Abstract
Purpose
We examined whether prospective molecular characterization of advanced metastatic disease can reveal grade and/or histology-specific differences to inform diagnosis and facilitate enrollment onto clinical trials.
Experimental Design
Patients with uterine sarcoma consented to a prospective study of next-generation sequencing (NGS). Clinical annotations were extracted from their medical record. Tumor and matched normal DNA were subjected to NGS, and the genomic landscape was explored for survival correlations and therapeutic targetability.
Results
Tumors from 107 women were sequenced and included leiomyosarcoma (uLMS, n=80), high-grade non-LMS (n=22), low-grade endometrial stromal sarcoma (LG-ESS, n=4), and smooth muscle tumor of uncertain malignant potential (STUMP, n=2). Genomic profiling influenced histologic diagnosis in three cases. Common uterine LMS (uLMS) alterations were loss-of-function mutations in TP53 (56%), RB1 (51%), and ATRX (31%). Homozygous deletions of BRCA2 were present in 5% of these patients. PTEN alteration frequency was higher in the metastases samples as compared to the primary samples. Genomes of low-grade tumors were largely silent, while 50.5% of high-grade tumors had whole genome duplication. Two metastatic uLMS cases were hypermutated. Both had prolonged disease-free survival. Potentially actionable mutations were identified in 48 patients (45%), eight (17%) of whom received matched therapy with two achieving clinical responses. Among uLMS patients with somatic BRCA2 alterations, sustained partial responses were observed with PARP inhibitor-containing therapy.
Discussion
Prospective genomic profiling can contribute to diagnostic precision and inform treatment selection in patients with uterine sarcomas. There was evidence of clinical benefit in uLMS patients with somatic BRCA2 alterations treated with PARP inhibitors.
Keywords: uterine cancer, sarcoma, genomic profiling, next-generation sequencing, precision medicine
Introduction
Uterine sarcomas are rare mesenchymal neoplasms that differ in histologic appearance and clinical behavior. Classification of uterine sarcoma has traditionally been based on histologic appearance including key features of cytologic atypia, tumor cell necrosis, mitotic rate, and select use of immunohistochemistry (IHC) to support tissue differentiation. The most common subtypes of uterine sarcomas are leiomyosarcoma (uLMS), low-grade endometrial stromal sarcoma (LG-ESS), and high-grade endometrial stromal sarcoma (HG-ESS). Immunophenotypes and molecular characterization of uterine sarcomas have increasingly been utilized to improve diagnostic classification and prognostication in uterine sarcomas. For example, LG-ESSs express the estrogen and progesterone receptors, have an indolent disease course, and are molecularly characterized by recurrent chromosomal translocations commonly involving JAZF1 (1). By comparison, a subset of HG-ESS harbor t(10;17)(q22;p13) translocations that result in expression of a YWHAE-NUTM2A/B fusion (2) and have an aggressive disease course (3) while others have BCOR alterations that are of unknown prognostic significance (4). As such, testing for YWHAE and BCOR fusions in histologically challenging high-grade sarcomas may facilitate a more precise diagnosis. Uterine leiomyosarcoma (uLMS), the most common subtype of uterine sarcoma, does not have a single defining molecular abnormality. Instead, these tumors often have multiple chromosomal abnormalities associated with chromothripsis, TP53 and RB1 inactivation, and whole genome duplication (WGD) (5).
To determine whether genomic profiling could improve diagnostic precision, inform prognosis, or aid in therapeutic selection, we prospectively characterized uterine sarcomas, enriching for high-risk subtypes that contribute to the greatest morbidity and mortality. We applied a clinically validated next-generation sequencing (NGS) platform to determine whether molecular profiling could enhance uterine sarcoma classification, provide prognostic information within histologic subtypes, and identify subsets of patients whose tumors harbor targetable mutations. We also evaluated whether patients with potentially actionable alterations received matched therapy, and if so, whether they derived clinical benefit.
Materials and Methods
Patients
Patients with a histologically confirmed uterine sarcoma (USARC, oncotree.mskcc.org) at Memorial Sloan Kettering Cancer Center (MSK) were consented to a prospective study using NGS under an Institutional Review Board (IRB)-approved protocol (ClinicalTrials.gov, NCT01775072). This study was conducted in accordance with International Ethical Guidelines for Biomedical Research Involving Human Subjects, Good Clinical Practice guidelines, the Declaration of Helsinki, and local laws, and with written consent of the subjects, where necessary.
Histologic diagnosis, stage, and grade
All consecutively sequenced uterine sarcomas and uterine mesenchymal tumors of uncertain malignant potential, regardless of histologic type or grade, successfully profiled between April 2014 and April 2017 were included in this cohort, with the exception of perivascular epithelioid tumors, which lack established standard diagnostic criteria. All tumors were reviewed and histopathologically confirmed to be uterine sarcoma by gynecologic sarcoma pathologists at MSK. To facilitate genomic and outcome analysis, we grouped cases into four diagnostic entities on the basis of histology: 1) uLMS, 2) high-grade non-LMS uterine sarcomas, 3) LG-ESS, and 4) smooth muscle tumor of uncertain malignant potential (STUMP). As grading of uLMS is not standardized and since most tumors meeting histologic criteria for a diagnosis of LMS at MSK are considered high grade, LMS cases were not further subdivided by grade. The two STUMP tumors were included, because both patients had an original diagnosis of STUMP in the uterus, followed by metastatic disease, which in one case was histologically shown to be LMS. In both cases, the genomic profiling was performed on the tissue that had been classified as STUMP. Tumors were staged according to the 2009 International Federation of Gynecology and Obstetrics (FIGO) staging system. Primary and metastatic sites were sequenced based on tissue availability.
