Although microbial colonization of the internal tissues of animals generally causes septicemia and death, various animals are persistently associated with benign or beneficial microorganisms in their blood or internal organs. The metabolic consequences of these persistent associations for the animal host are largely unknown. Our research on the facultative bacterium Hamiltonella, localized primarily to the hemolymph of pea aphids, demonstrated that although Hamiltonella imposed no major reconfiguration of the aphid metabolome, it did alter the metabolic relations between the aphid and its obligate intracellular symbiont, Buchnera. Specifically, Buchnera produced more histidine in Hamiltonella-positive aphids to support both Hamiltonella demand for histidine and Hamiltonella-induced increase in host demand. This study demonstrates how microorganisms associated with internal tissues of animals can influence specific aspects of metabolic interactions between the animal host and co-occurring microorganisms.
KEYWORDS: Buchnera, Hamiltonella, histidine, metabolism, symbiosis
ABSTRACT
Beneficial microorganisms associated with animals derive their nutritional requirements entirely from the animal host, but the impact of these microorganisms on host metabolism is largely unknown. The focus of this study was the experimentally tractable tripartite symbiosis between the pea aphid Acyrthosiphon pisum, its obligate intracellular bacterial symbiont Buchnera, and the facultative bacterium Hamiltonella which is localized primarily to the aphid hemolymph (blood). Metabolome experiments on, first, multiple aphid genotypes that naturally bear or lack Hamiltonella and, second, one aphid genotype from which Hamiltonella was experimentally eliminated revealed no significant effects of Hamiltonella on aphid metabolite profiles, indicating that Hamiltonella does not cause major reconfiguration of host metabolism. However, the titer of just one metabolite, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), displayed near-significant enrichment in Hamiltonella-positive aphids in both metabolome experiments. AICAR is a by-product of biosynthesis of the essential amino acid histidine in Buchnera and, hence, an index of histidine biosynthetic rates, suggesting that Buchnera-mediated histidine production is elevated in Hamiltonella-bearing aphids. Consistent with this prediction, aphids fed on [13C]histidine yielded a significantly elevated 12C/13C ratio of histidine in Hamiltonella-bearing aphids, indicative of increased (∼25%) histidine synthesized de novo by Buchnera. However, in silico analysis predicted an increase of only 0.8% in Buchnera histidine synthesis in Hamiltonella-bearing aphids. We hypothesize that Hamiltonella imposes increased host demand for histidine, possibly for heightened immune-related functions. These results demonstrate that facultative bacteria can alter the dynamics of host metabolic interactions with co-occurring microorganisms, even when the overall metabolic homeostasis of the host is not substantially perturbed.
INTRODUCTION
The function of beneficial microorganisms is traditionally interpreted in terms of one or a few well-defined services that enhance host fitness. For animal hosts, frequently reported services are promotion of host nutrition and protection against natural enemies (1). These services include microbial nutrient provisioning (e.g., B vitamins, essential amino acids), degradation of dietary plant polysaccharides that are intractable to host digestion, production of toxins that are active against pathogens, and promotion of host immunological defenses (2–7). Nevertheless, there is increasing evidence that beneficial microorganisms have pervasive effects on their hosts, influencing multiple physiological systems. Many of these effects cannot be explained adequately by formally described microbial services (8–11). For many associations, this functional complexity is compounded by two further factors: (i) a high diversity and variable composition of the microbial partners (12–15) and (ii) variable contributions of different microorganisms and among-microbe interactions to microbe-dependent host traits (16–19).
The basis of this study is that the bacterial symbiosis in the pea aphid Acyrthosiphon pisum offers a superb system to investigate host interactions with beneficial microorganisms for two reasons. The first is that the association is naturally of low diversity. All pea aphids bear a bacterial symbiont, Buchnera aphidicola (gammaproteobacteria, henceforth known as Buchnera), which is localized to specialized insect cells known as bacteriocytes (20). In addition, many pea aphids bear one or more additional facultative bacteria, generically known as secondary symbionts, which are localized to the hemolymph as well as aphid cells (21, 22). Both Buchnera and these facultative symbionts are vertically transmitted via the insect ovary, but they confer different services. Buchnera provides the insect with essential amino acids (EAAs), required for sustained growth and reproduction on the EAA-deficient diet of phloem sap (23, 24), while some facultative symbionts confer ecologically important benefits, including protection against parasitic wasps and fungal pathogens (25). The pea aphid gut generally bears minimal numbers of transient microorganisms (26–28).
The second valuable trait of the aphid-bacterial symbiosis is that several facultative symbionts are amenable to experimental manipulation. Most research has focused on Hamiltonella defensa (gammaproteobacteria, henceforth known as Hamiltonella), which can be eliminated from aphids by selective antibiotic treatment, administered to Hamiltonella-free aphids by feeding or injection, and can be cultured in insect cell cultures and cell-free medium (29–32). Many Hamiltonella strains confer aphid resistance against parasitic wasps, likely mediated by toxins coded by a prophage on the genomes of protective Hamiltonella strains (32–35). Other data, however, suggest that Hamiltonella may also influence host immunity and feeding behavior (36–38). Metabolite exchange between the whitefly Bemisia tabaci and the sister taxon of aphid Hamiltonella (also known as Hamiltonella defensa) has been inferred from genomic data and metabolic modeling (39–42), but the impact of Hamiltonella on aphid metabolism has not, to our knowledge, been investigated systematically.
Our specific purpose was to identify how Hamiltonella influences the metabolic function of the pea aphid and its obligate bacterial symbiont Buchnera. Initial comparisons of the metabolome of aphid lines that naturally bear or lack Hamiltonella and a single aphid genotype with or without Hamiltonella led us to focus on Buchnera-mediated synthesis of one EAA, histidine. Using metabolism experiments and metabolic modeling, we demonstrate that for the single genotype tested, Buchnera synthesizes histidine at elevated rates in aphids bearing Hamiltonella and that both the population of Hamiltonella and aphid host contribute to the increased demand for Buchnera-derived histidine.
RESULTS
Metabolite profiles of pea aphids bearing and lacking Hamiltonella.
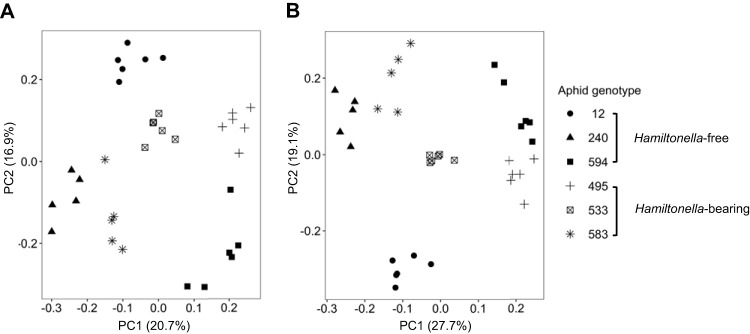
The metabolite profile of six pea aphid genotypes, three of which were naturally infected with Hamiltonella and three of which were naturally Hamiltonella-free, was measured by untargeted liquid chromatography-mass spectrometry (LC-MS) (see Data Sets S1 and S2 in the supplemental material). Principal-component analysis (PCA) of all detected features revealed clustering by aphid genotype, but not Hamiltonella infection status (Fig. 1). Initial analysis with individual t tests, as is standard for analysis of metabolomics data (43), identified 11 metabolites that differed significantly between Hamiltonella-bearing and Hamiltonella-free aphids (see Table S1 in the supplemental material). However, when these 11 metabolites were taken for more rigorous analysis that included the effect of aphid genotype and correction for multiple testing, no metabolites differed significantly between Hamiltonella-bearing and Hamiltonella-free aphids (Table S1).
