Highlights
-
•
We analysed over 450 references from all well-famed databases.
-
•
We provided a comprehensive survey on multimodal data fusion in neuroimaging.
-
•
This review encompassed current challenges & applications, strengths &limitations.
-
•
Fundamental fusion rules, and fusion quality assessment methods were reviewed.
-
•
Atlas-based fusion segmentation, quantification, & applications were reviewed.
Keywords: Multimodal data fusion, Neuroimaging, Magnetic resonance imaging, PET, SPECT, Fusion rules, Assessment, Applications, Partial volume effect
Abstract
Multimodal fusion in neuroimaging combines data from multiple imaging modalities to overcome the fundamental limitations of individual modalities. Neuroimaging fusion can achieve higher temporal and spatial resolution, enhance contrast, correct imaging distortions, and bridge physiological and cognitive information. In this study, we analyzed over 450 references from PubMed, Google Scholar, IEEE, ScienceDirect, Web of Science, and various sources published from 1978 to 2020. We provide a review that encompasses (1) an overview of current challenges in multimodal fusion (2) the current medical applications of fusion for specific neurological diseases, (3) strengths and limitations of available imaging modalities, (4) fundamental fusion rules, (5) fusion quality assessment methods, and (6) the applications of fusion for atlas-based segmentation and quantification. Overall, multimodal fusion shows significant benefits in clinical diagnosis and neuroscience research. Widespread education and further research amongst engineers, researchers and clinicians will benefit the field of multimodal neuroimaging.
1. Introduction
Neuroimaging has been playing pivotal roles in clinical diagnosis and basic biomedical research in the past decades. As described in the following section, the most widely used imaging modalities are magnetic resonance imaging (MRI), computerized tomography (CT), positron emission tomography (PET), and single-photon emission computed tomography (SPECT). Among them, MRI itself is a non-radioactive, non-invasive, and versatile technique that has derived many unique imaging modalities, such as diffusion-weighted imaging, diffusion tensor imaging, susceptibility-weighted imaging, and spectroscopic imaging. PET is also versatile, as it may use different radiotracers to target different molecules or to trace different biologic pathways of the receptors in the body.
Therefore, these individual imaging modalities (the use of one imaging modality), with their characteristics in signal sources, energy levels, spatial resolutions, and temporal resolutions, provide complementary information on anatomical structure, pathophysiology, metabolism, structural connectivity, functional connectivity, etc. Over the past decades, everlasting efforts have been made in developing individual modalities and improving their technical performance. Directions of improvements include data acquisition and data processing aspects to increase spatial and/or temporal resolutions, improve signal-to-noise ratio and contrast to noise ratio, and reduce scan time. On application aspects, individual modalities have been widely used to meet clinical and scientific challenges. At the same time, technical developments and biomedical applications of the concert, integrated use of multiple neuroimaging modalities are trending up in both research and clinical institutions. The driving force of this trend is twofold. First, all individual modalities have their limitations. For example, some lesions in MS can appear normal in T1-weighted or T2-weighted MR images but show pathological changes in DWI or SWI images [1]. Second, a disease, disorder, or lesion may manifest itself in different forms, symptoms, or etiology; or on the other hand, different diseases may share some common symptoms or appearances [2, 3]. Therefore, an individual image modality may not be able to reveal a complete picture of the disease; and multimodal imaging modality (the use of multiple imaging modalities) may lead to a more comprehensive understanding, identify factors, and develop biomarkers of the disease.
In the narrow sense, a multimodal imaging study would mean the use of multiple imaging devices such as PET and MRI scanners, different imaging modes such as structural MRI, diffusion-weighted imaging, and magnetic resonance spectroscopy, or even different contract mechanism such as with or without contract agents in a single examination or experiment of a subject. This practice has been widely used in clinical diagnosis and medical research. For example, a routine protocol of MRI examination of a stroke patient may include T1-weighted, T1-weighted high-resolution structural MRI scans, diffusion-weighted imaging, SWI, etc [4, 5]. A protocol of an MRI study of a psychiatric disorder may contain a combination of structural MRI, functional MRI, MR spectroscopic imaging, etc [6, 7].
In the broad sense, a multimodal imaging study may mean the use of multimodal imaging data obtained separately, from different subjects, and/or from different clinical or research sites. This practice offers the advantages of large and diverse datasets. However, it also comes with challenges of sophisticated models, complicated data normalization (that includes correction of errors and variations imbedded in data from different institutions), data fusion, and data integration [8, 9].
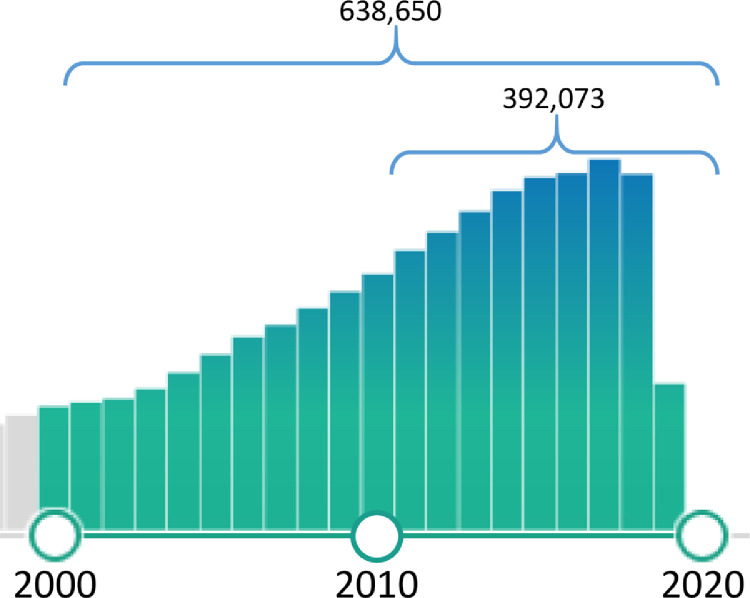
In recent years, the quantity of peer-reviewed journal articles on neuroimaging has been increasing steadily. A database (PubMed) query using the keywords in titles of “neuroimaging” OR “brain imaging” returned more than 39,000 articles from 2010 to the present time when this paper was drafted in Feb 2020 (Fig. 1 ). These publications include not only applications of multimodal neuroimaging in clinical examinations and biomedical research but also methodological studies in imaging processing and fusion of multimodal neuroimaging.
Fig. 1.
Numbers of peer-reviewed papers with the keywords of “neuroimaging” or “brain imaging” in titles (the numbers and the bar graph were generated by PubMed in Feb 2020).
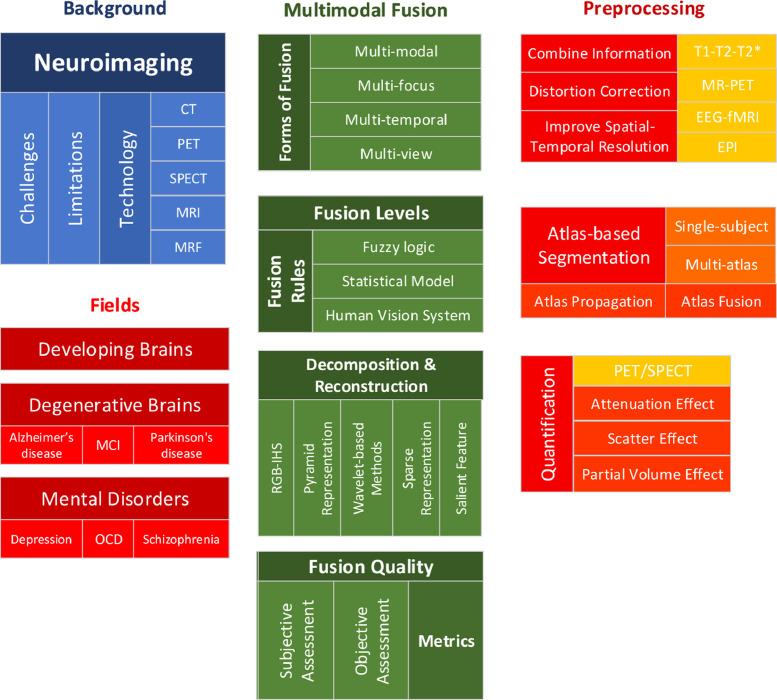
Therefore, this present paper will focus on the following two main aspects: (1) we will review some of the recent, typical papers that exhibit the strength and limitations of the neuroimaging modalities and the corresponding analysis methods, and in particular, the needs for improved image fusion methods and (2) we will review recent methodological development in data preprocessing and data fusion in multimodal neuroimaging. We note that although we tried to cover all neuroimaging modalities, we inevitably paid more attention to MRI modalities. This is not only due to the most practical application and versatility of the MRI but also due to the limitations of our expertise. Fig. 2 shows the taxonomy of this review.
Fig. 2.
Taxonomy of this review.
The main contents of the paper are organized as follows. Chapter 2 will give a brief introduction to neuroimaging, and challenges of multimodal imaging; Chapter 3 introduces the commonly used neuroimaging modalities, which include computerized tomography, positron emission tomography, single-proton emission computed tomography, and magnetic resonance imaging, which has many modalities in its own right. For each modality, we will concisely describe its signal source, energy level, spatial resolution, temporal resolution, and major applications; Chapter 4 describe applications of neuroimaging in three major areas: the developing brains, the degenerative brains, and mental disorders. In each part, we will first briefly describe what the clinical and/or biomedical problems are, we then review recent papers on how neuroimaging has been used to address these problems, and we point out what the unmet needs and challenges;
Chapters 5 to 9 are devoted to the multimodal neuroimaging fusion, covering some important procedures in data fusion. The topics are not necessarily complete and their order of presentation is not necessarily coherent with the pipeline of fusion processing. Chapter 5 reviews the fundamental methods, which covers types, rules, atlas-based segmentation, decomposition, reconstruction, and quantification; Chapter 6 reviews subjective and objective assessment of data fusion in multimodal neuroimaging; Chapter 7 reviews the advantages of data fusion in improving the spatial/temporal resolution, distortion correction, and contrast; it also reviews the benefits of these advantages in fusing structural and functional images; Chapter 8 reviews atlas-based segmentations in multimodal imaging fusion; Chapter 9 reviews the quantification in multimodal neuroimaging fusion. While the focus of this part is given to PET and SPECT, some of the approaches and principles discussed here, such as partial volume correction and attenuation (relaxation), can be applied to quantitative MRI modalities, such as DTI, ASL, quantitative susceptibility mapping (QSM), etc. Chapter 10 concludes the paper.
2. Multimodal imaging data fusion: challenges in neuroimaging
In this part, we will review the current challenges of neuroimaging, including limited spatial/temporal resolution, lack of quantification, and imaging distortions. These challenges often create fundamental limitations on individual modalities of neuroimaging, while some challenges also exist in current multi-modal neuroimaging. This part will mainly cover the challenges of individual neuroimaging modalities that led to the development and ongoing research of multimodal neuroimaging methods.
2.1. Individual modality imaging
Neuroimaging can be divided into structural imaging and functional imaging according to the imaging mode. Structural imaging is used to show the structure of the brain to aid the diagnosis of some brain diseases, such as brain tumors or brain trauma. Functional imaging is used to show how the brain metabolizes while carrying out certain tasks, including sensory, motor, and cognitive functions. Functional imaging is mainly used in neuroscience and psychological research, but it is gradually becoming a new way of clinical-neurological diagnosis [10].
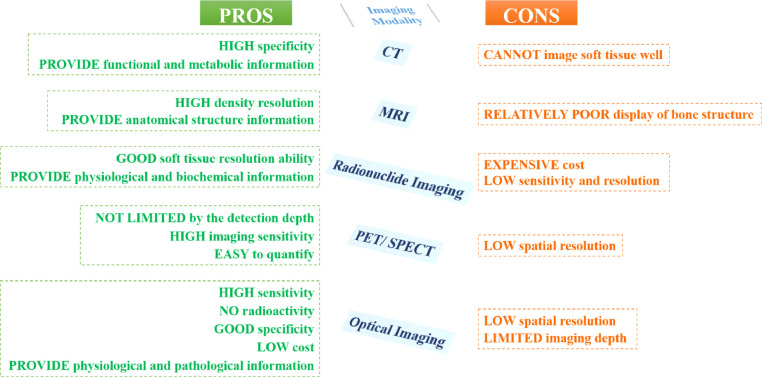
The amount of information obtainable through single-mode imaging is limited and often cannot reflect the complex specificity of organisms. For instance, although CT imaging is effective in identifying normal structures and abnormal diseased tissues according to their density and thus can provide clear anatomical structure information, it cannot image soft tissue well. Generally speaking, MRI imaging has good soft-tissue contrast resolution for most sequences, but its display of bone structure is relatively poor. PET imaging and SPECT imaging are not limited by the detection depth, with high imaging sensitivity, and are easy to quantify, but their spatial resolutions are low [11]. Optical imaging refers to the detection of fluorescence or bioluminescent dyes using the principle of light emission. This technique has high sensitivity, no radioactivity, good specificity, and low cost. Optical imaging allows dynamical monitoring of the replication process of virus bacteria in organisms. However, it has low spatial resolution and limited imaging depth [12, 13]. Therefore, it can be observed that various imaging technologies all have both benefits and drawbacks, as shown in Fig. 3 , and it is difficult to provide comprehensive and accurate information through utilizing individual modality imaging.
Fig. 3.
Advantages and disadvantages of various individual modality imaging.
2.2. Low spatial/temporal resolution
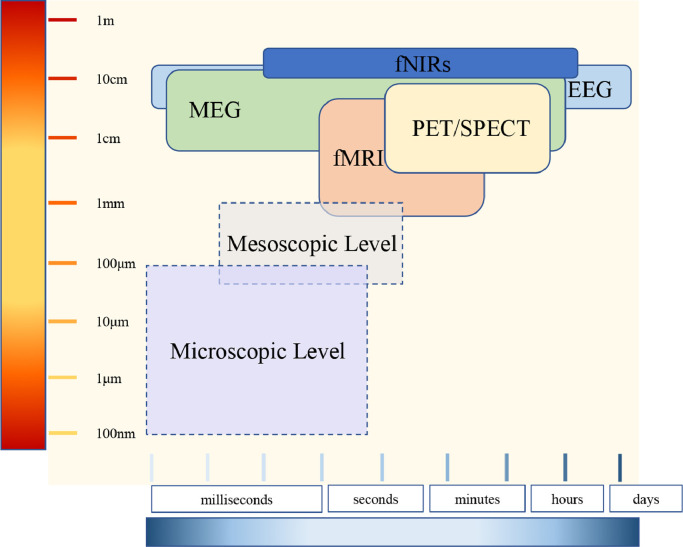
The nowadays most commonly used noninvasive functional imaging methods and their spatial and temporal range are illustrated in Fig. 4 . It can be distinctly observed that among these most advanced methods, functional MRI (fMRI) reaches the highest range of spatial resolution. fMRI can assess the whole brain and image the hemodynamic processes at the layered and columnar levels of the human cortex, under the condition of a high-intensity magnetic field (i.e., submillimeter level) [14]. However, it has a relatively lower temporal resolution in terms of imaging the neuronal population dynamics. Electroencephalogram (EEG) and Magnetoencephalography (MEG) can both measure electromagnetic changes in the scale of milliseconds. However, their spatial resolution/uncertainty is more than several millimeters [15, 16]. The microscopic level of neuroscience is often beyond the reach of noninvasive imaging techniques due to the requirement of high spatial or temporal resolution.
Fig. 4.
Functional neuroimaging modalities.
2.3. Non-quantitative
The majority of imaging modalities are non-quantitative and have to gain complement information from other data. This additional information allows the normalization of signals, acquirement of absolute units, and inter-subject comparison. As an example, the fMRI signal is a measure of neuronal activity incited hemodynamic changes caused by a combination of complex physical and physiological processes. In different subjects or brain areas, the same level of neuronal activity can evoke different corresponding fMRI signals. As a consequence, fMRI signals can only be considered as roughly proportional to the activity of neurons. Four years later, in 2008, the studies from Ances have found that the cerebral blood flow (CBF) is relevant to fMRI signal variations in individual's brain regions, patients’ age groups, and health conditions. This broad relevance brings fMRI signal high sensitivity [17]. Various approaches have been proposed to explain the ensuing sensitivity differences. Amongst these approaches, the so-called calibrated BOLD approach proposed and improved by Blockley, Chiarelli and Hoge over the years has been the most widely used [18], [19], [20]. However, the more reliable absolute quantitative results of CBF with improved spatial and temporal resolutions are provided by the Arterial spin labeling (ASL) technique by the UMICH fMRI lab [21].
2.4. Distortion
Some neuroimaging modalities are prone to geometric distortions. Echo‐planar imaging (EPI) is a fast imaging approach that could obtain the complete k‐space data set just in a single acquisition. Due to its unmatched acquisition speed, it has revolutionized the field of neuroimaging and has served as the standard readout module for most fMRI and dMRI acquisition. Nevertheless, EPI suffers from distortion and intensity loss mainly caused by field inhomogeneities, leading to relatively poor image quality [22]. In general, the imperfection of equipment may result in information loss, noise amplification and artifacts, resulting in the distortion of images.
To conclude, in practical application, utilizing individual modality imaging often has limitations, such as low sensitivity and specificity, low spatial/temporal/contrast resolution, distortion and so on. Because of these deficiencies, we need to introduce the usage of multimodal neuroimaging to eliminate those shortcomings in some degree.
3. Multimodal imaging data fusion: imaging technologies
Neuroimaging, more commonly known as brain imaging, is referred to as different types of technologies to display the function, pathology, and structure of the nervous system. There are mainly two types of neuroimaging: the functional imaging that directly or indirectly visualizes the processing of information by the central neural system of the brain and the structural imaging that shows the structure information of the brain. Neuroimaging is used for a patient who is found a neurological disorder by a physician to have a more in-depth investigation. There are different types of imaging modalities, such as Magnetic resonance imaging (MRI), Single-Photon Emission Computed Tomography, Computerized Tomography, Positron Emission Tomography, Pneumoencephalography, Functional Magnetic Resonance Imaging (fMRI). According to the types of possible diseases, patients will be investigated by different methods.
3.1. Computerized tomography
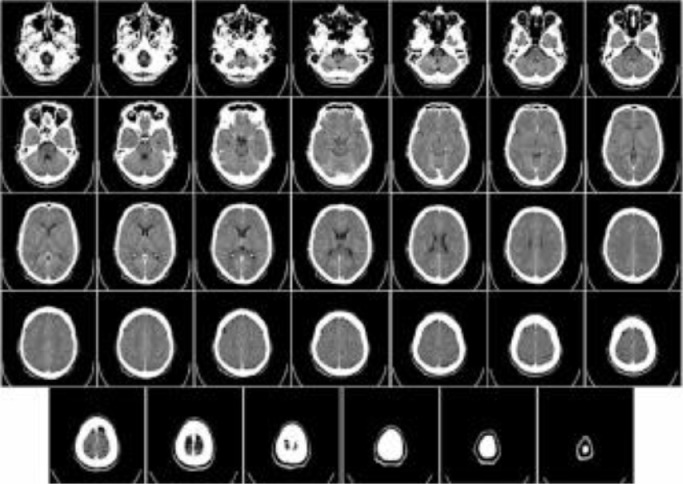
Computerized Tomography (CT), also known as computerized x-ray imaging, combines a series of X-ray signals obtained from multiple angles around the body and creates cross-sectional images by computer processing. Different from the conventional X-ray [23] that uses a fixed X-ray tube, the CT scanner uses a motorized x-ray source, which rotates around a gantry, a circular frame in a donut-shaped structure. CT scan images, therefore, can provide more information than conventional X-rays. CT can be recommended for disease or injury of various parts of the body, such as lesions or tumors of the abdomen, different types of heart disease, injuries, tumors or clots of the head [24], [25], [26]. Fig. 5 [27] shows an example of a CT scan of a brain.
Fig. 5.
An example of a CT brain scan.
3.2. Positron emission tomography
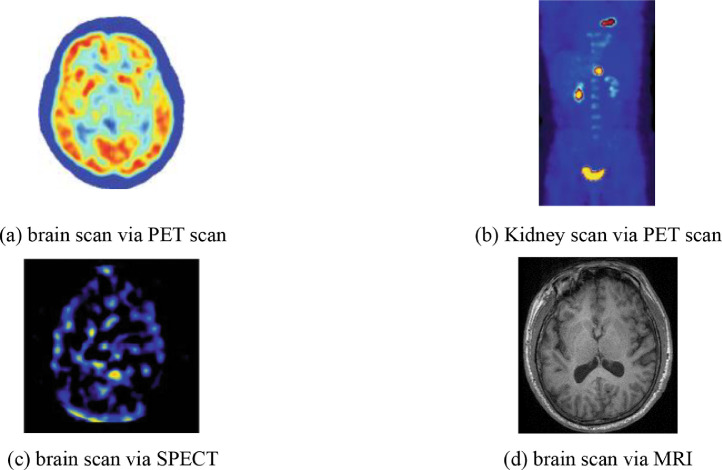
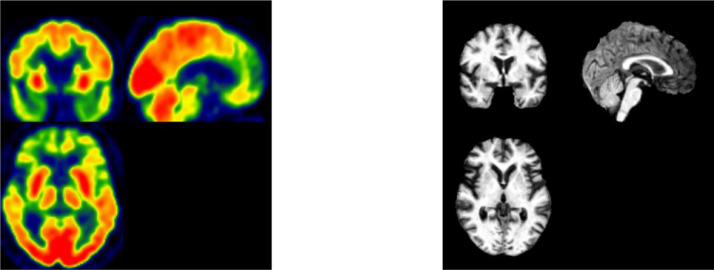
Positron Emission Tomography, shortened as PET, is a combination of nuclear medicine and biochemical analysis that is mostly used for the diagnosis of brain or heart conditions and cancer. Instead of detecting the amount of a radioactive substance existing in body tissues of a specific location to check the tissue's function, PET detects the biochemical changes within body tissues. The biochemical changes can reveal the onset of a disease process before other imaging processes can visualize the anatomical changes related to the disease. During PET studies, only a tiny amount of radioactive substance is needed for the examination of targeted tissues. PET scans not only can be used to detect the presence of disease or other conditions of organs or tissues but can also be used to evaluate the function of organs, like the heart or the brain. The most common application of PET scan is cancer detection and treatment. Fig. 6 (a) shows a PET image of the brain, and Fig. 6(b) shows a PET scan of the kidney [27].
Fig. 6.
Common scan modalities.
3.3. Single-photon emission computed tomography
Single-photon emission computed tomography, commonly known as SPECT, uses gamma rays as the tracer to detect blood flow in organs or tissue. Therefore, a gamma-emitting radioisotope, such as isotope gallium, should be injected into the bloodstream of the patient for SPECT. The computer collects the gamma rays from the tracer and shows it on the CT cross-section. It bears similarity with the traditional nuclear medicine planar imaging but provides 3D information as multiple images of cross-sectional slices through the patient. The information can be manipulated or reformatted freely according to diagnostic or research requirements. Besides detecting blood flow, SPECT scanning is also applied for the presurgical evaluation of medically controlled seizures. Fig. 6(c) [27] shows a SPECT scan of the brain
3.4. Magnetic resonance imaging
Magnetic resonance imaging (MRI) utilizes strong magnetic field, magnetic field gradients, and radio waves to generate pictures of the anatomy and the physiological processes of the body [28]. Different from PET or CT, MRI does not need the injection of ionizing radioisotopes or involve X-rays. As all radiation instances can cause ionization that leads to cancer, MRI, without exposing the body to radiation, becomes a better choice than CT and one of the safest medical procedures. MRI is widely used in hospitals and clinics for the medical diagnosis of different body regions, including the brain, spinal cord, bones and joints, breasts, heart and blood vessels, and other internal organs, such as the liver, womb or prostate gland [29], [30], [31]. Besides, MRI can also be used for non-living objects [32]. Fig. 6(d) shows an MRI brain image [33].
3.4.1. T1, T2, and proton density
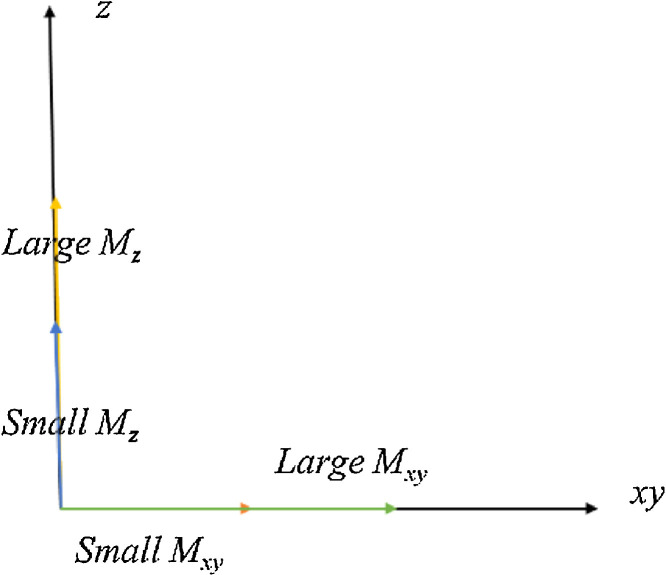
T1, T2 and Proton density (PD) are three basic types of MRI imaging. T1, T2 and PD, which all vary with sequence parameters, can simultaneously determine the contrast of the MR images [34]. Via selecting different pulse sequences with different timings, we can decide the contrast in the region being imaged. There are also other types of sequences, such as fluid attenuates inversion recovery (FLAIR) and short tau inversion recovery (STIR). In this section, we will only mention the three main types. Fig. 7 shows the relationship between Mz and Mxy, of which xy refers to plane [35].
Fig. 7.
The relationship between Mz and Mxy.
Higher Mz at the time of applying the 90° RF pulse brings the larger transverse signal (Mxy). The time to repetition (TR) is defined as the determent of the length of time between 90° RF pulses. The Echo time [26] is defined as the time between the excitation pulse and the peak of the signal.
T1, the longitudinal relaxation time, is defined as a time constant that stands for the magnetization to recover from 0 to 63% of their maximum Mz in a static magnetic field. T1 values of hydrogen nuclei are different for different molecules and different tissues. T1 relaxation is defined as the recovery of the longitudinal magnetization. T1 is commonly applied for detecting fatty tissue, general morphological information, and characterizing focal liver lesions.
T2, the transverse relaxation time, is defined as the time for transverse magnetization Mxy to decay to 37% of its initial Mxy. Similar to T1 values, we also have different T2 values of Hydrogen nuclei in different molecules and different tissues. T2 weighted imaging is suitable for revealing cerebral white matter lesions and assessing edema, inflammation and zonal anatomy in the prostate and uterus.
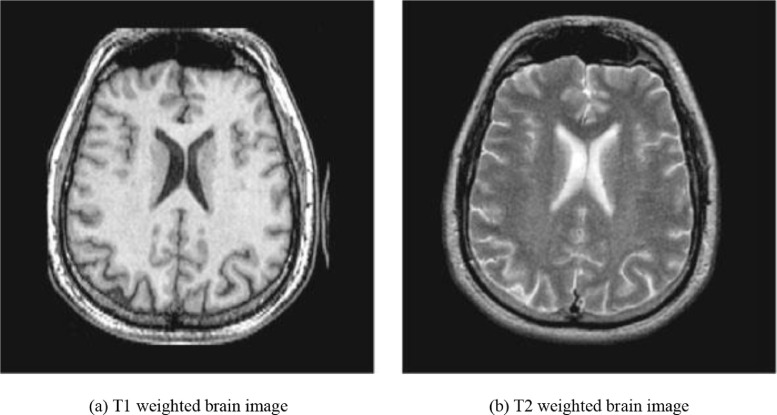
Different from T1 and T2, which mainly focuses on the magnetic characteristics of the hydrogen nuclei, PD is more related to the number of nuclei in the region being imaged. PD weighted images are obtained by a short echo time and a long repetition time, which can provide a more apparent distinction between the gray matter and white matter. PD weighted imaging is specifically useful for detecting joint disease and injury. Fig. 8 (a) and (b) show a T1 weighted brain image and T2 weighted brain image, respectively [27].
Fig. 8.
Two types of weighted images of the same brain.
3.4.2. Functional magnetic resonance imaging
Functional magnetic resonance imaging or functional MRI (fMRI) measures brain activity by detecting changes associated with blood flow. As it has been proved that when an area of the brain is in use, the blood flow to that area increases, which means that the neuronal activation and cerebral blood flow are matched. fMRI is a particular type of imaging technology used to map the neuron activities in the spinal cord and brain of humans or animals by visualizing the change in blood flow, which is related to the energy use by brain cells. fMRI also includes resting-state fMRI [36] or task-less fMRI, which can provide subjects’ baseline BOLD service [37].
3.4.3. Diffusion weighted/tensor imaging
Diffusion Weighted/Tensor Imaging (DWI) generates image contrast from the differences in the magnitude of diffusion of water molecules within the brain. Diffusion in biology is defined as the passive movement of molecules from a higher concentration region to a lower concentration region, which is also known as Brownian motion [38]. Diffusion within the brain is affected by many factors, such as temperature, type of molecule under investigation, and the microenvironmental architecture in which the diffusion takes place. Based on the MRI sequences of which diffusion is sensitive to, the image contrast can be generated according to the difference in diffusion rates. DWI is highly effective for the early diagnosis of ischemic tissue injury, even before the pathology can be shown by the traditional MR sequence. Therefore, DWI provides the time window for tissue salvaging interventions.
3.4.4. Perfusion and susceptibility weighted imaging
Perfusion weight imaging (PWI) is defined as a variety of MRI techniques that are able to provide insights into the perfusion of tissues by blood [39]. PWI can be used for the evaluation of ischaemic conditions, neoplasms, and neurodegenerative diseases. Perfusion MRI mainly has three main techniques: Dynamic susceptibility contrast (DSC), Dynamic contrast-enhanced (DCE), and Arterial spin labeling (ASL).
Susceptibility weighted imaging (SWI), previously known as BOLD venographic imaging, is a type of MRI sequence that is extremely sensitive to venous blood, hemorrhage, and iron storage. As an fMRI technique, SWI can explore the susceptibility differences between tissues and detect differences based on the phase image. An enhanced contrast magnitude image can be obtained by combing the magnitude and phase data. SWI is commonly used in traumatic brain injuries (TBI) and high-resolution brain venographies as it is sensitive to venous blood.
3.4.5. Magnetic resonance fingerprinting
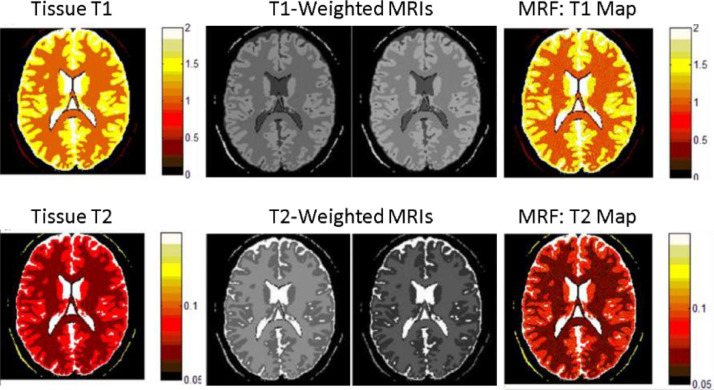
Magnetic resonance fingerprinting (MRF) [40] is new MRI technique that integrates MR physics theory and computer pattern recognition technology and realizes fast and multi-parameter parallel quantization imaging. The technique consists of three modules. First, the fingerprinting signals are excited and acquired from the subject in the MR scanner by the pseudorandom temporal varied pulse sequence to reflect the physiological property of tissue. Second, the evolution of fingerprinting signals with different physiological parameter combinations are predicted by the computer simulation using the Bloch equation; and a fingerprint dictionary indexed by the quantized parameters is constructed. Finally, the pattern recognition technology is applied to find the matched fingerprinting entries for the measured fingerprinting signals, so as to obtain the corresponding quantization parameters and realize quantization MR imaging. Different from most of the conventional MRI modalities, which provide qualitative contrast-based images that are determined not only by the tissue properties but also by experimental conditions, MRF provides quantitative images of tissue properties that reflects pathological conditions of the subject. Fig. 9 shows digital phantom experiments of conventional MRI and MRF. The upper row shows a digital brain with T1 values (left), T1-weighted MRIs with different experimental parameters (middle), and MRF reconstructed T1 map (right), respectively; The lower row shows a digital brain with T2 values (left), T2-weighted MRIs with different experimental parameters (middle), and MRF reconstructed T2 map, respectively. The experiments demonstrated that the contrasts of the conventionally “weighted” MRI images depend on both the tissue properties and experimental parameters, but MRF can reconstruct the parameter images independent of experimental parameters.
