Abstract
Spinocerebellar ataxia type 10 (SCA10) is a progressive genetic disorder caused by ATTCT pentanucleotide repeat expansions in intron 9 of the ATXN10 gene. ATTCT repeats have been reported to form unwound secondary structures which are likely linked to large-scale repeat expansions. In this study, we performed high-resolution nuclear magnetic resonance spectroscopic investigations on DNA sequences containing two to five ATTCT repeats. Strikingly, we found the first two repeats of all these sequences well folded into highly compact minidumbbell (MDB) structures. The 3D solution structure of the sequence containing two ATTCT repeats was successfully determined, revealing the MDB comprises a regular TTCTA and a quasi TTCT/A pentaloops with extensive stabilizing loop-loop interactions. We further carried out in vitro primer extension assays to examine if the MDB formed in the primer could escape from the proofreading function of DNA polymerase. Results showed that when the MDB was formed at 5-bp or farther away from the priming site, it was able to escape from the proofreading by Klenow fragment of DNA polymerase I and thus retained in the primer. The intriguing structural findings bring about new insights into the origin of genetic instability in SCA10.
INTRODUCTION
DNA repeat expansions in the human genome are known to cause >30 inherited neurological diseases (1,2). Among them, spinocerebellar ataxia type 10 (SCA10) is associated with ATTCT pentanucleotide repeat expansions in intron 9 of the ATXN10 gene on chromosome 22q13.3 (3). Clinical features of SCA10 involve cerebellar dysfunctions that usually start as poor balance and unsteady gait, followed by upper-limb ataxia, scanning dysarthria, dysphagia (4) and/or sleep disorders (5). A large difference in the number of ATTCT repeats between normal populations (∼10–22 repeats) and SCA10 patients (∼850–4500 repeats) has been reported, and a range from ∼280 to 850 repeats is considered to be the pre-mutation size (Figure 1) (3,6,7).
Figure 1.
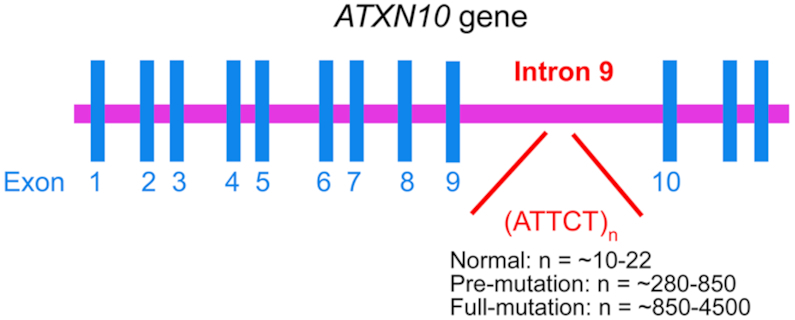
ATTCT repeat expansions in intron 9 of the ATXN10 gene are associated with SCA10.
Molecular mechanisms of repeat expansions have been studied extensively in the past three decades. The breakthrough in understanding the origin of repeat expansions is the realization that almost all expandable repeats are capable of forming unusual secondary structures such as the hairpin (8), triplex (9), G-quadruplex (10), i-motif (11) and dumbbell (12). It has been widely accepted that repeat expansions can occur via strand slippage, which is promoted by the formation of an unusual secondary structure in the nascent strand during DNA replication (13–15). ATTCT repeats have been reported to form unwound secondary structures which were associated with replication initiations (16). As a result, ATTCT repeat expansions have been proposed to occur via replication re-initiations (17) and template switching (18) pathways.
Recently, we have discovered that some pyrimidine-rich tetranucleotide repeats, including TTTA, CCTG and CTTG repeats, can form a new type of unusual DNA structure called minidumbbell (MDB) (19–23). All these three MDBs contain two adjacent type II tetraloops. A DNA tetraloop is commonly defined to have four residues in which the first (L1) and fourth (L4) residues form a loop-closing base pair. In a type II tetraloop, the second residue (L2) folds into the minor groove whereas the third residue (L3) stacks on L1–L4 (24,25). In the MDB structure, L1–L4 and L1′–L4′ form two loop-closing base pairs, L2 and L2′ fold into the minor groove, whereas L3 and L3′ stack on their nearby loop-closing base pairs, here L1, L2, L3, L4 represent residues of the 5′-type II tetraloop, and L1′, L2′, L3′ and L4′ represent residues of the 3′-type II tetraloop (Supplementary Figure S1). MDBs are distinguishable from larger dumbbell structures (12) by possessing loop–loop interactions between (i) the two loop-closing base pairs, and/or (ii) the two minor groove residues L2 and L2′. The TTTA, CCTG and CTTG MDBs formed stably at neutral pH with a melting temperature (Tm) of ∼18, 22 and 35°C, respectively (20,22,23). The relatively lower thermodynamic stabilities of the TTTA and CCTG MDBs probably facilitate their escape from the proofreading function of DNA polymerase and/or the repair functions of DNA repair systems. Thereby, their formations in nascent strands during DNA replication have been proposed to be the origins of TTTA and CCTG repeat expansions in Staphylococcus aureus pathogen and myotonic dystrophy type 2 (DM2) patients, respectively (22,23). No CTTG repeat expansion has ever been reported in any genomes.
As any structures with relatively low thermodynamic stabilities can easily be underdetermined, the pyrimidine-rich and expandable nature of ATTCT repeats brought us the question whether ATTCT repeats can also form any secondary structures like TTTA and CCTG repeats. We also noted that a folded structure with a transition temperature of ∼6°C was previously observed in a sequence containing nine ATTCT repeats, but the structural information was not known (26). A thorough understanding on the solution structural behaviors of ATTCT repeats will advance our knowledge of the origin of genetic instability in SCA10. Therefore, we decided to perform high-resolution nuclear magnetic resonance (NMR) spectroscopic investigations on ATTCT repeats starting from the simplest sequence containing two repeats, up to five repeats, of which unambiguous NMR assignments of the fingerprint regions of these sequences could still be obtained. Intriguingly, our NMR results reveal that at relatively low temperatures, the sequence containing two ATTCT repeats well folded into an MDB structure comprising a regular TTCTA pentaloop and a quasi TTCT/A pentaloop. The regular pentaloop has its first and fifth residues to form a loop-closing base pair, and every two adjacent loop residues sequentially connected by the phosphodiester backbone. The quasi pentaloop has a backbone discontinuous site between two adjacent loop residues (as indicated by the symbol of ‘/’) (27). When the ATTCT repeat length increased to three, four and five, the first two repeats at the 5′-termini of these sequences also formed similar MDB structures.
MATERIALS AND METHODS
DNA sequence design and sample preparation for NMR structural study
DNA sequences containing two, three, four and five ATTCT repeats were first investigated, and they were named as (ATTCT)2, (ATTCT)3, (ATTCT)4 and (ATTCT)5, respectively. To investigate the importance of a quasi pentaloop in the formation of MDB structure with two ATTCT repeats, we designed another sequence containing two TTCTA repeats which would potentially form an MDB containing two regular TTCTA pentaloops, and this sequence was named as (TTCTA)2. To investigate the effects of 3′ and/or 5′-partial length repeats on the MDB structure formed by two ATTCT repeats, we designed additional nine sequences including (i) four sequences by adding A, AT, ATT and ATTC at the 3′-end of two ATTCT repeats, (ii) four sequences by adding T, CT, TCT and TTCT at the 5′-end of two ATTCT repeats, and (iii) one sequence by adding an A at the 3′-end and a T at the 5′-end of two ATTCT repeats. These nine sequences were named as (ATTCT)2A, (ATTCT)2AT, (ATTCT)2ATT, (ATTCT)2ATTC, T(ATTCT)2, CT(ATTCT)2, TCT(ATTCT)2, TTCT(ATTCT)2 and T(ATTCT)2A, respectively. DNA sequences used for NMR structural study are shown in Table 1.
Table 1.
DNA sequences used for NMR structural study
| Name | Sequence |
|---|---|
| (ATTCT)2 | 5′-ATTCT ATTCT |
| (ATTCT)3 | 5′-ATTCT ATTCT ATTCT |
| (ATTCT)4 | 5′-ATTCT ATTCT ATTCT ATTCT |
| (ATTCT)5 | 5′-ATTCT ATTCT ATTCT ATTCT ATTCT |
| (TTCTA)2 | 5′-TTCTA TTCTA |
| (ATTCT)2A | 5′-ATTCT ATTCT A |
| (ATTCT)2AT | 5′-ATTCT ATTCT AT |
| (ATTCT)2ATT | 5′-ATTCT ATTCT ATT |
| (ATTCT)2ATTC | 5′-ATTCT ATTCT ATTC |
| T(ATTCT)2 | 5′-T ATTCT ATTCT |
| CT(ATTCT)2 | 5′-CT ATTCT ATTCT |
| TCT(ATTCT)2 | 5′-TCT ATTCT ATTCT |
| TTCT(ATTCT)2 | 5′-TTCT ATTCT ATTCT |
| T(ATTCT)2A | 5′-T ATTCT ATTCT A |
DNA samples were synthesized using an Applied Biosystems model 394 DNA synthesizer. They were purified by denaturing polyacrylamide gel electrophoresis (PAGE), diethylaminoethyl sephacel anion exchange column chromatography, and finally desalted using Amicon® Ultra-4 centrifugal filtering devices. For DNA quantification, the ultra-violet (UV) absorbance at 260 nm was measured. Unless otherwise mentioned, the NMR samples were prepared by dissolving ∼0.5 μmol purified DNA into 500 μl buffer solutions containing 10 mM sodium phosphate (NaPi) at pH 7.0 and 0.02 mM 2,2 dimethyl-2-silapentane-5-sulfonic acid (DSS).
