Abstract
In this study, the anti–severe acute respiratory syndrome coronavirus‐2 (anti‐SARS‐CoV‐2) activity of mycophenolic acid (MPA) and IMD‐0354 was analyzed. These compounds were chosen based on their antiviral activities against other coronaviruses. Because they also inhibit dengue virus (DENV) infection, other anti‐DENV compounds/drugs were also assessed. On SARS‐CoV‐2‐infected VeroE6/TMPRSS2 monolayers, both MPA and IMD‐0354, but not other anti‐DENV compounds/drugs, showed significant anti‐SARS‐CoV‐2 activity. Although MPA reduced the viral RNA level by only approximately 100‐fold, its half maximal effective concentration was as low as 0.87 µ m, which is easily achievable at therapeutic doses of mycophenolate mofetil. MPA targets the coronaviral papain‐like protease and an in‐depth study on its mechanism of action would be useful in the development of novel anti‐SARS‐CoV‐2 drugs.
Keywords: antiviral agents, IMD‐0354, mycophenolic acid, SARS‐CoV‐2, VeroE6/TMPRSS2
Abbreviations
- AMED
Japan Agency for Medical Research and Development
- DENV
dengue virus
- FDA
United States Food and Drug Administration
- HCoV
human coronavirus
- IKKβ
inhibitor of nuclear factor kappa‐B kinase subunit beta
- IRF3
interferon regulatory factor 3
- IκBα
inhibitor of nuclear factor kappa B
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- MPA
mycophenolic acid
- NF‐κB
nuclear factor kappa‐light‐chain‐enhancer of activated B cells
- PLpro
papain‐like protease
- SARS‐CoV
severe acute respiratory syndrome coronavirus
In the last two decades, three coronaviruses in the genus Betacoronavirus, namely, severe acute respiratory syndrome coronavirus (SARS‐CoV), Middle East respiratory syndrome coronavirus (MERS‐CoV), and SARS‐CoV‐2, have demonstrated their potential for high pathogenicity in humans. The clinical severity of the infection caused by SARS‐CoV‐2 (COVID‐19) varies greatly among patients, ranging from mild symptoms to severe pneumonia, and in some cases even being asymptomatic. 1 The COVID‐19 outbreak started in Wuhan, China, in December 2019, spreading rapidly and globally. On March 11, the World Health Organization declared COVID‐19 a pandemic to emphasize the urgent necessity of global countermeasures against this deadly disease. However, neither approved drugs nor vaccines are currently available for infections caused by coronaviruses, including those by COVID‐19; thus, their development is required urgently. In this study, we tested the antiviral activity of mycophenolic acid (MPA) and IMD‐0354 against SARS‐CoV‐2. Previous studies have shown that MPA has antiviral activity against MERS‐CoV, human coronavirus (HCoV)‐OC43, HCoV‐NL63, and mouse hepatitis virus, 2 , 3 and that IMD‐0354 has antiviral activity against porcine transmissible gastroenteritis virus. 4 We have previously demonstrated that both MPA and IMD‐0354 exhibit antiviral activity against dengue virus (DENV) as well. 5 Thus, in addition to MPA and IMD‐0354, 12 other compounds/drugs, with demonstrated antiviral activity against DENV (Supplementary Table 1), were also assessed in this study. 6 , 7 , 8
In this study a simple CPE method, which involves the observation of CPE as an indicator of virus growth, was used for roughly evaluating the antiviral activity of individual compounds/drugs against SARS‐CoV‐2 infection. The method could be established based on the recent discovery that VeroE6/TMPRSS2 cells are highly sensitive and show strong CPE upon SARS‐CoV‐2 infection. 9 SARS‐CoV‐2 and various concentrations of individual compounds/drugs were mixed and added onto the VeroE6/TMPRSS2 cell monolayer. If the CPE is strong, following the addition of a drug/compound, the infected cells detach from the culture plate, and the crystal violet staining is barely detected, suggesting cytotoxicity of that compound/drug. By contrast, if the compound/drug shows antiviral activity, the monolayers remain attached, and the crystal violet staining is clearly visible. In this study, cells treated with appropriate concentrations of IMD‐0354 or seven anti‐DENV compounds (CCT08159, PD102807, CD437, retinoic acid, indatraline, maprotiline, and quinacrine) remained attached with clearly detectable crystal violet staining (up to the concentration indicated with blue bars; Figure 1). However, poor crystal violet staining was observed with cells treated with high concentrations of the compounds/drugs, likely due to cytotoxicity (from the concentration above the level indicated with red bars; Figure 1). The assumption of cytotoxicity of compounds/drugs was confirmed by WST assay (Dojindo Molecular Technologies, Japan; Supplementary Figure 1). The results of the WST assay were consistent with the cell viability assessed with the CPE method. These results indicate that the effective concentration range (between blue and red bars; Figure 1), which shows antiviral activity with low cytotoxicity, was narrow for CCT08159, CD437, indatraline, maprotiline, and quinacrine. Thus, the anti‐SARS‐CoV‐2 activity of these five compounds/drugs remained uncertain. MPA showed no anti‐SARS‐CoV‐2 activity in this CPE assay. By contrast, the anti‐SARS‐CoV‐2 activity of IMD‐0354 and two other compounds (PD102807 and retinoic acid) was apparent, but the drug concentrations required for complete CPE inhibition was relatively high (32 µm) for both PD102807 and retinoic acid. Accordingly, the CPE assay suggested that IMD‐0354 could be a candidate for anti‐SARS‐CoV‐2 drug.
