Rtf1 is a conserved RNA polymerase II (RNAPII) elongation factor that promotes cotranscriptional histone modification, RNAPII transcript elongation, and mRNA processing. Rtf1 function requires the phosphorylation of Spt5, an essential RNAPII processivity factor. Spt5 is phosphorylated within its C-terminal domain (CTD) by cyclin-dependent kinase 9 (Cdk9), the catalytic component of positive transcription elongation factor b (P-TEFb). Rtf1 recognizes phosphorylated Spt5 (pSpt5) through its Plus3 domain.
KEYWORDS: Cdk9, Rtf1, Spt5, transcription elongation
ABSTRACT
Rtf1 is a conserved RNA polymerase II (RNAPII) elongation factor that promotes cotranscriptional histone modification, RNAPII transcript elongation, and mRNA processing. Rtf1 function requires the phosphorylation of Spt5, an essential RNAPII processivity factor. Spt5 is phosphorylated within its C-terminal domain (CTD) by cyclin-dependent kinase 9 (Cdk9), the catalytic component of positive transcription elongation factor b (P-TEFb). Rtf1 recognizes phosphorylated Spt5 (pSpt5) through its Plus3 domain. Since Spt5 is a unique target of Cdk9 and Rtf1 is the only known pSpt5-binding factor, the Plus3/pSpt5 interaction is thought to be a key Cdk9-dependent event regulating RNAPII elongation. Here, we dissect Rtf1 regulation by pSpt5 in the fission yeast Schizosaccharomyces pombe. We demonstrate that the Plus3 domain of Rtf1 (Prf1 in S. pombe) and pSpt5 are functionally distinct and that they act in parallel to promote Prf1 function. This alternate Plus3 domain function involves an interface that overlaps the pSpt5-binding site and that can interact with single-stranded nucleic acid or with the polymerase-associated factor (PAF) complex in vitro. We further show that the C-terminal region of Prf1, which also interacts with PAF, has a similar parallel function with pSpt5. Our results elucidate unexpected complexity underlying Cdk9-dependent pathways that regulate transcription elongation.
INTRODUCTION
Mechanisms regulating RNA polymerase II (RNAPII) transcription elongation are potential therapeutic targets in cancer, heart disease, and the pathogenesis of HIV (1–3). Although a number of conserved positive and negative regulators of elongation have been identified, their mechanisms of action remain poorly understood (4, 5). Rtf1 is a multifunctional elongation factor primarily implicated in promoting cotranscriptional histone modifications; it also has roles in RNAPII elongation and mRNA processing (4, 6, 7). Rtf1 is functionally linked to cyclin-dependent kinase 9 (Cdk9), the catalytic component of positive transcription elongation factor b (P-TEFb) and the key driver of RNAPII elongation in all eukaryotes (4). The most extensively characterized Cdk9 targets are Rpb1, the largest subunit of RNAPII, and Spt5, an essential RNAPII processivity factor, both of which are phosphorylated on repeated amino acid motifs that comprise their C-terminal domains (CTDs) (8, 9). The Spt5 CTD repeat is more variable than that of the Rpb1 CTD in size and sequence, both within and between species. A related repeat motif is conserved between Schizosaccharomyces pombe (consensus motif, TPAWNSKS) and human [consensus motif, TP(M/L)YGS(R/Q)], in which the Thr1 residue is the Cdk9 target (10–12). The roles of the Spt5 CTD and its phosphorylation in RNAPII elongation are mostly unknown, despite the fact that it is a primary and exclusive Cdk9 target both in vitro and in vivo (8–10, 13). In fact, the only established function of phosphorylated Spt5-T1 (pSpt5) is to create a binding site for Rtf1 (Prf1 in S. pombe) (14–16). Rtf1 recognizes pSpt5 through its conserved Plus3 domain, so named for three positively charged amino acids that are invariant among Rtf1 orthologs (17, 18). The Plus3 domain is essential for the localization of Rtf1 to transcribed genes (14, 17). Rtf1 also contains a highly conserved histone modification domain (HMD). The HMD directly stimulates the activity of the E2 ubiquitin-conjugating enzyme Rad6, leading to the monoubiquitylation of histone H2B (H2Bub1) (19, 20). H2Bub1, in turn, directly promotes the activity of the histone H3K79 methyltransferase Dot1 as well as the Set1 histone H3K4 methyltransferase complex and regulates chromatin dynamics during transcription (21–23). The C-terminal domain of Rtf1 interacts with the polymerase-associated factor (PAF) complex; this domain in human Rtf1 also stimulates RNAPII elongation in vitro (17, 24, 25). Therefore, the Rtf1 Plus3 domain interaction with pSpt5 is thought to be part of a key regulatory pathway linking Cdk9 activity to cotranscriptional histone modification.
A crystal structure of the Plus3 domain in complex with the phosphorylated Spt5 CTD has provided a high-resolution view of this interaction, and mutations that eliminate or decrease the interaction between the Plus3 domain and pSpt5 abrogate the association of Rtf1 with transcribed genes in vivo (15). Similarly, Spt5 CTD mutations that eliminate the Cdk9-dependent phosphorylation site also prevent the association of Rtf1 with chromatin and diminish H2Bub1 levels (14, 16, 26, 27), consistent with pSpt5 recognition by the Plus3 domain playing a central role in Rtf1 function. However, the Plus3 domain has also been shown to have other functions. For example, Plus3 contains a subdomain with structural similarity to the nucleic acid-binding PAZ domains found in Argonaute family proteins (18, 28). The Plus3 domain has been shown to interact with single-stranded DNA (ssDNA) in vitro. The physiological significance of the nucleic acid interaction is not understood, nor is the relationship between pSpt5 binding and nucleic acid binding. Previous biochemical studies argue that these two functions are likely to be separable, although this has not been formally tested (18, 28). In the fission yeast S. pombe, genetic ablation of H2Bub1 or of the Rtf1 ortholog Prf1 causes cell division and morphology phenotypes that are not caused by Spt5-T1 mutations (13, 16, 29, 30). These data call into question the idea of a simple, linear pathway connecting Cdk9 activity to H2Bub1 through Plus3 domain binding to pSpt5.
We have used the model eukaryote S. pombe to evaluate the physiological significance of the putative Cdk9-Spt5-Prf1 pathway. Surprisingly, our data suggest that both pSpt5 and the Prf1 Plus3 domain act independently to mediate Prf1 function in elongation. The additional Plus3 domain interaction involves an interface that overlaps the pSpt5-binding site, is necessary for Prf1 chromatin association, and shares function with a C-terminal region of Prf1 that interacts with the PAF complex. Our results suggest that the recruitment of Prf1/Rtf1 to sites of transcription involves multiple interactions that are modulated both directly and indirectly by Cdk9-dependent Spt5 phosphorylation.
RESULTS
Functional divergence of the Prf1 Plus3 domain and phosphorylated Spt5.
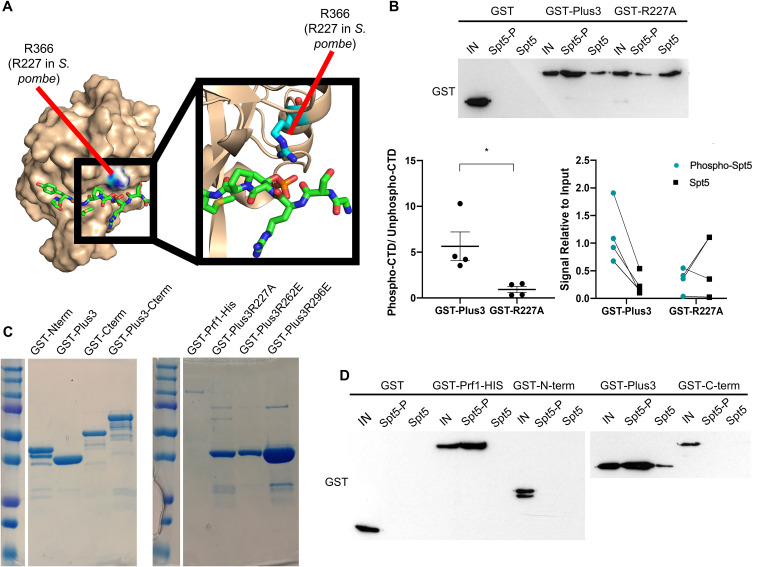
To examine the physiological significance of pSpt5 binding by the Plus3 domain in S. pombe, we introduced a point mutation at Arg227 of prf1+ that is predicted to disrupt the pSpt5-binding pocket (R227A) (Fig. 1A) (15, 18). We verified the effect on the pSpt5 interaction using immobilized peptide pulldown assays. Biotinylated peptides corresponding to either unmodified or phosphorylated Spt5 CTD repeats were immobilized on streptavidin beads and incubated with the purified recombinant Plus3 domain, and bound proteins were analyzed by immunoblotting. Quantification of the immunoblot signals showed that wild-type Plus3 preferentially bound to the pSpt5 peptide compared to the unmodified peptide. This preference was completely abrogated by the R227A mutation, as expected (Fig. 1B and C). We also verified that the HMD and C-terminal region of Prf1 did not contribute to pSpt5 binding (Fig. 1C and D).
FIG 1.
The prf1-R227A mutation abolishes pSpt5 binding. (A) PyMOL illustration mapping the location of Prf1 R227 on the crystal structure of the human Plus3 domain in complex with a pSpt5 peptide (PDB accession number 4L1U). R366 is the equivalent position in the human protein (15). (B) Immobilized peptide pulldown assays with the indicated Spt5 CTD peptides and the indicated recombinant GST fusion proteins. Binding reactions were analyzed by SDS-PAGE and immunoblotting with GST antibody. (Top) Representative GST immunoblot. “IN” denotes a 10% input. (Bottom left) Quantification of the ratio between the bound signals of phosphorylated Spt5 CTD and unphosphorylated Spt5 CTD peptides. Error bars denote standard errors of the means from 4 independent experiments. *, P ≤ 0.05 (two-sided t test). (Bottom right) Quantification of the bound signal relative to the input for each of the 4 independent experiments. Lines between the phosphorylated Spt5 CTD and unphosphorylated Spt5 CTD indicate corresponding signals within each experiment. (C) The indicated recombinant proteins were analyzed by SDS-PAGE and Coomassie staining. (D) Immobilized peptide pulldown assays with the indicated Spt5 CTD peptides and the indicated recombinant GST fusion proteins. Binding reactions were analyzed by SDS-PAGE and immunoblotting with GST antibody.
