Abstract
9-Hydroxyphenanthrene (9-OHPhe), the representative hydroxyl metabolite of phenanthrene, has generated increasing concern as it is potentially hazardous to organisms. Herein, multispectroscopic and molecular docking techniques were applied to investigate the molecular interaction of human serum albumin (HSA) with 9-hydroxyphenanthrene (9-OHPhe) under simulated physiological conditions. Steady-state fluorescence and time-resolved fluorescence spectral analysis showed that 9-OHPhe quenched HSA fluorescence through a mixed static and dynamic process. HSA can bind with 9-OHPhe to form a 1:1 complex, with binding constants of 1.28 × 105, 1.36 × 105, and 1.26 × 105 L·mol–1 at 298.15, 303.15, and 308.15 K, respectively. The strong binding between HSA and 9-OHPhe is spontaneous and entropy-driven. Molecular docking indicated that the optimal binding site of 9-OHPhe with HSA was located in the IA subdomain of HSA. Thermodynamic analysis and molecular docking results suggested that hydrophobic interactions and hydrogen bond force dominated the binding process of HSA with 9-OHPhe. Specifically, 9-OHPhe formed hydrophobic interactions with LEU134, LEU139, ILE142, LEU154, PHE157, ALA158, and TYR161 and formed a 1.86 Å hydrogen bond with LEU135. Circular dichroism spectral analysis showed that the α-helical content of HSA decreased from 52.3 to 50.9% after adding 9-OHPhe with a ratio of 1:1. The obtained results are hoped to provide basic data for understanding the potential effects of the hydroxyl metabolites of PAHs on functional biomacromolecules.
1. Introduction
Phenanthrene (Phe), as a typical polycyclic aromatic hydrocarbon (PAH), has been included in the list of Group 3 carcinogens by the International Agency for Research on Cancer since 2010.1 Like other PAHs, Phe enters into the environment primarily through the incomplete combustion of organic materials,2 posing a potential hazard to organisms. There are many routes by which humans are exposed to Phe or other PAHs, including diet, inhalation, and skin exposure.3 After entering the human body, Phe can be metabolized by the metabolic enzyme system into hydroxyl compounds or other metabolites, which are often detected in the urine of human beings.4,5 Therefore, the hydroxyl metabolites of Phe should not be ignored when evaluating the adverse effects of Phe on human beings.
In recent years, the binding between exogenous substances and biomacromolecules has attracted more and more attention,6−8 as the binding interactions are considered to be the beginning of the biological effects induced by exogenous substances.9 Human serum albumin (HSA), the most abundant carrier protein in human plasma, has the ability to bind a wide variety of endo- and exogenous substances.10 Moreover, it is commonly used as a representative model protein in clinical medicine, chemistry, and life sciences.11 The structure of HSA has been fully resolved, and it is a single polypeptide chain that contains 585 amino acid residues.12 HSA consists of three structurally homologous domains (i.e., I, II, and III), and each domain is further divided into two subdomains (i.e., A and B).13 The different subdomains have varying affinities to ligands. For instance, warfarin, indomethacin, and oxyphenbutazone preferentially bind to subdomain IIA, while diazepam and ibuprofen generally bind to the pocket in subdomain IIIA.14 This is due to the varying hydrophobicity and spatial structure of the pocket in different subdomains.15
Up to now, some research studies have focused on the molecular interactions between functional biomacromolecules and hydroxyl metabolites of PAHs. The data related to some biomacromolecules (i.e., DNA,16,17 bovine serum albumin,18,19 catalase,20,21 aryl hydrocarbon receptor,22 and estrogen receptors23) have been obtained through these studies, which demonstrated that the hydroxyl metabolites of PAHs could bind with these biomacromolecules and might induce different structural and/or physicochemical effects on them. However, until now, studies focusing on the interactions of the hydroxyl metabolites of PAHs with HSA and the corresponding effects on HSA have been insufficient.
In the present study, 9-hydroxyphenanthrene (9-OHPhe) was selected as the representative of monohydroxylated Phe, the molecular interaction of HSA with 9-OHPhe and the effects of 9-OHPhe on the structure of HSA were investigated using multispectroscopic and molecular docking techniques. Briefly, the fluorescence quenching mechanism of HSA by 9-OHPhe was determined through fluorescence spectra and fluorescence lifetime measurements. The number of binding sites was calculated using the Job plot method. The binding constants of HSA with 9-OHPhe under different temperatures were obtained through the mathematical analysis of the fluorescence spectral data. Thermodynamics analysis was conducted to determine the main binding forces that existed in the molecular interaction. The binding mode and detailed binding information of HSA with 9-OHPhe were investigated using molecular docking. Moreover, synchronous fluorescence and circular dichroism spectroscopy were utilized to analyze the effects of 9-OHPhe on the structural properties of HSA. Overall, the results obtained in this study can provide basic data for illustrating the molecular interaction mechanism between HSA and 9-OHPhe in vitro, which is helpful for understanding the biological effects of the hydroxyl metabolites of PAHs on functional biomacromolecules in vivo.
2. Materials and Methods
2.1. Materials
9-OHPhe (purity > 99.5%) and HSA (purity > 98%, fatty acid-free < 0.05%) were purchased from Sigma-Aldrich Chemical Co (St. Louis, MO). All of the other reagents used for experiments were of analytical reagent grade. Ultrapure water (18.2 MΩ·cm) was used throughout the experiments. The stock solution of HSA (5.0 × 10–5 mol·L–1) was prepared in 0.05 mol·L–1 Tris–HCl buffer (pH = 7.4, containing 0.01 mol·L–1 NaCl). The stock solutions of 9-OHPhe (2.0 × 10–3 mol·L–1) were prepared in ethanol. Tris–HCl buffer was used to keep the pH value constant and to maintain the ionic strength of the working solutions. The final volume of ethanol in the working solution was within 0.5%. All of the solutions were stored in the dark at 277.15 K.
2.2. Steady-State Fluorescence Spectroscopy Measurements
All of the fluorescence spectra of HSA without and with 9-OHPhe were obtained using an F-4600 fluorescence spectrophotometer (Hitachi, Japan) with a 10 × 10 mm2 quartz cuvette. The emission spectra of HSA in each system were scanned in the range of 305.0–575.0 nm with an excitation wavelength of 295.0 nm at three temperatures (298.15, 303.15, and 308.15 K), with a scan rate of 240 nm·min–1. The widths of excitation/emission slits were set at 5.0/5.0 or 5.0/10.0 nm as required. To avoid the inner-filter effect, all of the obtained spectral intensity values were corrected by multiplying a factor of 10(Aex+Aem)/2, where Aex and Aem are the differences in the absorption values of the sample after the addition of 9-OHPhe at the excitation and emission wavelengths, respectively.24
The synchronous fluorescence spectra of HSA in each system were also recorded, with wavelength intervals (Δλ) of 15 nm (excitation wavelength at 270.0–310.0 nm) and 60 nm (excitation wavelength at 220.0–330.0 nm) at 298.15 K. The widths of excitation/emission slits were set as 5.0/10.0 nm.
