Abstract
Colonization of the human stomach with Helicobacter pylori strains containing the cag pathogenicity island is a risk factor for development of gastric cancer. The cag pathogenicity island contains genes encoding a secreted effector protein (CagA) and components of a type IV secretion system (Cag T4SS). The molecular architecture of the H. pylori Cag T4SS is substantially more complex than that of prototype T4SSs in other bacterial species. In this review, we discuss recent discoveries pertaining to the structure and function of the Cag T4SS and its role in gastric cancer pathogenesis.
Keywords: type IV secretion system, bacterial protein secretion, bacterial nanomachines, macromolecular structures, cryo-electron microscopy, cryo-electron tomography, gastric cancer, gastric adenocarcinoma, peptic ulcer disease
Helicobacter pylori and gastric cancer
Helicobacter pylori, a Gram-negative spiral-shaped bacterium, persistently colonizes the stomach in about half of the global human population (1). While the majority never experience any adverse health consequences attributable to H. pylori, the presence of these bacteria in the stomach increases the risk for development of gastric adenocarcinoma and peptic ulcer disease (2). H. pylori infection is the strongest known risk factor for gastric cancer, and gastric cancer is the third leading cause of cancer-related death worldwide (3). Some H. pylori strains contain a 40-kb region of chromosomal DNA known as the cag pathogenicity island (cag PAI), whereas other strains do not (4-6). Individuals colonized with H. pylori strains containing the cag PAI have a higher risk of gastric adenocarcinoma and peptic ulcer disease compared to individuals colonized with H. pylori strains lacking the cag PAI (7-10). The cag PAI contains genes encoding a secreted effector protein (CagA) and multiple proteins required for delivery of CagA into host cells (11, 12). Several of the latter proteins exhibit low-level sequence relatedness to components of type IV secretion systems (T4SSs) in other bacterial species (Table 1)(12).
Table 1.
cag PAI-encoded proteins required for Cag T4SS activitya
| Gene numberb |
Protein name |
Localizationc | Predicted localizationd |
Structuree | Homologs | Putative function |
|---|---|---|---|---|---|---|
| Proteins with defined localization in T4SS apparatus | ||||||
| hp0528 | CagX | OMCC | 6OEG 5H3V |
VirB9 | ||
| hp0527 | CagY | OMCC | 6ODI | VirB10 | ||
| hp0532 | CagT | OMCC | 6OEE | VirB7 | ||
| hp0537 | CagM | OMCC | ||||
| hp0522 | Cag3 | OMCC | ||||
| hp0525 | Cagα | IMC | 2PT7 1G6O |
VirB11 | ATPase | |
| hp0524 | Cagβ | IMC | VirD4 | ATPase | ||
| hp0544 | CagE | IMC | VirB3/VirB4 | ATPase | ||
| Proteins for which localization in T4SS apparatus is not yet defined | ||||||
| hp0546 | CagC | IM, OM, S | VirB2 | |||
| hp0545 | CagDf | IM, PP, S | 3CWX | |||
| hp0543 | CagF | IM, C | CagA chaperone | |||
| hp0542 | CagGf | PP | ||||
| hp0541 | CagH | IM | ||||
| hp0540 | CagI | PP | ||||
| hp0539 | CagL | PP, S | 4CII 4YVM 3ZCI |
|||
| hp0538 | CagNf | PP, IM | ||||
| hp0531 | CagU | IM | ||||
| hp0530 | CagV | IM | 6IQT | VirB8 | ||
| hp0529 | CagW | IM | ||||
| hp0526 | CagZf | IM | 1S2X | |||
| hp0523 | Cag4 | PP | VirB1 | PG hydrolase | ||
| Secreted effector protein | ||||||
| hp0547 | CagA | C,S | 4DVY 4GOH |
Effector protein | ||
The proteins listed are required for CagA translocation into host cells (11, 12). Several proteins (for example, CagF and Cagβ) are required for CagA translocation into host cells but are not required for other T4SS-dependent phenotypes (such as stimulation of IL-8 production) (11, 43-47).
Gene number in H. pylori strain 26695
Location in T4SS apparatus based on biochemical analysis of extracted subassemblies, cryo-ET analysis or cryo-EM analysis. OMCC: outer membrane core complex. IMC: inner membrane complex.
Predicted localization based on features of protein sequences or fractionation experiments. OM, outer membrane; IM, inner membrane; C, cytoplasm; S, surface-exposed or supernatant; PP, periplasm.
Protein database (PDB) IDs for structures are listed.
Conflicting results about requirement for T4SS activity, or mutants reported to exhibit a partial reduction in T4SS activity.
Recent studies have elucidated T4SS-dependent activities relevant to gastric cancer pathogenesis in cell culture systems and animal models. Other recent studies have used cryo-electron tomography (cryo-ET) to view the Cag T4SS in situ in intact H. pylori (13, 14) as well as single particle cryo-electron microscopy (cryo-EM) to determine the structure of T4SS complexes extracted from H. pylori (15, 16). These studies reveal that the molecular architecture of the H. pylori Cag T4SS is substantially more complex than that of prototype T4SSs in other bacterial species (Text Box 1). In this review, we discuss recent discoveries pertaining to the structure and function of the H. pylori Cag T4SS and its role in gastric cancer pathogenesis.
Text Box 1. “Overview of bacterial type IV secretion systems”.
Bacteria utilize more than 10 different types of secretion systems to translocate proteins across the cell envelope (99). Type IV secretion systems (T4SSs) are complex and functionally diverse molecular machines found in both Gram-negative and Gram-positive bacteria as well as Archaea (61-64). In Gram-negative bacteria, T4SSs span both the inner and outer membrane. T4SSs can carry out a wide range of functions, including delivery of effector proteins into eukaryotic or bacterial target cells, delivery of DNA into target cells, and import (or export) of substrates from (or to) the extracellular environment (61-64, 100-102). Two of the most extensively studied examples of T4SSs are bacterial conjugation systems, which mediate DNA exchange between bacteria, and the Agrobacterium tumefaciens VirB/VirD4 T4SS, which delivers bacterial T-DNA into plant cells, resulting in tumor formation (crown gall disease). T4SS-mediated delivery of effector proteins into host cells is required for the pathogenicity of many bacterial species, including H. pylori, Legionella pneumophila, Bordetella pertussis, Coxiella, Brucella, and Bartonella. In most (but not all) cases, T4SS-mediated delivery of effector proteins into recipient cells is contact-dependent. T4SSs can be phylogenetically grouped into two main families: type IVA (exemplified by conjugation systems, the A. tumefaciens VirB/VirD4 T4SS, and the H. pylori Cag T4SS) and type IVB (exemplified by the Legionella Dot/Icm T4SS)(102). “Minimized” T4SSs, exemplified by conjugations systems and the A. tumefaciens VirB/VirD4 T4SS, have about 12 components (VirB1-11 and VirD4). In contrast, the H. pylori Cag T4SS and the Legionella Dot/Icm T4SS are more complex and have a substantially larger number of components.
The cag pathogenicity island
The cag PAI is a 40-kb chromosomal region containing about 27 genes. These include cagA (encoding a secreted effector protein), 17 genes required for Cag T4SS activity and several genes with no known functions (4-6, 11, 12). H. pylori is the only Helicobacter species known to harbor the cag PAI. The %GC content of the cag PAI is lower than that of the rest of the H. pylori chromosome, which suggests that it was horizontally acquired. Geographically distinct populations of H. pylori throughout the world harbor the cag PAI (6), which suggests that it was horizontally acquired by ancestral H. pylori in the distant past, prior to human migrations out of Africa.
