Abstract
Historically, medical imaging has been a qualitative or semi-quantitative modality. It is difficult to quantify what can be seen in an image, and to turn it into valuable predictive outcomes. As a result of advances in both computational hardware and machine learning algorithms, computers are making great strides in obtaining quantitative information from imaging and correlating it with outcomes. Radiomics, in its two forms “handcrafted and deep,” is an emerging field that translates medical images into quantitative data to yield biological information and enable radiologic phenotypic profiling for diagnosis, theragnosis, decision support, and monitoring. Handcrafted radiomics is a multistage process in which features based on shape, pixel intensities, and texture are extracted from radiographs. Within this review, we describe the steps: starting with quantitative imaging data, how it can be extracted, how to correlate it with clinical and biological outcomes, resulting in models that can be used to make predictions, such as survival, or for detection and classification used in diagnostics. The application of deep learning, the second arm of radiomics, and its place in the radiomics workflow is discussed, along with its advantages and disadvantages. To better illustrate the technologies being used, we provide real-world clinical applications of radiomics in oncology, showcasing research on the applications of radiomics, as well as covering its limitations and its future direction.
Introduction
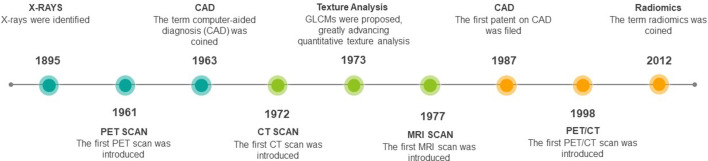
Medical imaging technologies in healthcare have expanded remarkably from the discovery of X-Rays 124 years ago to the use of CT, MRI, and positron emission tomography (PET), among others in modern-day clinical practice1 (Figure 1). These tools have become an integral part in detection and diagnosis for many diseases due to several factors, including: the minimally invasive nature of imaging, rapid technological developments, lower costs compared to alternatives, the high information density of images, and the hardware can be used for multiple diseases and sites.2,3
Figure 1.
Timeline highlighting key developments in medical imaging. CAD, computer-aided diagnosis; GLCM, grey level co-occurring matrix; PET, positron emission tomography.
Medical imaging in its infancy generated analogue images, which underwent subjective interpretation based on visual inspection and verbal communication. By the end of the 20th century, information technology has brought radiology to the digital world,4 although the interpretation of radiographs remained mostly qualitative. Humans excel at recognising patterns through visual inspection, however, they are often lacking when performing complex quantitative assessments.5,6 In the early 1960s, researchers started to focus on computerised quantitative analysis of medical data for aiding clinical diagnosis,7–9 what later came to be known as computer-aided diagnosis (CAD) systems. However, these systems were using a classical approach using statistical analysis and probability theories, and the volume of available data was low, so the results were often too inaccurate for clinical use. Later in the 1980s, further advances in theoretical computer science and digital imaging lead to the development of advanced machine learning and pattern recognition algorithms, which when integrated with CAD systems were able to generate clinically reliable results.10,11
In recent decades, simple quantitative image analysis (QIA) has been adopted by clinicians (e.g. RECIST12), and has been primarily focused on assisting qualitative observations.13 For instance, CAD systems can be found in healthcare worldwide, aiding radiologists and clinicians in making diagnostic and theragnostic decisions.14 One of the most typical applications of CAD systems is in recognising abnormalities during cancer screening.15 Notable contributions are in the area of lung and breast cancer research. For example, there are many CAD studies which focus on detecting and diagnosing lung nodules16,17 (as benign or malignant) on CT and chest radiographs. Similarly, many such studies have been conducted in breast mammography images for highlighting microcalcifications,18 architectural distortions, and the prediction of mass type.19,20
It is conceivable that the lack of quantitative information leads to increased follow-ups or invasive biopsies that would be deemed unnecessary given the unused information in medical images.21 Even though there have been various developments in QIA, traditionally radiologists are trained to understand the behaviour of the underlying disease through visual inspection of radiographic images.21 This partially explains why most of the developments in imaging technology are in optimising the visual representation of the generated images, with vendors competing to generate the highest quality images. With the exception of CT, with its semi-parametric calibrated Hounsfield Units, and some particular MRI sequences, individual voxel values do not correlate with the underlying biology without further calibration and modelling. Furthermore, qualitative analysis is not so dependent on reproducible voxel values, while machines on the other hand only process numerical values and rely on the standardisation of image acquisition and reconstruction to yield reproducible results. The lack of standardisation of medical images has been a major hurdle in the development of QIA in medical imaging.22–25 However, in recent years, quantitative imaging is becoming more popular with the advent of, e.g. quantitative fludeoxyglucose-PET26,27 or quantitative MRI28,29 for treatment response assessment.
The ubiquitous computer, vast amounts of data, and advanced algorithms have opened a new era in medical imaging. The high information density of images allows for many quantitative metrics since intricate pixel and voxel relationships can be captured by complex operations. Radiomics involves the process of extraction of quantifiable features from vast amounts of data that might correlate with the underlying biology or clinical outcomes using advanced machine learning analysis techniques.30,31 Radiomics has two main arms, based on how imaging information is transformed into mineable data: handcrafted radiomics and deep learning. Handcrafted features are formulas mostly based on intensity histograms, shape attributes, and texture, that can be used to fingerprint phenotypical characteristics of the radiograph32 while in deep learning a complex network “creates” its own features. Various statistical and machine learning models have been widely researched, and are envisioned to be complementary to best medical practice by aiding in making informed clinical decisions in both oncological and non-oncological diseases.33–36
Since the 1990s predictions were being made that genomics, spearheaded by the Human Genome Project, would completely transform therapeutic medicine, heralding precision medicine.37 Precision medicine, also termed personalised medicine, originally referred to the view that incorporating genomic information in the clinical workflow will lead to marked improvements in the prediction, diagnosis, and treatment of diseases. Recently, the scope of precision medicine has expanded to incorporate inputs beyond the genome.38 Radiomics and other “-omic” developments, such as metabolomics and proteomics, are contributing to this a paradigm shift in medicine, where the focus has changed from standard clinical protocols based on trial populations to a personalised treatment tailored not only to the disease and site but also the patient, further enabling precision medicine.
In this review, we provide a broad overview and update on the fast-growing field of quantitative imaging research, focussing on the two arms “handcrafted radiomics and deep learning” describing some of its caveats and giving examples of the budding clinical implementation, the stepping stones towards precision medicine.
Radiomics: from feature extraction to correlation with outcomes
Performing feature extraction of textures in medical imaging is nothing new and in fact serious research had begun in the early 1980s at Kurt Rossmann Laboratories for Radiologic Image Research in the Department of Radiology at the University of Chicago to develop CAD systems for the detection of lung nodules as well as detection of clustered microcalcifications in mammograms.39,40 The first CAD patent was filed all the way back in 1987 using a method of pixel thresholding and contiguous pixel area thresholding.40
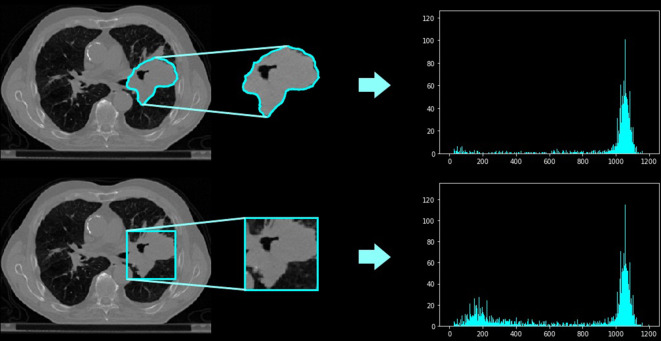
The radiomic workflow begins with the medical image, which can be represented in two, three, or four dimensions.32,41 Images contain quantitative data in the form of signals that are captured at different scales and variation across medical machines.42,43 Normalisation techniques are used to distribute pixel intensities evenly across a data set evenly and within a standardised range.42,43 44. Next, a region of interest (ROI) is defined so that only information related to the lesion can be extracted, and the useful information that can be extracted are called features. There are competing methods to extract features both in two-dimensional and three-dimensional. One such method is the manual segmentation of the lesion or the creation of a bounding box, as seen in Figure 2.45,46 This can also be performed using automated segmentation algorithms. Methods for automated segmentation include deep learning architectures such as U-Net, or semi-automatic methods like click-and-grow algorithms.45,46
Figure 2.
