Abstract
Although the physiological consequences of Notch signaling in hematopoiesis have been extensively studied, the differential effects of individual notch cleavage products remain to be elucidated. Given that ADAM10 is a critical regulator of Notch and that its deletion is embryonically lethal, we generated mice that overexpress ADAM10 (A10Tg) at early stages of lympho- and myeloid development. Transgene expression resulted in abrogated B cell development, delayed T cell development in the thymus and unexpected systemic expansion of CD11b+Gr-1+ cells, also known as myeloid-derived suppressor cells (MDSCs). Mixed bone marrow reconstitution assays demonstrated that transgene expression altered hematopoiesis via a cell intrinsic mechanism. Consistent with previously reported observations, we hypothesized that ADAM10 overexpression dysregulated Notch by uncoupling the highly regulated proteolysis of Notch receptors. This was confirmed using an in vitro model of hematopoiesis via culturing A10Tg hematopoietic Lineage−Sca-1+c-Kit+ cells (LSKs) with OP-9 stromal cells in the presence or absence of Delta-1 (OP9-DL1), a primary ligand for Notch. Blockade of the S2 and S3 cleavage of the Notch receptor demonstrated differential effects on hematopoesis. OP9-DL1 cultures containing the ADAM10 inhibitor (S2 cleavage site) enhanced and rescued B cell development from wild type and A10Tg LSKs, respectively. In contrast, blockade of γ-secretase at the S3 cleavage site induced accumulation of the S2 product and consequently prevented B cell development and resulted in myeloid cell accumulation. Collectively, these findings indicate that the differential cleavage of Notch into S2 and S3 products regulated by ADAM10 is critical to hematopoietic cell-fate determination.
Introduction.
Disintegrin and metalloproteinases (ADAMs) regulate cell signaling pathways by cleaving the extracellular domains of membrane-bound receptors and ligands. Consequently, these proteins serve as initiators for signaling pathways that require regulated intramembrane proteolysis (RIP) of receptor:ligand complexes. In vitro-based assays have revealed that ADAM10 is an important mediator of ectodomain shedding and RIP of numerous substrates, such as the low affinity IgE receptor, CD23, and the Notch ligand, Delta-1. Proteolytic processing of these substrates contributes to the pathogenesis of multiple disease states, including allergy, cancer and inflammation (1, 2). Accordingly, there is growing interest in ADAM10 as a pharmacological target for these conditions. However, determination of the physiologic consequences of ADAM10-mediated cleavage events has been limited by lethality of ADAM10-null murine embryos (3).
Production of ADAM10-deficient embryos and conditional knockout mice has demonstrated a critical role for ADAM10 in developmental pathways, including thymocyte and marginal zone B cell development (4, 5). Each report concluded that impaired development in the absence of ADAM10 was the result of diminished Notch signaling, which depends upon RIP for signal activation.
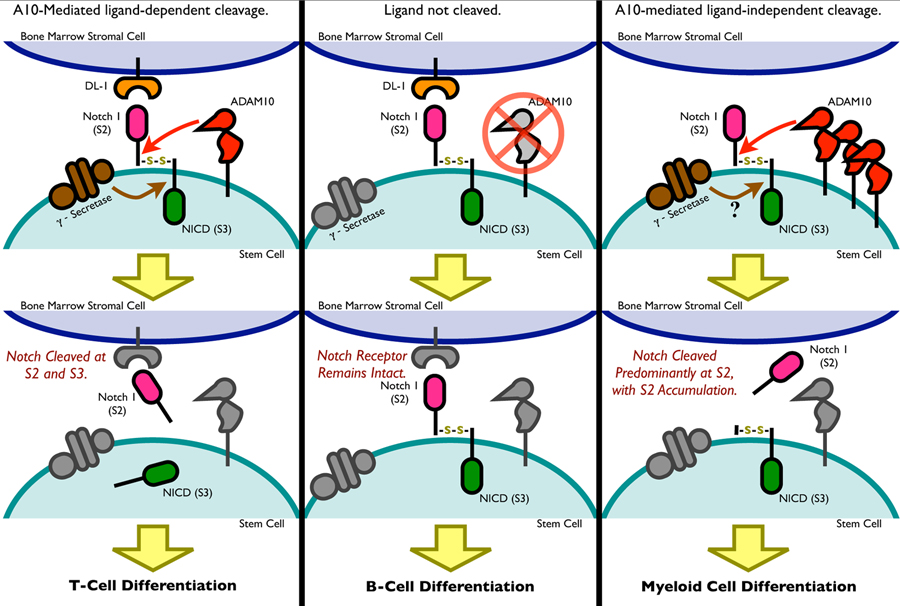
Many studies have demonstrated the importance of Notch signaling in lymphocyte development. The Notch signaling pathway is highly conserved, consisting of four families of receptors (Notch1–4) that interact with ligands (Jagged and Delta) expressed by neighboring cells(6–8). Following ribosomal synthesis, the Notch receptor undergoes a furin-mediated maturation at site 1 (S1) in the Golgi apparatus prior to trafficking to the cell surface. At the surface, Notch is expressed as an integral membrane protein, consisting of both extracellular (NEXT) and intracellular domains (NICD). Once engaged with its ligand, the extracellular domain undergoes an ADAM10-mediated cleavage at site 2 (S2). This event generates a substrate for γ-secretase complex to perform a final cleavage of Notch at site 3 (S3), releasing the transcriptionally active NICD (Figure 8) (9). Several studies have reported the accumulation of intact receptor and the S2 product as a result of ADAM10 and γ-secretase blockade, respectively (10, 11). Although inhibition of both enzymes prevents NICD activation, the consequences of accumulation of these different cleaved products on hematopoiesis remains to be determined.
Figure 8. A model of differential effects of Notch S2/S3 cleavage in hematopoietic differentiation.
In the presence of ligand, the Notch receptor undergoes both S2 and S3 cleavage, resulting in two cleaved products and ultimately promoting T cell development. In the absence of ligand or in the case of blocked S2 cleavage, the Notch receptor remains intact and B cell development results. However, S2 cleavage without S3 due to increased ADAM10 activity, results in myelopoiesis.
Notch is critical for T cell lineage commitment and maturation as well as marginal zone B cell development (6, 12). Notch1 signaling in common lymphoid progenitors (CLPs) is required for early thymocyte development (13), and enforced signaling enhances T cell development (12). In humans, mutations in the negative regulatory region (NRR) of the Notch1 receptor allow ligand-independent cleavage, resulting in excessive Notch1 signaling and the formation of T cell acute lymphocytic leukemia (T-ALL) (9). Several groups report the use of γ-secretase inhibitors (GSIs) as a means of suppressing the expansion of T-ALL cells, and thus as a potential method for treating T-ALL and other Notch-dependent lymphoproliferative disorders (14, 15). However, GSI treatment does not completely abrogate Notch signaling, as it does not affect the initiation of the signaling pathway mediated by ADAMs. GSI treatment results in the accumulation of the S2 cleavage product and its pathologic consequences remain to be elucidated (8, 16).
In contrast to T cell development, Notch signaling prevents commitment of CLPs to the B cell lineage (12). Furthermore, enforced expression of constitutively active NICD or Notch target genes in BM progenitors abrogates B lineage commitment and promotes non-cell autonomous expansion of CD11b+Gr-1+ myeloid cells in transplant recipients (17, 18).
