Abstract
Aim
Since RAP1B is critical for platelet functions, including hemostasis, this study was conducted to identify RAP1B regulating microRNAs (miRNAs) in ex vivo stored platelets.
Background
Previous studies with platelets identified factors affecting RAP1B activity but regulatory miRNAs that affect RAP1B protein expression have not been reported.
Objective
To understand the functional significance of miRNA mediated regulation of RAP1B in stored platelets, using microRNA, miR-181a as an example.
Methods
A Tagged RNA Affinity approach (MS2-TRAP) was employed to identify miRNAs that bound to the 3` untranslated region (3`UTR) of the RAP1B mRNA in HeLa cells as an assay system. And subsequently, the mRNA 3’UTR:miRNA interactions were verified in platelets through the ectopic expression of miR-181a mimic and appropriate controls. The interaction of such miRNAs with RAP1B mRNA was also validated by qRT-PCR and Western analysis.
Results
Sixty-two miRNAs from MS2 assay were then compared with already known 171 platelet abundant miRNAs to identify a common set of miRNAs. This analysis yielded six miRNAs (miR-30e, miR-155, miR-181a, miR-206, miR-208a and miR-454), which are also predicted to target RAP1B mRNA. From this pool, miR-181a was selected for further study since RAP1B harbors two binding sites for miR-181a in its 3′UTR. Ectopic expression of miR-181a mimic in platelets resulted in lowering the endogenous RAP1B at both mRNA and protein levels. Further, miR-181a ectopic expression reduced the surface expression of the platelet activation marker, P-selectin.
Conclusion
MicroRNA-181a can target RAP1B and this interaction has the potential to regulate platelet activation during storage.
Keywords: microRNA, miR-181a, MS2-TRAP, platelet activation, platelets, RAP1B
1. INTRODUCTION
Ras-related Protein 1 (RAP1) is a member of the Ras family of guanine triphosphatases. RAP1 exits in two isoforms, RAP1A and RAP1B, which are encoded by two separate genes. RAP1 cycles between inactive GDP bound form and active GTP bound form, which are positively regulated by Guanine-nucleotide Exchange Factors (GEFs) and negatively regulated by GTPase activating proteins (GAPs) [1, 2]. In circulating platelets, RAP1 activity is regulated via the actions of RAP-GAP, RASA3. Whereas, at the site of injury, RAP1 is activated by RAP-GEF, CalDAG-GEFI [1, 3]. For sustained activation of RAP1, RASA3 activity is regulated via ADP signaling [1].
RAP1B is the most abundant form of RAP1 in platelets where it plays a crucial role in agonist-induced platelet activation and hemostasis [4-6]. In addition to their redundant functions, both RAP1A and RAP1B also regulate specific functions of platelets [7, 8]. Once activated, RAP1B translocates to the cytoskeletal region in platelets to regulate different signaling pathways. Platelets from RAP1B deficient mice showed reduced platelet aggregation in response to different agonists (ADP, epinephrine, Collagen, and Convulxin), increased bleeding time and impaired integrin alpha-2 beta-3 activation [4]. RAP1B mediated integrin activation is important for hemostatic plug formation during hemostasis. During development, RAP1B is a critical regulator of vasculature formation as RAP1B deficient mice developed hemorrhages [7].
Several studies examined the roles of different molecules on RAP1B activation [1, 2, 9-11] but information on the miRNA-based regulation of RAP1B protein expression levels in platelets is starting to emerge only recently [12]. MicroRNAs are small regulatory non-coding RNAs involved in post-transcriptional regulation of gene expression. Several studies, including studies from our laboratories, show differ-ential expression of microRNAs in platelets during storage and the effect of microRNAs on gene regulation and platelet functions [12, 13]. MicroRNA mediated regulation of a target gene occurs via an RNA Induced Silencing Complex (RISC) where microRNA binds to its target sites present on a messenger RNA, followed by either messenger RNA degra-dation or translation inhibition [14-16].
