Niestroj et al. present findings from the largest investigation to date of copy number variants (CNVs) in epilepsy. The results reveal epilepsy subtype-specific differences in CNV burden and contribute to the understanding of the complex genetic architecture of epilepsies.
Keywords: copy number variation, epilepsy, genetic generalized epilepsy, developmental and epileptic encephalopathy, focal epilepsy
Abstract
Cytogenic testing is routinely applied in most neurological centres for severe paediatric epilepsies. However, which characteristics of copy number variants (CNVs) confer most epilepsy risk and which epilepsy subtypes carry the most CNV burden, have not been explored on a genome-wide scale. Here, we present the largest CNV investigation in epilepsy to date with 10 712 European epilepsy cases and 6746 ancestry-matched controls. Patients with genetic generalized epilepsy, lesional focal epilepsy, non-acquired focal epilepsy, and developmental and epileptic encephalopathy were included. All samples were processed with the same technology and analysis pipeline. All investigated epilepsy types, including lesional focal epilepsy patients, showed an increase in CNV burden in at least one tested category compared to controls. However, we observed striking differences in CNV burden across epilepsy types and investigated CNV categories. Genetic generalized epilepsy patients have the highest CNV burden in all categories tested, followed by developmental and epileptic encephalopathy patients. Both epilepsy types also show association for deletions covering genes intolerant for truncating variants. Genome-wide CNV breakpoint association showed not only significant loci for genetic generalized and developmental and epileptic encephalopathy patients but also for lesional focal epilepsy patients. With a 34-fold risk for developing genetic generalized epilepsy, we show for the first time that the established epilepsy-associated 15q13.3 deletion represents the strongest risk CNV for genetic generalized epilepsy across the whole genome. Using the human interactome, we examined the largest connected component of the genes overlapped by CNVs in the four epilepsy types. We observed that genetic generalized epilepsy and non-acquired focal epilepsy formed disease modules. In summary, we show that in all common epilepsy types, 1.5–3% of patients carry epilepsy-associated CNVs. The characteristics of risk CNVs vary tremendously across and within epilepsy types. Thus, we advocate genome-wide genomic testing to identify all disease-associated types of CNVs.
Introduction
Characterized by recurrent and unprovoked seizures, epilepsy is the third most common neurological disorder, affecting roughly 65 million individuals worldwide (Ngugi et al., 2010). The cause of epilepsy is unknown in many patients and can be the result of a variety of insults that perturb brain function. Along with acquired causes such as trauma, infectious diseases and autoimmune diseases, genetic variants play a major role in the disease aetiology (EpiPM Consortium, 2015). To date, ∼100 genes have been associated with epilepsy (EpiPM Consortium, 2015; Heyne et al., 2018).
The clinical representation of epilepsy is heterogeneous and subtype classification can be challenging. The epilepsies can be grouped into four major phenotypes (Scheffer et al., 2017): (i) genetic generalized epilepsies (GGE); (ii) focal epilepsies with non-acquired focal epilepsies (NAFE) and lesional focal epilepsies (LFE); (iii) developmental and epileptic encephalopathies (DEE); and (iv) unclassified epilepsies. Among all epilepsy phenotypes, the DEE group has the poorest prognosis (Berg et al., 2010; Scheffer et al., 2017).
In the past decade, many genetic studies have established that single nucleotide variants can confer risk or cause epilepsy (EpiPM Consortium, 2015; ILAE Consortium on Complex Epilepsies Consortium, 2018). Disease causing de novo variants have been reported in patients with DEE (Epi4K Consortium et al., 2013) and seizure susceptibility variants have been identified in GGE (Noebels, 2015). Focal epilepsies have been associated with germline, somatic and mosaic pathogenic variants in e.g. PCDH19 (Dibbens et al., 2008), LGI1, SCN1A and CHRNA4 (Helbig et al., 2016) and especially in genes associated with the mechanistic target of rapamycin (mTOR) pathway (Moller et al., 2016; Devinsky et al., 2018).
Additionally, rare copy number variants (CNVs) are strongly implicated in the aetiology of epilepsy. Around 4–8% of DEE patients carry pathogenic CNVs (Heinzen et al., 2010; Mefford et al., 2011) and CNVs at genomic hotspots such as 15q13.3, 15q11.2, 16p11.2, 16p13.11 and 22q11.2 have been associated with GGE (Dibbens et al., 2009; Helbig et al., 2009; de Kovel et al., 2010; Mefford et al., 2010; Mullen et al., 2013; Olson et al., 2014; Lal et al., 2015b; Perez-Palma et al., 2017). Rare genic CNVs were found in ∼10% of GGE patients (Mefford et al., 2010, 2011; Addis et al., 2016) and CNVs >1 Mb (megabase) were significantly enriched in patients compared to controls (Heinzen et al., 2010; Mefford et al., 2011; Striano et al., 2012; Lal et al., 2015b). Deletions at 15q13.3, 15q11.2 and 16p13.11 are rarely seen in patients with DEE, highlighting the notion that the major phenotypes of epilepsy have different genetic architectures (Mefford, 2014). Non-recurrent deletions in RBFOX1 have been additionally found in patients with focal epilepsies (Lal et al., 2015a) and the 16p13.11 deletion was found in a study including GGE, NAFE, and LFE patients combined (Heinzen et al., 2010). However, no significant CNV association has been identified to date with NAFE (Perez-Palma et al., 2017) and the role of CNVs in LFE has not been studied at large scale.
To date, all the current epilepsy CNV associations have been identified through candidate loci screens, as genome-wide scans were under-powered to confirm significant genetic associations of low frequency CNVs (<1%) with epilepsy. In addition, the vast majority of CNV association studies have focused on deletions and not duplications. Lastly, no large-scale study uniformly processed or analysed several types of epilepsy with the same genotyping platform and analysis protocol, which would enable robust comparisons across epilepsy phenotypes.
Here, we performed a large genome-wide analysis and the first CNV breakpoint association analysis of both deletions and duplications in four different epilepsy phenotypes (n = 10 712 cases and 6746 controls), to decipher epilepsy phenotype-specific patterns as well as to discover novel epilepsy-associated CNV loci.
Materials and methods
Sample ascertainment
Epilepsy patients and associated clinical data (n = 13 454) were ascertained from clinics distributed throughout Europe (37 sites), North America, Oceania and Asia as part of an ongoing collaborative effort by the Epi25 Consortium. Subjects were assessed for a diagnosis of DEE, GGE, NAFE and LFE.
DEE comprised subjects with severe refractory epilepsy of unknown aetiology with developmental plateauing or regression, no epileptogenic lesion on MRI, and with epileptiform features on EEG. As this is the group with the largest number of gene discoveries to date, we encouraged inclusion of those with negative epilepsy gene panel results, but we did not exclude those without prior testing. Diagnosis of GGE required a history of generalized seizure types (generalized tonic-clonic, absence, or myoclonus seizures) and generalized epileptiform discharges on EEG. We excluded cases with evidence of focal seizures, or with moderate to severe intellectual disability and those with an epileptogenic lesion on neuroimaging (although neuroimaging was not obligatory). If EEG was not available, then only cases with an archetypal clinical history as judged by the phenotyping committee (e.g. morning myoclonus and generalized tonic-clonic seizures) were accepted. Diagnosis of NAFE required a convincing history of focal seizure types, an EEG with focal epileptiform or normal findings, and neuroimaging showing no epileptogenic lesion or hippocampal sclerosis (MRI was preferred but CT was accepted). Exclusion criteria were a history of primarily generalized seizures or moderate to severe intellectual disability. LFE-compromised subjects with a convincing history of focal seizure types, an EEG with focal epileptiform or normal findings, and neuroimaging showing an epileptogenic lesion such as a low-grade brain tumour or a focal cortical dysplasia.
Patients who did not fulfil criteria for any of the aforementioned epilepsy phenotypes because of absence of critical data or conflicting data were excluded from the analyses. Patients or their legal guardians provided signed informed consent according to local national ethical requirements. This study was approved by the institutional review boards of all participating sites (Supplementary material). Samples had been collected over a 20-year period in some centres, so the consent forms reflected standards at the time of collection. Samples were only accepted if the consent did not exclude data sharing (see details in exome study using similar patient cohort in Epi25 Collaborative, 2019). Part of the dataset was published in dbGaP (phs001489.v1.p1).
