R-loop dynamics occur before changes in gene expression and are required for somatic cell reprogramming.
Abstract
R-loops modulate genome stability and regulate gene expression, but the functions and the regulatory mechanisms of R-loops in stem cell biology are still unclear. Here, we profiled R-loops during somatic cell reprogramming and found that dynamic changes in R-loops are essential for reprogramming and occurred before changes in gene expression. Disrupting the homeostasis of R-loops by depleting RNaseH1 or catalytic inactivation of RNaseH1 at D209 (RNaseH1D209N) blocks reprogramming. Sox2, but not any other factor in the Yamanaka cocktail, overcomes the inhibitory effects of RNaseH1 activity loss on reprogramming. Sox2 interacts with the reprogramming barrier factor Ddx5 and inhibits the resolvase activity of Ddx5 on R-loops and thus facilitates reprogramming. Furthermore, reprogramming efficiency can be modulated by dCas9-mediated RNaseH1/RNaseH1D209N targeting the specific R-loop regions. Together, these results show that R-loops play important roles in reprogramming and shed light on the regulatory module of Sox2/Ddx5 on R-loops during reprogramming.
INTRODUCTION
The induction of pluripotent stem cells (iPSCs) from fibroblasts by the Yamanaka cocktail—Oct4, Sox2, Klf4, and c-Myc (OSKM)—provides a powerful system for studying the mechanisms controlling cell fate. Somatic cell reprogramming must overcome roadblocks to cell-fate transitions that inhibit this process. Chromatin states and gene transcription occur in a highly phased process during reprogramming. RNA polymerase II plays an essential role in modulating gene transcription, and while it engages at pluripotency promoters, it pauses and is released during reprogramming (1).
During transcription, nascent RNAs exit from the RNA channel of RNA polymerase and inappropriately anneal to homologous genomic loci, generating a structure known as an R-loop, which contains one RNA strand that hybridizes to a complementary DNA strand and displaced single-stranded DNA (ssDNA). R-loops are present across a wide range of organisms, from bacteria to mammals, and they are believed to have multifaceted Yin and Yang effects in biology (2). R-loops not only lead to conflicts between transcription and replication causing genome instability but also promote DNA replication (3). R-loops regulate antisense transcription and the pausing and termination of transcription (4, 5). In addition, R-loops are tightly coupled to chromatin modifications (6).
R-loop formation likely requires specific proteins to control levels of R-loops in cells. Some enzymes can bind directly to R-loops and regulate R-loop levels, including members of the RNaseH family, which can degrade RNA in R-loops (7). Loss of RNaseH has been found to cause defect in development (8), suggesting the importance of maintaining correct levels of R-loops in organisms. In addition, R-loop–associated factors can also regulate the levels of R-loops and safeguard genomic integrity. DNA topoisomerases differentially modulate R-loops in the genomes of a large variety of species (9, 10). In addition, some chromatin remodelers, nuclear pore components, and DNA repair factors are involved in preventing the formation of harmful R-loops (11–13).
Although R-loops have important biological functions in gene regulation and are properly controlled by R-loop–associated factors, much less has been reported regarding the roles of R-loops in stem cell biology, especially during the reprogramming of somatic cells into PSCs. In this study, we showed that R-loops are dynamically regulated during reprogramming and accumulated at the iPSC stage, becoming highly enriched at specific genomic loci. The loss of RNaseH1 activity inhibits reprogramming in a culture medium–independent manner. Single amino acid mutations of enzymatic regions at D209 within RNaseH1 stabilize different fractions of R-loops, which are not the same as the normal accumulating R-loops in reprogramming, thereby blocking the reprogramming process. Factor selection indicates that Sox2, but not any other factor of the Yamanaka cocktail, is required for partially overcoming the inhibitory effects of harmful R-loops on reprogramming. Sox2, but not OKM, forms a complex with R-loop–associated complexes. Co-immunoprecipitation (co-IP) experiments for both Sox2 and the R-loop antibody S9.6 indicate that Ddx5 interacts with both Sox2 and R-loop complexes. We further found that Sox2 inhibits Ddx5-mediated R-loop–resolving capacity in vitro and during reprogramming. Site-specific modulating R-loops via dCas9 technology showed that targeting harmful R-loops by dCas9-RNaseH1 promoted reprogramming but inhibited reprogramming by dCas9-RNaseH1D209N. In consistent, our data indicated that targeting beneficial R-loops by dCas9-RNaseH1 blocked reprogramming but promoted reprogramming by dCas9-RNaseH1D209N, suggesting that RNaseH1D209N could stabilize R-loops in vivo. Together, these findings reveal that R-loops play a regulatory role during the reprogramming of somatic cells into pluripotent cells.
RESULTS
Genome-wide R-loops change dynamically during reprogramming
To investigate the genome-wide dynamics of R-loops during the reprogramming of mouse embryonic fibroblasts (MEFs) into iPSCs, we performed somatic cell reprogramming experiments in iCD1 medium by transfecting OG2 MEFs with OSKM (Fig. 1A, left); these MEFs bear an Oct4–green fluorescent protein (GFP) reporter whose expression reflects the establishment of pluripotency. Reprogramming intermediates of MEFs on days 0, 1, 3, 5, and 7, along with iPSCs, were harvested and subjected for RNA sequencing (RNA-seq) and ssDRIP-seq (ssDNA ligation–based library construction after DNA:RNA hybrid immunoprecipitation combined with next-generation sequencing) (14), a recently developed method for profiling the dynamics of R-loops, during reprogramming (Fig. 1A, right). After data processing, ssDRIP-seq in two biological replicates was basically identical except for some minor differences (fig. S1A). Principal component analysis (PCA) showed that gradual changes of R-loops occurred at the early stage of reprogramming, following large changes between days 3 and 5, and then R-loops changed markedly again once iPSCs were generated (fig. S1B). The PCA results of RNA-seq showed a dynamic pattern with data from ssDRIP-seq (fig. S1C). This similarity suggested that there might be a correlation between gene expression and R-loop dynamics during reprogramming. In summary, our data indicated that R-loops showed sharp changes at the initial and late stages of reprogramming but transient and subtle changes at the intermediate stage of reprogramming, leading to different R-loop patterns during iPSC generation.
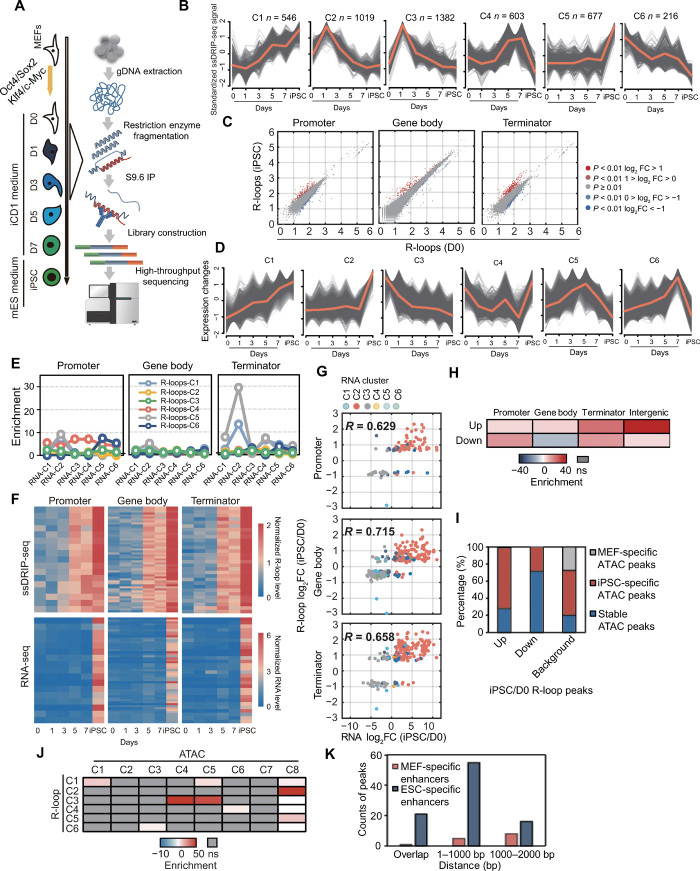
Fig. 1. Genome-wide detection of R-loops during reprogramming by ssDRIP-seq.
(A) Flowchart of ssDRIP-seq. (B) Fuzzy cluster analysis of ssDRIP-seq signals. Line plots show ssDRIP-seq signals, with individual loci (gray lines) and values of the cluster center (orange line). (C) Scatterplots of R-loop levels (iPSCs/D0). (D) Fuzzy cluster analysis of RNA-seq signals. Line plots show RNA-seq signal, with individual loci (gray lines) and values of the cluster center (orange line). (E) Colocalization analysis of R-loop clusters and different regions of RNA clusters. (F) Heatmaps of ssDRIP-seq and RNA-seq signals in different regions of RNA-C2 overlapped with R-loop-C1. (G) Scatterplots of RNA and R-loop (iPSCs/D0). The significantly changed genes are shown and colored by RNA clusters. (H) Colocalization analysis of ATAC peaks and differential R-loop regions (iPSCs/D0). (I) Percentages of differential R-loop regions (iPSCs/D0) overlapped with different ATAC peaks. MEF-specific, iPSC-specific, and stable ATAC peaks refer to peaks that were lost, gained, and unchanged during reprogramming, respectively. Background, the distribution of different ATAC peaks in all ATAC peaks. (J) Colocalization analysis of R-loop categories and ATAC clusters defined by Knaupp et al. (17). (K) Statistics of up-regulated R-loop regions (iPSCs/D0) with different distances from MEF- or ESC-specific enhancers. ns, not significant.
To investigate the potential roles of R-loops in reprogramming, we classified differential R-loop regions (fig. S1D; see Materials and Methods) and performed time-course fuzzy clustering using mFuzz (15). There were six clusters (C1 to C6) generated for differential R-loop regions (Fig. 1B). These six clusters fall into five major categories: first, R-loops are up-regulated persistently during the reprogramming process (C1); second, R-loops are instantaneously up-regulated after OSKM infection and down-regulated between days 1 and 7 (C2, C3); third, R-loops are gained at the late stage but reduced in the iPSC stage (C4); fourth, R-loops exhibit subtle changes during reprogramming but are greatly up-regulated in iPSCs (C5); and fifth, R-loops are gradually down-regulated during reprogramming (C6). These results indicated that R-loops are regulated dynamically and change along with reprogramming, suggesting that R-loops may be a vital feature in reprogramming.
We then analyzed the R-loop dynamics between iPSCs (final stage) and MEFs (beginning stage, day 0) by normalizing the ssDRIP-seq read counts on the promoter, gene body, and terminator regions with DESeq2 (16). The differential R-loop genes (DRGs) were defined by three types: promoter, gene body, and terminator (Prom-/Gb-/Term-) DRGs, which means a gene with differential R-loop level in its promoter, gene body, or terminator region (fig. S1E). Compared with MEFs, R-loops in iPSCs that resided in the promoter, gene body, and terminator showed variable changes, and more genes exhibited high level of R-loop in their terminator regions (Fig. 1C and fig. S1F). Moreover, most of the Term-DRGs were gained in the iPSC stage (fig. S1G). Together, these results indicate that R-loop occurs in waves during somatic cell reprogramming.