A well-established RNA affinity purification based method, known as MS2-TRAP [17] was applied to purify RAP1B mRNA 3’UTR bound miRNAs. In this report, one of the miRNAs which is abundant in platelets, miR-181a, identified by this method was evaluated for its regulatory interaction with RAP1B to understand the functional signi-ficance of this interaction in stored platelets.
2. MATERIALS AND METHODS
2.1. Plasmids and Cloning
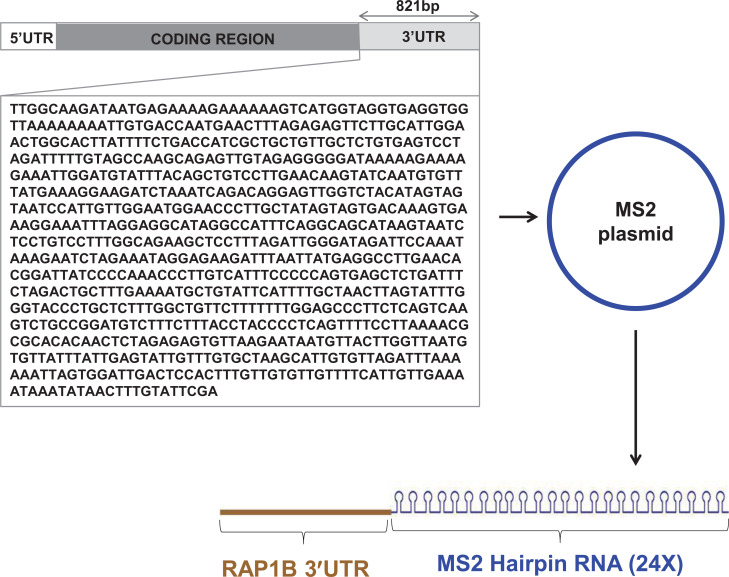
A plasmid expressing the chimeric RNA (RAP1B- 3' UTR-MS2 hairpin 24X) was constructed from plasmid pMS2-24X, which carries 24 cDNA copies of the MS2 hairpin loop. A copy of RAP1B-3’UTR (821bp) was cloned into pMS2-24X using ggaGAATTCTTGGCAAGATAATGAGAAAAG (Forward) and agaGCGGCCGCTCGAATACAAAGTTATATTT (Reverse) primers.
The plasmid expressing a chimeric protein MS2-GST (pMS2-GST) and pMS2-24X were gifts from Myriam Goro-spe (National Institute on Aging, NIH, Baltimore).
2.2. Pull Down of Ribonucleoprotein (RNP) Complexes
RNP complex was pulled down from HeLa cells lysates using the method described earlier [12]. Briefly, HeLa (ATCC® #CCL-2™) cells were obtained from the American Type Culture Collection (ATCC) and maintained in Eagle's Minimum Essential Medium (ATCC, USA) supplemented with 10% Fetal Bovine serum and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin). The pMS2-GST was co-transfected with control (pMS2-24X) or plasmid (p-RAP1B 3’UTR-MS2-24X) expressing chimeric RNA in HeLa cells. The cells lysates were prepared using a lysis buffer and the RNP complexes were purified using GSH beads which bind to GST protein associated with RNPs. The beads were subsequently washed and treated with DNAse I and proteinase K. Total RNA was extracted from the RNPs using Phenol chloroform method as previously described [12]. Briefly, 500ul water was added to the washed pellet. After thorough mixing, 500ul Acidic Phenol chloroform was added and vortexed. The sample was centrifuged at 10,000 rpm for 20 minutes at 4°C. The top aqueous phase (400ul) was transferred to a new tube and RNA was precipitated using 40ul 3M sodium acetate, 860ul 100% ethanol and 3ul Glycoblue (Life Technologies) followed by overnight incubation at -20°C. The next day, samples were centrifuged at 13000rpm for 30 minutes, washed with 75% ethanol and air-dried pellet was resuspended in 10ul RNAse-free water. RNA was quantified using NanoDrop (Life Technologies).