Although the effect of Notch signaling on lymphopoiesis has been well established, its role in myelopoiesis remains controversial. Several studies have shown that Notch signaling promotes expansion of undifferentiated myeloid cells (19), abrogates B cell development, and promotes immature myeloid cell formation (17). Additionally, mice deficient in downstream Notch effectors exhibit defective B cell and myeloid development (20). Collectively, these observations suggest that Notch-mediated alterations in lymphocyte development could serve to modulate myelopoiesis. This supposition is supported by recent data demonstrating that myeloid potential is retained in developing lymphocytes (21, 22). Nonetheless, other studies have either failed to detect myeloid alterations under the same conditions as the aforementioned studies (23) or have indicated that Notch1 and Notch2 are capable of inhibiting myeloid differentiation (24). Adding to this ambiguity is the observation that mice with diminished γ-secretase activity exhibit splenomegaly due to the accumulation of myeloid cells (25). The presence of multiple conflicting data suggests that myeloid differentiation may depend on both the strength and the temporal stage at which Notch signaling occurs.
Many disease states are characterized by the excessive production of ADAM10 cleavage products (2). Inspired by the successful exploitation of conditional knockout mice in previous studies, we generated a strain of ADAM10 transgenic mice (A10Tg) overexpressing ADAM10 at early stages of lympho- and myelopoiesis. The increased ADAM10 activity in these animals severely impaired B2 cell development and promoted the striking expansion of MDSCs via a cell-intrinsic mechanism. Lineage−Sca-1+c-Kit+ (LSK) hematopoietic stem cell culture assays indicated that selective blockade of S2 and S3 cleavage exerts a differential effect on Notch signaling and hematopoiesis in vitro. Furthermore, ADAM10 overexpression was found to alter hematopoiesis by dysregulating RIP-dependent Notch signaling. Taken together, these observations underscore the importance of ADAM10 in Notch-mediated signaling for both lympho and myelopoiesis.
Materials and Methods.
Mice
ADAM10 transgenic (A10Tg) mice were generated as described in the Supplemental methods. C57BL/6 and congenic CD45.1+ (B6-Ly5.2) mice were purchased from the National Cancer Institute. All mouse protocols were approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee.
Flow Cytometry and Cell Sorting
Cell isolation and labeling was conducted as previously described(5). Additionally, peritoneal fluid cells were obtained by flushing the peritoneal cavity with PBS (5mL). BM cells were isolated by flushing excised tibias and femurs with complete RPMI. Single cell suspensions of PLN cells, thymocytes, and splenocytes were created by disrupting inguinal, brachial, axillary lymph nodes, thymus, and spleens, respectively, with glass slides. Cells were labeled following red blood cell lysis and filtration through 40µM cell strainers. Antibodies included anti-mouse unlabeled 2.4G2, biotinylated CD135 (A2F10), FITC-conjugated IL7R (A7R34), CD44 (IM7), B220 (RA3–6B2), and Gr-1 (RB6–8C5); PE-conjugated B220, CD8 (53–6.7), Gr-1, Ter-119, Thy1.2, (30-H12) CD11b (M1/70), CD3ε (2C11); APC-labeled B220, CD4 (RM4–5), CD5 (53–7.3), CD45.2 (104) and c-kit (2B8); PE/Cy7-conjugated CD11b and sca-1 (D7), PE/Cy5 CD34 (A2F10), APC/Cy7-conjugated CD19 (6D5) and CD45.1 (A20), PerCP/Cy5.5-conjugated IgM (RMM-1) and IL7R from Biolegend; CD34-FITC (RAM34), Ly6G-FITC (1A8), c-kit-PE (2B8), Sca-1-PE/Cy7, and APC-conjugated CD3 (145–2C11),B220 (RA3–6B2), Ly6C/Ly6G (RB6–8C5), CD11b (M1/70), and TER-119 (TER-119) from BD Biosciences, and ADAM10-FITC (FAB946) from R&D Systems. Anti-mouse FcγRII/III (in-house) and anti-mouse Ly6D (49-H4) (BD Biosciences) were biotinylated with EZ-Link Sulfo-NHS-biotin (Pierce), followed by dialysis to remove free NHS-biotin. Streptavidin-ECD (Beckman Coulter) was used for secondary labeling of biotinylated-Ly6D and FcγRII/III labeled cells. Flow cytometric analysis was performed using a Canto or AriaII (BD Biosciences), and data analysis was conducted with FCS Express V3 software. Histogram overlays were generated in SigmaPlot 10.0. as line plots and smoothed using the SMOOTH transform. For fluorescence activated cell sorting (FACS) of LSKs, lineage positive cells were depleted with a lineage cell depletion kit (MACS; Miltenyi Biotec/ Stemcell Technologies). Remaining LSKs (Lin−IL7R−ckithisca-1hi) were sorted with an Aria II. Lineage positive cells include CD3ε, Gr-1, CD11b, B220, and Ter119 positive cells. In Figure 4, B220 was excluded from the lineage. Total spleen MDSCs (CD11b+Gr-1+) were sorted for Purity of sorted cells exceeded 95%. For photomicrographs of sorted spleen MDSCs, cells were cytospun on glass slides and stained with the HEMA 3 stain set (Fisher Scientific). Photographs were taken with a BIOQUANT NOVA camera attached to an Olympus BH-2 microscope.
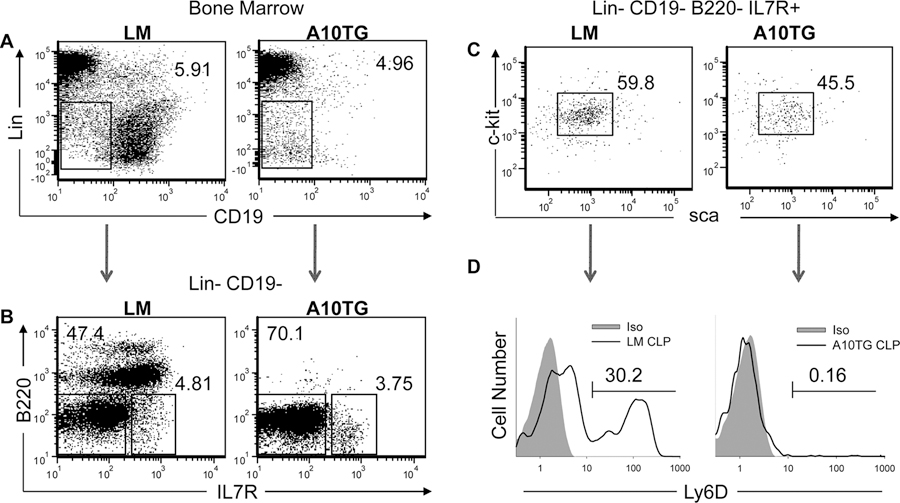
Figure 4. Overexpression of ADAM10 alters hematopoiesis prior to CLP commitment to the B cell lineage.
Flow cytometric analysis of (A) Lineage negative (Lin−) vs CD19 labeled BM cells, (B) B220 and IL-7 receptor (IL7R) expression by Lin−CD19− gated cells from (A), (C) c-kit and sca-1 expression of Lin−CD19−B220−IL7R+ cells from (B). (D) Expression of Ly6D by CLPs of LM and A10Tg mice. Numbers on dot plots and histograms indicate the percentage of gated cells. The cell lineage includes Ter-119, CD11b, Gr-1, and CD3ε positive cells. Plots are representative of 3 independent experiments. CLPs are defined as Lin−CD19−B220−IL7R+c-kitintsca-1int.