The difference between using (A) a contoured binary mask, and (B) using a bounding box.
Once the ROI is defined, the choice of features to be extracted depend on the information being sought. Shape features such as volume relate only to the definition of the ROI, and if this is manually created, suffer from inter- and intraobserver variability.47 First-order features give insight into the distribution of pixel intensities, e.g. histograms of pixel intensities are quantified by a large number of statistical methods, including variance, skewness, and kurtosis. These features, however, are unable to quantify how pixels are positioned in relation to each other. Second and higher-order features may capture this relationship, with second-order features obtained based on the average relationship between two pixels/voxels, and higher-order features for more than two pixels/voxels. An example of a second-order feature extraction method is the grey level co-occurring matrix (GLCM). GLCMs are co-occurring pixels in each defined direction (Figure 3) and are counted and recorded (Figure 4) into a matrix. Statistical analysis such as contrast, correlation, and homogeneity, as well as tailored formulae can then be applied on the GLCM to extract independent features.48 Features extracted in this manner are considered “handcrafted” features as they are features that are pre-defined by specially designed formulae.
Figure 3.
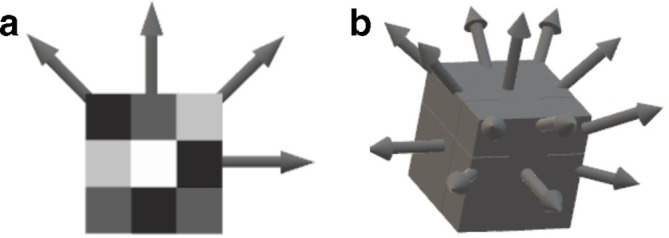
Possible angles for the calculation of co-occurrence matrices in two and three dimensions. (A) Shows the 4 possible directions in 2 dimensions while (B) shows the 13 possible directions in 3 dimensions.
Figure 4.
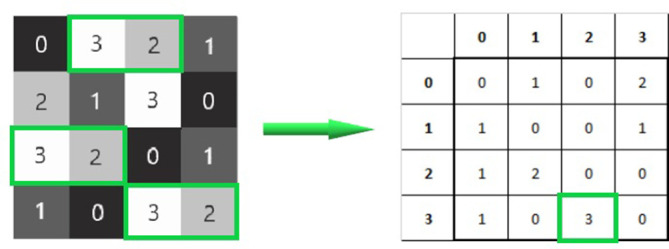
Calculating a GLCM for horizontal co-occurring pixel intensities. In total, 3 co-occurring pixel intensities of 3 and 2 that are next to each other on a horizontal plane can be totalled and tracked in the corresponding matrix. GLCM, grey level co-occurring matrix.
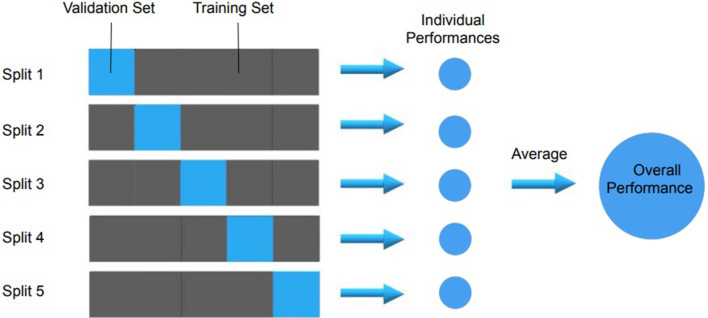
After features have been extracted from all the images in a database, a subset of features needs to be selected that go into the final model. To make a model generalisable, it is important to avoid finding spurious correlations in the data that do not generalise to other similar data sets, an occurrence termed overfitting.49–51 If a model has learned to recognise noise, outliers, or other kinds of variance, it is unlikely to perform well when presented with new data. The larger the number of predictors, the larger the chance to find spurious correlations, a major problem in the realm of machine learning.52 To detect overfitting, ideally, a model’s performance is validated in external data sets with similar population and outcome distributions, but from different centres—if the model performs significantly better on the training set than on the validation set, overfitting is likely.53,54 In the absence of an external validation data set, data can be split into different subsets, and the model trained in one group and validated on the other(s) in a process called cross-validation (Figure 5).55 During this process, the model hyperparameters (settings within the model itself, e.g. degree of polynomial fitting) can be further tuned to increase performance in the training and validation sets.56
Figure 5.
An example of fivefold cross-validation which can be used to evaluate machine learning models. Cross-validation gives the ability to test the result across the entirety of a data set, giving a better estimation of a model’s overall performance.
A method to overcome overfitting is to reduce the number of predictors, in this case, imaging features. Feature selection is the process of reducing the number of predictors while retaining the core important information that correlates with outcomes or the underlying biology.32 Many feature reduction methods exist, but none are known to work well on all kinds of data sets, and they can be combined in many ways.32 This remains an active field of research.57 Similar features can also be grouped to achieve dimensionality reduction, and methods such as principal component analysis and independent component analysis are employed to this end.58
Once features are selected, the task is to correlate these features—individually or in groups—to diagnostic and prognostic outcomes or to the underlying biology. There are numerous methods to find and test such models, from simple linear regression and curve-fitting to advanced machine learning methods such as decision trees, support vector machines, random forests, boosted trees, or neural networks.59 Ensembling is the combination of models that get trained on random samples of data from the training set called bags and then combined as a whole using a voting system. This is the basis for algorithms such as Random Forests, AdaBoost, and Gradient Boosting.60 An intuitive explanation is that even though the individual models can show a large amount of variance due to being trained on small subsets of the data, their averaging or voting smooths out the variance while improving the ability to better generalise.60
Once a generalisable model has been trained and externally validated, it might be desirable to expand the interoperability of the model to all hardware, acquisition, and reconstruction parameters found in general clinical practice. Instead of relying on the standardisation of images, the features themselves can be harmonised to a common frame-of-reference using combined batch methods such as ComBat,44,60,61 originally developed for similar problems encountered in gene sequencing assays.62
Deep learning for fully automated workflows
Artificial neural networks (ANNs) are a class of machine learning architecture that are loosely based on how biological brains work.63 With the exception of unsupervised learning (such as autoencoders), deep learning architectures usually rely on information regarding the outcome in order to craft their features, and unlike in handcrafted radiomics, feature extraction and correlation are intertwined.64 Also, unlike radiomics, there is generally no need for image segmentation, as the whole image can be presented to a deep learning model, both during training and in clinical routine.
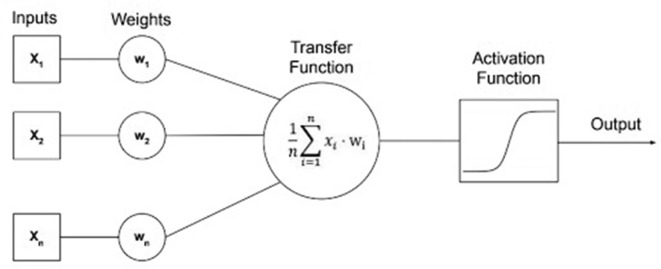
An ANN is able to use a collection of neurons and weights, one for each of the inputs preceding the neuron.65 These weights get continuously updated, or corrected, in steps called epochs that work together to create a very complex function able to make predictions. The weights are inputs for each neuron and are multiplied and averaged, resulting in a transfer function, which is converted to an output via a function called an activation function.66 These activation functions are often a sigmoidal function such as a hyperbolic tangent or sigmoid, or a function called a rectified linear unit that can be represented as the maximum of the product of the coefficient and zero or one. A representation of a single neuron, including the activation function, can be seen in Figure 6.67 Multiple neurons can then be stacked to create a single layer referred to as a “hidden layer” and hidden layers (were inputs and outputs all connect) can be stacked to create larger networks, see Figure 7. 65 The term deep learning is used to describe a neural network that has many layers, which is considered deep. For a binary classifier or regression, the final layer should contain only a single neuron and use a sigmoid activation function to make a prediction with a binary outcome (zero or one). If the problem is categorical, the network’s final layer should contain the same number of neurons as there are categories to be classified and the final activation will be a “softmax” function, which is the average of the exponentials of the inputs,68 yielding the probabilities of each category. Deep learning for image vision employs convolutional neural networks (CNNs) which are a type of ANN that have an automated feature extractor designed specifically for images.69 CNNs employ a filtering technique, which convolves the image with a kernel (sliding window), creating a new pixel/voxel value (and hence new image) by sliding a matrix of numbers over the image, see Figure 8. It is possible to make a variety of different filters using these types of convolutions, such as blurring, sharpening, edge detection, and gradient detection69,70, and CNNs are able to learn filters that are best suited to extracting features needed for making predictions.