Figure 1.
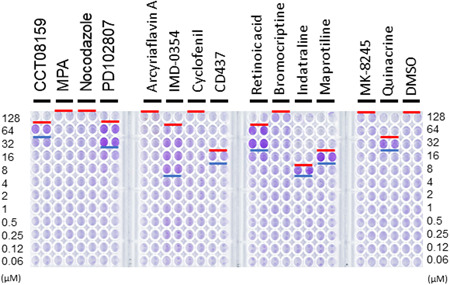
CPE assay. VeroE6/TMPRSS2 cell monolayers were cultured with the SARS‐CoV‐2 Wk521 strain 10 prepared in culture media containing different concentrations of drugs for 50 hr (MOI = 0.01). After the 50 hr incubation period, the cell monolayers were stained with crystal violet. Blue bars indicate the minimum drug concentration level by which viral CPE was blocked and cells remained attached after crystal violet staining (antiviral level). Red bars indicate the minimum drug concentration level by which the drugs showed cytotoxicity and poor crystal violet staining (cytotoxicity level)
Next, to evaluate the anti‐SARS‐CoV‐2 effect quantitatively and calculate the half maximal effective concentration (EC50) of these compounds/drugs, the level of SARS‐CoV‐2 RNA in the cells was analyzed using the same experimental conditions as in the CPE method. The compound/drug concentrations above the red bar in each graph (Figure 2) showed cytotoxicity in the CPE assay. The compound/drug concentrations above the blue bar in each graph showed anti‐SARS‐CoV activity in the CPE assay. Thus, the light purple area in each graph indicates the effective compound/drug concentration range, which is equivalent to the homogeneous crystal violet staining in the CPE assay. In agreement with the CPE assay data, the level of viral RNA was reduced by >10,000‐fold in cells treated with IMD‐0354 and seven identified compounds/drugs (CCT08159, PD102807, CD437, retinoic acid, indatraline, maprotiline, and quinacrine). The data from viral RNA and CPE assay revealed that a reduction of viral RNA to about 100‐fold (to the level of <106 copies of viral RNA) was necessary to inhibit the CPE to a level sufficient to keep cells attached after the crystal violet staining. MPA inhibited SARS‐CoV‐2 replication by 100‐fold; however, at high concentrations of MPA, when suppression was <100‐fold, CPE was observed (Figures 1 and 2). Importantly, the EC50 (dashed green lines in Figure 2) was the lowest (0.87 µm) for MPA among all the tested compounds/drugs. Indatraline and IMD‐0354, which showed strong inhibition activities for SARS‐CoV‐2 at moderate to high concentrations, showed the second and third lowest EC50 (1.57 and 1.74 µm, respectively) among the tested compounds/drugs (Figure 2).
Figure 2.