All of the spectra were scanned three times, and the data were analyzed using OriginPro 2018 software.
2.3. Fluorescence Lifetime Measurements
Fluorescence lifetime measurements were conducted at 298.15 K using an FS5 fluorescence spectrometer (Edinburgh Instruments, Livingston, U.K.) equipped with a 270 nm picosecond pulsed LED source using the time-correlated single-photon counting method as reported in previous work.19 The emission wavelength was set as 340.0 nm, and both excitation and emission slits were set as 5.0 nm. The instrumental response function (IRF) was detected by measuring the colloidal silica (Ludox AM-30, 30 wt % suspension in water) solution. The fluorescence decay plots were fitted with the biexponential decay law using Origin 7.5 software after deconvolution of the IRF.
2.4. Job’s Plot Analysis
Keeping the total concentration of HSA and 9-OHPhe as a constant (2.0 × 10–5 mol·L–1), sample solutions were prepared with varying molar fractions of HSA and 9-OHPhe. A control group was set up with the corresponding concentrations of HSA in the absence of 9-OHPhe. The fluorescence spectroscopy measurements of the samples were carried out as described in Section 2.2, and the fluorescence quenching data were obtained by subtracting the fluorescence intensity of the experimental sample from the corresponding control sample.
2.5. UV–Vis Absorption Spectra Measurements
The UV–vis absorption spectra were measured from 190.0 to 450.0 nm using a UV-8000S UV–vis spectrophotometer (Shanghai Yuanxi Instrument Co. Ltd., Shanghai, China). Quartz cuvettes with an optical path of 10.0 mm were used.
2.6. CD Spectral Measurements
The CD spectra of pure HSA and HSA-9-OHPhe complex were recorded using a Jasco-810 spectropolarimeter (Japan Spectroscopic Company, Japan). Each sample was scanned three times at 298.15 K in the range of 190.0–250.0 nm. The averaged spectrum of each sample was analyzed using the CDPro software (http://lamar.colostate.edu/~sreeram/CDPro/) coupled with the SELCON3 program to determine the secondary structure contents of HSA in different systems.
2.7. Molecular Docking Procedures
To verify the best binding mode of 9-OHPhe with HSA, blind docking procedures were performed using AutoDock 4.2.6 software.25 The crystal structure of HSA (PDB ID: 1AO6) was downloaded from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The molecular structure of 9-OHPhe was sketched using GaussView 5.08 software and then optimized using Gaussian 09 at the DFT/B3LYP/6-31G(d) level. Using AutoDockTools, the HSA and 9-OHPhe structures were prepared by adding polar hydrogen atoms, assigning ADT atom types, and adding partial charges. The grid box was set centered on the HSA molecule with dimensions of 126 × 126 × 126 Å3 and a spacing of 0.375 Å. After that, the Lamarckian genetic algorithm (LGA) was applied to seek the best binding site of 9-OHPhe in HSA with the default parameters. For each docking procedure, 25 conformations were output, of which the conformation with the lowest binding energy was selected and analyzed using the web service Protein–Ligand Interaction Profiler (projects.biotec.tu-dresden.de/plip-web).21 Finally, the docking results were further visualized and analyzed using PyMOL software.26
3. Results and Discussion
3.1. Fluorescence Quenching of HSA Induced by 9-OHPhe
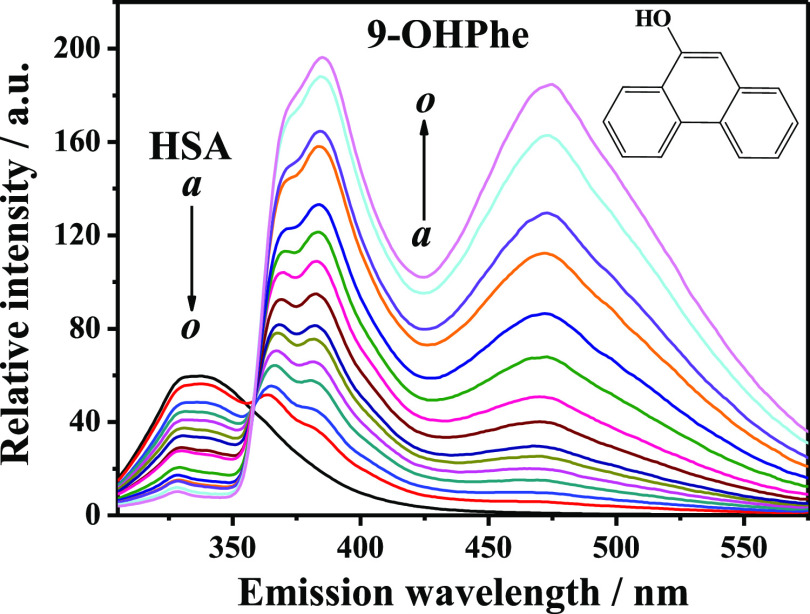
The intrinsic fluorescence of a protein is sensitive to the changes in the fluorophore surrounding environments when binding with small molecules, and thus, the fluorescence quenching method has been commonly used to study the interaction of proteins with ligands. Herein, the fluorescence quenching spectra of HSA in the presence of various concentrations of 9-OHPhe were measured at 298.15 K at an excitation wavelength of 295.0 nm (Figure 1).
Figure 1.
Fluorescence emission spectra of HSA in the presence of various concentrations of 9-OHPhe. T = 298.15 K; λex = 295.0 nm, ex./em. slit = 5.0/5.0 nm; a–o: [HSA] = 5.0 × 10–6 mol·L–1, [9-OHPhe] = (0, 1, 2, 3, 4, 5, 6, 8, 10, 15, 20, 25, 30, 40, and 50) × 10–6 mol·L–1.
Figure 1 shows that HSA has a fluorescence peak at around 338.0 nm, which is mainly attributed to the Trp residue. With the continuous addition of 9-OHPhe, regular quenching of HSA fluorescence was observed, indicating that an obvious interaction occurred between HSA and 9-OHPhe. Moreover, an isosbestic point was observed at around 358.0 nm in the HSA-9-OHPhe system, which suggested that the interaction between HSA and 9-OHPhe reached equilibrium.27,28
3.2. Fluorescence Quenching Mechanism of the HSA-9-OHPhe System
The fluorescence quenching of HSA may occur by different mechanisms that are usually classified as dynamic (the collisions between the protein at the excited state and the ligand in solution due to the diffusive motion) and static (the formation of a ground-state complex) quenching. To clarify the quenching mechanism of the HSA-9-OHPhe system, the fluorescence quenching data were analyzed using the well-known Stern–Volmer equation (eq S1).12 The fitting plots are shown in Figure S1; the Stern–Volmer quenching constant (Ksv) and the quenching rate constant (Kq) were calculated and are listed in Table 1. As can be seen, the Kq values of the HSA-9-OHPhe system at three different temperatures were much greater than the diffusion-limited quenching constant of biomolecules (2.0 × 1010 L·mol–1·s–1),21 which suggested that the static process played an important role in this interaction system.29
Table 1. Binding Parameters of HSA with 9-OHPhea.
