Significance
Mismatch repair is the process that corrects replication errors. Mutations in the proteins MutSα and MutLα that initiate MMR are responsible for Lynch syndrome, the most common hereditary cancer. We present results showing surprising stoichiometries and conformations of human MutSα and MutLα mismatch repair initiation complexes. Moreover, these multimeric MutSα–MutLα complexes reconfigure the DNA in the vicinity of the mismatch. These complexes may not only mark the position of the mismatch but also could protect it from nucleosome reloading and potentially remodel adjacent nucleosomes. These results contrast with current models in the field, which envision mobile 1:1 MutSα–MutLα complexes that leave the mismatch. Our results provide a framework for thinking about MMR initiation.
Keywords: DNA repair, AFM, MutS, MutL, DREEM
Abstract
DNA mismatch repair (MMR) corrects errors that occur during DNA replication. In humans, mutations in the proteins MutSα and MutLα that initiate MMR cause Lynch syndrome, the most common hereditary cancer. MutSα surveilles the DNA, and upon recognition of a replication error it undergoes adenosine triphosphate-dependent conformational changes and recruits MutLα. Subsequently, proliferating cell nuclear antigen (PCNA) activates MutLα to nick the error-containing strand to allow excision and resynthesis. The structure–function properties of these obligate MutSα–MutLα complexes remain mostly unexplored in higher eukaryotes, and models are predominately based on studies of prokaryotic proteins. Here, we utilize atomic force microscopy (AFM) coupled with other methods to reveal time- and concentration-dependent stoichiometries and conformations of assembling human MutSα–MutLα–DNA complexes. We find that they assemble into multimeric complexes comprising three to eight proteins around a mismatch on DNA. On the timescale of a few minutes, these complexes rearrange, folding and compacting the DNA. These observations contrast with dominant models of MMR initiation that envision diffusive MutS–MutL complexes that move away from the mismatch. Our results suggest MutSα localizes MutLα near the mismatch and promotes DNA configurations that could enhance MMR efficiency by facilitating MutLα nicking the DNA at multiple sites around the mismatch. In addition, such complexes may also protect the mismatch region from nucleosome reassembly until repair occurs, and they could potentially remodel adjacent nucleosomes.
Maintaining the integrity of the DNA genome is essential to all organisms, and the DNA mismatch repair (MMR) system plays a major role in mutation avoidance. MMR proteins identify and correct DNA synthesis errors that occur during replication, and they are involved in several other DNA transactions, including DNA damage-induced apoptosis, double-strand break repair, and recombination (1–5). Inactivation of MMR genes not only increases the frequency of mutations, it also decreases apoptosis, increases cell survival, and results in resistance to many chemotherapeutic agents (6–8). In humans, mutations in the MMR initiation proteins MutSα (MSH2-MSH6) or MutLα (MLH1-PMS2) cause Lynch syndrome, the most common hereditary cancer (3, 9–13).
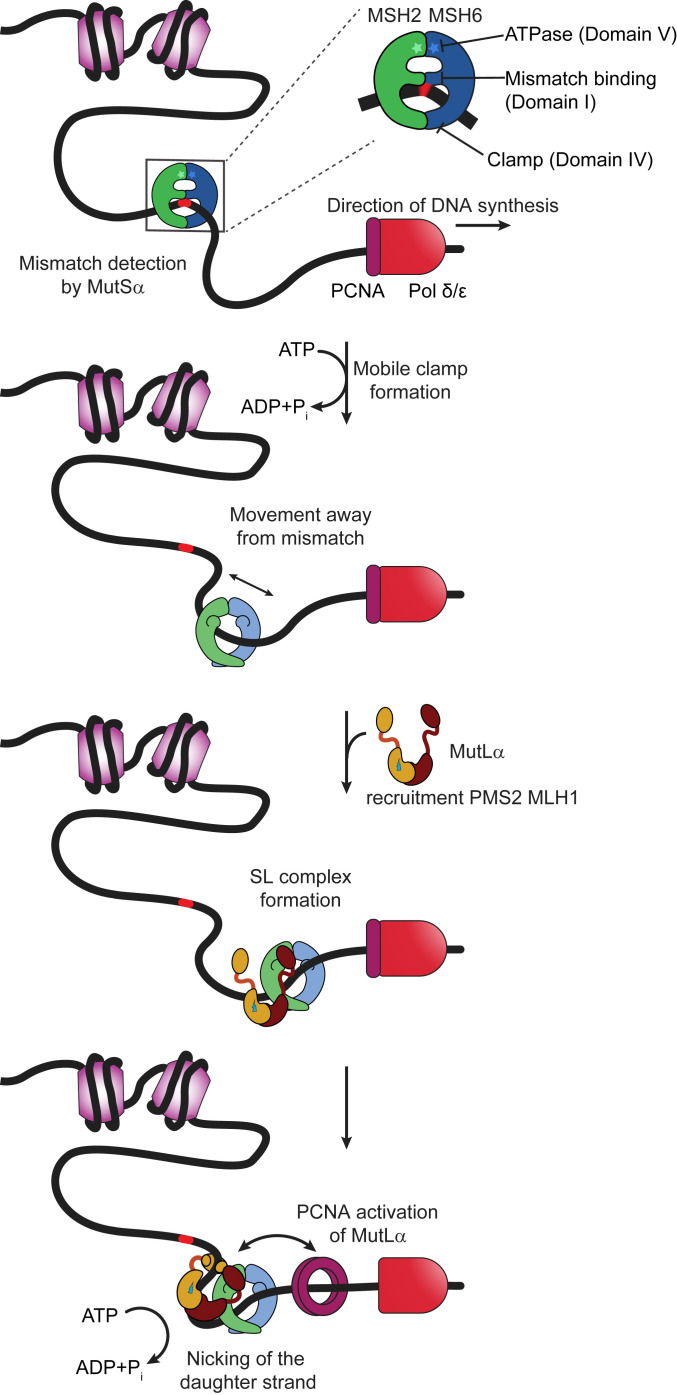
In all organisms, MMR is initiated by the highly conserved dimeric MutS and MutL homologs, which both contain DNA binding and ATPase activities (Fig. 1). Prokaryotic MutS or eukaryotic MutSα [collectively noted as MutS(α)] surveilles newly replicated DNA for mismatches and insertion–deletion loops (IDLs). After recognizing an error, MutS(α) undergoes adenosine triphosphate (ATP)-dependent conformational changes to form a clamp that can move along the DNA (1, 2, 4, 14–23). These conformational changes also promote its interaction with one or more MutL or MutLα proteins [collectively noted as MutL(α)] (1, 2, 4, 15, 16, 18, 22). In most organisms, with the exception of a few bacteria that utilize methyl-directed mismatch repair, such as Escherichia coli, MutL(α) contains a latent ATP-dependent endonuclease activity that is essential for repair (24–26). Subsequent interaction of the MutS(α)–MutL(α)–DNA (SL) complex with the mobile DNA polymerase processivity clamp (proliferating cell nuclear antigen [PCNA] in eukaryotes or β-clamp in prokaryotes) activates MutL(α) to preferentially nick the daughter strand in the region containing the error in an ATP-dependent manner (24, 27, 28). Once MutLα nicks the DNA 5′ to the mismatch, MutSα or MutLα activates the 5′-3′ exonuclease EXO1 to excise the DNA containing the incorrect nucleotide (29–33), or MutSα promotes POLδ/ε to initiate strand-displacement synthesis from the 5′-nick (34). Finally, DNA resynthesis is catalyzed by DNA polymerases δ or ε, and DNA ligase seals the nick (35–37).
Fig. 1.
Schematic of mammalian mismatch repair initiation showing mismatch recognition, mobile clamp formation, MutLα recruitment, and PCNA activation of MutLα. Further description is provided in the text. MutSα is depicted as a green (MSH2) and blue (MSH6) theta-like dimer, with the ATPase sites indicated by stars at the top of the theta. The middle DNA binding domain of MSH6 interacts specifically with the mismatch. Nucleosomes are represented as light purple cylinders, DNA polymerase as red bullet, and PCNA as a dark purple ring. MutLα is depicted showing the C-terminal dimerization domains of MLH1 (burgundy) and PMS2 (ochre) connected by long flexible linker arms to the N-terminal DNA binding and ATPase domains. The endonuclease site on PMS2 is shown as a lightning bolt.
