Abstract
Polycomb group (PcG) proteins are essential for maintenance of lineage fidelity by coordinating developmental gene expression programs. Polycomb group ring finger 6 (PCGF6) has been previously reported to repress expression of lineage-specific genes, especially germ cell–related genes in mouse embryonic stem cells (ESCs) via the noncanonical polycomb repressive complex PRC1.6. However, the molecular mechanism of this repression remains largely unknown. Here, using RNA-Seq, real-time RT-PCR, immunohistochemistry, immunoprecipitation, and ChIP analyses, we demonstrate that PCGF6 plays an essential role in embryonic development, indicated by the partially penetrant embryonic lethality in homozygous PCGF6 (Pcgf6−/−)-deficient mice. We also found that surviving Pcgf6-deficient mice exhibit reduced fertility. Using the Pcgf6-deficient mice, we observed that ablation of Pcgf6 in somatic tissues robustly derepresses germ cell–related genes. We further provide evidence that these genes are direct targets of PCGF6 in ESCs and that endogenous PCGF6 co-localizes with the histone-modifying proteins G9A histone methyltransferase (G9A)/G9a-like protein (GLP) and histone deacetylase 1/2 (HDAC1/2) on the promoters of the germ cell–related genes. Moreover, the binding of these proteins to their target genes correlated with methylation of Lys-9 of histone 3 and with the status of histone acetylation at these genes. Moreover, the recruitment of G9A/GLP and HDAC1/2 to target promoters depended on the binding of PCGF6. Our findings indicate that PCGF6 has a critical role in safeguarding lineage decisions and in preventing aberrant expression of germ cell–related genes.
Keywords: polycomb group ring finger 6 (Pcgf6), knockout mice, embryonic stem cells, germline gene silencing, G9A histone methyltransferase, G9a-like protein 1 (Glp), histone deacetylase 1/2 (Hdac1/2), PRC1.6, epigenetic regulation, gene regulation, polycomb, embryonic stem cell, histone deacetylase (HDAC), gene knockout, histone methylation, G9a/Glp, Pcgf6
PcG proteins act as evolutionarily conserved epigenetic mediators of transcription that have integral roles in stem cell identity and development by repressing key developmental genes (1–3). PcG proteins are categorized into at least two biochemically and functionally distinct multisubunit complexes, the polycomb repressive complex 1 (PRC1) and 2 (PRC2), which deposit monoubiquitination of histone H2A at Lys-119 (H2AK119ub1) and trimethylation at lysine 27 of histone H3 (H3K27me3), respectively (1, 2). In mammals, the PRC2 complex comprises the catalytic subunits Ezh1/Ezh2 and their interacting partners Eed and Suz12 (4). The canonical PRC1 complex comprises four core subunits: RING E3 ligase (Ring1A/B), Pcgf (Pcgf1–6), Cbx (polycomb; Cbx2/4/6/7/8), and Phc (polyhomeotic homologues; Phc1/2/3) (1, 2, 5). In noncanonical PRC1 complexes, Cbx and Phc proteins are replaced by Rybp/Yaf2 (5). Importantly, based on the exclusive association of one of the six Pcgf protein homologues, there exist six functionally distinct groups of mammalian PRC1 complexes named PRC1.1–1.6 (6).
PCGF6 was initially identified in a multimeric complex associated with the E2F6 transcription factor in HeLa cells, termed E2F6.com, that contains E2F6, DP1, MAX, MGA, L3MBTL2, RING1A/B, PCGF6, CBX3, YAF2, G9A (also known as EHMT2: euchromatic histone lysine N-methyltransferase 2), and GLP (G9A-like protein 1, also known as EHMT1) (7). Subsequent studies in other cell types, including ESCs (8), demonstrate that the major E2F6.com resembles PRC1.6 complex (6, 9). Essential roles for Pcgf6 in maintenance of ESC state has been demonstrated (10–12). Pcgf6 was identified as an essential self-renewal gene in ESCs by an unbiased genome-wide RNAi screen (13). More recent knockout studies revealed that Pcgf6 is required for ESC identity (10, 12). Notably, the transcriptional profile of Pcgf6-null ESCs, examined by microarray analysis or RNA-Seq, revealed the comprehensive repression of germline-specific genes by Pcgf6 (10, 12). Of interest, the expression of germ cell–specific genes is globally repressed in ESCs and all somatic cells in the body. Failure to appropriately express these genes in meiotic cells often results in a partial or complete loss of fertility (14), whereas illegitimate expression in somatic cells may lead to catastrophic consequences such as oncogenic transformation (15). However, the mechanisms that govern Pcgf6-mediated timely silencing of the germline-specific genes remain poorly understood. Intriguingly, the association of Pcgf6 with G9a/Glp (7), with major euchromatic H3K9me1 and H3K9me2 HKMTs (histone lysine methyltransferases), and with the histone deacetylase Hdac1/2 suggests that epigenetic modulations could play a role in silencing of germ line–expressed genes (8). Unfortunately, this issue has not been addressed experimentally.
In this study, we showed that disruption of the Pcgf6 gene led to partial embryonic lethality, and mutant male and female mice exhibited significantly impaired fertility. In the surviving Pcgf6-deficient mice, Pcgf6 was found to be essential for silencing germ cell–specific genes in somatic tissues, thereby suppressing the germ cell differentiation program. Furthermore, G9a/Glp and Hdac1/2 existed in a repressive complex assembled by Pcgf6. Pcgf6 directly bound to its silenced genes, and its binding highly correlated with G9a/Glp and Hdac1/2 in ESCs. Moreover, deletion of the Pcgf6 gene induced robust reduction of H3K9me1/2, accompanied with derepression of germ cell–specific genes. Our data genetically demonstrate mechanistic insight into how Pcgf6 acts as a dedicated repressor of genes associated with meiosis and germ cells, thereby preserving the identity of ESCs as well as somatic cells under physiological conditions.
Results
Pcgf6 null mice display partially penetrant embryonic lethality
To uncover the biological role of Pcgf6 in vivo, we generated mice with targeted disruption of the Pcgf6 locus (Fig. 1A). The conserved RING motif is located in exon 3 of the Pcgf6 gene (16); therefore, we designed a conditional targeting construct spanning 2 kb to disrupt exons 2, 3, 4, and 5. A loxP site (3′loxP) was cloned downstream of exon 5, and a PGKneo cassette flanked by FRT sites was cloned upstream of exon 2. The linearized targeting vector was introduced into C57BL/6J ESCs by electroporation, and cells were selected with G418. 96 G418-resistant ESC colonies were selected and screened for a recombination event using long-range PCR. The FRT-flanked Neo selection cassette was removed by subsequent FLPe-mediated recombination. Two ESC clones carrying one floxed Pcgf6 allele were microinjected into C57BL/6J blastocysts to generate chimeric animals. Both ESC clones successfully transmitted the Pcgf6-flox conditional allele through the germline. These mice were mated with a mouse strain that expresses Cre recombinase ubiquitously under the control of the cytomegalovirus promoter. Deletion of exons 2–5 of the Pcgf6 gene was confirmed by PCR analysis (Fig. 1B). Successful gene ablation of Pcgf6 in homozygous Pcgf6 (Pcgf6–/–)-deficient mice was also confirmed by the absence of its protein product in multiple tissues by Western blotting analyses (Fig. 1C). Heterozygous Pcgf6 mice were viable, fertile, and indistinguishable from WT littermates. When heterozygotes were intercrossed, we observed a significantly lower fraction of Pcgf6 homozygous mutant offspring than the 25% predicted by the Mendelian law (Fig. 1D), which is consistent with previous results (12). Additionally, surviving homozygotes appeared smaller in body size and lighter in weight than their WT or heterozygous littermates at the same age (Fig. 1, E and F). Notably, ablation of Pcgf6 did not affect the protein levels of other components of the PRC1.6 complex (Fig. 1C).
Figure 1.