Genomic sequencing
NGS of DNA extracted from formalin-fixed, paraffin embedded (FFPE) tumor and patient-matched blood was performed in the Clinical Laboratory Improvement Amendments (CLIA)-certified MSK Molecular Diagnostics Service Laboratory using MSK-IMPACT, an exon capture assay targeting all coding exons of 341 (n=44 samples), 410 (n=44 samples), or 468 (n=20 samples) cancer-associated genes, as previously described (6, 7). DNA was sequenced to an average of 646-fold sequence coverage. All somatic variant genomic results were reviewed by a molecular pathologist for quality and accuracy prior to adding the results into the patient’s medical record, as previously described (7). All patient-level clinical and genomic data are available at the cBioPortal (https://cbioportal.mskcc.org/study/summary?id=usarc_msk_2020).
Fusion detection
Fusion genes were identified either via MSK-IMPACT, which targets breakpoint-containing introns of known oncogenic fusion partners, or an RNA-based custom solid tumor fusion panel, MSK-Fusion (8), which was used as part of the diagnostic work-up for a subset of patients.
Microsatellite instability
The presence of microsatellite instability (MSI) was assessed genomically using MSIsensor (version 0.2) (9). MSIsensor assigns a numeric score based on the percentage of unstable microsatellite sites divided by the total number of microsatellite sites tested from aligned sequencing data. Based on the prior clinical validation of MSIsensor using MSK-IMPACT data, MSI status was defined as follows: <3: microsatellite stable (MSS); ≥3 and <10: MSI-indeterminate (MSI-I); and ≥10: MSI-high (MSI-H) (10). For tumor samples in which genomic sequencing yielded an MSIsensor score ≥10, the MSI phenotype was confirmed by IHC staining of mismatch repair (MMR) proteins (MLH1, MSH2, MSH6, and PMS2).
Mutational signature decomposition
To better characterize the mutational processes driving acquisition of somatic alterations, mutational signature decomposition analysis was performed for tumor samples with ten or more single nucleotide variant (SNV) somatic mutations, as previously described (11). For cases in which more than one signature was present, a weighted combination of signatures was calculated reflecting the proportion of mutations in the sample attributed to that signature.
Allele-specific copy number analysis
We performed FACETS analysis to determine allele-specific and absolute DNA copy number genome-wide in all patients (FACETS version 0.5.6, cval=100) (12). We used these allele-specific copy number data to estimate tumor purity and ploidy. Prior to further analysis, total copy number log ratios were corrected for ploidy and purity. Tumors with WGD were those in which greater than 50% of the autosomal genome had a major copy number ≥2, where major copy number is defined as the number of copies of the most prevalent allele present in the sample (13). Cancer cell fractions (CCFs) were calculated using a binomial distribution and maximum likelihood estimation normalized to produce posterior probabilities, and were used to infer the sequence/timing of mutations (14).
Germline analysis
Germline annotation for pathogenic or likely pathogenic variants was performed by MSK-IMPACT for 76 genes previously associated with cancer predisposition syndromes using a clinically validated platform (15, 16). In accordance with local IRB guidelines and protocol-mandated procedures, the germline variant annotation and the assessment of pathogenicity were performed after irreversible anonymization of patients. Histologic type and allele-specific absolute copy number were retained prior to irreversible anonymization, permitting subsequent determination of loss-of-heterozygosity (LOH). No other clinical data were retained following anonymization.
Programmed cell death protein 1 (PD-1) IHC
To confirm that deletions of PDCD1 (the gene that encodes PD-1) were associated with loss of protein expression in tumor samples with homozygous deletion of PDCD1, IHC with an anti–PD-1 monoclonal antibody (clone NAT105; Abcam, Cambridge, MA) using a Leica Bond-3 automated platform (Leica, Buffalo Grove, IL) was performed. A polymeric secondary kit (Refine, Leica) was used for the detection of the primary antibody.
Annotation of somatic alterations
To classify the individual identified somatic genomic variants, we utilized the OncoKB knowledgebase (OncoKB.org), which provides disease-specific levels of evidence for the actionability of individual mutant alleles, DNA copy number alterations, and translocations (17). A level 1 alteration is an FDA-recognized biomarker in the patient’s tumor type; a level 2 alteration is a biomarker routinely used to guide prescribing of an FDA-approved drug in the patient’s tumor type (2A) or another indication (2B); and a level 3 alteration has compelling clinical evidence to support its use as a biomarker predictive of treatment response. These annotations and integration with clinical data were performed as of October 2019.
Statistical analysis
We evaluated for enrichment of genomic alterations within this cohort across histologic subtypes and sample type (primary vs. metastasis), as well as between this cohort and other cohorts, including MSK non-uLMS and The Cancer Genome Atlas (TCGA) uLMS cohorts (18). Comparisons were conducted utilizing the Fisher Exact test, and nominal p-values were specified. Survival analyses were performed using univariate Cox proportional hazards model and Kaplan-Meier estimation of overall survival (OS) using the R ‘survival’ package (2.41–3).
