Abstract
Background:
Globally, toxic metal exposures are a well-recognized risk factor for many adverse health outcomes. DNA methylation-based measures of biological aging are predictive of disease, but have poorly understood relationships with metal exposures.
Objective:
We performed a pilot study examining the relationships of 24-hour urine metal concentrations with three novel DNA methylation-based measures of biological aging: DNAmAge, GrimAge, and PhenoAge.
Methods:
We utilized a previously established urine panel of five common metals [arsenic (As), cadmium (Cd), lead (Pb), manganese (Mn), and mercury (Hg)] found in a subset of the elderly US Veterans Affairs Normative Aging Study cohort (N = 48). The measures of DNA methylation-based biological age were calculated using CpG sites on the Illumina HumanMethylation450 BeadChip. Bayesian Kernel Machine Regression (BKMR) was used to determine metals most important to the aging outcomes and the relationship of the cumulative metal mixture with the outcomes. Individual relationships of important metals with the biological aging outcomes were modeled using fully-adjusted linear models controlling for chronological age, renal function, and lifestyle/environmental factors.
Results:
Mn was selected as important to PhenoAge. A 1 ng/mL increase in urine Mn was associated with a 9.93-year increase in PhenoAge (95%CI: 1.24, 18.61, p=0.03). The cumulative urine metal mixture was associated with increases in PhenoAge. Compared to a model where each metal in the mixture is set to its 50th percentile value, every one-unit increase of the cumulative mixture with each metal at its 70th percentile was associated with a 2.53-year increase in PhenoAge (95%CI: 0.10, 4.96, P<0.05).
Conclusion:
Our results add novel evidence that metals detected in urine are associated with increases in biological aging and suggest that these DNA methylation-based measures may be useful for identifying individuals at-risk for diseases related to toxic metal exposures. Further research is necessary to confirm these findings more broadly.
Keywords: Urine Metals, DNA Methylation Age, Aging, Epigenetics
1 |. Introduction
Across developed and developing communities worldwide, toxic metal exposures have become a well-recognized risk factor for many adverse health outcomes. It is estimated that in 2017, worldwide lead exposures were responsible for 1.06 million deaths and 24.4 million years of healthy life lost – particularly due to developmental intellectual disability1. Often, low- and middle-income countries bear a disproportionate burden of such exposures2, but toxic metal exposures are also prevalent in developed nations. An estimated 2.1 million people residing in the United States may be obtaining drinking water from wells with high concentrations of arsenic – a known neurotoxin and multi-system carcinogen3,4. Given the pervasiveness of toxic metal exposures and the gravity of their associated health consequences, methods of assessing the impact of these exposures on human health before the onset of clinical disease remain an area of active research5–7. Such tools would be particularly useful in cases of chronic insidious metal exposures that may wreak havoc on the human body for decades before coming to clinical attention. DNA methylation-based predictors of biological aging are a unique set of novel tools that offer some promise in addressing this important public health gap.
Measures of DNA methylation age are novel metrics of biological aging that can be calculated using DNA methylation values from specific combinations of age-correlated CpG dinucleotides. Presently, there are three leading DNA methylation age measures that are notable for demonstrating tissue independence and/or very robust disease relationships. The oldest of the three is DNAmAge and it is calculated using 353 CpGs8. DNAmAge maintains a very high predictive accuracy and has been associated with a host of disease processes9,10. The second of the three metrics, DNA Phenotypic Age (PhenoAge), was built using nine clinical variables (i.e. albumin, creatinine, glucose, C-reactive protein, lymphocyte percent, mean cell volume, red cell distribution width, alkaline phosphatase, and white blood cell count) and can be calculated using 513 CpGs (41 of which are shared with DNAmAge)11. PhenoAge is an even better predictor of lifespan and healthspan12–14. The final metric, DNA GrimAge, outperforms both DNAmAge and PhenoAge with respect to lifespan and healthspan15–17. In particular, GrimAge is unparalleled in its ability to predict all-cause time-to-death (Cox regression p=2.0 × 10−75)18. GrimAge was calculated from 1030 unique CpGs as well as DNA methylation surrogates of cigarette pack-years and 7 plasma protein markers [adrenomedullin (ADM), beta-2-microglobulim (B2M), cystatin C, GDF-15, leptin, plasminogen activator inhibitor-1 (PAI-1), and tissue inhibitor metalloproteinases 1 (TIMP-1)]18.
A growing body of literature has examined the relationships between DNA methylation age measures and toxic exposures19–23. Unfortunately, very little of this work has focused on metals. Of the two existing studies, the first examined the relationship of urinary cadmium with DNAmAge in 40 non-smoking women in Thailand24. The second examined the relationship of chronic serum cobalt/chromium levels from metal on metal hip replacements with DNAmAge25. Both found no associations between the metals and DNAmAge. In the present study, we reexamine some of these previously described relationships and investigate additional relationships of metals with DNA methylation age measures. Specifically, we examine the independent and cumulative relationships of 24-hour urine concentrations of five metals [arsenic (As), cadmium (Cd), lead (Pb), manganese (Mn), and mercury (Hg)] with DNAmAge, PhenoAge, and GrimAge. This five metal urinary panel was previously established in the VA Normative Aging Study (NAS) and we utilize the NAS cohort for the present analysis26,27. Our analyses also utilize Bayesian Kernel Machine Regression (BKMR), a Bayesian variable selection framework that also allows for the assessment of both individual and cumulative exposure relationships28.
2 |. Methods
2.1. Study Population
Individuals in the present analysis were active participants in the VA Normative Aging Study (NAS). The NAS is a longitudinal cohort study of aging, which recruited healthy male participants from the Greater Boston area beginning in 196329. The NAS is now a closed cohort of elderly community-dwelling men. Every 3–5 years since recruitment participants have returned for in-person, follow-up study visits. During these onsite follow-up visits, participants receive comprehensive outpatient medical evaluations, give detailed information about their diets/lifestyle factors that may affect their health, and provide bio-specimens including blood. In the NAS, dropout has been less than 1% per year and predominantly occurs when participants move out of the study area30. Our study sample consisted of participants who had all available exposure, outcome, and clinical data (N = 48). Each participant contributed one study visit for a total of 48 observations. All participants provided written informed consent to the VA Institutional Review Board (IRB), and human subjects approval was granted by the VA and Harvard T.H. Chan School of Public Health IRBs (protocol 14027–102).
