In December 2019, cases of a respiratory disease were reported in Hubei Province, China, caused by a positive-sense RNA virus from the family Coronaviridae (1). Subsequently, the disease was called coronavirus disease (COVID-19) and the virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The outcome of infection with SARS-CoV-2 is highly variable; on one hand, the virus has been responsible for more than 360,000 deaths worldwide, and on the other hand, there is a diverse range of clinical outcomes in different people (2). For any virus, infection depends on the ability to 1) enter, 2) evade cellular defenses, 3) hijack host machineries to express viral genes, 4) replicate new genomes, 5) assemble viral particles, and 6) exit. Virus tropism, the ability to infect particular cell types, is defined by the differential expression of host factors the virus subverts or evades during these processes. The earliest determinant is binding and entry via a cell surface receptor.
For SARS-CoV-2 entry, the primary receptor is ACE2 (angiotensin I–converting enzyme 2), which serves as receptor for SARS-CoV and a human seasonal coronavirus, human coronavirus NL63 (HCoV-NL63) (1). The physiological role of ACE2 is the regulation of the renin-angiotensin hormone system, regulating blood volume, systemic vascular resistance, and cardiovascular homeostasis (3). ACE2 is abundantly expressed in intestine, liver, kidney, and testis (proteinatlas.org). Because COVID-19 is primarily a respiratory disease with obvious virally induced lesions in the lung, there has been intense interest to characterize ACE2 expression in the respiratory tract.
In the current issue of the Journal, Zhang and colleagues (pp. 219–229) have analyzed a broad range of preexisting RNA expression microarray data from human trachea and small and large airway epithelium (SAE/LAE) (4). They confirm ACE2 expression in these tissues and report higher levels of ACE2 in the trachea and LAE as compared with SAE. Similarly, Sungnak and colleagues recently reported at a single-cell level that upper airway cell types, including ciliated cells, express ACE2 mRNA (5). Lee and colleagues confirmed this at the protein level, showing ACE2 expression on the motile cilia by immunofluorescent staining (6). Together, these findings imply that because of abundant ACE2 expression, respiratory cells in the upper respiratory tract, particularly ciliated cells, can be infected by SARS-CoV-2 and that they may be more susceptible to infection than those in deep lung. Indeed, Hou and colleagues employed an elegant reverse genetic approach in which recombinant SARS-CoV-2 viruses expressing GFP (green fluorescent protein) were used to infect cells from different levels of the respiratory tract and showed that the gradient of decreased expression of ACE2 from nose to alveolus is mirrored by a decrease in permissiveness to virus infection (7).
However, ACE2 expression may not be the only factor determining SARS-CoV-2 permissivity.
Not all cells that express ACE2 are susceptible to SARS-CoV-2 infection. Re-evaluating single-cell RNA sequencing allowed Zhang and colleagues to identify expression of ACE2 in all SAE cell types (even if at reduced expression relative to LAE), including club cells. Others have confirmed the presence of ACE2 protein and the surface activating protease TMPRSS2 (transmembrane protease, serine 2) in club cells (7). Nevertheless, club cells do not get productively infected by SARS-CoV-2 (7). Club cells have a stem cell–like function in the respiratory epithelium and potentially express intrinsically high levels of some antiviral IFN-stimulated genes, such as IFITMs (IFN-induced transmembrane proteins) and Ly6E (lymphocyte antigen 6E) (8), both described as coronavirus restriction factors (9, 10).
Just as expression of cell surface proteins used for SARS-CoV-2 entry does not always confer susceptibility to infection, different expression levels of ACE2 between individuals do not necessarily determine disease outcome. One key question in the field is why children are less affected by SARS-CoV-2 infection despite similar seroprevalence rates. Some studies have shown an age-dependent direct correlation between levels of ACE2 expression in nasal epithelium and age (11), but in other studies, this pattern did not hold up (6).