2.3. Microarray Analysis
Affymetrix Gene chip miRNA 3.0 arrays were used for microRNA profiling. After RNA quantification, 300ng RNA was polyadenylated (Poly A tail) and labeled with biotin using FlashTag Biotin HSR RNA labeling Kit (Affymetrix, Santa Clara, CA, USA), as per the manufacturer’s instructions. Biotin labeling was confirmed using Enzyme-Linked Oligosorbent Assays (ELOSA). The labeled RNA samples were hybridized to Affymetrix Gene chip miRNA microarray for 16hrs and incubated at 48C in Affymetrix Hybridization Oven-645, with shaking at 60rpm (Affymetrix). Following hybridization, the washing and staining of the microarrays were performed in Fluidics Station 450 and the chip was scanned with GeneChip Scanner 3000 7G (Thermo Scientific). Partek Genomic Suite software was used for data analysis, and the data were normalized using quantile normalization, which is based on the common distribution of intensities of all miRNAs within and across miRNA arrays.
2.4. Ectopic Expression and Quantification of miR-181a in Platelets
MiRNA control mimic and miR-181a mimic were obtained from Life Technologies. HeLa cells were transfected with the mimics using siPort Neo and incubated at 37°C in a CO2 incubator for 48hrs. Platelet samples collected by apheresis were obtained from the National Institutes of Health (NIH) blood bank under FDA-RIHSC approved protocol #03-120B. The samples were stored in a platelet incubator with agitation at 22°C. One day old platelets were transfected with the microRNA mimics using siPort Neo and incubated in the platelet incubator. After 48hrs, total RNA was extracted from both HeLa cells and platelets using TRIZOL (Invitrogen). Briefly, the cells and platelets were pelleted and each resuspended in 1ml TRIZOL and incubated at room temperature for 5 minutes on ice (at 4°C). After adding 200ul chloroform, samples were vortexed and centrifuged at 12000 rpm for 15 minutes at 4°C. The supernatant was transferred to a new tube, and RNA was precipitated using 0.5ml of 100% isopropanol and 2ul glycoblue (Invitrogen). After overnight incubation at -20°C, the samples were centrifuged at 13000 rpm for 30 minutes at 4°C. The pellet was washed with 75% ethanol, air-dried and resuspended in RNase-free water. miR-181a expression was determined by using TaqMan microRNA assay (Thermo Fisher) and the results were normalized with RNU6B as the endogenous control. For quantification of RAP1B mRNA in control mimic, miR-181a mimic treated cells and platelets were quantified using qRT-PCR [18]. DNAse I treated total RNA samples were reverse transcribed using SuperScript® III Reverse Transcriptase kit (Thermo Fisher, Waltham, MA, USA). The realtime PCR reaction was performed in QuantStudio™ 6 (Thermo Fisher) using SYBR Green Real-time PCR master mix (Thermo Fisher). Primers overlapping intron-exon boundaries were designed and synthesized at FDA core facility (Table 1). The RAP1B mRNA expression was estimated by the 2^ddCt method using GAPDH as a normalization control.
Table 1.
List of primers used for qPCR.