Native PAGE
To investigate the oligomeric states of the abovementioned sequences, i.e. whether the DNA adopted a monomeric conformation by a single strand or a multimeric conformation by two or more strands, PAGE experiments were performed using 20% native polyacrylamide gels. An electrophoresis buffer containing 89 mM Tris, 89 mM boric acid and 2 mM ethylenediaminetetraacetic acid (EDTA) at pH 7.5 was used. The DNA loading samples were prepared in the same buffer solutions as for NMR samples, that is, ∼1.0 mM DNA in 10 mM NaPi (pH 7.0). For the reference lane of DNA ladder, we prepared a mixture of seven DNA sequences containing two, three, four, five, six, seven and eight TTTA repeats, which correspond to 8, 12, 16, 20, 24, 28 and 32 nt, respectively. The monomeric states of these TTTA repeats have been demonstrated in our previous study (28). The concentration of each DNA sequence in the ladder was kept at ∼0.2 mM. The sequences were dissolved in a buffer solution containing 10 mM NaPi (pH 7.0), and 8 M urea was added into the ladder to further prevent the formation of multimeric structures. The electrophoresis experiments were conducted at ∼5°C in a fridge. DNA bands were visualized by staining the gels with stains-all solution.
Thermodynamic study
UV absorption melting experiments were performed to determine the thermodynamic stability of the MDB formed by (ATTCT)2. UV absorbance data at 260 nm were collected as a function of temperature using a Hewlett-Packard 8453 diode-array UV–visible spectrophotometer equipped with a thermostated temperature controller and sample holder. The sample holder temperature was set from 0 to 50°C at a heating rate of 1.0°C/min. The sample concentrations were kept at ∼5 μM in the same NMR buffer solution, and a 10 mm path length cuvette was used. The Tm values were extracted from curve fittings using a two-state transition model (29). Three replicate measurements were performed to determine the averaged Tm value and uncertainties.
NMR spectroscopy
All NMR experiments were performed using Bruker AV-500 and/or AV-700 spectrometer. For studying the labile protons, DNA samples were prepared in a 90% H2O/10% D2O buffer solution. One-dimensional (1D) imino, and two-dimensional (2D) nuclear Overhauser effect spectroscopy (NOESY) experiments were acquired using the excitation sculpting (30) or jump-return (31) pulse sequence to suppress the water signal. For studying the non-labile protons, the solvent was exchanged with a 99.96% D2O buffer solution, and a 2-s presaturation pulse was used to suppress the residual water signal. The NOESY spectra were acquired with a mixing time of 300 ms or 600 ms. To assign the adenine H2 resonances, 2D 1H–13C heteronuclear multiple bond coherence (HMBC) experiments were acquired with a delay of 65 ms for the evolution of long-range couplings (32,33). For the measurements of 3JH1′–H2′, 3JH4′–H5′ and 3JH4′–H5″ coupling constants of (ATTCT)2, double quantum filtered correlation spectroscopy (DQF-COSY) spectrum was acquired. All the NOESY, TOCSY and DQF-COSY spectra were acquired with a data set size of 4096 × 512. The NOESY and TOCSY spectra were zero-filled to give 4096 × 4096 spectra with a cosine window function applied to both dimensions, whereas the COSY spectrum was zero-filled to give an 8192 × 4096 spectrum with a sine window function applied to both dimensions. To assign the 31P resonances, 1H–31P heteronuclear single-quantum coherence (HSQC) spectroscopy was performed with the application of a Carr–Purcell–Meiboom–Gill (CPMG) pulse train during the periods of magnetization transfer between phosphorus and scalar coupled proton nuclei (34). The delays surrounding synchronous proton/phosphorus 180° refocusing pulses were set to ∼100 μs. The spectra were acquired with a data size of 4096 × 200 and zero-filled to give 4096 × 2048 spectra with a cosine window function applied to both dimensions. The 31P and 1H spectral widths were set to 9.0 and 11.0 ppm with the carrier frequencies positioned at –3.7 and 4.7 ppm, respectively. 31P and 13C chemical shifts were indirectly referenced to DSS using the derived nucleus-specific ratios of 0.404808636 and 0.251449530, respectively (35).
To compare the relative thermodynamic stabilities of (ATTCT)2 and (TTCTA)2, variable-temperature 1D 1H NMR experiments were performed from 0 to 75°C at a step size of 2.5°C.
Sequential resonance assignments of all sequences were made from NOESY H6/H8-H1′ fingerprint regions using standard methods (36,37) and they are shown in Supplementary Figures S2–S12. The adenine H2 resonance assignments were made based on the long-range couplings between H8/H2 and C4, and they are shown in Supplementary Figures S13–S16. The 31P resonance assignments were made by first assigning the H3′ resonances in the TOCSY spectra, then correlating each H3′ resonance of the ith residue with the 31P resonance of the i + 1th residue in 1H–31P HSQC spectra, and they are shown in Supplementary Figures S17–S23.
Extraction of NMR restraints
To extract the non-labile proton-proton distance restraints, 2D NOESY spectrum of the 99.96% D2O sample was acquired at 5°C with a mixing time of 300 ms. We classified the intensities of NOE cross-peaks into five categories including strong, strong or medium, medium, medium or weak, and weak. The corresponding distance restraints for each category were set as 1.8–4.0, 2.5–4.5, 3.0–5.0, 3.5–5.5 and 4.0–6.0 Å, respectively. In addition, a distance restraint of 1.8–6.0 Å was applied to seriously overlapping NOE cross-peaks of which the intensities could not be reliably determined. Due to the exchange between labile proton and water solvent, the intensities of NOE cross-peaks involving labile protons could not be reliably determined. Therefore, a distance restraint of 1.8–6.0 Å was also applied to these NOE cross-peaks. The T2 and T7 H3 signals appeared at 13.36 and 13.00 ppm, respectively, which fall into the chemical shift range of thymine imino involved in Watson–Crick base pairs (38). Observations of 1D NOEs of T2 H3-A6 H2 and T7 H3-A1 H2 further suggest that T2-A6 and T7-A1 adopted Watson–Crick base pairs. Therefore, we added Watson–Crick hydrogen bond restraints for T2-A6 and T7-A1 base pairs. Hydrogen bond distance restraints of 2.72–2.92 and 2.85–3.05 Å were applied to A N1-T N3 and A N6-T O4, respectively (39). A total of 284 NOE-derived distance restraints were finally obtained. Among them, 169 and 115 were inter-residue and intra-residue distance restraints, respectively. In addition, glycosidic torsion angles χ were obtained based on the intranucleotide H6/H8-H1′ NOE intensities in the D2O NOESY spectrum and adenine C8 chemical shifts. Restraints for backbone torsion angles γ were determined based on the analysis of 3JH4′–H5′ and 3JH4′–H5″ coupling constants (37). The H1′–C1′–C2′–H2′ sugar torsion angles were determined by the 3JH1′–H2′ coupling constants measured from the DQF-COSY spectrum and the Karplus equation (40). The H1′–C1′–C2′–H2′ sugar torsion angle restraints were obtained by adding ±15° to the calculated angle values.
In addition to the NMR-derived experimental restraints, we also generated the chirality restraints for C1′, C3′ and C4′ atoms of each residue in (ATTCT)2 using the script ‘makeCHIR_RST’ in AmberTools (41). A summary of the restraints used for calculating the solution structures of (ATTCT)2 is shown in Supplementary Table S1. The chemical shift information of (ATTCT)2 is shown in Supplementary Table S2.
Structural calculations
The solution structures of (ATTCT)2 were calculated via restrained molecular dynamics (rMD) and restrained energy minimization (rEM) using AMBER 16 with the ff14SB force field (42). The initial single-strand 5′-ATTCT ATTCT-3′ was obtained by manually removing the opposite strand from a classical double-helical B-DNA generated by Nucleic Acid Builder and then energy minimized. The in vacuo calculations were initiated by heating the system from 300 K to an annealing temperature in the first 15 ps, and then maintaining for 30 ps. After that, the temperature was gradually lowered to 300 K over the next 15 ps and maintained at 300 K for another 15 ps till the end of the rMD step. A set of structural coordinates was then saved for restrained energy minimization by 200 steps of steepest descent, followed by conjugated gradient minimization until the energy gradient difference between successive minimization steps was smaller than 0.1 kcal/mol·Å2. In order to assure that the annealing temperature was high enough to overcome bad local structural contacts and fulfill experimental restraints, we used ten different annealing temperatures from 400 to 1300 K with a step size of 100 K. Under each annealing temperature, 100 rMD trials were performed with random starting velocities. Therefore, a total of 1000 calculated structures were obtained. The 20 structures with the lowest total energies were selected to be the final representative ensemble of the refined structures.
Data analysis of NMR refined solution structures
For the identification of stabilizing interactions, a hydrogen bond was considered to exist if the distance between the hydrogen bond donor and acceptor was less than 3.2 Å. Hydrophobic interaction was considered to exist between two non-polar groups (e.g. thymine methyl and deoxyribose 2′-methylene) if their carbon-carbon distance fell within 3.8–6.5 Å (43). All figures showing the solution structures of (ATTCT)2 were generated using UCSF Chimera (44).
In vitro primer extension assays
To investigate if the MDB can form and retain in the nascent strand containing ATTCT repeats during DNA replication, we designed two primer-template models wherein the template contained (AGAAT)10 flanked by a few non-repeating residues, and its 3′-terminal was a three-carbon spacer (Glen Research) to avoid template extension or cleavage by DNA polymerase. The two primers were 5′-(ATTCT)5 and 5′-GC(ATTCT)5, respectively. For simplicity, we named these two primer-template models as P1-T and P2-T, respectively. The Klenow fragment (KF) of DNA polymerase I was used for primer extensions. The primer and template were first hybridized by annealing at 90°C for 2 min and then slowly cooling to room temperature. The mixture containing 0.02 mM DNA primer-template, 12 mM of each dNTPs (N = A/G/C/T) in 1× reaction buffer (50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris–HCl, pH 7.9) was incubated at 37°C for 20 min, and then 10 μl sample was collected as the 0-min timepoint. We then added 62 units of KF to the mixture to start the primer extensions at 37°C and collected every 10 μl reaction mixture after 30 min, 1 h, 3 h and 24 h. The reaction was quenched by adding 2 μl of 0.5 M EDTA to the collected sample and incubating the sample at 90°C for 5 min.