CagA effector protein
CagA was originally identified by detecting serum antibody responses to a high molecular mass (120-132 kDa) protein in humans colonized by H. pylori (17-20). Antibody responses to this protein are detected more commonly in individuals with gastric cancer or peptic ulcer disease than in asymptomatic H. pylori-positive individuals (7, 17, 18). The name “cytotoxin-associated gene A” (cagA) was originally assigned based on the observation that the 120-132 kDa antigenic protein and the gene encoding it are present more frequently in H. pylori strains with vacuolating toxin activity than in strains without detectable vacuolating toxin activity (19, 20). Subsequently it became clear that vacuolating toxin activity is mediated by the secreted VacA protein (21), which is encoded by a chromosomal locus distant from cagA, and cagA is not required for vacuolating toxin activity. Use of the term “cag” (cytotoxin-associated gene) when describing cagA, the cag PAI, and the Cag T4SS is based on the historical association of the vacuolating toxin phenotype with production of the 120-132 kDa antigenic protein.
Thus far, CagA is the only protein known to be secreted by the Cag T4SS. CagA is secreted and translocated into host cells by adherent H. pylori (Fig. 1), but it is not secreted into the extracellular space when the bacteria are cultured in vitro in the absence of host cells. The signals and regulatory mechanisms that trigger CagA secretion in response to H. pylori contact with host cells are not known. Upon entry into host cells, CagA undergoes phosphorylation by tyrosine kinases (Src and Abl) and interacts with at least 20 different host cell proteins, altering their activity (discussed in other reviews) (22-24).
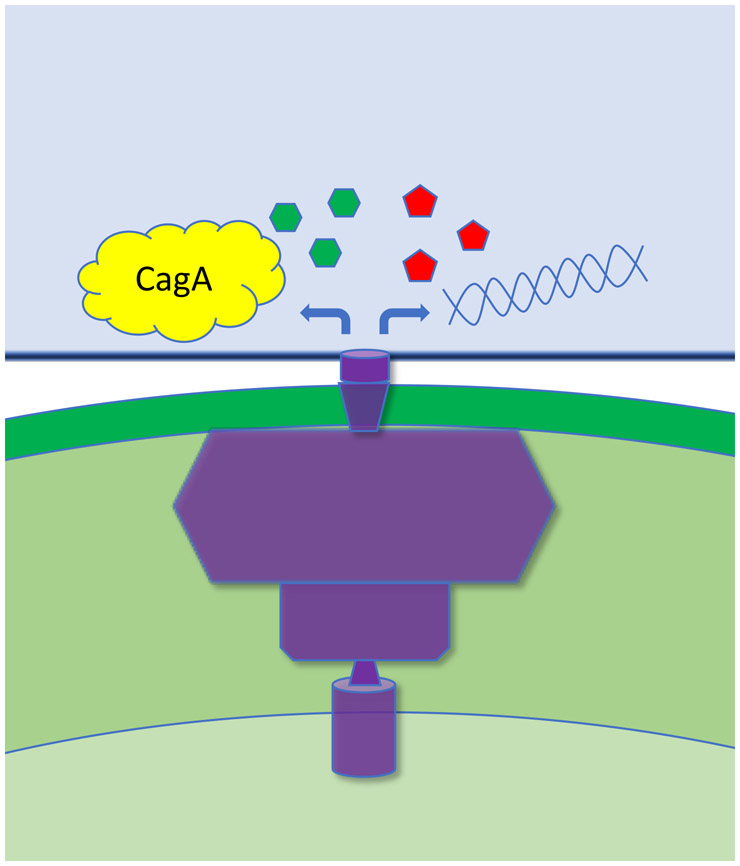
Fig. 1.
Schematic depicting an H. pylori bacterium adhering to a host cell. The Cag T4SS (purple) facilities delivery of the CagA effector protein into host cells, as well as intracellular entry of H. pylori LPS metabolites, peptidoglycan and DNA.
The CagA proteins produced by different strains of H. pylori vary from about 120 to 145 kDa in mass. Most H. pylori contain a single copy of cagA, but some strains contain multiple tandem copies. CagA is characterized by a structured N-terminal region and an intrinsically disordered C-terminal region (25, 26). Both a C-terminal 20-amino-acid translocation signal and the N-terminal domain are required for efficient secretion of CagA (27, 28). The disordered C-terminal portion of the protein contains multiple sites (EPIYA motifs) where the protein undergoes phosphorylation by tyrosine kinases within host cells (29-31).
Transgenic mice expressing CagA (in the absence of H. pylori infection) develop gastric epithelial hyperplasia, gastric polyps, and adenocarcinoma of the stomach and small intestine, as well as myeloid leukemia and B-cell lymphoma (32). Similarly, CagA expression in Drosophila promotes increased cell proliferation, and transgenic zebrafish expressing CagA exhibit increased rates of intestinal epithelial cell proliferation (33). CagA induces intestinal small cell carcinoma and adenocarcinoma in zebrafish with a loss-of-function allele of p53 (33), and CagA expression in epithelial cells enhances the growth and invasion of tumors in a Drosophila model of metastasis (34). Therefore, multiple lines of evidence indicate that CagA has an important role in gastric carcinogenesis, leading to its designation as a “bacterial oncoprotein” (22, 23).
Proteins required for Cag T4SS activity
To define the genes required for Cag T4SS activity, H. pylori genes within the cag PAI have been individually mutated and the resulting mutant strains have been tested in T4SS functional assays. About 17 genes in the cag PAI are required for CagA translocation into gastric cells, and mutagenesis of several additional cag PAI genes results in a partial reduction in CagA translocation (Table 1)(11, 12). At least two H. pylori genes in chromosomal loci outside the cag PAI, encoding an outer membrane protein (HopQ)(35) and hydrogenase (36), influence Cag T4SS activity but are not considered components of the Cag T4SS.
Role of the Cag T4SS in H. pylori-induced alterations in host cells
Co-culture of gastric epithelial cells with cag PAI-positive H. pylori but not Cag T4SS mutant strains results in an assortment of cellular alterations, many of which are attributable to the actions of CagA. T4SS-dependent cellular alterations attributable to actions of CagA include changes in cell shape (resulting from cytoskeletal changes), disruption of cell-cell junctions, altered cell polarity and cell adhesion, increased cell motility and cell migration, increased cell proliferation, β-catenin activation, and epithelial-mesenchymal transition (discussed in other reviews)(22-24).
H. pylori also causes T4SS-dependent changes in gastric epithelial cells that are not dependent on CagA. Among these, the most extensively studied phenomenon is stimulation of interleukin-8 (IL-8) production, which occurs through an NF-κB-dependent pathway. H. pylori-induced IL-8 production was initially attributed to the entry of peptidoglycan into host cells and recognition of peptidoglycan by Nod1 (Fig. 1)(37). More recently, this phenomenon has been attributed to the entry of lipopolysaccharide metabolites [heptose-1,7-bisphosphate (HBP) or ADP-glycero-β-D-manno-heptose (ADP-heptose), a derivative of HBP] into host cells and recognition of these molecules by tumor necrosis factor receptor-associated factor (TRAF)-interacting protein with forkhead-associated domain (TIFA), independent of Nod1 (38-41). Another T4SS-dependent alteration in host cells is activation of Toll-like receptor 9 (TLR9), a phenomenon attributed to the entry of H. pylori DNA into host cells (42). The cag PAI genes required for H. pylori-induced stimulation of IL-8 production in gastric epithelial cells or TLR9 activation are similar to those required for CagA translocation, but several genes required for CagA translocation [for example, CagF (a putative CagA chaperone) and Cagβ (an ATPase proposed to act as a coupling protein)] are not required for IL-8 production or TLR9 activation (11, 43-47).
