Abstract
Background & aim:
Genetic variability in drug absorption, distribution, metabolism and excretion (ADME) genes contributes to the high heterogeneity of drug responses. The present study investigated polymorphisms of ADME genes frequencies and compared the findings with populations from other continents, available in the 1000 Genome Project (1 KGP) and the Exome Aggregation Consortium (ExAC) databases.
Methodology & results:
We conducted a study of 100 patients in Brazil and a total of 2003 SNPs were evaluated by targeted next-generation sequencing in 148 genes, including Phase I enzymes (n = 50), Phase II enzymes (n = 38) and drug transporters (n = 60). Overall, the distribution of minor allele frequency (MAF) suggests that the distribution of 2003 SNPs is similar between Brazilian cohort, 1 KGP and ExAC; however, we found moderate SNP allele-frequency divergence between Brazilian cohort and both 1000 KGP and ExAC. These differences were observed in several relevant genes including CYP3A4, NAT2 and SLCO1B1.
Conclusion:
We concluded that the Brazilian population needs clinical assessment of drug treatment based on individual genotype rather than ethnicity.
Keywords: : ADME genes, admixture population, Brazilian, genetic diversity, next-generation sequencing, pharmacogenomics
Genetic variation in genes encoding drug absorption, distribution, metabolism and excretion (ADME) proteins is one of the factors that affects drug pharmacokinetics and contributes to variability of drug efficacy and safety [1]. Based on such genetic variability, pharmacogenetics of several drugs in current clinical practice has been described by several international consortia to predict adverse drug response and improve the treatment outcome. This is reflected by a growing number of drugs with pharmacogenetic information provided in their drug labels by the US FDA, so far issued 404 guidelines for 285 drugs, 86 biomarkers and 19 therapeutic areas (March 2020) [2].
Population studies have shown that interethnic variability occurs in the frequency of genetic variants [3] and that significant genetic differences in the ADME genes between different populations could result in therapeutic failure or adverse drug response. For example, the anticoagulant warfarin has the highest dose requirements in African–Americans, the lowest dose requirements in Asians and intermediate requirements in Caucasian populations [4]. Limdi et al. [5] showed that race-stratified analysis improves dose prediction in the USA. However, it is different in admixed populations, such as Brazil and other Latin American countries that predictive power of two such algorithms did not differ between white and black Brazilian: this was explained by the higher frequency of the rs9923231T allele in black Brazilians, as a result of the extensive European–African admixture [6].
Brazil, with more than 200 million people, is the most populous country in South America and has mainly a trichotomous ancestral contribution of the following distant parental populations: Europeans, Native Americans and Africans [7]. The Brazilian Institute of Geography and Statistics (IBGE) classifies individuals by self-reported color and they are categorized as white (48%), mixed ‘pardo’ (43%), black (7.6%), Asian descendant ‘amarelo’ (1.1%) and Native American (0.4%) [8]. In particular, the genetic admixture influences the genomic diversity of ADME genes and may cause high heterogeneity of drug responses in admixed populations such as Brazilians.
This brings up one major topic in pharmacogenetics studies: facing ethnic genetic difference [9]. Given the importance of developing drugs for patients worldwide and the increasing globalization of clinical drug development, identifying and quantifying all ADME genetic variations that contribute to interethnic differences in drug pharmacokinetics, efficacy and safety is of extreme interest [10]. Some initiatives have addressed genetic variation among populations. The 1000 Genome Project (1 KGP) created the largest public catalogue of human variation and sequencing data, aiming to identify most genetic variants with frequencies of at least 1% in the populations studied [11]. The Exome Aggregation Consortium (ExAC) grouped exome sequencing data from a number of disease-specific and population studies, making it publicly available [12]. However, there is an overrepresentation of European descendant populations in pharmacogenetic studies and the extrapolation of those findings to other populations are factors that undermine the strong evidence that supports the incorporation of precision medicine [13].
On the basis of this understanding, the objective of this study was to investigate similarities and differences in genetic polymorphisms in genes involved in drug ADME among Brazilian, 1 KGP and ExAC databases. To this end, a large number of genetic variants were assessed using the next-generation sequencing method. This study might help evaluate the difficulty of extrapolating clinical data between populations, mainly admixture populations. Also, we discussed the clinical implications of these genetic variations in drug safety and efficacy.
Materials & methods
Study design & participants
From December 2015 to December 2016, blood samples were collected from 100 patients with hepatitis C virus enrolled at the Department of Gastroenterology, Clinics Hospital of University of Sao Paulo, Brazil. The study protocol was approved by the local ethic committee of the Clinics Hospital (protocol no. 1142/09) and written informed consent was obtained from each participant. The study was conducted in accordance to the ethical guidelines of the 1975 Declaration of Helsinki.
ADME genes & polymorphisms selection
The design of the pharmacogenetics panel for all relevant ADME related genes (157 genes in total) included publicly available lists derived from the Pharmacogenomics Knowledge Base (PharmGKB; June 2016) [14], FDA table of pharmacogenomic biomarkers in drug labels (June 2016) [2] and PharmaADME.org core list [16,17]. In addition, we added pharmacogenetic genes derived from PubMed database searches using combinations of variant terms for drug response, ADME genes, genetic variations and polymorphisms (June 2016). For analysis, the genes were assorted into functional groups as follows: Phase I enzymes (n = 56), Phase II enzymes (n = 42) and drug transporters (n = 60), detailed in Supplementary Table 1.
Targeted ADME next-generation sequencing panel sequencing
Genomic DNA isolation from 200 μl blood and cell lines was performed using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration of DNA samples was measured by NanoDrop spectrophotometer (Thermo Fisher Scientific, MA, USA) and Qubit 2.0 (Invitrogen Life Technologies, CA, USA) as recommended. In brief, 4210 targets including exons, introns, UTR3′ and UTR5′ gene regions of 157 ADME genes, including Phases I and II drug-metabolizing enzymes and drug transporters, were specifically captured using SureSelectXT Target Enrichment System Kit for Illumina Paired-End Multiplexed Sequencing Library customized 1–499kb (Agilent Technologies, CA, USA) and sequenced using an Illumina NextSeq and MiSeq systems (Illumina Inc., CA, USA); see details in Supplementary Table 2.
Bioinformatics analyses
The raw data (fastq) was aligned to GRCh37/hg19 using BWA-MEM software [18] and ordered by the bamsort tool of the biobambam2 package. The call for variants was made by the freebayes software (v0.9.10-3-g47a713e) simultaneously, followed by filtering to exclude variants with low statistical support and strand bias (vcffilter tool of the vcflib package). Subsequently, the breakdown of the multiallelic variants in individual lines and normalization to the left of inDels was done by the vt software [19]. The annotation of the variants was done by ANNOVAR [20].