R-loop dynamics occur prior to changes in gene expression
To analyze the sequential order of R-loop dynamics and gene expression dynamics during reprogramming, we analyzed RNA-seq data and classified gene expression dynamics into six major clusters (RNA-C1 to RNA-C6; Fig. 1D). Then, we performed a permutation test to analyze the colocalization between gene expression and R-loop clusters. Our data indicated that R-loops in categories C1 and C5 showed strong colocalization with RNA-C2, while the colocalizations between R-loops in other categories and RNA clusters were weaker (Fig. 1E). In addition, a part of differential R-loop regions was mainly enriched at the promoter and terminator of differentially expressed genes (DEGs), respectively (Fig. 1E). The dynamic pattern of R-loops in category C5 was similar to the gene expression pattern of cluster RNA-C2 (Fig. 1, B and D). However, R-loop levels in category C1 exhibited a gradual increase (Fig. 1B), suggesting that there was a different relationship between C1 R-loops and RNA-C2, unlike the similar patterns seen for C5 R-loops and RNA-C2. With respect to dynamic R-loops and gene expression, our analysis demonstrated that R-loop levels at the promoter, gene body, and terminator DRGs changed earlier than the expression of colocalized genes during reprogramming (Fig. 1F), suggesting that the earlier R-loop changes might play an important role in regulating these genes. We further selected two genes belonging to cluster 1 of R-loops and performed nuclear run-on reverse transcription quantitative polymerase chain reaction (RT-qPCR) to measure nascent transcripts of these genes. Correlation analysis of nascent transcripts and R-loops (showing in Fig. 1B) indicated that R-loops also change earlier than nascent RNA transcription in Tdrd12 and Rif1 loci at different stages of reprogramming (fig. S1H). In addition, our results also indicated that there was a positive correlation between gene expression and R-loops (Fig. 1G), and most genes that increased in both R-loops and RNA level belong to the RNA-C2 category, while most genes with decreased R-loops and RNA level belong to the RNA-C3 category (fig. S1I). Together, our data indicate that R-loop dynamics correlate with the changes in gene expression, indicating that R-loops are functional changes and might be an important regulator for reprogramming.
Intergenic up-regulated R-loops are highly enriched at open chromatin and embryonic stem cell-specific enhancers during reprogramming
Genome-wide distribution analysis showed that about 50% of differential R-loop regions were located in the distal intergenic region (fig. S1J). It was reported that ~50% of total assay for transposase-accessible chromatin (ATAC) peaks and most dynamic ATAC peaks are located in the intergenic regions during reprogramming (17). Thus, we supposed that dynamic intergenic R-loops and ATAC peaks might be present at common chromatin regions. Our data showed that up-regulated intergenic R-loops were markedly enriched on open chromatin (Fig. 1H). We further investigated the colocalization between dynamic open chromatin and differential R-loop regions. Our results showed a high enrichment of up-regulated R-loops on iPSC-specific open chromatin and that down-regulated differential R-loop regions were enriched on stable ATAC peaks and a few differential R-loop regions were found to overlap with MEF-specific ATAC peaks (Fig. 1I). To gain further insight into R-loop dynamics and figure out the relationship between R-loop and chromatin accessibility during reprogramming, we intersected the R-loops from our ssDRIP-seq data with ATAC-seq clusters defined by Knaupp et al. (17). These data highlighted characteristics of R-loop and chromatin accessibility. R-loops in categories 1 and 3 were highly colocalized with iPSC-specific ATAC peaks (ATAC-C1, ATAC-C4 and ATAC-C5) (Fig. 1J and fig. S1K), which showed that R-loops were up-regulated during reprogramming. MEF-specific ATAC peaks in C6 and C7 showed a low level of R-loops and had no correlation with any R-loop category (Fig. 1J and fig. S1K). Furthermore, R-loops (C2) in stable ATAC clusters (ATAC-C8) exhibited gradual increase at the intermediate stage and reduction in iPSCs. Together, these results demonstrate that R-loops are closely related to chromatin accessibility during iPSC generation.
Enhancers were dynamically selected during reprogramming, which is accompanied by the silencing of somatic enhancers and the activation of pluripotent enhancers (18), and showed a differential dynamic relationship with chromatin accessibility (17). Considering that there is a dynamic relationship between open chromatin and enhancers, we further studied the relationship in the localization of differential R-loop regions and enhancers. The permutation test results showed that up-regulated differential R-loop regions were enriched on ESC-specific enhancers and depleted on MEF-specific enhancers (Fig. 1K). Besides overlapping directly, the up-regulated differential R-loop regions were also enriched in the nearby regions of ESC-specific enhancers (Fig. 1K). However, transient ATAC clusters (ATAC-C2 and ATAC-C3) composed of different enhancers coincided with a subtle gain of R-loops, which were down-regulated at the iPSC stage (fig. S1K). Previous reports indicated that enhancers can produce transcripts called enhancer RNA (eRNAs), and we supposed that enhancer RNA (eRNA) might be involved in enhancer function via R-loop. Permutation test and overlap analysis indicated that the genes with R-loop accumulation in enhancer regions showed strongly transcriptional up-regulation during reprogramming (fig. S1L). These data indicate that dynamic R-loops correlate well with dynamic enhancers in different cell types. Together, our data indicate that the dynamic intergenic R-loops show a positive relationship with open chromatin, and a part of these R-loops might be involved in enhancer functions.
Knockdown of RNaseH1 inhibits somatic cell reprogramming
R-loops can be cleaved directly by RNaseH1 (7). We noticed that RNaseH1 expression gradually increased during reprogramming (fig. S2A), suggesting that the R-loop cleavage activity of RNaseH1 may be needed during reprogramming. To investigate RNaseH1 function in reprogramming, we manipulated the activity of RNaseH1 in cells. First, we conducted two independent retroviral short hairpin RNA (shRNA) vectors to reduce RNaseH1 expression (fig. S2B). Then, we transduced OG2 MEFs with these shRNA vectors, together with OSKM, and found that RNaseH1 knockdown, along with overexpression of OSKM factors, led to a modest decrease in not only the number of alkaline phosphatase–positive (AP+) colonies but also the number of GFP-positive (GFP+) colonies (a late marker of reprogramming) (Fig. 2, A and B), indicating that RNaseH1 is required for reprogramming. To confirm these results, we performed reprogramming experiments by using another medium containing serum and vitamin C and found that RNaseH1 loss of function also led to a decrease in the number of both AP+ and GFP+ colonies (Fig. 2, C and D), suggesting that RNaseH1 loss of function inhibits reprogramming in a medium-independent manner.
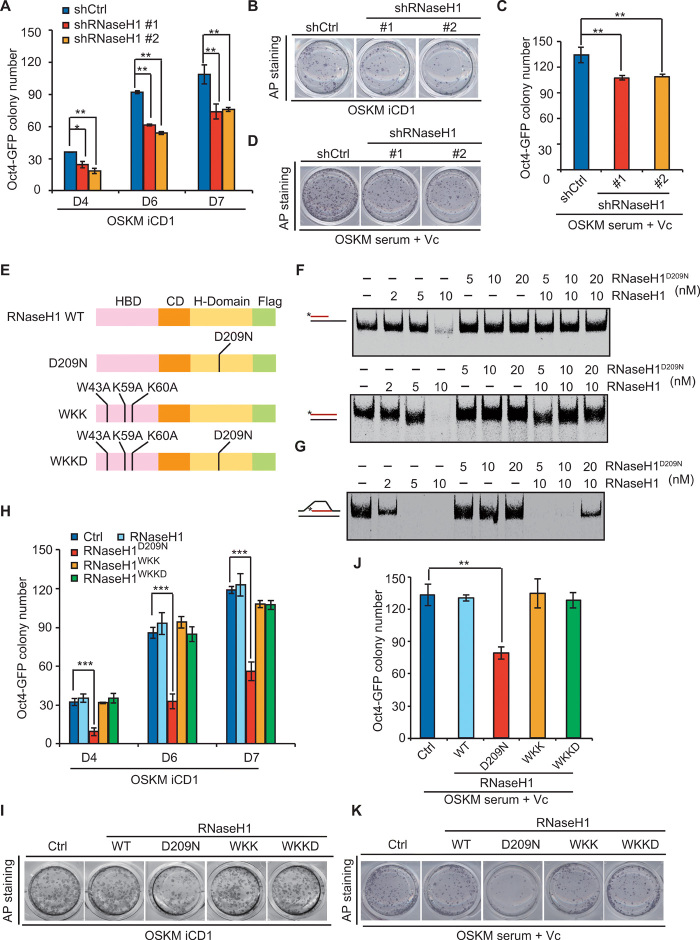
Fig. 2. Inhibition of RNaseH1 by catalytic-inactive mutant or knockdown impedes somatic cell reprogramming.
(A) Oct4-GFP colony number of MEFs reprogrammed with OSKM, shRNAs against control, or RNaseH1 in iCD1 medium. (B) AP staining corresponding to (A). (C) Oct4-GFP colony number of MEFs reprogrammed with OSKM, shRNAs against control, or RNaseH1 in serum + Vc medium. (D) AP staining corresponding to (C). (E) Schematic illustration of RNaseH1 and different mutants. HBD, RNA/DNA hybrid binding domain; CD, connection domain; H-Domain, catalytic domain. (F) RNA/DNA + ssDNA RNA/DNA substrate with Cy5-labeled (*) RNA (red) or DNA (black) was incubated with increasing concentrations of RNaseH1 and RNaseH1D209N. (G) R-loop substrate with Cy5-labeled (*) RNA (red) or DNA (black) was incubated with increasing concentrations of RNaseH1 and RNaseH1D209N. (H) Oct4-GFP colony number of MEFs reprogrammed with OSKM plus vehicle, RNaseH1, and the mutants in iCD1 medium. (I) AP staining corresponding to (H). (J) Oct4-GFP colony number of MEFs reprogrammed with OSKM, plus vehicle, RNaseH1, and the mutants in serum + Vc medium. (K) AP staining corresponding to (J). Error bars correspond to means ± SD (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, two-tailed Student’s t test).
The catalytic inactivation of RNaseH1 blocks reprogramming
Substitution of catalytic residue Asp210 with Asn abolishes the cleavage activity of human RNaseH1 (19). Sequence alignment of the catalytic domains of multiple RNaseH endonucleases in different species indicated that the catalytically conserved residue in mouse is Asp209 (fig. S2C). Therefore, we cloned mouse RNaseH1 and generated constructs with a mutation of amino acid 209 from Asp (D) to Asn (N) in the RNaseH1 catalytic domain (D209N), a mutation of amino acids [43 from Trp (W) to Ala (A), 59 from Lys (K) to Ala (A), and 60 from Lys (K) to Asn (A)] in the binding domain (WKK), which prevents RNaseH1 from binding to R-loops (20), and a mutation of both binding region (WKK to AAA) and catalytically conserved region (D209N) together (WKKD) (Fig. 2E). Our in vitro experiments showed that the mutation of D209N of RNaseH1 abolished the catalytic activity of RNaseH1 but protected both RNA/DNA hybrids (Fig. 2F) and R-loop structure (Fig. 2G), while RNaseH1WKK and RNaseH1WKKD had no effect on RNaseH1 activity (fig. S2D). We then investigated the effect of WT RNaseH1 and different mutations on reprogramming efficiency. We surprisingly found that RNaseH1 overexpression had no effect on reprogramming efficiency (Fig. 2, H and I and fig. S2E), likely because of redundancy of RNaseH1 in the endogenous level. Only RNaseH1D209N, but not other mutations, notably inhibited the formation of both AP+ and GFP+ colonies compared with the controls in iCD1 medium (Fig. 2, H and I). Furthermore, we examined the levels of endogenous RNaseH1 and exogenous RNaseH1D209N. The Western blot experiments showed that the level of RNaseH1D209N was about 20 times higher than the level of endogenous RNaseH1 (fig. S2F). Consistent with these results, RNaseH1D209N was also found to inhibit reprogramming in another reprogramming culture (serum + Vc medium system) (Fig. 2, J and K).