Figure 6.
The architecture of a single neuron with a transfer function and a sigmoid activation function visualised.
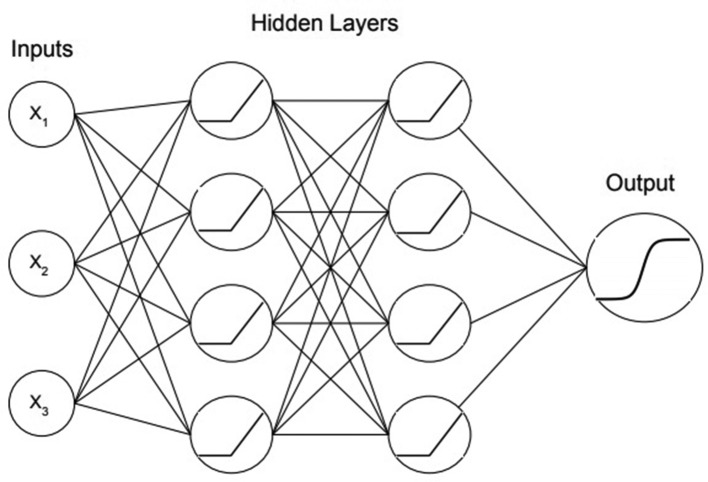
Figure 7.
A three layer neural network that is a binary classifier with three inputs. Nodes with Xn refer to inputs while other nodes refer to activation functions. The connecting lines between the nodes represent weights.
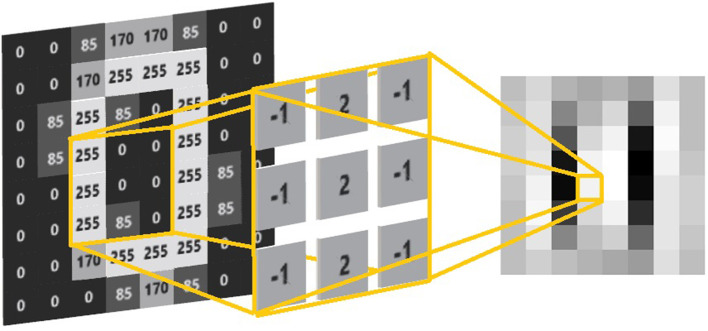
Figure 8.
A filter that is able to filter out vertical lines. The yellow lines represent the kernel or sliding window, while the image on the right is the result of performing convolutions across the entirety of the original image.
ANNs do have some drawbacks compared to using handcrafted features alongside other machine learning techniques. The main drawback is the intrinsic need for much larger datasets to train the models, since feature creation is contingent on the training data, as opposed to handcrafted radiomics. Another drawback to using ANNs is interpretability. ANNs build ultracomplex functions that can be extremely difficult for practitioners to make sense of. Although CNNs have performed very well in image recognition, they have been less successful learning texture features, since texture information inherently has a higher dimensionality compared to other types of data sets, making them more difficult for neural networks to master.69,71 According to Basu et al,71 a redesign of neural network architectures is required to extract features in a similar manner as GLCM and other features based on spatial correlation.
Currently, the main application of deep learning in the radiomics workflow still lies in the automated detection and localisation of organs and lesions, removing the major burden in data set curation. While there is no algorithm that can solve every problem, deep learning still has its place and is able to work as additional methods for delineation and feature extraction that compliments handcrafted radiomics. There is active research in combining both deep learning features and radiomics features that shows improved results.72–74
Potential clinical applications
Radiomics in oncology
Radiomics has been widely studied for application in diagnosis and treatment prognosis/selection in oncology, primarily due to the existence of large imaging data sets used for staging, often containing delineations of tumours and organs at risk necessary for radiation treatment planning. These data sets can be used to train diagnostic and prognostic models for a variety of cancer types and sites. Using clinical reports, pathology/histology, and genetic information along with radiomics analysis can give a global outlook on the biology of the disease.48 In this section, an overview of notable studies published in this area will be discussed.
Lung
Lung cancer is by far the leading cause of cancer-related deaths among both males and females worldwide.75 Recent studies have shown that radiomics can determine the risk of lung cancer from screening scans.76–78 Radiomic features found to have a strong association to decode tumour heterogeneity for risk stratification,79,80 concluding that patients with heterogeneous tumours tend to have a worse prognosis. In addition to that, Yoon et al were able to show the association of radiomic analysis with gene expression.81 Radiomic features were also found to correlate with TNM staging for lung and head-and-neck cancer.31,82 Later studies further validated the strong predictive power of radiomics for distant metastasis.83–85
Radiomics may also play a role in lung cancer treatment planning by evaluating tumour response to a specific treatment. Several studies focused on analysing the tumour response to radiation therapy.86,87 For instance, Mattonen et al developed a radiomics signature for treatment response to stereotactic ablative radiation therapy that was able to predict lung cancer recurrence post-therapy,86 while Fave et al used multiple time point information referred to as delta-radiomic analysis to evaluate the change of radiomic features as a predictor for tumour response to radiation therapy.87 The results suggest that δ radiomic features are in fact a good indicator of treatment response. Another interesting study by Mattonen et al found that radiomic analysis can identify features associated with local recurrence of lung cancer after radiation therapy,88 while physicians usually have great difficulty to distinguish local recurrence from radiation-induced sequelae.
Besides the traditional handcrafted feature extraction approach followed in the radiomics pipeline, deep learning radiomics is also gaining popularity among researchers. A deep learning-based approach followed by Shen et al yielded more accurate malignancy prediction of nodules compared to previous methods.89 Pham et al used a two-step deep learning approach for evaluating lymph node metastases with accurate cancer detection.90 Instead of using data from a single time point, deep recurrent convolutional network architectures can be used to analyse data from multiple time points to monitor treatment response.91
Brain
Brain tumours are usually graded based on clinical or pathological analysis to define their malignancy. Radiomics may be able to non-invasively perform grade assessment, as reported by Coroller et al in meningioma patients, suggesting a strong correlation between certain imaging features and histopathological grade.92 Zhang et al were able to classify between low-grade gliomas and high-grade gliomas with high accuracy.93 Chen et al investigated the prediction of brain metastases in T1 lung adenocarcinoma patients and found that the predictive performance for the radiomics model was significantly better compared to clinical models and could potentially be used for brain metastases screening.94 Fetit et al performed radiomic analysis for the classification of brain tumours in childhood suggesting that radiomics can aid in the classification of tumour subtype.95 However, the scalability of the techniques used in these studies needs to be assessed further by extensions to multicentric cohorts using different acquisition protocols and vendors.
Radiation therapy can lead to necrosis, which is difficult to distinguish from tumour recurrence on imaging. Larroza et al were able to develop a high classification accuracy model to distinguish between brain metastasis and radiation necrosis using radiomic analysis.96 Some radiomic studies successfully investigated the treatment response in recurrent glioblastoma patients with a radiomics approach.97–99 An iterative study by radiomic researchers found strong evidence of radiomic features in predicting survival and treatment response of patients with glioblastoma using pre-treatment imaging data.100–102
Deep learning has also made some other interesting contributions in this area. Chang et al used residual deep convolutional network for predicting the genotype in Grade II-IV glioma with high accuracy.103 Deep learning can also be used complementary to traditional handcrafted radiomics studies. For example, studies72,73 focused on using deep networks for segmentation, followed by radiomics analysis for survival prediction.