Quality control
Quality control tests were performed on data using PLINK v1.07 [21]. A total of 7065 SNPs passed through the first quality control filter with call rate of 0.95%. Then, 4231 SNPs were included for the evaluation of genetic frequency after considering the quality control filter that removes SNPs that were not in Hardy–Weinberg equilibrium 0.0001, minor allele frequency (MAF) 0.01 and chromosome X. After quality control, the final dataset was based on 2003 genetic markers in 148 genes, Phase I enzymes (n = 50), Phase II enzymes (n = 38) and drug transporters (n = 60) (Figure 1).
Figure 1. . Study overview.
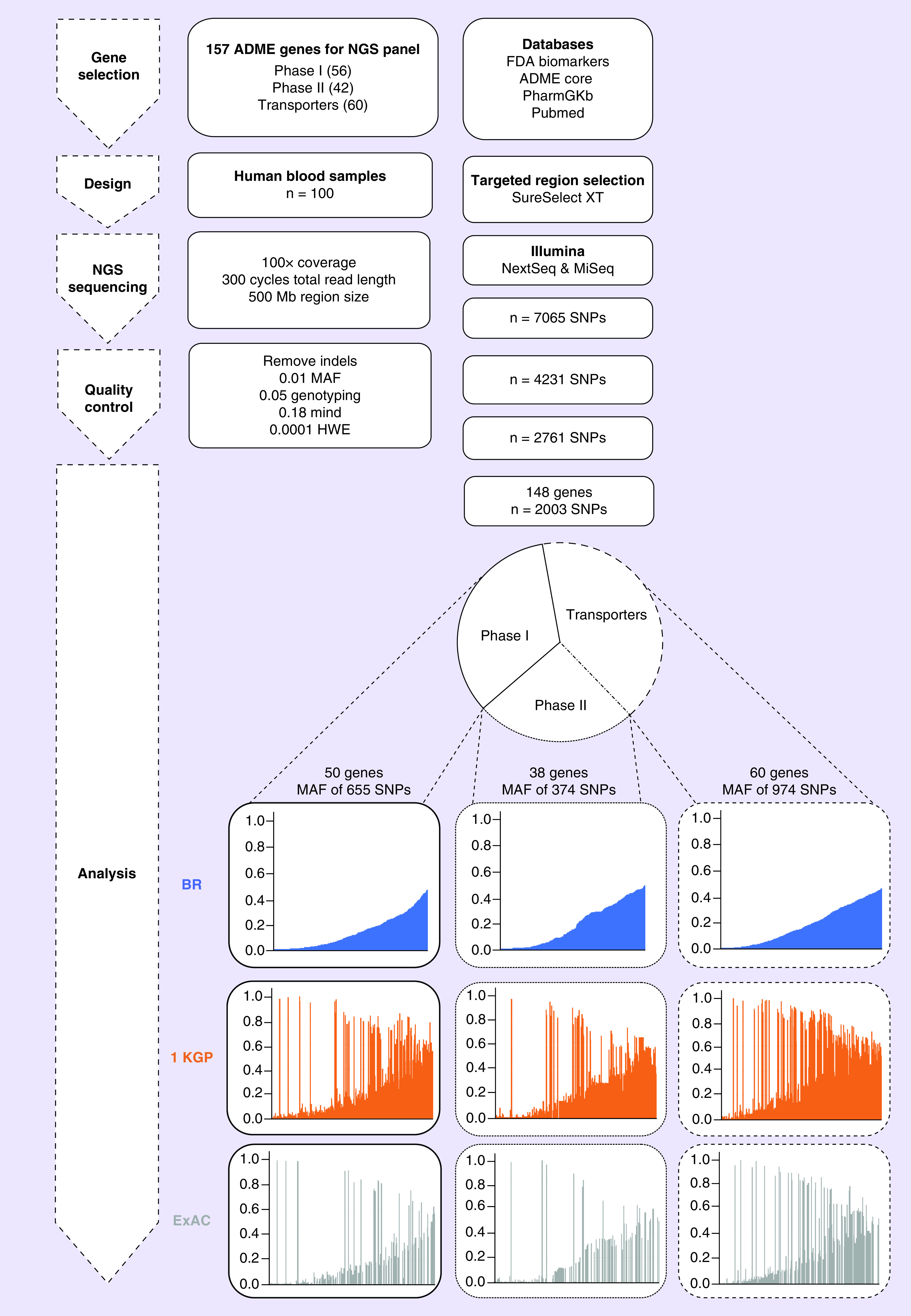
Schematic overview of the workflow for the ADME NGS panel sequencing. Composition of ADME NGS target genes displayed as Phase I, Phase I enzymes (n = 50); Phase II, Phase II enzymes (n = 38); and transporters (n = 60). For further details see Figures 2–4.
1 KGP: 1000 Genome Project; ADME: Absorption, distribution, metabolism and excretion; ExAC: Exome Aggregation Consortium; HWE: Hardy–Weinberg equilibrium; MAF: Minor allele frequency; Mind: Missingness per individual; NGS: Next-generation sequencing.
Results
Patient characteristics
The cohort of 100 patients was recruited from the Clinics Hospital of University of Sao Paulo. Most of the subjects were white (69%), male (63%) and had a median age of 59 years (range 34–74 years) (Table 1). The description of hepatitis C virus treatment is detailed in Supplementary Table 3.
Table 1. . Baseline characteristics in study population based in self-reported race.
| Characteristics | Number | White | Brown | Black | Asian |
|---|---|---|---|---|---|
| Overall | 100 | 69 | 15 | 8 | 8 |
| Age (range) | 59 (34–74) | 60 (34–74) | 54 (39–67) | 59 (43–70) | 64 (55–74) |
| Male | 63 | 45 | 11 | 5 | 2 |
| Female | 37 | 24 | 4 | 3 | 6 |
| BMI (range) | 26.9 (18.8–40.7) | 26.9 (18.8–38.5) | 28.6 (22.3–40.7) | 26.7 (19.4–29.6) | 23.6 (21.5–26.9) |
Impact of population admixture on pharmacogenomic implementation
Pharmacogenomic (PGx) studies focus on the genetic diversity patterns of functional genes, that is, the variants that play a significant role in drug response variability. In the present study, we used public databases such as PharmaADME.org and PharmGKB, to classify all genes into three categories as described Supplementary Table 1. We explored the possibility to estimate the frequencies of functional SNPs in the admixed population, which was sampled for deep sequencing to explore the polymorphic state of functional variants. From our dataset, we described the MAF of 2003 SNPs in 148 ADME-related genes in Brazilians and we compared with 1 KGP and ExAC as shown in Figure 1. Overall, the distribution of MAF suggests that the distribution of 2003 SNPs is similar between Brazilian cohort, 1 KGP and ExAC; however, we observed moderate SNP allele-frequency divergence between Brazilian cohort and both 1000 KGP and ExAC databases (Figures 2–4).