In view of the function of R-loop on DNA replication and DNA damage, we detected the expression of genes that are associated with DNA replication and examined γH2A.X level during reprogramming. Our data indicated that both overexpression of RNaseH1D209N and RNaseH1 knockdown resulted in little change on the expression of DNA replication–associated genes (fig. S2, G and H). Moreover, both RNaseH1 loss and RNaseH1D209N overexpression did not affect γH2A.X level (fig. S2, I and J). Moreover, cell apoptosis analysis showed that both RNaseH1 knockdown and RNaseH1D209N overexpression had no significant changes on cell death in contrast to the controls (fig. S2, K and L). In summary, we conclude that both RNaseH1 knockdown and RNaseH1 mutation (D209N) inhibit reprogramming independent of DNA replication, DNA damage, as well as cell apoptosis.
To confirm these results, we further performed reprogramming experiments by using the TetON lentivirus system (fig. S3A), which have four genes (Oct4, Sox2, Klf4, and c-Myc) constructed in one single vector. As expected, RNaseH1 knockdown remarkably reduced reprogramming efficiency (fig. S3, B and C). Overexpression of RNaseH1D209N abolished reprogramming, while other mutants of RNaseH1 caused a very subtle or no effect on reprogramming (fig. S3, D and E). Together, our data indicate that inhibiting the activity of RNaseH1 blocks reprogramming, which may be caused by dysregulation of R-loop balance in cells.
RNaseH1D209N induces R-loop accumulation and changes gene expression during reprogramming
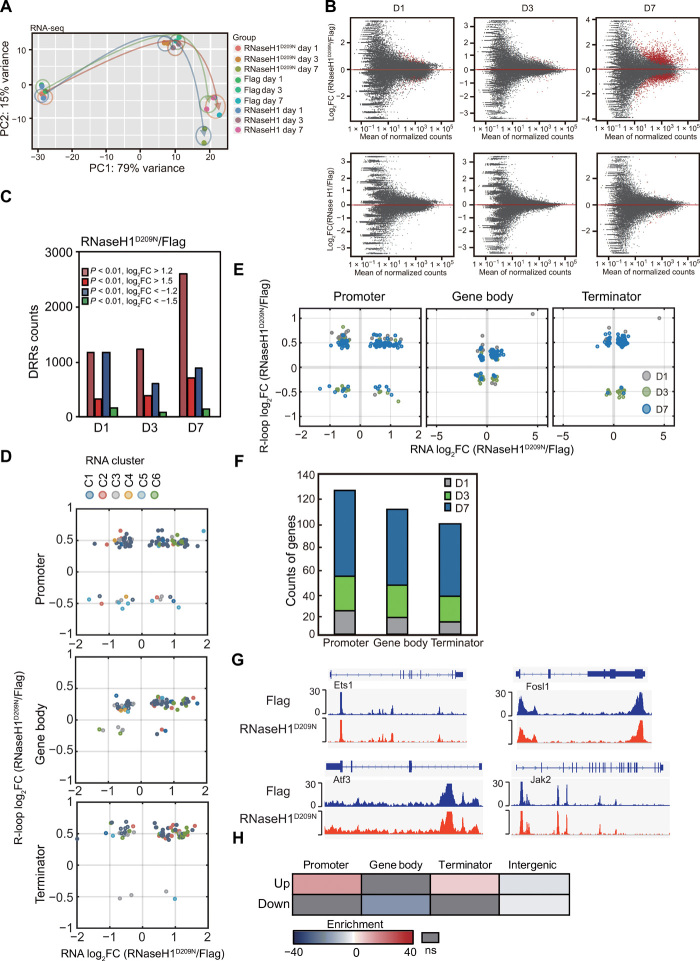
To examine the mechanism underlying RNaseH1D209N-mediated inhibition of reprogramming, we performed RNA-seq with OSKM plus Flag, RNaseH1, and RNaseH1D209N, respectively. RNA-seq data revealed that RNaseH1 plus OSKM had little effect on gene expression in contrast to OSKM plus Flag control at different stages of reprogramming (Fig. 3, A and B), consistent with the phenotype that RNaseH1 gain of function has no effect on reprogramming. Overexpression of RNaseH1D209N had a subtle effect on gene expression compared with Flag control at the early stage (D1 to D3) of reprogramming (Fig. 3, A and B). However, RNaseH1D209N led to a significant change in gene expression patterns compared with control at the late stage (D7) of reprogramming (Fig. 3, A and B). Similarly, the changes in gene expression patterns of RNaseH1D209N to RNaseH1 were identical to those of RNaseH1D209N to Flag (fig. S4A).
Fig. 3. RNaseH1D209N induces R-loop accumulation and changes gene expression during reprogramming.
(A) PCA of RNA-seq signals in cells transfected with OSKM and Flag, RNaseH1, or RNaseH1D209N during reprogramming. (B) MA plots showing the DEGs (red dots) of RNaseH1D209N/Flag and RNaseH1/Flag during reprogramming. (C) Statistics of up- or down-regulated differential R-loop regions (DRRs) of RNaseH1D209N compared with Flag during reprogramming. (D) Scatterplots of gene expression and R-loop log2 fold change (RNaseH1D209N/Flag, D7). (E) Scatterplots of RNA (x axis, D7) and promoter, gene body, or terminator R-loop (y axis, D1, D3, D7) of RNaseH1D209N compared with controls on concurrently changed genes. (F) Counts of Prom-/Gb-/Term-concurrently changed genes shown in (E) on D1, D3, and D7 during reprogramming. (G) Snapshots of ATAC-seq of MEFs reprogrammed with OSKM plus Flag and RNaseH1D209N on day 7 at representative genomic loci. (H) Colocalization analysis of total ATAC peaks (RNaseH1D209N + Flag, D7) and up- or down-regulated differential R-loop regions (RNaseH1D209N/Flag, D7) in promoter, gene body, terminator, and intergenic regions.
To study whether there is any correlation between gene expression and R-loop accumulation after RNaseH1D209N gain of function during reprogramming, we performed ssDRIP-seq experiments and profiled R-loop dynamics in the samples from OSKM plus Flag, RNaseH1, and RNaseH1D209N during reprogramming. We generated more than 40 million reads for each sample and found that RNaseH1D209N led to R-loop accumulation during reprogramming (Fig. 3C), except a part of R-loops was reduced. We then analyzed fold changes of R-loop peaks in the intergenic and genic regions of genes. Our data showed that RNaseH1D209N led to increases in both intergenic and genic R-loops (fig. S4B). Gene Ontology analysis showed that DRGs up-regulated by RNaseH1D209N were associated with a somatic cell state and were incompatible with reprogramming [e.g., oxidative phosphorylation and SUMOylation (fig. S4C)]. Down-regulated DRGs were mainly associated with ectodermal genes (fig. S4D). Together, these results suggest that RNaseH1D209N inhibits reprogramming by inducing the accumulation of aberrant R-loops of somatic cell-associated genes.
Furthermore, bioinformatics analysis showed that a large fraction (~60%) of genes were hyper-activated by RNaseH1D209N and most of these genes belong to the RNA-C6 category of genes (Fig. 3D and fig. S4E), which were normally activated at the early and middle stages but suppressed at the later stage of reprogramming (Fig. 1D). As our data suggested that R-loops change earlier than gene expression during reprogramming, we further examined the correlation between RNaseH1D209N-induced DRGs on days 1, 3, or 7 and DEGs on day 7. We found that RNaseH1D209N resulted in ~40% DEGs showing differential R-loop levels at the early stage of reprogramming (Fig. 3, E and F). These results suggest that a fraction of early R-loops regulate gene expression at the late stage of reprogramming.
As mentioned above, intergenic R-loop dynamics was strongly related with chromatin accessibility and enhancers during reprogramming; therefore, we wondered whether intergenic differential R-loop regions induced by RNaseH1D209N have similar epigenetic features. To address this question, we performed ATAC-seq experiments in both Flag and RNaseH1D209N reprogrammed cells. We surprisingly found that RNaseH1D209N had little effect on chromatin accessibility and dynamics of chromatin accessibility induced by RNaseH1D209N (~6% peaks are lost, ~7% peaks are gained) was much weaker than that during normal reprogramming (~40% ATAC peaks are lost and ~60% new ATAC peaks are gained) (Fig. 3G and fig. S4F). Permutation test results showed that differential R-loop regions induced by RNaseH1D209N were slightly enriched at promoter and terminator ATAC peaks (Fig. 3H) and that there was no significant colocalization between dynamic ATAC peaks and differential R-loop regions induced by RNaseH1D209N. These results indicate that RNaseH1D209N-accumulated R-loops could exist at open chromatin and that R-loop is not required for opening chromatin in cis. Together, our data indicate that RNaseH1D209N disrupts reprogramming by up-regulating aberrant R-loops on genes.
Inhibitory effect of RNaseH1 activity loss on reprogramming could be rescued by RNaseH1 overexpression
To investigate whether RNaseH1 could overcome the inhibitory effect of RNaseH1 activity loss on reprogramming, we performed several rescue experiments. Our data showed that ectopic expression of RNaseH1 could override the reprogramming-inhibiting effect by RNaseH1 knockdown, but the rescue reprogramming efficiency was similar to OSKM control, indicating that RNaseH1 is required but is not enough for promoting reprogramming (fig. S5, A and B). Furthermore, we found that overexpression of RNaseH1 could overcome the inhibitory effects of RNaseH1D209N on reprogramming (fig. S5, C and D). Together, these data suggest that restoring RNaseH1 could recover the blocking effect of RNaseH1 activity loss on reprogramming.