Control subjects (n = 12 857) were obtained from three external large-scale genetic studies, specifically selected because genotyping was performed on the same genotyping array (Illumina Infinium Global Screening Array) and at the same centre (Broad Institute) as the epilepsy cases. Controls provided as part of this study: (i) Genomic Psychiatry Cohort (GPC) controls; (ii) FINRISK controls; and (iii) Helmsley irritable bowel disease (IBD) cases and controls. The control subjects were not particularly checked for generalized epilepsy in childhood. For a detailed description see the Supplementary material.
Genotyping
Samples selected for this study were all genotyped on the GSA-MD v1.0 (Illumina, San Diego) in separate batches. A total of 688 032 markers were used for quality control (QC).
Genotype sample quality control
To correct for population stratification, we performed an initial round of QC based on single nucleotide polymorphism (SNP) genotype data for 13 420 epilepsy cases and 12 857 controls. Samples with a call rate <0.96 or discordant sex status were excluded. We filtered autosomal SNPs for low genotyping rate (>0.98), case-control difference in minor-allele frequency (>0.05), and deviation from Hardy-Weinberg equilibrium (HWE, P-value ≤ 0.001) before pruning SNPs for linkage disequilibrium (–indep-pairwise 200 100 0.2) using PLINK v1.9 (Chang et al., 2015) in order to perform principal component analysis (PCA) to assess for population stratification. Samples with non-European ancestry were excluded based on visual clustering of the PCA (Supplementary Fig. 1).
CNV calling
We focused only on autosomal CNVs because of higher quality of CNV calls from non-sex chromosomes (Pinto et al., 2011). We created GC wave-adjusted LRR (Log-R ratio) intensity files for all samples using PennCNV, and employed PennCNV’s CNV calling algorithms (Wang et al., 2007) to detect CNVs in our dataset. We generated a custom population B-allele frequency file before calling CNVs. Adjacent CNV calls were merged if the number of intervening markers between them was <20% of the total number when both segments were combined.
Intensity sample quality control
Intensity-based QC was conducted following established protocols for CNV calling (Huang et al., 2017) (Supplementary material). Following intensity-based QC, all samples had a LRR standard deviation (SD) of < 0.25, absolute value of waviness factor < 0.04, and a B-allele frequency drift < 0.007.
CNV load sample quality control
We performed a final round of sample QC by removing additional samples with excessively high CNV load based on the total number of CNV calls (>100), as suggested by PennCNV and Huang et al. (2017). This threshold was determined empirically by visual inspection of distributions across all datasets combined (Supplementary Fig. 2). Our final dataset after sample QC comprised 17 458 samples: 10 712 epilepsy cases and 6746 controls (DEE = 1308; GGE = 3643; LFE = 1263; NAFE = 4498).
Call filtering and delineation of rare CNVs
CNV calls were removed from the dataset if they spanned <20 markers, were <20 kb in length, had a SNP density <0.0001 (amount of markers/length of CNV) or overlapped by more than haf of their total length with regions known to generate artefacts in SNP-based detection of CNVs as described previously (Marshall et al., 2017) (for details see the Supplementary material). Additionally, all CNV calls spanning >20 markers and ≥1 Mb in length were included in the analysis even if the SNP density was <0.0001 (Huang et al., 2017; Marshall et al., 2017).
We assigned all CNV calls a specific frequency count using PLINK v.1.07 (Purcell et al., 2007), with the option –cnv-freq-method2 0.5. Here, the frequency count of an individual CNV is determined as 1 + the total number of CNVs overlap by at least 50% of its total length (in bp), irrespective of CNV type. We then filtered our callset for rare CNVs with a frequency of 186 or lower across all samples).
After CNV quality control, 11 826 of 17 992 (7425 cases and 4401 controls) QC-passed individuals had at least rare CNV.
CNV annotation
CNVs were annotated for gene content and recurrent deletion hotspots for epilepsy and neurodevelopmental disorders with various annotation files including gene name and the corresponding coordinates in hg19 assembly using in-house Perl scripts (available on request). We annotated 89 genes that were previously associated with epilepsy (EpiPM Consortium, 2015; Heyne et al., 2018), 93 genes associated with neurodevelopmental disorders (Deciphering Developmental Disorders Study, 2017), 2680 genes intolerant for protein truncating variants defined as probability of loss-of-function intolerance (pLI) score > 0.95) (Lek et al., 2016), >28 000 annotated regions from UCSC refseq genes, eight recurrent hotspot deletion regions for epilepsy and six recurrent hotspot regions for neurodevelopmental disorders (Carvill and Mefford, 2013). We only considered a CNV as ‘coding’ if it overlapped 80% of a gene (Coppola et al., 2019). We considered all other CNVs as ‘non-genic’.
Cytogenic testing is well-established for diagnostic evaluation of patients with neurodevelopmental disorders including epilepsies. We considered rare deletions (frequency ≤ 1%) overlapping known hotspot regions for epilepsy or rare duplications (frequency ≤ 1%) overlapping 16p11.2 or deletions overlapping epilepsy and/or neurodevelopmental disorder genes as ‘likely pathogenic’.
CNV burden analysis
We measured CNV burden for all four epilepsy phenotypes using four separate categories to evaluate relative contribution on epilepsy type risk: (i) the total length of all rare CNVs within an individual (CNV length); (ii) the carrier status of rare CNVs intersecting genes and neurodevelopmental- or epilepsy-associated CNVs hotspot regions; (iii) the carrier status of rare likely pathogenic CNVs; and (iv) the carrier status of rare deletions overlapping recurrent neurodevelopmental or epilepsy associated deletion hotspots. For length and CNV burden in different gene and hotspot lists, deletions and duplications were analysed separately. For likely pathogenic CNV burden duplications and deletions were analysed according to the definition of ‘likely pathogenic’ CNVs mentioned before. To assess for a CNV burden difference between epilepsy cases and controls, we fitted a logistic binomial regression model using the ‘glm’ function of the stats package (https://github.com/SurajGupta/r-source/tree/master/src/library/stats/R) in R for common and rare CNVs respectively (Huang et al., 2017):
| (1) |
where ‘y’ is a dichotomous outcome variable (epilepsy type = 1, control = 0); ‘sex’ is used as a covariate and ‘CNV burden’ represents one of the categories mentioned above. For all burden analyses, odds ratios, 95% confidence intervals (CIs), and significance were calculated. Odds ratios were calculated by taking the exponential of the logistic regression coefficient. Odds ratios above one indicate an increased risk for the specific epilepsy type per unit of CNV burden. Significance threshold was corrected for multiple testing using Bonferroni correction. Bonferroni multiple-testing threshold for significance was calculated combined for all epilepsy phenotypes and CNV types for all three categories [(i) CNV length burden P < 2.1 × 10−3; (ii) genome-wide burden P < 1.4 × 10−3; (iii) likely pathogenic CNV burden P < 0.0125; and (iv) deletion burden at recurrent hotspots P < 1.8 × 10−3].
Regression of potential confounds on case-control ascertainment
It is important to ensure that any bias in gender and ancestry does not drive spurious associations with epilepsy. To ensure the robustness of the analysis, CNV burden analyses included potential confounding variables as covariates in a logistic regression framework as previously described (Marshall et al., 2017) (for details see the Supplementary material).
CNV breakpoint level association
The CNV breakpoint level association was performed by quantifying the frequencies of case and control CNV carriers at all unique CNV breakpoint locations (i.e. the SNP probe defining the start and end of the CNV segment); the full set of CNV breakpoints represents the genome-wide space of CNV variation between cases and controls.