Adoptive Transfer and BM chimeras
Recipient CD45.2+ A10Tg (F240) and CD45.1+ WT mice were irradiated with 950 rads using a 137Cs source (Mark I , Model 68–0146; JL Shepherd & Associates). Donor Lineage−Sca-1+c-Kit+ (LSK) cells were isolated from mouse tibia, femur, and humerus by magnetic cell sorting with a lineage cell depletion kit. (MACS; Miltenyi Biotec). 24 hours after irradiation, recipient CD45.2+ A10Tg and CD45.1+ WT mice were injected i.v. with 2.5×106 CD45.1+ WT and 5×106 A10Tg LSK cells, respectively. For generation of mixed BM chimeras, a mixture of LSK cells from CD45.2+ A10Tg (2.5×106) and CD45.1+ WT (1.25×106) mice were injected i.v. into irradiated CD45.1+ WT mice. Cell populations were analyzed 42 and 63 days after reconstitution.
LSK cultures
BM-derived LSKs isolated via magnetic cell sorting and FACS were cultured in the presence of IL-7 (1 ng/mL, Peprotech) and Flt3L (5ng/mL, R&D Systems) as previously described(26). LSK differentiation was examined via flow cytometric analysis, and differentiated cells were passed onto freshly plated OP9 cells with additional cytokines every 4–5 days. OP9-GFP and OP9-DL1 cells were kindly provided by J.C. Zuniga-Pflucker (University of Toronto). Compound E (100nM, Alexis Biochemicals) and GI254023X (5µM, Glaxo Smith Kline) were used for blockade of γ-secretase and ADAM10 activity, respectively.
Statistical Analysis
P-values were calculated using unpaired two-tailed Student’s t-tests. Error bars represent the standard error of the mean between samples.
Results.
Generation of ADAM10 transgenic mice.
To examine the role of ADAM10 in hematopoiesis, we generated A10Tg mice that overexpress murine ADAM10 cDNA under control of the H-2Kb promoter and the IgH enhancer region (Fig. S1A). These transcriptional regulatory units were previously utilized to generate multiple transgenic mouse lines, including TCR, CD23, and bkl transgenics. Thus this vector allows expression in early lymphocyte progenitors (28–30). Inclusion of the IgH enhancer results in preferential expression on B lineage cells. Two founder lines, F240 and F258, were generated, and Southern blot analysis of genomic DNA from F2 progeny demonstrated that both lines contain similar copy numbers of the transgene (Fig. S1B). Because progeny of both lines have nearly identical phenotypes, the following data are presented from line F240, unless otherwise stated.
ADAM10 overexpression prevents B2 lymphocyte development and inhibits development of thymocyte progenitors.
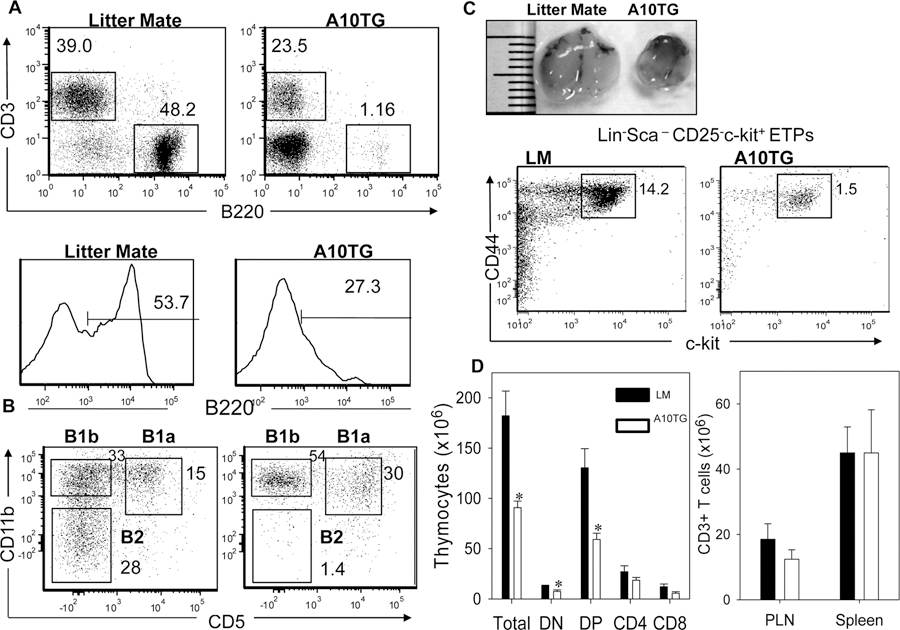
Western blot and flow cytometric analysis demonstrated that transgene expression resulted in elevated levels of ADAM10 in bone marrow (BM) cells, including pro/pre B cells (B220+IgM−) (Fig. S1C–E). Unexpectedly, overexpression markedly reduced the levels of pro/pre B cells and immature B cells (B220hiIgM+) in BM (Fig. S1C). This led to a near complete loss of peripheral B cells in peripheral organs including blood, lymph nodes and spleen (data not shown and Fig. 1A). Analysis of B cells from peritoneal fluid revealed that levels of B1a (B220intCD11b+CD5+) and B1b cells (B220intCD11b+CD5−) in A10Tg mice were not significantly altered compared to littermate (LM) controls, whereas B2 cells (B220hiCD11b−CD5−) were nearly absent (Fig. 1B). Thus, the block in B cell development was specific to bone marrow-derived B2 cells. Additionally, transgene expression suppressed development of thymocyte progenitors. A ten-fold reduction was observed in Lin−CD44+c-kit+ early thymocyte progenitors (ETPs) in A10Tg bone marrow compared to LM (Fig. 1C), which resulted in reduced levels of A10Tg thymic ETPs (data not shown) (31, 32). Accordingly, A10Tg mice have reduced levels of total, double negative, and double positive thymocytes and a small thymus. However, numbers of single positive thymocytes and peripheral T cells in PLN and the spleen were not altered in A10Tg mice (Fig. 1D).
Figure 1. ADAM10 overexpression prevents B2 cell development and suppresses development of thymocyte progenitors.
(A,B) Flow cytometric analysis of (A) T cells (CD3+) and B cells (B220+) in spleen (SPL); and (B) B cell subsets in peritoneal fluid. Lower two panels are gated on B220+ cells in upper histograms. B2 cells: B220hiCD11b−CD5−, B1a cells: B220intCD11b+CD5+, B1b cells: B220intCD11b+CD5−. (C) Representative thymi from indicated mice. Bone Marrow cells were analyzed for Lin−CD25−CD44+c-kit+ early thymocyte progenitors (ETPs). Lineage cocktail includes B220, Ter-119, CD11b, Gr-1, CD3,CD4, CD8. (D) Amount of thymocyte subsets and CD3+ T cells in the spleen and PLN; n=4, mean ± SEM, DN: CD4−CD8−, DP: CD4+CD8+, CD4: CD4+CD8−, CD8: CD4−CD8+. Dot plots and histograms are representative of 6 (A), 4(B), 3(C) independent experiments. Numbers on plots indicated percent of gated cells within box. * p<0.05.
MDSC accumulation in ADAM10Tg mice.