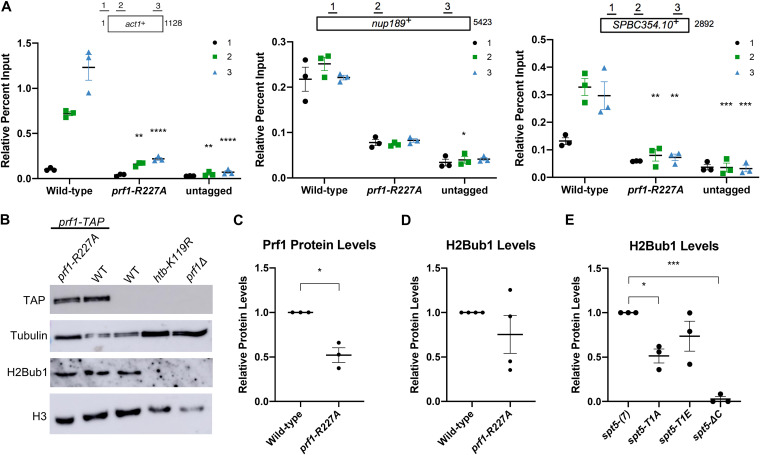
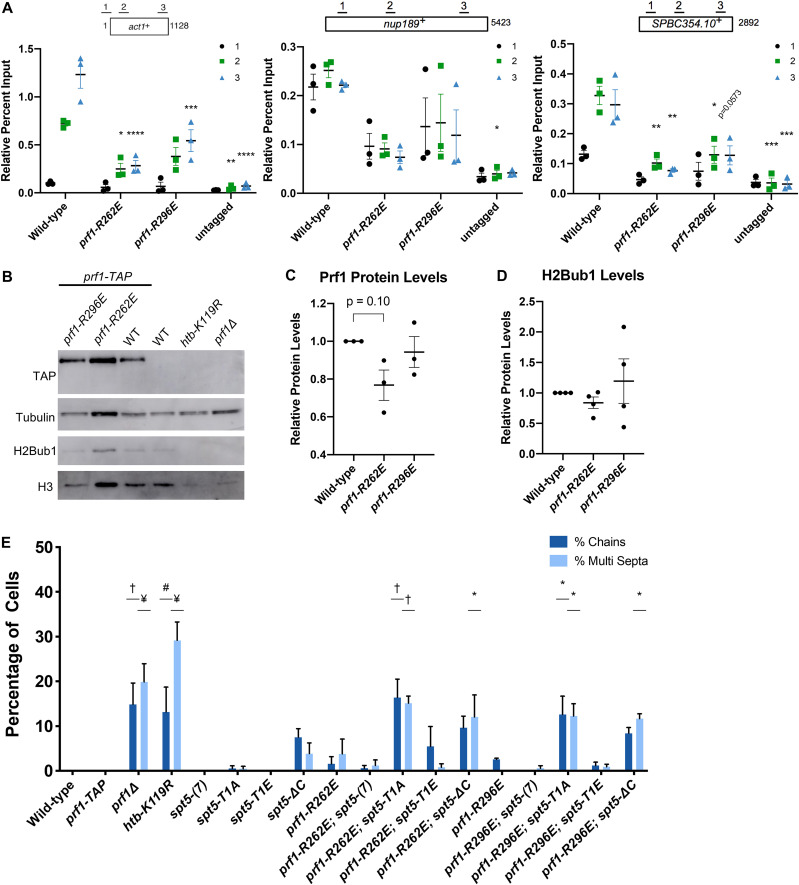
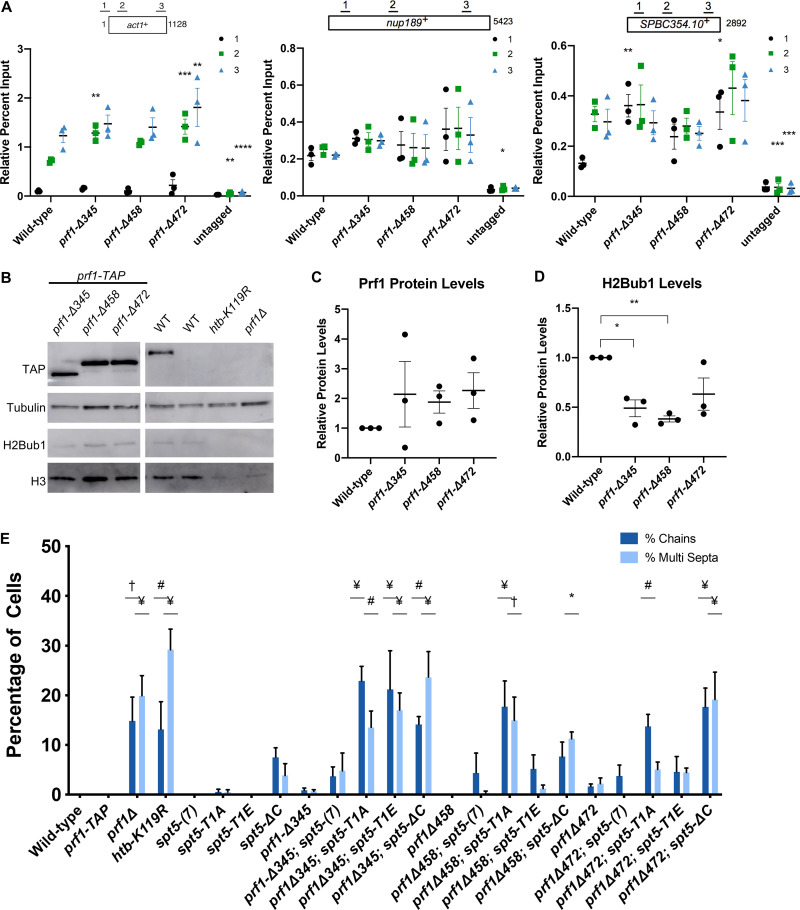
To examine the impact of pSpt5 binding on Prf1 function in vivo, we introduced the R227A mutation into the endogenous prf1+ locus and compared it to the effect of mutations in the Spt5 CTD that abolish all of the Cdk9-dependent phosphorylation sites. Spt5 CTD mutations (T1A or T1E) were engineered in the context of a truncated, 7-repeat spt5+ CTD whose function is comparable to that of the wild type [spt5(7)] (12, 16, 29). We also analyzed the spt5-ΔC mutant in which the entire CTD is deleted. In chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) assays, Prf1-R227A recruitment to transcribed genes was significantly decreased (up to 5-fold) throughout gene bodies compared to the wild type, to levels close to those obtained in the untagged control (Fig. 2A). A comparable effect on the Prf1 chromatin association was elicited by the spt5-T1A and spt5-T1E mutants (16). The prf1-R227A mutation caused a more modest, 2-fold reduction in Prf1 protein levels, which argues that the reduced chromatin occupancy reflects an impaired interaction (Fig. 2B and C). Despite the strong effects of Plus3 or Spt5 phosphorylation mutations on Prf1 chromatin occupancy, we observed relatively modest effects on H2Bub1 levels. Immunoblotting of whole-cell extracts indicated that H2Bub1 levels were not significantly affected by prf1-R227A and were reduced 2-fold by spt5-T1A; a complete loss was observed in the spt5-ΔC mutant (Fig. 2B, D, and E). These results suggest that the Plus3 domain/pSpt5 interaction is necessary for Prf1 chromatin binding but is only partially required for mediating Prf1 function and the effect of Cdk9 activity on H2Bub1.
FIG 2.
The prf1-R227A mutation abolishes chromatin association but preserves Prf1 function. (A) TAP tag ChIP was performed on the indicated strains and quantified by qPCR using the indicated primers in act1+, nup189+, and spb354+. Percent IP values were normalized using a primer pair in the S. cerevisiae PMA1 gene. The length of the gene (in base pairs) and positions of PCR amplicons are shown in the diagram at the top. Error bars denote standard errors of the means from 3 independent experiments. Two-way analysis of variance (ANOVA) was conducted followed by two-sided t tests with Bonferroni correction between each strain and the wild type within a specific primer pair. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (B) Immunoblots of whole-cell extracts from the indicated strains. Controls on each blot are the wild-type (WT) prf1-TAP strain (left) and an untagged strain (right). Antibodies are indicated on the left. (C) Quantification of immunoblots analyzing Prf1-TAP protein levels normalized to tubulin and then the wild type for the prf1-R227A strain. (D) Quantification of H2Bub1 levels normalized to total H3 levels and then the wild type for the prf1-R227A strain. (E) Quantifications of H2Bub1 levels normalized to total H3 levels in spt5 mutant strains. spt5(7) levels were set to 1. For panels C to E, error bars denote standard errors of the means from 3 independent experiments. A one-sample two-sided t test was conducted between each strain and its relative normalized wild type. *, P ≤ 0.05; ***, P ≤ 0.001.
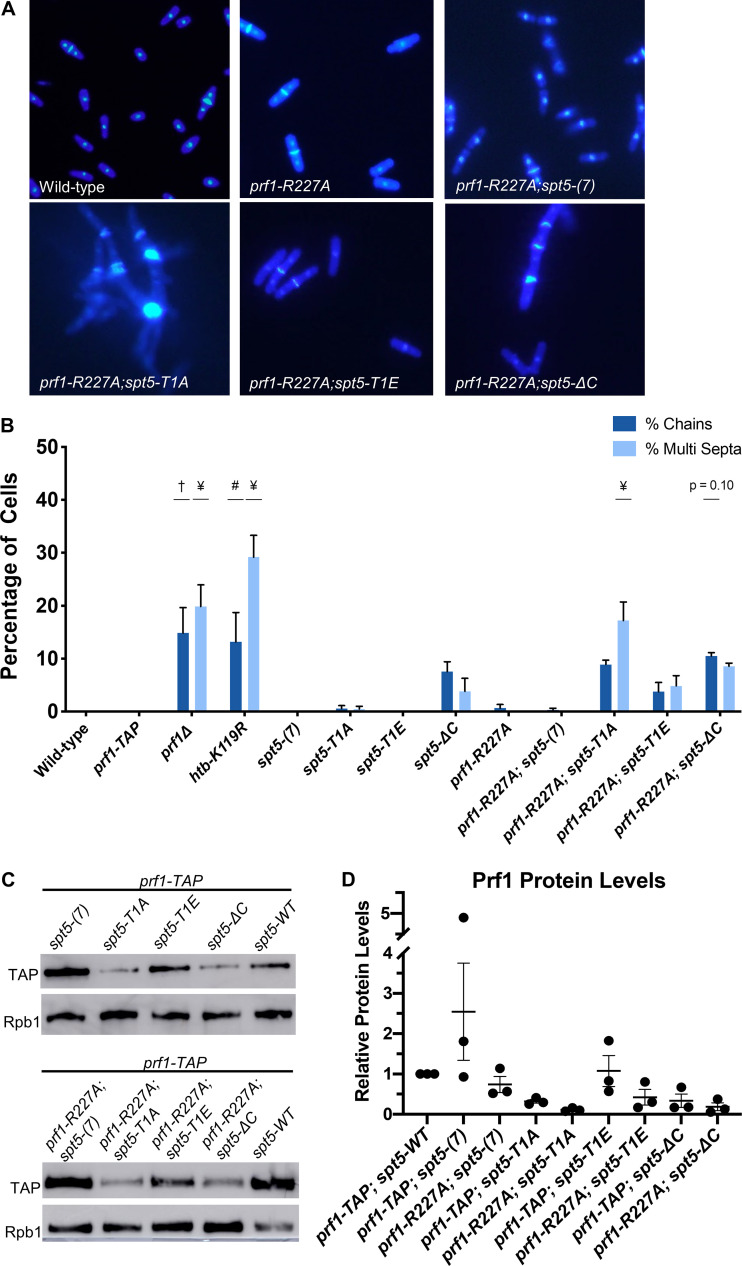
Genetic ablation of H2Bub1 (htb-K119R) or of Prf1 (prf1Δ) leads to cell division defects (13, 16, 30). These include unseparated chains of cells with division septa between each nucleus and “twinned” septa (multiple septa separating two nuclei) (13, 16). We examined prf1-R227A cells stained with 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor by fluorescence microscopy and saw no differences from the wild type (Fig. 3A; quantified in Fig. 3B). The spt5-T1A mutant was also similar to the wild type in these assays, consistent with the partial requirement of the Plus3 domain/pSpt5 interaction for H2Bub1 formation (Fig. 3B) (29).
FIG 3.
The Plus3 domain and pSpt5 function in parallel pathways. (A) The indicated strains were stained with DAPI and calcofluor and visualized by fluorescence microscopy. (B) Quantification of septation defects in the indicated strains normalized to the number of septated cells counted in each indicated strain. Error bars represent standard errors of the means from 3 independent experiments; at least 100 cells were counted for each strain per experiment. One-way ANOVA was conducted across all strains followed by two-sided t tests with Bonferroni correction between each strain and the wild-type prf1-TAP strain, for each specific morphology defect. #, P ≤ 0.01; †, P ≤ 0.001; ¥, P ≤ 0.0001. (C) Immunoblots of whole-cell extracts from the indicated strains. Antibodies are indicated on the left. (D) Quantification of Prf1-TAP protein levels in prf1-R227A spt5 double mutant strains and spt5 single mutant strains. Ratios of TAP/Rpb1 signals for each sample were normalized to that in prf1-TAP spt5+. Error bars denote standard errors of the means from 3 independent experiments. One-way ANOVA was conducted across all prf1 strains within an spt5 background followed by two-sided t tests with Bonferroni correction between each prf1 mutant strain and the wild-type prf1-TAP strain in the same spt5 background.