CNV breakpoint level association was run using the epilepsy residual phenotype as a quantitative variable, with significance determined through 1 million permutations of phenotype residual labels using PLINK v1.07 (Purcell et al., 2007). An additional z-scoring correction was used to efficiently estimate two-sided empirical P-values for highly significant loci. A fraction of our controls were patients from an IBD project, and therefore to rule out confounds, we ran the same CNV breakpoint level association for the ‘IBD-controls’ from the Helmsley dataset (since these represent IBD cases) and used them as cases to test association using the remaining controls as comparison group. IBD-related CNV breakpoints with P-values < 0.01 after genome wide correction were removed from the combined analysis (epilepsy cases versus all controls including the IBD fraction). Association tests were conducted for all CNV types, deletions, and duplications independently. CNVs spanning the centromere were removed from the analysis (in particular, at the centromere of chromosome 9). Bonferroni correction for 46 846 tests was used to identify significance threshold. Loci that surpassed genome-wide multiple testing correction in either test were followed up by manual CNV quality evaluation: B-allele frequency and LRR were manually investigated using Perl scripts provided by PennCNV and UCSC genome browser hg19 (https://genome.ucsc.edu/).
Phenotype analysis
The phenome-wide association study (PheWAS) design requires a good signal to noise ratio to discover novel CNV associations. To enrich for high confidence pathogenic CNVs, we tested the burden of big CNVs (>2 Mb) in patients with a specific phenotype among the different epilepsy phenotypes. Based on the data collected through the Epi25 consortium, we were able to include 43 different phenotype categories in the PheWAS (Supplementary material). P-values and odds ratios were obtained using a Fisher’s exact test (two-sided). Multiple testing correction for 161 tests results in a significant P-value < 3.1 × 10−4. We performed a meta-analysis for the association of GGE patients with big duplications (> 2 Mb) with febrile seizures to exclude a possible centre bias using the R package ‘metafor’ (https://cran.r-project.org/web/packages/metafor/metafor.pdf).
Network analysis
Network analysis was performed for all brain expressed protein coding genes covered by a CNV significantly enriched (P ≤ 0.05) in the four different epilepsy types than compared to controls. For details see the Supplementary material.
Data availability
The data that support the findings of this study are available from the Epi25 Consortium, upon reasonable request.
Results
Elevated epilepsy type-specific CNV burden in DEE and GGE patients
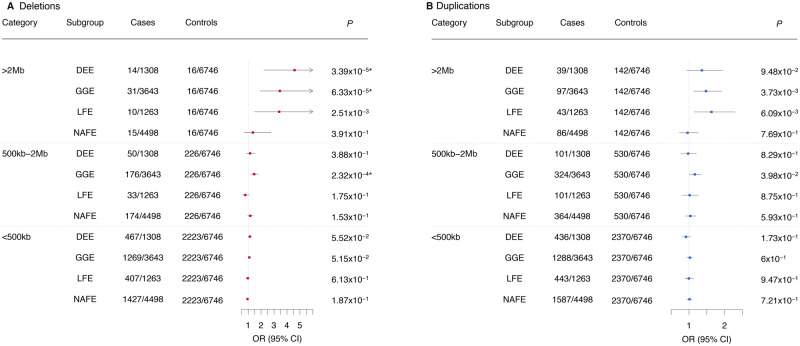
We applied logistic regression to investigate whether the four epilepsy phenotypes have on average a greater genomic region covered (combined CNV length) by either deletions or duplications. After correction for 24 tests, we found that patients with DEE and GGE showed independent enrichment for total deletions of an overall length of >2 Mb compared to controls [DEE: odds ratio (OR) 4.59 (95% CI 2.2–9.45), P = 3.39 × 10−5; GGE: OR 3.45 (95% CI 1.9–6.48), P = 6.33 × 10−5] (Fig. 1A). No epilepsy type showed a significant burden for duplications (Fig. 1B).
Figure 1.
Global burden of CNV by overall length across four epilepsy types. Rare CNV burden observed in the different epilepsy types is shown for (A) deletions and (B) duplications. Odds ratios (OR) and P-values were calculated using a binomial logistic regression for rare CNVs with sex as a covariate in three different categories (overall genomic sequence loss in one individual of >2 Mb, 500 kb–2 Mb and <500 kb). UE = unclassified epilepsies; *P-values surpassing the Bonferroni multiple testing for 30 tests cut-off (*P < 2.1 × 10−3).
Enrichment of gene-sets and CNV hotspots in DEE, GGE, and NAFE patients
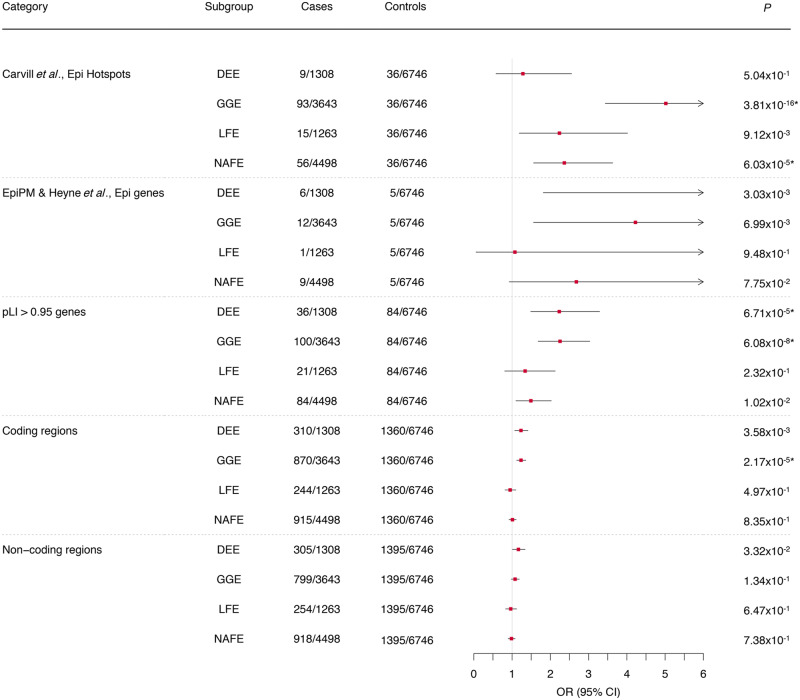
Next, we measured if the CNV burden was concentrated within defined sets of genes and known deletion hotspots for epilepsy and neurodevelopmental disorders. Compared to deletions identified in the controls, we found that the epilepsy hotspot list, genes intolerant for truncating variants, and coding regions were enriched for patient deletions (Fig. 2). DEE and GGE patients showed a significant burden of deletions in genes with pLI > 0.95 [DEE: OR 2.23 (95% CI 1.48–3.29), P = 6.7 × 10−5; GGE: OR 2.25 (95% CI 1.68–3.03), P = 6.08 × 10 − 08]. Additionally, GGE patients showed an enrichment of deletions at previously identified epilepsy hotspots [OR 5.02 (95% CI 3.44–7.49), P = 3.81 × 10−16] mostly driven by deletions at 15q13.3 [OR 36.04 (95% CI 7.49–647.45), P = 4.75 × 10−4] and 16p13.11 [OR 21.14 (95% CI 6.19–132.38), P = 3.8 × 10−5; Table 1]. Of the 30 GGE cases, 14 carry a >2 Mb deletion at one of the recurrent hotspot regions for epilepsy with four cases having a deletion at 16p13.11 and four at 22q11.2 whereas only one control carries a >2-Mb deletion at locus 22q11.2. An additional burden was observed for CNVs covering the coding regions [OR 1.14 (95% CI 1.06–1.23), P = 6.54 × 10−4] but no significant enrichment of known epilepsy genes. Furthermore, we detected a significant deletion enrichment in NAFE patients at previously identified epilepsy deletion hotspots [OR 2.37 (95% CI 1.56–3.63), P = 6.03 × 10−5]. Patients with LFE showed a significant deletion burden at 16p13.11 compared to controls [OR 4.98 (95% CI 2.14–9.66), P = 2.3 × 10−5; Table 1]. In contrast, no enrichment was observed in any gene list or loci tested when duplications were considered in any epilepsy phenotype (Supplementary Fig. 4). However, the duplication at locus 15q11.2 is recurrent in patients (13/259 patients with >2 Mb duplications) and absent from controls.
Figure 2.