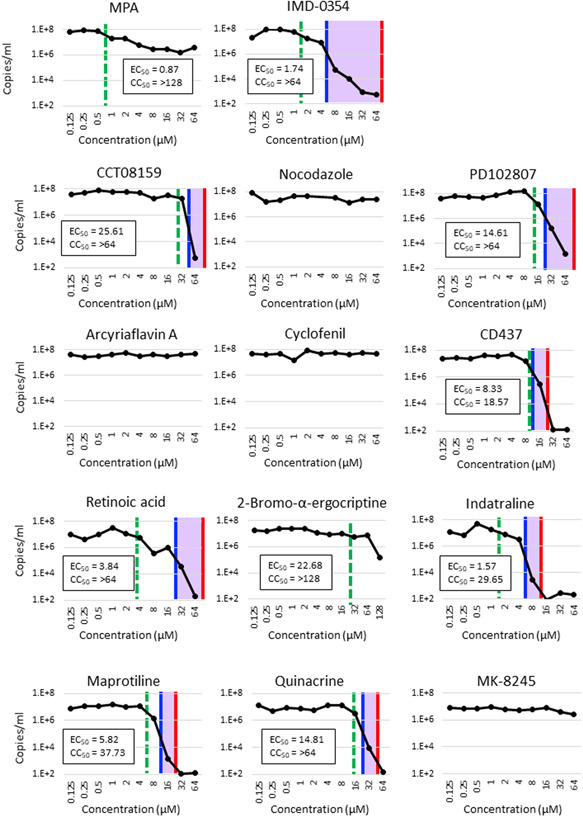
Effect of drugs on viral RNA synthesis. VeroE6/TMPRSS2 cell monolayers were cultured with the SARS‐CoV‐2 Wk521 strain 10 prepared in culture media containing different concentrations of drugs for 50 hr (MOI = 0.01). After the 50 hr incubation period, RNAs were extracted from the culture medium and SARS‐CoV‐2‐specific RNAs (E region) were quantified by RT‐quantitative PCR, as described previously. 11 , 12 Blue and red bars indicate the antiviral level and cytotoxicity level, respectively, indicated in Figure 1. CC50, 50% cytotoxic concentration
This study shows that IMD‐0354 inhibited SARS‐CoV‐2 replication. IMD‐0354 is an IKKβ (inhibitor of nuclear factor kappa‐B kinase subunit beta) inhibitor, which blocks IκBα (inhibitor of nuclear factor kappa B) phosphorylation and NF‐κB (nuclear factor kappa‐light‐chain‐enhancer of activated B cells) activation. Although NF‐κB is activated by coronavirus infection, stimulating production of proinflammatory cytokines and chemokines and contributing to the coronavirus pathogenesis, 10 , 13 , 14 the mechanism by which IMD‐0354 inhibits the coronavirus infection remains unclear because NF‐κB is a key transcription factor of the host innate immune responses and coronaviruses themselves inhibit the NF‐κB‐mediated pathway. 15 , 16 Furthermore, in this study, the antiviral effect of IMD‐0354 was evaluated using the VeroE6/TMPRSS2 cell line, which lacks type I IFN‐related genes. IMD‐0354 demonstrated antiviral activity, and therefore, its mechanism of action might be type I IFN independent. Thus, the underlying mechanisms for coronavirus inhibition by IMD‐0354 require further studies.
MPA is an inhibitor of inosine monophosphate dehydrogenases which play a key role in purine biosynthesis. Two FDA‐approved drugs, mizoribine and mycophenolic mofetil, are derivatives of MPA. Our study demonstrates the anti‐SARS‐CoV‐2 activity of MPA, similar to its reported activity against MERS‐CoV. 2 , 3 , 17 A previous study 17 has shown that MPA inhibits the MERS‐CoV papain‐like protease (PLpro), which proteolytically processes the viral nonstructural polyproteins for maturation. PLpro also contributes to the virus evasion from the host innate immunity by deubiquitination of interferon regulatory factor 3 (IRF3) and suppression of IFN‐β induction. 18 Importantly, for MERS‐CoV infection, MPA had a low EC50, which is easily achievable at therapeutic doses. Thus, MPA in combination with IFN‐β is proposed for the treatment of MERS‐CoV. This combination also increases the efficacy of MPA. Mycophenolate mofetil can be administered orally, and clinical and laboratory findings from patients with MERS suggest an increased survival rate when mycophenolate mofetil is used in their treatment. 19 Importantly, a mild inhibitory effect by MPA against SARS‐CoV infection was reported in cell culture and in vivo (mice) assays, 20 but our CPE assay failed to detect an anti‐SARS‐CoV‐2 effect for MPA, because the inhibition was not perfect even at high concentrations of MPA. However, it should be noted that the EC50 of MPA was as low as 0.87 µm, which is much lower than the MPA serum levels achieved during therapeutic use of mycophenolate mofetil. Thus, similar to the case of MERS‐CoV, it is worth considering the clinical use of FDA‐approved mycophenolate mofetil in combination with other effective drugs for COVID‐19 treatment.
In conclusion, this study demonstrates the anti‐SARS‐CoV‐2 activity of MPA and IMD‐0354. Our CPE assay was easily and efficiently used for screening anti‐SARS‐CoV‐2 compounds/drugs when the replication of SARS‐CoV‐2 is reduced by over 100‐fold. IMD‐0354 showed strong inhibitory effect and its EC50 was reasonably low (1.74 µm). Although the inhibition level of SARS‐CoV‐2 replication by MPA was less than 100‐fold, it still demonstrated significant anti‐SARS‐CoV‐2 activity at low concentrations (EC50 = 0.87 µm), a level that is easily achievable by the therapeutic dose of mycophenolate mofetil. Further studies on MPA, which is known to inhibit coronavirus PLpro, would be useful to develop novel anti‐SARS‐CoV‐2 drugs.