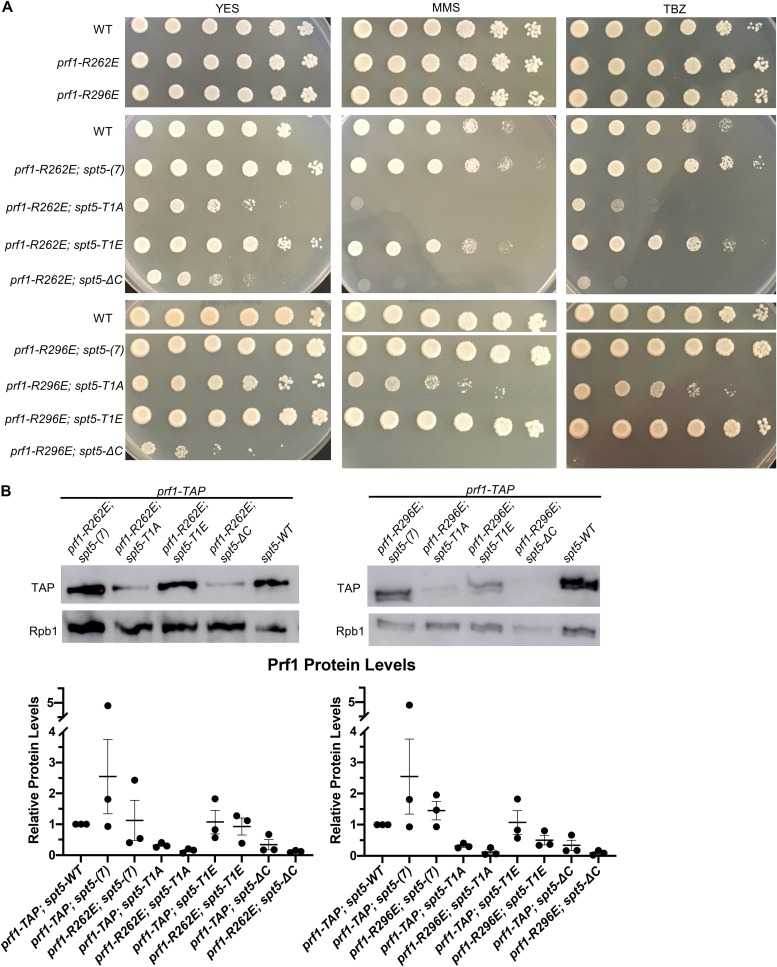
We also created double mutants harboring prf1-R227A in combination with each of the spt5 mutations. Given that the R227A mutation lies within a well-characterized binding site for pSpt5, we anticipated that any phenotypic effects would be due solely to the loss of pSpt5 binding, and thus, we predicted an epistatic relationship between prf1-R227A and spt5-T1A. Surprisingly, when the prf1-R227A mutation was combined with the spt5-T1A mutation, the resulting double mutants displayed a significant increase in the cell division phenotypes characteristic of prf1Δ and htb-K119R cells (Fig. 3A and B). This indicated a synthetic rather than an epistatic relationship between the mutations, which argues that they affect different pathways. No phenotype was observed in strains with prf1-R227A in combination with the spt5(7) allele. The modest synthetic effects observed in combination with the spt5-T1E or spt5-ΔC allele were not statistically significant, indicating that the synthetic effects were specifically related to the loss of pSpt5.
We performed immunoblot analyses to monitor Prf1 protein levels in single and double mutant strains. Interestingly, the spt5-T1A and spt5-ΔC mutations, but not spt5-T1E, resulted in decreased Prf1 protein levels in the wild-type prf1-TAP strain, supporting a direct or indirect functional link between pSpt5 and Prf1. However, the introduction of prf1-R227A into these strains did not significantly reduce Prf1 levels relative to those in wild-type prf1-TAP (Fig. 3C and D). We conclude that the synthetic phenotypes observed in prf1-R227A spt5-T1A double mutants are due to an impaired Prf1 interaction distinct from that with pSpt5.
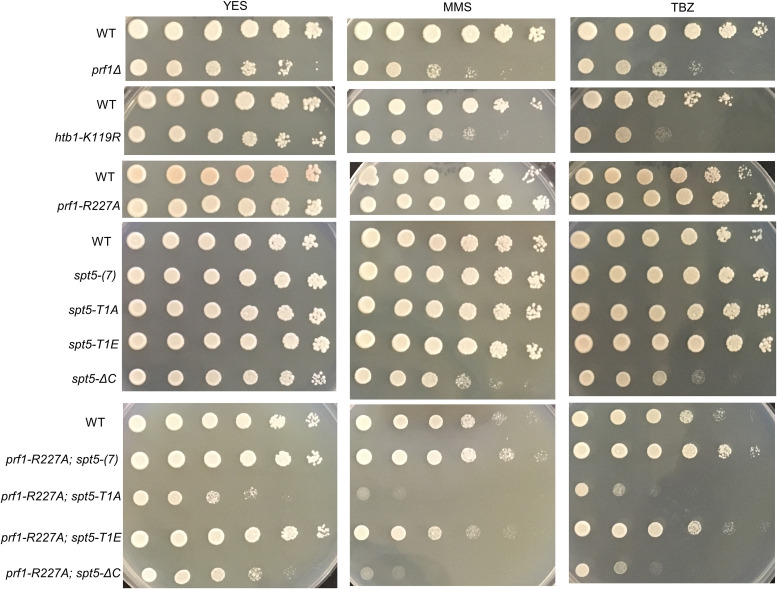
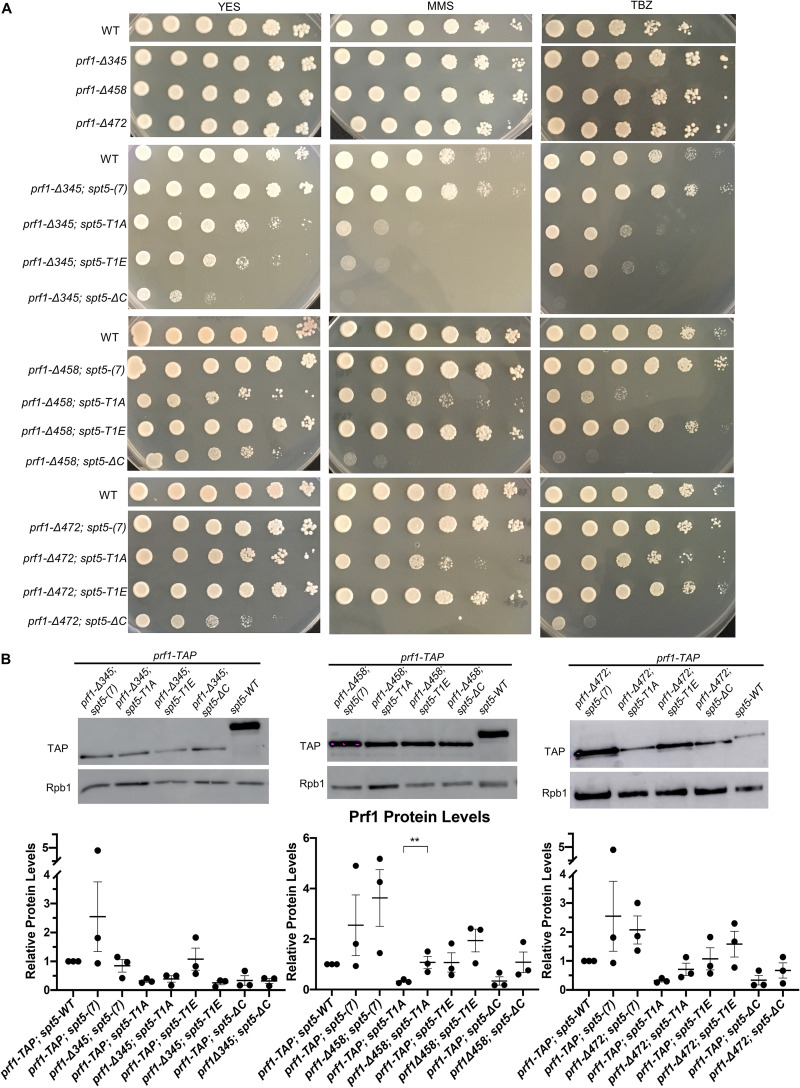
The prf1Δ and htb-K119R mutants are sensitive to thiabendazole (TBZ), a microtubule-destabilizing agent that perturbs mitotic chromosome segregation, and methyl methanesulfonate (MMS), a DNA-damaging agent (31–33) (Fig. 4). To determine whether these phenotypes were subject to similar synthetic effects, we assessed the growth of prf1-R227A and spt5 CTD single mutants and double mutants in the presence of TBZ or MMS. The double mutant strain with spt5-T1A, but not with spt5(7) or spt5-T1E, showed a marked decrease in growth on the control medium compared to either single mutant, consistent with the observed cell division phenotypes (Fig. 4). In the presence of either TBZ or MMS, the growth of this double mutant was specifically suppressed, whereas no effect on growth was observed for either single mutant or the other double mutant combinations. The spt5-ΔC mutant was sensitive to TBZ and MMS on its own, and this sensitivity was enhanced in combination with prf1-R227A. Together, these synthetic phenotypes establish an additional function for the pSpt5-binding surface of the Plus3 domain as well as a Plus3-independent function for pSpt5.
FIG 4.
Synthetic growth defects of prf1-R227A spt5 double mutants. Fivefold serial dilutions of strains were spotted onto agar plates containing control medium (YES), medium with 0.01% methyl methanesulfonate (MMS), or rich medium with 15 mg/ml thiabendazole (TBZ). Strains are indicated on the left. All experiments were repeated at least 3 times, and representative pictures are shown.
Nucleic acid-binding activity of the Plus3 domain is required for Prf1 recruitment to chromatin and is competitive with pSpt5 binding.
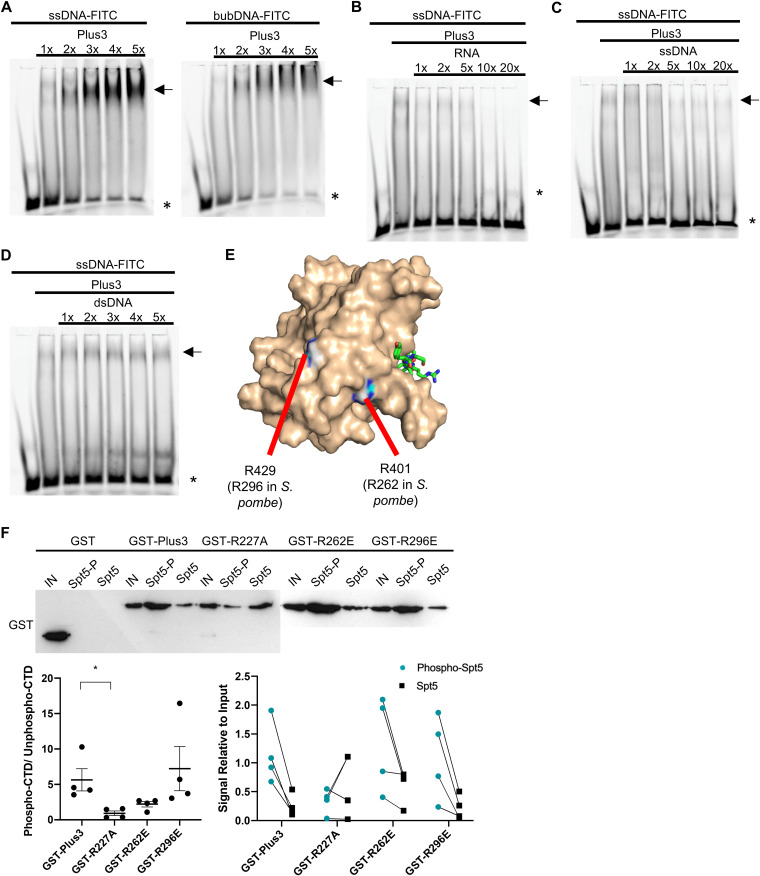
We investigated whether the additional function of the Plus3 domain could be attributed to its ability to bind nucleic acids. We first used electrophoretic mobility shift assays (EMSAs) to assess the binding of the recombinant Prf1 Plus3 domain to a fluorescently labeled ssDNA probe (30 deoxynucleotides in length). In the presence of increasing amounts of the Plus3 domain, the intensity of the band corresponding to the free probe diminished, and the intensity of a diffuse band close to the well increased; we also noted a general increase in the signal intensity throughout the lane (Fig. 5A, arrows). This pattern likely reflects the formation of heterogeneous protein-nucleic acid complexes, similar to what was previously observed for the Plus3 domain from human Rtf1 (18). The Prf1 Plus3 domain also bound a “bubble” DNA substrate (a double-stranded probe with a central region of noncomplementarity designed to mimic the transcription bubble) (Fig. 5A). Competition experiments showed no apparent binding to double-stranded DNA (dsDNA) but indicated that RNA competes for binding to the labeled ssDNA probe just as effectively as or more effectively than ssDNA (Fig. 5B to D). Thus, nucleic acid binding is a conserved property of the Plus3 domain.
FIG 5.
