Significance
We show that the combination of conductive polymer spray mass spectrometry (CPSI-MS) and machine learning provides a simple, fast, and affordable method for oral squamous cell carcinoma diagnosis with 86.7% accuracy. By using CPSI-MS, the direct, high-throughput metabolic profiling of saliva can be readily realized in a noninvasive manner. The self-conductive materials used in CPSI-MS for sample loading and ionization are clean, cheap, and consumable for cohort analysis. Wide coverage of chemical species provides not only the pools of possible metabolite signatures for molecular diagnosis but also the possibility of exploring metabolite function and oncological mechanism. The analysis of saliva samples from 373 individuals was completed within 4.5 h, satisfying the technical demand for point-of-care testing.
Keywords: point-of-care test, noninvasive diagnosis, conducting polymer spray ionization, salivary metabolomics, oral squamous cell carcinoma
Abstract
Saliva is a noninvasive biofluid that can contain metabolite signatures of oral squamous cell carcinoma (OSCC). Conductive polymer spray ionization mass spectrometry (CPSI-MS) is employed to record a wide range of metabolite species within a few seconds, making this technique appealing as a point-of-care method for the early detection of OSCC. Saliva samples from 373 volunteers, 124 who are healthy, 124 who have premalignant lesions, and 125 who are OSCC patients, were collected for discovering and validating dysregulated metabolites and determining altered metabolic pathways. Metabolite markers were reconfirmed at the primary tissue level by desorption electrospray ionization MS imaging (DESI-MSI), demonstrating the reliability of diagnoses based on saliva metabolomics. With the aid of machine learning (ML), OSCC and premalignant lesions can be distinguished from the normal physical condition in real time with an accuracy of 86.7%, on a person by person basis. These results suggest that the combination of CPSI-MS and ML is a feasible tool for accurate, automated diagnosis of OSCC in clinical practice.
Oral squamous cell carcinoma (OSCC) represents the most malignant neoplasm in oral cancer with a mortality rate of more than 50%. The OSCC is a multistep neoplasia initially developed from mild oral epithelial hyperplasia to dysplasia followed by carcinoma in situ (1). Oral leukoplakia, oral lichen planus, oral erythroplakia, and oral submucous fibrosis all belong to the premalignant lesion stage (PML) before aggressive OSCC (2). If necessary, intervention could be carried out before tumorigenesis; the currently maintained 50% 5-y survival rate could be improved (3). Unfortunately, there are still many cases not diagnosed until the advanced stage when metastases have happened (4), missing the best opportunity for treatment. Even for the OSCC patient who receives surgical resection, local recurrence is another cause for treatment failure (5). Therefore, a highly sensitive and specific screening approach is urgently needed for diagnosis and prognosis.
Currently, the visual inspection of the mouth combined with histopathology is still the gold standard method for oral cancer screening (6, 7). Although the oral cavity is relatively accessible, some asymptomatic cases at the early or postoperative stage are difficult to observe. Incisional biopsy not only causes second physical impairment but also faces the challenge of inaccurate sampling caused by tumor heterogeneity (8). As an approach complementary to routine visual examination, molecular diagnosis becomes indispensable, capturing the latent ongoing molecular phenotype changes before local recurrence, malignant transformation, or distant metastasis. Therefore, development of a cost-effective tool for multiplex molecular detection in the biological fluid is much desired.
For molecular diagnosis of OSCC, saliva is an ideal diagnostic fluid (2, 9, 10). Its collection procedure is noninvasive and more cost-effective than traditional intravenous or finger blood (2). The production amount of 500 –1,500 mL saliva per day is abundant enough for on-demand collection (11). Saliva has been widely reported to be a potential source of biomarkers owing to its diversity in components, including genome, transcriptome, proteome, microbiome, and metabolome (12–14). Considering that saliva is the natural pool closest to the primary carcinoma site, there must be locally expressed molecules representing the signs of OSCC tumorigenesis and malignant transformation.
The salivary metabolic profile has often been called the “mirror of the body” because it can provide a general outlook on significantly changed metabolites from aberrant enzymatic regulation, capture the oncometabolites originating from metabolic rewiring, and highlight those altered pathways during metabolic reprogramming (12, 15, 16). All of these can become ideal metabolite markers indicating the ongoing OSCC-associated changes without obvious pathological symptoms. Currently, more than 100 metabolites have been reported to be dysregulated with OSCC malignant progression, including choline, carnitine, lactate, glutamate, sialic acid, histidine, polyamines, pipecolic acid, and trimethylamine N-oxide (3, 17–22).
Practical molecular screening for clinical applications requires comprehensive consideration of several aspects: 1) single or panel of measurable oncomarkers with high specificity and sensitivity; 2) convenient collection of diagnostic fluid that minimizes the patient’s discomfort; 3) technical platform with affordable cost for routine examination; and 4) rapid information feedback for physician’s decision-making. Thus, a strategy for a point-of-care test (POCT) has been in high demand to satisfy these technical requirements (23).