Sox2 partially rescues RNaseH1D209N inhibitory effect on reprogramming
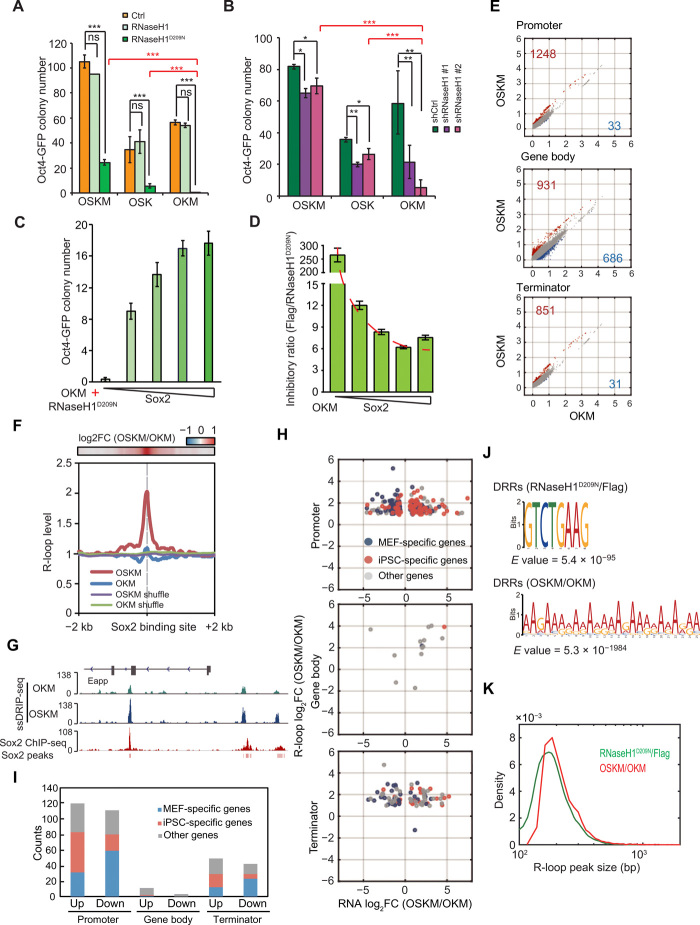
Each factor in the Yamanaka cocktail plays a different role in somatic cell reprogramming (21); therefore, we were curious to see whether the dysregulation of R-loops by RNaseH1D209N overexpression or RNaseH1 knockdown could be counteracted by any of the four transcription factors. In addition, down-regulated DRGs induced by RNaseH1D209N were mainly related to neuroectoderm lineage (fig. S4D); thus, we supposed that RNaseH1D209N might attenuate the function of Sox2 during reprogramming. To test this, we performed reprogramming experiments with either RNaseH1D209N overexpression or RNaseH1 knockdown, together with O/S/K/M in different combinations: OSKM/OSK/OKM/OSM (OSM generated very few GFP+ colonies in all treatments). Our data indicated that OKM plus RNaseH1D209N produced fewer GFP+ colonies than OKM alone or OKM plus RNaseH1, while a reprogramming system having Sox2 within OSKM or within OSK and RNaseH1D209N produced more iPSC colonies than a reprogramming system with OKM without Sox2 (Fig. 4A and fig. S6A). Alternatively, knockdown of RNaseH1 in the OKM-reprogramming system remarkably inhibited the generation of Oct4-GFP+ and AP+ colonies more than the reprogramming system with either OSKM or OSK plus RNaseH1 knockdown (Fig. 4B and fig. S6B).
Fig. 4. Sox2 partially rescues the inhibitory effect of aberrant R-loops on reprogramming.
(A) Oct4-GFP colony number of MEFs reprogrammed with OSKM, OSK, OKM plus Flag, RNaseH1, or RNaseH1D209N. (B) Oct4-GFP colony number of MEFs reprogrammed with OSKM, OSK, OKM, and shRNAs against control or RNaseH1. (C) Oct4-GFP colony number of MEFs reprogrammed with OKM, RNaseH1D209N, plus various concentrations of Sox2 virus (0, 50, 100, 150, and 200%). (D) Inhibitory ratio of reprogramming corresponding to (C). (E) Scatterplots of R-loop (OSKM/OKM, D9). The counts of up- or down-regulated DRGs are shown. (F) The metaplot of R-loop of MEFs, reprogrammed with OKM and OSKM, on Sox2 binding sites. (G) Snapshots of ssDRIP-seq of MEFs reprogrammed with OKM and OSKM on Sox2 binding peaks. (H) Scatterplots of RNA and R-loop (OSKM/OKM, D9). The concurrently changed genes are shown and classified as MEF-specific, iPSC-specific, or other genes. (I) Statistics of concurrently changed genes shown in (H). (J) DNA motif of differential R-loop regions of RNaseH1D209N/Flag and OSKM/OKM. (K) Peak size distribution of differential R-loop of RNaseH1D209N/Flag and OSKM/OKM. Error bars correspond to means ± SD (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, two-tailed Student’s t test).
To assess whether Sox2 could overcome the inhibitory effects of RNaseH1 activity loss on reprogramming, we gradually increased the amount of Sox2 during reprogramming MEFs toward iPSCs in the OKM plus RNaseH1D209N-inducing system. Our data indicated that the number of Oct4-GFP+ colonies increased as the amount of Sox2 used was gradually increased (Fig. 4C) and that the inhibitory ratio of RNaseH1D209N on reprogramming could be gradually overridden by increasing the amount of Sox2 added (Fig. 4D). To rule out that Oct4, Klf4, and c-Myc have no capacity to overcome the effect of RNaseH1D209N on reprogramming, we performed additional reprogramming experiments with Yamanaka factors plus RNaseH1D209N with the increasing amount of Oct4, Sox2, Klf4, or c-Myc via a doxycycline-induced dose-dependent expression system, respectively. Our results showed that only Sox2 but not Oct4, Klf4, and c-Myc continuously attenuated the inhibitive function of RNaseH1D209N on reprogramming (fig. S6, C and D). One possible explanation for these data is that Sox2 regulates R-loop and rescues the inhibitory effects of RNaseH1D209N on reprogramming.
Sox2 helps maintain beneficial R-loop levels during reprogramming
Because Sox2 partially neutralizes the inhibitory effect of RNaseH1D209N on reprogramming, we wondered whether Sox2 has an antagonistic role in RNaseH1D209N-mediated accumulation of R-loops. Therefore, we performed ssDRIP-seq and RNA-seq in OKM and OSKM cells to examine the genome-wide effects of Sox2 on R-loops in reprogramming. RNA-seq results showed marked differences in gene expression in cells reprogrammed with OSKM and OKM (fig. S6E), indicating that Sox2 induces a series of gene expression alternations for improving reprogramming. ssDRIP-seq results showed that there was a strong increase in R-loops on gene promoter and terminator regions in the OSKM system compared with the OKM system, while the decreased R-loops on gene body in the OSKM system are comparable with the increased R-loops in the OKM system (Fig. 4E). As Sox2 is a transcription factor, we wondered whether Sox2 could increase R-loop levels by binding directly to its target sites. Based on our analysis on ssDRIP-seq data and published Sox2 chromatin immunoprecipitation sequencing (ChIP-seq) data (17), we found that R-loops accumulated markedly at Sox2 binding sites in OSKM-reprogrammed MEFs compared with OKM-reprogrammed MEFs (Fig. 4, F and G).
OSKM-reprogrammed cells have more R-loops in DEGs compared with OKM-reprogrammed cells without Sox2 (Fig. 4H). In addition, we found that more pluripotent genes were up-regulated, while more MEF-specific genes were down-regulated in OSKM but not in OKM-reprogrammed cells (fig. S6F). In addition, differential R-loop regions of these DEGs were enriched at promoter and terminator regions (Fig. 4I). These results indicate that Sox2 induces reprogramming by maintaining a high level of R-loops at promoter or terminator regions of pluripotent genes.
The above results indicate that R-loops could be up-regulated by overexpressing RNaseH1D209N or Sox2. However, R-loops induced by RNaseH1D209N have a harmful effect on reprogramming, while Sox2-mediated R-loops facilitate reprogramming. Thus, we hypothesized that RNaseH1D209N and Sox2 might target different R-loops in the genome during reprogramming. The intersection results showed few overlapping DRGs between RNaseH1D209N and Sox2 (fig. S6G). We hypothesized that R-loop patterns induced by RNaseH1D209N and Sox2 were regulated by different mechanisms. To test this, we then searched for the motifs enriched in differential R-loop regions regulated by RNaseH1D209N and Sox2, respectively. AR (where R = purine G or A) motif was enriched in R-loops regulated by Sox2 (Fig. 4J), consistent with more R-loops in the promoter region and less in the gene body and intergenic region (fig. S6H). However, GTCTGAAC motif was found in RNaseH1D209N-regulated R-loops. Furthermore, Sox2 regulated larger R-loops than RNaseH1D209N (Fig. 4K and fig. S6I). The differences between the motif of OSKM/OKM and RNaseH1D209N/Flag might be the differential R-loop targets of RNaseH1 and Sox2. These results indicate that both RNaseH1D209N and Sox2 regulate a different fraction of R-loops that are required for reprogramming.
Sox2 blocks the activity of Ddx5 at R-loops
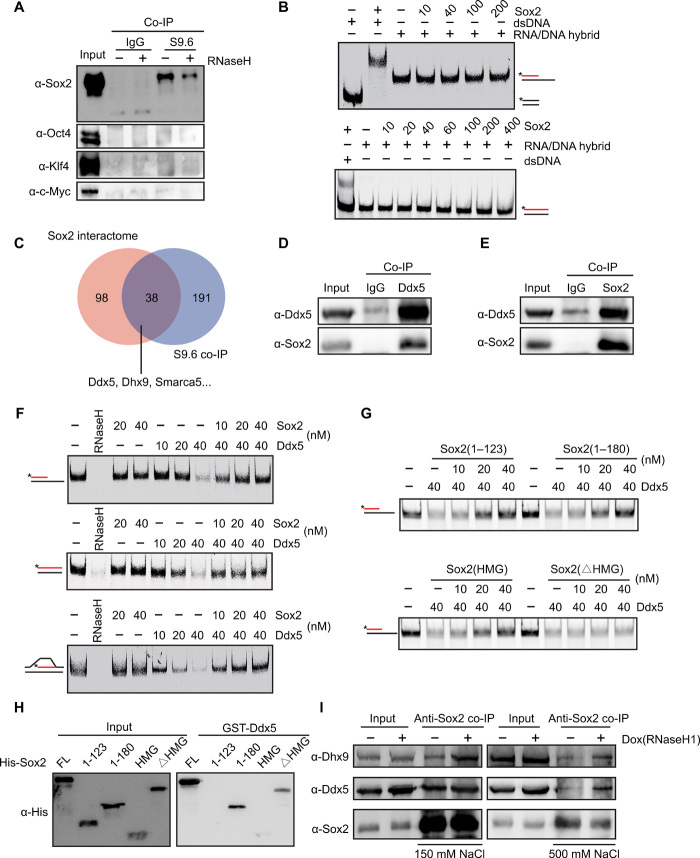
To determine the function of Sox2 on R-loops during reprogramming, co-IP experiments were carried out by using the S9.6 antibody. The results showed that Sox2, but not OKM, is associated with R-loop–associated protein complexes (Fig. 5A), suggesting that Sox2 is directly involved in the regulation of R-loops. It has been reported that Sox2 could bind to not only double-stranded DNA but also single-stranded RNA (22). We were curious to see whether Sox2 could bind to and regulate R-loops directly or indirectly. Electrophoretic mobility shift assay (EMSA) showed that Sox2 could not bind to RNA/DNA hybrid directly (Fig. 5B). It has been reported that R-loop could be regulated by R-loop-interacting cofactors (23); therefore, we wondered whether Sox2 might regulate R-loop by interacting with other factors that are involved in processing R-loop. To address this, we performed S9.6 RNA/DNA hybrid immunoprecipitation experiments followed by mass spectrometry to identify RNA/DNA hybrid–associated protein cofactors. By analyzing our RNA/DNA hybrid interactome and the Sox2 interactome reported previously (24, 25), we identified Ddx5 and Dhx9 as the potential cofactors of R-loops (Fig. 5C). To verify the data with mass spectrometry, we performed co-IP experiments and found that Sox2 interacted with Ddx5 (Fig. 5, D and E). These data support the previous finding that Ddx5 interacts with Sox2 (26). Similarly, the interaction between Sox2 and Dhx9 was confirmed by Western blotting (fig. S7A). It has been shown that Sox2 interacts with many RNA binding proteins, which are involved in R-loop metabolism (27, 28). Together, our results suggest that Sox2 might regulate R-loop via its chaperones.