| T/K | 10–5 Ksv (L·mol–1) | 10–13 Kq (L·mol–1·s–1) | R2 | 10–5 Kb (L·mol–1) | R2 | ΔH (kJ·mol–1) | TΔS (J·mol–1·K–1) | ΔG (kJ·mol–1) |
|---|---|---|---|---|---|---|---|---|
| 298.15 | 1.28 | 2.10 | 0.999 | 1.19 | 0.977 | 11.08 | 40.06 | –28.98 |
| 303.15 | 1.36 | 2.23 | 0.999 | 1.28 | 0.985 | 40.73 | –29.65 | |
| 308.15 | 1.26 | 2.07 | 0.998 | 1.38 | 0.988 | 41.40 | –30.32 |
R2 represents the correlation coefficient.
One additional method to ensure the presence of static quenching is by examination of the absorption spectra of the fluorophore.29 Thus, UV–vis absorption spectra of HSA in the presence of increasing amounts of 9-OHPhe were obtained by subtracting the corresponding spectrum of free 9-OHPhe from those of the HSA-9-OHPhe system (Figure S2). As can be seen, with gradual addition of 9-OHPhe, the absorbance intensity of HSA in the range of 240–278 nm decreased. This demonstrated again that a static quenching process existed in the HSA-9-OHPhe system.30 The UV–vis spectra of 9-OHPhe in the presence of increasing amounts of HSA were also measured and are shown in Figure S3. It can be seen from Figure S3 that 9-OHPhe has two absorption peaks at around 210 and 250 nm. Meanwhile, Figure S2 and previous studies31 indicated that HSA has two major absorption peaks located at 212 and 278 nm, while a new absorption band (from 375 to 420 nm) appeared in the HSA-9-OHPhe system, and the absorbance intensity of this peak increased with increasing concentrations of HSA. The appearance of this new absorption peak at longer wavelengths was considered to be ascribed to the formation of a ground-state complex (HSA-9-OHPhe complex), which had been verified above.32
Fluorescence lifetime analysis is a useful method to determine whether dynamic quenching exists in a quenching system or not.29 Herein, the fluorescence lifetime decay data of HSA in the absence and presence of 9-OHPhe were collected (Figure 2A) and fitted using a two-exponential decay function (Figure S4). As can be seen, the average fluorescence lifetime (τav) of HSA decreased slightly at increasing concentrations of 9-OHPhe (Table S1), indicating that a dynamic quenching process occurred in this system.29 When fitting a linear regression line between the τ0/τav values versus the concentration of 9-OHPhe (Figure 2B), the dynamic quenching constant was obtained to be 1.15 × 104 L·mol–1. Obviously, this value was less than one-tenth of the Stern–Volmer quenching constant Ksv calculated above (listed in Table 1). These results indicated that HSA fluorescence was quenched both by collisions and by complex formation with 9-OHPhe (mixed static and dynamic quenching processes) in the HSA-9-OHPhe system, while the static process played the main part.
Figure 2.
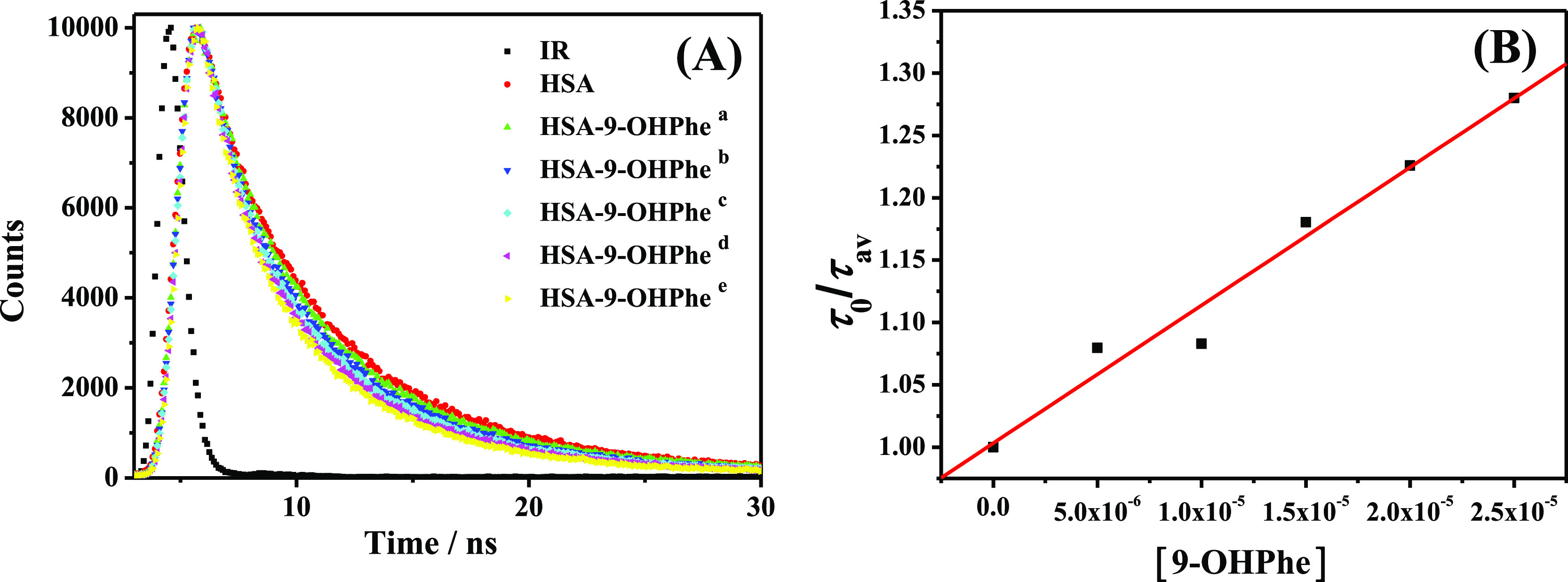
Fluorescence lifetime decay plots of the HSA-9-OHPhe system (A) and the fitting plots of τ0/τav versus the concentration of 9-OHPhe (B). τ0 and τav represent the average fluorescence lifetime of pure HSA and HSA in the presence of varying concentrations of 9-OHPhe, respectively. [HSA] = 5.0 × 10–6 mol·L–1; [9-OHPhe], a–e: (5.0, 10.0, 15.0, 20.0, 25.0) × 10–6 mol·L–1.