In addition to the Cag T4SS, three additional H. pylori T4SSs have been described (known as the ComB T4SS, TFSS3 and TFSS4) (48). The ComB T4SS allows H. pylori to take up exogenous DNA through natural transformation. The functions of the TFSS3 and TFSS4 systems are not yet well understood. Thus far, there is no evidence that genes encoding components of the ComB T4SS, TFSS3 and TFSS4 are required for CagA translocation or other phenotypes associated with the Cag T4SS.
Role of the Cag T4SS in animal models of H. pylori-induced gastric disease
Mongolian gerbils infected with wild-type cag PAI-positive H. pylori strains develop severe gastric inflammation, often associated with gastric ulceration or gastric cancer (49-53). Mutant H. pylori strains with defects in Cag T4SS activity are able to colonize the gerbil stomach but cause less severe gastric inflammation and a much lower rate of gastric cancer or gastric ulceration than wild-type strains (49-53). Similar to T4SS mutant strains, CagA mutant strains cause less severe gastric inflammation and a markedly reduced rate of gastric cancer or gastric ulceration in a Mongolian gerbil model compared to isogenic wild-type strains (53-56). Therefore, the relative inactivity of Cag T4SS mutant strains is attributable, at least in part, to the failure of these strains to secrete CagA. Whether Cag T4SS-dependent delivery of non-protein substrates also contributes to gastric inflammation and carcinogenesis remains unclear.
Mice experimentally infected with wild-type cag PAI-positive H. pylori strains develop mild gastric inflammation but rarely develop gastric ulceration or gastric cancer. Cag T4SS and CagA mutant strains cause less severe gastric inflammation in mice compared to isogenic wild-type cag PAI-positive strains (57-59), but the differences are less striking than those observed in the Mongolian gerbil model. Wild-type H. pylori strains with an intact Cag T4SS stimulate gastric stem cell expansion, resulting in increased cell proliferation and hyperplasia, whereas T4SS mutant bacteria do not (59, 60).
Molecular architecture of the Cag T4SS
Over the past decade, multiple models for the structural organization of the Cag T4SS have been proposed based on weak sequence relatedness of cag PAI-encoded proteins to components of T4SSs in other bacterial species (Table 1), localization of Cag proteins to specific subcellular fractions, experimentally identified protein-protein interactions among Cag proteins, and knowledge about T4SS architecture in other bacterial species. Generating models for the organization of the Cag T4SS has been challenging, however, because several Cag proteins do not exhibit sequence relatedness to components of T4SSs in other bacterial species (Table 1).
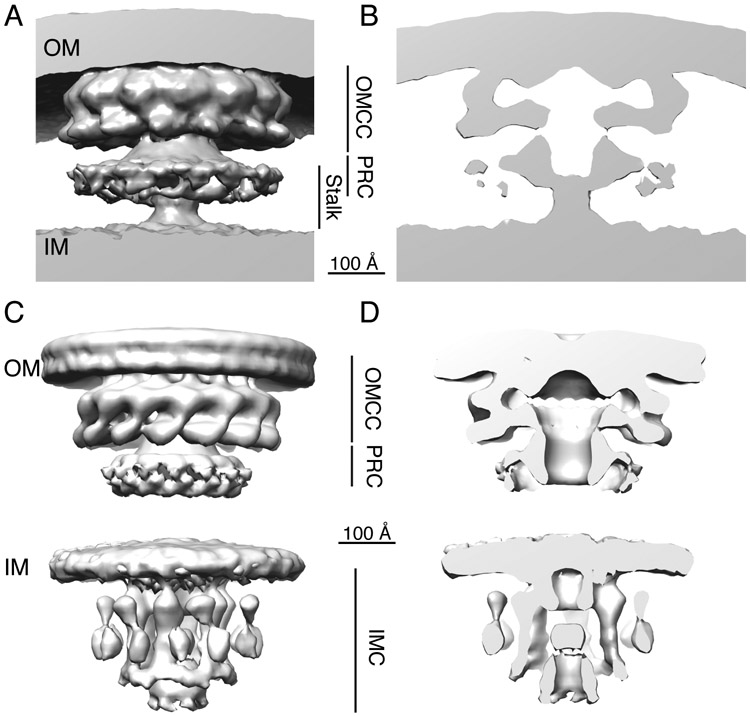
Two recent studies used cryo-electron tomography (cryo-ET) to reveal the structural organization of the Cag T4SS in intact bacteria (13, 14). When viewed by this approach, the Cag T4SS consists of a large mushroom-shaped structure that contacts the outer membrane, known as an outer membrane core complex (OMCC), as well as an inner membrane complex (IMC)(Fig. 2)(13, 14). The OMCC has 14-fold symmetry (13, 14). The IMC, consisting of three concentric rings surrounding a central channel, has six-fold symmetry (14). Mutant strains lacking individual ATPase components of the T4SS (CagE, Cagα and Cagβ) each exhibit detectable differences in IMC architecture compared to the wild-type strain (14).
Fig. 2.
Cryo-ET analyses of the H. pylori Cag T4SS. Spatial relationships between the T4SS, the bacterial outer membrane (OM) and inner membrane (IM) are shown. (A,B) Cryo-ET analysis of H. pylori attached to AGS gastric cells, showing the 3D structure and a central axial slice view of the 3D structure (EMD-7474)(13). Scale bar, 100 Å. (C,D). Cryo-ET analysis of H. pylori grown in pure culture (without AGS gastric cells) showing the 3D structure and a central axial slice view of the 3D structure (EMD- 0634 and 0635)(14). Scale bar, 100 Å. The outer membrane core complex (OMCC), periplasmic ring complex (PRC), stalk, and inner membrane complex (IMC) are labeled.
The mushroom-shaped OMCC has been successfully extracted from H. pylori, thereby allowing analysis of its composition and structure (15, 16). The extracted complex is composed of 5 main components (CagY, CagX, CagT, Cag3 and CagM) (15). Three of these components (CagY, CagX and CagT) are homologous to OMCC components of T4SSs in other bacterial species, whereas Cag3 and CagM are present exclusively in H. pylori (Table 1). CagY, CagX and CagM are each required for OMCC assembly or stability (14, 15). OMCCs produced by Cag3 or CagT mutants have a narrower diameter than wild-type complexes (14, 15), which suggests that Cag3 and CagT are localized in the periphery of the complex.
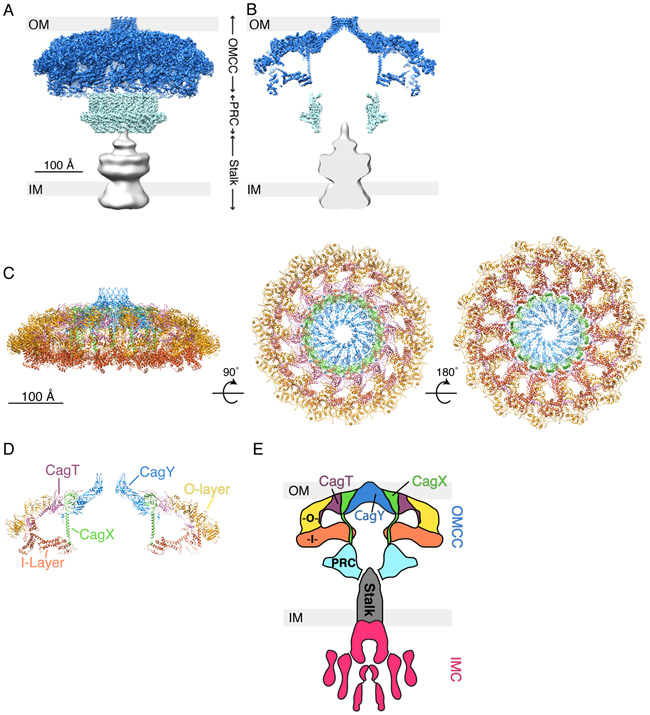
Recently a 3.5 Å three-dimensional structure of isolated Cag T4SS particles was determined by single particle cryo-EM analysis (16). The particles were described as three sub-assemblies: an OMCC with 41 nm diameter, a periplasmic ring complex (PRC) with 18.5 nm diameter, and a central stalk (Fig. 3A). The OMCC can be further subdivided into an O-layer and an I-layer. A large cavity about 270 Å wide is present within the structure, extending from the bottom of the PRC to the top of the OMCC, where the cavity tapers to a 35 Å opening (Fig. 3B). The overall structure of Cag T4SS particles extracted from H. pylori appears similar to the mushroom-shaped periplasmic structures observed by cryo-ET (13-16). Interestingly, the cryo-ET studies visualized a “plug” structure within the large central cavity between the PRC and OMCC, as well as peripheral periplasmic densities, which were not visible in the high-resolution 3D reconstruction resulting from single particle cryo-EM analysis of isolated T4SS particles.