FIG 1.
Principal-component analysis (PCA) of aphid metabolites quantified using untargeted liquid chromatography-mass spectrometry (LC-MS). (A) Positive mode. (B) Negative mode. Individual points represent biological replicates, and each symbol corresponds to an aphid genotype. The variance explained by each principal component axis is shown in parentheses.
Metabolites enriched in three naturally Hamiltonella-bearing compared to three naturally Hamiltonella-free aphid genotypes. Significant differences in metabolite abundances were detected using t tests and linear mixed-effects models. F statistics and P values are from the linear mixed-effects models where Hamiltonella status was treated as a fixed effect and aphid genotype was treated as a random effect. Benjamini-Hochberg (B to H) multiple testing corrections were performed on P values for fixed and random effects in the linear mixed-effects models. Download Table S1, DOCX file, 0.02 MB (16.1KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of isogenic lines bearing and lacking Hamiltonella.
Two factors may contribute to the lack of significant metabolic differences between Hamiltonella-bearing and Hamiltonella-free aphid genotypes: Hamiltonella may have a minimal effect on the metabolite pools of the aphids; or the influence of Hamiltonella on aphid metabolite profiles may be obscured by metabolic variation among the aphid-Buchnera genotypes. To control for genotype effects, we selectively eliminated Hamiltonella from aphids of genotype SC_583, yielding SC_583H- lacking Hamiltonella. (SC_583 was selected because it is amenable to Hamiltonella clearance using antibiotics, and it is one of the genotypes used in the initial metabolomics study.) Neither aphid performance nor the abundance and activity of their Buchnera populations differed significantly between the two lines (Table 1; see also Text S1A and B in the supplemental material). These data indicate that any metabolic differences between the two lines are likely direct effects of Hamiltonella on the metabolic function of the symbiosis, rather than a nonspecific consequence of Hamiltonella effects on aphid growth, development, or reproductive output or on the Buchnera population.
TABLE 1.
Performance and Buchnera symbiosis in isogenic pea aphid lines bearing Hamiltonella (SC_583) and experimentally deprived of Hamiltonella (SC_583H-)a
| Aphid line | Aphid performance |
Buchnera populationb
|
||
|---|---|---|---|---|
| Intrinsic rate of increase (rm) (aphids aphid−1 day−1) (10 replicates) |
Larval relative growth rate (mg mg−1 day−1) (15 replicates) |
Abundance (16S copies in gDNA/aphid ef1α copies) (6 replicates) |
Activity (16S copies in cDNA/gDNA) (6 replicates) |
|
| SC_583 | 0.351 ± 0.005 | 0.394 ± 0.006 | 0.77 + 0.035 | 1.24 + 0.060 |
| SC_583H- | 0.347 ± 0.010 | 0.377 ± 0.007 | 0.82 + 0.040 | 1.29 + 0.043 |
| t18 = 0.360, P = 0.723 | t28 = 1.90, P = 0.068 | t10 = 0.97, P = 0.356 | t10 = 0.58, P = 0.574 | |
The last row of the table shows the t value (test statistic with degrees of freedom indicated as subscript) and P value comparing the values for the SC_583 and SC_583H- aphid lines.
Determined by quantitative PCR (qPCR), described in Text S1A in the supplemental material. gDNA, genomic DNA.
Supplemental text. (A) Quantitative PCR (qPCR) analysis of Buchnera abundance and activity. (B) Estimation of biomass ratio of Buchnera/Hamiltonella. (C) Preparation of aphid samples for metabolomics analysis of aphid genotypes bearing and lacking Hamiltonella. (D) Preparation of aphid samples for metabolomics analysis of isogenic lines SC_583 and SC_583H-. (E) Preparation of protein hydrolysates for analysis of metabolism of dietary [13C6]histidine by isogenic aphid lines SC_583 and SC_583H-. (F) Diagnostic PCR for detection of Hamiltonella. (G) Metabolic model constraints and analysis. Download Text S1, DOCX file, 0.02 MB (23.4KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic profile of isogenic aphids bearing and lacking Hamiltonella.
To investigate the metabolic traits of the aphid lines SC_583 and SC_583H-, we applied LC-MS to measure the metabolite profiles of the 7-day-old aphid larvae that had been reared on chemically defined diets from day 2 (Table S2). Three metabolites, all with roles in phenylalanine metabolism differed significantly between the two lines after correction for multiple tests: hydroxyphenylpyruvate, prephenate, and phenyllactic acid (Table S3). These metabolites had not been identified as candidates in our initial analysis of naturally Hamiltonella-bearing and Hamiltonella-free clones (Table S1).
Metabolites identified by negative-mode LC-MS analysis of metabolite pools extracted from day 7 aphid larvae of isogenic lines bearing (SC_583) and lacking (SC_583H-) Hamiltonella reared on chemically defined diets. Download Table S2, DOCX file, 0.03 MB (34.1KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential metabolite abundance between isogenic lines bearing (SC_583) and lacking (SC_583H-) Hamiltonella. LC-MS analysis was performed on metabolite pools extracted from 7-day-old aphid larvae reared on chemically defined diets. Benjamini-Hochberg multiple testing corrections were performed on P values obtained from t tests on each metabolite. Download Table S3, DOCX file, 0.04 MB (44.6KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
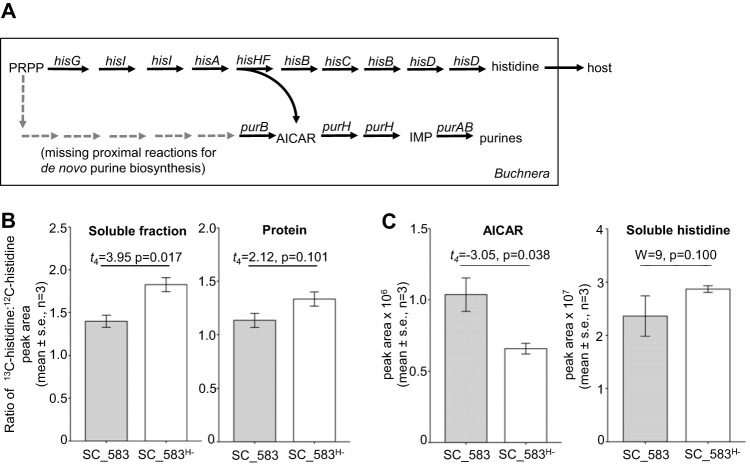
Further inspection of the metabolomics data yielded just one metabolite, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), which had a near-significant enrichment in Hamiltonella-bearing aphids (i.e., significant prior to adjustment for multiple tests) in both data sets (Table S1 and Table S3). AICAR is of considerable interest because its production is linked to the overproduction of the EAA histidine by Buchnera. Specifically, AICAR is a by-product of Buchnera histidine biosynthesis and, due to deletion of the proximal reactions for de novo purine biosynthesis, the sole Buchnera-derived substrate for Buchnera purine synthesis (Fig. 2A). It has been argued that Buchnera demand for AICAR to meet its purine requirements drives the overproduction of histidine, with the excess histidine delivered to the aphid host (44, 45). We hypothesized that, in Hamiltonella-bearing aphids, the metabolic demand for Buchnera-derived histidine exceeds Buchnera demand for purines, leading to the accumulation of AICAR.
FIG 2.
