Abstract
Poor adherence is associated with worse disease outcomes. Pharmacogenomics provides a possible intervention to address adherence. We hypothesized that pharmacogenomic-informed care could increase adherence. Patients in a prospective case-control study underwent preemptive pharmacogenomic genotyping with results available for provider use at the point-of-care; controls (not genotyped) were treated by the same providers. Over 6,000 e-prescriptions for 39 medications with actionable pharmacogenomic information were analyzed. Composite adherence, measured by modified proportion of days covered (mPDC), was compared between cases/controls and genomically-concordant vs genomically-higher-risk medications. Overall, 536 patients were included. No difference in mean mPDC was observed due to availability of pharmacogenomic guidance. However, case patients prescribed high-risk pharmacogenomic medications were more than twice as likely to have low mPDC for these medications compared to genomically-concordant prescriptions (odds ratio=2.4 (1.03–5.74), p<0.05). This study is the first to show that composite pharmacogenomic information predicts adherence.
Keywords: pharmacogenomics, adherence, implementation, precision medicine
Introduction
Poor adherence to medications has been associated with worsening disease progression, death and increased health care costs.1–7 At least a third of all medication-related hospital admissions in the United States are due to poor medication adherence, with a resultant cost of approximately 100 billion dollars a year.2 Poor adherence has been found to be particularly harmful for patients with chronic conditions. Diabetes, hypertension, hypercholesterolemia and chronic heart failure are all examples of conditions in which higher hospitalization rates were found in patients with low medication adherence.3 Thus, the full benefits of effective medications for these chronic diseases have not been realized, as approximately fifty percent of such patients do not take their medications as prescribed.6,8 Multiple randomized controlled trials have shown that past clinical interventions to improve adherence have had relatively low success rates, with only marginal improvements in the simplification of drug regimens and convenience of care.9,10
There is limited and mixed evidence that pharmacogenomic information may improve adherence. Research into pharmacogenomics and its clinical implementation aims to increase the efficacy of medications through personalized prescribing based on patient-specific genetic risk factors. Genetic variation among patients has been shown to have a significant impact on risk of efficacy and toxicity for many medications.11–13 The personal and interactive nature of pharmacogenomic-informed care may help address key factors that lead to poor medication adherence, such as perceived need/efficacy, cost, concerns about side effects and limited patient engagement in treatment decisions.14 This fostering of confidence and engagement can have a positive impact on medication-taking behavior.14 In fact, two studies looking at statin adherence saw improvements in patients provided with genetic information of a singular risk variant.15,16 However, there is currently no data on how pharmacogenomic information impacts patient adherence spanning multiple medical conditions. To increase access and use of personalized patient pharmacogenomic information, multiple hospitals have developed electronic medical record-embedded clinical decision support (CDS) tools that convey pharmacogenomic information and complement physician medication decision making. The University of Chicago’s Genomic Prescribing System (GPS) is an example of such a tool that has been used in both inpatient and outpatient settings.1,17 This tool1 delivers pharmacogenomic test results at the point-of-care (along with accompanying clinical decision support guidance) using a traffic light system such that genomically-congruent (favorable) medications are denoted by a green light, medications that are considered to be of moderate pharmacogenomic risk are denoted by a yellow light, and genomically-incongruent (high-risk) medications are denoted by a red light18. The number of medications with actionable pharmacogenomic results and decision supports in GPS has grown over time, and is based on those meeting a rigorous evidence evaluation standard19,20 along with consideration of and concordance with existing external guidance standards (e.g., Clinical Pharmacogenetics Implementation Consortium, PharmGKB, Dutch Pharmacogenetics Working Group)21.
This study aimed to show that the utilization of a healthcare delivery model that incorporates information on multiple risk variants to assist physician prescribing offers the potential to assess the possible larger impact of pharmacogenomics on medication adherence. We hypothesized that providing personalized, genomic-informed care—through the use of a pharmacogenomic CDS tool—would significantly increase patient composite adherence rates to prescribed medications, as compared with standard outpatient care.
Methods
Participants and Study Design
Patients for this study were identified from a large multi-center outpatient visit dataset collected from the already-established 1200 Patients Project (clinicaltrials.gov #NCT01280825). The 1200 Patients Project aimed to study the feasibility of incorporating pharmacogenomic testing into routine medical care and represented eight different medicine primary care and subspecialty clinics from two metropolitan outpatient locations.1 The project served as a prospective case-control institutional implementation project in which patient pharmacogenomic information was made available to enrolled physicians for clinical decision assistance. All enrolled case patients were genotyped according to the 1200 Patients Project clinical study protocol1 with use of a broad custom pharmacogenomic panel and a separate panel developed in conjunction with Hologic22 CYP2D6 assessment, plus ThermoFisher TaqMan copy number assay23. Control patients, who were not genotyped but received care from the same enrolled study providers, were concurrently enrolled. These cohorts provided populations in which medication adherence could be assessed in the contexts of pharmacogenomic-informed versus non-informed (usual) care. The study was approved by the Institutional Review Board of the University of Chicago.
Evaluable Encounters and Data Collection
Consented providers were trained on the use of the pharmacogenomic software tool, the GPS19 and agreed to have their prescribing decisions tracked/analyzed. GPS presents easily-decipherable patient-specific pharmacogenomic results using traffic light iconography to represent medications that are high-risk (red light), cautionary (yellow light), or favorable (green light) based on the patient’s genomics, along with CDS summaries.