Fig. 3.
3D structure of Cag T4SS particles extracted from H. pylori and determined by single particle cryo-EM. (A) 3D reconstruction of the Cag T4SS with the predicted position of the bacterial outer membrane (OM) and inner membrane (IM) shown (EMD-20023, 20020, and 20021) (16). Outer membrane core complex (OMCC), blue; periplasmic ring complex (PRC), cyan; and stalk, grey. (B) Central axial slice view of the 3D structure in panel A. (C) Secondary structure model of the Cag T4SS OMCC showing the positions of CagT (purple, PDB-6OEE), the C-terminal portion of CagX (green, PDB-6OEG), and the C-terminal portion of CagY (blue, PDB-6ODI), as well as unassigned outer-layer (O-layer, yellow, PDB-6OEF) and inner-layer (I-layer, orange, PDB-6OEH) densities (16). The structure is rotated 90° and then 180° around the X-axis. (D) Central slice view of the structure shown in panel C (left image). (E) Schematic summarizing our current understanding of Cag T4SS architecture, based on cryo-ET and single particle EM studies (13, 14, 16). The Cag T4SS is composed of an OMCC, periplasmic ring complex (PRC, cyan), stalk (grey), and inner membrane complex (IMC, magenta). The positions of CagT (purple) and portions of CagX (green) and CagY (blue) have been mapped into the OMCC. There are outer and inner layer densities (O, yellow and I, orange) that have yet to be assigned a specific T4SS component. The identities and position of T4SS components in the PRC and stalk also have not yet been structurally determined.
CagT, the C-terminal portion of CagY and the C-terminal portion of CagX are localized to the O-layer of the OMCC (Fig. 3). CagY localizes to the top of the OMCC, and a ring of CagY α-helices (two helices per protomer) is predicted to form a channel through the outer membrane. The structural features of putative transmembrane portions of CagY are markedly different from the typical β-barrel structures of most outer membrane proteins in Gram-negative bacteria. The exact protein composition of the I-layer, PRC, and central stalk has not yet been determined.
A recent model proposed that the Cag T4SS is built in a sequence beginning with assembly of a central cylinder (composed of CagY, CagX and maybe CagM), followed by incorporation of other OMCC components (CagT and Cag3), followed by assembly of the IMC (14). When viewed in cross-section, the OMCC has 14-fold symmetry, the PRC has 17-fold symmetry, and the IMC has 6-fold symmetry (14, 16). At present, very little is known about the protein-protein interactions that allow assembly of an apparatus with these multiple types of symmetry mismatch.
Structural comparisons of the H. pylori Cag T4SS and T4SSs in other bacterial species
Bacterial conjugation systems and the A. tumefaciens VirB/VirD4 T4SS are prototype T4SSs that have been studied extensively for several decades (Text Box 1)(61-64). In these systems, VirB7, VirB9 and VirB10 assemble into an OMCC, three ATPases and several other T4SS components localize to the inner membrane, and VirB2 and VirB5 localize to pilus structures (61-64). The OMCCs of conjugation systems encoded by pKM101 or R388 plasmids, the A. tumefaciens VirB/D4 system, and the Xanthomonas citri T4SS have been expressed in E. coli, extracted from the bacteria, and analyzed by single particle EM (electron microscopy) or crystallography (65-68). The OMCCs of these prototype T4SSs have outer and inner layers (O-layer and I-layer), each with 14-fold symmetry (65-68). VirB10 is predicted to span both the outer membrane and inner membrane. The outer membrane channel is formed by a portion of VirB10 (corresponding to a hydrophobic ring of two-helix bundles) (66). Lower resolution structures have been determined for a T4SS assembly encoded by the E. coli R388 conjugative plasmid, containing the OMCC connected by a stalk to an IMC (69). Within the IMC, 12 VirB4 ATPase subunits are organized as side-by-side hexameric barrels (69), and VirD4 localizes adjacent to VirB4 subunits (70).
The architecture of an F-plasmid-encoded T4SS (mediating bacterial conjugation) has been analyzed in intact bacteria using cryo-ET (71). Four distinct types of structures were visualized, presumably corresponding to different stages of T4SS biogenesis. The cryo-ET images provided an analysis of spatial relationships between the OMCC, IMC and pilus structures (71). In contrast to OMCCs of the prototype T4SSs described above, the OMCC of the F plasmid-encoded T4SS has 13-fold symmetry (71).
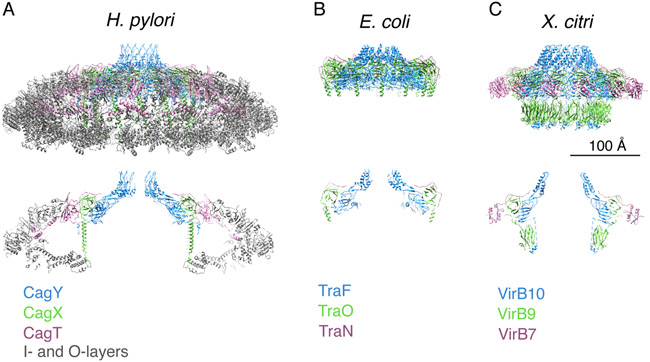
The H. pylori Cag T4SS exhibits structural similarities to the T4SSs described above, as well as notable differences (Fig. 4). Similarities include the overall shape, 14-fold symmetry of the OMCC, components with related sequences (Table 1), and conserved structural features of the portions of CagY or VirB10 homologs that span the outer membrane. Conversely, the diameter of the H. pylori Cag T4SS OMCC is roughly twice that of OMCC diameters in prototype T4SSs (Fig. 4), and the Cag T4SS OMCC contains two proteins (Cag3 and CagM) that are not present in prototype T4SSs (15, 16). The presence of a distinct periplasmic ring complex (PRC) and symmetry mismatch between the OMCC and PRC are additional features that differentiate the Cag T4SS from prototype T4SSs (16).
Fig. 4.
Structural comparison of the H. pylori Cag T4SS OMCC with minimized T4SSs from other bacterial species. (A-C) Upper panel, secondary structural models of the complexes. Lower panel, Central axial slice views of the secondary structural models. Scale bar, 100 Å. (A) OMCC from the H. pylori Cag T4SS (16). CagY (PDB-6ODI, blue), CagX (PDB-6OEG, green), CagT (PDB-6OEE, purple), and outer- and inner-layer proteins (PDB-6OEF and -6OEH, grey). (B) Conjugation system (encoded by pKM101) (66). TraF (PDB-3JQO, blue), TraO (PDB-3JQO, green), and TraN (PDB-3JQO, purple). (C) Xanthomonas citri T4SS (67). VirB10 (PDB-6GYB, blue), VirB9 (PDB-6GYB, green), and VirB7 (PDB-6GYB, purple).