Although little is known about the mechanisms by which the obligate repair complexes of MutS(α) and MutL(α) assemble on mismatch DNA, structural studies of MutS(α) and MutL(α) provide frameworks that guide models of MMR initiation. Crystal structures of MutS(α) mismatch recognition complexes show that MutS(α) forms a theta-like structure with two channels: one encircling and bending the DNA and the other empty. The ATPase domains are on the distal end from the DNA binding channel (Fig. 1). The DNA binding site is made up of four mobile domains (two on each subunit), with one of the middle domains (on MSH6) that separates the two channels interacting specifically with the mismatch (38–41). ATP binding drives large conformational changes in the four mobile domains, such that the middle domains open, resulting in a single larger channel for the DNA (17, 20). The structural rearrangements associated with opening this larger channel also create the binding site for MutL(α), establishing the structural mechanism by which ATP promotes the recruitment of MutL(α) to MutS(α) (20).
MutL(α) dimerizes via the C-terminal domains, which are linked to the N-terminal domains by long flexible linker arms (42–45). The N-terminal domains contain ATPase and DNA binding activities (46–48), and the endonuclease site resides in the C-terminal domain (in PMS2 in eukaryotes) (24–26, 49). In E. coli MutL, which does not have endonuclease activity, ATP promotes dimerization of the N-terminal domains (46); however, in organisms that do not utilize methyl-directed MMR, the N-terminal domains do not appear to undergo ATP-induced dimerization (50–52). Instead, ATP drives asymmetric condensation of the MLH1 and PMS2 linker arms, bringing the N- and C-terminal domains together in eukaryotes (42). These conformational changes result in the DNA binding domains and the endonuclease site being in close proximity (42), suggesting that these ATP-induced changes orient the DNA in the endonuclease site for cleavage. Importantly, specific nicking of the error-containing daughter strand requires that the mobile processivity factor, PCNA, associate with MutLα to activate and direct the endonucleolytic cleavage (24, 27, 28). A crucial precursor to activating MutLα’s endonuclease activity is the mismatch- and ATP-dependent formation of an SL complex; however, the nature of this complex remains enigmatic, with several disparate models being proposed.
The dominant model, which has been strongly motivated by studies of E. coli MMR, posits that MutL(α) joins MutS(α) to form hydrolysis-independent SL mobile clamps that diffuse on DNA in search of the strand discrimination signal, which is a hemimethylated dGATC site in E. coli (22, 53). Recent studies also suggest that the diffusive SL mobile clamps can separate to allow MutL alone to diffuse to the hemimethylated dGATC site (22, 54). Because the signal in non–methyl-directed MMR, PCNA, is itself mobile, such mobile SL complexes may not be necessary in this more widely employed MMR paradigm. In addition, the mobile SL-clamp model is challenged as the primary signaling mechanism in MMR by in vitro and in vivo studies that show MutS(α) and MutL(α) form multimeric assemblies on mismatch-containing DNA (16, 18, 55–58), that MutL(α) dramatically slows MutS(α) dissociation from DNA with free ends (18, 20, 22, 59), and that PCNA-induced MutLα nicking of the daughter strand occurs at preferential sites near the mismatch (24, 25). These results suggest an alternative pathway for MMR initiation involving SL complexes that do not freely diffuse on DNA, and three additional models have been proposed (1, 2): one which invokes ATP-hydrolysis–dependent movement (60, 61), one that suggests MutS(α) and MutL(α) remain at the mismatch and signal via DNA looping (55, 62), and one in which MutS(α) induces the polymerization of MutL(α) with varying stoichiometries around the mismatch (2, 15, 16, 18, 57, 63). This diversity of models highlights the complexity of MMR, which requires the coordinated assembly of transient dynamic complexes of multiple proteins on the DNA. The MMR cascade is particularly sensitive to stochastic variations because the behaviors and functions of MutS(α) and MutL(α) depend on the timing and sequence of their interactions with one another and with adenosine nucleotides, DNA, and other proteins in the pathway. Such diversity limits synchronization of the process and can lead to heterogeneous populations of complexes. Studies rich enough in information to unify all previous results with a single model are lacking, especially for eukaryotic MMR proteins.
Here, we use atomic force microscopy (AFM), which provides information about conformations and stoichiometries of proteins and protein–DNA complexes (64–69), to study the assembly of human MutSα and MutLα proteins on mismatch-containing DNA. Because AFM allows characterization of individual complexes, it is particularly powerful for revealing the properties of heterogeneous populations. Our experiments reveal the assembly of multimeric complexes containing multiple MutSα and MutLα proteins (complexes with three to eight proteins), with the majority of these complexes residing at or near the mismatch. Examining the properties of the complexes at different incubation times and protein concentrations provides a window into the assembly pathways. Our data suggest that, following the initial mismatch binding by MutSα, the complexes assemble stochastically in a stepwise fashion with one or two MutSα loading onto the DNA, followed by recruitment of one or more MutLα proteins. Unexpectedly, we also find that these complexes reconfigure and compact the DNA over time. These complexes may mark and protect the region around the mismatch, and they also may explain the observation of enhanced multiple MutLα nicking in proximity to the mismatch, which could enhance repair efficiency (15, 24).
Results
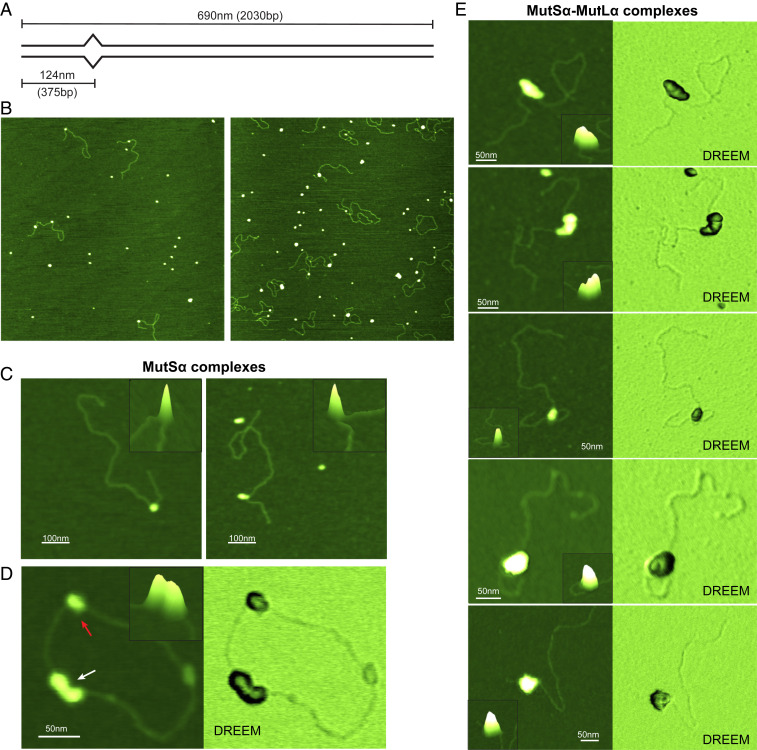
We used AFM to examine the properties of human MutSα and MutSα–MutLα (SL) complexes bound to 2-kbp DNA fragments that are perfectly paired (GC-DNA) or that contain a single GT mismatch 375 bp (124 nm) from one end (GT-DNA; Fig. 2A). We used a GT mismatch because it is well-characterized in MMR studies, especially in the repair reconstitution experiments using human proteins (4, 36, 70, 71). We incubated different concentrations of MutSα, MutLα, and adenosine diphosphate (ADP) or ATP with GC- or GT-DNA for varying lengths of time, cross-linked the complexes for 1 min with 0.85% glutaraldehyde, and deposited them on a mica surface for imaging (Methods and Fig. 2B). Representative images of GT-DNA deposited in the presence of MutSα or MutSα+MutLα show that the protein complexes are easily resolved on DNA (Fig. 2 and SI Appendix, Fig. S1). Because we know the position of the mismatch on the DNA, we can determine whether MutSα or the SL complexes are bound at the mismatch (specific complex) or at flanking homoduplex sites (nonspecific complex) by measuring the position of the complex relative to the ends of the DNA (Methods). In addition, comparison of the measured contour lengths of the free DNA versus protein-bound DNA reveals any compaction or DNA wrapping caused by the protein–DNA interactions (67, 69, 72). Finally, the volumes of the protein complexes in the AFM images provide an estimate of the number of proteins within each complex (65, 66, 68, 73).
Fig. 2.
