Significance
Influenza viruses (IV) replicate in the nucleus. Export of newly produced genomes, packaged in viral ribonucleoprotein (vRNP) complexes, relies on the nuclear CRM1 export pathway and appears to be timely controlled by virus-induced cellular signaling. However, the exact mechanism of the signaling-controlled complex assembly and export is enigmatic. Here we show that IV activates the Raf/MEK/ERK/RSK1 pathway, leading to phosphorylation at specific sites of the NP, which in turn, creates a docking site for binding of the M1 protein, an initial step in formation of vRNP export complexes. These findings are of broad relevance regarding the regulatory role of signaling pathways and posttranslational modifications in virus propagation and will strongly support ongoing development of an alternative anti-influenza therapy.
Keywords: influenza virus, Raf/MEK/ERK pathway, RSK
Abstract
Influenza viruses (IV) exploit a variety of signaling pathways. Previous studies showed that the rapidly accelerated fibrosarcoma/mitogen-activated protein kinase/extracellular signal-regulated kinase (Raf/MEK/ERK) pathway is functionally linked to nuclear export of viral ribonucleoprotein (vRNP) complexes, suggesting that vRNP export is a signaling-induced event. However, the underlying mechanism remained completely enigmatic. Here we have dissected the unknown molecular steps of signaling-driven vRNP export. We identified kinases RSK1/2 as downstream targets of virus-activated ERK signaling. While RSK2 displays an antiviral role, we demonstrate a virus-supportive function of RSK1, migrating to the nucleus to phosphorylate nucleoprotein (NP), the major constituent of vRNPs. This drives association with viral matrix protein 1 (M1) at the chromatin, important for vRNP export. Inhibition or knockdown of MEK, ERK or RSK1 caused impaired vRNP export and reduced progeny virus titers. This work not only expedites the development of anti-influenza strategies, but in addition demonstrates converse actions of different RSK isoforms.
Influenza viruses (IV) cause highly contagious respiratory infections with epidemic and pandemic potential and high morbidity and mortality (1). The currently licensed drugs against influenza directly targeting components of the virus are not very effective and lead to the emergence of resistant virus strains (2–4). Therefore, we urgently need alternative approaches to fight influenza.
IV are nuclear replicating viruses. During the viral life cycle the newly produced viral genome, that is packaged in viral ribonucleoprotein complexes (vRNP), must cross the nuclear-cytoplasmic barrier to be transported to the cell membrane and packaged in progeny virus particles. Using Leptomycin B (LMB), an inhibitor of Crm1/Exportin1, it was demonstrated that vRNPs are exported out of the nucleus via the Crm1-mediated nuclear export pathway (5, 6). The chain of events that orchestrates vRNP nuclear export complex is to date not understood. One putative model postulates the interaction of the viral nuclear export protein (NEP) with the viral polymerase complex to create a supporting binding site for the matrix protein 1 (M1) (7). The Crm1-interaction is mediated via the NEP N terminus (7, 8). However, since vRNP export does not take place in the absence of M1, while strongly reduced amounts of NEP do not influence this process, the exact contribution of the two proteins to vRNP export still remains elusive (9–12). It was shown that the vRNP export complex assembles at the dense chromatin to gain access to the cellular export machinery. This assembly takes place within RCC1 (Ran nucleotide exchange factor)-located regions, to ensure the direct interaction of the vRNPs with regenerated Crm1-RanGTP-complexes (13, 14). Furthermore, there is accumulating evidence that vRNP export does not occur constitutively but is regulated by cellular signaling pathways (15–17), to ensure a temporal control of vRNP migration to the cytoplasm in the later stages of the viral life cycle starting at around 5.0 to 6.0 h postinfection (p.i.) (18). However, the exact mechanisms are still enigmatic. Like any other virus, IV exploit many factors of the infected cell to replicate. Viral proteins are multifunctional and interact with a wide variety of cellular components. Thus, blockade of cellular factors that are required for viral propagation might not only inhibit replication on a broad antiviral scale but could also strongly reduce the emergence of resistant virus variants due to the inability of the virus to substitute for missing cellular functions (19–21). The Ras-dependent Raf/MEK/ERK mitogen-activated protein (MAP) kinase signaling pathway regulates important cellular functions involved in proliferation, differentiation, cell metabolism, and immune response (22). Downstream targets of the pathway can either be directly phosphorylated by the MAPK ERK or by ERK-activated protein kinases like the p90 ribosomal S6 kinases (RSKs), which are exclusively activated by ERK1/2 (23).
In previous studies we have shown that viral activation of the Raf/MEK/ERK pathway, induced by hemagglutinin (HA) accumulation in the cellular membrane, supports vRNP nuclear export (15, 24, 25). These findings indicated that vRNP export is a Raf/MEK/ERK signaling-induced event, ensuring timely regulation of the export late in the infection cycle when the pathway is activated. Accordingly, influenza virus infection triggers activation of the pathway in an unusual biphasic manner, with a very early phase directly after infection and a later phase that requires productive infection. By using a variety of inhibitors of the kinase MEK, which represents the bottleneck of the Raf/MEK/ERK cascade, it was shown that the MEK blockade not only suppressed both activation phases but, in addition, led to strongly decreased progeny virus titers correlating with a nuclear retention of newly synthesized vRNPs of both influenza A (IAV) and B viruses (IBV) (15, 24, 26–28). Accordingly, treatment also impaired viral replication in vivo (26, 27). Importantly, no escape mutants could be detected after multipassage use of the MEK inhibitor U0126 in contrast to treatment with virus-directed drugs such as Amantadine (24). In addition, Oseltamivir-resistant influenza strains are still fully sensitive to MEK inhibitor treatment (27). These findings indicate the inability of the virus to compensate for the missing cellular function, suggesting that MEK inhibition might be suitable as an antiviral strategy.
While it has been already known for quite a while that the Raf/MEK/ERK cascade triggers vRNP export, in the present study we have identified the full chain of events that lead to the signaling-driven nuclear export of vRNPs.
Results
Inhibition of the Raf/MEK/ERK Pathway Results in Retention of Progeny vRNPs at the Chromatin and Reduced Binding to the M1-Protein.