Results
Patient demographics
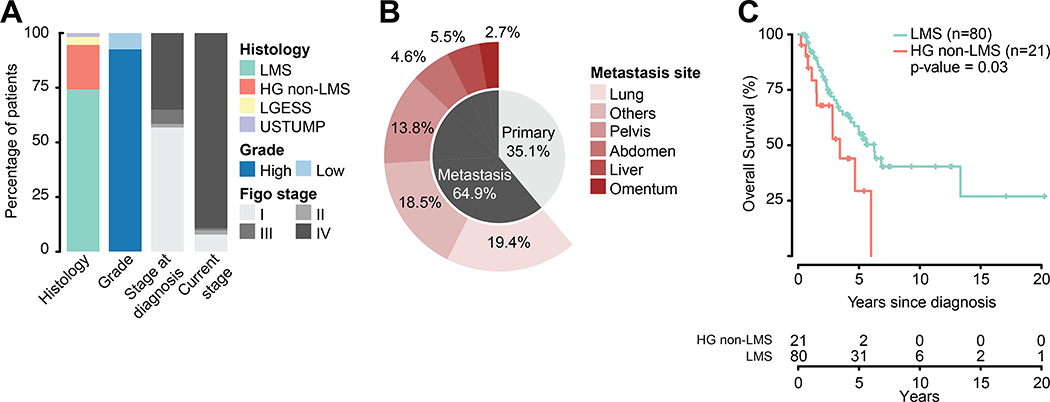
As part of the previously described MSK-IMPACT clinical sequencing cohort (6), which at the time of data freeze stood at 15816 patients for tumor types with at least 25 cases, the tumors of 107 women with uterine sarcoma were successfully sequenced. At the time of diagnosis, 57% (n=61) of patients had uterine-confined disease (FIGO stage I). At the time of sequencing, 89% (n=95) had recurrent/metastatic disease, Figure 1A. As expected for a cohort of patients with recurrent/metastatic uterine sarcoma, the majority of cases (94.3%, n=101) were histologically high grade. Histologic sarcoma subtypes included uLMS (n=80), high-grade non-LMS (n=21), LG-ESS (n=4), and STUMP (n=2). High-grade non-LMSs were further classified as: high-grade endometrial stromal sarcoma (n=7), undifferentiated (n=5), high-grade adenosarcoma with sarcomatous overgrowth (n=4), high-grade uterine sarcoma with heterologous elements (n=2) uterine sarcoma with focal dedifferentiation, high grade uterine sarcoma not otherwise specified, atypical myxoid neoplasm with differential including leiomyosarcoma or myofibroblastic sarcoma (one each). All patients are represented by a single sample, except for a high-grade non-LMS patient with two metastasis samples (pelvic and abdomen). In contrast to The Cancer Genome Atlas (TCGA), both primary (39%, n=42) and metastatic (61%, n=66) samples were profiled, Figure 1B. At the time of analysis, 45% of patients had died of disease. Median survival from diagnosis for the entire cohort was 5.6 years (range, 0.2 to 23.1 years). Survival for high-grade non-LMS patients was significantly worse than for patients with uLMS (3.4 years vs. 6.3 years, respectively; HR=0.48, CI=0.24–0.96, p=0.03), Figure 1C.
Figure 1.
A) Composition of the MSK uterine sarcoma cohort by histology, grade, and stage. B) Distribution of the biopsied primary and metastatic disease sites and sample numbers in the cohort. C) Overall survival of high grade uterine sarcoma cohort split by leiomyosarcoma (LMS) and high-grade non-leiomyosarcoma (HG non-LMS).
Genomic alterations and comparisons across uterine sarcoma histologies
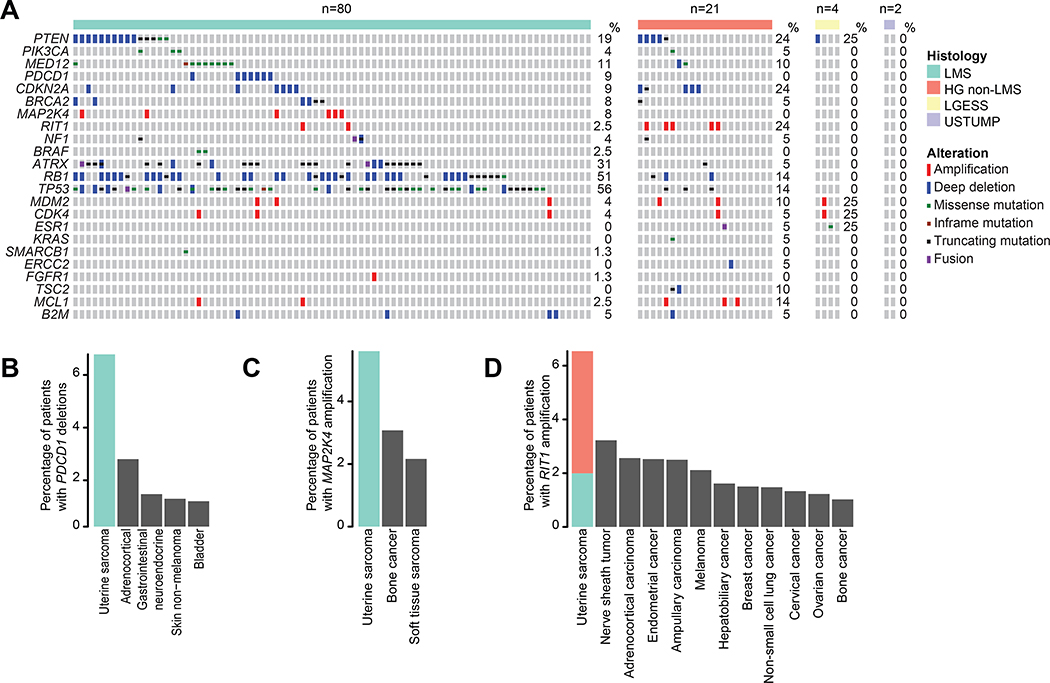
The most common alterations in uLMS were loss-of-function mutations or homozygous deletions in TP53 (56%), RB1 (51%), and ATRX (31%), Figure 2A. TP53, RB1, and ATRX alterations were less common in high-grade non-LMS cases (14%, 14%, and 5%, respectively; p<0.01 for all). Compared to other cancer types, uterine sarcomas have high frequency of homozygous deletions of BRCA2 (19), and these were found exclusively in uLMS, Figure 2A. uLMS was also characterized by recurrent homozygous deletions of PDCD1, which encodes PD-1. Indeed, uLMS had the highest rate of PDCD1 homozygous deletions among all cancer types with at least 25 cases in the contemporary MSK-IMPACT clinical series cohort of prospective sequenced cancers (n=15816), Figure 2B. To determine whether PDCD1 homozygous deletion was associated with loss of PDCD1 expression, we performed IHC in affected cases with sufficient archival material (n=2 of 7), which confirmed PDCD1 was negative in both cases (Figure S1). Similarly, amplifications of MAP2K4 and RIT1 were most common in uterine sarcomas compared to the same contemporary MSK-IMPACT clinical cohort, Figures 2C and 2D. Furthermore, RIT1 was amplified at a significantly higher frequency in high-grade non-LMSs at 24% (5/21), compared to only 2.5% (2/80) of uLMSs (p=0.004).