2.2. Urinary Metal Concentration Assessment
Concentrations for all five metals (As, Cd, Pb, Mn, and Hg) in urine were determined using methods previously described26. In short, a week before their scheduled study visit, participants received a 4-liter container for at-home 24-hour urine collection. Instructions and a questionnaire addressing sample collection time, missed collections, spillage, and medication regimens were also included with the container. Participants were instructed to collect urine after their first void in the morning through the first void of the next morning. Samples were returned during NAS study visits and stored at −20°C. Before the assay, they were thawed at room temperature, aliquoted, and digested with HNO3. Samples were then analyzed by plasma mass spectrometry (Sciex Elan 5000, Perkin-Elmer, Norwalk, CT) using standard operating and data processing procedures31. An average of five replicates were used to establish the final metal concentrations for each subject.
2.3. Measuring DNA Methylation and Calculating Measures of DNA Methylation Age
Processing of DNA methylation data has been previously described32. In brief, at each NAS follow-up visit, whole blood samples were collected from each participant. Bisulfite conversion (EZ-96 DNA Methylation Kit, Zymo Research, Orange, CA, USA) was performed on extracted DNA from the buffy coat of whole blood. Then, the Illumina Infinium HumanMethylation450 BeadChip was used to measure the DNA methylation of CpG probes. In an effort to minimize batch effects and achieve a similar age distribution across chips and plates, chips were randomized across plates. We then used a two-stage age-stratified algorithm to randomize samples. To achieve quality control, we removed samples where >5% of probes had a beadcount < 3 or > 1% of probes had a detection p-value >0.05. Probes mapping to single nucleotide polymorphisms, non-autosomal chromosomes, and cross-hybridizing probes were excluded32,33. After quality control, the remaining samples were preprocessed using the Illumina-type background correction34, dye-bias adjustment35, and BMIQ normalization36 to generate methylation beta values. Beta values represent the percentage of methylation for each of the ~480,000 CpG sites in the BeadChip array. Specifically, beta = intensity of the methylated signal (M)/[intensity of the unmethylated signal (U) + intensity of the methylated signal (M) + 100]. DNA methylation beta values range from 0 (completely unmethylated) to 1 (completely methylated). Previous analyses of this data demonstrated no evidence of batch effects after pre-processing37.
Measures of DNA Methylation Age were calculated using Horvath’s publicly available online calculator (http://dnamage.genetics.ucla.edu). Horvath’s initial DNAmAge was derived from an elastic net penalized regression run on multiple data sets of different cell and tissue types. After 21,369 CpG probes – shared by both the Illumina HumanMethylation27 and HumanMethylation450 BeadChip platforms – were regressed on a calibrated version of chronological age, the elastic net selected 353 CpGs that correlated with age (193 positively and 160 negatively). The model coefficients from these 353 CpGs were used by the calculator to predict the age of each DNA sample (i.e. DNAmAge). The calculator maintains predictive accuracy (age correlation 0.97, error = 3.6 years) across almost all body tissues including blood and brain8.
DNA phenotypic age (PhenoAge) was created by employing a Cox penalized model where the hazard of mortality was regressed on 42 clinical markers and chronological age11. From this penalized regression model, nine clinical variables (i.e. albumin, creatinine, glucose, C-reactive protein, lymphocyte percent, mean cell volume, red cell distribution width, alkaline phosphatase, and white blood cell count) and chronological age were selected to create a phenotypic age score. In test data, this score was highly correlated with chronological age (r=0.94) and every one-year increase in age score was associated with a 9% increase in all-cause mortality. The authors then used a different data set and elastic net penalized regression to determine methylation CpG sites related to their phenotypic score. Again they restricted their analyses to 20,169 CpGs shared across all three Illumina BeadChip platforms (27k, 450k, EPIC). In the end, they identified 513 CpGs to create the DNAm Phenotypic Age (PhenoAge) measure. 41 of these CpGs are shared with DNAmAge.
The final DNA methylation age metric used in our study is GrimAge. These authors wanted to first determine DNA methylation values that could be used to estimate plasm protein levels. They first regressed 88 plasma protein variables on chronological age, sex, and ~450,000 CpGs (shared between the 450K and EPIC platforms) in elastic net regression models. Only 12 out of the 88 plasma proteins demonstrated moderately high (r>0.35) correlations between their measured plasma levels and DNAm surrogate markers determined from the elastic net models. The authors also used 172 CpGs to establish a DNAm surrogate of cigarette smoking pack-years. They next regressed all-cause mortality time-to-death on: chronological age, sex, the pack-years DNAm surrogate marker, and the 12 plasma protein DNAm surrogate markers. The elastic net Cox regression model selected age, DNAm pack-years, sex, and 7 DNAm protein markers [adrenomedullin (ADM), beta-2-microglobulim (B2M), cystatin C, GDF-15, leptin, plasminogen activator inhibitor-1 (PAI-1), and tissue inhibitor metalloproteinases 1 (TIMP-1)]. After a linear transformation, the combination of these covariates was used to create DNAm GrimAge. Importantly, the CpGs used to create all DNAm surrogate markers are made up of 1030 unique CpGs. Out of all existing DNAm metrics, GrimAge is unparalleled in its ability to predict all-cause time-to-death (Cox regression p=2.0 × 10−75)18.
2.4. Statistical Analysis
Given that we had five metal exposures and three aging variables of interest, we sought a robust methodology for identifying biologically and statistically significant relationships. We were also cognizant that each metal as well as the entire exposure metal mixture could have unique relationships with the aging markers. Thus, we employed Bayesian Kernel Machine Regression (BKMR). BKMR functions as a Bayesian variable selection framework while also allowing for the assessment of both individual and cumulative metal relationships28.