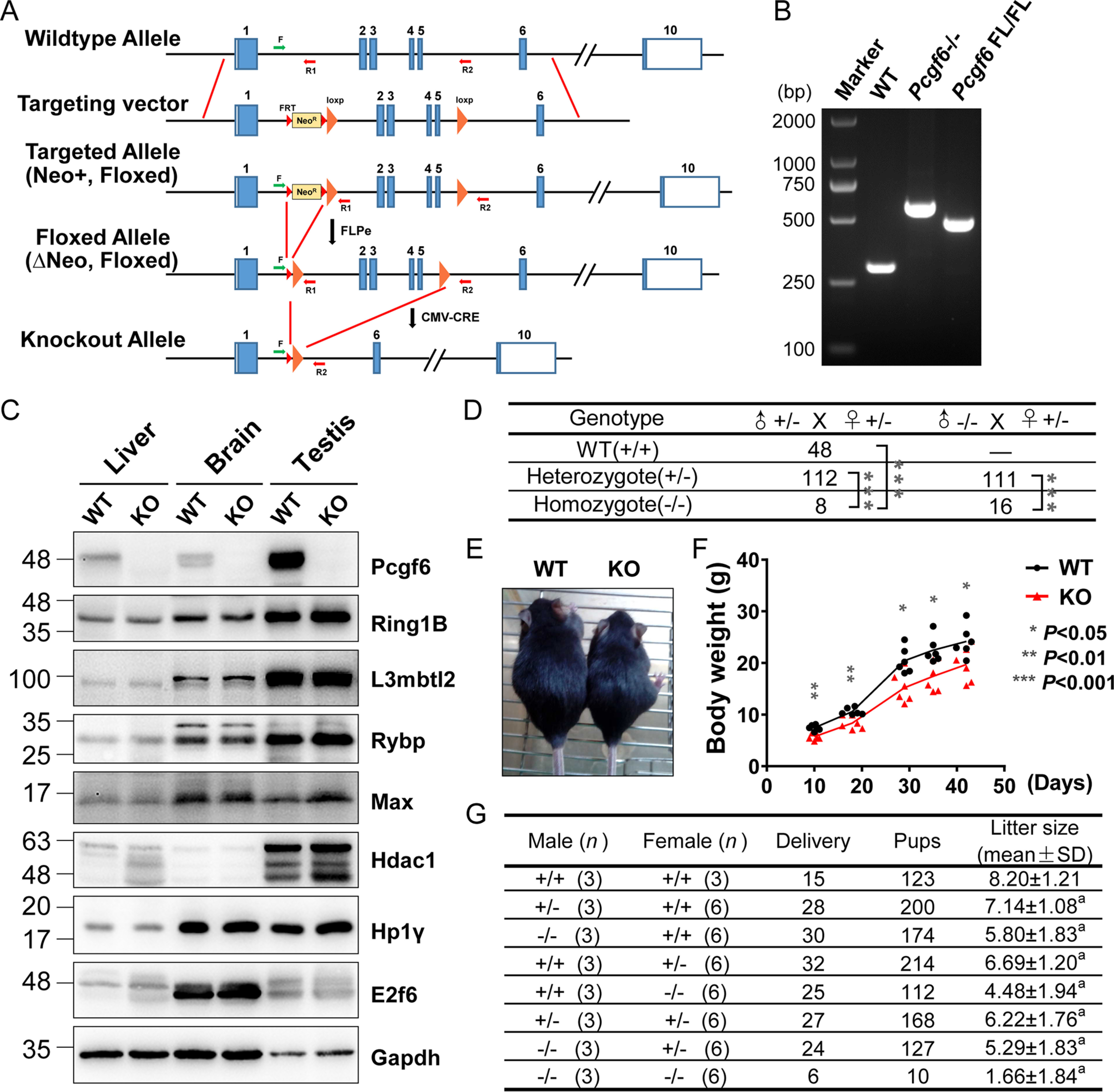
Pcgf6 null mice exhibit partially penetrant embryonic lethality. A, schematic of the gene targeting strategy used to generate Pcgf6 mutant mice. The WT Pcgf6 locus is shown at the top. The targeting vector contains loxP sites flanking exons 2–5 and a neomycin gene flanked by FRT sites. F, R1, R2 are primers used to determine genotypes. B, genomic PCR for analysis of Pcgf6 gene status. The primers F, R1, and R2 shown in A and DNA from toes of mice were used to determine genotypes. C, Western blotting demonstrating changes in the protein levels of the indicated genes in the liver, brain, and testis of indicated genotypes. Protein loading is 40 μg. Gapdh was used as a control. D, genotype distribution of progeny of different intercrosses. ***, p < 0.005 (χ2 test), indicating that the probability of conforming to the Mendelian law is <0.5%. E, body size of the Pcgf6+/+ and Pcgf6–/– mice at 6 weeks. F, body weight of Pcgf6−/− mice from birth to adulthood compared with WT mice. n = 6. G, fecundity of Pcgf6+/+, Pcgf6+/–, and Pcgf6–/– mice. n, number of mice. a, p < 0.01(t test) versus +/+ × +/+.
Mice deficient in Pcgf6 exhibit a strongly reduced fertility
qRT-PCR showed broad expression of Pcgf6 mRNA in various tissues of WT mice, among which the highest level of expression was detected in testis (Figs. S1 and S2A). Moreover, recent research has demonstrated that ablation of the genes encoding the unique subunits of PRC1.6 in ESCs is accompanied with robust derepression of germline-specific gene expression (8, 10, 13, 17, 18). Thus, the unique expression patterns of Pcgf6 prompted us to explore its potential regulation of reproduction and fertility in mice. When Pcgf6+/– males and females were crossed with their WT counterparts, the litter sizes (7.1 and 6.7 pups/litter, respectively) were slightly smaller than that after intercrossing the WT male and female mice (8.2 ± 1.2 pups) (Fig. 1G); however, crosses of Pcgf6–/– males and females with WT counterparts produced significantly smaller litters than did the controls (5.8 and 4.5 pups, respectively). It was noteworthy that mating of Pcgf6–/– mice produced an average litter size of only 1.7 pups (Fig. 1G). These data indicated that loss of Pcgf6 in both male and female mice caused a reduction in fecundity.
To evaluate fertility in the male Pcgf6 null mice, testicular development and morphology were analyzed. At 8 weeks postpartum, testes and epididymis from male Pcgf6–/– were smaller than those from WT littermates (Fig. 2A). When testes and epididymis weights were normalized to body weight, the ratios were significantly reduced in Pcgf6−/− mice compared with WT mice (Fig. 2B), suggesting an impairment of testicular function and/or spermatogenesis. Histological analysis of the testes by hematoxylin and eosin (H&E) staining indicated that there were no significant alterations in the interstitial spaces or seminiferous epithelium of testes from Pcgf6−/− mice (Fig. 2C). Meanwhile, as observed with the testis, H&E staining did not reveal any significant morphological differences in the ovaries of adult Pcgf6−/− mice compared with WT littermates (Fig. S2). However, compared with WT mice, Pcgf6−/− mice exhibited significant reductions in sperm motility, accompanied by more morphologically abnormal sperm than exhibited by WT littermates (Fig. 2, D–F). To gain insight into the molecular mechanisms underlying the observed phenotypic changes, we performed RNA-Seq on whole testes from Pcgf6–/– mice and their respective control. In Pcgf6–/– testes, 150 genes were deregulated (with 65 up-regulated and 85 down-regulated) on a -fold change cutoff of 2 and p value <0.05 (Fig. 2G and Table S3). Gene ontology (GO) analysis of down-regulated target genes scored them as being strongly related to sperm motility and differentiation (Fig. 2H), suggesting a particular role for Pcgf6 in sperm fate specification. In contrast, GO analysis of up-regulated genes did not show significant association with any biological process. Importantly, among down-regulated genes, our analysis revealed a set of genes, including Adam7, Crisp1, Rnase9, Spinkl, Spink8, Spink10, and Ceacam10 (Fig. 2I), which have been shown to have critical roles in testicular development and function (19–25). Spermatogenesis and fertility depend upon functional interactions between germ cells and somatic cells (Leydig and Sertoli cells). To have a more in-depth understanding of Pcgf6 in spermatogenetic events, we isolated multiple testicular cell populations (Sertoli, Leydig, and several spermatogenic cell populations) from testes. Interestingly, although Pcgf6 was expressed in Leydig and Sertoli cells, as well as in all types of germ cells of mouse testis, loss of the Pcgf6 gene affected its target gene expression (such as Crisp1, Rnase9, and Spinkl) in germ cells at the round and elongating spermatid stage but not in somatic cells or pachytene spermatocytes, raising the possibility that Pcgf6 functions in a stage-dependent manner during spermatogenesis (Fig. S3). Together, our studies suggest a previously unknown role for Pcgf6 in testicular development and spermatogenesis.
Figure 2.
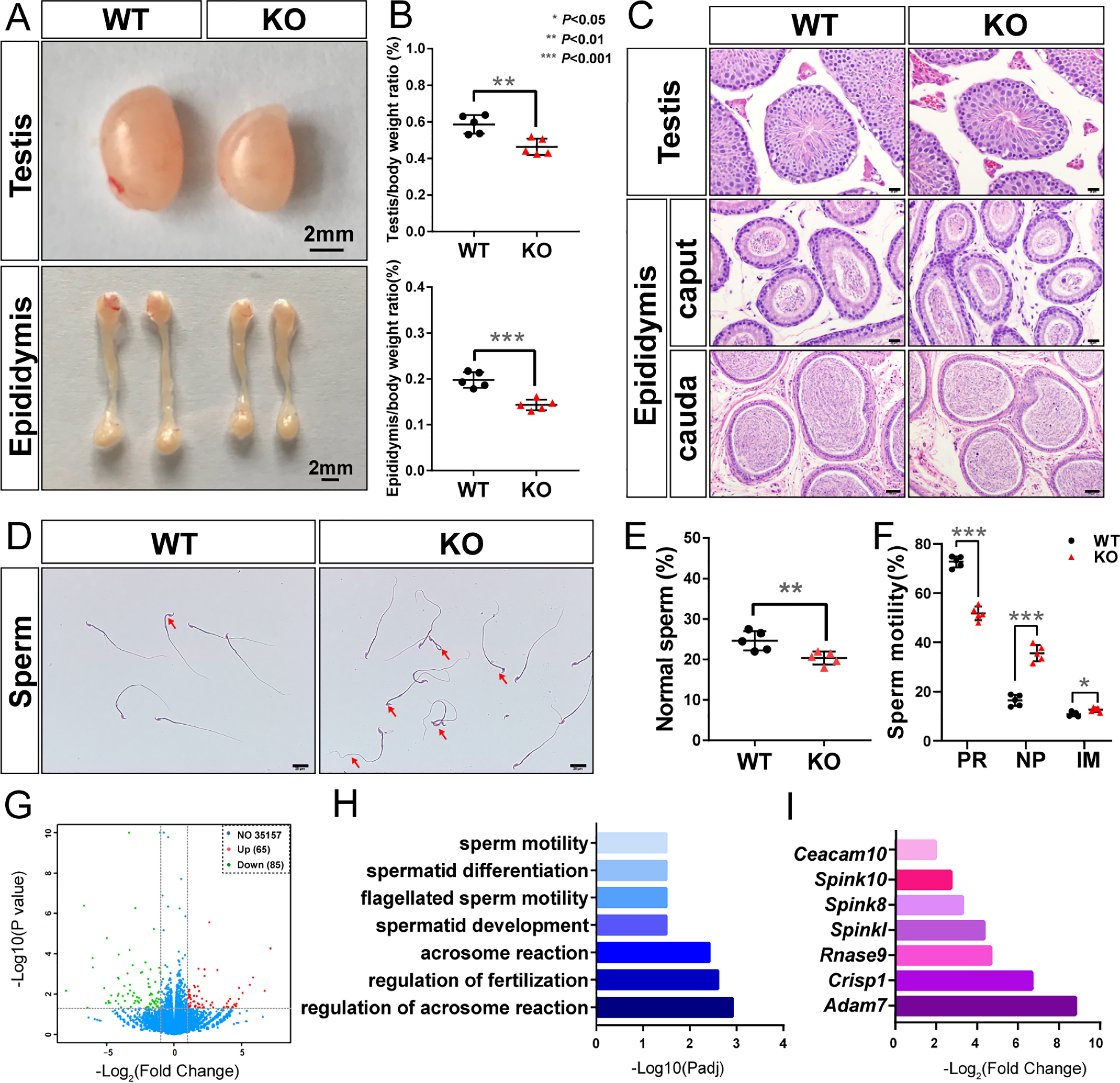
Pcgf6-deficient mice dramatically reduced in fertility. A, representative image of the testes and epididymides of the WT and Pcgf6−/− mice. Scale bars, 2 mm. B, testis and epididymis as a percentage of body weight of mice with the indicated genotype. C, H&E staining of representative sections of the testes and epididymis of the WT and Pcgf6−/− mice. Scale bars, 50 μm (cauda epididymis) or 20 μm (others). D, normal and abnormal sperms in WT and Pcgf6−/− mice at 8 weeks of age. Red arrows, defective sperms. E and F, the proportions of normal sperm and the sperm motility of mice with the indicated genotype at 8 weeks of age. Data are represented as the means ± S.D. (error bars) from five mice for each genotype. Sperm motility was measured using a CASA CEROS version 12 sperm analysis system (Hamilton Thorne). PR, progressively motile; NP, nonprogressively motile; IM, immotile. G, volcano plot of differentially expressed genes in testis between WT and Pcgf6−/− mice. H, GO analyses showing the biological functions of down-regulated genes in Pcgf6−/− testis. I, the log2(-fold change) of the several down-regulated genes related to sperm function in Pcgf6−/− testis.