To study if the MDB, which was pre-existing in the primer, can escape from the proofreading function of DNA polymerase, we designed four primer-template models in which the primer contained the MDB-forming sequence at 6, 5, 4 and 3-bp away from the priming site, respectively. For simplicity, these four primer-template models were named as MDB+6bp, MDB+5bp, MDB+4bp and MDB+3bp, respectively. The primer extension assays were performed following the abovementioned protocol except that the reaction mixture contained 0.05 mM DNA primer-template, 1.5 mM of each dNTPs and 5 units of KF, and the timepoints for sample collections were 15 min, 30 min, 1 h, 5 h and 24 h.
Primer extension products of P1-T, P2-T, MDB+6bp, MDB+5bp, MDB+4bp and MDB+3bp were examined by PAGE using 20% denaturing polyacrylamide gels. DNA bands were visualized by staining the gels with stains-all solution. The DNA sequences used for primer extension assays are summarized in Supplementary Table S3. The DNA synthesis and purification followed the same protocols as those in NMR structural study.
RESULTS AND DISCUSSION
Unusual secondary structures form at the 5′-termini of ATTCT repeats
We first performed variable-temperature 1H and 31P NMR investigations on the sequences containing two, three, four and five ATTCT repeats, which were named as (ATTCT)2, (ATTCT)3, (ATTCT)4 and (ATTCT)5, respectively. Interestingly, these sequences showed a similar pattern of unusually shifted NMR signals upon lowering the temperature to ∼5°C, including the downfield shifted methyl H7 and 31P signals of T3/T8 at around 1.9 to 2.0 ppm and –3.4 to –3.7 ppm, respectively (Figure 2). These chemical shifts were found to deviate from those of the corresponding residues in B-DNA (T3/T8 H7: ∼1.5 ppm) and random coil (T3/T8 H7/31P: ∼1.8/–4.0 ppm) predicted by the DSHIFT web server (45), suggesting that these sequences may form similar unusual secondary structures by their first two repeats at the 5′-termini. We also performed native PAGE experiments for these four sequences under the same NMR buffering condition at ∼5°C (Supplementary Figure S24), and the gel results reveal that they predominantly adopted monomeric conformations by a single strand instead of multimeric structures by two or more strands. To further investigate the structural details, we calculated the solution structure of the simplest repeating sequence, namely, (ATTCT)2.
Figure 2.
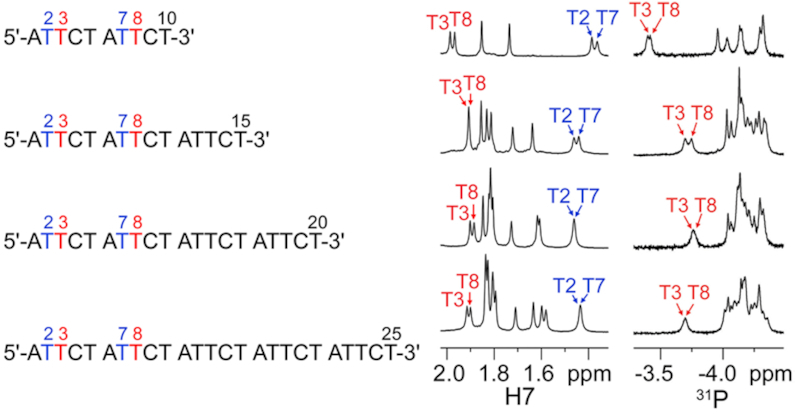
1D 1H and 31P NMR spectra of (ATTCT)2, (ATTCT)3, (ATTCT)4 and (ATTCT)5 at 5°C.
(ATTCT)2 forms an MDB structure containing a regular and a quasi pentaloops
Prior to extraction of NMR experimental restraints, we determined the thermodynamic stability of the folded structure of (ATTCT)2 from three replicative UV melting experiments. The Tm was calculated from curve fitting using a two-state model which defines the folded and unfolded states of macromolecules (29), and the Tm value was 17.0 ± 0.5°C (Supplementary Figure S25). The folded state was found to be predominantly populated at low temperatures, e.g. ∼85% and 15% populations of the folded and unfolded states, respectively, at 5°C. Therefore, we extracted the NMR experimental restraints at 5°C, including 284 NOE-derived distance restraints and 16 torsion angle restraints. Considering the presence of ∼15% unfolded state might have minor influences on the averaging NOE intensities, we used relatively loose NOE-derived distance ranges to avoid overestimations of the calculated structures (see ‘Extraction of NMR restraints’ in ‘Materials and Methods’).
Among the 1000 calculated structures, 20 with the lowest total energies were selected in the final representative ensemble (PDB ID: 6IY5) and their refinement statistics are summarized in Table 2. These structures well agree with NMR experimental restraints as there is no large restraint violation. The heavy-atom RMSD from mean structure is 0.8 ± 0.2 Å, representing one conformation adopted by these 20 structures. We realized that the heavy-atom RMSD value of (ATTCT)2 is much larger than that of the stable CTTG MDB (0.4 ± 0.1 Å) (19), reflecting the calculated MDB solution structures with relatively low thermodynamic stabilities are more dynamic, as there is a minor contribution of the unfolded state to the averaged NOEs. In this ensemble of structures, (ATTCT)2 folds into a highly compact MDB structure containing a regular TTCTA pentaloop formed by the loop residues L1 to L5, i.e. T2, T3, C4, T5 and A6, and a quasi TTCT/A pentaloop formed by the loop residues L1′ to L5′, i.e. T7, T8, C9, T10 and A1 (Figure 3A). For simplicity, we named it as the ATTCT-Q/L5′ MDB where ‘Q/L5′’ indicates in the quasi-loop, the backbone discontinuous site is right before the L5′ residue.
Table 2.
NMR refinement statistics of the ATTCT-Q/L5′ MDB formed by (ATTCT)2
| Experimental restraints | |
| Distance restraints | |
| Inter-residue | 169 |
| Intra-residue | 115 |
| Hydrogen bond | 4 |
| Torsion angle restraints | |
| Glycosidic (χ) | 10 |
| Backbone (γ) | 3 |
| Sugar torsion H1′–C1′–C2′–H2′ | 3 |
| Restraint satisfaction | |
| Distance restraints (Å) | |
| Number of violations > 0.2 Å | 0 |
| Maximum violation | 0.006 |
| Average violation | 0.003 ± 0.001 |
| Torsion angle restraints (°) | |
| Number of violations > 5° | 0 |
| Maximum violation | 0.4 |
| Average violation | 0.2 ± 0.1 |
| Bonds (Å) | 0.0092 ± 0.0004 |
| Angles (°) | 2.7 ± 0.1 |
| Heavy-atom RMSD (Å) a | |
| Average pairwise RMSD | 1.2 ± 0.2 |
| RMSD from mean structure | 0.8 ± 0.2 |
aRMSD values were calculated among 20 refined structures for all residues.
Figure 3.
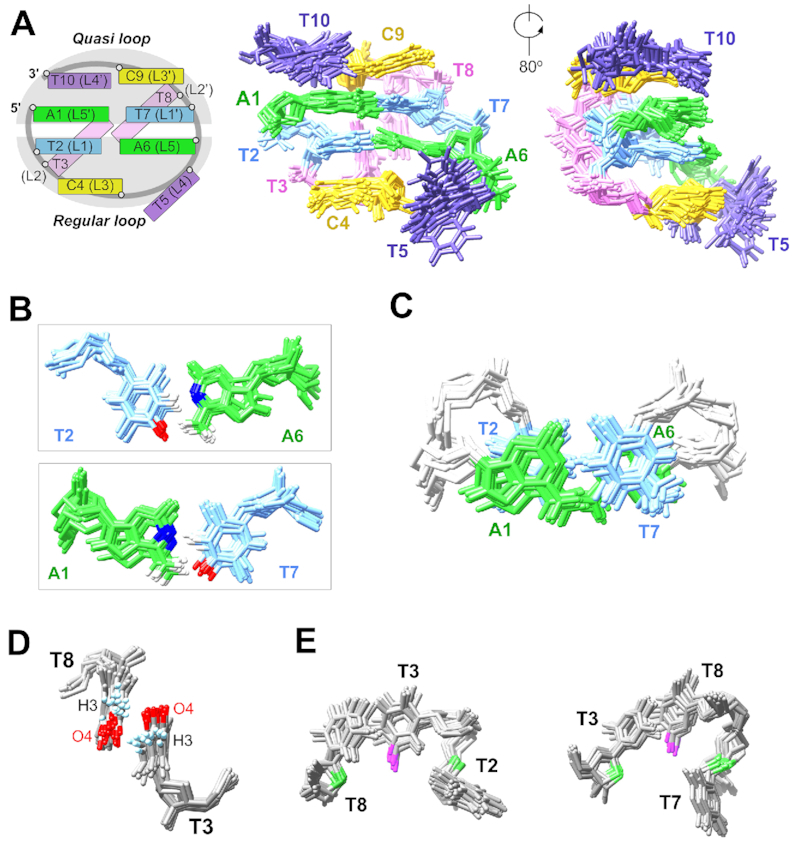
(A) Schematic of the ATTCT-Q/L5′ MDB and the superimposed 20 solution NMR structures (PDB ID: 6IY5). (B) T2-A6 and T7-A1 form Watson–Crick base pairs. (C) T2-A6 and T7-A1 base pairs show extensive base-base stackings. (D) T3 and T8 stack with each other, providing an orientation for favorable electrostatic interactions between T3 H3 and T8 O4, and between T3 O4 and T8 H3. (E) Dual hydrophobic cores formed by T3 methyl (magenta) and T2/T8 2′-methylene groups (green), and T8 methyl and T3/T7 2′-methylene groups.
In the ATTCT-Q/L5′ MDB, T2-A6 and T7-A1 form Watson–Crick loop-closing base pairs (Figure 3B), as suggested by the 1D NOE on A6 H2 upon selectively saturating T2 H3 at 13.36 ppm, and the 1D NOE on A1 H2 upon selectively saturating T7 H3 at 13.00 ppm (Supplementary Figure S26A). In addition, T2-A6 and T7-A1 base pairs show base-base stackings (Figure 3C), which agree with the observation of T2 H6-A1 H8 and T7 H6-A6 H8 NOEs (Supplementary Figure S26B).