| S. No. | Name | Sequence |
|---|---|---|
| 1. | RAP1B Forward | GAGGAGGTTGTGGTGGATGA |
| 2. | RAP1B Reverse | CTCCATTAACTGCCGAATGAT |
| 3. | GAPDH Forward | GCACCACCAACTGCTTAGCA |
| 4. | GAPDH Reverse | GTCTTCTGGGTGGCAGTGATG |
| 5. | RAP1B 3’UTR Forward | CTGACCATCGCTGCTGTTG |
| 6. | RAP1B 3’UTR Reverse | CTACTATGTAGACCAACTCCC |
2.5. Western Blot Analysis
Platelets transfected with control mimic and miR-181a mimic were lysed in SDS lysis buffer [62.5 mM Tris-HCl (pH 6.8), 10% glycerol, and 2% SDS] after 48hr incubation. After quantification of proteins by using the BCA assay kit (Pierce, Rockford, IL), samples were run on 10-20% Tris-Glycine gels and transferred on to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). To avoid non-specific binding of the antibody, membranes were blocked in 5% nonfat dry milk, washed in TBST (Tris-buffered saline, 0.1% Tween 20) and incubated with Mouse monoclonal anti-RAP1B (Abcam) or rabbit anti-GAPDH (Protein Tech) antibodies. The next day, membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibody (anti-mouse or anti-rabbit IgG, 1:10,000; Amersham Biosciences Corp., Piscataway, NJ) for 1hr at room temperature, washed with TBST and developed using chemiluminescence kit (ECL; Amersham Biosciences Corp.). Protein band intensities were quantified using the Image J program developed at the National Institutes of Health.
2.6. MicroRNA Binding Site Analysis
Target Scan algorithms was used to identify miR-181a binding sites in RAP1B 3’UTR (http://www.targetscan.org/vert_72/).
2.7. Flow Cytometry
Expression of platelet surface marker was determined using FITS or PE-conjugated antibodies against CD41 and CD62. CD41 and Isotype IgG-FITC antibodies were obtained from Beckman Coulter whereas CD62 and Isotype IgG-PE were obtained from Becton Dickinson (Becton Dickinson, San Jose, CA). Platelet samples (control and test) were washed with PBS supplemented with 0.1% human albumin (PBS/albumin) and resuspended in PBS supplemented with 0.1% human albumin. Saturating concentrations of FITC- or PE-conjugated monoclonal antibodies against CD41, CD62 and isotype control antibodies were added to 50ul of diluted platelets (2x105). After 20 minutes of incubation in the dark at room temperature, platelets were washed with PBS/albumin, resuspended in 200ul PBS/albumin, and analyzed using a FACSCalibur (Becton Dickinson, San Jose, CA) and CellQuestPro software. Data analysis was performed using FlowJo software (Becton Dickinson, San Jose, CA).
3. RESULTS
3.1. MS2-TRAP Method Enriched RAP1B 3`UTR Interacting microRNAs
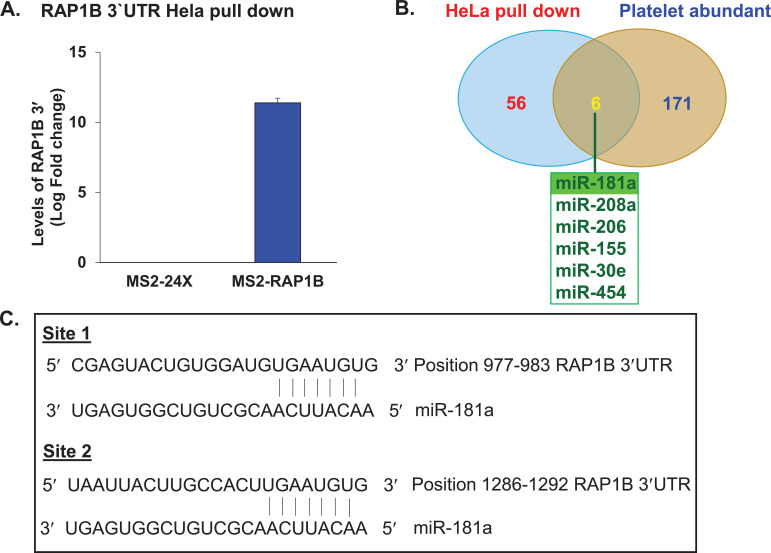
To enrich microRNAs that are associated with RAP1B 3`UTR, a plasmid expressing chimeric RNA containing 24 copies of RNA hairpin loop attached to RAP1B 3`UTR was generated (Fig. 1). HeLa cells were co-transfected with pMS2-GST/ control (pMS2-24X) or pMS2-GST/ p-RAP1B 3’UTR-MS2 (plasmid expressing chimeric RNA containing RAP1B 3`UTR). Cell lysates were prepared and subjected to pull down to extract native RNP complexes. First, the levels of RAP1B 3`UTR in control and test samples after pull-down were tested. There was a significant enrichment of RAP1B 3`UTR in RNP complexes isolated from test samples expressing chimeric RNA compared to the control samples (Fig. 2A).