Germline-specific genes are aberrantly expressed in Pcgf6 null adult somatic tissues
Given that genetic ablation of Pcgf6 in ESCs results in illegitimate expression of germ cell– and meiosis–related genes, we wondered whether Pcgf6 is functionally required for the silencing of germline genes in somatic tissues (10, 12). To this end, we compared the expression profiles of liver and brain (organs that show the high Pcgf6 mRNA expression levels (Fig. S1)) from Pcgf6 knockout and littermate WT control mice by RNA-Seq analysis. A total of 160 versus 61 genes were considerably up-regulated in Pcgf6-deficient liver and brain, respectively, and 50 versus 56 down-regulated genes in Pcgf6-deficient liver and brain, respectively, compared with their corresponding controls (Fig. 3 (A and C) and Tables S4 and S5). To gain insight into the physiological functions of Pcgf6-mediated gene regulation in somatic tissues, we performed gene ontology (GO) analyses, which revealed that the up-regulated genes are significantly enriched in germ cell– and meiosis–related functions (Fig. 3B). Strikingly, analysis of genes commonly up-regulated in Pcgf6-deficient liver and brain revealed highly significant overlap between the two RNA-Seq datasets, and GO analysis of these overlapping genes again demonstrated a strong enrichment of terms involved in meiosis and germ cell development (Fig. 3, C and D). By contrast, no significant association to any pathways or biological processes was identified in the down-regulated gene group. Together, these data revealed a previously unappreciated function of Pcgf6 for suppressing genes associated with germ cells and meiosis in cell types other than ESCs. Interestingly, we examined the proximal promoter regions of 41 up-regulated germ cell–specific genes and found specific enrichment of the binding motifs for E2f6/Dp1 (GCGGGAA) (26) and Mga/Max (the E-box, CACGTG) (27) (Fig. 3, E and F). The presence of centrally enriched E-box and E2f6/Dp1 motifs in these germ cell promoters suggests that Pcgf6 represses germ cell gene expression in the context of a PRC1.6 complex.
Figure 3.
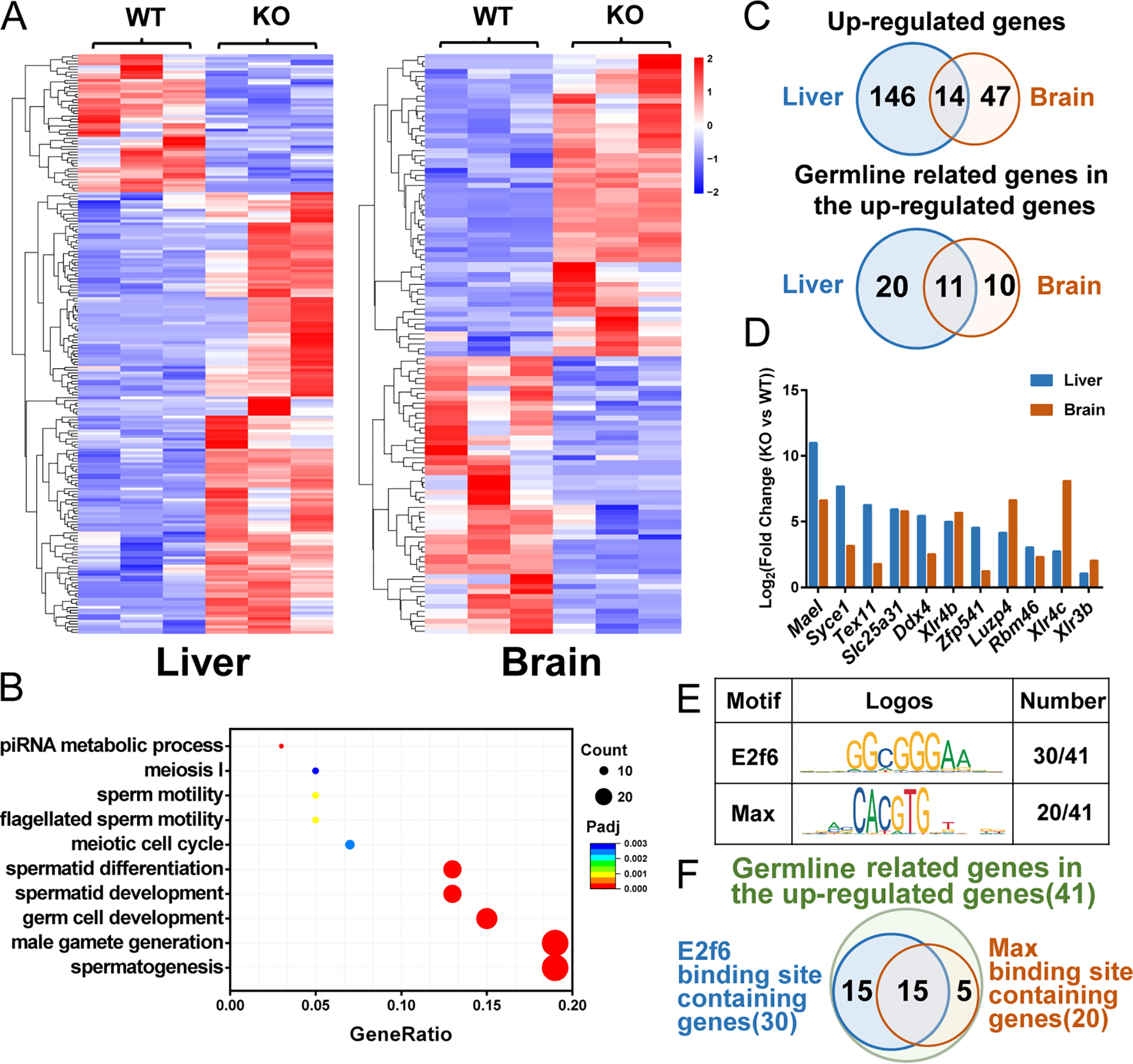
Pcgf6 represses the expression of germline-related genes with binding sites of Max and E2f6 in the liver and brain. A, RNA-Seq heat map of transcripts with 2-fold expression differences and p < 0.05 in liver and brain comparing WT mice with Pcgf6−/− mice. B, GO analysis of biological functions of 207 up-regulated genes in the liver and brain. C, Venn diagrams of up-regulated target genes and germline-related genes in the liver and brain of Pcgf6−/− mice. D, log2(-fold changes (KO versus WT)) in the expression of the 11 germline-related genes up-regulated in both liver and brain. E, motif analysis of 41 germline-related genes in up-regulated genes of liver and brain. F, the overlap between E2f6-binding sites containing genes and Max-binding sites containing genes.