Fig. 5. Sox2 blocks the activity of Ddx5 at R-loops.
(A) Western blot to examine the interactions between Yamanaka factors and R-loop–associated complexes via S9.6 co-IP. (B) RNA/DNA + ssDNA or DNA/DNA substrate (top, 25 nM), or RNA/DNA or DNA/DNA substrate (bottom, 25 nM) with Cy5-labeled (*) RNA (red) or DNA (black) was incubated with increasing concentrations of Sox2. (C) Overlap between Sox2 interactome and RNA/DNA hybrid interactome in mESCs. (D) Co-IP to detect the interaction between Ddx5 and Sox2 in mESCs by using anti-Ddx5 antibody. (E) Co-IP to detect the interaction between Ddx5 and Sox2 in mESCs by using anti-Sox2 antibody. (F) RNA/DNA + ssDNA (top), RNA/DNA (middle), or R-loop substrate (bottom) was incubated with RNaseH, Sox2, or Ddx5 with different concentrations. (G) RNA/DNA + ssDNA substrate (25 nM) was incubated with increasing concentrations of different Sox2 deletions in the presence of Ddx5. (H) Glutathione S-transferase (GST)–pull-down analysis of the interaction between GST-Ddx5 and different Sox2 deletions. (I) Co-IP analysis of interaction between Sox2 and Ddx5 or Dhx9 in mESCs before and after inducing RNaseH1 expression by doxycycline. Experiments were performed under mild conditions (wash with 150 mM KCl) or stringent conditions (wash with 500 mM KCl). IgG, immunoglobulin G.
Our previous data indicated that Ddx5 functions as a barrier to reprogramming (29). To investigate whether and how Sox2 and Ddx5 regulate R-loops, we purified His-Ddx5 and His-Sox2 fusion proteins (fig. S7B), synthesized several different types of RNA/DNA hybrids and R-loop structures, and then performed in vitro RNA/DNA resolution assays. Our biochemical data further indicated that Sox2 alone could not resolve either RNA/DNA hybrids or R-loops, but that Ddx5 has a strong capacity to resolve these structures in vitro (Fig. 5F). To test whether Sox2 has any effect on Ddx5-resolving R-loops, in vitro experiments indicated that the activity of Ddx5 in resolving both RNA/DNA hybrids and R-loops was blocked by Sox2 in a concentration-dependent manner (Fig. 5F). In addition, our data showed that Sox2 could also prevent Dhx9 from resolving R-loop (fig. S7C). Together, these results suggest that Sox2 promotes iPSC generation at least partially by maintaining those R-loops that are beneficial for reprogramming.
To identify the specific domain of Sox2 (30) that inhibits the resolving activity of Ddx5 at R-loops, we generated different Sox2 deletions (fig. S7, D and E) and tested for their ability to protect R-loops via in vitro RNA/DNA resolution. Our data indicated that the HMG domain of Sox2, but not other domains, is required for reducing the activity of Ddx5 on RNA/DNA hybrids (Fig. 5G). Furthermore, domain-mapping experiments showed that the connection domain, but not any other domain of Sox2, is necessary for Sox2 binding to Ddx5 (Fig. 5H). These results indicated that the connection domain of Sox2 is required for the interaction of Sox2 with Ddx5, but that the HMG domain of Sox2 is essential for inhibition of Ddx5 activity at R-loops. It has been shown that the serine-rich domain (amino acids 207 to 254) and C-terminal end (amino acids 206 to 319) of Sox2 are required for its transactivation activity (31); therefore, we investigated whether the N-terminal domain (1 to 180 amino acids, S180) of Sox2 (without transactivation activity) could override the inhibitory effect of RNaseH1D209N on reprogramming. Our reprogramming data showed that OKMS180 achieved higher reprogramming efficiency compared with OKM in the presence of RNaseH1D209N, though lower than OSKM plus RNaseH1D209N (fig. S7F). On the basis of these results, our data suggested that Sox2 might corporate with R-loop independent of its function as a transcription factor.
To explore the interaction model of Sox2 and Ddx5 or Dhx9 responding to R-loop in vivo, we generated doxycycline-inducible mESC lines and modulated R-loop levels via adding doxycycline into the culture to induce the expression of RNaseH1. Our data indicated that RNaseH1 overexpression in mES cells markedly increased Sox2 binding affinity to both Ddx5 and Dhx9 after loss of R-loop (Fig. 5I), but has little effect on the protein level of Sox2, Ddx5/Dhx9 (fig. S7G). Together, R-loop is an important regulator in mediating Sox2-Ddx5 interaction. A high level of R-loop impedes but a low level of R-loop strongly facilitates Sox2-Ddx5 interaction.
Sox2 maintains R-loop levels at key pluripotent genes to promote reprogramming
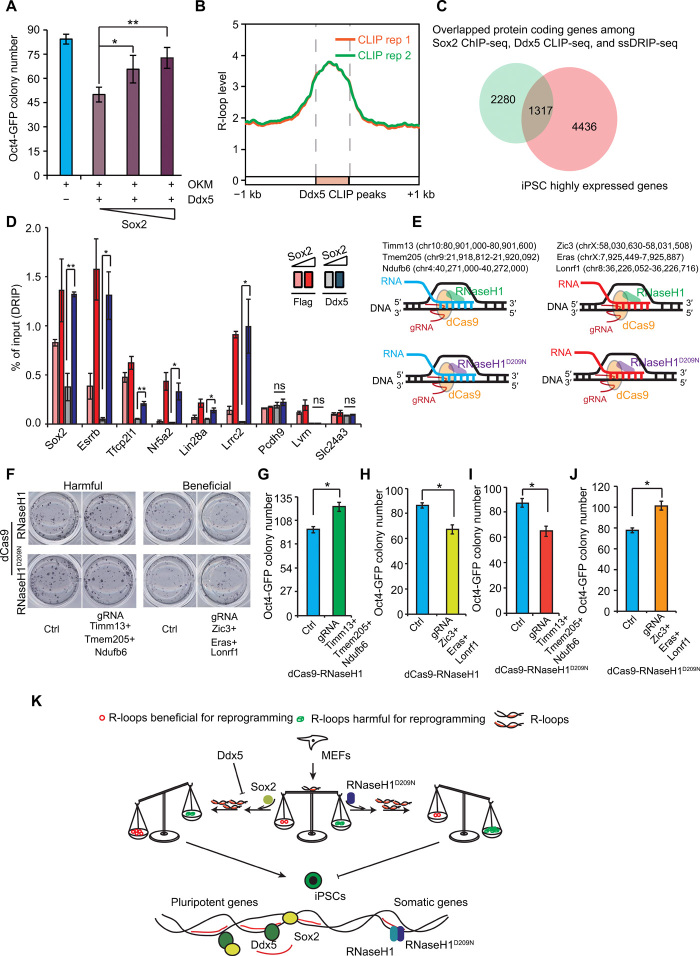
On the basis of the above data, we postulated that Sox2 could rescue the inhibition of reprogramming mediated by Ddx5. Our experiments indeed indicated that a gradual increase of Sox2 rescued the inhibitory phenotype of Ddx5 on reprogramming (Fig. 6A). To elucidate the binding targets of Ddx5, we performed Ddx5 cross-linking immunoprecipitation with highthroughput sequencing (CLIP-seq) experiments and found that Ddx5 peaks were highly enriched at R-loops in iPSCs (Fig. 6B). We then identified the co-binding genes among the data from the Sox2 ChIP-seq, Ddx5 CLIP-seq, and ssDRIP-seq experiments. Venn diagrams showed that many pluripotent genes were among the co-binding genes (Fig. 6C). Furthermore, we assessed whether Sox2 could maintain the R-loops of these pluripotent genes by inhibiting the activity of Ddx5 to promote reprogramming in cells. To address this, we performed DRIP-qPCR experiments by using reprogrammed cells treated with OKM plus Ddx5 in addition to different amounts of Sox2 on day 7 of reprogramming and examined R-loop levels at selected key pluripotent genes and somatic genes. Our data indicated that Ddx5 inhibited R-loop levels of all pluripotent genes, consistent with the resolving activity of Ddx5 at R-loops in vitro (Fig. 5F). Sox2 could notably rescue Ddx5-resolving capacity on R-loops and maintain high levels of R-loops at the promoters of these key pluripotent genes, with little effect on somatic genes (Fig. 6D). These data suggest that stabilization of R-loops by Sox2 is required for reprogramming somatic cells into pluripotent cells.
Fig. 6. Sox2 and Ddx5 balance the R-loops at key pluripotency genes for reprogramming.
(A) Oct4-GFP colony number of MEFs reprogrammed with OKM, Ddx5, plus various concentrations of Sox2. (B) The metaplot of R-loop in iPSCs on Ddx5 binding peaks. (C) Intersection of the overlapping genes of Sox2 ChIP-seq, Ddx5 CLIP-seq, and ssDRIP-seq and iPSC highly expressed genes. (D) DRIP-qPCR monitoring R-loop levels at different pluripotent genes on day 7 of the reprogramming. (E) Schematic of strategy to modulate specific R-loop regions. The selected harmful R-loops (left) and beneficial R-loops (right). (F) AP staining of MEFs reprogrammed with OSKM, guide RNA (gRNA) targeting harmful or beneficial R-loops, plus dCas9-RNaseH1 or dCas9-RNaseH1D209N. (G) Oct4-GFP colony number of MEFs reprogrammed with OSKM, gRNA targeting harmful R-loops, plus dCas9-RNaseH1. (H) Oct4-GFP colony number of MEFs reprogrammed with OSKM, gRNA targeting beneficial R-loops, plus dCas9-RNaseH1. (I) Oct4-GFP colony number of MEFs reprogrammed with OSKM, gRNA targeting harmful R-loops, plus dCas9-RNaseH1D209N. (J) Oct4-GFP colony number of MEFs reprogrammed with OSKM, gRNA targeting beneficial R-loops, plus dCas9-RNaseH1D209N. (K) A proposed working model. Error bars correspond to means ± SD (*P ≤ 0.05, **P ≤ 0.01, two-tailed Student’s t test).
The effects of dCas9-mediated site-specific R-loop editing on reprogramming
On the basis of these findings, we proposed that some R-loops were harmful for reprogramming, the aberrant accumulation of which might inhibit reprogramming, while another fraction of R-loops were beneficial for reprogramming. To validate this hypothesis, we took advantage of dCas9 technology and made MEFs transduced with guide RNAs (gRNAs) targeting the selected R-loops sites either harmful (Timm13 + Tmem205 + Ndufb6) or beneficial (Zic3 + Eras + Lonrf1) to reprogramming (Fig. 6E). Then, we further investigated whether the reprogramming efficiency would be affected after site-specific R-loops were modulated by dCas9-RNaseH1 or dCas9-RNaseH1D209N. Our data indicated that the reprogramming efficiency was increased when dCas9-RNaseH1 targeted harmful R-loop regions at Timm13 + Tmem205 + Ndufb6 loci (Fig. 6, F and G, and fig. S7H) but was inhibited when dCas9-RNaseH1 targeted beneficial R-loop regions at Zic3 + Eras + Lonrf1 (Fig. 6, F and H, and fig. S7H). Furthermore, accumulation of the harmful R-loops induced by dCas9-RNaseH1D209N inhibited reprogramming and accumulation of the beneficial R-loops resulted in the opposite phenotype (Fig. 6, F, I, and J, and fig. S7I). Together, these data demonstrate that R-loop plays important roles in reprogramming.