Histidine biosynthesis in isogenic aphid lines bearing Hamiltonella (SC_583) and lacking Hamiltonella (SC_583H-). (A) AICAR is a metabolic by-product of histidine biosynthesis in Buchnera. Reactions for de novo purine biosynthesis are absent (dashed arrows indicate genes missing from the Buchnera genome), and purines are instead synthesized from AICAR, which is a by-product of histidine biosynthesis in Buchnera. Abbreviations: PRPP, phosphoribosyl pyrophosphate; 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR); IMP, inosine monophosphate. (B) Incorporation of dietary [13C6]histidine, determined as the ratio of [13C]histidine to [12C]histidine into aphid soluble pools and hydrolyzed protein pools. (C) Corrected peak area of AICAR and total soluble histidine (all measurable isotopes combined) of aphids. Statistical tests applied the critical probability of 0.025, following Bonferroni correction for two tests. s.e., standard error.
Buchnera-mediated histidine biosynthesis.
To investigate the effect of Hamiltonella on Buchnera-mediated histidine biosynthesis, we raised larvae of the lines SC_583 and SC_583H- on chemically defined diet with histidine supplied exclusively as [13C6]histidine for 5 days and then measured the 13C/12C ratio of free histidine and protein-bound histidine (Table S4). [12C]histidine is derived from Buchnera-mediated synthesis (Hamiltonella lacks the genetic capacity to synthesize histidine [46], and reduced [13C]histidine/[12C]histidine is indicative of increased contribution of Buchnera-derived histidine).
Histidine isotopes and AICAR identified in soluble metabolite and hydrolyzed protein pools by negative-mode LC-MS analysis of isogenic lines SC_583 and SC_583H- reared on chemically defined diets containing 2mM [13C6]histidine. Download Table S4, DOCX file, 0.01 MB (15.4KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Consistent with our prediction that histidine synthesis is increased in Hamiltonella-bearing aphids, the 13C/12C ratio in the soluble histidine pool was significantly lower in SC_583 than SC_583H- aphids at P = 0.025 threshold (Bonferroni correction for two tests) (Fig. 2B). The equivalent data for histidine in the protein fraction showed the same trend of reduced 13C/12C in SC_583, but the effect was not significant (Fig. 2B). 13C was predominantly recovered from fully labeled [13C]histidine (His M+6), accounting for 53 to 63% of total histidine in the soluble fraction and 47 to 55% in the protein fraction, and the equivalent values for fully unlabeled [12C]histidine (His M+0) were 32 to 42% and 39 to 48%, respectively (see Fig. S1 in the supplemental material).
Relative proportions of histidine isotopes. (A) Hydrolyzed protein pools. (B) Soluble pools of histidine. Extracted from isogenic 7-day-old line SC_583 (bearing Hamiltonella) and SC_583H- (Hamiltonella-free) larvae reared on diets containing [13C]histidine from day 2 to day 7 of larval development. Error bars show standard deviations for three biological replicates per treatment and time point. Download FIG S1, PDF file, 0.3 MB (267.6KB, pdf) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Further analysis of this data set showed that AICAR content, but not histidine content, was significantly elevated in SC_583 relative to SC_583H- (Fig. 2C), recapitulating the results of the previous experiments (Tables S1 and S3).
The metabolic determinants of AICAR content.
We hypothesized that the relationship between increased Buchnera production of histidine (as revealed by the 13C/12C ratio of histidine) and increased AICAR content of aphids bearing Hamiltonella could be explained by the metabolic link between the synthesis of histidine and purines in Buchnera (Fig. 2A). Specifically, AICAR is predicted to accumulate under conditions where the total symbiosis demand for Buchnera-derived histidine exceeds the Buchnera demand for AICAR as the substrate for purines.
To investigate whether Hamiltonella demand for extra histidine creates an overflow of AICAR from Buchnera, we compared the metabolic flux in a two-compartment metabolic model, comprising Buchnera and the aphid host, and three-compartment models that also included Hamiltonella with Buchnera/Hamiltonella biomass ratios ranging from 10:1 to 1:5 (Fig. 3). We applied flux balance analysis to quantify how AICAR production varies with histidine synthesis, as determined by flux through the HisD reaction. In the Hamiltonella-free model, Buchnera releases no AICAR (Fig. 3A). In the three-compartment model with Buchnera/Hamiltonella biomass ratio greater than one, Buchnera exhibits modest increase in histidine production (Fig. 3B), supporting Hamiltonella demand for this EAA, accompanied by AICAR overflow (Fig. 3A). As the Hamiltonella biomass exceeds that of Buchnera, the model predicts further increase in Buchnera histidine production and AICAR overflow. At the most extreme Buchnera/Hamiltonella biomass ratio tested of 1:5 (equivalent to 1:130 cell number ratio), HisD flux is increased by 35% and AICAR overflow is more than doubled.
FIG 3.
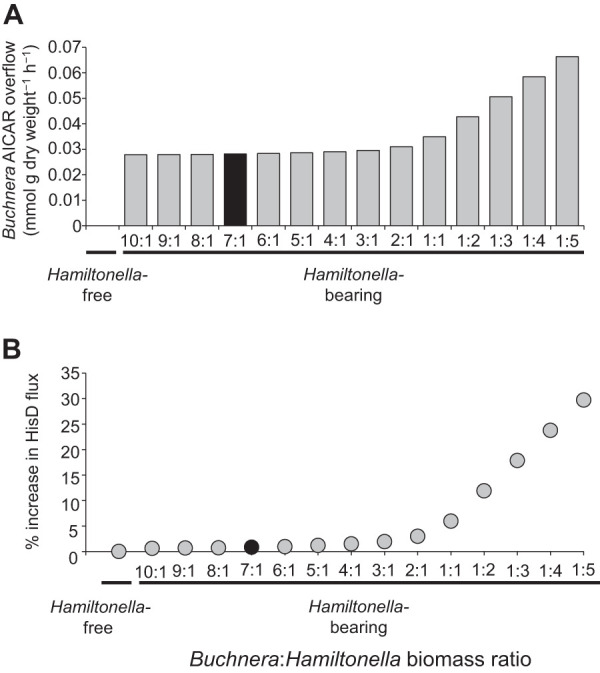
Histidine and AICAR production by Buchnera predicted from flux balance analysis of the Hamiltonella-free model comprising Buchnera and the aphid, and three-compartment models with increasing biomass of Hamiltonella. (A) AICAR overflow. (B) Flux through HisD reaction, as an index of total histidine production. Values corresponding to the empirically determined Buchnera/Hamiltonella ratio are indicated by a black bar and black circle.
The relative cell number of Buchnera/Hamiltonella in line SC_583 was determined empirically, at 1:3.9 (standard error [s.e.] = 0.3, n = 18), equivalent to the biomass ratio of 6.7:1 (Text S1B). Under these conditions, the predicted increase in histidine biosynthesis to support the Hamiltonella population is 0.8%, and the predicted AICAR overflow is 0.03 mmol g Buchnera biomass−1 h−1 (Fig. 3).
The modest increase in histidine yield (0.8%) predicted by our models compared to the 25% increase in histidine pools observed empirically suggests that an increase in Hamiltonella demand for histidine alone may not fully account for the increased Buchnera histidine production observed in Hamiltonella-bearing line SC_583. Altogether, our empirical and modeling data indicate that Hamiltonella induces additional demands for Buchnera histidine production by a third player, the aphid host.
DISCUSSION
Animals that naturally house a few microbial taxa are powerful systems to investigate the metabolic interactions among microbial taxa and the host. Here, we leveraged the tripartite symbiosis between the pea aphid, its obligate nutritional symbiont Buchnera, and a facultative defensive symbiont Hamiltonella to investigate how Hamiltonella affects host-Buchnera metabolic function. We demonstrated that, in the presence of Hamiltonella, Buchnera increases production of the EAA histidine, and we inferred that the extra Buchnera-derived histidine meets both Hamiltonella demand and Hamiltonella-induced increase in host demand for this EAA. Here, in the Discussion, we explore, in turn, how our three approaches, metabolomics, metabolic experiments, and metabolic modeling, contribute to our understanding of the metabolic consequences of Hamiltonella and some implications for metabolic interactions between other symbiotic microorganisms and their animal hosts.
