The diversified NF-κB transcription factor family has been extensively characterized in organisms ranging from flies to humans. However, homologs of NF-κB and many upstream signaling components have recently been characterized in basal phyla, including Cnidaria (sea anemones, corals, hydras, and jellyfish), Porifera (sponges), and single-celled protists, including Capsaspora owczarzaki and some choanoflagellates. Herein, we review what is known about basal NF-κBs and how that knowledge informs on the evolution and conservation of key sequences and domains in NF-κB, as well as the regulation of NF-κB activity.
KEYWORDS: NF-kappaB, development, evolution, innate immunity, signal transduction
ABSTRACT
The diversified NF-κB transcription factor family has been extensively characterized in organisms ranging from flies to humans. However, homologs of NF-κB and many upstream signaling components have recently been characterized in basal phyla, including Cnidaria (sea anemones, corals, hydras, and jellyfish), Porifera (sponges), and single-celled protists, including Capsaspora owczarzaki and some choanoflagellates. Herein, we review what is known about basal NF-κBs and how that knowledge informs on the evolution and conservation of key sequences and domains in NF-κB, as well as the regulation of NF-κB activity. The structures and DNA-binding activities of basal NF-κB proteins resemble those of mammalian NF-κB p100 proteins, and their posttranslational activation appears to have aspects of both canonical and noncanonical pathways in mammals. Several studies suggest that the single NF-κB proteins found in some basal organisms have dual roles in development and immunity. Further research on NF-κB in invertebrates will reveal information about the evolutionary roots of this major signaling pathway, will shed light on the origins of regulated innate immunity, and may have relevance to our understanding of the responses of ecologically important organisms to changing environmental conditions and emerging pathogen-based diseases.
INTRODUCTION
The nuclear factor κB (NF-κB) superfamily comprises a group of related transcription factors that have been intensively studied for their involvement in development and immunity since their nearly simultaneous discovery in a retrovirus, flies, and mice almost 35 years ago (1–5). Indeed, there are now approximately 100,000 papers on NF-κB, with the vast majority focusing on NF-κB proteins from mammals, flies, and viruses. However, with the proliferation of genomic and transcriptomic sequencing over the past decade, it has been discovered that many organisms ostensibly less complex than insects have NF-κB-like genes (6–13). Herein, we critically review what is known about NF-κB in these more basal organisms, namely, cnidarians, poriferans, and protists.
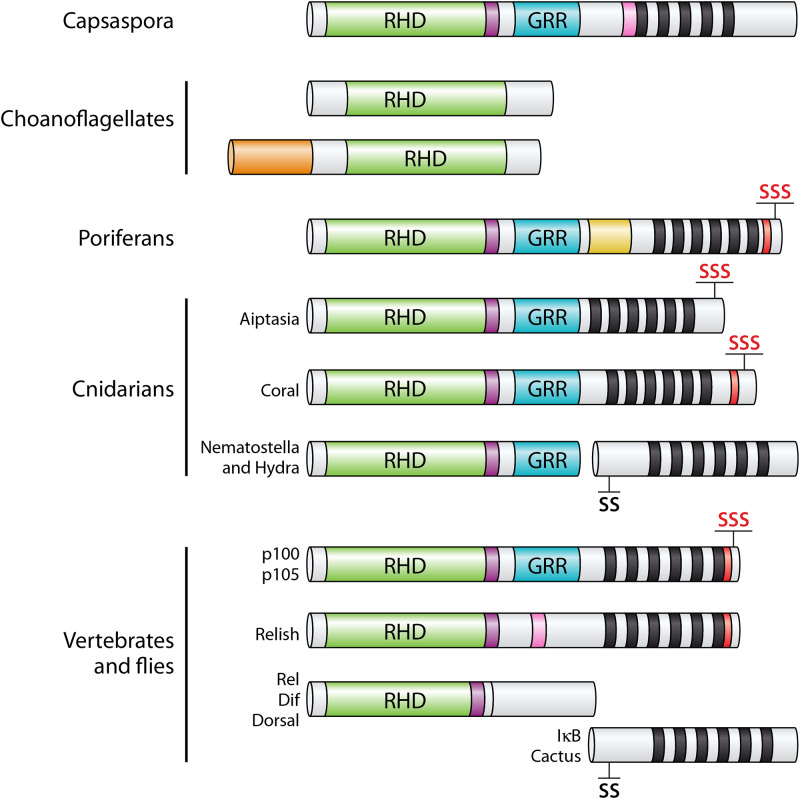
All NF-κB proteins are related by an N-terminal DNA-binding and dimerization region, called the Rel homology domain (RHD), containing a nuclear localization sequence (NLS), and the RHD allows them to enter the nucleus, bind to specific DNA sites (κB sites), and activate or repress the transcription of target genes for specified biological outcomes (14). In vertebrates and flies, the NF-κB superfamily can be divided into two subfamilies: the NF-κB proteins, which consist of vertebrate p100 and p105 and Drosophila Relish, and the Rel proteins, which include vertebrate RelA, RelB, and c-Rel, as well as Drosophila Dif and Dorsal (14). Thus, flies and vertebrates all contain multiple NF-κB proteins that, for the most part, show complete combinatorial diversity by forming homodimers and heterodimers, which have distinct DNA target site specificities. The two subfamilies can be distinguished phylogenetically by sequence alignment of their RHDs and by sequences C terminal to the RHD. That is, NF-κB proteins contain C-terminal inhibitory ankyrin (ANK) repeat domains, and Rel proteins contain C-terminal transactivation domains (Fig. 1). The ANK repeats, either within the NF-κB proteins themselves or in a separate family of NF-κB inhibitors (IκBs), regulate the subcellular localization of NF-κBs by binding to the RHD and sequestering them in the cytoplasm. Activation of the pathway by an appropriate upstream signal results in the degradation of the ANK repeat inhibitor, thus allowing the NF-κB dimer to enter the nucleus and bind DNA (14, 15). The NF-κB p100 and p105 proteins also contain a C-terminal death domain (DD) that is important for protein-protein interactions with other members of the DD superfamily, which serve as adaptors in signaling pathways and/or to recruit other proteins into signaling complexes (15).
FIG 1.
Structures of NF-κBs in organisms basal to Bilateria are most similar to human NF-κB proteins rather than Rel proteins. The basic domain structures of homologs of NF-κB in Capsaspora, choanoflagellates, poriferans (sponges), and cnidarians are compared to those of the fly and vertebrate NF-κBs (Relish and p100/p105), Rels (Dif, Dorsal, RelA, RelB, and c-Rel), and IκB (Cactus). Green, Rel homology domain (RHD); purple, nuclear localization signal (NLS); blue, glycine-rich region (GRR); pink, caspase cleavage site; black bars, ankyrin repeats; red, death domain; SSS/SS, conserved serines important for phosphorylation and degradation of the protein (the red SSSs indicate homology to p100); orange, N-terminal domains of choanoflagellates not seen in other NF-κB proteins.
Many evolutionarily conserved receptors can elicit downstream signals to activate NF-κB translocation to the nucleus. These receptors, which include Toll-like receptors (TLRs) and interleukin-1 receptors (IL-1Rs), interact with cytoplasmic adaptor proteins (e.g., MYD88 and MAL) to initiate downstream signaling (15). Once the pathway is activated, a series of phosphorylation events leads to the degradation of the ANK repeat inhibitor sequences, freeing NF-κB to translocate from the cytoplasm to the nucleus. In vertebrates, activation of NF-κB generally happens in one of two ways. In the canonical pathway, activation of an IκB kinase β (IKKβ) complex results in the phosphorylation of an independent IκB protein, and phosphorylated IκB then undergoes ubiquitination and proteasomal degradation to liberate the NF-κB dimer. In the noncanonical pathway, activation of the related kinase IKKα leads to the phosphorylation of a serine cluster located C terminal to the ANK repeats on the p100 protein (16). These phosphorylations result in proteasomal processing of the inhibitory ANK repeats on p100 until the proteasome reaches a glycine-rich region (GRR). At the GRR, the proteasome falls off, generating p52, a C-terminally truncated version of p100 that can enter the nucleus (16). Similar pathways exist in flies, except that the p100 homolog Relish has no GRR and the truncated, active form of Relish is generated by a site-specific proteolytic cleavage that removes the ANK repeat region (17; for more in-depth information on basal TLR-to-NF-κB pathways, we refer readers to references 6 and 18).