The inhibition of the Raf/MEK/ERK pathway by specific MEK inhibitors, such as U0126 (15), Trametinib (28), or CI-1040 (27) led to a reduction of progeny viral titers, concomitant with the retention of newly synthesized vRNPs in the nuclei of infected cells. The aim of the present study was to unravel the molecular chain of events that links virus-induced activation of the kinase pathway to the nuclear export of vRNPs. Inhibitors might have off-target effects; therefore, we first aimed to confirm by genetic means that the antiviral action of MEK inhibitors is indeed due to inhibition of the kinase pathway. The kinases ERK1 and 2 are the only known direct downstream targets for MEK (29). Thus, we knocked down expression of ERK1/2 with specific small interfering RNAs (siRNAs). We tested the knockdown efficiency in A549 cells and chose a concentration of 100 nM siRNA for further experiments (SI Appendix, Fig. S1B). Indeed, progeny virus titers of Wilson-Smith neurotropic (WSN), which we used as a model IAV strain, were significantly decreased after a total infection time of 24 h in the ERK1/2 knockdown cells compared to control (siCtrl) (SI Appendix, Fig. S1C). These reduced virus titers correlated well with a nuclear retention of viral RNA (vRNA) and viral NP, polymerase acidic protein (PA), and M1 proteins (Fig. 1A and C and SI Appendix, Fig. S1 D, F, H, and J), that are all constituents of vRNP complexes. Furthermore, we analyzed the proteins associated with low-soluble dense chromatin in an in situ fractionation assay. First, soluble proteins were extracted from the cytoplasm and the nucleoplasm, followed by a chromatin digestion with DNase I and an extraction step using 250 mM NaCl. Proteins associated with dense packaged chromatin cannot be extracted with this concentration (14, 30). This in situ fractionation revealed higher immunofluorescence signals of vRNA, NP, M1, and PA at the remaining dense chromatin (Fig. 1 B and D and SI Appendix, Fig. S1 E, G, I, and K). After this confirmation of MEK/ERK pathway involvement in vRNP export, we decided to inhibit MEK by chemical means for further experiments using the MEK inhibitor CI-1040, which is highly specific for MEK1/2 due to its non-adenosine triphosphate (ATP) competitive action (31). This strategy allows a more precise timing to manipulate the pathway during viral infection, compared to a knockdown or knockout. CI-1040 was originally developed as an anti-tumor drug and, while it was not significantly effective on the tumor target, it very efficiently inhibited MEK and was well tolerated in humans in clinical trials (32, 33). Therefore CI-1040 might be a suitable drug for an antiviral therapy.
Fig. 1.
ERK1/2 knockdown and MEK-inhibition result in chromatin retention of progeny vRNPs. (A) Cellular localization of WSN vRNA 7 h p.i. after an ERK1/2 knockdown. See also SI Appendix, Fig. S1D. (B) Dense chromatin analysis of PB2-vRNA 7 h p.i. after an ERK1/2 knockdown. Same laser and detector settings were used. See also SI Appendix, Fig. S1E. (C) Cellular localization of WSN vRNPs (NP) and M1 7 h p.i. after an ERK1/2 knockdown. See also SI Appendix, Fig. S1J. (D) Dense chromatin analysis of vRNPs (NP) and M1 7 h p.i. after an ERK1/2 knockdown. Same laser and detector settings were used. See also SI Appendix, Fig. S1K. (E) Cellular localization of WSN vRNA 7 h p.i. after CI-1040 (10, 15 µM) and LMB (5 nM) treatment 3 h p.i. DMSO (0.1%) and MeOH (0.1%) served as negative controls. See also SI Appendix, Fig. S1L. (F) Dense chromatin analysis of PB2-vRNA 7 h p.i. after CI-1040 (10, 15 µM) and LMB (5 nM) treatment. DMSO (0.1%) and MeOH (0.1%) served as negative controls. Same laser and detector settings were used. See also SI Appendix, Fig. S1M. (G) Cellular localization of WSN vRNPs (NP) and M1 7 h p.i. after CI-1040 (10 µM) and LMB (5 nM) treatment 3 h p.i. DMSO (0.1%) and MeOH (0.1%) served as negative controls. See also SI Appendix, Fig. S1R. (H) Dense chromatin analysis of vRNPs (NP) and M1 7 h p.i. after CI-1040 (10 µM) and LMB (5 nM) treatment. Same laser and detector settings were used. See also SI Appendix, Fig. S1S. (A–H) Representative images of three independent experiments. Dashed squares indicate zoom-in areas. (Scale bar, 20 µm.)
As it is already known that the Crm1/exportin 1 inhibitor LMB leads to a nuclear retention of vRNPs at the chromatin (14, 34), we used LMB as a control treatment. The incubation of infected cells with either CI-1040 or LMB, starting 3.0 h p.i., led to nuclear retention of vRNPs (Fig. 1 E and G and SI Appendix, Fig. S1 L, N, P, and R). In addition, higher immunofluorescence signals of vRNA, NP, M1, and PA could also be detected at the chromatin (Fig. 1 F and H and SI Appendix, Fig. S1 M, O, Q, and S), confirming the findings of the ERK1/2 knockdown. In addition, stochastic optical reconstruction microscopy (STORM) was used to determine nuclear NP and M1 protein spatial distribution at high resolution (Fig. 2 A and B and SI Appendix, Fig. S2 G and H). We found areas of NP and M1 colocalization in total nuclei and at the chromatin in CI-1040- and LMB-treated cells, indicating that although NP and M1 are localized to the chromatin at a late step in the replication cycle, assembly of the export complex or its release from the chromatin is impaired under inhibitor treatment. Furthermore, nuclear localization of NP in LMB compared to CI-1040-treated cells appeared to differ (Fig. 2A and SI Appendix, Fig. S2G). While in CI-1040-treated cells NP seems to be distributed evenly in the nucleus, the protein appears to accumulate in proximity to the nuclear membrane in LMB-treated cells. This was a first indication that, although both compounds lead to vRNP retention, their molecular mechanism of antiviral action might be different.
Fig. 2.