Our first metabolomics analysis revealed a far greater effect of aphid genotype than Hamiltonella on the metabolome of aphids naturally bearing and lacking Hamiltonella (Fig. 1). These results show that our methodology was appropriate to detect substantial metabolomics differences and suggest that Hamiltonella may not cause a major reconfiguration of the host metabolome. Additionally, the clustering of the metabolomics data by genotype is indicative of large-scale intraspecific variation in metabolic function of the pea aphid. This striking pattern complements published evidence for significant among-genotype variation in aphid utilization of sucrose and amino acids (29, 47–49), the chief carbon and nitrogen sources in the aphid diet of plant phloem sap. Furthermore, this variation has been causally linked to specific genes of the aphid and Buchnera, as well as variation in Buchnera population size (47, 50–52). These patterns are also fully consistent with evidence from other animals that host genotype can strongly influence metabolic traits linked to microbiome function (53–57).
Although Hamiltonella does not perturb the global metabolic homeostasis of its aphid host, our metabolomics data sets included one metabolite, AICAR, with elevated titer in Hamiltonella-bearing aphids for both the comparisons between genotypes that naturally harbor and lack Hamiltonella and between a single genotype bearing and experimentally deprived of Hamiltonella. The correspondence between the presence of Hamiltonella, increased AICAR, and increased Buchnera-mediated histidine production, as determined by our 13C metabolic experiments, confirmed our hypothesis that the additional AICAR was likely of Buchnera origin and linked to increased demand for Buchnera-derived histidine in Hamiltonella-bearing aphids.
Aphids bearing Hamiltonella are expected to have an elevated histidine requirement because Hamiltonella is auxotrophic for this EAA (46, 58). Hamiltonella has the genetic capacity to synthesize just two EAAs, threonine and lysine, and it is expected to be a sink for the other eight EAAs (including histidine), all of which are synthesized by Buchnera (24). These predicted EAA fluxes from Buchnera to Hamiltonella do not translate into significant effects on the titers of histidine or other EAAs in the aphid metabolome, presumably because of homeostatic controls over metabolite pool sizes. Hamiltonella may impose a greater demand for histidine than other EAAs; alternatively, increased flux through other Buchnera EAA biosynthetic pathways in Hamiltonella-bearing aphids may have gone undetected in our study because it did not result in the accumulation of unique by-product(s) equivalent to AICAR for histidine synthesis. An indication that Hamiltonella may alter the metabolism of a second EAA, phenylalanine, comes from the significant underrepresentation of three intermediates in the phenylalanine biosynthetic and catabolic pathways (hydroxyphenylpyruvate, prephenate and phenyllactic acid) in Hamiltonella-bearing isogenic aphids (see Table S3 in the supplemental material).
Interestingly, the nutritional requirements of Hamiltonella resulted in no discernible reduction of aphid fitness, despite increased EAA demand. Possible contributory factors were that we used naturally occurring aphid-Hamiltonella combinations and a susceptible plant cultivar for insect culture. In published studies, the effect of Hamiltonella on aphid performance varies with aphid and bacterial genotype (59–61) and can be particularly deleterious for aphids reared on partially resistant plants (36, 62).
An important inference from this study is that Hamiltonella demand for histidine is unlikely to account fully for the difference in histidine production between Hamiltonella-bearing and Hamiltonella-free lines. The chief evidence came from metabolic models that assumed fixed aphid demand for histidine; when Hamiltonella at the empirically determined biomass was added to the aphid-Buchnera model, the computed flux of histidine synthesis increased by just 0.8%, substantially less than the observed 25% difference in Buchnera-derived histidine production between aphids bearing and lacking Hamiltonella. A possible factor contributing to this discrepancy may have been the simplifying assumptions required to construct the flux balance models (63, 64), although the model equations are not discernibly biased to underestimate Hamiltonella demand for histidine. It is most probable that increased host demand for histidine in Hamiltonella-bearing aphids contributes much of the discrepancy between the empirical data (Fig. 2B) and model data (Fig. 3).
Why might Hamiltonella increase host demand for histidine and possibly other Buchnera-derived EAAs? Two processes may be involved. First, Hamiltonella has been demonstrated to alter the cellular immunity of pea aphids, specifically by increasing the population of hemocytes (37). This effect would increase the host sink for histidine because as for animals generally (65, 66), immune cell proliferation in aphids is metabolically demanding and requires metabolic resources, including EAAs. Second, aphid feeding, including probing behavior and food ingestion can be altered by Hamiltonella (36). These feeding traits can substantially affect dietary EAA supply to the aphid by influencing aphid choice of feeding site and food consumption rates. Previous research has demonstrated that rearing aphids on diets lacking a specific EAA results in increased synthesis of that EAA by Buchnera (67). This suggests that increased production of histidine by Buchnera could arise from feeding changes in aphids bearing Hamiltonella that resulted in reduced uptake of dietary histidine.
In conclusion, this study has revealed that the tripartite relationship between two bacterial symbionts and their aphid host is metabolically interactive. Specifically, the nutritional requirements of one bacterium, Hamiltonella, can modify the metabolic function of a second symbiont, Buchnera, for increased histidine production without altering the Buchnera population size, and the Buchnera response to support Hamiltonella is likely compounded by the Hamiltonella-induced increase in host demand for histidine. These findings raise two general questions. The first relates to the role of Hamiltonella as a defensive symbiont that protects the aphid host against parasitic wasps (33). Although this defensive function has been attributed to toxins encoded by a prophage on the Hamiltonella genome (35), future research should consider the possible contribution to parasitoid resistance of Hamiltonella-induced changes to aphid metabolism and immunity. The second question is the incidence of multiway metabolic interactions in symbioses. On the one hand, the substantial among-genotype differences in the aphid metabolome identified in this study suggests that further insights can be gleaned from analysis of intraspecific variation in these interactions. This avenue would be a productive extension of recent research on variation in aphid-Buchnera interactions (50, 52) and idiosyncratic effects of facultative symbionts on host phenotype (59–61). On the other hand, the broad principle of multiway interactions may be general to multiple taxa localized to the hemolymph and cells of insects (68), including other facultative symbionts of aphids and bacteria with a broad distribution in arthropods, e.g., Wolbachia, Spiroplasma. As in this study, these future investigations will be facilitated by the combined application of metabolomics, isotope tracer experiments, and metabolic modeling.
MATERIALS AND METHODS
Experimental aphids.
The experiments were conducted on six genotypes of the pea aphid Acyrthosiphon pisum collected from an alfalfa field in Ithaca, NY, USA, in May 2015. All genotypes bore the vertically transmitted bacterial symbiont Buchnera; genotypes SC_12, SC_240 and SC_594 bore no previously described facultative symbionts, also known as secondary symbionts, SC_533 and SC_583 bore Hamiltonella, and SC_495 had both Hamiltonella and Spiroplasma (50).
The aphids were maintained on Vicia faba cv. Windsor at 20°C with 16-h light/8-h dark light cycle. To generate age-synchronized larvae, adult apterous females were allowed to larviposit for 24 h on V. faba plants or excised leaves and then removed. The deposited larvae were left to develop for a further day (to day 2) for use in experiments.
Metabolomics (LC-MS) analysis.