Fig. (1).
Construction of a chimeric plasmid. A cDNA copy of RAP1B 3′UTR (821bp) was cloned into a plasmid carrying 24 cDNA copies of MS2 hairpin RNA. This plasmid produces chimeric RNA, RAP1B 3'UTR-MS2 Hairpin RNA. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (2).
Relative expression levels of (A) RAP1B 3′UTR in HeLa cells after pull-down, enriched microRNAs and miRNAs binding sites. RAP1B 3′UTR expression levels in pRAP1B 3′UTR-MS2 transfected cells compared to the control pMS2 transfected cells following pull-down. (B). Comparison of microRNAs enriched in HeLa cells expressing chimeric RNA (RAP1B 3'UTR-MS2 Hairpin RNA) with microRNAs known to be abundant in platelets. (C). MicroRNA-181a binding sites in RAP1B 3′UTR as predicted by Target Scan algorithm. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Microarray analysis of the RNA extracted from control and test samples showed enrichment of 62 microRNAs in test samples compared to the control. The comparison of these enriched microRNAs with 171 microRNAs that are known to be abundant in platelets as previously reported in the literature [13, 18-20] has identified 6 microRNAs (miR-181a, miR-208a, miR-206, miR-155, miR-30e and miR-454, shown in Fig. 2B). Based on the Target Scan prediction, all 6 microRNAs have their potential target sites on RAP1B 3`UTR. Since miR-181a is one of the six common microRNAs among the two cell lines tested in the previous report [12], and in-silico analysis identified the presence of two miR-181a binding sites in RAP1B 3`UTR (Fig. 2C), miR-181a was selected for further study.
3.2. MiR-181a Targets RAP1B Messenger RNA and Downregulates Endogenous RAP1B Protein Expression
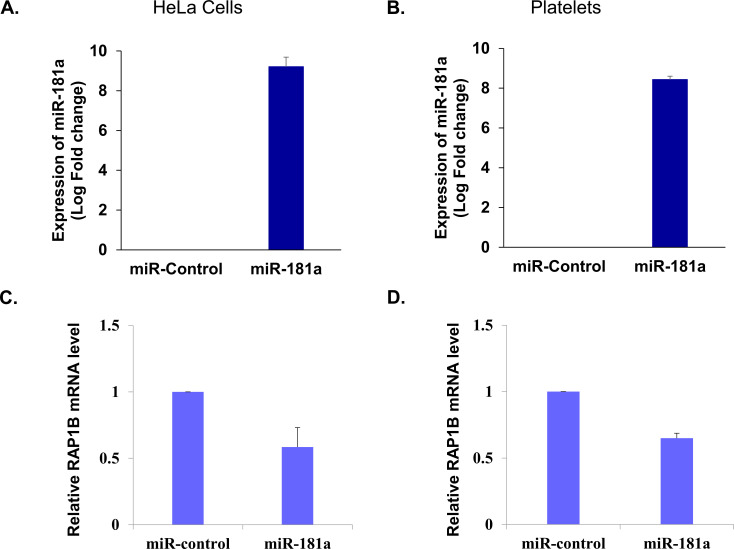
The mechanism of miR-181a mediated regulation of RAP1B was examined. The effect of this miRNA on RAP1B messenger RNA in Hela cells and platelets was tested by transfecting both the cells with miRNA control mimic and
miR-181a (test group). Total RNA extracted from the control and test group was quantified for miR-181a expression and endogenous RAP1B messenger respectively by RT-qPCR. Both HeLa cells and platelets showed increased expression of miR-181a in miRNA-181a mimic transfected samples compared to their control counterparts (Figs. 3A and 3B). This ectopic expression of miR-181a in both HeLa cells and platelets was able to reduce endogenous RAP1B messenger RNA by more than 40% (Figs. 3C and 3D), suggesting a regulatory interaction of miRNA-181a with RAP1B messenger RNA.