Figure 2. . The minor allele frequency of 64 Phase I enzyme genes variants.
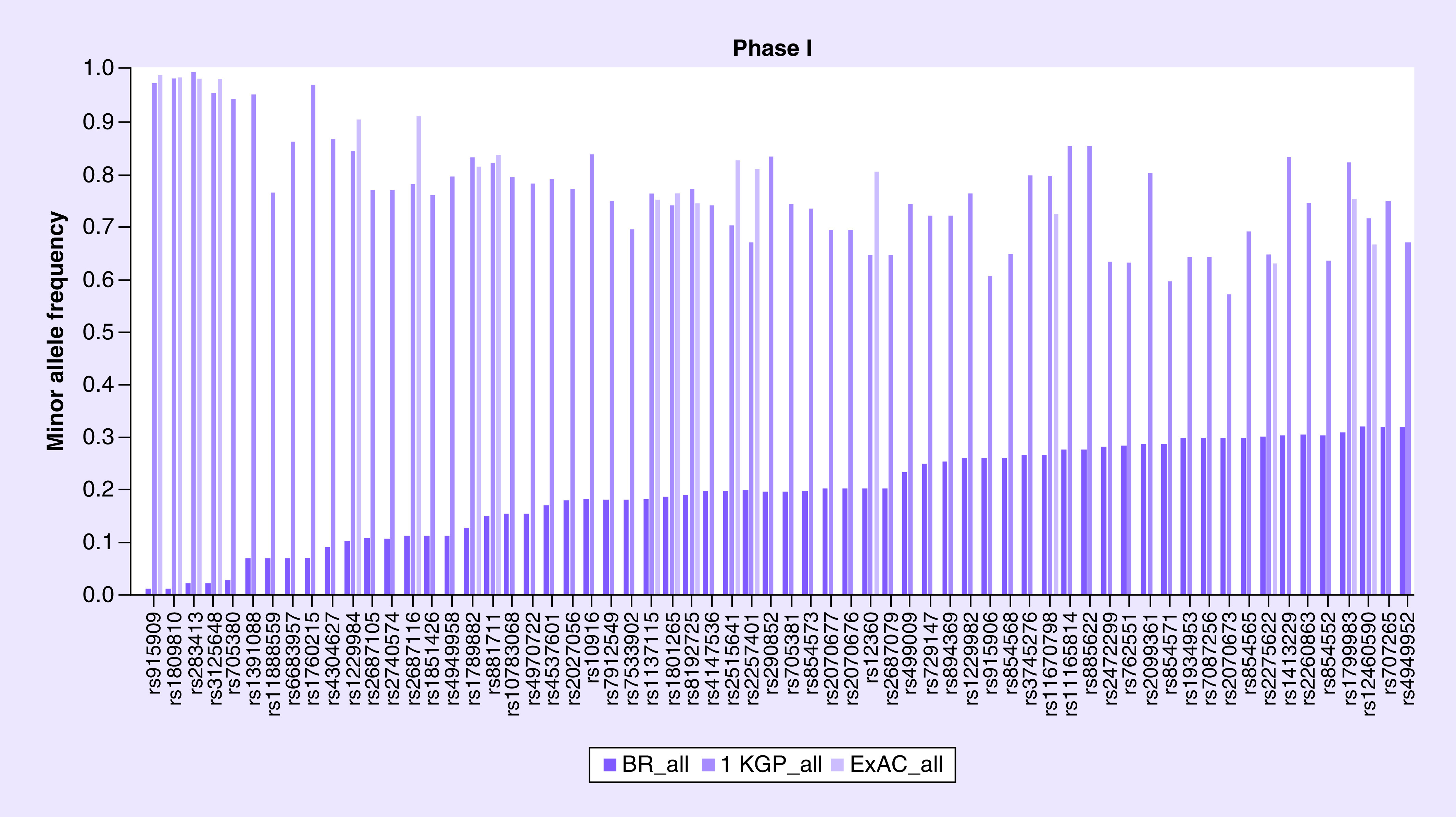
1 KGP: 1000 Genome Project; ExAC: Exome Aggregation Consortium.
MAF profiling of common functional polymorphisms of Phase I enzymes genes
The MAF was described for 655 polymorphisms of 50 genes of Phase I and compared with 1 KGP and ExAC databases, as shown in Supplementary Table 4. Our study showed that several polymorphisms such as rs4079369 of CYP2A6 have similar MAF (6%) among the same ethnic groups of 1 KGP (6%) and ExAC (8%). However, there are several variants such as rs915909 (CYP2E1) and rs1809810 (CYP2A6) which have MAF of 1%, while 1 KGP and ExAC shows a frequency of 98 and 99%, respectively (Figure 2).
When comparing populations of similar ethnicities, such as African population, for variant rs1799853 (CYP2C9), our results showed that MAF was 13% for black, ExAC shows 2% for African (AFR) and another Brazilian cohort described 7% for black [22]. On the other hand, populations with less admixture, such as the Asian population, have a similar frequency. For instance, our results showed a MAF of 43% for black and 25% for Asian, while ExAC shows 73% for African and 26% for East Asian for this variant rs2242480 located in CYP3A4 gene.
MAF profiling of common functional polymorphisms of Phase II enzymes genes
The MAF was described for 374 polymorphisms of 38 genes of Phase II and compared with 1 KGP and ExAC databases, as shown in Supplementary Table 4. Several variants such as rs650985 (GSTM4) have MAF of 1%, while 1 KGP and ExAC shows a frequency of 98 and 96%, respectively (see Figure 3). Our study identified that polymorphism rs1800822 (FMO3) presented a similar minor allele T frequency among white, black and Asian when compared with 1 KGP and ExAC databases. For the variant rs1208 of the NAT2 gene, the frequency of the minor allele G is similar between Caucasian populations, such as white Brazilian (46%), European 1 KGP (44%) and Finnish European ExAC (43%). On the other hand, the frequency of rs4680 variant (COMT) is different between our white population (37%) and the Caucasian populations European and Finnish European (50 and 53%). It should be noted that the polymorphism rs28365062 of the UGT2B7 has a similar minor allele frequency among Brazilian white, mixed and black, but it has a different frequency between Asian descendant (0%), Japanese 1 KGP (10%) and East Asian ExAC (5%).
Figure 3. . The minor allele frequency of 37 Phase II enzyme genes variants.