Mass spectrometry (MS) is a powerful platform for proteomics/metabolomics and has become prevalent in biomedical research owing to its in-depth coverage of various molecular species. Versatile MS platforms such as liquid chromatography mass spectrometry (LC-MS) (24), gas chromatography mass spectrometry (GC-MS) (18, 25), capillary electrophoresis mass spectrometry (CE-MS) (26), two-dimensional electrophoresis mass spectrometry (2DE-MS) (27), matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF) (28), and surface enhanced laser desorption ionization time-of-flight mass spectrometry (SELDI-TOF) (29) have received increasing biomedical and clinical applications. Unfortunately, most of these approaches require time-consuming pretreatment of a complex biological sample before a single run. This limitation becomes nonnegligible when the cohort sample size goes beyond hundreds or thousands in size, which consumes at least several weeks or months for analysis, restricting the use of these platforms for salivary POCT.
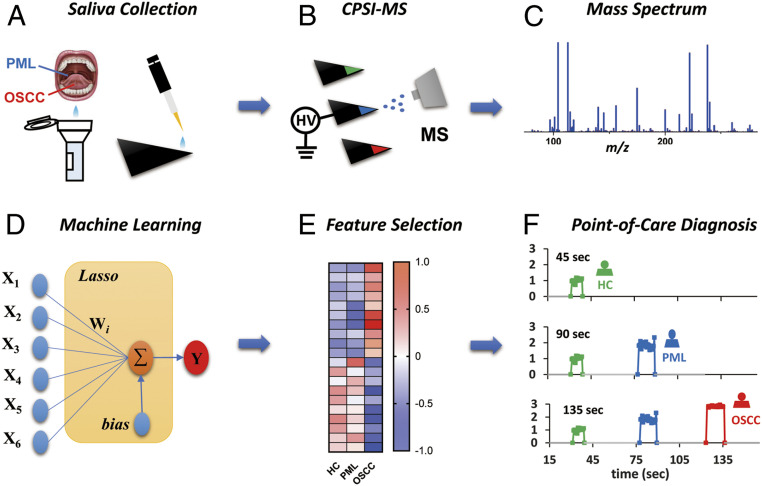
Paper spray ionization has become popular because it integrates sample loading, storage, filtering, and ionization into one cost-effective substrate. Assisted with continuous solvent supply, enhanced ionization can be realized for target drugs, metabolites, glycans, and proteins detection (30). However, the paper’s impurities interference and its porous nature strongly suppress desorption and ionization especially for hydrophilic metabolites in biofluids. To overcome this drawback, we have introduced an ambient ionization-based multiplex molecular screening method, conductive polymer ionization mass spectrometry (CPSI-MS) (31), which we believe meets all of the above-mentioned criteria for a POCT. Fig. 1 shows the analytical pipeline for CPSI-MS analysis, which is able to identify hundreds of metabolites directly from trace saliva under atmospheric conditions and allows in the future the mass spectrometer to go from the laboratory to the bedside. The analysis of each sample requires only a few seconds by using a self-conductive polymer as the ionization probe. The introduction of machine learning (ML) combined with CPSI-MS has allowed us to successfully translate the metabolomics into actual practice in clinic diagnostics. With the aid of the CPSI-MS/ML approach, we believe that a noninvasive salivary diagnosis can be realized to give a quick, accurate, cost-effective diagnosis of OSCC progression, as will be described in what follows.
Fig. 1.
Schem3atic illustration of the salivary diagnosis workflow. (A) Noninvasive collection of saliva mixed with solvent and internal standard. Load saliva onto the tip of conductive polymer to form the dried saliva spot for further analysis. (B) Different groups of saliva samples are sequentially tested by CPSI-MS by applying trace extraction solvent and high voltage on the tip to trigger each component’s desorption and ionization into the MS inlet. (C) The corresponding mass spectrum is acquired and recorded. (D) An ML model is trained by recording salivary mass spectra with known types. (E) Those important metabolite peaks that had significant changes and contributed to the classification are searched for as input features. (F) By combining CPSI-MS with ML, POC diagnosis can be carried out in nearly real time.
Results
Salivary Metabolic Profiling.
The collected 373 saliva cases (124 healthy contrast [HC], 124 PML, and 125 OSCC) were divided into two batches for CPSI-MS data acquisition (see SI Appendix, Table S1 for more details). From the first batch of 193 saliva cases, 627 common peaks were selected to characterize the global metabolic profiles of different groups (HC, PML, and OSCC). Orthogonal partial least-squares discriminant analysis (OPLS-DA) was introduced to extract two orthogonal latent features from these 627 variables. In the constructed space of the two features, saliva cases from the same group were well clustered, while those samples from different groups were separated (SI Appendix, Fig. S1A). This initial work demonstrated that metabolic profiles acquired by CPSI-MS contain underlying bioinformation that can differentiate HC, PML, and OSCC.
To further investigate the reproducibility of the metabolic profile acquired with CPSI-MS, another 180 saliva cases were evenly divided to acquire metabolomics information over three successive days. It was surprising to find that mass spectrum profiles from different groups were in high agreement. There were at least 30 discriminant peaks that were identified in both batches (SI Appendix, Fig. S2 A and B). Taking the Pearson correlation coefficient as the metric, the similarities of average MS profiles acquired from the two batches were 95% (HC), 90% (PML), and 86% (OSCC) (SI Appendix, Fig. S2C). Most saliva samples in each day can be accurately projected onto the corresponding clusters in the OPLS-DA model that was previously developed (SI Appendix, Fig. S1 B–D). The cluster distributions in feature space for the second batch (SI Appendix, Fig. S1E) were similar to those of the first batch (SI Appendix, Fig. S1A). The pattern recognition results for the two batches can be overlaid (SI Appendix, Fig. S1F). The ion intensities of the internal standard (IS) spiked in the pooled saliva samples (as quality control, QC) behaved quite stably in the three successive days, with the average interday relative standard deviation (RSD%) being less than 15% (SI Appendix, Fig. S3). These results gave us confidence that the CPSI-MS system has robust performance for acquiring reproducible metabolic profiles.