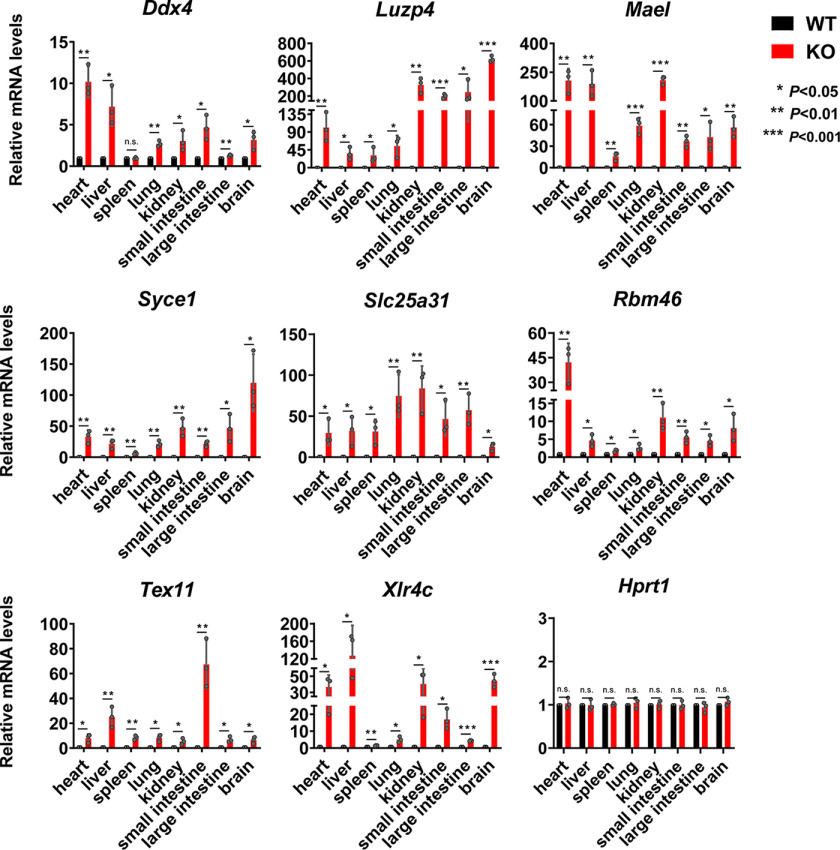
To build on this observation, we next chose to focus our efforts on the set of germline-specific genes showing aberrant expression in adult somatic tissues derived from Pcgf6 null mice. qRT-PCR analysis of Ddx4, Luzp4, Mael, Syce1, Slc25a31, Rbm46, Tex11, and Xlr4c mRNA confirmed that expression of these genes was undetectable in WT somatic tissues, whereas they were derepressed in liver and brain but also in all of the tissues we tested derived from Pcgf6 null adult mice (Fig. 4). From these results, we concluded that Pcgf6 is primarily involved in suppression of the germ cell lineage differentiation program in somatic cells and tissues.
Figure 4.
Pcgf6 represses the expression of germline-related genes almost in all somatic organs. Relative expression of the indicated germline-related genes in various tissues of Pcgf6−/− mice determined by qRT-PCR. Actin was used as a control for normalization. Hprt1 was used as a negative control. Data are from three biological replicates each with three technical replicates. Error bars, S.D. n.s., nonsignificant (two-tailed Student's t test).
Because Pcgf6 is highly expressed in ESCs and contributes to the repression of germline genes in somatic cells, we postulated that Pcgf6 may be required for somatic cell reprogramming. To explore this possibility, we reprogrammed mouse embryonic fibroblasts (MEFs) derived from the Pcgf6−/− mice using the classical Yamanaka's method (28). As shown in Fig. S4, Pcgf6 deficiency in MEFs dramatically reduced the formation of alkaline phosphatase–positive colonies in Oct4-, Sox2-, Klf4-, and c-Myc (OSKM)-driven reprogramming. Importantly, reprogramming efficiency of Pcgf6−/− MEFs could be rescued by transduction of Pcgf6 (Fig. S4). The requirement for Pcgf6 in somatic cell reprogramming was also reported in recent studies using an shRNA-mediated knockdown approach (11). Taken together, these studies reveal that Pcgf6 is specifically implicated in somatic cell reprogramming.
DNA methylation is largely dispensable for Pcgf6-mediated germline gene silencing
Promoter CpG island DNA methylation is generally associated with gene silencing (29). A cluster of X-linked germ line–specific genes has been reported to be derepressed in cells deficient in DNA methyltransferases (30). Therefore, we asked whether the up-regulation of germ cell–specific genes in Pcgf6 null cells would be accompanied by changes in promoter DNA methylation. The content in methylated cytosines in the proximal promoters of the four selected genes, which were significantly up-regulated in Pcgf6 null somatic tissues (Fig. 4), was determined by conventional Sanger sequencing bisulfite-assisted genomic DNA. As shown in Fig. S5, following Pcgf6 inactivation, the promoter-proximal CpG islands of Ddx4, Mael, and Slc25a31 underwent marginal DNA demethylation, whereas Syce1 showed no differences from WT liver. By contrast, Pcgf6 deficiency in testis had no detectable effect on DNA methylation in the proximal CpG islands of these four genes, consistent with the unaltered mRNA levels in testis after deletion of Pcgf6 (Fig. S5). Together, these results suggest that maintenance of DNA methylation patterns is not essential for Pcgf6-mediated repression of germ cell–specific gene expression.
Pcgf6 associates with G9a/glp and Hdac1/2 and directs them to its target promoters to repress germ cell–related genes
Generally, repressors recruit multiple cofactors involved in histone deacetylation and methylation or chromatin remodeling to mediate concerted transcriptional silencing via multiple repression pathways (29). Interestingly, Hdac1/2 and G9a/Glp have repeatedly been identified as additional subunits of PRC1.6 (6, 8,9, 12). The presence of Hdac1/2 and G9a/Glp in Pcgf6-PRC1 complex prompted us to assess whether these epigenetic regulators associate with Pcgf6 to cooperatively repress germ cell–related genes. We explored this possibility via biochemical and genetic analyses. First, we examined interactions between Pcgf6 and these four epigenetic regulators in mouse ESCs by co-immunoprecipitation assays. As shown in Fig. 5A, both the Hdac1/2 and the G9a/Glp as well as L3mbtl2 and Ring1B proteins were co-immunoprecipitated with anti-Pcgf6 antibody, whereas deletion of Pcgf6 resulted in undetectable G9a/Glp or Hdac1/2 in the precipitates. In contrast, the interaction of Pcgf6 with Ezh2 and Cbx8, a histone H3K27 methyltransferase and a canonical PRC1 subunit, respectively, was not detected, indicating that interaction between Pcgf6 and the individual G9a/Glp and Hdac1/2 is specific. To further explore whether Pcgf6 could interact directly with G9a/Glp and Hdac1/2, we purified recombinant GSH S-transferase (GST)-tagged Pcgf6, His-Ring1B, His-G9a/Glp, and His-Hdac1/2 and measured their direct interactions using a GST pulldown assay. We found that Pcgf6 selectively recognized Ring1B and did not exhibit detectable binding to G9a/Glp or Hdac1/2, likely suggesting that Pcgf6 interacts with G9a/Glp and Hdac1/2 through other PRC1.6 subunits (Fig. S6).
Figure 5.
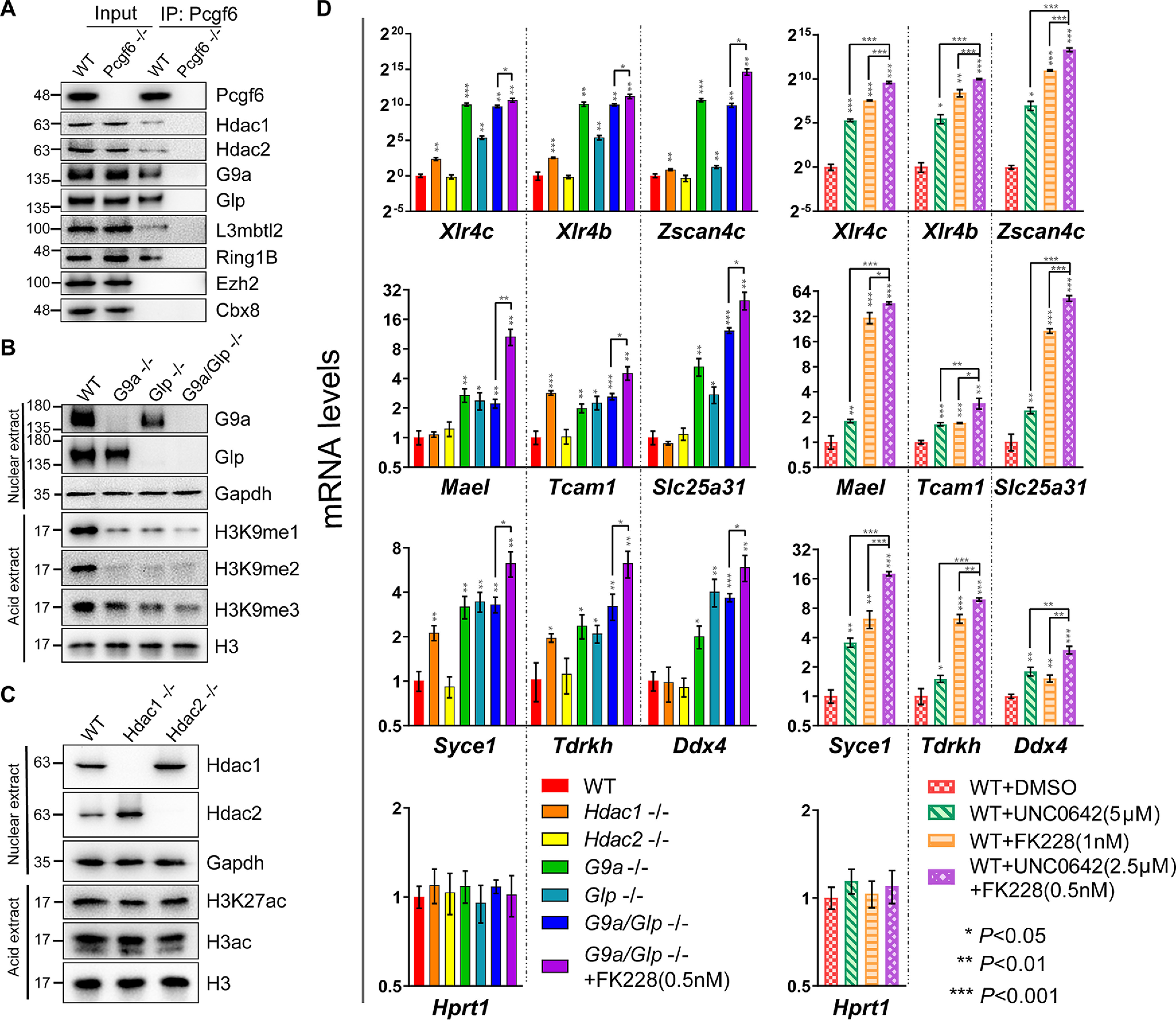
G9a/Glp and Hdac1/2 repress the expression of Pcgf6 targeted germline genes. A, IP–Western blot analysis showing physical interaction of Pcgf6, Hdac1/2, and G9a/Glp in cell extracts from WT and Pcgf6−/− ESCs. B, Western blot analysis showing the methylation levels of H3K9 in G9a−/−, Glp−/−, and G9a/Glp−/− ESCs. C, Western blot analysis showing the acetylation levels of H3 in Hdac1−/− and Hdac2−/− ESCs. D, the mRNA levels of germline-related genes in ESCs of the indicated genotype or in ESCs treated with the indicated inhibitors. All cells were treated with inhibitors for 72 h. DMSO is the solvent control. Error bars, S.D. of three independent experiments. The asterisks arranged vertically refer to significant differences compared with WT ESCs. Horizontally arranged asterisks indicate significant differences between groups.