Treatment of infected cells with CI-1040 results in chromatin retention of progeny vRNPs and decreased vRNP-binding to the M1 protein. (A) STORM analysis of WSN vRNPs (NP) and M1 localization after CI-1040 (10 µM) and LMB (5 nM) treatment. DMSO (0.1%) and MeOH (0.1%) served as negative controls. See also SI Appendix, Fig. S2G. (B) STORM analysis of dense chromatin after WSN infection and CI-1040 (10 µM) or LMB (5 nM) treatment. See also SI Appendix, Fig. S2H. (A and B) Dashed lines mark the nuclear periphery. Squares (Epi) indicate high-resolution areas. (Scale bar, 500 nm.) (C) Fractionation of WSN infected and CI-1040 (10 µM) (+) treated A549 cells. DMSO (0.1%) (−) served as negative control. Total infection times are indicated. Results of one out of three independent experiments for each time point are shown. See also SI Appendix, Fig. S2 A, C, and D. (D) Total protein amounts of WSN infected and CI-1040 (10 µM) (+) treated A549 cells. DMSO (0.1%) (−) served as negative control. Total infection times are indicated. Results of one out of three independent experiments are shown. See also SI Appendix, Fig. S2B. (E and F) Experiments were conducted as in C using Strep-PB2-WSN. Fractionation and vRNP purification were performed after the indicated time points. (E) Results of one out of three independent experiments. See also SI Appendix, Fig. S2F. (F) Total Western blot images were cropped to show the ch150 fraction. Indicated time points were analyzed once. Total analysis is shown in SI Appendix, Fig. S2E.
According to previous reports, the influenza vRNP nuclear export complex assembles at the chromatin where it enters the nuclear export machinery (14, 35). To validate the retention of the viral proteins NP and M1 at the chromatin (shown in Figs. 1H and 2B and SI Appendix, Figs. S1S and S2H), both essential components in the formation of the vRNP export complex, we first aimed to estimate their protein amounts in different cellular/nuclear compartments in presence or absence of the MEK inhibitor. We chose a time window of 6.5 h to 8.0 h p.i. because this is the prime time period when nuclear export takes place with our model virus (11). Infected treated or untreated cells were separated into cytoplasmic (cyt), nucleoplasmic (nuc), and two fractions of dense chromatin with different salt solubilities (ch150: 150 mM NaCl-extractable chromatin; ch500: 500 mM NaCl-extractable chromatin) (Fig. 2C and SI Appendix, Fig. S2 C and D). Inhibition of the Raf/MEK/ERK pathway resulted in an increased amount of NP in the ch500-subnuclear fraction, which indicates a retention of vRNPs at the chromatin (Fig. 2C and SI Appendix, Fig. S2A). Interestingly, the distribution of the M1 protein showed a more dynamic pattern, as higher subnuclear amounts compared to the control were only found up to 7.0 h p.i. (Fig. 2C). These differential localization patterns of NP and M1 under CI-1040 treatment were not due to overall changes in total viral protein accumulation in CI-1040-treated cells (Fig. 2D and SI Appendix, Fig. S2B), confirming earlier findings with other MEK inhibitors (15, 28).
To further analyze the composition of the vRNP export complex in the presence or absence of CI-1040, we infected cells with a recombinant WSN virus containing a C-terminally Strep-tagged PB2. Strep-purification after cell fractionation allowed us to purify vRNPs from the different fractionation lysates. This was confirmed by detection of copurified polymerase basic protein 1 (PB1) and PA as markers for the trimeric polymerase complex, as well as copurified NP, the main viral protein in the vRNP complex. While these vRNP proteins could be detected in all fractions regardless of whether MEK was inhibited or not, we found striking differences with regard to associated M1 protein (Fig. 2 E and F and SI Appendix, Fig. S2E). vRNP-associated M1 could only be detected in the chromatin fraction extracted with 150 mM NaCl. Furthermore, M1 association was virtually abolished if cells were treated with CI-1040, a pattern that was robustly detected over an observation period from 6.0 h to 7.5 h p.i. (Fig. 2 E and F and SI Appendix, Fig. S2 E and F). This clearly indicates that MEK inhibition alters the interaction of M1 with vRNPs and thus results in a subsequent block in the assembly of the export complex at a particular chromatin fraction.
Raf/MEK/ERK Pathway-Dependent Phosphorylation of a Specific Motif within the Nucleoprotein.
The data so far indicate that the Raf/MEK/ERK pathway promotes vRNP export by facilitating M1 association to the vRNP export complex at the chromatin. Since on one hand the Raf/MEK/ERK kinase cascade transmits signals within the cell via timely regulated sequential phosphorylation (36), and on the other hand the viral NP is long known to be differentially phosphorylated throughout the replication cycle (37), we hypothesized that posttranslational phosphorylation of the NP may be the decisive signal for M1 association. To identify phosphorylation sites within the viral NP that are controlled via MEK/ERK, HEK293T cells were infected with the recombinant Strep-PB2-WSN virus. At 3.0 h p.i., the infected cells were treated with dimethylsulfoxide (DMSO) or CI-1040. At 7.0 h p.i., vRNPs were Strep-purified from the total protein lysates. Phospho-modification patterns in the vRNP proteins purified from the cells treated with DMSO and CI-1040 were analyzed by mass spectrometry. We found peptides corresponding to two phospho-serine residues at S269 and S392, which were absent after CI-1040 treatment, indicating that these sites are sensitive to inhibitor treatment.
The validity of the mass spectrometry analysis is reflected by the fact that we additionally found already described phosphorylation sites (S402, S403, S457) (38, 39) (SI Appendix, Fig. S3B), which, however, were not sensitive to CI-1040 treatment. The crystallographic structure of a vRNP bound to RNA and monomeric NP revealed that S269 and S392 are located in close proximity to each other. Furthermore, S269 localizes within the nuclear export signal 2 (NES2) and S392 is located near the NES2 and NES3 of the nucleoprotein (40). A positively charged RNA-binding groove and a loop formed by the viral RNA surrounds the two residues (Fig. 3 A–C and SI Appendix, Fig. S3C).
Fig. 3.