Fig. 9.
Digital phantom experiments of conventional T1 weighted and T2 weighted MRIs, and MRF.
Currently, the applications of MRF have been limited to biomedical research and the fusion of MRF with other neuroimaging modalities has not been reported. Given its parametric and quantitative features, the MRF technique will play an important role not only in neuroimaging but also in fusion of multimodal neuroimaging.
3.5. Comparison of imaging methods
Table 1 lists the main advantages, disadvantages and applications of each neuroimaging technology.
Table 1.
Comparison of various imaging methods.
| Imaging methods | Advantages | Disadvantages | Applications |
|---|---|---|---|
| Computerized Tomography (CT) | • Painless, noninvasive and accurate | • Radiation | • Brain tumors. |
| • Blood clots and blood | |||
| • Image bone, soft tissue | vessel defects. enlarged ventricles | ||
| and blood vessels all at the same time | |||
| • Not recommended for pregnant women | • Abnormalities in the | ||
| nerves or muscles of the eye | |||
| • Fast and simple | |||
| Positron Emission Tomography (PET) | • Double the diagnostic clarity compared to CT | • Not recommended for pregnant women | • Cancer |
| • Heart disease | |||
| • Diabetics require certain precautions. | • Brain disorders | ||
| • Easy,Nondisruptive | |||
| Single-photon Emission Computed Tomography (SPET) | • More available and widely used | • Long scan times | • Functional brain imaging |
| • Low-resolution and prone to artifacts and attenuation | |||
| • Functional cardiac imaging | |||
| • Less expensive than PET | |||
| Magnetic Resonance Imaging (MRI) | • No radiation | • Expensive | • Anomalies of the brain and spinal cord |
| • Apparent, detailed images of soft-tissue structures compared to other imaging techniques | • Cannot find all cancers | ||
| • Tumors, cysts, and other anomalies in various parts of the body | |||
| • Cannot always distinguish between malignant or benign tumors | |||
| • Breast cancer screening for women who face a high risk of breast cancer | |||
| • Injuries or abnormalities of the joints, such as the back and knee | |||
| • Certain types of heart conditions | |||
| • Diseases of the liver and other abdominal organs | |||
| • The evaluation of pelvic pain in women, with causes including fibroids and endometriosis | |||
| • Suspected uterine anomalies in women undergoing infertility evaluation | |||
3.6. Databases
In this section, we listed some public databases, as shown in Table 2 . The International Cat Association (TICA) database is an extensive database that contains different types of medical images of cancers, including lung cancer, breast cancer, and kidney cancer. ATLAS is a public database of Harvard University, which mainly contains image data of Cerebrovascular Disease, Neoplastic Disease, Degenerative Disease, Inflammatory or Infectious Disease. CTisus has numerous MRI, CT, X-rays of different organs and tissues. The Open Access Series of Imaging Studies (OASIS) dataset contains 2000 MR sessions, which includes: T1 weighted image, T2 weighted image, FLAIR, ASL, SWI, time of flight, resting-state BOLD and DTI sequences, PET images from three types of traces, PIB, AV45 and FDG. The Alzheimer's Disease Neuroimaging Initiative (ADNI) database contains several types of data like MR, PET from a group of volunteers and dementia patients. The Federal Interagency Traumatic Brain Injury Research (FITBIR) shares the data for Traumatic Brain Injury (TBI) research.
Table 2.
Public datasets.
| Database Name | Web Address |
|---|---|
| TCIA | http://www.cancerimagingarchive.net/ |
| ATLAS | http://www.med.harvard.edu/aanlib/home.html |
| CTisus | http://www.ctisus.com/ |
| OASIS | https://www.oasis-brains.org/ |
| ADNI | http://adni.loni.usc.edu/ |
| FITBIR | https://fitbir.nih.gov/ |
4. Multimodal imaging data fusion: diseases
In this part, we will review recent advancements in the application of multimodal neuroimaging in some clinical and research areas such as early brain development, neurodegenerative diseases, psychiatric disorders, and neurological diseases. It is not our intention to cover all aspects or provide a complete review of these areas. Instead, we focus on the aspects related to the development and applications of the multimodal neuroimaging techniques that meet the expectations and challenges of biomedicine. As such, each of the areas will begin with a brief description of background information such as clinical features, pathology, diagnosis, treatment of the diseases; then a general introduction of the roles, applications, and current status of the medical imaging techniques to the disease; the major part will be a review of recent papers that used one or more imaging modalities and used image fusion in multiple imaging modalities.
4.1. Developing brains
Recent studies show that the human brain experiences a rapid development in the first eight years and continues to develop and change into adulthood. During this long period, the brain develops in size, neuroanatomy, and functions. This period is significant for a person's physical and mental health, intellectual and emotional development, and learning, working, and life success [41], [42], [43].
Many factors have influences on the brain development of young children, which will have an impact on cognitive abilities and mental health in later life. These influencing factors include genes, maternal stress, and drug abuse, exposure to toxic environments, infectious diseases, socioeconomic status of the family, etc. Approximately one-third of genes in the human genome are expressed primarily in the brain and will affect brain development. Many psychiatric and mental disorders, such as autism, ADHD, bipolar, and schizophrenia, are highly heritable or have genetic risk factors. Maternal stress and drug abuse are associated with preterm birth and low birth weight and increased risk of neurodevelopmental disorders and mental disorders in children [44]. The nutritional status of a child, which is affected by the socioeconomic status of the family, has a significant impact on neurocognitive development [45].
Neuroimaging techniques have been used to study normal and/or abnormal development of the brain, enhancing our understanding of neuroanatomy, connectivity, and functionality of the brain. These techniques also reveal the etiological associations of abnormal brain development with risk factors and contribute to the development of intervention procedures for diseased children [42]. Young children are more sensitive to radiation than adults are, so the use of PET and CT is limited. Thanks to the in vivo nature and versatility of MRI, not only young children but also newborn babies can be imaged, offering the opportunity to study white matter development and cognition in babies [46], [47], [48]. MRI has become the most important pediatric neuroimaging modality and has been widely used to study normal and abnormal brain development, allowing repeated longitudinal observation of the changes of brains of the same individuals before and after birth [49]. In the following, our review will focus on the major MRI modalities in pediatric imaging, which include structural, functional, and diffusion tensor imaging.
Early pediatric brain MRI studies focused on the anatomical aspects using T1-weighted and T2-weighted images. Qualitative studies provided information about changing patterns of gray matter and white matter differentiation and myelination in the first months of birth [50] and early childhood [51]. Quantitative studies also revealed the changes in water contents, T1, and T2 relaxation times in both gray matter and white matter; age-related changes in gray matter, white matter, and CSF volumes. All these reflect ongoing maturation and remodeling of the central nervous system [52, 53].
Compared with adult cohorts, brain MR imaging of young children is challenging because of several factors. Young children are less cooperative than adults with scanning procedures, which can be long, noisy, and uncomfortable when lying still for long; the images are often plagued with motion artifacts. The brain changes rapidly with age in early life after birth; the brain is not well myelinated; the contrast between gray matter and white matter is low. These pose difficulties to the optimized parameters for data acquisition protocol and also the standard parameters or criteria for the postprocessing procedures, such as the segmentation of the brain to determine cortical thickness [54]. As a result, the physical properties, such as relaxation times, water content, diffusion coefficients, of the developing brain are not very well characterized. Other technical challenges exist to scan young children [55].
Knowledge of the variations of biophysical properties, such as T1 and T2 relaxation times, water contents in GM and WM during the early life of children, is of critical importance to the understanding of neurodevelopment of young children and also to the development of diagnostic protocols of abnormal brain development. The measurement of these biophysical properties is challenging due to the prolonged scan time. Recent technical development of magnetic resonance fingerprinting (MRF) allows rapid and quantitative analysis of multiple tissue properties [40]. For example, MRF can provide T1, T2, and proton density maps of the brain in contrast to the conventional T1-weighted, T2-weighted, or proton density-weighted images. A recent paper reported an application of the MRF to study the T1, T2, and MWF of children aged from 0 to 5 years old [56]. This study was able to record different patterns of variations of tissue biophysical parameters over different age stages. MRF techniques were also used to parametrically characterize brain tumors in children and young adults [57]. In a broad sense, the parametric information in MRF opened doors to studies of the correlations between brain tissue properties and brain development, impairment, and physiopathology. Techniques of image fusion can play an essential role in the processing, interpretation, and application of MRF data.
4.2. Degenerative brains
Degenerative brain diseases are caused by the decline of neuronal function and the reduction of numbers of neurons in the central nervous system (CNS). Known degenerative brain diseases include mild cognition impairment, Alzheimer's disease (AD), Parkinson's disease, etc. The patients of these diseases suffer from losses of functions in memory, speech, movement, etc. Most of these diseases (except for some mild cognitive impairment subtypes) are progressive, i.e. the symptoms deteriorate as the brains age. As the population is rapidly aging, degenerative brain diseases post enormous impacts on individuals, families, and society. The etiology of these diseases is still unknown, and there is currently no cure. In the following sessions, we will review advances of neuroimaging on MCI, ADs, and PDs.
4.2.1. Mild cognitive impairment
Mild cognitive impairment (MCI) is a clinical transition between normal aging and dementia or Alzheimer's disease (AD), in which individuals have memory or other cognitive impairments beyond their age, but not to the extent of dementia. Patients with MCI often only have minor difficulties in functional ability.
In studies based on people older than 65 years of age, the incidence of MCI is estimated to be at 10–20% [58], and the Mayo clinical study on aging shows an 11.1% incidence of amnestic MCI (aMCI) and 4.9% incidence of non-amnestic MCI (naMCI) in undiagnosed patients aged 70-89 years [59]. Several longitudinal studies have shown that most MCI patients have a significantly higher risk of developing to dementia compared to the general U.S. population (1-2%/ year) [60], the community population (5-15%/ year), and the clinical patients (10-15%/ year) [61], [62], [63]. The latter data suggest that cognitive impairment tends to develop more rapidly for the patients that display serious symptoms. Although some studies have shown that the incidence of MCI reversals to normal cognitive function is as high as 25-30%, recent studies suggest that the incidence may be lower. In addition, cognitive reversals over a short period of follow-up study showed that they did not prevent subsequent disease progression.
Magnetic resonance imaging
Magnetic resonance imaging (MRI) techniques have been used in the clinical identification of MCI and various types of dementia to predict the progression of MCI to dementia. For MRI measurements of brain structure, linear, area, or volume measurements can be used. The results showed that the area of MCI brain atrophy was consistent with AD, but to a lesser extent, between the normal elderly (control group) and AD patients [64], [65], [66], [67]. Similar results were found using voxel-based measurement and analysis, with abnormal changes in not only gray matter but also white matter [68, 69]. The previous diagnosis of AD by structural MRI was mainly based on the degree of brain atrophy, especially in the medial temporal lobe. The structural MRI studies showed the atrophy along the hippocampal pathway (entorhinal cortex, hippocampus and posterior cingulate cortex), which was consistent with the loss of early memory. As the disease progresses, the temporal, frontal, and apical lobes shrink with neuronal loss, causing abnormalities in language, practice, vision, and behavior [70, 71]. However, no definitive biomarkers have been identified by structural MRI alone to distinguish MCI and AD, to stage MCI, and to predict MCI conversion to AD or not [72, 73].
MRI-based functional imaging has been applied to the understanding of and to the discrimination between AD and MCI. These techniques include perfusion-weighted imaging (PWI), diffusion-weighted imaging (DWI), diffusion tensor imaging (DTI), and blood oxygen-dependent fMRI (including task execution and resting state) [72]. Functional MRI allows the delineation of microstructural brain changes, which is complementary to structural MRI that can depict the global changes of the brain in MCI. An MRI-based functional imaging study that employed PWI, DTI and proton MRS showed significant abnormalities in parameters derived from the three imaging modalities for AD patients. PWI and DTI parameters showed a significant, but a lower degree of abnormalities in some areas for MCI patients. fMRI has also been used to distinguish AD and MCI and to predict the transition from cognitive normal to MCI and from MCI to AD. Recent studies show that BOLD-fMRI can detect changes in brain function before MCI progresses to AD, making it an important technique to study the neural mechanism of MCI [74, 75].
Proton magnetic resonance spectroscopy (1HMRS) is a noninvasive imaging method that can detect biochemical and metabolic changes in brain tissue in vivo and conduct quantitative analysis. Early MRS studies show abnormal concentrations of N-acetylaspartate (NAA), creatine, and choline are associated with the status of memory and cognition impairment and have a promise for assessing cognitive status, evaluating response to medicine, and monitoring progression during treatment [76], [77], [78]. In recent years, with advances in the technical development of MR hardware and pulse sequences, the roles of glutamate, the excitatory neurotransmitter, and GABA, the inhibitory neurotransmitter, in MCI patients became the main focus [79], [80], [81]. For example, with ultra-high field 7 Tesla MR scanner, abnormal concentrations of GABA, glutamate, NAA, glutathione, and myo-inositol (mI) in different brain regions were detected [82]. The manifestations of 1HMRS in MCI patients were mainly shown in decreased NAA/Cr ratio and increased mI/Cr ratio. The pathological results showed neuronal deletion and glial proliferation, and the changes in metabolite concentration were consistent with the pathological results [82, 83].
Multimodal imaging
PET and SPECT provide insight into blood perfusion and metabolism in tissues and organs, as well as explore changes in function. The nuclear medical images of aMCI patients showed decreased perfusion and metabolism in the hippocampus, temporoparietal lobe, and posterior cingulate gyrus. Studies using PET, SPECT, and MRI have shown that glucose metabolism in the hippocampus, glucose metabolism rate in the bilateral temporal-parietal lobe, and blood perfusion in patients with aMCI are lower than those in normal elderly. These studies have also shown that low glucose metabolism in the temporal-parietal lobe is a reliable indicator of conversion to AD [84], [85], [86]. Excessive deposition of β-amyloid peptide in the brain and the cascade reaction caused by it are the early onset of AD. Therefore, early detection of β-amyloid peptide in the brain can help identify patients with aMCI, and monitor the progression of the disease and treatment effect. It was found that the 11C-PiB-PET could attach to Aβ in the brain. PET imaging showed the amount and location of Aβ deposition in the brain, which was expected to be an early diagnostic method for AD [87], [88], [89].
Multimodal imaging techniques involving MRI-based imaging and PET-based imaging have been frequently used for prediction, characterization, and classification of MCI [90, 91]. In facilitating these complex tasks, imaging fusion methods based on artificial intelligence, neural network, deep learning and graph theory have been used [92], [93], [94]. Brain network studies based on multimodal MRI and graph theory analysis have found that the topological properties of AD and aMCI affected brain networks have undergone abnormal changes, which mainly manifested as the imbalance between functional differentiation and integration. This approach provided a new way to reveal topological mechanisms and pathophysiological mechanisms of brain networks [93, 95]. In addition, the combination of graph theory analysis and classification analysis suggests that the brain network topology attribute can be used as an imaging marker of AD and has a good clinical application prospect.
4.2.2. Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disorder and the most common cause of dementia. AD is characterized by progressive memory loss, aphasia, loss of use, loss of recognition, impairment of visual-spatial skills, executive dysfunction, and personality and behavior changes [96, 97]. It has become one of the major diseases that seriously threaten the health and quality of life of the elderly [98]. The onset of AD is slow or insidious, with patients and their families often unable to tell when it starts. It is more common in the elderly over the age of 70 (the average male is 73, and the average female is 75 years old), with more females than males (female to male ratio of 3:1) [99].
There is currently no cure for AD, but large numbers of novel compounds are currently under development that have the potential to modify the course of the disease and to assess the efficacy of these proposed treatments. There is a pressing need for imaging biomarkers to improve understanding of the disease and to assess the efficacy of these proposed treatments.
Magnetic resonance imaging
Structural MRI (sMRI) is the most widely used imaging modality for the study of AD. The techniques for analyzing sMRI are classified into volume-based and surface-based methods [100]. Previous studies have shown that hippocampal volume atrophy and whole-brain atrophy independently predicted the progression of AD [101]. Hippocampal damage or atrophy occurs in the early stage of AD, which is an important structural basis for the clinical manifestations of AD. Although global hippocampal atrophy in AD was well accepted, the differences were often detected large sample-size studies [102].
De Winter et al. studied 48 elderly AD patients with depression and 52 healthy control elderly people and examined all the subjects with sMRI and neuropsychology [103]. They found that there was no significant difference in the positive rate of Aβ between the depression group and the healthy control group. However, the hippocampal volume in the depression group was significantly smaller than that in the healthy control group. There is significant hippocampal atrophy in elderly depression patients, and hippocampal atrophy has nothing to do with Aβ, which challenges the reliability of hippocampal atrophy in the clinical diagnosis of AD. It is suggested that hippocampal atrophy not only occurs in AD but also in senile depression. The study of sMRI indicates that the brain atrophy shown by brain morphology and structure has reference value for the diagnosis of AD. However, the diagnosis of AD still needs to be confirmed by combining clinical manifestations, neuropsychological assessments, and other examination methods. It also indicates that follow-up is needed for suspected depression in patients with AD. The above studies showed the limitations of structural MRI and the necessity of the multimodal approach in the study of AD [104].
Other MRI modalities, including functional MRI, DWI, PWI, have also been widely used in the study of neurodegenerative diseases. We will review recent advances of the resting-state functional magnetic resonance imaging (rs-fMRI) as an example. As opposed to the conventional task-based fMRI, rs-fMRI does not require the subject to perform any task or be subjected to any external excitation. The rs-fMRI captures the low-frequency oscillations signals that are related to the spontaneous neural activity of the brain by analyzing the brain blood oxygen level dependent (BOLD) signal. Sophisticated methods of analysis of the rs-fMRI data depict the functional connectivity of the brain. The rs-fMRI has been used to reveal how the networks of the functional connectivity are correlated to the brain functions of individuals with cognitive impairment. Zamboni et al. found that the recognition task of AD patients was related to the increased activation of the lateral prefrontal area, which also overlapped with the functional connection enhancement area indicated by the rs-fMRI [105]. Zhou et al. predicted the pathological changes of AD by using the calculation model of resting brain function network and studied five different brain regions vulnerable to neurodegenerative diseases through the use of task state fMRI [106]. They found that the brain network of AD patients may have the phenomenon of weak functional connectivity and their ability to transmit information of functional brain network decline. Wang et al. found that the functional brain network of MCI patients had different degrees of functional connectivity disorder. The evaluation of overall functional brain connectivity of patients plays an important role in the early diagnosis and treatment of AD [107]. Abnormal brain connectivity can be a biomarker of the disease.
Many neuropsychiatric diseases and dementia can change the default mode network (DMN) of the brain. Identification of the change in the connectivity of DMN is constructive for the early recognition of AD. Jin et al. collected 8 patients with aMCI and 8 healthy people to analyze rs-fMRI data by independent component analysis (ICA) [108]. They found that the functional activities of the lateral prefrontal cortex, left medial temporal lobe, left middle temporal gyrus and right angular gyrus in aMCI patients decreased, while the activity of the middle and medial prefrontal cortex and the left parietal cortex increased. Further studies found that the functional activities of the left lateral prefrontal cortex, left middle temporal gyrus and right angular gyrus were positively correlated with memory, especially delayed memory [109]. Although there was no significant difference between the two groups in the degree of medial temporal lobe atrophy, the functional activities of the left medial temporal lobe decreased. This decrease suggests that the functional changes of DMN may occur in the early stage of AD, i.e. aMCI, and the functional changes may occur before the obvious change of brain structure.
Multimodal imaging
Due to severe overlap in symptoms and findings of individual imaging modalities of the neurodegenerative diseases, it is difficult to identify the biomarkers that could be used to differentiate the types of these diseases and/or to stage the progress of a disease. Therefore, multimodal neuroimaging techniques are used to overcome the challenges [110]. As pointed out in [111], individual modalities of MRI and EEG lack precision in AD diagnosis and staging. By employing both imaging modalities, with the MRI measuring the cortical thickness and the EEG measuring the rhythmic activities, the authors found joint markers that identified the subjects of Alzheimer's disease with an accuracy of 84.7%, a significant increase from those of individual modalities. While some studies of multimodal imaging confirmed correlations of findings among individual modalities as in a study of sMRI and fMRI [112], multimodal imaging studies can also be used to dissociate the tau deposition and brain atrophy in early ADs using PET and MRI. The study found that the tau load had little effect on the gray matter atrophy, and this might imply that tau protein deposit precedes and predicts brain atrophy. The multimodal imaging studies require statistical and analytical models, advanced computing algorithms, and especially, novel data fusion methods [113], [114], [115], [116], which will be reviewed in detail in the following sections.
4.2.3. Parkinson's disease
Parkinson's disease (PD) is a chronic progressive degenerative disease of the central nervous system, which is commonly seen in elderly patients. Typical clinical manifestations of PD include static tremor, myotonia, bradykinesia, and abnormal posture and pace [117]. With the continuous increase of the aging population, the incidence and disability rates of the population are also increasing year by year. The results of epidemiological surveys indicate that the prevalence of PD in people over 65 is about 1.7%, and the prevalence of PD in people over 80 is as high as 4% [118], [119], [120]. PD is more and more harmful to the health of the middle-aged and elderly, especially involving the central nervous system.
Due to the lack of objective basis and diagnostic criteria for the diagnosis of PD, the previous clinical diagnosis of PD was mainly based on the clinical symptoms, resulting in a low coincidence rate between the clinical diagnosis and pathology of PD and in a significant lag behind the pathological changes of brain microstructure. With the increasingly standardized diagnosis and treatment of PD, neuroimaging examination has become an indispensable part of the diagnosis. This differential diagnosis of PD can help identify different movement disorders, locate anatomical dysfunction sites, and determine the causes of the lesion, which will improve clinical evaluation and prognosis [121, 122].
Magnetic resonance imaging
Structural cranial MRI can distinguish white matter from gray matter by setting different imaging parameters while avoiding radiation. It is better than cranial CT in revealing white matter lesions, small infarcts, subacute intracerebral hemorrhage, and lesions in the brain stem, subcortical regions, and posterior fossa. On structural MRI such as T1, T2-weighted, and fluid-attenuated inversion recovery (FLAIR) images, PD patients usually exhibit broadening of the ventricles (caused by extrapyramidal atrophy) and widened sulci (diffuse brain cortical atrophy) [123]. The quantitative measurements of cortical atrophy can be measured based on voxel morphometric assessment. When the compact belt of the substantia nigra shrinks and the short T2 signal of the substantia nigra disappears, the width of the dense belt of the substantia nigra, the ratio of the width of the dense belt of the substantia nigra to the diameter of the midbrain, the caudex nucleus, the putamen nucleus, the thalamus and other areas of interest are measured. In evaluating the extent of atrophy, physiological changes such as age increase and relevant clinical supporting evidence should be taken into account [124].
Neuromelanin-sensitive MRI is used to detect neuromelanin, a surrogate biomarker for the PD. Neuromelanin is a dark pigment found in neurons in the substantia nigra pars compacta. The concentration of neuromelanin increases with age but is found to be around 50% higher in PD patients compared with age-matched non-PD subjects, due to the death of cells in the substantia nigra. Neuromelanin-sensitive MRI allows the visualization of the neuromelanin-containing neurons in the substantia nigra, pars compacta. With the use of morphological analysis and signal intensity (contrast to noise ratio), the width and CNR of the lateral and central substantia nigra were found to be significantly lower in the PD subjects than in the control group and untreated essential tremor (ET) group [125, 126]. Therefore, this imaging technique can be potentially used as a biomarker to differentiate ET from the de novo tremor-dominant PD subtype. The neuromelanin levels were quantitatively assessed using neuromelanin-sensitive MRI and quantitative susceptibility mapping (QSM) [127], an MRI modality for measuring the absolute concentrations of iron, calcium, and other substances in tissues based on changes of local susceptibility [128]. While the neuromelanin imaging found significantly lower neuromelanin levels in the PD group than the health controls (HC), which is in agreement with the neuromelanin MRI only study, the QSM values were significantly higher in the PS group than in the HC group. This result suggested the usefulness of QSM in detecting PD [127].
Resting functional magnetic resonance imaging (fMRI) is a technique that collects the blood oxygen level dependent signal changes of patients in the awake and resting states to obtain the functional activity level of the brain in the baseline state. In recent years, fMRI has been widely used in clinical studies of all motor disorders or neurodegenerative diseases, including PD [129], [130], [131]. Resting-state functional MRI (rs-fMRI) can calculate a variety of brain activity attributes, such as local consistency, range of low-frequency fluctuation, and amplitude of low-frequency fluctuation etc. By observing the correlation between time-dependent signals of blood oxygen levels in different voxels or areas of interest, we can further evaluate the synchronization of functional activity in different brain areas, i.e. functional connectivity [129, 132, 133]. In recent years, calculation methods based on independent component analysis, Granger causality analysis and Graph Theory can help to find complex pattern changes in the brain network of PD patients.
Other imaging modalities and multimodal imaging
Other imaging modalities, including PET, SPECT, EEG, CT, have been applied to study the functional and structural abnormalities and changes of PD patients, and they provide much complementary information to MR-based imaging modalities mentioned earlier. PET studies investigated cerebral glucose metabolism with or without medications, with or without brain stimulations [134], [135], [136]. Metabolic and brain chemical changes related to dopamine neurons in PD patients were also studied using SPECT, which cannot be assessed by other MRI modalities including proton magnetic resonance spectroscopy (MRS) [137]. By jointly applying SPECT and DTI, this study identified regions and connections of the brain that differentiate PD patients and healthy controls. Different from the imaging modalities in that study, a recent study employed PET scans with two different tracers and rs-fMRI to investigate variations of metabolism and functional connectivity of the PD patients [138]. It identified correlations between motor impairments with hypometabolism and hypoconnectivity in multiple brain regions. With the use of different modalities under similar aims, results from these studies can provide complementary information for the impaired regions. The data from them can be integrated and analyzed using data fusion like the work in [139], in which data of anatomical MRI, rs-fMRI, and DTI were analyzed for more accurate and reliable biomarkers of PD.
4.3. Mental disorders
Mental disorders are conditions that affect a person's thinking, mode, behavior, relationship with others, and functions of daily life and work. Major psychiatric disorders include depressive disorders, bipolar disorders, obsessive-compulsive disorders, schizophrenic disorders, autistic spectrum disorders, attention deficit, and hyperactivity disorder. It is estimated that nearly one-fifth of adults aged 18 or older in the United States live with a psychiatric disorder [140]. The World Health Organization estimates that mental disorders affect one-fourth of the worldwide population [141]. The high prevalence of mental disorders have a significant impact on the wellbeing of societies and the development of the world economy [142], [143], [144]
Unlike the diagnosis of other diseases, such as cancer and diabetes, there are currently no medical tests that can determine mental illness. The diagnosis of mental illness is determined by a psychiatrist using official criteria such as The Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5) according to the feeling, symptoms, and behaviors of the patient. However, neuroimaging techniques have been used to detect, identify, differentiate, and understand the abnormalities, differences, etiologies, and biomarkers of psychiatric disorders [145], [146], [147], [148], [149].
4.3.1. Depression
Depression is a common mood disorder, which can be caused by a variety of reasons. The main clinical feature is marked with persistent depression of mood, which is incompatible with the situation. In severe cases, suicidal thoughts and behaviors may occur. Most cases tend to show recurrence; and most can be relieved each time, while some may have residual symptoms or progress to chronic depression. At least 10% of clinical depression patients also show manic episodes and should be diagnosed as bipolar disorder [150, 151]. What we commonly call depression is clinical or major depression, which affects 16% of the population at some point in their lives [152]. In addition to the severe emotional and social costs of depression, the economic costs are also enormous. According to the World Health Organization, depression has become the fourth most serious disease in the world and is expected to become the second most serious disease after coronary heart disease by 2020 [153].
So far, the etiology and pathogenesis of depression are not clear, and there are no obvious signs or laboratory indicators of abnormality. Although there have been many basic and clinical studies on depression, no critical breakthrough has been made in the three most important clinical problems: pathogenesis, objective diagnosis, and efficient treatment. A key breakthrough in these issues is to find and establish a stable biological marker from gene to clinical phenotype and then further study its pathogenesis, establish objective diagnostic methods and develop efficient clinical therapy.
Magnetic resonance imaging
Up to now, in the clinical research field of mental illness, especially depression, the most sought after biological markers may potentially be provided by the study of neuroimaging, especially brain MRI. Brain MRI examination have characteristics of good clinical applicability, non-invasive, simple operation, universal, relatively stable results and easy to repeat, but its sensitivity and specificity need to be improved. Brain MRI research has become an intermediate mechanism from molecular research to clinical phenotype. Through this mechanism research, we can not only explore how genes, molecules, and proteins affect the brain structure and function of patients with depression but also use MRI as an objective diagnostic tool for the most urgent clinical needs. Over the past 20 years, the application of multi-mode MRI technology to study the brain structural and functional characteristics of depression, especially to establish clear biological marker targets around the characteristics of emotional circuits, has become one of the major scientific frontiers in the basic and clinical research of neuroscience.
Many studies have found that patients with depression have abnormalities in brain structure and function of emotional circuits, as well as in neurotransmitters associated with these circuits [149, 154]. MRI studies in recent years found that the depressive mood is associated with three brain regions, namely in the amygdala and the ventral striatum as the primary mood areas, the orbital gyrus, medial prefrontal cortex and cingulate gyrus as the emotional auto-regulation areas, and the dorsolateral and ventrolateral prefrontal cortex as the center of the active emotional regulation area [154], [155], [156].
Multimodal MRI
Multimodal MRI techniques used in mental disorders seek to find correlated, complementary, and/or converging image features from multiple image modalities and applied sophisticated analytical methods to identify robust biomarkers for the types of depression.
A study employed DTI, magnetic resonance spectroscopy (MRS), rs-fMRI, and magnetoencephalography (MEG) and revealed patterns of abnormalities of patients with major depressive disorders. These patterns included factors in the neurotransmitters (glutamate concentration), white matter fibers (fractional anisotropy), and functional excitations (fMRI) [157]. A multimodal MRI study involves structural MRI and ASL to assess grey matter volume and regional cerebral blood flow in MDD patients. This multimodal study revealed negative correlations between the extent of depressive symptoms and CBF in the bilateral para-hippocampus and between depressive symptoms and CBF in the right middle frontal cortex [158]. In addition to confirming the correlations among findings of individual modalities, some multimodal MRI studies, however, found disrelations among individual findings in the MDD group [159]. Further multimodal data analysis involving MRI imaging data and clinical, neurobiological metrics of the patients may resolve the disparities.