DISCUSSION
It has been suggested that R-loops may act as epigenetic markers, by being involved in binding alteration of transcription factors, chromatin modification, DNA methylation, and chromatin interaction. In this study, we mapped the landscapes of R-loops during OSKM-mediated somatic cell reprogramming and showed that there is a dynamic association between R-loop and the process of somatic cell reprogramming. Our data suggested that R-loop causes sharp changes at both the early and late stages of reprogramming, but it has transient and subtle changes at the intermediate stage, which shows a similar pattern of chromatin opening, DNA methylation, and gene expression during reprogramming (17, 32). The cross-talk between R-loops and other epigenetic events, such as chromatin modification and binding of transcription factors, needs to be further investigated. We surprisingly found that a part of R-loops changes before RNA output and that these R-loops accumulate at terminator regions and are positively associated with gene expression during reprogramming (Fig. 1F). In addition, most R-loops reside in intergenic regions and overlap with ESC-specific enhancers. Furthermore, transient transcriptome analysis has been recently used as a tool for monitoring the dynamics of enhancer landscapes and transcription programs during cellular responses and differentiation (33). On the basis of data from both previous transient transcriptome analysis and our R-loops at ESC-specific enhancers, we concluded that both R-loops and transient transcripts produced during transcription might exist together at enhancers or that R-loop–enriched enhancer RNAs are short-lived transient transcripts. In line with these discoveries, R-loop might be an indicator of epigenomic signatures for somatic cell reprogramming.
After defining the profile of R-loops, we would like to understand how R-loops affect stem cell fate and reprogramming. Our results suggested that the balance of R-loops influences the reprogramming of somatic cells into iPSCs. Disrupting this balance could impede somatic cell reprogramming (Fig. 6K). Our data uncovered that high levels of R-loops are closely related to open chromatin and enhancers (Fig. 1, H and I) but that R-loops could not open chromatin in cis (Fig. 3G). After chromatin is opened, R-loops may recruit epigenetic factors and trigger active epigenetic markers.
The balance of R-loops during reprogramming can be disrupted by either depleting RNaseH1, resulting in over-accumulation of R-loops, or overexpressing RNaseH1D209N, resulting in stabilization of R-loops. Aberrant unbalanced R-loops abolish reprogramming (Fig. 6K). Despite no major transcriptional differences between control and RNaseH1D209N-treated reprogrammed cells at the early stage of reprogramming, RNaseH1D209N caused remarkable inhibition of reprogramming, supported by the marked changes in R-loop dynamics observed throughout reprogramming (Fig. 1). Accumulation of R-loops at the early stage of reprogramming might prevent the redistribution of transcription factors, in line with the importance of stage-specific binding patterns of transcription factors for efficient reprogramming, thereby suggesting that R-loops play a crucial role in cell-fate transitions.
Our S9.6 co-IP experiments revealed that only Sox2, but not any other factor of the Yamanaka cocktail, forms a complex with R-loops. Coincidentally, of these four factors, only Sox2 can overcome the inhibitory effect of both RNaseH1 loss of function and Ddx5 gain of function on reprogramming. From these two aspects, we conclude that Sox2 plays an important role in reprogramming by ensuring the maintenance of R-loops (Fig. 6K). Ddx5 acts as a barrier for reprogramming through microRNA-125b–based repression of RYBP. This function of Ddx5 is focused on the cross-talk between Ddx5 and H2AK119ub1 in regulating reprogramming (29). However, as a DEAD-box RNA helicase, the helicase activity of Ddx5 remains to be further investigated. This study demonstrates that Sox2 inhibits the helicase activity of Ddx5 on R-loop. Our data suggest that, during reprogramming, Ddx5 maintains the activation of somatic genes via repressing H2AK119ub1 and silences the expression of pluripotent genes via its activity on R-loop. These findings support and reflect bivalent functions of Ddx5 in regulating reprogramming.
On the basis of our data from biochemical and reprogramming experiments, we believe that Sox2 itself does not resolve R-loops but that it prevents Ddx5/Dhx9 from resolving R-loops. Sox2 protein purification followed by mass spectrometry showed that Sox2 interacts with many RNA binding proteins and helicases, which are involved in R-loop metabolism. These raise the possibility that Sox2 might function in many biological processes via regulating R-loop by interacting with R-loop associated factors. Together, the results of this study point to the importance of balancing R-loop dynamics in reprogramming. In addition, they demonstrate that Sox2 is not only a transcription factor that induces transcription but also an essential regulator that maintains the balance of R-loops and further promotes reprogramming together with R-loop–resolving factors.
MATERIALS AND METHODS
Cell culture and plasmids
NIH3T3 and Plat-E cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM)/high-glucose media (Hyclone) supplemented with 10% fetal bovine serum (FBS) (Natocor). OG2 MEFs were derived from E13.5 mouse embryos from crossing male Oct4-GFP transgenic allele–carrying mice (CBA/CaJ × C57BL/6J) with 129Sv/Jae female mice. MEFs were cultured in DMEM/high-glucose media (Hyclone) supplemented with 10% FBS (GIBCO), 1 mM nonessential amino acids (GIBCO), and 1× GlutaMAX (GIBCO). mESCs and iPSCs were maintained in feeder-free or feeder-coated ESC medium containing DMEM/high-glucose media (Hyclone), 15% FBS (GIBCO), 1 mM sodium pyruvate (GIBCO), 1 mM nonessential amino acids (GIBCO), 1× GlutaMAX (GIBCO), 0.1 mM 2-mercaptoethanol (GIBCO), leukemia inhibitory factor (LIF; 1000 U/ml) (Millipore), and the 2i inhibitors: 3 mM CHIR99021 (Selleck) and 1 mM PD0325901 (Selleck).
Two independent shRNA oligos targeting RNaseH1 were designed and constructed into the pSUPER plasmid. The sequences of shRNA oligos are listed in table S1. Individual gRNA was ligated onto Lenti-guide-puro vector that contained a U6 promoter (U6) linearized by Bsmb I. Primers used to construct individual small guide RNA (sgRNAs) are shown in table S1.
Reprogramming experiments
OG2 MEFs at passage 2 were plated at 10,000 cells per well in 12-well plates or 5000 cells per well in 24-well plates precoated with 0.1% gelatin; then, they were infected twice with retrovirus generated from Plat-E cells. Infected cells were cultured in iCD1 medium or ESC medium containing DMEM/high-glucose media (Hyclone), 15% FBS (GIBCO), 1 mM sodium pyruvate (GIBCO), 1 mM nonessential amino acids (GIBCO), 1× GlutaMAX (GIBCO), 0.1 mM 2-mercaptoethanol (GIBCO), LIF (1000 U/ml; Millipore), and vitamin C (50 μg/ml; Sigma-Aldrich). The reprogramming system takes 7 days in iCD1 medium or 12 days in serum medium. iPSC colonies were picked and then maintained as mESCs. For overexpression or knockdown experiments in reprogramming, to ensure high transfection efficiency, we transfected virus supernatant into MEFs twice to make sure that more than 95% of MEFs were transfected via adjusting multiplicity of infection.
Western blot
Cells were collected and lysed in cell lysis buffer [50 mM tris-HCl (pH 7.6), 1% Triton X-100, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, and protein inhibitor cocktail]. After centrifugation at 12,000g for 10 min, soluble protein mixtures were quantified. After SDS–polyacrylamide gel electrophoresis (PAGE), proteins were transferred onto polyvinylidene fluoride membrane. The membrane was incubated with the corresponding primary antibody and secondary antibodies. The antibodies used in this paper are as follows: anti–RNase H1 (Abcam ab56560), anti–β-Actin (Abcam ab8227), anti–Histone H3 (Abcam ab1791), anti–Histone H2A.X (Abcam ab11175), anti–Histone-γH2A.X (Abcam, ab81299), anti-Sox2 (Abcam ab79351), anti-Klf4 (R&D Systems AF3158), anti–c-Myc (Abcam ab32072), anti–Flag M2 (Sigma-Aldrich F1804), anti-Oct3/4 (Santa Cruz sc-5279), anti-Dhx9 (Proteintech 17721-1-AP), and anti-Ddx5 (Abcam ab21696).
Quantitative RT-qPCR analysis
RNA extraction was performed with RaPure Total RNA Kit (Magen, R4011). One microgram of total RNA was then reverse-transcribed with HiScript II Q RT SuperMix for qPCR (Vazyme, R123-01). The primers used are listed in table S2. All the experiments were repeated three times.
AP staining
AP staining was performed with BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime, C3206) following the manufacturer’s instructions.
S9.6 co-IP
S9.6 co-IP was performed as described previously (28), with several modifications. Briefly, genomic DNA was extracted and treated with or without 5 U RNase H per microgram of DNA after the DNA was fragmented with 50 U of enzymes, Mse I, Dde I, Alu I, and Mbo I. DNA (2 μg) treated with or without RNase H was bound to 2 μg of S9.6 antibody or immunoglobulin G (IgG) and 25 μl of Dynabeads Protein A or Protein G (Invitrogen) in IP buffer [20 mM tris-HCl (pH 7.4), 150 mM KCl, 0.1% Triton X-100, and 5 mM EDTA (pH 8.0)] at 4°C for 4 hours. Beads were washed three times with IP buffer and incubated with nuclear extraction pretreated with RNase A at 4°C for 4 hours. The beads were then washed three times and eluted for the Western blot. The interactors are listed in table S3.
Immunoprecipitation and pull-down experiments
For co-IP experiments, nuclear protein extracts were incubated with 1 μg antibody and 20 μl of Dynabeads Protein A or Protein G (Invitrogen) at 4°C overnight. Beads were then washed five times with wash buffer [20 mM tris-HCl (pH 7.4), 250 mM KCl, and 0.1% Triton X-100]. Beads were boiled in 1× SDS loading buffer for the Western blot. For pull-down experiments, purified proteins were incubated with glutathione resin (GenScript) at 4°C overnight. The resin was washed five times with wash buffer [20 mM tris-HCl (pH 7.4), 500 mM KCl, and 0.1% Triton X-100] and then boiled in 1× SDS loading buffer for the Western blot.
Protein expression and purification
The bacteria expression plasmids pET-28a containing cDNAs of different Sox2 deletions, Ddx5, and RNaseH1 fusing to N-terminal tagged 6× Histidine were expressed into transetta (DE3) chemically competent cells (Transgene Biotech, CD801). Transformed cells were grown at 37°C to a density of 0.6 to 0.8 at OD600 (optical density at 600 nm) and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 6 hours. The cells were collected and resuspended in lysis buffer [25 mM tris-HCl (pH 7.5), 150 mM NaCl, 5% glycerol, and 20 mM imidazole]. The cells were lysed and protein supernatants were allowed to flow through a Ni-NTA Sefinose Column (Sangon Biotech, C600791). The columns were washed with lysis buffer and 100 mM imidazole. The proteins were eluted with elution buffer [25 mM tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, and 300 mM imidazole]. To remove imidazole, the eluted fraction was dialyzed in dialysis buffer [25 mM tris-HCl (pH 7.5), 150 mM NaCl, and 10% glycerol] and stored at −80°C.