Figure 2.
A) Oncoprint of genomic alterations in the cohort split by histology. Alterations represented were selected by the following criteria: 1) All actionable alterations (OncoKB); 2) All genes with oncogenic alterations (OncoKB) in at least 5% of cases; 3) An alteration type in a given gene was found to be most frequent in uterine sarcomas when compared to the contemporary MSK-IMPACT clinical series cohort of prospectively sequenced cancers (n=15816). B) Frequency of PDCD1 homozygous deletions in the MSK-IMPACT clinical sequencing cohort compared to other cancer types with at least 25 cases and 1% altered cases. C) and D) Frequency of MAP2K4 and RIT1 amplifications in the MSK-IMPACT clinical sequencing cohort compared to other cancer types with at least 25 cases and 1% altered cases.
There were no significant differences in mutation patterns between primary and metastatic samples except for PTEN alterations which were more frequent in the metastasis samples (p=0.046). This difference remained significant within the uLMS subset (p=0.04). Only 11% of uLMS patients harbored MED12 mutations, an alteration present in an estimated 70% of benign uterine leiomyomas (20). The genomic landscape of LG-ESS (n=4) and uterine STUMP cases (n=2) had few oncogenic alterations, with only four identified likely oncogenic alterations across three patients, Figure 2A. 50.5% (50/99) of the high grade uterine sarcomas (uLMS and high grade non-LMS combined) displayed evidence of WGD, Figure S2. In contrast, none of the low-grade tumors displayed (WGD), supporting the association between WGD and high-grade uterine sarcomas (p=0.04).
We identified two extreme outliers in terms of tumor mutational burden (TMB) among the high-grade uLMS cases, hitherto referred to as hypermutated, Figure S3A. Both possessed microsatellite instability (MSI) based upon MSIsensor analysis, Figure S3B. Using an orthogonal approach, mutational signature decomposition confirmed both cases as possessing MSI/MMR-D mutational signatures, Figure S3C. IHC identified loss of MSH2 protein expression in one case and MSH6 in the other. Neither case had somatic oncogenic alterations in these genes that would explain the MSI phenotype, implying that the cause of the MSI phenotype could be epigenetic or germline. Both patients had distant metastatic disease, were managed with both systemic cytotoxic and local therapies, achieved complete responses, and have remained disease free 5.9 and 13 years from initial diagnosis. Neither received treatment with immune checkpoint inhibitors. Excluding the two MSI-H hypermutated cases, tumor mutation burden was higher for the remaining uLMSs compared to high-grade non-LMSs (2.5 mutations/Mb, range 0–7.9 and 1.1 mutations/Mb range 0–10.6, respectively; p=0.05). Following permanent anonymization (excluding the two MSI phenotype patients), two patients were found to be carriers of pathogenic germline TP53 mutations, one of which was accompanied by somatic biallelic inactivation of TP53. One of these patients was known to have Li-Fraumeni syndrome. No other known or likely pathogenic germline variants were identified in the anonymized analysis.