The prf1-R262E and prf1-R296E mutations affect nucleic acid-binding activity but not pSpt5 binding. (A) EMSAs using an FITC-labeled ssDNA (left) or bubble DNA (bubDNA) (right) probe and 1× to 5× molar equivalents of the Prf1 Plus3 domain. (B and C) Competition EMSAs using an ssDNA probe, a 1× molar equivalent of the Prf1 Plus3 domain, and either unlabeled RNA of the same sequence (left) or unlabeled ssDNA of the same sequence (right) at 1× to 20× molar ratios to the probe. (D) Competition experiments using an ssDNA probe, a 1× molar equivalent of the Prf1 Plus3 domain, and unlabeled dsDNA of the same sequence added at 1× to 5× molar ratios to the probe. For panels A to D, the predominant shifted band is denoted with an arrow. * indicates the free probe. All experiments were repeated at least 3 independent times, and representative images are shown. (E) PyMOL illustration mapping the location of Prf1 R262 and R296 on the human Plus3 domain/pSpt5 crystal structure (PDB code 4L1U). Conservation of the equivalent positions in the human protein is indicated (15). (F) Immobilized peptide pulldown assays with the indicated Spt5 CTD peptides and the indicated recombinant GST fusion proteins. Binding reactions were analyzed by SDS-PAGE and immunoblotting with GST antibody. (Top) Representative GST immunoblot (the blot for GST, GST-Plus3, and GST-R227A is from Fig. 1B). “IN” denotes a 10% input. The left half of this blot is identical to that in Fig. 1B. (Bottom left) Quantification of the ratio between the bound signals of phosphorylated Spt5 CTD and unphosphorylated Spt5 CTD peptides. A two-sided t test was conducted between each Plus3 mutant and the Plus3 wild-type signal ratios. Error bars denote standard errors of the means from 4 independent experiments. *, P ≤ 0.05. (Bottom right) Quantification of the bound signal relative to the input for each of the 4 independent experiments. Lines between the phosphorylated Spt5 CTD and unphosphorylated Spt5 CTD indicate corresponding signals within each experiment.
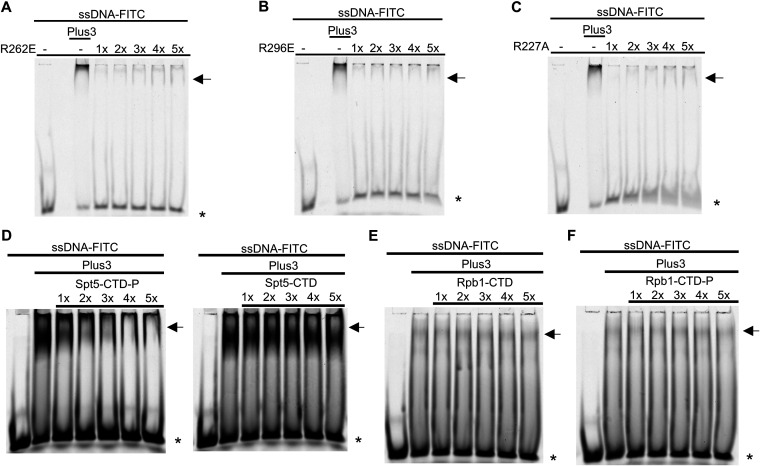
Previous studies showed that residues in the predicted PAZ subdomain, distant from the pSpt5-binding pocket, were critical for ssDNA binding (15, 18). We replaced two equivalent positions in the Prf1 Plus3 domain, Arg262 and Arg296, with glutamates (Fig. 5E). These substitutions had a minimal effect on the interaction with pSpt5 in immobilized peptide pulldown assays, although the R262E mutation decreased (but did not eliminate) the phospho-binding preference (Fig. 5F). As was found for human Plus3, both mutations abolished the nucleic acid binding of Prf1 Plus3 (Fig. 6A and B). Interestingly, the R227A mutation also dramatically decreased the Plus3 domain’s nucleic acid-binding activity, although some mobility shift could be discerned at higher protein concentrations (Fig. 6C). This suggests that pSpt5-binding and nucleic acid-binding functions may reside in overlapping regions of the Plus3 domain. To confirm this, we performed EMSAs with the wild-type Plus3 domain in the presence of either phosphorylated or unphosphorylated Spt5 CTD peptides. We found that the pSpt5 peptide, but not the unphosphorylated peptide or a phosphorylated Rpb1 CTD peptide, effectively competed for binding to ssDNA (Fig. 6D to F). Therefore, pSpt5 and nucleic acid interact with the Plus3 domain on overlapping binding surfaces in a mutually exclusive manner.
FIG 6.
Nucleic acid-binding activity of the Plus3 domain is mutually exclusive with pSpt5 binding. (A to C) EMSAs using an FITC-labeled ssDNA probe and 1× to 5× molar equivalents of the indicated Prf1 Plus3 domain. The lane marked “Plus3” contains the wild-type Plus3 domain at a 1× concentration. (D) Competition experiments using an ssDNA probe, a 1× molar equivalent of the Prf1 Plus3 domain, and the Spt5 CTD peptide (either phosphorylated or unphosphorylated) added at 1× to 5× molar ratios to the probe. (E and F) Competition experiments using an ssDNA probe, a 1× molar equivalent of the Prf1 Plus3 domain, and the Rpb1 CTD peptide (either unphosphorylated or phosphorylated at serines 5 and 2) added at 1× to 5× molar ratios to the probe. For all experiments, the predominant shifted band is denoted with an arrow. * indicates the free probe. All experiments were repeated at least 3 independent times, and representative images are shown.
To determine the physiological relevance of nucleic acid binding, we conducted an in vivo examination of R262E and R296E mutants. Both the prf1-R262E and prf1-R296E mutations significantly decreased the recruitment of Prf1 to chromatin in ChIP-qPCR assays; prf1-R262E conferred an ∼5-fold decrease, similar to prf1-R227A, whereas prf1-R296E conferred a more modest ∼2-fold decrease (Fig. 7A). Neither of the prf1-R262E and prf1-R296E mutations significantly affected Prf1 protein or H2Bub1 levels (Fig. 7B to D). These data define nucleic acid binding as a biochemical activity distinct from pSpt5 binding that is required for Prf1 association with transcribed genes.
FIG 7.
Disruptions of Plus3 domain nucleic acid binding and pSpt5 binding have similar phenotypic outcomes. (A) TAP tag ChIP was performed on the indicated strains and quantified by qPCR using the indicated primers in act1+, nup189+, and spb354+. Percent IP values were normalized using a primer pair in the S. cerevisiae PMA1 gene. The length of the gene (in base pairs) and positions of PCR amplicons are shown in the diagram at the top. Error bars denote standard errors of the means from 3 independent experiments. Two-way ANOVA was conducted followed by two-sided t tests with Bonferroni correction between each strain and the wild type within a specific primer pair. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (B) Immunoblots of whole-cell extracts from the indicated strains. Controls on each blot are the wild-type prf1-TAP strain (left) and an untagged strain (right). Antibodies are indicated on the left. (C) Quantification of immunoblots analyzing Prf1-TAP protein levels normalized to tubulin and then the wild type of the indicated strains. (D) Quantifications of H2Bub1 levels normalized to total H3 levels and then the wild type of the indicated strains. For panels C and D, error bars denote standard errors of the means from 3 independent experiments. A one-sample two-sided t test was conducted between each strain and its relative normalized wild type. (E) Quantification of septation defects in the indicated strains normalized to the number of septated cells counted in each indicated strain. At least 100 cells were counted for each strain per experiment. Error bars denote standard errors of the means from 3 independent experiments. One-way ANOVA was conducted across all strains followed by two-sided t tests with Bonferroni correction between each strain and the wild-type prf1-TAP strain, for each specific morphology defect. *, P ≤ 0.05; #, P ≤ 0.01; †, P ≤ 0.001; ¥, P ≤ 0.0001.
The prf1-R262E and prf1-R296E mutants displayed phenotypic profiles that were very similar to that of prf1-R227A: they did not show any cell division/morphology deficits or drug sensitivity on their own but showed strong synthetic phenotypes in combination with spt5-T1A (Fig. 7E and Fig. 8A). The prf1-R296E mutation had milder synthetic effects with spt5-T1A on drug sensitivity than either of the other Plus3 domain mutations, although it interacted strongly with spt5-ΔC (Fig. 8A). This may be a reflection of its milder effect on Prf1 function in the ChIP assay (Fig. 7A). Prf1 protein levels in the prf1-R262E spt5 and prf1-R296E spt5 double mutants were not significantly different in comparison to Prf1 levels from the respective spt5 single mutants (Fig. 8B). These results show that the nucleic acid-binding surface of the Plus3 domain is important for the in vivo function of Prf1 independently of the pSpt5 interaction.
FIG 8.
Synthetic growth defects of prf1-R262E spt5 and prf1-R296E spt5 double mutants. (A) Fivefold serial dilutions of strains were spotted onto agar plates containing control medium (YES), medium with 0.01% MMS, or rich medium with 15 mg/ml TBZ. Strains are indicated on the left. All experiments were repeated at least 3 times, and representative pictures are shown. (B) Immunoblots of whole-cell extracts from the indicated strains (top). Ratios of TAP/Rpb1 signals for each sample were normalized to that in prf1-TAP spt5+. Error bars denote standard errors of the means from 3 independent experiments (bottom). One-way ANOVA was conducted across all prf1+ strains within an spt5+ background followed by two-sided t tests with Bonferroni correction between each prf1+ mutant strain and the prf1-TAP strain in the same spt5+ background.
Evidence that the Plus3 domain and the Prf1 C terminus share a function.
In an effort to characterize other functional regions of S. pombe Prf1, we analyzed a series of truncations of the Prf1 C terminus, a region of the protein previously implicated in binding to the PAF complex (17, 24, 25). C-terminal truncation mutants terminating at amino acid (aa) 345, 458, or 472 (prf1-Δ345, prf1-Δ458, and prf1-Δ472) were still recruited to transcribed genes by ChIP-qPCR at levels similar to or even higher than those for the wild type (Fig. 9A). The increased ChIP-qPCR signals correlated with increases in Prf1 protein levels by immunoblotting (Fig. 9B and C). However, H2Bub1 levels were decreased in all three mutants (Fig. 9D). Thus, C-terminally truncated Prf1 proteins are functionally impaired in a manner distinct from that of Prf1 mutants that disrupt the Plus3 domain.
FIG 9.
The Prf1 C-terminal region and the Plus3 domain have a shared function. (A) TAP tag ChIP was performed on the indicated strains and quantified by qPCR using the indicated primers in act1+, nup189+, and spb354+. Percent IP values were normalized using a primer pair in the S. cerevisiae PMA1 gene. The length of the gene (in base pairs) and positions of PCR amplicons are shown in the diagram at the top. Error bars denote standard errors of the means from 3 independent experiments. Two-way ANOVA was conducted followed by two-sided t tests with Bonferroni correction between each strain and the wild type within a specific primer pair. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (B) Immunoblots of whole-cell extracts from the indicated strains. Controls on each blot are the wild-type prf1-TAP strain (left) and an untagged strain (right). Antibodies are indicated on the left. The right half of this blot is identical to the one in Fig. 7B. (C) Quantification of immunoblots analyzing Prf1-TAP protein levels normalized to tubulin and then wild type of the indicated strains. (D) Quantifications of H2Bub1 levels normalized to total H3 levels and then the wild type of the indicated strains. For panels C and D, error bars denote standard errors of the means from 3 independent experiments. A one-sample two-sided t test was conducted between each strain and its relative normalized wild type. *, P ≤ 0.05; **, P ≤ 0.01. (E) Quantification of septation defects in the indicated strains normalized to the number of septated cells counted in each strain. At least 100 cells were counted for each strain per experiment. Error bars denote standard errors of the means from 3 independent experiments. One-way ANOVA was conducted across all strains followed by two-sided t tests with Bonferroni correction between each strain and the wild-type prf1-TAP strain, for each specific morphology defect. *, P ≤ 0.05; #, P ≤ 0.01; †, P ≤ 0.001; ¥, P ≤ 0.0001.
The Prf1 C-terminal truncations did not give rise to cell growth and morphology phenotypes on their own (Fig. 9E). However, like Plus3 domain mutations, they exhibited synthetic phenotypes in combination with mutations in the Spt5 CTD. For the prf1-Δ472 and prf1-Δ458 double mutant strains, the synthetic phenotypes were observed for all assays in combination with spt5-ΔC. Double mutants with spt5-T1A exhibited assay-dependent effects: prf1-Δ472 caused septation defects and MMS sensitivity, whereas prf1-Δ458 caused septation defects, TBZ sensitivity, and MMS sensitivity (Fig. 9E and Fig. 10A). Modest septation phenotypes were observed in double mutants with spt5-T1E, but these were not statistically significant, suggesting that the function of this C-terminal portion of Prf1 is related to the loss of Spt5 CTD phosphorylation (Fig. 9E and Fig. 10A). The largest prf1 truncation mutation, prf1-Δ345, displayed cell division/morphology deficits and drug sensitivity with the spt5-T1A, -T1E, and -ΔC mutants (Fig. 9E and Fig. 10A). The fact that alanine and glutamate substitutions at the T1 position were similarly deleterious in this background suggests that the larger truncation impinges on a function that is either stringently dependent on T1 or dependent on phosphorylated T1 in a way that is not compensated for by the negatively charged side chain. Levels of the C-terminally truncated Prf1 proteins were unchanged or increased compared to those of wild-type Prf1 in the respective spt5 mutant backgrounds (Fig. 10B). Taken together, these results suggest that amino acids 459 to 562 of Prf1 participate in an interaction that functions in parallel with Spt5-T1 phosphorylation, similar to the Plus3 domain, and that amino acids 345 to 458 of Prf1 participate in an additional function that is more generally sensitive to the Spt5 CTD structure.