Breast
Among females, breast cancer is the second leading cause of death for cancer worldwide.75 However, earlier diagnosis can lead to a better prognosis. Radiomics in the field of breast cancer has been applied to several imaging modalities including (PET)-MRI, (contrast-enhanced) mammography, ultrasound, and digital breast tomosynthesis focusing on tumour classification, molecular subtypes, tumour response prediction to neoadjuvant systemic therapy (NST), lymph node metastasis, overall survival, and recurrence risks. For example, a large number of radiomics studies have been used for the prediction of malignant breast cancers.104–107 Besides the prediction of tumour malignancy, several radiomics studies examined the prediction of breast cancer molecular subtypes with the aim of leaving out liquid biopsies in the future.108–111 Lymph node metastasis identification is an important prognostic factor and often determines treatment. In all clinically node negative patients, a sentinel lymph node procedure is the basis of the axillary treatment.112 Dong et al was able to provide an alternative to this invasive approach by successfully applying radiomics for the prediction of lymph node metastasis in the sentinel lymph node using imaging data.113
In addition to the prediction of breast tumour malignancy, tumour molecular subtypes and sentinel lymph node metastasis identification, radiomics studies have also made some significant contributions to treatment planning. Chan et al investigated the power of radiomics to discriminate between patients with low and high treatment failure risk on pre-treatment imaging data.114 There are multiple studies that predict tumour response to NST using radiomic analysis. For instance, Braman et al found a combination of intratumoral and peritumoral radiomics features as a robust and strong indicator for pathologic complete tumour response using pre-treatment imaging data.115 Two other studies116,117 found similar evidence on serial imaging data containing follow-up scans. The use of multiparametric MRI for the prediction of tumour response to NST showed promising results.118,119
Deep learning approaches have also been adopted in breast cancer research. The study of Huynh et al investigated tumour classification capacity of deep features extracted from convolutional networks trained on a different data set to analytically extracted features.120 The results suggested a higher performance of deep features. Similarly, another study,121 used deep learning for risk assessment and found higher performance compared to conventional texture analysis.
Other sites and diseases
While cancers of the lung, brain, and breast have received wide attention from the radiomics research community, any site is open to QIA research. Diagnostic and prognostic radiomics research is ongoing for cancers of the head-and-neck,122 ovaries,38 prostate,123 kidney,124 liver,125 colon and rectum,126 and many other sites. The main requirements for a radiomics study are the presence of a radiologic phenotype which allows for the clustering of patients based on differences within that phenotype or some correlation to the underlying biology, and the availability of imaging and clinical data. While not nearly as prevalent,127 this has meant that non-oncological diseases which require medical imaging as part of the standard of care have also been the subject of radiomics analysis, such as in the fields of neurology,35 ophthalmology,128 and dentistry.129
Limitations of radiomics and future directions towards precision medicine
While radiomics facilitates new possibilities in the field of personalised medicine, some challenges remain. One of the primary obstacles is the lack of big and standardised clinical data. Although large amounts of medical imaging data are stored, these data are dispersed across different centres and acquired using different protocols. Access for research purposes is highly restricted by law and ethics. An exhaustive data curation and harmonisation process is still necessary to make it usable for research. Radiomics will potentially enable imaging-based clinical decision support systems, however, the current black box approach, particularly in deep learning, makes it less acceptable for clinical application. In certain cases, handcrafted radiomic features have already been correlated with biological processes,130–132 but it is essential to work further in the direction of interpretable artificial intelligence (AI) to make it more accessible for clinical implementation.33
In recent years, various countries have already adopted many measures to control variability in clinical trial protocols, data acquisition, and analysis.133,134 For example, across Europe consistent protocol guidance was adopted with the help of European Association of Nuclear Medicine.135 The Quantitative Imaging Biomarker Alliance initiative also aims to achieve the same task in a much broader level.136,137 On the other hand, algorithmically, developments in deep learning allow for automated quality check, clustering of data, and automated detection and contouring of organs and lesions, vastly improving data curation times. Generative adversarial networks open up the possibility of generating synthetic data138 or domain adaptive algorithms139,140 might be able to deal with the shortage of standardised data. Techniques like distributed learning provide the ability to train machine learning models using distributed data without the data ever leaving their original locations. Distributed learning has already been applied across several medical institutions to build predictive and segmentation models.141–144 Furthermore, this approach can be coupled with other technologies such as blockchain to trace back data provenance and monitor the use of the final models.145 Various techniques to visualise deep features have already been put forward by researchers to generate an intuitive understanding. A completely new research area of AI called explainable AI aims to track the decisions made by the intelligent algorithms so that it can be better understood by humans. Companies like Google, IBM, Microsoft and Facebook are at the forefront in this research. This will not only help to build trust of AI systems among medical professionals but also unlocks new possibilities in understanding a disease.146,147
The implementation of precision medicine itself has its own limitations and has drawn criticism due to the lack of a “transformation in therapeutic medicine” in the last two decades.148 So far, life expectancies or other public health measures have not shown any dramatic improvements, regardless of the vast amounts of precision medicine research being conducted. Contentious points remain such as excessive costs (e.g. gene therapy), although new developments such as radiomics promise to reduce costs in the long run. Furthermore, the diagnostic and prognostic power of complex “omics-driven” models is still to be determined in specific populations, and evidence needs to be produced that such methods improve health outcomes.149 Precision medicine is likely to mature and translate to clinical workflows over the next decade and will change the way health services are delivered and evaluated. Healthcare systems will need to adjust their methods and processes to accommodate for these changes.
Conclusion
Radiomics, whether handcrafted or deep, is an emerging field that translates medical images into quantitative data to give biological information and enable phenotypic profiling for diagnosis, theragnosis, decision support, and monitoring. Radiomics, in essence, allows personalised care by identifying features or signatures correlated with a disease or a treatment response with high precision and in a non-invasive way. Recent developments in genomics and deep learning have pushed radiomics researchers to focus more on extracting deep features and explore new possibilities in AI modelling. In the future, radiomics will be a valued addition to precision medicine workflows by facilitating earlier and more accurate diagnosis, providing prognostic information, aiding in treatment choice, monitoring disease and treatment non-invasively, and enabling routine dynamic treatment based on individual responses. But the road to this vision is long, and many technical, regulatory, and ethical problems still need to be solved.
Footnotes
Funding: Authors acknowledge financial support from ERC advanced grant (ERC-ADG-2015, no 694812 - Hypoximmuno), ERC-2018-PoC (no 81320 – CL-IO). This research is also supported by the Dutch Technology Foundation STW (grant no P14-19 Radiomics STRaTegy), which is the applied science division of NWO, and the Technology Programme of the Ministry of Economic Affairs. Authors also acknowledge financial support from SME Phase 2 (RAIL - no 673780), EUROSTARS (DART, DECIDE), the European Program H2020-2015-17 (BD2Decide - PHC30-689715, ImmunoSABR - no 733008, PREDICT - ITN - no 766276), TRANSCAN Joint Transnational Call 2016 (JTC2016 “CLEARLY”- no UM 2017–8295), Interreg V-A Euregio Meuse-Rhine (“Euradiomics”). This work was supported by the Dutch Cancer Society (KWF Kankerbestrijding), Project number 12085/2018–2
Conflict of interest: Dr Philippe Lambin reports, within and outside the submitted work, grants/sponsored research agreements from Varian medical, Oncoradiomics, ptTheragnostic, Health Innovation Ventures and DualTpharma. He received an advisor/presenter fee and/or reimbursement of travel costs/external grant writing fee and/or in-kind manpower contribution from Oncoradiomics, BHV, Merck and Convert pharmaceuticals. Dr Lambin has shares in the company Oncoradiomics SA and Convert pharmaceuticals SA and is co-inventor of two issued patents with royalties on radiomics (PCT/NL2014/050248, PCT/NL2014/050728) licensed to Oncoradiomics and one issue patent on mtDNA (PCT/EP2014/059089) licensed to ptTheragnostic/DNAmito, three non-patentable invention (software) licensed to ptTheragnostic/DNAmito, Oncoradiomics and Health Innovation Ventures.
Dr Woodruff has (minority) shares in the company Oncoradiomics.
William Rogers and Sithin Thulasi Seetha have contributed equally to this study and should be considered as co-first authors.
Contributor Information
William Rogers, Email: w.rogers@maastrichtuniversity.nl.
Sithin Thulasi Seetha, Email: s.thulasiseetha@maastrichtuniversity.nl.
Turkey A. G. Refaee, Email: t.refaee@maastrichtuniversity.nl.
Relinde I. Y. Lieverse, Email: relinde.lieverse@maastrichtuniversity.nl.
Renée W. Y. Granzier, Email: r.granzier@maastrichtuniversity.nl.
Abdalla Ibrahim, Email: a.ibrahim@maastrichtuniversity.nl.
Simon A. Keek, Email: s.keek@maastrichtuniversity.nl.
Sebastian Sanduleanu, Email: s.sanduleanu@maastrichtuniversity.nl.
Sergey P. Primakov, Email: s.primakov@maastrichtuniversity.nl.
Manon P. L. Beuque, Email: m.beuque@maastrichtuniversity.nl.