Nucleoprotein residues serine 269 and 392 phosphorylation upon Raf/MEK/ERK activation. (A) Cryo-electron reconstruction of a helical part of A/Wilson-Smith/1933 (H1N1) ribonucleoprotein obtained from Protein Data Bank (PDB) ID 4BBL (41). (B) Localization of S269 and S392 in vRNP, surrounded by a vRNA (orange) loop. Green: Nuclear export signals (NES). Purple: RNA-binding groove. See also SI Appendix, Fig. S3C. (C) Localization of S269 and S392 in monomeric NP. Green: NES. Purple: RNA-binding groove. PDB ID 2IQH (42). (D) Multireplication cycle analysis of WSN wild type (wt) or mutants in A549 cells. Titers were determined after the indicated time points. Data represents mean ± SD of four independent experiments, each performed in triplicates. Data passed a one-way ANOVA test followed by Dunnett’s multiple comparison test for each time point individually (**P ≤ 0.01, ***P ≤ 0.001).
To analyze whether the two serine residues play a functional role in virus replication, WSN mutants with nonphosphorylatable amino acids at the positions 269 and 392 (S269A, S392A, S269A/S392A) were generated. Notably, phospho-mimicking mutants could not be rescued, indicating that permanent negative charges at these positions are not tolerated. The replication efficiency of the S392A-mutants was decreased within multicycle replication experiments by up to two log10. The S269A-mutant showed only a slight increase in the replication efficiency, especially at earlier time points (Fig. 3D). These data indicate a strong virus-supportive contribution of the phosphoacceptor residue S392 on the viral life cycle.
The Kinase RSK1 Links Activation of the Raf/MEK/ERK Pathway to the Control of vRNP Export.
The amino acid sequences adjacent to the identified CI-1040 sensitive phosphorylation sites (L-I-L-R-G-S269-V, A-I-R-T-R-S392-G) lack similarity to the consensus sequence of the ERK phosphorylation motive (P-X-S/T-P) (SI Appendix, Fig. S3D) (43). Therefore it appears unlikely that the identified serine residues are directly phosphorylated by ERK. Instead, the consensus target sequences of the ERK-downstream kinase 90 kDa ribosomal S6 kinase (RSK) (R/L-X-R-X-X-S/T; R-R-X-S/T) showed much higher identities (SI Appendix, Fig. S3D) (44). To explore whether RSK is the link between the activation of the Raf/MEK/ERK signaling pathway and the phosphorylation of NP, which subsequently drives nuclear export of newly synthesized vRNPs, RSK activation during the viral life cycle was analyzed. Indeed, in later stages of the infection, not only ERK but also RSK as well as the RSK downstream target glycogen synthase kinase GSK-3β were increased in their phosphorylation (Fig. 4A and SI Appendix, Fig. S4A). This activation could be blocked by incubation with the MEK inhibitor CI-1040, clearly indicating that virus-induced RSK activation is mediated by the Raf/MEK/ERK pathway (SI Appendix, Fig. S4B). To test for a functional involvement of RSK activation, we used a specific inhibitor of RSK, BI-D1870 (45), which led to a concentration-dependent reduction of GSK-3β phosphorylation, confirming its inhibitory effect on RSK activation during the viral life cycle (Fig. 4B and SI Appendix, Fig. S4C). While, similar to MEK inhibitors (Fig. 2D), BI-D1870 did not affect the synthesis and accumulation of viral proteins, we interestingly found an increase of ERK activation after the inhibition of RSK (Fig. 4B and SI Appendix, Fig. S4C). This is indicative of a BI-D1870-mediated inhibition of a negative feedback loop that under normal conditions would prevent the overactivation of the pathway (45, 46). To test whether RSK inhibition would also lead to impaired export of viral RNPs, BI-D1870 was compared side by side with CI-1040 on their impact on localization of the vRNA and viral proteins NP, M1, PA, and NEP (Figs. 4C and 5D and SI Appendix, Figs. S4 D–K and S5D). Similar to CI-1040, treatment with BI-D1870 resulted in a strong impairment of vRNP nuclear export. The effect on nuclear retention correlates with a strong antiviral activity of BI-D1870 (SI Appendix, Fig. S4 L and M), with a 50% effective concentration (EC50) of 2.8 μM (SI Appendix, Fig. S4M) and a selectivity index (SI) of 157.98 (SI Appendix, Fig. S4N). As a control, a second RSK inhibitor, SL0101-1 was used, resulting in similar antiviral effects (SI Appendix, Fig. S4 O–Q). This demonstrates a strong anti-IAV activity of RSK inhibitors in the absence of any toxic side effects in the effective concentrations. Furthermore, if BI-D1870 and CI-1040 were used in a combinational treatment, there were no significant additive effects compared to CI-1040 treatment alone, again demonstrating that RSK acts within the same pathway and is directly activated by the Raf/MEK/ERK kinase cascade (Fig. 4D).
Fig. 4.