CPSI-MS Robustness Despite Salivary Component Variation.
Saliva from nine healthy volunteers, treated as negative controls, were collected at six different times, before and after food consumption, over three successive days. After CPSI-MS data acquisition, the two representative features were extracted from the salivary metabolic profile, based on the previously developed OPLS-DA model. The clustering results suggest that the salivary metabolic profile may be more susceptible to diet and individual differences (SI Appendix, Fig. S4 A and B) than to interday or intertime variation (SI Appendix, Fig.S4 C and D), especially when the oral cavity was not fully rinsed after eating. Nevertheless, all of these negative control saliva cases can still be projected onto the same cluster as the other samples from HC group members, demonstrating the metabolic profiling method’s robustness to the fluctuation in salivary components.
Dysregulated Metabolites and Metabolic Pathways.
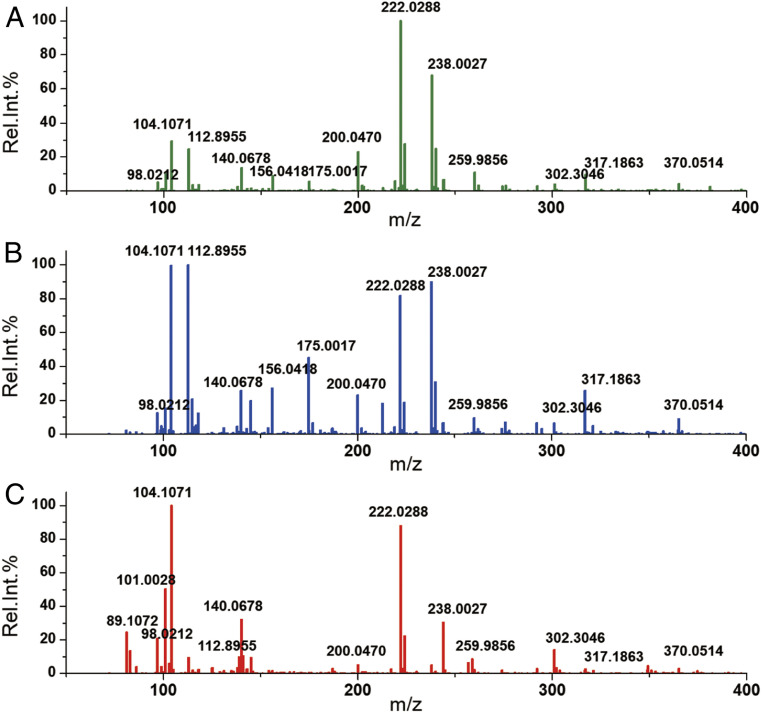
After CPSI-MS data acquisition of the first batch, the metabolite ions were tentatively assigned according to exact m/z values, isotope distribution, and adduct ion type (SI Appendix, Table S2). Fig. 2 displays the average mass spectra acquired from HC, PML, and OSCC. Compared with the IS peak (m/z 222.0288, [M+Na]+), several obvious changes in relative abundance can be directly observed in peaks such as m/z 89.1072 (putrescine, [M+H]+), 98.0212 (glycine, [M+Na]+), 104.1068 (choline, [M+H]+), 140.0678, and 156.0418 (betaine, [M+H]+ and [M+Na]+); 175.0017 (hypoxanthine, [M+H]+), 259.9856 (phosphocholine, [M+2K-H]+), 302.3046 (sphinganine, [M+H]+), 317.1863 (linoleic acid, [M+K]+), and 370.0514 (adenosine monophosphate, [M+Na]+).
Fig. 2.
Average CPSI mass spectra of saliva collected from different groups: (A) healthy contrast, (B) premalignant lesion, and (C) oral squamous cell carcinoma.
To discover more underlying dysregulated metabolites, Student’s t test and multivariate analysis were employed to search for those significantly changed metabolite ions (fold change > 2.0 or < 0.5, P < 0.05) that contributed more to the classification in OPLS-DA. Considering only the identified metabolites, there were 58 that significantly changed during progression from HC to PML. In contrast, during development from PML to OSCC, 116 metabolites were discovered to be significantly elevated or decreased (SI Appendix, Tables S3 and S4). Table 1 lists the top 10 metabolites that were up-regulated or down-regulated during malignant progression from HC to OSCC. These top 10 and other representative metabolites were further validated by MS/MS fragmentation (SI Appendix, Figs. S5 and S6). For the already collected OSCC cases, we tried to investigate whether these metabolites also have significant difference among different stage (from stage I to stage IV). Unfortunately, no one among these metabolites was discovered to have a significant difference during staging from I to IV using carried out the one-way analysis of variance with multiple comparisons.
Table 1.