All metals were log transformed to achieve uniformity/normal distributions before beginning any analyses. Subsequent Rosner’s tests did not reveal any outliers in the metal concentration levels. Metal exposures were also standardized using Z scores before running BKMR models. Using the R “bkmr” package, we first created BKMR selection models for each of the three aging metrics28. In previously published works, 10,000 iterations by Markov Chain Monte Carlo algorithm per BKMR model were deemed appropriate for selection38. However, because of our smaller sample, we ran 100,000 iterations per selection model. Specifically, each of the five metals and the covariates were modeled as predictors of DNAmAge, PhenoAge, and GrimAge respectively. We also examined traceplots from each model to ensure that each model demonstrated good convergence. Important covariates were determined a priori from the relevant literature20,24,26,27. Covariates included chronological age (continuous), season of visit (spring [March-May], Summer [June-August], Fall [September-November], and Winter [December-February]), body mass index (BMI) (lean [<25], overweight [25–30], obese [>30]), alcohol intake (yes/no ≥ 2 drinks daily), cumulative cigarette pack-years (continuous), smoking status (never, former, or current), education (<12 years, 12–16 years, >16 years), glomerular filtration rate (GFR) (continuous), white blood cell proportions (continuous), diabetes (yes/no), hypertension (yes/no), and ischemic heart disease (yes/no). The BKMR model was as follows:
,where the function h() is a dose-response function containing nonlinear and/or interactions between exposure metals, and Z = Z1, …, Zk are k potential confounders. Each model produced posterior inclusion probabilities (PIPs), which are a measure of how important each metal was for each of the aging outcomes. Although a PIP >0.50 threshold has been utilized in previously published studies39,40; given our small sample size, we decided to employ a more rigorous threshold (PIP >0.60) for determining important predictor metals. We then used linear regression models (using the lm function from the “stats” R package) to determine the independent association of selected metal exposures with their respective DNA methylation aging metrics. From the same BKMR models, we were able to plot the cumulative effects of the five urine metals by comparing the estimate value of the exposure-response function when all exposures are at a particular quantile. We considered the hypothesis that disease diagnoses may actually diminish the relationship between our exposures and outcomes. Hence, both our independent and cumulative association analyses were first run without the age-related disease covariates of diabetes, hypertension, and ischemic heart disease. We then repeated the models in sensitivity analyses adjusting for diabetes, hypertension, and ischemic heart disease. As an additional sensitivity analysis, all models were replicated using DNA methylation age acceleration (AgeAccel) as an outcome. AgeAccel values are the residuals from models where each methylation age variable was regressed on chronological age.
All statistical analyses were performed using R Version 3.6.3 (R Core Team, Vienna, Austria). We considered a p-value < 0.05 to be statistically significant. To improve the interpretability of the results, the logarithmic transformations of metals, where applicable, were reversed when conveying the results and creating the final tables.
3|. Results
3.1. Study Sample Characteristics and Descriptive Statistics
The demographic, biomarker, and clinical descriptive statistics for the study participants are presented in Table 1. The mean (SD) chronological age of the participants was 76.9 (5.3) years. Participants had a mean (SD) GrimAge, DNAmAge, and PhenoAge of 74.2 (6.4), 78.5 (7.3), and 71.6 (9.2) years respectively. The mean (SD) glomerular filtration rate (GFR) was 69.5 (13.4) mL/min/1.73 m2. 75% of participants consumed less than two alcoholic beverages a day. The majority of participants had study visits in the summer months (33%), were former smokers (67%), had a lean BMI (38%), and had an education of at least 12 years (69%). In this study sample, the prevalence of diabetes, hypertension, and ischemic heart disease were 29%, 67%, and 31%. Table 2 presents the descriptive statistics for the urine metal concentrations across study subjects. Mean (SD) levels for As, Cd, Hg, Mn, and Pb were 40.9 (57.2), 0.8 (0.4), 1.6 (1.5), 1.4 (0.4), and 1.7 (1.0) ng/mL respectively. Table 2 also lists the urine quintile values for each metal as well as the current EPA safe drinking water levels for each metal. 75% of urine samples exceeded the As drinking water standards, and 25% of urine samples exceeded the Hg standards. Urine samples for all other metals were within the safe drinking water standard levels.
Table 1.
Characteristics of Study Subjects (N = 48)
| Age Variables | ||
|---|---|---|
| Chronological Age (years), mean (SD) | 76.9 (5.3) | |
| DNAmAge (years), mean (SD) | 78.5 (7.3) | |
| GrimAge (years), mean (SD) | 74.2 (6.4) | |
| PhenoAge (years), mean (SD) | 71.6 (9.2) | |
| Health and Lifestyle Variables | ||
| Glomerular Filtration Rate (mL/min/1.73 m2), mean (SD) | 69.5 (13.4) | |
| Pack-years, mean (SD) | 17.5 (21.7) | |
| Alcohol Consumption, N (%) | ||
| < 2 drinks/day | 36 (75) | |
| ≥ 2 drinks/day | 12 (25) | |
| BMI, N (%) | ||
| Healthy/Lean | 18 (38) | |
| Overweight | 17 (35) | |
| Obese | 13 (27) | |
| Diabetes, N (%) | ||
| Yes | 14 (29) | |
| No | 34 (71) | |
| Education, N (%) | ||
| ≤ 12 years | 15 (31) | |
| 12 – 16 years | 20 (42) | |
| > 16 years | 13 (27) | |
| Hypertension, N (%) | ||
| Yes | 32 (67) | |
| No | 16 (33) | |
| Ischemic Heart Disease, N (%) | ||
| Yes | 15 (31) | |
| No | 33 (69) | |
| Season, N (%) | ||
| Spring | 8 (17) | |
| Summer | 16 (33) | |
| Fall | 13 (27) | |
| Winter | 11 (23) | |
| Smoking Status, N (%) | ||
| Current | 1 (02) | |
| Former | 32 (67) | |
| Never | 15 (31) | |
Table 2.
24-Hour Urine Metal Concentrations Across Study Subjects (N = 48) and EPA Drinking Water Guidelines
| Metal | Measures (ng/mL) | ||||||
|---|---|---|---|---|---|---|---|
| mean ± SD | 0% | 25% | Quantiles 50% | 75% | 100% | EPA Drinking Water Standards | |
| As | 40.9 ± 57.2 | 4.35 | 13.61 | 20.69 | 39.53 | 303.16 | 10 |
| Cd | 0.8 ± 0.4 | 0.23 | 0.52 | 0.66 | 0.88 | 2.05 | 5 |
| Hg | 1.6 ± 1.5 | 0.30 | 0.58 | 1.21 | 1.97 | 8.47 | 2 |
| Mn | 1.4 ± 0.4 | 0.45 | 1.22 | 1.41 | 1.64 | 2.20 | 50 |
| Pb | 1.7 ± 1.0 | 0.39 | 1.06 | 1.43 | 1.99 | 6.19 | 15 |
We also present the Pearson correlation matrices and data distributions of age variables (Figure 1A) and urine metal concentrations (Figure 1B) across all participants in the study sample (N = 48). GrimAge and PhenoAge shared the weakest correlation (r=0.53). GrimAge and DNAmAge shared the strongest correlation (r=0.71). Correlation coefficients between urine metal concentrations ranged from 0.12 to 0.48. The weakest correlation was between Hg and Pb while the strongest was between Hg and As.
Figure 1 |. orrelation Matrices of Age and Metal Variables.