In both of the regular TTCTA and quasi TTCT/A pentaloops, the second loop residues T3 and T8 fold into the minor groove (Figure 3A), and their minor groove locations agree with the NOEs between T3/T8 H7/H2′/H2″ and A1/A6 H2 as these adenine H2 protons point to the minor groove (Supplementary Figure S26C). T3 and T8 partially stack with each other, providing an optimized orientation for favorable electrostatic interactions between T3 H3 and T8 O4, and between T3 O4 and T8 H3 (Figure 3D). The distances from T3 H3 to T8 O4, and from T3 O4 to T8 H3 were measured to be 2.9 ± 0.4 and 3.4 ± 0.2 Å, respectively. In addition, there are dual hydrophobic cores (19) formed by (i) the methyl group of T3 and the 2′-methylene groups of T2/T8, and (ii) the methyl group of T8 and the 2′-methylene groups of T3/T7 (Figure 3E). The distances from T3 C7 to T2/T8 C2′, and from T8 C7 to T3/T7 C2′ were 5.1 ± 0.2/4.5 ± 0.3 and 5.8 ± 0.4/5.1 ± 0.2 Å, respectively, which fall into the range of non-polar carbon-carbon distances for favorable hydrophobic interactions (3.8–6.5 Å) (43).
The bases of the third loop residues C4 and C9 well stack on the bases of T2 and T7, respectively (Figure 4A), which also agrees with the base-base NOEs of C4 H5-T2 H6 and C9 H5-T7 H6, and the NOEs between C4/C9 H6 and T2/T7 H1′/H2′/H2″ also suggest C4/C9 are close to T2/T7 (Supplementary Figure S26D). The sugar rings of C4 and C9 were found to stack on the bases of A6 and A1, respectively, as supported by the sugar-base NOEs of C4/C9 H4′/H5′/H5″-A6/A1 H2 (Figure 4A and Supplementary Figure S26D). The stackings of C4/C9 sugars on the loop-closing base pairs make most of the C4/C9 sugar protons locate in a more shielding chemical environment, and thus their sugar proton chemical shifts including H1′, H2′, H2″ and H4′ are much more upfield than those of other residues (Supplementary Table S2).
Figure 4.
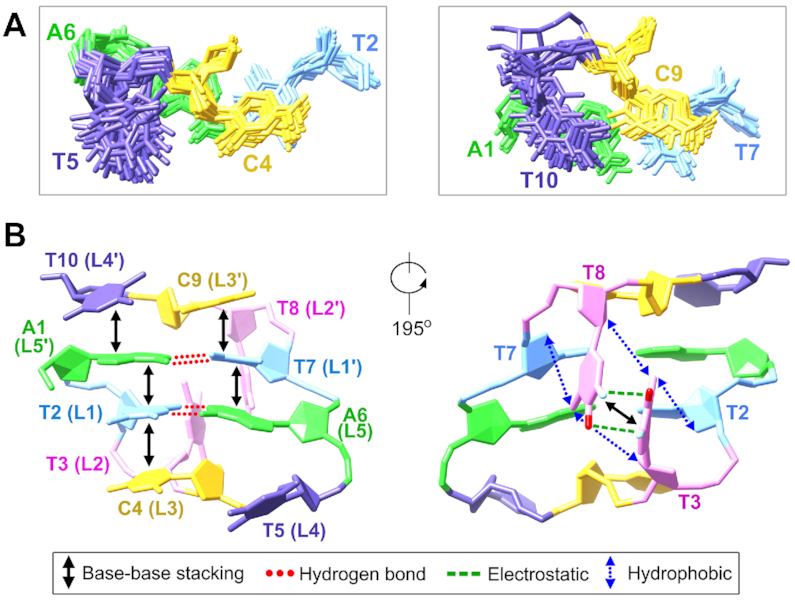
(A) In the ATTCT-Q/L5′ MDB, C4 and C9 stack on T2-A6 and T7-A1 base pairs, respectively. T5 does not stack with any residue, whereas T10 well stacks on A1. (B) A summary of the stabilizing forces in the ATTCT-Q/L5′ MDB.
The fourth loop residues T5 and T10 in the ATTCT-Q/L5′ MDB were found to locate in the major groove, showing some different structural behaviors. T5 in the regular loop appears to be more dynamical comparing to T10 in the quasi loop. Among the 20 structures, 17 of them show T5 is far from A6, three show T5 is nearly perpendicular to A6 (Supplementary Figure S27). None of them shows base-base stacking between T5 and A6. However, T10 stacks well with A1 in all 20 structures, forming favorable 3′-5′ terminal stacking to stabilize the MDB structure (Figures 3A and 4A). The well stacking between T10 and A1 agrees with the observation of multiple NOEs between T10 H6 and A1 H2/H8, and between T10 H7/H2′/H2″ and A1 H8 (Supplementary Figure S26E).
Terminal stacking between L4′ and L5′ in the quasi pentaloop provides substantial stabilizations to the MDB
Structural flexibilities provided by quasi tetraloops have been frequently observed in DNA three-way junctions (46–48). In the ATTCT-Q/L5′ MDB, the backbone discontinuity between terminal T10 and A1 in the quasi pentaloop provides a structural flexibility for T10 to stack well on A1, bringing about substantial stabilizations to the quasi pentaloop and thus the ATTCT-Q/L5′ MDB. Meanwhile, T5 in the regular TTCTA pentaloop of the MDB shows no base-base stacking with any other residue, behaving similarly to the fourth loop residue in the previously reported regular CTTTG pentaloop in hairpins (49,50). In order to consolidate the importance of the quasi pentaloop in providing substantial 3′–5′ terminal stacking to the formation of MDB, we designed another sequence, 5′-TTCTA TTCTA-3′ which was named as (TTCTA)2, to replace the quasi TTCT/A pentaloop in the ATTCT-Q/L5′ MDB with a regular TTCTA pentaloop. As suggested by the (i) downfield shifted H7/31P signals of T2/T7, (ii) upfield shifted H7 signals of T1/T6, (iii) base-base NOEs of C3 H5-T1 H6 and C8 H5-T6 H6 and (iv) 3′-5′ terminal base-base NOEs between A10 and T1 (Supplementary Figure S28), (TTCTA)2 shows a tendency to form an MDB containing two regular TTCTA pentaloops (herein we called it as the TTCTA MDB). However, the variable-temperature chemical shift profiles suggest that the thermodynamic stability of the TTCTA MDB was much lower than that of the ATTCT-Q/L5′ MDB (Figure 5), revealing that a quasi pentaloop is more favorable than a regular pentaloop in facilitating the formation of MDBs.
Figure 5.
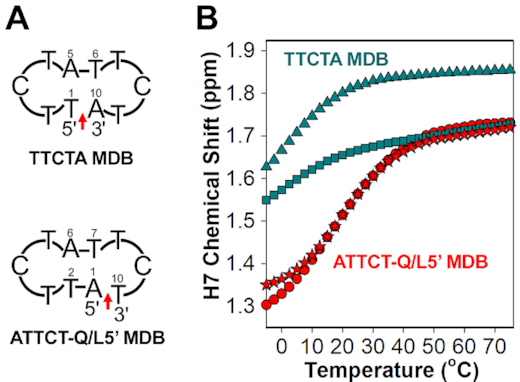
(A) Schematics of the TTCTA MDB containing two regular TTCTA pentaloops, and the ATTCT-Q/L5′ MDB containing a regular TTCTA pentaloop and a quasi TTCT/A pentaloop. Red arrows indicate the backbone discontinuous sites. (B) The variable-temperature L1/L1′ H7 chemical shift profiles of the TTCTA MDB (T1/T6: square/triangle in green) and ATTCT-Q/L5′ MDB (T2/T7: circle/star in red).
The critical stabilizing forces identified in the formation of the ATTCT-Q/L5′ MDB are summarized in Figure 4B. These include (i) the hydrogen bonds within T2-A6 and T7-A1 Watson–Crick loop-closing base pairs, (ii) base-base stackings between A1 and T2, A6 and T7, T2 and C4, T7 and C9, A1 and T10, and T3 and T8, (iii) electrostatic interactions between T3 H3/O4 and T8 O4/H3 and (iv) hydrophobic interactions from the dual hydrophobic cores formed by T3 methyl and 2′-methylene groups of T2/T8, and T8 methyl and 2′-methyle groups of T3/T7. The recognition of these stabilizing forces enhances our understanding on the formation of such a highly compact MDB structure with only two ATTCT pentanucleotide repeats.
NMR solution structures of the ATTCT-Q/L5′ MDB were determined under a relatively low ionic strength of 10 mM NaPi at 5°C to maximize the population of the folded state. To examine if the MDB can also be formed under a more physiologically relevant ionic condition, we acquired NMR spectra for the ATTCT-Q/L5′ MDB under 150 mM NaCl. Consistent NMR spectral features suggest that the ATTCT-Q/L5′ MDBs formed under 10 mM NaPi and 150 mM NaCl should have similar structures and thermodynamic stabilities (Supplementary Figure S4B). The population of the folded ATTCT-Q/L5′ MDB at 37°C was determined to be ∼9% from the fitted UV melting curves (Supplementary Figure S25) using a two-state transition model (29), suggesting that the ATTCT-Q/L5′ MDB may have a chance to form under physiological ionic condition and temperature.