The global burden of deletions across different gene sets, hotspot regions and non-coding regions in four different epilepsy phenotypes. Common deletion burden was elucidated for epilepsy hotspot regions (Carvill and Mefford, 2013) and rare (<1% frequency) deletion burden was elucidated for all other gene lists (Category). Odds ratios (OR) and P-values were calculated using a binomial regression for common CNVs and a binomial regression for rare CNVs with sex as a covariate. CNVs are defined as ‘genic’ if they overlap 80% of a gene. Notably, not all individuals carry a CNV. (Results of CNV burden in neurodevelopmental disorder hotspots and genes are not shown because of very small sample sizes and no significance; results of duplication burden are shown in Supplementary Fig. 4.) When they exceed a specified limit, 95% CIs are clipped to arrows. *P-values surpassing the Bonferroni multiple testing for 36 tests cut-off (*P < 1.4 × 10−3).
Table 1.
Deletion burden of recurrent CNVs across four different epilepsy phenotypes
| Recurrent hotspot | Subgroup | Cases | Controls | OR | P-value |
|---|---|---|---|---|---|
| 1q21.1 | DEE | 0/1308 | 1/6746 | NA | NA |
| GGE | 4/3643 | 1/6746 | 7.84 | 6.6 × 10−2 | |
| LFE | 0/1263 | 1/6746 | NA | NA | |
| NAFE | 0/4498 | 1/6746 | NA | NA | |
| 15q11.2 | DEE | 6/1308 | 23/6746 | 1.35 | 0.52 |
| GGE | 29/3643 | 23/6746 | 2.4 | 1.9 × 10−3 | |
| LFE | 5/1263 | 23/6746 | 1.16 | 0.77 | |
| NAFE | 30/4498 | 23/6746 | 1.97 | 1.5 × 10−2 | |
| 15q13.3 | DEE | 1/1308 | 1/6746 | 4.83 | 0.27 |
| GGE | 20/3643 | 1/6746 | 36.04 | 4.7 × 10−4* | |
| LFE | 0/1263 | 1/6746 | NA | NA | |
| NAFE | 2/4498 | 1/6746 | 3.1 | 0.36 | |
| 16p11.2 | DEE | 0/1308 | 2/6746 | NA | NA |
| GGE | 6/3643 | 2/6746 | 6.19 | 2.6 × 10−2 | |
| LFE | 1/1263 | 2/6746 | 2.66 | 0.42 | |
| NAFE | 6/4498 | 2/6746 | 4.52 | 6.5 × 10−2 | |
| 16p12.1 | DEE | 0/1308 | 3/6746 | NA | NA |
| GGE | 9/3643 | 3/6746 | 5.52 | 1.1 × 10−2 | |
| LFE | 1/1263 | 3/6746 | 1.78 | 0.62 | |
| NAFE | 6/4498 | 3/6746 | 3.04 | 0.12 | |
| 16p13.11 | DEE | 2/1308 | 2/6746 | 5 | 0.11 |
| GGE | 21/3643 | 2/6746 | 21.14 | 3.8 × 10−5* | |
| LFE | 7/1263 | 2/6746 | 18.91 | 2.5 × 10−4* | |
| NAFE | 11/4498 | 2/6746 | 8.36 | 5.8 × 10−3 | |
| 22q11.2 | DEE | 0/1308 | 1/6746 | NA | NA |
| GGE | 4/3643 | 1/6746 | 7.31 | 7.6 × 10−2 | |
| LFE | 0/1263 | 1/6746 | NA | NA | |
| NAFE | 0/4498 | 1/6746 | NA | NA |
Odds ratios (ORs) and P-values were calculated using a binomial regression for rare deletions with sex as a covariate. NA = not available.
P-values surpassing the Bonferroni multiple testing for 28 tests cut-off (*P < 1.8 × 10−3).
Enrichment of likely pathogenic CNVs in all epilepsy types
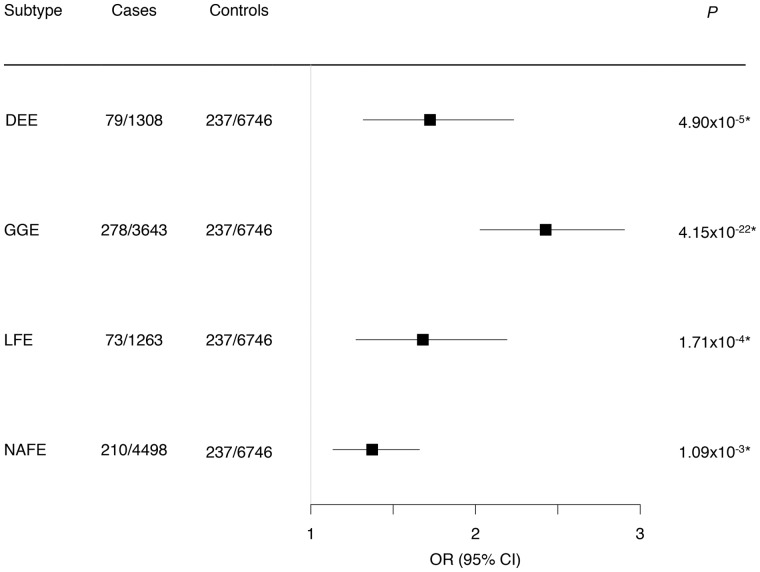
For our next category, we evaluated the combined burden of the CNVs that are considered as ‘likely pathogenic’ (see ‘Materials and methods’ section for selection criteria) in the four studied epilepsy phenotypes. Likely pathogenic CNVs were identified in 1.15% of DEEs, 2.88% of GGEs, 1.19% of LFEs and in 1.49% of NAFEs. However, likely pathogenic CNVs were also present in 0.58% of controls. Nevertheless, in a direct comparison with the controls, we observed a significant enrichment of likely pathogenic CNVs in all epilepsy types (Fig. 3). The likely pathogenic CNV effect size was greatest in patients with GGE [OR 2.43 (95% CI 2.03–2.9), P = 4.1 × 10−22; Fig. 3], mainly driven by deletions overlapping recurrent hotspot regions (Fig. 2).
Figure 3.
Global burden of likely pathogenic CNVs across four different epilepsy phenotypes. Likely pathogenic CNVs were defined as rare deletions in neurodevelopmental disorders and epilepsy genes and in recurrent epilepsy hotspots and duplications of 16p11.2, odds ratios (OR) and P-values were calculated using a binomial logistic regression for rare CNVs with sex as a covariable. Genic CNVs are defined as those that overlap 80% of any exon of a known protein-coding gene. *P values surpassing the Bonferroni multiple testing for four tests cut-off (*P < 0.0125).
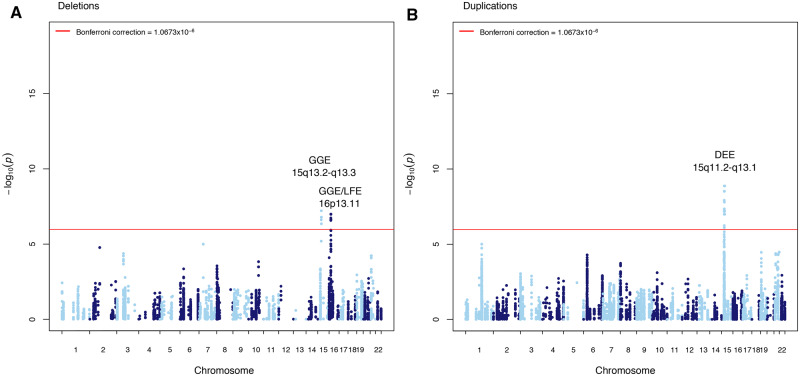
Genome-wide CNV breakpoint association reveals significant loci for DEE, GGE, and LFE
Three independent CNV loci in three epilepsy phenotypes surpassed genome-wide significance; all loci have been previously reported in association with GGE (Helbig et al., 2009; de Kovel et al., 2010; Mefford et al., 2010; Lal et al., 2015b). For two of the identified CNV loci we extended the phenotypic spectrum by identifying novel epilepsy phenotype associations. In line with previous results from candidate loci studies, our analysis showed that patients with GGE were most significantly enriched for deletions overlapping hotspot loci on chromosomes 15q13.2-q13.3 (P = 6.06 × 10−8) and 16p13.11 (P = 1.06 × 10−7; Fig. 4A). The DEE analysis revealed a genome-wide significant duplication locus overlapping the recurrent region on chromosome 15q11.2-q13.1 also known as the Prader-Willi/Angelman critical region (P = 1.35 × 10−9; Fig. 4B). No locus was significantly enriched in the NAFE cohort. Deletions in LFE patients were enriched at epilepsy hotspot 16p13.11 (P = 1.86 × 10−7; Fig. 4A).