In vitro RNA/DNA hybrid resolution assay
The procedure for the in vitro RNA/DNA hybrid resolution assay was performed as described previously (34), with some modifications. In brief, the indicated protein was incubated in buffer A [25 mM tris-HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, bovine serum albumin (BSA; 50 mg/ml), and 50 mM KCl] on ice for 10 min. Then, RNA/DNA hybrids were added to the reaction and incubated at 37°C. The reaction was halted by adding stop buffer [proteinase K (5 mg/ml), 0.05 M EDTA, and 5% SDS]. The reaction products were analyzed by using 10% native polyacrylamide gel at 4°C. Gels were scanned with Fujifilm FLA-7000.
The list of oligonucleotide sequences used in this study is as follows:
RNA oligo (CCUACUACCUGUGCCUUUAAGUACCUUUUCUGUCUUCUA)
DNA oligo (TAGAAGACAGAAAAGGTACTCTAAAGGCACAGGTAGTAGG)
DNA + ssDNA oligo (TAGAAGACAGAAAAGGTACTTAAAGGCACAGGTAGTAGGTTTGCCCACCTGCAGGTTCACCTCGTCCCTGGC)
For the R-loop substrate, the RNA oligo was annealed with oligo 1 and 2:
Oligo 1 (GCCAGGGACGAGGTGAACCTGCAGGTGGGCGGCTACGTCATGACTGTCATAGATGTGTCACATCACGAGGCTTATTGGTAGAATTCGGCAGCGTCATGCGACGGC)
Oligo 2 (GCCGTCGCATGACGCTGCCGAATTCTACCACGCTAGAAGACAGAAAAGGTACTTAAAGGCACAGGTAGTAGGTTTGCCCACCTGCAGGTTCACCTCGTCCCTGGC)
Electrophoretic mobility shift assay
EMSA was performed as previously described (34). The indicated proteins were mixed with RNA/DNA hybrids containing 5′-cy5–labeled RNA in buffer B [25 mM tris-HCl (pH 7.5), 1 mM DTT, 10 mM EDTA, BSA (50 mg/ml), and 50 mM KCl] at room temperature for 30 min. The reaction products were analyzed with 10% native polyacrylamide gel at 4°C. Gels were scanned with Fujifilm FLA-7000.
Nuclear run-on assay
Briefly, cells during reprogramming were washed with ice-cold phosphate-buffered saline (PBS) and resuspended in NP-40 lysis buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.5% IGEPAL CA-630]. Cell suspension was incubated on ice for 5 min before nuclei were centrifuged at 400g for 4 min at 4°C. Pellets containing nuclei were washed carefully with ice-cold NP-40 lysis buffer and collected by centrifugation (400g, 4 min, 4°C). Nuclei were added to nuclei storage buffer [50 mM tris-HCl (pH 8.3), 0.1 mM EDTA, 5 mM MgCl2, and 40% (v/v) glycerol] and 2× transcription buffer [20 mM tris-HCl (pH 8.3), 5 mM MgCl2, 300 mM KCl, and 4 mM DTT] plus Ribonucleoside triphosphate set and bromouridine 5′-triphosphate, and incubated for 30 min at 30°C. Nascent RNA was extracted using TRIzol and precipitated in ethanol. Extracted nascent RNA was purified using 2 μg of anti–5-bromo-2′-deoxyuridine monoclonal antibody and 30 μl of protein G Dynabeads. Nuclear run-on assay transcripts were quantified by RT-qPCR. The primers used are listed in table S2.
RNA-seq and bioinformatics analysis
Total RNA was extracted with TRIzol. An RNA library was generated with Vazyme mRNA-seq V3 Library Prep Kit for Illumina (Vazyme, NR611). Raw RNA-seq reads were stripped to remove adaptor sequences and low-quality bases using Trimmomatic (35). The processed reads with lengths greater than 35 nt were defined as clean reads. The reads were mapped to mm10 (mouse) reference genomes using Spliced Transcripts Alignment to a Reference (STAR) (36). Only uniquely mapped reads with mapping quality more than or equal to 20 were kept for subsequent analysis. Transcript identification and counting were processed by HTSeq (37). The DEseq2 (16) package was used to analyze DEGs. The RNA-seq reads on exons were counted and used to perform differential analysis. A DEG was defined as a gene with q value < 0.05 and |log2FC| > 1.
ATAC-seq and bioinformatics analysis
In brief, 50,000 cells were harvested and washed once with 50 μl of cold PBS. Then, the cells were resuspended in 50 μl of lysis buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.2% (v/v) IGEPAL CA-630]. Then, the suspension of nuclei was centrifuged at 500g at 4°C for 10 min. The pellet was resuspended by adding 50 μl of transposition reaction mix (10 μl of TD buffer, 5 μl of Tn5 transposase, and 35 μl of nuclease-free H2O) and incubated at 37°C for 30 min. Last, DNA was isolated using MinElute PCR Purification Kit (QIAGEN). ATAC-seq libraries were constructed and purified with AMPure XP beads (Beckman Coulter).
After trimming adaptor sequence with Cutadapt, ATAC-seq reads were aligned to mm10 genome by using Bowtie2 with default parameters. MACS2 was used to call ATAC peaks with ≥10 enrichment and with q value ≤ 0.001. MEF-specific (or iPSC-specific) ATAC peaks were defined as the ATAC peaks observed in MEFs (or iPSCs) only, without any overlapping with ATAC peaks in iPSCs (or MEFs).
ssDRIP-seq and DRIP-qPCR
DRIP was performed as described previously (5, 14), with several modifications. In brief, 1 × 107 cells were resuspended in TE buffer, followed by adding 10% SDS (final concentration: 0.5%) and proteinase K (Thermo Fisher Scientific, 25530015), and then incubated in a constant-temperature shaker at 37°C. After adding 5 M KAc, the tubes were mixed and placed on ice for 20 min. Phenol:chloroform:isoamyl alcohol (25:24:1) was added, mixed well by vortex, and centrifuged at 12,000g at 4°C for 10 min. The supernatant was transferred to a new tube, and DNA was precipitated by adding 0.7 volume of isopropanol (Sigma-Aldrich). DNA was washed with 70% ethanol and air-dried, and then the DNA pellet was resuspended with TE buffer. RNase H treatment: half of the genomic DNA (gDNA) was transferred to a new tube, and 1/10 volume of 10× RNase H buffer and RNase H (NEB) was added, followed by incubation at 37°C overnight and then purification by phenol:chloroform:isoamyl alcohol (25:24:1) extraction as described above. gDNA was digested with Mse I, Dde I, Alu I, and Mbo I (NEB; final concentration: 100 U/ml for each enzyme) at 37°C. After purifying with phenol:chloroform:isoamyl alcohol (25:24:1) extraction, fragmented gDNA was resuspended in TE buffer and quantified by Qubit 3.0 (Invitrogen). For each sample, input DNA was immunoprecipitated with 1× DRIP binding buffer [10 mM NaPO4 (pH 7.0), 140 mM NaCl, and 0.05% Triton X-100] and S9.6 antibody (American Type Culture Collection, HB-8730) in a shaker at 4°C overnight. After adding Dynabeads Protein G (Invitrogen, 10004D) and incubating at 4°C, the beads with antibody were washed four times with 1× DRIP binding buffer. Elution buffer [50 mM tris-HCl (pH 8.0) and 10 mM EDTA] containing proteinase K was added into beads/antibody complexes, and the tube was incubated in an Eppendorf ThermoMixer at 55°C. Then, the reaction mixtures were purified with phenol/chloroform extraction, and the supernatant was moved into a new tube. A 1/10 volume of 3 M NaAc, 1 μl GlycoBlue (Invitrogen), and 1 volume isopropanol was added, and the DNA was precipitated at −20°C. The DRIPed DNA pellet was washed with 70% ethanol, air-dried, resuspended in TE buffer [10 mM tris-HCl (pH 8.0) and 0.1 mM EDTA], and used for next-generation sequencing (NGS) library construction or qPCR.
ssDRIP-seq library was constructed by using the Accel-NGS 1S Plus DNA Library Kit (Swift Biosciences). Briefly, the DRIPed DNA sample was fragmented to an average size of 250 base pairs (bp) with an S220 Focused-ultrasonicator (Covaris, Woburn, MA, USA). The sonicated DNA was denatured into ssDNA at 98°C for 2 min and placed on ice immediately for another 2 min. The truncated adapter 1 was ligated to the 3′ end of ssDNA first, followed by extension. The truncated adapter 2 was ligated to the 5′ end. After PCR amplification, libraries were purified with AMPure XP beads (Beckman Coulter). The quality of each library was evaluated with an Agilent Bioanalyzer (Palo Alto, CA, USA), and each library was sequenced on an Illumina HiSeq X Ten platform (San Diego, CA, USA).
qPCR was performed and the signal intensity plotted was the relative abundance of RNA/DNA hybrids immunoprecipitated in each region and normalized to the input. The primers used in the DRIP-qPCR assays are listed in table S2.
ssDRIP-seq data analysis
Adaptor sequences were trimmed off for all raw reads. Then, 10 bases from the beginning and the ends of both R1 and R2 were also trimmed to remove the library tails. Reads that were less than 50 bases in length were discarded. All the above processes are implemented through Cutadapt. The remaining reads were aligned to the mm10 (mouse) reference genomes using Bowtie2, with default parameters. Only uniquely mapped reads with mapping quality score ≥20 were kept for the subsequent analysis for each sample using Samtools. MACS2 was used to call narrow R-loop peaks with ≥10 enrichment and with q value ≤ 0.001. Promoter and terminal regions were defined as −2000 to +2000 bp around TSS or TTS, respectively. Gene body was defined as from TSS to TTS.
Differential R-loop analysis
To reduce biases from algorithms, differential R-loop regions, and DRG, two different methods were used to assess the R-loop difference among the samples. For differential R-loop regions analysis, briefly, total R-loop regions were merged from the R-loop peaks of all samples analyzed. After identifying a common set of peaks (total R-loop regions), the reads in each peak across samples were counted. Differential analysis was performed using the DEseq2 package (16). The differential R-loop regions analysis method covers all the genomic regions including intergenic regions but ignores the R-loop signal out of R-loop peaks. For DRG analysis, the ssDRIP-seq reads in the promoter, gene body, or terminator region of each gene were counted, and the differential analysis was performed using the DEseq2 package (16). Prom-DRG, Gb-DRG, or Term-DRG was defined as a gene with a significant R-loop difference in the promoter, gene body, or terminator, respectively. DRG analysis method covers all the genic R-loop signals and assigns a unique value to the promoter, gene body, or terminator of each gene but ignores intergenic R-loops. A concurrently changed gene was defined as a gene with a differential RNA level [false discovery rate (FDR) < 0.05] and R-loop level (P < 0.01 or prob. >0.9). Prom-/Gb-/Term-concurrently changed genes: differential RNA level [FDR0) < 0.05] and differential promoter-/gene body-/terminator R-loop level (P < 0.01 or prob. >0.9). For motif analysis of differential R-loop regions, we used MEME suit with default parameters.