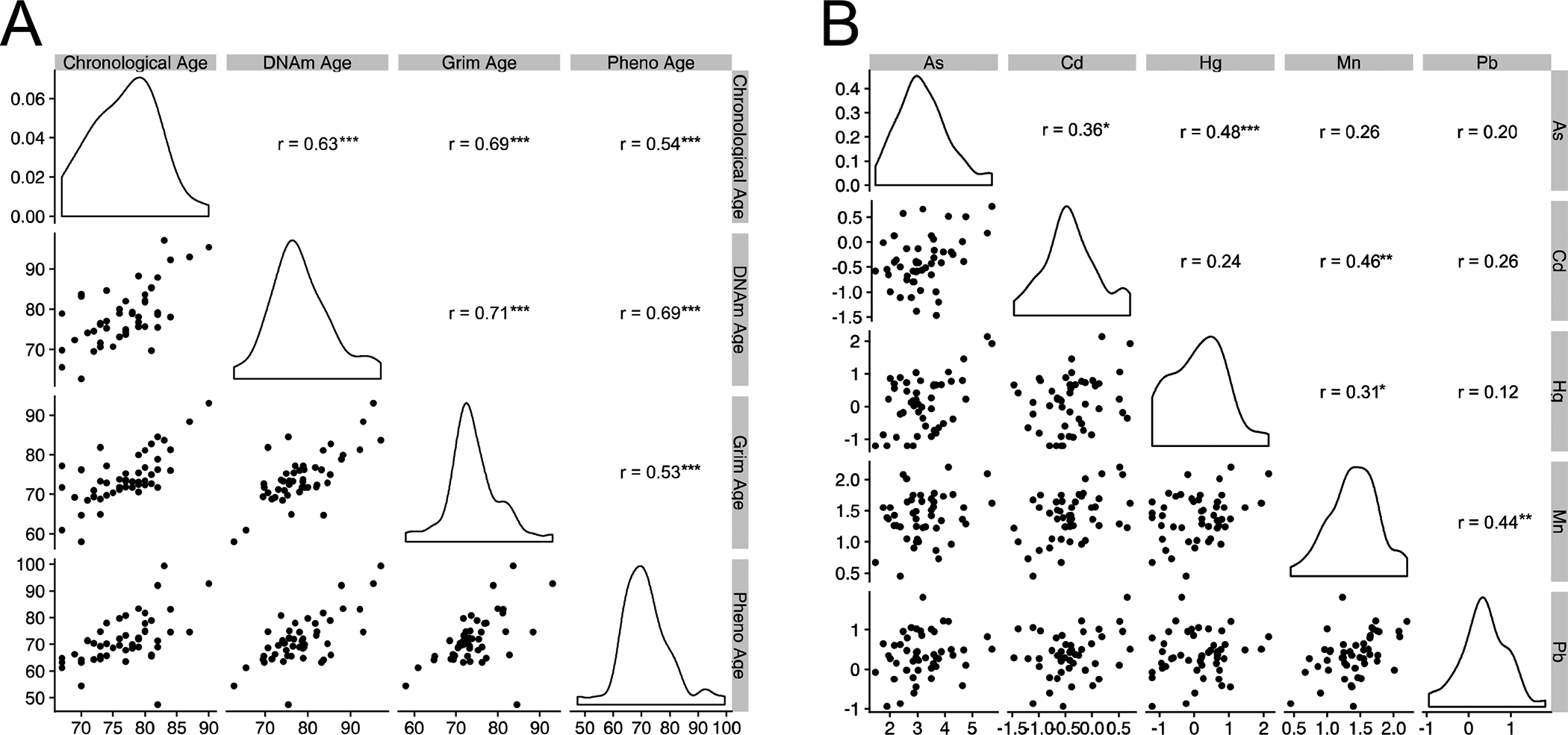
Pearson correlation matrices of [A] age variables and [B] urine metal concentrations across participants in the study sample. All correlations are based on the full cohort (N = 48). For the correlation matrix, all metals with the exception of Mn were log transformed to achieve distributions closer to normal. For uniformity, all metals were log transformed in the BKMR analyses. Subsequent Rosner’s tests did not reveal any outliers in the metal concentration levels. Asterisks denote statistical significance of correlations:
* = P < 0.05, ** = P < 0.01, *** = P < 0.001.
3.2. Bayesian Kernel Machine Regression (BKMR) Selection
Table 3 details the posterior inclusion probabilities (PIPs) derived from BKMR selection models. PIPs operate as a selection tool by representing the degree to which data supports each metal being included in the final regression model. PIPs for DNAmAge ranged from 0.35 (Hg) to 0.47 (As). No metals in the DNAmAge BKMR models met the 0.60 PIP threshold to be included in the final model. PIPs for GrimAge ranged from 0.36 (Pb) to 0.55 (Cd). Again, no metals met the threshold to be included in the final GrimAge regression model. PIPs for PhenoAge ranged from 0.39 (Pb) to 0.61 (Mn). Mn met the threshold for inclusion in the final PhenoAge regression model. BKMR selection results for all AgeAccel measures mirrored the results of their respective DNA methylation age BKMR selection models.
Table 3.
Posterior Inclusion Probabilities (PIPs) for Bayesian Kernel Machine Regression (BKMR) Fully-Adjusted Models
| Metal | DNAmAge | DNAm AgeAccel | GrimAge | Grim AgeAccel | PhenoAge | Pheno AgeAccel |
|---|---|---|---|---|---|---|
| As | 0.47 | 0.48 | 0.44 | 0.43 | 0.47 | 0.44 |
| Cd | 0.46 | 0.45 | 0.55 | 0.53 | 0.46 | 0.46 |
| Hg | 0.35 | 0.35 | 0.41 | 0.40 | 0.45 | 0.43 |
| Mn | 0.40 | 0.42 | 0.49 | 0.47 | 0.61ϕ | 0.62 ϕ |
| Pb | 0.37 | 0.40 | 0.36 | 0.34 | 0.39 | 0.36 |
In these analyses, metal PIPs 0.60 are marked with a “ϕ” and represent those selected as the important predictors of the outcome in the fully-adjusted BKMR model.