Figure 4.
Genome-wide CNV breakpoint association. Manhattan plot displaying the −log10 deviance P-value for (A) genome-wide deletion breakpoint association for DEE, GGE, LFE, and NAFE; and (B) genome-wide duplication breakpoint association for DEE, GGE, LFE, and NAFE. P-value cut-offs corresponding to correction for 46 846 tests at 1.0673 × 10−6 are highlighted in red. Loci significant after multiple test correction in the appropriate epilepsy type are labelled.
Phenome-wide association study analysis reveals enrichment of large CNVs in epilepsy subtypes
We performed a phenome-wide association study (PheWAS) to identify an association between large effect CNVs and a large number of different phenotypes. We analysed whether the CNV burden is enriched in any clinical phenotype within the four different epilepsy phenotypes. After multiple testing correction for 161 applied tests, we identified two significant associations. We observed a 3.25-fold enrichment of large duplications (>2 Mb) in patients with GGE and febrile seizures when comparing to GGE patients without febrile seizures [OR 3.25 (95% CI 1.8–5.92), P = 4.07 × 10−5; Supplementary Table 2]. Further, a 2.72-fold enrichment of large duplications was detected for focal epilepsy patients with structural abnormalities versus without [OR 2.72 (95% CI 1.57–4.56), P = 2.33 × 10−4; Supplementary Table 2]. An evaluation of types of lesions in this group showed that pathogenic CNVs are not specific to a single lesion type but found in patients with five different lesion types (Supplementary Fig. 6).
GGE and NAFE form disease modules
Finally, we performed a network analysis, to explore the largest connected component of the genes covered by CNVs and enriched in the different epilepsy types. Among the four epilepsy phenotypes, GGE and NAFE formed modules (Supplementary Fig. 7A and B), while DEE and LFE do not. The largest connected component versus total number of genes of each phenotype was as follows: GGE, 62/167 (P = 0.041); NAFE, 41/129 (P = 0.056); DEE, 3/72 (P = 0.787); and LFE, 2/23 (P = 0.389). The largest connected component networks for GGE and NAFE are shown in Supplementary Fig. 7C and D. GGE had three hub genes: APP, SUMO3, and UBE3A. NAFE had APP, SUMO3, but not UBE3A, which was not enriched in NAFE patients.
The network proximity analysis showed that DEE and LFE have a small distance (Supplementary Table 4; z-score = −1.65, P = 0.046), although the number of overlapping genes was only three (Supplementary Table 5). The z-score for the proximity of GGE and NAFE was −1.49 (P = 0.062; Supplementary Table 4), and there was a large number of overlapping genes (n = 101; Supplementary Table 5). DEE and NAFE also had a small distance (z-score = −1.43, P = 0.063; Supplementary Table 4).
Discussion
In this study, we identify several novel CNV-epilepsy associations using a case-control approach with >17 000 individuals genotyped on the same platform and analysed with the same CNV calling, quality control, and analysis pipeline. We observe an increased burden of CNVs in different epilepsy phenotypes, report novel risk loci that surpass genome-wide multiple testing correction, and show that also LFE can be associated with an increased CNV burden. Consistent with results from genetic studies in other neurodevelopmental disorders, we show that novel risk loci lay at the ultra-rare end of the CNV frequency spectrum. Thus, larger samples will be needed to identify additional risk loci at convincing levels of statistical evidence (Huang et al., 2017; Marshall et al., 2017).
CNV burden
We and others have previously shown a burden of deletions overlapping genes associated with neurodevelopmental processes in patients with GGE, and that the signal was particularly concentrated within epilepsy hotspot loci (Dibbens et al., 2009; Helbig et al., 2009; de Kovel et al., 2010; Mefford et al., 2010; Mullen et al., 2013; Olson et al., 2014; Lal et al., 2015b; Perez-Palma et al., 2017). In the present study, we were able to replicate the original GGE signal with a significant enrichment for deletions at epilepsy-associated hotspots, which gives confidence about the reliability of the results from this study. Previously, in cohort studies the recurrent CNVs at 15q11.2, 15q13.3, 16p13.11, and 22q11.2, have been associated with GGE or GGE and focal epilepsy combined (Heinzen et al., 2010; Mullen et al., 2013; Lal et al., 2015b). For DEEs no significant association for recurrent CNVs has been identified suggesting that recurrent CNVs are rather associated with the more common and milder forms of epilepsy (Mefford et al., 2012). In our study we replicated findings for 15q13.3 and 16p13.11 with GGE. However, we could not replicate the previous associations of GGE with 15q11.2 and 22q11.2. Our study represents the largest CNV investigation so far. Based on our results, we suggest interpreting CNVs at 15q11.2 and 22q11.2 in epilepsy patients with caution. Additionally, we observed a significant deletion burden in genes intolerant for protein truncating variants in the general population, which has been suggested recently in a smaller cohort of 160 generalized, 32 focal, and six unclassified epilepsy patients (Monlong et al., 2018). Consistent with the well-established role of rare, large effect CNVs in the aetiology of the severe and early onset DEEs (Mefford et al., 2011), we identified a significant deletion enrichment covering genes intolerant for truncating variants in the general population. Previous studies did not find significant differences between focal epilepsy patients and controls within hotspot loci, most likely due to the small sample size (Perez-Palma et al., 2017). Here, we detect deletions overlapping epilepsy hotspot regions enriched in patients with NAFE. Although epilepsy-associated brain lesions have mainly been associated with somatic variants, which affect the mechanistic target of rapamycin (mTOR) pathway (Moller et al., 2016; Devinsky et al., 2018) also germline variants in DEPDC5 have been identified as risk factors for lesional epilepsies. Here, we show that CNVs play a role in the aetiology of LFE. The detected pathogenic CNVs were not specific to a single brain lesion, suggesting that the CNVs confer risk to epilepsy rather than to the lesion itself. An alternative hypothesis of how these CNVs could confer risk for epilepsy-associated brain lesions could be a second hit scenario, in which patients carry in addition to the germline CNV a somatic variant or the non-mutated allele is lost (loss of heterozygosity). Such a disease mode has recently been shown for patients with focal cortical dysplasia and variants in DEPDC5 (Baldassari et al., 2019).
CNVs are present in most individuals and usually represent benign genetic variation without clinical significance (Zarrei et al., 2015). Therefore, we concentrated on the burden of likely pathogenic CNVs that were 1.37–2.43-fold enriched in epilepsy patients. Although we used state-of-the-art criteria to support the categorization as ‘likely pathogenic’ CNV, the modest enrichment indicates that also population controls carry similar types of CNVs. This observation is in accordance with the presence of recurrent CNVs in epilepsy hotspot loci in healthy controls, suggesting an incomplete penetrance for epilepsy risk (Dibbens et al., 2009; Crawford et al., 2019). Additionally, detection of large gene-disrupting CNVs and epilepsy-associated gene deletions does not imply causality but rather increased susceptibility or incomplete penetrance. Many CNV hotspots and large-gene disrupting CNVs are known to be co-morbid with other disorders like intellectual disability (Mullen et al., 2013) and autism (Weiss et al., 2008; Glessner et al., 2009; Levy et al., 2011; Sanders et al., 2011), but we did not observe an enrichment of likely pathogenic CNVs in patients with these comorbidities in our cohort (data not shown). Interestingly, we found an enrichment of large duplications (>2 Mb) in GGE patients with febrile seizures compared to GGE patients without febrile seizures (Supplementary Table 2, Supplementary Fig. 5). Additional comorbidities in GGE patients with CNVs have been reported before (Mullen et al., 2013). Large duplications at 1q21.1, 22q11.2, and 16p11.2 are known to be enriched in syndromic epilepsies (Mefford and Eichler, 2009; Mefford and Mulley, 2010; Mefford et al., 2012), suggesting that those GGE patients carry additional phenotypic co-morbidities.
