Abstract
The complete plastome sequences of six species were sequenced to better understand the evolutionary relationships and mutation patterns in the chloroplast genome of the genus Colobanthus. The length of the chloroplast genome sequences of C. acicularis, C. affinis, C. lycopodioides, C. nivicola, C. pulvinatus and C. subulatus ranged from 151,050 to 151,462 bp. The quadripartite circular structure of these genome sequences has the same overall organization and gene content with 73 protein-coding genes, 30 tRNA genes, four rRNA genes and five conserved chloroplast open reading frames. A total of 153 repeat sequences were revealed. Forward repeats were dominant, whereas complementary repeats were found only in C. pulvinatus. The mononucleotide SSRs composed of A/T units were most common, and hexanucleotide SSRs were detected least often. Eleven highly variable regions which could be utilized as potential markers for phylogeny reconstruction, species identification or phylogeography were identified within Colobanthus chloroplast genomes. Seventy-three protein-coding genes were used in phylogenetic analyses. Reconstructed phylogeny was consistent with the systematic position of the studied species, and the representatives of the same genus were grouped in one clade. All studied Colobanthus species formed a single group and C. lycopodioides was least similar to the remaining species.
Subject terms: Plant genetics, Comparative genomics, Next-generation sequencing
Introduction
The genus Colobanthus in the family Caryophyllaceae contains 26 species1. Most species are found in the Southern Hemisphere, and the greatest diversity is observed in New Zealand2. Colobanthus species are low-growing perennials with a cushion growth habit, narrow and dense leaves, and inconspicuous, solitary, greenish flowers without petals, but with four to six prominent sepals3,4. The relationships within the family Caryophyllaceae are not easy to elucidate, partly due to arbitrarily and poorly defined genera and difficulties in determining phylogenetically useful morphological characters5. Colobanthus is one of the least studied genera, and it is sometimes confused with the related genus Sagina (Caryophyllaceae)6. Many areas where Colobanthus species occur are under protection, and some of them, such as the cold-temperate South Pacific Islands, have world heritage status7. Moreover, in contrast to the widespread species of C. quitensi, C. affinis and C. apetalus, taxa such as C. strictus, C. squarrosus and C. curtisiae (recorded in only three Tasmanian populations)8–10 or C. nivicola (endemic to the alpine tract of the Mt Kosciusko area in New South Wales, Australia)11 are extremely rare and are on the Australian list of rare or threatened plant species12. In many areas of Australia, where the habitats of Colobanthus species overlap, species such as C. nivicola and C. pulvinatus may be difficult to distinguish in the field11. Therefore, a precise identification of these species is necessary.
The members of the genus Colobanthus are extremely rarely studied, excluding Antarctic pearlwort (Colobanthus quitensis (Kunth) Bartl) which rose to fame as the only native representative of Magnoliopsida in the maritime Antarctic13. Colobanthus quitensis has been intensively studied to explore the traits responsible for its high tolerance to extreme Antarctic conditions14–19. Despite the above, our knowledge of the genetic diversity of this species and the entire genus Colobanthus remains limited20–24. Only two papers presented the complete sequence of the chloroplast genomes of C. quitensis25 and C. apetalus26, whereas the size of the nuclear genome in C. quitensis was estimated by flow cytometry in only one study27.
In recent years, chloroplast genome sequences attracted significant interest in plant phylogenetics, phylogeography and molecular evolution research28. Chloroplast genome sequences have numerous advantages, including low molecular weight, simple structure, uniparental (generally maternal) mode of inheritance, haploidy, highly conserved structure and a slower evolutionary rate of change than nuclear genomes. For this reason, chloroplast genomes constitute valuable data that are relatively easy to handle with source molecular data, support the validation of complex evolutionary relationships and detailed phylogenetic analyses at group, family or even genus level29–32. Chloroplast sequences also have numerous applications in biotechnology33,34 and the development of molecular markers for identifying species and distinguishing morphologically similar species35–37. A high number of new chloroplast genomes have been reported ever since the complete chloroplast genome sequences of Nicotiana tabacum38 and Marchantia polymorpha39 were published in 1986. This phenomenal progress was made possible by the development of new high-throughput genome sequencing technologies which enable scientists to obtain high quality cp genome sequences in a more convenient and relatively inexpensive way. As a result, around 3,500 chloroplast genomes have been deposited in the database of the National Center for Biotechnology Information (NCBI). It has been recently proposed that the whole chloroplast genome sequence should be used as a universal super-barcode in the identification of plant species. This approach may overcome the limitations of the traditional two-locus barcode based mainly on sequence variation within two plastome regions (rbcL and matK) that is not always sufficient for species discrimination40. In Colobanthus, the development of cpDNA markers has so far been restricted to the following loci: matK, trnK (C. masonae, C. affinis41), matK, trnK, trnL, trnL–trnF, trnF, rps16 (C. muscoides; unpublished data, available in NCBI) and ndhF (C. brevisepalus42). Complete chloroplast genome sequences are available only for C. quitensis25 and C. apetalus26. The published data indicate that the Colobanthus cp genome is typical for angiosperms in terms of size (151,276 bp for C. quitensis and 151,228 bp for C. apetalus) and composition (112 genes). This genome has a conserved quadripartite circular structure with a large single copy (LSC) region, a small single copy (SSC) region and two copies of inverted repeat (IR) regions43,44. The complete sequences of the cp genome of C. quitensis and C. apetalus provide molecular data that are essential for advanced genomic studies. However, the genomic data for other members of the genus Colobanthus are still limited.
The complete chloroplast genomes of six Colobanthus species have been sequenced and annotated for the first time in this study. The comparative study of these six chloroplast genomes as well as the previously published cp genomes for C. quitensis and C. apetalus had the following goals: (1) to determine the size and structure of Colobanthus cp genomes, (2) to identify genomic repeats, including forward, reverse, palindromic and complementary sequences among Colobanthus genomes, (3) to compare the variation of simple sequence repeats (SSRs) among Colobanthus cp genomes, (4) to verify the phylogenetic relationships among Colobanthus species and other Caryophyllaceae species for which complete chloroplast genomes are available.
Results
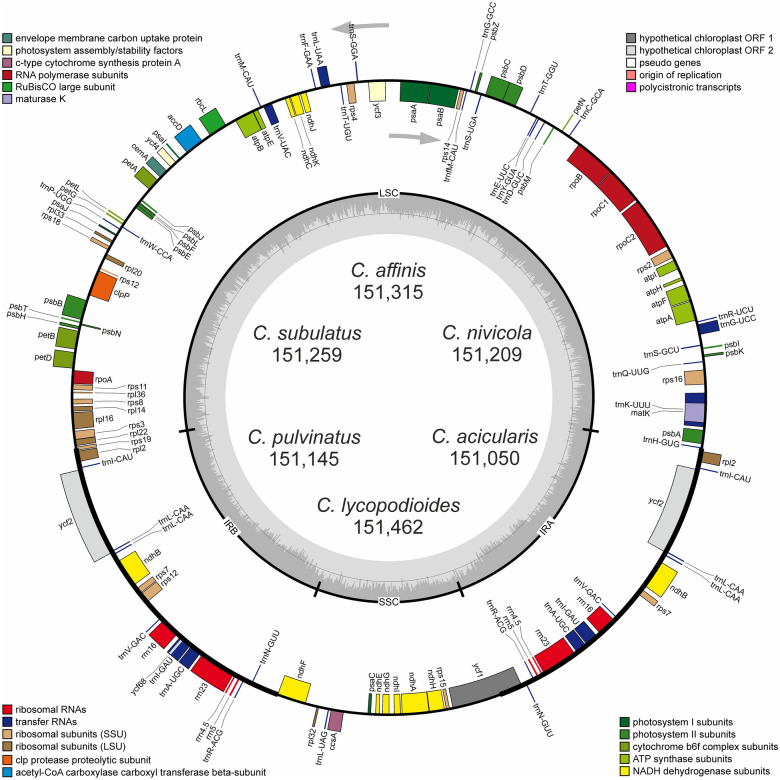
Organization of chloroplast genomes
Six Colobanthus species were sequenced to produce 1,986,760–7,849,218 raw reads (150 bp for average read length) which were mapped separately to the reference genome of C. quitensis. A total of 210,882–414,160 reads were ultimately mapped with 202.7× to 396.5 × coverage (Table 1). The six Colobanthus cp genome sequences were deposited in GenBank under the following accession numbers: MN273320 for C. acicularis, MN273318 for C. affinis, MN273317 for C lycopodioides, MN273316 for C. nivicola, MN273315 for C. pulvinatus, and MN273319 for C. subulatus. The chloroplast genome sequences described in this study ranged from 151,050 (C. acicularis) to 151,462 bp (C. lycopodioides). Each chloroplast genome was assembled into a single circular, double-stranded DNA sequence. All plastomes displayed a typical quadripartite structure with a pair of IRs (25,309–25,326 bp) separated by SSC (17,177–17,256 bp) and LSC (83,198–83,645 bp) regions (Fig. 1). The overall GC content was 36.61–36.66% and it was nearly identical in all Colobanthus cp genomes (Table 1). All of the analyzed Colobanthus cp genomes contained an identical set of 112 genes composed of 73 protein-coding genes, 30 tRNA genes, four rRNA genes and five conserved chloroplast ORFs (ycf1, ycf2, ycf3, ycf4, ycf68) (Table 2). Fifty-eight protein-coding genes, 22 tRNA genes and 2 conserved chloroplast ORFs (ycf3 and ycf4) are located in LSC, whereas the SSC region contained 11 protein-coding genes and one tRNA gene. The IR region contained four rRNA genes, seven tRNA genes and eight protein-coding genes, including ycf2, ycf68 and ycf1 on the border between IRA/IRB and SSC. The full ycf1 sequence is located on the IRA/SSC border, and its incomplete copy on the IRB/SSC border acts as a pseudogene. Fifteen genes contained one intron (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, trnI-GAU, trnA-UGC, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, ycf3), and two genes consisted of three exons (rps12 and clpP). The first exon of rps12 (5′ end of the sequence) was found in the LSC region, and the remaining two exons were located in the IR region. This unique feature supported the identification of rps12 as a trans-spliced gene. The introns of the two remaining genes, trnK-UUU and trnI-GAU, include coding sequences for matK and ycf68, respectively.