Top 10 metabolites with marked up-regulation or down-regulation during malignant progression from HC to OSCC
| Metabolites | PML vs. HC | OSCC vs. PML | OSCC vs. HC | |||
| FDR adjusted P value* | Fold change† | FDR adjusted P value* | Fold change† | FDR adjusted P value* | Fold change† | |
| Putrescine | 0.0075 | 2.22 | 0.0000 | 2.50 | 0.0000 | 5.53 |
| Cadaverine | 0.0006 | 4.10 | 0.0056 | 5.51 | 0.0010 | 22.56 |
| Thymidine | 0.0671 | 2.55 | 0.0000 | 4.12 | 0.0000 | 10.51 |
| Adenosine | 0.2160 | 3.74 | 0.0000 | 12.96 | 0.0000 | 48.47 |
| 5-aminopentoate | 0.6101 | 1.21 | 0.0000 | 6.60 | 0.0000 | 7.97 |
| Hippuric acid | 0.1266 | 0.58 | 0.0006 | 0.35 | 0.0008 | 0.20 |
| Phosphocholine | 0.0000 | 0.48 | 0.0000 | 0.36 | 0.0000 | 0.17 |
| Glucose | 0.0558 | 0.39 | 0.0091 | 0.11 | 0.0002 | 0.04 |
| Serine | 0.3830 | 0.79 | 0.0000 | 0.07 | 0.0000 | 0.05 |
| Adrenic acid | 0.0832 | 0.54 | 0.0000 | 0.07 | 0.0000 | 0.04 |
FDR: false discovery rate, which was calculated according to the Benjamini–Hochberg method.
Fold changes were calculated by the mean intensities ratio of PML/HC, OSCC/PML, and OSCC/HC.
The dysregulated metabolic pathways during both stages of premalignancy and malignant progression were further investigated by inputting those metabolites into the open-source platform MetaboAnalyst (32). They were mainly focused on five representative pathways including aminoacyl tRNA biosynthesis, arginine/proline metabolism (proline, putrescine, spermine, spermidine, N-acetyl putrescine, N-acetyl spermidine), arginine biosynthesis (arginine, citrulline, ornithine, urea), lysine degradation (lysine, cadaverine, piperidine, pipecolate, 5-pentanoate), and histidine metabolism (histidine, methyl-histidine, urocanic acid) (SI Appendix, Tables S5 and S6). The impact value and the significance (-log10P) of these five pathways increased with the aggressive progression from PML to OSCC, reflecting the stepped-up exacerbation in metabolic dysregulation (Fig. 3).
Fig. 3.
Altered metabolites and their underlying metabolic pathways during cancerous progression: (A) significantly changed metabolic pathways during both premalignancy and malignancy stages; and (B) heatmap visualization of the metabolite’s expression within HC, PML, and OSCC groups. The circle diameter in the bubble graph represents the number of hits in certain pathways.
Validation of the Discovered Metabolic Changes.
The second batch of 180 saliva samples was collected for external validation. After the same CPSI-MS data acquisition and statistical screening, there were 52 and 97 metabolites dysregulated during premalignant and malignant stages, respectively. By comparing with previously discovered metabolites from the first batch, 42 out of 52 and 94 out of 97 metabolites were reconfirmed with confidence (SI Appendix, Tables S3 and S4). Among these, there were 30 metabolites aberrant in both the PML and OSCC stages (SI Appendix, Fig. S7). Desorption electrospray ionization MS imaging (DESI-MSI) was employed to further validate these discovered metabolites at the level of primary oncological tissue sites. It was revealed that the polyamines (e.g., spermidine, spermine) and amino acids (e.g., arginine, lysine, histidine, glutamine, leucine) that evolved in the arginine biosynthesis/metabolism, lysine degradation, and histidine metabolism go through aberrant metabolic regulation. Energy metabolism-related metabolites (e.g., glucose, creatine, creatinine) and purine metabolites (e.g., inosine, hypoxanthine) were also found to change in abundance in the tumor region (SI Appendix, Fig. S8).
Features Selection and Lasso Regression for Cancer Stage Prediction.
As there were a variety of metabolites that differ among HC, PML, and OSCC, it is likely a combination of metabolites can effectively give a more accurate prediction of tumor development. For this purpose, Lasso regression (33) was introduced to construct an ML model for OSCC prediction, where a sparse set of metabolites that are most informative in the classification are selected. The two batches of saliva samples were arranged as training (193 cases) and validation (180 cases) datasets, respectively. Each dataset comprises the measured relative abundances of 627 detected metabolite ions and the diagnostic class of each individual’s saliva, namely HC, PML, or OSCC. To select for the tuning parameter lambda that optimizes the model performance, we selected the model via 20-fold cross-validation on the training set (SI Appendix, Fig. S14). The 20-fold cross-validation was carried out by stratifying the training dataset into training folds (184 cases, 95%) and testing folds (9 cases, 5%). Based on this approach, the Lasso model with lowest lambda achieves 90% accuracy on the held-out test fold and selects 62 out of 627 detected metabolite ions (SI Appendix, Table S7) across all 193 saliva cases in the training dataset (SI Appendix, Table S8 and Fig. S9). When evaluating the model performance on the second batch of external validation samples (180 cases), the model achieves an accuracy of 86.7% (SI Appendix, Table S8). This gives us a sense that the model generalizes well to new patients and achieves desirable accuracy without overfitting.