Cross-linking immunoprecipitation followed by sequencing and bioinformatics analysis
CLIP-seq was performed as previously described (38), with changing the 3′ RNA linker into a preA-L3-IR800-biotin DNA adaptor. Briefly, mESCs were irradiated at 400 mJ/cm2 with 254-nm ultraviolet light. The cross-linked cells were lysed and 15 μg of anti-DDX5–specific antibody was applied to pull down protein-RNA complexes. After micrococcal nuclease treatment and 3′ DNA adaptor ligation, the immunoprecipitated complexes were fractionated on a 4 to 12% NuPAGE bis-tris gel and transferred onto a nitrocellulose membrane. The DDX5-specific smear bands were excised with scalpels and treated with proteinase K (Thermo Fisher Scientific, 25530015) before extraction of the respective RNA by phenol and chloroform. A 5′ RNA linker was then added to the isolated RNA, and the RNA was reverse-transcribed by superscript reverse transcriptase III (Life Technologies, 18080-051). The cDNA strands were PCR-amplified to produce libraries for deep sequencing. L3-IR800-biotin DNA adaptor: 5′OH-ATC TCG TAT GCC GTC TTC TGC TTG TAA AAA AAA AAA A/iAzideN/A AAA AAA AAA AA/3Bio/-3′, 5′ RNA linker: 5′-(OH) rArCrArCrGrArCrGrCrUrCrUrUrCrCrGrArUrCrU(rN)(rN) (rN)rU(OH)-3′. For CLIP-seq data analysis, only R1 was aligned to mm10, and other data processing was the same as for the RNA-seq analysis described above. PARCLIP peaks were called with PEAKachu (https://github.com/tbischler/PEAKachu/, v0.0.1alpha2) using default parameters. Peaks were filtered by |log2FC| > 5. IgG control was used as peak calling background.
Colocalization analysis
Bedtools (39) and regioneR (permutation test) (40) were used to assess the association between different types of peaks. Using regioneR, the random peaks were generated by shuffling in the mm10 genome regions with 1000 times, the permutated values were determined from the 1000 random peaks, and the observed values were the real overlaps between the two peaks, in which colocalization analysis is required. The differences between the observed value and the permutation value represent the colocalization.
ssDRIP-seq and RNA-seq fuzzy cluster analysis
Fuzzy cluster analysis was performed by using mFuzz (15). For RNA cluster analysis, all DEGs between any two samples were analyzed. For R-loop cluster analysis, all differential R-loop regions between any two samples were analyzed. The normalized RNA-seq or ssDRIP-seq read counts on exons of DEGs or differential R-loop regions were analyzed with a cluster membership threshold of 0.7.
Supplementary Material
Acknowledgments
We thank the support from the Guangzhou Branch of the Supercomputing Center of the Chinese Academy of Sciences. Funding: This work was supported by the National Key R&D Program of China (2016YFA0100400 to H.Y.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010502 to H.Y.), the Ministry of Science and Technology of the People’s Republic of China (2016YFA0500800 to Q.S.), the National Natural Science Foundation of China (31925009 to H.Y. and 91740105 and 31822028 to Q.S.), the National Mega-project of China for Innovative Drugs (2018ZX09201002-005 to M.Y.), the Science and Technology Planning Project of Guangdong Province, China (2019B020234004, 2017B030314056, and 2016B030229006 to H.Y.), the Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR110104007 to H.Y.), and the Science and Technology Program of Guangzhou, China (201807010101 and 201707020042 to H.Y. and 201906010096 to M.Y.). The Sun Lab was supported by the Tsinghua University Initiative Scientific Research Program, the Tsinghua-Peking Joint Center for Life Sciences, and the 1000 Young Talent Program of China. W.X. was supported by a postdoctoral fellowship from Tsinghua-Peking Joint Center for Life Sciences. The Schöler Lab was supported by the Max Planck Society as well as the EU grant Prometheus I0D: 669168 “Novel Cells for Organ Repair” funded under the program H2020-EU.1.1.–Excellent Science–European Research Council (ERC) for the period 1 August 2015 to 31 July 2020. Author contributions: H.Y. and Y.L. conceived and designed the experiments of reprogramming. Q.S. and W.X. conceived and managed the ssDRIP-seq and bioinformatics analysis. Y.L. and Y.S. conducted somatic cell reprogramming, RNA-seq, ATAC-seq, and the molecular and biochemical experiments. Q.L. performed the ssDRIP-seq. W.X. and Q.L. performed the bioinformatics analysis with assistance from K.L. X.W., J.W., Z.L., S.V., R.Y., Q.X., L.W., R.G., X.D., Z.Z., Y.D., H.L., M.Y., Y.X., and H.R.S. contributed to the work. H.Y., Y.L., and W.X. wrote the manuscript. H.Y. and Q.S. conceived and supervised the entire study. H.Y. approved the final version. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The data for enhancer analysis are from GSE90895. The ATAC-seq, ssDRIP-seq, CLIP-seq, and RNA-seq data described in this paper are deposited at NCBI GEO: GSE125644.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/24/eaba0777/DC1
REFERENCES AND NOTES
- 1.Liu L. Q., Xu Y., He M. H., Zhang M., Cui F. G., Lu L. N., Yao M. Z., Tian W. H., Benda C., Zhuang Q., Huang Z. J., Li W. J., Li X. C., Zhao P., Fan W. X., Luo Z. W., Li Y., Wu Y. S., Hutchins A. P., Wang D. Y., Tse H. F., Schambach A., Frampton J., Qin B. M., Bao X. C., Yao H. J., Zhang B. L., Sun H., Pei D. Q., Wang H. T., Wang J., Esteban M. A., Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell 15, 574–588 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Costantino L., Koshland D., The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 34, 39–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamperl S., Bocek M. J., Saldivar J. C., Swigut T., Cimprich K. A., Transcription-replication conflict orientation modulates R-Loop levels and activates distinct DNA damage responses. Cell 170, 774–786.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K. M., Eaton M. L., Kellis M., Hill S. J., Parmigiani G., Proudfoot N. J., Livingston D. M., BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Q., Csorba T., Skourti-Stathaki K., Proudfoot N. J., Dean C., R-Loop stabilization represses antisense transcription at the arabidopsis FLC locus. Science 340, 619–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skourti-Stathaki K., Kamieniarz-Gdula K., Proudfoot N. J., R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahba L., Amon J. D., Koshland D., Vuica-Ross M., RNase H and multiple RNA biogenesis factors cooperate to prevent RNA: DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerritelli S. M., Frolova E. G., Feng C., Grinberg A., Love P. E., Crouch R. J., Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11, 807–815 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Manzo S. G., Hartono S. R., Sanz L. A., Marinello J., De Biasi S., Cossarizza A., Capranico G., Chedin F., DNA topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 19, 100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Hage A., French S. L., Beyer A. L., Tollervey D., Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salas-Armenteros I., Perez-Calero C., Bayona-Feliu A., Tumini E., Luna R., Aguilera A., Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 36, 3532–3547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Benítez F., Gaillard H., Aguilera A., Physical proximity of chromatin to nuclear pores prevents harmful R loop accumulation contributing to maintain genome stability. Proc. Natl. Acad. Sci. U.S.A. 114, 10942–10947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatia V., Barroso S. I., Garcia-Rubio M. L., Tumini E., Herrera-Moyano E., Aguilera A., BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Xu W., Xu H., Li K., Fan Y., Liu Y., Yang X., Sun Q., The R-loop is a common chromatin feature of the arabidopsis genome. Nat. Plants 3, 704–714 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Kumar L., E Futschik M., Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2, 5–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knaupp A. S., Buckberry S., Pflueger J., Lim S. M., Ford E., Larcombe M. R., Rossello F. J., de Mendoza A., Alaei S., Firas J., Holmes M. L., Nair S. S., Clark S. J., Nefzger C. M., Lister R., Polo J. M., Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell 21, 834–845.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K., Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H., Lima W. F., Crooke S. T., Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 276, 23547–23553 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Chen L., Chen J.-Y., Zhang X., Gu Y., Xiao R., Shao C., Tang P., Qian H., Luo D., Li H., Zhou Y., Zhang D.-E., Fu X.-D., R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell 68, 745–757.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi K., Schöler H. R., Directing reprogramming to pluripotency by transcription factors. Curr. Opin. Genet. Dev. 22, 416–422 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Ng S. Y., Bogu G. K., Soh B. S., Stanton L. W., The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Gorthi A., Romero J. C., Loranc E., Cao L., Lawrence L. A., Goodale E., Iniguez A. B., Bernard X., Masamsetti V. P., Roston S., Lawlor E. R., Toretsky J. A., Stegmaier K., Lessnick S. L., Chen Y., Bishop A. J. R., EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 555, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang X., Yoon J. G., Li L., Tsai Y. S., Zheng S., Hood L., Goodlett D. R., Foltz G., Lin B., Landscape of the SOX2 protein-protein interactome. Proteomics 11, 921–934 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Gao Z., Cox J. L., Gilmore J. M., Ormsbee B. D., Mallanna S. K., Washburn M. P., Rizzino A., Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors. J. Biol. Chem. 287, 11384–11397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding J., Huang X., Shao N., Zhou H., Lee D. F., Faiola F., Fidalgo M., Guallar D., Saunders A., Shliaha P. V., Wang H., Waghray A., Papatsenko D., Sanchez-Priego C., Li D., Yuan Y., Lemischka I. R., Shen L., Kelley K., Deng H., Shen X., Wang J., Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell 16, 653–668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang I. X., Grunseich C., Fox J., Burdick J., Zhu Z., Ravazian N., Hafner M., Cheung V. G., Human proteins that interact with RNA/DNA hybrids. Genome Res. 28, 1405–1414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristini A., Groh M., Kristiansen M. S., Gromak N., RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Lai P., Jia J., Song Y., Xia Q., Huang K., He N., Ping W., Chen J., Yang Z., Li J., Yao M., Dong X., Zhao J., Hou C., Esteban M. A., Gao S., Pei D., Hutchins A. P., Yao H., RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell 20, 462–477.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Cox J. L., Mallanna S. K., Luo X., Rizzino A., Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PLOS One 5, e15486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowling T. K., Johnson L. R., Wiebe M. S., Rizzino A., Identification of the transactivation domain of the transcription factor Sox-2 and an associated co-activator. J. Biol. Chem. 275, 3810–3818 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Sardina J. L., Collombet S., Tian T. V., Gomez A., Di Stefano B., Berenguer C., Brumbaugh J., Stadhouders R., Segura-Morales C., Gut M., Gut I. G., Heath S., Aranda S., Di Croce L., Hochedlinger K., Thieffry D., Graf T., Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell 23, 727–741.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Schwalb B., Michel M., Zacher B., Fruhauf K., Demel C., Tresch A., Gagneur J., Cramer P., TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Nguyen H. D., Yadav T., Giri S., Saez B., Graubert T. A., Zou L., Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65, 832–847.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S., Pyl P. T., Huber W., HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y. S., Zhang C., Yeo G., Black D. L., Sun H., Fu X. D., Zhang Y., Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell 36, 996–1006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gel B., Díez-Villanueva A., Serra E., Buschbeck M., Peinado M. A., Malinverni R., regioneR: An R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/24/eaba0777/DC1