Table 1.
Summary of chloroplast genome characteristic of Colobanthus.
| Genome features | C. acicularis | C. affinis | C. lycopodioides | C. nivicola | C. pulvinatus | C. subulatus |
|---|---|---|---|---|---|---|
| Raw data reads no | 5,263,336 | 4,940 388 | 5,780,642 | 1,986 760 | 5,473,888 | 7,849,218 |
| Mapped reads no | 352,843 | 224 899 | 366,573 | 210,882 | 233,901 | 414 160 |
| Percent of chloroplast genome reads (%) | 6.70 | 4.55 | 6.34 | 10.61 | 4.27 | 5.28 |
| Mean coverage (x) | 338.0 | 216.6 | 355.5 | 202.7 | 220.6 | 396.5 |
| Size (bp) | 151,050 | 151 315 | 151,462 | 151,209 | 151,145 | 151,259 |
| LSC length (bp) | 83,198 | 83,419 | 83,645 | 83,351 | 83,306 | 83,456 |
| SSC length (bp) | 17,224 | 17,256 | 17,177 | 17,206 | 17,188 | 17,185 |
| IR length (bp) | 25,314 | 25,320 | 25,320 | 25,326 | 25,326 | 25,309 |
| Number of unique genes | 112 | 112 | 112 | 112 | 112 | 112 |
| Protein-coding genes | 78 | 78 | 78 | 78 | 78 | 78 |
| tRNA genes | 30 | 30 | 30 | 30 | 30 | 30 |
| rRNA genes | 4 | 4 | 4 | 4 | 4 | 4 |
| Number of genes duplicated in IR | 19 | 19 | 19 | 19 | 19 | 19 |
| Overall GC content (%) | 36.66 | 36.65 | 36.61 | 36.65 | 36.66 | 36.66 |
Species arranged alphabetically.
Figure 1.
Gene map of the six Colobanthus chloroplast genomes. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise (indicated by arrows). Differential functional gene groups are color-coded. GC content variations is shown in the middle circle. Gene map was generated with the OrganellarGenomeDRAW (OGDRAW) 1.3.1. (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html.)
Table 2.
Genes present in six chloroplast genomes of Colobanthus species.
| Category | Group of gene | Name of genes |
|---|---|---|
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cytochrome complex | petA, petB, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| NADH dehydrogenase | ndhA, ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Large subunit of RUBISCO | rbcL | |
| DNA replication and protein synthesis | Ribosomal RNA | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) |
| Small subunit ribosomal proteins | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 (×2), rps14, rps15, rps16, rps18, rps19b | |
| Large subunit ribosomal proteins | rpl2 (×2), rpl14, rpl16, rpl20, rpl22, rpl32, rpl33, rpl36 | |
| RNA polymerase subunits | rpoA, rpoB, rpoC1, rpoC2 | |
| Transfer RNA | trnA-UGC (×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU (×2), trnI-GAU (×2), trnK-UUU, trnL-CAA (×4), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), trnV-UAC, trnW-CCA, trnY-GUA | |
| Other genes | Conserved hypothetical chloroplast ORF | ycf1b, ycf2 (×2), ycf3a, ycf4a, ycf68 (×2) |
| Other proteins | accD, ccsA, cemA, clpP, matK |
Genes list arranged alphabetically.
aGenes associated with Photosystem I.
bGene with its pseudogene copy at IRB/LSC and IRA/SSC border: ψrps19 and ψycf1, respectively.
The total number of codons for all protein-coding genes in the cp genomes of six Colobanthus species ranged from 25,159 to 26,162. The most and least abundant codons (excluding these associated with the initiation and termination of translation) were ATT (4.33%) and TGC (0.25%), respectively (Supplementary Table S1 online). Furthermore, leucine appeared as the dominant amino acid (10.7%), whereas cysteine was less frequently encountered (1.2%). Since the data for codon usage were not available for the previously published plastomes of C. quitensis and C. apetalus, these species were included in the analysis. Both species shared the same pattern of codon usage and amino acid frequency.
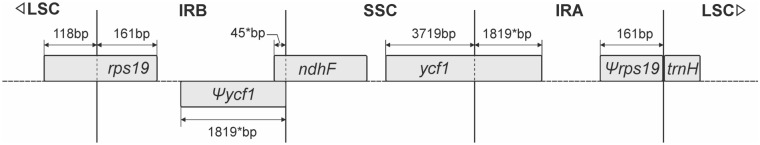
The boundaries between IR and SSC/LSC regions in all Colobanthus cp genomes were identified (Fig. 2). The IRA/SSC junction was found within the ycf1 gene (1819 bp from its 5′ end), and the boundary between IRB and LSC region was identified within the rps19 gene (161 bp from its 5′ end). Consequently, the full ycf1 sequence is located only on the IRA/SSC border, and its incomplete copy on IRB/SSC border acts as a pseudogene (ψycf1). A similar situation was observed for rps19, for which the complete sequence can be found on the IRB/LSC border, whereas the ψrps19 pseudogene is located in the IRA region. The IRB/SSC boundary was identified within the ndhF gene, and a 45 bp string from its 3′-end overlapped ψycf1 within IRB. The IRA/LSC junction was adjacent to the trnH gene. The boundaries between the IR and SSC/LSC regions of six Colobanthus species were generally found in the same positions and within the same genomic elements. One base shift in the IRA/SSC and IRB/SSC border position was found only in C. subulatus (Fig. 2).
Figure 2.
Comparison of LSC, SSC, and IR boundaries of six Colobanthus chloroplast genomes. Asterisk represents the location of one base shift in IRA/SSC and SSC/IRB boundary position; 45 and 1,819 bp values should be replaced by 44 and 1,818 bp for C. subulatus.