Genomic and transcriptomic sequencing data strongly suggest that NF-κB was also pervasive through early evolution, based on the presence of NF-κB-like homologs in many extant organisms basal to flies and vertebrates. NF-κB homologs have been identified in single-celled premetazoans, such as the protist Capsaspora owczarzaki (11), a symbiont that lives in the hemolymph of the freshwater snail Biomphalaria glabrata, and choanoflagellates (12), aquatic unicellular colonial organisms which have been proposed to be the closest living relatives to multicellular animals. NF-κBs have also been identified in a variety of multicellular basal marine metazoans, including poriferans (sponges) and cnidarians (corals, sea anemones, hydras, and jellyfish) (7–10, 13). In most of these basal organisms, there are single NF-κB proteins, which, as discussed below, most closely resemble NF-κB subfamily proteins, such as Relish and p100. Curiously, no NF-κB homologs have been found in nematodes (e.g., Caenorhabditis elegans and Caenorhabditis briggsae) or ctenophores, where the pathway appears to have been lost.
As discussed in this review, only recently have several studies investigated how the transcription factor NF-κB and its upstream and downstream pathways function in early-branching organisms. We provide insight into how this highly conserved transcription factor and its regulatory pathway arose, describe domains that have appeared at pivotal points of evolution and biological processes likely controlled by NF-κB, and discuss areas of basal NF-κB knowledge that remain unanswered.
STRUCTURES OF BASAL NF-κB PROTEINS
Many basal eukaryotes have NF-κB proteins that are structurally and phylogenetically most similar to vertebrate NF-κB proteins rather than Rel proteins. In most cases, these organisms have single p100-like RHD-ANK repeat NF-κB proteins that often possess C-terminal serine residues known in vertebrates to be required for processing of p100 via the noncanonical NF-κB pathway (see “Activity and regulation of basal NF-κBs” below). For example, the protist Capsaspora, the sponge Amphimedon queenslandica, and several cnidarians all have NF-κBs with an RHD and C-terminal ANK repeats on the same transcript (8, 19–24). Many ANK repeat-containing proteins are found throughout the genomes of archaea, bacteria, and all plants and animals (25, 26). Accordingly, it is likely that during the evolution of eukaryotes there arose a primitive RHD-only protein which developed the ability to interact with a preexisting ANK repeat-containing protein. At some point, these interacting RHD and ANK repeat genes fused to create the more modern single RHD-ANK repeat protein.
Nevertheless, some basal organisms have NF-κB-like proteins without C-terminal ANK repeat domains. Among protists, several choanoflagellates have NF-κB-like proteins that do not have C-terminal ANK repeat domains (12). Choanoflagellates comprise a sister group to all metazoans and are part of a diverse group of single-celled, colony-forming organisms that live in waters around the world. Although the genomes of the classically studied choanoflagellates Monosiga brevicollis and
Salpingoeca rosetta do not contain NF-κB homologs (27), a large-scale transcriptomic analysis of 19 choanoflagellates (12) showed that 12 had transcripts that encode RHD-containing NF-κB-like sequences (12). However, none of these choanoflagellate NF-κB-like proteins appears to have C-terminal ANK repeat domains (Fig. 1). Moreover, several choanoflagellate NF-κB-like proteins also have extended N-terminal domains that are not present in any other NF-κB proteins (Fig. 1; see also Table S1 in the supplemental material). Overall, these 19 choanoflagellate transcriptomes encode zero to three NF-κB-like proteins, none of which has C-terminal ANK repeats. Indeed, it is not yet known whether choanoflagellate NF-κBs are regulated by an ANK repeat-containing IκB-like inhibitor. The lack of ANK repeats in choanoflagellate NF-κB proteins and the wide diversification of NF-κBs among choanoflagellates are likely a result of 600 million years of independent evolution among the diverse species of choanoflagellates. Furthermore, this gene expansion mirrors the increasing number and complexity of NF-κBs seen throughout metazoans. In contrast, the protist Capsaspora does contain a bipartite RHD-ANK protein (11). Thus, the earliest NF-κB with an RHD-ANK fusion likely arose in the protist lineage that led to Capsaspora but was excluded or lost in the lineage that led to choanoflagellates.
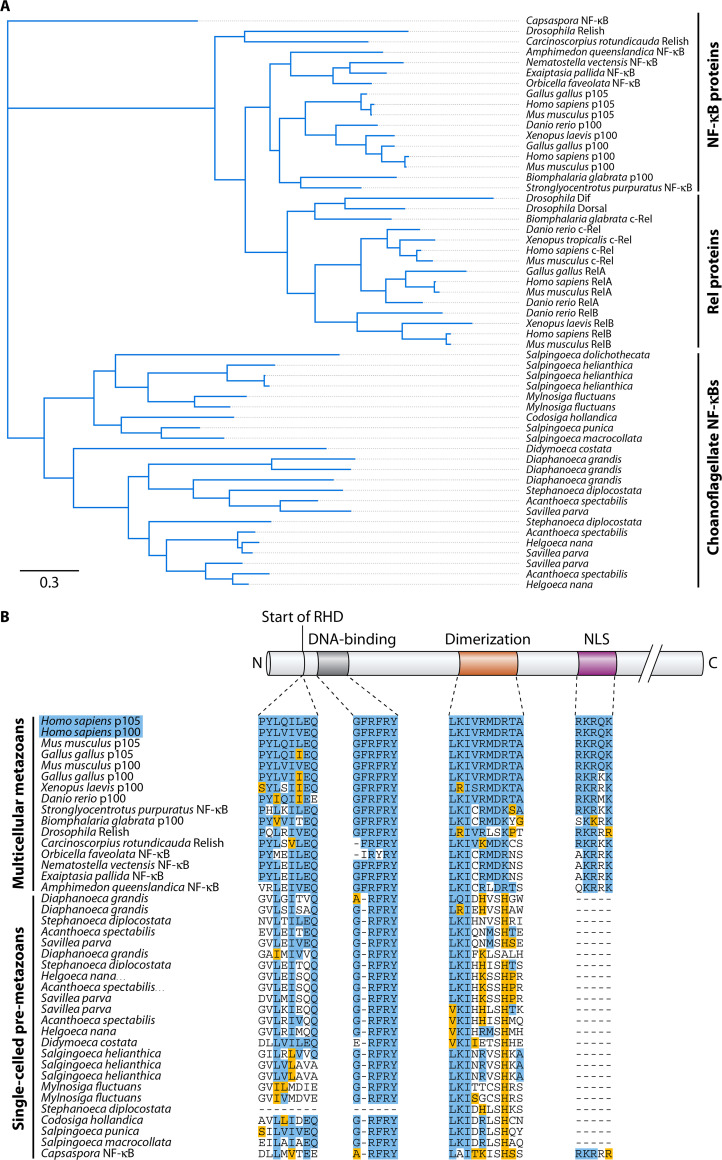
Although the choanoflagellate and Capsaspora NF-κBs differ from one another in their overall structures (i.e., especially the absence of a C-terminal ANK repeat region in choanoflagellates), they share two motifs within their RHDs (Fig. S1, motifs 7 and 10 in teal and yellow) that are not present in any metazoan NF-κBs. Among choanoflagellate NF-κBs, there is a great deal of sequence diversity, which is consistent with the overall genetic differences among choanoflagellates, wherein the average phylogenetic distance between any pair of choanoflagellates is greater than the phylogenetic distance between sponges and mammals (12). Nevertheless, a Bayesian tree analysis of the choanoflagellate RHDs identified to date clusters them as an outgroup to several fly and vertebrate NF-κBs and Rels, which demonstrates a clear divergence of the choanoflagellate NF-κBs from the metazoan NF-κB superfamily (Fig. 2A). In general, the NF-κBs within a single choanoflagellate species that has multiple NF-κB proteins (e.g., Salpingoeca rosetta) are highly related (Fig. 2A), suggesting that they arose by gene duplication events. However, it is important to note that some RHD subdomains are not present in the single-celled premetazoan lineages. For example, Capsaspora and choanoflagellates have a low homology in the dimerization sequence, and choanoflagellates have no NF-κB-like NLS. All metazoans and premetazoan NF-κB sequences contain a highly conserved sequence that is important for DNA binding (Fig. 2B).