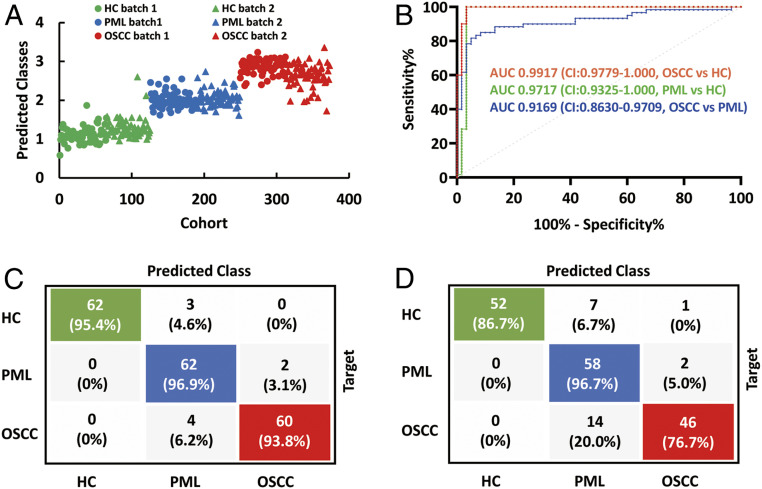
The final prediction results for all of the 373 cases are shown in Fig. 4A. The training and validation set accuracies of the Lasso model for the two batches are 95.3% and 86.7%, respectively (SI Appendix, Table S9). The area under the curve (AUC) values from the receiver operating characteristic (ROC) curves that differentiate PML from HC, OSCC from PML, and OSCC from HC were 0.9717 (CI: 0.9325–1.000), 0.9169 (CI: 0.8630–0.9709), and 0.9917 (CI: 0.9779–1.000) (Fig. 4B), showing excellent diagnostic performance in the external validation dataset. The confusion matrix specifies the classification results among the first batch of 193 cases plus the second batch of 180 cases (Fig. 4 C and D).
Fig. 4.
Lasso-based ML results for differentiating HC, PML, and OSCC. (A) The prediction results for HC, PML, OSCC from training dataset and validation dataset. (B) ROC curves and corresponding AUC values to characterize general diagnosis accuracy of OSCC vs. PML, OSCC vs. HC, and HC vs. PML in the validation dataset (second batch). (C and D) Confusion matrices of the HC, PML, and OSCC prediction results for the training and validation dataset.
Simulating POC Salivary Diagnosis Based on the Lasso Model.
For each saliva case, there will be at least 15 scans acquired with CPSI-MS to generate the average mass spectrum characterizing its general metabolic profile. Considering the developed Lasso model performed well in distinguishing different groups of saliva, this model can be applied to predict the health of each person at the single-MS scan level. To this end, the pretrained Lasso model was constructed by Simulink to observe the scenario of real-time molecular diagnosis. In the actual CPSI-MS analysis, all of the saliva cases were arranged as one test sequence with the data recorded in one .raw file. After a rapid (within a few seconds) format conversion into a .cdf file, all of the mass peaks will immediately be read and processed by a self-written MATLAB script. Meanwhile, the extracted metabolite ion features will be put into the predeployed Simulink Lasso model to provide nearly real-time prediction at the single-scan level (SI Appendix, Fig. S10). The final diagnosis for certain cases depends on the number of diagnoses of different scans. As a result, almost all of the saliva cases can be processed around 10 s (SI Appendix, Fig. S11).
Discussion
Several DNA, messenger RNA (mRNA), and proteins were reported to be associated with OSCC progression, such as B7-H3, MMP1, KNG1, ANXA2, HSPA5, DUSP1, H3F3A, OAZ1, M2BP, MRP14, Prolifin, Ki67, Actin, and Myosin (1, 4, 34, 35). The current gene- or protein-based clinic diagnosis mainly relies on the use of several immunoassays that introduce the hybrid probe or antibody as specific recognition elements. This multiplex detection is inevitably restricted by spectral signal overlap. The analytical period and economic cost also increases with the introduction of more biorecognition probes. In contrast, ambient ionization MS-based metabolite profiling has the advantage of wide coverage, high speed, label-free detection that can be performed with comparable accuracy and at little cost.
The first advantage of the above-described CPSI-MS procedure is that it is high throughput and therefore well-suited to big cohort analysis. No additional enrichment or purification was required before sample loading to form a dried biofluid spot. Both desorption and ionization can be triggered by applying an extraction solvent and high voltage, which is as simple as operating an on/off switch. Thus, CPSI-MS is quite suitable for rapid, direct metabolic profiling of complex biological fluids such as saliva, tears, sweat, urine, serum, plasma, or even whole blood. Direct metabolic profiling of single case only takes few seconds for collecting enough MS data (SI Appendix, Fig. S11). The total analytical period for these saliva samples collected from volunteers took 4.5 h, which means that CPSI-MS can analyze ∼1,000 samples within 12 h, satisfying the practical requirement for a daily bedside POCT. In addition, the conductive polymer composite multi-walled carbon nanotube/polymethyl methacrylate (MWCNT/PMMA) used in CPSI-MS is cheap, clean, and consumable, which facilitates the economic testing of large-scale samples.