3.3. Energy Transfer from HSA to 9-OHPhe
According to Förster’s resonance energy transfer theory,33 energy transfer will occur under the following conditions: (1) the donor can produce fluorescence; (2) the fluorescence emission spectrum of the donor overlaps with the absorption spectrum of the acceptor; and (3) the distance between the donor and the acceptor is lower than 8 nm. As shown in Figure S5, the absorption spectrum of 9-OHPhe and the fluorescence emission spectrum of HSA sufficiently overlap. The average distance between HSA and 9-OHPhe was calculated using eqs S2–S4, and the following results were obtained: E = 38.3%, J = 3.42 × 10–14 cm3 L·mol–1, R0 = 3.1 nm, and r = 3.4 nm. The average distance (r) between HSA and 9-OHPhe was less than 8 nm and R0 < r < 1.5 R0, suggesting that the energy transfer from HSA to 9-OHPhe occurred with great possibility.34
3.4. Binding Site Number (n) of HSA with 9-OHPhe
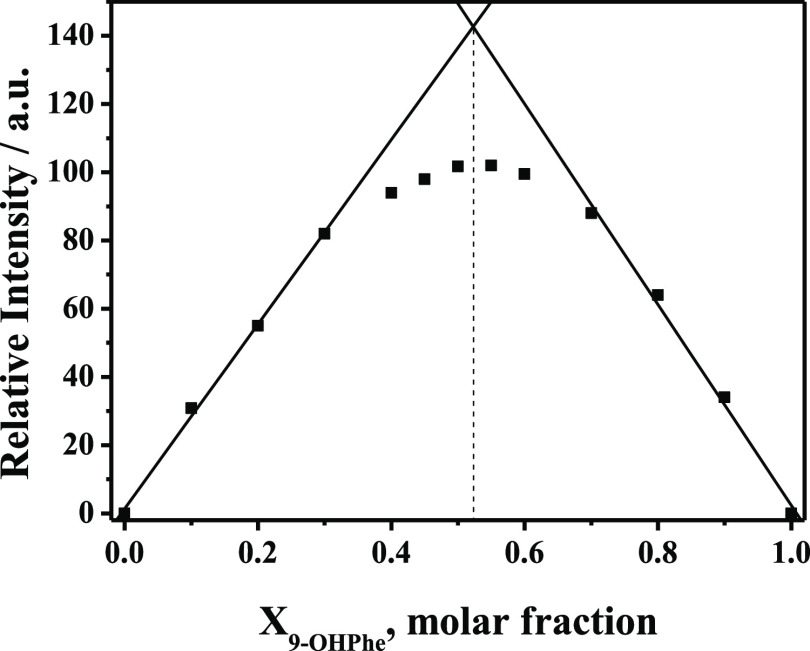
To obtain the binding affinity between HSA and 9-OHPhe, the n value of HSA with 9-OHPhe should be determined first. Herein, a Job’s plot analysis35 was carried out to calculate the binding stoichiometry of the HSA-9-OHPhe complex, and the fluorescence emission spectra related to the Job’s plot analysis are shown in Figure S6. The plots of the values of fluorescence quenching in the HSA-9-OHPhe system versus the molar fraction of 9-OHPhe are shown in Figure 3. As can be seen, two linear regression fittings were made based on the first and last four points, respectively. The crossing point of the two straight lines appeared at a molar fraction (X9-OHPhe) of about 0.52. This value was approximately equal to 0.5, which suggested that the binding stoichiometry of HSA with 9-OHPhe was 1:1 (n = 1).
Figure 3.
Job’s plot for fluorescence quenching of the HSA-9-OHPhe system with a total concentration of 2.0 × 10–5 mol·L–1 ([HSA] + [9-OHPhe]). Ex. = 295.0 nm and Em. = 338.0 nm.
3.5. Binding Constant (Kb) of HSA with 9-OHPhe at Different Temperatures
Herein, the fluorescence quenching spectra of HSA in the presence of various concentrations of 9-OHPhe were measured at three temperatures (298.15, 303.15, and 308.15 K) with an excitation wavelength of 295.0 nm. For the 1:1 binding interaction, the Kb values can be calculated using the most generally valid equation, eq 1.24
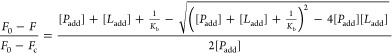 |
1 |
In this equation, F0 and F represent the fluorescence intensity of HSA in the absence and presence of 9-OHPhe, respectively. Fc is the residual fluorescence intensity of HSA when fully bonded with 9-OHPhe. [Padd] is the concentration of HSA, and [Ladd] is the concentration of added 9-OHPhe.
Based on this equation, the fitting plots for the fluorescence spectral data at different temperatures were obtained (Figure S7), and the calculated Kb values are shown in Table 1. The Kb values at three temperatures were all in the order of magnitude of 105 L·mol–1, indicating a strong interaction between HSA and 9-OHPhe. This suggested that 9-OHPhe could be carried by HSA and transported through blood circulation to other organs in vivo.
3.6. Thermodynamic Analysis
To have a better understanding of the interaction between HSA and 9-OHPhe, thermodynamic parameters should be obtained. As has been reported,36 the molecular forces contributed to the interaction between HSA and ligands mainly due to noncovalent interactions, including van der Waals interactions, hydrogen bonds, hydrophobic interactions, electrostatic interactions, salt bridges, π–π effects, etc. The major type of binding forces can be estimated through the sign of the values of enthalpy (ΔH) and entropy (ΔS).37 Thus, the thermodynamic parameters (Gibbs free energy (ΔG), ΔH, and ΔS) were calculated using the van’t Hoff equation (eqs S5 and S6), and the corresponding plots of ln Kb versus T–1 are shown in Figure S8.
As shown in Table 1, the negative ΔG values indicated that 9-OHPhe could bind to HSA spontaneously. Meanwhile, as |TΔS| > |ΔH|, it suggested that the formation of the HSA-9-OHPhe complex was an entropy-driven process.38 Moreover, the positive values of both ΔH and ΔS demonstrated that hydrophobic interactions dominated the binding of HSA with 9-OHPhe.