Another example is the effect of smoking. When the pandemic first started, smoking was considered a risk factor for COVID-19, as it is for many other respiratory virus infections. Zhang and colleagues were able to categorize their analysis of SAEs according to the smoking status and identified that male smokers had an increased expression of ACE2. This is complementary with other studies that have reported the same observations at the protein level (12). Strikingly, numerous epidemiological reports have found that smokers are actually underrepresented for COVID-19 complications (2, 13). Notably, a study with 1,099 individuals showed that smokers represented only 12.6% of COVID-19 cases while representing 30% of the Chinese population (2). These observations are inconsistent with the increased expression of the virus receptor and emphasize that receptor abundance is not the only factor important for severe disease progression.
Understanding the wide spectrum in severity of COVID-19 disease in different individuals infected by SARS-CoV-2 is important but challenging because disease outcome is determined by a combination of exposure levels, virus, and host responses. A first and crucial step is to understand how expression levels of genes we know to be involved in virus replication might vary within and between hosts. The paper from Zhang and colleagues makes an important contribution to this body of knowledge and underscores again the key role of ACE2 as the virus receptor. However, taken in the wider context of accumulating cell biological and epidemiological information, the correlates appear to be less clear cut than expected (Figure 1). Other host genes that correlate with cell permissivity to SARS-CoV-2 infection now need to be defined in an unbiased manner. In addition, coupling epidemiological observations with this knowledge, for example to understand why different cohorts have different risks of disease progression, might reveal a novel Achilles heel for the virus.
Figure 1.
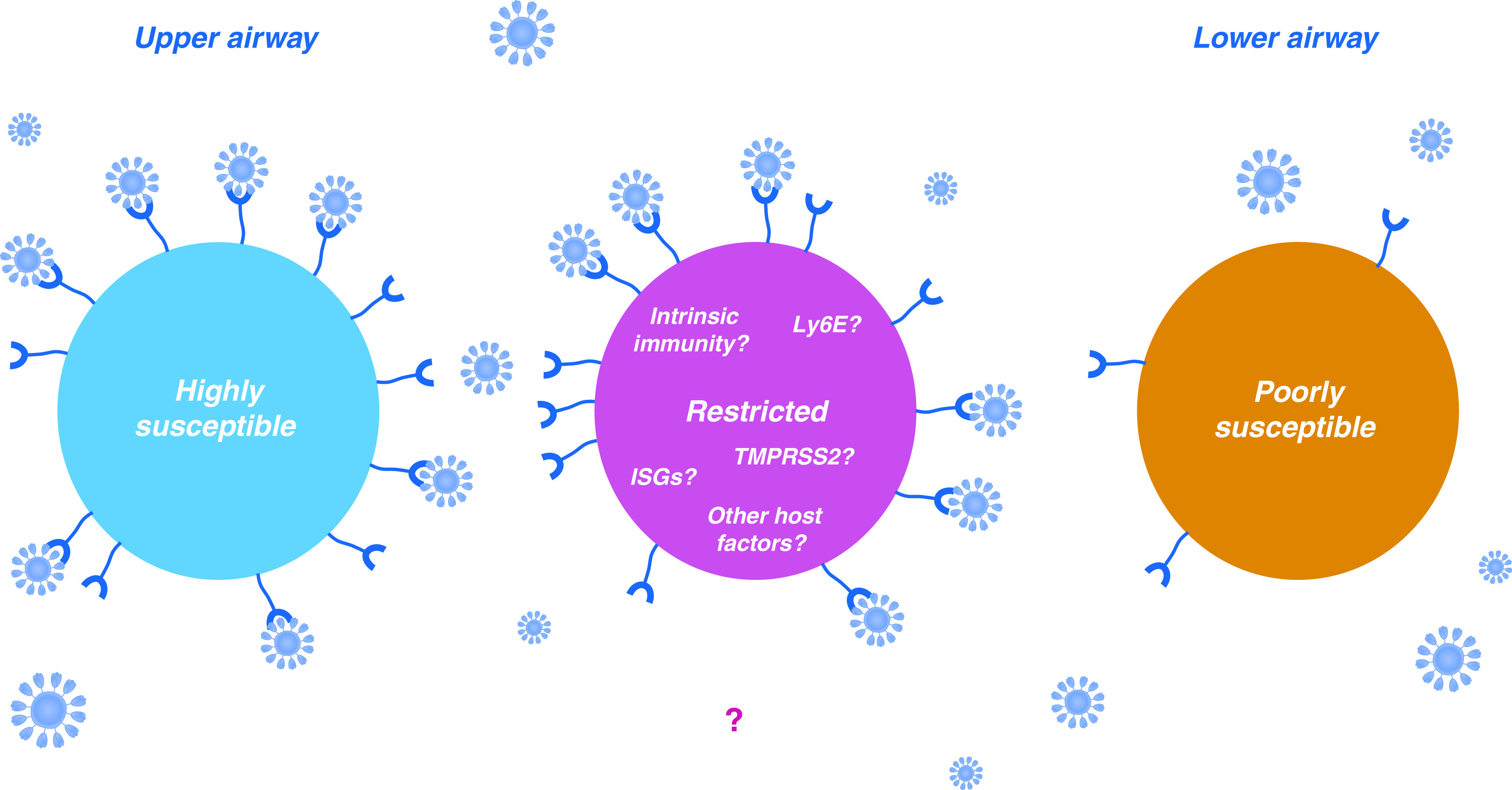
In some cells, ACE2 expression is higher (left cell), whereas in other cells, ACE2 expression is lower (right cell). In the case of the airways, this renders the more highly expressing upper airway cells inherently more permissive to infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Nevertheless, in other cells (central cell), ACE2 expression is high, and yet the population remains resistant to SARS-CoV-2 infection. This phenotype is potentially dictated by the expression levels of additional proviral factors, or restrictive host factors such as IFN-stimulated genes (ISGs).