FIG 2.
Phylogenetic analysis places choanoflagellate NF-κBs as an outgroup of vertebrate and fly NF-κBs. (A) Bayesian analysis of holozoan RHDs, including the recently identified choanoflagellate NF-κBs. MEME analysis was performed on each sequence to identify shared motifs. NF-κB proteins and Rel proteins cluster separately from each other, and choanoflagellate NF-κBs cluster as a single outgroup. (B) A multiple-sequence alignment of important NF-κB subdomains across multicellular metazoans and single-celled premetazoans. Exact amino acid matches to either p100 or p105 are highlighted in blue across phyla, and residues with conserved functional groups are highlighted in yellow. All multicellular metazoans contain a defined start to the Rel homology domain (RHD), dimerization sequence, and nuclear localization sequence (NLS). Single-celled premetazoans have a less defined RHD start, low homology in the dimerization sequence, and no canonical NLS (with the exception of Capsaspora). All metazoan and premetazoan NF-κB sequences contain a highly conserved DNA-binding sequence.
Among poriferans, the fully sequenced genome of the demosponge Amphimedon queenslandica encodes a single full-length NF-κB protein that contains clear features of the human p100 NF-κB protein: that is, it has an RHD, an NLS, a GRR, six ANK repeats, and a DD (22, 28) (Fig. 1). However, between the GRR and the ANK repeats, A. queenslandica NF-κB contains a region that has no known function and no homology to any other protein (22, 28) (Fig. 1, yellow). More limited sequencing data indicate that other sponges also contain NF-κB-like transcripts, which, to date, contain only RHD sequences (29). Many of these sponge NF-κB-like proteins appear much shorter than the prototypical NF-κB proteins, but this may simply be due to a lack of sequence coverage; for example, Corticium candelabrum is reported to have a transcript for NF-κB that consists of only 86 amino acids that are similar to the beginning of the RHD (29).
All characterized cnidarians appear to contain single NF-κB proteins, but the overall structures of these NF-κB proteins have diverged, in some cases, likely by gene-splitting events. Thus, many cnidarians the anemones (Edwardsiella lineata [24] and Exaiptasia pallida [23] and the corals Acropora digitifera, Stylophora pistillata, Orbicella faveolata, and Pocillopora damicornis [19–21]) have prototypical RHD-ANK repeat bipartite proteins, whereas others have separate RHD and ANK repeat proteins. In the best-studied example of the latter, the sea anemone Nematostella vectensis has a single 440-amino-acid NF-κB-like protein that is most similar to the processed vertebrate p50/p52 NF-κB proteins, based on phylogenetic analysis of RHD amino acid sequences, the intron-exon structure of the RHD, and the presence of a GRR after the RHD (30). Moreover, a separate N. vectensis gene encodes an IκB-like ANK repeat protein with substantial homology to C-terminal sequences of the ANK repeat domains of vertebrate p100/p105 (30). That the separate N. vectensis NF-κB and IκB genes arose due to a gene-splitting event is supported by four lines of evidence: (i) the existence of RHD-ANK repeat bipartite proteins in more basal organisms (e.g., Capsaspora and A. queenslandica), (ii) the presence of remnant GRR sequences at the C terminus of N. vectensis NF-κB, (iii) the homology of the separate N. vectensis IκB protein to mammalian NF-κB protein C-terminal ANK repeats, and (iv) the presence of an intact RHD-ANK protein in the closely related anemone E. lineata (24). Similar to N. vectensis, the hydras Hydractinia symbiolongicarpus (31) and Hydra magnipapillata (32) and the jellyfish Aurelia (13) have separate RHD-only NF-κB proteins and IκB-like genes, all of which likely came about due to gene-splitting events, which are common in cnidarians (33, 34).
ACTIVITY AND REGULATION OF BASAL NF-κBs
The activity and regulation of NF-κB proteins in vertebrates are now known in great detail. Many of these properties are conserved in basal NF-κBs; however, there are clear exceptions.
Although not formally shown either biochemically or structurally, it is likely that all basal NF-κB-like proteins bind as dimers to extremely similar DNA sites. Nevertheless, the absence of a clearly defined dimerization sequence in the premetazoan NF-κBs does raise the possibility that they bind DNA as monomers, similar to what has been found for the NFAT proteins (35). However, consistent with the basal NF-κBs binding as dimers, the NF-κB κB site protein-DNA complexes seen in electrophoretic mobility shift assays using cnidarian and sponge NF-κB proteins migrate in a manner suggesting that the NF-κB proteins are dimers (19, 23, 28, 36). Furthermore, based on protein-binding microarrays (PBMs), the NF-κB proteins of Capsaspora, the sponge A. queenslandica, and two sea anemones bind to a set of κB sites that are more similar to the sites bound by mammalian NF-κB proteins than to those bound by Rel proteins (23, 37), a biochemical finding that is consistent with their overall structural organization and phylogenetic data.
Basal NF-κB proteins appear to be activators of κB site-containing promoters. That is, the RHD sequences of sponge and cnidarian NF-κB proteins function as activators of transcription when expressed in reporter gene assays in Saccharomyces cerevisiae and human cells (19, 23, 28, 36). The ability of these basal NF-κBs to function as independent activators of transcription is more similar to that of Relish, which can act as a homodimeric transcriptional activator (17), than to that of mammalian p50/p52 proteins, which are generally activators of transcription only when in heterodimers with a Rel protein (14, 15). No DNA-binding or transcriptional regulatory studies have been performed with choanoflagellate NF-κBs.
The most extensive research on basal NF-κB activity has been done with the NF-κB protein of the sea anemone N. vectensis (Nv-NF-κB). Curiously, it was found that wild populations of these anemones have two major alleles of Nv-NF-κB that differ at 10 residues, 6 of which are in the RHD (38). Two of these variable residues are ones that are predicted to contact DNA, and, as a consequence, one of the allelic Nv-NF-κB proteins (Nv-NF-κB-C) binds DNA with an approximately 2-fold higher affinity than the other variant (Nv-NF-κB-S) (39). Despite its reduced DNA-binding activity, Nv-NF-κB-S activates transcription in reporter gene assays in human cells more effectively than Nv-NF-κB-C (39). Overall, the PBM-based binding site profiles of both Nv-NF-κB alleles still largely resemble the profile of mammalian p50 (23, 37). Interestingly, the DNA-binding site profiles of Nv-NF-κB-C and Nv-NF-κB-S are as different from each other as each profile is different from the profile of human p50 (23), suggesting that within a given species (i.e., N. vectensis) there can be considerable flexibility in DNA-binding site recognition that does not have an obvious effect on the organism’s overall developmental phenotype. However, anemones harboring the distinct NF-κB alleles may have differences in other properties, such as immunity.