Evaluable encounters included all visits in which an enrolled patient saw an enrolled provider. Among case patients, inclusion criteria to define an evaluable patient for this analysis also required that pharmacogenomic results were available for consideration through GPS (i.e., the genotyping results could not be pending) and that the provider accessed the GPS for at least one clinic visit for that patient. Click log data detailing provider navigations into and within GPS were recorded. These data included login date/times and alert colors of prescribed drugs at each visit.
Available health records including medication data were obtained for this study from the University of Chicago Clinical Research Data Warehouse for enrolled 1200 Patient Project patients between October 1, 2012 and June 1, 2017. This dataset included patient demographics and e-prescription records. The e-prescription records detailed the medications and dosages prescribed, prescription start and end dates, amounts of medications dispensed, number of refills prescribed, prescription instructions, and ordering physician e-signatures. Inclusion criteria for this study required patients to have e-prescription data for at least one of the 39 pharmacogenomically-actionable chronic medications available in GPS (Table S1; and Figure 1 for study overview). The process for inclusion of specific medications as ‘pharmacogenomically actionable’ and deployed in GPS has been extensively previously described19,20, but the incremental implementation of certain medications also had a practical purposefulness in that the medications most relevant to our initial 1200 Patients Project cohort were prioritized first. This means that some known pharmacogenomically actionable medications (e.g., psychiatry medications, transplant medications, pediatric oncology medications) were not implemented in GPS at the time of the clinical evaluation of the cohort evaluated in this study because those populations were not eligible for genotyping. To account for this and further enhance the generalizability of our current findings, we have performed additional analyses focused on medications in GPS that also have CPIC Level A or B designations (as described below).
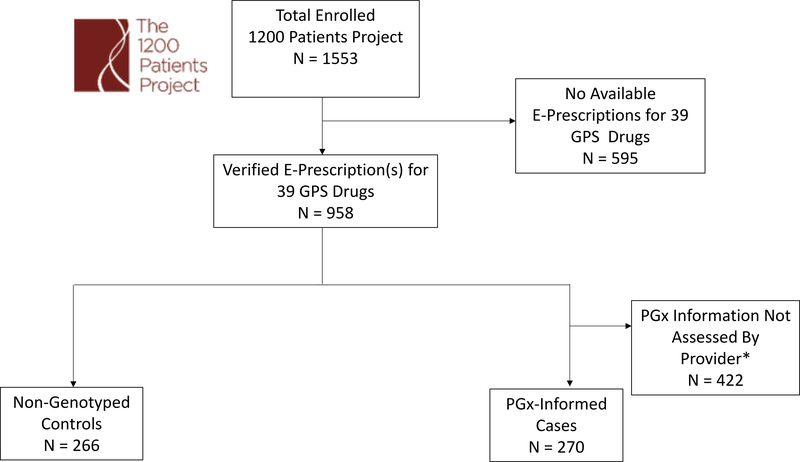
Figure 1: CONSORT Diagram Showing Patient Inclusion into the Study.
E-Prescription data were obtained for all patients enrolled in the 1200 Patients Project1. Patients without e-prescriptions for at least one of the 39 pharmacogenomic actionable medications were excluded. Genotyped cases were further excluded if their physician never accessed their pharmacogenomic information. *As part of the 1200 Patients project, providers were not required to access patient pharmacogenomic results at clinic visits.
Assessment of Medication Adherence
Medication adherence was assessed using e-prescription records and was based on proportion of days covered (PDC), or the total number of days covered with medication (starting from the e-prescription index date) divided by the total time period measured (in days). PDC has been previously utilized extensively as a measure of adherence and has been cited as the most commonly applied method for assessing adherence retrospectively using electronic medical record data of those with chronic conditions.24–27 PDC provides a more conservative estimate of adherence compared to medication possession ratio (MPR), as it takes into account oversupply instances where additional prescribing occurs before previous prescription supply has run out.28
For our analyses, we used a modified PDC calculation (mPDC):
** If the number of medication dispensed was greater than the length of its given e-prescribe order and a subsequent prescription for the given medication was e-prescribed, the number of medication dispensed was set to the length of the e-prescription order. In periods of time in between e-prescription orders, any medication supplies that were numerically possibly remaining from a previous prescription order were included in the mPDC calculation.
Individual patient medication-specific mPDC values were calculated and then averaged in order to obtain an overall composite mPDC that described a given patient’s total medication adherence for the measured medications.
Statistical Analyses
mPDC were compared between cases and controls, and between patients prescribed genomically-concordant (green light) versus genomically-incongruent (yellow/red light, i.e. higher risk) medications. mPDC was also assessed prior to the pharmacogenomic intervention for case patients and was compared to mPDC after implementation. Multivariate logistic regression evaluated the relationship between adherence, pharmacogenomic intervention, and genetic risk. Our study’s primary outcome was overall composite mPDC for the 39 pharmacogenomically-actionable medications available through our institutional implementation program via the GPS. Secondary analyses were conducted assessing mPDC for pharmacogenomic medications in GPS that also had existing CPIC Level A or B designations (https://cpicpgx.org/genes-drugs/; accessed October 1, 2019).