Among the methods of the multimodal MRI image fusion in depressive disorders, support vector machine (SVM) [160] and linked independent component analysis [161] was recently used, respectively, to identify biomarkers for the classification and prediction of symptom loads of heterogeneous MDD cohorts. Imaging data and neurobiological data were included in the data fusion. In both studies, the results did not show strong support for the hypothesis and did not provide sufficient evidence for the sought biomarkers [160, 161].
4.3.2. Obsessive-compulsive disorder
Obsessive-compulsive disorder (OCD) is a group of neuropsychiatric disorders with obsessive thinking and compulsive behavior as the main clinical manifestations. It is characterized by the co-existence of conscious compulsion and anti-compulsion, and the repeated intrusion of thoughts or impulses into the daily life of patients that are often meaningless and involuntary. Although patient perceives that these thoughts or impulses are their own and resist them to the utmost degree, he or she is still unable to control them. The intense conflict between the two causes the patient great anxiety and pain, which affects his other study, work, interpersonal communication, and even daily life.
Magnetic resonance imaging
Voxel-based morphometry was widely used in the sMRI studies of OCD. These studies measure the structures and volumes of regions of interest in various OCD groups and healthy control groups. OCD patients were found to have lower grey matter volumes in specific regions of the brain. For children with OCD, these regions include the bilateral frontal lobe, cingulate cortex, and temporal-parietal junction [162]. For adults with OCD, these regions are the left and right orbitofrontal cortex [67]. Lower volumes were also seen in white matter in the cingulate and occipital cortex, right frontal and parietal and left temporal regions [162] and in a small area of the parietal cortex for patients with OCD [163]. fMRI studies can provide information about the pathophysiology of OCD [164, 165]. However, whether this information from single fMRI modality alone could be of clinical value in the diagnosis of individual patients is not clear [166]. The pathophysiological feature of OCD, as revealed by fMRI studies, suggest that abnormal brain metabolites may be implied in OCDs. Abnormalities in brain metabolite concentrations in patients with OCD were investigated using proton MRS [167], [168], [169], [170]. Among about ten detectable metabolites, glutamate, glutamine, and GABA are of particular interest, as they are involved in neurotransmission. Recent studies show that OCD patients had an elevated GABA level and a higher GABA/glutamate ratio in the anterior cingulate cortex [171], and they had lower GABA concentration in the prefrontal lobe, as compared to healthy control groups [172]. The roles of glutamate and glutamine in OCD are in the focus of research interest, but the findings lacked reasonable consistency [173], [174], [175]. The heterogeneity of structural neuroimaging findings of OCD may reflect the heterogeneity of the disease itself.
Multimodal MRI
Multimodal MRI studies in OCD provide complementary, correlated and/or integrated information of findings from individual modalities [176, 177]. Early structural MRI study suggested that the volume reduction of superior temporal gyrus (STG) is associated with the pathophysiology of OCD [178]. A functional MRI study found increased low-frequency fluctuations in neural activities in STG [179]. A correlation between these findings was found in a combined structural MRI and fMRI study, which shows that the volume of the superior temporal sulcus is strongly correlated with functional connectivity between several brain regions that may form a neuro-network [177, 180]. The simultaneous 1H-MRS and DTI study, to investigate metabolic and white matter integrity alterations in OCD, found that the level of Glx to Cr ratio in the anterior cingulate cortex was higher in the OCD group than the healthy control group [181]. The study also found from DTI analysis that the FA values in the left cingulate bundle of the OCD group were significantly higher than the healthy controls. A limitation of this study is that the Glx level, which is a combination of glutamate and glutamine, was measured instead of measuring glutamate and glutamine individually. It has been recognized that it is difficult to distinguish these two structurally similar metabolites using 3 Tesla scanners and ultra-high magnetic field (e.g. 7T) scanners are required.
4.3.3. Schizophrenia
Schizophrenia is a group of serious psychosis with unknown etiology, which usually starts slowly or sub-acute in the young and middle-aged individuals [182]. Clinically, it often manifests as a syndrome with different symptoms, including abnormalities in sensory perception, thinking, emotion, and behavior, as well as uncoordinated mental activities [183]. Schizophrenia is a multifactor disease [184]. Although the understanding of etiology is not clear at present, effects of the susceptible quality of individual psychology and the adverse factors of external social environment on the occurrence and development of the disease have been widely recognized. Both susceptible quality and external adverse factors may lead to the occurrence of disease through the joint action of internal biological factors. The course of schizophrenia is generally protracted, showing repeated attack, aggravation, or deterioration. Some patients eventually show recession and mental disability, but some patients can maintain recovery or basic recovery after treatment [185].
Magnetic resonance imaging
Structural MRI was widely used to study the morphology and volumetry of the brains of schizophrenia patients. Studies found that the average brain volume of schizophrenia patients was smaller than that of healthy people [186, 187]. The abnormal volume and structure of white matter usually appear before the onset of the disease, and these abnormalities tend to be stable during the development of the disease [188]; the change of gray matter volume is more evident after the onset of the disease and decreases progressively over time [189]. According to a longitudinal study, gray matter deficiency in schizophrenia mainly occurs in the first five years [190].
Quiet complement to the structural MRI, DTI reveals the abnormalities of white matter microstructure of schizophrenia patients [191]. Decreased fractional anisotropy in white matter tracts, different cortical regions, and subcortical regions was found in schizophrenia patients in some studies. However, controversial findings were also reported [192, 193]. These inconsistencies might be attributed, in part, to small sample sizes. A large-scale DTI study involving more 4,000 subjects found widespread white matter microstructural differences between schizophrenia patients and healthy controls [194]. Significantly reduced fractional anisotropy values were found in 20 of the 25 investigated regions within the white matter. Furthermore, significantly higher mean diffusivity and radial diffusivity were also observed in schizophrenia patients than in healthy controls.
Functional MRI techniques are used to detect the deficits in neural networks of patients with schizophrenia [195]. Brain network studies show that the functional connectivity of the default mode network (DMN) in schizophrenic patients has changed. Although the research structures are inconsistent, most studies show that DMN functional connectivity is enhanced in schizophrenia, and functional connectivity in the prefrontal cortex is weakened (especially in the prefrontal cortex) [196]. In addition, the functional connections of auditory/linguistic networks and basal nuclei are related to auditory hallucinations and delusional symptoms. The study of brain structure networks found that the number of frontal and temporal core nodes decreased and the average shortest path increased, indicating decrease in global efficiency. Wang et al constructed a network of DTI images of 79 schizophrenia patients and 96 age-matched normal subjects [197]. They found that: compared with the normal subjects, the global efficiency of the schizophrenic group decreased; the local efficiency of the core nodes distributed in the frontal cortex, the paralimbic system, the limbic system and the left putamen decreased; and the global efficiency of the network was negatively correlated with the PANSS score. Research shows that the change in brain structure network started at the beginning of the disease. More severe symptoms indicate lower the global or local efficiency of the network, and the slower the speed of information integration.
Magnetic resonance spectroscopy studies found that the brain metabolism of schizophrenia patients was abnormal [198], [199], [200]. The levels of N-acetyl-aspartic acid (NAA) in the hippocampus, frontal lobe, temporal lobe, and thalamus of schizophrenic patients decreased; the levels of NAA in the thalamus of high-risk groups also decreased, while the level of NAA in temporal lobe decreased. It was also found that the increase of glutamate level in the hippocampus and medial temporal lobe was related to the decrease of executive function.
Multimodal imaging
The heterogeneities of findings of individual imaging modalities have been driving the multimodal neuroimaging approach to the search of the more consistent and precise biomarkers for the deficits, abnormalities in functions of schizophrenia patients. A concerted use of three MRI modalities, namely, resting-state of fMRI, structural MRI, and diffusion MRI, was able to simultaneously reveal abnormalities from these three kinds of MRI images and, thereby, identify the cortico-striato-thalamic circuits that might be related the cognitive impairments in schizophrenia [201]. In addition to the multimodal imaging investigation of structural and functional brain abnormalities in schizophrenia, proton MRS has also been used in combination with fMRI to investigate cognitive impairment in schizophrenia at both neurometabolic and functional levels [202]. The combined proton MRS and fMRI study are particularly useful for short-term longitudinal studies on the effects of medication, which is invariant to brain structural change. In this study [202], the relationship between Glx/Cr levels and BOLD response significantly changed after six weeks of medication for schizophrenia patients, although factors that confound interpretations of the results remain. More examples of multimodal imaging studies on schizophrenia are given in recent review articles [203, 204].
5. Multimodal imaging data fusion: methodology
Multimodal fusion has gradually entered the center of research interest as an approach to tackle the challenges of neuroimaging. The first main reason is that there exists a great complementarity between different imaging modes. For instance, the images obtained by positron emission tomography imaging (PET) or single-photon emission tomography imaging (SPECT) do not contain high resolution, three-dimensional anatomical information. On the other hand, high-resolution structural images can be obtained via the use of CT and/or MRI. These images complement each other to provide a complete picture of the targeted organs’ anatomy, physiology, and pathology. The fusion of these images is of great significance for the relevant clinical and pre-clinical studies.
Another outstanding merit of utilizing multimodal fusion is that it efficiently enhances the spatial and temporal resolution in the characterization of brain processes. In other words, multimodal imaging may allow the combination of the hyper-temporal resolution of one imaging mode with the hyper-spatial resolution of another, taking advantage of the spatial-temporal complementarity. Take a study in 2014 as an example, Ke Zhang et al have successfully measured the cerebral blood flow with a combination of Arterial Spin Labeling (ASL), MRI and PET [205]. Apart from this, utilizing EEG together with fMRI to improve spatial and temporal resolution has also been studied by many scientists in neuroscience [206], [207], [208], [209]
It is worth mentioning that, in both narrow and wide senses, multimodal data fusion has a high capacity of generalization. A typical instance is the alignment of functional MRI, EEG, and fNIRS images to an anatomical coordinate system. The coordinate system can act as a template to standardize reported results. The alignment of the images to a single coordinate system not only allows comparison with other studies but also allows the combination of functional and structural information [210].
PET/CT combination is a multimodal absolute quantification approach where CT provides structural data information on bones, which also serves as the main absorber for the γ-rays in PET. This combination allows decay correction, in which the accumulation of radioactive isotopes in human tissues becomes apparent, and the amount of radioactive decay can be absolutely quantified [211, 212].
Multimodal imaging also has the benefit of utilizing data from one modality to improve the data quality of another modality, such as correcting the geometric distortions of EPI images by acquiring a B0 field map or obtaining EPI with different parameters [213, 214]. Another classic case is in a combined MR-PET study, where the motion information provided by high-temporal resolution MRI data was used to help the reconstruction of the PET data [10].
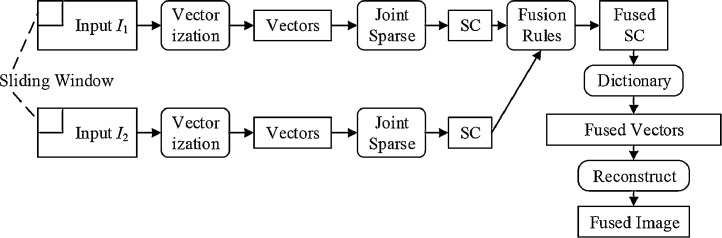
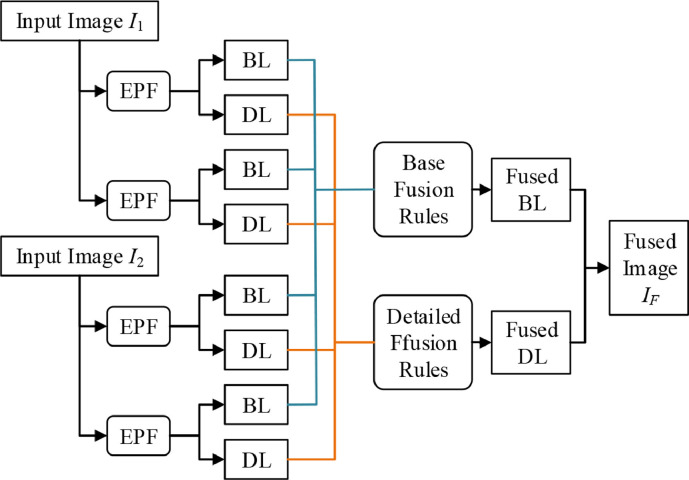
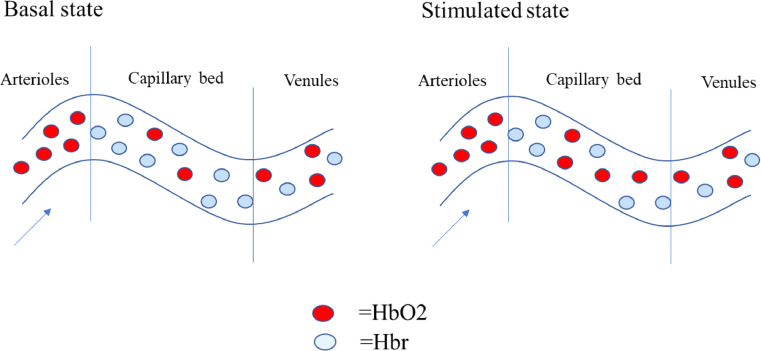
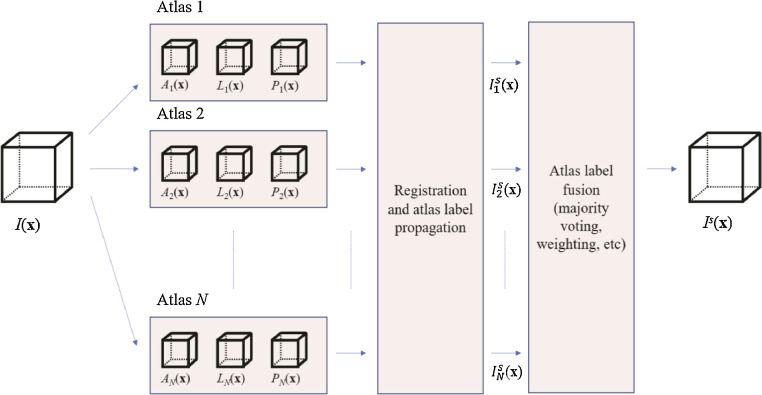
5.1. Forms of multimodality fusion
The long history of neuroimaging has led to the development of an assortment of imaging technologies and modalities, as seen in Section 3. The existing research and apparatus provided a solid foundation for multimodal fusion, leading to the rapid development of many fusion techniques. In this part, we classify reviewed methodologies into four primary forms: multimodal, multi-focus, multi-temporal, and multi-view.
5.1.1. Multimodal fusion
Modern medical imaging methods aim at revealing possible dysfunctions in patients. For example, neuroimaging methods are often used to image the structure of nervous system on a macroscopic level, which in turn helps explore the neuroglial basis of behavior and cognition of patients. However, how to combine different medical imaging methods to provide images with better quality or clearer structures remains to be the heated research interest in the field. Therefore, multimodal image fusion is proposed to minimize the gap.
In a narrow sense, multimodal image fusion, a technique to improve the interpretation of the structure and functions of target organ or region, generally combines two or even more images collected from different imaging instruments. In order to achieve the goal of simultaneous acquisition, researchers have developed particular instrumentations to allow data of one modality to be acquired with low or neglectable inference from another modality. For instance, the EEG-fMRI combination fuses data acquired from EEG instruments like amplifiers and MRI scanners. Also, the novel instrumentations range from simple arrangements to relatively complex technological innovations. However, in some combinations, simultaneous acquisition of data was impossible due to the physical interactions of the imaging devices.
Multimodal image fusion, in a broader sense, also combines data but collected with the same instrument. In this case, MRI is widely used due to its versatility in the generation of different tissue contrasts with the well-studied phenomenon of magnetic resonance. There is also research that combines two and more contrasts in the same acquisition, which has been routinely used for many years. Multiple contrasts can also be acquired by PET (Positron emission tomography) when radioactive compounds injected are different.
5.1.2. Multi-focus fusion
Multi-focus fusion, which keeps interested objects staying in focus, is a technique to fuse images acquired with different focal length. Generally, regions containing objects of interest were segmented from individual images, and then fusion is applied to form fused images. Many multi-focus image fusion methods have been developed during the past decades. These methods, however, can be classified into two categories: spatial domain and frequency domain.
Spatial domain methods work on image pixels. Methods like Intensity Hue Saturation [215] and Principle Component Analysis (PCA) belong to the category of the spatial domain [216]. A method based on Discrete Cosine Transform proposed by Phamila and Amutha [217] turns out to be more efficient compared to other existing DCT methods [218]. In the proposed method, original images are divided into 8*8 blocks, while DCT coefficients of each block are calculated. AC coefficients, together with DC coefficients, are two components that form DCT coefficients. The blocks with higher AC coefficient values are selected for fusion since higher AC coefficient value indicates higher variance and more fine-grained images. The analysis of performance is based on metrics of mean square error (MSE), Petrovic's metric, and Peak Signal to Noise Ratio (PSNR). The reason why this method is so efficient is likely because it does not require complex floating-point operations.
A region-based method, which incorporated segmentation into the fusion process, was proposed by S. Li in [219]. The algorithm is composed of three steps: segmentation, clarity measurement, and fusion of images. Prior to these steps, images are first fused by averaging. Objects of interest are then extracted by normalization-based cuts segmentation. The normalized criterion measures similarity and dissimilarity between different images. After measuring the spatial frequency, the fusion of corresponding regions of source images can be performed. In order to evaluate the performance, mutual information and the Petrovic metric are considered. The Petrovic metric examines the quality of edge-based information transferred from source images to fused images. Compared to pixel-based fusion methods, which suffer from problems including blurring effect, sensitivity to noise and pixel misregistration, the proposed method has lower complexity while being more robust and reliable.
Based on the scale-invariant feature transform (SIFT), Yu et al. proposed that local image features such as SIFT can be used for image fusion [220]. The characteristic of SIFT is the robustness to spatial variations, including scale, rotation and translation. Generally, there are two phases in SIFT: feature points detection and feature point description. The proposed algorithm acquired the initial decision map by dense SIFT descriptor, which was used to measure the activity level of the source image patches. The decision map is further improved with feature matching and local focus measure comparison. It was pointed out that local features such as dense SIFT can also be used to match pixels that were misregistered between multiple source images.
In methods based on frequency, images are transformed into the frequency-domain for fusion. Wavelet-based methods, including Discrete Wavelet Transform (DWT) [221], Haar wavelet [222] and pyramid-based methods such as the Laplacian pyramid [223], fall under the category of frequency-domain methods. A DWT based method was proposed in [224]. Principal component analysis (PCA) was used for approximating coefficients of input images. The principal components were evaluated to obtain multiscale coefficients. The weights for the fusion rule were then acquired by averaging those components. Besides the promising performance, the proposed method was widely applied in medical image fusion for CT and MRI images. In [225], the authors proposed the Daubechies complex wavelet transform (DCxWT) to fuse multimodal medical image. By using the maximum selection rule, the complex wavelet coefficients of source images are fused. Source images at different levels are decomposed by DCxWT, followed by Inverse DCxWT to form the fused image. Compared to the other five methods, including Dual tree complex wavelet transform (DTCWT) based fusion and PCA based fusion, the proposed method achieved the best performance in terms of five measurements, including standard deviation, entropy, edge strength, fusion symmetry, and fusion factor. It was proved that the proposed method was robust to noises, including speckle, salt and pepper, while maintaining the property of shift-invariance.
5.1.3. Multi-temporal fusion and data acquisition
Multi-temporal fusion is to fuse images taken at different times but of the same modality. Multi-temporal fusion enables easy detection of changes in images by subtracting one or multiple images from another. Data acquisition consists of separate recording and simultaneous recording. The choice between separate or simultaneous acquisition should be cautiously considered. Compared to separate recordings, where an image from each modality is acquired individually, simultaneous acquisitions have relatively lower data quality and more artifacts. For example, EEG-fMRI simultaneously acquires cardio-ballistic artifacts and MRI gradients. Data acquisition consists of separate recording and simultaneous recording [226]. In the MR-PET scenario, components of the MRI scanner would trigger degradation in PET scanning results. Therefore, the costs of simultaneous acquisition are higher than separate acquisition due to subject discomfort and long set-up time. However, there are also many cases in which costs are of less concern, given the benefits of simultaneous acquisitions.
5.1.4. Multi-view fusion
In multi-view fusion, images of the same modality are taken under different conditions at the same time. This fusion technique is applied to increase the amount of information for the fused images, while source images are taken under different conditions.
The choice between asymmetric and symmetric data fusion makes a significant difference in the integration and joint analysis of multimodal data. For integration methods falling under the asymmetric category, information from different modalities is assigned with different weights so that information from one modality could be treated as a constraint on the other modality. For instance, fMRI contrast maps limit the source localization of EEG/MEG. Modalities in symmetric data fusion, however, are treated equally in terms of spatial and temporal resolution as well as the uncertainty in the possible indirect relation to neural activity. Hypothesis-driven approaches and data-driven approaches are the two main categories of symmetric fusion approaches. Hypothesis-driven approaches, also called model-driven approaches, are usually in model-based setting, while data-driven approaches belong to blind source separation methods [227]. Examples of different fusion methods are shown in Table 3 .
Table 3.
Examples of different fusion methods.
5.2. Fusion rules
Image fusion rules are applied to highlight the features of interest in images while suppressing the unimportant features. Generally, fusion rules are mainly comprised of four components: activity-level measurement, coefficient grouping, coefficient combination, and consistency verification [230].
5.2.1. Components of fusion rules
The activity-level measurement rule, as can be subdivided into window-based activity (WBA), coefficient-based activity (CBA) and region-based activity (RBA), characterizes coefficients at different scales. In the coefficient grouping component, there are mainly three typical groupings, including single-scale grouping (SG), multi-scale grouping (MG), and no-grouping (NG). SG indicates that the same strategy is applied to fuse different coefficients between sub-images on the same scale.
The coefficient combination component mainly comprises of maximum rules (MR), weighted rules (WAR), and average rules (AR). MR can be given as:
| (1) |
where CF indicates the combined coefficient, and are the coefficients of two input images at the level of j. For WAR, and are combined by multiplying different weights and , that is
| (2) |
AR, which has the coefficient of input images averaged, is a special case of WAR.
| (3) |
The consistency verification component ensures that the same rules are applied to fuse the coefficients in the neighborhood.
5.2.2. Fusion levels
There are three different levels of image fusion: pixel level, feature level, and decision level. This categorization can be seen in Table 4 and Fig. 10 . Pixel level rules directly deal with the information acquired from each pixel of source images and then generates pixel values for the fused image correspondingly. Feature level rules focus on regional information and features such as texture and salient features. The fused image in the decision level is acquired through rules of fuzzy logic and statistics. Before rules in feature level and decision level apply to the source images, segmentation of the source images is needed. Compared to the pixel level fusion, feature and decision level fusion shows more advantages are less affected by noises and misregistration. Feature and decision level fusion also shows better contrast and lower complexity [231]. In the following sections, we will introduce fusion rules for multi-model image fusion, followed by validation metrics.
Table 4.
Three fusion levels.
| Decision level | Feature level | Pixel level |
|---|---|---|
| • Image segmentation | • Image segmentation | • Information acquisition of each pixel |
| • Fusion based on initial object detection and classification | • Fusion based on the properties | • Fusion of each pixel based on the information |
Fig. 10.
pixel-based and window-based fusion.
5.2.3. Fuzzy logic
Fuzzy logic-based rules belong to decision level fusion. These rules are usually used to solve challenges in blurry fused images. Mamdani and T-S models are two fuzzy logic models. The difference between them lies in the consequence parts. T-S models linearly mapped input variables into functions to form the consequence parts, while Mamdani models used fuzzy sets. The T-S model is more advantageous than the Mamdani model in regard to number of rules and accuracy.
In general, feature extraction through fuzzy logic algorithms is performed prior to fusion, which generates pixel-wise features C 1 and C 2 from two input images. The fusion procedure can be divided into three steps. In the first step, the fuzzy logic, which is usually comprised of four conditional rules, is used to label the individual pixels as following.
| (4) |
Then the new pixel-wise feature values can be calculated through:
| (5) |
where 1, 2, 3 corresponds to low, medium and high components, respectively. α and β are the mean and variance of each component. By incorporating the center average defuzzifier to process fuzzy outputs, the weight of fuzzy logic can be obtained.
5.2.4. Statistics model
The essence in statistics-based methods lies in the data-driven technique and high order statistics that can reveal the underlying pattern across multiple modes of data. Principal component analysis (PCA) [232], [233], [234], [235] together with Hidden Markow Tree (HMT) [236], [237], [238] are two typical examples of statistic methods in the field of multi-modal medical image fusion.
PCA is an orthogonal linear transformation that reveals the most valuable components of the input images. Let C 1 and C 2 be the two coefficients of the input images that can be denoted as:
| (6) |
| (7) |
where and (1 ≤ j ≤ M) are column vectors of two coefficients C 1 and C 2. The importance of components is related to the eigenvalues in the covariance matrix between C 1 and C 2. The covariance matrix can be computed through:
| (8) |
where E is the expectation of vectors, and corresponds to the average of C 1 and C 2 respectively, that is
| (9) |
| (10) |
Given the eigenvalues obtained from the covariance matrix as Y, the normalized weights w 1 and w 2 for C 1 and C 2 can be denoted as:
| (11) |
| (12) |
Therefore, the fused coefficient CF can be the combination of two input images:
| (13) |
The two-state HMT method, unlike PCA methods, can be deployed to model the coefficients. Two mixed Gaussian random distributions, as well as the hidden states, depict Intra-coefficients. The hidden states here refer to the one parent and four children coefficients. Given each coefficient denoted as C, the coefficient is obtained by probability density function:
| (14) |
when the coefficient Ci in the state n (n=0, 1), is the probability density function correspondingly. The fused coefficient is then given by:
| (15) |
5.2.5. Human vision system
Methods based on the Human visual system (HVS) aim at solving the fusion problem in the way of image recognition and comprehension. The system includes components such as visibility, smallest univalve segment assimilating nucleus (SUSAN) [239], and retina-inspired model (RIM) [240, 241].
The sharpness of an image can be quantified by visibility. Therefore, the images with higher visibility show lower blurriness. Given an image I of size h × w, then the visibility of the I can be mathematically expressed as:
| (16) |
where μ is acquired by calculating the mean grey value of the image I and α, the visual constant varying from 0.6 to 0.7.
SUSAN, proposed in [242], is a feature extraction algorithm inspired by HVS. SUSAN computes the feature of a pixel by considering a circular mask around the pixel. In the mask, the area consisting of pixels that have similar brightness to the nucleus, or the central pixel, is selected and is called Univalue Segment Assimilating Nucleus (USAN). Let the input image to be I and the circular mask with the radius , then simplest USAN can be given as:
| (17) |
where r 0 is the central pixel, r is a nominal depicting surrounding pixels. I(r)is the pixel value at r and L is the brightness difference threshold. The value of L, which specifies the range of pixel values to be considered, must be carefully chosen because extracted features are sensitive to it. In order to lower the sensitiveness, the distance between pixels is also taken into consideration, and the extended USAN function is:
| (18) |
where ρ is the distance scaling factor.
For components in intensity-hue saturation (IHS) decomposition methods, the RIM model can be used as the fusion rules. There are five layers in RIM. The first cone layer outputs an array of high-resolution cone photoreceptors with high resolution. The second layer is the extractor for spatial feature, while the third layers are horizontal cells. The fourth and fifth layers, which combine features, are bipolar and ganglion cells The RIM based image fusion rule can be demonstrated as:
| (19) |
where C 1 and C 2 stand for intensity components of two source images. h 1 and h 2 are the filters of feature extractors. The filter h 1, a high-scale spatial feature extractor, calculates the spatial difference between high-resolution and low-resolution. Filter h 2 combines the output of horizontal cells.
5.2.6. Validation metrics
Objective evaluation metrics are used to evaluate the efficacy of image fusion rules on improving the quality of the fused image. Widely used metrics includes spatial frequency (SF) [243], the ratio of spatial frequency error (rSFe) [244], wavelet entropy (WE) [245], Signal noise ratio (SNR) [246], mutual information (MI) [247] and directive contrast (DC) [248].
SF metric, which measures images’ activity, is generally used for PCA integrated HIS methods in multimodal image fusion. SF can be defined as
| (20) |
where CF and RF stand for column frequency and row frequency, respectively.
Based on SF, rSFe compares the of the fused image with and of the input images where SF’ can be extended as:
| (21) |
where MDF and SDF are the main diagonal SF and the secondary diagonal SF. rSFe can then be formulated as:
| (22) |
Calculated by multi-scale entropy, WE can be given by:
| (23) |
where i is the resolution level, pi is the density distribution derived from the energy of the detail signal Ei and the total energy ET
| (24) |
In [246], the authors proposed SNR-based image fusion rules. The proposed fusion method can be formulated in the following form.
| (25) |
for a specific region k. A(t) is the activity level of that, while Mt is the total number of pixels. The probability of pixel activity pl can be calculated through:
| (26) |
where w is the weight of SNR from the image, N is the number of decomposition levels and di(j 1, j 2) is the detailed wavelet coefficient.
MI is deployed as the fusion rule for WT-based multi-modal medical image fusion. MI between the input images I 1 , I 2 and the fused image I F, is maximized to give CF, which is acquired by:
| (27) |
DC measures the difference between the pixel and its neighbors. The ratio between the high-frequency intensity I H and the low-frequency intensity I L is the intensity contrast DC.
| (28) |
5.3. Decomposition and reconstruction
For the multi-modal image fusion of neuroimaging, the selection of various decomposition and reconstruction methods influence the fusion procedure and outcomes. In this survey, we shall discuss seven popular methods: (i) RGB-IHS; (ii) Pyramid representation; (iii) wavelet-based approach and its variants; (iv) multi-resolution analysis; (v) sparse representation; and (vi) salient features.
5.3.1. RGB-IHS
The intensity-hue-saturation (IHS) model [249] helps transform the original image in RGB color space to hue, saturation, and intensity channels. This RGB to IHS procedure is calculated by simple equations. Besides, the reconstruction is carried out by inversed transformation (IHS to RGB).
For the RGB->IHS procedure, the intensity of each input image was estimated by the following equations:
| (29) |
Then, we consider three conditions: C 1: B<R,G; C 2: R<B,G; C 3: G<R,B.
The hue value is obtained by
| (30) |
The saturation value was yielded from the equation
| (31) |
On the other hand, after the revision was performed on the I, H, and S components, we can convert from IHS color space to original RGB space based on three various conditions:
For C 1, we have
| (32) |
For C 2, we have
| (33) |
For C 3, we have
| (34) |
5.3.2. Pyramid representation
Pyramid representation (PR) is commonly employed in image fusion. PR is a typical multi-scale signal representation approach that can be used for 1D signals, 2D images, etc. Fig. 11 shows a toy example using the cameraman picture. There are two pyramids commonly seen in practice: lowpass pyramid and bandpass pyramid. The former smooths the image and then under-samples the smoothed image. The latter generates the difference between images at adjacent levels and carry out image interpolation between neighboring levels of resolution.
Fig. 11.
A toy example showing pyramid representation.
Let us assume we have input image I. We have a series of pyramid representations of this input image I as
| (35) |
Suppose Fk is the filter and down-sampling, PR can be obtained by
| (36) |
| (37) |
where rk is the residual image of I at i-th level, and k is in the range of
| (38) |
where K is the maximum iteration number.
After modification on the PR of input images, the fused image IF is obtained by inverse pyramid transformation (IPT) as
| (39) |
The process of PR based fusion is shown in Fig. 12 .
Fig. 12.
Diagram of two-level pyramid representation based fusion.