Repetitive sequences and SSRs
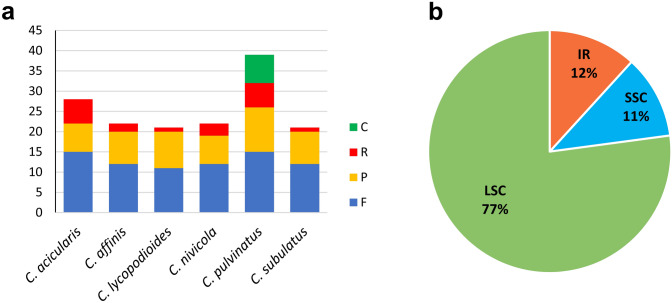
A total of 153 repeats were observed in the plastomes of six Colobanthus species. The number of repeats was highest (39) in C. pulvinatus and lowest (21) in C. lycopodioides and C. subulatus (Supplementary Table S2A–F online). Forward repeats dominated in the identified repetitive sequences (from 38.5% in C. pulvinatus to 57.1% in C. subulatus), followed by palindromic (from 25% in C. acicularis to 42.9% in C. lycopodioides) and reverse repeats (from 4.8% in C. lycopodioides and C. subulatus to 21.4% in C. acicularis). Complementary repeats were found only in the cp genome of C. pulvinatus with a frequency of 17.9% (Fig. 3A). Most repeat sequences (77.1%) were detected in the LSC region, followed by IR (11.8%) and SSC regions (11.1%) (Fig. 3B). These sequences were found predominantly within intergenic spacers and introns with a frequency of 79.5% (C. pulvinatus) to 57.1% (C. subulatuc and C. lycopodioides). Our study also revealed that many repeats shared the same locus in all Colobanthus cp genomes. Fourteen such loci were identified: psaA, psaA–ycf3, psaB, psaI–ycf4, petN–psbM, trnG-GCC, trnG-UCC, trnS-GCU, trnS-GGA and trnS-UGA in the LSC region, ndhA and ycf1 in the SSC region, and trnV-GAC-rps7 and ycf2 in the IR region.
Figure 3.
Number of repeat types and distribution of repeats in six Colobanthus species. (A) Types of repeats, (B) Location of repeat sequences. F, P, R and C represent forward, palindromic, reverse and complementary repeats.
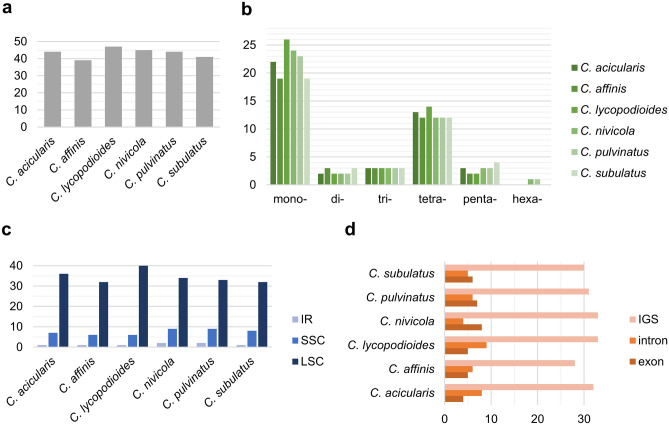
The Phobos analysis supported the identification of 39–47 SSRs in six cp genomes (Fig. 4A), including mono-, di-, tri-, tetra-, penta- and hexanucleotides (Supplementary Table S3A–F online). The mononucleotide SSRs were most common with a frequency ranging from 46.3% in C. subulatus to 55.3% in C. lycopodioides (Fig. 4B). All mononucleotide SSRs were composed of A/T repeat units. Motifs composed of adenine and thymine were also predominant in di- and trinucleotide SSRs, where only AT/TA and AAT/TTA motifs were observed, respectively. Tetranucleotide SSRs were the second most frequent repeats that ranged from 26.7% in C. nivicola to 30.8% in C. affinis. The frequency of di-, tri- and pentanucleotide SSRs did not exceed 9.8%. Hexanucleotide SSRs were detected only in C. nivicola and C. pulvinatus with a frequency of 2.2% and 2.3%, respectively. The majority of SSRs were located in the LSC region (from 75% in C. pulvinatus to 85.1% in C. lycopodioides), followed by SSC (from 12.8% in C. lycopodioides to 20.5% in C. pulvinatus) and IR regions (from 2.1% in C. lycopodioides to 4.5% in C. pulvinatus) (Fig. 4C). Furthermore, SSRs were identified predominantly within intergenic spacers (from 70.2% in C. lycopodioides to 73.3% in C. nivicola), whereas the remaining SSRs were distributed in various proportions between exons and introns (Fig. 4D).
Figure 4.
The distribution and type of simple sequence repeats (SSRs) in cp genomes of six Colobanthus species. (A) Number of different SSRs types. (B) Location of different SSRs in IR, SSC and LSC regions. (C) Distribution of SSR motifs in different repeat class types. (D) Partition of SSRs among IGS, introns and exons.
Sequence divergence
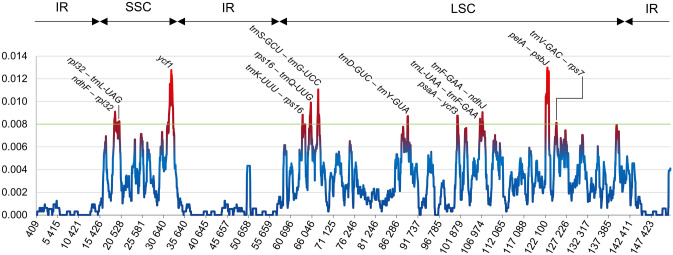
The overall sequence identity and divergent regions in the cp genomes of C. acicularis, C. affinis, C. lycopodioides, C. nivicola, C. pulvinatus, C. subulatus and the previously published plastomes of C. quitensis and C. apetalus25,26 were determined in MAUVE and DnaSP programs. MAUVE results are shown in Supplementary Figure S1 online. Rearrangements (inversions or translocations) were not detected in any of the eight chloroplast genome sequences. The high sequence similarity points to the conservative character of all eight cp genomes. In the DnaSP, nucleotide diversity (π) in the cp genomes of Colobanthus species was determined at 0.00262. The most variable regions were identified in sliding window analysis, i.e. regions for which π values exceeded 0.008 (Fig. 5). Divergence was generally higher in non-coding regions. In the coding region, differences were found only in the ycf1 locus. In non-coding regions, the highest divergence and the highest π value (0.01299) were observed for ndhF–rpl32, rpl32–trnL-UAG, trnK-UUU–rps16, rps16–trnQ-UUG, trnS-GCU–trnG-UCC, trnD-GUC–trnY-GUA, psaA–ycf3, trnL-UAA–trnF-GAA, trnF-GAA–ndhJ, petA–psbJ and trnV-GAC–rps7 (Supplementary Table S4 online). The majority of highly variable regions (9) were identified in LSC. There were three such regions in SSC, and none in the IR region.
Figure 5.
Sliding window analysis of the eight Colobanthus complete chloroplast genome sequences (window length: 800 bp; step size 50 bp). The Y-axis presents nucleotide diversity of each window, while the X-axis represents position of the midpoint.
Synonymous (Ks) and non-synonymous (Ka) substitution rate analysis
The substitution rate varied widely across plastome genes in each functional group, and the values of Ka and Ks were determined in the range of 0–0.0117 and 0–0.152, respectively (Supplementary Table S5 online). The highest average value of Ks (0.0111) was noted for coding sequences associated with the large subunit of ribosome. The average value of Ks was lowest in genes related to the cytochrome b/f complex (0.0008), RubisCO large subunit (0.0015), Photosystem II (0.0022) and Photosystem I (0.0026). The genes associated with Photosystem II, Photosystem I and the cytochrome b/f complex were also characterized by the lowest average values of Ka (0.0003, 0.0005 and 0.0005, respectively), and the highest average value of Ka (0.0025) was noted in the RubisCO large subunit. In general, no differences were observed in the sequences of 18 plastome genes (Ka = 0, Ks = 0) of the studied Colobanthus species. The remaining 60 genes shared 99% similarity, but only synonymous substitutions (Ka = 0) were observed in 33 of those genes. The Ka/Ks ratio was less than 1 in all genes, excluding rpoC2 (1.5238 for C. affinis, and 1.381 for C. nivicola and C. pulvinatus) and matK (1.1333 for C. lycopodioides). The Ka/Ks ratio exceeded 1 in rpoC2 and matK, which could suggest that in the above species, these genes had undergone positive selection (adaptation to a specific environment). A Ka/Ks ratio of less than 1 points to the influence of purifying selection on the remaining 76 genes.