Supplementary Material
Footnotes
Originally Published in Press as DOI: 10.1164/rccm.202006-2151ED on June 10, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu W, Tao ZW, Wang L, Yuan ML, Liu K, Zhou L, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl) 2020;133:1032–1038. doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang W, McKinnie SM, Farhan M, Paul M, McDonald T, McLean B, et al. Angiotensin-converting enzyme 2 metabolizes and partially inactivates Pyr-Apelin-13 and Apelin-17: physiological effects in the cardiovascular system. Hypertension. 2016;68:365–377. doi: 10.1161/HYPERTENSIONAHA.115.06892. [DOI] [PubMed] [Google Scholar]
- 4. Zhang H, Rostami MR, Leopold PL, Mezey JG, O’Beirne SL, Strulovici-Barel Y, et al. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med. 2020;202:219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al. HCA Lung Biological Network. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee I, Nakayama T, Wu CT, Goltsev Y, Jiang S, Gall PA, et al. Robust ACE2 protein expression localizes to the motile cilia of the respiratory tract epithelia and is not increased by ACE inhibitors or angiotensin receptor blockers [preprint] MedRxiv; 2020 [accessed 2020 May 12]. Available from: https://www.medrxiv.org/content/10.1101/2020.05.08.20092866v1.
- 7. Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. doi: 10.1016/j.cell.2020.05.042. [online ahead of print] 27 May 2020; DOI: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–438, e25. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfaender S, Mar KB, Michailidis E, Kratzel A, Hirt D, V’kovski P, et al. LY6E impairs coronavirus fusion and confers immune control of viral disease [preprint] BioRxiv 2020 [accessed 2020 Apr 10]. Available from: https://www.biorxiv.org/content/10.1101/2020.03.05.979260v1. [DOI] [PMC free article] [PubMed]
- 10. Huang I-C, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. doi: 10.1001/jama.2020.8707. [online ahead of print] 20 May 2020; DOI: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leung JM, Yang CX, Tam A, Shaipanich T, Hackett TL, Singhera GK, et al. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J. 2020;55:2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontanet A, Tondeur L, Madec Y, Grant R, Besombes C, Jolly N, et al. Cluster of COVID-19 in northern France: a retrospective closed cohort study [preprint] MedRxiv; 2020 [accessed 2020 Apr 23]. Available from: https://www.medrxiv.org/content/10.1101/2020.04.18.20071134v1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