From the aspect of analytical performance, another advantage of CPSI-MS is its wide coverage of different metabolite species. It is estimated that CPSI-MS under positive mode can successfully detect several hundreds of salivary metabolites including amino acids, carbohydrates, fatty acids, nucleosides, nucleotides, acyl carnitines, carboxylic acids, polyamines, glycerolipids, glycerophospholipids, sphingolipids, etc. It is worth noting that only a trace volume of biofluid dissolved in the methanol–water extraction solvent (7:3, vol/vol, 3 μL) is needed, which minimizes the hazard of chemical or biological exposure. The wide coverage of endogenous metabolite species provides not only a candidate pool for metabolite marker discovery but also the possibility of deep insight into the underlying metabolism pathways during tumorigenesis.
It has been recognized that saliva has great potential as an ideal noninvasive diagnostic fluid. However, practical objections to its usage mainly arise from the need to have a standardized collection protocol and to do QC of the endogenous components’ stability and variation. Failure to consider these factors may lead to misleading correlations between discovered markers and disease progression (2, 4). Therefore, a strict protocol has been followed throughout the entire analytical procedure, including prerinsing and unstimulated saliva collection, IS incorporation, and QC insertion. All individuals tested are selected based on the statement that they have no other diseases to their knowledge to avoid the influence of other factors of nonoral cancer diseases. Among the OSCC patients compared with PML and HC volunteers, poor oral hygiene conditions were prevalent because many cannot brush their teeth every day. Therefore, fully prerinsing with pure water was critical to reduce the oral hygiene differences before saliva collection. We specially investigated to what extent dietary interference, sample collection times, or individual differences will cause variation in the salivary composition and metabolic-profile-based classification accuracy. Although diet is found to contribute more to the metabolic profile perturbation than circadian rhythm, this factor had little influence on the final classification results. This is partly because the prerinsing cleanup (SI Appendix, Fig. S12) and the metabolite ion features we selected rule out interfering peaks from diet variation. Additionally, the fluctuation of the CPSI-MS system and the variation of the salivary metabolite components can be effectively monitored (SI Appendix, Fig. S3) and compensated for within a controllable range by normalization and QC sample calibration.
Among the discovered metabolites, there were 53 (more than 50%) previously reported metabolite markers also found in PML or OSCC, proving the reliability of our proposed CPSI-MS approach in metabolite marker discovery. Uncontrolled tumor growth is the hallmark of aggressive carcinoma, which relies on overconsumption of nitrogen sources, carbon sources, and energy to support dysregulated cancer cell proliferation (36, 37). Five representative altered pathways involved three basic amino acids (histidine, lysine, and arginine), which covered the urea cycle and polyamine metabolism (SI Appendix, Tables S5 and S6). These pathways not only provided an abundant source of nitrogen but also promoted hyperproliferative disorders in various cancers (38, 39). The significant change in aminoacyl transfer RNA (tRNA) biosynthesis pathway hinted at the dysregulated in mRNA translation for protein synthesis (40, 41). The up-regulated expression of ketoleucine, indole-acetate, and 3-hydroxyphenylacetate and the down-regulated desaminotyrosine revealed abnormal metabolism of leucine, tryptophan, phenylalanine, and tyrosine, respectively (SI Appendix, Fig. S13). Cancer’s “glutamine addiction” (42) promoted the decrease in glutamine and the concomitant elevation of glutamate by glutaminolysis (43). Decreased glucose and elevated lactate indicated ongoing increased glycolysis (the Warburg effect) (44). The energy fueling was also observed by changes in creatine metabolism (e.g., creatine, creatinine), acyl carnitine (e.g., acetyl carnitine, butyryl carnitine), free fatty acids (e.g., oleic acid, linoleic acid) and monoacyl glycerol [e.g., MG(20:4)] for mitochondrion β-oxidation. In addition, purine and pyrimidine metabolism pathways became abnormal in PML, as seen by 10 metabolites that were observed to be significantly changed (e.g., inosine, hypoxanthine, adenosine, thymidine, uridine, guanosine, cytosine), which are building blocks for nucleotide replication in cell proliferation (SI Appendix, Table S6) (45, 46). The abnormal choline (e.g., choline, phosphocholine) and sphingolipid metabolism (e.g., sphingosine, phytosphingosine) hint at cell proliferation dysregulation (47, 48). These dysregulated metabolite markers were indicative not only for OSCC diagnosis, but also for evaluating the outcome of OSCC therapy.
Although salivary metabolomics can generally reflect the in situ metabolic status within the primary OSCC location, the metabolite concentration was inevitably diluted to some extent because of continuous saliva secretion, mainly by the parotid gland, submandibular gland, and sublingual gland (49). Therefore, we also investigated the expression of these metabolite markers in the tumor tissue by DESI-MSI. Six of the top 10 most altered metabolites also showed abundance differences in tumors and positive margin regions compared with normal tissue (SI Appendix, Fig. S8). It is also worth noting that certain metabolites in saliva and tumor tissue changed their abundance in opposite directions. However, we believe that these differences arise because of many complex factors, like the increased/decreased uptake of cancer cells in the tumor tissue (19), the body fluid dilution, and the disagreement between population-based statistical tendency and personal-based dispersion. These results not only provide direct evidence that the salivary metabolites can indeed reflect the ongoing metabolic dysregulation within primary carcinoma sites but also support the feasibility of CPSI-MS as a noninvasive salivary diagnostic for PML and OSCC.