The dimensions and number of components in the H. pylori Cag T4SS most closely resemble corresponding features of the Legionella pneumophila Dot/Icm T4SS (72, 73). The Legionella Dot/Icm potentially contains more than 20 components. Cryo-ET studies indicate that the OMCC of the Dot/Icm T4SS is about 42 nm in diameter (72, 73), which is similar to dimensions of the Cag T4SS OMCC and much larger than dimensions of OMCCs in prototype T4SSs. The Dot/Icm OMCC is predicted to contain at least 5 proteins (DotH, DotC, DotD, DotF and DotF). Cryo-ET analysis of the Dot/Icm T4SS suggests that it contains a subassembly similar to the PRC observed in the H. pylori Cag T4SS (72, 73), and there are also similarities in the structural organization of IMCs in the H. pylori Cag T4SS and the Legionella Dot/Icm T4SS (14, 72). In contrast to the 14-fold symmetry of the H. pylori Cag T4SS, the Legionella OMCC has 13-fold symmetry (72, 73). While the Cag T4SS and Dot/Icm T4SS exhibit multiple structural similarities, the sequences of individual proteins in these systems are highly divergent. One region of conserved sequence among OMCC components of the Legionella Dot/Icm T4SS, H. pylori Cag T4SS and prototype T4SSs is within the C-terminal portions of DotG/CagY/VirB10/TraF, which likely form a pore in the outer membrane (16).
“Pilus” structures associated with the H. pylori Cag T4SS
In response to contact with gastric epithelial cells, H. pylori produces extracellular filamentous structures (55, 74-82). An initial study reported that the structures were membrane-sheathed and produced more abundantly by bacteria co-cultured with cultured gastric epithelial cells than by bacteria cultured in the absence of epithelial cell contact (74). The structures were labeled “pili” based on the view that they were analogous to pilus components of conjugation systems and the A. tumefaciens VirB/VirD4 T4SS. Several studies reported that the pili are not produced by H. pylori strains lacking the cag PAI or harboring mutations in various genes encoding Cag T4SS components (74-76, 79, 80, 83). The dimensions of the structures described in various studies vary considerably (ranging from about 14 nm to 70 nm in width)(74, 76-79). Some studies reported that the structures are produced only when H. pylori is in contact with gastric epithelial cells, whereas other studies reported that the structures are also produced by bacteria in the absence of epithelial cell contact. A recent study reported that the structures form primarily when H. pylori contacts the basolateral surface of polarized epithelial cells, but not the apical surface (82). Several components of the Cag T4SS, including CagY, CagT and CagL, have been localized to the structures using immunogold EM (74, 76), and CagA has been localized to the tip of the structures (76, 78). The structures have never been isolated, so their biochemical composition has not been defined.
Cryo-ET methods were used recently to analyze H. pylori in contact with gastric epithelial cells, and membranous tube-like structures (about 37 nm in diameter) protruding from the outer membrane were visualized (13). The structures were produced by a wild-type strain but not a cag PAI mutant strain (13), and were observed only when the bacteria were co-cultured with gastric epithelial cells but not in bacteria grown in pure culture. Interestingly, the membranous tubes visualized by cryo-ET were not detected in direct association with T4SS OMCCs.
While many studies have concluded that the extracellular structures are pilus components of the Cag T4SS, several gaps in knowledge and apparent inconsistencies need to be addressed before this view can be fully accepted. First, a major pilin subunit has not been conclusively identified. Second, the lack of close proximity between the filaments and T4SS OMCCs in cryo-ET images (13) raises questions about whether there is any relationship between these structures. Finally, it is notable that H. pylori strains containing deletion or insertion mutations in several genes required for Cag T4SS activity (cagC, cagH and cagY) produce extracellular pilus structures, similar to a wild-type strain (79, 80, 83). CagY (a VirB10 homolog) would likely be required for export of pilus components (80, 83). In future studies, it will be important to define the composition of the pili, filaments or tubes, clarify if one or more than one type of structure is produced, and more rigorously determine the genetic requirements for production of these structures.
H. pylori Cag T4SS interactions with the surface of host cells
Relatively little is known about interactions between the Cag T4SS and host cells. CagL, a T4SS component localized to pilus structures and the bacterial surface, is recognized by TLR5 on host cells, resulting in flagellin-independent activation of TLR5 and NF-κB activation (84). In addition, multiple components of the T4SS (CagL, CagY and CagI) as well as CagA interact with integrins (α5β1 as well as other types), and it has been proposed that integrins might be host cell receptors for the T4SS (26, 76, 78, 85, 86). Interactions of the T4SS with integrins could potentially be mediated by either Cag proteins localized on the surface of H. pylori or by proteins localized to pilus structures. A ring of CagY α-helices localized at the apex of the OMCC is predicted to form a pore through the H. pylori outer membrane (16) and would likely be accessible for interactions with integrins or other host cell receptors. Integrins are typically localized on the basolateral surface of gastric epithelial cells, so it has been suggested that secreted H. pylori proteases may allow the bacteria to breach cell-cell junctions and gain access to integrins on the basolateral surface of cells (82). While multiple studies have detected interactions between the components of the Cag T4SS and integrins, one study reported that integrins were not required for CagA translocation, and instead, CEACAM proteins (receptors for the H. pylori HopQ protein) were required (87).
Whether components of the T4SS insert into the plasma membrane of host cells to form a channel for translocation of CagA and other H. pylori constituents is unknown. If a component of the T4SS inserts into the plasma membrane (analogous to the translocon of T3SSs), the protein responsible for such an action has not yet been identified. One model proposed that adherence of H. pylori to gastric epithelial cells stimulates externalization of phosphatidylserine to the outer leaflet of the plasma membrane, and proposed that binding of CagA to phosphatidylserine then triggers internalization of CagA through an energy-dependent process distinct from receptor-mediated endocytosis (88). In this model, CagA translocation through the bacterial envelope is proposed to be mediated by the Cag T4SS, and CagA internalization into host cells is proposed to occur without a requirement for T4SS-dependent permeabilization of the plasma membrane (88, 89).
Concluding remarks and future perspectives
Type IV secretion systems are complex molecular machines found in many bacterial species. Our current understanding of these machines has come mainly from studies of conjugation systems and the A. tumefaciens VirB/VirD4 T4SS. Although multiple features of these prototype T4SSs are broadly conserved, there is extensive variation in the structure and function of T4SSs from different bacterial species. The H. pylori Cag T4SS is of particular interest because of its role in the pathogenesis of gastric cancer. Over the past 25 years since the potential existence of a secretion system encoded by the cag PAI was first recognized (4, 5, 90), there has been a steady increase in our understanding of the Cag T4SS. Cryo-ET analyses have provided low resolution models of the Cag T4SS OMCC and IMC in the context of intact H. pylori cells (13, 14), and single particle cryo-EM analysis has provided a high resolution model of the OMCC and PRC, as well as a molecular model for the positions of CagY, CagX and CagT in the OMCC (16) (Fig. 3). These structural studies combined with biochemical and functional analyses provide a strong foundation for further investigation (see “Outstanding Questions”).
Outstanding questions.
How does the T4SS apparatus assemble? Where are individual proteins localized with the T4SS and what are their functions? If the Cag T4SS is a dynamic structure, how many structural states can be identified?
T4SSs found in H. pylori and Legionella are larger in size and more complex than prototype T4SSs. In what ways do increased size and complexity contribute to the function of these systems?
How are CagA and non-protein H. pylori constituents (LPS metabolites, peptidoglycan and DNA) recruited to the T4SS?