Fig. (3).
Expression of miR-181a and its effect on RAP1B messenger RNA. Expression levels of miR-181a in (A) HeLa cells and (B) Platelets transfected with control miRNA mimic and miR-181a mimic. Effect of miR-181a ectopic expression on (C) RAP1B messenger RNA levels in HeLa cells and (D) Platelets transfected with control miRNA mimic and miR-181a mimic. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
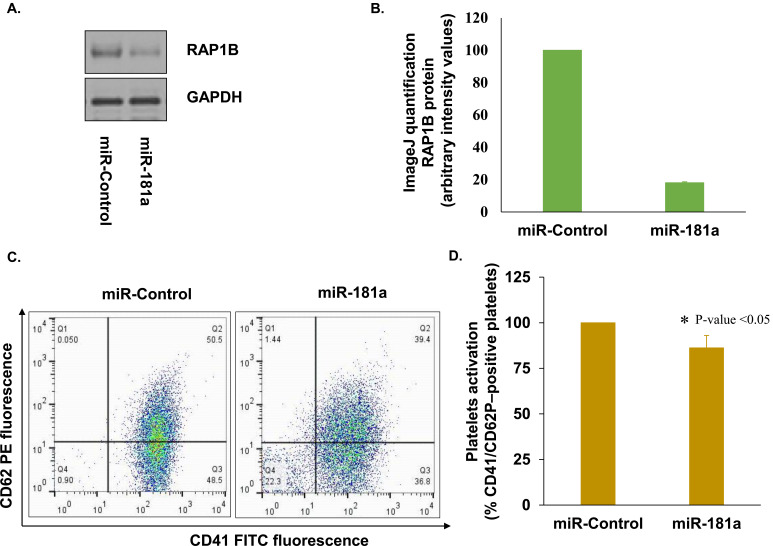
After observing the regulatory effect of microRNA-181a on RAP1B mRNA, the effect of miRNA-181a on the expression of endogenous RAP1B was tested. Levels of RAP1B proteins were determined by western analysis in platelets transfected with miRNA control mimic and miR-181a (test group). Increased expression of miR-181a significantly reduced (>81%) the levels of endogenous RAP1B in platelets, confirming that miRNA-181a downregulates RAP1B expression in platelets (Figs. 4A and 4B).
Fig. (4).
Effect of miR-181a on RAP1B mRNA, protein and platelet activation. (A) Effect of miR-181a ectopic expression on RAP1B protein in platelets by western blot; (B) Quantification of western bands using ImageJ analysis; (C) Scatter plot of platelets transfected with control miRNA mimic and miR-181a mimic; (D) Effect of increased levels of miR-181a on platelet activation. Surface expression of CD41 and CD62 were examined in platelets transfected with the control miRNA mimic and miR-181a mimic by Flow cytometry. Data is represented as percentage of CD41 and CD62P–positive platelets. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.3. Increased Expression of miR-181a Reduces Platelet Activation
To understand the functional significance of miRNA-181a and RAP1B mRNA interaction in platelets, the effect of ectopic expression of miRNA-181a in platelets as a function of platelet activation was tested. The activation was measured by evaluating the levels of P-selectin (CD62P) surface expression on CD41 positive platelets by FACS analysis. Platelets transfected with miR-181a mimic exhibited reduced surface expression of P-selectin (CD62P) compared to platelets transfected with miRNA control mimic by more than 14% (Figs. 4C and 4D).