Second, to test the requirement of these histone-modifying enzymes for repression of germ cell–related genes, we generated single or double knockout of Hdac1/2 and G9a/Glp ESCs using CRISPR/Cas9 technology (31–33) (Figs. S7 and S8). The successful knockout of these enzymes was confirmed by genomic PCR and Western blotting (Fig. 5 (B and C) and Figs. S7 and S8). Consistent with previous reports, G9a or Glp single deficient ESCs lacked nearly all of the H3K9me2 and showed a significant decrease in H3K9me1, whereas H3K9me3 levels were mildly affected (Fig. 5B). Interestingly, double knockout of G9a/Glp further reduced the methylation level of H3K9me3 but not H3K9me1 or H3K9me2, suggesting that G9a and Glp generally cannot compensate for the function of each other (34, 35). As reported previously (36), individual Hdac1/2 disruption in ESCs was not associated with changes in chromatin modifications altered by these enzymes (i.e. histone 3 acetylation (H3ac) and H3K27ac) (Fig. 5C). As expected, the expression of the germline genes, including Xlr4c, Xlr4b, Zscan4c, Mael, Tcam1, Slc25a31, Syce1, Tdrkh, and Ddx4, in ESCs with single deficiency of G9a/Glp was significantly up-regulated, as evidenced by qRT-PCR. Comparing expression changes of germ cell–related genes in single mutant cells with those in cells with combined inactivation of G9a/Glp, we observed no synergistic effects (Fig. 5D). In agreement with these results, the G9a/Glp inhibitor UNC0642-treated WT ESCs displayed a significant induction of germ cell–related gene expression. Interestingly, despite the fact that Hdac1, but not Hdac2, single knockout displayed a mild effect on derepression of germ cell–related genes, Hdac1/2-selective inhibitor FK228 significantly induced the expression of these genes, suggesting functional redundancy between these two Hdacs (Fig. 5D). In addition, Hdac1/2 inhibitor treatment of G9a/Glp double null ESCs further enhanced the expression of germline genes. Likewise, FK228 in combination with UNC0642 in ESCs showed strong synergistic effects (Fig. 5D). Together, these data suggest a possible synergistic cooperation between the two enzymatic activities.
The above results establish a role for G9a/Glp and Hdac1/2 in the repression of germ cell–related genes; however, the molecular mechanism by which they are targeted to a small subset of promoters is not understood. Given that Pcgf6 interacts with G9a/Glp and Hdac1/2, we wondered whether it is directly involved in recruitment of these proteins. We first examined the association between Pcgf6 and the promoters of known target genes (10) (Fig. 6A) via ChIP assays by anti-Pcgf6 antibodies (Fig. 6B). We found that the Pcgf6 was enriched at the promoters of selected germ cell–related genes—including Mael, Tcam1, Slc25a31, Syce1, Tdrkh, and Ddx4—whereas enrichment of Pcgf6 was not observed at the promoter of Hprt1, whose expression in ESCs was not changed by absence of Pcgf6 (Fig. 6B). Importantly, this was accompanied by considerable enrichment of Ring1B, G9a/Glp, and Hdac1/2, suggesting that Pcgf6 is involved in the recruitment of the PRC1.6 complex (Fig. 6, B–D). Accordingly, we observed association of H3K9me1 and H3K9me2 with the indicated target genes. Moreover, enrichment of Ring1B, G9a/Glp, Hdac1/2, H3K9me1, and H3K9me2 (but not H3K9me3) at the target promoters was indeed markedly depleted in the absence of Pcgf6 (Fig. 6 (B–D) and Fig. S9), whereas their global levels remained largely unperturbed following loss of Pcgf6 (Fig. 5A and Fig. S10). ChIP analysis of histone 3 and histone 4 acetylation (H3ac and H4ac) revealed that histone acetylation at Pcgf6-bound promoters increased dramatically after disruption of Pcgf6 (Fig. 6D), consistent with reduced local activity of Hdac1/2 (Fig. 6D). Notably, consistent with our previous report (10, 37), H2AK119ub1 enrichment at these targets did not show considerable changes in the Pcgf6 null ESCs despite substantially depleted Ring1B occupancy (Fig. 6B). This finding agrees with the observation that Pcgf1-PRC1 complex is essential for shaping normal H2AK119ub1 and Pcgf6-PRC1 complex is a weak ubiquitin ligase in ESCs in vivo (38). Together, these results suggest that Pcgf6 coordinates epigenetic repression of germ cell–related genes by recruitment of G9a/Glp and Hdac1/2 (Fig. 6E).
Figure 6.
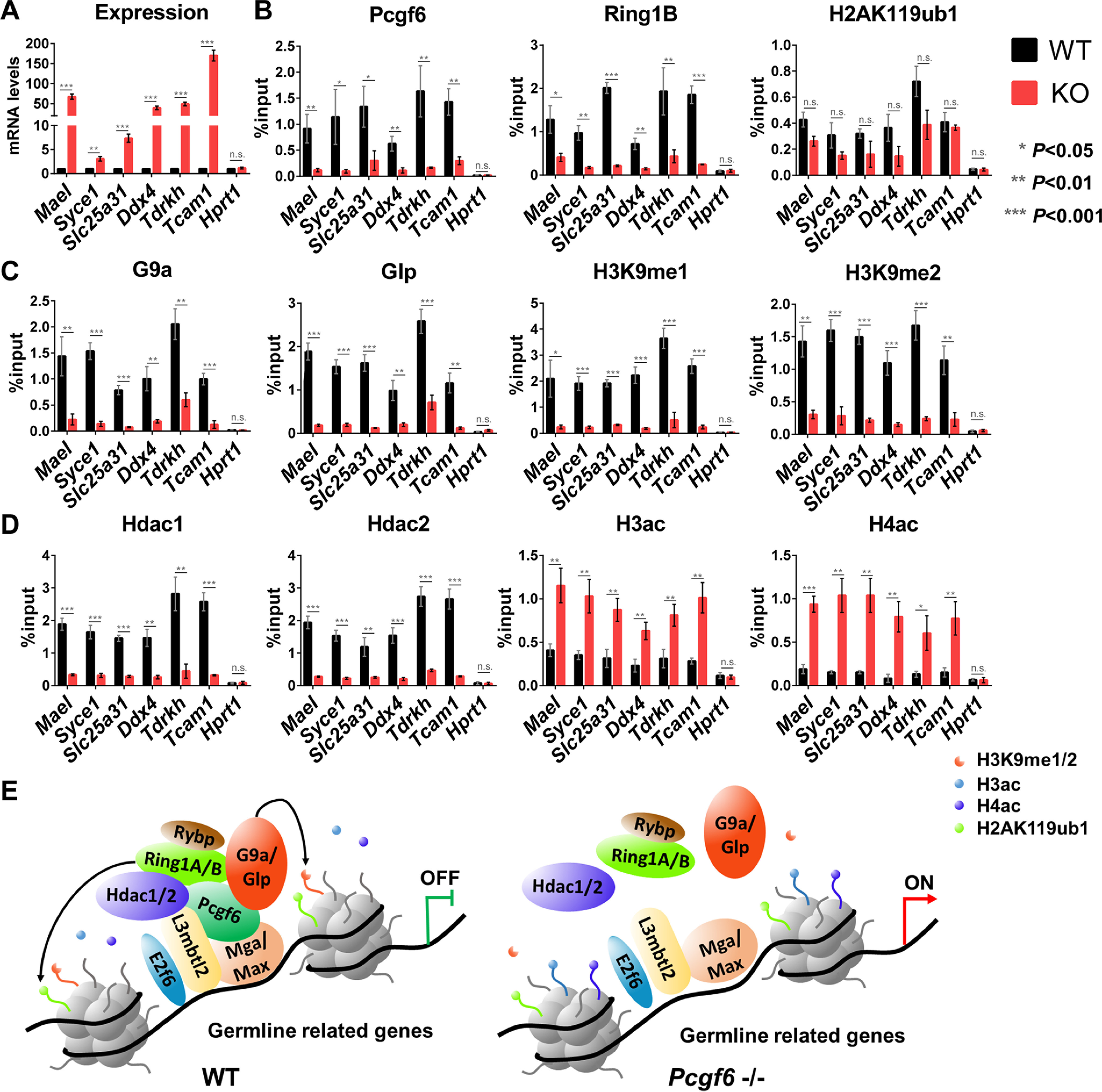
Pcgf6 plays a key role in recruiting G9a/Glp and Hdac1/2 to germline-related genes. A, mRNA expression analysis of the selected genes in ESCs of the indicated genotype. B–D, ChIP-qPCR analysis showing enrichment of the indicated proteins on the selected gene promoters in WT and Pcgf6−/− ESCs. Error bars, S.D. of three independent repeats. E, a schematic model showing that recruiting Hdac1/2 and G9a/Glp is an important mechanism used by Pcgf6 to modify histone tails and repress the expression of germline-related genes (see “Discussion” for a detailed description).