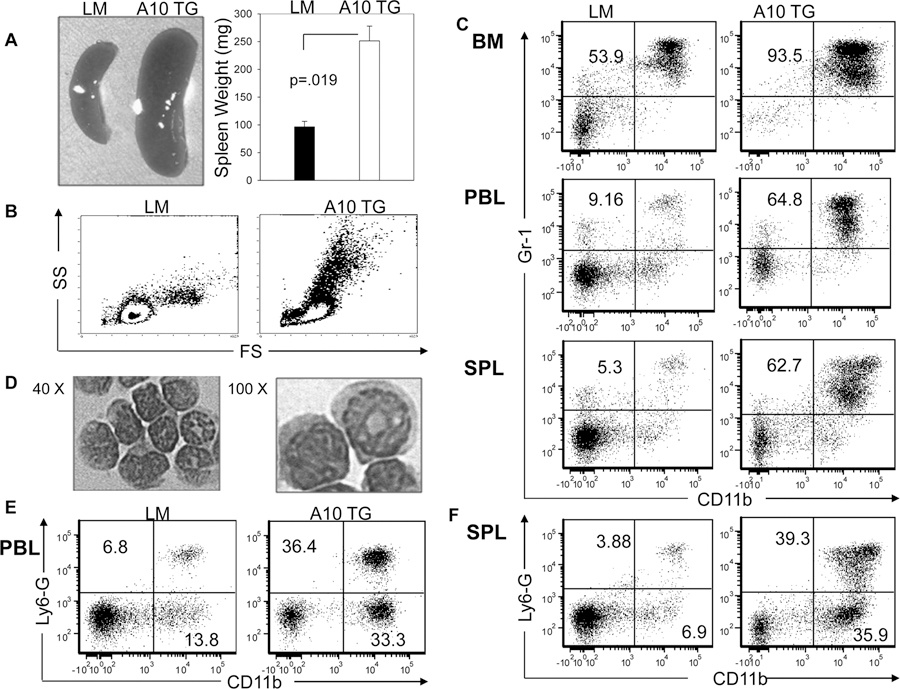
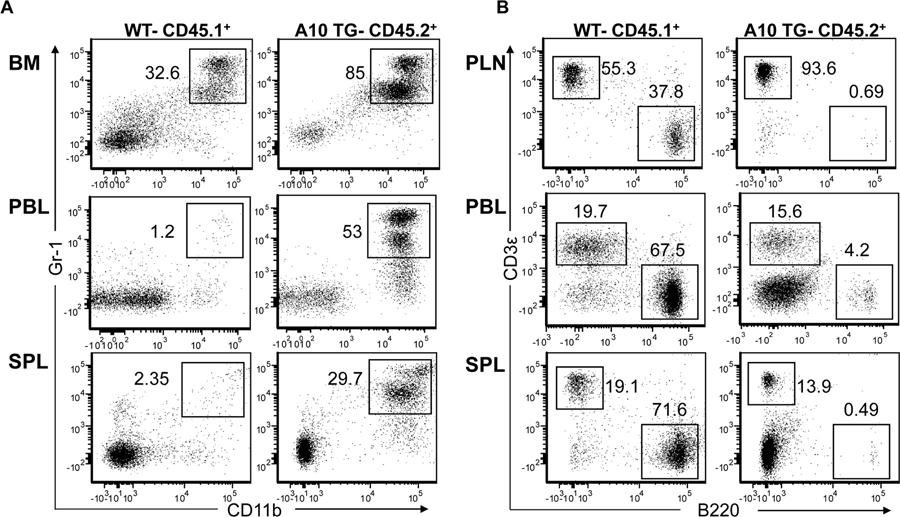
The blockade of B2 cell development was expected to result in reduced levels of total splenocytes. However, the spleens of A10Tg mice were noticeably enlarged, weighing an average of 2.5 fold more than LM spleens (Fig. 2A), containing twice as many nucleated cells (data not shown). The forward and side scatter pattern of A10Tg splenocytes indicated the presence of large granular myeloid cells (Fig. 2B). Further analysis, as shown in Figure 2C, revealed that approximately 63% of transgenic splenocytes were CD11b+Gr-1+ cells, compared to 5.3% of LM splenocytes. While the majority of wild type (WT) CD11b+Gr-1+ cells differentiate into mature myeloid cells prior to exiting the BM, A10Tg CD11b+Gr-1+ cells expanded in BM, constituting 93.5% of BM cells, and entered the spleen and PBL at dramatically high levels (Fig. 2C). CD11b+Gr-1+ cells outside the bone marrow are classified as myeloid derived suppressor cells (MDSCs), consisting of monocytic (CD11b+Gr-1intLy6G−) and granulocytic subsets (CD11b+Gr-1hiLy6G+) (33). Light micrographs of sorted A10Tg CD11b+Gr-1+ splenocytes and flow cytometry confirmed that A10Tg mice contain both monocytic and granulocytic MDSCs in PBL and spleen (Fig. 2D–F).
Figure 2. ADAM10 overexpression causes the expansion of myeloid-derived suppressor cells.
(A) Representative spleens and average spleen weight of indicated mice, n=4, mean ± SEM. Flow cytometric analysis of (B) forward scatter (FS) vs. side scatter (SS) of splenocytes, the percentage of (C) CD11b+Gr-1+ MDSCs present in the bone marrow (BM), PBL, and spleen (SPL), and the percentage of (E,F) CD11b+Ly6-G+ and CD11b+Ly6-G− MDSCs in (E) PBL and (F) spleen of indicated mice. (D) 40X and 100X photomicrographs of sorted CD11b+Gr-1+ splenocytes from A10Tg mice. Flow cytometry plots and photomicrographs are representative of 4 independent experiments. Numbers on plots indicate the percentage of cells in the UR (C) and UR,LR quadrants (E) and (F).
Hematopoietic expression of ADAM10 alters development via a cell-intrinsic mechanism.
To confirm that the observed phenotype was the result of transgene expression by hematopoietic cells and not a dysregulation in the BM stromal environment, mixed bone marrow assays were conducted. As controls, irradiated CD45.2+ A10Tg hosts were reconstituted with LSK cells from WT CD45.1+ congenic mice, and irradiated WT CD45.1+ congenic hosts were reconstituted with CD45.2+ A10Tg LSK BM cells. Despite being in a WT host, A10Tg BM recapitulated the observed altered hematopoiesis of A10Tg mice. Additionally, WT BM cells demonstrated normal cell differentiation even in an A10Tg host (data not shown). This finding indicated that altered cell differentiation in A10Tg mice was due to alterations in signaling pathway(s) within hematopoietic cells rather than the microenvironment. Mixed bone marrow chimera studies further confirmed this, as irradiated CD45.1+ WT hosts were reconstituted with a mixture of LSK BM cells from CD45.2+ A10Tg and CD45.1+ WT donors. This resulted in similar reconstitution of host BM by A10Tg and WT donor cells 42 and 63 days after cell transfer. However, development from BM into peripheral lymphoid organs was less efficient in A10Tg cells, as approximately 66% ± 5.1 (S.E.) and 81% ± 6.2 of recipient spleen and PLN cells, respectively, developed from WT BM at Day 42. The selective development of WT thymocytes was most striking, as 98.1% ± 1.0 of thymocytes at day 63 were of WT origin (data not shown). This result supports the diminished thymocyte development in A10Tg mice (Fig. 1C,D). Despite these differences, hematopoietic development of CD45.1+ WT cells was similar to development in LM control mice, and development of CD45.2+ A10Tg cells closely mimicked development in A10Tg mice. MDSCs only expanded from A10Tg BM cells, and B lineage cells predominantly differentiated from WT BM (Fig. 3). These results demonstrate that ADAM10-mediated MDSC expansion is not the indirect result of abrogated B cell development, trans-cleavage of BM stromal cell ligands, nor cytokine secretion, which would cause WT MDSC expansion. They also illustrate that ADAM10 overexpression on hematopoietic cells causes MDSC expansion via an intrinsic cell autonomous mechanism.
Figure 3. ADAM10 alters hematopoiesis by a cell-autonomous intrinsic mechanism.