Does CagA transit through the central chamber within the T4SS? Is it translocated in a folded or unfolded state?
What are the stimuli associated with bacteria-host cell contact that trigger CagA secretion, and what are the corresponding regulatory mechanisms?
By what mechanisms do CagA and non-bacterial constituents transit the plasma membrane of host cells?
What is the role of extracellular structures (pili or membranous tubes) in T4SS-mediated processes?
What benefits are conferred to H. pylori by the Cag T4SS?
The Cag T4SS contributes to gastric cancer and peptic ulcer disease in patients colonized with H. pylori. Do H. pylori strains producing the Cag T4SS also confer health benefits to humans?
Assembly of the Cag T4SS is energetically costly and would be unfavorable unless the secretion system conferred substantial benefits to the bacteria. The Cag T4SS is not required for H. pylori colonization of the human stomach but it likely confers competitive advantages. In vitro studies suggest that CagA enhances the capacity of H. pylori to extract iron and other nutrients directly from polarized gastric epithelial cells (91, 92) or access nutrients by disrupting intercellular junctions (93), thereby promoting bacterial growth. Gastric inflammation stimulated by T4SS-dependent processes may also enhance availability of nutrients utilized by H. pylori. CagA and the Cag T4SS inhibit expression of β-defensin 1 and β-defensin 3, thereby allowing H. pylori to evade innate host defenses (94, 95). Enhanced ability to obtain nutrients and resist host defenses are two possible factors contributing to an increased density of gastric colonization observed with CagA-positive H. pylori strains compared to CagA-negative strains (96). Actions of the Cag T4SS might allow H. pylori to colonize expanded gastric niches (for example, the gastric corpus)(50). In addition, T4SS-dependent actions can potentially lead to alterations in the gastric microbiome or intestinal microbiome (97, 98). Collectively, actions of the Cag T4SS are likely to enhance H. pylori growth in the stomach and enhance transmission of H. pylori to new hosts. In future studies, it will be important to further delineate mechanisms by which the Cag T4SS confers benefits to H. pylori. In addition, it will be important to develop a clearer understanding of not only the mechanisms by which Cag T4SS-positive H. pylori strains contribute to the pathogenesis of gastric cancer and peptic ulcer disease, but also mechanisms by which these strains potentially confer health benefits to human hosts.
Highlights.
The Cag T4SS has a key role in the pathogenesis of H. pylori-associated gastric cancer.
Recent advances in molecular imaging have facilitated investigation of H. pylori Cag T4SS structure.
The H. pylori Cag T4SS contains multiple components unrelated to components of T4SSs in other bacterial species, and the molecular architecture of the H. pylori Cag T4SS is substantially more complex than that of prototype T4SSs.
T4SSs are a heterogenous group of secretion systems with diverse functions. Despite a low level of sequence relatedness among corresponding components of T4SSs from different bacterial species, there are shared structural features among all T4SSs analyzed thus far.
Acknowledgments:
Supported by the National Institutes of Health (AI118932, CA116087, AI039657) and the Department of Veterans Affairs (1I01BX004447).
Glossary
- Helicobacter pylori
Gram negative bacteria that colonize the human stomach
- cag pathogenicity island (PAI)
a region of chromosomal DNA, present in some H. pylori strains but not others, which contains genes encoding CagA and components of a T4SS
- T4SS (type IV secretion system)
a diverse family of bacterial secretion systems with multiple possible functions (for example, delivery of effector proteins into target cells or bacterial conjugation)
- Cag T4SS
a T4SS composed of proteins encoded by genes in the H. pylori cag PAI
- CagA
the only known effector protein transported by the Cag T4SS, designated as a bacterial oncoprotein
- Interleukin-8 (IL-8)
a proinflammatory cytokine
- Dot/Icm T4SS
a type of T4SS present in Legionella pneumophila (causative agent of Legionnaire’s disease)
- VirB/VirD4 T4SS
a prototype form of T4SS present in Agrobacterium tumefaciens (which delivers T-DNA into plant cells, resulting in crown gall disease)
- Conjugation systems
class of T4SSs (composed of Tra components) that mediate DNA exchange between bacteria
- Outer membrane core complex (OMCC)
a T4SS subassembly localized within the periplasm, in contact with the outer membrane
- Periplasmic ring complex (PRC)
a T4SS subassembly localized within the periplasm
- Inner membrane complex (IMC)
a T4SS subassembly containing ATPases, localized in association with the inner membrane and extending into the cytoplasm
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-analysis. Gastroenterology doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. [DOI] [PubMed] [Google Scholar]
- 4.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci 93:14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Molecular Microbiology 28:37–53. [DOI] [PubMed] [Google Scholar]
- 6.Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. 2010. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet 6:e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res 55:2111–5. [PubMed] [Google Scholar]
- 8.Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, Capelinha AF, Quint W, Caldas C, van Doorn LJ, Carneiro F, Sobrinho-Simoes M. 2002. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Natl Cancer Inst 94:1680–7. [DOI] [PubMed] [Google Scholar]
- 9.Plummer M, van Doorn LJ, Franceschi S, Kleter B, Canzian F, Vivas J, Lopez G, Colin D, Munoz N, Kato I. 2007. Helicobacter pylori cytotoxin-associated genotype and gastric precancerous lesions. J Natl Cancer Inst 99:1328–34. [DOI] [PubMed] [Google Scholar]
- 10.Cover TL. 2016. Helicobacter pylori Diversity and Gastric Cancer Risk. MBio 7:e01869–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol 42:1337–48. [DOI] [PubMed] [Google Scholar]
- 12.Fischer W 2011. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J 278:1203–12. [DOI] [PubMed] [Google Scholar]
- 13.Chang YW, Shaffer CL, Rettberg LA, Ghosal D, Jensen GJ. 2018. In Vivo Structures of the Helicobacter pylori cag Type IV Secretion System. Cell Rep 23:673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, Khara P, Song L, Lin AS, Frick-Cheng AE, Harvey ML, Cover TL, Christie PJ. 2019. In Situ Molecular Architecture of the Helicobacter pylori Cag Type IV Secretion System. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL. 2016. Molecular and Structural Analysis of the Helicobacter pylori cag Type IV Secretion System Core Complex. MBio 7:e02001–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung JM, Sheedlo MJ, Campbell AM, Sawhney N, Frick-Cheng AE, Lacy DB, Cover TL, Ohi MD. 2019. Structure of the Helicobacter pylori Cag type IV secretion system. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cover TL, Dooley CP, Blaser MJ. 1990. Characterization of and human serologic response to proteins in Helicobacter pylori broth culture supernatants with vacuolizing cytotoxin activity. Infect Immun 58:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crabtree JE, Taylor JD, Wyatt JI, Heatley RV, Shallcross TM, Tompkins DS, Rathbone BJ. 1991. Mucosal IgA recognition of Helicobacter pylori 120 kDa protein, peptic ulceration, and gastric pathology. Lancet 338:332–5. [DOI] [PubMed] [Google Scholar]
- 19.Tummuru MK, Cover TL, Blaser MJ. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun 61:1799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci 90:5791–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cover TL, Blanke SR. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol 3:320–32. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama M 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer 4:688–94. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama M 2014. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15:306–16. [DOI] [PubMed] [Google Scholar]