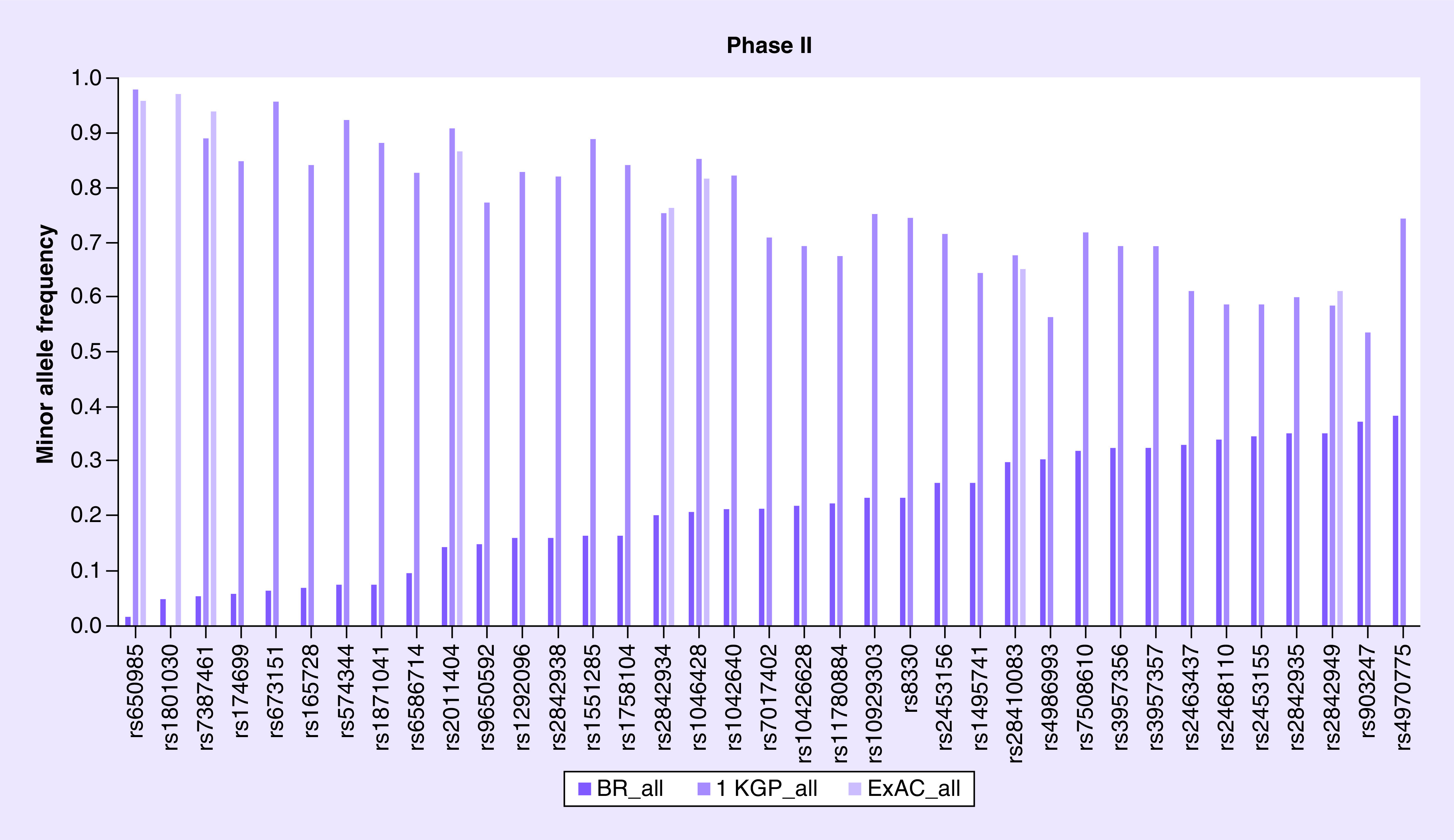
1 KGP: 1000 Genome Project; ExAC: Exome Aggregation Consortium.
Allele frequency distribution of common functional polymorphisms of drug transporter genes
The MAF was described for 974 polymorphisms of 60 genes of drug-transporter genes and compared with 1 KGP and ExAC databases, as shown in Supplementary Table 4. Several variants such as rs9524765 (ABCC4) have MAF of 1%, while 1 KGP and ExAC shows a frequency of 93 and 95%, respectively (Figure 4). The polymorphism rs717620 (ABCC2) affects the response of antiepileptic drugs [23], influences the metabolism of erythromycin [24] and is associated with toxicity among patients treated with fluorouracil, leucovorin and oxaliplatin [25]. Our study shows that this polymorphism presents a similar lower allele frequency among the ethnic groups when comparing the Brazilian population with 1 KGP and ExAC databases. However, there are several variants such as rs2306283 (SLCO1B1) which has a general lower allele frequency of 46%, whereas 1 KGP and ExAC showed an allele frequency of less than 38 and 52%, respectively. The variant rs1867351 (SLC22A1) has a frequency of the allele less than 19% for black Brazilians, whereas 1 KGP and ExAC showed a frequency lower than 33 and 30%. On the other hand, the ABCG2 variant rs2231142 (G>T) had a frequency of the minor allele T of 15, 10, 6 and 25% among white, mixed, black and Asian, respectively. We observed a similar frequency when compared with 1 KGP (9, 14, 1 and 32%, respectively) and ExAC (10, 24, 3 and 30%, respectively). This polymorphism is associated with increased plasma concentration of rosuvastatin [26,27].
Figure 4. . The minor allele frequency of 62 transporters genes variants.