3.7. Molecular Docking
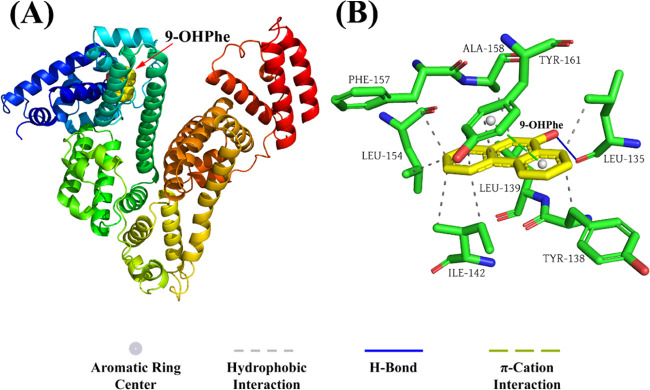
To further reveal the binding information of HSA with 9-OHPhe, the molecular docking method was performed using AutoDock 4.2.6 software to investigate the specific binding mode of the HSA-9-OHPhe complex. Out of the 25 obtained docking conformations, the conformation with the lowest free binding energy was selected and analyzed. As illustrated in Figure 4A, 9-OHPhe inserted into the IA subdomain of HSA with a free binding energy of −32.09 kJ·mol–1. The calculated binding constant value (4.19 × 105 L·mol–1) was larger than that obtained from the fluorescence quenching analysis, which may be due to the differences in crystal structures of HSA used in the fluorescence quenching test and those employed in docking calculations.39Figure 4B and Table S2 display the specific binding information of 9-OHPhe with the surrounding amino acids located in the binding site. The results showed that 9-OHPhe formed hydrophobic interactions with LEU134, LEU139, ILE142, LEU154, PHE157, ALA158, and TYR161 and formed a 1.86 Å hydrogen bond with LEU135. Besides, π-stacking interactions were detected between 9-OHPhe and TYR161 with a distance of 4.19 Å and an angle of 19.75°. Combined with the experimental results obtained in Section 3.6, it can be concluded that the intermolecular forces that dominated the stability of the HSA-9-OHPhe complex were mainly hydrophobic interactions and hydrogen bonds.
Figure 4.
Binding modes of 9-OHPhe to HSA. (A) Binding site of 9-OHPhe in HSA. (B) Detailed illustration of the binding forces of 9-OHPhe with the surrounding amino acid residues.
3.8. Conformational Changes of HSA
For proteins with some flexibility, the intermolecular interactions with ligands may commonly have an effect on the molecular conformation of the protein. Herein, to verify the conformational changes of HSA induced by 9-OHPhe, synchronous fluorescence, EEM fluorescence, and CD spectra were recorded.
3.8.1. Synchronous Fluorescence Spectral Analysis
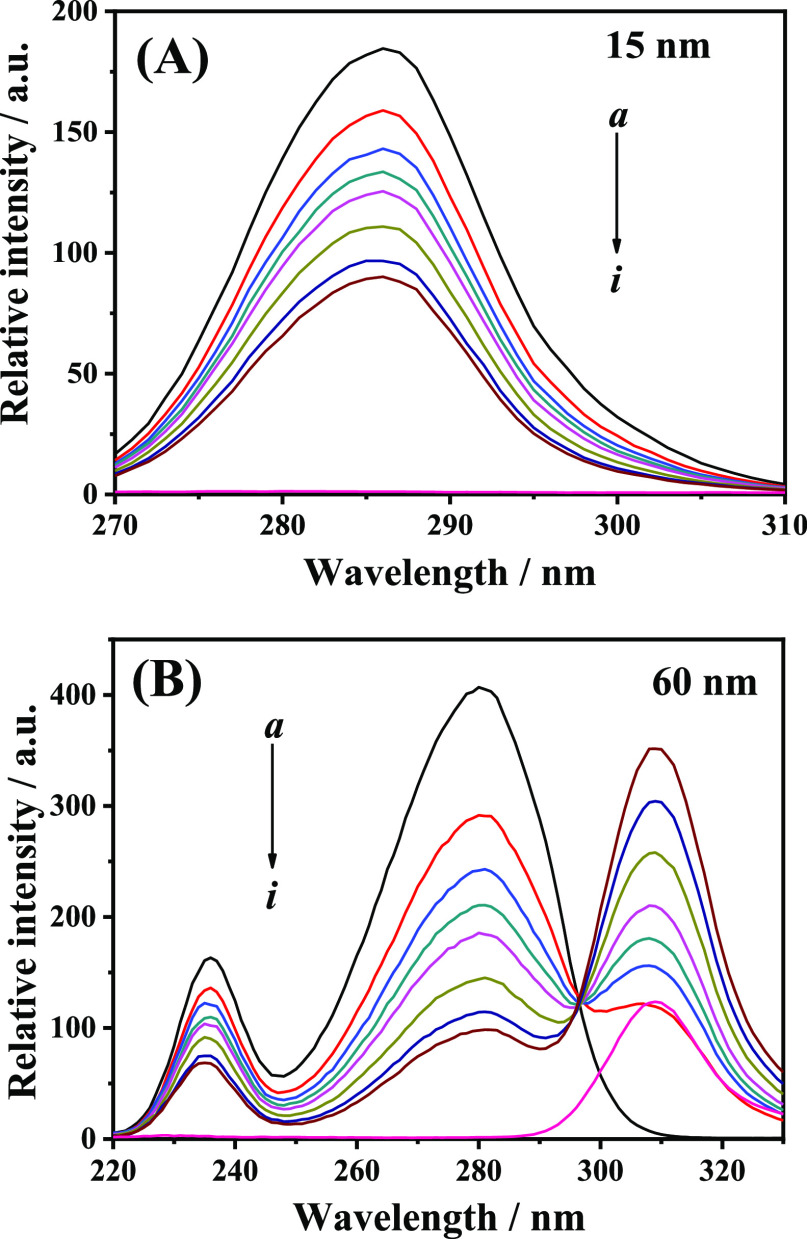
Synchronous fluorescence spectroscopy can provide useful information about the microenvironmental changes around fluorophore functional groups, and thus, it was widely applied to investigate the influence of ligands on the conformation of proteins.40 When setting Δλ at 15 and 60 nm, the synchronous fluorescence spectra of HSA represent the characteristic information of Tyr and Trp residues, respectively.41 The synchronous fluorescence spectra of HSA with various concentrations of 9-OHPhe were measured and are shown in Figure 5.
Figure 5.
Synchronous fluorescence spectra of the HSA-9-OHPhe system for Δλ = 15 nm (A) and Δλ = 60 nm (B). T = 298.15 K; λex = 295.0 nm, ex./em. slit = 5.0/10.0 nm; a–h: [HSA] = 5.0 × 10–6 mol·L–1, [9-OHPhe] = (0, 4.0, 6.0, 8.0, 10.0, 15.0, 20.0, 25.0) × 10–6 mol·L–1; i: [9-OHPhe] = 4.0 × 10–6 mol·L–1.
As can be seen in Figure 5, 9-OHPhe has no obvious fluorescence in the range of 270.0–310.0 nm at Δλ = 15 nm and 220.0–290.0 nm at Δλ = 60 nm. As 9-OHPhe can affect the synchronous fluorescence spectra of Trp residues in the range of 290.0–330.0 nm, this part of the spectra should not be used to characterize the fluorescence information of Trp residues in this system. With increasing concentrations of 9-OHPhe, the synchronous fluorescence intensities of both Trp and Tyr residues decreased and the fluorescence peaks of Trp and Tyr residues showed no obvious shift. This indicated that 9-OHPhe did not perturb the microenvironment around Trp and Tyr residues.41 Moreover, the Ksv values for Tyr and Trp residues in the HSA-9-OHPhe system were calculated to be 4.28 × 104 and 1.29 × 105 L·mol–1, respectively, which suggested that Trp residues played a more important role in the interaction of HSA with 9-OHPhe than Tyr.