Patient demographics were reported as means and standard deviations for continuous data and percentages for categorical data. Statistical comparisons for continuous variables were performed using two-sided t-tests, and comparisons for categorical variables were analyzed using Pearson’s Chi-squared or Fisher’s exact test. Variables found to be statistically different between the genotyped and control groups or variables that have previously been reported to affect medication adherence15,16 were considered for multivariate logistic regression models. These covariates included age, gender, race, ethnicity, education attainment, and total number of medications. P-values were considered significant if less than 0.05. Analyses were performed using RStudio’s software package (Version 0.98.1091, RStudio Inc., Boston, MA, USA).
Results
Patient Characteristics
Among the 1553 patients enrolled from the University of Chicago institutional pharmacogenomic implementation project known as The 1200 Patients Project (clinicaltrials.gov #NCT01280825), 958 (62%) had e-prescription data for at least one of the 39 pharmacogenomically-actionable medications available in GPS. Of these patients, 703 (73%) were from the study’s pharmacogenomic-informed cohort (“cases”) and 266 (27%) patients were from the non-genotyped cohort (“controls”). The control cohort was comprised of patients who, while not preemptively genotyped, were treated by the same study providers. These patients with available e-prescription data reflected the composition of the overall 1200 Patients Project population (1150 [74%] cases and 403 [26%] controls). Since the purpose of this study was to examine the potential influence of delivered pharmacogenomic results/decision-supports, the pre-specified study exclusion criteria required 422 genotyped patients to be excluded from analysis. This group included a majority who never returned for a second clinic visit (i.e., they were enrolled at a first clinic visit but then never sought care at our medical center again), an additional large number whose pharmacogenomic results were pending at the time of any subsequent visit(s), and patients for whom results were available but whose providers never accessed their GPS results during the study period. It is notable that the exclusion of these patients from the primary and secondary adherence analyses of this study is justified because most of these patients were not longitudinally followed (i.e., most were only seen at one visit and thus adherence data over time are unavailable). This resulted in 270 pharmacogenomic-informed case patients and 266 non-genotyped control patients for the final analysis (CONSORT diagram, Figure 1).
Cases and controls were demographically similar (Table 1). There were no significant differences in gender, race, ethnicity, or education level attained. Cases were, on average, older than controls (68.8 versus 64.1 years, respectively, p<0.05). Cases were also taking slightly more total medications compared to controls (4.6 versus 3.9, respectively, p<0.05), but fewer medications with actionable pharmacogenomic information (1.8 versus 2.1, respectively, p<0.05).
Table 1.
Patient Demographics
| Characteristic | PGx-Informed Cases (n = 270) (%) | Non-Genotyped Controls (n = 266) (%) | p value |
|---|---|---|---|
| Age in years, mean (SD) | 68.8 (13.3) | 64.1 (14.9) | < 0.05 |
| Male, n (%) | 130 (48.1) | 136 (51.1) | 0.49 |
| Race | 0.53 | ||
| White | 141 (52.2) | 143 (53.8) | |
| Black or African American | 116 (43.0) | 105 (39.5) | |
| Asian/Mideast Indian | 8 (3.0) | 8 (3.0) | |
| Other | 5 (1.8) | 10 (3.2) | |
| Ethnicity | 0.36 | ||
| Not Hispanic or Latino | 259 (95.9) | 254 (95.5) | |
| Hispanic or Latino | 10 (3.7) | 8 (3.0) | |
| Not answered/unknown | 1 (0.4) | 4 (1.5) | |
| Education attainment | 0.33 | ||
| < High School | 8 (3.0) | 10 (3.8) | |
| High School/GED | 42 (15.6) | 37 (13.9) | |
| Some College | 57 (21.1) | 65 (24.4) | |
| College Graduate | 57 (21.1) | 68 (25.6) | |
| Graduate School | 102 (37.8) | 79 (29.7) | |
| Unknown | 4 (1.4) | 7 (2.6) | |
| No. of total medications, mean (SD) | 4.6 (2.3) | 3.9 (2.5) | < 0.05 |
| No. of PGx medications, mean (SD) | 1.8 (1.0) | 2.1 (1.2) | < 0.05 |
| No. of patients with low adherence | 33 | 45 |
GED, general education diploma; PGx, Pharmacogenomics
Medication Adherence Measures
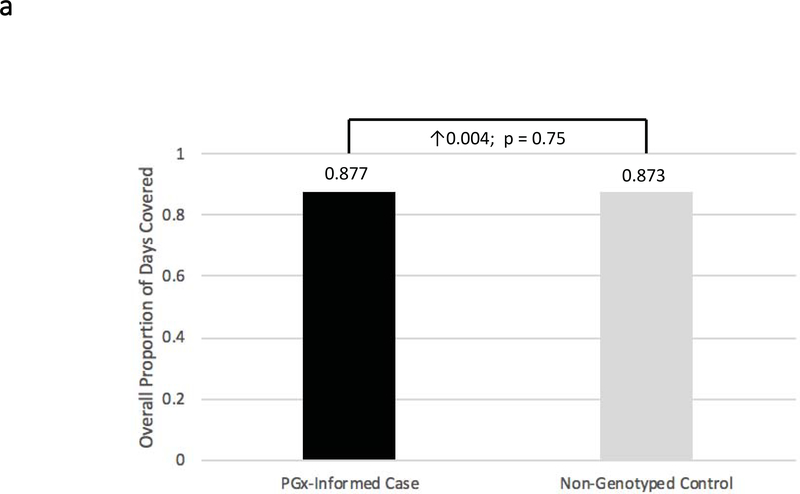
With respect to the primary study endpoint, we found both the pharmacogenomic-informed cases and non-genotyped controls had very high overall adherence throughout the study. The mean overall mPDCs were 0.88 ± 0.14 and 0.87 ± 0.16, respectively (Figure 2a) (p=NS). Further, in order to assess whether patient activation due to pharmacogenomic testing alone is a potential confounder, the adherence of case patients prior to the pharmacogenomic intervention was compared to that of case patients after genomic results became available. Prior to the intervention, mean overall mPDC was 0.86 ± 0.15, which was not statistically different from the mean overall mPDC during the study period.