4. DISCUSSION
In this study, a well-established pull-down approach was applied to identify microRNAs that can regulate RAP1B. Six microRNAs (miR-181a, miR-208a, miR-206, miR-155, miR-30e and miR-454) that were enriched in the pull-down experiments are also known to be abundant in platelets. Based on the prediction by the Target Scan algorithm [21], RAP1B 3`UTR carries microRNA binding sites for all six platelet abundant miRNAs identified in this study [18, 20-23]. Subsequently, enrichment of the same miRNAs through the pull-down assay tool further validates that these miRNAs are true targets for RAP1B 3’UTR.
Further studies in platelets were conducted using one selected microRNA, miR-181a from the 6 miRNAs identified in the analysis. Two miR-181a binding sites were identified in the RAP1B 3’UTR with perfect complementarity in the seed region (2-8 base pair from microRNA 5’ end). It has been known that miRNAs can regulate target mRNA by either degradation or by interfering with translation, or both [14-16, 24, 25]. The ectopic expression experiments with miR-181a resulted in decreased RAP1B mRNA expression in both, HeLa cells and platelets. This further suggests that miR-181a binding sites in RAP1B 3`UTR are functional, and the protein regulation is via RNA degradation.
Platelets transfected with miR-181a showed a significant decrease in RAP1B protein levels as well (>80%) compared to control miRNA transfected platelets. Recently, the regulatory role of miR-320c on RAP1B protein expression was reported [12]. Interestingly, while both miRNAs target the same 3’UTR, miR-181a and miR-320c differ in how they mediate downregulation of RAP1B. As reported here, miRNA-181a downregulates RAP1B mRNA and consequently the protein also. On the other hand, microRNA-320c mainly regulates RAP1B via translational inhibition without affecting the levels of RAP1B messenger RNA [12]. Both miR-181a as reported here and miR-320c as reported previously [12], reduce platelet activation as assessed by the surface expression of P-selectin. Following the platelet activation, P-selectin translocates from internal granules to the platelet surface [26].
conclusion
Together, these studies indicate that miRNAs are also emerging as regulators of ex vivo stored platelet functions. Since RAP1B signaling plays an important role in platelet’s function, understanding RAP1B regulation further will provide necessary insights on the mechanisms that control RAP1B in platelets in circulation, during quiescent state and in hemostasis at the site of injury.
Acknowledgements
We would like to acknowledge Dr. Myriam Gorospe for proving the pMS2 plasmids used in this study.
List of Abbreviations
- CBER
Center for Biologics Evaluation and Research
- RAP1B
Ras-related Protein 1B
- 3′UTR
3′ Untranslated Region
- ELOSA
Enzyme-Linked Oligosorbent Assays
- RISC
RNA-Induced Silencing Complex
- MS2-TRAP
MS2-Tagged RNA Affinity Purification
Authors’ Contributions
ND conceived and designed the study, performed the experiments, analyzed the data, and wrote the manuscript for this study. CDA provided training to ND, provided overall guidance for the study and participated in designing, writing and editing the manuscript.
Ethics Approval and Consent to Participate
The study prootocol was approved by FDA-RIHSC, USA protocol #03-120B.
Human and Animal Rights
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Materials
The data and materials will be available upon sending a request to Dr. Chintamani Atreya (corresponding author of this manuscript).