Images of MutSα and MutSα–MutLα complexes bound to GT-DNA. (A) Schematic view of the GT-DNA substrate used in this study. The length of the DNA fragment and the position of the mismatch (depicted as bulge) in base pairs and in nanometers from the nearest end are shown. (B) Representative 2-μm × 2-μm top-view images of MutSα (Left) or MutSα+MutLα (Right) deposited in the presence of GT-DNA and ATP. (C) Zoomed 3D topographic images of MutSα–GT–DNA complexes containing one (Left) or two (Right) MutSα proteins. (D) Topographic (Left) and DREEM (Right) images of MutSα on circular GT-DNA showing complexes containing one (red arrow) and three MutSα (white arrow) proteins in the complex. DREEM images show the DNA passing through the MutSα proteins. (E) Topographic and DREEM images of MutSα–MutLα–GT-DNA complexes showing different sizes and increasingly compacted structures. Control experiments show that the larger complexes seen in the presence of MutSα, MutLα, and ATP do not form on GC-DNA and that MutLα alone with ATP exhibits no significant binding to DNA (SI Appendix, Figs. S2H and S4). Scale bars are shown in white. (Insets, C–E) Zoomed-in 3D topographic views of the complexes. Cartoons depicting possible complex conformations are shown in Fig. 5. Additional images are shown in SI Appendix, Figs. S1 and S5 A and B.
ATP Promotes MutSα–MutSα Interactions on Mismatched DNA.
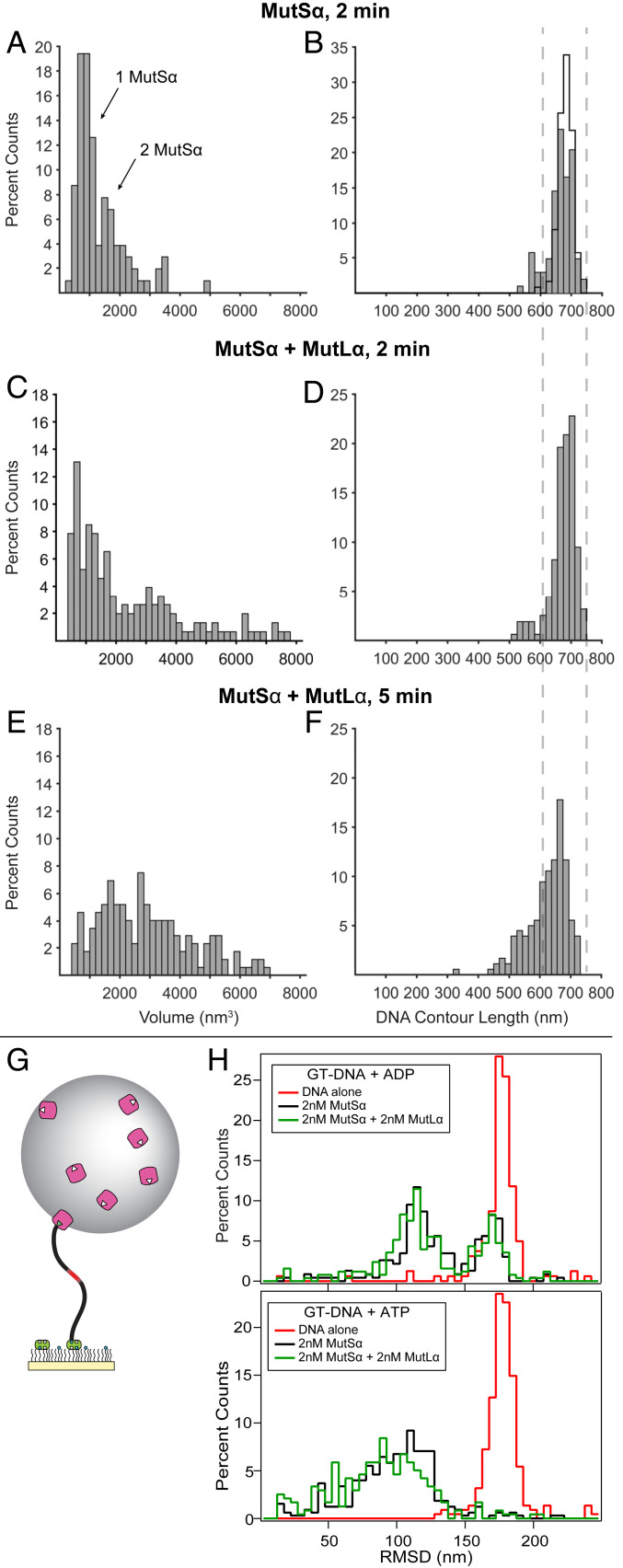
On GC-DNA in the presence of ADP or ATP, MutSα exhibits a random distribution of positions on the DNA (SI Appendix, Fig. S2 A, Right), as expected. In contrast, on GT-DNA in the presence of ADP, the position distribution of MutSα shows a sharp peak at the position of the mismatch (∼120 nm; SI Appendix, Fig. S2 A, Left), indicating that a MutSα binds the mismatch with high specificity, as expected based on DNA binding studies (71, 74). ATP moderately decreases the percentage of specific MutSα complexes relative to ADP (SI Appendix, Fig. S2A), consistent with DNA binding measurements that show an ∼10-fold lower affinity for a GT mismatch in ATP compared to ADP (71, 74). The distributions of measured volumes of MutSα–DNA complexes on GT-DNA in the presence of ADP and on GC-DNA in the presence of ADP or ATP each show a single peak at ∼800 nm3 (SI Appendix, Fig. S2 B and C; data not shown), which is similar to the volume of free MutSα (SI Appendix, Fig. S3A) and thereby establishes the volume of a single MutSα bound to DNA (Methods).
Interestingly, in the presence of ATP, many of the MutSα–GT–DNA complexes, both specific and nonspecific, appear to contain more than one MutSα (Fig. 2 B–D and SI Appendix, Fig. S1A and Table S1), and the volume distribution exhibits two main peaks at ∼800 nm3 and ∼1,600 nm3, consistent with one and two MutSα per complex, respectively (Fig. 3A and SI Appendix, Fig. S2D). Similar results are seen using nicked plasmid (circular) GT-DNA (Fig. 2D and SI Appendix, Fig. S2E), although it is not possible to distinguish between specific and nonspecific complexes. Some of these of these complexes are globular, with the two proteins indistinguishable; however, many follow the contour of the DNA molecule, with each protein clearly interacting with the DNA (Fig. 2 C and D and SI Appendix, Fig. S1A). This latter result suggests that the MutSα–MutSα complexes are formed via sequential rounds of mismatch binding and ATP-induced mobile clamp formation by MutSα, followed by collision of the mobile clamps on the DNA (or a mobile clamp and a second MutSα bound at the mismatch). These results further suggest that the mobile clamp state also facilitates MutSα–MutSα interactions. Pertinent to this observation, the C-terminal domain of prokaryotic MutS is known to promote oligomerization, and mutations in this domain in MutSα are associated with hereditary nonpolyposis colorectal cancers (HNPCCs) (ref. 3; http://insight-database.org/).
Fig. 3.
MutSα–MutLα complexes compact GT-DNA. (A–F) Distributions of AFM volumes (left column) and total DNA contour lengths (right column) for MutSα and MutSα–MutLα complexes bound to GT-DNA. Volumes (A, C, and E) and lengths (B, D, and F) of protein–DNA complexes for MutSα incubated in the presence of GT-DNA and ATP for 2 min (A and B; n = 103) and for MutSα+MutLα incubated in the presence of GT-DNA and ATP for 2 min (C and D; n = 199) and 5 min (E and F; n = 173). Both specific and nonspecific complexes are included in the analyses. Cityscape in B shows the length distribution for free DNA. Dashed lines across B, D, and F show ±1 SD of the measured free DNA length. Additional data using nicked plasmid GT-DNA as a substrate are included in SI Appendix, Figs. S2E and S5. (G) Cartoon of tethered particle motion assay showing mismatched DNA tethered to a surface with bead attached (not to scale). (H) Histograms of the RMSDs of many DNA molecules using the tethered particle motion assay with 550-bp GT-DNA. (Upper) Experiments with 2 mM ADP without protein (red; n = 161), with 2 nM MutSα (black; n = 231), and with 2 nM each of MutSα+MutLα (green; n = 252). (Lower) Experiments with 2 mM ATP without protein (red; n = 224), with 2 nM MutSα (black; n = 326), and with 2 nM each of MutSα+MutLα (green; n = 238). No significant changes in bead motion were observed with GC-DNA for either MutSα alone or MutSα and MutLα in the presence of ATP (SI Appendix, Fig. S6).
MutSα and MutLα Form Multimeric Complexes on Mismatched DNA.