RNA-editing
The results of the PREP prediction revealed 49 editing sites in 18 protein coding genes in the plastomes of Colobanthus species, excluding C. acicularis and C. lycopodioides where 48 such elements were found (Supplementary Table S6 online). One RNA editing site within the ndhF gene was missing in C. acicularis, and one editing site within the ndhB sequence was missing in C. lycopodioides. All editing events involved C to U conversion. Fifteen non-synonymous mutations were found at the first position of the codon, 34 mutations were identified at the second position, and none were found at the third position. Serine (S) to leucine (L) changes accounted for nearly a third (32.6%) of the identified mutations, whereas arginine (R) to tryptophan (W) and serine (S) to phenylalanine (F) changes were least frequently observed (4.1% for both). Each RNA editing site in the corresponding genes of the eight Colobanthus species was generally found at the same nucleotide position. Three base shifts were identified in only two RNA editing sites within the rpoB sequence in the cp genome of C. quitensis.
Phylogenetic analysis
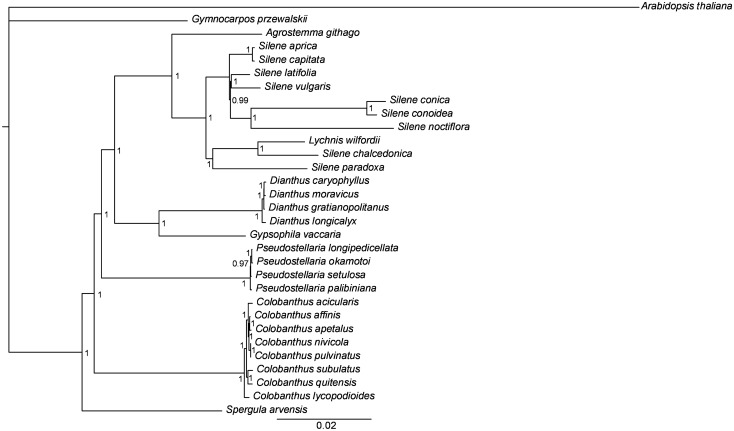
The phylogenetic trees generated by BI and ML had a consistent topology. In the BI tree, Bayesian posterior probability reached 1.0 in 92.6% of the nodes (25 out of 27). The reconstructed phylogeny is consistent with the taxonomic position of the studied species, and it revealed the following relationships: all studied Colobanthus species formed one clade; the second clade grouped four Pseudostellaria species; the third clade consisted of four Dianthus species and a solitary branch of Gysophilla vacaria; all members of the genus Silene and Lychnis wilfordii formed the fourth clade in the proximity of a separate branch of Agrostemma githago; the most divergent branches were formed by Gymnocarpos przewalski and Spergula arvensis (Fig. 6, Table 3).
Figure 6.
Phylogenetic tree based on sequences of sheared 73 protein-coding genes from eight Colobanthus species and 22 other Caryophyllaceae representatives using Bayesian posterior probabilities (PP). Bayesian PP are given at each node.
Table 3.
GenBank accession numbers and references for cp genomes used in this study.
| Species | Accession number | Length (bp) | Reference |
|---|---|---|---|
| Agrostemma githago | NC_023357 | 151,733 | Sloan et al.54 |
| Arabidopsis thaliana | NC_000932 | 154,478 | Sato et al.103 |
| Colobanthus acicularis | MN273320 | 151,050 | This study |
| Colobanthus affinis | MN273318 | 151,315 | This study |
| Colobanthus apetalus | NC_036424 | 151,228 | Androsiuk et al.26 |
| Colobanthus lycopodioides | MN273317 | 151,462 | This study |
| Colobanthus nivicola | MN273316 | 151,209 | This study |
| Colobanthus pulvinatus | MN273315 | 151,145 | This study |
| Colobanthus subulatus | MN273319 | 151,259 | This study |
| Colobanthus quitensis | NC_028080 | 151,276 | Kang et al.25 |
| Dianthus caryophyllus | NC_039650 | 147,604 | Chen et al. (2018)a |
| Dianthus gratianopolitanus | LN877389 | 149,735 | Michling et al. (2018)a |
| Dianthus longicalyx | KM668208 | 149,539 | Gurusamy et al.104 |
| Dianthus moravicus | LN877396 | 149,524 | Michling et al. (2018)a |
| Gymnocarpos przewalskii | NC_036812 | 150,636 | Yang (2017)a |
| Gypsophila vaccaria | NC_040936 | 150,042 | Yao et al.105 |
| Pseudostellaria longipedicellata | NC_039454 | 149,626 | Kim et al. (2018)a |
| Pseudostellaria okamotoi | NC_039974 | 149,653 | Kim et al. (2019)a |
| Pseudostellaria palibiniana | NC_041166 | 149,668 | Kim and Park (2019)a |
| Pseudostellaria setulosa | MK172842 | 149,479 | Kim and Park (2019)a |
| Lychnis wilfordii | NC_035225 | 152,320 | Kang et al.69 |
| Silene aprica | NC_040934 | 150,293 | Yao et al.105 |
| Silene capitata | NC_035226 | 150,224 | Kang et al.69 |
| Silene chalcedonica | NC_023359 | 148,081 | Sloan et al.54 |
| Silene conica | NC_016729 | 147,208 | Sloan et al.53 |
| Silene conoidea | NC_023358 | 147,896 | Sloan et al.54 |
| Silene latifolia | NC_016730 | 151,736 | Sloan et al.53 |
| Silene noctiflora | NC_016728 | 151,639 | Sloan et al.53 |
| Silene paradoxa | NC_023360 | 151,632 | Sloan et al.54 |
| Silene vulgaris | NC_016727 | 151,583 | Sloan et al.53 |
| Spergula arvensis | NC_041240 | 152,703 | Yao et al.105 |
Species list arranged alphabetically.
aDirect submission to NCBI, unpublished.
Discussion
The length of the complete sequences of the six new chloroplast genomes of Colobanthus species ranged from 151,050 (C. acicularis) to 151,462 bp (C. lycopodioides), it was very similar to the previously sequenced plastomes of C. quitensis (151,276 bp)25 and C. apetalus (151,228 bp)26, and was within the size range of cp genomes of other angiosperms45. A comparison of all Colobanthus cp genomes that have been sequenced to date also revealed considerable similarities in genome composition—all eight species had the same gene content and order. Moreover, their protein-coding sequences were characterized by low variation. Consequently, the differences in the size and organization of intergenic spacers were most probably responsible for the observed variations in the size of Colobanthus cp genomes. As previously described46, the variation in the size of cp genomes in different plant lineages could also be attributed to the expansion and contraction of IR regions. Similarly to most angiosperms, IR boundaries were found within ycf1 and rps19 genes in the presented cp genomes of Colobanthus47. The location of IR boundaries was identical in all Colobanthus species. One base shift in the position of IRA/SSC and IRB/SSC borders was found only in C. subulatus. The size of IR regions was highly similar in all Colobanthus cp genomes that have been sequenced to date, ranging from 25,303 (C. quitensis) to 25,326 bp (C. nivicola and C. pulvinatus), which corresponds to the values reported in other dicotyledons48, 49. Somewhat greater differences were observed in Colobanthus species when the length of LSC and SSC regions was considered. The differences in the length of LSC and SSC between the longest and the shortest element were determined at 447 bp (C. lycopodioides vs. C. acicularis) and 79 bp (C. affinis vs. C. lycopodioides), respectively.