In the mammalian noncanonical pathway, human p100 is phosphorylated by IKKα at a cluster of serine residues that are located C terminal to the ANK repeats, and this phosphorylation induces proteasomal processing of the C-terminal sequences up to the GRR (16). This cluster of serine residues is conserved in many sequenced cnidarian NF-κBs, including those of the corals Pocillopora damicornis, Orbicella faveolata, Stylophora pistillata, and Acropora millepora and the sea anemones Actinia tenebrosa, Aulactinia veratra, and Aiptasia, as well as one sponge (A. queenslandica) (7, 19–23, 28, 40, 41). Furthermore, phosphorylation and proteasomal processing of NF-κBs from two cnidarians (Aiptasia and O. faveolata) and the sponge A. queenslandica can be induced by coexpression of human IKKα and IKKβ or the single Aiptasia or O. faveolata IKK (19, 23, 28), suggesting that IKK-dependent processing of cnidarian and sponge NF-κBs can occur in their natural settings. However, no IKK homolog has yet been identified in the sponge A. queenslandica. On the other hand, the Capsaspora NF-κB protein clearly has C-terminal ANK repeats and a GRR, but it does not have any apparent C-terminal IKK target site serine residues, and no IKK can be found in the Capsaspora genome. Intriguingly, Capsaspora does have at least one possible caspase cleavage site, similar to what is seen in the Drosophila protein Relish (Fig. 1; Fig. S2). Overall, the emergence of IKK proteins appears to be generally coincident with the presence of regulatory serine residues in the ANK repeat domains of basal NF-κBs. Taken together, these findings suggest that phosphorylation-dependent processing of the RHD-ANK repeat proteins was first invented in sponges and that a different form of regulated processing occurs with Capsaspora NF-κB. Of note, the Drosophila Relish protein also lacks C-terminal serine residues and a GRR and is processed by a site-specific protease cleavage event (17), and under some conditions, shortened human p105 NF-κB proteins can be generated by ribosomal stalling (42).
For the N. vectensis NF-κB protein, which lacks C-terminal ANK repeats, it was shown that the independent N. vectensis IκB (Nv-IκB) could directly bind to Nv-NF-κB and that this Nv-IκB could be phosphorylated by an N. vectensis IKK (Nv-IKK)-related kinase (36). Thus, aspects of canonical signaling may also be present in cnidarians.
Proteasome-mediated processing has not been directly demonstrated to induce the nuclear translocation of any basal NF-κB in its natural setting. Furthermore, while the ubiquitin-proteasome system is present in basal phyla, regulated processing has not been experimentally demonstrated for any protein (43). However, in a reconstituted mammalian cell system, IKK-induced processing of the anemone Aiptasia NF-κB can be blocked by a proteasome inhibitor (23). In addition, when expressed in chicken tissue culture cells, the full-length RHD-ANK proteins of the coral O. faveolata, the anemone Aiptasia, and the sponge A. queenslandica are located in the cytoplasm, and deletion of the ANK repeat domains of these NF-κBs causes them to localize to the nucleus (19, 23, 28), suggesting that C-terminal processing would do the same in their host organisms. Similarly, in the same assays, the naturally truncated anemone Nv-NF-κB protein localizes to the nucleus, but it is sequestered in the cytoplasm when Nv-IκB is coexpressed (36). Of note, although the C-terminal ANK repeat sequences block nuclear translocation and reporter gene activation by the sponge NF-κB protein, the ANK repeats do not inhibit NF-κB’s in vitro DNA-binding activity (28), unlike what is seen with NF-κBs of cnidarians and other higher metazoans. Nothing is known about the subcellular localization or regulation of the choanoflagellate NF-κB proteins, which lack a clear NLS as well as C-terminal ANK repeats.
Notwithstanding the above-described reconstitution experiments conducted with cnidarian and sponge NF-κBs in vertebrate cell systems, it is not known whether or how regulated processing of basal NF-κBs occurs in any natural setting. Indeed, in both the anemone Aiptasia (23) and one demosponge (28), most NF-κB protein is largely processed and nuclear in the absence of any known stimulus. Furthermore, certain treatments (e.g., bleaching in Aiptasia and lipopolysaccharide [LPS] treatment of O. faveolata coral tissue) may induce NF-κB activation by inducing the transcriptional upregulation of NF-κB pathway genes rather than C-terminal processing (19, 23).
BASAL NF-κBs LIKELY HAVE ROLES IN DEVELOPMENT AND IMMUNITY
One of the most important questions regarding NF-κB in basal metazoans is what biological processes are regulated by this transcription factor. At least in cnidarians and sponges, there is evidence that NF-κB has roles in both early development and adult immunity (Table 1).
TABLE 1.
Functions of NF-κB in basal organisms
| Organism | NF-κB structure | Biological function(s) | Data available | Reference(s) |
|---|---|---|---|---|
| Aiptasia | RHD-ANK | Immunity | Protein, transcriptomic, mRNA, DNA-binding | 23, 56, 57, 68 |
| N. vectensis | RHD only | Immunity (?) and development | Protein, transcriptomic, mRNA, DNA-binding | 36, 45, 49, 69 |
| Hydra | RHD only | Immunity | Transcriptomic, protein | 31–33 |
| Corals | RHD-ANK | Immunity | Transcriptomic | 19, 40, 59–62 |
| Sponge | RHD-ANK | Immunity and development | Protein, transcriptomic, mRNA, DNA-binding | 22, 28, 53, 54 |
| Capsaspora | RHD-ANK | ? | Genomic | 11 |
| Choanoflagellates | RHD only | ? | Transcriptomic | 12 |
Role of NF-κB in sponge and cnidarian development.
In the sponge A. queenslandica and several cnidarians, NF-κB transcripts are expressed in the early embryo. In A. queenslandica, fluorescent in situ staining has shown that NF-κB transcripts are broadly expressed throughout the embryo after cleavage and are particularly strong in granular cells, which eventually become the outer layer of the developing embryo (22). Similarly, some cnidarians express NF-κB transcripts in the early embryo and juvenile larvae (44–49). In the anemone N. vectensis, NF-κB mRNA expression is seen as early as 1 h postfertilization (hpf) (46–49), and nuclear NF-κB protein was detected as early as the late gastrula stage (45). Morpholino-based knockdown of NF-κB in the developing N. vectensis embryo led to a failure to develop cnidocytes at the juvenile polyp stage (45). Cnidocytes are a Cnidaria-specific cell type that is involved in a variety of sensing, prey capture, and, perhaps, defense roles (50). In addition, morpholino knockdown of the single NF-κB-inducing Toll-like receptor (TLR) transcript also led to an early developmental defect in N. vectensis (51).
Role of NF-κB in juvenile and adult sponges and cnidarians.
NF-κB transcripts and proteins have also been detected in several cell types in developed sponge tissues. Whole-mount in situ hybridization of juvenile sponge tissue showed that NF-κB transcripts are present in flask cells, which are large ciliated cells that express a range of genes whose orthologues play roles in eumetazoan neurons (22). Two recent studies (52, 53) performed single-cell sequencing on different adult sponge cell types, and they reported that NF-κB transcripts were primarily expressed in two cell types: choanocytes, which are cells that reside in the chambers that allow the sponge to filter feed, and archeocytes, which are motile phagocytic cells that are inside the mesohyl (a gelatinous matrix between the internal and external layers of sponges). In a black encrusting sponge (Cliona sp.), nuclear NF-κB protein was detected in several scattered cells throughout the animal (28). Furthermore, anti-NF-κB Western blots of whole-tissue extracts from this Cliona sp. contained proteins similar in size to the NF-κB proteins of A. queenslandica. Treatment of this sponge tissue with lipopolysaccharide (LPS; a potent TLR-to-NF-κB inducer in mammals) produced further processing of the putative full-length NF-κB protein and an increase in κB-site binding activity. Similarly, treatment of two Mediterranean species of sponges, Aplysina aerophoba and Dysidea avara, with an immunogenic cocktail of LPS and peptidoglycan led to the upregulation of immune-related receptors involved in signaling to NF-κB (54).