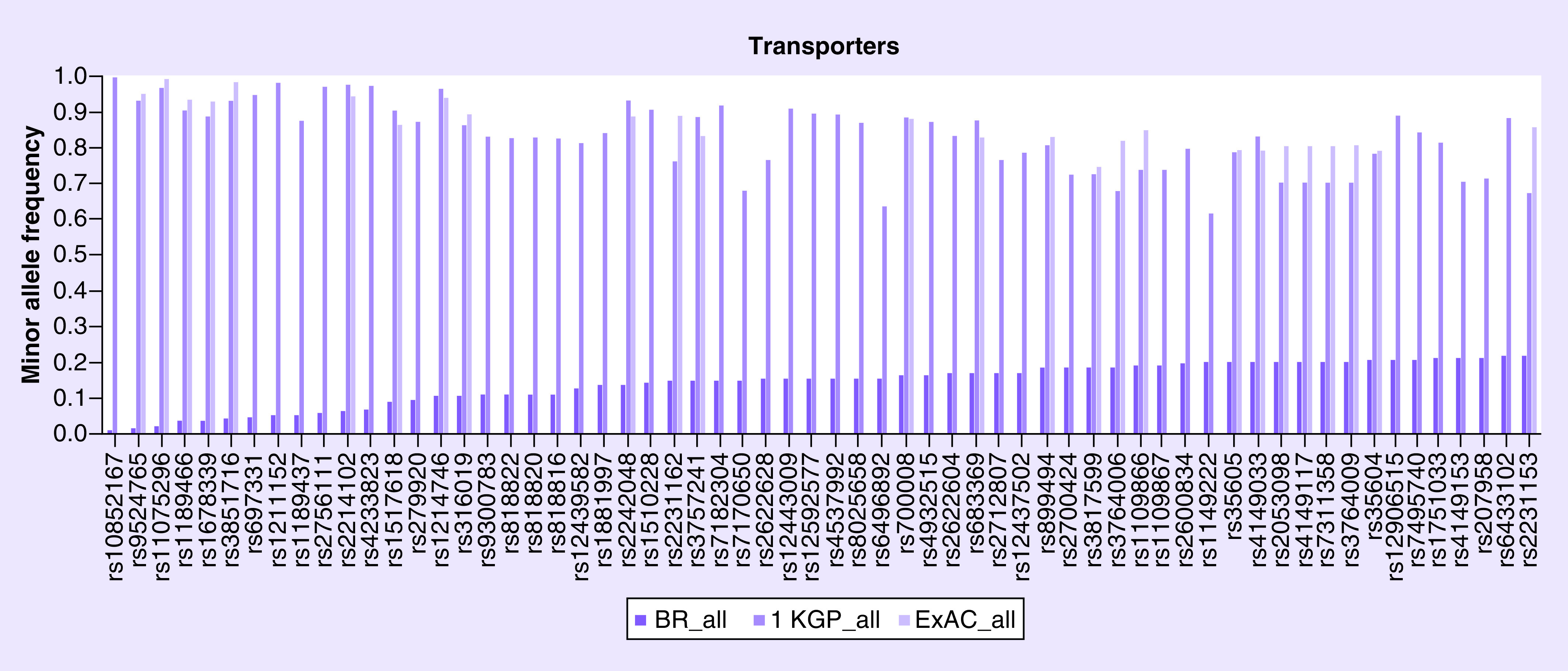
1 KGP: 1000 Genome Project; ExAC: Exome Aggregation Consortium.
Discussion
Pharmacogenetics studies have been conducted primarily on study cohorts consisting of European non-Hispanic whites. A collapse of the genetic diversity associated with the first human colonization of Europe during the Paleolithic period, followed by the recent mixture of African, European and Native American ancestors resulted in different ethnic groups with varying degrees of genetic diversity [28]. There is robust evidence from clinical trials for different medical conditions that show that individuals from different ethnic groups experience varying responses to specific therapeutic agents [21].
CYP2C19 *2 (rs4244285) and *3 (rs4986893) are known to affect the metabolism and responses of several commonly prescribed medications, including antidepressants [29], antiplatelets [30] and antiulcer drugs [31]. The most commonly mutated allele is CYP2C19 *2 in slow metabolizing Caucasians [32], while CYP2C19 *3 is rare among Caucasian individuals [33]. In the present study, we found the same results that the incidence of CYP2C19 *2 among white Brazilians (18%) was similar to that found in other Caucasian populations, 1 KGP (15%) and ExAC (15%), while CYP2C19 *3 is also rare in the Brazilian white population (0%).
CYP2C8 is a Phase I metabolizing enzyme that plays an integral role in the biotransformation of structurally diverse xenobiotic and endogenous compounds. CYP2C8 *2 is common in African (19%), but it is rare in white and Asian [34]. The results are similar for the frequency in our Brazilian black population of 13% and rare for white and Asian Brazilians, 1 and 0%, respectively. In vitro, CYP2C8 *2 was associated with a decrease in enzyme activity and a lower intrinsic clearance of paclitaxel, ibuprofen and repaglinide compared with the wild-type enzyme [35].
The rs1057910 (A) at CYP2C9 most commonly encodes the amino acid isoleucine at position 359 and the resulting allele is also known as CYP2C9 *1. The rs1057910 (C) encodes a leucine at this same position and the resulting allele is termed CYP2C9 *3 (Ile359Leu). Individuals with this SNP may be at increased risk of developing acute gastrointestinal bleeding during the use of NSAIDs, such as, celecoxib, diclofenac and ibuprofen [36]. In addition, there is a risk of severe skin reactions when taking phenytoin, an antiepileptic drug [37]. According to 1 KGP, the CYP2C9 *3 allele has a frequency of 7% in European, 4% in American, 0% in African and 2% in Japanese. Our results corroborate allelic frequencies of 5, 7, 0 and 0% in Brazilian white, mixed, black and Asian individuals, respectively. This finding is similar to that of other populations in which the frequency of the CYP2C9 *3 allele. Lower frequencies have been reported for the CYP2C9 *3 variant in East Asian populations; Japanese (1.1–2.1%) [38] and Korean [39] and the highest frequency of CYP2C9 *3 was reported in Tamil Sri Lankans living in England (0.5%) [11].
Nakamura et al. [40] suggest that instabilities and reduction of intrinsic clearance by the protein encoded by the rs1065852 (T) allele are the main reasons why Asian have lower metabolic activities than Caucasian for drugs metabolized by CYP2D6, since this (T) allele occurs more frequently in Asians. In Korean women with metastatic breast cancer, the CYP2D6 *10 genotype is associated with significantly lower steady-state plasma concentrations of endoxifen (the active metabolite of tamoxifen). It is found more frequently among non-responders [41], although another large randomized study (BIG 1–98) involving mainly white patients (>98%) found no such association between CYP2D6 variants and disease control [42]. The allelic frequency of CYP2D6 *10 (rs1065852) varies among different ethnic groups: 21% in white, 20% in mixed, 13% in black and 50% in Asian.
CYP4F2 regulates the bioavailability of vitamin E and vitamin K, a critical co-factor for blood clotting. Variations in CYP4F2 that affect the bioavailability of vitamin K also affect the dosage of vitamin K antagonists, such as warfarin or acenocoumarol [43]. A study in three separate Caucasian populations was the first to report that the T allele in rs2108622 on CYP4F2 was associated with an increase in the dose of warfarin. The study calculated that each T allele in rs2108622 is associated with a 4–12% increase in the warfarin dose, so that a patient with TT genotype could require an approximate dose of 1 mg/day of warfarin compared with a patient with CC genotype [32]. Our results show that the frequency of rs2108622 was the closest to that reported for Caucasian populations (30%), European 1 KGP (29%) and ExAC (29%).
The variants in COMT, the metabolizing enzyme of Phase II, have been associated with psychiatric disorders, including schizophrenia, perception of pain mediated by opioid receptors and breast cancer. COMT is one of the main pathways of dopamine degradation and COMT Val/Met polymorphisms are associated with enzyme activity which is related to the involvement of dopamine in the process of nicotine addiction. This meta-analysis found that patients with AA genotype who are treated with nicotine replacement therapy have an increased likelihood of smoking cessation compared with patients with GG genotype [44]. The variant rs4680 in the COMT has an allele frequency of less than approximately 50% in Caucasian, in our study population, it was 37%.