Nuclear retention of progeny vRNPs upon RSK inhibition and RSK1 knockdown. (A) ERK1/2, RSK1, and GSK-3β activation after WSN infection in A549 cells 7 h and 9 h p.i. Results of one out of three independent experiments are shown. See also SI Appendix, Fig. S4A. (B) Increased ERK1/2 and decreased GSK-3β phosphorylation after WSN infection and BI-D1870 treatment in A549 cells 7 h p.i. DMSO (0.1%) served as negative control. Results of one out of four independent experiments are shown. See also SI Appendix, Fig. S4C. (C) Nuclear retention of vRNPs after BI-D1870 (15 µM) treatment in A549 cells 9 h p.i. DMSO (0.1%) served as negative control, CI-1040 (10 µM) as positive control. Localization of vRNPs (NP) and M1 was analyzed. Representative images of three independent experiments. Dashed squares indicate zoom-in areas (Scale bar, 20 µm.) See also SI Appendix, Fig. S4 J and K. (D) Titer reduction of WSN in A549 cells after BI-D1870 (10 µM), CI-1040 (10 µM), and a combinational treatment (each 10 µM) 24 h p.i. DMSO (0.2%) served as negative control. Data represent means ± SD of four independent experiments, each performed in duplicates. Data passed a one-way ANOVA test followed by Tukey´s multiple comparison test for each time point separately (***P ≤ 0.001). (E) Cellular localization of WSN vRNPs (NP) and M1 9 h p.i. after RSK1 or RSK2 were knocked down in A549 cells. Representative images of three independent experiments. Dashed squares indicate zoom-in areas (Scale bar, 20 µm.) See also SI Appendix, Fig. S4 V and W. (F and G) Titers of WSN 24 h p.i. after RSK1 or RSK2 knockdown in A549 cells. Shown are means ± SD of three independent experiments, each performed in triplicates. Data passed a paired two-tailed t test (**P ≤ 0.01; ***P ≤ 0.001). See also SI Appendix, Fig. S4 X and Y. (H and I) Titers of WSN wt or mutants after BI-D1870 (10 µM) treatment in A549 cells 24 h p.i. DMSO (0.1%) served as negative control. Shown are means ± SD of three independent experiments, each performed in triplicates. Statistics: plaque-forming unit (PFU)/mL: Data passed an unpaired two-tailed t test with Welch-correction for each virus individually, Percentage: Data passed a paired two-tailed t test for each virus individually (ns > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). (J–O) Titers of WSN wt or mutants after knockdown of ERK1/2 (J and K), RSK1 (L and M), or RSK2 (N and O) in A549 cells 24 h p.i. Shown are means ± SD of three independent experiments, each performed in triplicates. Statistics: PFU/mL: Data passed an unpaired two-tailed t test with Welch-correction for each virus individually. Percentage: Data passed a paired two-tailed t test for each virus individually (ns > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Fig. 5.
BI-D1870 treatment results in chromatin retention of progeny vRNPs and decreased binding rates to the M1 protein. (A) Fractionation and Strep-purification of Strep-PB2-vRNP (WSN) after BI-D1870 (10 µM) (+) treatment 7 h p.i. DMSO (−) served as negative control. See also SI Appendix, Fig. S5A. (B) Total protein amounts of the fractionated cell lysates from A. (C) Protein amounts of total cell lysates from A. (D) Cellular localization of WSN vRNA 7 h p.i. after BI-D1870 (10, 15 µM) treatment 3 h p.i. DMSO (0.1%) served as negative control. See also SI Appendix, Fig. S5D. (E) Dense chromatin analysis of PB2-vRNA 7 h p.i. after BI-D1870 (10, 15 µM) treatment. DMSO (0.1%) served as negative control. Same laser and detector settings were used. See also SI Appendix, Fig. S5E.
To exclude off-target effects of the RSK inhibitors, the two isoforms RSK1 and RSK2 were knocked down with specific siRNAs and effects on the viral life cycle were analyzed (Fig. 4 E–G and SI Appendix, Fig. S4 R–Y). RSK1 knockdown led to a nuclear retention of newly synthesized vRNPs whereas the RSK2 knockdown had no effect on the nuclear export (Fig. 4E and SI Appendix, Fig. S4 S–W). In multicycle replication analysis, RSK1 knockdown resulted in a strong decrease of viral titers, confirming the data obtained with the inhibitors (Fig. 4F and SI Appendix, Fig. S4X). This was in clear contrast to the RSK2 knockdown, which seemed to have a proviral effect (Fig. 4G and SI Appendix, Fig. S4Y). This supportive effect of RSK2 knockdown on viral replication was already described by Kakugawa and colleagues in 2009, who showed that RSK2 is specifically involved in mounting an antiviral response (47). While we could fully confirm this finding here, our data also unravel isoform-specific functions of RSK1 and RSK2, leading to opposite effects on the influenza virus life cycle. Since the RSK inhibitors BI-D1870 and SL0101-1 are described to block both isoforms, we can also conclude that the virus-supportive function of RSK1 is dominant over the antiviral function of RSK2. If RSK would phosphorylate the identified serine residues in NP, the viral mutants lacking these phosphorylation sites should be less susceptible to the inhibition or knockdown of RSK1. Indeed, the inhibitory effect of BI-D1870 on the viral life cycle was decreased. This effect was more prominent for the S392A-mutants, supporting the previous findings (Fig. 4 H and I). Comparable effects were found after RSK1 or ERK1/2 was knocked down (Fig. 4 J–M). The proviral effect of the RSK2 knockdown was found for the wild-type virus and the mutants, whereby it was not as prominent for the S392A-mutants as for the wild type and the S269A-mutans (Fig. 4 N and O).
To further study whether RSK1 is the downstream effector of MEK/ERK in the control of vRNP export, we infected cells with Strep-PB2-WSN virus in the absence or presence of BI-D1870 and performed chromatin fractionation assays as described in Fig. 2E. BI-D1870 treatment led to a similar vRNP chromatin retention and impairment of vRNP-M1 interaction (Fig. 5 A, B, and E and SI Appendix, Fig. S5 A, B, and E) as previously shown for CI-1040 (Fig. 2 E and F and SI Appendix, Fig. S2 E and F). The total viral protein amounts again were not affected by the inhibitor (Fig. 5C), while in these lysates again a strong induction of the ERK1/2 phosphorylation could be detected, most likely as a consequence of an inhibited negative feedback loop. From these data we could also conclude that RSK1 is the sole effector kinase of ERK in the vRNP export-regulating activity of the pathway since enhanced ERK activity in the presence of the RSK inhibitor would not lead to enhanced export but rather the opposite.
MEK and RSK Inhibitors Do Not Generally Block Crm1-Mediated Nuclear Export.
LMB blocks the general Crm1/exportin1-mediated nuclear export pathway by alkylating and inhibiting Crm1 (48). Since Crm1-mediated export is an essential process for cell homeostasis and LMB acts irreversibly, the compound is toxic and not suitable as a drug. The question now arises, whether MEK and RSK inhibitors would also generally affect Crm1-mediated export, which would disqualify them as possible drug candidates. As a control cargo for the cellular Crm1 export pathway, we used the Ran-binding protein RanBP1, which is known to be exported to the cytoplasm in a Crm1-pathway-dependent manner (49). We compared LMB to the MEK inhibitor CI-1040 and the RSK inhibitor BI-D1870 regarding their effect on the Crm1 pathway. Cells were infected with WSN and treated with the respective inhibitors. Quantification of immunofluorescence stainings 9.0 h p.i. revealed nuclear retention of the viral NP for all tested inhibitors. In contrast, nuclear retention of RanBP1 was only detected in the LMB-treated samples (Fig. 6 A and B). Comparable results were found for other MEK and RSK inhibitors, such as ATR-002 and SL0101-1, respectively (SI Appendix, Fig. S5 F and G). This strongly indicates that the inhibition of the Raf/MEK/ERK/RSK pathway has no general effect on the Crm1 export but specifically controls export of the influenza vRNPs.