FIG 10.
Synthetic growth defects of prf1-Δ345 spt5, prf1-Δ458 spt5, and prf1-Δ472 double mutants. (A) Fivefold serial dilutions of strains were spotted onto agar plates containing control medium (YES), medium with 0.01% MMS, or rich medium with 15 mg/ml TBZ. Strains are indicated on the left. All experiments were repeated at least 3 times, and representative pictures are shown. (B, top) Immunoblots of whole-cell extracts from the indicated strains. (Bottom) Ratios of TAP/Rpb1 signals for each sample were normalized to that in prf1-TAP spt5+. Error bars denote standard errors of the means from 3 independent experiments. One-way ANOVA was conducted across all prf1+ strains within an spt5+ background followed by two-sided t tests with Bonferroni correction between each prf1+ mutant strain and the prf1-TAP strain in the same spt5+ background. **, P ≤ 0.01.
Both the Prf1 Plus3 domain and C-terminal region interact with the PAF complex.
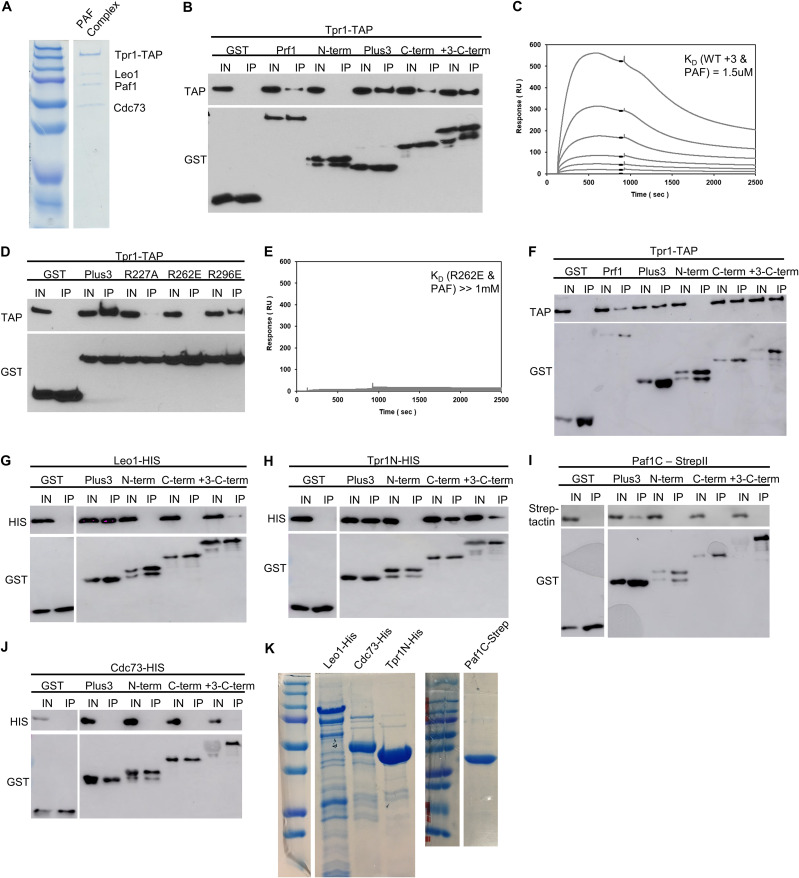
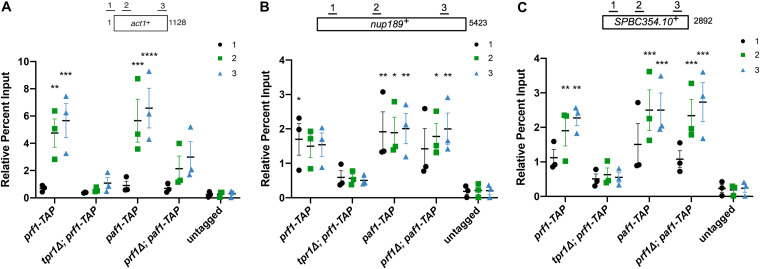
We hypothesized that the Prf1 Plus3 domain and C-terminal region may share a physical interactor that accounts for their shared function. This is unlikely to be nucleic acid, as we have not detected any nucleic acid binding by the Prf1 C-terminal region (data not shown). The C-terminal region also has no affinity for the Spt5 CTD (Fig. 1C and D). Given that the PAF complex has previously been shown to interact with the C-terminal regions of human and Saccharomyces cerevisiae Rtf1, we investigated the interaction between Prf1 and the PAF complex using purified proteins (17, 24, 25). We observed that full-length Prf1, the Plus3 domain, and the C-terminal region (amino acids 345 to 562), produced as recombinant glutathione S-transferase (GST) fusion proteins (Fig. 1C), associated with the native S. pombe PAF complex (purified via the tandem affinity purification [TAP] method) (Fig. 11A) in GST pulldown experiments (Fig. 11B). The N-terminal HMD did not pull down PAF, indicating that PAF interacts specifically with the Plus3 domain and C-terminal region in vitro (Fig. 11B). As an interaction between the Plus3 domain and PAF has not been previously reported, we used surface plasmon resonance (SPR) to confirm it. Specific dose-dependent binding between the PAF complex and the Plus3 domain was also apparent in SPR experiments (Fig. 11C). Importantly, the R227A, R262E, and R296E mutations in the Plus3 domain all reduced this interaction (Fig. 11D and E). As these mutations all affect nucleic acid binding with the Plus3 domain, we tested whether the interaction between the Plus3 domain and PAF is nucleic acid dependent. We observed a similar interaction between the Plus3 domain and PAF in the presence of ethidium bromide, suggesting that it reflects a direct protein-protein interaction (Fig. 11F). Indeed, the addition of exogenous ssDNA reduced the efficiency of the Plus3 domain-PAF interaction in GST pulldowns (data not shown). Thus, the interaction with the PAF complex is an additional Plus3 domain function that may operate in parallel with pSpt5. We have not yet identified genetic interactions between PAF complex mutations and spt5 CTD mutations that support this (data not shown). This is likely due to the fact that the interaction of the Plus3 domain with the PAF complex seems to involve multiple individual PAF complex subunits based on our in vitro characterization, including Leo1, the N-terminal half of Tpr1 (Tpr1N), and the C-terminal half of Paf1 (Paf1C) (Fig. 11G to K). However, ChIP-qPCR assays showed a significant decrease in prf1-TAP chromatin occupancy at the act1+, spbc354.10+, and nup189+ genes in a tpr1Δ strain compared to the wild type (Fig. 12). As tpr1Δ is predicted to eliminate the PAF complex, this indicated that PAF, like the Plus3 domain and pSpt5, is necessary for the Prf1 chromatin association. PAF chromatin occupancy showed a locus-specific dependence on Prf1, as paf1-TAP recruitment was affected by prf1Δ at act1+ but not at the other two loci; this may reflect locus-specific functions for the Prf1-PAF interaction (Fig. 12A). These data suggest that a direct Prf1-PAF interaction, mediated in part by the Plus3 domain, promotes Prf1 function in conjunction with pSpt5.
FIG 11.
Prf1 interacts with the PAF complex through its Plus3 domain and C-terminal region. (A) The native PAF complex purified from a tpr1-TAP strain was analyzed by SDS-PAGE and Coomassie staining. Subunits of the complex are labeled on the right; size markers are indicated on the left. “Tpr1-CBP” refers to Tpr1 fused to the calmodulin-binding peptide that is present after TAP purification. (B) GST pulldown of the native PAF complex (purified via Tpr1-TAP) with full-length, recombinant GST-Prf1 or the indicated domains tagged with GST was analyzed by SDS-PAGE and immunoblotting with the indicated antibodies (left). (C) Representative SPR sensorgrams displaying a 2-fold serial titration (0 to 540 nM) of either the wild-type Plus3 domain or the immobilized PAF complex (925 RU). Steady-state binding responses (black bars) were plotted against the concentration to determine the apparent equilibrium dissociation constant (KD). (D) Same as panel B, with the indicated GST-Plus3 domain fusions. (E) Same as panel C, with the GST-R262E mutant. (F) GST pulldown assays of the native PAF complex (Tpr1-TAP) with the full-length protein in the presence of ethidium bromide (0.1%). Recombinant GST-Prf1 or the indicated domains tagged with GST were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (G to J) GST pulldown containing recombinantly expressed subunits of the PAF complex tagged with either 6×His or Strep-tag II and GST-tagged Prf1 fusions. “Paf1C” denotes the C-terminal half of the Paf1 subunit (Paf1C-Strep). “Tpr1N” denotes the N-terminal half of the Tpr1 subunit. Immunoblotting was performed with Strep-Tactin–HRP or the indicated antibodies. For panels B, D, and F to J, “+3-C-term” denotes a fragment of Prf1 consisting of both the Plus3 domain and the C terminus. “I” denotes the input (5%); “IP” denotes the bound fraction (50%). All experiments were repeated at least 3 independent times, and representative blots are shown. (K) SDS-PAGE/Coomassie staining of recombinant PAF complex subunits.
FIG 12.
The PAF complex is necessary for Prf1 recruitment to chromatin. TAP tag ChIP was performed on the indicated strains and quantified by qPCR using the indicated primers in act1+ (A), nup189+ (B), and spb354+ (C); percent IP values were normalized to the input for each corresponding strain and primer pair. The length of the gene (in base pairs) and positions of PCR amplicons are shown in the diagram at the top. Error bars denote standard errors of the means from 3 independent experiments. Two-way ANOVA was conducted followed by two-sided t tests with Bonferroni correction between each strain and the untagged control within a specific primer pair. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
DISCUSSION
This study provides novel insights into the function of the Rtf1 Plus3 domain and its relationship to the Spt5 CTD. Previous studies have centered on the direct interaction between the Plus3 domain and Spt5 CTD repeats phosphorylated at the conserved T1 position and have emphasized its importance for the recruitment of Rtf1 and the PAF complex to transcribed genes (14, 15). Our genetic and biochemical analyses strongly argue that (i) the Plus3 domain engages in an additional interaction, exclusive of that with pSpt5, that is critical for Prf1/Rtf1 function and recruitment in vivo and (ii) pSpt5 promotes Prf1/Rtf1 function in parallel through another factor.
We observed broad phenotypic overlap between prf1-R227A, which abolishes pSpt5 recognition, and both prf1-R262E and prf1-R296E, which retain pSpt5 binding. The phenotypic effects of these mutations were strongest in the spt5-T1A and spt5-ΔC genetic backgrounds and absent or weak in combination with spt5-T1E, indicating that the introduction of a negative charge at the T1 position is important for Prf1 function when the Plus3 domain is compromised. The fact that phenotypic enhancement was observed with spt5-ΔC (albeit to various extents) negates the possibility that another CTD phosphorylation site is bound by the mutant Plus3 domains or that Plus3 domain binding to the unmodified CTD drives the phenotypic effects. We cannot exclude the possibility that a physical interaction occurs between Prf1 and Spt5 that is independent of the Spt5 CTD altogether but that requires the Plus3 domain. This would be an interaction entirely different from that suggested by previous work in budding yeast (14).
All of these mutations reduce Prf1 occupancy on gene coding regions by ChIP. This effect is particularly pronounced for prf1-R227A and prf1-R262E, both of which exhibit occupancy levels close to the background. Thus, the pSpt5-independent interaction of the Plus3 domain is important for Prf1 recruitment to chromatin, consistent with the role of the Plus3 domain previously defined in S. cerevisiae (14, 17). The spt5-T1A mutation reduces Prf1 chromatin occupancy as well as Prf1 protein levels, although we argue that these effects are not solely attributable to the interaction with Prf1 (16). Thus, Prf1 chromatin occupancy requires both Plus3 domain function and pSpt5, but Prf1 function can be maintained in the absence of either one. These findings suggest that Prf1 function does not require its stable association with chromatin and is compatible with more dynamic associations that are not captured by ChIP (19).