Genomic comparison of uterine vs. non-uterine LMS
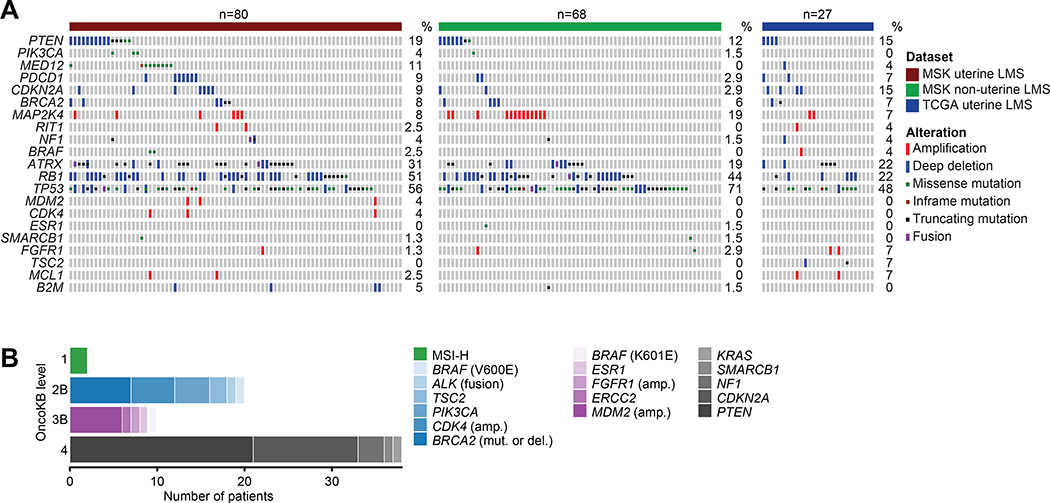
We next sought to determine how the genomic landscape of uLMSs in our cohort (n=80) differed from non-uterine LMSs sequenced at MSK (n=68), Figure 3A. Alterations in TP53 were significantly more common in non-uterine LMS, 71% (52/68) compared to uLMS 56% (45/80), p=0.01. Similarly, MAP2K4 was amplified at a higher frequency in non-uterine LMS 19% (13/68) compared to uLMS at 8% (6/80), p=0.06. uLMSs had a significantly higher frequency of MED12 mutations, 11% (9/80) compared to non-uterine LMSs where MED12 alterations are absent, p=0.01. Furthermore, in a comparison of uLMSs from this study to those of a small cohort (n=27) from TCGA, RB1 alterations were significantly more frequent in the MSK cohort, 51% (41/80) vs. 22% (6/27), p=0.02.
Figure 3.
A) Comparison of genomic alterations across MSK uterine-LMS, MSK non-uterine LMS, and TCGA uterine-LMS. B) All clinically actionable alterations identified via MSK-IMPACT or MSK-Fusion, split by OncoKB levels of evidence.
Genomic alterations not prognostic for OS
In order to determine whether any of the alterations detected were prognostic for OS, we performed univariate Cox regression analysis on all USARC-relevant genes with oncogenic (or likely oncogenic) alterations in at least five of the LMS and high-grade non-LMS cases (LG-ESS and STUMP tumors were excluded from the survival analysis due to small sample size (n=6) and their known long-survival prognoses). Notably, none of the alterations in these genes, nor broader somatic features like WGD were associated with OS in this study cohort.
Therapeutic actionability and diagnostic utility
In addition to IMPACT sequencing, seven patients received MSK-Fusion (a targeted multiplex RNA sequencing assay (8)) as part of their diagnostic work up, Table S1. Overall, from IMPACT and MSK-Fusion we identified potentially therapeutically actionable alterations in 45% of all patients profiled (n=48 of 107, Figure 3B). Among these was a tumor with an LBH-ALK fusion, which was initially missed by IMPACT DNA-based sequencing as the breakpoint was located in an intron not commonly targeted by ALK fusions found in lung cancers. Previously classified as an atypical uLMS, pathology re-review revealed a myxoid spindle cell neoplasm with moderate-to-severe nuclear pleomorphism, with IHC positive for smooth muscle actin and negative for desmin. The ALK fusion prompted revision of the diagnosis to inflammatory myofibroblastic tumor (IMT), a rare sarcoma subtype in which ALK fusions are characteristic (21). This patient was subsequently treated with an ALK inhibitor, crizotinib, and had radiographic stabilization of disease for 30+ months. Subsequent to the data freeze, a second uLMS was found to harbor an ALK fusion via IMPACT sequencing, again prompting review and reclassification as an IMT. This patient was treated with crizotinib, achieving a brief radiographic response followed by progression. The patient further progressed on the second-generation ALK inhibitor ceritinib. One patient had a BCOR-ZC3H7B fusion identified by IMPACT sequencing. This patient was originally diagnosed with a metastatic high-grade myxoid uLMS and survived for 3.4 years. BCOR mutations have been described as diagnostic markers of a specific subset of high-grade endometrial stromal sarcomas, and as histologic mimickers of myxoid LMS (4, 22). In this case, genomic profiling led to reclassification of the tumor as a BCOR-mutated high-grade stromal sarcoma, which has implications for therapeutic choices since certain sarcoma chemotherapy agents are approved only for LMS, and clinical trial eligibility may be histology-specific. Additional diagnostically relevant fusions were identified in three more patients by MSK-Fusion - YWHAE/NUTM2 rearrangements in two high-grade, non-pleomorphic stromal sarcomas (3) and one JAZF1-SUZ12 in a LG-ESS (23).