Funding
This work is supported by US-FDA Intramural Research Funding.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
references
- 1.Stefanini L., Paul D.S., Robledo R.F., et al. RASA3 is a critical inhibitor of RAP1-dependent platelet activation. J. Clin. Invest. 2015;125(4):1419–1432. doi: 10.1172/JCI77993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stefanini L., Bergmeier W. RAP1-GTPase signaling and platelet function. J. Mol. Med. (Berl.) 2016;94(1):13–19. doi: 10.1007/s00109-015-1346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefanini L., Roden R.C., Bergmeier W. CalDAG-GEFI is at the nexus of calcium-dependent platelet activation. Blood. 2009;114(12):2506–2514. doi: 10.1182/blood-2009-04-218768. [DOI] [PubMed] [Google Scholar]
- 4.Chrzanowska-Wodnicka M., Smyth S.S., Schoenwaelder S.M., Fischer T.H., White G.C., II Rap1b is required for normal platelet function and hemostasis in mice. J. Clin. Invest. 2005;115(3):680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardi B., Guidetti G.F., Campus F., et al. The small GTPase Rap1b regulates the cross talk between platelet integrin alpha2beta1 and integrin alphaIIbbeta3. Blood. 2006;107(7):2728–2735. doi: 10.1182/blood-2005-07-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., Holly S.P., Larson M.K., et al. Rap1b is critical for glycoprotein VI-mediated but not ADP receptor-mediated alpha2beta1 activation. J. Thromb. Haemost. 2009;7(4):693–700. doi: 10.1111/j.1538-7836.2009.03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chrzanowska-Wodnicka M., White G.C., II, Quilliam L.A., Whitehead K.J. Small GTPase Rap1 is essential for mouse development and formation of functional vasculature. PLoS One. 2015;10(12):e0145689. doi: 10.1371/journal.pone.0145689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefanini L., Lee R.H., Paul D.S., et al. Functional redundancy between RAP1 isoforms in murine platelet production and function. Blood. 2018;132(18):1951–1962. doi: 10.1182/blood-2018-03-838714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woulfe D., Jiang H., Mortensen R., Yang J., Brass L.F. Activation of Rap1B by G(i) family members in platelets. J. Biol. Chem. 2002;277(26):23382–23390. doi: 10.1074/jbc.M202212200. [DOI] [PubMed] [Google Scholar]
- 10.Guidetti G.F., Torti M. The small GTPase Rap1b: a bidirectional regulator of platelet adhesion receptors. J. Signal Transduct. 2012;2012:412089. doi: 10.1155/2012/412089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G., Xiang B., Ye S., et al. Distinct roles for Rap1b protein in platelet secretion and integrin αIIbβ3 outside-in signaling. J. Biol. Chem. 2011;286(45):39466–39477. doi: 10.1074/jbc.M111.239608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahiya N., Atreya C.D. RAP1 Downregulation by miR-320c reduces platelet activation in ex-vivo storage. MicroRNA. 2019;8(1):36–42. doi: 10.2174/2211536607666180521094532. [DOI] [PubMed] [Google Scholar]
- 13.Nagalla S., Shaw C., Kong X., et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103(11):4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwakawa H.O., Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25(11):651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Yoon J.H., Srikantan S., Gorospe M. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012;58(2):81–87. doi: 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry P., Plante I., Ouellet D.L., Perron M.P., Rousseau G., Provost P. Existence of a microRNA pathway in anucleate platelets. Nat. Struct. Mol. Biol. 2009;16(9):961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahiya N., Sarachana T., Kulkarni S., et al. miR-570 interacts with mitochondrial ATPase subunit g (ATP5L) encoding mRNA in stored platelets. Platelets. 2017;28(1):74–81. doi: 10.1080/09537104.2016.1203405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osman A., Fälker K. Characterization of human platelet microRNA by quantitative PCR coupled with an annotation network for predicted target genes. Platelets. 2011;22(6):433–441. doi: 10.3109/09537104.2011.560305. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal V., Bell G.W., Nam J., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. http://www.targetscan.org/vert_72/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plé H., Landry P., Benham A., Coarfa C., Gunaratne P.H., Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahiya N., Kulkarni S., Atreya C.D. The repertoire and features of human platelet microRNAs. PLoS One. 2016;7:e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu S., Kay M.A. How do miRNAs mediate translational repression? Silence. 2010;1(1):11. doi: 10.1186/1758-907X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng Y., Yi R., Cullen B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA. 2003;100(17):9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merten M., Thiagarajan P. P-selectin expression on platelets determines size and stability of platelet aggregates. Circulation. 2000;102(16):1931–1936. doi: 10.1161/01.CIR.102.16.1931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials will be available upon sending a request to Dr. Chintamani Atreya (corresponding author of this manuscript).