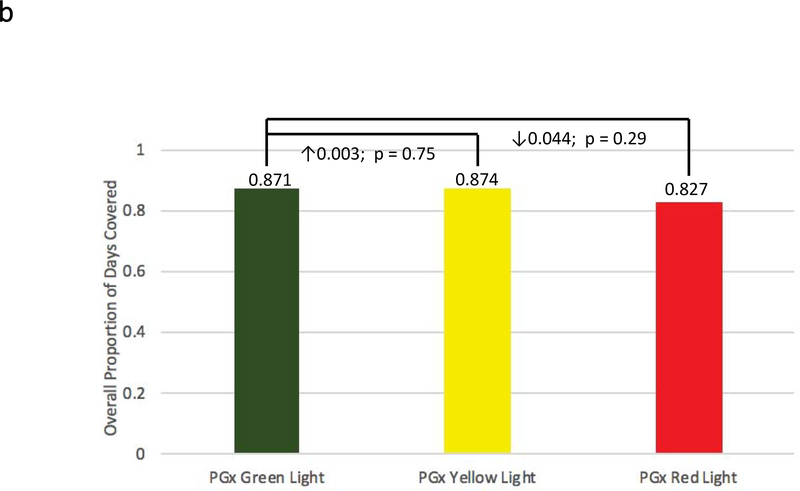
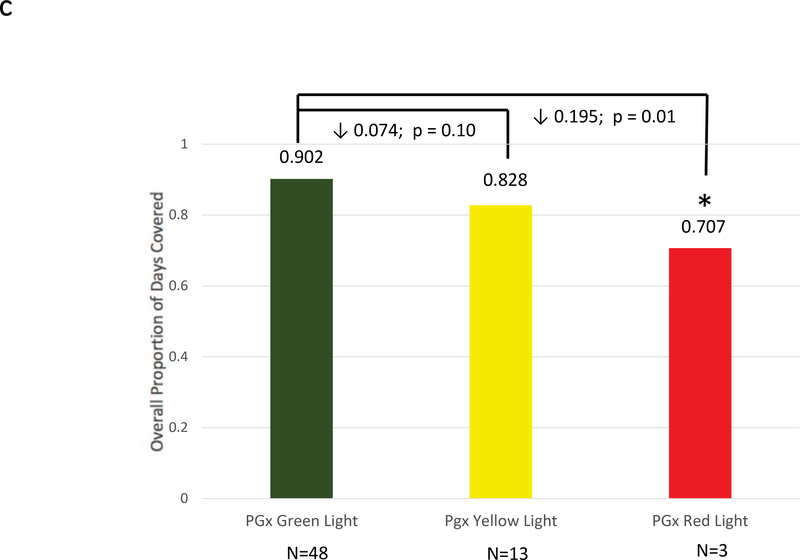
Figure 2: Adherence Comparisons between Genotyped Cases and Non-Genotyped Controls.
Adherence was defined as the mean composite modified proportion of days covered (mPDC) for all evaluable medications. (a) Comparisons were made between pharmacogenomic informed case patients (N = 270) and non-genotyped control patients (N = 266) using all medications reported within GPS for which e-prescriptions were written. (b) Comparisons were made between cases that were prescribed genomically-concordant (green light), cautionary (yellow light), and genomically-incongruent, high-risk (red light) medications, using all medications reported within GPS. (c) Comparisons were made between cases that were prescribed genomically-concordant (green light), cautionary (yellow light), and genomically-incongruent, high-risk (red light) medications, using all medications reported within GPS that have CPIC A or CPIC B designations. GPS = Genomic Prescribing System; CPIC = Clinical Pharmacogenetics Implementation Consortium.
Next, we examined the pharmacogenomic-informed case group based on genetic risk for 39 medications with pharmacogenomic information available through our institutional GPS. Interestingly, patients prescribed genomically-concordant (low genomic risk medications; green lights) had a mean overall mPDC of 0.87 ± 0.18. Those prescribed genomically-cautionary (yellow light) medications had mean overall mPDC values of 0.87 ± 0.18. However, patients prescribed genomically-incongruent, high-risk (red light) medications had a mean mPDC of 0.83 ± 0.24, a relative decrease in adherence within a clinically relevant range4, although this difference did not reach statistical significance (Figure 2b). Prior to the pharmacogenomic intervention, mean overall mPDC for prescribed green, yellow, and red light medications were 0.87 ± 0.15, 0.86 ± 0.16, and 0.85 ± 0.17, respectively (p=NS).