Images and volume analyses from depositions containing MutSα, MutLα, and GT-DNA in the presence of ATP reveal larger complexes on DNA than are seen with MutSα alone (Figs. 2 and 3 A and C and SI Appendix, Fig. S1). In control experiments, we rarely observe MutLα bound to DNA under any conditions used (<3% compared to >50% for SL complexes; SI Appendix, Fig. S4). To capture different stages of the SL complex assembly, we varied MutSα, MutLα, and ATP concentrations and incubation times. The distributions of complex sizes shift to larger volumes with higher protein concentrations and longer incubation times (Fig. 3 C and E and SI Appendix, Fig. S2 F and G). In all conditions, the volumes range from ∼800 nm3 (single MutSα) to ∼8,000 nm3 (6 to 10 proteins; Methods), with the majority having volumes <4,000 nm3 (<5 to 6 proteins). Notably, the number of proteins in the SL complexes is similar to that which we determined for Thermus aquaticus (Taq) SL complexes using photobleaching (18) and is consistent with surface plasmon resonance (SPR) studies on eukaryotic proteins (25, 59). Multimeric complexes of E. coli MutS and MutL have also been detected with AFM on mismatched DNA (75). The larger complexes detected in our study may correlate to the foci of fluorescent-protein fusions of MMR proteins observed in live cells, which appeared to contain ∼6 to 11 MutLα proteins (57).
At 50 nM of each MutSα and MutLα, ∼30% of the complexes exhibit volumes consistent with SL complexes, but their sizes are smaller (2,000 nm3 to 4,000 nm3; SI Appendix, Fig. S2F) than those at 125 nM of each MutSα and MutLα (2,000 nm3 to 8,000 nm3; Fig. 3 C and E and SI Appendix, Fig. S2G). In addition, at 125 nM concentrations, the population with volumes consistent with MutSα alone (<2,000 nm3) decreases over time (2 min vs. 5 min), with a concomitant increase in the population with volumes indicative of SL complexes (2,000 to 8,000 nm3; Fig. 3 C and E). Notably, the SL complexes do not grow without bound, and the largest complexes are limited to volumes of ∼8,000 nm3. The limited size of these complexes suggests that MutLα may be joining and leaving. This suggestion is consistent with previous studies showing dynamic SL complexes (59, 76) and with our observation that 10-fold dilution of these samples without cross-linking before deposition results in their dissociation in less than ∼1 min (Methods). Together, these data suggest that the majority of MutSα–GT–DNA complexes formed in the presence of ATP will eventually convert to SL complexes.
The MutSα and the MutSα+MutLα data, together with previous experimental studies, lead us to propose that these complexes likely contain one or two MutSα proteins and varying numbers of MutLα proteins. Several observations support this proposal. Single-molecule fluorescence studies showed that both human MutLα and Taq MutL limit multiple loading of MutS(α) onto mismatched DNA to one to three MutS(α), in contrast to the up to six in the absence of MutL(α) (18, 77). In addition, SPR measurements find superstoichiometric responses for complexes formed on mismatched DNA when using MutSα+MutLα compared to MutSα alone (25, 59). Finally, live cell studies with fluorescent-protein fusions of MMR proteins find foci that contain more MutL(α) than MutS(α) (57, 58).
Our series of experiments also allowed us to visualize the conformations of different assembly states of MutSα and MutLα on GT-DNA. For those complexes with volumes large enough to contain both MutSα and MutLα (>2,000 nm3), the conformational states can be categorized into three classes: complexes that are assembled linearly along the DNA, globular complexes in which individual protein peaks are indistinguishable, and complexes that are intermediate between these two, exhibiting assembly along the DNA but with bent conformations (Fig. 2E and SI Appendix, Fig. S1B). We observe these three classes of classes of conformations in all conditions; however, the linear and bent species are more common at short incubation times (1 min) and low protein concentrations, while, at longer times (2 min and 5 min), the globular species become dominant. Interestingly, we also observe protein-mediated DNA looping (Fig. 2E and SI Appendix, Fig. S1B) in ∼10% of complexes in each condition, with ∼95% of the loops involving the mismatch. Loops are rarely observed (<1%) with MutSα alone on linear DNA, suggesting MutLα is important for their formation. Experiments with E. coli MMR proteins have observed loops mediated by MutS alone as well as MutS and MutL (60, 75, 78). As discussed later, these SL complex shapes may reflect different steps in the assembly of MutSα and MutLα after mismatch recognition by MutSα.
MutSα–MutLα Interactions Compact Mismatched DNA.
A striking finding is that, after formation, the SL complexes appear to undergo reorganization over time, leading to compaction of the DNA within the complex (Fig. 3 C–F). Specifically, at 2 min, the distribution of DNA contour lengths exhibits a major peak that overlaps with the distributions of both free DNA and MutSα–DNA complexes, with a small shoulder at shorter lengths (Fig. 3 B and D). At 5 min, the shoulder peak height increases, and both the shoulder and main peaks shift to shorter lengths relative to 2 min (Fig. 3F vs. Fig. 3 B and D). The shorter DNA lengths suggest that some SL complexes can contain 50 to 300 bp in a compacted configuration. To glean qualitative information about the conformation of DNA within these complexes, we examined a few SL complexes using dual resonance frequency-enhanced electrostatic force microscopy (DREEM), which is sensitive to electrostatic force gradients and can reveal the DNA path within protein–DNA complexes (64, 79–82) (Fig. 2 D and E). In the linear SL complexes (or MutSα alone), the DNA appears to pass through the center of the proteins. In the bent complexes and the rare, larger globular complexes, the DNA appears to be folded inside (Fig. 2E). This DNA folding within the complex appears to account for the DNA shortening measured from the topographic images (Fig. 3F).
As a control, we examined MutSα–MutLα assembly on nicked plasmid (circular) GT-DNA (SI Appendix, Fig. S5), which mimics substrates that are used for in vitro DNA mismatch repair assays (4, 71), although it is not possible to distinguish between specific and nonspecific complexes. We observe a broader distribution of complex sizes, with an increase in larger complexes and a greater protein-induced DNA shortening on nicked plasmid GT-DNA relative to linear GT-DNA (SI Appendix, Fig. S5 C and D); however, the overall conformations and properties of the complexes are similar to those seen on linear GT-DNA (Fig. 2 B and E and SI Appendix, Figs. S1B and S5 A and B). The larger complexes likely result from the stable loading of multiple MutSα proteins onto nicked plasmid GT-DNA, which in turn may promote the recruitment of more MutLα. Linear DNA not only allows the distinction between specific and nonspecific complexes but also allows the observation of early events in the assembly of MutSα–MutLα complexes on mismatched DNA.
Given the finding of DNA shortening from the AFM experiments, we sought independent in-solution evidence for MutSα-MutLα–induced DNA compaction using tethered particle motion (TPM) experiments (83–85). For the TPM experiments, we use a 550-bp DNA substrate with a single central GT mismatch (or a GC for homoduplex DNA) tethered to a surface by one end and with a bead attached to the other end (Fig. 3G). The Brownian motion of the bead correlates with the length and/or flexibility of the DNA and reports its configurations in solution (Methods and SI Appendix, Methods). The bead motion is characterized by the root mean square displacement (RMSD) of the excursions around its center attachment point. For DNA in the absence of protein, the distributions of RMSDs for many molecules exhibits a single peak centered around 180 nm (Fig. 3H and SI Appendix, Fig. S6 A and B). In the presence of ADP, addition of MutSα results in a new peak in the RMSD distribution at ∼120 nm (Fig. 3H), indicating reduced Brownian motion of the bead-bound GT-DNA end. The decreased RMSD is consistent with MutSα-induced DNA bending (41, 83, 86). A peak in the RMSD distribution at the position of free DNA remains detectable, which indicates that not all of the DNA is bound by MutSα. This partial occupancy is expected based on the MutSα concentration used (2 nM) and the KD we previously determined (8.9 ± 8.8 nM) (71). The RMSD distribution does not significantly change upon including MutLα with MutSα in the presence of ADP (Fig. 3H), which likely reflects the ATP requirement for MutSα to recruit MutLα (1, 2).