Fig. 6.
The Raf/MEK/ERK/RSK pathway inhibitors CI-1040 and BI-D1870 act specifically on the nuclear export of viral proteins. (A) Cellular localization of WSN vRNPs (NP) and RanBP1 in A549 cells after treatment with LMB (5 nM), CI-1040 (10 µM), or BI-D1870 (15 µM) 9 h p.i. MeOH (0.1%) and DMSO (0.1%) served as negative controls. Representative images of three independent experiments. Dashed squares indicate zoom-in areas (Scale bar, 20 µm.) See also SI Appendix, Fig. S5F. (B) Quantification of the intracellular protein localization of NP and RanBP1. Results are depicted as means ± SD of three independent experiments. Data passed a one-way ANOVA test followed by Tukey´s multiple comparison test (***P ≤ 0.001). See also SI Appendix, Fig. S5G.
Broad Anti-Influenza Activity of BI-D1870.
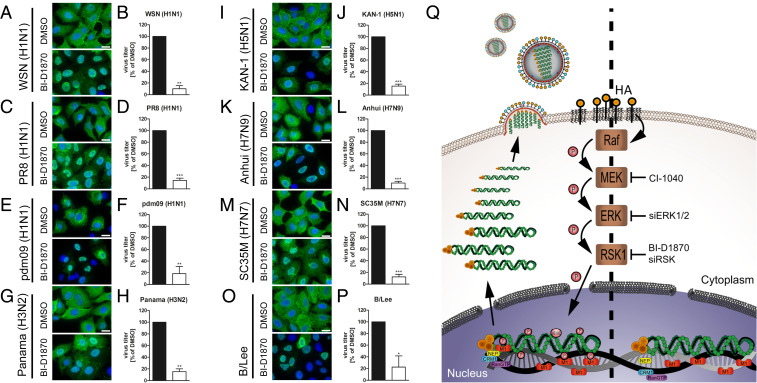
To confirm that the effect of RSK inhibition on viral replication is not strain specific, we used a range of different IAV subtypes, including a seasonal H3N2 virus, a swine origin H1N1 pandemic (pdm) virus from 2009, highly pathogenic avian viruses of subtype H5 and H7, as well as an IBV strain to determine the broad anti-influenza virus activity. All tested viruses showed both nuclear retention of newly synthesized vRNPs and a reduction of progeny viral titers of around one log10 in the presence of the RSK inhibitor (Fig. 7). These results confirm a broad dependence of IAV and IBV on the Raf/MEK/ERK/RSK pathway (26–28) and show that RSK is the critical mediator that links the pathway to vRNP export.
Fig. 7.
RSK inhibition affects vRNP nuclear export and progeny virus titers of a range of different influenza viruses. Cellular localization (IF) and titers of human IAV (A–H), avian-derived IAV (I–L), IAV/SC35M/H7N7 (M and N), or IBV (O and P) in A549 cells (A–O) or MDCKII cells (P). Cells were treated with BI-D1870 (15 µM) or DMSO (0.1%). At 9 h p.i. (IAV) or 12 h p.i. (IBV), localization of vRNPs (NP) was analyzed. At 24 h p.i., titers were analyzed. Shown are means ± SD of three independent experiments. Each experiment was performed in triplicates. Data passed a paired two-tailed t test (*P ≤ 0.05; **P ≤ 0.01, ***P ≤ 0.001) (Scale bar, 20 µm.) (Q) Hypothetical mechanism of the RSK-induced nuclear export of newly synthesized vRNPs.
In summary, we have identified the complete chain of events that link the nuclear export of newly synthesized vRNPs with viral activation of the Raf/MEK/ERK/RSK pathway (Fig. 7Q). As inhibitors targeting the pathway are not toxic, they may serve as promising candidates for the development of broadly active anti-influenza drugs.
Discussion
Influenza viruses are nuclear-replicating viruses. Hence, the viral genome needs to be exported out of the nucleus at late stages of the replication cycle to be transported to the cellular membrane and packaged in progeny viruses. Since at early and intermediate stages of replication new genomes are needed as templates for the amplification of RNA replication and transcription, vRNP export should not be constitutive but regulated and preferentially promoted in late stages of infection. We showed previously that late-stage viral activation of the Raf/MEK/ERK pathway by accumulation of HA in the host membrane and protein kinase C (PKCα) activation is linked to vRNP nuclear export and may represent such a regulatory principle (15, 25). However, the exact mechanism of how the pathway supports this process was completely enigmatic for a long time. By the use of siRNA knockdown approaches as well as kinase inhibitors we could now not only unravel the complete chain of events how the Raf/MEK/ERK kinase pathway controls the nuclear export of newly produced vRNPs, but at the same time dissect the antiviral mode of action of the MEK inhibitor CI-1040. A derivate of CI-1040, ATR-002 has now successfully passed a phase I clinical trial (clinicaltrials.gov: NCT04385420) and is under further clinical development as an anti-influenza drug.
It is known that the vRNP export complex assembles at the chromatin, where it can interact with the cellular export protein Crm1 (14). The inhibition of MEK or RSK during the viral life cycle led to a retention of the newly synthesized vRNPs at the chromatin and reduced binding abilities to the viral M1 protein, which is essential for the nuclear export of the viral genome (Figs. 1 E–H, 2, 4, and 5 and SI Appendix, Figs. S1, S2, S4, and S5) (11, 12).