5.3.3. Wavelet-based method
Wavelet transform (WT) based fusion is one of the multi-scale analysis methods. The idea is simple, as shown in Fig. 13 . WT will decompose the input images into low-frequency (LF) and high-frequency (HF) subbands. The corresponding image fusion rules will be applied to fuse LF and HF subbands. Finally, the fused image is yielded by the WT reconstruction technique [250].
Fig. 13.
Diagram of wavelet-based fusion methods.
Continuous wavelet transform (CWT) decomposes a square-integrable function S(t) as follows
| (40) |
where
| (41) |
Here, the wavelet Φc, b(t) is calculated from the mother wavelet Φ(t) by translation and dilation. The dilation factor c and translation factor b are all positive numbers.
On the other hand, the discrete wavelet transform (DWT) captures both spatial information along x and y axes, by discretizing above two equations
| (42) |
| (43) |
Then, suppose a low-pass filter (LPF) and a high-pass filter (HPF) are created, we have
| (44) |
| (45) |
where n is the discrete version of time t, e represents the coefficients, a and d correspond to approximation and detail, respectively. The symbol ↓ represents the downsampling operation.
| (46) |
The above decomposition process can be iterated with successive approximations being decomposed in turn, so that one signal is broken down into various levels of resolution.
When I(n) is extended to be a 2D brain image I(m, n), the 1D-DWT is applied to row and column directions separately. The approximation (a) and detail (d) subbands now expand to four subbands: approximation subband (a), horizontal subband (h), vertical subband (v), and diagonal subband (d), as shown in Fig. 14 .
| (47) |
| (48) |
| (49) |
| (50) |
Fig. 14.
Diagram of 2D-DWT.
2D-DWT can even be generalized to 3D-DWT. A straightforward implementation is to apply 1D-DWT to row, column, and slice directions, respectively. Fig. 15 shows an example of carrying out 3D-DWT to a cubic image. Instead of using a, h, v, and d, we now use LLL, LLH, LHL, LHH, HLL, HLH, HHL, and HHH to represent the eight subbands in 3D-DWT [251]. Here L and H represent the result after LPF and HPF, respectively.
Fig. 15.
Illustration of performing 3D-DWT on a cubic image.
5.3.4. Variants of wavelet-based analysis
The DWT can provide better performances than traditional signal processing techniques, but it lacks translation-invariance and directional selectivity. Hence, new variants of wavelet-based techniques have been applied to multi-modality image fusion.
The stationary wavelet transform (SWT) can solve the shift-variance problem by getting rid of the downsampling operation from ordinary DWT. SWT can provide more details and texture information than DWT. Prakash and Khare [252] fused CT and MR images based on SWT by Modulus Maxima. Pawar and Kadam [253] used SWT and convolutional sparse representation for multi-modal image fusion.
The DWT calculates each decomposition level by passing only the previous approximation coefficients to quadrature mirror filters (QMF). Nevertheless, the discrete wavelet packet transform (DWPT) [254, 255] passes all coefficients (both approximation and detail) through QMF to create a full binary tree. Sreekala and Kuncheria [256] used WPT to implement misaligned image fusion. Shah, Merchant [257] combined curvelet wavelet and the WPT method to carry out image fusion.
To increase directional selectivity, the dual-tree complex wavelet transform (DTCWT) used two separate two-channel filter banks. The scaling and wavelet filters in the dual-tree cannot be selected arbitrarily. In one tree, the wavelet and scaling filters produce a wavelet and scaling function, which are approximate Hilbert transforms of those generated by another tree. At each level of 2D DTCWT, it produces a total of six directionally selective subbands (±15°, ±45°, ±75°). Fig. 16 presented a comparison of DWT, SWT, DWPT, and DTCWT.
Fig. 16.
Comparison of different wavelet-based decomposition techniques.
Lift scheme (LS) is a solution to reduce computation time and accelerate the design and application of DWT [258]. The idea of LS is to factorize DWT with finite filters, into a sequence of ordinary convolution operators, which are called “lifting steps” [259]. This procedure can reduce the arithmetic operations by almost two. Mathematically, the analysis filters can be formulated in the form of polyphase matrix
| (51) |
This polyphase matrix P(z) is a 2 × 2 matrix, which contains the analysis LPF and HPF, each split up into their even and odd polynomial coefficients and normalized.
| (52) |
P(z) is then factored into a series of 2 × 2 upper triangular matrices (UTM) and lower triangular matrices (LTM), each with diagonal entries equal to 1. UTM contains the coefficients for prediction, while LTM contains the coefficients for updates. Prakash, Park [260] used LS based biorthogonal wavelet transform to realize the multi-scale fusion of multimodal medical images. Haouam, Beladgham [261] used the level-set method and LS-based CDF wavelet to compress magnetic resonance images.
5.3.5. Other multi-resolution analysis
Apart from the wavelet analysis, scholars have also proposed other multi-resolution analysis (MRA) methods for multimodal image fusion. The wavelet does not work well in detecting smoothness along the edges, and it lacks directional resolution because it only has three high-frequency subbands. The contourlet transform (CT) utilizes the contour segments to capture the geometrical structures of the input images. The procedure is two-stage: First, the Laplacian pyramid (LP) performs multi-scale decomposition, capturing point discontinuities. Second, directional filter bank (DFB) yields directional information and forms those point discontinuities into a linear structure. The flowchart of CT is shown in Fig. 17 .
Fig. 17.
Diagram of contour transform.
Similar to stationary wavelet transform developed from discrete wavelet transform, nonsubsampled contourlet transform (NSCT) [262] was also developed due to the shift-variance of CT. Ramlal, Sachdeva [263] proposed an improved multimodal medical image fusion scheme, via a hybrid combination technique of NSCT and SWT. Li, Wang [264] presented a new practical medical image enhancement based on NSCT. Wang, Zhao [265] used NSCT and simplified-spatial frequency-pulse coupled neural network to develop a multi-modal functional/anatomical medical image fusion framework.
Wavelets also fail to capture the geometric regularity along the singularities of surfaces, because of their isotropic support. Shearlet transform (ST) is one of the best sparse directional image representation methods. Fig. 18 showed the ST coefficients, where the input image is Fig. 16(a). Li, Wang [266] proposed a novel medical image fusion approach based on nonsubsampled ST (NSST). Vishwakarma and Bhuyan [267] offered a new image fusion framework via adjustable NSST. Akbarpour, Shamsi [268] suggested a novel combination of NSST and principal component averaging.
Fig. 18.
Example of coefficients of shearlet transform.
5.3.6. Sparse representation
Different from standard multi-scale analytic methods, the sparse representation (SR) assumes that both high-frequency and low-frequency components share the same set of sparse coefficients [269]. SR based fusion method root from compressed sensing. Nowadays, there are some famous variants of SR, such as group sparse representation and joint sparse representation (JSP) [270], the diagram of which is shown in Fig. 19 .
Fig. 19.
Diagram of JSP based fusion method.
From Fig. 19 we can observe the detailed steps of JSP [271]. First, both input images I 1 and I 2 are transformed into vectors via sliding window
| (53) |
where SW represents the sliding window method. An over-complete dictionary sparsely represents the vectors of both images
| (54) |
where represents the intersection of vectors of images I 1 and I 2, and represents their differences. The two variances and can be obtained by
| (55) |
| (56) |
where E stands for the over-complete dictionary, and TC and TU denote the sparse coefficients (SC) of and .
Afterward, SCs from both input images are combined using fusion rules as
| (57) |
where TF stands for the fused SC. The over-complete dictionary can generate the fused vectors
| (58) |
where stands for the fused vector.
Finally, we can transform to image space and get the fused image IF.
5.3.7. Salient feature
Salient feature based fusion approaches are a type of novel methods, with the benefits of shift-invariance, low-cost computation, and saliency-feature preservation. The edge-preserving filter (EPF) is an important research method among all salient feature fusion approaches. Scholars have proposed many EPF approaches, such as local extrema scheme [272], multi-scale edge-preserving decomposition [273], edge-preserving smoothing pyramid [274].
Fig. 20 shows the diagram of the EPF-based image fusion approach, where BL and DL represent the base layer and detailed layer, respectively. The procedures of EPF-based image fusion approach are listed below:
Fig. 20.
Diagram of EPF based fusion approach.
First, both input images are decomposed into base layer (BL) and detailed layer (DL) by edge-preserving filters. The BL of each input image at different scales is obtained by
| (59) |
where k ∈ [1, 2] means the index of two input images, and j is the level of decomposition, and Fj represents the EPF at j-th level. Similarly, the DLs are obtained by
| (60) |
Afterward, the BL and DL fusion rules and are applied to corresponding BLs and DLs, respectively;
| (61) |
| (62) |
where Bf and Df are the fused BL and DL, and J is the maximum decomposition level. Finally, the fused image IF is recovered by the summation below or other weighted equations.
| (63) |
6. Multimodal imaging data fusion: assessment
Assessment is an essential component of multimodal data fusion that provides perspective into the quality of fusion results. There are two main forms of qualitative assessment: subjective quality assessment and objective quality assessment. In this part, we will review the conventional techniques and metrics applied in both forms.
6.1. Subjective quality assessment
Subjective assessments are established as a reliable form of quality evaluation in image fusion [275]. Professional subjective assessments based on medical and radiology expertise are widely applied in neuroimaging studies. Conventional subjective quality assessments are in the form of surveys with a set predesignated of questions and solutions, which represents a non-linear mapping to quantitative quality metrics [276]. Typical questions in neuroimaging fusion are the number of artifacts or distortion, while typical solutions can be in the form of basic descriptors (e.g. none, minimal, some, substantial) associated with continuous scores. Subjects are required to answer questions with respect to a set of images. It is ideal for image sets between subjects to overlap in order to ensure multiple surveys for the same image. With a set of high-quality fused images as references, the survey scores are then converted to difference scores with respect to scores of reference images. The difference scores are then converted to Z-scores and rescaled to the required range for analysis [276]. The double stimulus method is commonly applied to account for the potential misalignment in quality scales between different image sets or different assessment sessions. In the double stimulus method, both surveys and references are randomly included in the test set [277]. Conventional quality metrics obtained using these methods are the mean opinion score (MOS) and difference mean opinion score (DMOS). Data noise elimination and outlier removal for images & subjects are then performed after obtaining the scores. Subjective quality assessments are vulnerable to both intra-expert and inter-expert variability, while the requirement for expertise significantly increases the cost of assessments. For extensive studies, considerable efforts are required to standardize and maintain assessment protocols for optimal consistency [278].
6.2. Objective quality assessment
In contrast to subjective Quality Assessment (QA), which involves a complex organization of observers and strict tests, objective QA only requires the computation of a single numerical score. Objective QA should be consistent with subjective QA, and often relies on statistical properties of the images, or even stochastic modeling of the Human Visual System (HVS) [279].
Objective QA can be classified according to the reference (distortion-free) image II used in the score computation. The best-case scenario is the full-reference QA [280], in which II, with the same resolution as the fused image IF, is known. This approach is, by far, the most widely extended. However, this full-reference is rarely available in practical applications. For example, in multi-spectral satellite imaging or medical imaging, some image modalities are acquired at a lower resolution and then fused with higher-resolution gray-level images. This is known as the reduced-reference case, where II is sometimes available at a coarser resolution and sometimes just a set of extracted features, and the QA is performed in this feature space. Finally, a third case is when no reference is available at all [281]. In this case, IF is considered by itself, and measures like entropy and contrast are computed using only the information contained within.
Depending on the type of quantification used, the metrics can be classified into “signal distortion” and “salient feature” categories. The former uses strict mathematical theory to assess quality, e.g. with entropy, standard deviation, etc. The later is grounded on a modeling of the HVS to assess salient feature transferred from II to IF.
6.2.1. Signal distortion based metrics
The first signal distortion metric is a commonly used statistic, the standard deviation (STD) between the input image II and IF. Let us consider the images as a function II(x, y) and IF(x, y) defined in the range and , with M the number of rows and N the number of columns in the image. The STD is computed as:
| (64) |
with being the mean intensity of the fused image.
Similar is the Root Mean Squared Error (RMSE), also widely used in many applications, included objective QA:
| (65) |
An additional measure based only on pixel intensities is the Sharpness (SP), that reflects the level of detail transferred to IF:
| (66) |
And finally, the Peak Signal-to-Noise Ratio (PSNR), which is perhaps the most widely used objective quality assessment:
| (67) |
where L is the number of intensity levels, typically 256 for 8-bit images.
Now there are many other measures based on information theory and entropy [282]. To perform this computation, let us note P(i∨I) as the ratio of pixels with gray value equal to i over the total number of pixels N × M of image I. We define the Entropy (EN) as:
| (68) |
The difference of entropy (DEN) [282] quantifies the differences between IF and II. The smaller, the better:
| (69) |
Another possibility when a reference image is available is to compute the Cross-Entropy (CE) between the reference II and the fused image IF:
| (70) |
From CE we can derive the Overall Cross-Entropy (OCE), which measures the entropy between several input images and the fused image. The formula below refers to K images, but the most common case is the one that uses just two images, I 1 and I 2:
| (71) |
In Sheikh and Bovik [276], the authors propose the Visual Information Fidelity (VIF) measure. This measure quantifies how much of the information at II can be extracted from IF. It is based on a Gaussian Scale Mixture (GSM) random field source model and a modelling of the HVS as a distortion channel. It uses the information MI(X, Y) between two given images, and sums over all subbands as in:
| (72) |
where and represent the information that could ideally be extracted by the brain in the reference and fusion images. , , represent N elements of the GSM model for the visual model (C), reference image (II) and fused image (IF) in subband j. A more detailed explanation of the computation of these coefficients is found at Sheikh and Bovik [276].
However, Hossny, Nahavandi [283] propose a new formulation of mutual information (MI), based on the joint statistical distribution of two random variables. To do so, we note the mutual entropy as:
| (73) |
where P(i∨X) and P(j∨Y) are the probability of intensity i and j on the images X and Y respectively, and P(i, j∨X, Y) is their joint probability. Note that , identical to Eq. (68).
The Mutual Information (MI) is then obtained in [284] as:
| (74) |
with to obtain them with no reference images. However, the H(IF, I 1) and H(IF, I 2) are not guaranteed to be on the same scale. The solution is the Normalized Mutual Information, devised in [284] and re-formulated in [283] for the fusion of two source images I 1, I 2 as:
| (75) |
The Spatial Frequency (SF) [285] is often used to measure the overall clarity of the fused images. It is obtained from the row frequency (FR) and column frequency (FC) of the image I as follows:
| (76) |
| (77) |
and the SP is obtained by the harmonic mean of these two measures.
| (78) |
6.2.2. Salient feature based metrics
Salient feature metrics assess whether the salient features of the source images have passed to the fused image, following the hypothesis that the HVS is highly adapted for the perception of structural information. The most widely used measure in this category is the Structural Similarity (SSIM) index [281], which is based on the degradation of structural information.
The SSIM compares local patterns of pixel intensities, based on luminance l, contrast c and structure s:
| (79) |
weighted by some exponential variables that were set to in the original paper [281]. Several expressions are provided for s(X, Y), l(X, Y) and c(X, Y), with certain constraints, and after substituting on the previous equation, it becomes:
| (80) |
where C 1 and C 2 are two constraints of the model, expressed by , and K 1 ≪ 1, K 2 ≪ 1. The notation μI and σI corresponds to the mean intensity and unbiased standard deviation of the intensities of I, as in previous equations, and is the covariance matrix of the intensities of II and IF.
Finally, Mittal et al. proposed the Natural Image Quality Evaluator (NIQE) [286], a QA model based on the construction of a Multivariate Gaussian Model (MGM) from a corpus of undistorted images with mean μM and covariance matrix σM, N(μM, σM). These features are “quality aware”, and the quality of IF is estimated as a distance between the statistics of the model and the fused image.
| (81) |
with and the mean vector and covariance matrix of the MGM of IF.
7. Multimodal imaging data fusion: Benefits
Multimodal neuroimaging and the fusion of multimodal data tackle the challenges of neuroimaging and the fundamental limitations of individual modalities, and therefore provide significant benefits to the overarching aim to achieve higher image quality and reveal brain physiology. This part will review some of the main benefits with specific fusion examples.
7.1. Combination of physiological aspects of brain structures and processes
7.1.1. MR-PET
MR-PET refers to a functional metabolic and molecular multimodal imaging method integrated through a combination of MRI and PET. It has the potential to achieve the maximum complementary advantages with the examination function of both PET and MRI [287].
MRI can not only display structural details through multi-parameter sequences but also perform a variety of functional imaging. In other words, it can be seen as a means of anatomical imaging. However, compared with PET, MRI still has certain limitations in metabolite imaging. PET imaging can show trace amounts of radiolabeled molecules, but its image resolution is poor and the anatomical structure is not clear. It is the complementary characteristics of MRI and PET that led to the birth of MR-PET imaging, which not only has high soft-tissue contrast and resolution but also can provide valuable functional information [288].
At the radiological society of North America meeting in November 2006, Siemens presented the first MR-PET images of the brain from their diagnostic machine. During the imaging process of MR-PET, PET and MR imaging can be performed simultaneously with minimal interference; the PET scan detects the accumulation of fluorodeoxyglucose, while multiple sequences of MR images were obtained [289].
7.1.2. Combination of different contrast images
In neuroimaging studies of MRI, the common approach tends to acquire T1- and T2-weighted anatomical, T2*-weighted functional data within the same session and then make a combination of these different contrasts for further exploration and research. Here, T1 refers to the recovery time of longitudinal magnetization, and T2 refers to the decay time of transverse magnetization. Both of them are special values associated with the spin of the nucleus in the tissues.
In humans, T1 and T2 values of diseased tissue and normal tissue are different, so diseases can be diagnosed by nuclear magnetic resonance imaging [290].
Magnetic resonance images are presented in different shades of gray, reflecting differences in the intensity of magnetic resonance signals, or in the length of relaxation times T1 and T2.
Pure T2 dephasing is intrinsic to the sample. While T2* dephasing is relevant with true T2, field inhomogeneity (T2M) and tissue susceptibility (T2MS) as shown in Eq. (82).
| (82) |
According to the common neuroimaging protocols, T1-weighted scans have good resolution and gray-white matter contrast, so it is better for observation of anatomical structure. T2-weighted scans perform well in showing histologic lesions, so it is often used for checking permanent brain injury. T2* is mostly used in scanning brain activity. An increase in T2* weighted signal between baseline and an active condition is associated with brain activation in studies using blood oxygenation-level dependent (BOLD) fMRI. For instance, as displayed in Fig. 21 , brain regions become oxygen-rich after activity, which leads to a decrease of the Hbr/HbrO2 ratio and the increase of fMRI signal. Consequently, for T1, T2 and T2*, each method provides a physiologically and physically filtered view on one or more brain processes of interest [291].
Fig. 21.
Comparison of Hbr/HbrO2 ratio in brain capillaries before and after activity.
Thus, combining different contrasts has the general merit of getting a more comprehensive physiological view on brain processes than utilizing just one imaging method alone.
7.2. Improving temporal/spatial resolution
In Fig. 4, the spatial and temporal ranges for the most widely used non-invasive functional imaging methods are presented. Functional MRI, as the non-invasive functional imaging method, has the highest spatial resolution though the temporal resolution is relatively low compared to neuronal population dynamics. MEG and EEG can measure magnetic and electrical changes on millisecond time scales while the spatial resolution is beyond seven millimeters [15]. Ultra high-field MRI has been widely used for the mesoscopic level of neuroscience in humans [292], [293], [294]. As a consequence, the Spatio-temporal resolution can be improved by combining different imaging methods, especially through the combination of one modality that has a higher temporal resolution with another modality of superior spatial resolution. The fused images are called validation when Spatio-temporal resolutions are similar.
7.2.1. Nominal resolution and effective resolution
When considering improving spatio-temporal resolution, the resolution of each modality is not of the biggest concern. Each modality's effective resolution and the additional effect after the combination of data from different modalities should be considered instead. While the nominal resolution is generated according to fixed physical parameters of instruments, the effective resolution depends on the information of the data instead. In fMRI, field-of-view and the k-space acquisition matrix determine the nominal spatial resolution, while the effective resolution is largely affected by fMRI sequences and spread of the hemodynamic response [295]. Because of the slow development of hemodynamic response, the effective resolution is lower than nominal temporal resolution in fMRI, while the latter resolution is determined by the repetition timeTR.
The nominal temporal resolution of electrophysiological methods is usually in the millisecond range. The effective resolution, however, can vary from tens of milliseconds to hundreds of milliseconds due to the slow evolution of neuronal field potentials and statistical detectability. Also, many repetitions have to be implemented before a detectable signal comes up for the evoked potentials in EEG, while a detectable hemodynamic response can be acquired through a single stimulus in fMRI.
7.2.2. EEG-fMRI
EEG-fMRI, as the combination of EEG and fMRI, turns out to be the typical example of improving spatio-temporal resolution through multimodal neuroimaging. Epilepsy research facilitated the development of simultaneous EEG-fMRI recording [296]. In the case of localizing epileptic foci, simultaneously recorded fMRI help by connecting epileptic foci with interictal epileptiform discharges (IED) acquired by EEG. Simultaneous EEG-fMRI can also be utilized as neuronal oscillations in neuroscience [209, 297]. In these studies, the process of analyzing the recorded fMRI data takes power in the frequency domain of the EEG signal as a variable.
7.3. Distortion correction
An MRI machine uses a static, homogenous, external magnetic field B0, a radio-frequency magnetic field B1, and three orthogonal gradient fields. However, magnetic field variation may result from imperfections of the main magnetic field coil, the exciting gradient coils, and susceptibility of the tissues [298]. Field inhomogeneity is one of the causes of blurring or spatial shifts in MRI images. A typical region of distortion is the borderline between tissue and air, where a difference in magnetic susceptibilities exists. A standard process in neuroimage processing and fusion is the registration of multimodal images to standardized templates. Hellier and Barillot [299] introduced 3D non-rigid registration of multimodal images by mapping the deformation through cost optimization. The registration process provides the foundation for higher quality fusion with less distortion.
The fusion of the main imaging sequence with supplementary sequences is also widely employed for distortion correction. As mentioned previously, a well-studied example is EPI, a high temporal resolution MRI technique that can collect multiple images per second. The high imaging frequency of EPI acquisition results in geometric distortions to its images. Acquisition of the static Bo fieldmap before or after EPI can provide additional information on local static field inhomogeneity and magnetic susceptibility. Holland, Kuperman [300] developed an efficient non-linear registration method by acquiring EPI of opposite phase encoding polarities, while Oh, Chung [213] applied PSF modeling to map the distortion field.
Another example is the fusion of MRI and PET images in MR-PET systems. In these systems, apart from the combination of structural and metabolic information, the fusion of MRI and PET also allows enhanced distortion correction. In simultaneous MP-PET systems, motion distortions of PET can be corrected with the volumetric information provided by continuous EPI acquisitions or navigator sequences [301, 302]. Joint reconstruction of MRI and PET utilizes dependence between the two modalities for image reconstruction of one mode based on the other. Reconstructed images show sharper edges and less distortion [303]. This entire process can also be formulated as a single optimization problem [301].
8. Multimodal imaging data fusion: atlas-based segmentation
Segmentation has been one of the main challenges faced by neuroimaging experts during the last decades. It refers to the process of tagging image pixels or voxels with biologically meaningful labels, such as anatomical structures and tissue types [304]. The segmentation task and data analysis are very often guided in most of the neuroimaging experiments by atlases that provide standardized, or stereotaxic, 3D coordinate systems for statistical data analysis and report of findings [305]. One of the earliest works in anatomical brain standardization was by Talairach, Tournoux [306], who developed a 3D coordinate space for the brain to assist surgical techniques. This work was later updated by a full printed three-dimensional atlas of the human brain, which was especially beneficial for clinical studies, electroencephalographic investigations, and statistical computations [307]. Tzourio-Mazoyer, Landeau [308] developed an anatomical parcellation approach with the spatially normalized single-subject high-resolution T1 volume provided by the Montreal Neurological Institute (MNI) [309]. These developments have contributed to better define the relationships between brain structures and their functions in modern human neuroscience research, as well as to reduce the variability found between different subjects.
The traditional approach for brain image segmentation is the manual annotation or delimitation of the regions of interest (ROIs) by a trained expert [304]. This process is subjective and strongly influenced by expert performance. It is then of limited applicability since it is time-consuming, prone to error and difficult to reproduce.
Automated segmentation methods have been developed during the past decades. These methods can be classified into basic tissue classification and anatomical segmentation procedures [310]. Tissue classification methods segment a 3D image of the brain into different tissue types (Grey Matter, White Matter, CSF, etc.), while also correcting for spatial intensity variations (also known as bias field or RF inhomogeneities). These methods normally incorporate probabilistic tissue atlases to improve their performance and have been successfully automated for fMRI studies as well as integrated into several neuroimaging toolkits such as FSL FMRIB's Automated Segmentation Tool (FAST) [311] and Statistical Parametric Mapping (SPM) [312]. However, anatomical segmentation procedures are much more difficult to automate, since different anatomical structures that consist of different tissue types may exhibit similar signal properties. This observation is the reason why automated anatomical segmentation of the brain into non-homogeneous regions needs to be guided by atlases.
8.1. Single-subject atlas-based segmentation
For atlas-based segmentation, an atlas is defined as the combination of a brain volume (atlas template) and its corresponding coregistered segmented volume (atlas labels). The atlas-based segmentation method registers the atlas template to the target image, and then the atlas labels are propagated to the target image using an effective image warping method [313].
Let I(x) be the volume to be segmented, with representing a 3D voxel coordinate vector. For the purpose of single-subject atlas-based segmentation, a grey level atlas volume A(x), a labeled volume
| (83) |
where L is the number of anatomical regions or labels defined in the atlas and, optionally, a probabilistic atlas P(x), all within the same spatial coordinates (the atlas space), are required. The segmentation process consists of a registration step and a label propagation step.
In the registration step, the atlas volume A(x) is registered to the input image I(x) by means of a spatial transformation T(x), which optimizes a given cost function. In the label propagation step, labels defined in the tagged volume L(x) are assigned to each of the voxels of the input image I(x) by applying the normalization transformation T(x) to L(x), thus spatially aligning the atlas to the input image I(x). The atlas probability maps P(x) are normally used for MRI since they enable to improve the segmentation accuracy of T1-weighted MRI into grey matter, white matter and cerebrospinal fluid tissues with higher accuracy when the regional tissue densities need to be quantified and compared for between different groups of subjects [314].
Medical image registration is routinely used in neuroimaging. It aims at determining the spatial alignment between images of the same or different subjects by means of optimizing a cost function, which measures the similarity between the transformed input image and the reference image (template) [315]. The registration process involves a spatial transformation being global rigid and affine transformations usually enough for intra-subject image registration [313]. However, the atlas-based segmentation task requires intersubject matching or registration of an input image to an atlas image and, as a consequence of the variability of the anatomical structure across different subjects, non-rigid registration algorithms are widely used.
8.2. Multi-atlas segmentation
Single-subject atlas-based segmentation approaches suffer from a reduced ability to capture the variability of the spatial distribution of anatomical structures across different subjects. Multi-atlas segmentation was introduced in some pioneering works [310, 316, 317] to address this problem and to offer superior segmentation accuracy.
Fig. 22 shows a block diagram of a multi-atlas segmentation method. Instead of a model-based average atlas representation, a number of expert-annotated image volumes of different subjects are required. The input image is coregistered to each one of the different atlases available, and label propagation is performed. The final label is obtained for each voxel of the input image through a label fusion technique. In a multi-atlas segmentation approach, each atlas is used in the parcellation of the input image separately, since they are not summarized in a single probabilistic model.
Fig. 22.
Block diagram of multi-atlas segmentation based on image registration, label propagation, and label fusion.
8.2.1. Atlas propagation
For the purposes of reviewing the existing methods for atlas propagation and atlas fusion in multi-atlas based segmentation, a similar notation to the adopted ones in [318, 319] will be used. These works have addressed the atlas-based segmentation problem as a classification task where the atlas is the training set, and the training is associated with the process of computing the registration between the image and the atlas.
Let I(x) be a 3D image to be segmented into L different classes belonging to the atlas label set . Multi-atlas segmentation methods use K 3D atlas images Ak(x) and their corresponding atlas labels
| (84) |
The coordinate transformation Tk : R3 → R3 defines a mapping from the coordinates of the k atlas Ak(x) to the target image I(x). Once the registration transformation is obtained, the input image I(x) is automatically segmented for each one of the K atlases available by applying a label propagation technique, thus obtaining K different segmentations
| (85) |
These candidate segmentations must be combined later into a final estimated segmentation Is(x) ∈ Λ by applying a label fusion technique, as shown in Fig. 22.
Once each of the K atlas images has been coregistered to the input image, the labels are then propagated to the space of the input image in a process called label propagation. Label propagation applies the registration transformations to map each of the available atlas volumes Lk(x) to the input image, and an image warping technique preserves the discrete nature of the labels. Note that the transformations Tk: applied to the atlas label volumes Lk(x) are 3D continuous transformations that do not match grid points in the atlas space to grid points in the target input image. The purpose of label propagation is to perform the interpolation of the transformed atlas label volume for grid points in the target image space [320].
Among the different label propagation techniques, the most widely used are nearest-neighbor interpolation [310] and linear interpolation [316, 320]. Nearest-neighbor provides the unique label of the nearest atlas grid point. Partial volume interpolation, a technique that was first introduced in [321] for image registration, is a more sophisticated method for label propagation that takes into account the contribution of adjacent grip points to the final propagated label. During the last decade, several improvements have been proposed and reviewed for label propagation in multi-atlas segmentation [304]. Most of them incorporate additional information to improve the performance by augmenting the information with a tissue consistency map in nearest-neighbor interpolation [322] or introducing signed distance maps of the original atlas labels [323, 324].
8.2.2. Atlas fusion
The final step in atlas-based segmentation is atlas fusion. After registration and label propagation for each of the K available atlases, a final unique segmentation Is(x) ∈ Λ is obtained by merging the information provided by each of the individual atlas-based segmentations into a single segmented image. This approach has been shown to be more accurate than an individual atlas segmentation [316] in the same manner as a combination of classifiers is generally more accurate than a single classifier in many pattern recognition scenarios [325], [326], [327], [328], [329], [330].
The conventional method for combining individual segmentations is an equally weighted majority voting framework [319]. A more sophisticated approach estimates the performances of the individual atlas segmentations and combines them by weighting them according to their estimated performance. Both approaches are described and discussed in this subsection.
Majority voting atlas fusion.
The combined multi-atlas segmentation output Is(x) ∈ Λ for a voxel sample x given the set of K single atlas segmentations , with , is obtained through the vote rule decision function
| (86) |
where the Q function is defined to be
| (87) |
Atlas label fusion based on majority voting considers all the segmentations equally accurate, and no prior knowledge of segmentation performance is required.
Performance weighting atlas fusion.
Majority voting equally weighs the individual atlas segmentation without using a segmentation performance model. In order to improve the performance of the multi-atlas segmentation, the combined segmentation output Is(x) ∈ Λ should be the class maximizing the probability, given all the individual segmentations , with and some available segmentation performance model P [319]:
| (88) |
Using the Bayes rule, and assuming independence of the individual classifiers, simplifies the problem and enables us to find an optimal segmentation based on a model of segmentation performance. Rohlfing, Russakoff [319] proposed different classifier performance models and used an expectation-maximization (EM) algorithm that simultaneously estimated the performance parameters of the segmentations and provided an estimation of the unknown ground truth. This novel strategy enabled us to learn classifier performance parameters and to adopt weighted atlas combination for atlas label fusion in multi-atlas segmentation following a novel supervised training stage [331, 332].