Additional analyses were conducted to assess adherence to medications with CPIC guidelines. Specifically, those medications in GPS with CPIC Level A or B recommendations (clopidogrel, esomeprazole, lansoprazole, omeprazole, rabeprazole, rosuvastatin, simvastatin, and warfarin) were included for sub-analysis. For these medications specifically, the mean overall mPDC for pharmacogenomic-informed cases versus non-genotyped controls were 0.85 ± 0.18 and 0.83 ± 0.20, respectively (p=NS). However, case patients showed a stepwise decrease in adherence for CPIC A/B medications as genomic risk increased. Patients prescribed genomically-concordant green light medications had a mean overall mPDC of 0.90 ± 0.12, compared to 0.83 ± 0.20 for yellow light medications and only 0.71 ± 0.17 for genomically-incongruent red light medications (p=0.01 for comparison of red versus green; Figure 2c).
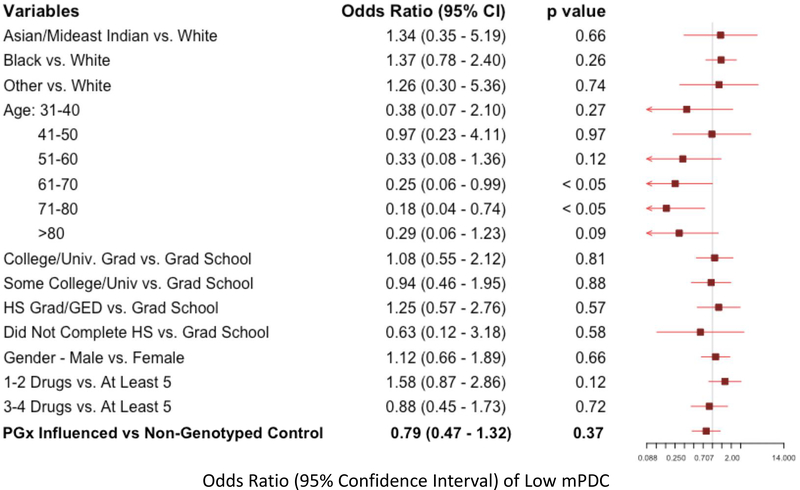
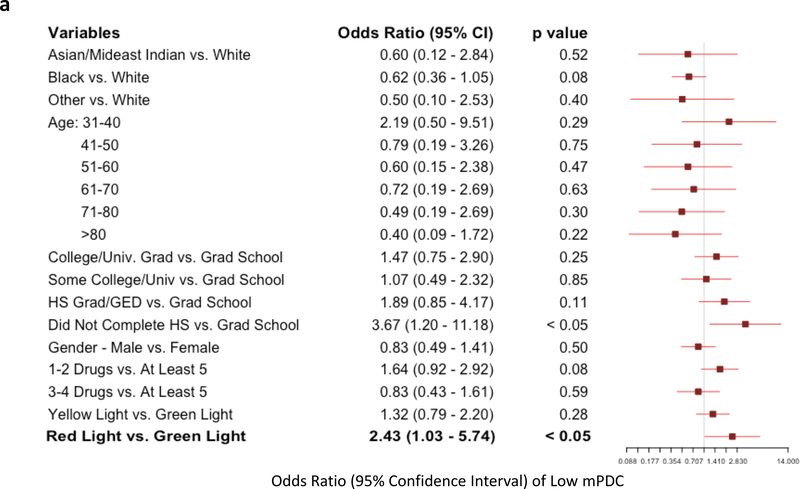
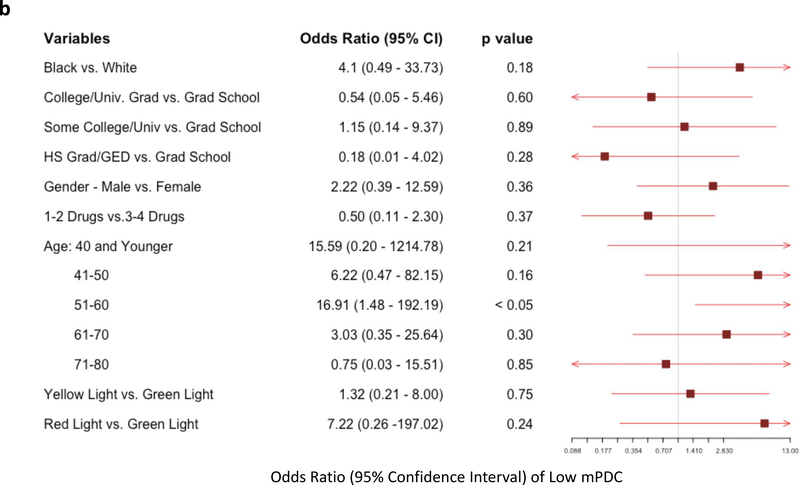
Multivariate logistic regression models were used to assess the likelihood of having very low adherence, defined as one standard deviation below the cohort mean composite mPDC (“low mPDC”). Individuals with low mPDC represent those with the worst cohort-specific adherence patterns. The first comparison of low mPDC between pharmacogenomic-informed cases and control patients again showed that the mere presence of pharmacogenomic results did not influence likelihood of adherence (odds ratio [OR]: 0.79, p=NS; Figure 3). However, the comparison between genomically-concordant and genomically-incongruent medications intriguingly revealed a significant increase in likelihood of low mPDC in patients prescribed high-risk (red light) medications. Specifically, we found that patients receiving genomically-incongruent, high-risk (red light) medications had significantly increased odds of having a low mPDC compared to genomically-concordant (green light) medications (OR: 2.43; p<0.05; Figure 4a). While age and education level also significantly affected adherence, medications conferring a high genomic risk remained an independent predictor of having a low mPDC. Similar results (including an even greater magnitude of effect, in the same direction) were obtained when only examining the subset of CPIC A/B medications, although these findings did not reach statistical significance probably because of the relatively smaller number of patients in this analysis (Figure 4b). Overall, these findings suggest that medications that are genomically more likely to pose toxicity risk or be ineffectual are indeed less likely to be reliably taken/refilled by patients, even without apparent specific patient knowledge about genomic risk classification.