The accumulation of point mutations in the form of synonymous and non-synonymous nucleotide substitutions is one of the key mechanisms of gene evolution50. In this study, synonymous nucleotide substitutions were more frequently observed; therefore, the analyzed Colobanthus species have maintained a high degree of sequence conservation, especially in genes involved in photosynthesis. However, considerable variation was observed in several genes, including rps16 and ycf1 which are characterized by the highest number of non-synonymous nucleotide substitutions. These two elements of chloroplast genomes are very often found among the most variable chloroplast loci of numerous genera51, including Silene (Caryophyllaceae)52–54. The Ka/Ks ratio was examined to determine selection pressure on protein-coding genes. The six Colobanthus cp genomes exhibited highly conserved organization, but positive selection pressure (Ka/Ks > 1) was observed in rpoC2 (C. affinis, C. nivicola and C. pulvinatus) and matK (C. lycopodioides), which suggests that these loci are undergoing essential adaptation to environmental conditions. In C. lycopodioides, C. nivicola and C. pulvinatus, the Ka/Ks ratio for the rbcL gene was only somewhat lower than 1 (0.965), which could indicate that positive selection played some role in the acceleration of the substitution rate for that locus. All three genes have been also previously reported to undergo positive selection in other plant species. Gene rbcL, which encodes the large subunit of RuBisCO, appears to undergo positive selection most often, in up to 75–88% of terrestrial plants55. According to the literature, rbcL could be a chloroplast region that was positively selected during the evolutionary processes associated with adaptation to temperature55–58, CO2 concentration55,59,60 or water deficiency57,60. The matK gene encoding the maturase enzyme which catalyzes the removal of a nonautocatalytic intron from premature RNAs61 was also found to undergo positive selection in 32 plant groups62. Despite its important function, the rpoC2 gene encoding the β subunit of plastid-encoded plastid RNA polymerase (RNA polymerase type I) is also a relatively rapidly evolving chloroplast sequence63 that undergoes positive selection in various groups of plants, including Lamiaceae64, Orobanchaceae65 and Annonaceae66. The fact that traces of positive selection were detected in such different functional gene classes, including genes encoding photosynthesis (rbcL), transcription and transcript processing (matK and rpoC2), could indicate that natural selection targets different chloroplast functions. A higher rate of substitutions in these Colobanthus genes could be indicative of continuous fine-tuning to specific environmental conditions.
Repeat regions within genomes play an important role in sequence divergence and rearrangement, which is why they have to be identified, and their number and distribution has to be determined in genomic studies67,68. In the six reported here plastomes of Colobanthus species, most repeat regions were identified in intergenic regions and introns (57.1–79.5%), which is highly consistent with the values previously reported in C. apetalus (76.7%) and C. quitensis (53.3%)26, as well as for other Caryophyllaceae such as Silene capitata (56.0%) and Lychnis wilfordii (69.2%)69. Our study also demonstrated that repeat regions are not randomly distributed within Colobanthus cp genomes, and they were identified mainly within highly divergent regions of rpl32–trnL-UAG, ycf1, trnK-UUU–rps16, psaA–ycf3 and petA–psbJ in the LSC.
Simple sequence repeats, also known as microsatellites, are important molecular markers with many applications, including species identification, population genetics and phylogenetic studies70–72. Mononucleotide SSRs were identified most frequently (51% on average) among the microsatellites of the six analyzed cp genomes of Colobanthus species, with A/T as the prevalent motif type. The frequency of mononucleotide SSRs was highly similar in C. apetalus and C. quitensis (48.8% and 54.2%, respectively)26 as well as in other plant species, including members of the family Caryophyllaceae69,73,74. In turn, hexanucleotide SSRs were least abundant, and only one such element was identified in the cp genomes of C. nivicola and C. pulvinatus. Moreover, the majority of SSRs were located within intergenic spacers and introns, while only 13.5% (on average) were positioned in the exons of 11 genes. Four of these loci (ycf1, rpoC2, rrn23, atpA) harbored SSRs in all six Colobanthus species, whereas four other loci (petB, petD, ndhA, rpoA) contained SSRs in only one, specific species.
In terrestrial plants, cytidines are systematically converted to uridines (C to U editing) in both mitochondrial and plastid mRNA transcripts to restore conserved codons75. This important post-transcriptional process that had appeared in early stages of flowering plant evolution76 is considered to be functionally significant in chloroplasts77. In our work, potential RNA editing sites were identified in 18 out of the 34 analyzed chloroplast protein-coding genes. These sites were highly conserved in the analyzed species, excluding two sites within the rpoB gene in the cp genome of C. quitensis. In this species, the three base shift appear to be a consequence of the insertion of three bases (CAG) which added glutamine in position 636 of the rpoB amino acid sequence. This study also demonstrated that leucine codons, including those that have potentially emerged from RNA editing, are heavily used in the cp genomes of all eight Colobanthus species. Previous research revealed a high demand for leucine biosynthesis in chloroplasts and suggested that this amino acid plays an important role in photosynthesis-related metabolism78,79.
Colobanthus is one of the 86 genera within the family Caryophyllaceae42. Genus Colobanthus contains around 26 confirmed species and 24 taxa whose status has not been resolved1. Therefore, a taxonomic inventory of the genus Colobanthus is still needed. However, this is a challenging task due to the extensive geographic distribution of the species and high variation within morphological traits. The DNA barcode has recently emerged as an effective biological tool for accurate species identification80. Despite the above, the molecular data for Colobanthus are highly limited, and species-specific barcode sequences are not available. In the eight Colobanthus species with completely sequenced cp genomes, the genetic variation in the chloroplast regions of matK and rbcL that are widely used in the barcoding of terrestrial plants was lower than expected. However, genome-wide comparative analyses based on nucleotide diversity (π) supported the identification of 12 highly variable regions (π > 0.008) that could be utilized as a source of potential markers for species identification and reconstruction of the phylogenetic relationships within this plant group: ndhF–rpl32, rpl32–trnL-UAG, ycf1, trnK-UUU–rps16, rps16–trnQ-UUG, trnS-GCU–trnG-UCC, trnD-GUC–trnY-GUA, psaA–ycf3, trnL-UAA–trnF-GAA, trnF-GAA–ndhJ, petA–psbJ and trnV-GAC–rps7. High nucleotide diversity (π) in regions ycf1, rpl32–trnL, trnS–trnG, petA–psbJ and rps16–trnQ was also reported in other plants51. Moreover, the ndhF–rpl32–trnL-UAG region has been widely used in phylogenetic studies81,82. Two of these highly variable regions (rps16–trnQ-UUG and trnL-UAA–trnF-GAA) were among the 5 chloroplast sequences that were analyzed by Greenberg and Donoghue83 to explore the phylogenetic relationships within the family Caryophyllaceae.
It has recently been postulated that complete chloroplast genome sequences can be used as a super barcode for identifying plant species84. This approach is particularly useful for distinguishing between closely related taxa where limited sequence variation has resulted from a low rate of genome evolution or a relatively short time since the divergence event85,86. Considering the still unresolved status of many Colobanthus species, a phylogeny reconstruction based on whole plastome sequences seems to be highly desired for this genus. In this study, chloroplast genome sequences were used to resolve the phylogenetic relationships within the genus Colobanthus and the Caryophyllaceae family, but the analysis involved only species with completely elucidated plastome sequences. The phylogeny reconstructed based on 73 concatenated protein-coding gene sequences appeared to be consistent with the taxonomic position of the studied species and previous phylogenies of the Caryophyllaceae41,83,87. However, in the genus Colobanthus, this was an initial step towards resolving the phylogenetic relationships within that group of plants. The phylogenetic tree with highly supported nodes revealed that all eight Colobanthus species were grouped in one clade, where C. pulvinatus and C. nivicola, C. apetalus and C. affinis as well as C. subulatus and C. quitensis formed three pairs of most similar species, whereas C. lycopodioides appeared to be most different.
Conclusion
The chloroplast genomes of C. acicularis, C. affinis, C lycopodioides, C. nivicola, C. pulvinatus and C. subulatus were sequenced and characterized for the first time. Their plastomes have a typical quadripartite circular structure and share the same overall organization and gene content. The information regarding sequence variation, distribution and characteristics of SSR loci within the studied cp genomes could be useful in future studies on the population genetics, phylogenetics and evolution of Colobanthus. Nevertheless, further research is required to investigate whether highly variable regions or complete chloroplast genome sequences could be used as reliable and effective DNA barcodes for Colobanthus species.