9. Multimodal imaging data fusion: quantification
The application and improvement of the quantification in medical image allows to increase characteristics such as sensitivity and specificity, among others, obtaining more accurate patient's diagnoses. To perform this analysis, the emission computed tomography (ECT) is used, since it is the most important medical imaging modality in nuclear medicine. More specifically, the two image modalities analyzed are positron emission tomography (PET) and single-photon emission computed tomography (SPECT), which differ in the radiotracer used and the nature of the emission measurement. It should be noted that the factors associated with quantification that affect PET and SPECT are also extrapolated to multimodal images. Furthermore, multimodal images can be of great interest to improve quantification. Both, PET and SPECT, have proven to be effective imaging techniques for the diagnosis and monitoring of treatments in different medical applications, offering information about biological processes, which is very important for the study of brain activity [333]. The level of intensity in these types of images is bound to physical parameters, such as cerebral blood flow, glucose or receptor binding among others [334, 335].
Since these modalities were conceived in the 50’s and 60’s Jones and Townsend [336], several improvements have been developed in terms of quantification. One of the most relevant improvements in this field is due to the development of dual-modality imaging in the 90′s [337]. This type of imaging allows the combination of complementing functional information (PET and SPECT) with structural information (computed tomography (CT) and magnetic resonance imaging (MRI)), leading to increases in sensitivity and specificity. The combination of different characteristics that can be observed from each type of image allows us to obtain a better understanding of the structure and function of the human body. For example, PET/MRI combination offers remarkable advantages compared to the use of PET/CT since CT radiation dose is avoided and soft tissue images from MR could be acquired at the same time as the PET ones [338].
PET offers higher resolution and better quality than SPECT, particularly in PET/CT. Furthermore, it allows for easier quantitative measurements [335]. For example, sensitivity could be up to two orders of magnitude greater with comparable axial fields of view [339]. Nevertheless, the spatial resolution that can be obtained in both techniques tends to be low, due to its dependence on the radiation dose administrated to the patients. Also, ECT images are usually degraded by several factors, such as photon attenuation, partial volume effect or scatter radiation [337]. Moreover, spatial resolutions associated with structural images continue to be higher (≈ 1 mm) than in functional images (≈ 4–6 mm) [340].
Regarding quantification, an increasing number of relevant research aims to propose new quantification techniques or improve previously existing ones is observed in recent years. The possibility of quantification in nuclear medicine imaging is one of its successes. Due to the increasing use of nuclear images in therapies, the way of measuring functional images is changing. While historically, the use of relative and semiquantitative measures was typical, the recent application of absolute quantification is gaining support. One of the first steps was considered activity concentration and normalized uptake using the standard uptake value (SUV) [341]. Nevertheless, to reach quantitative imaging as a useful and potential tool, several factors must be addressed. As Zaidi and Hasegawa [339] mentioned among these factors are the system sensitivity and spatial resolution, dead-time and pulse pile-up effects, the linear and angular sampling intervals of the projections, the size of the object, photon attenuation, scatter, partial volume effects, patient motion, kinetic effects, and filling of excretory routes (e.g., the bladder) by the radiopharmaceutical.
In neuroimaging, PET and SPECT are a popular option to detect biomarkers associated with degenerative diseases. For Alzheimer's disease (AD), the accumulation of Amyloid-β plaques and tau aggregates can be detected [342, 343], while an increased diffusivity in the striatum and thalamus can be observed in Parkinson's disease (PD) patients. A well-defined quantification of these biomarkers would allow more precise and reliable diagnoses.
This section is organized as follows. Firstly, quantification both in PET and SPECT is analyzed considering the associated effects and their correction, especially those related to photon attenuation, scatter, and partial volume effects. Then, novel developments focused on cerebral medical imaging are highlighted.
9.1. PET and SPECT quantification
Even though PET scans could be evaluated qualitatively through the visual examination of the tracer uptake in cortical regions by a trained radiologist [344], the best way to analyze these images is quantitative. For that, automated or semi-automated localization methods can be used to evaluate regional levels of tracer uptake [345, 346]. Within automated studies, an essential step is to use spatial normalization to register subjects' brain to a standardized template space, so that all subjects in the study can be compared [347]. Another procedure of great importance is intensity normalization, which is the object of study in this analysis.
As Foster, Bagci [348] mentioned, several semi-quantitative and quantitative parameters that can be considered for intensity normalization exist, such as standardized uptake value (SUV), fractional uptake rate (FUR), tumor-to-background ratio (TBR), nonlinear regression techniques, total lesion evaluation (TLE) or the Patlak-derived methods. Nevertheless, the most widely used is SUV, which can also be used in SPECT [348]. The use of body weight (BW) in SUV has been discussed in the literature, and the general advice is to use more reliable measures such as body surface area (BSA) or lean body mass (LBM) [349, 350]. Moreover, a decay factor that depends on the particular radiotracer used may also be considered [351].
Several physiological and physical factors can influence the standardized uptake value obtained [348, 352]. Regarding physiological factors, some of them are weight and fat of the subject or blood glucose concentration, etc. Physical factors include partial volume effect (PVE), image manipulation (reconstruction, smoothing), and artifacts related to involuntary movements of the patient. The relevance of these factors lies in the variability of SUV. Studies carried out in the literature suggest that these factors may alter SUV in the range of 10%–30% [353, 354]. Nevertheless, the importance of correcting these effects depends on the clinical trial performed, since it can be a complex task and is not always worthwhile [355].
Moreover, these factors affect not only semi-quantitative measurements but also affect absolute quantification. There are several methods to consider within the absolute quantification, such as image reconstruction, effects correction or calibration to obtain the measured activity distribution [356, 357].
Among the mentioned factors associated with imaging quantification, we analyzed three effects and their respective corrections, since they are popular bias sources for PET and SPECT quantification. These effects are PVE, scatter, and photon attenuation effects.
9.1.1. Partial volume effect
Regarding the quantitative accuracy of PET image, partial volume effect (PVE) is the most relevant effect [358]. PVE is related to the finite spatial resolution of PET scanners and its discrete nature. That discrete nature causes a voxel to could be composed of more than one tissue, which leads to a final signal that is an averaged mix of signals, which is also called tissue fraction effect. Moreover, the larger the voxel size, the more tissues can coexist in it. This effect, along with the point-spread effect (or spillover), is the cause of image blur [354, 359]. Also, it is an effect that often causes confusion among clinicians and researchers when analyzing the image, since it is necessary to distinguish between loss of radioactivity due to PVE and the true loss of tissue.
Strategies to address this effect are called partial volume correction (PVC) methods. Distribution of both signal and noise are the main factors to consider for the selection of algorithms for PVC to apply [359]. Several PVC techniques have been proposed in the literature for improving image quality and quantitative accuracy in PET [360], [361], [362], [363]. Historically, these techniques have been classified in several forms. In this work, these methods are grouped as region-based (RB) and voxel-based (VB) techniques, based the most commonly used classifications [340, 355]. The most relevant differences between them are that VB methods produce PVE-recovered images while RB methods do not, and RB methods are associated with regions of interest (ROIs) while VB methods are applied at a voxel level considering the recovery of the spatial resolution of the system. Usually, RB techniques tend to be used more frequently than VB ones since they include regional homogeneity assumptions, simplifying the problem [359].
Table 5 shows a summary of the different existing techniques. Among the several PVC methods, the one associated with recovery coefficients (RC) is the simplest and most popular in clinical trials. Nevertheless, the most common technique used in neuroimaging is PVE correction based on anatomical images, which was developed for neuroimaging. The first technique proposed by Videen, Perlmutter [364] in 2D and later extended to 3D by Meltzer, Leal [365] was a segmentation in anatomy into two classes: brain and non-brain. Mullergartner, Links [366] proposed an improved version of this technique (known as GM algorithm) to use three instead of two classes: grey matter (GM), white matter (WM) and cerebrospinal fluid (CSF). Meltzer, Zubieta [367] added a fourth class to the previous algorithms in order to compensate for the real heterogeneity in GM tissue. The importance of this method is that PVE can be corrected between high and low intensity brain structures. Regarding the iterative reconstruction algorithms associated to VB methods, as V. Bettinardi, Castiglioni [340] noted, some of the best known and used is the Van Cittert (VC) algorithm, the re-blurred Van Cittert (R-VC) and the Richardson–Lucy (RL) algorithm. For broader coverage, readers are referred to reviews related to PVE [340] and iterative reconstruction methods [368].
Table 5.
Classification of PVC methods (RB = region-based; VB = voxel-based).
| Category | Method | Definition | Benefits | Limitations | Examples |
|---|---|---|---|---|---|
| RB | Recovery coefficients (RC) | They are numerical calculated in the image domain as a ratio of measured radioactivity concentration and actual concentration using spheres filled with a known radioactivity concentration | Fairly simple Practical Popular within clinical trials | More related to oncology | [369,370] |
| PET raw data | Instead of using the image domain, the sinogram is used | Low computational cost | Some methods require segmentation | [371], [372], [373] | |
| Geometric transfer matrix (GTM) method | Average uptake is estimated considering multiple ROIs using co-registered anatomical images | Commonly used Good accuracy | Requires segmentation Bias | [374], [375], [376], [377] | |
| VB | Image reconstruction | Spatial resolution is recovered within the image reconstruction process. It is highly recommended to use iterative reconstruction algorithms. The most relevant drawback of this is the high number of iterations needed | Increase the quality of reconstruction | Computational cost | [378], [379], [380], [381] |
| Image deconvolution | Spatial resolution is recovered by using a post-reconstruction restoration technique (deconvolution) that applies the point spread function (PSF). Iterative algorithms are commonly used | Compensation of spill-over effects | Computational cost | [382] | |
| Multi-resolution approach | Spatial resolution is recovered by using information from spatially coregistered high-resolution anatomical images, which allows transfer high spatial frequencies from anatomical to functional images | Adjust of radioactivity concentration Used in brain imaging | In poorly correlated areas the amount of artifacts increases | [383] | |
| Anatomical images | First, a segmentation of a coregistered anatomical image is done. Then, PVE correction is applied to the set of voxels associated with each region/class (tissue) | Highly used in brain imaging | Need segmentation | Two classes: [364,365] Three classes: [366] Four classes: [367] |
Finally, data extracted from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu) is used in order to show some PVE correction methods. This data consists of a patient diagnosed with Alzheimer's disease from which FDG-PET and T1 weighted MR images were taken, as shown in Fig. 23 . Both modalities are registered to respective templates of T1 and PET, and PET image was not normalized in intensity due to use only one subject in this manuscript. PVC techniques are applied by using PVElab [384, 385], a platform developed by the EU sponsored PVEOut project. PVElab can facilitate PVE correction through a graphical interface with several steps, including registration, segmentation, reslicing, optional application of an atlas (none in our case), and PVE correction. Three methods discussed above are those analyzed: 3D Meltzer technique [365] and GM algorithm [366] regarding VB methods, and the RB method proposed by Rousset, Ma [374].
Fig. 23.
Original PET (left) and MR (right) images.
The large difference between methods is shown in Fig. 24 a. The difference in the result between considering two (Fig. 24a) and three tissues (Fig. 24b) is highlighted. The method related to GTM (Fig. 24c) reflects intermediate results between the two images already commented.
Fig. 24.
Evolution of PVC techniques.
9.1.2. Attenuation effect
Another relevant effect to analyze is photon attenuation. This phenomenon occurs due to the interaction between the photon associated to the radiotracer and elements in the body such as tissue. Normally, this interaction leads to a scatter in the photon radiation. The probability that a photon experiences an interaction is represented by the linear attenuation coefficient [386]. The situation of this phenomenon differs between PET and SPECT imaging. In PET, two antiparallel photons are detected in the collimator, so the total tissue thickness crossed by them is the same body thickness that intersected the straight line between the two detections (line of response). Nevertheless, in SPECT imaging, the attenuation depends on the total tissue thickness crossed and its type (e.g., soft tissue, bone), which varies depending on where the points of emission and detection are [386]. Traditionally, this correction was much more commonly performed in PET imaging than in SPECT. Nowadays, it is done in both to be able to achieve much more accurate quantitative results, although it is more difficult to apply in SPECT.
Attenuation correction is necessary in order to obtain accurate quantitative results and is widely implemented. For this, an attenuation map is needed, which Zaidi and Hasegawa [386] defined as a representation of the spatial distribution of linear attenuation coefficients and delineates the body structures located in the image. Once the attenuation map is done, the reconstruction algorithm can be implemented with more information. Attenuation correction techniques can be classified into two major groups: transmission-less approaches and the ones based on transmission scanning. While the first group considers the measured emission data to develop the attenuation map or assumes a uniform distribution for the coefficients, the second one uses transmission data of external sources such as CT and MRI, i.e., the use of anatomical data. The second group offers a more accurate solution. Nevertheless, there are situations in which the correction made by the first group is sufficient, without the need to complicate the applied technique. A brief introduction to the various existing methods for performing the attenuation map is shown in Table 6 . For a more extensive reading of this topic, readers are referred to reviews about attenuation correction [386], [387], [388].
Table 6.
Classification of attenuation correction methods.
| Category | Method | Definition | Benefit | Limitations | Examples |
|---|---|---|---|---|---|
| Transmission-less based | Uniform fit-ellipse method (UFEM) | Object outline is approximated as an ellipse around its edges. In order to generate the attenuation map, uniform attenuation is assigned Then, uniform attenuation is assigned inside the figure. | Quick and easy Functional for brain studies | Low precision Limited to homogeneous areas | [389] |
| Automated contour detection method (ACDM) | Edge-detection algorithms are used to generate the shape of the object. This allows convex shapes | It is independent of the specialist Functional for brain studies | Low precision but higher than UFEM Only for homogeneous areas | [390,391] | |
| Other methods | Several techniques fit here, such as algebraic reconstruction–based techniques (MLAA or MLACF) or machine-learning techniques | No anatomical information is needed | Possibility of cross-talk between emission data and attenuation map Virtually not used in clinical trials | [392], [393], [394], [395], [396] | |
| Transmission-based | Radionuclide transmission | Apply an external source (PET, SPECT or SPECT/PET) interleaving transmission and emission scanning | Available in most systems. Existence of complementary methods to reduce noise | Need to modify the obtained attenuation coefficients due to be energy-dependent Possibility of errors due to cross-talk of data | [397], [398], [399], [400] |
| CT transmission | It can be addressed by segmenting the CT regions and assigning linear attenuation coefficients to each tissue or transforming the CT image to the attenuation map associated with the radiotracer photon's energy (or a combination of both methods using different scale factors for bone and tissues) | Quick Low noise Good spatial resolution Functional for brain studies | Misregistration due to respiratory movements Erroneus uptake due to patient's possession of unnatural materials (e.g. prostheses) CT photonic energy usually differs from that of the radionuclide used for emission scanning | [401], [402], [403], [404], [405] | |
| MRI transmission | Segmentation-based techniques, where first the PET and MRI images are co-registered and then a segmentation technique is applied. Usually fuzzy is used to divide the image from two to five tissues, assigning to each tissue some attenuation coefficients Atlas-based techniques, where an MR template is used instead of multistep segmentation procedures | High precision Popular for brain studies | Total dependence on co-registration success in the patient's image | [370,[405], [406], [407], [408], [409] |
Transmission-less based methods are easily applied in brain imaging because the brain is considered a practically homogeneous region composed mainly of soft-tissue. Moreover, these methods allow results regarding the skull [390] and optical tracking systems to be used to obtain head contours [391]. Despite being a typology that has not been used frequently, research on these methods has increased over the last years due to shift away from anatomical information. One of the first methods was proposed by Nuyts, Dupont [392], which consists of a maximum likelihood approach known as MLAA (maximum likelihood reconstruction of attenuation and activity). It is commonly used in non-time-of-flight (TOF) PET images, and several studies have tried to improve or test it [393, 395, 396]. Another popular algorithm is MLACF (maximum likelihood attenuation correction factors) [394], mostly used in TOF PET, which jointly estimates the attenuation sinogram and the activity image. Less popular than the previous one, this method has also been tested in other studies [410]. Moreover, machine learning techniques for attenuation correction are emerging, such as deep convolutional encoder-decoders [334] or deep convolutional neural networks [411] have also been applied in attenuation correction. For this last type of technique, there is considerable interest in this field of research, and new algorithms are constantly presented.
Regarding transmission-based techniques, the added difficulty for MRI systems to obtain attenuation maps compared to CT systems should be highlighted. The reason for this is the connection between the attenuation coefficients and the electron density of the tissue. In CT, the data is associated with electron density and photon attenuation properties of the tissues. In contrast, MRI data is correlated to proton density and magnetic relaxation properties of the tissues [387]. As a challenge related to PET/MRI (or SPECT/MRI) systems, several methods have been proposed in literature regarding attenuation estimation. The MRI-based methods can be classified into two groups: segmentation-based and atlas-based methods.
Of the two groups, the segmentation-based one tends to be used more on the literature. One of the first methods published in this area was that of Le Goff-Rougetet, Frouin [412]. This method aims to reduce the patient dose associated with PET imaging without affecting the accurate quantification. This technique is based on a surface matching technique for coregistration of PET and MR images. Another relevant proposal was based on registered T1-weighted MRI. Zaidi, Montandon [407] used a supervised fuzzy C-means clustering segmentation technique, which depends on the density and composition of five tissues: air, skull, brain tissue, and nasal sinuses. Similar to the latter case, Wagenknecht, Kops [408] presented a method where tissue classification is done with neural networks. First, a voxel classification with five tissues (GM, WM, CFS, adipose tissue (AT) and background (BG)) is made, then a second classification is done depending on the previous classes to detect extracerebral tissue and finally, segmentation is obtained. The main drawback of this method is the possibility of mis-segmentation or over-segmentation of bones, especially in the presence of abnormal anatomy or pathology.
In order to improve bone detection, ultrashort echo time (UTE) MRI began to be investigated. Firstly, dual-echo ultra-short echo time and three tissue classification (bone, soft tissue and air) were used [370, 413]. In parallel, methods related to the Dixon technique were raised. Dixon-Water–Fat-segmentation (DWFS) allows the separation of soft and adipose tissues. The method was applied DWFS for the whole body in [414]. Considering both techniques (UTE and DWFS), Berker, Franke [415] proposed a method trying to reduce the drawbacks of these two techniques: time-consuming and complex image registration. This new technique applies UTE sampling for bone detection and uses gradient echoes for water-fat separation, obtaining a four-class PET attenuation map. An example of DWFS application in neuroimaging is the study conducted by Andersen, Ladefoged [416], where this technique is used to generate the attenuation maps. Taking into account the time-consuming problem with UTE, short echo-time (STE) has been tested as a method to obtain attenuation maps of three (cortical bone, air and soft tissue) [417] and four (cortical bone, air, soft tissue and fat tissue) tissues [418], both in combination with fuzzy C-means (FCM) clustering method. Besides, the second study also used two-point Dixon sequences in image acquisition. The latest research in this field has been the use of zero echo time (ZTE), initially proposed by Wiesinger, Sacolick [419] as part of a segmentation method for cranial bone structures. All studies found in the literature, specially the ones related to ZTE, indicate an improvement in results compared to the use of atlas-based methods [420], [421], [422], [423].
The basis of atlas-based approaches is the use of an MR template instead of multistep segmentation procedures, so atlas-based methods use a general map while segmentation-based methods generate a map for each individual. This template can be obtained from two sources: an image labeled as if it were a segmentation of the different tissues or a coregistered attenuation map from a PET or CT scan with continuous attenuation values [354]. As usual, it is obtained from an average of co-registered normal patients, and it has to be warped to the target patient image volume. In any case, most of the studies related to this approach combine atlas-based and segmentation-based methods since the use of an atlas can be an important source of information and can reduce computational cost [424], [425], [426]. Despite this, other studies have proposed methods purely based on atlas images [405, 409], such as the one developed by Johanson, Garpebring [427] where linear attenuation coefficients are predicted from a Gaussian mixture regression algorithm.
Once the attenuation map is generated, attenuation correction is applied in the process. For that, two techniques are mainly used [386]. The first one multiplies the attenuation coefficients obtained by PET image data in the sinogram (or projection) space [428]. The other procedure is performed when PET image reconstruction is performed with an iterative algorithm, using attenuation coefficients as data weighting. While PET images can apply both methods, only iterative methods are used for SPECT [429], [430], [431].
9.1.3. Scatter effect
Scatter in PET and SPECT is another relevant effect for absolute quantification, especially in SPECT, and which is usually associated with Compton scattering. It consists of an energy loss and a change of direction of a photon after an interaction with surrounding atoms. Nevertheless, this effect is not highly relevant in the clinical environment, according to the literature. The reason for this is that the techniques implemented have little impact on the final result, and since the photon change of direction is practically zero, the energy loss is minimal. However, several researchers are convinced that in order to obtain a high accuracy quantitative image, this artifact should be removed [432, 433].
Several methods have been proposed for scatter correction, but most of them were developed many years ago and are inefficient. However, a few recent methods are getting remarkable results. Regarding the categorization of these techniques, a similar classification could be made for both PET and SPECT, considering that PET techniques began to be developed much earlier. Following the review of Zaidi and Montandon [432], the classification would consist of five groups: hardware approaches using coarse septa or beam stoppers, multiple-energy-window approaches, convolution/deconvolution-based approaches, approaches based on direct estimation of scatter distribution and approaches based on statistical reconstruction. Due to the low clinical implementation and the diversity of existing methods, this work will only highlight the most innovative and relevant methods. Therefore, readers interested in this effect are advised to read the following reviews [389, 433, 434]. Table 7 summarizes the different existing categories indicating some examples.
Table 7.
Classification of scatter correction methods.
| Category | Methods Description | Benefit | Limitations | Examples |
|---|---|---|---|---|
| Hardware-based techniques | If coarse septa or beam stoppers are used. lines of response intercepted by the septa can be used to determine the scatter component | No noise increase | Unused | [442,443] |
| Multiple-energy window techniques | The energy spectrum is estimated by using windows below and above the photopeak window | Highly used Simple | Noise | [444,445] |
| Convolution and deconvolution-based techniques | In this case, the standard energy acquisition window is used. Data collected in it helps to estimate the distribution of scatter | Good image contrast Good accuracy | Not commonly used | [446], [447], [448] |
| Direct calculation techniques | Extract information from emission data, or a combination of emission and transmission data for estimating scatter distribution. Monte Carlo technique and ToF information can achieve great progress | The most popular High accuracy | Computational cost | [449], [450], [451], [452] |
| Iterative reconstruction-based scatter-correction techniques | Scatter distribution is obtained and used during image reconstruction | Parallel processing High contrast Low noise | Computational cost | [368,[453], [454], [455] |
The first method proposed to narrow the photopeak energy window to avoid the acceptance of scattered photons. However, this technique has significant drawbacks, such as the elimination of unscattered photons in the process, and therefore, a loss of intensity appears in the image [434, 435]. Thus, multiple-energy-window became much more popular, and it is one of the simplest and most used approaches. The techniques have been developed from two [436, 437] three [438, 439] or even multiple energy windows [440, 441]. Decades have passed since it is known that Monte Carlo techniques are ideal for scatter correction [433], [434], [435]. Nevertheless, it has recently become a viable solution in the clinical environment, as the computational cost is reducing and techniques are faster. Moreover, the Monte Carlo technique can be applied to several methods, either iterative reconstruction based or direct calculation methods.
Finally, another factor to consider in the quantification of functional images is the voluntary and involuntary movement of the patient (e.g. breathing). However, since it does not have high relevance in neuroimaging, its analysis has been dismissed in this study.
9.2. PET and SPECT in neurology
Functional imaging is very useful for the diagnosis of neurodegenerative diseases, such as Alzheimer's disease or Parkinson's disease due to differences in brain activity which can be observed in temporal and parietal lobes (AD) or the striatum (PD) with respect to healthy subjects. Therefore, the systems must allow a correct quantification in order to offer an accurate diagnosis of the patient's condition.
As already mentioned, the correction of the aforementioned effects improves the quantification of the images. For example, some studies associated with attenuation correction demonstrate this improvement. Delso, Kemp [421] achieved a bias reduction of −0.5% using a CT-based correction instead of a regular ZTE attenuation correction in a PET/MR image. Also associated with PET/MR images, Berker, Franke [415] compared 4-class tissue segmentation and 3-class tissue segmentation, concluding that better results are obtained with a 4-class tissue segmentation, and these results are also very similar to those that would be obtained with a PET/CT system. In the study developed by Sousa, Appel [422], a comparison between using an attenuation correction method based on ZTE and atlas-based correction shows that the former produces less variability than the latter. However, bias correction is similar in analyzed brain regions. For example, the correlation coefficient associated with anterior cortical regions is 0.99 for ZTE-based correction and 0.92 for atlas-based correction. Similar results are obtained in the study by Sgard, Khalife [423]. Other interesting results are produced by those that apply machine learning methods. For example, the deep convolutional neural network used by Yang, Park [411] for both attenuation and scatter correction designed for situations where it is difficult to use a combined CT or transmission source. The results were similar to the ones obtain with an CT-based scatter and attenuation correction.
PVC methods are also used in brain images. For example, regarding PD studies, Du, Tsui [376] proved than using a modified GTM method in brain SPECT images, the underestimation of striatal activities could be reduced to 1.2% from an initial value of 30%.
Finally, a correct normalization of the images can also highly increase the accuracy of the study. Salas-Gonzalez, Gorriz [456] proposed a method for intensity normalization of FP-CIT SPECT brain image based on α-stable distribution. This method was tested by Castillo-Barnes, Arenas [457], showing significant differences between the images before and after normalizing. For more information on intensity normalization, especially associated with PD, we recommend reading [458].
In conclusion, the possibility of improving the techniques exposed in this manuscript to reduce unwanted effects on images is highlighted. Despite being applied in clinical systems, they are not considered of high relevance due to the limited improvement they provide or, sometimes, the increase in noise they produce. Therefore, the area of absolute quantification must continue to be investigated for more accurate and faster solutions.
10. Conclusion
This review presents an overview of multimodal data fusion in the field of neuroimaging, including current developments and challenges. We first outlined the fundamental limitations of individual modalities, which can include distortion, non-quantitative nature, and limited temporal/spatial resolutions. These limitations are the general motivators for the development of multimodal neuroimaging and fusion. Multimodal neuroimaging provides more comprehensive information on pathology.
We have summarised the individual benefits and limitations of the current imaging technologies and modalities, including CT, PET, SPECT, MRI, fMRI, DWI, PWI, and MRF. Building upon the available individual techniques, we summarised current development and application of multimodal neuroimaging and fusion in terms of neurological disorders and brain diseases, with a focus in three areas: developing brains, degenerative brains, and psychiatric disorders. The utilization of multiple modalities helps in clinical diagnosis, prevention of misdiagnosis, progression analysis, and research-oriented studies that allow us to gain a more in-depth understanding of human brain pathologies. Nowadays the effects of COVID in the human body are not well-known and maybe we could find in the future some people affected in the brain structure (micro ictus or ischemia). Nevertheless, the fusion techniques and strategies discussed in this survey may be transferred to COVID-19 multimodality image analysis [459]. AI can contribute to the information fusion [460].
The forms of multimodality fusion include multi-modal, multi-focus, multi-temporal, and multi-view. These forms combine images from different instruments/acquisitions, acquisition focal lengths, time of acquisitions and conditions, respectively. Fusion rules were specified with respect to its components and three levels of fusion and theoretical foundation, i.e. rules derived from fuzzy logic, statistic models, and the human vision system. Then we summarised conventional and novel image decomposition and reconstruction methods for the fusion process, including methods based on RGB-IHS, pyramid representations, multi-resolution analysis, sparse representation, and salient features. In addition, we summarised both subjective and objective methods for fusion quality assessment.
The major benefits of multimodal data fusion in neuroimaging include distortion correction, higher temporal/spatial resolution, the combination of structural and functional information. We summarised these benefits with the current applications of multimodal fusion, e.g. MR-PET, EEG-fMRI, and EPI correction. This review also adds a particular focus on the application of multimodal image fusion in standardization, via atlas-based anatomical brain segmentation and the use of multi-atlas fusion. In addition, we summarised the effect of multimodal data fusion on the shift of neuroimaging diagnosis from qualitative analysis to quantitative evaluation, with an example of the effect of multimodal data in photon attenuation, scatter, and partial volume effects of PET and SPECT quantification.
Modern neuroimaging has seen significant improvements in acquisition quality and a constant increase in the abundance of imaging modalities. The fusion of modalities combines complementary information, expands resolution limits, provides standardization, and improves data quality.
It is expected that the effect of multimodal imaging fusion could also effectively scale the amount and quality of information accessible to radiologists for more both precise diagnosis and higher quality research in multiple aspects.
CRediT authorship contribution statement
Yu-Dong Zhang: Conceptualization, Project administration, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing. Zhengchao Dong: Validation, Writing - original draft, Writing - review & editing. Shui-Hua Wang: Methodology, Validation, Writing - original draft, Writing - review & editing. Xiang Yu: Validation, Writing - original draft, Writing - review & editing. Xujing Yao: Validation, Writing - original draft, Writing - review & editing. Qinghua Zhou: Validation, Writing - original draft, Writing - review & editing. Hua Hu: Validation, Writing - original draft, Writing - review & editing. Min Li: Validation, Writing - original draft, Writing - review & editing. Carmen Jiménez-Mesa: Validation, Writing - original draft, Writing - review & editing. Javier Ramirez: Validation, Writing - original draft, Writing - review & editing. Francisco J. Martinez: Validation, Writing - original draft, Writing - review & editing. Juan Manuel Gorriz: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Acknowledgments
This study was partly supported by Royal Society International Exchanges Cost Share Award, UK (RP202G0230); Medical Research Council Confidence in Concept Award, UK (MC_PC_17171); Hope Foundation for Cancer Research, UK (RM60G0680); British Heart Foundation Accelerator Award, UK; the MINECO/ FEDER under the RTI2018-098913-B100 and A-TIC-080-UGR18 projects; FPU predoctoral grant (FPU 18/04902) from Ministerio de Universidades, Spain; Fundamental Research Funds for the Central Universities (CDLS-2020-03); Key Laboratory of Child Development and Learning Science (Southeast University), Ministry of Education; Guangxi Key Laboratory of Trusted Software (kx201901).