We could identify two new phosphorylation residues within the viral NP, the major constituent of vRNPs (serine 269 and serine 392) (Fig. 3 and SI Appendix, Fig. S3). The sites are located in the loop region (264 to 276; 452 to 463) and the body domain (21 to 149; 273 to 396; 464 to 489) of the protein, respectively, and are located in close proximity to each other. Crystallographic structures of the vRNP helix bound to vRNA revealed that these two serine residues are surrounded by a specifically formed vRNA loop (Fig. 3B and SI Appendix, Fig. S3C) (40, 41).
Signaling pathways are often controlled by negative feedback loops. This is also true for the Raf/MEK/ERK pathway that is negatively controlled by the active downstream kinase RSK (46). If RSK is inhibited, its negative regulating mode of action is missing and the kinases upstream of RSK are over activated (Figs. 4B and 5C and SI Appendix, Fig. S4C). Together with other findings in the present study, this observation allows the conclusion that RSK1 is the predominant downstream effector of ERK that mediates vRNP export. The hyperactivation of ERK in the presence of the RSK inhibitor does not lead to enhanced vRNP export but rather the opposite, indicating that the signal is not further transmitted if RSK is blocked.
Kakugawa and colleagues already previously investigated the role of RSK2 in influenza virus-infected cells (47). They could show via knockdown experiments that RSK2 has an antiviral function as the knockdown resulted in increased IAV and IBV replication. This effect was explained by a role of RSK2 in the innate immune response due to the activation and expression regulation of nuclear factor kB (NF-κB), interferon β (IFNβ), and protein kinase R (PKR). We could fully confirm the antiviral function of RSK2 in our experiments, since RSK2 knockdown led to higher viral titers (Fig. 4G and SI Appendix, Fig. S4Y). In clear contrast, RSK1 knockdown resulted in strongly decreased viral titers and the nuclear retention of newly synthesized vRNPs (Fig. 4 F, L, and M and SI Appendix, Fig. S4X). These findings point to opposite roles of the two RSK subtypes during influenza virus infection. It was shown that messenger RNA (mRNA) and protein expression levels of RSK1 are much higher than RSK2 levels in human and mice lungs, supporting a more prominent role of RSK1 compared to RSK2 (44, 50). The fact that two downstream effectors of the same pathway exhibit opposite roles allows several conclusions. First, it can be concluded that activation of the Raf/MEK/ERK pathway most likely occurs as part of the antiviral response of the cell to drive RSK2-mediated innate immune responses. Second, it seems that IV have acquired the capability to misuse this supposedly antiviral activity for their own replication in a dominant fashion. Third, this concept well explains the observation that viral signaling antagonists such as NS1, that readily inhibits other antiviral signaling pathways, would not block Raf/MEK/ERK activation (25).
In summary, we have established the mechanism by which IV misuses the cellular Raf/MEK/ERK/RSK pathway to control a particular step in the virus life cycle in a spatiotemporal manner. In turn, this represents an Achilles heel of virus replication that may be used for an antiviral approach. The fact that the inhibitors of MEK and RSK used in this study are not toxic and exhibit a high selectivity-index (SI Appendix, Fig. S4 L–Q) or were even shown to be well tolerated in mice and humans (CI-1040) (27, 32) suggests that they may be considered as lead compounds for a generation of cell-directed antivirals in a repurposing approach.
Material and Methods
A549, MDCKII, or HEK293 cells were infected with different viruses (SI Appendix, Table S1) for 30 min at 37 °C, washed once with phosphate-buffered saline (PBS), and incubated in infection Dulbecco's modified Eagle medium (DMEM) or minimum essential medium (MEM). For multicycle replication experiments, cells were treated with the inhibitors or DMSO diluted in infection medium 30 min p.i. for the indicated time points. For single-cycle replication experiments, cells were treated 3.0 h p.i. for the indicated time points.
The ERK1/2 and RSK1,2 knockdown was introduced in A549 cells with the aid of Lipofectamine 2000 (Invitrogen) 48 h preinfection, according to the manufacturer’s protocol.
Infected and inhibitor-treated cells were fractionated in a cytoplasmic (cyt), nucleoplasmic (nuc), and two chromatin fractions (ch150; ch500). The PB2-Strep-tagged WSN virus was used to purify the vRNPs out of the individual fractionations.
Protein expression rates, amounts, and phosphorylation states of viral and cellular proteins were analyzed by Western blot (SI Appendix, Table S2).
Infected and inhibitor-treated cells were in situ fractionated and chromatin-associated vRNA and viral proteins were analyzed by indirect immunofluorescence staining or the dSTORM reporter-only method with conventional antibodies (SI Appendix, Table S2).
Strep-PB2 purified WSN-vRNPs were analyzed by mass spectrometry in a Proxeon Easy-nLC in combination with a Q-Exactive HF mass spectrometer. Data were analyzed with MaxQuant V1.6.6.0 (Max Planck Institute).
Detailed material and methods are described in SI Appendix, Material and Methods.
Data Availability.
The data supporting the findings of this study are available within the paper and in SI Appendix. Raw datasets from mass spectrometry proteomics are publicly available on the Proteomics Identifications Database (PRIDE Archive) at http://www.ebi.ac.uk/pride/archive/projects/PXD016638.
Supplementary Material
Acknowledgments
We thank Atriva Therapeutics GmbH, Tuebingen, Germany, for providing the CI-1040 and ATR-002 inhibitors. This work was supported by the German Research Foundation (DFG) (grants SFB1009B02 and KFO342) as well as by the innovative medical research (IMF) and the interdisciplinary center of clinical research (IZKF) of the Medical Faculty, Westfaelische Wilhelms Universitaet Muenster.
Footnotes
Competing interest statement: S.L. is cofounder and head of the board of Atriva Therapeutics GmbH, Tuebingen, Germany.
This article is a PNAS Direct Submission.
Data deposition: Raw datasets from mass spectrometry proteomics can be publicly accessed on the Proteomics Identifications Database (PRIDE Archive) at http://www.ebi.ac.uk/pride/archive/projects/PXD016638.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002828117/-/DCSupplemental.