Figure 3: Multivariate Logistic Regression for Likelihood of Low Adherence Based on Pharmacogenomic-Guided vs Usual Care Prescribing.
Forest plot showing variables evaluated as possible predictors of likelihood of low adherence (modified proportion of days covered (mPDC) < one standard deviation below the composite mean cohort mPDC). A total of 45 patients met the low adherence threshold. Patients receiving pharmacogenomic-informed prescriptions were less likely to be less adherent, although the difference was not statistically significant compared to non-genotyped patients.
Figure 4: Multivariate Logistic Regression for Likelihood of Low Adherence in Patients Taking Genomically-Incongruent (High Risk; Red Light) Medications vs Genomically-Concordant Medications (Green Light).
Forest plot showing variables evaluated as possible predictors of likelihood of low adherence (modified proportion of days covered (mPDC) < one standard deviation below the composite mean cohort mPDC). A total of 33 patients met the low adherence threshold. (a) Multivariate analysis including all 39 GPS medications. (b) Multivariate analysis focused on CPIC A/B medications only. In both analyses, patients who were prescribed medications that were genomically-incongruent (high risk; red lights) were more likely to have low adherence to those prescriptions compared to prescriptions for genomically-concordant (green light) medications.
Discussion
The results of this study provide the first indication that comprehensive pharmacogenomic testing with clinical delivery of results could potentially impact the complex problem of poor adherence in multi-drug regimens. Patients prescribed pharmacogenomic high-risk (genomically-incongruent) medications were more than twice as likely to have low adherence compared to those prescribed genomically-concordant medications. Bolstering the credibility of our results, in multivariate analyses we found that age and education level—known variables identified by prior studies15,16—also affected adherence, however genomic high risk (i.e. red light results) remained as an independent predictor in our study. Similar results were obtained when limiting analyses to medications with CPIC guidance, enhancing the generalizability and robustness of our findings. These data justify the future prospective examination of whether preemptive pharmacogenomic testing with the ability to predict high-risk (genomically-incongruent) medications may prevent poor adherence.
Prior literature has provided limited and mixed evidence as to whether adherence might be influenced by the clinical use of pharmacogenomic results. Some studies have demonstrated positive effects (increased adherence using pharmacogenomics).15,16 These studies, while encouraging, were focused solely on one medication class (statins) for a single medical condition. Another study was unable to demonstrate the same impact with statins29 and a separate study failed to suggest an increase in adherence in another treatment area (nicotine replacement therapy30), although both of these investigations were limited by very small sample sizes. In related work, our group previously showed that use of pharmacogenomic results during prescribing significantly increased patient recall of physician medication recommendations, a potential intermediate step towards increasing adherence with a prescribed regimen.31 This idea is concordant with the prior survey-based findings of Olson et al., who found in a large Mayo Clinic population that patients with imperfect drug adherence were highly inclined to report being more likely to use a medication as prescribed if pharmacogenomic information was used during medication selection.32 While these data (and ours) are encouraging, it remains possible that pharmacogenomic testing itself may be a total patient ‘activator’ (i.e., inducing behavior change irrespective of the genotypic findings), so future follow-on studies need to account for this important variable (ideally in a randomized way).
Previous research focused on pharmacogenomic effects on adherence for a single medication also does not reflect an accurate depiction of a typical patient’s medical regimen, and adherence is likely to be related in complex ways to polypharmacy.33,34 A 2002 U.S. survey revealed that about 25% of the overall population takes five or more medications per week.35 Our study provides the first attempt at assessing the impact of pharmacogenomics on composite adherence, integrating comprehensive genomic results for multiple medications simultaneously. We thought it important to incorporate multiple medications into our analysis, as it more fully characterizes the potential impact pharmacogenomics has on overall medication adherence for multi-drug regimens. Nevertheless, we found that the mere presence of pharmacogenomic results does not itself impact adherence. This is perhaps not surprising since providers preferentially react to1 – and will communicate to patients31 – only those pharmacogenomic results that are actionable (i.e., those that necessitate a possible regimen alteration). In line with this, our results importantly indicated that the patients most likely to be impacted and benefitted are those being prescribed genomically-incongruent (high risk) medications. In the absence of prior knowledge of genomic congruity, and presumably by trial (and error), patients seem to be less adherent to medications that are more likely to pose a toxicity risk or be ineffectual.