Methods
Plant material, DNA extraction and chloroplast genome sequencing
Fresh leaves of five Colobanthus species were sampled from plants grown from seeds in a greenhouse of the Department of Plant Physiology, Genetics and Biotechnology at the University of Warmia and Mazury in Olsztyn, Poland. The seeds of C. affinis were obtained from the Royal Botanic Gardens, Victoria, Australia. The seeds of C nivicola and C. pulvinatus were acquired from the Australian National Botanic Gardens, Canberra. The seeds of C. subulatus originated from the Royal Botanic Gardens, Kew, United Kingdom. The seeds of C. lycopodioides were collected in the region of Mendoza, Andes, Argentina, at an altitude of 4,024 m a.s.l., (33°10′ S; 69°50′ W). Only in C. acicularis, DNA was extracted from dried tissue of one individual, supplied in silica gel by the Royal Botanic Garden, Edinburgh, UK. Plant material was formally identified before the analyses. Professor Irena Giełwanowska performed morphological and anatomical analyses of both vegetative and generative organs harboring characteristic traits for the identification of Colobanthus species1,88,89. Voucher specimens of each studied species have been deposited in the Vascular Plants Herbarium of the Department of Botany and Nature Protection at the University of Warmia and Mazury in Olsztyn, Poland (OLS), under the following numbers: C. acicularis (No. OLS 33824), C. affinis (No. OLS 33825), C lycopodioides (No. OLS 33826), C. nivicola (No. OLS 33827), C. pulvinatus (No. OLS 33828) and C. subulatus (No. OLS 33829). Total genomic DNA was extracted from fresh/dry tissue of a single plant using the Maxwell16LEV Plant DNA Kit (Promega, Madison, WI). The quality of DNA was verified on 1% (w/v) agarose gel stained with 0.5 µg/ml ethidium bromide. The concentration and purity of DNA samples were assessed spectrophotometrically.
Genome libraries were prepared using the Nextera XT kit (Illumina Inc., San Diego, CA, USA), and genomes were sequenced on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) with a 150 bp paired-end read.
Annotation and genome analysis
The FastQC tool was used to check the quality of raw reads. Raw reads were trimmed (5 bp of each read end, regions with more than 5% probability of error per base) and mapped to the reference chloroplast genome of C. quitensis (NC_028080) in Geneious v.R7 software90 with default medium–low sensitivity settings. Subsequent steps of annotation and genome analysis were described in our previous paper26. The gene maps of the annotated cp genomes were developed with the OrganellarGenome DRAW tool91.
Genomic repeats and SSR analysis
The size and location of genomic repeats, including forward, reverse, palindromic and complementary sequences within the analyzed chloroplast genomes, were identified using REPuter software92 with the following settings: (1) hamming distance of 3, (2) sequence identity ≥ 90%, and (3) minimum repeat size ≥ 30 bp. Phobos v.3.3.1293 was used to detect chloroplast simple sequence repeats (SSRs). Only perfect SSRs with a motif size of one to six nucleotide units were considered, with standard thresholds for chloroplast SSRs identification: ≥ 12 repeat units for mononucleotide SSRs, ≥ 6 repeat units for dinucleotide SSRs, ≥ 4 repeat units for trinucleotide SSRs, and ≥ 3 repeat units for tetra-, penta- and hexanucleotide SSRs94. A single IR region was used to eliminate the influence of doubled IR regions. Redundant results in REPuter were deleted manually.
Comparative chloroplast genome analysis
Genome synteny analysis of the eight Colobanthus plastomes (six genomes reported in this paper, and C. quitensis and C. apetalus that were previously characterized and deposited in NCBI25,26) was performed with the use of MAUVE v.1.1.195. The sequences were aligned in MAFFT v.7.31096 to perform sliding window analysis and evaluate nucleotide diversity (π) in cp genomes using DnaSP v.6.10.0497. The step size was set to 50 base pairs, and window length was set to 800 base pairs.
The evolutionary rate of the plastome genes identified in all Colobanthus species (C. acicularis, C. affinis, C. apetalus, C lycopodioides, C. nivicola, C. pulvinatus, C. quitensis and C. subulatus) was analyzed. A total of 78 genes were selected to estimate the ratio of non-synonymous (Ka) to synonymous (Ks) substitutions. Colobanthus quitensis was the reference species. These genes were extracted and aligned separately using MAFFT v7.310. The values of Ka and Ks in the shared genes were calculated in DnaSP v.6.10.04. Genes with non-applicable (NA) Ka/Ks ratios were changed to zero.
The chloroplast genome borders of LSC, SSC, and IRs were identified and compared based on their annotations. The data on the distribution of codon usage was acquired from the Geneious v.7 statistics panel.
Potential RNA editing sites in the protein-coding genes of chloroplast genomes were predicted using the Predictive RNA Editor for Plants (PREP) suite98. The cutoff value for the analyzed Colobanthus species was set at 0.8, and 34 out of the 35 reference genes in PREP were used. rpl23 was not included in the analysis because it was not identified within the chloroplast genomes of the studied Colobanthus species. Two previously sequenced cp genomes of C. quitensis and C. apetalus25,26 were also included in this analysis.
Phylogenetic analysis
The available (24) complete chloroplast genomes representing Caryophyllaceae lineages and the cp genome of Arabidopsis thaliana as an outgroup were downloaded from the NCBI database to investigate the phylogenetic relationships among the studied representatives of the genus Colobanthus and the genera in the family Caryophyllaceae. The cp genomes used in phylogenetic analyses are presented in Table 3. The sequences of 73 shared protein coding genes were extracted using custom R script, and they were aligned in MAFFT v.7.310. Finally, 73 concatenated protein-coding gene sequences where used for phylogeny reconstruction by Bayesian Inference (BI) and Maximum-Likelihood (ML) method. The best-fit model of sequence evolution was identified in MEGA v.799, and the GTR + G + I model was selected. The BI analysis was performed in MrBayes v.3.2.6100,101, and the ML analysis was conducted in PhyML v.3.0102. Parameter settings were previously described by Androsiuk et al.26.
Supplementary information
Acknowledgements
Authors are grateful to Royal Botanic Gardens, Victoria, Australia; Australian National Botanic Gardens, Canberra; Royal Botanic Gardens, Kew, UK and Royal Botanic Garden, Edinburgh, UK for sharing the research material representing genus Colobanthus. This research was supported by National Science Centre, Poland (No. 2018/02/X/NZ8/02243). The funding agency had no role in the design of the experiment, analysis, and interpretation of data and in writing the manuscript.
Author contributions
P.A. conceived and designed this study, acquired the research material, performed the repeat and phylogenetic analyses, conducted the comparative analyses and interpreted the data, wrote the manuscript; J.P.J., K.M. and Ł.P.: performed genome assembly and annotation, assisted in comparative analyses, were responsible for figures preparation and GenBank submissions, drew the cp genome map; A.O. and A.P.: performed DNA extraction, were responsible for genome library preparation and DNA sequencing; K.J.C. collected C. lycopodioides, assisted in data interpretation, revised all versions of the manuscript; R.G. provided valuable comments on manuscript development and revised the manuscript; I.G. was responsible for seeds germination and plant growth in the greenhouse, assisted in data interpretation, revised all versions of the manuscript. All authors have read and approved the final manuscript.
Data availability
The complete chloroplast genomes of the six Colobanthus species have been submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/https://www.ncbi.nlm.nih.gov/) under the accession numbers: MN273320 for C. acicularis, MN273318 for C. affinis, MN273317 for C. lycopodioides, MN273316 for C. nivicola, MN273315 for C. pulvinatus and MN273319 for C. subulatus.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-68563-5.
References
- 1.The Plant List 2013. Version 1.1.https://www.theplantlist.org/browse/A/Caryophyllaceae/Colobanthus. (2019).
- 2.West JG. Colobanthus curtisiae (Caryophyllaceae), a new species from eastern Tasmania, Australia. R. Soc. Tasm. Hobart. 1991;124(2):75–78. [Google Scholar]
- 3.Giełwanowska I, et al. Biology of generative reproduction of Colobanthus quitensis (Kunth) Bartl. from King George Island, South Shetland Islands. Pol. Polar Res. 2011;32:139–155. [Google Scholar]
- 4.The AGS online Plant Encyclopaedia. Alpine Garden Society. http://encyclopaedia.alpinegardensociety.net/plants/Colobanthus. (2019).