References
- 1.Trip S.A. Imaging in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. 2005;76(Suppl 3):iii11–iii18. doi: 10.1136/jnnp.2005.073213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levenson R.W. Emotional and behavioral symptoms in neurodegenerative disease: a model for studying the neural bases of psychopathology. Annu. Rev. Clin. Psychol. 2014;10:581–606. doi: 10.1146/annurev-clinpsy-032813-153653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram L. The genetic epidemiology of neurodegenerative disease. J. Clin. Invest. 2005;115(6):1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C. MR image features predicting hemorrhagic transformation in acute cerebral infarction: a multimodal study. Neuroradiology. 2015;57(11):1145–1152. doi: 10.1007/s00234-015-1575-8. [DOI] [PubMed] [Google Scholar]
- 5.Macintosh B.J. Magnetic resonance imaging to visualize stroke and characterize stroke recovery: a review. Front. Neurol. 2013;4:60. doi: 10.3389/fneur.2013.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ercan E. A multimodal MRI approach to identify and characterize microstructural brain changes in neuropsychiatric systemic lupus erythematosus. Neuroimage Clin. 2015;8:337–344. doi: 10.1016/j.nicl.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astley S.J. Functional magnetic resonance imaging outcomes from a comprehensive magnetic resonance study of children with fetal alcohol spectrum disorders. J. Neurodev. Disord. 2009;1(1):61–80. doi: 10.1007/s11689-009-9004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calhoun V.D. Multimodal fusion of brain imaging data: a key to finding the missing link(s) in complex mental illness. Biol. Psychiatry Cognit. Neurosci, Neuroimaging. 2016;1(3):230–244. doi: 10.1016/j.bpsc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tulay E.E. Multimodal neuroimaging: basic concepts and classification of neuropsychiatric diseases. Clin. EEG Neurosci. 2019;50(1):20–33. doi: 10.1177/1550059418782093. [DOI] [PubMed] [Google Scholar]
- 10.Uludağ K. General overview on the merits of multimodal neuroimaging data fusion. Neuroimage. 2014;102:3–10. doi: 10.1016/j.neuroimage.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Tang T. Design and Applications of Nanoparticles in Biomedical Imaging. Springer; 2017. PET/SPECT/MRI multimodal nanoparticles; pp. 205–228. [Google Scholar]
- 12.Hu Z. From PET/CT to PET/MRI: advances in instrumentation and clinical applications. Mol. Pharm. 2014;11(11):3798–3809. doi: 10.1021/mp500321h. [DOI] [PubMed] [Google Scholar]
- 13.Luker G.D. Optical imaging: current applications and future directions. J. Nucl. Med. 2008;49(1):1–4. doi: 10.2967/jnumed.107.045799. [DOI] [PubMed] [Google Scholar]
- 14.Weiskopf N. Principles of a brain-computer interface (BCI) based on real-time functional magnetic resonance imaging (fMRI) IEEE Trans. Biomed. Eng. 2004;51(6):966–970. doi: 10.1109/TBME.2004.827063. [DOI] [PubMed] [Google Scholar]
- 15.Hari R. Magnetoencephalography: from SQUIDs to neuroscience: neuroimage 20th anniversary special edition. Neuroimage. 2012;61(2):386–396. doi: 10.1016/j.neuroimage.2011.11.074. [DOI] [PubMed] [Google Scholar]
- 16.Michel C.M. Towards the utilization of EEG as a brain imaging tool. Neuroimage. 2012;61(2):371–385. doi: 10.1016/j.neuroimage.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 17.Ances B.M. Regional differences in the coupling of cerebral blood flow and oxygen metabolism changes in response to activation: implications for BOLD-fMRI. Neuroimage. 2008;39(4):1510–1521. doi: 10.1016/j.neuroimage.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blockley N.P. A review of calibrated blood oxygenation level‐dependent (BOLD) methods for the measurement of task‐induced changes in brain oxygen metabolism. NMR Biomed. 2013;26(8):987–1003. doi: 10.1002/nbm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiarelli P.A. A calibration method for quantitative BOLD fMRI based on hyperoxia. Neuroimage. 2007;37(3):808–820. doi: 10.1016/j.neuroimage.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 20.Hoge, R.D., Calibrated fMRI. NeuroImage, 2012. 62(2): p. 930–937. [DOI] [PubMed]
- 21.Borogovac A. Arterial spin labeling (ASL) fMRI: advantages, theoretical constrains and experimental challenges in neurosciences. Int. J. Biomed. Imaging. 2012:2012. doi: 10.1155/2012/818456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng H. Image distortion correction in EPI: comparison of field mapping with point spread function mapping. Mag. Reson. Med. 2002;48(1):137–146. doi: 10.1002/mrm.10200. [DOI] [PubMed] [Google Scholar]
- 23.Musalar E. Conventional vs invert-grayscale X-ray for diagnosis of pneumothorax in the emergency setting. Am. J. Emerg. Med. 2017;35(9):1217–1221. doi: 10.1016/j.ajem.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 24.Liu S. Application of high-resolution CT images information in complicated infection of lung tumors. J. Infect. Public Health. 2019 doi: 10.1016/j.jiph.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhao S. Application of CT combined with electrocardiographic gating in hypertensive patients with brain and nerve diseases. World Neurosurg. 2020 doi: 10.1016/j.wneu.2019.12.161. [DOI] [PubMed] [Google Scholar]
- 26.Stepniak K. Novel 3D printing technology for CT phantom coronary arteries with high geometrical accuracy for biomedical imaging applications. Bioprinting. 2020:e00074. [Google Scholar]
- 27.Wang S.-H. Springer; Germany: 2018. Pathological Brain Detection; p. 222. [Google Scholar]
- 28.Warman Chardon J. MYO-MRI diagnostic protocols in genetic myopathies. Neuromuscul. Disord. 2019;29(11):827–841. doi: 10.1016/j.nmd.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H. Comparison of the clinical application value of mo-targeted X-ray, color doppler ultrasound and MRI in preoperative comprehensive evaluation of breast cancer. Saudi J. Biol. Sci. 2019;26(8):1973–1977. doi: 10.1016/j.sjbs.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X. Magnetic Fe3O4@PVP nanotubes with high heating efficiency for MRI-guided magnetic hyperthermia applications. Mater. Lett. 2020;262 [Google Scholar]
- 31.Kazemivalipour E. Reconfigurable MRI technology for low-SAR imaging of deep brain stimulation at 3T: application in bilateral leads, fully-implanted systems, and surgically modified lead trajectories. Neuroimage. 2019;199:18–29. doi: 10.1016/j.neuroimage.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van As H. MRI of plants and foods. J. Magn. Reson. 2013;229:25–34. doi: 10.1016/j.jmr.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 33.Wang, S.-H., et al., Unilateral sensorineural hearing loss identification based on double-density dual-tree complex wavelet transform and multinomial logistic regression. 2019. 26(4): p. 411–426.
- 34.Lee D.H. Mechanisms of contrast enhancement in magnetic resonance imaging. Can. Assoc. Radiol. J. 1991;42(1):6–12. [PubMed] [Google Scholar]
- 35.Wang X. Magnetic properties and magnetization reversal process in (Pt/CoFe/MgO)10 multilayers at low temperature. J. Magn. Magn. Mater. 2019;499 [Google Scholar]
- 36.Parsons N. Single-subject manual independent component analysis and resting state fMRI connectivity outcomes in patients with juvenile absence epilepsy. Magn. Reson. Imaging. 2020;66:42–49. doi: 10.1016/j.mri.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Angenstein F. The role of ongoing neuronal activity for baseline and stimulus-induced BOLD signals in the rat hippocampus. Neuroimage. 2019;202 doi: 10.1016/j.neuroimage.2019.116082. [DOI] [PubMed] [Google Scholar]
- 38.Magdziarz M. Lamperti transformation of scaled Brownian motion and related Langevin equations. Commun. Nonlinear Sci. Numer. Simul. 2020;83 [Google Scholar]
- 39.Xie D. Denoising arterial spin labeling perfusion MRI with deep machine learning. Magn. Reson. Imaging. 2020 doi: 10.1016/j.mri.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma D. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson M.H. Functional brain development in humans. Nat. Rev. Neurosci. 2001;2(7):475–483. doi: 10.1038/35081509. [DOI] [PubMed] [Google Scholar]
- 42.Gogtay N. Dynamic mapping of human cortical development during childhood through early adulthood. Proc. Natl. Acad. Sci. U. S. A. 2004;101(21):8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegel D.J. third ed. Guilford Press; New York: 2020. The Developing Mind: How Relationships and the Brain Interact to Shape Who We Are. [Google Scholar]
- 44.O'Connor T.G. Maternal antenatal anxiety and behavioural/emotional problems in children: a test of a programming hypothesis. J. Child Psychol. Psychiatry. 2003;44(7):1025–1036. doi: 10.1111/1469-7610.00187. [DOI] [PubMed] [Google Scholar]
- 45.Nyaradi A. Diet in the early years of life influences cognitive outcomes at 10 years: a prospective cohort study. Acta Paediatr. 2013;102(12):1165–1173. doi: 10.1111/apa.12363. [DOI] [PubMed] [Google Scholar]
- 46.O'Muircheartaigh J. White matter development and early cognition in babies and toddlers. Hum. Brain Mapp. 2014;35(9):4475–4487. doi: 10.1002/hbm.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dean D.C., 3rd Modeling healthy male white matter and myelin development: 3 through 60months of age. Neuroimage. 2014;84:742–752. doi: 10.1016/j.neuroimage.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulikova S. Multi-parametric evaluation of the white matter maturation. Brain Struct. Funct. 2015;220(6):3657–3672. doi: 10.1007/s00429-014-0881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine D. Central nervous system abnormalities assessed with prenatal magnetic resonance imaging. Obstet. Gynecol. 1999;94(6):1011–1019. doi: 10.1016/s0029-7844(99)00455-x. [DOI] [PubMed] [Google Scholar]
- 50.Barkovich A.J. Techniques and methods in pediatric magnetic resonance imaging. Semin. Ultrasound CT MR. 1988;9(3):186–191. [PubMed] [Google Scholar]
- 51.Holland B.A. MRI of normal brain maturation. AJNR Am. J. Neuroradiol. 1986;7(2):201–208. [PMC free article] [PubMed] [Google Scholar]
- 52.Reiss A.L. Brain development, gender and IQ in children. A volumetric imaging study. Brain. 1996;119(Pt 5):1763–1774. doi: 10.1093/brain/119.5.1763. [DOI] [PubMed] [Google Scholar]
- 53.Jernigan T.L. Late childhood changes in brain morphology observable with MRI. Dev. Med. Child Neurol. 1990;32(5):379–385. doi: 10.1111/j.1469-8749.1990.tb16956.x. [DOI] [PubMed] [Google Scholar]
- 54.Phan T.V. Processing of structural neuroimaging data in young children: bridging the gap between current practice and state-of-the-art methods. Dev. Cognit. Neurosci. 2018;33:206–223. doi: 10.1016/j.dcn.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thieba C. Factors associated with successful MRI scanning in unsedated young children. Front. Pediat.r. 2018;6:146. doi: 10.3389/fped.2018.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y. MR fingerprinting enables quantitative measures of brain tissue relaxation times and myelin water fraction in the first five years of life. Neuroimage. 2019;186:782–793. doi: 10.1016/j.neuroimage.2018.11.038. [DOI] [PubMed] [Google Scholar]
- 57.de Blank P. Magnetic resonance fingerprinting to characterize childhood and young adult brain tumors. Pediatr. Neurosurg. 2019;54(5):310–318. doi: 10.1159/000501696. [DOI] [PubMed] [Google Scholar]
- 58.Langa K.M. The diagnosis and management of mild cognitive impairment: a clinical review. JAMA. 2014;312(23):2551–2561. doi: 10.1001/jama.2014.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petersen R.C. Prevalence of mild cognitive impairment is higher in men. The Mayo Clinic Study of Aging. Neurology. 2010;75(10):889–897. doi: 10.1212/WNL.0b013e3181f11d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jack C.R., Jr. Alzheimer disease: new concepts on its neurobiology and the clinical role imaging will play. Radiology. 2012;263(2):344–361. doi: 10.1148/radiol.12110433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu-Ambrose T.Y. Increased risk of falling in older community-dwelling women with mild cognitive impairment. Phys. Ther. 2008;88(12):1482–1491. doi: 10.2522/ptj.20080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ackl N. Hippocampal metabolic abnormalities in mild cognitive impairment and Alzheimer's disease. Neurosci. Lett. 2005;384(1-2):23–28. doi: 10.1016/j.neulet.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 63.Petersen R.C. Current concepts in mild cognitive impairment. Arch. Neurol. 2001;58(12):1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 64.Apostolova L.G. Use of magnetic resonance imaging to identify mild cognitive impairment: who should be imaged? CNS Spectr. 2008;13(10 Suppl 16):18–20. doi: 10.1017/s1092852900026997. [DOI] [PubMed] [Google Scholar]
- 65.Bartos A. Brain volumes and their ratios in Alzheimer s disease on magnetic resonance imaging segmented using Freesurfer 6.0. Psychiatry Res. Neuroimaging. 2019;287:70–74. doi: 10.1016/j.pscychresns.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Basiratnia R. Hippocampal volume and hippocampal angle (a more practical marker) in mild cognitive impairment: a case-control magnetic resonance imaging study. Adv. Biomed. Res. 2015;4:192. doi: 10.4103/2277-9175.166153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atmaca M. Volumetric MRI study of orbito-frontal cortex and thalamus in obsessive-compulsive personality disorder. J. Clin. Neurosci. 2019;64:89–93. doi: 10.1016/j.jocn.2019.03.062. [DOI] [PubMed] [Google Scholar]
- 68.Bilello M. Correlating cognitive decline with white matter lesion and brain atrophy magnetic resonance imaging measurements in alzheimer's disease. J. Alzheimers Dis. 2015;48(4):987–994. doi: 10.3233/JAD-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang L. Inhibition of eukaryotic initiation factor 3B suppresses proliferation and promotes apoptosis of chronic myeloid leukemia cells. Adv. Clin. Exp. Med. 2019 doi: 10.17219/acem/110323. [DOI] [PubMed] [Google Scholar]
- 70.Saka E. Linear measures of temporal lobe atrophy on brain magnetic resonance imaging (MRI) but not visual rating of white matter changes can help discrimination of mild cognitive impairment (MCI) and Alzheimer's disease (AD) Arch. Gerontol. Geriatr. 2007;44(2):141–151. doi: 10.1016/j.archger.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 71.Shen Q. Volumetric and visual rating of magnetic resonance imaging scans in the diagnosis of amnestic mild cognitive impairment and Alzheimer's disease. Alzheimers Dement. 2011;7(4):e101–e108. doi: 10.1016/j.jalz.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandra A. Magnetic resonance imaging in Alzheimer's disease and mild cognitive impairment. J. Neurol. 2019;266(6):1293–1302. doi: 10.1007/s00415-018-9016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu L. Prediction of progressive mild cognitive impairment by multi-modal neuroimaging biomarkers. J. Alzheimers Dis. 2016;51(4):1045–1056. doi: 10.3233/JAD-151010. [DOI] [PubMed] [Google Scholar]
- 74.Chiti A. Functional magnetic resonance imaging with encoding task in patients with mild cognitive impairment and different severity of leukoaraiosis. Psychiatry Res. Neuroimaging. 2018;282:126–131. doi: 10.1016/j.pscychresns.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 75.Forouzannezhad P. A survey on applications and analysis methods of functional magnetic resonance imaging for Alzheimer's disease. J. Neurosci. Methods. 2019;317:121–140. doi: 10.1016/j.jneumeth.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 76.Frederick B. Brain proton magnetic resonance spectroscopy in Alzheimer disease: changes after treatment with xanomeline. Am. J. Geriatr. Psychiatry. 2002;10(1):81–88. [PubMed] [Google Scholar]
- 77.Modrego P.J. Conversion from mild cognitive impairment to probable Alzheimer's disease predicted by brain magnetic resonance spectroscopy. Am. J. Psychiatry. 2005;162(4):667–675. doi: 10.1176/appi.ajp.162.4.667. [DOI] [PubMed] [Google Scholar]
- 78.Garcia Santos J.M. Magnetic resonance spectroscopy performance for detection of dementia, Alzheimer's disease and mild cognitive impairment in a community-based survey. Dement. Geriatr. Cognit. Disord. 2008;26(1):15–25. doi: 10.1159/000140624. [DOI] [PubMed] [Google Scholar]
- 79.Jahng G.H. Glutamine and glutamate complex, as measured by functional magnetic resonance spectroscopy, alters during face-name association task in patients with mild cognitive impairment and alzheimer's disease. J. Alzheimers Dis. 2016;52(1):145–159. doi: 10.3233/JAD-150877. [DOI] [PubMed] [Google Scholar]
- 80.Vijayakumari A.A. Glutamatergic response to a low load working memory paradigm in the left dorsolateral prefrontal cortex in patients with mild cognitive impairment: a functional magnetic resonance spectroscopy study. Brain Imaging Behav. 2019 doi: 10.1007/s11682-019-00122-7. [DOI] [PubMed] [Google Scholar]
- 81.Wong D. Reduced hippocampal glutamate and posterior cingulate N-acetyl aspartate in mild cognitive impairment and alzheimer's disease is associated with episodic memory performance and white matter integrity in the cingulum: a pilot study. J. Alzheimers Dis. 2020 doi: 10.3233/JAD-190773. [DOI] [PubMed] [Google Scholar]
- 82.Oeltzschner G. Neurometabolites and associations with cognitive deficits in mild cognitive impairment: a magnetic resonance spectroscopy study at 7 Tesla. Neurobiol. Aging. 2019;73:211–218. doi: 10.1016/j.neurobiolaging.2018.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kantarci K. Proton MRS in mild cognitive impairment. J. Magn. Reson. Imaging. 2013;37(4):770–777. doi: 10.1002/jmri.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coutinho A.M.N. Analysis of the posterior cingulate cortex with [18F]FDG-PET and Naa/mI in mild cognitive impairment and Alzheimer's disease: correlations and differences between the two methods. Dement. Neuropsychol. 2015;9(4):385–393. doi: 10.1590/1980-57642015DN94000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vannini P. Anosognosia for memory deficits in mild cognitive impairment: insight into the neural mechanism using functional and molecular imaging. Neuroimage Clin. 2017;15:408–414. doi: 10.1016/j.nicl.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bailly M. Precuneus and cingulate cortex atrophy and hypometabolism in patients with alzheimer's disease and mild cognitive impairment: MRI and (18)F-FDG PET quantitative analysis using freesurfer. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/583931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marcus C. Brain PET in the diagnosis of Alzheimer's disease. Clin. Nucl. Med. 2014;39(10):e413–e422. doi: 10.1097/RLU.0000000000000547. quiz e423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cohen A.D. Early detection of Alzheimer's disease using PiB and FDG PET. Neurobiol. Dis. 2014;72(Pt A):117–122. doi: 10.1016/j.nbd.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murphy M.P. Alzheimer's disease and the amyloid-beta peptide. J. Alzheimers Dis. 2010;19(1):311–323. doi: 10.3233/JAD-2010-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang F. Prediction and characterization of protein-protein interaction networks in swine. Proteome Sci. 2012;10(1):2. doi: 10.1186/1477-5956-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer's disease. Neuroimage. 2012;59(2):895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu H. The impact of marine shipping and its DECA control on air quality in the Pearl River Delta, China. Sci. Total Environ. 2018;625:1476–1485. doi: 10.1016/j.scitotenv.2018.01.033. [DOI] [PubMed] [Google Scholar]
- 93.Kim D. A graph-based integration of multimodal brain imaging data for the detection of early mild cognitive impairment (E-MCI) Multimodal. Brain Image Anal. 2013;2013(8159):159–169. doi: 10.1007/978-3-319-02126-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young J. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin S.Y. Multiparametric graph theoretical analysis reveals altered structural and functional network topology in Alzheimer's disease. Neuroimage Clin. 2019;22 doi: 10.1016/j.nicl.2019.101680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tromp D. Episodic memory in normal aging and Alzheimer disease: insights from imaging and behavioral studies. Ageing Res. Rev. 2015;24(Pt B):232–262. doi: 10.1016/j.arr.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 97.Werheid K. Are faces special in Alzheimer's disease? Cognitive conceptualisation, neural correlates, and diagnostic relevance of impaired memory for faces and names. Cortex. 2007;43(7):898–906. doi: 10.1016/s0010-9452(08)70689-0. [DOI] [PubMed] [Google Scholar]
- 98.Cass S.P. Alzheimer's disease and exercise: a literature review. Curr. Sports Med. Rep. 2017;16(1):19–22. doi: 10.1249/JSR.0000000000000332. [DOI] [PubMed] [Google Scholar]
- 99.Alzheimer's A. 2014 Alzheimer's disease facts and figures. Alzheimers Dement. 2014;10(2):e47–e92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 100.Tucholka A. An empirical comparison of surface-based and volume-based group studies in neuroimaging. Neuroimage. 2012;63(3):1443–1453. doi: 10.1016/j.neuroimage.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 101.Henneman W.J. Hippocampal atrophy rates in Alzheimer disease: added value over whole brain volume measures. Neurology. 2009;72(11):999–1007. doi: 10.1212/01.wnl.0000344568.09360.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kerchner G.A. Hippocampal CA1 apical neuropil atrophy in mild Alzheimer disease visualized with 7-T MRI. Neurology. 2010;75(15):1381–1387. doi: 10.1212/WNL.0b013e3181f736a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Winter F.L. No association of lower hippocampal volume with alzheimer's disease pathology in late-life depression. Am. J. Psychiatry. 2017;174(3):237–245. doi: 10.1176/appi.ajp.2016.16030319. [DOI] [PubMed] [Google Scholar]
- 104.Chen J. Can multi-modal neuroimaging evidence from hippocampus provide biomarkers for the progression of amnestic mild cognitive impairment? Neurosci. Bull. 2015;31(1):128–140. doi: 10.1007/s12264-014-1490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zamboni G. Resting functional connectivity reveals residual functional activity in Alzheimer's disease. Biol. Psychiatry. 2013;74(5):375–383. doi: 10.1016/j.biopsych.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 106.Zhou J. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73(6):1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J. Disrupted functional brain connectome in individuals at risk for Alzheimer's disease. Biol. Psychiatry. 2013;73(5):472–481. doi: 10.1016/j.biopsych.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 108.Jin M. Aberrant default mode network in subjects with amnestic mild cognitive impairment using resting-state functional MRI. Magn. Reson. Imaging. 2012;30(1):48–61. doi: 10.1016/j.mri.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bayram E. Current understanding of magnetic resonance imaging biomarkers and memory in Alzheimer's disease. Alzheimers Dement. 2018;4:395–413. doi: 10.1016/j.trci.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Promteangtrong C. Multimodality imaging approach in Alzheimer disease. Part I: Structural MRI, functional MRI, diffusion tensor imaging and magnetization transfer imaging. Dement. Neuropsychol. 2015;9(4):318–329. doi: 10.1590/1980-57642015DN94000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Waser M. Neuroimaging markers of global cognition in early Alzheimer's disease: a magnetic resonance imaging-electroencephalography study. Brain Behav. 2019;9(1):e01197. doi: 10.1002/brb3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harman P. Technical note: can resting state functional MRI assist in routine clinical diagnosis? BJR Case Rep. 2018;4(4) doi: 10.1259/bjrcr.20180030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Basheera S. Convolution neural network-based Alzheimer's disease classification using hybrid enhanced independent component analysis based segmented gray matter of T2 weighted magnetic resonance imaging with clinical valuation. Alzheimers Dement. 2019;5:974–986. doi: 10.1016/j.trci.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hirjak D. Multimodal Magnetic Resonance Imaging Data Fusion Reveals Distinct Patterns of Abnormal Brain Structure and Function in Catatonia. Schizophr. Bull. 2020;46(1):202–210. doi: 10.1093/schbul/sbz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chabiniok R. Multiphysics and multiscale modelling, data-model fusion and integration of organ physiology in the clinic: ventricular cardiac mechanics. Interface Focus. 2016;6(2) doi: 10.1098/rsfs.2015.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Adali T. Multi-modal data fusion using source separation: two effective models based on ICA and IVA and their properties. Proc. IEEE Inst. Electr. Electron Eng. 2015;103(9):1478–1493. doi: 10.1109/JPROC.2015.2461624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marino B.L.B. Parkinson's disease: a review from the pathophysiology to diagnosis, new perspectives for pharmacological treatment. Mini. Rev. Med. Chem. 2019 [Google Scholar]
- 118.Driver J.A. Incidence and remaining lifetime risk of Parkinson disease in advanced age. Neurology. 2009;72(5):432–438. doi: 10.1212/01.wnl.0000341769.50075.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee J.H. The incidence rates and risk factors of Parkinson disease in patients with psoriasis: a nationwide population-based cohort study. J. Am. Acad. Dermatol. 2019 doi: 10.1016/j.jaad.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 120.Van Den Eeden S.K. Incidence of Parkinson's disease: variation by age, gender, and race/ethnicity. Am. J. Epidemiol. 2003;157(11):1015–1022. doi: 10.1093/aje/kwg068. [DOI] [PubMed] [Google Scholar]
- 121.Bharti K. Neuroimaging advances in Parkinson's disease with freezing of gait: a systematic review. Neuroimage Clin. 2019;24 doi: 10.1016/j.nicl.2019.102059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Al-Radaideh A.M. The role of magnetic resonance imaging in the diagnosis of Parkinson's disease: a review. Clin. Imaging. 2016;40(5):987–996. doi: 10.1016/j.clinimag.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 123.Alegret M. MRI atrophy parameters related to cognitive and motor impairment in Parkinson's disease. Neurologia. 2001;16(2):63–69. [PubMed] [Google Scholar]
- 124.Prasad S. Three-dimensional neuromelanin-sensitive magnetic resonance imaging of the substantia nigra in Parkinson's disease. Eur. J. Neurol. 2018;25(4):680–686. doi: 10.1111/ene.13573. [DOI] [PubMed] [Google Scholar]
- 125.Wang J. Neuromelanin-sensitive MRI of the substantia nigra: an imaging biomarker to differentiate essential tremor from tremor-dominant Parkinson's disease. Parkinsonism Relat. Disord. 2019;58:3–8. doi: 10.1016/j.parkreldis.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 126.Jin L. Combined visualization of nigrosome-1 and neuromelanin in the substantia nigra using 3T MRI for the differential diagnosis of essential tremor and de novo Parkinson's disease. Front. Neurol. 2019;10:100. doi: 10.3389/fneur.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takahashi H. Comprehensive MRI quantification of the substantia nigra pars compacta in Parkinson's disease. Eur. J. Radiol. 2018;109:48–56. doi: 10.1016/j.ejrad.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 128.Wang Y. Quantitative susceptibility mapping (QSM): decoding MRI data for a tissue magnetic biomarker. Magn. Reson. Med. 2015;73(1):82-101. doi: 10.1002/mrm.25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Burciu R.G. Imaging of motor cortex physiology in Parkinson's disease. Mov. Disord. 2018;33(11):1688–1699. doi: 10.1002/mds.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Niethammer M. Functional neuroimaging in Parkinson's disease. Cold Spring Harb. Perspect. Med. 2012;2(5) doi: 10.1101/cshperspect.a009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Evangelisti S. L-dopa modulation of brain connectivity in parkinson's disease patients: a pilot EEG-fMRI study. Front. Neurosci. 2019;13:611. doi: 10.3389/fnins.2019.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tessitore A. Sensorimotor connectivity in Parkinson's disease: the role of functional neuroimaging. Front. Neurol. 2014;5:180. doi: 10.3389/fneur.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Amboni M. Resting-state functional connectivity associated with mild cognitive impairment in Parkinson's disease. J. Neurol. 2015;262(2):425–434. doi: 10.1007/s00415-014-7591-5. [DOI] [PubMed] [Google Scholar]
- 134.Borghammer P. Glucose metabolism in small subcortical structures in Parkinson's disease. Acta Neurol. Scand. 2012;125(5):303–310. doi: 10.1111/j.1600-0404.2011.01556.x. [DOI] [PubMed] [Google Scholar]
- 135.Hilker R. Functional imaging of deep brain stimulation in idiopathic Parkinson's disease. Nervenarzt. 2010;81(10):1204–1207. doi: 10.1007/s00115-010-3027-3. [DOI] [PubMed] [Google Scholar]
- 136.Berding G. Resting regional cerebral glucose metabolism in advanced Parkinson's disease studied in the off and on conditions with [(18)F]FDG-PET. Mov. Disord. 2001;16(6):1014–1022. doi: 10.1002/mds.1212. [DOI] [PubMed] [Google Scholar]
- 137.Son S.J. Imaging analysis of Parkinson's disease patients using SPECT and tractography. Sci. Rep. 2016;6:38070. doi: 10.1038/srep38070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ruppert M.C. Network degeneration in Parkinson's disease: multimodal imaging of nigro-striato-cortical dysfunction. Brain. 2020 doi: 10.1093/brain/awaa019. [DOI] [PubMed] [Google Scholar]
- 139.Bowman F.D. Multimodal imaging signatures of Parkinson's disease. Front. Neurosci. 2016;10:131. doi: 10.3389/fnins.2016.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mental Illness . 2020. Available from: https://www.nimh.nih.gov/health/statistics/mental-illness.shtml. [Google Scholar]
- 141.Mental Disorders Affect One in Four People . 2001. https://www.who.int/whr/2001/media_centre/press_release [cited 2020 9/March]; Available from. [Google Scholar]
- 142.Rehm J. Global burden of disease and the impact of mental and addictive disorders. Curr. Psychiatry Rep. 2019;21(2):10. doi: 10.1007/s11920-019-0997-0. [DOI] [PubMed] [Google Scholar]
- 143.Eaton W.W. The burden of mental disorders. Epidemiol. Rev. 2008;30:p. 1-14. doi: 10.1093/epirev/mxn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mental Illness Will Cost the World $16 USD Trillion by 2030 . 2018. https://www.psychiatrictimes.com/mental-health/mental-illness-will-cost-world-16-usd-trillion-2030 [cited 2020 9/3]; Available from. [Google Scholar]
- 145.Silbersweig D.A. Neuroimaging in psychiatry: a quarter century of progress. Harv. Rev. Psychiatry. 2017;25(5):195–197. doi: 10.1097/HRP.0000000000000177. [DOI] [PubMed] [Google Scholar]
- 146.Cannon D.M. Neuroimaging in psychiatry. Ir. J. Psychol. Med. 2007;24(3):86–88. doi: 10.1017/S0790966700010363. [DOI] [PubMed] [Google Scholar]
- 147.Wibawa P. Understanding MRI in clinical psychiatry: perspectives from neuroimaging psychiatry registrars. Aust. Psychiatry. 2019;27(4):396–403. doi: 10.1177/1039856219842647. [DOI] [PubMed] [Google Scholar]