3.8.2. CD Spectral Analysis
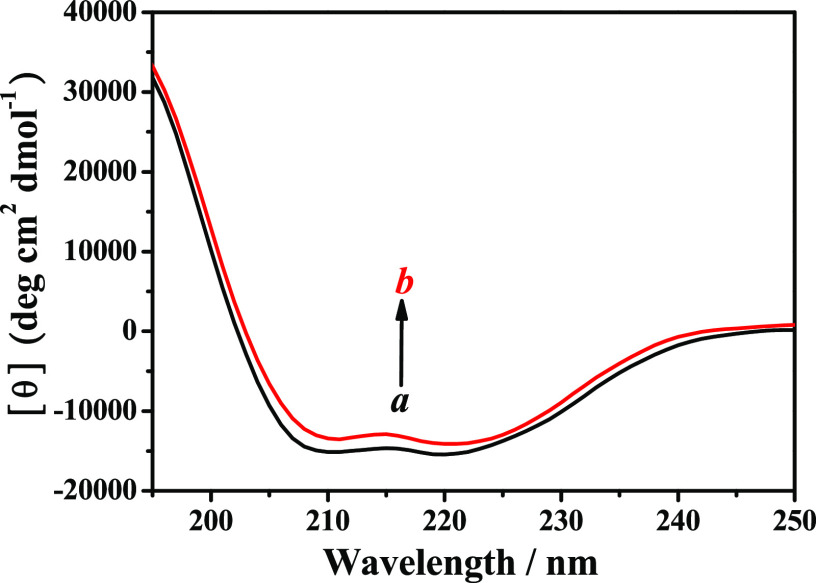
To further obtain the information on the secondary structural changes of HSA induced by 9-OHPhe, CD spectroscopy was employed here. The CD spectra of pure HSA show two negative bands at approximately 208.0 and 220.0 nm (Figure 6), characterizing the α-helical structure of HSA, which are caused by n → π* transfer for the peptide bond of the α-helix.42 After the addition of 9-OHPhe, the CD spectra of HSA changed significantly with a decrease in its band intensity. The CD spectral data were analyzed using the CDPro software package, and the quantitative data on the secondary structural changes of HSA were obtained and are shown in Table 2.
Figure 6.
CD spectra of HSA in the absence and presence of 9-OHPhe. [HSA] = 5.0 × 10–8 mol·L–1; a, b: [9-OHPhe] = 0 and 5.0 × 10–8 mol·L–1.
Table 2. Secondary Structural Content of HSA in the Absence and Presence of 9-OHPhe.
| system | α-helix (%) | β-sheets (%) | turns (%) | random coils (%) |
|---|---|---|---|---|
| HSA | 52.3 | 9.3 | 13.9 | 24.0 |
| HSA-9-OHPhe | 50.9 | 10.4 | 13.0 | 24.2 |
As shown in Table 2, the α-helical content of HSA decreased from 52.3 to 50.9% after adding 9-OHPhe with a ratio of 1:1. Meanwhile, the β-sheets of HSA increased from 9.3 to 10.4%. It may be because the interactions of 9-OHPhe with certain amino acid residues in HSA destroyed the hydrogen-bonding networks of HSA and induced the conversion of helical structures to sheets.21 Meanwhile, the decrease of the α-helical content of HSA indicated that 9-OHPhe could cause unfolding of the HSA skeleton.20 As the structure of a protein is closely related to its biological function, the structural changes of HSA caused by 9-OHPhe suggested that 9-OHPhe might induce adverse effects on the biological function of HSA, which would be harmful to the human body.
4. Conclusions
The molecular interaction between HSA and 9-OHPhe has been investigated using multispectroscopic and molecular docking methods in the present work. The results showed that (1) both dynamic and static quenching processes occurred in the HSA-9-OHPhe system, while the static process played the major role; (2) HSA could bind with 9-OHPhe in 1:1 mode with a binding constant of 1.28 × 105 L·mol–1 at 298.15 K; (3) hydrophobic interactions and hydrogen bond were considered to be the main interaction forces involved in the binding process between HSA and 9-OHPhe; (4) 9-OHPhe was located in the IA subdomain of HSA in the HSA-9-OHPhe complex, and 9-OHPhe formed hydrophobic interactions with 7 amino acid residues around it and formed a 1.86 Å hydrogen bond with LEU135; and (5) 9-OHPhe could induce unfolding of the structure of HSA.
Overall, this work revealed the interaction mechanism of HSA with 9-OHPhe and the corresponding effects on the structure of HSA. The obtained results will be useful for understanding the potential toxic effects of hydroxylated PAHs on the transport protein in organisms. However, it is worth noting that the molecular interactions between hydroxylated PAHs and functional biomacromolecules are still far from being fully understood. For instance, little information is known about the interactions of hydroxylated PAHs with some other biomacromolecules like superoxide dismutase, glutathione peroxidase, estrogen receptor, etc. Moreover, as the structural difference of hydroxylated PAHs may affect their interactions with biomacromolecules, it is quite necessary to further investigate the interactions between biomacromolecules and hydroxylated PAHs with varying substituent groups or varying number of benzene rings. In addition, molecular dynamics simulations have been well applied in the previous studies for obtaining more useful binding information based on molecular docking,43,44 which can be further used in our subsequent relevant studies.
Acknowledgments
We gratefully acknowledge the financial support from the Natural Science Foundation of Fujian Province [grant number 2018J05024] and the Program for Prominent Young Talents in Fujian Province University, China [grant number [2018]47]. We are also grateful to Linfeng Chen for assistance with the experiments and valuable discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02031.
Stern–Volmer equation (Eq S1); Stern–Volmer plots for the quenching of HSA by 9-OHPhe at three temperatures (Figure S1); The UV–visible absorption spectra of HSA in the presence of 9-OHPhe at different concentrations (Figure S2); The UV–visible absorption spectra of 9-OHPhe in the presence of increasing concentrations of HSA (Figure S3); The fitting lines for the fluorescence lifetime decay plots of the HSA-9-OHPhe system (Figure S4); The fluorescence lifetime of HSA in the presence of various concentrations of 9-OHPhe (Table S1); The overlap of the fluorescence emission spectrum of HSA and the absorption spectrum of 9-OHPhe (Figure S5); Equations for the calculation of the binding distance between HSA and 9-OHPhe (Eqs S2–S4); Fluorescence emission spectra of HSA in the presence (A) and absence (B) of 9-OHPhe (Figure S6); The fitting plots for the fluorescence quenching of HSA by 9-OHPhe at three temperatures (Figure S7); van’t Hoff equations (Eqs S5 and S6); Fitting line of the van’t Hoff equation for the 9-OHPhe–HSA system (Figure S8); Specific information on the interaction forces between 9-OHPhe and HSA (Table S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- International Agency for Research on Cancer. Agents Classified by the IARC Monographs, Volumes 1–125. https://monographs.iarc.fr/agents-classified-by-the-iarc/ (accessed Jan 18, 2020).