MDBs also form in sequences containing three, four and five ATTCT repeats
As the first two repeats in (ATTCT)3, (ATTCT)4 and (ATTCT)5 show similar NMR spectral features to (ATTCT)2 (Figure 2), we expected that the first two repeats of these three sequences also folded into ATTCT-Q/L5′ MDBs. To confirm the formation of MDBs in these longer sequences, we performed further NMR analyses and the results revealed that (ATTCT)3 adopted a major MDB conformer and a minor dumbbell conformer (Figure 6A). For the major conformer, the first two repeats folded into an ATTCT-Q/L5′ MDB, leaving the third repeat as a 3′-overhang. T2-A6 and T7-A1 formed Watson-Crick base pairs, as suggested by (i) the relatively stronger 1D NOEs on A6/A1 H2 upon saturating the thymine imino signals at ∼12.8 ppm (Figure 6B), and (ii) the upfield shifted H7 signals of T2 and T7 (Figure 2). T3 and T8 folded into the minor groove, as suggested by the relatively downfield shifted H7/31P signals of T3/T8 (Figure 2). C4 and C9 stacked on T2-A6 and T7-A1 base pairs, respectively, which is supported by the base-base NOEs of C4 H5-T2 H6, C4 H6-A6 H2, C9 H5-T7 H6 and C9 H6-A1 H2 (Figure 6C). Furthermore, multiple NOEs between T10 H6/H7/H1′ and A1 H2/H8 (Figure 6D) support the existence of base-base stacking between terminal T10 and A1, consolidating the formation of the MDB structure.
Figure 6.
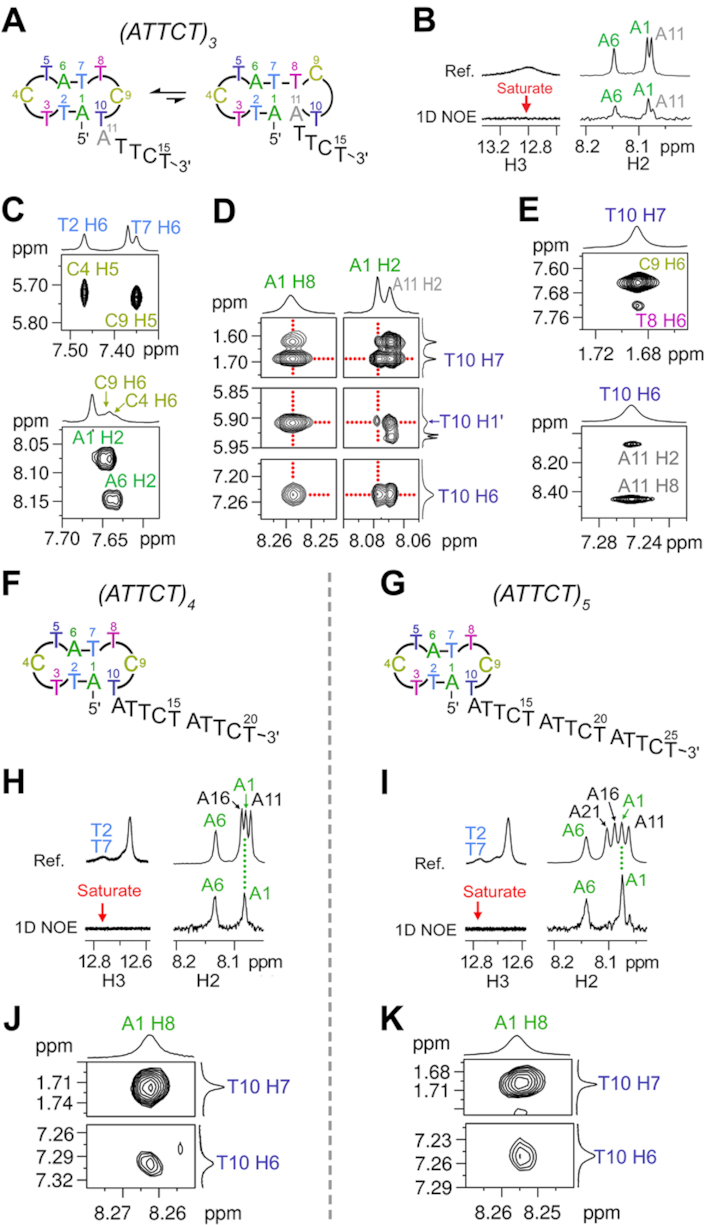
(A) (ATTCT)3 forms a major MDB conformer and a minor dumbbell conformer. The major MDB conformer is supported by the (B) 1D NOEs on A1/A6 H2 upon saturating thymine H3 at ∼12.8 ppm, (C) base-base NOEs between C4 and T2-A6, and between C9 and T7-A1, and (D) NOEs between A1 and T10. (E) NOEs between T10 and T8-A11 suggest the formation of TCTA type II tetraloop in the minor dumbbell conformer. (F–G) (ATTCT)4 and (ATTCT)5 form ATTCT-Q/L5′ MDBs by their first two repeats, leaving a two-repeat and three-repeat 3′-overhang, respectively. For (ATTCT)4 and (ATTCT)5, (H-I) 1D NOE signals on A1/A6 H2 suggest the formation of T2-A6 and T7-A1 Watson-Crick base pairs, and (J-K) base-base NOEs between T10 and A1 suggest T10 stacks on A1. The 1D NOE and 2D NOESY spectra were acquired at 0 and 5°C, respectively.
From the 1D NOE spectra of (ATTCT)3, we also realized that upon saturating the thymine imino signals at ∼12.8 ppm, there was a relatively weaker 1D NOE on A11 H2 (Figure 6B), suggesting A11 may also be involved in T-A Watson–Crick base pair formation. In addition, we observed unusual NOEs of T10 H7-T8 H6 and T10 H6-A11 H8/H2 (Figure 6E), which agree with the structural features of TCTA type II tetraloop wherein the L3 residue stacks on L1–L4 (21,51). Therefore, we believe that there was also a minor dumbbell conformer containing a TTCTA pentaloop, a T7-A1 base pair in the stem, a TCTA tetraloop in which T10 stacks on T8-A11 loop-closing base pair, and a 3′-TTCT overhang (Figure 6A, right). The T7-A1 base pair lengthens the distance between the two loops, thus avoiding the two minor groove residues T3 and C9 to interact.
Similar MDB conformers were also found to predominantly form by the first two repeats in (ATTCT)4 and (ATTCT)5 (Figure 6F, G). In the 1D NOE experiments, upon selectively saturating the thymine imino signals at ∼13.6 ppm, the 1D NOEs were mainly observed on A1 H2 and A6 H2 (Figure 6H, I). Together with the upfield shifted T2 and T7 H7 signals at ∼1.4 ppm (Figure 2), we concluded that T2-A6 and T7-A1 formed Watson-Crick base pairs. The base-base NOEs of C4 H5-T2 H6 and C4 H6-A6 H2, and the NOEs of C9 H5-T7 H6 and C9 H6-A1 H2 (Supplementary Figure S29) suggest C4 and C9 stacked on T2-A6 and T7-A1 base pairs, respectively. More importantly, the NOEs of T10 H6/H7-A1 H8 (Figure 6J-K) support the base-base stacking between T10 and A1, further consolidating the formation of predominantly MDBs by the first two repeats in (ATTCT)4 and (ATTCT)5. Based on the solution structural behaviors of two to five ATTCT repeats, we believe that similar MDBs will also form in longer ATTCT repeats at their 5′-termini.
Effects of 3′ and/or 5′-partial length repeats on the MDB formation
The above results reveal the formation of MDBs at the 5′-termini of integral ATTCT repeats. During DNA replication, the transiently dissociated part of the nascent strand may also contain non-integral ATTCT repeats, i.e. with 3′ and/or 5′-partial length repeats (Figure 7A). Therefore, we also studied if ATTCT-Q/L5′ MDBs would still form in the presence of (i) 3′-partial length repeats, (ii) 5′-partial length repeats and (iii) both 3′ and 5′-partial length repeats. Firstly, we studied the sequences of (ATTCT)2A, (ATTCT)2AT, (ATTCT)2ATT and (ATTCT)2ATTC to investigate the effects of 3′-partial length repeats. Our NMR results reveal that (ATTCT)2A adopted a major ATTCT-Q/L5′ MDB conformer, and a minor dumbbell conformer (Figure 7B, left). The major MDB conformer is suggested by (i) the upfield shifted T2/T7 H7 signals at 1.40 ppm, (ii) the downfield shifted T3/T8 H7 signals at 2.00/1.90 ppm (Figure 7B, right), (iii) the downfield shifted T3/T8 31P signals at -3.50/-3.60 ppm (Supplementary Figure S30A), and (iv) the base-base NOEs of C4 H6-T2 H6 and C9 H5-T7 H6 (Supplementary Figure S30B), which are consistent with the NMR spectral features of the ATTCT-Q/L5′ MDB formed by two ATTCT repeats. For the minor dumbbell conformer, the TCTA type II tetraloop is supported by (i) the downfield shifted 31P signal of C9 (Supplementary Figure S30A), (ii) the NOEs of T10 H7-T8 H6 and T10 H6-A11 H2/H8, and (iii) the NOEs of A11 H1′/H2′/H2″-A1 H8 (Supplementary Figure S30C-D), which suggest C9 folded into the minor groove, T10 stacked on T8-A11 base pair, and A11 and A1 were close in space, respectively. Because T8 pairs up with A11 in the dumbbell conformer, T8 H7 became obviously less downfield shifted than T3 H7 in (ATTCT)2A (Figure 7B). Interestingly, such minor dumbbell conformer was not observed in the sequences of (ATTCT)2AT, (ATTCT)2ATT and (ATTCT)2ATTC as no downfield shifted C9 31P signal was observed (Supplementary Figure S30E). They were found to form predominantly ATTCT-Q/L5′ MDBs with a 3′-AT, ATT or ATTC overhang, as their T8 H7 signals were found to be close to their downfield shifted T3 H7 signals at ∼1.95 ppm (Figure 7C). In addition, their downfield shifted T3/T8 31P signals at around –3.5 ppm, and the NOEs of C4 H5-T2 H6 and C9 H5-T7 H6 also agree with the NMR features of the ATTCT-Q/L5′ MDBs (Supplementary Figure S30E, F).
Figure 7.