In the presence of ATP and MutSα, the free DNA peak in the RMSD distribution is absent, which is expected based on the 2-min incubation time before measurement and the observation that MutSα forms long-lived mobile clamps on end-blocked, mismatch-containing DNA (71, 77) (Methods). The protein–DNA peak is broadened with a shoulder extending to shorter lengths compared to ADP (Fig. 3H). This broader RMSD distribution peak likely reflects the multiple types of MutSα–DNA complexes, with varying numbers and conformations of MutSα that were observed by AFM (Figs. 2 C and D and 3A and SI Appendix, Fig. S1A). In the presence of both MutSα and MutLα with ATP, the DNAs convert to protein complexes, with further reduction of bead motion beyond MutSα alone (Fig. 3H). Specifically, statistical analysis comparing the distributions of MutSα alone to MutSα+MutLα using the two-sample Kolmogorov–Smirnov test (SI Appendix, Methods) confirms that the RMSD distributions for MutSα+MutLα are shifted to significantly shorter lengths in the presence of ATP (P = 0.001) but not ADP (P = 0.035), consistent with the DNA shortening that we see in our AFM experiments (Figs. 3F and 4 and SI Appendix, Fig. S5D). The breadth of the RMSD distributions is also consistent with our AFM experiments in which we observe a population of complexes with a broad range of sizes (Fig. 3 C and E and SI Appendix, Fig. S5C). In control experiments with homoduplex DNA and ATP, the RMSD distributions are unchanged from DNA alone when MutSα or MutSα+MutLα are included (SI Appendix, Fig. S6A). Together, these TPM results bolster the AFM observation of SL-induced DNA shortening in the presence of ATP. Our observation of the unexpected formation of dynamic, multimeric SL complexes that can compact DNA over time is supported by prior in vitro and in vivo studies that found recruitment of multiple MutL(α) and MutS(α) proteins to mismatched DNA (57–59, 76) and protection of significantly larger segments of DNA in the presence of both E. coli MutS and MutL than MutS alone (55, 56).
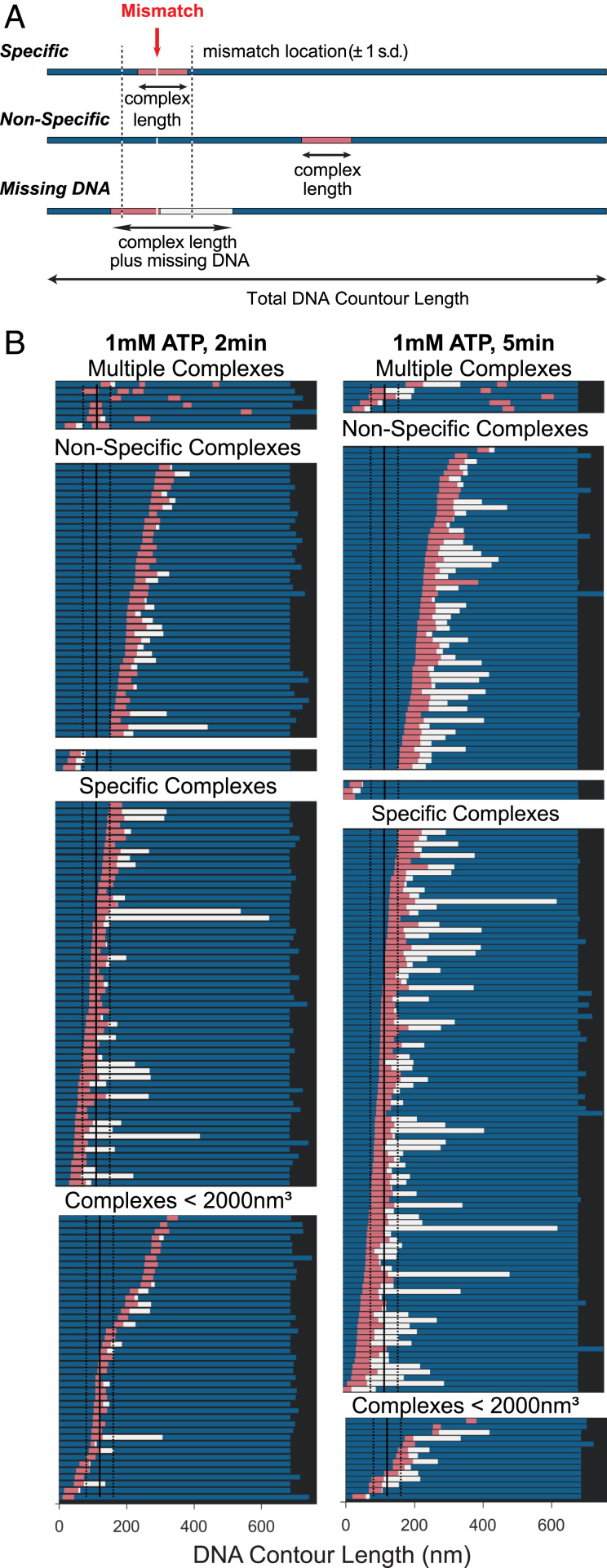
Fig. 4.
Position analysis of MutSα–MutLα complexes bound to GT-DNA. (A) Schematics of DNA molecules (represented as bars) showing location of MutSα–MutLα complex and extent of DNA contained within the complex. Schematics for three types of complexes are shown: specific, mismatch within complex (Top); nonspecific, complex away from the mismatch (Middle); and missing DNA, complexes where the DNA is shorter than ±1 SD of the distribution of DNA contour length (Bottom). For all, the bar denotes the total DNA contour length and the pink section is the length of the complex on the DNA. The blue sections represent the length of the DNA observed on either side of the complex. The white section (in missing DNA example) represents the missing DNA length for molecules with lengths less than 1 SD of free DNA. The total amount of DNA that the protein complex covers is the summation of the pink and white bars. On the top example, a red arrow points to the mismatch site, and dashed black lines on either side indicate ±1 SD of the measured position of the mismatch on the DNA. (B) Positions of MutSα–MutLα complexes on GT-DNA in the presence of 1 mM ATP incubated for 2 min (Left) or 5 min (Right). Complexes are first separated into two groups: complex volumes <2,000 nm3 consistent with one or two MutSα (Bottom) and >2,000 nm3 consistent with MutSα–MutLα complexes (Upper). Those complexes with volumes >2,000 nm3 (SL complexes) are separated into specific and nonspecific complexes. Each group is sorted from top to bottom based on the position of the complex relative to the nearest end to the mismatch, and those with multiple complexes are shown on the top. DNA is only considered to be compacted if the contour length is shorter than the 1 SD in the length measurements. These plots are generated from the same data as in Fig. 3 C–F.
MutSα–MutLα Complexes Reside Both at the Mismatch and at Adjacent Sites.
Shortening of the DNA complicates determination of whether the SL complexes in the AFM images encompass the mismatch. To address this challenge, we generated a series of bar graphs that display both the positions of SL complexes on DNA and the length of DNA contained within individual complexes, including the DNA that is absent from the measured contour and is presumed to be buried within the complexes (Fig. 4A). Each bar denotes the total DNA contour length. The pink section is the length of the complex on the DNA, and the blue sections represent the length of the DNA observed on either side of the complex. For molecules with lengths shorter than free DNA, the white section represents the missing DNA length. The total amount of DNA that the protein complex covers is the summation of the pink and white bars. Fig. 4B shows data for the 2-min (left column) and 5-min (right column) incubations of MutSα and MutLα in the presence of ATP. The few DNAs that have multiple complexes bound are grouped together (Fig. 4 B, Top). For the remaining complexes, the data for each condition are then grouped based on the protein complex volume to separate MutSα–MutLα–DNA complexes (>2,000 nm3; Fig. 4 B, Middle) from potential MutSα–DNA complexes (<2,000 nm3; Fig. 4 B, Bottom). Those complexes with volumes consistent with SL complexes are then separated into specific and nonspecific complexes. Comparison of the data for the 2-min and 5-min incubations shows increased “missing DNA” (Fig. 4, white bars) with increased incubation time, as is also revealed in the DNA length distribution plots (Fig. 3 D and F). In addition, these data show that SL complexes are located within the region of the mismatch and at nonspecific sites for both the 2-min and 5-min incubations, with the majority encompassing the mismatch (Fig. 4B). The observation of nonspecific SL complexes indicates that (i) MutLα can assemble into SL complexes with MutSα mobile clamps that have moved away from mismatch, (ii) SL complexes formed at a mismatch can move away, or both. Studies showing that MutL can stop MutS at a mismatch (18) and stop or dramatically slow MutS mobile clamps (20, 22, 87) support the former mechanism, and our observation of dynamic SL complexes that reconfigure mismatched DNA (Fig. 3 D and F) supports the latter suggestion. Notably, these mechanisms of nonspecific complex formation are not mutually exclusive, and both may occur in our experiments and in vivo.
Discussion
Unifying Model of Stochastic Pathways to Assemble MutSα–MutLα Complexes That Compact Mismatched DNA.
Following MutSα recognition of a mismatch and recruitment of MutLα, a key step in DNA MMR is the ATP- and PCNA-dependent activation of MutLα to nick error-containing daughter strands (24). The fundamental importance of this step is evidenced by the observation that inactivation of MutLα’s ATPase or endonuclease activity completely abrogates repair (25, 26, 44, 47, 88, 89). Despite the importance of ATP- and mismatch-dependent eukaryotic SL complexes, virtually nothing is known about their assembly states or the conformations. Using AFM and other single-molecule techniques, we provide a picture of the dynamic assembly and the configurations of human SL MMR complexes on mismatched DNA.