Glutathione S-transferases, also Phase II metabolizing enzymes, are responsible for the detoxification of a number of drugs and potential carcinogens through glutathione conjugation. Patients with advanced rectal colon cancer with a homozygous variant genotype for GSTP1 were more likely to discontinue FOLFOX (folinic acid, fluorouracil and oxaliplatin) because of neurotoxicity (24 vs 10%; p = 0.01) [45]. The GSTP1 rs4147581 high frequency in Asians of 1 KGP (64%) and ExAC (69%) showed a higher chance of adverse events compared with the Brazilian Asian (25%).
N-acetyltransferase encodes enzymes genes involved in acetylation of aromatic amines and heterocyclic compounds [46]. The rs1799930 AA genotype is associated with an increased risk of hepatotoxicity when treated with isoniazid in subjects with tuberculosis compared with the GG genotype [47]. Women with ovarian neoplasms with C allele and T allele (CT) compared with TT of rs1801280 present an increased anemia risk when treated with cisplatin and cyclophosphamide [48]. The rs1801280 has a high frequency (40%) in Brazilian individuals, but low in Asians (6%). This result corroborates with other populations of 1 KGP and ExAC.
TPMT encodes S-methyltransferase thiopurine, which catalyzes the S-methylation of thiopurine drugs and aromatic and heterocyclic sulfhydryl compounds. TPMT variation may lead to thiopurine toxicity [49]. Polymorphism rs1800462 occurred with an overall frequency of 1% in white Brazilian subjects, the same as the 1% in Europeans and 0% in other ethnicities.
UGT1A1 is the only enzyme responsible for bilirubin glucuronidation, allowing its excretion. It is also responsible for glucuronidation of other xenobiotics, such as SN-38, the active metabolite of irinotecan [50]. Epileptic patients with G/G genotype in UGT2B7 *2 and *3, rs7439366 and rs12233719, respectively, are associated with increased concentrations of valproic acid compared with T/T genotype. Therefore, patients with epilepsy with these genotypes may need to increase (or decrease) the dose of valproic acid to ensure its therapeutic effect [49]. Individuals homozygous for the present allele have 32% of normal enzyme activity and may present with Gilbert’s syndrome [51].
ABCB1 is one of the most important genes involved in transport including antidepressants, antipsychotics, antihypertensive and analgesics. The rs1045642 showed a frequency of 47% in white, 43% in mixed, 25% in black and 38% in Asian and the frequency of these polymorphisms in 1 KGP and ExAC were similar to those found in our study. The A allele of this polymorphism is associated with a decreased risk of hepatotoxicity when treated with nevirapine in HIV-positive individuals compared with G allele [52]. The polymorphism rs1128503 of the ABCB1 gene is associated with an increase in the overall survival period from 0.34 to 0.44 among oxaliplatin-treated patients for colorectal neoplasms [53]. On the other hand, rs10276036 is associated with an increased risk of death in patients with osteosarcoma after chemotherapy [54]. Rs1045642 is associated with increased serum concentrations of digoxin and hepatotoxicity induced by nevirapine [55,56] and rs212091 of virologic failure in antiretroviral therapy [57]. In the present study, we observed that the general frequency of ABCB1 gene polymorphisms among Brazilians was similar to that found in other general populations, in 1 KPG and ExAC. Larger frequencies of variant rs1128503 were reported for Asian populations in the three databases.
The rs22273697, also known as c1249G>A or p.V471I, is a nonsynonymous polymorphism of the ABCC2 transporter gene. From an initial case–control study of 146 patients with epilepsy, followed by replication in another 279 patients, the allele rs2273697 (A) was associated with neurological adverse reactions to the use of carbamazepine (p = 0.001). Functional studies have shown that this SNP selectively reduced the transport of carbamazepine through the cell membrane [58]. In addition, rs2273697 influences the pharmacokinetics of talinolol and irinotecan [59,60]. The frequency of the Brazilian population corroborates with 1 KGP and ExAC databases for the four ethnic groups. Another rs3740066 in the ABCC2 is reported to be associated with an increased risk of developing intrahepatic cholestasis of pregnancy based on a study of approximately 70 Argentinean patients [61]. ABCC4 variant rs1751034 (C > T) allele of less than 25% for white, 23% for mixed, 6% for black and 25% for Asian. The rs1751034 CC and CT genotypes are associated with increased intracellular concentrations of tenofovir when treated in HIV-infected individuals compared with the TT genotype [62].
The rs4149056, found in the SLCO1B1, which encodes the polypeptide protein transport of organic anions. This protein, found mainly in the liver, regulates the absorption of countless drugs and natural compounds. The variant rs4149056 (C) defines the SLCO1B1 *5 allele which gives rise to an amino acid change (from valine to alanine at residue 174) which has reduced uptake and transport activity. Therefore, drugs metabolized by SLCO1B1 tend to accumulate higher circulating concentrations [63]. SLCO1B1 *5 is associated with a high risk of muscle disease when treated with simvastatin. Other drugs associated with *5 variant include cerivastatin, pravastatin and rosuvastatin [64,65]. According to 1 KPG, the frequency of the rs4149056 variant of the SLCO1B1 gene is reported as different between Caucasian (16%) and African (1%) populations. However, in the Brazilian white population (14%) and black (13%) this difference was not observed. Another SNP related to adverse events is the rs628031 variant of the SLC22A1 gene that is associated with gastrointestinal side effects when treated with metformin [66].
There were also some limitations to our analysis. Our dataset is a small sample size and it is representative of individuals with hepatitis C virus. Despite these challenges, such data are important to assess and improve the current strategies. We evaluated and described the minor allele frequency of 2003 polymorphisms of 148 ADME-related genes. Our findings demonstrated that the Brazilian population exhibited substantial functional SNPs differences at some ADME genes compared with the 1 KGP and ExAC database. Then, we suggest population differences should be carefully considered in pharmacogenetics studies, designing clinical trials of new drugs and treatments across continents and personalization of drug treatment based on the individual genotype rather than ethnicity may be the most appropriate. Further studies are warranted to accomplish this major goal. The success of the Personalized Medicine paradigm will depend on our capabilities to recognize the genetic variability existing between and within different ethnic groups.
Summary points.
Absorption, distribution, metabolism and excretion genes
Genetic variation in genes encoding drug absorption, distribution, metabolism and excretion (ADME) proteins is one of the factors that affects the drug pharmacokinetics and contributes to variability of drug efficacy and safety.
The 1000 Genome Project & Exome Aggregation Consortium
There is an overrepresentation of European descendant populations in pharmacogenetic studies.
Extrapolation of those findings to other populations are factors that undermine the strong evidence that supports the incorporation of precision medicine.
Minor allele-frequency profiling
The minor allele frequency of 2003 SNPs in 148 ADME genes was described in Brazilian cohort and compared with the 1000 Genome Project (1 KGP) and Exome Aggregation Consortium (ExAC).