Damiënne Marcus, Email: d.marcus@maastrichtuniversity.nl.
Alexander M. A. van der Wiel, Email: a.vanderwiel@maastrichtuniversity.nl.
Fadila Zerka, Email: f.zerka@maastrichtuniversity.nl.
Cary J. G. Oberije, Email: c.oberije@maastrichtuniversity.nl.
Henry C. Woodruff, Email: h.woodruff@maastrichtuniversity.nl.
Philippe Lambin, Email: philippe.lambin@maastrichtuniversity.nl.
REFERENCES
- 1. Scatliff JH, Morris PJ. From roentgen to magnetic resonance imaging: the history of medical imaging. N C Med J 2014; 75: 111–3. doi: 10.18043/ncm.75.2.111 [DOI] [PubMed] [Google Scholar]
- 2. Giakos GC, Pastorino M, Russo F, Chowdhury S, Shah N, Davros W. Noninvasive imaging for the new century [Internet]. Vol. 2. IEEE Instrumentation & Measurement Magazine 1999; 49: 32–5. [Google Scholar]
- 3. Prince J, Links J. Medical Imaging: Signals and Systems (Prince, J.L. and Links, J.M.; 2006) [Book Review. IEEE Signal Process Mag 2008; 25: 152–3. doi: 10.1109/MSP.2008.4408454 [DOI] [Google Scholar]
- 4. Kesner A, Laforest R, Otazo R, Jennifer K, Pan T. Medical imaging data in the digital innovation age. Med Phys 2018; 45: e40–52. doi: 10.1002/mp.12794 [DOI] [PubMed] [Google Scholar]
- 5. Miller GA. The magical number seven plus or minus two: some limits on our capacity for processing information. Psychol Rev 1956; 63: 81–97. doi: 10.1037/h0043158 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y-XJ, Ng CK. The impact of quantitative imaging in medicine and surgery: charting our course for the future. Quant Imaging Med Surg 2011; 1: 1–3. doi: 10.3978/j.issn.2223-4292.2011.09.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lodwick GS, Haun CL, Smith WE, Keller RF, Robertson ED. Computer diagnosis of primary bone tumors. Radiology 1963; 80: 273–5. doi: 10.1148/80.2.273 [DOI] [Google Scholar]
- 8. Meyers PH, Nice CM. Automated computer analysis of radiographic images. Arch Environ Health 1964; 8: 774–5. doi: 10.1080/00039896.1964.10663755 [DOI] [PubMed] [Google Scholar]
- 9. Winsberg F, Elkin M, Macy J, Bordaz V, Weymouth W. Detection of radiographic abnormalities in mammograms by means of optical scanning and computer analysis. Radiology 1967; 89: 211–5. doi: 10.1148/89.2.211 [DOI] [Google Scholar]
- 10. Summers RM. Road maps for advancement of radiologic computer-aided detection in the 21st century. Radiology 2003; 229: 11–13. doi: 10.1148/radiol.2291030010 [DOI] [PubMed] [Google Scholar]
- 11. Doi K. Computer-Aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph 2007; 31(4-5): 198–211. doi: 10.1016/j.compmedimag.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. . New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1. Eur J Cancer 2009; 45: 228–47. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 13. Zheng B. Identifying and testing new quantitative image, an analysis based clinical markers to predict breast cancer risk and prognosis. OMICS Journal of Radiology 2016; 05. doi: 10.4172/2167-7964.C1.009 [DOI] [Google Scholar]
- 14. Kobayashi T, Xu XW, MacMahon H, Metz CE, Doi K. Effect of a computer-aided diagnosis scheme on radiologists' performance in detection of lung nodules on radiographs. Radiology 1996; 199: 843–8. doi: 10.1148/radiology.199.3.8638015 [DOI] [PubMed] [Google Scholar]
- 15. Halalli B, Makandar A. Computer Aided Diagnosis - Medical Image Analysis Techniques [Internet. Breast Imaging 2018;. [Google Scholar]
- 16. The Robust Computer Aided Diagnostic System for Lung Nodule Diagnosis [Internet. International Journal of Recent Technology and Engineering. 2019; 8: 5670–5. [Google Scholar]
- 17. Ziyad SR, Radha V, Vayyapuri T. Overview of computer aided detection and computer aided diagnosis systems for lung nodule detection in computed tomography. Curr Med Imaging Rev 2020; 16: 16–26. doi: 10.2174/1573405615666190206153321 [DOI] [PubMed] [Google Scholar]
- 18. Rizzi M, D'Aloia M, Castagnolo B. Computer aided detection of microcalcifications in digital mammograms adopting a wavelet decomposition. Integr Comput Aided Eng 2009; 16: 91–103. doi: 10.3233/ICA-2009-0306 [DOI] [Google Scholar]
- 19. Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced Mr images of the breast. Magn Reson Med 2003; 50: 92–8. doi: 10.1002/mrm.10496 [DOI] [PubMed] [Google Scholar]
- 20. Murakami R, Kumita S, Tani H, Yoshida T, Sugizaki K, Kuwako T, et al. . Detection of breast cancer with a computer-aided detection applied to full-field digital mammography. J Digit Imaging 2013; 26: 768–73. doi: 10.1007/s10278-012-9564-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Z, Wang S, Dong D, Wei J, Fang C, Zhou X, et al. . The applications of Radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 2019; 9: 1303–22. doi: 10.7150/thno.30309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hunter LA, Krafft S, Stingo F, Choi H, Martel MK, Kry SF, et al. . High quality machine-robust image features: identification in nonsmall cell lung cancer computed tomography images. Med Phys 2013; 40: 121916. doi: 10.1118/1.4829514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One 2016; 11: e0166550. doi: 10.1371/journal.pone.0166550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016; 6: 34921. doi: 10.1038/srep34921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Timmeren JE, Leijenaar RTH, van Elmpt W, Wang J, Zhang Z, Dekker A, et al. . Test-Retest data for Radiomics feature stability analysis: generalizable or Study-Specific? Tomography 2016; 2: 361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, et al. . Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit 2009; 42: 1162–71. doi: 10.1016/j.patcog.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ulaner GA. Measuring Treatment Response on FDG PET/CT [Internet. Fundamentals of Oncologic PET/CT. 2019;: 225–9. [Google Scholar]
- 28. Xu Q-G, Xian J-F. Role of quantitative magnetic resonance imaging parameters in the evaluation of treatment response in malignant tumors. Chin Med J 2015; 128: 1128–33. doi: 10.4103/0366-6999.155127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Degnan AJ, Chung CY, Shah AJ. Quantitative diffusion-weighted magnetic resonance imaging assessment of chemotherapy treatment response of pediatric osteosarcoma and Ewing sarcoma malignant bone tumors. Clin Imaging 2018; 47: 9–13. doi: 10.1016/j.clinimag.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 30. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. . Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, et al. . Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. doi: 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. . Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–62. doi: 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 33. Sanduleanu S, Woodruff HC, de Jong EEC, van Timmeren JE, Jochems A, Dubois L, et al. . Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother Oncol 2018; 127: 349–60. doi: 10.1016/j.radonc.2018.03.033 [DOI] [PubMed] [Google Scholar]