Taken together, the above-described studies suggest that NF-κB plays a role in a select number of cells involved in sponge immunity. In one sponge, the NF-κB protein appears to be constitutively in its processed active state and in the nucleus of select cells (28). Whether this constitutive nuclear localization of NF-κB is the result of a chronic processing of this protein in the sponge or represents a naturally truncated isoform that is preferentially expressed has not been determined.
Several studies have suggested that NF-κB also has a role in immunity in adult cnidarians. In the anemone N. vectensis, cytoplasmic NF-κB and IκB are expressed in a subset of cnidocytes in the body column of juvenile and adult anemones (44). NF-κB protein is also highly expressed in cnidocytes, which are present in circulating multicellular bodies called nematosomes, which also express high levels of TLR and c-GAS-STING innate immune signaling components, which are upstream of NF-κB (50, 51). Those results, along with the ability of nematosomes to take up bacteria (50, 51), suggest that the nematosome is a primitive circulating immune organ in some anemones.
Aiptasia is a tropical anemone that has been used as a model for cnidarian symbiosis with algal dinoflagellates in the family Symbiodiniaceae (55). Colonization of both larval and adult Aiptasia anemones with algal symbionts of certain strains results in the downregulation of NF-κB transcripts, protein, and DNA-binding activity (23, 56, 57). Conversely, induction of loss of symbiosis with either chemical or heat treatment results in increased levels of NF-κB expression and activity (23), and Aiptasia anemones lacking symbionts are more resistant to bacterial infection than ones with symbionts (57). These results have led to the hypothesis that the suppression of NF-κB-directed immunity is required for the establishment of algal symbiosis in some cnidarians, similar to what has been found in amphibians (58). It is noteworthy that the symbiont-modulated changes in NF-κB activity in Aiptasia occurred primarily at the level of expression (23) and not by posttranslational induction, as is generally seen in vertebrate systems. That is, both symbiotic and aposymbiotic Aiptasia anemones had nuclear NF-κB-staining cells, but the number of NF-κB-positive cells was substantially increased in Aiptasia anemones in which symbiont loss had occurred (23).
The research on NF-κB in Aiptasia is of high interest because corals host the same family of algal symbionts hosted by Aiptasia and because the ocean warming-induced loss of symbiosis (bleaching), as well as microbial pathogen infections, is causing a large-scale loss of coral reef health (55). Several studies have also investigated NF-κB and NF-κB pathway gene expression in corals, and such studies further suggest a role for NF-κB in immunity and symbiosis/dysbiosis (Table 1). For example, transcripts encoding NF-κB and its signaling components have been identified in transcriptomes from bleached O. faveolata coral (59), and treatment of O. faveolata tissue with LPS resulted in the differential expression of genes in a manner that indicated activation of the NF-κB pathway (19). Furthermore, NF-κB mRNA was substantially upregulated in the coral Acropora palmata following extended exposure to an elevated water temperature (60), while another study found that NF-κB transcripts were only transiently induced shortly after heat treatment in the coral Acropora hyacinthus (61). Thus, NF-κB may be affected in different ways in different cnidarians undergoing heat stress-induced changes. Additionally, some studies have shown an increase in transcripts encoding members of the NF-κB pathway in corals with microbial diseases (19, 62), further suggesting that NF-κB has an immune-related role in cnidarians.
Several lines of evidence suggest that TLR and TLR-like pathways are upstream activators of NF-κB in cnidarians. First, homologs of TLR-to-NF-κB pathway signaling are present in most cnidarians (6–8, 13, 19, 20, 30, 33, 36). Second, Hydra and N. vectensis TLR-like proteins have a conserved ability to activate NF-κB when ectopically expressed in human cells (32, 51). Third, as mentioned above, TLR and NF-κB proteins are expressed in many of the same cnidocytes in adult N. vectensis anemones (51). Fourth, treatment of O. faveolata coral tissue with the mammalian TLR ligand LPS can induce NF-κB pathway gene expression (19). Indeed, TLR-to-NF-κB signaling has been proposed to play a role in embryonic development (44, 51), immunity (19, 32, 51, 59, 62), and regeneration (63) in cnidarians. Nevertheless, TLR is not the sole receptor that can signal to NF-κB in mammals that is also present in cnidarians. For example, cnidarians also possess homologs to tumor necrosis factor (TNF) receptors and their downstream components (30), which are extensively characterized activators of NF-κB in vertebrates. However, the ability of cnidarian TNF receptors to activate NF-κB in cnidarians has not been investigated.
The biological role of NF-κB in single-celled organisms remains unknown (Table 1). However, one can speculate that the biological processes controlled by NF-κB in protists are quite different from those controlled by NF-κB in multicellular animals, given that protists have no innate or adaptive immunity in the traditional sense of specialized immune cells. Thus, if NF-κB plays a role in immunity in protists, it may do so in an unexpected manner. Or, in the case of Capsaspora, which is a symbiont of the snail B. glabrata, NF-κB may modulate either protist or host immunity to facilitate symbiosis, similar to what has been proposed for Aiptasia-Symbiodiniaceae symbiosis. Alternatively, NF-κB could play a developmental role in the solitary versus colony states seen in many protists, including choanoflagellates. Finally, NF-κB may be involved in some protist-specific process. For example, one species of choanoflagellate has been shown to be induced to sexually reproduce upon stimulation with bacterial components (64).
CONCLUSIONS AND FUTURE PROSPECTS
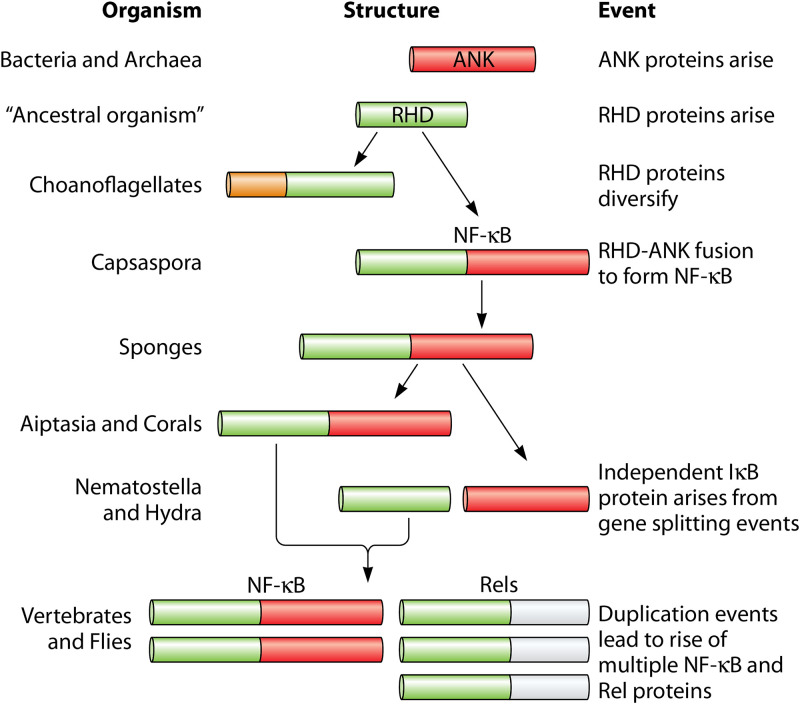
Since its discovery, NF-κB has been studied as a prominent player in many biological processes in vertebrate and fly systems. Nonetheless, until a decade ago, NF-κB was not known to be present in the genomes of organisms basal to insects. The discovery of NF-κBs in single-celled protists has led to improved hypotheses for how this transcription factor evolved (Fig. 3). That is, we propose that prior to the rise of holozoan life, ANK repeats were present in bacteria and archaea genomes. Some ancestral organism developed a primordial RHD-only protein that eventually fused to an ANK repeat protein, resulting in the modern-day bipartite NF-κB protein. RHDs then diversified in choanoflagellates, while the metazoan lineage retained the full-length NF-κB fusion protein and also developed separate NF-κB/Rel and IκB proteins, likely from gene-splitting and duplication events (Fig. 3).