- 5.Harbaugh DT, et al. A new lineage-based tribal classification of the family Caryophyllaceae. Int. J. Plant Sci. 2010;171(2):185–198. [Google Scholar]
- 6.Timaná ME. Sagina diffusa (Hook.f.) Timaná, comb. Nov. (Caryophyllaceae), a new combination for the flora of île St. Paul (Southern Indian Ocean), with some historical notes. Adansonia. 2018;40(3):47–53. [Google Scholar]
- 7.Cooper, J., Cuthbert, R. J., Gremmen, N. J. M., Ryan, P. G. & Shaw, J. D. Earth, fire and water: Applying novel techniques to eradicate the invasive plant, procumbent pearlwort Sagina procumbens, on Gough Island, a World Heritage Site in the South Atlantic. International Conference on Island Invasives, Auckland, New Zealand (Gland, Switzerland: IUCN 2011).
- 8.West, J. G. Colobanthus curtisiae (Caryophyllaceae), a new species from Eastern Australia. In Aspects of Tasmanian Botany: A Tribute to Winifred Curtis (eds. Banks, M. R., Smith, S. J., Orchard, A. E. & Kantvilas, G.) (Royal Society of Tasmania, 1991).
- 9.Sneddon BV. The taxonomy and breeding system of Colobanthus squarrosus (Caryophyllaceae) N. Z. J. Bot. 1999;37:195–204. [Google Scholar]
- 10.Gilfedder L, Kirkpatrick JB. The distribution, ecology and conservation needs of Colobanthus curtisiae west. Pap. Proc. R. Soc. Tasmania. 1996;130(1):25–30. [Google Scholar]
- 11.Gray M. Miscellaneous notes on Australian plants. 3. Craspedia, Gnaphalium, Epacris, Tasmannia, Colobanthus and Deyeuxia. Contr. Herbarium Australiense. 1976;26:1–11. [Google Scholar]
- 12.Briggs JD, Leigh JH. Rare or Threatened Australian Plants. Clayton: CSIRO Publishing; 1996. [Google Scholar]
- 13.Skottsberg C. Antarctic flowering plants. Svensk Bot. Tidskr. 1954;51:330–338. [Google Scholar]
- 14.Bravo LA, et al. Effect of cold acclimation on the photosynthetic performance of two ecotypes of Colobanthus quitensis (Kunth) Bartl. J. Exp. Bot. 2007;58:3581–3590. doi: 10.1093/jxb/erm206. [DOI] [PubMed] [Google Scholar]
- 15.Giełwanowska I, Pastorczyk M, Lisowska M, Wegrzyn M, Gorecki RJ. Cold stress effects on organelle ultrastructure in polar Caryophyllaceae species. Pol. Polar Res. 2014;35:627–646. [Google Scholar]
- 16.Bascunan-Godoy L, et al. Cold-acclimation limits low temperature induced photoinhibition by promoting a higher photochemical quantum yield and a more effective PSII restoration in darkness in the Antarctic rather than the Andean ecotype of Colobanthus quitensis Kunt Bartl (Cariophyllaceae) BMC Plant Biol. 2012;12:114. doi: 10.1186/1471-2229-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarrete-Gallegos AA, Bravo LA, Molina-Montenegro MA, Corcuera LJ. Antioxidant responses in two Colobanthus quitensis (Caryophyllaceae) ecotypes exposed to high UV-B radiation and low temperature. Rev. Chil. Hist. Nat. 2012;85:419–433. [Google Scholar]
- 18.Pastorczyk M, Giełwanowska I, Lahuta LB. Changes in soluble carbohydrates in polar Caryophyllaceae and Poaceae plants in response to chilling. Acta Physiol. Plant. 2014;36:1771–1780. [Google Scholar]
- 19.Cuba-Díaz M, Castel K, Acuña D, Machuca Á, Cid I. Sodium chloride effect on Colobanthus quitensis seedling survival and in vitro propagation. Antarct. Sci. 2017;29:45–46. [Google Scholar]
- 20.Lee DW, Postle RL. Isozyme variation in Colobanthus quitensis (Kunth) Bartl.: Methods and preliminary analysis. Brit. Antarct. Surv. B. 1975;41–42:133–137. [Google Scholar]
- 21.Gianoli E, et al. Ecotypic differentiation in morphology and cold resistance in populations of Colobanthus quitensis (Caryophyllaceae) from the Andes of Central Chile and the Maritime Antarctic. Arct. Antarct. Alp. Res. 2004;36:484–489. [Google Scholar]
- 22.Acuña-Rodríguez IS, Oses R, Cortés-Vasquez J, Torres-Díaz C, Molina-Montenegro MM. Genetic diversity of Colobanthus quitensis across the Drake Passage. Plant Genet. Resour. C. 2014;12:147–150. [Google Scholar]
- 23.Androsiuk P, Chwedorzewska KJ, Szandar K, Giełwanowska I. Genetic variability of Colobanthus quitensis from King George Island (Antarctica) Pol. Polar Res. 2015;36:281–295. [Google Scholar]
- 24.Koc J, et al. Range-wide pattern of genetic variation in Colobanthus quitensis. Polar Biol. 2018;41:2467–2479. [Google Scholar]
- 25.Kang Y, et al. The complete chloroplast genome of Antarctic pearlwort Colobanthus quitensis (Kunth) Bartl. Mitochondrial DNA A. 2016;27:4677–4678. doi: 10.3109/19401736.2015.1106498. [DOI] [PubMed] [Google Scholar]
- 26.Androsiuk P, et al. The complete chloroplast genome of Colobanthus apetalus (Labill.) Druce: Genome organization and comparison with related species. PeerJ. 2018;6:e4723. doi: 10.7717/peerj.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuba-Díaz M, Cerda G, Rivera C, Gómez A. Genome size comparison in Colobanthus quitensis populations show differences in species ploidy. Polar Biol. 2017;40:1475–1480. [Google Scholar]
- 28.Dong W, Xu C, Cheng T, Zhou S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE. 2013;8:e77965. doi: 10.1371/journal.pone.0077965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988;75(10):1443–1458. [Google Scholar]
- 30.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008;49:827–831. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabir J, et al. Evolutionary and biotechnology implications of plastid genome variation in the inverted repeat lacking clade of legumes. Plant Biotechnol. J. 2014;12(6):743–754. doi: 10.1111/pbi.12179. [DOI] [PubMed] [Google Scholar]
- 34.Ruhlman. T. & Jansen, R. The plastid genomes of flowering plants. In Chloroplast Biotechnology (ed. Maglia, P.) 3–38 (Humana Press, 2014). [DOI] [PubMed]
- 35.Nock CJ, et al. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 2011;9:328–333. doi: 10.1111/j.1467-7652.2010.00558.x. [DOI] [PubMed] [Google Scholar]
- 36.Kane N, et al. Ultrabarcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 2012;99:320–329. doi: 10.3732/ajb.1100570. [DOI] [PubMed] [Google Scholar]
- 37.Park I, Yang S, Choi G, Kim WJ, Moon BC. The complete chloroplast genome sequences of Aconitum pseudolaeve and Aconitum longecassidatum, and development of molecular markers for distinguishing species in the Aconitum subgenus Lycoctonum. Molecules. 2017;22:E2012. doi: 10.3390/molecules22112012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. [Google Scholar]
- 40.Hollingsworth PM, et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dillenberger MS, Kadereit JW. Maximum polyphyly: Multiple origins and delimitation with plesiomorphic characters require a new circumscription of Minuartia (Caryophyllaceae) Taxon. 2014;63(1):64–88. [Google Scholar]
- 42.Smissen RD, Clement JC, Garnock-Jones PJ, Chambers GK. Subfamilial relationships within Caryophyllaceae as inferred from 5' ndhF sequences. Am. J. Bot. 2002;89(8):1336–1341. doi: 10.3732/ajb.89.8.1336. [DOI] [PubMed] [Google Scholar]
- 43.Palmer JD. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 44.Jansen RK, Ruhlman TA. Plastid Genomes of Seed Plants. Berlin: Springer Press; 2012. [Google Scholar]
- 45.Dong W, Xu C, Cheng T, Lin K, Zhou S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013;5(5):989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Messing J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE. 2011;6(9):e24670. doi: 10.1371/journal.pone.0024670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 48.Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. Analysis of the Amborella trichopoda chloroplast genome sequence suggest that amborella is not a basal angiosperm. Mol. Biol. Evol. 2003;20(9):1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- 49.Hupfer H, et al. Complete nucleotide sequence of the Oenothera elata plastid genome representing plastome I of the five distinguishable Euoenothera plastomes. Mol. Gen. Genet. 2000;263(4):581–585. doi: 10.1007/pl00008686. [DOI] [PubMed] [Google Scholar]