- 34. Deist TM, Dankers FJWM, Valdes G, Wijsman R, Hsu I-C, Oberije C, et al. . Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med Phys 2018; 45: 3449–59. doi: 10.1002/mp.12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ibrahim A, Vallières M, Woodruff H, Primakov S, Beheshti M, Keek S, et al. . Radiomics analysis for clinical decision support in nuclear medicine. Semin Nucl Med 2019; 49: 438–49. doi: 10.1053/j.semnuclmed.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 36. Refaee T, Wu G, Ibrahim A, Halilaj I, Leijenaar RTH, Rogers W, et al. . The emerging role of Radiomics in COPD and lung cancer. Respiration 2020; 99: 99–107. doi: 10.1159/000505429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collins FS. Shattuck lecture--medical and societal consequences of the Human Genome Project. N Engl J Med 1999; 341: 28–37. doi: 10.1056/NEJM199907013410106 [DOI] [PubMed] [Google Scholar]
- 38. Nougaret S, Tardieu M, Vargas HA, Reinhold C, Vande Perre S, Bonanno N, et al. . Ovarian cancer: an update on imaging in the era of radiomics. Diagn Interv Imaging 2019; 100: 647–55. doi: 10.1016/j.diii.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 39. Giger ML, Doi K, MacMahon H, Dwyer SJ III, Schneider RH. Computerized Detection Of Lung Nodules In Digital Chest Radiographs [Internet. Medical Imaging 1987;. [Google Scholar]
- 40. Giger ML, Doi K, MacMahon H. Image feature analysis and computer-aided diagnosis in digital radiography. 3. automated detection of nodules in peripheral lung fields. Med Phys 1988; 15: 158–66. doi: 10.1118/1.596247 [DOI] [PubMed] [Google Scholar]
- 41. Larue RTHM, Van De Voorde L, van Timmeren JE, Leijenaar RTH, Berbée M, Sosef MN, et al. . 4DCT imaging to assess radiomics feature stability: an investigation for thoracic cancers. Radiother Oncol 2017; 125: 147–53. doi: 10.1016/j.radonc.2017.07.023 [DOI] [PubMed] [Google Scholar]
- 42. Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ. On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers. Med Phys 2017; 44: 1755–70. doi: 10.1002/mp.12188 [DOI] [PubMed] [Google Scholar]
- 43. Haga A, Takahashi W, Aoki S, Nawa K, Yamashita H, Abe O, et al. . Standardization of imaging features for radiomics analysis. J Med Invest 2019; 66(1.2): 35–7. doi: 10.2152/jmi.66.35 [DOI] [PubMed] [Google Scholar]
- 44. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8: 118–27. doi: 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 45. Kalpathy-Cramer J, Mamomov A, Zhao B, Lu L, Cherezov D, Napel S, et al. . Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography 2016; 2: 430-437. doi: 10.18383/j.tom.2016.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. . Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014; 9: e102107. doi: 10.1371/journal.pone.0102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leijenaar RTH, Carvalho S, Velazquez ER, van Elmpt WJC, Parmar C, Hoekstra OS, et al. . Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 2013; 52: 1391–7. doi: 10.3109/0284186X.2013.812798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–77. doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. . Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2018; 2: 36. doi: 10.1186/s41747-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vial A, Stirling D, Field M, Ros M, Ritz C, Carolan M, et al. . The role of deep learning and radiomic feature extraction in cancer-specific predictive modelling: a review. Transl Cancer Res 2018; 7: 803–16. doi: 10.21037/tcr.2018.05.02 [DOI] [Google Scholar]
- 51. Fan J, Shao Q-M, Zhou W-X. Are discoveries spurious? distributions of maximum spurious correlations and their applications. Ann Stat 2018; 46: 989–1017. doi: 10.1214/17-AOS1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lever J, Krzywinski M, Altman N. Model selection and overfitting [Internet]. Vol. 13. Nature Methods 2016;: 703–4. [Google Scholar]
- 53. Parmar C, Barry JD, Hosny A, Quackenbush J, Aerts HJWL. Data analysis strategies in medical imaging. Clin Cancer Res 2018; 24: 3492–9. doi: 10.1158/1078-0432.CCR-18-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chatterjee A, Vallières M, Dohan A, Levesque IR, Ueno Y, Bist V, et al. . An empirical approach for avoiding false discoveries when applying high-dimensional Radiomics to small datasets. IEEE Transactions on Radiation and Plasma Medical Sciences 2019; 3: 201–9. doi: 10.1109/TRPMS.2018.2880617 [DOI] [Google Scholar]
- 55. Wong T-T. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit 2015; 48: 2839–46. doi: 10.1016/j.patcog.2015.03.009 [DOI] [Google Scholar]
- 56. Duarte E, Wainer J. Empirical comparison of cross-validation and internal metrics for tuning SVM hyperparameters. Pattern Recognit Lett 2017; 88: 6–11. doi: 10.1016/j.patrec.2017.01.007 [DOI] [Google Scholar]
- 57. Solorio-Fernández S, Ariel Carrasco-Ochoa J, Martínez-Trinidad JF. A review of unsupervised feature selection methods [Internet. Artificial Intelligence Review 2019;. [Google Scholar]
- 58. Zhang D, Zou L, Zhou X, He F. Integrating feature selection and feature extraction methods with deep learning to predict clinical outcome of breast cancer. IEEE Access 2018; 6: 28936–44. doi: 10.1109/ACCESS.2018.2837654 [DOI] [Google Scholar]
- 59. Choudhary R, Gianey HK. Comprehensive Review On Supervised Machine Learning Algorithms [Internet. International Conference on Machine Learning and Data Science 2017;. [Google Scholar]
- 60. Ren Y, Zhang L, Suganthan PN. Ensemble Classification and Regression-Recent Developments, Applications and Future Directions [Review Article. IEEE Computational Intelligence Magazine 2016; 11: 41–53. doi: 10.1109/MCI.2015.2471235 [DOI] [Google Scholar]
- 61. Lovinfosse P, Visvikis D, Hustinx R, Hatt M. Fdg PET radiomics: a review of the methodological aspects. Clinical and Translational Imaging 2018; 6: 379–91. doi: 10.1007/s40336-018-0292-9 [DOI] [Google Scholar]
- 62. Lucia F, Visvikis D, Vallières M, Desseroit M-C, Miranda O, Robin P, et al. . External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging 2019; 46: 864–77. doi: 10.1007/s00259-018-4231-9 [DOI] [PubMed] [Google Scholar]
- 63. Kriegeskorte N. Deep neural networks: a new framework for modeling biological vision and brain information processing. Annu Rev Vis Sci 2015; 1: 417–46. doi: 10.1146/annurev-vision-082114-035447 [DOI] [PubMed] [Google Scholar]
- 64. Bengio Y, Delalleau O, Le Roux N. The curse of highly variable functions for local kernel machines. Adv Neural Inf Process Syst 2005; 18: 107–14. [Google Scholar]
- 65. Hinton GE. Learning multiple layers of representation. Trends Cogn Sci 2007; 11: 428–34. doi: 10.1016/j.tics.2007.09.004 [DOI] [PubMed] [Google Scholar]
- 66. LeCun Y. Deep learning & convolutional networks [Internet. 2015 IEEE Hot Chips 27 Symposium 2015.;. [Google Scholar]
- 67. LeCun Y, Bengio Y, Hinton G. Deep learning [Internet. Nature 2015; 521: 436–44. [DOI] [PubMed] [Google Scholar]
- 68. Aggarwal CC. Neural networks and deep learning: a textbook. Springer 2018; 497. [Google Scholar]
- 69. Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: a review. Neurocomputing 2016; 187: 27–48. doi: 10.1016/j.neucom.2015.09.116 [DOI] [Google Scholar]
- 70. Tyagi V. Introduction to Digital Image Processing [Internet. Understanding Digital Image Processing. 2018;: 1–12. [Google Scholar]
- 71. Basu S, Mukhopadhyay S, Karki M, DiBiano R, Ganguly S, Nemani R, et al. . Deep neural networks for texture classification-A theoretical analysis. Neural Netw 2018; 97: 173–82. doi: 10.1016/j.neunet.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 72. Yogananda CGB, Nalawade SS, Murugesan GK, Wagner B, Pinho MC, Fei B, et al. . Fully Automated Brain Tumor Segmentation and Survival Prediction of Gliomas using Deep Learning and MRI [Internet..