FIG 3.
Evolution of NF-κB. Prior to the rise of holozoan life, ankyrin (ANK) repeats (red) were present in bacteria and archaea. The appearance of an RHD-containing protein (green) likely led to RHD-only proteins that diversified in choanoflagellates and to an RHD-ANK fusion in an ancestor of Capsaspora. Metazoans generally retained the full-length NF-κB fusion protein, although in some cnidarians (e.g., Hydra and Nematostella) there were gene-splitting events that created separate RHD and ANK repeat proteins. Eventually, a series of duplication events gave rise to the multiple NF-κB and Rel proteins that are present in vertebrates and flies.
Several studies have now demonstrated that the NF-κB proteins of evolutionarily basal organisms have many of the same structural features, activities, modes of regulation, and biological effects that are found in the expanded set of NF-κB and Rel proteins of more complex organisms. For example, ANK repeat regulation of the NF-κB RHD is even found in protists, and NF-κB is likely to have dual roles in development and immunity in organisms ranging from sponges to humans. Nevertheless, there are clear differences in basal NF-κBs. Particularly intriguing is the finding that NF-κB proteins appear to be constitutively processed and active in some sponges and anemones and that induced activation of the NF-κB pathway may be at the transcriptional level in some of the same basal organisms. It is interesting to speculate that continuous exposure to pathogens or other activating ligands induces NF-κB processing in these situations and results in a constant heightened immunological state. We note that this concept of heightened immunity has been suggested in more complex organisms as well; for example, some species of bats appear to have constitutively active immune systems due to their microbe-rich habitats (65, 66).
Overall, it is clear that research on the evolution of NF-κB has only scratched the surface of what is yet to be discovered. Future studies will likely seek to identify the most primitive biological processes controlled by NF-κB in protists. Moreover, the identification of NF-κB target genes and the development of genetic systems to study gene function will certainly reveal much about the function of NF-κB in cnidarians and sponges. In particular, the identification of the immune response gene targets of NF-κB in basal organisms may provide insights into novel antimicrobials, as well as reveal information about the molecular processes underlying global ecological crises of marine invertebrates, such as coral bleaching and microbial pathogenesis. Looking backwards at the evolutionary history of NF-κB will no doubt bring further knowledge to our understanding of extant immunological systems, especially in organisms that are environmentally sensitive and that are situated at the base of multicellular life.
PHYLOGENETIC ANALYSIS
For the phylogenetic analysis, the RHD sequences of NF-κB proteins from cnidarians, poriferans, and choanoflagellates were compared to the sequences of annotated vertebrate NF-κB and Rel proteins and rooted with the predicted RHD of Capsaspora owczarzaki NF-κB. Details on the databases and sequence acquisition can be found in Table S2 in the supplemental material. Conserved motifs from MEME analysis were truncated based on motif predictions (Table S2) and were aligned by use of the Clustal Omega program (67) for sequence alignment and Bayesian analysis (Fig. 2A). Other phylogenetic analyses showed similar results (data not shown).
Supplementary Material
ACKNOWLEDGMENTS
We thank Anvitha Addanki for help with the analysis of choanoflagellate NF-κB proteins.
Our research on the evolution and basal functions of NF-κB was supported by the following National Science Foundation grants (to T.D.G.): MCB-0924749, IOS-1557804, and IOS-1937650. L.M.W. was supported by an NSF Graduate Research Fellowship.
We declare no conflict of interest.
Biographies

Leah M. Williams is a Ph.D. candidate in cell and molecular biology at Boston University (BU). She received her bachelor of arts from Wheaton College (Massachusetts) and then worked for 2 years at Harvard Medical School doing proteomic pathway discovery before starting her graduate career. Ms. Williams’s doctoral research seeks to understand immunity in evolutionarily distant and ecologically significant organisms, and she has published seminal papers on basal immunity in sponges and cnidarians. Her research has been supported by multiple internal BU grants and external grants, including an NSF GRFP. Ms. Williams has been a mentor for a dozen undergraduate and high school students, including NSF-REU students, and she has served on panels for science fairs and volunteered in various scientific teaching settings for students in K-12 through college. Ms. Williams intends to pursue postdoctoral research combining molecular and systems approaches to answer evolutionary and ecological questions.

Thomas D. Gilmore is a professor of biology at Boston University (BU). He received his Ph.D. from the University of California, Berkeley, performed postdoctoral research at the University of Wisconsin, and has been on the faculty at BU since 1987. Dr. Gilmore’s lab has made key contributions to many aspects of NF-κB biology, starting with his postdoctoral studies on the v-Rel oncoprotein with Howard Temin. Dr. Gilmore lab’s longstanding research interest has been in the area of NF-κB signaling and diseases, including human B-cell diseases, and disease pathologies of marine invertebrates, such as corals and sea anemones. This research has been supported by grants from the NIH, NSF, and the American Cancer Society, among others. Dr. Gilmore has written numerous reviews on NF-κB, and his lab hosts the website nf-kb.org. Dr. Gilmore has carried out research primarily with an army of student trainees, including 27 Ph.D. students, 41 M.A. students, and over 100 undergraduates. Dr. Gilmore has received BU’s highest awards for teaching and research.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gilmore TD, Temin HM. 1986. Different localization of the product of the v-rel oncogene in chicken fibroblasts and spleen cells correlates with transformation by REV-T. Cell 44:791–800. doi: 10.1016/0092-8674(86)90845-7. [DOI] [PubMed] [Google Scholar]
- 2.Gilmore TD. 1990. NF-κB, KBF1, dorsal, and related matters. Cell 62:841–843. doi: 10.1016/0092-8674(90)90257-f. [DOI] [PubMed] [Google Scholar]
- 3.Steward R. 1987. Dorsal, an embryonic polarity gene in Drosophila, is homologous to the vertebrate proto-oncogene, c-rel. Science 238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh S, Gifford AM, Riviere LR, Tempst P, Nolan GP, Baltimore D. 1990. Cloning of the p50 DNA binding subunit of NF-κB: homology to rel and dorsal. Cell 62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- 5.Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban MB, Kourilsky P, Baeuerle PA, Israël A. 1990. The DNA binding subunit of NF-κB is identical to factor KBF1 and homologous to the rel oncogene product. Cell 62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 6.Gilmore TD, Wolenski FS. 2012. NF-κB: where did it come from and why? Immunol Rev 246:14–35. doi: 10.1111/j.1600-065X.2012.01096.x. [DOI] [PubMed] [Google Scholar]