References
- 1.Girard M. P., Cherian T., Pervikov Y., Kieny M. P., A review of vaccine research and development: Human acute respiratory infections. Vaccine 23, 5708–5724 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussain M., Galvin H. D., Haw T. Y., Nutsford A. N., Husain M., Drug resistance in influenza A virus: The epidemiology and management. Infect. Drug Resist. 10, 121–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omoto S. et al., Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 8, 9633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam M. I. et al., Verapamil has antiviral activities that target different steps of the influenza virus replication cycle. J. Antivir. Antiretrovir. 8, 121–130 (2016). [Google Scholar]
- 5.Elton D. et al., Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75, 408–419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe K. et al., Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 77, 31–42 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Brunotte L. et al., The nuclear export protein of H5N1 influenza A viruses recruits Matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export. J. Biol. Chem. 289, 20067–20077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S. et al., A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 87, 767–778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolstenholme A. J., Barrett T., Nichol S. T., Mahy B. W. J., Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins. J. Virol. 35, 1–7 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith D. B., Inglis S. C., Regulated production of an influenza virus spliced mRNA mediated by virus-specific products. EMBO J. 4, 2313–2319 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin K., Helenius A., Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell 67, 117–130 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Bui M., Wills E. G., Helenius A., Whittaker G. R., Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74, 1781–1786 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemergut M. E., Mizzen C. A., Stukenberg T., Allis C. D., Macara I. G., Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Chase G. P. et al., Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog. 7, e1002187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleschka S. et al., Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3, 301–305 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Nencioni L. et al., Bcl-2 expression and p38MAPK activity in cells infected with influenza A virus: Impact on virally induced apoptosis and viral replication. J. Biol. Chem. 284, 16004–16015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrhardt C. et al., The NF-κB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell. Microbiol. 15, 1198–1211 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Eisfeld A. J., Kawakami E., Watanabe T., Neumann G., Kawaoka Y., RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85, 6117–6126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig S., Disruption of virus-host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections. Biol. Chem. 392, 837–847 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Müller K. H. et al., Emerging cellular targets for influenza antiviral agents. Trends Pharmacol. Sci. 33, 89–99 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Li C. C., Wang X. J., Wang H. R., Repurposing host-based therapeutics to control coronavirus and influenza virus. Drug Discov. Today 24, 726–736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cargnello M., Roux P. P., Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz M., Amit I., Yarden Y., Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta 1773, 1161–1176 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig S. et al., MEK inhibition impairs influenza B virus propagation without emergence of resistant variants. FEBS Lett. 561, 37–43 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Marjuki H. et al., Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J. Biol. Chem. 281, 16707–16715 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Droebner K., Pleschka S., Ludwig S., Planz O., Antiviral activity of the MEK-inhibitor U0126 against pandemic H1N1v and highly pathogenic avian influenza virus in vitro and in vivo. Antiviral Res. 92, 195–203 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Haasbach E. et al., The MEK-inhibitor CI-1040 displays a broad anti-influenza virus activity in vitro and provides a prolonged treatment window compared to standard of care in vivo. Antiviral Res. 142, 178–184 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Schräder T. et al., The clinically approved MEK inhibitor Trametinib efficiently blocks influenza A virus propagation and cytokine expression. Antiviral Res. 157, 80–92 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Pearson G. et al., Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 22, 153–183 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Mirsky A. E., Burdick C. J., Davidson E. H., Littau V. C., The role of lysine-rich histone in the maintenance of chromatin structure in metaphase chromosomes. Proc. Natl. Acad. Sci. U.S.A. 61, 592–597 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen L. F., Sebolt-Leopold J., Meyer M. B., CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK). Semin. Oncol. 30 (suppl. 16), 105–116 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Lorusso P. M. et al., Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J. Clin. Oncol. 23, 5281–5293 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Barrett S. D. et al., The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg. Med. Chem. Lett. 18, 6501–6504 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Ma K., Roy A. M. M., Whittaker G. R., Nuclear export of influenza virus ribonucleoproteins: Identification of an export intermediate at the nuclear periphery. Virology 282, 215–220 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Takizawa N., Watanabe K., Nouno K., Kobayashi N., Nagata K., Association of functional influenza viral proteins and RNAs with nuclear chromatin and sub-chromatin structure. Microbes Infect. 8, 823–833 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Yoon S., Seger R., The extracellular signal-regulated kinase: Multiple substrates regulate diverse cellular functions. Growth Factors 24, 21–44 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Kistner O., Müller K., Scholtissek C., Differential phosphorylation of the nucleoprotein of influenza A viruses. J. Gen. Virol. 70, 2421–2431 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Hutchinson E. C. et al., Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8, e1002993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mondal A., Potts G. K., Dawson A. R., Coon J. J., Mehle A., Phosphorylation at the homotypic interface regulates nucleoprotein oligomerization and assembly of the influenza virus replication machinery. PLoS Pathog. 11, e1004826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng A. K., Wang J. H., Shaw P. C., Structure and sequence analysis of influenza A virus nucleoprotein. Sci. China C Life Sci. 52, 439–449 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Arranz R. et al., The structure of native influenza virion ribonucleoproteins. Science 338, 1634–1637 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Ye Q., Krug R. M., Tao Y. J., The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444, 1078–1082 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez F. A., Raden D. L., Davis R. J., Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266, 22159–22163 (1991). [PubMed] [Google Scholar]
- 44.Romeo Y., Zhang X., Roux P. P., Regulation and function of the RSK family of protein kinases. Biochem. J. 441, 553–569 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Sapkota G. P. et al., BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem. J. 401, 29–38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha M. et al., RSK phosphorylates SOS1 creating 14-3-3-docking sites and negatively regulating MAPK activation. Biochem. J. 447, 159–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakugawa S. et al., Mitogen-activated protein kinase-activated kinase RSK2 plays a role in innate immune responses to influenza virus infection. J. Virol. 83, 2510–2517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kudo N. et al., Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plafker K., Macara I. G., Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol. Cell. Biol. 20, 3510–3521 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeniou M., Ding T., Trivier E., Hanauer A., Expression analysis of RSK gene family members: The RSK2 gene, mutated in Coffin-Lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum. Mol. Genet. 11, 2929–2940 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and in SI Appendix. Raw datasets from mass spectrometry proteomics are publicly available on the Proteomics Identifications Database (PRIDE Archive) at http://www.ebi.ac.uk/pride/archive/projects/PXD016638.