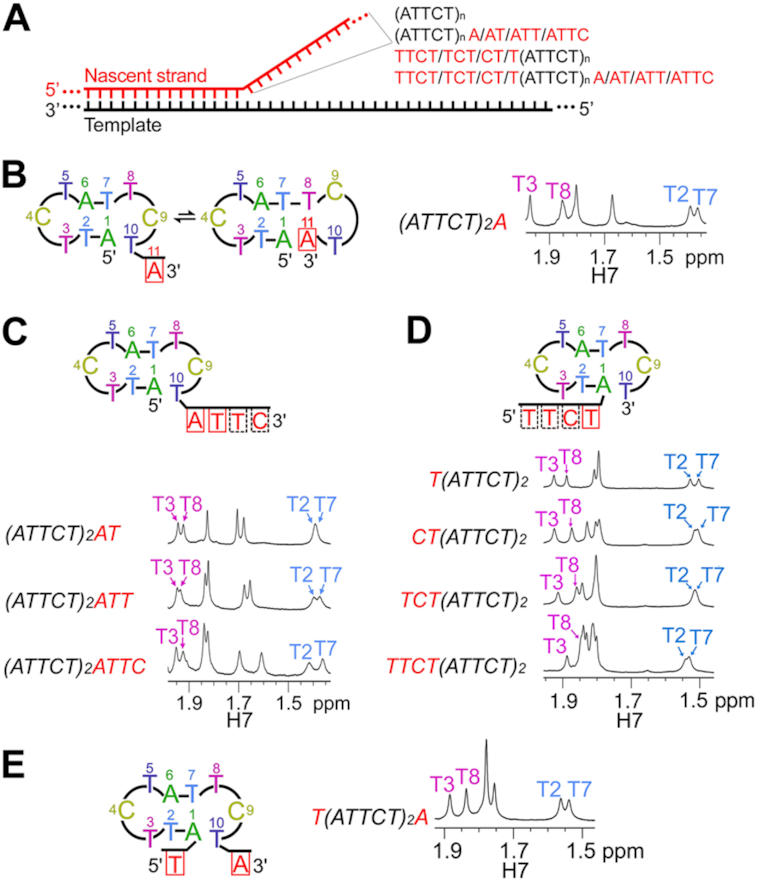
(A) During DNA replication, the momentarily dissociated nascent strand may contain integral repeats (ATTCT)n, integral repeats flanked by 3′ or 5′-partial length repeats, or both 3′ and 5′-partial length repeats. Solution structural behaviors and methyl H7 NMR signals of (B) (ATTCT)2A, (C) (ATTCT)2AT, (ATTCT)2ATT and (ATTCT)2ATTC, (D) T(ATTCT)2, CT(ATTCT)2, TCT(ATTCT)2 and TTCT(ATTCT)2, and (E) T(ATTCT)2A at 0°C.
Secondly, we studied four other sequences, including T(ATTCT)2, CT(ATTCT)2, TCT(ATTCT)2 and TTCT(ATTCT)2, to investigate the effects of 5′-partial length repeats. All of these sequences showed similar NMR spectral features of (ATTCT)2, including (i) the upfield shifted T2/T7 H7 signals at ∼1.5 ppm, (ii) the relatively downfield shifted T3/T8 H7 signals at ∼1.9 ppm (Figure 7D), (iii) the downfield shifted T3/T8 31P signals (Supplementary Figure S31A), and (iv) the base-base NOEs of C4 H5-T2 H6 and C9 H5-T7 H6 (Supplementary Figure S31B), revealing the propensities of these sequences to form ATTCT-Q/L5′ MDBs with a 5′-T, CT, TCT or TTCT overhang.
Comparing the T3/T8 and T2/T7 H7 chemical shifts of two ATTCT repeats flanked with same length of 3′ and 5′-overhangs, i.e. (ATTCT)2AT versus CT(ATTCT)2, (ATTCT)2ATT versus TCT(ATTCT)2, and (ATTCT)2ATTC versus TTCT(ATTCT)2, the chemical shifts of MDBs with 3′-overhangs are more downfield shifted (Figure 7C-D), suggesting higher formation propensities of the MDBs with 3′-overhangs than the MDBs with 5′-overhangs. We have shown that a quasi ATTC/T pentaloop is much more stabilizing than a regular TTCTA pentaloop for the MDB formation (Figure 5). Therefore, the 3′-overhanging residues attached to the quasi pentaloop are expected to have less destabilizing effect on the MDB formation than the 5′-overhanging residues attached to the regular TTCTA pentaloop, and this also explains the preferential formations of MDBs at the 5′-termini in (ATTCT)3, (ATTCT)4 and (ATTCT)5.
Thirdly, we studied the solution structural behaviors of the sequence T(ATTCT)2A to see if an ATTCT-Q/L5′ MDB can still form in the presence of both 5′ and 3′-partial length repeats. Our NMR results show that a ATTCT-Q/L5′ MDB with a 5′ and 3′-overhangs was formed as suggested by (i) the upfield shifted T2/T7 H7 signals at ∼1.5 ppm, (ii) the downfield shifted T3/T8 H7 signals at ∼1.9 ppm (Figure 7E), (iii) the downfield shifted T3/T8 31P signals (Supplementary Figure S32A), and (iv) the NOEs of C4 H5-T2 H6 and C9 H5-T7 H6 (Supplementary Figure S32B), which are similar to the NMR spectral features of the ATTCT-Q/L5′ MDB.
The above structural results show that ATTCT-Q/L5′ MDBs will form not only in sequences containing integral ATTCT repeats, but also in sequences containing partial ATTCT repeats. Therefore, whenever the 3′-terminal of the nascent strand containing ATTCT repeats with integral or partial repeats momentarily dissociates from the template during DNA replication, it is likely that the ATTCT-Q/L5′ MDB will form, thus promoting strand slippage which possibly accounts for the unstable nature of ATTCT repeats in SCA10.
In vitro primer extension assays reveal the ability of ATTCT-Q/L5′ MDB to escape from the proofreading function of KF
With the advancement in DNA sequencing technology, more and more repeat expansion diseases are being found (2,52). A thorough understanding on the origins of these genetic instabilities may benefit the development of disease therapeutics. For repeat expansions occurring via the unusual structure-mediated pathway during DNA replication, the unusual structure formed in the nascent strand must be able to escape from (i) the proofreading function of DNA polymerase, and (ii) the subsequent DNA repair process (1,28,53,54). Recently, there are increasing evidences showing that DNA repair proteins, which are supposed to fix replication errors, indeed promote trinucleotide repeat expansions (15,55). Therefore, escaping from the proofreading function of DNA polymerase becomes a crucial step to bring about repeat expansions. A previous primer extension study showed that the hairpin structure formed with at least two 3′-neighboring base pairs in the nascent (CTG)n or (CAG)n strand could escape from the proofreading of the high fidelity DNA polymerase (54). Yet, little has been known for pentanucleotide repeat expansions.
To investigate if the MDB can form and retain in the nascent strand harbouring ATTCT repeats during DNA replication, we first designed two simplified primer-template models. The template contained (AGAAT)10 flanked by a few non-repeating residues, and its 3′-terminal was a three-carbon spacer ‘C3s’ to avoid template cleavage or extension by KF of DNA polymerase I. The first primer was a 25-nt sequence containing five ATTCT repeats, i.e. 5′-(ATTCT)5. In case that this primer hybridized with the template at multiple sites, we also prepared a second primer which was a 27-nt sequence, i.e. 5′-GC(ATTCT)5. For simplicity, we named these two primer-template models as P1-T and P2-T, respectively (Figure 8A). In vitro primer extensions were performed using the KF at 37°C. Results show that approximately half population of the primer was fully extended and there remained some incomplete extended primer products after 24 h in P1-T and P2-T (Figure 8A). However, no expanded primer product was observed, which may be attributed to stable hybridizations between the primer and template, and no occurrence of strand slippage during primer extensions. This may also because our in vitro primer extension conditions are different from the cellular environments such as the DNA replication machinery.
Figure 8.
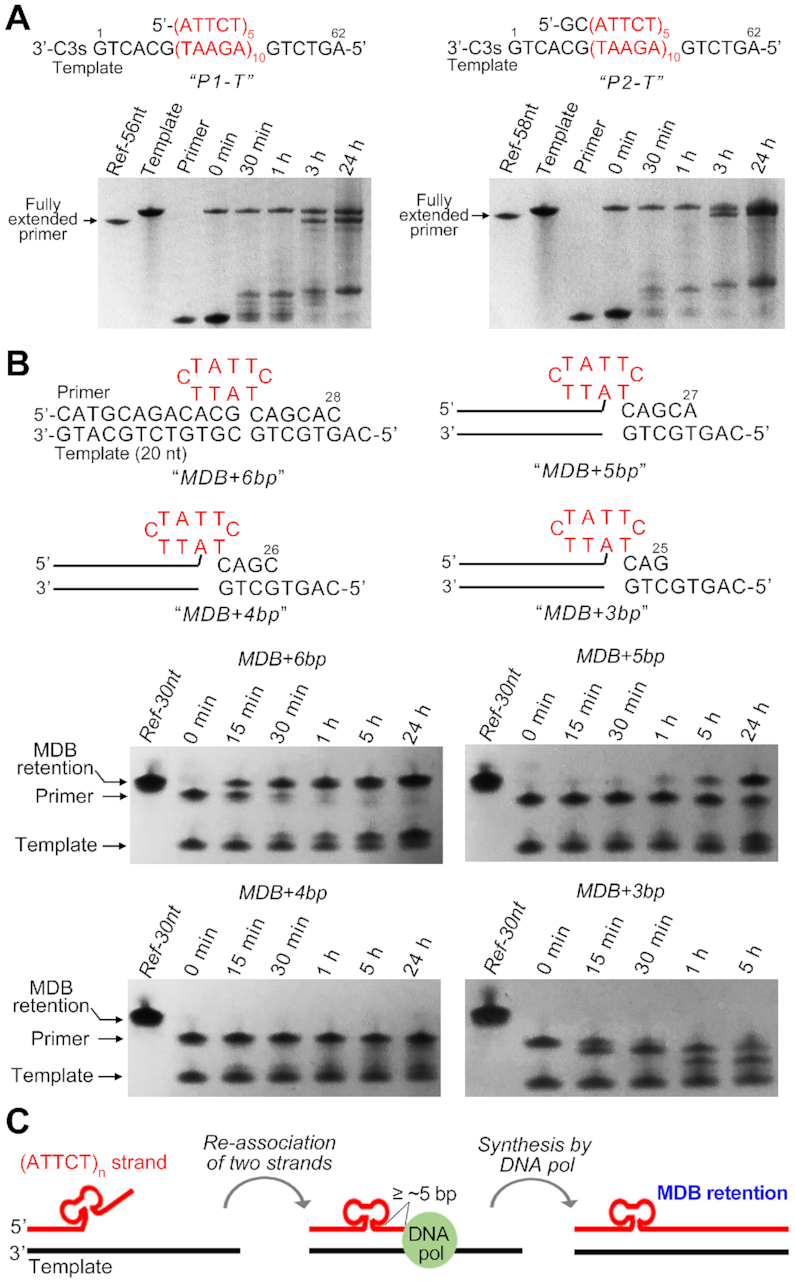
Denaturing PAGE show the in vitro primer extension products of (A) P1-T and P2-T, and (B) MDB+6bp, MDB+5bp, MDB+4bp and MDB+3bp. (C) A proposed model for MDB-mediated ATTCT repeat expansions. The 3′ slippage within ATTCT repeats in the nascent strand leads to the formation of an MDB near the 5′ end. If there are five or more base pairs as the 3′ neighbors of the MDB after re-association between the nascent and template strands, the MDB is able to escape from the proofreading by DNA polymerase, resulting in the MDB retention in the nascent strand.