- 50.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- 51.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erixon P, Oxelman B. Reticulate or tree-like chloroplast DNA evolution in Sileneae (Caryophyllaceae)? Mol. Phylogenet. Evol. 2008;48:313–325. doi: 10.1016/j.ympev.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 53.Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol. Evol. 2012;4:294–306. doi: 10.1093/gbe/evs006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sloan DB, et al. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae) Mol. Phylogenet. Evol. 2014;72:82–89. doi: 10.1016/j.ympev.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Kapralov MV, Filatov DA, et al. Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol. Biol. 2007;7:73. doi: 10.1186/1471-2148-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapralov MV, Filatov DA. Molecular adaptation during adaptive radiation in the Hawaiian endemic genus Schiedea. PLoS ONE. 2006;1:e8. doi: 10.1371/journal.pone.0000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iida S, et al. Molecular adaptation of rbcL in the heterophyllous aquatic plant Potamogeton. PLoS ONE. 2009;4:e4633. doi: 10.1371/journal.pone.0004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kapralov MV, Smith JAC, Filatov DA. Rubisco evolution in C4 eudicots: An analysis of Amaranthaceae sensu lato. PLoS ONE. 2012;7:e52974. doi: 10.1371/journal.pone.0052974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christin PA, et al. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Mol. Biol. Evol. 2008;25:2361–2368. doi: 10.1093/molbev/msn178. [DOI] [PubMed] [Google Scholar]
- 60.Galmes J, et al. Environmentally driven evolution of Rubisco and improved photosynthesis and growth within the C3 genus Limonium (Plumbaginaceae) New Phytol. 2014;203:989–999. doi: 10.1111/nph.12858. [DOI] [PubMed] [Google Scholar]
- 61.Vogel J, Borner T, Hess W. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hao DC, Chen SL, Xiao PG. Molecular evolution and positive Darwinian selection of the 520 chloroplast maturase matK. J. Plant Res. 2010;123:241–247. doi: 10.1007/s10265-009-0261-5. [DOI] [PubMed] [Google Scholar]
- 63.Logacheva MD, Penin AA, Samigullin TH, Vallejo-Roman CM, Antonov AS. Phylogeny of flowering plants by the chloroplast genome sequences: In search of a “Lucky Gene”. Biochemistry. 2007;72:1324–1330. doi: 10.1134/s0006297907120061. [DOI] [PubMed] [Google Scholar]
- 64.Krawczyk K, Sawicki J. The uneven rate of the molecular evolution of gene sequences of DNA-dependent RNA polymerase I of the genus Lamium L. Int. J. Mol. Sci. 2013;14:11376–11391. doi: 10.3390/ijms140611376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng S, et al. The complete chloroplast genome sequences of six Rehmannia species. Genes. 2017;8:103. doi: 10.3390/genes8030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blazier JC, et al. Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci. Rep. 2016;6:24595. doi: 10.1038/srep24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cavalier-Smith T. Chloroplast evolution: Secondary symbiogenesis and multiple losses. Curr. Biol. 2002;12:R62–R64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- 68.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of diverged regions and categorization of shared repeats. Am. J. Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- 69.Kang JS, Lee BY, Kwak M. The complete chloroplast genome sequences of Lychnis wilfordii and Silene capitata and comparative analyses with other Caryophyllaceae genomes. PLoS ONE. 2017;12(2):e0172924. doi: 10.1371/journal.pone.0172924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang AH, Zhang JJ, Yao XH, Huang HW. Chloroplast microsatellite markers in Liriodendron tulipifera (Magnoliaceae) and cross-species amplification in L. chinense. Am. J. Bot. 2011;98(5):e123–e126. doi: 10.3732/ajb.1000532. [DOI] [PubMed] [Google Scholar]
- 71.Jiao Y, et al. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra) BMC Genomics. 2012;13:201. doi: 10.1186/1471-2164-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, et al. The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuang DY, et al. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implications for DNA barcoding and population genetics. Genome. 2011;54(8):663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- 74.Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK. Genome-wide distribution and organization of microsatellites in plants: An insight into marker development in Brachypodium. PLoS ONE. 2011;6(6):e21298. doi: 10.1371/journal.pone.0021298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knoop V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol. Life Sci. 2011;68:567–586. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsunewaki K, Matsuoka Y, Yamazaki Y, Ogihara Y. Evolutionary dynamics of wheat mitochondrial gene structure with special remarks on the origin and effects of RNA editing in cereals. Genes Genet. Syst. 2008;83(4):301–320. doi: 10.1266/ggs.83.301. [DOI] [PubMed] [Google Scholar]
- 77.Sasaki Y, Kozaki A, Ohmori A, Iguchi H, Nagano Y. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem. 2001;276(6):3937–3940. doi: 10.1074/jbc.M008166200. [DOI] [PubMed] [Google Scholar]
- 78.Knill T, Reichelt M, Paetz C, Gershenzon J, Binder S. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 2009;71(3):227–239. doi: 10.1007/s11103-009-9519-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He Y, et al. A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 2009;60(4):679–690. doi: 10.1111/j.1365-313X.2009.03990.x. [DOI] [PubMed] [Google Scholar]
- 80.Yu XQ, Drew BT, Yang JB, Gao LM, Li DZ. Comparative chloroplast genomes of eleven Schima (Theaceae) species: Insights into DNA barcoding and phylogeny. PLoS ONE. 2017;12:e0178026. doi: 10.1371/journal.pone.0178026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaw J, et al. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- 82.Song Y, et al. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 2015;6:662. doi: 10.3389/fpls.2015.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greenberg AK, Donoghue MJ. Molecular systematics and character evolution in Caryophyllaceae. Taxon. 2011;60(6):1637–1652. [Google Scholar]
- 84.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: Diversity, evolution and applications in genetic engineering. Genome Biol. 2016;17(1):134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tonti-Filippini J, Nevill PG, Dixon K, Small I. What can we do with 1000 plastid genomes? Plant J. 2017;90(4):808–818. doi: 10.1111/tpj.13491. [DOI] [PubMed] [Google Scholar]
- 87.Fior S, Karis PO, Casazza G, Minuto L, Sala F. Molecular phylogeny of the Caryophyllaceae (Caryophyllales) inferred from chloroplast matK and nuclear rDNA ITS sequences. Am. J. Bot. 2006;93:399–411. doi: 10.3732/ajb.93.3.399. [DOI] [PubMed] [Google Scholar]
- 88.West, J. G. & Cowley, K. J. Colobanthus. In Flora of Victoria. Vol. 3, Dicotyledons Winteraceae to Myrtaceae (ed. Walsh, N. G. & Entwisle, T. J.) (Inkata Press, 1996).
- 89.Mantowani A, Vieira RC. Leaf micromorphology of Antarctic pearlwort Colobanthus quitensis (Kunth) Bartl. Polar Biol. 2000;23:531–538. [Google Scholar]
- 90.Kearse M, et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 92.Kurtz S, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mayer, C. Phobos Version 3.3.11. A tandem repeat search program. 2006–2010. https://www.rub.de/spezzoo/cm/cm_phobos.htm (2019).
- 94.Sablok G, et al. ChloroMitoSSRDB 2.00: More genomes, more repeats, unifying SSRs search patterns and on-the-fly repeat detection. Database. 2015;2015:av084. doi: 10.1093/database/bav084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rozas J, et al. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 98.Mower JP. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 101.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 102.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 103.Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6(5):283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- 104.Gurusamy R, Lee DH, Park S. The complete chloroplast genome sequence of Dianthus superbus var. longicalycinus. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27(3):2015–2017. doi: 10.3109/19401736.2014.974167. [DOI] [PubMed] [Google Scholar]
- 105.Yao G, et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenet. Evol. 2019;134:74–86. doi: 10.1016/j.ympev.2018.12.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete chloroplast genomes of the six Colobanthus species have been submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/https://www.ncbi.nlm.nih.gov/) under the accession numbers: MN273320 for C. acicularis, MN273318 for C. affinis, MN273317 for C. lycopodioides, MN273316 for C. nivicola, MN273315 for C. pulvinatus and MN273319 for C. subulatus.