- Han F. L.; Guo H.; Hu J. L.; Zhang J.; Ying Q.; Zhang H. L. Sources and health risks of ambient polycyclic aromatic hydrocarbons in China. Sci. Total Environ. 2020, 698, 134229 10.1016/j.scitotenv.2019.134229. [DOI] [PubMed] [Google Scholar]
- Ma Y. N.; Harrad S. Spatiotemporal analysis and human exposure assessment on polycyclic aromatic hydrocarbons in indoor air, settled house dust, and diet: A review. Environ. Int. 2015, 84, 7–16. 10.1016/j.envint.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Cao L. M.; Wang D. M.; Wen Y. H.; He H.; Chen A. L.; Hu D.; Tan A. J.; Shi T. M.; Zhu K. J.; Ma J. X.; Zhou Y.; Chen W. H. Effects of environmental and lifestyle exposures on urinary levels of polycyclic aromatic hydrocarbon metabolites: A cross-sectional study of urban adults in China. Chemosphere 2020, 240, 124898 10.1016/j.chemosphere.2019.124898. [DOI] [PubMed] [Google Scholar]
- Lu S. Y.; Yu Z. Q.; Yuan J. Investigation of polycyclic aromatic hydrocarbon exposure levels in farmers in an area of China. J. Environ. Health 2010, 27, 611–614. [Google Scholar]
- Xu M. C.; Wan J. Q.; Niu Q. G.; Liu R. T. PFOA and PFOS interact with superoxide dismutase and induce cytotoxicity in mouse primary hepatocytes: A combined cellular and molecular methods. Environ. Res. 2019, 175, 63–70. 10.1016/j.envres.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Ding K. K.; Kong X. T.; Wang J. P.; Lu L. P.; Zhou W. F.; Zhan T. J.; Zhang C. L.; Zhuang S. L. Side chains of parabens podulate antiandrogenic activity: In vitro and molecular docking studies. Environ. Sci. Technol. 2017, 51, 6452–6460. 10.1021/acs.est.7b00951. [DOI] [PubMed] [Google Scholar]
- Zhuang S. L.; Wang H. F.; Ding K. K.; Wang J. Y.; Pan L. M.; Lu Y. L.; Liu Q. J.; Zhang C. L. Interactions of benzotriazole UV stabilizers with human serum albumin: Atomic insights revealed by biosensors, spectroscopies and molecular dynamics simulations. Chemosphere 2016, 144, 1050–1059. 10.1016/j.chemosphere.2015.09.085. [DOI] [PubMed] [Google Scholar]
- Ghosh D.; Chattopadhyay N. Equilibrium and dynamic effects on ligand binding to biomacromolecules and biomimetic model systems. Int. Rev. Phys. Chem. 2013, 32, 435–466. 10.1080/0144235X.2013.782683. [DOI] [Google Scholar]
- Mohammadzadeh-Aghdash H.; Akbari N.; Esazadeh K.; Dolatabadi J. E. N. Molecular and technical aspects on the interaction of serum albumin with multifunctional food preservatives. Food Chem. 2019, 293, 491–498. 10.1016/j.foodchem.2019.04.119. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Wang L. J.; Zhu M. Q.; Xue J. Y.; Hua R. M.; Li Q. X. Comparative studies on biophysical interactions between gambogic acid and serum albumin via multispectroscopic approaches and molecular docking. J. Lumin. 2019, 205, 210–218. 10.1016/j.jlumin.2018.09.005. [DOI] [Google Scholar]
- Razzak M. A.; Lee J. E.; Choi S. S. Structural insights into the binding behavior of isoflavonoid glabridin with human serum albumin. Food Hydrocolloids 2019, 91, 290–300. 10.1016/j.foodhyd.2019.01.031. [DOI] [Google Scholar]
- Bagheri M.; Fatemi M. H. Fluorescence spectroscopy, molecular docking and molecular dynamic simulation studies of HSA-Aflatoxin B1 and G1 interactions. J. Lumin. 2018, 202, 345–353. 10.1016/j.jlumin.2018.05.066. [DOI] [Google Scholar]
- Ghuman J.; Zunszain P. A.; Petitpas I.; Bhattacharya A. A.; Otagiri M.; Curry S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- Fasano M.; Curry S.; Terreno E.; Galliano M.; Fanali G.; Narciso P.; Notari S.; Ascenzi P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- Wang L. R.; Wang Y.; Chen J. W.; Guo L. H. A structure-based investigation on the binding interaction of hydroxylated polycyclic aromatic hydrocarbons with DNA. Toxicology 2009, 262, 250–257. 10.1016/j.tox.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Li F.; Li X. H.; Liu X. L.; Zhang L. B.; You L. P.; Zhao J. M.; Wu H. F. Noncovalent interactions between hydroxylated polycyclic aromatic hydrocarbon and DNA: Molecular docking and QSAR study. Environ. Toxicol. Pharmacol. 2011, 32, 373–381. 10.1016/j.etap.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Wu T. Q.; Wu Q.; Guan S. Y.; Su H. X.; Cai Z. J. Binding of the environmental pollutant naphthol to bovine serum albumin. Biomacromolecules 2007, 8, 1899–1906. 10.1021/bm061189v. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Chen W. X.; Tang B. W.; Zhang W.; Chen L. F.; Duan Y.; Zhu Y. X.; Zhu Y. X.; Zhang Y. Interactions of 1-hydroxypyrene with bovine serum albumin: insights from multi-spectroscopy, docking and molecular dynamics simulation methods. RSC Adv. 2016, 6, 23622–23633. 10.1039/C6RA00981F. [DOI] [Google Scholar]
- Chen L. F.; Zhang J.; Zhu Y. X.; Zhang Y. Molecular interactions of 1-hydroxypyrene with catalase revealed by spectroscopic methods combined with molecular docking. Chem. J. Chin. Univ. 2015, 36, 2394–2401. [Google Scholar]
- Zhang J.; Chen L. F.; Zhu Y. X.; Zhang Y. Study on the molecular interactions of hydroxylated polycyclic aromatic hydrocarbons with catalase using multi-spectral methods combined with molecular docking. Food Chem. 2020, 309, 125743 10.1016/j.foodchem.2019.125743. [DOI] [PubMed] [Google Scholar]
- Ohura T.; Kurihara R.; Hashimoto S. Aryl hydrocarbon receptor activities of hydroxylated polycyclic aromatic hydrocarbons in recombinant yeast cells. Toxicol. Environ. Chem. 2010, 92, 737–742. 10.1080/02772240903109043. [DOI] [Google Scholar]
- Sievers C. K.; Shanle E. K.; Bradfield C. A.; Xu W. Differential action of monohydroxylated polycyclic aromatic hydrocarbons with estrogen receptors alpha and beta. Toxicol. Sci. 2013, 132, 359–367. 10.1093/toxsci/kfs287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakar K. A.; Feroz S. R. A critical view on the analysis of fluorescence quenching data for determining ligand-protein binding affinity. Spectrochim. Acta, Part A 2019, 223, 117337 10.1016/j.saa.2019.117337. [DOI] [PubMed] [Google Scholar]
- Zhang L. L.; Liu Y. C.; Wang Y. M. Interaction between an (-)-epigallocatechin-3-gallate-copper complex and bovine serum albumin: Fluorescence, circular dichroism, HPLC, and docking studies. Food Chem. 2019, 301, 125294 10.1016/j.foodchem.2019.125294. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.3r1; Schrödinger, LLC: New York, USA, 2010.