Concerning the ML model investigation, we systematically evaluated various classification and regression models including Lasso regression, artificial neural network (ANN), K nearest neighbor, decision tree, supporting vector machine, and Naive Bayesian. The performances were compared both in the training and validation datasets with the general accuracy and mean-squared error as the metrics. Although Lasso ranked in the second place behind ANN in terms of performance in accuracy (SI Appendix, Table S11), the resulting model with 62 features is much simpler and more informative compared to ANN, which introduces nonlinear activation function and complex hidden layers. For example, selected Lasso metabolites include N6,N6,N6-trimethyl-l-lysine and symmetric dimethylarginine, suggesting their role in OSCC dysregulated transcription. The advantages of Lasso for identifying specific metabolic changes related to disease phenotypes have also been widely shown in our previous applications of DESI-MSI in cancer diagnosis (50–52).
With the well-trained Lasso model, we envisioned that this CPSI-MS/Lasso approach might give automatic, real-time feedback of diagnosis. Therefore, we made a simulation for the complete analytical process from the acquired MS data to the final prediction about the tumor progression status in the MATLAB/Simulink platform (SI Appendix, Fig. S10). The results turned out to be successful both in accurate prediction and nearly real-time feedback. It illustrated the future potential of CPSI-MS/ML in realizing on-demand, bedside diagnosis, although further development in data acquisition software for synchronized data transfer to the ML model is required for ultimate real-time prediction.
Conclusion
The salivary metabolic profile can reflect oral cancer development. Most discovered metabolites in saliva were found to be highly linked to their expression levels within the primary oncological site of oral cavity tissues, demonstrating the potential of saliva for in vitro molecular diagnosis of OSCC. By cohort analysis using CPSI-MS, slight metabolic changes were found to start from aminoacyl t-RNA biosynthesis, arginine biosynthesis and metabolism, lysine degradation, histidine metabolism, and proline metabolism during the precancerous stage. When the tumor has developed into the aggressive stage, more metabolites involved in the above pathways were found to be increasingly up-regulated or down-regulated. These findings provide potential clinical markers for indicating OSCC tumorigenesis. It has been demonstrated that CPSI-MS, as a promising ambient MS tool, shares cost-effective performance in monitoring hundreds of salivary metabolites without any laborious sample pretreatment. The combination of CPSI-MS with ML enabled excellent molecular diagnosis (86.7% accuracy) for the external held-out validation set. In addition, the potential of CPSI-MS/ML for automated, on-demand diagnosis of OSCC in real time has also been shown in concept by dynamic simulation. All of these findings indicate that CPSI-MS/ML can be a very useful tool to provide a simple, fast, affordable noninvasive diagnosis for OSCC.
Materials and Methods
The full study protocol was approved by the medical ethics committee of the Nanjing Stomatology Hospital. All patients were informed and signed consent forms. The CPSI-MS/ML procedure is similar to the reporting recommendations for tumor marker prognostic studies (REMAKRS) (53). The raw data, processing code, and machine learning models can be accessed at the following link: https://osf.io/nv32d/. Additional details about technical descriptions of the methods may be found in SI Appendix, Material and Methods: Saliva Collection and Pretreatment; CPSI-MS and DESI-MSI method; CPSI-MS and DESI-MSI Data Preprocessing; Statistical Analysis; Machine Learning and Dynamic Simulation; Metabolite Identification and Metabolic Pathway Searching.
Supplementary Material
Acknowledgments
X.S. thanks the China Postdoctoral Science Foundation (2019M651337) and the National Natural Science Foundation of China (81903575) for support of this study. This work was also supported by the Scientific Research Startup Foundation (IDH1615113) from Fudan University and the National Natural Science Foundation of China (81772880).
Footnotes
The authors declare no competing interest.
Data Deposition: Raw data, processing code, and machine learning models related to this article can be accessed at https://osf.io/nv32d/.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001395117/-/DCSupplemental.