Flow cytometric analysis of (A) myeloid and (B) lymphocyte differentiation in mixed BM chimeras generated as described in the Methods 42 days after cell transfer. CD45.1+ and CD45.2+ gated cells differentiated from WT and A10Tg LSK BM, respectively. Data are representative of 3 independent experiments, except PBL data is representative of 6 independent experiments; numbers on dot plots indicate the percent of CD45.1 or CD45.2 gated cells within boxes.
ADAM10 prevents commitment of CLPs to the B cell lineage.
Expansion of MDSCs in conjunction with blockade of B2 cell development indicated that ADAM10 regulates the commitment of BM progenitors to myeloid or lymphoid lineages. According to the classical model of hematopoiesis, hematopoietic stem cells (HSCs), which are Lineage−Sca-1+c-Kit+ (LSKs) in the BM, develop into common myeloid progenitors (CMPs) or common lymphoid progenitors (CLPs), giving rise to early thymocyte precursors or pro-B cells (34). Figure S1C illustrates that a small percentage of B220+ cells was present in A10Tg BM. However, further analysis revealed that the few B220+CD19+ BM cells in A10Tg mice also expressed the myeloid markers, CD11b and Gr-1 (data not shown), indicating that alterations in hematopoiesis occur prior to the pro-B cell stage. Thus, to determine the stage at which ADAM10 overexpression alters hematopoiesis, levels of LSKs, CLPs, and CMPs were examined. Analysis of BM lineage positive cells (Ter119, CD3ε, CD11b, Gr-1) and CD19+ cells demonstrated the near absence of Lin−CD19+ B cells in A10Tg mice. However, the percentage of Lin−CD19− cells was similar to LM levels (Fig. 4A). Lin−CD19− LM cells contain B220+ B cell precursors, which were absent from A10Tg BM (Fig. 4B). Analysis of IL-7R+ cells revealed a modest decrease in CLPs (Lin−CD19−B220−IL-7R+c-kitintscaint) in A10Tg BM (Fig. 4C). Additionally, Inlay et al. recently demonstrated that Ly6D+ CLPs are committed to the B cell lineage, whereas Ly6D− CLPs are uncommitted lymphoid progenitors (35). Accordingly, 30% of LM CLPs expressed high levels of Ly6D (Fig. 4D). However, only 0.16% of A10Tg CLPs were Ly6D+. These findings indicate that hematopoietic alterations in A10Tg mice occur prior to the commitment of CLPs to the B cell lineage and explains the near absence of B cell precursors in the bone marrow.
ADAM10 alters myeloid but not LSK Development.
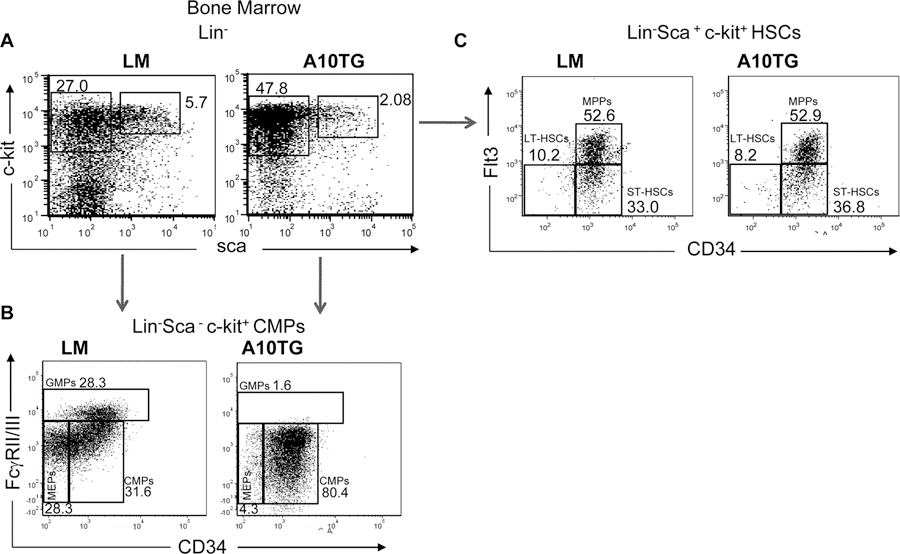
To further determine that state at which A10 overexpression altered cell differentiation, we analyzed numerous subsets of LSKs and myeloid precursors. Multiple studies have shown that BM LSKs can be subdivided into various distinct populations based upon CD34 and fms-related tyrosine kinase 3 (Flt3) expression: CD34−Flt3− long-terms HSCs (LT-HSCs), CD34+Flt3− short-term HSCs (ST-HSCs), and finally CD34+Flt3+ multipotent progenitor populations (MPPs)(36–38). We therefore analyzed these distinct populations within the LSK compartment in A10Tg mice.
Although the percentage of LSKs was slightly lower in A10Tg BM (Fig. 5A), we did not observe any differences in the levels of LT-HSC, ST-HSCs nor MPPs between LM and A10Tg mice (Fig. 5C). However, the percentage of myeloid progenitors (Lin−c-kithisca−) in A10Tg mice was approximately 2-fold greater than the level in LM mice. It was previously shown that myeloid progenitors can also be subdivided into three distinct populations based upon CD34 and low affinity IgG Fc receptors (FcγRII/RIII). These include CD34+FcγRII/IIIlo Common Myeloid Progenitors (CMP), CD34+FcγRII/IIIhi Granulocyte-Macrophage Progenitors (GMP), and CD34−FcγRII/IIIlo Megakaryocyte-Erythroid Progenitors (MEP)(36). As shown in Figure 5B, there is a striking difference in the myeloid compartment of A10Tg and LM BM. GMP and MEP populations are nearly absent from A10Tg mice. Additionally, approximately 80% of the A10Tg myeloid progenitors are in the CMP stage compared to 31.6% in the LM. This finding may account for the systemic expansion of CD11b+Gr-1+ MDSCs in A10Tg mice.
Figure 5. ADAM10 overexpression alters development of myeloid and T cell progenitors, but not LSK subsets.
Flow cytometric analysis of (A) Lin− bone marrow cells. (B) CD34 and FcγRII/III expression on CMP gate from (A). (C) CD34 and Flt3 expression in the Lineage−Sca-1+c-kit+ (LSK) gated cells from (A). (D) Bone marrow cells were also analyzed for early thymocyte progenitors (ETPs). For D, CD25 was added to the lineage negative cocktail. Numbers on dot plots indicate the percentage of gated BM cells. Cell lineage cocktail includes B220, Ter-119, CD11b, Gr-1, and CD3 positive cells. Plots are representative of 3 independent experiments. LSKs were gated as Lin−sca-1hic-kit+ and subdivided to Long-term Hematopoietic Stem Cells (LT-HSCs): CD34−Flt3−, Short-term Hematopoietic Stem Cells ST-HSCs: CD34+Flt3−, and Multipotent Progenitors (MPPs): CD34+Flt3+. The c-kit+ myeloid cells were gated and defined as Common Myeloid Progenitors (CMPs): CD34+FcgRII/IIIlo, Granulocyte-Macrophage Progenitors (GMPs): CD34+FcgRII/IIIhi, and Megakaryocyte-Erythroid Progenitors (MEPs): CD34−FcgRII/IIIlo.
ADAM10 overexpression alters hematopoiesis by dysregulating Notch signaling.