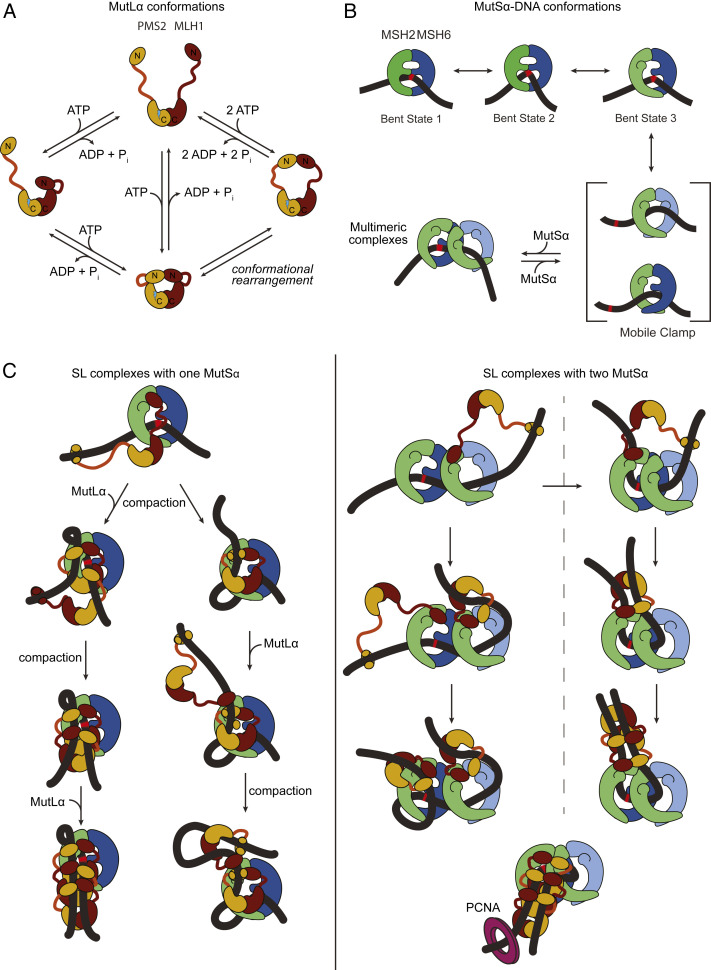
Taking our results on MutSα together with those from MutSα+MutLα suggests that the SL complexes are formed in a stepwise fashion with one or two MutSα loading onto the DNA, followed by recruitment of one or more MutLα proteins as diagramed in Fig. 5 B and C. This idea is consistent with single-molecule fluorescence studies on Taq and human MMR proteins, which showed that MutL(α) limits loading of MutS(α) to one to three proteins (18, 77). The SL complexes observed in our AFM experiments appear to assemble “linearly” along the DNA and, over time, evolve to more globular forms (in which individual protein peaks are indistinguishable) that can reconfigure the DNA (Fig. 2E). This DNA reconfiguration involves compaction of DNA within the protein complexes and, in some cases, loop formation (Fig. 2E and SI Appendix, Fig. S1B). MutLα’s abilities to simultaneously interact with two double strands of DNA via its N-terminal domains (47, 90, 91) and to undergo large ATP-induced asymmetric conformational changes (Fig. 5A) (42) may promote this reorganization. For example, DNA reconfiguration will result if one of the MutLα N-terminal domains binds distally on the DNA with that arm in an extended state (as in Fig. 5C), followed by nucleotide-induced retraction of that arm toward the C-terminal domains containing the endonuclease site. This process is stochastic, and the specific DNA location where the MutLα N-terminal domain binds will determine the details of the final compacted state. Reaching nearby will result in a compacted complex where the DNA is within the complex, whereas reaching far away will result in a loop where the DNA is exposed. The higher probability of binding adjacent DNA compared to binding distant sites may be reflected in the small population of loops that we observe. MutSα may also contribute to the DNA reconfiguration, given our finding that ATP-activated MutSα can self-associate (Figs. 3A and 5B).
Fig. 5.
Example models of potential pathways to the formation of compacted MutSα–MutLα–GT-DNA complexes. (A) Previously identified conformational states of MutLα (42). MLH1 and PMS2 are shown in burgundy and ochre, respectively. The C-terminal dimerization domains are connected to the N-terminal ATPase and DNA binding domains by a flexible linker. The endonuclease site of PMS2 is shown as a lightning bolt. (B) Model for MutSα recognition, mobile clamp formation, and association of multiple MutSα. The early states (bent states 1 to 3) are from studies of Taq MutS (19), and the two mobile clamp conformations are based on the crystal structure of E. coli MutS mobile clamp in complex with the N-terminal domains of MutL (20), in conjunction with our recent single-molecule fluorescence studies that indicate that Taq MutS mobile clamps can exist in two conformations (87). (C) Example models for formation of SL complexes containing a single MutSα (Left) or two MutSα (Right). The Discussion includes a detailed description. Complexes are shown at the mismatch, but they can also occur at nonspecific sites on GT-DNA. These models also suggest that loop formation and DNA compaction could both result from the MutLα N-terminal domain binding distally on the DNA with one of its arms in an extended state (as in A), followed by nucleotide-induced retraction of that arm toward the C-terminal domain containing the endonuclease site. The lower right model depicts how PCNA (purple ring) could interact with an SL complex with antiparallel DNA strands to activate MutLα to nick on either side of the mismatch depending on the orientation of MutLα (15). The conformations of MutSα and MutLα depicted in the model are based on the conformations shown in A and B, and the location of the interactions between MutSα and MutLα are derived from the crystal structure of E. coli MutS sliding clamp in complex with the N-terminal domains of MutL (20) and from hydrogen/deuterium exchange mass spectrometry studies of E. coli MutS and MutL coupled with functional studies of yeast MutSα and MutLα (92). Models are only representative and are not intended to imply specific pathways or any structural details, but rather to provide ideas of how MutSα–MutLα complexes could compact the DNA.
MutSα–MutLα Complexes May Mark and Protect the Mismatch until Repair Occurs.
For repair to occur, complexes of MutSα and MutLα need to form on the DNA before nucleosomes reassemble behind the replication fork because DNA that is packaged in nucleosomes is refractory to repair (93, 94). Furthermore, PCNA-induced MutLα nicking in the vicinity of the mismatch should enhance repair efficiency by localizing the excision tracts to the mismatch-containing region. Our observations provide a mechanistic framework that successfully interprets previous studies of MMR and establishes a framework for thinking about how MutSα and MutLα initiate repair (Fig. 5).
The picture of SL complex assembly that emerges from our studies commences with MutSα binding a mismatch. Upon mismatch recognition, MutSα undergoes ATP-induced conformational changes that license MutLα interaction and mobile clamp formation. Interaction of MutLα with MutSα on the pathway to mobile clamp formation results in a mismatch-localized SL complex (18). Subsequent interaction with PCNA can activate MutLα within such a complex to nick the daughter strand locally to the mismatch, thereby allowing excision and repair to commence. If MutLα does not associate with MutSα quickly enough to trap it at the mismatch, MutSα can convert into a mobile clamp and free the mismatch for additional MutSα loading. Repetitive rebinding of the mismatch by MutSα mobile clamps, similar to that recently observed for Taq MutS mobile clamps (87), would increase the residence time of MutSα near the mismatch and the probability that SL complexes (and nicking) are localized near the mismatch. Any of the complexes depicted in Fig. 5C, including a single MutSα–MutLα complex, are expected to be competent for activation by PCNA (15, 25, 28).
In the absence of interactions with downstream repair proteins such as PCNA or EXO1, the SL complexes may progress toward more complicated forms, with some of these reorganizing to compact DNA within the complex. Such dynamic assemblies on the exposed duplex between the replication fork and nucleosomes could have multiple functions. They could serve to prevent nucleosome assembly over mismatch-containing DNA or possibly even move nucleosomes off the mismatch. Consistent with these suggestions, studies have shown that active MMR inhibits nucleosome loading preferentially near mismatches (95, 96). Assembling SL complexes in the vicinity of the mismatch, coupled with their observed longevity, may mark and protect the mismatch until repair can proceed. Supporting this idea, SL complexes have been observed in yeast far from the replication fork (57). Marking and protection could be particularly important for late repair, such as the small fraction of repair events associated with RNase H2 nicking at ribonucleotides in the leading strand (15). We also speculate that folding of DNA by SL complexes may facilitate MutLα nicking the daughter strand on both sides of the mismatch (24, 25) by generating antiparallel configurations that bring PCNA in proximity to both sides (15) (Fig. 5C). Finally, the dynamic DNA folding properties of SL complexes could promote multiple MutLα-induced nicks in DNA around the mismatch and amplify the nicking signal (15, 24), helping efficient recruitment of EXO1 or a strand-displacing polymerase to start the next stage of DNA repair (4, 24).