References
- 1.Yu J. S. et al., Saliva protein biomarkers to detect oral squamous cell carcinoma in a high-risk population in Taiwan. Proc. Natl. Acad. Sci. U.S.A. 113, 11549–11554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikkonen J. J. et al., Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J. Periodontal Res. 51, 431–437 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Wei J. et al., Salivary metabolite signatures of oral cancer and leukoplakia. Int. J. Cancer 129, 2207–2217 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y. S., Rees T., Wright J., A review of research on salivary biomarkers for oral cancer detection. Clin. Transl. Med. 3, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.-T. et al., Glycoprotein B7-H3 overexpression and aberrant glycosylation in oral cancer and immune response. Proc. Natl. Acad. Sci. U.S.A. 112, 13057–13062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell L. H., Reply to Galvão-Moreira and da Cruz: Saliva biomarkers to complement the visualization-based oral cancer detection. Proc. Natl. Acad. Sci. U.S.A. 114, E111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galvão-Moreira L. V., da Cruz M. C., Saliva protein biomarkers and oral squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 114, E109–E110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakob M., Fuentes L., Wang M. B., Abemayor E., Wong D. T., Salivary biomarkers for detection of oral squamous cell carcinoma - current state and recent advances. Curr. Oral Health Rep. 1, 133–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa S. et al., Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 6, 31520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay I., Panda M., Recent trends of saliva omics biomarkers for the diagnosis and treatment of oral cancer. J. Oral Biosci. 61, 84–94 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Cuevas-Córdoba B., Santiago-García J., Saliva: A fluid of study for OMICS. OMICS 18, 87–97 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Rai V., Mukherjee R., Ghosh A. K., Routray A., Chakraborty C., “Omics” in oral cancer: New approaches for biomarker discovery. Arch. Oral Biol. 87, 15–34 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Zhang A., Sun H., Wang X., Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl. Biochem. Biotechnol. 168, 1718–1727 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Esteves C. V. et al., Diagnostic potential of saliva proteome analysis: A review and guide to clinical practice. Braz. Oral Res. 33, e043 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Shin J. M., Kamarajan P., Fenno J. C., Rickard A. H., Kapila Y. L., Metabolomics of head and neck cancer: A mini-review. Front. Physiol. 7, 526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward P. S., Thompson C. B., Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohavanichbutr P. et al., Salivary metabolite profiling distinguishes patients with oral cavity squamous cell carcinoma from normal controls. PLoS One 13, e0204249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enomoto Y. et al., Exploring a novel screening method for patients with oral squamous cell carcinoma: A plasma metabolomics analysis. Kobe J. Med. Sci. 64, E26–E35 (2018). [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Yu D., Metabolomics study of oral cancers. Metabolomics 15, 22 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa S. et al., Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis. 26, 35–42 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Sridharan G., Ramani P., Patankar S., Vijayaraghavan R., Evaluation of salivary metabolomics in oral leukoplakia and oral squamous cell carcinoma. J. Oral Pathol. Med. 48, 299–306 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Yang X. H. et al., Amino acids signatures of distance-related surgical margins of oral squamous cell carcinoma. EBioMedicine 48, 81–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schafer C. A. et al., Saliva diagnostics: Utilizing oral fluids to determine health status. Monogr. Oral Sci. 24, 88–98 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Wang Q., Gao P., Wang X., Duan Y., The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 4, 6802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye G. et al., Study of induction chemotherapy efficacy in oral squamous cell carcinoma using pseudotargeted metabolomics. J. Proteome Res. 13, 1994–2004 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto M., Wong D. T., Hirayama A., Soga T., Tomita M., Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6, 78–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu S. et al., Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics 5, 1714–1728 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Ploypetch S. et al., Salivary proteomics of canine oral tumors using MALDI-TOF mass spectrometry and LC-tandem mass spectrometry. PLoS One 14, e0219390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schipper R. et al., SELDI-TOF-MS of saliva: Methodology and pre-treatment effects. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 45–53 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Riboni N. et al., Solvent-assisted paper spray ionization mass spectrometry (SAPSI-MS) for the analysis of biomolecules and biofluids. Sci. Rep. 9, 10296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song X., Chen H., Zare R. N., Conductive polymer spray ionization mass spectrometry for biofluid analysis. Anal. Chem. 90, 12878–12885 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Chong J., Wishart D. S., Xia J., Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics 68, e86 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Tibshirani R., Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B 58, 267–288 (1996). [Google Scholar]
- 34.Lorenzo-Pouso A. I. et al., Protein-based salivary profiles as novel biomarkers for oral diseases. Dis. Markers 2018, 6141845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rathy Ravindran D. M. G., Salivary tumour markers in oral cancer: Brief review. Oral Maxillofac. Pathol. J. 2, 238–244 (2012). [Google Scholar]
- 36.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J., Thompson C. B., Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 20, 436–450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casero R. A. Jr., Murray Stewart T., Pegg A. E., Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 18, 681–695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobieszczuk-Nowicka E. et al., Polyamines–A new metabolic switch: Crosstalk with networks involving senescence, crop improvement, and mammalian cancer therapy. Front. Plant. Sci. 10, 859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S., You S., Hwang D., Aminoacyl-tRNA synthetases and tumorigenesis: More than housekeeping. Nat. Rev. Cancer 11, 708–718 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Park S. G., Schimmel P., Kim S., Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perera R. M., Bardeesy N., Pancreatic cancer metabolism: Breaking it down to build it back up. Cancer Discov. 5, 1247–1261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altman B. J., Stine Z. E., Dang C. V., From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X. et al., Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 20, 661–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin J. et al., Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 9, 1697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogretmen B., Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 18, 33–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glunde K., Penet M. F., Jiang L., Jacobs M. A., Bhujwalla Z. M., Choline metabolism-based molecular diagnosis of cancer: An update. Expert Rev. Mol. Diagn. 15, 735–747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Kaczor-Urbanowicz K. E., Wong D. T., Salivary biomarkers in cancer detection. Med. Oncol. 34, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eberlin L. S. et al., Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc. Natl. Acad. Sci. U.S.A. 111, 2436–2441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margulis K. et al., Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc. Natl. Acad. Sci. U.S.A. 115, 6347–6352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee S. et al., Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. U.S.A. 114, 3334–3339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altman D. G., McShane L. M., Sauerbrei W., Taube S. E., Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. PLoS Med. 9, e1001216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.