Genome-wide CNV breakpoint association
Several recurrent CNVs have been previously associated with epilepsy (Helbig et al., 2009; de Kovel et al., 2010); however, all have been identified in candidate loci studies. In this study, our sample size and uniform CNV calling pipeline allowed us to test CNV loci at genome-wide scale with adequate power at the CNV breakpoint level. Here, we performed the first genome-wide CNV breakpoint association analysis to identify associated loci among different epilepsy phenotypes. We replicated three of seven previously published locus-associations with epilepsy types at genome-wide significance level (15q11.2, 15q13.3 and 16p13.11) (Helbig et al., 2009; de Kovel et al., 2010; Mefford et al., 2010; Lal et al., 2015b), whereas 1q21.1, 16p11.2, 16p12, and 22q11.2 only reached suggestive significance (P-value < 0.05), suggesting that larger datasets are needed to reach genome-wide significance. The majority of these previously established loci are co-morbid with other neurodevelopmental disorders such as schizophrenia, psychotic disorder, autism or intellectual disability (Brunetti-Pierri et al., 2008; Coe et al., 2012; Marshall et al., 2017). Notably, our previous GGE CNV study re-evaluated clinical records of GGE patients carrying a 22q11.2 deletion, revealing additional congenital and developmental features (Lal et al., 2015b). Possibly in this study, we used more stringent sample inclusion criteria with a smaller fraction of patients with comorbidities. This may explain why four of seven recurrent loci were not significantly enriched in our analysis. Nonetheless, we show a significant association of deletions in 16p13.11 with LFE. Previously, deletions of 16p13.11 were found to be enriched in candidate loci studies of GGE and CECTS (childhood epilepsy with centrotemporal spikes) along with autism, intellectual disability, schizophrenia and additionally in non-lesional focal epilepsies (de Kovel et al., 2010; Mefford et al., 2010). The signal of non-lesional focal epilepsies could have been driven by misdiagnosed patients with small lesions undetectable by neuroimaging so that a lesional focal epilepsy might not have been confidently ruled out in these patients.
Network analysis
Using the human interactome, we examined the largest connected component of the genes covered by CNVs and enriched in the different epilepsy types to see if the genes form disease modules. GGE and NAFE were the only epilepsy types forming disease modules both with the hub genes APP and SUMO3. APP is suggested to be involved in the migration of neurons during early development and constitutive mutations and duplications are believed to cause rare forms of familial Alzheimer’s disease and Alzheimer’s disease neuropathology in Down syndrome (Murrell et al., 1991; Hooli et al., 2014; Wiseman et al., 2015). SUMO3 plays a role in a number of cellular processes such as nuclear transport, DNA replication and repair, mitosis and signal transduction. SUMO3 immunoreactivity is predominantly detected in neurons in brains from Alzheimer’s disease, Down syndrome, and non-demented humans providing a regulatory mechanism in APP amyloidogenesis. Therefore, it has been suggested that components in the sumoylation pathway may be critical in Alzheimer’s disease onset or progression (Li et al., 2003). Additionally, GGE has a third hub gene UBE3A, which plays a critical role in the normal development and function of the nervous system. UBE3A has been suggested to regulate the proteostasis at synapses (Greer et al., 2010) and CNVs at this gene are well known to be implicated in neurodevelopmental disorders such as Angelman syndrome and autism spectrum disorder (for a review see Khatri and Man, 2019).
The three ‘Hub’ genes are not related to genes already known to cause epilepsy; however, these genes are brain expressed and show neurological phenotypes when truncated. Additionally, UBE3A has been suggested to regulate the proteostasis at synapses (Greer et al., 2010) and also genes that play a role in ion channel and synaptic function have been associated with epilepsy including SCN1A, KCNQ2, and GRIN2A (for a review see McTague et al., 2016).
Based on the literature, CNVs in all three genes lead to a brain phenotype assuming that further studies will help to unravel the importance of these genes specifically for GGE and NAFE.
Study limitations
It is important to note that CNV breakpoints in the current study are estimated from genotyped SNPs around the true breakpoint, and these breakpoint estimates are limited by the resolution of the genotyping platform. Last, we recognize that especially small structural variants are not detectable with current genotyping platforms (Sudmant et al., 2015). New technologies for whole-genome sequencing will ultimately enable the assessment of the contribution of a wider array of rare variants, including balanced rearrangements, small CNVs (Brandler et al., 2016) and short tandem repeats (Gymrek et al., 2016).
Conclusion
Large-scale collaborations in epilepsy genetics have greatly advanced discovery through genome-wide association studies. Here, we have extended this framework to rare CNVs in four different epilepsy phenotypes including stringent ancestry and data quality control criteria, after generating the data under the same genotype array and calling pipeline for each subject. Our results help to refine the list of promising candidate CNVs associated with specific epilepsy types and extend the phenotypic spectrum for identified loci. We are confident that the application of this framework to even larger datasets has the potential to advance the discovery of loci and identification of the relevant genes and functional elements.
Funding
This work is part of the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI). CCDG-funded Epi25 research activities at the Broad Institute, including genomic data generation in the Broad Genomics Platform, are supported by NHGRI grant UM1 HG008895 (PIs: Eric Lander, Stacey Gabriel, Mark Daly, Sekar Kathiresan). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
- CNV =
copy number variation
- DEE =
developmental and epileptic encephalopathies
- GGE =
-
genetic generalized epilepsies
IBD = Helmsley irritable bowel disease
- LFE =
lesional focal epilepsies
- NAFE =
non-acquired focal epilepsies
- SNP =
single nucleotide polymorphism
Appendix 1
Epi Consortium
Full details are provided in the Supplementary material.