3.3. Associations of BKMR Individual Selected Urinary Metals and DNA Methylation-Based Measures of Biological Age
Table 4 summarizes the results of linear models where BKMR selected urinary metals were modeled as predictors of DNA methylation-based measures of biological age. No metals achieved selection criteria for a final DNAmAge or GrimAge model. A 1 ng/mL increase in urinary Mn was associated with a 9.93-year increase in PhenoAge (95%CI: 1.24, 18.61, P=0.03). This finding was similar to the results of a sensitivity analysis model including the covariates diabetes, hypertension, and ischemic heart disease; however, these results did not meet statistical significance (β = 10.20, 95%CI: −0.07, 20.47, P=0.05). A 1 ng/mL increase in urinary Mn was also significantly associated with a 9.93-year increase in Pheno Age Accel (95%CI: 1.24, 18.61, P=0.03). Urine Mn findings from the AgeAccel sensitivity analysis model, including the covariates diabetes, hypertension, and ischemic heart disease, were again. not statistically significant (β = 10.20, 95%CI: −0.07, 20.47, P=0.05).
Table 4.
Bayesian Kernel Machine Regression (BKMR) Selected 24-Hour Urinary Metal Concentrations as Predictors of DNA Methylation Age
| Models | Difference in DNA Methylation Age (95% CI) | P |
|---|---|---|
| PhenoAge | ||
| Mna (ng/mL) | 9.93 (1.24, 18.61) | 0.03 |
| Mnb (ng/mL) with Diseases | 10.20 (−0.07, 20.47) | 0.05 |
| Pheno AgeAccel | ||
| Mna (ng/mL) | 9.93 (1.24, 18.61) | 0.03 |
| Mnb (ng/mL) with Diseases | 10.20 (−0.07, 20.47) | 0.05 |
Main model adjusted for chronological age, year of visit, pack years, smoking status, season, alcohol consumption, education, GFR, BMI, and white blood cell proportions.
Sensitivity analysis model adjusted for Model “a” covariates and diabetes, hypertension, and ischemic heart disease.
All metals with the exception of Mn were log transformed to achieve normal distributions for the analyses. P values < 0.05 are italicized and considered statistically significant in these analyses.
3.4. Associations of Cumulative Metal Mixtures with DNA Methylation-Based Measures of Biological Age
Figure 2 demonstrates the results of analyses where cumulative metal exposures are modeled as a predictor of DNAmAge, GrimAge, PhenoAge, and their AgeAccel measures respectively. All models are compared to a model where each urine metal in the mixture is set to its 50th percentile value. For DNAmAge, PhenoAge, and their AgeAccel measures, we observe a trend of increasing methylation age as each metal in the mixture increases from its 10th to 90th percentile. These results were consistently statistically significant for PhenoAge and Pheno AgeAccel when each metal in the mixture is at its 70th percentile (Figure 2A). Here, every one-unit increase of the cumulative mixture with each metal at its 70th percentile was associated with a 2.53-year increase in PhenoAge (95%CI: 0.10, 4.96, P<0.05) and a 2.47-year increase in Pheno AgeAccel (95%CI: 0.18, 4.76, P<0.05). In cumulative mixture sensitivity analysis models including the covariates diabetes, hypertension, and ischemic heart disease, we observe trends similar to our main analyses (Figure 2B). Again, results were statistically significant for PhenoAge. However, this time, significance was met when each metal in the mixture was at its 90th percentile. Every one-unit increase of the cumulative mixture with each metal at its 90th percentile was associated with a 6.10-year increase in PhenoAge (95% CI: 0.59, 11.61, P<0.05) and a 6.18-year increase in Pheno AgeAccel (95% CI: 0.77, 11.59, P<0.05). For comparison, every one-unit increase of the cumulative mixture with each metal at its 90th percentile in the main analysis was associated with a 5.50-year increase in PhenoAge (95%CI: 0.09, 10.91, P<0.05) and a 5.68-year increase in PhenoAge (95%CI: 0.17, 11.19, P<0.05).
Figure 2 |. Relationships between Cumulative Metal Mixtures and DNA Methylation Age Variables.
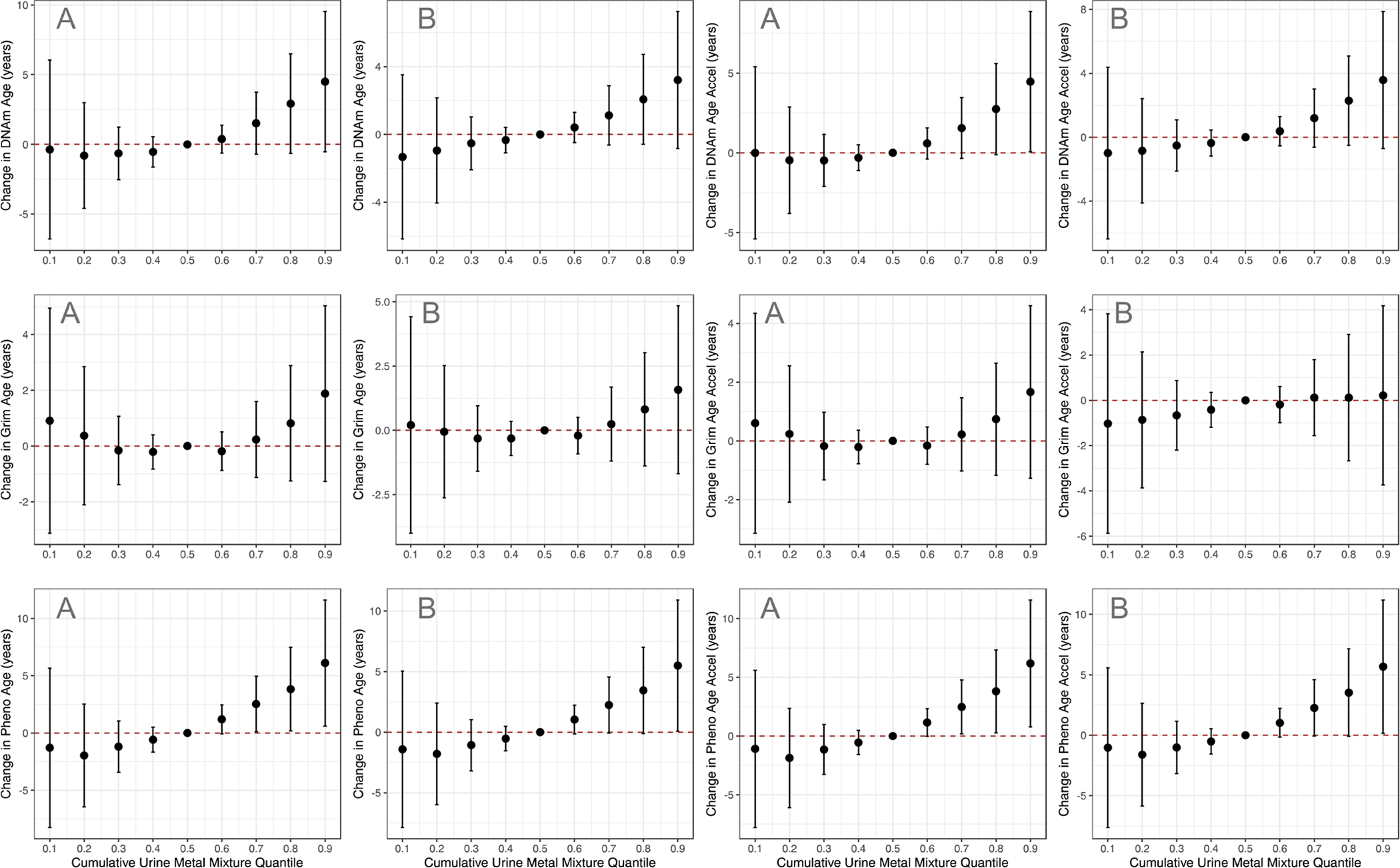
Difference in DNA methylation age variables (95% CI) of cumulative urine metal mixture exposure in the [A] main analysis models, and [B] sensitivity analysis models including the diseases diabetes, hypertension, and ischemic heart disease. In each BKMR model, all urine metals at the specified percentiles (x-axis) were compared to a model with all urine metals at their 50th percentile.