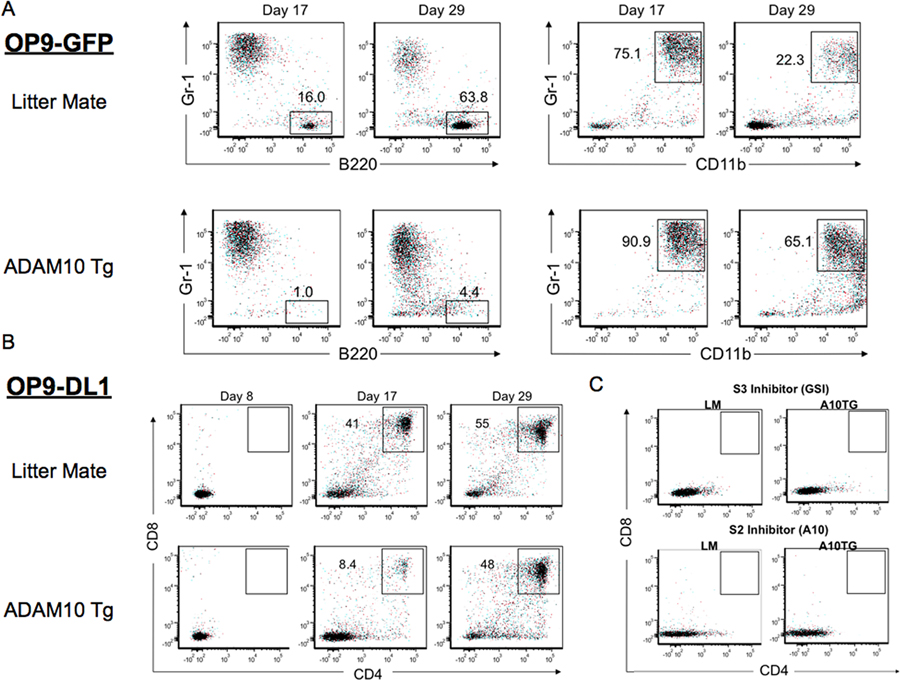
Although the mechanism by which ADAM10 regulates hematopoiesis remains ambiguous, recent studies have demonstrated its essential role in performing S2 cleavage of Notch receptors (see model in Figure 8) during embryonic, thymocyte, and marginal zone B cell development (3–5). Thus, to determine the effect of ADAM10 overexpression on Notch signaling in cell differentiation, we cultured purified LSKs on OP9 stromal cells that either did not express Notch ligands (OP9-GFP) or expressed a primary Notch ligand, Delta-1, (OP9-DL1). The addition of IL-7 and Flt3L promote LSK differentiation into T cells on OP9-DL1 cells and B cells on OP9-GFP cells (26). OP9-GFP cultures showed that while LM LSKs developed into B cells, A10Tg LSKs only generated CD11b+Gr-1+ myeloid cells (Fig. 6A). OP9-DL1 cultures demonstrated that A10Tg LSKs are capable of T cell differentiation (Fig. 6B). However, their development was delayed compared to LM LSKs. This further supports the adverse effect of ADAM10 overexpression on development of B cell and thymocyte progenitors. Interestingly, high expression of Delta-1 on OP9-DL1 cells prevented myeloid expansion of A10Tg LSKs (Fig. 7A). This result suggests that ADAM10 overexpression may cause myeloid development by dysregulating Notch signaling in the absence of sufficient ligand.
Figure 6. Inhibition of γ-secretase or ADAM10 activity prevents Notch-dependent T cell development.
(A,B) Flow cytometric analysis of differentiated LSKs co-cultured with (A) OP9-GFP or (B) OP9-DL1 stromal cells for 8, 17, and 29 days; representative of 4 independent experiments. (C) T cell development of LSKs co-cultured with OP9-DL1 cells for 29 days in the presence of a γ-secretase inhibitor, Compound E, or an ADAM10 inhibitor, GI254023X; representative of 2 independent experiments.
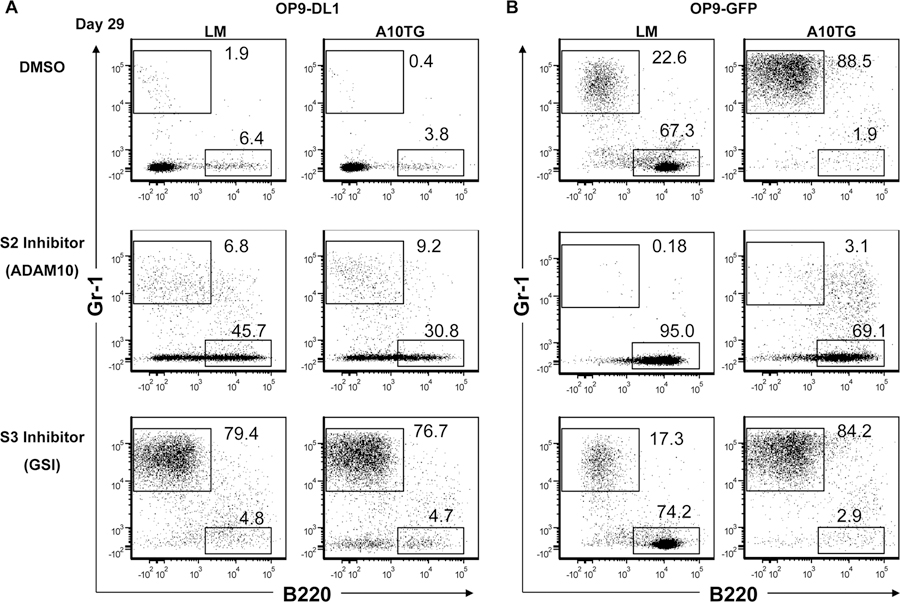
Figure 7. ADAM10 overexpression and γ-secretase inhibition disrupt Notch signaling and direct myeloid cell development.
Flow cytometric analysis of LSK differentiation after 29 days of LSK co-culture with (A) OP9-DL1 or (B) OP9-GFP stromal cells as described in the Methods. Compound E, GI254023X, or control DMSO was added to media to inhibit γ-secretase or ADAM10 activity, respectively. DMSO-treated plots are representative of 4 independent experiments; plots of inhibitor-treated cultures are representative of 2 independent experiments. Numbers on plots indicate the percentage of gated live cells based upon forward and side scatter.
To further test whether ADAM10 overexpression deregulates Notch signaling, we examined the effects of inhibiting Notch receptor cleavage in LM and A10Tg LSKs. Inhibition of Notch signaling with the ADAM10 inhibitor (GI254023X) or the γ-secretase inhibitor (GSI, Compound E) (14) in OP9-DL1 cultures prevented LM and A10Tg T cell development (Fig. 6C). In accordance with reports of ADAM10’s critical role in Notch S2 cleavage in other cell types (3–5, 8, 16, 39), this result indicates that ADAM10 also mediates S2 cleavage in developing hematopoietic precursors. Intriguingly, while both inhibitors blocked Notch-dependent T cell development, their effects on development of myeloid and B lineage cells were distinct. ADAM10 inhibition in OP9-DL1 cultures resulted in B cell development of LM and A10Tg LSKs. Additionally, ADAM10 inhibition in OP9-GFP cultures not only resulted in enhanced LM B cell development, but it also rescued B cell development of A10Tg cells and prevented myeloid differentiation.
In contrast to ADAM10 inhibition, GSI treatment of OP9-DL1 cultures caused myeloid development that was strikingly similar to myeloid differentiation of A10Tg cells in OP9-GFP cultures (Fig. 7). Additionally, GSI treatment of LM OP9-GFP cultures had no effect on LSK development. This demonstrates that GSI treatment only directs myeloid development following Notch ligand binding and ADAM-mediated S2 cleavage, resulting in accumulation of the S2 cleavage product. Thus, both inhibition of the protease responsible for degrading the S2 product (γ-secretase) and overexpression of the protease that produces the S2 product (ADAM10) caused myeloid development. Collectively, these results indicate that ADAM10 overexpression may also direct myeloid development by generating excessive S2 product.