The recombinant Plus3 domain binds to the purified, native PAF complex in a manner that is also sensitive to prf1-R227A, prf1-R262E, and prf1-R296E mutations. The Prf1 interaction with the PAF complex also involves its C-terminal region, the truncation of which leads to synthetic phenotypes in combination with spt5-T1A. A direct interaction between Prf1 and PAF was previously demonstrated using purified components and was primarily attributed to the C-terminal region of Prf1 (25, 34). Our finding that PAF can also directly interact with the Plus3 domain is also consistent with cross-linking mass spectrometry analyses of the S. cerevisiae Rtf1/PAF complex, in which both Plus3 and C-terminal regions could be cross-linked to PAF (34). These results support a model in which both the Plus3 domain and the C-terminal region interact with the PAF complex, thereby promoting Prf1 function in parallel with pSpt5. This idea is further supported by the fact that the PAF complex is necessary for Prf1 chromatin occupancy. However, given that Prf1 function is maintained in cases where its chromatin occupancy is greatly reduced, how the interaction with PAF contributes to Prf1 function remains unclear. Greater insight into the significance of this interaction will require the identification of additional pSpt5-binding factors, as the genetics argues that pSpt5 contributes in parallel to PAF’s function in this context. We detect interactions between multiple PAF subunits and the Plus3 domain in vitro, but interactions between Prf1 and the PAF complex are weak or undetectable in extracts (as is the case in metazoans), further complicating efforts to dissect the function of the interaction in vivo (16, 35). A more detailed picture of the molecular basis for the cell division and morphology phenotypes of prf1Δ will also be critical for understanding the significance of Prf1 interactions.
The prf1-R227A, prf1-R262E, and prf1-R296E mutations all impair a nucleic acid-binding activity of the Plus3 domain. This activity prefers ssDNA over dsDNA, as has been demonstrated previously for the human Plus3 domain (18). We also show that the affinity of the Prf1 Plus3 domain for RNA is similar to that for ssDNA. This is consistent with studies showing interactions of S. cerevisiae Rtf1 with RNA in vitro and in vivo (36, 37). Whether or not nucleic acid is a physiologically relevant binding partner for the Plus3 domain in vivo remains to be determined. It is clear, however, that the binding of the Plus3 domain to pSpt5 and binding to nucleic acid are mutually exclusive, because (i) prf1-R227A abrogates both and (ii) pSpt5 competes with nucleic acid for Plus3 domain binding. Nucleic acid also competes with the PAF complex for Plus3 binding (data not shown). The differential effects of R262E and R296E on nucleic acid binding (and PAF complex binding) versus pSpt5 binding suggest that the interaction interface for the former may be larger. Nonetheless, the results of the competition experiments suggest that the Plus3 domain can interact with multiple partners through a common interface (or distinct but overlapping interfaces). We suggest that multiple Plus3 domain interactions could occur in the context of an extended Spt5 CTD with multiple phosphorylated repeats. Whereas Prf1 may directly bind to pSpt5 at some repeats, alternate modes of association may predominate at others. Our data also suggest that all modes of interaction are needed to observe a stable association of Prf1 with chromatin but that this apparent plasticity could explain how function is maintained when either pSpt5 or the Plus3 domain is compromised. Determining whether this plasticity might be regulated, and what the functional consequences might be for transcription, is an important avenue for future study.
Our results point to additional Spt5 CTD interactors that are regulated by CTD phosphorylation and that promote the function of Prf1/Rtf1. Few direct interactions with the Spt5 CTD have been described previously, and the Plus3/CTD interaction is the only one known to be phospho-specific. Factors involved in 5′ and 3′ mRNA processing also interact with the Spt5 CTD (38–40). The phospho-specificity of cleavage and polyadenylation factor interactions with the Spt5 CTD has not been determined, whereas the capping enzyme interaction is blocked by T1 phosphorylation (38). Interestingly, we observed that T1 phosphorylation also blocks the interaction of the PAF complex with the Spt5 CTD, although the physiological relevance of this interaction is not known (16). Further investigation of the functional relationship between the Spt5 CTD and Prf1/Rtf1 may uncover novel mechanisms linking Spt5 CTD phosphorylation to RNAPII elongation control.
MATERIALS AND METHODS
Yeast strains.
S. pombe strains used in this study are listed in Table 1. All genetic manipulations were conducted using standard techniques as previously described (41). Standard YES medium (5 g/liter yeast extract, 30 g/liter d-glucose, 250 mg/liter each of histidine, leucine, adenine, and uracil) and a temperature of 30°C were used for the growth of all liquid cultures.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| JT204 | h− ade6-M216 | 30 |
| JT202 | h− prf1-TAP::kanMX6 ade6 | 13 |
| JT362 | h+ leu1-32 ura4-D18 his3-D1 ade6-M210 | R. Fisher |
| JT98-1 | h− htb1-K119R::kanMX6 ade6 leu1-32 ura4-D18 | 30 |
| JT113-2 | h− prf1Δ::hphMX4 ade6 | 16 |
| JT435 | h− tpr1-TAP::kanMX6 | 16 |
| JT924 | h− prf1-R227A-TAP::kanMX6 ade6-M216 | This study |
| JT925 | h− prf1-R296E-TAP::kanMX6 ade6-M216 | This study |
| JT926 | h− prf1-R262E-TAP::kanMX6 ade6-M216 | This study |
| JT921 | h− prf1-Δ345-TAP::kanMX6 ade6-M216 | This study |
| JT923 | h− prf1-Δ458-TAP::kanMX6 ade6-M216 | This study |
| JT933 | h− prf1-Δ472-TAP::kanMX6 ade6-M216 | This study |
| JT956 | h? prf1-Δ345-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT957 | h? prf1-Δ345-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT958 | h? prf1-Δ345-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT959 | h? prf1-Δ345-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT960 | h? prf1-Δ458-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT961 | h? prf1-Δ458-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT962 | h? prf1-Δ458-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT965 | h? prf1-Δ458-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT977 | h? prf1-R296E-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32 his3-D1 | This study |
| JT978 | h? prf1-R296E-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216? ura4-D18 leu1-32 his3-D1 | This study |
| JT979 | h? prf1-R296E-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216? ura4-D18 leu1-32 his3-D1 | This study |
| JT980 | h? prf1-R296E-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216? ura4-D18 leu1-32 his3-D1 | This study |
| JT981 | h? prf1-R262E-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT982 | h? prf1-R262E-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT983 | h? prf1-R262E-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT984 | h? prf1-R262E-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT966 | h? prf1-Δ472-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT986 | h? prf1-Δ472-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT987 | h? prf1-Δ472-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT988 | h? prf1-Δ472-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT997 | h? prf1-R227A-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT998 | h? prf1-R227A-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT999 | h? prf1-R227A-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT1000 | h? prf1-R227A-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216? ura4-D18 leu1-32? his3-D1? | This study |
| JT1040 | h? prf1-TAP::kanMX6 spt5(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT1041 | h? prf1-TAP::kanMX6 spt5-T1A(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT1042 | h? prf1-TAP::kanMX6 spt5-T1E(7)::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT1043 | h? prf1-TAP::kanMX6 spt5-ΔC::ura4+ ade6-M216 ura4-D18 leu1-32? his3-D1? | This study |
| JT340 | h− spt5(7)::ura4+ leu1-32 ura4-D18 his3-D1 ade6-M210 | This study |
| JT341 | h− spt5-T1A(7)::ura4+ leu1-32 ura4-D18 his-D1 ade6 | This study |
| JT342 | h− spt5-T1E(7)::ura4+ leu1-32 ura4-D18 his-D1 ade6 | This study |
| JT343 | h− spt5-ΔC::ura4+ leu1-32 ura4-D18 his3-D1 ade6 | This study |
| JT771 | h+ spt5(7)::ura4+ leu1-32 ura4-D18 his3-D1 ade6-M210 | 12 |
| JT772 | h+ spt5-T1A(7)::ura4+ leu1-32 ura4-D18 his-D1 ade6-M210? | 12 |
| JT773 | h+ spt5-T1E(7)::ura4+ leu1-32 ura4-D18 his-D1 ade6-M210? | 12 |
| JT774 | h+ spt5-ΔC::ura4+ leu1-32 ura4-D18 his3-D1 ade6-M210? | 12 |
| JT483 | h? paf1-TAP::kanMX6 | This study |
| JT501 | h? prf1Δ::hphMX4; paf1-TAP::kanMX6 | This study |
| JT625 | h? tpr1Δ::kanMX5; prf1-TAP::kanMX6 ade6-M216 ura4-D18 leu1-32 his− | This study |
To integrate C-terminal truncation mutations into the chromosomal prf1+ locus, primers were designed to amplify the C-terminal TAP tag from pJT9 (pFA6a-kanMX6-CTAP2) as described previously (42). PCR products were transformed into competent JT204 S. pombe cells as described previously (43). To integrate Plus3 domain point mutations into the chromosomal prf1+ locus, EagI-XhoI digests of plasmid pJT161, pJT162, or pJT163 (described below) were transformed into competent JT204 cells as described above. Positive transformants were verified by sequencing and Western blotting.
Plasmids.
Plasmids used in this study are listed in Table 2. Full-length Prf1, the N-terminal region (amino acids 1 to 213), the Plus3 domain (amino acids 214 to 345), and the C-terminal region (amino acids 346 to 563) were PCR amplified from S. pombe cDNA and cloned into pGEX-6P-1. For the full-length protein, a C-terminal six-histidine tag was introduced by PCR. Plus3 domain point mutations were introduced using the Phusion site-directed mutagenesis kit (Thermo Fisher Scientific) and verified by sequencing.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pJT9 | pFA6a-kanMX6-CTAP2 | 48 |
| pJT48 | pGEX-6P-1 | Addgene |
| pJT118 | pET StrepII tag LIC cloning vector (2RT) | Addgene |
| pJT123 | pET His6 TEV tag LIC cloning vector (1B) | Addgene |
| pJT124 | pGEX-6P-1-Prf1-His6 (expresses GST-Prf1-His, inserted at BamHI/EcoRI) | This study |
| pJT125 | pGEX-6P-1-N-Terminal domain of Prf1 (expresses GST–N-terminal domain of Prf1, inserted at BamHI/XhoI) | This study |
| pJT126 | pGEX-6P-1-plus3 domain of Prf1 (expresses GST-Plus3 domain of Prf1, inserted at BamHI/XhoI) | This study |
| pJT127 | pGEX-6P-1-C-Terminal domain of Prf1 (expresses GST–C-terminal domain of Prf1, inserted at BamHI/XhoI) | This study |
| pJT129 | pGEX-6P-1-(+3+C) domains of Prf1 (expresses GST-+3+C fragment of Prf1, inserted at BamHI/XhoI) | This study |
| pJT130 | pGEX-6P-1-(+3 domainR227A of Prf1) (expresses GST-Plus3 domain R227A point mutant, inserted at BamHI/XhoI) | This study |
| pJT131 | pGEX-6P-1-(+3 domainR262E of Prf1) (express GST-Plus3 domain R262E point mutant, inserted at BamHI/XhoI) | This study |
| pJT132 | pGEX-6P-1-(+3 domainR296E of Prf1) (expresses GST-Plus3 domain R296E point mutant, inserted at BamHI/XhoI) | This study |
| pJT150 | pGex-6p-1-Rtf1-TAP-kanMX6 (inserted at XhoI/EagI) | This study |
| pJT152 | pET His6 TEV LIC-Cdc73 (expresses His-Cdc73) | This study |
| pJT153 | pET His6 TEV LIC-Leo1 (expresses His-Leo1) | This study |
| pJT154 | pET His6 TEV LIC-Tpr1N (1-400aminoacids) (expresses His-Tpr1N) | This study |
| pJT161 | pGex-6p-1-Rtf1-R227A-TAP-kanMX6 (inserted at XhoI/EagI) | This study |
| pJT162 | pGex-6p-1-Rtf1-R262E-TAP-kanMX6 (inserted at XhoI/EagI) | This study |
| pJT163 | pGex-6p-1-Rtf1-R296E-TAP-kanMX6 (inserted at XhoI/EagI) | This study |
| pJT175 | pET Strep-tag vector (p118) C-terminus of Paf1 (aa 260–456) (expresses Strep-tag II–Paf1C) | This study |
To integrate mutations into the chromosomal prf1+ locus, an ∼4.5-kb region spanning the locus and including ∼250 bp of 5′ and 3′ homology was PCR amplified from strain JT202 (prf1-TAP::kanMX6) and cloned into pGEX-6P-1 to create pJT150. Plus3 domain mutations were introduced by site-directed mutagenesis as described above. Wild-type and mutant prf1-TAP::kanMX6 constructs were verified by sequencing.