- Ghosh S.; Mitra A. K.; Pal U.; Basu S.; Saha C. Evidence of two structurally related solvatochromic probes complexed with beta-cyclodextrin by using spectroscopic methods. J. Mol. Struct. 2017, 1130, 810–817. 10.1016/j.molstruc.2016.10.097. [DOI] [Google Scholar]
- Song Y. M.; Kang J. W.; Zhou J.; Wang Z. H.; Lu X. Q.; Wang L. F.; Gao J. Z. Study on the fluorescence spectra and electrochemical behavior of ZnL2 and Morin with DNA. Spectrochim. Acta, Part A 2000, 56, 2491–2497. 10.1016/S1386-1425(00)00340-1. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R.Quenching of Fluorescence. In Principles of Fluorescence Spectroscopy, 3rd ed.; Lakowicz J. R., Ed.; Springer Science and Business Media: New York, 2006; pp 278–327. [Google Scholar]
- Lyu S. L.; Wang W. Spectroscopic methodologies and computational simulation studies on the characterization of the interaction between human serum albumin and astragalin. J. Biomol. Struct. Dyn. 2020, 1–12. 10.1080/07391102.2020.1758213. [DOI] [PubMed] [Google Scholar]
- Parsekar S. U.; Velankanni P.; Sridhar S.; Haldar P.; Mate N. A.; Banerjee A.; Antharjanam P. K. S.; Koley A. P.; Kumar M. Protein binding studies with human serum albumin, molecular docking and in vitro cytotoxicity studies using HeLa cervical carcinoma cells of Cu(II)/Zn(II) complexes containing a carbohydrazone ligand. Dalton Trans. 2020, 49, 2947–2965. 10.1039/C9DT04656A. [DOI] [PubMed] [Google Scholar]
- Birla L.; Cristian A. M.; Hillebrand M. Absorption and steady state fluorescence study of interaction between eosin and bovine serum albumin. Spectrochim. Acta, Part A 2004, 60, 551–556. 10.1016/S1386-1425(03)00261-0. [DOI] [PubMed] [Google Scholar]
- Förster T.Delocalized Excitation and Excitation Transfer; Florida State University, 1965. [Google Scholar]
- Almutairi F. M.; Ajmal M. R.; Siddiqi M. K.; Amir M.; Khan R. H. Multi-spectroscopic and molecular docking technique study of the azelastine interaction with human serum albumin. J. Mol. Struct. 2020, 1201, 127147 10.1016/j.molstruc.2019.127147. [DOI] [Google Scholar]
- Shree G. J.; Sivaraman G.; Siva A.; Chellappa D. Anthracene- and pyrene-bearing imidazoles as turn-on fluorescent chemosensor for aluminum ion in living cells. Dyes Pigm. 2019, 163, 204–212. 10.1016/j.dyepig.2018.11.061. [DOI] [Google Scholar]
- Ross P. D.; Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry 1981, 20, 3096–3102. 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- Liu F. R.; Zhang Y. Y.; Yu Q. Y.; Shen Y. Z.; Zheng Z.; Cheng J. S.; Zhang W. C.; Ye Y. W. Exploration of the binding between ellagic acid, a potentially risky food additive, and bovine serum albumin. Food Chem. Toxicol. 2019, 134, 110867 10.1016/j.fct.2019.110867. [DOI] [PubMed] [Google Scholar]
- Wang L.; Sun J. P.; Zhang H. H.; Shen Z. Q.; Xu L.; Liu G. J. Solubility and thermodynamic analysis of methyleneaminoacetonitrile in binary solvents from T = (278.15 to 323.15) K. J. Mol. Liq. 2019, 283, 462–471. 10.1016/j.molliq.2019.03.069. [DOI] [Google Scholar]
- Sharma A. S.; Anandakumar S.; Ilanchelian M. A combined spectroscopic and molecular docking study on site selective binding interaction of Toluidine blue O with Human and Bovine serum albumins. J. Lumin. 2014, 151, 206–218. 10.1016/j.jlumin.2014.02.009. [DOI] [Google Scholar]
- Ren C.; Xiong W. F.; Li B. Binding interaction between beta-conglycinin/glycinin and cyanidin-3-O-glucoside in acidic media assessed by multi-spectroscopic and thermodynamic techniques. Int. J. Biol. Macromol. 2019, 137, 366–373. 10.1016/j.ijbiomac.2019.07.004. [DOI] [PubMed] [Google Scholar]
- Abdelhameed A. S.; Alanazi A. M.; Bakheit A. H.; Hassan E. S.; Herqash R. N.; Almutairi F. M. Novel BTK inhibitor acalabrutinib (ACP-196) tightly binds to site I of the human serum albumin as observed by spectroscopic and computational studies. Int. J. Biol. Macromol. 2019, 127, 536–543. 10.1016/j.ijbiomac.2019.01.083. [DOI] [PubMed] [Google Scholar]
- Mu H. T.; Chen S. H.; Liu F. Y.; Xiao J. B.; Huang H.; Zhang Y. H.; Sun Y. M.; Gao X. Y.; Lei H. T.; Yuan X. W. Stereoselective interactions of lactic acid enantiomers with HSA: Spectroscopy and docking application. Food Chem. 2019, 270, 429–435. 10.1016/j.foodchem.2018.07.135. [DOI] [PubMed] [Google Scholar]
- Lu L. P.; Zhan T. J.; Ma M.; Xu C.; Wang J. P.; Zhang C. L.; Liu W. P.; Zhuang S. L. Thyroid Disruption by Bisphenol S Analogues via Thyroid Hormone Receptor beta: in Vitro, in Vivo, and Molecular Dynamics Simulation Study. Environ. Sci. Technol. 2018, 52, 6617–6625. 10.1021/acs.est.8b00776. [DOI] [PubMed] [Google Scholar]
- Wu H.; Lu L. P.; Chen J. Y.; Zhang C. L.; Liu W. P.; Zhuang S. L. Inhibited Nitric Oxide Production of Human Endothelial Nitric Oxide Synthase by Nitrated and Oxygenated Polycyclic Aromatic Hydrocarbons. Environ. Sci. Technol. 2020, 54, 2922–2930. 10.1021/acs.est.9b07163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.