To analyze aphid genotypes that naturally harbored and lacked Hamiltonella, five or six replicate pools of mixed-age aphids of each genotype were collected from routine culture on plants, with 25 mg fresh weight per replicate. Following sample preparation (see Text S1C in the supplemental material), 5 μl of each sample was injected onto an AB SCIEX 5600 TripleTOF (triple time of flight) liquid chromatography-mass spectrometry (LC-MS) for analysis in positive and negative electrospray ionization (ESI) mode, with method blanks and a pooled quality control (QC) sample. Samples were separated by reverse-phase high-performance liquid chromatography (HPLC) using Prominence 20 UFLCXR system (Shimadzu) with a BEH C18 column (100 mm × 2.1 mm; 1.7 μm particle size; Waters) maintained at 55°C and a 20-min aqueous acetonitrile gradient (flow rate 250 μl min−1). The initial conditions were 97% solvent A (HPLC grade water with 0.1% formic acid) and 3% solvent B (HPLC grade acetonitrile with 0.1% formic acid), increasing to 45% solvent B at 10 min, 75% solvent B at 12 min where it was held until 17.5 min before returning to initial conditions. The eluate was delivered into a 5600 (QTOF) TripleTOF using a Duospray ion source (AB SCIEX). The capillary voltage was set at 5.5 kV in positive ion mode and 4.5 kV in negative ion mode, with declustering potential of 80 V. The mass spectrometer was operated in information-dependent acquisition mode with 100-ms survey scan from 100 to 1200 m/z, and up to 20 MS/MS (tandem MS) product ion scans (100 ms) per duty cycle using a collision energy of 50 V with a 20-V spread.
Instrument raw data files were converted into mzML format using Proteowizard (69) and analyzed using MS-DIAL (70). Mass spectrometry tolerances for MS1 and MS2 were set to 0.01 Da and 0.05 Da, corresponding to the resolution of the TripleTOF instrument. For smoothing extracted-ion chromatography, the linear weighted moving average was applied with a smoothing level of 3, and the minimum peak height was set to 3,000 for noise signal. Compound identification used MS/MS similarity to the curated public library in MS-DIAL with 80% similarity threshold and peak areas normalized using the internal standard chlorpropamide (Santa Cruz Biotech).
Metabolomics analysis of isogenic lines SC_583 and SC_583H- used three groups of 10 2-day-old larvae administered diet with 2 mM histidine to mimic V. faba phloem sap (71). Five days later, the larvae (7-days-old, approximately 25 mg fresh aphid material per sample) were snap-frozen in liquid nitrogen and stored at –80°C. Following extraction of metabolites (protocol in Text S1D), metabolites were separated using a Waters XSelect HSS T3 column (100 Å, 5 μm, 2.1 mm × 100 mm) fitted with a Restex UltraShield 0.2-μm precolumn filter and analyzed on a Thermo Exactive Plus Orbitrap (72–74). Blanks and a pooled QC sample were included in the analysis. Raw data files were converted to mzXML format and processed in MAVEN (75) using an in-house targeted metabolite library (72). Compounds were identified based on m/z (±10 ppm) and retention time (±0.5 min) tolerances to standards. Peak areas were normalized against the total ion chromatogram to account for analytical drift, followed by blank subtraction.
For analysis of dietary [13C]histidine metabolism by isogenic aphid lines SC_583 and SC_583H-, six replicate groups of 2-day-old aphids per line were raised on diets containing 2 mM 13C6-labeled histidine for 5 days (to day 7). Each replicate, comprising 30 larvae (approximately 25 mg fresh aphid material), was snap-frozen in liquid nitrogen and stored at –80°C. Three replicates per line were analyzed for soluble metabolites and extracted as described above, and protein hydrolysates were prepared as described previously (76) (Text S1E). Samples were analyzed on a Thermo Exactive Plus Orbitrap as described above. For histidine isotopes, all isotopic peaks were picked manually using the expected isotopic mass and the tolerances m/z (±10 ppm) and retention time (±0.5 min).
Antibiotic treatment of pea aphid genotype SC_583 to eliminate Hamiltonella.
The larvae of genotype SC_583 were treated with antibiotics as in reference 29. Three cages, each with five 2-day-old larvae of genotype SC_583, were maintained on a chemically defined diet (formulation in reference 77 with 0.5 M sucrose and 0.15 M amino acids), supplemented with the antibiotics gentamicin, cefotaxime, and ampicillin (Sigma) each at 50 μg ml−1 diet for 5 days (to day 7), when they were transferred individually to plants and allowed to larviposit.
A single offspring (generation 2) per aphid was tested for Hamiltonella by diagnostic PCR assay (50) (Text S1F). Up to 10 siblings of individuals that tested negative were transferred individually to fresh plants, and five of their progeny (generation 3) were tested by PCR for Hamiltonella. This was repeated to generation 10. Where an aphid tested positive for Hamiltonella, all codescendants from the generation 2 aphid were discarded. By this procedure, we generated the Hamiltonella-free line SC_583H-. As expected, hemolymph samples from leg bleeds of genotype SC_583 bore many bacterial cells of the morphology predicted for Hamiltonella (21, 31,) but hemolymph samples from line SC_583H- were bacteria free.
Aphid performance assays.
To determine larval relative growth rate (RGR) of aphid lines SC_583 and SC_583H-, 15 replicate groups of five 2-day-old aphids were weighed (±1 μg) and confined to clip cages on V. faba plants. On day 7, the surviving larvae in each clip cage were counted and weighed. RGR was calculated as loge (day 7 weight per aphid/day 2 weight per aphid)/5 days. To determine the intrinsic rate of increase (rm), larvae deposited by adults over 24 h on V. faba plants were allowed to develop for 6 days, when they were in the final larval stadium, and then individually transferred to the underside of a leaf of a fresh plant in a clip cage. Aphids were then examined daily until the first offspring was deposited, to give the time from larviposition to onset of reproduction (d). Progeny were subsequently counted and removed every several days until 2d, yielding the total number of progeny per aphid (Md). The formula rm = 0.745(loge Md)/d (78) was applied, with 10 replicate individuals per line.
Metabolic model reconstruction and analysis.
A genome scale metabolic model of Hamiltonella defensa was generated by combining two draft model reconstructions. The first identified gene orthologs in the Hamiltonella defensa genome (NCBI JAABOV000000000) and Escherichia coli strain K-12 substrain MG1655 by reciprocal BLAST searches and then manually extracted reactions encoded by these genes from E. coli strain K-12 substrain MG1655 metabolic model iML1515 (79) to create a draft model, as previously described (41). The second draft reconstruction was generated from the automated reconstruction pipeline ModelSEED (80) using a RAST (81) reannotated Hamiltonella defensa genome as input. The two draft models were then integrated and manually curated to remove redundant reactions and ensure correct reaction gene association, directionality, stoichiometry, and mass-charge balance. Hamiltonella-specific features and genes encoding metabolic reactions absent in the E. coli iML1515 metabolic model were identified by literature review and searches of the BioCyc, KEGG, EcoCyc, BiGG, and BRENDA databases (82–86) and then added to the draft model.
The Buchnera metabolic model was updated from the published model (44, 87) by removing two reactions, adding 62 reactions (Data Set S3A) and updating the biomass equation (see Data Set S3B for details).