Yen-Chen Anne Feng, Daniel P. Howrigan, Liam E. Abbott, Katherine Tashman, Felecia Cerrato, Dennis Lal, Claire Churchhouse, Namrata Gupta, Benjamin M. Neale, Samuel F. Berkovic, Holger Lerche, David B. Goldstein, Daniel H. Lowenstein, Gianpiero L. Cavalleri, Patrick Cossette, Chris Cotsapas, Peter De Jonghe, Tracy Dixon-Salazar, Renzo Guerrini, Hakon Hakonarson, Erin L. Heinzen, Ingo Helbig, Patrick Kwan, Anthony G. Marson, Slavé Petrovski, Sitharthan Kamalakaran, Sanjay M. Sisodiya, Randy Stewart, Sarah Weckhuysen, Chantal Depondt, Dennis J. Dlugos, Ingrid E. Scheffer, Pasquale Striano, Catharine Freyer, Roland Krause, Patrick May, Kevin McKenna, Brigid M. Regan, Susannah T. Bellows, Costin Leu, Brigid M. Regan, Caitlin A. Bennett, Susannah T. Bellows, Esther M.C. Johns, Alexandra Macdonald, Hannah Shilling, Rosemary Burgess, Dorien Weckhuysen, Melanie Bahlo, Terence J. O’Brien, Patrick Kwan, Slavé Petrovski, Marian Todaro, Sarah Weckhuysen, Hannah Stamberger, Peter De Jonghe, Chantal Depondt, Danielle M. Andrade, Tara R. Sadoway, Kelly Mo, Heinz Krestel, Sabina Gallati, Savvas S. Papacostas, Ioanna Kousiappa, George A. Tanteles, Katalin Šterbová, Markéta Vlcková, Lucie Sedlácková, Petra Laššuthová, Karl Martin Klein, Felix Rosenow, Philipp S. Reif, Susanne Knake, Wolfram S. Kunz, Gábor Zsurka, Christian E. Elger, Jürgen Bauer, Michael Rademacher, Manuela Pendziwiat, Hiltrud Muhle, Annika Rademacher, Andreas van Baalen, Sarah von Spiczak, Ulrich Stephani, Zaid Afawi, Amos D. Korczyn, Moien Kanaan, Christina Canavati, Gerhard Kurlemann, Karen Müller-Schlüter, Gerhard Kluger, Martin Häusler, Ilan Blatt, Johannes R. Lemke, Ilona Krey, Yvonne G. Weber, Stefan Wolking, Felicitas Becker, Christian Hengsbach, Sarah Rau, Ana F. Maisch, Bernhard J. Steinhoff, Andreas Schulze-Bonhage, Susanne Schubert-Bast, Herbert Schreiber, Ingo Borggräfe, Christoph J. Schankin, Thomas Mayer, Rudolf Korinthenberg, Knut Brockmann, Gerhard Kurlemann, Dieter Dennig, Rene Madeleyn, Reetta Kälviäinen, Pia Auvinen, Anni Saarela, Tarja Linnankivi, Anna-Elina Lehesjoki, Mark I. Rees, Seo-Kyung Chung, William O. Pickrell, Robert Powell, Sanjay M. Sisodiya, Natascha Schneider, Simona Balestrini, Sara Zagaglia, Vera Braatz, Anthony G. Marson, Michael R. Johnson, Pauls Auce, Graeme J. Sills, Patrick Kwan, Larry W. Baum, Pak C. Sham, Stacey S. Cherny, Colin H.T. Lui, Nina Barišic, Gianpiero L. Cavalleri, Norman Delanty, Colin P. Doherty, Arif Shukralla, Mark McCormack, Hany El-Naggar, Laura Canafoglia, Silvana Franceschetti, Barbara Castellotti, Tiziana Granata, Pasquale Striano, Federico Zara, Michele Iacomino, Francesca Madia, Maria Stella Vari, Maria Margherita Mancardi, Vincenzo Salpietro, Francesca Bisulli, Paolo Tinuper, Laura Licchetta, Tommaso Pippucci, Carlotta Stipa, Lorenzo Muccioli, Raffaella Minardi, Antonio Gambardella, Angelo Labate, Grazia Annesi, Lorella Manna, Monica Gagliardi, Elena Parrini, Davide Mei, Annalisa Vetro, Claudia Bianchini, Martino Montomoli, Viola Doccini, Carla Marini, Toshimitsu Suzuki, Yushi Inoue, Kazuhiro Yamakawa, Birute Tumiene, Ruta Mameniskiene, Algirdas Utkus, Ruta Praninskiene, Jurgita Grikiniene, Ruta Samaitiene, Lynette G. Sadleir, Chontelle King, Emily Mountier, S. Hande Caglayan, Mutluay Arslan, Zuhal Yapici, Uluc Yis, Pinar Topaloglu, Bulent Kara, Dilsad Turkdogan, Asli Gundogdu-Eken, Nerses Bebek, Sibel Ugur-Iseri, Betül Baykan, Baris Salman, Garen Haryanyan, Emrah Yücesan, Yesim Kesim, Çigdem Özkara, Beth R. Sheidley, Catherine Shain, Annapurna Poduri, Russell J. Buono, Thomas N. Ferraro, Michael R. Sperling, Dennis J. Dlugos, Warren Lo, Michael Privitera, Jacqueline A. French, Patrick Cossette, Steven Schachter, Hakon Hakonarson, Ruben I. Kuzniecky, Dennis J. Dlugos, Orrin Devinsky, Ruben I. Kuzniecky, Jacqueline A. French, Manu Hegde, Pouya Khankhanian, Katherine L. Helbig, Colin A. Ellis, Gianfranco Spalletta, Fabrizio Piras, Federica Piras, Tommaso Gili, Valentina Ciullo.
Contributor Information
the Epi25 Collaborative:
Yen-Chen Anne Feng, Daniel P Howrigan, Liam E Abbott, Katherine Tashman, Felecia Cerrato, Dennis Lal, Claire Churchhouse, Namrata Gupta, Benjamin M Neale, Samuel F Berkovic, Holger Lerche, David B Goldstein, Daniel H Lowenstein, Gianpiero L Cavalleri, Patrick Cossette, Chris Cotsapas, Peter De Jonghe, Tracy Dixon-Salazar, Renzo Guerrini, Hakon Hakonarson, Erin L Heinzen, Ingo Helbig, Patrick Kwan, Anthony G Marson, Slavé Petrovski, Sitharthan Kamalakaran, Sanjay M Sisodiya, Randy Stewart, Sarah Weckhuysen, Chantal Depondt, Dennis J Dlugos, Ingrid E Scheffer, Pasquale Striano, Catharine Freyer, Roland Krause, Patrick May, Kevin McKenna, Brigid M Regan, Susannah T Bellows, Costin Leu, Brigid M Regan, Caitlin A Bennett, Susannah T Bellows, Esther C Johns, Alexandra Macdonald, Hannah Shilling, Rosemary Burgess, Dorien Weckhuysen, Melanie Bahlo, Terence J O’Brien, Patrick Kwan, Slavé Petrovski, Marian Todaro, Sarah Weckhuysen, Hannah Stamberger, Peter De Jonghe, Chantal Depondt, Danielle M Andrade, Tara R Sadoway, Kelly Mo, Heinz Krestel, Sabina Gallati, Savvas S Papacostas, Ioanna Kousiappa, George A Tanteles, Katalin Šterbová, Markéta Vlcková, Lucie Sedlácková, Petra Laššuthová, Karl Martin Klein, Felix Rosenow, Philipp S Reif, Susanne Knake, Wolfram S Kunz, Gábor Zsurka, Christian E Elger, Jürgen Bauer, Michael Rademacher, Manuela Pendziwiat, Hiltrud Muhle, Annika Rademacher, Andreas van Baalen, Sarah von Spiczak, Ulrich Stephani, Zaid Afawi, Amos D Korczyn, Moien Kanaan, Christina Canavati, Gerhard Kurlemann, Karen Müller-Schlüter, Gerhard Kluger, Martin Häusler, Ilan Blatt, Johannes R Lemke, Ilona Krey, Yvonne G Weber, Stefan Wolking, Felicitas Becker, Christian Hengsbach, Sarah Rau, Ana F Maisch, Bernhard J Steinhoff, Andreas Schulze-Bonhage, Susanne Schubert-Bast, Herbert Schreiber, Ingo Borggräfe, Christoph J Schankin, Thomas Mayer, Rudolf Korinthenberg, Knut Brockmann, Gerhard Kurlemann, Dieter Dennig, Rene Madeleyn, Reetta Kälviäinen, Pia Auvinen, Anni Saarela, Tarja Linnankivi, Anna-Elina Lehesjoki, Mark I Rees, Seo-Kyung Chung, William O Pickrell, Robert Powell, Sanjay M Sisodiya, Natascha Schneider, Simona Balestrini, Sara Zagaglia, Vera Braatz, Anthony G Marson, Michael R Johnson, Pauls Auce, Graeme J Sills, Patrick Kwan, Larry W Baum, Pak C Sham, Stacey S Cherny, Colin H T Lui, Nina Barišic, Gianpiero L Cavalleri, Norman Delanty, Colin P Doherty, Arif Shukralla, Mark McCormack, Hany El-Naggar, Laura Canafoglia, Silvana Franceschetti, Barbara Castellotti, Tiziana Granata, Pasquale Striano, Federico Zara, Michele Iacomino, Francesca Madia, Maria Stella Vari, Maria Margherita Mancardi, Vincenzo Salpietro, Francesca Bisulli, Paolo Tinuper, Laura Licchetta, Tommaso Pippucci, Carlotta Stipa, Lorenzo Muccioli, Raffaella Minardi, Antonio Gambardella, Angelo Labate, Grazia Annesi, Lorella Manna, Monica Gagliardi, Elena Parrini, Davide Mei, Annalisa Vetro, Claudia Bianchini, Martino Montomoli, Viola Doccini, Carla Marini, Toshimitsu Suzuki, Yushi Inoue, Kazuhiro Yamakawa, Birute Tumiene, Ruta Mameniskiene, Algirdas Utkus, Ruta Praninskiene, Jurgita Grikiniene, Ruta Samaitiene, Lynette G Sadleir, Chontelle King, Emily Mountier, S Hande Caglayan, Mutluay Arslan, Zuhal Yapici, Uluc Yis, Pinar Topaloglu, Bulent Kara, Dilsad Turkdogan, Asli Gundogdu-Eken, Nerses Bebek, Sibel Ugur-Iseri, Betül Baykan, Baris Salman, Garen Haryanyan, Emrah Yücesan, Yesim Kesim, Çigdem Özkara, Beth R Sheidley, Catherine Shain, Annapurna Poduri, Russell J Buono, Thomas N Ferraro, Michael R Sperling, Dennis J Dlugos, Warren Lo, Michael Privitera, Jacqueline A French, Patrick Cossette, Steven Schachter, Hakon Hakonarson, Ruben I Kuzniecky, Dennis J Dlugos, Orrin Devinsky, Ruben I Kuzniecky, Jacqueline A French, Manu Hegde, Pouya Khankhanian, Katherine L Helbig, Colin A Ellis, Gianfranco Spalletta, Fabrizio Piras, Federica Piras, Tommaso Gili, and Valentina Ciullo
References
- Addis L, Rosch RE, Valentin A, Makoff A, Robinson R, Everett KV, et al. Analysis of rare copy number variation in absence epilepsies. Neurol Genet 2016; 2: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassari S, Ribierre T, Marsan E, Adle-Biassette H, Ferrand-Sorbets S, Bulteau C, et al. Dissecting the genetic basis of focal cortical dysplasia: a large cohort study. Acta Neuropathol 2019; 138: 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia 2010; 51: 676–85. [DOI] [PubMed] [Google Scholar]