We then redesigned primer-template models to see if the ATTCT-Q/L5′ MDB, which was pre-existing in the primer as a mimic of MDB formation upon strand slippage, could escape from the proofreading function of DNA polymerase. We constructed four primer-template models in which the primer contained the ATTCT-Q/L5′ MDB forming sequence at a position of 6, 5, 4 and 3-bp away from the priming site. These four primer-template models were named as MDB+6bp, MDB+5bp, MDB+4bp and MDB+3bp, respectively (Figure 8B). NMR spectra of MDB+6bp under 50 mM NaCl and 10 mM MgCl2 were acquired at 37°C. Characteristic spectral features of the ATTCT-Q/L5′ MDB, i.e. the unusually downfield shifted T15 and T20 methyl H7 signals at 2.04 and 1.98 ppm (Supplementary Figure S33), suggest the formation of ATTCT-Q/L5′ MDB under an ionic and temperature condition similar to that of primer extension assays. In vitro primer extension assays were then performed using the KF at 37°C, and the results are shown in Figure 8B. The MDB formed in MDB+6bp escaped from the proofreading by KF and the primer was almost fully extended in 1 h. When reducing the number of 3′-neighboring base pairs, MDB+5bp showed a tiny extended primer product in 1 h, and the extended primer product became predominant after 24 h, suggesting that the MDB formed at 5-bp away from the priming site could still escape from the cleavage by KF, but it slowed down the DNA synthesis by KF. The primer of MDB+4bp was not extended nor cleaved after 24 h, suggesting that the MDB formed at 4-bp away from the priming site might stall primer extensions. For MDB+3bp, a major product of primer cleavage by 1 nt was observed after 30 min, and another product of primer cleavage by ∼2 nt appeared after 1 h. The result of MDB+3bp suggests that KF could sense the MDB and remove a few 3′-neighboring nucleotide(s), but the MDB structure was still refractory to the cleavage activity of KF and stalled primer extensions. We also noticed that the template was extended by 1 nt in these four primer-templates, and the literature has reported that KF tended to add one more nucleotide at the 3′ of a blunt end (56).
Based on our NMR structural findings and the results of in vitro primer extension assays on MDB+6bp to MDB+3bp, we proposed an alternative model for ATTCT repeat expansions in addition to the previously reported replication re-initiations (17) and template switching (18) models. During DNA replication, when the dissociated part of the nascent strand contains more than three ATTCT repeats, an MDB structure is likely to form at the 5′ end as suggested by the structural behaviors of (ATTCT)3 to (ATTCT)5. After re-association between the nascent and templating strands, the MDB will have five or more 3′-neighboring base pairs and escape from the proofreading function of DNA polymerase, causing the MDB retention in the nascent strand (Figure 8C). In contrast, If the MDB is formed closer to the priming site, it may interfere with DNA polymerase activity and result in replication stalling. Notably, mismatch repair (MMR) is a post-replication repair system to maintain the fidelity of DNA replication. The eukaryotic MSH2-MSH3 complex recognizes replication errors including some base-base mismatches and internal loops up to ∼17 nucleotides, and such loops can form as a result of strand slippage during replication of repetitive DNA sequences (57,58). The internal MDB formed by ATTCT repeats may likely be recognized and removed by MSH2-MSH3. If the MDB can further escape from the MMR, a small scale of repeat expansion will occur.
It should be noted that the pathogenetic number of ATTCT repeats ranges from ∼850 to 4500 (3,6,7), and the replication re-initiations (17) and template switching (18) models were proposed for large-scale ATTCT repeat expansions. The formation of an MDB is expected to cause a step size of two-repeat expansion, and the formation of MDBs at multiple sites, if occurred, can bring about larger-size expansions. The MDB-mediated repeat expansions are in a much smaller scale comparing to hundreds or thousands of repeats. We were aware that the expandable CCTG tetranucleotide repeats, of which expansions are also known to be in a large scale, could generate one to two-repeat expansions during in vitro primer extensions (59), and the formation of a mini-loop or MDB structure by one or two CCTG repeats has been proposed to be possible structural intermediates accounting for small-size expansions (23). Although the occurrence of small-scale ATTCT repeat expansions has not been reported at current stage, the MDB structure discovered here provides an alternative pathway for genetic instabilities in SCA10.
Intriguingly, the MDB formed by ATTCT repeats shows similarities in structure and thermodynamic stability to our previously reported TTTA and CCTG MDBs, which have been suggested to associate with TTTA and CCTG tetranucleotide repeat expansions in Staphylococcus aureus pathogen and DM2 patients, respectively (22,23). A previous study on CCTG repeat expansions showed that the sequences which formed thermodynamically less stable secondary structures had more expanded products whereas the sequences that could form the most stable secondary structures did not show any expanded products (59). These suggest that less stable unusual structures indeed can form during DNA replication and escape from the proofreading by DNA polymerase. Coincidently, the newly discovered MDBs formed by expandable tetra- and pentanucleotide repeats all have relatively low thermodynamic stabilities (Tm of ∼20°C). During DNA replication, we cannot exclude the possibility of their formations in the presence of other factors such as supercoiling stresses and macromolecular crowding.
CONCLUSIONS
In this study, the solution structural behaviors of ATTCT pentanucleotide repeats have been investigated by NMR, revealing that ATTCT repeats form well folded MDB structures which comprise a regular TTCTA pentaloop and a quasi TTCT/A pentaloop. Results of in vitro primer extension assays suggest that the MDB is able to escape from the proofreading function of DNA polymerase and thus results in the MDB retention in the nascent strand. Discovery of the MDB formed by ATTCT repeats provides an alternative pathway for repeat expansions in SCA10. In addition to our previously reported MDBs formed by expandable TTTA and CCTG tetranucleotide repeats, the MDB formed by ATTCT repeats here elevates the potential importance of MDBs in causing genetic instabilities.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported ATTCT-Q/L5′ MDB NMR solution structures have been deposited to the Protein Data Bank under the accession number of 6IY5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Xi Zhang for some preliminary works of this project.
Contributor Information
Pei Guo, School of Biology and Biological Engineering, South China University of Technology, Guangzhou, Guangdong 510006, China.