Positive-mode LC-MS analysis of six aphid genotypes naturally bearing and lacking Hamiltonella. (A) Metabolites identified by fragmentation of precursor ions (MS2). (B) Metabolites identified from precursor ions (MS1). (C) Precursor ions that could not be identified as known metabolites. (D) Sample key. Download Data Set S1, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Negative-mode LC-MS analysis of six aphid genotypes naturally bearing and lacking Hamiltonella. (A) Metabolites identified by fragmentation of precursor ions (MS2). (B) Metabolites identified from precursor ions (MS1). (C) Precursor ions that could not be identified as known metabolites. (D) Sample key. Download Data Set S2, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic model data. (A) Updates to published Buchnera model iSM199. (B) Objective function components for Buchnera, Hamiltonella, and aphid metabolic models. (C) Aphid hemolymph growth medium used for simulations. Download Data Set S3, XLSX file, 0.02 MB (26.2KB, xlsx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A genome scale metabolic model for the aphid host was generated as previously described (41), using aphid reactions involved in primary metabolism identified from publicly available Acyrthosiphon pisum genome data (NCBI: GCA_000142985.2). Additional reactions to generate or consume dead-end metabolites were identified and incorporated into the aphid host draft reconstruction. Individual bacterial and host metabolic models were integrated into a three-compartment model using previously described methods (41, 88). Model testing was conducted in COBRA Toolbox version 3.0 (89) run in Matlab 2015b (The MathWorks Inc.), using the Gurobi version 6.5.0 solver (Gurobi Optimization 2016). Details of model analysis and constraints applied are provided in Text S1G.
Statistical analyses.
All statistical analyses were performed in R (90). Data sets were inspected for normality and homoscedasticity using Shapiro-Wilk and Kolomogorov-Smirnov tests and Bartlett’s test, respectively. Differences in data with normal distributions and homogenous variances were tested with Student’s t test, and data sets with nonnormal distributions and/or heterogeneous variances were tested with Mann-Whitney U test. Statistically significant differences in metabolite abundance detected by LC-MS analysis of six aphid genotypes were calculated using a linear mixed-effects model (LMM) using the “lmer” function in the lme4 package, version 1.1-19 (91). Hamiltonella status was treated as a fixed effect, and aphid genotype was treated as a random effect. Residuals were visually inspected for normality, and an analysis of variance (ANOVA) was performed using the car package, version 3.0-2 (92). P values were adjusted for multiple testing using the Benjamini-Hochberg method.
Data availability.
The Hamiltonella defensa genome assembly and raw sequencing reads used to generate the genome scale metabolic model are available in the GenBank repository under accession number PRJNA602159 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA602159). The multicompartment model is provided in three formats—SBML (.xml), MATLAB (.mat), and Excel (.xls)—and deposited in GitHub (https://github.com/na423/Aphid_symbiosis). An SBML file of the models is also available in the BioModels database (93) with the identifier MODEL2001310002. All other data are provided in supplemental files.
ACKNOWLEDGMENTS
We thank Mihee Choi for assistance with aphid maintenance and Mariyam Masood for help with aphid sample preparation for DNA sequencing.
This study was funded by NSF grant IOS-1354743 and foundational grants 2015-67013-23421 and 12216941 from the National Institute of Food and Agriculture.
F.B., N.Y.D.A., and A.E.D. designed the study. F.B. and N.C. performed the experiments. N.Y.D.A. conducted the metabolic modeling. E.L.A., Q.L. and M.A. prepared samples for LC-MS. I.K. and E.L.A. analyzed LC-MS data. A.D.P. oversaw LC-MS experiments. F.B. and A.E.D. wrote the first draft of the manuscript. All authors reviewed the manuscript.
Footnotes
Citation Blow F, Ankrah NYD, Clark N, Koo I, Allman EL, Liu Q, Anitha M, Patterson AD, Douglas AE. 2020. Impact of facultative bacteria on the metabolic function of an obligate insect-bacterial symbiosis. mBio 11:e00402-20. https://doi.org/10.1128/mBio.00402-20.
Contributor Information
Yoshitomo Kikuchi, National Institute of Advanced Industrial Science and Technology.
Edward G. Ruby, University of Hawaii at Manoa.
REFERENCES
- 1.Douglas AE. 2010. The symbiotic habit. Princeton University Press, Princeton, NJ. [Google Scholar]
- 2.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerardo NM, Parker BJ. 2014. Mechanisms of symbiont-conferred protection against natural enemies: an ecological and evolutionary framework. Curr Opin Insect Sci 4:8–14. doi: 10.1016/j.cois.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Granato ET, Meiller-Legrand TA, Foster KR. 2019. The evolution and ecology of bacterial warfare. Curr Biol 29:R521–R537. doi: 10.1016/j.cub.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Lapebie P, Lombard V, Drula E, Terrapon N, Henrissat B. 2019. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat Commun 10:2043. doi: 10.1038/s41467-019-10068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 7.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 8.Smith K, McCoy KD, Macpherson AJ. 2007. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas AE. 2014. Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harbor Perspect Biol 6:a016113. doi: 10.1101/cshperspect.a016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. 2017. Dysbiosis and the immune system. Nat Rev Immunol 17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd-Price J, Abu-Ali G, Huttenhower C. 2016. The healthy human microbiome. Genome Med 8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garud NR, Pollard KS. 2020. Population genetics in the human microbiome. Trends Genet 36:53–67. doi: 10.1016/j.tig.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adair KL, Douglas AE. 2017. Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol 35:23–29. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Cosetta CM, Wolfe BE. 2019. Causes and consequences of biotic interactions within microbiomes. Curr Opin Microbiol 50:35–41. doi: 10.1016/j.mib.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Trosvik P, de Muinck EJ. 2015. Ecology of bacteria in the human gastrointestinal tract–identification of keystone and foundation taxa. Microbiome 3:44. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7:e01250-16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol 17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 20.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 21.Moran NA, Russell JA, Koga R, Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol 71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry LM, Peccoud J, Simon J-C, Hadfield JD, Maiden MJC, Ferrari J, Godfray HCJ. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol 23:1713–1717. doi: 10.1016/j.cub.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akman Gunduz E, Douglas AE. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci 276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 25.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 26.Grenier AM, Nardon C, Rahbe Y. 1994. Observations on the micro-organisms occurring in the gut of the pea aphid Acyrthosiphon pisum. Entomol Exp Appl 70:91–96. doi: 10.1111/j.1570-7458.1994.tb01762.x. [DOI] [Google Scholar]
- 27.Jing X, Wong AC, Chaston JM, Colvin J, McKenzie CL, Douglas AE. 2014. The bacterial communities in plant phloem-sap-feeding insects. Mol Ecol 23:1433–1444. doi: 10.1111/mec.12637. [DOI] [PubMed] [Google Scholar]
- 28.Gauthier JP, Outreman Y, Mieuzet L, Simon JC. 2015. Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS One 10:e0120664. doi: 10.1371/journal.pone.0120664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douglas AE, Francois CLMJ, Minto LB. 2006. Facultative ‘secondary’ bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol Entomol 31:262–269. doi: 10.1111/j.1365-3032.2006.00516.x. [DOI] [Google Scholar]