4 |. Discussion
In this study conducted in a cohort of community-dwelling older men, we demonstrate a novel positive association of urine metal concentrations with a DNA methylation-based measure of biological age. Using BKMR selection models, we identified 24-hour urine Mn concentrations as an important predictor of PhenoAge and Pheno AgeAccel. In further analyses using linear regression models adjusted for chronological age and lifestyle factors, we demonstrate that 24-hour urine Mn levels are significantly associated with higher PhenoAges and higher Pheno AgeAccel. Additionally, returning to BKMR models, we demonstrate that our cumulative 5-metal exposure mixture (As, Cd, Hg, Pb, and Mn) was also significantly associated with increases in PhenoAge and Pheno AgeAccel.
We hypothesized that measures of DNA methylation age could be biomarkers of subclinical toxic metal exposures. A critical component to testing this hypothesis was to use a cohort with subclinical toxic metal exposures. 24-hour urine analysis allows for the immediate detection of short-term exposures, and in our cohort, average urine concentrations with the exception of 75% of As samples and 25% of Hg levels were within drinking water standards permitted by the US Environmental Protection Agency (EPA)41–45. Given this descriptive data, we further hypothesized that As and Hg would be metals with important associations with our DNA methylation age metrics.
Arsenic (As) exposure can occur via inhalation, skin absorption, and ingestion (e.g. drinking water and food). Importantly, As intake is usually higher in food than in liquids like drinking water46. Seafood is the richest source of As, but As in food often exists as non-toxic organic compounds like (arsenobentaine and arsenocholine). Although these compounds may result in raised serum concentrations of As, they are rapidly excreted in urine. Studies have demonstrated that individuals who consumed seafood in the last 24-hours had higher total urine As levels than individuals who abstained for seafood in the prior day47. Seafood consumption may partially explain the elevated concentrations of As in the samples from our New England-based cohort48. Acute As toxicity manifests itself as abdominal pain, nausea, profuse vomiting, and diarrhea. Chronic toxicity can manifest as peripheral neuropathy, encephalopathy, and multisystem malignancy4. Existing research has demonstrated significant associations between As and aging processes including telomere shortening49 and cellular senescence50. Similar to As, a substantial proportion of Hg exposure occurs through the ingestion of seafood. Nonetheless, inhalation of Hg vapors is also a source of exposure. Depending on the route of exposure, acute Hg toxicity can present as shortness of breath, pleuritic chest pain, abdominal pain, vomiting, and bloody stools. Chronic toxicity is usually characterized by neurological, renal, and cutaneous symptoms51. The literature examining relationships of Hg with aging biomarkers is very limited and reaches mixed conclusions52,53
Despite these hypotheses, Mn was selected by our BKMR models as important to PhenoAge and Pheno AgeAccel respectively. Arriving at similar – if not identical – results in our PhenoAge and AgeAceel models gave us additional confidence in our findings. Mn exposure primarily occurs through inhalation and oral routes. Both natural processes (e.g. rock erosion and plant decomposition) and human activities (e.g. gasoline combustion) contribute to environmental Mn levels. Acute exposures can result in respiratory and neurobehavioral changes54, while chronic exposures result in neurotoxic symptoms that mimic Parkinson’s disease55. Very little evidence directly supports relationships of Mn with biological aging markers. A number of studies have, however, described links between Mn levels and aging-related neurological diseases56,57. A previous study in the NAS described relationships between high dietary Mn consumption and DNA methylation of inflammatory biomarker producing genes58. In our study sample, the relationship of Mn with PhenoAge and Pheno AgeAccel was statistically significant in the main analysis linear models, but the effect sizes were much larger than anything observed in previous DNA methylation age studies in the NAS20,59. One hypothesis explaining why the sensitivity analysis model loses statistical significance is that some of the effect of Mn on PhenoAge and Pheno AgeAccel could be mediated by age-related disease processes like diabetes or heart disease. Given that there is evidence for this60,61, models without these variables likely reveal the true relationship of Mn with PhenoAge and Pheno AgeAccel. Future work beyond this pilot will be important for better elucidating this relationship.
Ultimately, we expected to observe relationships with more metals and more DNA methylation biomarkers. Reasons for this lack of evidence may include our small pilot study sample and our exposure matrix. With the exception of Cd62, 24-hour urine metal levels give a short-term snapshot of an individual’s exposures. Much of the existing environmental exposure studies where robust relationships have been demonstrated in this area use long-term exposures of air pollution19,59. We assumed that in cases of ongoing environmental exposures, these short-term urine samples would be reflective of participant’s long-term metal exposures, but this may not be the case. Another explanation is that most of the individual metals truly do not have independent relationships with measures of DNA methylation age. The entire mixture, however, could have a statistically and biologically significant relationship with our outcomes. Unique relationships of metal mixtures with other outcomes have previously been reported in the literature63,64. Still, we only observed a significant relationship of the entire mixture with PhenoAge and Pheno AgeAccel in our study sample. Additional work exploring long-term metal relationships in urine and other exposure matrices will be useful for clarifying if many of these relationships are indeed null. We were also hoping to observe some trends across the three biomarkers of aging. Our study sample and others demonstrate that these markers are highly correlated; however, there are some notable differences11,18. These differences include largely unique component CpGs to calculate each metric as well as distinct abilities to predict mortality and other diseases. Hence, it is plausible to observe different metal-biomarker relationships, especially if the metrics have distinct physiological pathways of toxicity.