- 148.Todeva-Radneva A. The value of neuroimaging techniques in the translation and trans-diagnostic validation of psychiatric diagnoses - selective review. Curr. Top. Med. Chem. 2020 doi: 10.2174/1568026620666200131095328. [DOI] [PubMed] [Google Scholar]
- 149.Lai C.H. Promising neuroimaging biomarkers in depression. Psychiatry Investig. 2019;16(9):662–670. doi: 10.30773/pi.2019.07.25.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kessing L.V. Rate and predictors of conversion from unipolar to bipolar disorder: a systematic review and meta-analysis. Bipolar Disord. 2017;19(5):324–335. doi: 10.1111/bdi.12513. [DOI] [PubMed] [Google Scholar]
- 151.Vieta E. Early intervention in bipolar disorder. Am. J. Psychiatry. 2018;175(5):411–426. doi: 10.1176/appi.ajp.2017.17090972. [DOI] [PubMed] [Google Scholar]
- 152.Kessler R.C. Epidemiology of women and depression. J. Affect. Disord. 2003;74(1):5–13. doi: 10.1016/s0165-0327(02)00426-3. [DOI] [PubMed] [Google Scholar]
- 153.Andrews G. Why does the burden of disease persist? Relating the burden of anxiety and depression to effectiveness of treatment. Bull. World Health Organ. 2000;78(4):446–454. [PMC free article] [PubMed] [Google Scholar]
- 154.Schmaal L. Brain structural signatures of adolescent depressive symptom trajectories: a longitudinal magnetic resonance imaging study. J. Am. Acad. Child Adolesc. Psychiatry. 2017;56(7):593-601 e9. doi: 10.1016/j.jaac.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 155.Vassilopoulou K. A magnetic resonance imaging study of hippocampal, amygdala and subgenual prefrontal cortex volumes in major depression subtypes: melancholic versus psychotic depression. J. Affect. Disord. 2013;146(2):197–204. doi: 10.1016/j.jad.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 156.Sacchet M.D. Myelination of the brain in major depressive disorder: an in vivo quantitative magnetic resonance imaging study. Sci. Rep. 2017;7(1):2200. doi: 10.1038/s41598-017-02062-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nugent A.C. Multimodal imaging reveals a complex pattern of dysfunction in corticolimbic pathways in major depressive disorder. Hum. Brain Mapp. 2019;40(13):3940–3950. doi: 10.1002/hbm.24679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vasic N. Baseline brain perfusion and brain structure in patients with major depression: a multimodal magnetic resonance imaging study. J. Psychiatry Neurosci. 2015;40(6):412–421. doi: 10.1503/jpn.140246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Finkelmeyer A. Altered hippocampal function in major depression despite intact structure and resting perfusion. Psychol. Med. 2016;46(10):2157–2168. doi: 10.1017/S0033291716000702. [DOI] [PubMed] [Google Scholar]
- 160.Yang J. Development and evaluation of a multimodal marker of major depressive disorder. Hum. Brain Mapp. 2018;39(11):4420–4439. doi: 10.1002/hbm.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maglanoc L.A. Multimodal fusion of structural and functional brain imaging in depression using linked independent component analysis. Hum. Brain Mapp. 2020;41(1):241–255. doi: 10.1002/hbm.24802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen J. Widespread decreased grey and white matter in paediatric obsessive-compulsive disorder (OCD): a voxel-based morphometric MRI study. Psychiatry Res. 2013;213(1):11–17. doi: 10.1016/j.pscychresns.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 163.Lazaro L. Brain changes in children and adolescents with obsessive-compulsive disorder before and after treatment: a voxel-based morphometric MRI study. Psychiatry Res. 2009;172(2):140–146. doi: 10.1016/j.pscychresns.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 164.Qiu L. Abnormal regional spontaneous neuronal activity associated with symptom severity in treatment-naive patients with obsessive-compulsive disorder revealed by resting-state functional MRI. Neurosci. Lett. 2017;640:99–104. doi: 10.1016/j.neulet.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 165.Lazaro L. Cerebral activation in children and adolescents with obsessive-compulsive disorder before and after treatment: a functional MRI study. J. Psychiatr. Res. 2008;42(13):1051–1059. doi: 10.1016/j.jpsychires.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 166.Bu X. Investigating the predictive value of different resting-state functional MRI parameters in obsessive-compulsive disorder. Transl. Psychiatry. 2019;9(1):17. doi: 10.1038/s41398-018-0362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Park S.E. Metabolic abnormality in the right dorsolateral prefrontal cortex in patients with obsessive-compulsive disorder: proton magnetic resonance spectroscopy. Acta Neuropsychiatr. 2017;29(3):164–169. doi: 10.1017/neu.2016.48. [DOI] [PubMed] [Google Scholar]
- 168.Fan S. Abnormalities in metabolite concentrations in tourette's disorder and obsessive-compulsive disorder-A proton magnetic resonance spectroscopy study. Psychoneuroendocrinology. 2017;77:211–217. doi: 10.1016/j.psyneuen.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 169.Tukel R. Proton magnetic resonance spectroscopy in obsessive-compulsive disorder: evidence for reduced neuronal integrity in the anterior cingulate. Psychiatry Res. 2014;224(3):275–280. doi: 10.1016/j.pscychresns.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 170.Brennan B.P. A critical review of magnetic resonance spectroscopy studies of obsessive-compulsive disorder. Biol. Psychiatry. 2013;73(1):24–31. doi: 10.1016/j.biopsych.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li Y. Investigation of anterior cingulate cortex gamma-aminobutyric acid and glutamate-glutamine levels in obsessive-compulsive disorder using magnetic resonance spectroscopy. BMC Psychiatry. 2019;19(1):164. doi: 10.1186/s12888-019-2160-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang Z. Brain gamma-aminobutyric acid (GABA) concentration of the prefrontal lobe in unmedicated patients with obsessive-compulsive disorder: a research of magnetic resonance spectroscopy. Shanghai Arch. Psychiatry. 2016;28(5):263–270. doi: 10.11919/j.issn.1002-0829.216043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rosenberg D.R. Reduced anterior cingulate glutamate in pediatric major depression: a magnetic resonance spectroscopy study. Biol. Psychiatry. 2005;58(9):700–704. doi: 10.1016/j.biopsych.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 174.Lazaro L. Proton magnetic resonance spectroscopy in pediatric obsessive-compulsive disorder: longitudinal study before and after treatment. Psychiatry Res. 2012;201(1):17–24. doi: 10.1016/j.pscychresns.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 175.Whiteside S.P.H. The effect of behavior therapy on caudate N-acetyl-l-aspartic acid in adults with obsessive-compulsive disorder. Psychiatry Res. 2012;201(1):10–16. doi: 10.1016/j.pscychresns.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 176.Pico-Perez M. Modality-specific overlaps in brain structure and function in obsessive-compulsive disorder: multimodal meta-analysis of case-control MRI studies. Neurosci. Biobehav. Rev. 2020;112:83–94. doi: 10.1016/j.neubiorev.2020.01.033. [DOI] [PubMed] [Google Scholar]
- 177.Moreira P.S. The neural correlates of obsessive-compulsive disorder: a multimodal perspective. Transl. Psychiatry. 2017:7. doi: 10.1038/tp.2017.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Choi J.S. Morphometric alterations of anterior superior temporal cortex in obsessive-compulsive disorder. Depress. Anxiety. 2006;23(5):290–296. doi: 10.1002/da.20171. [DOI] [PubMed] [Google Scholar]
- 179.Fan J. Spontaneous neural activity in the right superior temporal gyrus and left middle temporal gyrus is associated with insight level in obsessive-compulsive disorder. J. Affect. Disord. 2017;207:203–211. doi: 10.1016/j.jad.2016.08.027. [DOI] [PubMed] [Google Scholar]
- 180.Bruin W. Diagnostic neuroimaging markers of obsessive-compulsive disorder: initial evidence from structural and functional MRI studies. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;91:49–59. doi: 10.1016/j.pnpbp.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 181.de Salles Andrade J.B. An MRI study of the metabolic and structural abnormalities in obsessive-compulsive disorder. Front. Hum. Neurosci. 2019;13:186. doi: 10.3389/fnhum.2019.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.McCutcheon R.A. Schizophrenia-an overview. JAMA Psychiatry. 2019:1–10. doi: 10.1001/jamapsychiatry.2019.3360. [DOI] [PubMed] [Google Scholar]
- 183.Davies G. A meta-analytic review of the relationship between neurocognition, metacognition and functional outcome in schizophrenia. J. Ment. Health. 2018:1–11. doi: 10.1080/09638237.2018.1521930. [DOI] [PubMed] [Google Scholar]
- 184.Zamanpoor M. Schizophrenia in a genomic era: a review from the pathogenesis, genetic and environmental etiology to diagnosis and treatment insights. Psychiatr. Genet. 2020;30(1):1–9. doi: 10.1097/YPG.0000000000000245. [DOI] [PubMed] [Google Scholar]
- 185.Tandon R. Schizophrenia, "Just the Facts": what we know in 2008 part 1: overview. Schizophr. Res. 2008;100(1-3):4–19. doi: 10.1016/j.schres.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 186.van Erp T.G. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry. 2016;21(4):585. doi: 10.1038/mp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cahn W. Brain volume changes in first-episode schizophrenia: a 1-year follow-up study. Arch. Gen. Psychiatry. 2002;59(11):1002–1010. doi: 10.1001/archpsyc.59.11.1002. [DOI] [PubMed] [Google Scholar]
- 188.De Peri L. Brain structural abnormalities at the onset of schizophrenia and bipolar disorder: a meta-analysis of controlled magnetic resonance imaging studies. Curr. Pharm. Des. 2012;18(4):486–494. doi: 10.2174/138161212799316253. [DOI] [PubMed] [Google Scholar]
- 189.Vita A. Progressive loss of cortical gray matter in schizophrenia: a meta-analysis and meta-regression of longitudinal MRI studies. Transl. Psychiatry. 2012;2:e19. doi: 10.1038/tp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Thompson P.M. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 2001;98(20):11650–11655. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Karlsgodt K.H. Diffusion imaging of white matter in schizophrenia: progress and future directions. Biol. Psychiatry Cognit. Neurosci. Neuroimaging. 2016;1(3):209–217. doi: 10.1016/j.bpsc.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Peters B.D. White matter fibertracking in first-episode schizophrenia, schizoaffective patients and subjects at ultra-high risk of psychosis. Neuropsychobiology. 2008;58(1):19–28. doi: 10.1159/000154476. [DOI] [PubMed] [Google Scholar]
- 193.Price G. The corpus callosum in first episode schizophrenia: a diffusion tensor imaging study. J. Neurol. Neurosurg. Psychiatry. 2005;76(4):585–587. doi: 10.1136/jnnp.2004.042952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kelly S. Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the ENIGMA Schizophrenia DTI Working Group. Mol. Psychiatry. 2018;23(5):1261–1269. doi: 10.1038/mp.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Birur B. Brain structure, function, and neurochemistry in schizophrenia and bipolar disorder-a systematic review of the magnetic resonance neuroimaging literature. NPJ Schizophr. 2017;3:15. doi: 10.1038/s41537-017-0013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fox J.M. Default mode functional connectivity is associated with social functioning in schizophrenia. J. Abnorm. Psychol. 2017;126(4):392–405. doi: 10.1037/abn0000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang Q. Anatomical insights into disrupted small-world networks in schizophrenia. Neuroimage. 2012;59(2):1085–1093. doi: 10.1016/j.neuroimage.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 198.Tarumi R. Levels of glutamatergic neurometabolites in patients with severe treatment-resistant schizophrenia: a proton magnetic resonance spectroscopy study. Neuropsychopharmacology. 2020;45(4):632–640. doi: 10.1038/s41386-019-0589-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Iwata Y. Glutamatergic neurometabolite levels in patients with ultra-treatment-resistant schizophrenia: a cross-sectional 3T proton magnetic resonance spectroscopy study. Biol. Psychiatry. 2019;85(7):596–605. doi: 10.1016/j.biopsych.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 200.Brugger S. Proton magnetic resonance spectroscopy and illness stage in schizophrenia–a systematic review and meta-analysis. Biol. Psychiatry. 2011;69(5):495–503. doi: 10.1016/j.biopsych.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 201.Sui J. In search of multimodal neuroimaging biomarkers of cognitive deficits in schizophrenia. Biol. Psychiatry. 2015;78(11):794–804. doi: 10.1016/j.biopsych.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cadena E.J. A longitudinal multimodal neuroimaging study to examine relationships between resting state glutamate and task related BOLD response in schizophrenia. Front. Psychiatry. 2018;9:632. doi: 10.3389/fpsyt.2018.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Isobe M. Multimodal neuroimaging as a window into the pathological physiology of schizophrenia: current trends and issues. Neurosci. Res. 2016;102:29–38. doi: 10.1016/j.neures.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 204.Aine C.J. Multimodal neuroimaging in schizophrenia: description and dissemination. Neuroinformatics. 2017;15(4):343–364. doi: 10.1007/s12021-017-9338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang K. Comparison of cerebral blood flow acquired by simultaneous [15O] water positron emission tomography and arterial spin labeling magnetic resonance imaging. J. Cereb. Blood Flow Metab. 2014;34(8):1373–1380. doi: 10.1038/jcbfm.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rosenkranz K. Present and future of simultaneous EEG-fMRI. Magnetic resonance materials in physics. Biol. Med. 2010;23(5-6):309–316. doi: 10.1007/s10334-009-0196-9. [DOI] [PubMed] [Google Scholar]
- 207.Goldman R.I. Simultaneous EEG and fMRI of the alpha rhythm. Neuroreport. 2002;13(18):2487. doi: 10.1097/01.wnr.0000047685.08940.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Laufs H. EEG-correlated fMRI of human alpha activity. Neuroimage. 2003;19(4):1463–1476. doi: 10.1016/s1053-8119(03)00286-6. [DOI] [PubMed] [Google Scholar]
- 209.Ritter P. simultaneous EEG–fMRI. Neurosci. Biobehav. Rev. 2006;30(6):823–838. doi: 10.1016/j.neubiorev.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 210.Eickhoff S.B. Assignment of functional activations to probabilistic cytoarchitectonic areas revisited. Neuroimage. 2007;36(3):511–521. doi: 10.1016/j.neuroimage.2007.03.060. [DOI] [PubMed] [Google Scholar]
- 211.Acton P.D. Quantification in PET. Radiol. Clin. 2004;42(6):1055–1062. doi: 10.1016/j.rcl.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 212.Zeeberg B.R. Accuracy of in vivo neuroreceptor quantification by PET and review of steady-state, transient, double injection, and equilibrium models. IEEE Trans. Med. Imaging. 1988;7(3):203–212. doi: 10.1109/42.7783. [DOI] [PubMed] [Google Scholar]
- 213.Oh S.H. Distortion correction in EPI at ultra‐high‐field MRI using PSF mapping with optimal combination of shift detection dimension. Magn. Reson. Med. 2012;68(4):1239–1246. doi: 10.1002/mrm.23317. [DOI] [PubMed] [Google Scholar]
- 214.Andersson J.L. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20(2):870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 215.Choi M. A new intensity-hue-saturation fusion approach to image fusion with a tradeoff parameter. IEEE Trans. Geosci. Remote Sens. 2006;44(6):1672–1682. [Google Scholar]
- 216.Kaur G. 2016 International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT) IEEE; 2016. Survey on multifocus image fusion techniques; pp. 1420–1424. [Google Scholar]
- 217.Phamila Y.A.V. Discrete Cosine Transform based fusion of multi-focus images for visual sensor networks. Signal Process. 2014;95:161–170. [Google Scholar]
- 218.Cao L. Multi-focus image fusion based on spatial frequency in discrete cosine transform domain. IEEE Signal Process Lett. 2014;22(2):220–224. [Google Scholar]
- 219.Li S. Multifocus image fusion using region segmentation and spatial frequency. Image Vision Comput. 2008;26(7):971–979. [Google Scholar]
- 220.Liu Y. Multi-focus image fusion with dense SIFT. Inf. Fusion. 2015;23:139–155. [Google Scholar]
- 221.Pu T. Contrast-based image fusion using the discrete wavelet transform. Opt. Eng. 2000:39. [Google Scholar]
- 222.Singh G. MHWT-a modified haar wavelet transformation for image fusion. Int. J. Comput. Appl. 2013;79(1) [Google Scholar]
- 223.Burt P. The Laplacian pyramid as a compact image code. IEEE Trans. Commun. 1983;31(4):532–540. [Google Scholar]
- 224.Vijayarajan R. Discrete wavelet transform based principal component averaging fusion for medical images. AEU-Int. J. Electron. Commun. 2015;69(6):896–902. [Google Scholar]
- 225.Singh R. Fusion of multimodal medical images using Daubechies complex wavelet transform–a multiresolution approach. Inf. Fusion. 2014;19:49–60. [Google Scholar]
- 226.Vulliemoz S. Simultaneous intracranial EEG and fMRI of interictal epileptic discharges in humans. Neuroimage. 2011;54(1):182–190. doi: 10.1016/j.neuroimage.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 227.Sui J. A review of multivariate methods for multimodal fusion of brain imaging data. J. Neurosci. Methods. 2012;204(1):68–81. doi: 10.1016/j.jneumeth.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zuzhang X. 2020. new_image_fusion. [cited 2020 2.23] [Google Scholar]
- 229.Johnson K.A. 2020. Brain Image.http://www.med.harvard.edu/AANLIB/cases/case9/mr1-tc1/020.html [cited 2020 02.24]; Available from. [Google Scholar]
- 230.Shen R. Cross-scale coefficient selection for volumetric medical image fusion. IEEE Trans. Biomed. Eng. 2012;60(4):1069–1079. doi: 10.1109/TBME.2012.2211017. [DOI] [PubMed] [Google Scholar]
- 231.Lewis J.J. Pixel-and region-based image fusion with complex wavelets. Inf. Fusion. 2007;8(2):119–130. [Google Scholar]
- 232.Nandi D. Principal component analysis in medical image processing: a study. Int. J. Image Min. 2015;1(1):65–86. [Google Scholar]
- 233.Vijayarajan R. Iterative block level principal component averaging medical image fusion. Optik. 2014;125(17):4751–4757. [Google Scholar]
- 234.Wang H.-q. Multi-mode medical image fusion algorithm based on principal component analysis. 2009 International Symposium on Computer Network and Multimedia TechnologyIEEE. 2009:1–4. [Google Scholar]
- 235.Krishn, A., et al., Medical Image Fusion Using Combination of PCA and Wavelet Analysis.
- 236.Wang L. EGGDD: an explicit dependency model for multi-modal medical image fusion in shift-invariant shearlet transform domain. Inf. Fusion. 2014;19:29–37. [Google Scholar]
- 237.Yang J. IEEE 60th Vehicular Technology Conference. IEEE; 2004. Image fusion using the expectation-maximization algorithm and a hidden Markov model; pp. 4563–4567. VTC2004-Fall. 2004. 2004. [Google Scholar]
- 238.Yang S. Contourlet hidden Markov Tree and clarity-saliency driven PCNN based remote sensing images fusion. Appl. Soft Comput. 2012;12(1):228–237. [Google Scholar]
- 239.Bhatnagar G. Human visual system inspired multi-modal medical image fusion framework. Expert Syst. Appl. 2013;40(5):1708–1720. [Google Scholar]
- 240.Daneshvar S. MRI and PET image fusion by combining IHS and retina-inspired models. Inf. Fusion. 2010;11(2):114–123. [Google Scholar]
- 241.Jang J.H. Contrast-enhanced fusion of multisensor images using subband-decomposed multiscale retinex. IEEE Trans. Image Process. 2012;21(8):3479–3490. doi: 10.1109/TIP.2012.2197014. [DOI] [PubMed] [Google Scholar]
- 242.Smith S.M. SUSAN—a new approach to low level image processing. Int. J. Comput. Vision. 1997;23(1):45–78. [Google Scholar]
- 243.He C. Multimodal medical image fusion based on IHS and PCA. Proc. Eng. 2010;7:280–285. [Google Scholar]
- 244.Zheng Y. A new metric based on extended spatial frequency and its application to DWT based fusion algorithms. Inf. Fusion. 2007;8(2):177–192. [Google Scholar]
- 245.Wencang Z. 2008 IEEE International Conference on Automation and Logistics. IEEE; 2008. Medical image fusion method based on wavelet multi-resolution and entropy; pp. 2329–2333. [Google Scholar]
- 246.Garg S. 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference. IEEE; 2006. Multilevel medical image fusion using segmented image by level set evolution with region competition; pp. 7680–7683. [DOI] [PubMed] [Google Scholar]
- 247.Li X. Wavelet Analysis and Applications. Springer; 2006. Medical image fusion by multi-resolution analysis of wavelets transform; pp. 389–396. [Google Scholar]
- 248.Bhatnagar G. Directive contrast based multimodal medical image fusion in NSCT domain. IEEE Trans. Multimed. 2013;15(5):1014–1024. [Google Scholar]
- 249.Zhu X.L. Investigation of remote sensing image fusion strategy applying PCA to wavelet packet analysis based on IHS transform. J. Indian Soc. Remote Sens. 2019;47(3):413–425. [Google Scholar]
- 250.Deepa B. An intensity factorized thresholding based segmentation technique with gradient discrete wavelet fusion for diagnosing stroke and tumor in brain MRI. Multidimen. Syst. Signal Process. 2019;30(4):2081–2112. [Google Scholar]
- 251.Phillips P. Detection of Alzheimer's disease and mild cognitive impairment based on structural volumetric MR images using 3D-DWT and WTA-KSVM trained by PSOTVAC. Biomed. Signal Process. Control. 2015;21:58–73. [Google Scholar]
- 252.Prakash O. CT and MR images fusion based on stationary wavelet transform by modulus maxima. In: Sethi I.K., editor. Computational Vision and Robotics. Springer-Verlag; Berlin: Berlin: 2015. pp. 199–204. [Google Scholar]
- 253.Pawar G.A. Computing, Communication and Signal Processing (ICCASP) Springer International Publishing Ag; Cham: 2019. Multi-focal image fusion with convolutional sparse representation and stationary wavelet transform; pp. 865–873. [Google Scholar]
- 254.Li Y. Detection of dendritic spines using wavelet packet entropy and fuzzy support vector machine. CNS Neurol. Disord. 2017;16(2):116–121. doi: 10.2174/1871527315666161111123638. [DOI] [PubMed] [Google Scholar]
- 255.Yang J. Preclinical diagnosis of magnetic resonance (MR) brain images via discrete wavelet packet transform with Tsallis entropy and generalized eigenvalue proximal support vector machine (GEPSVM) Entropy. 2015;17(4):1795–1813. [Google Scholar]
- 256.Sreekala K. International Conference on Circuit, Power and Computing Technologies. IEEE; Karnataka, India: 2016. Wavelet packet transform based fusion of misaligned images. [Google Scholar]
- 257.Shah P. Fusion of surveillance images in infrared and visible band using curvelet, wavelet and wavelet packet transform. Int. J. Wavel. Multiresol. Inf. Process. 2010;8(2):271–292. [Google Scholar]
- 258.Choubey A. Novel data-access scheme and efficient parallel architecture for multi-level lifting 2-D DWT. Circ. Syst. Signal Process. 2018;37(10):4482–4503. [Google Scholar]
- 259.Shiralashetti S.C. Wavelet-based lifting scheme for the numerical solution of some class of nonlinear partial differential equations. Int. J. Wavel. Multiresol. Inf. Process. 2018;16(5):14. 1850046. [Google Scholar]
- 260.Prakash O. Multiscale fusion of multimodal medical images using lifting scheme based biorthogonal wavelet transform. Optik. 2019;182:995–1014. [Google Scholar]
- 261.Haouam I. International Conference on Signal, Image, Vision and Their Applications. IEEE; Guelma, Algeria: 2018. MRI image compression using level set method and biorthogonal CDF wavelet based on lifting scheme. [Google Scholar]
- 262.Zemouri E.T. Nonsubsampled contourlet transform and k-means clustering for degraded document image binarization. J. Electron. Imaging. 2019;28(4):19. Article ID. 043021. [Google Scholar]
- 263.Ramlal S.D. An improved multimodal medical image fusion scheme based on hybrid combination of nonsubsampled contourlet transform and stationary wavelet transform. Int. J. Imaging Syst. Technol. 2019;29(2):146–160. [Google Scholar]
- 264.Li L.L. A practical medical image enhancement algorithm based on nonsubsampled contourlet transform. J. Med. Imaging Health Inform. 2019;9(5):1046–1056. [Google Scholar]
- 265.Wang C. Multi-modality anatomical and functional medical image fusion based on simplified-spatial frequency-pulse coupled neural networks and region energy-weighted average strategy in non-sub sampled contourlet transform domain. J. Med. Imaging Health Inform. 2019;9(5):1017–1027. [Google Scholar]
- 266.Li L.L. A novel medical image fusion approach based on nonsubsampled shearlet transform. J. Med. Imaging Health Inform. 2019;9(9):1815–1826. [Google Scholar]
- 267.Vishwakarma A. Image fusion using adjustable non-subsampled shearlet transform. IEEE Trans. Instrum. Meas. 2019;68(9):3367–3378. [Google Scholar]
- 268.Akbarpour T. Medical image fusion based on nonsubsampled shearlet transform and principal component averaging. Int. J. Wavel. Multiresol. Inf. Process. 2019;17(4):21. Article ID. 1950023. [Google Scholar]
- 269.Yang B. Pixel-level image fusion with simultaneous orthogonal matching pursuit. Inf. Fusion. 2012;13(1):10–19. [Google Scholar]
- 270.Li S. Multimodal image fusion with joint sparsity model. Opt. Eng. 2011;50(6) [Google Scholar]
- 271.Yu N. Image features extraction and fusion based on joint sparse representation. IEEE J. Sel. Top. Signal Process. 2011;5(5):1074–1082. [Google Scholar]
- 272.Xu Z.P. Medical image fusion using multi-level local extrema. Inf. Fusion. 2014;19:38–48. [Google Scholar]
- 273.Zhu H.R. Infrared and visible image fusion based on contrast enhancement and multi-scale edge-preserving decomposition. J. Electron. Inf. Technol. 2018;40(6):1294–1300. [Google Scholar]
- 274.Kou F. Edge-preserving smoothing pyramid based multi-scale exposure fusion. J. Visual Commun. Image Represent. 2018;53:235–244. [Google Scholar]
- 275.Petrović V. Subjective tests for image fusion evaluation and objective metric validation. Inf. Fusion. 2007;8(2):208–216. [Google Scholar]
- 276.Sheikh H.R. Image information and visual quality. IEEE Trans. Image Process. 2006;15(2):430–444. doi: 10.1109/tip.2005.859378. [DOI] [PubMed] [Google Scholar]
- 277.Yang Y. User models of subjective image quality assessment on virtual viewpoint in free-viewpoint video system. Multimed. Tools Appl. 2016;75(20):12499–12519. [Google Scholar]
- 278.Du J. An overview of multi-modal medical image fusion. Neurocomputing. 2016;215:3–20. [Google Scholar]
- 279.Du J. An overview of multi-modal medical image fusion. Neurocomputing. 2016;215:3–20. [Google Scholar]
- 280.Yang Y. Contourlet-based image quality assessment for synthesised virtual image. Electron. Lett. 2010;46(7):492–493. [Google Scholar]
- 281.Wang Z. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 2004;13(4):600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 282.Miao Q.G. A novel algorithm of image fusion using shearlets. Opt. Commun. 2011;284(6):1540–1547. [Google Scholar]
- 283.Hossny M. Comments on 'Information measure for performance of image fusion'. Electron. Lett. 2008;44(18):1066–1067. [Google Scholar]
- 284.Horibe Y. Entropy and correlation. IEEE Trans. Syst. Man Cybern. 1985;SMC-15(5):641–642. [Google Scholar]
- 285.Eskicioglu A.M. Image quality measures and their performance. IEEE Trans. Commun. 1995;43(12):2959–2965. [Google Scholar]
- 286.Mittal A. Making a "completely blind" image quality analyzer. IEEE Signal Process. Lett.. 2013;20(3):209–212. [Google Scholar]
- 287.Herzog H. The current state, challenges and perspectives of MR-PET. Neuroimage. 2010;49(3):2072–2208. doi: 10.1016/j.neuroimage.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 288.Schlemmer H.-P.W. Simultaneous MR/PET imaging of the human brain: feasibility study. Radiology. 2008;248(3):1028–1035. doi: 10.1148/radiol.2483071927. [DOI] [PubMed] [Google Scholar]
- 289.Grazioso R. APD-based PET for combined MR-PET imaging. Proc. Intl. Soc. Mag. Reson. Med. 2005:408. [Google Scholar]
- 290.Hamilton B.E. Comparative analysis of ferumoxytol and gadoteridol enhancement using T1-and T2-weighted MRI in neuroimaging. Am. J. Roentgenol. 2011;197(4):981–988. doi: 10.2214/AJR.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Just M. Tissue characterization with T1, T2, and proton density values: results in 160 patients with brain tumors. Radiology. 1988;169(3):779–785. doi: 10.1148/radiology.169.3.3187000. [DOI] [PubMed] [Google Scholar]
- 292.Xie S. Alcoholism identification based on an AlexNet transfer learning model. Front.Psychiatry. 2019;10 doi: 10.3389/fpsyt.2019.00205. Article ID. 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Dawood Y. Novel imaging techniques to study postmortem human fetal anatomy: a systematic review on microfocus-CT and ultra-high-field MRI. Eur. Radiol. 2020;30(4):2280–2292. doi: 10.1007/s00330-019-06543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Tuzzi E. Ultra-high field mri in Alzheimer's disease: effective transverse relaxation rate and quantitative susceptibility mapping of human brain in vivo and ex vivo compared to histology. J. Alzheimers Dis. 2020;73(4):1481–1499. doi: 10.3233/JAD-190424. [DOI] [PubMed] [Google Scholar]
- 295.Buxton R.B. Cambridge University Press; 2009. Introduction to Functional Magnetic Resonance Imaging: Principles and Techniques. [Google Scholar]
- 296.Rosenkranz K. Present and future of simultaneous EEG-fMRI. Magn. Reson. Mater. Phys. Biol. Med, 2010;23(5):309–316. doi: 10.1007/s10334-009-0196-9. [DOI] [PubMed] [Google Scholar]
- 297.Laufs H. A personalized history of EEG–fMRI integration. Neuroimage. 2012;62(2):1056–1067. doi: 10.1016/j.neuroimage.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 298.Medič J. Off-resonance frequency filtered magnetic resonance imaging. Magn. Reson. Imaging. 2010;28(4):527–536. doi: 10.1016/j.mri.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 299.Hellier P. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2000. Multimodal non-rigid warping for correction of distortions in functional MRI; pp. 512–520. [Google Scholar]
- 300.Holland D. Efficient correction of inhomogeneous static magnetic field-induced distortion in Echo Planar Imaging. Neuroimage. 2010;50(1):175–183. doi: 10.1016/j.neuroimage.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chen Z. From simultaneous to synergistic MR‐PET brain imaging: a review of hybrid MR‐PET imaging methodologies. Hum. Brain Mapp. 2018;39(12):5126–5144. doi: 10.1002/hbm.24314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ullisch M.G. MR-based PET motion correction procedure for simultaneous MR-PET neuroimaging of human brain. PLoS ONE. 2012;7(11):e48149. doi: 10.1371/journal.pone.0048149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ehrhardt M.J. Joint reconstruction of PET-MRI by exploiting structural similarity. Inverse Problems. 2014;31(1) [Google Scholar]