- 30.Koga R, Tsuchida T, Sakurai M, Fukatsu T. 2007. Selective elimination of aphid endosymbionts: effects of antibiotic dose and host genotype, and fitness consequences. FEMS Microbiol Ecol 60:229–239. doi: 10.1111/j.1574-6941.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- 31.Darby AC, Chandler SM, Welburn SC, Douglas AE. 2005. Aphid-symbiotic bacteria cultured in insect cell lines. Appl Environ Microbiol 71:4833–4839. doi: 10.1128/AEM.71.8.4833-4839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandt JW, Chevignon G, Oliver KM, Strand MR. 2017. Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proc Biol Sci 284:866. doi: 10.1098/rspb.2017.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degnan PH, Moran NA. 2008. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol 74:6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver KM, Degnan PH, Hunter MS, Moran NA. 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leybourne DJ, Valentine TA, Bos JIB, Karley AJ. 2020. A fitness cost resulting from Hamiltonella defensa infection is associated with altered probing and feeding behaviour in Rhopalosiphum padi. J Exp Biol 223:jeb207936. doi: 10.1242/jeb.207936. [DOI] [PubMed] [Google Scholar]
- 37.Laughton AM, Garcia JR, Gerardo NM. 2016. Condition-dependent alteration of cellular immunity by secondary symbionts in the pea aphid, Acyrthosiphon pisum. J Insect Physiol 86:17–24. doi: 10.1016/j.jinsphys.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz A, Anselme C, Ravallec M, Rebuf C, Simon J-C, Gatti J-L, Poirié M. 2012. The cellular immune response of the pea aphid to foreign intrusion and symbiotic challenge. PLoS One 7:e42114. doi: 10.1371/journal.pone.0042114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao D, Hoffmann AA, Zhang Z, Niu H, Guo H. 2018. Interactions between facultative symbionts Hamiltonella and Cardinium in Bemisia tabaci (Hemiptera: Aleyrodoidea): cooperation or conflict? J Econ Entomol 111:2660–2666. doi: 10.1093/jee/toy261. [DOI] [PubMed] [Google Scholar]
- 40.Shan HW, Luan JB, Liu YQ, Douglas AE, Liu SS. 2019. The inherited bacterial symbiont Hamiltonella influences the sex ratio of an insect host. Proc Biol Sci 286:20191677. doi: 10.1098/rspb.2019.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ankrah NYD, Luan J, Douglas AE. 2017. Cooperative metabolism in a three-partner insect-bacterial symbiosis revealed by metabolic modeling. J Bacteriol 199:e00872-16. doi: 10.1128/JB.00872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao Q, Rollat-Farnier P-A, Zhu D-T, Santos-Garcia D, Silva FJ, Moya A, Latorre A, Klein CC, Vavre F, Sagot M-F, Liu S-S, Mouton L, Wang X-W. 2015. Genome reduction and potential metabolic complementation of the dual endosymbionts in the whitefly Bemisia tabaci. BMC Genomics 16:226. doi: 10.1186/s12864-015-1379-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia J, Wishart DS. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 44.Thomas GH, Zucker J, Macdonald SJ, Sorokin A, Goryanin I, Douglas AE. 2009. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst Biol 3:24. doi: 10.1186/1752-0509-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsey JS, MacDonald SJ, Jander G, Nakabachi A, Thomas GH, Douglas AE. 2010. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol 19(Suppl 2):241–248. doi: 10.1111/j.1365-2583.2009.00945.x. [DOI] [PubMed] [Google Scholar]
- 46.Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A 106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel KJ, Moran NA. 2011. Sources of variation in dietary requirements in an obligate nutritional symbiosis. Proc Biol Sci 278:115–121. doi: 10.1098/rspb.2010.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacDonald SJ, Thomas GH, Douglas AE. 2011. Genetic and metabolic determinants of nutritional phenotype in an insect-bacterial symbiosis. Mol Ecol 20:2073–2084. doi: 10.1111/j.1365-294X.2011.05031.x. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava PN, Gao Y, Levesque J, Auclair JL. 1985. Differences in amino acid requirements between two biotypes of the pea aphid, Acyrthosiphon pisum. Can J Zool 63:603–606. doi: 10.1139/z85-087. [DOI] [Google Scholar]
- 50.Chung SH, Parker BJ, Blow F, Brisson JA, Douglas AE. 2020. Host and symbiont genetic determinants of nutritional phenotype in a natural population of the pea aphid. Mol Ecol 29:848–858. doi: 10.1111/mec.15355. [DOI] [PubMed] [Google Scholar]
- 51.Chong RA, Moran NA. 2016. Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proc Natl Acad Sci U S A 113:13114–13119. doi: 10.1073/pnas.1610749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith TE, Moran NA. 2020. Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc Natl Acad Sci U S A 117:2113–2121. doi: 10.1073/pnas.1916748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, Westmiller S, Wong AC-N, Clark AG, Lazzaro BP, Douglas AE. 2015. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun 6:6312. doi: 10.1038/ncomms7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 55.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AAM, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A. 2016. The effect of host genetics on the gut microbiome. Nat Genet 48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 56.Kolde R, Franzosa EA, Rahnavard G, Hall AB, Vlamakis H, Stevens C, Daly MJ, Xavier RJ, Huttenhower C. 2018. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med 10:6. doi: 10.1186/s13073-018-0515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, Sogin ML. 2017. The microbiome and human biology. Annu Rev Genomics Hum Genet 18:65–86. doi: 10.1146/annurev-genom-083115-022438. [DOI] [PubMed] [Google Scholar]
- 58.Chevignon G, Boyd BM, Brandt JW, Oliver KM, Strand MR. 2018. Culture-facilitated comparative genomics of the facultative symbiont Hamiltonella defensa. Genome Biol Evol 10:786–802. doi: 10.1093/gbe/evy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez AJ, Doremus MR, Kraft LJ, Kim KL, Oliver KM. 2018. Multi-modal defences in aphids offer redundant protection and increased costs likely impeding a protective mutualism. J Anim Ecol 87:464–477. doi: 10.1111/1365-2656.12675. [DOI] [PubMed] [Google Scholar]
- 60.Weldon SR, Russell JA, Oliver KM. 2019. More is not always better: coinfections with defensive symbionts generate highly variable outcomes. Appl Environ Microbiol 86:e02537-19. doi: 10.1128/AEM.02537-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russell JA, Moran NA. 2006. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc Biol Sci 273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chandler SM, Wilkinson TL, Douglas AE. 2008. Impact of plant nutrients on the relationship between a herbivorous insect and its symbiotic bacteria. Proc Biol Sci 275:565–570. doi: 10.1098/rspb.2007.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kauffman KJ, Prakash P, Edwards JS. 2003. Advances in flux balance analysis. Curr Opin Biotechnol 14:491–496. doi: 10.1016/j.copbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Orth JD, Thiele I, Palsson BO. 2010. What is flux balance analysis? Nat Biotechnol 28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ardia DR, Gantz JE, Schneider BC, Strebel S. 2012. Costs of immunity in insects: an induced immune response increases metabolic rate and decreates antimicrobial activity. Funct Ecol 26:732–739. doi: 10.1111/j.1365-2435.2012.01989.x. [DOI] [Google Scholar]
- 66.Goldszmid RS, Trinchieri G. 2012. The price of immunity. Nat Immunol 13:932–938. doi: 10.1038/ni.2422. [DOI] [PubMed] [Google Scholar]
- 67.Russell CW, Poliakov A, Haribal M, Jander G, van Wijk KJ, Douglas AE. 2014. Matching the supply of bacterial nutrients to the nutritional demand of the animal host. Proc Biol Sci 281:20141163. doi: 10.1098/rspb.2014.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blow F, Douglas AE. 2019. The hemolymph microbiome of insects. J Insect Physiol 115:33–39. doi: 10.1016/j.jinsphys.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 69.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. 2015. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sasaki T, Aoki T, Hayashi H, Ishikawa H. 1990. Amino acid composition of the honeydew of symbiotic and aposymbiotic pea aphids Acyrthosiphon pisum. J Insect Physiol 36:35–40. doi: 10.1016/0022-1910(90)90148-9. [DOI] [Google Scholar]