Additional potentially actionable alterations included seven BRCA2 alterations (four homozygous deletions (all high-grade uLMS) three somatic BRCA2 mutations (two high-grade uLMS, one high-grade non-LMS), two BRAF activating mutations (V600E and K601E; both uLMS), two MSI tumors, and one ESR1 Y537S ligand-binding domain mutation. Overall, 17% (8/48) of patients received therapy matched to their potentially actionable genomic alteration. Two patients achieved radiographic responses, one achieved prolonged stable disease (the ALK fusion patient), and five experienced progression. One of the two patients who achieved a radiologic response to matched therapy had metastatic LG-ESS, treated with anti-estrogen therapy for more than 20 years. Biopsy at progression confirmed JAZF1 fusion-positive LG-ESS, and IMPACT detected an ESR1 ligand-binding domain mutation (Y537S). Since ESR1 mutations have been described in the context of estrogen receptor-positive breast cancer to confer ligand-independent signaling and thus resistance to aromatase inhibitors (24, 25), her hormonal therapy was changed to the selective estrogen modulator fulvestrant, resulting in tumor regression. The second patient to achieve radiographic response had high-grade uLMS with somatic BRCA2 deletion. She was treated on a clinical trial with a Poly (ADP-ribose) polymerase (PARP) inhibitor, achieving an objective partial response lasting over 6 months.
Both BRAF-mutant patients received matched therapy. The BRAF V600-mutant patient was treated with an oral BRAF inhibitor on a basket trial, experiencing progression within 4 weeks. The patient with the BRAF K601E mutation, a class two dimer-dependent BRAF mutant which has been previously biologically characterized and is resistant to first-generation BRAF inhibitors (26), was enrolled on a clinical trial of a pan-RAF inhibitor. Her best response was stable disease lasting 4 months. As detailed above, both MSI-H high-grade LMS patients remained disease-free following multimodality management of metastatic disease and have not received immune checkpoint inhibitor treatment.
Given the activity of PARP inhibitors in BRCA-mutated advanced breast and ovarian cancers (27, 28), we sought to validate the actionability of BRCA2 alterations that our analysis identified as common in uLMS patients. Post data freeze, five subsequent high-grade uLMS patients with BRCA2 alterations were identified by routine clinical MSK-IMPACT testing (three with somatic biallelic inactivation and one each with a somatic and germline truncating mutations accompanied by LOH). Four received PARP inhibitor-containing therapy as part of various clinical trials, and the remaining patient harboring a germline BRCA2 mutation received PARP inhibitor off-label. As with the BRCA2-altered uLMS patient from the study cohort, all five had at least some radiographic regression, including one patient who achieved a complete radiographic response with treatment durations ranging from 6 months to 28 months (two of five patients remain on therapy at the time of submission). These results demonstrate the potential actionability of BRCA2 alterations in uLMS and the potential durability of responses.
Discussion
Uterine sarcomas are a histologically and clinically heterogenous group of tumors. A subset of uterine sarcomas have distinct molecular diagnostic characteristics (e.g., fusion rearrangements involving JAZF1 in LG-ESS and YWHAE/NUTM2 rearrangements in high-grade, non-pleomorphic stromal sarcomas). However, LMS, the most common histologic subtype of uterine sarcoma, has complex karyotypes with numerous structural aberrations and lacks a characteristic translocation or single driving or defining mutation (29, 30). Three core molecular mechanisms in sarcomagenesis have been described for soft tissue sarcomas. These include DNA copy number alterations, somatic mutations in key signaling pathways, and transcriptional dysregulation from chimeric transcriptional factors (31). In our cohort of uterine sarcomas, we found evidence to support these three molecular mechanisms for sarcomagenesis.
Findings of clinical utility included BCOR rearrangement for more precise diagnosis, detection of potentially actionable mutations in ALK, BRAF, ESR1, and BRCA2, and identification of MSI-H tumors, each of which could guide selection of FDA-approved therapies or influence clinical trial eligibility. The discovery of the BCOR fusion illustrates that prospective molecular characterization can lead to refinements in histologic classifications or distinguish among challenging histologic appearances such as true myxoid or epithelioid LMS from high-grade stromal sarcomas. Moreover, the further sub-classification of high-grade endometrial stromal sarcomas into either YWHAE or BCOR altered cases will ultimately facilitate studies to determine their prognostic or therapeutic implications. Prolonged disease stabilization with an ALK inhibitor in the patient with an ALK fusion sarcoma is consistent with recent results of a phase two trial of crizotinib in IMT in which 50% of ALK-positive patients achieved an objective response (32). The frequency of MED12 mutations in uLMS patients (11%) was much lower than one would expect if uLMS commonly evolved from antecedent benign leiomyomas, which harbor MED12 mutations in ~70% of cases. This finding suggests that uLMS may not evolve from antecedent benign leiomyomas, but rather arise independently as de novo cancers. This observation may have potential clinical implications, since women with stable-appearing leiomyomas on imaging may be reassured; however, new and growing uterine masses may be considered more concerning for harboring malignancy.
In total, prospective sequencing yielded diagnostic and therapeutic information with clinical utility for at least 17% of sequenced patients. Although actionable mutations were identified in 45% of patients only 17% (7% of the whole cohort) of those received matched therapy, so the broader therapeutic relevance of these data will require further research. Given the complex chromosomal abnormalities characteristic of uterine sarcomas, it will be a continuing challenge to characterize mutations to be either likely drivers or passengers.