- Brandler WM, Antaki D, Gujral M, Noor A, Rosanio G, Chapman TR, et al. Frequency and complexity of de novo structural mutation in autism. Am J Hum Genet 2016; 98: 667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet 2008; 40: 1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, Mefford HC.. Microdeletion syndromes. Curr Opin Genet Dev 2013; 23: 232–9. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Girirajan S, Eichler EE.. A genetic model for neurodevelopmental disease. Curr Opin Neurobiol 2012; 22: 829–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola A, Cellini E, Stamberger H, Saarentaus E, Cetica V, Lal D, et al. Diagnostic implications of genetic copy number variation in epilepsy plus. Epilepsia 2019; 60: 689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K Bracher-Smith M Owen D Kendall KM Rees E Pardiñas AF, et al. . Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J Med Genet 2019; 56: 131–8. [DOI] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain 2010; 133: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devinsky O, Vezzani A, O’Brien TJ, Jette N, Scheffer IE, de Curtis M, et al. Epilepsy. Nat Rev Dis Primers 2018; 4: 18024. [DOI] [PubMed] [Google Scholar]
- Dibbens LM, Mullen S, Helbig I, Mefford HC, Bayly MA, Bellows S, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet 2009; 18: 3626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet 2008; 40: 776–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, Berkovic SF, Cossette P, Delanty N, et al. De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi25 Collaborative. Ultra-rare genetic variation in the epilepsies: a whole-exome sequencing study of 17,606 individuals. Am J Hum Genet 2019; 105: 267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EpiPM Consortium. A roadmap for precision medicine in the epilepsies. Lancet Neurol 2015; 14: 1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009; 459: 569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 2010; 140: 704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 2016; 48: 22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, Need AC, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet 2010; 86: 707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Heinzen EL, Mefford HC, Commission IG.. Primer Part 1-The building blocks of epilepsy genetics. Epilepsia 2016; 57: 861–8. [DOI] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 2009; 41: 160–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyne HO, Singh T, Stamberger H, Abou Jamra R, Caglayan H, Craiu D, et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet 2018; 50: 1048–53. [DOI] [PubMed] [Google Scholar]
- Hooli BV, Kovacs-Vajna ZM, Mullin K, Blumenthal MA, Mattheisen M, Zhang C, et al. Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Mol Psychiatry 2014; 19: 676–81. [DOI] [PubMed] [Google Scholar]
- Huang AY, Yu D, Davis LK, Sul JH, Tsetsos F, Ramensky V, et al. Rare copy number variants in NRXN1 and CNTN6 increase risk for tourette syndrome. Neuron 2017; 94: 1101–11 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ILAE Consortium on Complex Epilepsies Consortium. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun 2018; 9: 5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri N, Man HY.. The Autism and Angelman Syndrome protein Ube3A/E6AP: the gene, E3 ligase ubiquitination targets and neurobiological functions. Front Mol Neurosci 2019; 12: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, Pernhorst K, Klein KM, Reif P, Tozzi R, Toliat MR, et al. Extending the phenotypic spectrum of RBFOX1 deletions: sporadic focal epilepsy. Epilepsia 2015. a; 56: e129–33. [DOI] [PubMed] [Google Scholar]
- Lal D, Ruppert AK, Trucks H, Schulz H, de Kovel CG, Kasteleijn-Nolst TD, et al. Burden analysis of rare microdeletions suggests a strong impact of neurodevelopmental genes in genetic generalised epilepsies. PLoS Genet 2015. b; 11: e1005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 2011; 70: 886–97. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang H, Wang S, Quon D, Liu YW, Cordell B.. Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci USA 2003; 100: 259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 2017; 49: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol 2016; 15: 304–16. [DOI] [PubMed] [Google Scholar]
- Mefford HC. CNVs in epilepsy. Curr Genet Med Rep 2014; 2: 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Batshaw ML, Hoffman EP.. Genomics, intellectual disability, and autism. N Engl J Med 2012; 366: 733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE.. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev 2009; 19: 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet 2010; 6: e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Mulley JC.. Genetically complex epilepsies, copy number variants and syndrome constellations. Genome Med 2010; 2: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Yendle SC, Hsu C, Cook J, Geraghty E, McMahon JM, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol 2011; 70: 974–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller RS, Weckhuysen S, Chipaux M, Marsan E, Taly V, Bebin EM, et al. Germline and somatic mutations in the MTOR gene in focal cortical dysplasia and epilepsy. Neurol Genet 2016; 2: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlong J, Girard SL, Meloche C, Cadieux-Dion M, Andrade DM, Lafreniere RG, et al. Global characterization of copy number variants in epilepsy patients from whole genome sequencing. PLoS Genet 2018; 14: e1007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen SA, Carvill GL, Bellows S, Bayly MA, Trucks H, Lal D, et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology 2013; 81: 1507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell J, Farlow M, Ghetti B, Benson MD.. A mutation in the amyloid precursor protein associated with hereditary Alzheimer’s disease. Science 1991; 254: 97–9. [DOI] [PubMed] [Google Scholar]
- Ncbi Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2016; 44: D7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi AK, Bottomley C, Kleinschmidt I, Sander JW, Newton CR.. Estimation of the burden of active and life-time epilepsy: a meta-analytic approach. Epilepsia 2010; 51: 883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels JL. Single-gene determinants of epilepsy comorbidity. Cold Spring Harb Perspect Med 2015; 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson H, Shen Y, Avallone J, Sheidley BR, Pinsky R, Bergin AM, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol 2014; 75: 943–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Palma E, Helbig I, Klein KM, Anttila V, Horn H, Reinthaler EM, et al. Heterogeneous contribution of microdeletions in the development of common generalised and focal epilepsies. J Med Genet 2017; 54: 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 2011; 29: 512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 2011; 70: 863–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017; 58: 512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striano P, Coppola A, Paravidino R, Malacarne M, Gimelli S, Robbiano A, et al. Clinical significance of rare copy number variations in epilepsy: a case-control survey using microarray-based comparative genomic hybridization. Arch Neurol 2012; 69: 322–30. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015; 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007; 17: 1665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 2008; 358: 667–75. [DOI] [PubMed] [Google Scholar]
- Wiseman FK, Al-Janabi T, Hardy J, Karmiloff-Smith A, Nizetic D, Tybulewicz VL, et al. A genetic cause of Alzheimer disease: mechanistic insights from Down syndrome. Nat Rev Neurosci 2015; 16: 564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW.. A copy number variation map of the human genome. Nat Rev Genet 2015; 16: 172–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the Epi25 Consortium, upon reasonable request.