DISCLOSURE
The authors declare that there are no conflict of interests.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supporting information
Supporting information
Supporting information
ACKNOWLEDGEMENTS
This study was supported by Grant‐in Aid from the Japan Agency for Medical Research and Development (AMED) under Grant number JP19fk0108111j0001 and from the Japan Society for the Promotion of Science under Grant number 20K16020.
Kato F, Matsuyama S, Kawase M, Hishiki T, Katoh H, Takeda M. Antiviral activities of mycophenolic acid and IMD‐0354 against SARS‐CoV‐2. Microbiology and Immunology. 2020;64:635–639. 10.1111/1348-0421.12828
REFERENCES
- 1. Siordia JA Jr. Epidemiology and clinical features of COVID‐19: a review of current literature. J Clin Virol. 2020;127:104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shen L, Niu J, Wang C, et al. High‐throughput screening and identification of potent broad‐spectrum inhibitors of coronaviruses. J Virol. 2019;93:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hart BJ, Dyall J, Postnikova E, et al. Interferon‐beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell‐based assays. J Gen Virol. 2014;95:571‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang CW, Lee YZ, Hsu HY, et al. Targeting coronaviral replication and cellular JAK2 mediated dominant NF‐kappaB activation for comprehensive and ultimate inhibition of coronaviral activity. Sci Rep. 2017;7:4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kato F, Nio Y, Yagasaki K, et al. Identification of inhibitors of dengue viral replication using replicon cells expressing secretory luciferase. Antiviral Res. 2019;172:104643. [DOI] [PubMed] [Google Scholar]
- 6. Hishiki T, Kato F, Nio Y, et al. Stearoyl‐CoA desaturase‐1 is required for flavivirus RNA replication. Antiviral Res. 2019;165:42‐6. [DOI] [PubMed] [Google Scholar]
- 7. Kato F, Ishida Y, Oishi S, et al. Novel antiviral activity of bromocriptine against dengue virus replication. Antiviral Res. 2016;131:141‐7. [DOI] [PubMed] [Google Scholar]
- 8. Tohma D, Tajima S, Kato F, et al. An estrogen antagonist, cyclofenil, has anti‐dengue‐virus activity. Arch Virol. 2019;164:225‐34. [DOI] [PubMed] [Google Scholar]
- 9. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS‐CoV‐2 by TMPRSS2‐expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001‐03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X, Wu K, Wang D, et al. Nucleocapsid protein of SARS‐CoV activates interleukin‐6 expression through cellular transcription factor NF‐kappaB. Virology. 2007;365:324‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuyama S, Shirato K, Kawase M, et al. Middle East Respiratory Syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J Virol. 2018;92:e00683‐11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25:2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dediego ML, Nieto‐Torres JL, Regla‐Nava JA, et al. Inhibition of NF‐kappaB‐mediated inflammation in severe acute respiratory syndrome coronavirus‐infected mice increases survival. J Virol. 2014;88:913‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanzawa N, Nishigaki K, Hayashi T, et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF‐kappaB activation. FEBS Lett. 2006;580:6807‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canton J, Fehr AR, Fernandez‐Delgado R, et al. MERS‐CoV 4b protein interferes with the NF‐kappaB‐dependent innate immune response during infection. PLoS Pathog. 2018;14:e1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Sun A, Sun Y, et al. Porcine transmissible gastroenteritis virus inhibits NF‐kappaB activity via nonstructural protein 3 to evade host immune system. Virol J. 2019;16:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng KW, Cheng SC, Chen WY, et al. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain‐like protease of Middle East respiratory syndrome coronavirus. Antiviral Res. 2015;115:9‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang X, Chen X, Bian G, et al. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain‐like protease. J Gen Virol. 2014;95:614‐26. [DOI] [PubMed] [Google Scholar]
- 19. Al Ghamdi M, Alghamdi KM, Ghandoora Y, et al. Treatment outcomes for patients with Middle Eastern Respiratory Syndrome Coronavirus (MERS CoV) infection at a coronavirus referral center in the Kingdom of Saudi Arabia. BMC Infect Dis. 2016;16:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barnard DL, Day CW, Bailey K, et al. Enhancement of the infectivity of SARS‐CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antiviral Res. 2006;71:53‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