Our study possesses a number of strengths including the incorporation of three novel biomarkers of aging, the application of a robust selection/modeling technique, and the utilization of participant data from a well-established longitudinal cohort. Despite these strengths, we still have a few notable limitations. First, we utilized a panel of urine metals that was collected for other purposes. This practice is not unique for major longitudinal cohort studies, and helps make pilot studies feasible. Nevertheless, it does limit the number of metals that were considered in our analysis. Despite this limitation, we still were able to explore relationships of As, Pb, Hg, Cd, and Mn which are among the leading toxic metal exposures worldwide65. Secondly, this is a relatively small cross-sectional analysis conducted in a cohort of elderly Caucasian males who now reside in the United States. As we noted previously, much of the developing world bears the burden of toxic environment exposures2. Hence, additional studies with a larger number of participants, in other environments, and involving other demographic groups are needed to broadly confirm our findings. Lastly, even though our study sample is taken from a longitudinal cohort with a breadth of data on participant demographic/lifestyle factors and we determined important covariates a priori from the relevant literature20,24,26,27, we cannot rule out the possibility of unknown or residual confounding in our analyses.
In conclusion, our study offers some evidence that DNA methylation aging biomarkers have important relationships with toxic metal exposures. Our data specifically suggests that PhenoAge may capture some of the physiological impacts of Mn exposures in individuals not explicitly presenting with clinical signs of Mn poisoning. Future research aimed at broadening the understanding of Mn and other metals with DNA methylation age measures will be crucial for defining the public health utility of these methylation-based tools as biomarkers of subclinical metal toxicity.
Highlights:
We examined relationships of five urine metals with three markers of DNA methylation age.
Urine Mn was identified as important to PhenoAge..
The cumulative urine metal mixture (As, Cd, Hg, Mn, and Pb) was associated with increases in PhenoAge.
Our results add novel evidence that metals detected in urine are associated with increases in biological aging.
Funding Information
This work was supported by National Institutes of Health (R01ES025225, R01ES021733, R01ES027747, P30ES009089); The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center, Boston, Massachusetts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: None declared
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
5 | References
- 1.Lead poisoning and health. https://www.who.int/news-room/fact-sheets/detail/lead-poisoning-and-health. [Google Scholar]
- 2.Alidadi H et al. Health risk assessments of arsenic and toxic heavy metal exposure in drinking water in northeast Iran. Environ. Health Prev. Med 24, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayotte JD, Medalie L, Qi SL, Backer LC & Nolan BT Estimating the High-Arsenic Domestic-Well Population in the Conterminous United States. Environ. Sci. Technol 51, 12443–12454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratnaike RN Acute and chronic arsenic toxicity. Postgrad. Med. J 79, 391–396 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keil DE, Berger-Ritchie J & McMillin GA Testing for Toxic Elements: A Focus on Arsenic, Cadmium, Lead, and Mercury. Lab. Med 42, 735–742 (2011). [Google Scholar]
- 6.Pizzorno J Conventional Laboratory Tests to Assess Toxin Burden. Integr. Med. Clin. J 14, 8–16 (2015). [PMC free article] [PubMed] [Google Scholar]
- 7.Andrade V, Mateus M, Batoréu M, Aschner M & Marreilha dos Santos A Lead, arsenic and manganese metal mixture exposures: focus on biomarkers of effect. Biol. Trace Elem. Res 166, 13–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath S et al. Perinatally acquired HIV infection accelerates epigenetic aging in South African adolescents. Aids 32, 1465–1474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhoeven JE et al. Epigenetic Age in Male Combat-Exposed War Veterans: Associations with Posttraumatic Stress Disorder Status. Mol. Neuropsychiatry 4, 90–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine ME et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thurston RC et al. Vasomotor Symptoms and Accelerated Epigenetic Aging in the Women’s Health Initiative (WHI). J. Clin. Endocrinol. Metab 105, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X et al. Association between sleep disordered breathing and epigenetic age acceleration: Evidence from the Multi-Ethnic Study of Atherosclerosis. EBioMedicine 50, 387–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T et al. Dysfunctional epigenetic aging of the normal colon and colorectal cancer risk. Clin. Epigenetics 12, 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillary RF et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol. Psychiatry (2019) doi: 10.1038/s41380-019-0616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arpón A et al. Interaction Among Sex, Aging, and Epigenetic Processes Concerning Visceral Fat, Insulin Resistance, and Dyslipidaemia. Front. Endocrinol 10, 496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezwan FI et al. Association of adult lung function with accelerated biological aging. Aging 12, 518–542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu AT et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhingra R, Nwanaji-Enwerem JC, Samet M & Ward-Caviness CK DNA Methylation Age-Environmental Influences, Health Impacts, and Its Role in Environmental Epidemiology. Curr. Environ. Health Rep 5, 317–327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nwanaji-Enwerem JC et al. Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: The VA normative aging study. Environ. Int 102, 57–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward-Caviness CK et al. Long-term exposure to air pollution is associated with biological aging. Oncotarget 7, 74510–74525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis SW et al. Environmental exposure to polybrominated biphenyl (PBB) associates with an increased rate of biological aging. Aging 11, 5498–5517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X et al. Effect of tobacco smoking on the epigenetic age of human respiratory organs. Clin. Epigenetics 11, 183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demanelis K et al. Cadmium exposure and age-associated DNA methylation changes in non-smoking women from northern Thailand. Environ. Epigenetics 3, dvx006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinberg J, Shah KM, Gartland A, Zeggini E & Wilkinson JM Effects of chronic cobalt and chromium exposure after metal-on-metal hip resurfacing: An epigenome-wide association pilot study. J. Orthop. Res. Off. Publ. Orthop. Res. Soc 35, 2323–2328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsaih SW et al. The independent contribution of bone and erythrocyte lead to urinary lead among middle-aged and elderly men: the normative aging study. Environ. Health Perspect 107, 391–396 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lampe BJ et al. Association between 24-hour urinary cadmium and pulmonary function among community-exposed men: the VA Normative Aging Study. Environ. Health Perspect 116, 1226–1230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bobb JF et al. Bayesian kernel machine regression for estimating the health effects of multi-pollutant mixtures. Biostat. Oxf. Engl 16, 493–508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell B, Rose CL & Damon A The Veterans Administration Longitudinal Study of Healthy Aging. The Gerontologist 6, 179–184 (1966). [DOI] [PubMed] [Google Scholar]