Discussion.
ADAM10-mediated S2 cleavage is required for the initiation of the canonical Notch signaling pathway. Accordingly, we hypothesized that ADAM10 activity would also regulate differentiation of early hematopoietic progenitors. Our experimental observations demonstrate that ADAM10 overexpression attenuates the development of thymocytes, abrogates B2 cell development, and promotes expansion of functional MDSCs via a cell-intrinsic mechanism. Furthermore, the S2 and S3 cleavage products of Notch are shown to exert a differential effect on hematopoietic cell fate determination.
Although effects of Notch signaling in B and T cell lineage commitment has been extensively described, the activity of ADAM10 in cell differentiation and early B lineage commitment was previously uncharacterized. We demonstrate that overexpression of ADAM10 prevented B2 cell commitment from CLPs, and that ADAM10 inhibitors in OP9 cultures enhanced B cell differentiation of LM and A10Tg LSKs. Thus, ADAM10 exerts adverse effects on B cell development. Further analysis of LSK, myeloid and lymphocyte compartments demonstrates the stage at which ADAM10 overexpression alters hematopoiesis. Given that the LSK compartment is not affected, ADAM10 overexpression influences differentiation post LSK subset differentiation but prior to mature lineage commitment (B cells, Ly6D+ CLPs, T cells (ETPs), CMPs, GMPs and MEPs), which are dramatically affected in the A10Tg mice. The precise stage at which ADAM10 overexpression affects LSK commitment is likely to be very transient, and remains to be determined. Although our data shows a generalized overexpression of ADAM10 in BM, this overexpression is limited to B220+ cells. Thus, this transient phase may include a fraction of B220+ cells. Following differentiation of these B220+ cells into non-B cells, the transgene is no longer overexpressed, likely because the IgH enhancer is not utilized by other non-B cells.
The shift in favor of CMP development (Fig. 5B) could explain the robust accumulation of MDSCs in the periphery. Additionally, because CMPs have been shown to retain erythromyeloid potential, it is not surprising that despite the decreased MEP and GMP compartments, erythropoieis remains unaffected. Furthermore, the presence of comparable Natural Killer (NK) cells (data not shown) and T cells in the periphery of A10Tg mice (Fig. 1D) suggests that while CLPs are unable to give rise to B cells, their ability to differentiate into ETPs and NK cells is retained.
It should be noted that upon lineage depletion, comparable bone marrow cell numbers were recovered from both A10Tg and LM mice. Therefore, the percentage of ETPs obtained is reflective of absolute numbers of thymocyte precursors in the BM. Although peripheral T cell levels are comparable, the observed reduction in ETPs is nonetheless very intriguing and consistent with the diminished thymus seeding of A10Tg BM cells. This explains the reduced thymus size, relative inability of A10Tg thymocyte precursors to develop in mixed BM chimeras, and delayed T cell development in OP-DL1 cultures.
The classical model of hematopoiesis describes the initial dichotomous differentiation of LSKs into CLPs or CMPs. However, recent studies demonstrating that early thymocyte progenitors possess myeloid potential have challenged this model (21, 22). Additional studies demonstrated that B cell progenitors, including CLPs, also retain myeloid potential (40, 41), while other progenitors that lack T cell potential can develop into B cells or macrophages (42). For this reason, a myeloid-based model of LSK development is beginning to emerge (34, 43). In A10Tg mice, the moderate effects on thymocyte development in combination with the more pronounced effects on B lineage commitment and myeloid expansion indicate that B2 cells and expanded myeloid cells develop from common progenitor(s), whereas thymocytes may develop from a unique precursor.
The data presented is consistent with the hypothesis that excess ADAM10 causes ligand independent S2 cleavage of Notch. It is possible that, in the absence of ligand, γ-secretase complex is not recruited to the cell membrane; thus leading to reduced S3 cleavage and accumulation of the S2 product. This would then result in enhanced myeloid development concurrent with diminished numbers of T cell progenitors. Given the high ligand expression levels in OP9-DL1 cells, we expected proper T cell differentiation in A10Tg LSKs as any excess S2 product would be processed by recruited γ -secretase. Indeed, even though T cell development in A10Tg LSKs was delayed, it was comparable to LM at day 29. Additionally, complete γ-secretase inhibition of LM and A10Tg OP9-DL1 cultures, a condition previously shown to cause S2 product accumulation(8, 16), mimicked myeloid development of A10Tg LSKs in the absence of ligand.
These findings support a model through which Notch cleavage mediates cell fate determination (Figure 8). In the presence of ligand, the Notch receptor undergoes cleavage at both S2 and S3 sites, resulting in two products and ultimately promoting T cell development. In the absence of Notch ligand or in the case of blocked S2 cleavage, the Notch receptor remains intact and B cell development results. However, S2 cleavage without S3 cleavage, as observed during ADAM10 overexpression or γ-secretase inhibition, induces myelopoiesis and delayed T cell development. This is in agreement with other studies which have shown γ-secretase blockade to result in accumulation of the S2 product (8, 16), that could direct myeloid development. Indeed, diminished presenilin (PS) dependent γ-secretase activity in PS1+/−PS2−/− mice results in myeloproliferative disease, characterized by accumulation of Mac-1/CD11b+Gr-l+ cells, causing splenomegaly (25).
The differential effects of S2 and S3 blockade on WT Notch signaling has significant implications for the treatment of Notch-related diseases. Many reports have proposed the use of GSIs for the treatment of T-ALL and B cell lymphoma (14, 44), however; our findings indicate that GSI treatment could cause MDSC expansion that would ultimately induce immunosuppression and enhance tumor growth. Thus, studies of GSI treatment in mice and clinical trials should include careful monitoring of myeloid cell development. This study indicates that pharmacologic blockade of S2 cleavage with ADAM10 inhibitors may be a more advantageous strategy.
Our data demonstrates that the proteolytic activity of ADAM10 regulates the lineage commitment of B2 cells and the expansion of functional MDSCs in a cell-intrinsic manner. Moreover, the data affords an in vivo model for further examination of MDSC expansion and MDSC-mediated immune suppression in the absence of confounding tumor-derived factors. Given that these factors also regulate immune responses (45), A10Tg animals provide a tool with which specific pathways leading to MDSC expansion may be elucidated in a controlled fashion. The singular expression of B1 cells in the absence of B2 cells likewise makes this mouse useful for the study of B1-mediated humoral immunity. Furthermore, our observations support the developing myeloid-based model of hematopoiesis, leading us to propose a novel mechanism through which the Notch S2 and S3 cleavage products differentially regulate cell fate determination.
Supplementary Material
Acknowledgements.
The authors thank S. Barbour, H. Bear, M. Manjili and J. Tew for their comments, L. Graham, J. Elenesky and J. Gomez-Arroyo for their technical assistance, and the VCU Transgenic/ Knockout Mouse Core Facility for the generation of the mice and the Flow cytometry facility for both flow analysis and sorting. Paul Saftig (Christian-Albrechts-Universität zu Kiel, Germany) is acknowledged for providing the ADAM10 cDNA.
Supported by NIAID/NIH (RO1AI18697 and U19AI077435 to D.H.C.), the American Heart Association (0815066E to D.R.G) and NIH-NCI Cancer Center Support Grant P30 CA016059 for VCU transgenic and flow cytometry facility.