- 73. Sun L, Zhang S, Chen H, Luo L. Brain tumor segmentation and survival prediction using multimodal MRI scans with deep learning. Front Neurosci 2019; 13: 810. doi: 10.3389/fnins.2019.00810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jochems A, Leijenaar RTH, Bogowicz M, Hoebers FJP, Wesseling F, Huang SH, et al. . PO-0932: combining deep learning and radiomics to predict HPV status in oropharyngeal squamous cell carcinoma. Radiotherapy and Oncology 2018; 127: S504–5. doi: 10.1016/S0167-8140(18)31242-8 [DOI] [Google Scholar]
- 75. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, et al. . Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 76. Hawkins S, Wang H, Liu Y, Garcia A, Stringfield O, Krewer H, et al. . Predicting malignant nodules from screening CT scans. J Thorac Oncol 2016; 11: 2120–8. doi: 10.1016/j.jtho.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kumar D, Chung AG, Shaifee MJ, Khalvati F, Haider MA, Wong A. Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction [Internet. Lecture Notes in Computer Science. 2017;: 54–62. [Google Scholar]
- 78. Liu Y, Balagurunathan Y, Atwater T, Antic S, Li Q, Walker RC, et al. . Radiological image traits predictive of cancer status in pulmonary nodules. Clin Cancer Res 2017; 23: 1442–9. doi: 10.1158/1078-0432.CCR-15-3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol 2012; 22: 796–802. doi: 10.1007/s00330-011-2319-8 [DOI] [PubMed] [Google Scholar]
- 80. Yoon HJ, Sohn I, Cho JH, Lee HY, Kim J-H, Choi Y-L, et al. . Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a Radiomics approach. Medicine 2015; 94: e1753. doi: 10.1097/MD.0000000000001753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yoon HJ, Sohn I, Cho JH, Lee HY, Kim J-H, Choi Y-L, et al. . Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a Radiomics approach. Medicine 2015; 94: e1753. doi: 10.1097/MD.0000000000001753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Parmar C, Leijenaar RTH, Grossmann P, Rios Velazquez E, Bussink J, Rietveld D, et al. . Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci Rep 2015; 5: 11044. doi: 10.1038/srep11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RTH, Hermann G, et al. . Ct-Based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015; 114: 345–50. doi: 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wu J, Aguilera T, Shultz D, Gudur M, Rubin DL, Loo BW, et al. . Early-Stage Non-Small Cell Lung Cancer: Quantitative Imaging Characteristics of (18)F Fluorodeoxyglucose PET/CT Allow Prediction of Distant Metastasis. Radiology 2016; 281: 270–8. doi: 10.1148/radiol.2016151829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhou H, Dong D, Chen B, Fang M, Cheng Y, Gan Y, et al. . Diagnosis of distant metastasis of lung cancer: based on clinical and radiomic features. Transl Oncol 2018; 11: 31–6. doi: 10.1016/j.tranon.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mattonen SA, Tetar S, Palma DA, Senan S, Ward AD. Automated texture analysis for prediction of recurrence after stereotactic ablative radiation therapy for lung cancer. Int J Radiat Oncol Biol Phys 2015; 93: S5–6. doi: 10.1016/j.ijrobp.2015.07.019 [DOI] [Google Scholar]
- 87. Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, et al. . Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep 2017; 7: 588. doi: 10.1038/s41598-017-00665-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mattonen SA, Palma DA, Johnson C, Louie AV, Landis M, Rodrigues G, et al. . Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys 2016; 94: 1121–8. doi: 10.1016/j.ijrobp.2015.12.369 [DOI] [PubMed] [Google Scholar]
- 89. Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, et al. . Multi-crop Convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit 2017; 61: 663–73. doi: 10.1016/j.patcog.2016.05.029 [DOI] [Google Scholar]
- 90. Pham HHN, Futakuchi M, Bychkov A, Furukawa T, Kuroda K, Fukuoka J. Detection of lung cancer lymph node metastases from Whole-Slide histopathologic images using a two-step deep learning approach. Am J Pathol 2019; 189: 2428–39. doi: 10.1016/j.ajpath.2019.08.014 [DOI] [PubMed] [Google Scholar]
- 91. Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, et al. . Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res 2019; 25: 3266–75. doi: 10.1158/1078-0432.CCR-18-2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Coroller TP, Bi WL, Huynh E, Abedalthagafi M, Aizer AA, Greenwald NF, et al. . Radiographic prediction of meningioma grade by semantic and radiomic features. PLoS One 2017; 12: e0187908. doi: 10.1371/journal.pone.0187908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang X, Yan L-F, Hu Y-C, Li G, Yang Y, Han Y, et al. . Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget 2017; 8: 47816–30. doi: 10.18632/oncotarget.18001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen A, Lu L, Pu X, Yu T, Yang H, Schwartz LH, et al. . Ct-Based Radiomics model for predicting brain metastasis in category T1 lung adenocarcinoma. AJR Am J Roentgenol 2019;: 1–6. doi: 10.2214/AJR.18.20591 [DOI] [PubMed] [Google Scholar]
- 95. Fetit AE, Novak J, Peet AC, Arvanitits TN. Three-Dimensional textural features of conventional MRI improve diagnostic classification of childhood brain tumours. NMR Biomed 2015; 28: 1174–84. doi: 10.1002/nbm.3353 [DOI] [PubMed] [Google Scholar]
- 96. Larroza A, Moratal D, Paredes-Sánchez A, Soria-Olivas E, Chust ML, Arribas LA, et al. . Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging 2015; 42: 1362–8. doi: 10.1002/jmri.24913 [DOI] [PubMed] [Google Scholar]
- 97. Kickingereder P, Götz M, Muschelli J, Wick A, Neuberger U, Shinohara RT, et al. . Large-Scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for Stratifying anti-angiogenic treatment response. Clin Cancer Res 2016; 22: 5765–71. doi: 10.1158/1078-0432.CCR-16-0702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chang K, Zhang B, Guo X, Zong M, Rahman R, Sanchez D, et al. . Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol 2016; 18: 1680–7. doi: 10.1093/neuonc/now086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Grossmann P, Narayan V, Chang K, Rahman R, Abrey L, Reardon DA, et al. . Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol 2017; 19: 1688–97. doi: 10.1093/neuonc/nox092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pérez-Beteta J, Molina D, Martínez-González A, Arregui E, Asenjo B, Iglesias L, et al. . P09.43 novel geometrical imaging biomarkers predict survival and allow for patient selection for surgery in glioblastoma patients. Neuro Oncol 2017; 19(suppl_3): iii80. doi: 10.1093/neuonc/nox036.299 [DOI] [Google Scholar]
- 101. Pérez-Beteta J, Molina-García D, Ortiz-Alhambra JA, Fernández-Romero A, Luque B, Arregui E, et al. . Tumor surface regularity at MR imaging predicts survival and response to surgery in patients with glioblastoma. Radiology 2018; 288: 218–25. doi: 10.1148/radiol.2018171051 [DOI] [PubMed] [Google Scholar]
- 102. Pérez-Beteta J, Molina-García D, Martínez-González A, Henares-Molina A, Amo-Salas M, Luque B, et al. . Correction to: morphological MRI-based features provide pretreatment survival prediction in glioblastoma. Eur Radiol 2019; 29: 2729. doi: 10.1007/s00330-018-5870-8 [DOI] [PubMed] [Google Scholar]
- 103. Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. . Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin Cancer Res 2018; 24: 1073–81. doi: 10.1158/1078-0432.CCR-17-2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bickelhaupt S, Paech D, Kickingereder P, Steudle F, Lederer W, Daniel H, et al. . Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J Magn Reson Imaging 2017; 46: 604–16. doi: 10.1002/jmri.25606 [DOI] [PubMed] [Google Scholar]
- 105. Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. npj Breast Cancer 2017; 3. doi: 10.1038/s41523-017-0045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bickelhaupt S, Jaeger PF, Laun FB, Lederer W, Daniel H, Kuder TA, et al. . Radiomics based on adapted diffusion Kurtosis imaging helps to clarify most mammographic findings suspicious for cancer. Radiology 2018; 287: 761–70. doi: 10.1148/radiol.2017170273 [DOI] [PubMed] [Google Scholar]
- 107. Whitney HM, Taylor NS, Drukker K, Edwards AV, Papaioannou J, Schacht D, et al. . Additive benefit of Radiomics over size alone in the distinction between benign lesions and luminal a cancers on a large clinical breast MRI dataset. Acad Radiol 2019; 26: 202–9. doi: 10.1016/j.acra.2018.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Guo W, Li H, Zhu Y, Lan L, Yang S, Drukker K, et al. . Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging 2015; 2: 041007. doi: 10.1117/1.JMI.2.4.041007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, et al. . Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2016; 211 05 2016. doi: 10.1038/npjbcancer.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Fan M, Li H, Wang S, Zheng B, Zhang J, Li L. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS One 2017; 12: e0171683. doi: 10.1371/journal.pone.0171683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ma W, Zhao Y, Ji Y, Guo X, Jian X, Liu P, et al. . Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol 2019; 26: 196–201. doi: 10.1016/j.acra.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Dong Y, Feng Q, Yang W, Lu Z, Deng C, Zhang L, et al. . Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol 2018; 28: 582–91. doi: 10.1007/s00330-017-5005-7 [DOI] [PubMed] [Google Scholar]