Discussion
We reported here that targeted disruption of polycomb protein Pcgf6 in mice resulted in partially penetrant embryonic lethality, and surviving mutants displayed markedly impaired fertility. Taking advantage of Pcgf6-deficient mice, we showed that Pcgf6 targets were enriched for meiosis– and germ cell–related genes in liver and brain and that such genes were dramatically derepressed in all Pcgf6 null tissues we examined. Mechanically, Pcgf6 recruited G9a/Glp and Hdac1/2 to modify the promoter region of these germline genes targeted for repression by Pcgf6. Our work thus shows a link between target promoters tethering by Pcgf6 with transcriptional silencing of germ cell–specific genes by G9a/Glp and Hdac1/2.
Histone H3K9 methylation is generally linked to gene repression, and G9a/Glp is reported to be targeted to selected gene promoters by sequence-specific DNA-binding factors (7). Additionally, G9a has been shown to associate with transcription factors CDP/cut, PRDI-BF1, Gfi1, and SHP and is recruited to their correspondingly targeted sites (39–42). Importantly, each of these G9a-interacting transcription factors also associates with Hdacs. Here, we have shown that Pcgf6 targets the action of both G9a/Glp and Hdac1/2 to the promoters of germline genes to dictate the transcriptional silencing of germline genes in ESCs, even though the precise molecular mechanisms by which it exerts this function still need further investigation. Of note, we recently reported that efficient recruitment of PRC1.6 depends on functional cooperation among unique subunits (L3mbtl2, Max, Mga, and Pcgf6) in the complex (37). Consistently, knockout of the genes encoding these unique subunits of PRC1.6 is accompanied with induction of germline gene expression (8, 10, 12, 17, 18). Intriguingly, deletion of Max in ESCs results in marked cytological changes reminiscent of meiotic cell division (17, 43). Therefore, Pcgf6 ablation–mediated selective derepression of germline genes reflects the impairment of PRC1.6 in which Pcgf6 is one of the important components. In further support of this scenario, our analysis of the proximal promoters of derepressed germline genes following genetic ablation of the Pcgf6 gene allowed the identification of putative signature motifs for Pcgf6 target genes, with the general enrichment of the E2f6/Dp1- and Mga/Max-binding sites (26, 27). It is worth mentioning that Ring1B, which is a major common subunit of PRC1, has been reported to be required for repression of Stra8 and other germline genes driving meiosis (44). This strongly indicates the existence of additional machineries for germline gene repression. Of particular interest, at selected Pcgf6 target promoters, Ring1B occupancy coincides with low or undetectable H2AK119ub1 levels, and the H2AK119ub1 signals were not compromised by loss of Pcgf6, suggesting that Pcgf6 complex could play a role at these promoters that is largely independent from its ability to modify histone H2A. It has been reported, however, that in ESCs from the conditional targeting, H2AK119ub1 occupancy is reduced in the absence of Pcgf6, implying the contribution of H2AK119ub1 in Pcgf6-mediated gene repression (12). This discrepancy might arise from differences in gene targeting approaches (conditional versus constitutive) used. Notably, our observation is in line with a recent report demonstrating a pivotal role of Pcgf1-PRC1 in shaping normal H2AK119ub1 in ESCs (38). Further studies are needed to explore the potential link between H2AK119ub1 and Pcgf6-mediated gene silencing. On the basis of these findings, we propose that Pcgf6 forms a complex with Hdac1/2 and G9a/Glp in undifferentiated ESCs as well as specified cell types and represses the expression of germline genes via repressive histone modification, namely histone deacetylation and H3K9 methylation (Fig. 6E).
PcG proteins are traditionally described as transcriptional repressors (1, 3). However, they can also bind to active promoters, where they mediate positive regulation of gene expression (45–47). A substantial number of expression changes with germ genes being down-regulated in Pcgf6 null testes may imply an additional role for Pcgf6 in germline-specific gene regulation. Pcgf6 may in fact play dual roles as both an activator and a repressor. Its functions may be determined by cell/tissue type and by differences in the complexes with which Pcgf6 is associated. For instance, in somatic tissues and ESCs, Pcgf6 represses transcription by acting as a platform upon which multiple factors that mediate gene silencing are assembled. Most notably, these include transcriptional repressors Hdac1/2 and G9a/Glp, thereby contributing to silencing of active chromatin. In meiotic cells, Pcgf6 may instead be recruiting an activation complex to meiosis-specific genes. We hypothesize that deregulated expression of these germline genes upon Pcgf6 ablation contributes to the observed phenotypes in spermatogenesis. Indeed, Pcgf6 and L3mbtl2 knockout mice exhibited a severely impaired spermatogenesis (Fig. 2) (48), supporting this notion. The function of Pcgf6 for key steps of spermatogenesis needs to be further elucidated. Given that the functionality of the PRC depends on its diverse biochemical composition, Pcgf6-containing PRC1 may consist of two or more subsets of complexes, thus allowing it to act as both an activator and a repressor. Further proteomic investigation of Pcgf6 protein interactome is required to gain deeper insights into the diverse functions of Pcgf6 in ESCs and multiple types of specialized cells.
Experimental procedures
Generation of Pcgf6−/− mice
Pcgf6-targeting vectors containing loxP sites flanking exons 2–5 and a Neo flanked with FRT sites were linearized and then introduced into C57BL/6 ESCs and selected by G418. The Neo cassette was deleted by transfecting a vector expressing FLPe recombinase. Chimeras were generated by injecting correctly targeted ESCs into C57BL/6J blastocysts. The consequent male chimeric mice were bred with female C57BL/6 mice, and germ line transmission of the Pcgf6-targeted (Pcgf6FL/+) allele was examined in the offspring. Conditional single allele mice were mated with cytomegalovirus-Cre transgenic mice to generate heterozygous mice for the Pcgf6 deficiency. The homozygous knockout mice (KO, Pcgf6−/−) were produced by intercrossing of Pcgf6+/– male and female mice. Genotyping primers are given in Table S1. The experimental animal facility has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, and the institutional animal care and use committee of the Model Animal Research Center of Nanjing University approved all animal protocols used in this study.
Generation of mutant cell lines and cell culture
Construction and transfection of Cas9 vector with a single guide RNA (sgRNA) were performed as described previously (10). Briefly, sgRNAs of the genes of interest were designed by the CRISPR design tool (RRID:SCR_015935) (33) and cloned into PX459 vector. pSpCas9(BB)-2A-Puro (PX459) V2.0 was a gift from Feng Zhang (Addgene plasmid 62988; RRID:Addgene_62988). After a 24-h transfection with the plasmids expressing a pair of sgRNAs and a puromycin resistance plasmid by Lipofectamine 2000 (Thermo Fisher Scientific), mouse ESCs were selected by puromycin for 2 days and then seeded on mitomycin-irradiated feeder cells in ESC medium. The null ESC clones were identified by genomic PCR analysis, qRT-PCR, and Western blotting. As previously described, ESCs were cultured in Dulbecco's modified Eagle's medium high glucose (Gibco) with 15% fetal calf serum (Gibco), l-glutamine (Gibco), nonessential amino acids (Gibco), β-mercaptoethanol (Sigma), 1000 units/ml leukemia inhibitory factor, and penicillin/streptomycin (Gibco).
Histology and immunohistochemistry
Tissues were harvested and fixed with 4% paraformaldehyde overnight at 4 °C and then dehydrated and embedded in paraffin. 5-μm-thick paraffin sections were stained with H&E or used for immunohistochemistry. Images were obtained by BX53 semi-electric fluorescence microscope (Olympus).
Fertility and fecundity assessment
Fertility and fecundity assessment was performed as described previously (49, 50). Briefly, Pcgf6+/– and Pcgf6−/− littermate males (>8 weeks old) were placed with WT fertile females of 2–6 months of age, respectively. Pcgf6+/– and Pcgf6−/− littermate females were caged with mature WT males for a similar period. The presence of a copulatory plug among females was checked over the first 4 days. Females with copulatory plug were either left with the male or removed to another cage and then checked again at 3 weeks for pregnancy. The number of offspring from each pregnancy was recorded.
Sperm collection, motility and morphology test
To assess the effect of Pcgf6 deficiency on sperms, the left and right caudal epididymitis of sexually mature mice were dissected out, minced in prewarmed TYH medium at 37 °C, and incubated for 120 min. The sperm suspension was filtered through a 70-µm nylon cell strainer to remove tissue debris. Sperm motility was then measured using a CASA CEROS version 12 sperm analysis system (Hamilton Thorne). The analysis setting described previously was used (51). To evaluate sperm abnormalities in Pcgf6-deficient mice, we made a thin smear of sperm air-dried on a premarked cleaned glass slide. After staining with a Diff-Quik stain kit (Solarbio, G2572) according to the instructions, 200 spermatozoa were examined per mouse using bright field illumination at ×400 magnification for normal and abnormal. All slides were coded for avoiding scorer biasness. Sperm normality was expressed as a percentage.
Transcriptome sequencing and analysis
For RNA-Seq, total RNA was extracted from the indicated tissues of WT and Pcgf6–/– mice, respectively, by the TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions and sequenced by Novogene (Beijing, China). Briefly, sequencing libraries were constructed using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (New England Biolabs) and assessed on the Agilent Bioanalyzer 2100 system. The library preparations were sequenced on an Illumina Hiseq platform. For data analysis, genes with a p value of <0.05 and 2-fold expression differences found by DESeq2 were considered as differentially expressed. GO enrichment analysis of differentially expressed genes was accomplished by the cluster Profiler R package. GO terms with corrected p value <0.05 were considered significantly enriched by differential expressed genes. We performed motif-scanning analysis using the online tool FIMO (RRID:SCR_001783). RNA-Seq data were available from the Gene Expression Omnibus under accession number GSE143206.