- 30.Mehta AJ et al. Associations between Changes in City and Address Specific Temperature and QT Interval - The VA Normative Aging Study. PLOS ONE 9, e106258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viczián M, Lásztity A, Wang X & Barnes RM On-line isotope dilution and sample dilution by flow injection and inductively coupled plasma mass spectrometry. J. Anal. At. Spectrom 5, 125–133 (1990). [Google Scholar]
- 32.Colicino E et al. Blood DNA methylation biomarkers of cumulative lead exposure in adults. J. Expo. Sci. Environ. Epidemiol (2019) doi: 10.1038/s41370-019-0183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW & Siegmund KD Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res 41, e90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis S, Du P, Bilke S, Triche T & Bootwalla M methylumi: Handle Illumina methylation data. R Package Version 2, (2015). [Google Scholar]
- 36.Teschendorff AE et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinforma. Oxf. Engl 29, 189–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nwanaji-Enwerem JC et al. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ. Epigenetics 2, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashrap P et al. Maternal blood metal and metalloid concentrations in association with birth outcomes in Northern Puerto Rico. Environ. Int 138, 105606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coker E et al. Association between prenatal exposure to multiple insecticides and child body weight and body composition in the VHEMBE South African birth cohort. Environ. Int 113, 122–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y et al. Association between exposure to a mixture of phenols, pesticides, and phthalates and obesity: Comparison of three statistical models. Environ. Int 123, 325–336 (2019). [DOI] [PubMed] [Google Scholar]
- 41.US EPA, O. Chemical Contaminant Rules. US EPA https://www.epa.gov/dwreginfo/chemical-contaminant-rules (2015). [Google Scholar]
- 42.US EPA, O. Drinking Water Criteria Document for Manganese. US EPA https://www.epa.gov/wqc/drinking-water-criteria-document-manganese (2018). [Google Scholar]
- 43.ATSDR. Cadmium (Cd) Toxicity: What Are the U.S. Standards for Cadmium Exposure? | ATSDR -Environmental Medicine & Environmental Health Education - CSEM https://www.atsdr.cdc.gov/csem/csem.asp?csem=6&po=7. [Google Scholar]
- 44.US EPA, O. National Primary Drinking Water Regulations. US EPA https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations (2015). [Google Scholar]
- 45.ATSDR. Lead (Pb) Toxicity: What Are the U.S. Standards for Lead Levels? | ATSDR -Environmental Medicine & Environmental Health Education - CSEM https://www.atsdr.cdc.gov/csem/csem.asp?csem=34&po=8. [Google Scholar]
- 46.Thomas KW, Pellizzari ED & Berry MR Population-based dietary intakes and tap water concentrations for selected elements in the EPA region V National Human Exposure Assessment Survey (NHEXAS). J. Expo. Anal. Environ. Epidemiol 9, 402–413 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Navas-Acien A, Francesconi KA, Silbergeld EK & Guallar E Seafood intake and urine concentrations of total arsenic, dimethylarsinate and arsenobetaine in the US population. Environ. Res 111, 110–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stavely KWF & Fitzgerald K America’s Founding Food: The Story of New England Cooking (Univ of North Carolina Press, 2004). [Google Scholar]
- 49.Alegría-Torres JA, Pérez-Rodríguez RY, García-Torres L, Costilla-Salazar R & Rocha-Amador D Exposure to arsenic and lead in children from Salamanca México, effects on telomeric lengthening and mitochondrial DNA. Environ. Sci. Pollut. Res. Int 27, 6420–6428 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Chung Y-P, Chen Y-W, Weng T-I, Yang R-S & Liu S-H Arsenic induces human chondrocyte senescence and accelerates rat articular cartilage aging. Arch. Toxicol 94, 89–101 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Broussard LA, Hammett-Stabler CA, Winecker RE & Ropero-Miller JD The Toxicology of Mercury. Lab. Med 33, 614–625 (2002). [Google Scholar]
- 52.Vriens A et al. Exposure to Environmental Pollutants and Their Association with Biomarkers of Aging: A Multipollutant Approach. Environ. Sci. Technol 53, 5966–5976 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Yeates AJ et al. PUFA Status and Methylmercury Exposure Are Not Associated with Leukocyte Telomere Length in Mothers or Their Children in the Seychelles Child Development Study. J. Nutr 147, 2018–2024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhoeven WM, Egger JI & Kuijpers HJ Manganese and acute paranoid psychosis: a case report. J. Med. Case Reports 5, 146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Neal SL & Zheng W Manganese Toxicity Upon Overexposure: a Decade in Review. Curr. Environ. Health Rep 2, 315–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genoud S et al. Subcellular compartmentalisation of copper, iron, manganese, and zinc in the Parkinson’s disease brain. Met. Integr. Biometal Sci 9, 1447–1455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huat TJ et al. Metal Toxicity Links to Alzheimer’s Disease and Neuroinflammation. J. Mol. Biol 431, 1843–1868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kresovich JK et al. The Inflammatory Potential of Dietary Manganese in a Cohort of Elderly Men. Biol. Trace Elem. Res 183, 49–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, Koutrakis P, Gao X, Baccarelli A & Schwartz J Associations of annual ambient PM2.5 components with DNAm PhenoAge acceleration in elderly men: The Normative Aging Study. Environ. Pollut. Barking Essex 1987 258, 113690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shan Z et al. U-Shaped Association between Plasma Manganese Levels and Type 2 Diabetes. Environ. Health Perspect 124, 1876–1881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y & Zheng W Cardiovascular Toxicities Upon Manganese Exposure. Cardiovasc. Toxicol 5, 345–354 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suwazono Y et al. Biological half-life of cadmium in the urine of inhabitants after cessation of cadmium exposure. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem 14, 77–81 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Valeri L et al. The Joint Effect of Prenatal Exposure to Metal Mixtures on Neurodevelopmental Outcomes at 20–40 Months of Age: Evidence from Rural Bangladesh. Environ. Health Perspect 125, 067015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Mukherjee B & Park SK Associations of cumulative exposure to heavy metal mixtures with obesity and its comorbidities among U.S. adults in NHANES 2003–2014. Environ. Int 121, 683–694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mamtani R, Stern P, Dawood I & Cheema S Metals and Disease: A Global Primary Health Care Perspective. J. Toxicol 2011, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