The overall distribution suggests that it is similar between Brazilian cohort, 1 KGP and ExAC. However, we observed moderate SNP allele-frequency divergence between our cohort and both databases.
Admixture population
The genetic admixture influences the genomic diversity of ADME genes and may cause high heterogeneity of drug responses in admixed populations such as Brazilians.
Our study concludes that the Brazilian population needs clinical assessment of drug treatment based on individual genotype rather than ethnicity.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/pgs-2020-0013
Financial & competing interests disclosure
V Kim would like to thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES – 1399290) and Programa de Doutorado Sanduíche no Exterior (PDSE 88881.132608/2016-01). SK Ono would like to thank Fundação de Amparo à pesquisa de São Paulo (FAPESP 2015/16291-0) and Brazilian Council for Development of Science and Technology (CNPq) for grant PQ 308609/2018-2. Y Hoshida would like to thank the US NIH/NIDDK R01 DK099558, European Commission ERCE2014EAdGE671231, Department of Defense W81XWHE16E1E0363 and the Cancer Prevention and Research Institute of Texas (CPRIT) RR180016. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Sim SC, Kacevska M, Ingelman-Sundberg M. Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J. 13(1), 1–11 (2013). [DOI] [PubMed] [Google Scholar]
- 2.US FDA. Table of pharmacogenomic biomarkers in drug labeling with labeling text. (2020). https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm
- 3.Rosenberg NA. A population-genetic perspective on the similarities and differences among worldwide human populations. Hum. Biol. 83(6), 659–684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang MT, Hambleton J, Kayser SR. The influence of ethnicity on warfarin dosage requirement. Ann. Pharmacother. 39(6), 1008–1012 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Limdi NA, Wadelius M, Cavallari L. et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 115(18), 3827–3834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifaz-Pena V, Contreras AV, Struchiner CJ. et al. Exploring the distribution of genetic markers of pharmacogenomics relevance in Brazilian and Mexican populations. PLoS ONE 9(11), e112640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima-Costa MF, Rodrigues LC, Barreto ML. et al. Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative). Sci. Rep. 5, 9812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.IBGE. Censo Demográfico. Rio de Janeiro: Instituto Brasileiro de Geografia e Estatística. (2011). https://biblioteca.ibge.gov.br/visualizacao/periodicos/93/cd_2010_caracteristicas_populacao_domicilios.pdf
- 9.Maisano Delser P, Fuselli S. Human loci involved in drug biotransformation: worldwide genetic variation, population structure and pharmacogenetic implications. Hum. Genet. 132(5), 563–577 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Hovelson DH, Xue Z, Zawistowski M. et al. Characterization of ADME gene variation in 21 populations by exome sequencing. Pharmacogenet. Genomics 27(3), 89–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auton A, Brooks LD, Durbin RM. et al. A global reference for human genetic variation. Nature 526(7571), 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616), 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos E, Callier SL, Rotimi CN. Why personalized medicine will fail if we stay the course. Per Med. 9(8), 839–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whirl-Carrillo M, McDonagh EM, Hebert JM. et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92(4), 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown AM, Renaud Y, Ross C. et al. Development of a broad-based ADME panel for use in pharmacogenomic studies. Pharmacogenomics 15(9), 1185–1195 (2014). [DOI] [PubMed] [Google Scholar]
- 17.PharmaADME.org. Montreal Heart Institute Pharmacogenomics Center. (2016). http://pharmaadme.org/joomla/
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14), 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan A, Abecasis GR, Kang HM. Unified representation of genetic variants. Bioinformatics 31(13), 2202–2204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16), e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dries DL, Exner DV, Gersh BJ, Cooper HA, Carson PE, Domanski MJ. Racial differences in the outcome of left ventricular dysfunction. N. Engl. J. Med. 340(8), 609–616 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues-Soares F, Kehdy FSG, Sampaio-Coelho J. et al. Genetic structure of pharmacogenetic biomarkers in Brazil inferred from a systematic review and population-based cohorts: a RIBEF/EPIGEN-Brazil initiative. Pharmacogenomics J. 18(6), 749–759 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Qu J, Zhou BT, Yin JY. et al. ABCC2 polymorphisms and haplotype are associated with drug resistance in Chinese epileptic patients. CNS Neurosci. Ther. 18(8), 647–651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franke RM, Lancaster CS, Peer CJ. et al. Effect of ABCC2 (MRP2) transport function on erythromycin metabolism. Clin. Pharmacol. Ther. 89(5), 693–701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KH, Chang HJ, Han SW. et al. Pharmacogenetic analysis of adjuvant FOLFOX for Korean patients with colon cancer. Cancer Chemother. Pharmacol. 71(4), 843–851 (2013). [DOI] [PubMed] [Google Scholar]
- 26.DeGorter MK, Tirona RG, Schwarz UI. et al. Clinical and pharmacogenetic predictors of circulating atorvastatin and rosuvastatin concentrations in routine clinical care. Circ. Cardiovasc. Genet. 6(4), 400–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HK, Hu M, Lui S, Ho CS, Wong CK, Tomlinson B. Effects of polymorphisms in ABCG2, SLCO1B1, SLC10A1 and CYP2C9/19 on plasma concentrations of rosuvastatin and lipid response in Chinese patients. Pharmacogenomics 14(11), 1283–1294 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J. Allergy Clin. Immunol. 133(1), 16–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Vos A, van der Weide J, Loovers HM. Association between CYP2C19*17 and metabolism of amitriptyline, citalopram and clomipramine in Dutch hospitalized patients. Pharmacogenomics J. 11(5), 359–367 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Kim HS, Chang K, Koh YS. et al. CYP2C19 poor metabolizer is associated with clinical outcome of clopidogrel therapy in acute myocardial infarction but not stable angina. Circ. Cardiovasc. Genet. 6(5), 514–521 (2013). [DOI] [PubMed] [Google Scholar]
- 31.de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J. Biol. Chem. 269(22), 15419–15422 (1994). [PubMed] [Google Scholar]
- 32.Caldwell MD, Awad T, Johnson JA. et al. CYP4F2 genetic variant alters required warfarin dose. Blood 111(8), 4106–4112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol. Pharmacol. 46(4), 594–598 (1994). [PubMed] [Google Scholar]