Quantitative RT-PCR
The indicated tissues of mice and ESCs were harvested and lysed in TRIzol reagent (Thermo Fisher Scientific). Total RNA was extracted as described previously (10). Gene expression was quantified in triplicates on a Roche Light Cycler using PowerUpTM SYBR Green Master Mix (Thermo Fisher Scientific) with the primers shown in Table S1. Relative changes in transcript levels were analyzed using the 2−ΔΔCt method. Values were normalized to β-actin mRNA content.
Conventional bisulfite sequencing
Genomic DNA was extracted from the liver and testis of WT and Pcgf6–/– mice using a Qiagen DNeasy blood and tissue kit and bisulfite-converted using the Qiagen Epitect bisulfite kit according to the provided instructions. The forward and reverse primers for the forward strand were designed by the online primer design tool Methprimer (RRID:SCR_010269), shown in Table S1 (52). The targeted regions were PCR-amplified from bisulfite-converted DNAs using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, P505). The purified PCR products were subsequently cloned into pBluescript II KS(−) vectors using ClonExpress II (Vazyme). To reveal the methylation status of individual CpG sites in the promoter of the selected genes, we sequenced 20 single clones using conventional Sanger sequencing.
Whole-cell lysates, histone extraction, and Western blotting
Whole-cell lysates and histone extraction were performed as described previously (10). Briefly, ESCs were washed with ice-cold PBS containing 5 mm sodium butyrate twice, lysed with a Triton extraction buffer (PBS, 0.5% Triton, 2 mm phenylmethylsulfonyl fluoride, 0.02% NaN3), and centrifuged. The pellet was resuspended in 0.2 n HCl after removing the supernatant. The histone was extracted overnight at 4 °C, and the supernatant was saved after centrifuging for 10 min Proteins were separated via SDS-PAGE and analyzed by immunoblotting. The antibodies used in this study are shown in Table S2.
GST pulldown assay
Expression and purification of GST or His tag fusion proteins were done as described previously (37). The GST pulldown assay was performed using the PierceTM GST Protein Interaction Pull-Down Kit (Thermo Fisher Scientific, catalog no. 21516) according to the manufacturer's protocol. Aliquots of 50 µl of beads immobilized with GST or GST-Pcgf6 fusion proteins were incubated in 1 ml of binding buffer for 3 h at 4 °C. Then the GST-bound beads were washed three times with binding buffer. The His tag fusion protein was added to the beads, and after 3 h of gentle rocking motion on a rotating platform at 4 °C, the tube was centrifuged and washed three times with the wash buffer. The bait- and prey-bound proteins were separated by SDS-PAGE and detected by Western blotting with the indicated antibodies.
MEF isolation and iPS reprogramming
To analyze the effects on Pcgf6 deficiency for iPS reprogramming, MEF isolation and iPS reprogramming were performed as described previously (28). Briefly, MEFs were isolated from embryonic day 13.5 WT and Pcgf6–/– embryos, respectively, and cultured in MEF medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, 100 units/ml penicillin/streptomycin, and 1 mm sodium pyruvate). MEFs were reprogrammed to iPS cells by lentiviral transduction. Lentivirus encoding Oct4, Sox2, Klf4, or c-Myc were generated by transfecting 293T cells as described previously (8). Viral supernatants from different transfections were equally mixed. WT and Pcgf6–/– MEFs were incubated with virus mixtures in the presence of Polybrene (Sigma; 8 μg/ml) for 24 h, and then supernatant was exchanged for MEF medium. 24 h later, infected cells were seeded onto mitomycin-irradiated WT MEFs and cultured in ESC medium. 3 weeks after transduction, the cells were stained using the Alkaline Phosphatase Stain Kit (Yeasen, 40749ES60) according to the manufacturer's instructions, and iPS reprogramming efficiencies were determined as frequencies of alkaline phosphatase colonies relative to input cell numbers.
Isolation of Sertoli, Leydig, and spermatogenic cells
Various testicular cell types were isolated from the whole-mouse testis using rigorous double enzymatic digestion with fraction enrichment on a Percoll (Sigma) gradient or a discontinuous BSA density gradient as described previously (53, 54). In brief, testes were collected from 8-week-old mice. Leydig cells were separated from collagenase-treated decapsulated testes by gravity sedimentation through a Percoll cushion. The dispersed seminiferous tubules were then digested with trypsin and DNase I to single-cell suspensions. Next, Sertoli cells were separated from germ cells by filtration through a 40-µm cell strainer and by adhesion to lectin-coated culture plates. Different populations of germ cells were separated by using a manually prepared 0.5–5% discontinuous BSA density gradient for velocity sedimentation sediment. After sedimentation, enriched fractions of the three germ cell types (pachytene spermatocyte, round spermatid, and elongating spermatid) were manually collected. RNA of purified somatic cell and germ cell fractions was reverse-transcribed and then analyzed by a Roche Light Cycler using primers set specific to germ cell, Sertoli cell, and Leydig cell marker genes (Table S1).
Immunoprecipitation
IP analysis was performed as described previously (8, 10). Briefly, cells were lysed and centrifuged. The supernatant was incubated with appropriate primary antibodies and protein A/G–Sepharose overnight at 4 °C. After they were washed three times with buffer containing 500 mm NaCl, immunoprecipitates were boiled with 5× loading buffer for 5 min and then subjected to Western blotting using the appropriate primary and second antibodies. All of the antibodies used in this study are listed in Table S2.
ChIP
ChIP analysis was performed as described previously (8, 10). Briefly, ESCs were cross-linked in 1% fresh formaldehyde solution followed by quenching with 0.125 m glycine. To produce chromatin fragments of 200–1000 bp, nuclear extracts were prepared in lysis buffer and sonicated using the Bioruptor (Diagenode). For immunoprecipitation, chromatin solution was mixed with appropriate antibodies and protein A/G–Sepharose beads in ChIP dilution buffer, rotated overnight at 4 °C. After they were washed twice by low-salt buffer and high-salt buffer, respectively, immunocomplexes were eluted from beads. The cross-linking of immunocomplexes was reversed, and the DNAs were purified using a DNAgel extraction kit (Axygen). Quantitative PCRs were performed using PowerUpTM SYBR Green Master Mix (Thermo Fisher Scientific) on a Roche Light Cycler. The percentage of input was calculated as 2−ΔCt, where ΔCt = Ct(CHIP) − Ct(input). ChIP primers are listed in Table S1.
Data availability
The RNA-Seq data are located in the Gene Expression Omnibus under accession number GSE143206.
Supplementary Material
Acknowledgments
We thank Ting Su for crucial assistance with RNA-Seq analysis. We are indebted to Drs Yun Shi and Zhaoyu Lin for helpful suggestions and for kindly providing reagents. We thank members of our laboratory for helpful discussion.
This article contains supporting information.
Author contributions—M. L. data curation; M. L. software; M. L., Y. Z., F. X., S. L., and J. Q. formal analysis; M. L. and J. Q. validation; M. L., Y. Z., F. X., S. L., and J. Q. methodology; M. L. and J. Q. writing-original draft; Y. X., Q. J., and J. Q. supervision; Y. X., Q. J., and J. Q. investigation; Y. X. and J. Q. project administration; J. Q. conceptualization; J. Q. resources; J. Q. funding acquisition; J. Q. writing-review and editing.
Funding and additional information—This work was supported by National Natural Science Foundation of China Grants 31671532 and 31970810 (to J. Q.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PcG
- polycomb group
- ESC
- embryonic stem cell
- PRC1
- polycomb repressive complex 1
- PRC2
- polycomb repressive complex 2
- IP
- immunoprecipitation
- sgRNA
- single guide RNA
- H2AK119ub1
- monoubiquitination of lysine 119 in histone H2A
- H3K27me3
- trimethylation of lysine 27 on histone H3
- H3K9me1/2
- mono- and dimethylation of histone H3 lysine 9
- qRT-PCR
- quantitative RT-PCR
- H&E
- hematoxylin and eosin
- GO
- gene ontology
- MEF
- mouse embryo fibroblast
- GST
- GSH S-transferase
- KO
- knockout
- GLP
- G9a-like protein
- HDAC
- histone deacetylase.