This study had limitations. Both the pharmacogenomic-informed cases and non-genotyped controls had overall mean mPDC values that were well above expected national values based on previous studies in the outpatient setting15,16. This could mean that our use of the modified PDC as an adherence measure might have over-estimated actual adherence, although we used the PDC because it is inherently more conservative and we took several steps to limit over-inflation of mPDC values when evaluating e-prescribing and refill patterns. It is more likely that our study population—derived from a relatively healthy, relatively highly-educated outpatient cohort of participants who had agreed to be part of a larger institutional precision medicine effort—were generally more adherent than typical patients, which may limit the generalizability of our findings. Despite this, and worthy of consideration on its own, the very high baseline adherence levels for our overall study population likely decreased our ability to detect differences that might have resulted from our primary intervention (point-of-care pharmacogenomic results/decision support availability). This potentially means that future studies may be able to identify additional impacts for pharmacogenomic availability in other patient groups that are less inherently adherent. Further, we acknowledge that it is unknown whether the patients actually took the prescribed medications or whether patients were prescribed additional medications (outside of our e-prescribing records, or from outside physicians) not captured in our analysis. It could also be the case that extraneous factors, including but not limited to route of refill, drug class, disease state, and insurance coverage, may have influenced mPDC, although it seems unlikely that any of these variables would correlate with pharmacogenomic genotype.
In conclusion, we found that pharmacogenomic congruency predicts medication adherence in patients receiving multiple medication prescriptions for common conditions. Although most previous clinical interventions to improve adherence (focused on simplification of regimens and convenience of care) have been unsuccessful at providing lasting impact, our results suggest that a focus on a personalized approach that allows potential avoidance of high risk medications may allow pharmacogenomic-informed care to have a different impact. Whether this will be true deserves prospective evaluation to understand whether knowledge of pharmacogenomics via preemptive testing can positively and meaningfully influence adherence for composite prescribing.
Supplementary Material
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Poor medication adherence is associated with worse health outcomes and increased healthcare costs. Pharmacogenomics provides a potential tool to combat poor adherence as it can address key contributing factors such as perceived medication efficacy and patient engagement.
WHAT QUESTION DID THIS STUDY ADDRESS?
Previous attempts to characterize the potential impact of pharmacogenomics on adherence have focused on individual medications rather than comprehensive pharmacogenomic testing with broad result availability and clinical decision support. We hypothesized that pharmacogenomic-informed care—through use of a point-of-care pharmacogenomic clinical decision support (CDS) tool—would significantly increase composite patient adherence rates to prescribed medications, as compared to standard outpatient care.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE?
Patient groups receiving pharmacogenomic-informed care and non-genotyped controls in this study both had high overall baseline adherence, with no detectable difference based on use of the pharmacogenomic CDS tool. However, patients prescribed genomically high-risk (incongruent) medications were more than twice as likely to have low adherence compared to those prescribed genomically-concordant medications.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
This study is the first to show that pharmacogenomics impacts composite adherence. The ability to predict high-risk (genomically-incongruent) medications using preemptive pharmacogenomic testing warrants prospective study as a means to prevent poor adherence.
Acknowledgments
Funding Sources: This research was supported by National Institutes of Health (NIH) K23 GM 100288-01A1 (P.H.O.), The University of Chicago Comprehensive Cancer Center support grant (P.H.O), The University of Chicago Bucksbaum Institute for Clinical Excellence Pilot Award (P.H.O.), the Central Society for Clinical and Translational Research - Early Career Development Award (P.H.O.), The Benjamin McAllister Scholar Program (C.C.), NIH 5T35DK062719-28 for the University of Chicago Pritzker School of Medicine Summer Research Program (as training support for C.C.), and The William F. O’Connor Foundation (M.J.R.).
Footnotes
Conflict of Interest: M.J.R. is a coinventor holding patents related to pharmacogenetic diagnostics and receives royalties related to UGT1A1 genotyping, although no royalties were received from the genotyping performed in this study. All other authors declared no competing interests for this work.
References
- 1.O’Donnell PH, Wadhwa N, Danahey K, Borden BA, Lee SM, Hall JP, et al. Pharmacogenomics-Based Point-of-Care Clinical Decision Support Significantly Alters Drug Prescribing. Clin Pharmacol Ther 102, 859–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osterberg L, Blaschke T Adherence to Medication. New England Journal of Medicine 353, 487–97 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Sokol MC, Mcguigan KA, Verbrugge RR, Epstein RS Impact of Medication Adherence on Hospitalization Risk and Healthcare Cost. Medical Care 43, 521–30 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Lam WY, Fresco P Medication Adherence Measures: An Overview. BioMed Research International 2015, 217047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iuga AO, McGuire MJ Adherence and health care costs. Risk Management and Healthcare Policy 7, 35–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkhart PV, Sabaté E Adherence to long-term therapies: evidence for action. Journal of Nursing Scholarship: An Official Publication of Sigma Theta Tau International Honor Society of Nursing 35, 207 (2003). [PubMed] [Google Scholar]
- 7.Berg JS, Dischler J, Wagner DJ, Raia JJ, Palmer-Shevlin N Medication compliance: a healthcare problem. The Annals of Pharmacotherapy 27, S1–24 (1993). [PubMed] [Google Scholar]