The SL complexes may have a role in both activating and regulating excision of the mismatch given that interaction with either MutSα or MutLα is sufficient to enable EXO1-dependent repair (33). Multimeric SL complexes, such as those that we observe in our AFM experiments, could be activated to nick the daughter strand by PCNA and subsequently provide a scaffold that recruits and regulates EXO1 activity. Consistent with this idea, in vitro, MutSα increases EXO1 processivity, but MutLα restricts the excision tracts to shorter lengths (77). We speculate that the dynamic nature of the SL complexes coupled with their interaction with EXO1 allows them to reconfigure during excision, perhaps with MutLα dissociating. This reconfiguration could, in turn, promote the dissolution of the SL complex, dissociation of EXO1, and termination of excision. Given that a majority of the SL complexes span the mismatch in our experiments (Fig. 4), we would expect nicking and excision to be directed preferentially to the vicinity of the mismatch. Furthermore, because PCNA could promote nicking on both sides of the SL complex, EXO1 can load onto a nick, processively excise DNA from 5′-3′, and terminate at the next nick it encounters. If that terminating nick is before the mismatch, SL complexes should still be present to activate further excision. After the mismatch has been excised successfully, MutSα will no longer be recruited to the DNA, limiting further excision activity. In summary, the view of MMR initiation that emerges from our studies shifts the focus from a mobile signaling complex that leaves the mismatch to a dynamic signaling complex that remains in the vicinity of the mismatch, localizing the excision tracks and resynthesis reactions where they are required.
Materials and Methods
DNA Substrate Preparation and Protein Expression and Purification.
Human MutSα and human MutLα were purified as previously described (71, 97) and also provided by Paul Modrich (Duke University, Durham, NC). We modified a pSCW02 plasmid to make the GT-DNA substrates that were used for AFM as done previously (71, 97). To create linear GT-DNA or linear GC-DNA (using unmodified pSCW02 DNA), the plasmid was cut with Xmn1 (New England Biolabs), which results in linear DNA with the mismatch 375 bp (124 nm) from one end. Nicked plasmid DNA was generated by cutting pSCW02 with Nt.BspQ1 (New England Biolabs).
Sample Preparation and Deposition.
Freshly cleaved ruby mica discs (Spruce Pine Mica Company) were placed in a desiccator next a piece of Parafilm containing 30 μL of (3-aminopropyl) triethoxysilane (APTES) or ethanolamine for 15 min to modify the mica surface to facilitate DNA deposition. For experiments with MutSα alone, MutSα was diluted to a concentration of 125 nM with 100 μM ADP, 100 μM ATP, 500 μM ATP, or 1 mM ATP incubated with 1 ng/μL of the DNA substrate for 2 or 5 min at room temperature in imaging buffer [25 mM Hepes, pH 7.5, 100 mM NaOAc, 10 mM Mg(OAc)2, 1 mM DTT, 5% glycerol] in a total volume of 20 μL. For experiments with both MutSα and MutLα, the concentrations of MutSα, MutLα, and ATP and the length of incubation prior to cross-linking are as indicated in the figure legends. The protein–DNA samples were cross-linked with 0.85% glutaraldehyde for 30 to 60 s, and the cross-linking was stopped either by quenching with Tris or diluting 10-fold and depositing on the mica. Cross-linking conditions were optimized to minimize artifacts, and non–cross-linked control experiments with MutSα alone were conducted (SI Appendix, Fig. S3). Control experiments with MutLα and ATP confirm that it has minimal binding to GC- or GT-DNA under any of the conditions used in the SL experiments (<3% DNAs have a bound MutLα; SI Appendix, Fig. S4), and control experiments with MutSα, MutLα, ATP, and GC-DNA do not show any higher-order oligomers, with only ∼15% of the of the observed complexes having volumes >2,000 nm3 and <2% having volumes >2,800 nm3 (SI Appendix, Fig. S2H). Together, these results indicate that cross-linking does not promote nonspecific protein assemblies on the DNA. In addition, to confirm that our cross-linking conditions do not promote higher-order oligomers of MutSα or MutLα, we used SDS polyacrylamide electrophoresis to examine the cross-linking products. After cross-linking in the presence of ATP or ATP+GC-DNA or ATP+GT-DNA, the positions of the dominant band on the gel for each condition are consistent with heterodimer of MSH2-MSH6 (MutSα) or MLH1-PMS2 (MutLα), as seen previously (91, 98). A faint band with slower migration is seen for MutSα (but not MutLα) and may represent a dimer of MutSα (SI Appendix, Fig. S3). These results demonstrate that our cross-linking protocol efficiently stabilizes heterodimers, as seen in other studies (91, 98), but does not promote formation of higher-order oligomers.
In the AFM experiments, variable extents of protein–DNA cross-linking were observed for each experiment, but the relative populations of species were found to be independent of cross-linking efficiency, which is estimated by the prevalence of protein–DNA complexes in comparison to the total DNA. Cross-linking was most important in experiments incubating MutSα and MutLα with the DNA substrate to observe a surface free from excess proteins. Additionally, without cross-linking, dilution of the samples to reduce the protein concentrations for imaging results in very few complexes observed on the surface, suggesting that complexes dissociate prior to deposition and imaging. This result is consistent with SPR studies that show that the MutSα–MutLα-GT mismatch ternary complex dissociates rapidly in the presence of ATP (76). The cross-linked samples were either filtered through a 4% agarose bead gel filtration column prior to deposition to remove excess free proteins or diluted 10-fold in imaging buffer prior to deposition. Fractions collected from the filtration column or the diluted samples were deposited onto the APTES- or ethanolamine-treated mica, rinsed with water, wicked dry with filter paper, and then dried under a stream of nitrogen before imaging. The experiments were conducted by multiple researchers with multiple protein preparations from two different labs. Results were independent of whether the cross-linked complexes were filtered or diluted, the choice of surface treatment, the protein preparation, or the researcher conducting the experiments.
Imaging and Image Analysis.
Details of the imaging and image analysis are in the SI Appendix. Briefly, topographic and DREEM images were captured in air with a Nanoscope IIIa (Digital Instruments), an Asylum Research MFP-3D, or a JPK NanoWizard 4 microscope in tapping mode and analyzed as described previously (65, 66, 82, 86). We found that the cross-linking significantly increased the heights of the MutSα and MutLα on the surface (SI Appendix, Fig. S3). As discussed in the SI Appendix, the number of proteins indicated in the text and figures for SL complexes are rough estimates based on the volume of MutSα alone. Extrapolation of these estimates to the larger complexes with “missing” DNA is particularly difficult due to contributions from the DNA and the effect that shape and height have on the apparent volumes measured from AFM images (99) (SI Appendix, Methods, and legend to SI Appendix, Fig. S3B). The position distributions for MutSα or MutSα–MutLα complexes on the DNA were generated as described in the SI Appendix.
Tethered Particle Motion Assay.
Tethered particle motion (TPM) experiments measure changes in DNA configurations that result in changes in the DNA end-to-end distances, such as protein-induced DNA bending, DNA looping, or wrapping, by monitoring the Brownian motion of a bead on the end of a surface-tethered DNA fragment (83–85). Experimental details of the TPM measurements are in the SI Appendix. Briefly, our TPM experiments were performed in chambers with surfaces passivated by PEG, with 550-bp, double-stranded DNA with a biotin on one end, a centrally located GT mismatch (or GC for control), and a bead attached to the other end via digoxigenin–antidigoxigenin interactions. The beads were SPHERO protein G polystyrene particles with 0.84-μm diameter. Due to nonspecific interactions of the MutSα and MutLα proteins with the surface and beads, these experiments were conducted at lower protein concentrations than the AFM experiments, such that no nonspecific interactions were observed with homoduplex DNA. These experiments also differ from the AFM experiments on linear DNA in that the DNA end is blocked, so MutSα mobile clamps cannot slide off the end of the DNA, similar to our control AFM experiment with nicked plasmid DNA (SI Appendix, Fig. S5). Data analysis used custom MATLAB codes.
Data and Code Availability.
All primary data and computer codes for this paper are available in the Carolina Digital Repository (https://cdr.lib.unc.edu/) at https://doi.org/10.17615/wqe7-t470. The custom image analysis program ImageMetrics is freely available at http://imetrics.app.