Expression and purification of recombinant Prf1.
GST fusion proteins were expressed in Escherichia coli BL21. Log-phase cultures (500 ml) were induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and grown at 16°C for 12 to 16 h. The cells were then harvested by centrifugation and resuspended in 25 ml of lysis buffer (20 mM Tris [pH 7.5], 200 mM NaCl, 20% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM benzamidine, protease inhibitor cocktail [Roche Applied Sciences]) with 2.5 mg of lysozyme. After 30 min on ice, the cell extract concentration was adjusted to 350 mM NaCl and 0.5% Triton X-100 and then sonicated with the Misonix 3000 sonicator (30 s on/off for 14 rounds; output, 5.0). All subsequent steps were conducted at 4°C. The suspension was centrifuged for 10 min at 25,000 × g, and the lysate supernatant was incubated for 3 h with 1 ml of glutathione-Sepharose beads (GE Healthcare) prewashed in lysis buffer. The beads were collected, transferred to a small column (Bio-Rad), and washed with 20 ml of wash buffer (20 mM Tris [pH 7.5], 350 mM NaCl, 20% glycerol, 1 mM EDTA, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine). The beads were eluted with 10× 0.5-ml volumes of felution buffer (20 mM Tris [pH 7.5], 350 mM NaCl, 20% glycerol, 1 mM EDTA, 100 mM reduced glutathione, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine). Peak fractions were pooled and dialyzed overnight against 2 liters of dialysis buffer {20 mM HEPES (pH 7.6), 20% glycerol, 0.15 M potassium acetate (KOAc), 10 mM magnesium acetate [Mg(OAc)2], 1 mM EDTA, 1 mM DTT}. For full-length Prf1, a second purification step was performed before dialysis. Briefly, eluates were supplemented with imidazole at a final concentration of 10 mM and incubated for 2 h with 200 μl of Ni-nitrilotriacetic acid (NTA)–agarose beads (Qiagen) that were prewashed in buffer C (20 mM HEPES [pH 7.6], 150 mM KCl, 5% glycerol, 10 mM imidazole, 0.1% NP-40, 1 mM PMSF, 1 mM β-mercaptoethanol). The beads were collected, washed four times with 1 ml of buffer C, and eluted into 1 ml elution buffer 2 (20 mM Tris [pH 7.5], 150 mM KCl, 5% glycerol, 200 mM imidazole, 0.1% NP-40, 1 mM PMSF, 1 mM β-mercaptoethanol). The eluate was then dialyzed overnight as described above.
Cloning of PAF complex subunits.
Full-length and truncated PAF complex subunits were PCR amplified from S. pombe cDNA and introduced into the pET StrepII-tag or pET His6 tobacco etch virus (TEV) protease tag expression vector by ligation-independent cloning (LIC) (Table 2) (QB3 Macrolab, UC Berkeley [https://macrolab.qb3.berkeley.edu/]). Transformants were screened by PCR and sequencing.
Immobilized peptide binding assays.
Spt5 CTD peptides (16) were synthesized as described previously (44). Ten micrograms of either the phosphorylated or unphosphorylated peptide was immobilized on 15 μl of prewashed streptavidin Dynabeads (Invitrogen) in 200 μl 1× phosphate-buffered saline (PBS). After a 3-h incubation at room temperature on a rocking platform, beads were collected on a magnet and washed twice with wash buffer (20 mM HEPES [pH 7.6], 5% glycerol, 0.1% Triton X-100, 1 mM EDTA, 350 mM KOAc, 10 mM β-glycerophosphate, 1 mM PMSF). Fifty nanograms of purified protein was added to beads, and the volume was made to 200 μl with binding buffer (20 mM HEPES [pH 7.6], 0.1% Triton X-100, 50 mM KOAc, 10 mM β-glycerophosphate, 1 mM PMSF, 1 mg/ml bovine serum albumin [BSA]). The reaction mixture was incubated at 4°C for 1 h with rocking. The beads were collected, washed four times with 1 ml of wash buffer, and resuspended in 20 μl of 1× SDS sample buffer. All samples (5% input and 50% beads) were boiled at 95°C for 2 min, centrifuged, and analyzed by SDS-PAGE and immunoblotting.
GST pulldown.
GST fusion proteins and purified factors were added in equimolar amounts (approximately 20 nM) in a 200-μl binding reaction mixture containing 20 mM HEPES (pH 7.6), 0.1% Triton X-100, 50 mM KOAc, 1 mM PMSF, 10 mM β-glycero-3-phosphate, and 0.1 mg/ml BSA. Binding reaction mixtures were incubated for 2 h at 4°C on a rocking platform. GST fusions were recovered by the addition of 25 μl of glutathione-Sepharose beads (prewashed twice with 400 μl of binding buffer) and incubation for a further 1 h at 4°C with rocking. The beads were collected and then washed four times with wash buffer (20 mM HEPES [pH 7.6], 5% glycerol, 0.1% Triton X-100, 1 mM EDTA, 350 mM KOAc, 10 mM β-glycerophosphate, 1 mM PMSF). The beads were then resuspended with 25 μl of 1× SDS sample buffer. All samples (5% input and 50% beads) were boiled at 95°C for 2 min, centrifuged, and analyzed by SDS-PAGE and immunoblotting.
Immunoblotting.
Whole-cell extracts prepared in trichloroacetic acid or purified proteins were analyzed by SDS-PAGE and immunoblotting as described previously (13). The following commercial antibodies were used: TAP tag (catalog number PICAB1001; Thermo Fisher Scientific), Rpb1 (clone 8WG16, catalog number MMS-126R-200; Covance), histone H3 (catalog number ab1791; Abcam), ubiquityl-histone H2B (catalog number 05-1312-I; Millipore), GST tag (catalog number 8-326; Thermo Fisher Scientific), His tag (catalog number H1029; Sigma-Aldrich), and Strep-Tactin–horseradish peroxidase (HRP) (catalog number 21130; Thermo Fisher Scientific). Alpha-tubulin antibody (TAT-1) was provided by Keith Gull (45). Images were acquired on an Amersham 600 imager (GE Healthcare) or on film. Images were processed using ImageJ software for quantification.
Electrophoretic mobility shift assays.
Reaction mixtures contained 0.1 μM fluorescein isothiocyanate (FITC)-labeled deoxynucleotide probe (5′-CCGCCCCGCC-T10-CCCGCCGCCC-FITC), 10 mM Tris (pH 7.5), 10% glycerol, 100 mM NaCl, 0.1 mg/ml BSA, and 0.1 to 0.5 μM recombinant GST-Plus3 protein in a final volume of 20 μl. For reaction mixtures containing the “bubble” probe, the probe described above was first hybridized to 5′-GGGCGGCGGG-T10-GGCGGGGCGG. After a 20-min incubation on ice, reaction mixtures were briefly centrifuged and loaded onto native 5% polyacrylamide gels. Gels were prepared and run in 0.5× Tris-borate-EDTA at 100 V for 1 h at 4°C. Images were acquired on the Typhoon Trio variable-mode imaging system (GE Healthcare). Rpb1 peptides used in competition experiments were synthesized as described previously (43), with the sequence biotin-PSYSPTSPSYSPTSPS (unphosphorylated) or biotin-PSYSPTS*PSYS*PTSPS (phosphorylated) (asterisks follow phosphoserines).
TAP-tagged protein purification.
Tpr1-TAP was purified from whole-cell extracts as described previously (16, 46). Ten microliters of the purified material was analyzed by SDS-PAGE alongside BSA standards, followed by Coomassie or silver staining.
His-tagged protein purification.
His fusion proteins were expressed in E. coli BL21. Log-phase cultures (500 ml) were induced with 1 mM IPTG and grown at 16°C for 12 to 16 h. Cells were harvested by centrifugation and resuspended in 10 ml of cold lysis buffer (50 mM NaPO4 [pH 8], 100 mM NaCl, 10% glycerol, 25 mM imidazole, 1 mM PMSF, 2 mM benzamidine), with the cell extract concentration adjusted to 250 mM NaCl and 0.5% Triton X-100. The cell extract was then sonicated with the Misonix 3000 sonicator (30 s on/1 min off for 12 rounds; output, 3.0) and spun down at 15,000 rpm for 20 min at 4°C. All subsequent steps were also conducted at 4°C. The lysate supernatant was incubated for 1 h with 1 ml of Ni-NTA–agarose beads (Qiagen) prewashed with wash buffer (50 mM NaPO4 [pH 8], 500 mM NaCl, 10% glycerol, 25 mM imidazole, 0.1% Triton X-100, 1 mM PMSF, 2 mM benzamidine). The beads were collected and washed with 20 ml wash buffer. The beads were eluted with 10× 0.3-ml volumes of elution buffer (50 mM NaPO4 [pH 8], 500 mM NaCl, 10% glycerol, 200 mM imidazole, 1 mM PMSF, 2 mM benzamidine). Peak aliquot fractions were pooled and dialyzed overnight against 2 liters of dialysis buffer (50 mM NaPO4 [pH 8], 100 mM NaCl, 30% glycerol, 1 mM EDTA, 1 mM DTT).
Strep-tag II protein purification.
Strep-tag II fusion proteins were expressed in E. coli BL21. Log-phase cultures (500 ml) were induced with 1 mM IPTG and grown at 16°C for 12 to 16 h. Cells were harvested by centrifugation and resuspended in 5 ml of cold lysis buffer (100 mM Tris [pH 8], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, 1 mM DTT, protease inhibitor cocktail [Roche Applied Sciences]) with 0.5 mg of lysozyme. After 30 min on ice, the cell extract concentration was adjusted to 0.5% Triton X-100 and then sonicated with the Misonix 3000 sonicator (30 s on/off for 10 rounds; output, 3.0). The suspension was spun down at 13,000 rpm for 15 min at 4°C. A small column (Bio-Rad) was packed with 1 ml of Strep-Tactin–Sepharose beads (IBA) and equilibrated with 2 ml of wash buffer (100 mM Tris [pH 8], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, 1 mM DTT). The supernatant from the cleared lysate was added to the column, and after it had completely entered the column, the column was washed stepwise with 5× 1 ml of wash buffer. Following this, 6× 500 μl of elution buffer (IBA) (100 mM Tris [pH 8], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, 1 mM dithiothreitol, 2.5 mM desthiobiotin) was added stepwise to elute the beads. Peak fractions were pooled and dialyzed overnight against 2 liters of dialysis buffer (20 mM HEPES [pH 7.5], 150 mM potassium acetate [KAc], 20% glycerol, 10 mM magnesium acetate [MgAc], 1 mM EDTA, 1 mM DTT).
Fluorescence microscopy.
DAPI and calcofluor staining was conducted as previously described, with minor changes (13). Cells were viewed using a Leica DM 5000b microscope with Lumenera’s Infinity 3-1UR camera with a 40× objective. Images were processed and cells were counted using ImageJ software. Phenotypes scored were unseparated chains of cells (with septa between each nucleus) and “twinned” septa (multiple septa separating two nuclei) (13, 16). Each strain was scored three times based on images of >100 cells.
Yeast spot tests.
Each strain was grown in 2 ml YES to an optical density at 600 nm (OD600) of 0.3 to 0.6. One milliliter of the culture was removed and adjusted to an OD600 of 2 with sterile water. Fivefold serial dilutions were performed, and 3 μl of each dilution was spotted onto the appropriate plates. The plates were then incubated at 30°C for 2 to 5 days. Methyl methanesulfonate (MMS) was used at 0.01%, and thiabendazole (TBZ) was used at 15 mg/ml.
Chromatin immunoprecipitation.
TAP-chromatin immunoprecipitation (ChIP) was conducted as previously described (47), with some modifications. Data from ChIP experiments were normalized to spiked-in S. cerevisiae chromatin (prepared from strain JTY41 [genotype, MATa his3Δ1 leu2Δ0 LYS2 ura3Δ0 RPB1-TAP::kanMX6]), unless otherwise indicated. S. pombe chromatin was prepared as described above using 1.5 × 107 cells per cross-linked sample in a final volume of 1 ml. S. cerevisiae chromatin was prepared using the same protocol but with 3.0 × 107 cells per cross-linked sample. Fifty microliters of S. cerevisiae spike-in chromatin was added to each S. pombe chromatin sample prior to conducting the immunoprecipitation (IP) step. A 100-μl sample of the input was then taken from each sample. IP was conducted by adding 20 μl of IgG-Sepharose beads (GE Healthcare) to each IP sample for a 4-h incubation at 4°C. Following IP, the beads were eluted and then washed with 150 μl of Tris-EDTA (TE). For DNA purification, all samples were incubated with 0.5 μl of RNase A (10 mg/ml) and 1 μl of glycogen (20 mg/ml) for an hour at 37°C, followed by 1.25 μl of proteinase K (20 mg/ml) for 3 h at 37°C. Samples were then extracted with 250 μl of phenol-chloroform-isoamyl alcohol and 250 μl of chloroform as described above. Primers for qPCR analysis are listed in Table 3. A primer pair in the coding region of the S. cerevisiae PMA1 gene was used for normalization.