Sik Lok Lam, Department of Chemistry, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong SAR, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
General Research Fund [CUHK14302915]; Special Equipment Grant [SEG/CUHK09] from the Research Grants Council of the Hong Kong Special Administrative Region, a Faculty Strategic Funding (Scheme II) and Direct Grant [4053194] from the Faculty of Science of The Chinese University of Hong Kong; Fund for Double First-Class University Plan from South China University of Technology [x2sw-K5190420]. Funding for open access charge: South China University of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007; 447:932–940. [DOI] [PubMed] [Google Scholar]
- 2. Hannan A.J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 2018; 19:286–298. [DOI] [PubMed] [Google Scholar]
- 3. Matsuura T., Yamagata T., Burgess D.L., Rasmussen A., Grewal R.P., Watase K., Khajavi M., McCall A.E., Davis C.F., Zu L. et al.. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000; 26:191–194. [DOI] [PubMed] [Google Scholar]
- 4. Lin X., Ashizawa T.. SCA10 and ATTCT repeat expansion: clinical features and molecular aspects. Cytogenet. Genome Res. 2003; 100:184–188. [DOI] [PubMed] [Google Scholar]
- 5. London E., Camargo C.H.F., Zanatta A., Crippa A.C., Raskin S., Munhoz R.P., Ashizawa T., Teive H.A.G.. Sleep disorders in spinocerebellar ataxia type 10. J. Sleep Res. 2018; 27:e12688. [DOI] [PubMed] [Google Scholar]
- 6. Schule B., McFarland K.N., Lee K., Tsai Y.C., Nguyen K.D., Sun C., Liu M., Byrne C., Gopi R., Huang N. et al.. Parkinson's disease associated with pure ATXN10 repeat expansion. NPJ Parkinsons Dis. 2017; 3:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alonso I., Jardim L.B., Artigalas O., Saraiva-Pereira M.L., Matsuura T., Ashizawa T., Sequeiros J., Silveira I.. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology. 2006; 66:1602–1604. [DOI] [PubMed] [Google Scholar]
- 8. Chi L.M., Lam S.L.. Structural roles of CTG repeats in slippage expansion during DNA replication. Nucleic Acids Res. 2005; 33:1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mariappan S.V., Catasti P., Silks L.A. 3rd, Bradbury E.M., Gupta G.. The high-resolution structure of the triplex formed by the GAA/TTC triplet repeat associated with Friedreich's ataxia. J. Mol. Biol. 1999; 285:2035–2052. [DOI] [PubMed] [Google Scholar]
- 10. Brcic J., Plavec J.. NMR structure of a G-quadruplex formed by four d(G4C2) repeats: insights into structural polymorphism. Nucleic Acids Res. 2018; 46:11605–11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kovanda A., Zalar M., Sket P., Plavec J., Rogelj B.. Anti-sense DNA d(GGCCCC)n expansions in C9ORF72 form i-motifs and protonated hairpins. Sci. Rep. 2015; 5:17944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lam S.L., Wu F., Yang H., Chi L.M.. The origin of genetic instability in CCTG repeats. Nucleic Acids Res. 2011; 39:6260–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt M.H., Pearson C.E.. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst.). 2016; 38:117–126. [DOI] [PubMed] [Google Scholar]
- 14. McMurray C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010; 11:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iyer R.R., Pluciennik A., Napierala M., Wells R.D.. DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 2015; 84:199–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu G., Bissler J.J., Sinden R.R., Leffak M.. Unstable spinocerebellar ataxia type 10 (ATTCT)·(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol. Cell Biol. 2007; 27:7828–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Potaman V.N., Bissler J.J., Hashem V.I., Oussatcheva E.A., Lu L., Shlyakhtenko L.S., Lyubchenko Y.L., Matsuura T., Ashizawa T., Leffak M. et al.. Unpaired structures in SCA10 (ATTCT)n·(AGAAT)n repeats. J. Mol. Biol. 2003; 326:1095–1111. [DOI] [PubMed] [Google Scholar]
- 18. Cherng N., Shishkin A.A., Schlager L.I., Tuck R.H., Sloan L., Matera R., Sarkar P.S., Ashizawa T., Freudenreich C.H., Mirkin S.M.. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo P., Lam S.L.. Unprecedented hydrophobic stabilizations from a reverse wobble T.T mispair in DNA minidumbbell. J. Biomol. Struct. Dyn. 2020; 38:1946–1953. [DOI] [PubMed] [Google Scholar]
- 20. Liu Y., Guo P., Lam S.L.. Formation of a DNA mini-dumbbell with a quasi-type II loop. J. Phys. Chem. B. 2017; 121:2554–2560. [DOI] [PubMed] [Google Scholar]
- 21. Guo P., Lam S.L.. Minidumbbell: a new form of native DNA structure. J. Am. Chem. Soc. 2016; 138:12534–12540. [DOI] [PubMed] [Google Scholar]
- 22. Guo P., Lam S.L.. Unusual structures of TTTA repeats in icaC gene of Staphylococcus aureus. FEBS Lett. 2015; 589:1296–1300. [DOI] [PubMed] [Google Scholar]
- 23. Guo P., Lam S.L.. New insights into the genetic instability in CCTG repeats. FEBS Lett. 2015; 589:3058–3063. [DOI] [PubMed] [Google Scholar]
- 24. Antao V.P., Tinoco I. Jr. Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992; 20:819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ippel H.H., van den Elst H., van der Marel G.A., van Boom J.H., Altona C.. Structural similarities and differences between H1- and H2-family DNA minihairpin loops: NMR studies of octameric minihairpins. Biopolymers. 1998; 46:375–393. [Google Scholar]
- 26. Handa V., Yeh H.J., McPhie P., Usdin K.. The AUUCU repeats responsible for spinocerebellar ataxia type 10 form unusual RNA hairpins. J. Biol. Chem. 2005; 280:29340–29345. [DOI] [PubMed] [Google Scholar]
- 27. Wu B., Girard F., van Buuren B., Schleucher J., Tessari M., Wijmenga S.. Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold. Nucleic Acids Res. 2004; 32:3228–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo P., Lam S.L.. The competing mini-dumbbell mechanism: new insights into CCTG repeat expansion. Signal Transduct. Target. Ther. 2016; 1:16028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006; 1:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stott K., Stonehouse J., Keeler J., Hwang T.L., Shaka A.J.. Excitation sculpting in high-resolution nuclear magnetic resonance spectroscopy: application to selective NOE experiments. J. Am. Chem. Soc. 1995; 117:4199–4200. [Google Scholar]
- 31. Plateau P., Gueron M.. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982; 104:7310–7311. [Google Scholar]
- 32. van Dongen M.J., Wijmenga S.S., Eritja R., Azorin F., Hilbers C.W.. Through-bond correlation of adenine H2 and H8 protons in unlabeled DNA fragments by HMBC spectroscopy. J. Biomol. NMR. 1996; 8:207–212. [DOI] [PubMed] [Google Scholar]
- 33. Bax A., Summers M.F.. Proton and carbon-13 assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J. Am. Chem. Soc. 1986; 108:2093–2094. [Google Scholar]
- 34. Luy B., Marino J.P.. 1H-31P CPMG-correlated experiments for the assignment of nucleic acids. J. Am. Chem. Soc. 2001; 123:11306–11307. [DOI] [PubMed] [Google Scholar]
- 35. Markley John L., Bax A., Arata Y., Hilbers C.W., Kaptein R., Sykes Brian D., Wright Peter E., Wüthrich K.. Recommendations for the presentation of NMR structures of proteins and nucleic acids (IUPAC Recommendations 1998). Pure Appl. Chem. 1998; 70:117–142. [Google Scholar]
- 36. Feigon J., Wright J.M., Leupin W., Denny W.A., Kearns D.R.. Use of two-dimensional NMR in the study of a double-stranded DNA decamer. J. Am. Chem. Soc. 1982; 104:5540–5541. [Google Scholar]
- 37. Wijmenga S.S., van Buuren B.N.M.. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998; 32:287–387. [Google Scholar]
- 38. Lam S.L., Chi L.M.. Use of chemical shifts for structural studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 2010; 56:289–310. [DOI] [PubMed] [Google Scholar]
- 39. Saenger W. Principles of Nucleic Acid Structure. 1984; NY: Springer-Verlag; 122–123. [Google Scholar]
- 40. Altona C., Sundaralingam M.. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972; 94:8205–8212. [DOI] [PubMed] [Google Scholar]
- 41. Case D.A., Betz R.M., Cerutti D.S., Cheatham T., Darden T., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., Homeyer N. et al.. AMBER 2016. 2016; San Francisco: University of California. [Google Scholar]
- 42. Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C.. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015; 11:3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Onofrio A., Parisi G., Punzi G., Todisco S., Di Noia M.A., Bossis F., Turi A., De Grassi A., Pierri C.L.. Distance-dependent hydrophobic-hydrophobic contacts in protein folding simulations. Phys. Chem. Chem. Phys. 2014; 16:18907–18917. [DOI] [PubMed] [Google Scholar]
- 44. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 45. Lam S.L. DSHIFT: a web server for predicting DNA chemical shifts. Nucleic Acids Res. 2007; 35:W713–W717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Buuren B.N., Overmars F.J., Ippel J.H., Altona C., Wijmenga S.S.. Solution structure of a DNA three-way junction containing two unpaired thymidine bases. Identification of sequence features that decide conformer selection. J. Mol. Biol. 2000; 304:371–383. [DOI] [PubMed] [Google Scholar]
- 47. Overmars F.J., Pikkemaat J.A., van den Elst H., van Boom J.H., Altona C.. NMR studies of DNA three-way junctions containing two unpaired thymidine bases: the influence of the sequence at the junction on the stability of the stacking conformers. J. Mol. Biol. 1996; 255:702–713. [DOI] [PubMed] [Google Scholar]
- 48. Welch J.B., Walter F., Lilley D.M.. Two inequivalent folding isomers of the three-way DNA junction with unpaired bases: sequence-dependence of the folded conformation. J. Mol. Biol. 1995; 251:507–519. [DOI] [PubMed] [Google Scholar]
- 49. Baouendi M., Cognet J.A., Ferreira C.S., Missailidis S., Coutant J., Piotto M., Hantz E., Herve du Penhoat C.. Solution structure of a truncated anti-MUC1 DNA aptamer determined by mesoscale modeling and NMR. FEBS J. 2012; 279:479–490. [DOI] [PubMed] [Google Scholar]
- 50. Pakleza C., Cognet J.A.. Biopolymer Chain Elasticity: A novel concept and a least deformation energy principle predicts backbone and overall folding of DNA TTT hairpins in agreement with NMR distances. Nucleic Acids Res. 2003; 31:1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blommers M.J., van de Ven F.J., van der Marel G.A., van Boom J.H., Hilbers C.W.. The three-dimensional structure of a DNA hairpin in solution two-dimensional NMR studies and structural analysis of d(ATCCTATTTATAGGAT). Eur. J. Biochem. 1991; 201:33–51. [DOI] [PubMed] [Google Scholar]
- 52. Ishiura H., Doi K., Mitsui J., Yoshimura J., Matsukawa M.K., Fujiyama A., Toyoshima Y., Kakita A., Takahashi H., Suzuki Y. et al.. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 2018; 50:581–590. [DOI] [PubMed] [Google Scholar]
- 53. Guo J., Gu L., Leffak M., Li G.M.. MutSbeta promotes trinucleotide repeat expansion by recruiting DNA polymerase beta to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis. Cell Res. 2016; 26:775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chan N.L., Guo J., Zhang T., Mao G., Hou C., Yuan F., Huang J., Zhang Y., Wu J., Gu L. et al.. Coordinated processing of 3′ slipped (CAG)n/(CTG)n hairpins by DNA polymerases beta and delta preferentially induces repeat expansions. J. Biol. Chem. 2013; 288:15015–15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao X.N., Usdin K.. The repeat expansion diseases: the dark side of DNA repair. DNA Repair (Amst.). 2015; 32:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clark J.M., Joyce C.M., Beardsley G.P.. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J. Mol. Biol. 1987; 198:123–127. [DOI] [PubMed] [Google Scholar]
- 57. Kunkel T.A., Erie D.A.. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015; 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jensen L.E., Jauert P.A., Kirkpatrick D.T.. The large loop repair and mismatch repair pathways of Saccharomyces cerevisiae act on distinct substrates during meiosis. Genetics. 2005; 170:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heidenfelder B.L., Topal M.D.. Effects of sequence on repeat expansion during DNA replication. Nucleic Acids Res. 2003; 31:7159–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported ATTCT-Q/L5′ MDB NMR solution structures have been deposited to the Protein Data Bank under the accession number of 6IY5.