References.
- 1.Blobel CP 2005. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 6:32–43. [DOI] [PubMed] [Google Scholar]
- 2.Crawford HC, Dempsey PJ, Brown G, Adam L, and Moss ML. 2009. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des 15:2288–2299. [DOI] [PubMed] [Google Scholar]
- 3.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, and Saftig P. 2002. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet 11:2615–2624. [DOI] [PubMed] [Google Scholar]
- 4.Tian L, Wu X, Chi C, Han M, Xu T, and Zhuang Y. 2008. ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int Immunol 20:1181–1187. [DOI] [PubMed] [Google Scholar]
- 5.Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, and Conrad DH. 2010. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med 207:623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, and Hirai H. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18:675–685. [DOI] [PubMed] [Google Scholar]
- 7.Radtke F, Fasnacht N, and Macdonald HR. 2010. Notch signaling in the immune system. Immunity 32:14–27. [DOI] [PubMed] [Google Scholar]
- 8.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, and Vooijs M. 2009. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem 284:31018–31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopan R, and Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, and Kopan R. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell 5:197–206. [DOI] [PubMed] [Google Scholar]
- 11.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, and Israel A. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell 5:207–216. [DOI] [PubMed] [Google Scholar]
- 12.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, and Pear WS. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299–308. [DOI] [PubMed] [Google Scholar]
- 13.Tanigaki K, and Honjo T. 2007. Regulation of lymphocyte development by Notch signaling. Nat Immunol 8:451–456. [DOI] [PubMed] [Google Scholar]
- 14.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, Meijerink J, Aifantis I, Basso G, Cordon-Cardo C, Ai W, and Ferrando A. 2009. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med 15:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanda T, Li X, Gutierrez A, Ahn Y, Neuberg DS, O’Neil J, Strack PR, Winter CG, Winter SS, Larson RS, von Boehmer H, and Look AT. 2010. Interconnecting molecular pathways in the pathogenesis and drug sensitivity of T-cell acute lymphoblastic leukemia. Blood 115:1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bozkulak EC, and Weinmaster G. 2009. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol 29:5679–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamata S, Du C, Li K, and Lavau C. 2002. Notch1 perturbation of hemopoiesis involves non-cell- autonomous modifications. J Immunol 168:1738–1745. [DOI] [PubMed] [Google Scholar]
- 18.Kawamata S, Du C, Li K, and Lavau C. 2002. Overexpression of the Notch target genes Hes in vivo induces lymphoid and myeloid alterations. Oncogene 21:3855–3863. [DOI] [PubMed] [Google Scholar]
- 19.Schroeder T, and Just U. 2000. Notch signalling via RBP-J promotes myeloid differentiation. EMBO J 19:2558–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder T, Kohlhof H, Rieber N, and Just U. 2003. Notch signaling induces multilineage myeloid differentiation and up-regulates PU.1 expression. J Immunol 170:5538–5548. [DOI] [PubMed] [Google Scholar]
- 21.Bell JJ, and Bhandoola A. 2008. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452:764–767. [DOI] [PubMed] [Google Scholar]
- 22.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, and Kawamoto H. 2008. Adult T-cell progenitors retain myeloid potential. Nature 452:768–772. [DOI] [PubMed] [Google Scholar]
- 23.Stier S, Cheng T, Dombkowski D, Carlesso N, and Scadden DT. 2002. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood 99:2369–2378. [DOI] [PubMed] [Google Scholar]
- 24.Bigas A, Martin DI, and Milner LA. 1998. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol 18:2324–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qyang Y, Chambers SM, Wang P, Xia X, Chen X, Goodell MA, and Zheng H. 2004. Myeloproliferative disease in mice with reduced presenilin gene dosage: effect of gamma-secretase blockage. Biochemistry 43:5352–5359. [DOI] [PubMed] [Google Scholar]
- 26.de Pooter R, and Zuniga-Pflucker JC. 2007. T-cell potential and development in vitro: the OP9-DL1 approach. Curr Opin Immunol 19:163–168. [DOI] [PubMed] [Google Scholar]
- 27.Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, Broadway N, Hartmann D, Saftig P, Umland S, Sehara-Fujisawa A, Black RA, Ludwig A, Becherer JD, Conrad DH, and Blobel CP. 2006. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol 7:1293–1298. [DOI] [PubMed] [Google Scholar]
- 28.Pircher H, Mak TW, Lang R, Ballhausen W, Ruedi E, Hengartner H, Zinkernagel RM, and Burki K. 1989. T cell tolerance to Mlsa encoded antigens in T cell receptor V beta 8.1 chain transgenic mice. EMBO J 8:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malek SN, Dordai DI, Reim J, Dintzis H, and Desiderio S. 1998. Malignant transformation of early lymphoid progenitors in mice expressing an activated Blk tyrosine kinase. Proc Natl Acad Sci U S A 95:7351–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payet ME, Woodward EC, and Conrad DH. 1999. Humoral response suppression observed with CD23 transgenics. J Immunol 163:217–223. [PubMed] [Google Scholar]
- 31.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, and Bhandoola A. 2005. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol 6:663–670. [DOI] [PubMed] [Google Scholar]
- 32.von Boehmer H 2005. Notch in lymphopoiesis and T cell polarization. Nat Immunol 6:641–642. [DOI] [PubMed] [Google Scholar]
- 33.Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, De Baetselier P, and Van Ginderachter JA. 2008. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood 111:4233–4244. [DOI] [PubMed] [Google Scholar]
- 34.Kawamoto H, and Katsura Y. 2009. A new paradigm for hematopoietic cell lineages: revision of the classical concept of the myeloid-lymphoid dichotomy. Trends Immunol 30:193–200. [DOI] [PubMed] [Google Scholar]
- 35.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, and Weissman IL. 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23:2376–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, and Bryder D. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1:428–442. [DOI] [PubMed] [Google Scholar]
- 37.Wilson A, and Trumpp A. 2006. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol 6:93–106. [DOI] [PubMed] [Google Scholar]
- 38.Luc S, Buza-Vidas N, and Jacobsen SE. 2007. Biological and molecular evidence for existence of lymphoid-primed multipotent progenitors. Ann N Y Acad Sci 1106:89–94. [DOI] [PubMed] [Google Scholar]
- 39.Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, Bartsch U, Weskamp G, Blobel CP, Glatzel M, De Strooper B, and Saftig P. 2010. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci 30:4833–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balciunaite G, Ceredig R, Massa S, and Rolink AG. 2005. A B220+ CD117+ CD19− hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol 35:2019–2030. [DOI] [PubMed] [Google Scholar]
- 41.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, and Hardy RR. 2006. Lineage specification and plasticity in CD19− early B cell precursors. J Exp Med 203:675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montecino-Rodriguez E, Leathers H, and Dorshkind K. 2001. Bipotential B-macrophage progenitors are present in adult bone marrow. Nat Immunol 2:83–88. [DOI] [PubMed] [Google Scholar]
- 43.Chi AW, Bell JJ, Zlotoff DA, and Bhandoola A. 2009. Untangling the T branch of the hematopoiesis tree. Curr Opin Immunol 21:121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan K, Murakami M, Bajaj B, Kaul R, He Z, Gan R, Feldman M, and Robertson ES. 2009. Inhibition of KSHV-infected primary effusion lymphomas in NOD/SCID mice by gamma-secretase inhibitor. Cancer Biol Ther 8:2136–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrand-Rosenberg S, and Sinha P. 2009. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol 182:4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.