- 24.Tegtmeyer N, Neddermann M, Asche CI, Backert S. 2017. Subversion of host kinases: a key network in cellular signaling hijacked by Helicobacter pylori CagA. Mol Microbiol 105:358–372. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi T, Senda M, Morohashi H, Higashi H, Horio M, Kashiba Y, Nagase L, Sasaya D, Shimizu T, Venugopalan N, Kumeta H, Noda NN, Inagaki F, Senda T, Hatakeyama M. 2012. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe 12:20–33. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan-Turkoz B, Jimenez-Soto LF, Dian C, Ertl C, Remaut H, Louche A, Tosi T, Haas R, Terradot L. 2012. Structural insights into Helicobacter pylori oncoprotein CagA interaction with beta1 integrin. Proc Natl Acad Sci U S A 109:14640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hohlfeld S, Pattis I, Puls J, Plano GV, Haas R, Fischer W. 2006. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol 59:1624–37. [DOI] [PubMed] [Google Scholar]
- 28.Schindele F, Weiss E, Haas R, Fischer W. 2016. Quantitative analysis of CagA type IV secretion by Helicobacter pylori reveals substrate recognition and translocation requirements. Mol Microbiol 100:188–203. [DOI] [PubMed] [Google Scholar]
- 29.Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA 96:14559–14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497–500. [DOI] [PubMed] [Google Scholar]
- 31.Stein M, Rappuoli R, Covacci A. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci U S A 97:1263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M, Yamada G, Azuma T, Hatakeyama M. 2008. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A 105:1003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neal JT, Peterson TS, Kent ML, Guillemin K. 2013. H. pylori virulence factor CagA increases intestinal cell proliferation by Wnt pathway activation in a transgenic zebrafish model. Dis Model Mech 6:802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wandler AM, Guillemin K. 2012. Transgenic expression of the Helicobacter pylori virulence factor CagA promotes apoptosis or tumorigenesis through JNK activation in Drosophila. PLoS Pathog 8:e1002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N, Salama N, Churin Y, Meyer TF. 2013. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol 15:1896–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G, Romero-Gallo J, Benoit SL, Piazuelo MB, Dominguez RL, Morgan DR, Peek RM Jr., Maier RJ. 2016. Hydrogen Metabolism in Helicobacter pylori Plays a Role in Gastric Carcinogenesis through Facilitating CagA Translocation. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5:1166–74. [DOI] [PubMed] [Google Scholar]
- 38.Stein SC, Faber E, Bats SH, Murillo T, Speidel Y, Coombs N, Josenhans C. 2017. Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLoS Pathog 13:e1006514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gall A, Gaudet RG, Gray-Owen SD, Salama NR. 2017. TIFA Signaling in Gastric Epithelial Cells Initiates the cag Type 4 Secretion System-Dependent Innate Immune Response to Helicobacter pylori Infection. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmermann S, Pfannkuch L, Al-Zeer MA, Bartfeld S, Koch M, Liu J, Rechner C, Soerensen M, Sokolova O, Zamyatina A, Kosma P, Maurer AP, Glowinski F, Pleissner KP, Schmid M, Brinkmann V, Karlas A, Naumann M, Rother M, Machuy N, Meyer TF. 2017. ALPK1- and TIFA-Dependent Innate Immune Response Triggered by the Helicobacter pylori Type IV Secretion System. Cell Rep 20:2384–2395. [DOI] [PubMed] [Google Scholar]
- 41.Pfannkuch L, Hurwitz R, Traulsen J, Sigulla J, Poeschke M, Matzner L, Kosma P, Schmid M, Meyer TF. 2019. ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J 33:9087–9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varga MG, Shaffer CL, Sierra JC, Suarez G, Piazuelo MB, Whitaker ME, Romero-Gallo J, Krishna US, Delgado A, Gomez MA, Good JA, Almqvist F, Skaar EP, Correa P, Wilson KT, Hadjifrangiskou M, Peek RM. 2016. Pathogenic Helicobacter pylori strains translocate DNA and activate TLR9 via the cancer-associated cag type IV secretion system. Oncogene 35:6262–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couturier MR, Tasca E, Montecucco C, Stein M. 2006. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun 74:273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattis I, Weiss E, Laugks R, Haas R, Fischer W. 2007. The Helicobacter pylori CagF protein is a type IV secretion chaperone-like molecule that binds close to the C-terminal secretion signal of the CagA effector protein. Microbiology 153:2896–909. [DOI] [PubMed] [Google Scholar]
- 45.Jurik A, Hausser E, Kutter S, Pattis I, Prassl S, Weiss E, Fischer W. 2010. The coupling protein Cagbeta and its interaction partner CagZ are required for type IV secretion of the Helicobacter pylori CagA protein. Infect Immun 78:5244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonsor DA, Weiss E, Iosub-Amir A, Reingewertz TH, Chen TW, Haas R, Friedler A, Fischer W, Sundberg EJ. 2013. Characterization of the translocation-competent complex between the Helicobacter pylori oncogenic protein CagA and the accessory protein CagF. J Biol Chem 288:32897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin AS, Dooyema SDR, Frick-Cheng AE, Harvey ML, Suarez G, Loh JT, McDonald WH, McClain MS, Peek RM Jr., Cover TL. 2020. Bacterial Energetic Requirements for Helicobacter pylori Cag Type IV Secretion System-Dependent Alterations in Gastric Epithelial Cells. Infect Immun 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohrer S, Holsten L, Weiss E, Benghezal M, Fischer W, Haas R. 2012. Multiple pathways of plasmid DNA transfer in Helicobacter pylori. PLoS One 7:e45623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogura K, Maeda S, Nakao M, Watanabe T, Tada M, Kyutoku T, Yoshida H, Shiratori Y, Omata M. 2000. Virulence factors of Helicobacter pylori responsible for gastric diseases in mongolian gerbil. J Exp Med 192:1601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rieder G, Merchant JL, Haas R. 2005. Helicobacter pylori cag-type IV secretion system facilitates corpus colonization to induce precancerous conditions in Mongolian gerbils. Gastroenterology 128:1229–42. [DOI] [PubMed] [Google Scholar]
- 51.Shibata W, Hirata Y, Maeda S, Ogura K, Ohmae T, Yanai A, Mitsuno Y, Yamaji Y, Okamoto M, Yoshida H, Kawabe T, Omata M. 2006. CagA protein secreted by the intact type IV secretion system leads to gastric epithelial inflammation in the Mongolian gerbil model. J Pathol 210:306–14. [DOI] [PubMed] [Google Scholar]
- 52.Wiedemann T, Loell E, Mueller S, Stoeckelhuber M, Stolte M, Haas R, Rieder G. 2009. Helicobacter pylori cag-Pathogenicity island-dependent early immunological response triggers later precancerous gastric changes in Mongolian gerbils. PLoS ONE 4:e4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM Jr. 2008. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res 68:379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM Jr., Algood HM, Cover TL. 2013. High Dietary Salt Intake Exacerbates Helicobacter pylori-Induced Gastric Carcinogenesis. Infect Immun 81:2258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC, Tan S, Morgan DR, Wilson KT, Bravo LE, Correa P, Cover TL, Amieva MR, Peek RM Jr. 2013. Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J Clin Invest 123:479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mimuro H, Suzuki T, Nagai S, Rieder G, Suzuki M, Nagai T, Fujita Y, Nagamatsu K, Ishijima N, Koyasu S, Haas R, Sasakawa C. 2007. Helicobacter pylori dampens gut epithelial self-renewal by inhibiting apoptosis, a bacterial strategy to enhance colonization of the stomach. Cell Host Microbe 2:250–63. [DOI] [PubMed] [Google Scholar]
- 57.Fox JG, Wang TC, Rogers AB, Poutahidis T, Ge Z, Taylor N, Dangler CA, Israel DA, Krishna U, Gaus K, Peek RM Jr. 2003. Host and microbial constituents influence Helicobacter pylori-induced cancer in a murine model of hypergastrinemia. Gastroenterology 124:1879–90. [DOI] [PubMed] [Google Scholar]