- 304.Iglesias J.E. Multi-atlas segmentation of biomedical images: a survey. Med. Image Anal. 2015;24(1):205–219. doi: 10.1016/j.media.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Evans A.C. Brain templates and atlases. Neuroimage. 2012;62(2):911–922. doi: 10.1016/j.neuroimage.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 306.Talairach J. Masson; 1957. Atlas d'anatomie Stereotaxique du Telencephale: Etudes Anatomo-Radiologiques. [Google Scholar]
- 307.Talairach J., Tournoux . 3-Dimensional Proportional System: An Approach to Cerebral Imaging. 1988. Co-planar stereotaxic atlas of the human brain. 1988. [Google Scholar]
- 308.Tzourio-Mazoyer N. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 309.Collins D.L. Design and construction of a realistic digital brain phantom. IEEE Trans. Med. Imaging. 1998;17(3):463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 310.Heckemann R.A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 311.Zhang Y.Y. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 312.Ashburner J. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 313.Cabezas M. A review of atlas-based segmentation for magnetic resonance brain images. Comput. Methods Programs Biomed. 2011;104(3):E158–E177. doi: 10.1016/j.cmpb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 314.Ashburner J. Voxel-based morphometry - the methods. Neuroimage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 315.Hill D.L.G. Medical image registration. Phys. Med. Biol. 2001;46(3):R1–R45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- 316.Rohlfing T. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004;21(4):1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 317.Klein A. Mindboggle: automated brain labeling with multiple atlases. BMC Med. Imaging. 2005;5:7. doi: 10.1186/1471-2342-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Artaechevarria X. Combination strategies in multi-atlas image segmentation: application to brain MR Data. IEEE Trans. Med. Imaging. 2009;28(8):1266–1277. doi: 10.1109/TMI.2009.2014372. [DOI] [PubMed] [Google Scholar]
- 319.Rohlfing T. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans. Med. Imaging. 2004;23(8):983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- 320.Rohlfing T. Multi-classifier framework for atlas-based image segmentation. Pattern Recognit. Lett. 2005;26(13):2070–2079. [Google Scholar]
- 321.Maes F. Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imaging. 1997;16(2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 322.Sdika M. Combining atlas based segmentation and intensity classification with nearest neighbor transform and accuracy weighted vote. Med. Image Anal. 2010;14(2):219–226. doi: 10.1016/j.media.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 323.Gholipour A. Multi-atlas multi-shape segmentation of fetal brain MRI for volumetric and morphometric analysis of ventriculomegaly. Neuroimage. 2012;60(3):1819–1831. doi: 10.1016/j.neuroimage.2012.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Gorthi S. Weighted shape-based averaging with neighborhood prior model for multiple atlas fusion-based medical image segmentation. IEEE Signal Process Lett. 2013;20(11):1036–1039. [Google Scholar]
- 325.Garcia-Pedrajas N. An empirical study of binary classifier fusion methods for multiclass classification. Inf. Fusion. 2011;12(2):111–130. [Google Scholar]
- 326.Nweke H.F. Data fusion and multiple classifier systems for human activity detection and health monitoring: review and open research directions. Inf. Fusion. 2019;46:147–170. [Google Scholar]
- 327.Yilmaz M.B. Score level fusion of classifiers in off-line signature verification. Inf. Fusion. 2016;32:109–119. [Google Scholar]
- 328.Viswanath P. Fusion of multiple approximate nearest neighbor classifiers for fast and efficient classification. Inf. Fusion. 2004;5(4):239–250. [Google Scholar]
- 329.Castillo-Barnes D. Robust ensemble classification methodology for I123-Ioflupane SPECT images and multiple heterogeneous biomarkers in the diagnosis of Parkinson's disease. Front. Neuroinform. 2018;12:16. doi: 10.3389/fninf.2018.00053. Article ID. 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ramirez J. Ensemble of random forests One vs. Rest classifiers for MCI and AD prediction using ANOVA cortical and subcortical feature selection and partial least squares. J. Neurosci. Methods. 2018;302:47–57. doi: 10.1016/j.jneumeth.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 331.Lam L. Optimal combinations of pattern classifiers. Pattern Recognit. Lett. 1995;16(9):945–954. [Google Scholar]
- 332.Woods K. Combination of multiple classifiers using local accuracy estimates. IEEE Trans. Pattern Anal. Mach. Intell. 1997;19(4):405–410. [Google Scholar]
- 333.Sedvall G. Imaging of neurotransmitter receptors in the living human-brain. Arch. Gen. Psychiatry. 1986;43(10):995–1005. doi: 10.1001/archpsyc.1986.01800100089012. [DOI] [PubMed] [Google Scholar]
- 334.Shiri I. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC) Eur. Radiol. 2019;29(12):6867–6879. doi: 10.1007/s00330-019-06229-1. [DOI] [PubMed] [Google Scholar]
- 335.Sarikaya I. PET studies in epilepsy. Am. J. Nucl. Med. Mol. Imaging. 2015;5(5):416–430. [PMC free article] [PubMed] [Google Scholar]
- 336.Jones T. History and future technical innovation in positron emission tomography. J Med. Imaging. 2017;4(1):17. doi: 10.1117/1.JMI.4.1.011013. Article ID. 011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Hasegawa B.H. Dual-modality imaging: more than the sum of its components. In: Zaidi H., editor. Quantitative Analysis in Nuclear Medicine Imaging. Springer US; Boston, MA: 2006. pp. 35–81. Editor. [Google Scholar]
- 338.Lillington J. PET/MRI attenuation estimation in the lung: a review of past, present, and potential techniques. Med. Phys. 2020;47(2):790–811. doi: 10.1002/mp.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zaidi H. Overview of nuclear medical imaging: physics and instrumentation. In: Zaidi H., editor. Quantitative Analysis in Nuclear Medicine Imaging. Springer US; Boston, MA: 2006. pp. 1–34. Editor. [Google Scholar]
- 340.Bettinardi V. PET quantification: strategies for partial volume correction. Clin. Transl. Imaging. 2014;2(3):199–218. [Google Scholar]
- 341.Dickson J. Quantitative SPECT: the time is now. Ejnmmi Phys. 2019;6:7. doi: 10.1186/s40658-019-0241-3. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Nordberg A. The use of PET in Alzheimer disease. Nat. Rev. Neurol. 2010;6(2):78–87. doi: 10.1038/nrneurol.2009.217. [DOI] [PubMed] [Google Scholar]
- 343.Okamura N. Brain imaging: applications of tau PET imaging. Nat. Rev. Neurol. 2017;13(4):197–198. doi: 10.1038/nrneurol.2017.38. [DOI] [PubMed] [Google Scholar]
- 344.Seibyl J. Impact of training method on the robustness of the visual assessment of 18F-Florbetaben PET scans: results from a phase-3 study. J. Nucl. Med. 2016;57(6):900–906. doi: 10.2967/jnumed.115.161927. [DOI] [PubMed] [Google Scholar]
- 345.Joshi A.D. A semiautomated method for quantification of F 18 florbetapir PET images. J. Nucl. Med. 2015;56(11):1736–1741. doi: 10.2967/jnumed.114.153494. [DOI] [PubMed] [Google Scholar]
- 346.Marcoux A. An automated pipeline for the analysis of PET data on the cortical surface. Front. Neuroinform. 2018;12:13. doi: 10.3389/fninf.2018.00094. Article ID. 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Tahmi M. A fully automatic technique for precise localization and quantification of amyloid-beta PET scans. J. Nucl. Med. 2019;60(12):1771–1779. doi: 10.2967/jnumed.119.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Foster B. A review on segmentation of positron emission tomography images. Comput. Biol. Med. 2014;50:76–96. doi: 10.1016/j.compbiomed.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zasadny K.R. Standardized uptake values of normal tissues at PET with 2-[fluorine-18]-fluoro-2-deoxy-D-glucose: variations with body weight and a method for correction. Radiology. 1993;189(3):847–850. doi: 10.1148/radiology.189.3.8234714. [DOI] [PubMed] [Google Scholar]
- 350.Kim C.K. Standardized uptake values of FDG: body surface area correction is preferable to body weight correction. J. Nucl. Med. 1994;35(1):164–167. [PubMed] [Google Scholar]
- 351.Basu S. Quantitative techniques in PET-CT imaging. Curr. Med. Imaging Reviews. 2011;7(3):216–233. [Google Scholar]
- 352.Huang S.-C. Anatomy of SUV. Nucl. Med. Biol. 2000;27(7):643–646. doi: 10.1016/s0969-8051(00)00155-4. [DOI] [PubMed] [Google Scholar]
- 353.Fahey F.H. Variability in PET quantitation within a multicenter consortium. Med. Phys. 2010;37(7):3660–3666. doi: 10.1118/1.3455705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Holman B.F. Improved correction for the tissue fraction effect in lung PET/CT imaging. Phys. Med. Biol. 2015;60(18):7387–7402. doi: 10.1088/0031-9155/60/18/7387. [DOI] [PubMed] [Google Scholar]
- 355.Rahmim A. Resolution modeling in PET imaging: theory, practice, benefits, and pitfalls. Med. Phys. 2013;40(6) doi: 10.1118/1.4800806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Bailey D.L. Quantitative SPECT/CT: SPECT joins PET as a quantitative imaging modality. Eur. J. Nucl. Med. Mol. Imaging. 2014;41:S17–S25. doi: 10.1007/s00259-013-2542-4. [DOI] [PubMed] [Google Scholar]
- 357.Ritt P. Absolute quantification in SPECT. Eur. J. Nucl. Med. Mol. Imaging. 2011;38(1):69–77. doi: 10.1007/s00259-011-1770-8. [DOI] [PubMed] [Google Scholar]
- 358.Yang J.R. Partial volume correction for PET quantification and its impact on brain network in Alzheimer's disease. Sci. Rep. 2017;7:14. doi: 10.1038/s41598-017-13339-7. Article ID. 13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Aston J.A.D. Positron emission tomography partial volume correction: estimation and algorithms. J. Cereb. Blood Flow Metab. 2002;22(8):1019–1034. doi: 10.1097/00004647-200208000-00014. [DOI] [PubMed] [Google Scholar]
- 360.Soret M. Partial-volume effect in PET tumor imaging. J. Nucl. Med. 2007;48(6):932–945. doi: 10.2967/jnumed.106.035774. [DOI] [PubMed] [Google Scholar]
- 361.Rousset O.G. Correction for partial volume effects in emission tomography. In: Zaidi H., editor. Quantitative Analysis in Nuclear Medicine Imaging. Springer US; Boston, MA: 2006. pp. 236–271. [Google Scholar]
- 362.Rousset O. Partial volume correction strategies in PET. PET Clin. 2007;2(2):235–249. doi: 10.1016/j.cpet.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 363.Erlandsson K. A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Phys. Med. Biol. 2012;57(21):R119–R159. doi: 10.1088/0031-9155/57/21/R119. [DOI] [PubMed] [Google Scholar]
- 364.Videen T.O. Regional correction of positron emission tomography data for the effects of cerebral atrophy. J. Cereb. Blood Flow Metab. 1988;8(5):662–670. doi: 10.1038/jcbfm.1988.113. [DOI] [PubMed] [Google Scholar]
- 365.Meltzer C.C. Correction of PET data for partial volume effects in human cerebral-cortex by MR imaging. J. Comput. Assist. Tomogr. 1990;14(4):561–570. doi: 10.1097/00004728-199007000-00011. [DOI] [PubMed] [Google Scholar]
- 366.Mullergartner H.W. Measurement of radiotracer concentration in brain gray-matter using positron emission tomography - MRI-based correction for partial volume effects. J. Cereb. Blood Flow Metab. 1992;12(4):571–583. doi: 10.1038/jcbfm.1992.81. [DOI] [PubMed] [Google Scholar]
- 367.Meltzer C.C. MR-based correction of brain PET measurements for heterogeneous gray matter radioactivity distribution. J. Cereb. Blood Flow Metab. 1996;16(4):650–658. doi: 10.1097/00004647-199607000-00016. [DOI] [PubMed] [Google Scholar]
- 368.Hutton B.F. Iterative reconstruction methods. In: Zaidi H., editor. Quantitative Analysis in Nuclear Medicine Imaging. Springer US; Boston, MA: 2006. pp. 107–140. [Google Scholar]
- 369.Srinivas S.M. A recovery coefficient method for partial volume correction of PET images. Ann. Nucl. Med. 2009;23(4):341–348. doi: 10.1007/s12149-009-0241-9. [DOI] [PubMed] [Google Scholar]
- 370.Catana C. PET/MRI for neurologic applications. J. Nucl. Med. 2012;53(12):1916–1925. doi: 10.2967/jnumed.112.105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Huesman R.H. A new fast algorithm for the evaluation of regions of interest and statistical uncertainty in computed-tomography. Phys. Med. Biol. 1984;29(5):543–552. doi: 10.1088/0031-9155/29/5/007. [DOI] [PubMed] [Google Scholar]
- 372.Muzic R.F. A method to correct for scatter, spillover, and partial volume effects in region of interest analysis in PET. IEEE Trans. Med. Imaging. 1998;17(2):202–213. doi: 10.1109/42.700732. [DOI] [PubMed] [Google Scholar]
- 373.Carson R.E. A maximum likelihood method for region-of-interest evaluation in emission tomography. J. Comput. Assist. Tomogr. 1986;10(4):654–663. doi: 10.1097/00004728-198607000-00021. [DOI] [PubMed] [Google Scholar]
- 374.Rousset O.G. Correction for partial volume effects in PET: principle and validation. J. Nucl. Med. 1998;39(5):904–911. [PubMed] [Google Scholar]
- 375.Frouin V. Correction of partial-volume effect for PET striatal imaging: fast implementation and study of robustness. J. Nucl. Med. 2002;43(12):1715–1726. [PubMed] [Google Scholar]
- 376.Du Y. Partial volume effect compensation for quantitative brain SPECT imaging. IEEE Trans. Med. Imaging. 2005;24(8):969–976. doi: 10.1109/TMI.2005.850547. [DOI] [PubMed] [Google Scholar]
- 377.Sattarivand M. Symmetric geometric transfer matrix partial volume correction for PET imaging: principle, validation and robustness. Phys. Med. Biol. 2012;57(21):7101–7116. doi: 10.1088/0031-9155/57/21/7101. [DOI] [PubMed] [Google Scholar]
- 378.Sureau F.C. Impact of image-space resolution modeling for studies with the high-resolution research tomograph. J. Nucl. Med. 2008;49(6):1000–1008. doi: 10.2967/jnumed.107.045351. [DOI] [PubMed] [Google Scholar]
- 379.Akamatsu G. Improvement in PET/CT image quality with a combination of point-spread function and time-of-flight in relation to reconstruction parameters. J. Nucl. Med. 2012;53(11):1716–1722. doi: 10.2967/jnumed.112.103861. [DOI] [PubMed] [Google Scholar]
- 380.Andersen F.L. Clinical evaluation of PET image reconstruction using a spatial resolution model. Eur. J. Radiol. 2013;82(5):862–869. doi: 10.1016/j.ejrad.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 381.Bowen S.L. Influence of the partial volume correction method on F-18-fluorodeoxyglucose brain kinetic modelling from dynamic PET images reconstructed with resolution model based OSEM. Phys. Med. Biol. 2013;58(20):7081–7106. doi: 10.1088/0031-9155/58/20/7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Sibarita J.-B. Deconvolution microscopy. In: Rietdorf J, editor. Microscopy Techniques. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. pp. 201–243. Editor. [Google Scholar]
- 383.Boussion N. A multiresolution image based approach for correction of partial volume effects in emission tomography. Phys. Med. Biol. 2006;51(7):1857–1876. doi: 10.1088/0031-9155/51/7/016. [DOI] [PubMed] [Google Scholar]
- 384.Quarantelli M. Integrated software for the analysis of brain PET/SPECT studies with partial-volume-effect correction. J. Nucl. Med. 2004;45(2):192–201. [PubMed] [Google Scholar]
- 385.Svarer C. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage. 2005;24(4):969–979. doi: 10.1016/j.neuroimage.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 386.Zaidi H. Attenuation correction strategies in emission tomography. In: Zaidi H., editor. Quantitative Analysis in Nuclear Medicine Imaging. Springer US; Boston, MA: 2006. pp. 167–204. [Google Scholar]
- 387.Mehranian A. Vision 20/20: magnetic resonance imaging-guided attenuation correction in PET/MRI: challenges, solutions, and opportunities. Med. Phys. 2016;43(3):1130–1155. doi: 10.1118/1.4941014. [DOI] [PubMed] [Google Scholar]
- 388.Hofmann M. Towards quantitative PET/MRI: a review of MR-based attenuation correction techniques. Eur. J. Nucl. Med. Mol. Imaging. 2009;36:93–104. doi: 10.1007/s00259-008-1007-7. [DOI] [PubMed] [Google Scholar]
- 389.Zaidi H. Attenuation compensation in cerebral 3D PET: effect of the attenuation map on absolute and relative quantitation. Eur. J. Nucl. Med. Mol. Imaging. 2004;31(1):52–63. doi: 10.1007/s00259-003-1325-8. [DOI] [PubMed] [Google Scholar]
- 390.Weinzapfel B.T. Automated PET attenuation correction model for functional brain imaging. J. Nucl. Med. 2001;42(3):483–491. [PubMed] [Google Scholar]
- 391.Watabe H. Acquisition of attenuation map for brain PET study using optical tracking system. In: Seibert J.A., editor. vols 1-4. Ieee; New York: 2002. pp. 1458–1461. (IEEE Nuclear Science Symposium, Conference Records). Editor. [Google Scholar]
- 392.Nuyts J. Simultaneous maximum a posteriori reconstruction of attenuation and activity distributions from emission sinograms. IEEE Trans. Med. Imaging. 1999;18(5):393–403. doi: 10.1109/42.774167. [DOI] [PubMed] [Google Scholar]
- 393.Nuyts J. Completion of a truncated attenuation image from the attenuated PET emission data. IEEE Trans. Med. Imaging. 2013;32(2):237–246. doi: 10.1109/TMI.2012.2220376. [DOI] [PubMed] [Google Scholar]
- 394.Rezaei A. ML-reconstruction for TOF-PET with simultaneous estimation of the attenuation factors. IEEE Trans. Med. Imaging. 2014;33(7):1563–1572. doi: 10.1109/TMI.2014.2318175. [DOI] [PubMed] [Google Scholar]
- 395.Benoit D. Optimized MLAA for quantitative non-TOF PET/MR of the brain. Phys. Med. Biol. 2016;61(24):8854–8874. doi: 10.1088/1361-6560/61/24/8854. [DOI] [PubMed] [Google Scholar]
- 396.Ladefoged C.N. A multi-centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage. 2017;147:346–359. doi: 10.1016/j.neuroimage.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Bailey D.L. Transmission scanning in emission tomography. Eur. J. Nucl. Med. 1998;25(7):774–787. doi: 10.1007/s002590050282. [DOI] [PubMed] [Google Scholar]
- 398.Ichihara T. Evaluation of SPET quantification of simultaneous emission and transmission imaging of the brain using a multidetector SPET system with the TEW scatter compensation method and fan-beam collimation. Eur. J. Nucl. Med. 1996;23(10):1292–1299. doi: 10.1007/BF01367583. [DOI] [PubMed] [Google Scholar]
- 399.Van Laere K. Nonuniform transmission in brain SPECT using 201Tl, 153Gd, and 99mTc static line sources: anthropomorphic dosimetry studies and influence on brain quantification. J. Nucl. Med. 2000;41(12):2051–2062. [PubMed] [Google Scholar]
- 400.Brown S. Investigation of the relationship between linear attenuation coefficients and CT Hounsfield units using radionuclides for SPECT. Appl. Radiat. Isot. 2008;66(9):1206–1212. doi: 10.1016/j.apradiso.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 401.Patton J.A. Image fusion using an integrated, dual-head coincidence camera with x-ray tube-based attenuation maps. J. Nucl. Med. 2000;41(8):1364–1368. [PubMed] [Google Scholar]
- 402.Kamel E.M. Impact of metallic dental implants on CT-based attenuation correction in a combined PET/CT scanner. Eur. Radiol. 2003;13(4):724–728. doi: 10.1007/s00330-002-1564-2. [DOI] [PubMed] [Google Scholar]
- 403.Kinahan P.E. X-ray-based attenuation correction for positron emission tomography/computed tomography scanners. Semin. Nucl. Med. 2003;33(3):166–179. doi: 10.1053/snuc.2003.127307. [DOI] [PubMed] [Google Scholar]
- 404.Carney J.P.J. Method for transforming CT images for attenuation correction in PET/CT imaging. Med. Phys. 2006;33(4):976–983. doi: 10.1118/1.2174132. [DOI] [PubMed] [Google Scholar]
- 405.Wollenweber S.D. Evaluation of an atlas-based PET head attenuation correction using PET/CT & MR patient data. IEEE Trans. Nucl. Sci. 2013;60(5):3383–3390. [Google Scholar]
- 406.Stodilka R.Z. Scatter and attenuation correction for brain SPECT using attenuation distributions inferred from a head atlas. J. Nucl. Med. 2000;41(9):1569–1578. [PubMed] [Google Scholar]
- 407.Zaidi H. Magnetic resonance imaging-guided attenuation and scatter corrections in three-dimensional brain positron emission tomography. Med. Phys. 2003;30(5):937–948. doi: 10.1118/1.1569270. [DOI] [PubMed] [Google Scholar]
- 408.Wagenknecht G. Knowledge-based segmentation of attenuation-relevant regions of the head in T1-weighted MR images for attenuation correction in MR/PET systems. In: Yu B., editor. 2009 IEEE Nuclear Science Symposium Conference Record, vols 1-5. Ieee; New York: 2009. p. 3338. [Google Scholar]
- 409.Yang J. Quantitative evaluation of atlas-based attenuation correction for brain PET in an integrated time-of-flight PET/MR imaging system. Radiology. 2017;284(1):169–179. doi: 10.1148/radiol.2017161603. [DOI] [PubMed] [Google Scholar]
- 410.Bal H. Evaluation of MLACF based calculated attenuation brain PET imaging for FDG patient studies. Phys. Med. Biol. 2017;62(7):2542–2558. doi: 10.1088/1361-6560/aa5e99. [DOI] [PubMed] [Google Scholar]
- 411.Yang J. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain (18)F-FDG PET. Phys. Med. Biol. 2019;64(7) doi: 10.1088/1361-6560/ab0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Le Goff-Rougetet R. Segmented MR images for brain attenuation correction in PET. Med. Imaging. 1994;2167 SPIE. [Google Scholar]
- 413.Keereman V. MRI-based attenuation correction for pet/mri using ultrashort echo time sequences. J. Nucl. Med. 2010;51(5):812–818. doi: 10.2967/jnumed.109.065425. [DOI] [PubMed] [Google Scholar]
- 414.Martinez-Moller A. Tissue classification as a potential approach for attenuation correction in whole-body PET/MRI: evaluation with PET/CT data. J. Nucl. Med. 2009;50(4):520–526. doi: 10.2967/jnumed.108.054726. [DOI] [PubMed] [Google Scholar]
- 415.Berker Y. MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echo-time/dixon MRI sequence. J. Nucl. Med. 2012;53(5):796–804. doi: 10.2967/jnumed.111.092577. [DOI] [PubMed] [Google Scholar]
- 416.Andersen F.L. Combined PET/MR imaging in neurology: MR-based attenuation correction implies a strong spatial bias when ignoring bone. Neuroimage. 2014;84:206–216. doi: 10.1016/j.neuroimage.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 417.Kazerooni A.F. Generation of MR-based attenuation correction map of PET images in the brain employing joint segmentation of skull and soft-tissue from single short-TE MR imaging modality. In: Gao F., Shi K., Li S., editors. Computational Methods for Molecular Imaging. Springer International Publishing: Cham; 2015. pp. 139–147. Editors. [Google Scholar]
- 418.Khateri P. Generation of a four-class attenuation map for MRI-based attenuation correction of PET data in the head area using a novel combination of STE/Dixon-MRI and FCM clustering. Mol. Imaging Biol. 2015;17(6):884–892. doi: 10.1007/s11307-015-0849-1. [DOI] [PubMed] [Google Scholar]
- 419.Wiesinger F. Zero TE MR bone imaging in the head. Magn. Reson. Med. 2016;75(1):107–114. doi: 10.1002/mrm.25545. [DOI] [PubMed] [Google Scholar]
- 420.Yang J. Evaluation of sinus/edge-corrected zero-echo-time-based attenuation correction in brain PET/MRI. J. Nucl. Med. 2017;58(11):1873–1879. doi: 10.2967/jnumed.116.188268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Delso G. Improving PET/MR brain quantitation with template-enhanced ZTE. Neuroimage. 2018;181:403–413. doi: 10.1016/j.neuroimage.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 422.Sousa J.M. Evaluation of zero-echo-time attenuation correction for integrated PET/MR brain imaging-comparison to head atlas and (68)Ge-transmission-based attenuation correction. EJNMMI Phys. 2018;5(1):20. doi: 10.1186/s40658-018-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Sgard B. ZTE MR-based attenuation correction in brain FDG-PET/MR: performance in patients with cognitive impairment. Eur. Radiol. 2020;30(3):1770–1779. doi: 10.1007/s00330-019-06514-z. [DOI] [PubMed] [Google Scholar]
- 424.Roy S. PET attenuation correction using synthetic CT from ultrashort echo-time MR imaging. J. Nucl. Med. 2014;55(12):2071–2077. doi: 10.2967/jnumed.114.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Poynton C.B. Probabilistic atlas-based segmentation of combined T1-weighted and DUTE MRI for calculation of head attenuation maps in integrated PET/MRI scanners. Am. J. Nucl. Med. Mol. Imaging. 2014;4(2):160–171. [PMC free article] [PubMed] [Google Scholar]
- 426.Delso G. Cluster-based segmentation of dual-echo ultra-short echo time images for PET/MR bone localization. EJNMMI Phys. 2014;1(1):7. doi: 10.1186/2197-7364-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Johanson A. Improved quality of computed tomography substitute derived from magnetic resonance (MR) data by incorporation of spatial information - potential application for MR-only radiotherapy and attenuation correction in positron emission tomography. Acta Oncol. (Madr) 2013;52(7):1369–1373. doi: 10.3109/0284186X.2013.819119. [DOI] [PubMed] [Google Scholar]
- 428.Chang L.-T. A method for attenuation correction in radionuclide computed tomography. IEEE Trans. Nucl. Sci. 1978;25(1):638–643. [Google Scholar]
- 429.Shepp L.A. Maximum likelihood reconstruction for emission tomography. IEEE Trans. Med. Imaging. 1982;1(2):113–122. doi: 10.1109/TMI.1982.4307558. [DOI] [PubMed] [Google Scholar]
- 430.Lange K. EM reconstruction algorithms for emission and transmission tomography. J. Comput. Assist. Tomogr. 1984;8(2):306–316. [PubMed] [Google Scholar]
- 431.Gullberg G.T. An attenuated projector-backprojector for iterative SPECT reconstruction. Phys. Med. Biol. 1985;30(8):799–816. doi: 10.1088/0031-9155/30/8/004. [DOI] [PubMed] [Google Scholar]
- 432.Zaidi H. Scatter compensation techniques in PET. PET Clin. 2007;2(2):219–234. doi: 10.1016/j.cpet.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 433.Hutton B.F. Review and current status of SPECT scatter correction. Phys. Med. Biol. 2011;56(14):R85–R112. doi: 10.1088/0031-9155/56/14/R01. [DOI] [PubMed] [Google Scholar]
- 434.Zaidi H. Scatter correction strategies in emission tomography. In: Zaidi H., editor. Quantitative Analysis in Nuclear Medicine Imaging. Springer US; Boston, MA: 2006. pp. 205–235. Editor. [Google Scholar]
- 435.Kupferschlaeger J. Absolute quantification in SPECT - a phantom study. Eur. J. Nucl. Med. Mol. Imaging. 2015;42:S148–S149. [Google Scholar]
- 436.Jaszczak R.J. Improved SPECT quantification using compensation for scattered photons. J. Nucl. Med. 1984;25(8):893–900. [PubMed] [Google Scholar]
- 437.Grootoonk S. Correction for scatter in 3D brain PET using a dual energy window method. Phys. Med. Biol. 1996;41(12):2757–2774. doi: 10.1088/0031-9155/41/12/013. [DOI] [PubMed] [Google Scholar]
- 438.Ichihara T. Compton scatter compensation using the triple-energy window method for single- and dual-isotope SPECT. J. Nucl. Med. 1993;34(12):2216–2221. [PubMed] [Google Scholar]
- 439.Shao L. Triple energy window scatter correction technique in PET. IEEE Trans. Med. Imaging. 1994;13(4):641–648. doi: 10.1109/42.363104. [DOI] [PubMed] [Google Scholar]
- 440.Koral K.F. SPECT Compton-scattering correction by analysis of energy spectra. J. Nucl. Med. 1988;29(2):195–202. [PubMed] [Google Scholar]
- 441.Bentourkia M. Energy dependence of scatter components in multispectral PET imaging. IEEE Trans. Med. Imaging. 1995;14(1):138–145. doi: 10.1109/42.370410. [DOI] [PubMed] [Google Scholar]
- 442.Hasegawa T. A Monte Carlo simulation study on coarse septa for scatter correction in 3-D PET. IEEE Trans. Nucl. Sci. 2002;49(5):2133–2138. [Google Scholar]
- 443.Chuang K.S. Novel scatter correction for three-dimensional positron emission tomography by use of a beam stopper device. Nucl. Instrum. Methods Phys. Res.h Section A. 2005;551(2-3):540–552. [Google Scholar]
- 444.Chen H.T. A fast, energy-dependent scatter reduction method for 3D PET imaging. In: Metzler S.D., editor. IEEE Nuclear Science Symposium, Conference Record, Vols 1-5. IEEE; Portland, OR: 2004. pp. 2630–2634. [Google Scholar]
- 445.Popescu L.M. PET energy-based scatter estimation and image reconstruction with energy-dependent corrections. Phys. Med. Biol. 2006;51(11):2919–2937. doi: 10.1088/0031-9155/51/11/016. [DOI] [PubMed] [Google Scholar]
- 446.Bailey D.L. A convolution-subtraction scatter correction method for 3d PET. Phys. Med. Biol. 1994;39(3):411–424. doi: 10.1088/0031-9155/39/3/009. [DOI] [PubMed] [Google Scholar]
- 447.Meikle S.R. A transmission-dependent method for scatter correction in SPECT. J. Nucl. Med. 1994;35(2):360–367. [PubMed] [Google Scholar]
- 448.Lubberink M. Non-stationary convolution subtraction scatter correction with a dual-exponential scatter kernel for the Hamamatsu SHR-7700 animal PET scanner. Phys. Med. Biol. 2004;49(5):833–842. doi: 10.1088/0031-9155/49/5/013. Article ID. Pii s0031-9155(04)69622-9. [DOI] [PubMed] [Google Scholar]
- 449.Bendriem B. A technique for the correction of scattered radiation in a PET system using time-of-flight information. J. Comput. Assist. Tomogr. 1986;10(2):287–295. doi: 10.1097/00004728-198603000-00021. [DOI] [PubMed] [Google Scholar]
- 450.Levin C.S. A Monte-Carlo correction for the effect of compton-scattering in 3-d PET brain imaging. IEEE Trans. Nucl. Sci. 1995;42(4):1181–1185. [Google Scholar]
- 451.Watson C.C., New, faster, image-base d scatter correction for 3D PET. IEEE Trans. Nucl. Sci. 2000;47(4):1587–1594. [Google Scholar]
- 452.Accorsi R. Optimization of a fully 3D single scatter simulation algorithm for 3D PET. Phys. Med. Biol. 2004;49(12):2577–2598. doi: 10.1088/0031-9155/49/12/008. Article ID. PII s0031-9155(04)70798-8. [DOI] [PubMed] [Google Scholar]
- 453.Beekman F.J. Efficient fully 3-D iterative SPECT reconstruction with Monte Carlo-based scatter compensation. IEEE Trans. Med. Imaging. 2002;21(8):867–877. doi: 10.1109/TMI.2002.803130. [DOI] [PubMed] [Google Scholar]
- 454.Cot A. Absolute quantification in dopaminergic neurotransmission SPECT using a Monte Carlo-based scatter correction and fully 3-dimensional reconstruction. J. Nucl. Med. 2005;46(9):1497–1504. [PubMed] [Google Scholar]
- 455.Lazaro D. Fully 3D Monte Carlo reconstruction in SPECT: a feasibility study. Phys. Med. Biol. 2005;50(16):3739–3754. doi: 10.1088/0031-9155/50/16/006. [DOI] [PubMed] [Google Scholar]
- 456.Salas-Gonzalez D. Linear intensity normalization of FP-CIT SPECT brain images using the alpha-stable distribution. Neuroimage. 2013;65:449–455. doi: 10.1016/j.neuroimage.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 457.Castillo-Barnes D. On a heavy-tailed intensity normalization of the parkinson's progression markers initiative brain database. In: Vicente J.M.F., editor. Natural and Artificial Computation for Biomedicine and Neuroscience, Pt I. Springer International Publishing Ag; Cham: 2017. pp. 298–304. [Google Scholar]
- 458.Brahim A. Comparison between different intensity normalization methods in 123I-Ioflupane imaging for the automatic detection of parkinsonism. PLoS ONE. 2015;10(6) doi: 10.1371/journal.pone.0130274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.D'Andrea A. The role of multimodality imaging in COVID-19 patients: from diagnosis to clinical monitoring and prognosis. Giornale Italiano Di Cardio Logia. 2020;21(5):345–353. doi: 10.1714/3343.33132. [DOI] [PubMed] [Google Scholar]
- 460.Górriz J.M. Artificial intelligence within the interplay between natural and artificial Computation: advances in data science, trends and applications. Neurocomputing. 2020 doi: 10.1016/j.neucom.2020.05.078. [DOI] [Google Scholar]