Our data, albeit limited, indicate that clinical response to BRAF inhibition in BRAF-mutant uLMS may be modest. In contrast, the prolonged disease control achieved with PARP inhibition among the six patients with IMPACT detected BRCA2 alterations, which occur in 8% of high-grade uLMSs, suggests that BRCA2 may be a therapeutically relevant target. Recent work has highlighted the importance of lineage specificity of BRCA dependency in relation to PARP response (19), and there is growing evidence to ascribe uterine sarcomas as a BRCA-related tumor type. In addition to relatively high incidences of homozygous deletions in BRCA2, BRCA1 promoter hypermethylation as a potential mechanism of BRCA1 downregulation in uLMS (33) and other hallmarks of “BRCAness” such as homologous recombination deficiency (HRD) mutational signatures (5) have been identified in uterine sarcomas. It seems likely that a subset of uterine sarcomas, in particular uLMSs, may be driven by HRD, and given the responses seen in BRCA-positive tumors to PARP inhibition, BRCA testing on all patients seems justified. However, whether BRCA1 promotor hypermethylated uterine sarcoma tumors would respond to PARP inhibition requires further investigation, not least identifying whether hypermethylation of the BRCA1 promotor leads to an HRD phenotype in uLMS tumors. Unlike other established BRCA-related tumor types, uterine sarcomas seem to almost exclusively harbor BRCA2 DNA alterations.
Genomic alterations such as RIT1 and MAP2K4 amplifications, despite their association with poor prognosis and other phenotypes in other cancer types (34–37), were not associated with prognostic differences in our cohort. None of the common oncogenic or likely oncogenic genomic alterations in uterine sarcoma genes were associated with prognosis in the study cohort. Similarly, although WGD was present in more than half of high-grade sarcomas, WGD was not associated with OS. In a recent study of undifferentiated uterine sarcomas, high DNA copy number variation was found in 62% (25/40 cases), but did not have statistically significant association with poorer OS (38). By contrast, the two patients with MSI-H disease both achieved complete responses with multi-modality interventions and have had long disease-free survivals, suggesting that MSI-H may be prognostic for chemotherapy sensitivity and/or disease-free survival in uLMS. Furthermore, our cohort identified PDCD1 deletions as more common in uLMS than in non-uterine LMS. Such discrepancies may be relevant given the paucity of objective responses seen in phase II trials of immunotherapeutic agents in uLMS (39, 40).
Our study illustrates the complementary value of specialized gynecologic pathology review coupled with molecular characterization of patient tumors. Detecting rare or pathognomonic fusions can refine uterine sarcoma diagnoses, as previous reports have indicated for high-grade endometrial sarcomas and myxoid mesenchymal uterine tumors (41, 42). However, incorporating the iterative changes in pathologic classification as new molecular diagnostic technologies become available will be an ongoing challenge. Key considerations in terms of the generalizability of our findings include the fact that all patients in the cohort had to be alive in order to enroll in the study and have their tumors sequenced. Thus, the cohort may over-represent patients with a more favorable survival who were able to seek consultation and care at a comprehensive cancer center. The absence of prognostically significant genomic alterations may be due to the clinical characteristics of the study cohort, which is likely biased to include patients with favorable survival. Patients who die very shortly after diagnosis may be under-represented, and their tumors may have a different genomic profile.
In conclusion, analysis of this large cohort of patients with uterine sarcomas demonstrates that genomic profiling can provide clinical utility for patients, contributing to genome-driven diagnostic precision and elucidating potential novel treatment options for a subset of patients.
Supplementary Material
Translational Relevance.
Prospective genomic profiling can contribute to diagnostic precision and inform treatment selection in a subset of patients with uterine sarcomas. Potentially actionable mutations were identified in 45% of patients. There was evidence of clinical benefit in uterine leiomyosarcoma patients with somatic BRCA2 alterations treated with PARP inhibitors. These data support the development of clinical trials for uterine sarcomas that incorporate genomic findings and test treatments matched to potential therapeutic targets.
Acknowledgements
Funding: This work is supported in part by the National Institutes of Health/National Cancer Institute MSK Cancer Center Support Grant P30 CA008748 (institutional support covering all MSK faculty).
Footnotes
Conflict of Interest Statement: DBS has consulted with/accepted honoraria from Pfizer, Loxo Oncology, Lilly Oncology, Vivideon Therapeutics, Illumina and QED Therapeutics. Over the past 2 years, MLH has served on advisory boards for Tesaro and Lilly; has received author royalties from UpToDate; and has served as faculty speaker for Research to Practice. Also, her spouse is employed at Sanofi. B.S.T. reports advisory board activities for Boehringer Ingelheim and honoraria and research funding from Genentech.