- 7.Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, Gough J, Weis VM, Aranda M, Pringle JR, Voolstra CR. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A 112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, Fujiyama A, Miller DJ, Satoh N. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- 9.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sánchez-Pons N, Torruella G, Derelle R, Manning G, Lang BF, Russ C, Haas BJ, Roger AJ, Nusbaum C, Ruiz-Trillo I. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun 4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter DJ, Fozouni P, Eisen MB, King N. 2018. Gene family innovation, conservation and loss on the animal stem lineage. Elife 7:e34226. doi: 10.7554/eLife.34226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold DA, Katsuki T, Li Y, Yan X, Regulski M, Ibberson D, Holstein T, Steele RE, Jacobs DK, Greenspan RJ. 2019. The genome of the jellyfish Aurelia and the evolution of animal complexity. Nat Ecol Evol 3:96–104. doi: 10.1038/s41559-018-0719-8. [DOI] [PubMed] [Google Scholar]
- 14.Gilmore TD. 2006. Introduction to NF-κB: players, pathways, perspectives. Oncogene 25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 15.Hayden MS, Ghosh S. 2008. Shared principles in NF-κB signaling. Cell 132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Sun S-C. 2011. Non-canonical NF-κB signaling pathway. Cell Res 21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stöven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engström Y, Maniatis T, Hultmark D. 2003. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc Natl Acad Sci U S A 100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan JJ, Gilmore TD. 2018. Evolutionary origins of Toll-like receptor signaling. Mol Biol Evol 35:1576–1587. doi: 10.1093/molbev/msy050. [DOI] [PubMed] [Google Scholar]
- 19.Williams LM, Fuess LE, Brennan JJ, Mansfield KM, Salas-Rodriguez E, Welsh J, Awtry J, Banic S, Chacko C, Chezian A, Dowers D, Estrada F, Hsieh Y-H, Kang J, Li W, Malchiodi Z, Malinowski J, Matuszak S, McTigue T, Mueller D, Nguyen B, Nguyen M, Nguyen P, Nguyen S, Njoku N, Patel K, Pellegrini W, Pliakas T, Qadir D, Ryan E, Schiffer A, Thiel A, Yunes SA, Spilios KE, Pinzón C JH, Mydlarz LD, Gilmore TD. 2018. A conserved Toll-like receptor-to-NF-κB signaling pathway in the endangered coral Orbicella faveolata. Dev Comp Immunol 79:128–136. doi: 10.1016/j.dci.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 20.Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot J-F, Tambutté S, Allemand D, Aranda M. 2017. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep 7:17583. doi: 10.1038/s41598-017-17484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N. 2018. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci Rep 8:16134. doi: 10.1038/s41598-018-34459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gauthier M, Degnan BM. 2008. The transcription factor NF-κB in the demosponge Amphimedon queenslandica: insights on the evolutionary origin of the Rel homology domain. Dev Genes Evol 218:23–32. doi: 10.1007/s00427-007-0197-5. [DOI] [PubMed] [Google Scholar]
- 23.Mansfield KM, Carter NM, Nguyen L, Cleves PA, Alshanbayeva A, Williams LM, Penvose AR, Crowder C, Finnerty JR, Gilmore TD, Siggers TW, Weis VM. 2017. Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci Rep 7:16025. doi: 10.1038/s41598-017-16168-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefanik DJ, Lubinski TJ, Granger BR, Byrd AL, Reitzel AM, DeFilippo L, Lorenc A, Finnerty JR. 2014. Production of a reference transcriptome and transcriptomic database (EdwardsiellaBase) for the lined sea anemone, Edwardsiella lineata, a parasitic cnidarian. BMC Genomics 15:71. doi: 10.1186/1471-2164-15-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. 2010. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol 18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jernigan KK, Bordenstein SR. 2014. Ankyrin domains across the Tree of Life. PeerJ 2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmeyer TT, Burkhardt P. 2016. Choanoflagellate models—Monosiga brevicollis and Salpingoeca rosetta. Curr Opin Genet Dev 39:42–47. doi: 10.1016/j.gde.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Williams LM, Inge MM, Mansfield KM, Rasmussen A, Afghani J, Agrba M, Albert C, Andersson C, Babaei M, Babaei M, Bagdasaryants A, Bonilla A, Browne A, Carpenter S, Chen T, Christie B, Cyr A, Dam K, Dulock N, Erdene G, Esau L, Esonwune S, Hanchate A, Huang X, Jennings T, Kasabwala A, Kehoe L, Kobayashi R, Lee M, LeVan A, Liu Y, Murphy E, Nambiar A, Olive M, Patel D, Pavesi F, Petty CA, Samofalova Y, Sanchez S, Stejskal C, Tang Y, Yapo A, Cleary JP, Yunes SA, Siggers T, Gilmore TD. 2020. Transcription factor NF-κB in a basal metazoan, the sponge, has conserved and unique sequences, activities, and regulation. Dev Comp Immunol 104:103559. doi: 10.1016/j.dci.2019.103559. [DOI] [PubMed] [Google Scholar]
- 29.Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol 31:1102–1120. doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan JC, Kalaitzidis D, Gilmore TD, Finnerty JR. 2007. Rel homology domain-containing transcription factors in the cnidarian Nematostella vectensis. Dev Genes Evol 217:63–72. doi: 10.1007/s00427-006-0111-6. [DOI] [PubMed] [Google Scholar]
- 31.Zárate-Potes A, Ocampo ID, Cadavid LF. 2019. The putative immune recognition repertoire of the model cnidarian Hydractinia symbiolongicarpus is large and diverse. Gene 684:104–117. doi: 10.1016/j.gene.2018.10.068. [DOI] [PubMed] [Google Scholar]
- 32.Franzenburg S, Fraune S, Künzel S, Baines JF, Domazet-Loso T, Bosch TCG. 2012. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci U S A 109:19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, Disbennett K, Pfannkoch C, Sumin N, Sutton GG, Viswanathan LD, Walenz B, Goodstein DM, Hellsten U, Kawashima T, Prochnik SE, Putnam NH, Shu S, Blumberg B, Dana CE, Gee L, Kibler DF, Law L, Lindgens D, Martinez DE, Peng J, Wigge PA, Bertulat B, Guder C, Nakamura Y, Ozbek S, Watanabe H, Khalturin K, Hemmrich G, Franke A, Augustin R, Fraune S, Hayakawa E, Hayakawa S, Hirose M, Hwang JS, Ikeo K, Nishimiya-Fujisawa C, Ogura A, Takahashi T, Steinmetz PRH, et al. 2010. The dynamic genome of Hydra. Nature 464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gacesa R, Chung R, Dunn SR, Weston AJ, Jaimes-Becerra A, Marques AC, Morandini AC, Hranueli D, Starcevic A, Ward M, Long PF. 2015. Gene duplications are extensive and contribute significantly to the toxic proteome of nematocysts isolated from Acropora digitifera (Cnidaria: Anthozoa: Scleractinia). BMC Genomics 16:774. doi: 10.1186/s12864-015-1976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroud JC, Chen L. 2003. Structure of NFAT bound to DNA as a monomer. J Mol Biol 334:1009–1022. doi: 10.1016/j.jmb.2003.09.065. [DOI] [PubMed] [Google Scholar]
- 36.Wolenski FS, Garbati MR, Lubinski TJ, Traylor-Knowles N, Dresselhaus E, Stefanik DJ, Goucher H, Finnerty JR, Gilmore TD. 2011. Characterization of the core elements of the NF-κB signaling pathway of the sea anemone Nematostella vectensis. Mol Cell Biol 31:1076–1087. doi: 10.1128/MCB.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryzhakov G, Teixeira A, Saliba D, Blazek K, Muta T, Ragoussis J, Udalova IA. 2013. Cross-species analysis reveals evolving and conserved features of the nuclear factor κB (NF-κB) proteins. J Biol Chem 288:11546–11554. doi: 10.1074/jbc.M113.451153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan JC, Wolenski FS, Reitzel AM, French CE, Traylor-Knowles N, Gilmore TD, Finnerty JR. 2009. Two alleles of NF-κB in the sea anemone Nematostella vectensis are widely dispersed in nature and encode proteins with distinct activities. PLoS One 4:e7311. doi: 10.1371/journal.pone.0007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolenski FS, Chandani S, Stefanik DJ, Jiang N, Chu E, Finnerty JR, Gilmore TD. 2011. Two polymorphic residues account for the differences in DNA binding and transcriptional activation by NF-κB proteins encoded by naturally occurring alleles in Nematostella vectensis. J Mol Evol 73:325–336. doi: 10.1007/s00239-011-9479-7. [DOI] [PubMed] [Google Scholar]