- 72.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. 2010. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem 82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allman EL, Painter HJ, Samra J, Carrasquilla M, Llinas M. 2016. Metabolomic profiling of the malaria box reveals antimalarial target pathways. Antimicrob Agents Chemother 60:6635–6649. doi: 10.1128/AAC.01224-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klee SM, Sinn JP, Finley M, Allman EL, Smith PB, Aimufua O, Sitther V, Lehman BL, Krawczyk T, Peter KA, McNellis TW. 2019. Erwinia amylovora auxotrophic mutant exometabolomics and virulence on apples. Appl Environ Microbiol 85:e00935-19. doi: 10.1128/AEM.00935-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics Chapter 14:Unit14.11. doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haribal M, Jander G. 2015. Stable isotope studies reveal pathways for the incorporation of non-essential amino acids in Acyrthosiphon pisum (pea aphids). J Exp Biol 218:3797–3806. doi: 10.1242/jeb.129189. [DOI] [PubMed] [Google Scholar]
- 77.Douglas AE, Minto LB, Wilkinson TL. 2001. Quantifying nutrient production by the microbial symbionts in an aphid. J Exp Biol 204:349–358. [DOI] [PubMed] [Google Scholar]
- 78.Wyatt IJ, White PF. 1977. Simple estimation of intrinsic increase rates for aphids and tetranychid mites. J Appl Ecol 14:757–766. doi: 10.2307/2402807. [DOI] [Google Scholar]
- 79.Monk JM, Lloyd CJ, Brunk E, Mih N, Sastry A, King Z, Takeuchi R, Nomura W, Zhang Z, Mori H, Feist AM, Palsson BO. 2017. iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol 35:904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol 28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 81.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. 2008. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res 36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Schröder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schellenberger J, Park JO, Conrad TM, Palsson BO. 2010. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 11:213. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. 2004. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res 32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. 2012. The central role of the host cell in symbiotic nitrogen metabolism. Proc Biol Sci 279:2965–2973. doi: 10.1098/rspb.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ankrah NYD, Chouaia B, Douglas AE. 2018. The cost of metabolic interactions in symbioses between insects and bacteria with reduced genomes. mBio 9:e01433-18. doi: 10.1128/mBio.01433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V, Magnusdóttir S, Ng CY, Preciat G, Žagare A, Chan SHJ, Aurich MK, Clancy CM, Modamio J, Sauls JT, Noronha A, Bordbar A, Cousins B, El Assal DC, Valcarcel LV, Apaolaza I, Ghaderi S, Ahookhosh M, Ben Guebila M, Kostromins A, Sompairac N, Le HM, Ma D, Sun Y, Wang L, Yurkovich JT, Oliveira MAP, Vuong PT, El Assal LP, Kuperstein I, Zinovyev A, Hinton HS, Bryant WA, Aragón Artacho FJ, Planes FJ, Stalidzans E, Maass A, Vempala S, Hucka M, Saunders MA, Maranas CD, Lewis NE, Sauter T, Palsson BØ, Thiele I, Fleming RMT. 2019. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc 14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 91.Bates D, Machler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme-4. J Stat Software 67:1–48. [Google Scholar]
- 92.Fox J, Weisberg S. 2010. An R companion to applied regression. SAGE Publications Inc, Thousand Oaks, CA. [Google Scholar]
- 93.Chelliah V, Juty N, Ajmera I, Ali R, Dumousseau M, Glont M, Hucka M, Jalowicki G, Keating S, Knight-Schrijver V, Lloret-Villas A, Natarajan KN, Pettit J-B, Rodriguez N, Schubert M, Wimalaratne SM, Zhao Y, Hermjakob H, Le Novère N, Laibe C. 2015. BioModels: ten-year anniversary. Nucleic Acids Res 43:D542–D548. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolites enriched in three naturally Hamiltonella-bearing compared to three naturally Hamiltonella-free aphid genotypes. Significant differences in metabolite abundances were detected using t tests and linear mixed-effects models. F statistics and P values are from the linear mixed-effects models where Hamiltonella status was treated as a fixed effect and aphid genotype was treated as a random effect. Benjamini-Hochberg (B to H) multiple testing corrections were performed on P values for fixed and random effects in the linear mixed-effects models. Download Table S1, DOCX file, 0.02 MB (16.1KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental text. (A) Quantitative PCR (qPCR) analysis of Buchnera abundance and activity. (B) Estimation of biomass ratio of Buchnera/Hamiltonella. (C) Preparation of aphid samples for metabolomics analysis of aphid genotypes bearing and lacking Hamiltonella. (D) Preparation of aphid samples for metabolomics analysis of isogenic lines SC_583 and SC_583H-. (E) Preparation of protein hydrolysates for analysis of metabolism of dietary [13C6]histidine by isogenic aphid lines SC_583 and SC_583H-. (F) Diagnostic PCR for detection of Hamiltonella. (G) Metabolic model constraints and analysis. Download Text S1, DOCX file, 0.02 MB (23.4KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolites identified by negative-mode LC-MS analysis of metabolite pools extracted from day 7 aphid larvae of isogenic lines bearing (SC_583) and lacking (SC_583H-) Hamiltonella reared on chemically defined diets. Download Table S2, DOCX file, 0.03 MB (34.1KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential metabolite abundance between isogenic lines bearing (SC_583) and lacking (SC_583H-) Hamiltonella. LC-MS analysis was performed on metabolite pools extracted from 7-day-old aphid larvae reared on chemically defined diets. Benjamini-Hochberg multiple testing corrections were performed on P values obtained from t tests on each metabolite. Download Table S3, DOCX file, 0.04 MB (44.6KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Histidine isotopes and AICAR identified in soluble metabolite and hydrolyzed protein pools by negative-mode LC-MS analysis of isogenic lines SC_583 and SC_583H- reared on chemically defined diets containing 2mM [13C6]histidine. Download Table S4, DOCX file, 0.01 MB (15.4KB, docx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative proportions of histidine isotopes. (A) Hydrolyzed protein pools. (B) Soluble pools of histidine. Extracted from isogenic 7-day-old line SC_583 (bearing Hamiltonella) and SC_583H- (Hamiltonella-free) larvae reared on diets containing [13C]histidine from day 2 to day 7 of larval development. Error bars show standard deviations for three biological replicates per treatment and time point. Download FIG S1, PDF file, 0.3 MB (267.6KB, pdf) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Positive-mode LC-MS analysis of six aphid genotypes naturally bearing and lacking Hamiltonella. (A) Metabolites identified by fragmentation of precursor ions (MS2). (B) Metabolites identified from precursor ions (MS1). (C) Precursor ions that could not be identified as known metabolites. (D) Sample key. Download Data Set S1, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Negative-mode LC-MS analysis of six aphid genotypes naturally bearing and lacking Hamiltonella. (A) Metabolites identified by fragmentation of precursor ions (MS2). (B) Metabolites identified from precursor ions (MS1). (C) Precursor ions that could not be identified as known metabolites. (D) Sample key. Download Data Set S2, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic model data. (A) Updates to published Buchnera model iSM199. (B) Objective function components for Buchnera, Hamiltonella, and aphid metabolic models. (C) Aphid hemolymph growth medium used for simulations. Download Data Set S3, XLSX file, 0.02 MB (26.2KB, xlsx) .
Copyright © 2020 Blow et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The Hamiltonella defensa genome assembly and raw sequencing reads used to generate the genome scale metabolic model are available in the GenBank repository under accession number PRJNA602159 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA602159). The multicompartment model is provided in three formats—SBML (.xml), MATLAB (.mat), and Excel (.xls)—and deposited in GitHub (https://github.com/na423/Aphid_symbiosis). An SBML file of the models is also available in the BioModels database (93) with the identifier MODEL2001310002. All other data are provided in supplemental files.