- 34.Dai D, Zeldin DC, Blaisdell JA. et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 11(7), 597–607 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Yu L, Shi D, Ma L, Zhou Q, Zeng S. Influence of CYP2C8 polymorphisms on the hydroxylation metabolism of paclitaxel, repaglinide and ibuprofen enantiomers in vitro. Biopharm. Drug Dispos. 34(5), 278–287 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Agundez JA, Garcia-Martin E, Martinez C. Genetically based impairment in CYP2C8- and CYP2C9-dependent NSAID metabolism as a risk factor for gastrointestinal bleeding: is a combination of pharmacogenomics and metabolomics required to improve personalized medicine? Expert Opin. Drug Metab. Toxicol. 5(6), 607–620 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Chung WH, Chang WC, Lee YS. et al. Genetic variants associated with phenytoin-related severe cutaneous adverse reactions. JAMA 312(5), 525–534 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Kimura M, Ieiri I, Mamiya K, Urae A, Higuchi S. Genetic polymorphism of cytochrome P450s, CYP2C19 and CYP2C9 in a Japanese population. Ther. Drug Monit. 20(3), 243–724 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Yoon YR, Shon JH, Kim MK. et al. Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br. J. Clin. Pharmacol. 51(3), 277–280 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura K, Ariyoshi N, Yokoi T. et al. CYP2D6.10 present in human liver microsomes shows low catalytic activity and thermal stability. Biochem. Biophys. Res. Commun. 293(3), 969–973 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J. Clin. Oncol. 25(25), 3837–3845 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Tan SH, Lee SC, Goh BC, Wong J. Pharmacogenetics in breast cancer therapy. Clin. Cancer Res. 14(24), 8027–8041 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Alvarellos ML, Sangkuhl K, Daneshjou R, Whirl-Carrillo M, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for CYP4F2. Pharmacogenet. Genomics 25(1), 41–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi HD, Shin WG. Association between catechol-O-methyltransferase (COMT) Val/Met genotype and smoking cessation treatment with nicotine: a meta-analysis. Pharmacogenomics 16(16), 1879–1885 (2015). [DOI] [PubMed] [Google Scholar]
- 45.McLeod HL, Sargent DJ, Marsh S. et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J. Clin. Oncol. 28(20), 3227–3233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley LA, Sim E. Update on the pharmacogenetics of NATs: structural considerations. Pharmacogenomics 9(11), 1673–1693 (2008). [DOI] [PubMed] [Google Scholar]
- 47.An HR, Wu XQ, Wang ZY, Zhang JX, Liang Y. NAT2 and CYP2E1 polymorphisms associated with antituberculosis drug-induced hepatotoxicity in Chinese patients. Clin. Exp. Pharmacol. Physiol. 39(6), 535–543 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Khrunin AV, Khokhrin DV, Moisseev AA, Gorbunova VA, Limborska SA. Pharmacogenomic assessment of cisplatin-based chemotherapy outcomes in ovarian cancer. Pharmacogenomics 15(3), 329–337 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Wang P, Lin XQ, Cai WK. et al. Effect of UGT2B7 genotypes on plasma concentration of valproic acid: a meta-analysis. Eur. J. Clin. Pharmacol. 74(4), 433–442 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Barbarino JM, Haidar CE, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for UGT1A1. Pharmacogenet. Genomics 24(3), 177–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto K, Sato H, Fujiyama Y, Doida Y, Bamba T. Contribution of two missense mutations (G71R and Y486D) of the bilirubin UDP glycosyltransferase (UGT1A1) gene to phenotypes of Gilbert's syndrome and Crigler–Najjar syndrome Type II. Biochim. Biophys. Acta 1406(3), 267–273 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Haas DW, Kwara A, Richardson DM. et al. Secondary metabolism pathway polymorphisms and plasma efavirenz concentrations in HIV-infected adults with CYP2B6 slow metabolizer genotypes. J. Antimicrob. Chemother. 69(8), 2175–2182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H, Kang H, Liu Y. et al. Association of ABCB1 genetic polymorphisms with susceptibility to colorectal cancer and therapeutic prognosis. Pharmacogenomics 14(8), 897–911 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Caronia D, Patino-Garcia A, Perez-Martinez A. et al. Effect of ABCB1 and ABCC3 polymorphisms on osteosarcoma survival after chemotherapy: a pharmacogenetic study. PLoS ONE 6(10), e26091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aarnoudse AJ, Dieleman JP, Visser LE. et al. Common ATP-binding cassette B1 variants are associated with increased digoxin serum concentration. Pharmacogenet. Genomics 18(4), 299–305 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Ciccacci C, Borgiani P, Ceffa S. et al. Nevirapine-induced hepatotoxicity and pharmacogenetics: a retrospective study in a population from Mozambique. Pharmacogenomics 11(1), 23–31 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Coelho AV, Silva SP, de Alencar LC. et al. ABCB1 and ABCC1 variants associated with virological failure of first-line protease inhibitors antiretroviral regimens in Northeast Brazil patients. J. Clin. Pharmacol. 53(12), 1286–1293 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Kim WJ, Lee JH, Yi J. et al. A nonsynonymous variation in MRP2/ABCC2 is associated with neurological adverse drug reactions of carbamazepine in patients with epilepsy. Pharmacogenet. Genomics 20(4), 249–256 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Haenisch S, May K, Wegner D, Caliebe A, Cascorbi I, Siegmund W. Influence of genetic polymorphisms on intestinal expression and rifampicin-type induction of ABCC2 and on bioavailability of talinolol. Pharmacogenet. Genomics 18(4), 357–365 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Fujita K, Nagashima F, Yamamoto W. et al. Association of ATP-binding cassette, sub-family C, number 2 (ABCC2) genotype with pharmacokinetics of irinotecan in Japanese patients with metastatic colorectal cancer treated with irinotecan plus infusional 5-fluorouracil/leucovorin (FOLFIRI). Biol. Pharm. Bull. 31(11), 2137–2142 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Sookoian S, Castano G, Burgueno A, Gianotti TF, Pirola CJ. Association of the multidrug-resistance-associated protein gene (ABCC2) variants with intrahepatic cholestasis of pregnancy. J. Hepatol. 48(1), 125–132 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Kiser JJ, Aquilante CL. anderson PL, King TM, Carten ML, Fletcher CV. Clinical and genetic determinants of intracellular tenofovir diphosphate concentrations in HIV-infected patients. J. Acquir. Immune Defic. Syndr. 47(3), 298–303 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Pasanen MK, Backman JT, Neuvonen PJ, Niemi M. Frequencies of single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide 1B1 SLCO1B1 gene in a Finnish population. Eur. J. Clin. Pharmacol. 62(6), 409–415 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Marciante KD, Durda JP, Heckbert SR. et al. Cerivastatin, genetic variants and the risk of rhabdomyolysis. Pharmacogenet. Genomics 21(5), 280–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho RH, Choi L, Lee W. et al. Effect of drug transporter genotypes on pravastatin disposition in European– and African–American participants. Pharmacogenet. Genomics 17(8), 647–656 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarasova L, Kalnina I, Geldnere K. et al. Association of genetic variation in the organic cation transporters OCT1, OCT2 and multidrug and toxin extrusion 1 transporter protein genes with the gastrointestinal side effects and lower BMI in metformin-treated type 2 diabetes patients. Pharmacogenet. Genomics 22(9), 659–666 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