References
- 1.Nucci MR, Harburger D, Koontz J, Dal Cin P, Sklar J. Molecular analysis of the JAZF1-JJAZ1 gene fusion by RT-PCR and fluorescence in situ hybridization in endometrial stromal neoplasms. The American journal of surgical pathology. 2007;31:65–70. [DOI] [PubMed] [Google Scholar]
- 2.Lee CH, Ou WB, Marino-Enriquez A, Zhu M, Mayeda M, Wang Y, et al. 14–3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:929–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CH, Marino-Enriquez A, Ou W, Zhu M, Ali RH, Chiang S, et al. The clinicopathologic features of YWHAE-FAM22 endometrial stromal sarcomas: a histologically high-grade and clinically aggressive tumor. The American journal of surgical pathology. 2012;36:641–53. [DOI] [PubMed] [Google Scholar]
- 4.Lewis N, Soslow RA, Delair DF, Park KJ, Murali R, Hollmann TJ, et al. ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2018;31:674–84. [DOI] [PubMed] [Google Scholar]
- 5.Chudasama P, Mughal SS, Sanders MA, Hubschmann D, Chung I, Deeg KI, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nature communications. 2018;9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature medicine. 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:4712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics (Oxford, England). 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Middha S, Zhang L, Nafa K, Jayakumaran G, Wong D, Kim HR, et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO precision oncology. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic acids research. 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nature genetics. 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Science translational medicine. 2015;7:283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng DT, Prasad M, Chekaluk Y, Benayed R, Sadowska J, Zehir A, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC medical genomics. 2017;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandelker D, Zhang L, Kemel Y, Stadler ZK, Joseph V, Zehir A, et al. Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. Jama. 2017;318:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO precision oncology. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell. 2017;171:950–65.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science (New York, NY). 2011;334:252–5. [DOI] [PubMed] [Google Scholar]
- 21.Mohammad N, Haimes JD, Mishkin S, Kudlow BA, Leong MY, Chew SH, et al. ALK Is a Specific Diagnostic Marker for Inflammatory Myofibroblastic Tumor of the Uterus. The American journal of surgical pathology. 2018;42:1353–9. [DOI] [PubMed] [Google Scholar]
- 22.Hoang LN, Aneja A, Conlon N, Delair DF, Middha S, Benayed R, et al. Novel High-grade Endometrial Stromal Sarcoma: A Morphologic Mimicker of Myxoid Leiomyosarcoma. The American journal of surgical pathology. 2017;41:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koontz JI, Soreng AL, Nucci M, Kuo FC, Pauwels P, van Den Berghe H, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardia A, Iafrate JA, Sundaresan T, Younger J, Nardi V. Metastatic Breast Cancer With ESR1 Mutation: Clinical Management Considerations From the Molecular and Precision Medicine (MAP) Tumor Board at Massachusetts General Hospital. The oncologist. 2016;21:1035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fribbens C, O’Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34:2961–8. [DOI] [PubMed] [Google Scholar]
- 26.Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017;548:234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oza AM, Tinker AV, Oaknin A, Shapira-Frommer R, McNeish IA, Swisher EM, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: Integrated analysis of data from Study 10 and ARIEL2. Gynecologic oncology. 2017;147:267–75. [DOI] [PubMed] [Google Scholar]
- 28.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. The New England journal of medicine. 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 29.Gibault L, Perot G, Chibon F, Bonnin S, Lagarde P, Terrier P, et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. The Journal of pathology. 2011;223:64–71. [DOI] [PubMed] [Google Scholar]
- 30.Halbwedl I, Ullmann R, Kremser ML, Man YG, Isadi-Moud N, Lax S, et al. Chromosomal alterations in low-grade endometrial stromal sarcoma and undifferentiated endometrial sarcoma as detected by comparative genomic hybridization. Gynecologic oncology. 2005;97:582–7. [DOI] [PubMed] [Google Scholar]
- 31.Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nature reviews Cancer. 2011;11:541–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoffski P, Sufliarsky J, Gelderblom H, Blay JY, Strauss SJ, Stacchiotti S, et al. Crizotinib in patients with advanced, inoperable inflammatory myofibroblastic tumours with and without anaplastic lymphoma kinase gene alterations (European Organisation for Research and Treatment of Cancer 90101 CREATE): a multicentre, single-drug, prospective, non-randomised phase 2 trial. The Lancet Respiratory medicine. 2018;6:431–41. [DOI] [PubMed] [Google Scholar]
- 33.Xing D, Scangas G, Nitta M, He L, Xu X, Ioffe YJ, et al. A role for BRCA1 in uterine leiomyosarcoma. Cancer research. 2009;69:8231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finegan KG, Tournier C. The mitogen-activated protein kinase kinase 4 has a pro-oncogenic role in skin cancer. Cancer research. 2010;70:5797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JT, Liu W, Kuang ZH, Chen HK, Li DJ, Feng QS, et al. [Amplification of RIT1 in hepatocellular carcinoma and its clinical significance]. Ai zheng = Aizheng = Chinese journal of cancer. 2003;22:695–9. [PubMed] [Google Scholar]
- 36.Pavese JM, Ogden IM, Voll EA, Huang X, Xu L, Jovanovic B, et al. Mitogen-activated protein kinase kinase 4 (MAP2K4) promotes human prostate cancer metastasis. PloS one. 2014;9:e102289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu F, Sun S, Yan S, Guo H, Dai M, Teng Y. Elevated expression of RIT1 correlates with poor prognosis in endometrial cancer. International journal of clinical and experimental pathology. 2015;8:10315–24. [PMC free article] [PubMed] [Google Scholar]
- 38.Binzer-Panchal A, Hardell E, Viklund B, Ghaderi M, Bosse T, Nucci MR, et al. Integrated Molecular Analysis of Undifferentiated Uterine Sarcomas Reveals Clinically Relevant Molecular Subtypes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:2155–65. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Ami E, Barysauskas CM, Solomon S, Tahlil K, Malley R, Hohos M, et al. Immunotherapy with single agent nivolumab for advanced leiomyosarcoma of the uterus: Results of a phase 2 study. Cancer. 2017;123:3285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tawbi HA, Burgess M, Bolejack V, Van Tine BA, Schuetze SM, Hu J, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. The Lancet Oncology. 2017;18:1493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busca A, Parra-Herran C. Myxoid Mesenchymal Tumors of the Uterus: An Update on Classification, Definitions, and Differential Diagnosis. Advances in anatomic pathology. 2017;24:354–61. [DOI] [PubMed] [Google Scholar]
- 42.Hoang L, Chiang S, Lee CH. Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology. 2018;50:162–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.