- 58.Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, Muller A. 2011. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology 140:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wroblewski LE, Choi E, Petersen C, Delgado AG, Piazuelo MB, Romero-Gallo J, Lantz TL, Zavros Y, Coffey RJ, Goldenring JR, Zemper AE, Peek RM Jr. 2019. Targeted mobilization of Lrig1(+) gastric epithelial stem cell populations by a carcinogenic Helicobacter pylori type IV secretion system. Proc Natl Acad Sci U S A 116:19652–19658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, Passarelli B, Camorlinga M, Bouley DM, Alvarez G, Nusse R, Torres J, Amieva MR. 2015. Helicobacter pylori Activates and Expands Lgr5(+) Stem Cells Through Direct Colonization of the Gastric Glands. Gastroenterology 148:1392–404 e21. [DOI] [PubMed] [Google Scholar]
- 61.Grohmann E, Christie PJ, Waksman G, Backert S. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107:455–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galan JE, Waksman G. 2018. Protein-Injection Machines in Bacteria. Cell 172:1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waksman G 2019. From conjugation to T4S systems in Gram-negative bacteria: a mechanistic biology perspective. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li YG, Hu B, Christie PJ. 2019. Biological and Structural Diversity of Type IV Secretion Systems. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sgro GG, Costa TRD, Cenens W, Souza DP, Cassago A, Coutinho de Oliveira L, Salinas RK, Portugal RV, Farah CS, Waksman G. 2018. Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri. Nat Microbiol 3:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gordon JE, Costa TRD, Patel RS, Gonzalez-Rivera C, Sarkar MK, Orlova EV, Waksman G, Christie PJ. 2017. Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol Microbiol 105:273–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Redzej A, Ukleja M, Connery S, Trokter M, Felisberto-Rodrigues C, Cryar A, Thalassinos K, Hayward RD, Orlova EV, Waksman G. 2017. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J 36:3080–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu B, Khara P, Christie PJ. 2019. Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli. Proc Natl Acad Sci U S A 116:14222–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chetrit D, Hu B, Christie PJ, Roy CR, Liu J. 2018. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol 3:678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghosal D, Jeong KC, Chang YW, Gyore J, Teng L, Gardner A, Vogel JP, Jensen GJ. 2019. Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat Microbiol 4:1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49:219–34. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka J, Suzuki T, Mimuro H, Sasakawa C. 2003. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol 5:395–404. [DOI] [PubMed] [Google Scholar]
- 76.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862–6. [DOI] [PubMed] [Google Scholar]
- 77.Couturier MR, Stein M. 2008. Helicobacter pylori produces unique filaments upon host contact in vitro. Can J Microbiol 54:537–48. [DOI] [PubMed] [Google Scholar]
- 78.Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. 2009. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog 5:e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog 7:e1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, Cariaga TA, Suarez G, Peek RM Jr., Cover TL, Solnick JV. 2013. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog 9:e1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaddy JA, Radin JN, Loh JT, Piazuelo MB, Kehl-Fie TE, Delgado AG, Ilca FT, Peek RM, Cover TL, Chazin WJ, Skaar EP, Scott Algood HM. 2014. The Host Protein Calprotectin Modulates the Helicobacter pylori cag Type IV Secretion System via Zinc Sequestration. PLoS Pathog 10:e1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT, Asche CI, Boehm M, Loessner H, Figueiredo C, Naumann M, Palmisano R, Solcia E, Ricci V, Backert S. 2017. Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery. Cell Host Microbe 22:552–560 e5. [DOI] [PubMed] [Google Scholar]
- 83.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. 2014. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun 82:3457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pachathundikandi SK, Tegtmeyer N, Arnold IC, Lind J, Neddermann M, Falkeis-Veits C, Chattopadhyay S, Bronstrup M, Tegge W, Hong M, Sticht H, Vieth M, Muller A, Backert S. 2019. T4SS-dependent TLR5 activation by Helicobacter pylori infection. Nat Commun 10:5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koelblen T, Berge C, Cherrier MV, Brillet K, Jimenez-Soto L, Ballut L, Takagi J, Montserret R, Rousselle P, Fischer W, Haas R, Fronzes R, Terradot L. 2017. Molecular dissection of protein-protein interactions between integrin alpha5beta1 and the Helicobacter pylori Cag type IV secretion system. FEBS J 284:4143–4157. [DOI] [PubMed] [Google Scholar]
- 86.Skoog EC, Morikis VA, Martin ME, Foster GA, Cai LP, Hansen LM, Li B, Gaddy JA, Simon SI, Solnick JV. 2018. CagY-Dependent Regulation of Type IV Secretion in Helicobacter pylori Is Associated with Alterations in Integrin Binding. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao Q, Busch B, Jimenez-Soto LF, Ishikawa-Ankerhold H, Massberg S, Terradot L, Fischer W, Haas R. 2018. Integrin but not CEACAM receptors are dispensable for Helicobacter pylori CagA translocation. PLoS Pathog 14:e1007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murata-Kamiya N, Kikuchi K, Hayashi T, Higashi H, Hatakeyama M. 2010. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe 7:399–411. [DOI] [PubMed] [Google Scholar]
- 89.Tohidpour A, Gorrell RJ, Roujeinikova A, Kwok T. 2017. The Middle Fragment of Helicobacter pylori CagA Induces Actin Rearrangement and Triggers Its Own Uptake into Gastric Epithelial Cells. Toxins (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tummuru MK, Sharma SA, Blaser MJ. 1995. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol 18:867–76. [DOI] [PubMed] [Google Scholar]
- 91.Tan S, Tompkins LS, Amieva MR. 2009. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog 5:e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan S, Noto JM, Romero-Gallo J, Peek RM Jr., Amieva MR. 2011. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog 7:e1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bauer B, Pang E, Holland C, Kessler M, Bartfeld S, Meyer TF. 2012. The Helicobacter pylori virulence effector CagA abrogates human beta-defensin 3 expression via inactivation of EGFR signaling. Cell Host Microbe 11:576–86. [DOI] [PubMed] [Google Scholar]
- 95.Patel SR, Smith K, Letley DP, Cook KW, Memon AA, Ingram RJ, Staples E, Backert S, Zaitoun AM, Atherton JC, Robinson K. 2013. Helicobacter pylori downregulates expression of human beta-defensin 1 in the gastric mucosa in a type IV secretion-dependent fashion. Cell Microbiol 15:2080–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Atherton JC, Tham KT, Peek RM Jr., Cover TL, Blaser MJ. 1996. Density of Helicobacter pylori infection in vivo as assessed by quantitative culture and histology. J Infect Dis 174:552–6. [DOI] [PubMed] [Google Scholar]
- 97.Jones TA, Hernandez DZ, Wong ZC, Wandler AM, Guillemin K. 2017. The bacterial virulence factor CagA induces microbial dysbiosis that contributes to excessive epithelial cell proliferation in the Drosophila gut. PLoS Pathog 13:e1006631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noto JM, Zackular JP, Varga MG, Delgado A, Romero-Gallo J, Scholz MB, Piazuelo MB, Skaar EP, Peek RM Jr. 2019. Modification of the Gastric Mucosal Microbiota by a Strain-Specific Helicobacter pylori Oncoprotein and Carcinogenic Histologic Phenotype. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–59. [DOI] [PubMed] [Google Scholar]
- 100.Gonzalez-Rivera C, Bhatty M, Christie PJ. 2016. Mechanism and Function of Type IV Secretion During Infection of the Human Host. Microbiol Spectr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Christie PJ. 2016. The Mosaic Type IV Secretion Systems. EcoSal Plus 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Christie PJ, Gomez Valero L, Buchrieser C. 2017. Biological Diversity and Evolution of Type IV Secretion Systems. Curr Top Microbiol Immunol 413:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]