References
- 1. Simon J. A., and Kingston R. E. (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- 2. Di Croce L., and Helin K. (2013) Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- 3. Brand M., Nakka K., Zhu J., and Dilworth F. J. (2019) Polycomb/trithorax antagonism: cellular memory in stem cell fate and function. Cell Stem Cell 24, 518–533 10.1016/j.stem.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Margueron R., and Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turner S. A., and Bracken A. P. (2013) A “complex” issue: deciphering the role of variant PRC1 in ESCs. Cell Stem Cell 12, 145–146 10.1016/j.stem.2013.01.014 [DOI] [PubMed] [Google Scholar]
- 6. Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., and Reinberg D. (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., and Nakatani Y. (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136 10.1126/science.1069861 [DOI] [PubMed] [Google Scholar]
- 8. Qin J., Whyte W. A., Anderssen E., Apostolou E., Chen H. H., Akbarian S., Bronson R. T., Hochedlinger K., Ramaswamy S., Young R. A., and Hock H. (2012) The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11, 319–332 10.1016/j.stem.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trojer P., Cao A. R., Gao Z., Li Y., Zhang J., Xu X., Li G., Losson R., Erdjument-Bromage H., Tempst P., Farnham P. J., and Reinberg D. (2011) L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42, 438–450 10.1016/j.molcel.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao W., Tong H., Huang Y., Yan Y., Teng H., Xia Y., Jiang Q., and Qin J. (2017) Essential role for polycomb group protein Pcgf6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity. J. Biol. Chem. 292, 2773–2784 10.1074/jbc.M116.763961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zdzieblo D., Li X., Lin Q., Zenke M., Illich D. J., Becker M., and Müller A. M. (2014) Pcgf6, a polycomb group protein, regulates mesodermal lineage differentiation in murine ESCs and functions in iPS reprogramming. Stem Cells 32, 3112–3125 10.1002/stem.1826 [DOI] [PubMed] [Google Scholar]
- 12. Endoh M., Endo T. A., Shinga J., Hayashi K., Farcas A., Ma K. W., Ito S., Sharif J., Endoh T., Onaga N., Nakayama M., Ishikura T., Masui O., Kessler B. M., Suda T., et al. (2017) PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife 6 10.7554/eLife.27970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu G., Kim J., Xu Q., Leng Y., Orkin S. H., and Elledge S. J. (2009) A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23, 837–848 10.1101/gad.1769609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matzuk M. M., and Lamb D. J. (2002) Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 4, s41–s49 10.1038/ncb-nm-fertilityS41 [DOI] [PubMed] [Google Scholar]
- 15. Simpson A. J., Caballero O. L., Jungbluth A., Chen Y. T., and Old L. J. (2005) Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 5, 615–625 10.1038/nrc1669 [DOI] [PubMed] [Google Scholar]
- 16. Akasaka T., Takahashi N., Suzuki M., Koseki H., Bodmer R., and Koga H. (2002) MBLR, a new RING finger protein resembling mammalian Polycomb gene products, is regulated by cell cycle-dependent phosphorylation. Genes Cells 7, 835–850 10.1046/j.1365-2443.2002.00565.x [DOI] [PubMed] [Google Scholar]
- 17. Maeda I., Okamura D., Tokitake Y., Ikeda M., Kawaguchi H., Mise N., Abe K., Noce T., Okuda A., and Matsui Y. (2013) Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 4, 1754 10.1038/ncomms2780 [DOI] [PubMed] [Google Scholar]
- 18. Pohlers M., Truss M., Frede U., Scholz A., Strehle M., Kuban R. J., Hoffmann B., Morkel M., Birchmeier C., and Hagemeier C. (2005) A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr. Biol. 15, 1051–1057 10.1016/j.cub.2005.04.060 [DOI] [PubMed] [Google Scholar]
- 19. Choi H., Han C., Jin S., Kwon J. T., Kim J., Jeong J., Kim J., Ham S., Jeon S., Yoo Y. J., and Cho C. (2015) Reduced fertility and altered epididymal and sperm integrity in mice lacking ADAM7. Biol. Reprod. 93, 70 10.1095/biolreprod.115.130252 [DOI] [PubMed] [Google Scholar]
- 20. Da Ros V. G., Maldera J. A., Willis W. D., Cohen D. J., Goulding E. H., Gelman D. M., Rubinstein M., Eddy E. M., and Cuasnicu P. S. (2008) Impaired sperm fertilizing ability in mice lacking cysteine-rich secretory protein 1 (CRISP1). Dev. Biol. 320, 12–18 10.1016/j.ydbio.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Westmuckett A. D., Nguyen E. B., Herlea-Pana O. M., Alvau A., Salicioni A. M., and Moore K. L. (2014) Impaired sperm maturation in RNASE9 knockout mice. Biol. Reprod. 90, 120 10.1095/biolreprod.113.116863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin M. H., Lee R. K., Hwu Y. M., Lu C. H., Chu S. L., Chen Y. J., Chang W. C., and Li S. H. (2008) SPINKL, a Kazal-type serine protease inhibitor-like protein purified from mouse seminal vesicle fluid, is able to inhibit sperm capacitation. Reproduction 136, 559–571 10.1530/REP-07-0375 [DOI] [PubMed] [Google Scholar]
- 23. Jalkanen J., Kotimäki M., Huhtaniemi I., and Poutanen M. (2006) Novel epididymal protease inhibitors with Kazal or WAP family domain. Biochem. Biophys. Res. Commun. 349, 245–254 10.1016/j.bbrc.2006.08.023 [DOI] [PubMed] [Google Scholar]
- 24. Jeong J., Lee B., Kim J., Kim J., Hong S. H., Kim D., Choi S., Cho B. N., and Cho C. (2019) Expressional and functional analyses of epididymal SPINKs in mice. Gene Expr. Patterns 31, 18–25 10.1016/j.gep.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 25. Li S. H., Lee R. K., Hsiao Y. L., and Chen Y. H. (2005) Demonstration of a glycoprotein derived from the Ceacam10 gene in mouse seminal vesicle secretions. Biol. Reprod. 73, 546–553 10.1095/biolreprod.105.039651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaubatz S., Wood J. G., and Livingston D. M. (1998) Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. U.S.A. 95, 9190–9195 10.1073/pnas.95.16.9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hurlin P. J., Steingrìmsson E., Copeland N. G., Jenkins N. A., and Eisenman R. N. (1999) Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 18, 7019–7028 10.1093/emboj/18.24.7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 29. Fuks F. (2005) DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15, 490–495 10.1016/j.gde.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 30. Fouse S. D., Shen Y., Pellegrini M., Cole S., Meissner A., Van Neste L., Jaenisch R., and Fan G. (2008) Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2, 160–169 10.1016/j.stem.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., and Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., and Shinkai Y. (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 10.1101/gad.1284005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y., and Allis C. D. (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 10.1016/S1097-2765(03)00479-9 [DOI] [PubMed] [Google Scholar]
- 36. Dovey O. M., Foster C. T., and Cowley S. M. (2010) Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 8242–8247 10.1073/pnas.1000478107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang Y., Zhao W., Wang C., Zhu Y., Liu M., Tong H., Xia Y., Jiang Q., and Qin J. (2018) Combinatorial control of recruitment of a variant PRC1.6 complex in embryonic stem cells. Cell Rep. 22, 3032–3043 10.1016/j.celrep.2018.02.072 [DOI] [PubMed] [Google Scholar]
- 38. Fursova N. A., Blackledge N. P., Nakayama M., Ito S., Koseki Y., Farcas A. M., King H. W., Koseki H., and Klose R. J. (2019) Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol. Cell 74, 1020–1036.e8 10.1016/j.molcel.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boulias K., and Talianidis I. (2004) Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 32, 6096–6103 10.1093/nar/gkh947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gyory I., Wu J., Fejér G., Seto E., and Wright K. L. (2004) PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5, 299–308 10.1038/ni1046 [DOI] [PubMed] [Google Scholar]
- 41. Nishio H., and Walsh M. J. (2004) CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. U.S.A. 101, 11257–11262 10.1073/pnas.0401343101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duan Z., Zarebski A., Montoya-Durango D., Grimes H. L., and Horwitz M. (2005) Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25, 10338–10351 10.1128/MCB.25.23.10338-10351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki A., Hirasaki M., Hishida T., Wu J., Okamura D., Ueda A., Nishimoto M., Nakachi Y., Mizuno Y., Okazaki Y., Matsui Y., Izpisua Belmonte J. C., and Okuda A. (2016) Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat. Commun. 7, 11056 10.1038/ncomms11056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yokobayashi S., Liang C. Y., Kohler H., Nestorov P., Liu Z., Vidal M., van Lohuizen M., Roloff T. C., and Peters A. H. (2013) PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 495, 236–240 10.1038/nature11918 [DOI] [PubMed] [Google Scholar]
- 45. Cohen I., Zhao D., Bar C., Valdes V. J., Dauber-Decker K. L., Nguyen M. B., Nakayama M., Rendl M., Bickmore W. A., Koseki H., Zheng D., and Ezhkova E. (2018) PRC1 fine-tunes gene repression and activation to safeguard skin development and stem cell specification. Cell Stem Cell 22, 726–739.e7 10.1016/j.stem.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao W., Huang Y., Zhang J., Liu M., Ji H., Wang C., Cao N., Li C., Xia Y., Jiang Q., and Qin J. (2017) Polycomb group RING finger proteins 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 292, 21527–21537 10.1074/jbc.M117.804054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van den Boom V., Maat H., Geugien M., Rodríguez López A., Sotoca A. M., Jaques J., Brouwers-Vos A. Z., Fusetti F., Groen R. W., Yuan H., Martens A. C., Stunnenberg H. G., Vellenga E., Martens J. H., and Schuringa J. J. (2016) Non-canonical PRC1.1 targets active genes independent of H3K27me3 and is essential for leukemogenesis. Cell Rep. 14, 332–346 10.1016/j.celrep.2015.12.034 [DOI] [PubMed] [Google Scholar]
- 48. Meng C., Liao J., Zhao D., Huang H., Qin J., Lee T. L., Chen D., Chan W. Y., and Xia Y. (2019) L3MBTL2 regulates chromatin remodeling during spermatogenesis. Cell Death Differ. 26, 2194–2207 10.1038/s41418-019-0283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harris T., Marquez B., Suarez S., and Schimenti J. (2007) Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the sun domain-containing family of putative RNA methyltransferases1. Biol. Reprod. 77, 376–382 10.1095/biolreprod.106.058669 [DOI] [PubMed] [Google Scholar]
- 50. Sapiro R., Kostetskii I., Olds-Clarke P., Gerton G. L., Radice G. L., and Strauss Iii J. F. (2002) Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell Biol. 22, 6298–6305 10.1128/mcb.22.17.6298-6305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goodson S. G., Zhang Z., Tsuruta J. K., Wang W., and O'Brien D. A. (2011) Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol. Reprod. 84, 1207–1215 10.1095/biolreprod.110.088989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li L. C., and Dahiya R. (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 10.1093/bioinformatics/18.11.1427 [DOI] [PubMed] [Google Scholar]
- 53. Chang Y. F., Lee-Chang J. S., Panneerdoss S., MacLean J. A. 2nd, and Rao M. K. (2011) Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. BioTechniques 51, 341–344 10.2144/000113764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Da Ros M., Lehtiniemi T., Olotu O., Meikar O., and Kotaja N. (2019) Enrichment of pachytene spermatocytes and spermatids from mouse testes using standard laboratory equipment. J. Vis. Exp. 10.3791/60271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data are located in the Gene Expression Omnibus under accession number GSE143206.