- 40.Anderson DA, Walz ME, Weil E, Tonellato P, Smith MC. 2016. RNA-Seq of the Caribbean reef-building coral Orbicella faveolata (Scleractinia-Merulinidae) under bleaching and disease stress expands models of coral innate immunity. PeerJ 4:e1616. doi: 10.7717/peerj.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying H, Hayward DC, Cooke I, Wang W, Moya A, Siemering KR, Sprungala S, Ball EE, Forêt S, Miller DJ. 2019. The whole-genome sequence of the coral Acropora millepora. Genome Biol Evol 11:1374–1379. doi: 10.1093/gbe/evz077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L, DeMartino GN, Greene WC. 1998. Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell 92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 43.Fort P, Kajava AV, Delsuc F, Coux O. 2015. Evolution of proteasome regulators in eukaryotes. Genome Biol Evol 7:1363–1379. doi: 10.1093/gbe/evv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siboni N, Abrego D, Motti CA, Tebben J, Harder T. 2014. Gene expression patterns during the early stages of chemically induced larval metamorphosis and settlement of the coral Acropora millepora. PLoS One 9:e91082. doi: 10.1371/journal.pone.0091082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolenski FS, Bradham CA, Finnerty JR, Gilmore TD. 2013. NF-κB is required for cnidocyte development in the sea anemone Nematostella vectensis. Dev Biol 373:205–215. doi: 10.1016/j.ydbio.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Warner JF, Guerlais V, Amiel AR, Johnston H, Nedoncelle K, Röttinger E. 2018. NvERTx: a gene expression database to compare embryogenesis and regeneration in the sea anemone Nematostella vectensis. Development 145:dev162867. doi: 10.1242/dev.162867. [DOI] [PubMed] [Google Scholar]
- 47.Fischer AH, Mozzherin D, Eren AM, Lans KD, Wilson N, Cosentino C, Smith J. 2014. SeaBase: a multispecies transcriptomic resource and platform for gene network interface. Integr Comp Biol 54:250–263. doi: 10.1093/icb/icu065. [DOI] [PubMed] [Google Scholar]
- 48.Tulin S, Aguiar D, Istrail S, Smith J. 2013. A quantitative reference transcriptome for Nematostella vectensis early embryonic development: a pipeline for de novo assembly in emerging model systems. EvoDevo 4:16. doi: 10.1186/2041-9139-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helm RR, Siebert S, Tulin S, Smith J, Dunn CW. 2013. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics 14:266. doi: 10.1186/1471-2164-14-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babonis LS, Martindale MQ. 2014. Old cell, new trick? Cnidocytes as a model for the evolution of novelty. Integr Comp Biol 54:714–722. doi: 10.1093/icb/icu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brennan JJ, Messerschmidt JL, Williams LM, Matthews BJ, Reynoso M, Gilmore TD. 2017. Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-κB signal transduction, and development. Proc Natl Acad Sci U S A 114:E10122–E10131. doi: 10.1073/pnas.1711530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sogabe S, Hatleberg WL, Kocot KM, Say TE, Stoupin D, Roper KE, Fernandez-Valverde SL, Degnan SM, Degnan BM. 2019. Pluripotency and the origin of animal multicellularity. Nature 570:519–522. doi: 10.1038/s41586-019-1290-4. [DOI] [PubMed] [Google Scholar]
- 53.Musser JM, Schippers KJ, Nickel M, Mizzon G, Kohn AB, Pape C, Hammel JU, Wolf F, Liang C, Hernández-Plaza A, Achim K, Schieber NL, Francis WR, Sv R, Kling S, Renkert M, Feuda R, Gaspar I, Burkhardt P, Bork P, Beck M, Kreshuk A, Wörheide G, Huerta-Cepas J, Schwab Y, Moroz LL, Arendt D. 2019. Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. bioRxiv doi: 10.1101/758276. [DOI] [PMC free article] [PubMed]
- 54.Pita L, Hoeppner M, Ribes M, Hentschel U. 2018. Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns. Sci Rep 8:16081. doi: 10.1038/s41598-018-34330-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weis VM. 2008. Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J Exp Biol 211:3059–3066. doi: 10.1242/jeb.009597. [DOI] [PubMed] [Google Scholar]
- 56.Wolfowicz I, Baumgarten S, Voss PA, Hambleton EA, Voolstra CR, Hatta M, Guse A. 2016. Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci Rep 6:32366–32312. doi: 10.1038/srep32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansfield KM, Cleves PA, Vlack EV, Kriefall NG, Benson BE, Camacho DJ, Hemond O, Pedroza M, Siggers T, Pringle JR, Davies SW, Gilmore TD. 2019. Varied effects of algal symbionts on transcription factor NF-κB in a sea anemone and a coral: possible roles in symbiosis and thermotolerance. bioRxiv doi: 10.1101/640177. [DOI]
- 58.Burns JA, Zhang H, Hill E, Kim E, Kerney R. 2017. Transcriptome analysis illuminates the nature of the intracellular interaction in a vertebrate-algal symbiosis. Elife 6:e22054. doi: 10.7554/eLife.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pinzón JH, Kamel B, Burge CA, Harvell CD, Medina M, Weil E, Mydlarz LD. 2015. Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R Soc Open Sci 2:140214. doi: 10.1098/rsos.140214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeSalvo MK, Sunagawa S, Voolstra CR, Medina M. 2010. Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar Ecol Prog Ser 402:97–113. doi: 10.3354/meps08372. [DOI] [Google Scholar]
- 61.Traylor-Knowles N, Rose NH, Sheets EA, Palumbi SR. 2017. Early transcriptional responses during heat stress in the coral Acropora hyacinthus. Biol Bull 232:91–100. doi: 10.1086/692717. [DOI] [PubMed] [Google Scholar]
- 62.Fuess LE, Pinzón C JH, Weil E, Grinshpon RD, Mydlarz LD. 2017. Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc Biol Sci 284:20170771. doi: 10.1098/rspb.2017.0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wenger Y, Buzgariu W, Reiter S, Galliot B. 2014. Injury-induced immune responses in Hydra. Semin Immunol 26:277–294. doi: 10.1016/j.smim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 64.Woznica A, Gerdt JP, Hulett RE, Clardy J, King N. 2017. Mating in the closest living relatives of animals is induced by a bacterial chondroitinase. Cell 170:1175–1183.e11. doi: 10.1016/j.cell.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brook CE, Boots M, Chandran K, Dobson AP, Drosten C, Graham AL, Grenfell BT, Müller MA, Ng M, Wang LF, van Leeuwen A. 2020. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 9:e48401. doi: 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banerjee A, Rapin N, Bollinger T, Misra V. 2017. Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci Rep 7:2232. doi: 10.1038/s41598-017-01513-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobovitz MR, Rupp S, Voss PA, Gornik SG, Guse A. 2019. Dinoflagellate symbionts escape vomocytosis by host cell immune suppression. bioRxiv doi: 10.1101/864579. [DOI] [PMC free article] [PubMed]
- 69.Babonis LS, Martindale MQ, Ryan JF. 2016. Do novel genes drive morphological novelty? An investigation of the nematosomes in the sea anemone Nematostella vectensis. BMC Evol Biol 16:114. doi: 10.1186/s12862-016-0683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.