- 113. Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. . Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin Cancer Res 2018; 24: 1073–81. doi: 10.1158/1078-0432.CCR-17-2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chan HM, van der Velden BHM, Loo CE, Gilhuijs KGA. Eigentumors for prediction of treatment failure in patients with early-stage breast cancer using dynamic contrast-enhanced MRI: a feasibility study. Phys Med Biol 2017; 62: 6467–85. doi: 10.1088/1361-6560/aa7dc5 [DOI] [PubMed] [Google Scholar]
- 115. Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. . Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 2017; 19: 57. doi: 10.1186/s13058-017-0846-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Henderson S, Purdie C, Michie C, Evans A, Lerski R, Johnston M, et al. . Interim heterogeneity changes measured using entropy texture features on T2-weighted MRI at 3.0 T are associated with pathological response to neoadjuvant chemotherapy in primary breast cancer. Eur Radiol 2017; 27: 4602–11. doi: 10.1007/s00330-017-4850-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Parikh J, Selmi M, Charles-Edwards G, Glendenning J, Ganeshan B, Verma H, et al. . Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology 2014; 272: 100–12. doi: 10.1148/radiol.14130569 [DOI] [PubMed] [Google Scholar]
- 118. Liu Z, Li Z, Qu J, Zhang R, Zhou X, Li L, et al. . Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res 2019; 25: 3538–47. doi: 10.1158/1078-0432.CCR-18-3190 [DOI] [PubMed] [Google Scholar]
- 119. Xiong Q, Zhou X, Liu Z, Lei C, Yang C, Yang M, et al. . Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy [Internet].. Clinical and Translational Oncology. 2019;. [DOI] [PubMed] [Google Scholar]
- 120. Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging 2016; 3: 034501. doi: 10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Li H, Giger ML, Huynh BQ, Antropova NO. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full-field digital mammograms. J Med Imaging 2017; 4: 1. doi: 10.1117/1.JMI.4.4.041304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wong AJ, Kanwar A, Mohamed AS, Fuller CD. Radiomics in head and neck cancer: from exploration to application. Transl Cancer Res 2016; 5: 371–82. doi: 10.21037/tcr.2016.07.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Toivonen J, Montoya Perez I, Movahedi P, Merisaari H, Pesola M, Taimen P, et al. . Radiomics and machine learning of multisequence multiparametric prostate MRI: towards improved non-invasive prostate cancer characterization. PLoS One 2019; 14: e0217702. doi: 10.1371/journal.pone.0217702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kocak B, Ates E, Durmaz ES, Ulusan MB, Kilickesmez O. Influence of segmentation margin on machine learning-based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas. Eur Radiol 2019; 29: 4765–75. doi: 10.1007/s00330-019-6003-8 [DOI] [PubMed] [Google Scholar]
- 125. Saini A, Breen I, Pershad Y, Naidu S, Knuttinen MG, Alzubaidi S, et al. . Radiogenomics and Radiomics in Liver Cancers. Diagnostics (Basel) [Internet. 2018; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Badic B, Hatt M, Durand S, Jossic-Corcos CL, Simon B, Visvikis D, et al. . Radiogenomics-based cancer prognosis in colorectal cancer. Sci Rep 2019; 9: 9743. doi: 10.1038/s41598-019-46286-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sollini M, Antunovic L, Chiti A, Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging 2019; 46: 2656–72. doi: 10.1007/s00259-019-04372-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tian Y, Liu Z, Tang Z, Li M, Lou X, Dong E, et al. . Radiomics analysis of DTI data to assess vision outcome after intravenous methylprednisolone therapy in neuromyelitis optic neuritis. J Magn Reson Imaging 2019; 49: 1365–73. doi: 10.1002/jmri.26326 [DOI] [PubMed] [Google Scholar]
- 129. Bianchi J, Gonçalves JR, Ruellas ACdeO, Vimort J-B, Yatabe M, Paniagua B, et al. . Software comparison to analyze bone radiomics from high resolution CBCT scans of mandibular condyles. Dentomaxillofac Radiol 2019; 48: 20190049. doi: 10.1259/dmfr.20190049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Panth KM, Leijenaar RTH, Carvalho S, Lieuwes NG, Yaromina A, Dubois L, et al. . Is there a causal relationship between genetic changes and radiomics-based image features? an in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol 2015; 116: 462–6. doi: 10.1016/j.radonc.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 131. Leijenaar RT, Bogowicz M, Jochems A, Hoebers FJ, Wesseling FW, Huang SH, et al. . Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br J Radiol 2018; 91: 20170498. doi: 10.1259/bjr.20170498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ou D, Blanchard P, Rosellini S, Levy A, Nguyen F, Leijenaar RTH, et al. . Predictive and prognostic value of CT based radiomics signature in locally advanced head and neck cancers patients treated with concurrent chemoradiotherapy or bioradiotherapy and its added value to human papillomavirus status. Oral Oncol 2017; 71: 150–5. doi: 10.1016/j.oraloncology.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 133. Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med 2009; 50 Suppl 1: 122S–50. doi: 10.2967/jnumed.108.057307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Fukukita H, Senda M, Terauchi T, Suzuki K, Daisaki H, Matsumoto K, et al. . Japanese guideline for the oncology FDG-PET/CT data acquisition protocol: synopsis of version 1.0. Ann Nucl Med 2010; 24: 325–34. doi: 10.1007/s12149-010-0377-7 [DOI] [PubMed] [Google Scholar]
- 135. Boellaard R, O’Doherty MJ, Weber WA, Mottaghy FM, Lonsdale MN, Stroobants SG, et al. . Fdg PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging 2010; 37: 181–200. doi: 10.1007/s00259-009-1297-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kinahan PE, Perlman ES, Sunderland JJ, Subramaniam R, Wollenweber SD, Turkington TG, et al. . The QIBA profile for FDG PET/CT as an imaging biomarker measuring response to cancer therapy. Radiology 2020;: 191882.. doi: 10.1148/radiol.2019191882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mankoff DA. Quantitative imaging as cancer biomarker [Internet. Medical Imaging 2015: Physics of Medical Imaging 2015;. [Google Scholar]
- 138. Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification [Internet. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). 2018;. [Google Scholar]
- 139. Mahmood F, Chen R, Durr NJ. Unsupervised reverse domain adaptation for synthetic medical images via Adversarial training. IEEE Trans Med Imaging 2018; 37: 2572–81. doi: 10.1109/TMI.2018.2842767 [DOI] [PubMed] [Google Scholar]
- 140. Wang Q, Milletari F, Nguyen HV, Albarqouni S, Jorge Cardoso M, Rieke N, et al. . Domain adaptation and representation transfer and medical image learning with less labels and imperfect data: first MICCAI workshop, dart 2019, and first International workshop, MIL3ID 2019, Shenzhen, held in conjunction with MICCAI 2019, Shenzhen, China, October 13 and 17, 2019, proceedings. Springer Nature 2019; 254. [Google Scholar]
- 141. Jochems A, Deist TM, El Naqa I, Kessler M, Mayo C, Reeves J, et al. . Developing and validating a survival prediction model for NSCLC patients through distributed learning across 3 countries. Int J Radiat Oncol Biol Phys 2017; 99: 344–52. doi: 10.1016/j.ijrobp.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Deist TM, Jochems A, van Soest J, Nalbantov G, Oberije C, Walsh S, et al. . Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin Transl Radiat Oncol 2017; 4: 24–31. doi: 10.1016/j.ctro.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Brisimi TS, Chen R, Mela T, Olshevsky A, Paschalidis IC, Shi W. Federated learning of predictive models from federated electronic health records. Int J Med Inform 2018; 112: 59–67. doi: 10.1016/j.ijmedinf.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Sheller MJ, Anthony Reina G, Edwards B, Martin J, Bakas S. Multi-institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation [Internet. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 2019;: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lugan S, Desbordes P, Tormo LXR, Legay A, Macq B. Secure Architectures Implementing Trusted Coalitions for Blockchained Distributed Learning (TCLearn) [Internet]. 2019. Available from: http://arxiv.org/abs/1906.07690 [cited 2019 Oct 17].
- 146. Holzinger A. Explainable AI (ex-AI) [Internet. Informatik-Spektrum. 2018; 41: 138–43. [Google Scholar]
- 147. Khedkar S, Subramanian V, Shinde G, Gandhi P. Explainable AI in Healthcare [Internet. SSRN Electronic Journal.. [Google Scholar]
- 148. Joyner MJ, Promises PN. promises, and precision medicine [Internet]. Vol. 129. Journal of Clinical Investigation 2019;: 946–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Saracci R. Epidemiology in Wonderland: big data and precision medicine. Eur J Epidemiol 2018; 33: 245–57. doi: 10.1007/s10654-018-0385-9 [DOI] [PubMed] [Google Scholar]