- 8.Brown MT, Bussell JK Medication adherence: WHO cares? Mayo Clinic Proceedings 86, 304–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kripalani S, Yao X, Haynes RB Interventions to enhance medication adherence in chronic medical conditions: a systematic review. Archives of Internal Medicine 167, 540–50 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Nieuwlaat R, Wilczynski N, Navarro T, Hobson N, Jeffery R, Keepanasseril A, et al. Interventions for enhancing medication adherence. The Cochrane Database of Systematic Reviews, CD000011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein TE, Altman RB PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Pharmacogenomics J 4, 1 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Relling MV, Evans WE Pharmacogenomics in the clinic. Nature 526, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman RB Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clin Pharmacol Ther 89, 348–50 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Haga SB, LaPointe NMA The potential impact of pharmacogenetic testing on medication adherence. The Pharmacogenomics Journal 13, 481–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JH, Joy SV, Haga SB, Orlando LA, Kraus WE, Ginsburg GS, et al. Genetically guided statin therapy on statin perceptions, adherence, and cholesterol lowering: a pilot implementation study in primary care patients. Journal of Personalized Medicine 4, 147–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charland SL, Agatep BC, Herrera V, Schrader B, Frueh FW, Ryvkin M, et al. Providing patients with pharmacogenetic test results affects adherence to statin therapy: results of the Additional KIF6 Risk Offers Better Adherence to Statins (AKROBATS) trial. The Pharmacogenomics Journal 14, 272–80 (2014). [DOI] [PubMed] [Google Scholar]
- 17.O’Donnell PH, Danahey K, Jacobs M, Wadhwa NR, Yuen S, Bush A, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care--initial results of the University of Chicago “1,200 Patients Project”. American Journal of Medical Genetics Part C, Seminars in Medical Genetics 166C, 68–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 92, 446–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danahey K, Borden BA, Furner B, Yukman P, Hussain S, Saner D, et al. Simplifying the use of pharmacogenomics in clinical practice: Building the genomic prescribing system. J Biomed Inform 75, 110–21 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Kaufman AL, Spitz J, Jacobs M, Sorrentino M, Yuen S, Danahey K, et al. Evidence for Clinical Implementation of Pharmacogenomics in Cardiac Drugs. Mayo Clin Proc 90, 716–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pharm GKB . Clinical Guideline Annotations [Available from: https://www.pharmgkb.org/guidelineAnnotations.
- 22.Fang H, Liu X, Ramírez J, Choudhury N, Kubo M, Im HK, et al. Establishment of CYP2D6 Reference Samples by Multiple Validated Genotyping Platforms. The pharmacogenomics journal 14, 564–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung EKY, Agolini E, Melis R, McMillin G, Friedman PN, Peterson P, Danahey K, O’Donnell PH, Yeo KTJ Validation of an extensive CYP2D6 Assay Panel based on Invader and TaqMan Copy Number Assays. J Appl Lab Med 01, 471–82 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Bijlsma MJ, Janssen F, Hak E Estimating time-varying drug adherence using electronic records: extending the proportion of days covered (PDC) method. Pharmacoepidemiol Drug Saf 25, 325–32 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Martin BC, Wiley-Exley EK, Richards S, Domino ME, Carey TS, Sleath BL Contrasting measures of adherence with simple drug use, medication switching, and therapeutic duplication. Ann Pharmacother 43, 36–44 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Tamargo C, Sando K, Prados Y, Cowart K Change in Proportion of Days Covered for Statins Following Implementation of a Pharmacy Student Adherence Outreach Program. J Manag Care Spec Pharm 25, 588–92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard K, Cowles B, McCall K 3rd, Henningsen RM, O’Toole M, Tu C Impact of medication synchronization programs on proportion of days covered (PDC) scores and Medicare Part D medication-related adherence metrics. J Am Pharm Assoc (2003) 59, 343–8 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Raebel MA, Schmittdiel J, Karter AJ, Konieczny JL, Steiner JF Standardizing Terminology and Definitions of Medication Adherence and Persistence in Research employing Electronic Databases. Medical care 51, S11–S21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peyser B, Perry EP, Singh K, Gill RD, Mehan MR, Haga SB, et al. Effects of Delivering SLCO1B1 Pharmacogenetic Information in Randomized Trial and Observational Settings. Circ Genom Precis Med 11, e002228 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Wright AJ, Sutton S, Armstrong D, Aveyard P, Kinmonth AL, Marteau TM Factors influencing the impact of pharmacogenomic prescribing on adherence to nicotine replacement therapy: A qualitative study of participants from a randomized controlled trial. Transl Behav Med 8, 18–28 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Borden BA, Lee SM, Danahey K, Galecki P, Patrick-Miller L, Siegler M, et al. Patient-provider communications about pharmacogenomic results increase patient recall of medication changes. Pharmacogenomics J DOI: 101038/s41397-019-0076-2, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson JE, Rohrer Vitek CR, Bell EJ, McGree ME, Jacobson DJ, St Sauver JL, et al. Participant-perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time). Genet Med 19, 819–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calip GS, Xing S, Jun DH, Lee WJ, Hoskins KF, Ko NY Polypharmacy and Adherence to Adjuvant Endocrine Therapy for Breast Cancer. J Oncol Pract 13, e451–e62 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Ulley J, Harrop D, Ali A, Alton S, Fowler Davis S Deprescribing interventions and their impact on medication adherence in community-dwelling older adults with polypharmacy: a systematic review. BMC Geriatr 19, 15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nisly NL, Gryzlak BM, Zimmerman MB, Wallace RB Dietary supplement polypharmacy: an unrecognized public health problem? Evid Based Complement Alternat Med 7, 107–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.