TABLE 3.
Primers used for qPCR analysis in this study
| Primer | Sequence |
|---|---|
| Act15′Fw | GGTTGCTCAATGTTATCCGTTTC |
| Act15′Rv | TGATAAAGCCACACACAGCGTTA |
| Act1geneFw | CTCAAAGCAAGCGTGGTATTT |
| Act1geneRv | TCTTTTCCATATCATCCCAGTTG |
| Act13′Fw | CCACTATGTATCCCGGTATTGC |
| Act13′Rv | CAATCTTGACCTTCATGGAGCT |
| Nup1895′Fw | TGGTGGCTTGTTTGGTTCTCA |
| Nup1895′Rv | GGTCGTAGCGGGTGTAGTGGA |
| Nup189geneFw | GCGCCGATGCTTTTACTCC |
| Nup189geneRv | CGTTGCCATTTTTCACACCAT |
| Nup1893′Fw | CCCAGTTGGAAGCATCAGG |
| Nup1893′Rv | CTAATCCCCCACGAAGGTT |
| Spb3545′Fw | TCGGCCAAAGAAGGTCGGAAATA |
| Spb3545′Rv | TTGGCGGAAGAGGATGAAG |
| Spb354geneFw | GGGCTTTCCAACATCTCTT |
| Spb354geneRv | TAACGACGGGGAATCTATCA |
| Spb3543′Fw | AGTACTTTGGGTGGTTTGTCTT |
| Spb3543′Rv | AATGGCGTTTGTTGAGGAGGTAA |
| Pma1Fw | ACCCCAGCTAGTTAAAGAAAATCA |
| Pma1Rv | CGTCATCGTCAGAAGATTCAGATG |
Surface plasmon resonance.
Binding interactions of GST (negative control) or the GST-Plus3 domain (46-kDa wild type or mutants) with PAF (intact 280-kDa complex) were examined by label-free, real-time surface plasmon resonance (SPR) using the Biacore T200 system (GE Healthcare Bio-Sciences AB, Upsala, Sweden) (Control software v2.0 and Evaluation software v1.0). The S-series CM5 sensors and the Biacore amine-coupling kit were obtained from GE Healthcare. Protein-grade detergents were obtained from Calbiochem (cetyltrimethylammonium bromide [CTAB] [catalog number 219374]) or Anatrace (Tween 20 [catalog number APT020] and Empigen [catalog number D350]), Pierce gentle elution (PGE) buffer was obtained from Thermo (catalog number 21027), and all other chemicals were of reagent-grade quality. All SPR experiments were performed using low-ionic-strength running buffer (10 mM HEPES [pH 7.4], 75 mM NaCl, 3 mM EDTA, 0.05% [vol/vol] Tween 20). To ensure that the PAF complex remained intact, it was amine coupled to CM5 sensors at neutral pH as recommended by the manufacturer (e.g., 13 μg/ml PAF in running buffer [pH 7.4] containing 1.4 mM CTAB; final density of 850 to 950 reference units); reference surfaces (i.e., dextran-only surfaces) were amine coupled at neutral pH in a similar manner. GST (negative control) or GST fusions (wild-type Plus3 domain or R262E mutant) were titrated in tandem over reference and active surfaces in multicycle mode (25 μl/min with 10 min of association and 30 min of dissociation). Between sample injections, the surfaces were regenerated at 50 μl/min using two 30-s pulses of solution I (running buffer containing 1 M NaCl and 0.1% [vol/vol] Empigen) and solution II (PGE buffer containing 0.1% [vol/vol] Empigen). Due to the inherent heterogeneity of the PAF complex (i.e., sensorgrams deviated from a simple 1:1 binding model), the “steady-state affinity” model (BIAevaluation software) was used to predict the apparent equilibrium dissociation constants (KDs).
ACKNOWLEDGMENTS
We thank K. Gull for kindly providing TAT1 monoclonal antibody against alpha-tubulin, R. Fisher, B. Schwer, and S. Shuman for S. pombe strains, F. Robert for S. cerevisiae expressing RPB1-TAP, and members of the Tanny lab for helpful discussions.
This work was supported by the Canadian Institutes for Health Research (MOP-130362 to J.C.T. and MOP-142184 to D.C.) and the Natural Sciences and Engineering Research Council of Canada (RGPIN 03661-15 to J.C.T. and RGPIN-2015-04848 to D.C.). J.C.T. and J.J.C. acknowledge support from the Fonds de Recherche santé Quebec (chercheur boursier 33115; doctorat boursier 259846). The McGill SPR-MS Facility is supported by the Canada Foundation for Innovation.
REFERENCES
- 1.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krystof V, Chamrad I, Jorda R, Kohoutek J. 2010. Pharmacological targeting of CDK9 in cardiac hypertrophy. Med Res Rev 30:646–666. doi: 10.1002/med.20172. [DOI] [PubMed] [Google Scholar]
- 3.Baker A, Gregory GP, Verbrugge I, Kats L, Hilton JJ, Vidacs E, Lee EM, Lock RB, Zuber J, Shortt J, Johnstone RW. 2016. The CDK9 inhibitor dinaciclib exerts potent apoptotic and antitumor effects in preclinical models of MLL-rearranged acute myeloid leukemia. Cancer Res 76:1158–1169. doi: 10.1158/0008-5472.CAN-15-1070. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanny JC. 2014. Chromatin modification by the RNA polymerase II elongation complex. Transcription 5:e988093. doi: 10.4161/21541264.2014.988093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Oss SB, Cucinotta CE, Arndt KM. 2017. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci 42:788–798. doi: 10.1016/j.tibs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi Y, Shibata H, Handa H. 2013. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim Biophys Acta 1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Jeronimo C, Collin P, Robert F. 2016. The RNA polymerase II CTD: the increasing complexity of a low-complexity protein domain. J Mol Biol 428:2607–2622. doi: 10.1016/j.jmb.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 11.Pei Y, Shuman S. 2003. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J Biol Chem 278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- 12.Schneider S, Pei Y, Shuman S, Schwer B. 2010. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol 30:2353–2364. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanso M, Lee KM, Viladevall L, Jacques PE, Page V, Nagy S, Racine A, St Amour CV, Zhang C, Shokat KM, Schwer B, Robert F, Fisher RP, Tanny JC. 2012. A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet 8:e1002822. doi: 10.1371/journal.pgen.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayekar MK, Gardner RG, Arndt KM. 2013. The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol Cell Biol 33:3259–3273. doi: 10.1128/MCB.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wier AD, Mayekar MK, Heroux A, Arndt KM, VanDemark AP. 2013. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci U S A 110:17290–17295. doi: 10.1073/pnas.1314754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mbogning J, Nagy S, Page V, Schwer B, Shuman S, Fisher RP, Tanny JC. 2013. The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet 9:e1004029. doi: 10.1371/journal.pgen.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warner MH, Roinick KL, Arndt KM. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE. 2008. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 16:149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. 2012. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci U S A 109:10837–10842. doi: 10.1073/pnas.1116994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Oss SB, Shirra MK, Bataille AR, Wier AD, Yen K, Vinayachandran V, Byeon IL, Cucinotta CE, Heroux A, Jeon J, Kim J, VanDemark AP, Pugh BF, Arndt KM. 2016. The histone modification domain of Paf1 complex subunit Rtf1 directly stimulates H2B ubiquitylation through an interaction with Rad6. Mol Cell 64:815–825. doi: 10.1016/j.molcel.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu PL, Shi H, Leonen C, Kang J, Chatterjee C, Zheng N. 2019. Structural basis of H2B ubiquitination-dependent H3K4 methylation by COMPASS. Mol Cell 76:712–723.e4. doi: 10.1016/j.molcel.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. 2011. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worden EJ, Hoffmann NA, Hicks CW, Wolberger C. 2019. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176:1490–1501.e12. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao QF, Yamamoto J, Isobe T, Tateno S, Murase Y, Chen Y, Handa H, Yamaguchi Y. 2015. Characterization of the human transcription elongation factor Rtf1: evidence for nonoverlapping functions of Rtf1 and the Paf1 complex. Mol Cell Biol 35:3459–3470. doi: 10.1128/MCB.00601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Guermah M, Roeder RG. 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A 106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng P, Zhou Y, Jiang J, Li H, Tian W, Cao Y, Qin Y, Kim J, Roeder RG, Patel DJ, Wang Z. 2018. Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology. Proc Natl Acad Sci U S A 115:9998–10003. doi: 10.1073/pnas.1812256115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mbogning J, Page V, Burston J, Schwenger E, Fisher RP, Schwer B, Shuman S, Tanny JC. 2015. Functional interaction of Rpb1 and Spt5 C-terminal domains in co-transcriptional histone modification. Nucleic Acids Res 43:9766–9775. doi: 10.1093/nar/gkv837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. 2007. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page V, Chen JJ, Durand-Dubief M, Grabowski D, Oya E, Sanso M, Martin RD, Hebert TE, Fisher RP, Ekwall K, Tanny JC. 2019. Histone H2B ubiquitylation regulates histone gene expression by suppressing antisense transcription in fission yeast. Genetics 213:161–172. doi: 10.1534/genetics.119.302499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadeghi L, Siggens L, Svensson JP, Ekwall K. 2014. Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat Struct Mol Biol 21:236–243. doi: 10.1038/nsmb.2776. [DOI] [PubMed] [Google Scholar]
- 33.Zeng M, Ren L, Mizuno K, Nestoras K, Wang H, Tang Z, Guo L, Kong D, Hu Q, He Q, Du L, Carr AM, Liu C. 2016. CRL4(Wdr70) regulates H2B monoubiquitination and facilitates Exo1-dependent resection. Nat Commun 7:11364. doi: 10.1038/ncomms11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Bernecky C, Lee C-T, Maier KC, Schwalb B, Tegunov D, Plitzko JM, Urlaub H, Cramer P. 2017. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat Commun 8:15741. doi: 10.1038/ncomms15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. 2015. The BioPlex Network: a systematic exploration of the human interactome. Cell 162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dermody JL, Buratowski S. 2010. Leo1 subunit of the yeast Paf1 complex binds RNA and contributes to complex recruitment. J Biol Chem 285:33671–33679. doi: 10.1074/jbc.M110.140764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battaglia S, Lidschreiber M, Baejen C, Torkler P, Vos SM, Cramer P. 2017. RNA-dependent chromatin association of transcription elongation factors and Pol II CTD kinases. Elife 6:e25637. doi: 10.7554/eLife.25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doamekpor SK, Sanchez AM, Schwer B, Shuman S, Lima CD. 2014. How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes. Genes Dev 28:1323–1336. doi: 10.1101/gad.242768.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. 2012. The Spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol 32:1321–1331. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen Y, Shatkin AJ. 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev 13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 42.Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-López M, Cotobal C, Fernández-Sánchez O, Borbarán Bravo N, Oktriani R, Abendroth H, Uka D, Hoti M, Wang J, Zaratiegui M, Bähler J. 2017. A CRISPR/Cas9-based method and primer design tool for seamless genome editing in fission yeast. Wellcome Open Res 1:19. doi: 10.12688/wellcomeopenres.10038.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamine A, Poujol de Molliens M, Letourneau M, Hebert TE, Vaudry D, Fournier A, Chatenet D. 2019. The amidated PACAP1-23 fragment is a potent reduced-size neuroprotective agent. Biochim Biophys Acta 1863:129410. doi: 10.1016/j.bbagen.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci 93(Part 3):491–500. [DOI] [PubMed] [Google Scholar]
- 46.Racine A, Page V, Nagy S, Grabowski D, Tanny JC. 2012. Histone H2B ubiquitylation promotes activity of the intact Set1 histone methyltransferase complex in fission yeast. J Biol Chem 287:19040–19047. doi: 10.1074/jbc.M112.356253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mbogning J, Tanny JC. 2017. Chromatin immunoprecipitation of histone modifications in fission yeast. Methods Mol Biol 1528:199–210. doi: 10.1007/978-1-4939-6630-1_12. [DOI] [PubMed] [Google Scholar]
- 48.Tasto JJ, Carnahan RH, McDonald WH, Gould KL. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]