Supplementary Material
Acknowledgments
We thank Paul Modrich, Tom Kunkel, Jackie Bower, Hong Wang, and Manju Hingorani for critical reading of the manuscript. We thank Paul Modrich for providing human MutSα and MutLα proteins. We also thank Yan Yan, David Dunlap, and Laura Finzi from Emory University for advice on conducting and interpreting the TPM studies. Funding was provided by National Institute of General Medical Sciences (NIGMS) grants R01 GM109832 to D.A.E. and K.R.W., R01 GM079480 and R35 GM127151 to D.A.E., and R01 GM132263 to K.R.W.; and the Division of Intramural Research, National Institute of Diabetes and Digestive and Kidney Diseases, NIH, to P.H.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.K.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918519117/-/DCSupplemental.
References
- 1.Kunkel T. A., Erie D. A., DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L., DNA mismatch repair: Functions and mechanisms. Chem. Rev. 106, 302–323 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Hsieh P., Yamane K., DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 129, 391–407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modrich P., Mechanisms in E. coli and human mismatch repair (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8490–8501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty U., Alani E., Understanding how mismatch repair proteins participate in the repair/anti-recombination decision. FEMS Yeast Res. 16, fow071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta D., Heinen C. D., The mismatch repair-dependent DNA damage response: Mechanisms and implications. DNA Repair (Amst.) 78, 60–69 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Bignami M., Casorelli I., Karran P., Mismatch repair and response to DNA-damaging antitumour therapies. Eur. J. Cancer 39, 2142–2149 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Fedier A., Fink D., Mutations in DNA mismatch repair genes: Implications for DNA damage signaling and drug sensitivity (review). Int. J. Oncol. 24, 1039–1047 (2004). [PubMed] [Google Scholar]
- 9.Martín-López J. V., Fishel R., The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam. Cancer 12, 159–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boland C. R., Lynch H. T., The history of Lynch syndrome. Fam. Cancer 12, 145–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinen C. D., Mismatch repair defects and Lynch syndrome: The role of the basic scientist in the battle against cancer. DNA Repair (Amst.) 38, 127–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebbink J. H., Drost M., de Wind N., DNA mismatch repair: From biophysics to bedside. DNA Repair (Amst.) 38, 1–2 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Gros J. et al., Post-licensing specification of eukaryotic replication origins by facilitated Mcm2-7 sliding along DNA. Mol. Cell 60, 797–807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradia S. et al., hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Kunkel T. A., Erie D. A., Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes G. X., Schmidt T. T., Kolodner R. D., Hombauer H., New insights into the mechanism of DNA mismatch repair. Chromosoma 124, 443–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu R. et al., Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 31, 2528–2540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu R. et al., MutL traps MutS at a DNA mismatch. Proc. Natl. Acad. Sci. U.S.A. 112, 10914–10919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeBlanc S. J. et al., Coordinated protein and DNA conformational changes govern mismatch repair initiation by MutS. Nucleic Acids Res. 46, 10782–10795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groothuizen F. S. et al., MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 4, e06744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong C. et al., MutS switches between two fundamentally distinct clamps during mismatch repair. Nat. Struct. Mol. Biol. 18, 379–385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J. et al., Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 539, 583–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendillo M. L. et al., Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. J. Biol. Chem. 285, 13170–13182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Kadyrov F. A. et al., Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C. E. et al., Dominant mutations in S. cerevisiae PMS1 identify the Mlh1-Pms1 endonuclease active site and an exonuclease 1-independent mismatch repair pathway. PLoS Genet. 9, e1003869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pluciennik A. et al., PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 107, 16066–16071 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genschel J. et al., Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 114, 4930–4935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szankasi P., Smith G. R., A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267, 1166–1169 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Tishkoff D. X. et al., Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. U.S.A. 94, 7487–7492 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran P. T., Simon J. A., Liskay R. M., Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 9760–9765 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genschel J., Bazemore L. R., Modrich P., Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 277, 13302–13311 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Goellner E. M. et al., Identification of Exo1-Msh2 interaction motifs in DNA mismatch repair and new Msh2-binding partners. Nat. Struct. Mol. Biol. 25, 650–659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadyrov F. A. et al., A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 106, 8495–8500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longley M. J., Pierce A. J., Modrich P., DNA polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem. 272, 10917–10921 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Constantin N., Dzantiev L., Kadyrov F. A., Modrich P., Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 280, 39752–39761 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y. et al., Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Lamers M. H. et al., The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407, 711–717 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Obmolova G., Ban C., Hsieh P., Yang W., Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407, 703–710 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Natrajan G. et al., Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: A common recognition mode for diverse substrates. Nucleic Acids Res. 31, 4814–4821 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren J. J. et al., Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell 26, 579–592 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Sacho E. J., Kadyrov F. A., Modrich P., Kunkel T. A., Erie D. A., Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol. Cell 29, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang Q., Prolla T. A., Liskay R. M., Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol. 17, 4465–4473 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran P. T., Liskay R. M., Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol. Cell. Biol. 20, 6390–6398 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guarné A. et al., Structure of the MutL C-terminal domain: A model of intact MutL and its roles in mismatch repair. EMBO J. 23, 4134–4145 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ban C., Junop M., Yang W., Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell 97, 85–97 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Hall M. C., Shcherbakova P. V., Kunkel T. A., Differential ATP binding and intrinsic ATP hydrolysis by amino-terminal domains of the yeast Mlh1 and Pms1 proteins. J. Biol. Chem. 277, 3673–3679 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Groothuizen F. S., Sixma T. K., The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst.) 38, 14–23 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Pillon M. C. et al., Structure of the endonuclease domain of MutL: Unlicensed to cut. Mol. Cell 39, 145–151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iino H. et al., Small-angle X-ray scattering analysis reveals the ATP-bound monomeric state of the ATPase domain from the homodimeric MutL endonuclease, a GHKL phosphotransferase superfamily protein. Extremophiles 19, 643–656 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Arana M. E. et al., Functional residues on the surface of the N-terminal domain of yeast Pms1. DNA Repair (Amst.) 9, 448–457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guarné A., Junop M. S., Yang W., Structure and function of the N-terminal 40 kDa fragment of human PMS2: A monomeric GHL ATPase. EMBO J. 20, 5521–5531 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acharya S., Foster P. L., Brooks P., Fishel R., The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 12, 233–246 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Mardenborough Y. S. N. et al., The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. Nucleic Acids Res. 47, 11667–11680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schofield M. J., Nayak S., Scott T. H., Du C., Hsieh P., Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 276, 28291–28299 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Grilley M., Welsh K. M., Su S. S., Modrich P., Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem. 264, 1000–1004 (1989). [PubMed] [Google Scholar]
- 57.Hombauer H., Campbell C. S., Smith C. E., Desai A., Kolodner R. D., Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147, 1040–1053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elez M., Radman M., Matic I., Stoichiometry of MutS and MutL at unrepaired mismatches in vivo suggests a mechanism of repair. Nucleic Acids Res. 40, 3929–3938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendillo M. L., Mazur D. J., Kolodner R. D., Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J. Biol. Chem. 280, 22245–22257 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Allen D. J. et al., MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 16, 4467–4476 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blackwell L. J., Martik D., Bjornson K. P., Bjornson E. S., Modrich P., Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 273, 32055–32062 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Junop M. S., Obmolova G., Rausch K., Hsieh P., Yang W., Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7, 1–12 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Modrich P., DNA mismatch correction. Annu. Rev. Biochem. 56, 435–466 (1987). [DOI] [PubMed] [Google Scholar]
- 64.LeBlanc S. et al., Using atomic force microscopy to characterize the conformational properties of proteins and protein-DNA complexes that carry out DNA repair. Methods Enzymol. 592, 187–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ratcliff G. C., Erie D. A., A novel single-molecule study to determine protein–Protein association constants. J. Am. Chem. Soc. 123, 5632–5635 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Yang Y., Wang H., Erie D. A., Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods 29, 175–187 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Dame R. T., Wyman C., Goosen N., Insights into the regulation of transcription by scanning force microscopy. J. Microsc. 212, 244–253 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Verhoeven E. E., Wyman C., Moolenaar G. F., Goosen N., The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 21, 4196–4205 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beerens N., Hoeijmakers J. H., Kanaar R., Vermeulen W., Wyman C., The CSB protein actively wraps DNA. J. Biol. Chem. 280, 4722–4729 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Genschel J., Modrich P., Functions of MutLalpha, replication protein A (RPA), and HMGB1 in 5′-directed mismatch repair. J. Biol. Chem. 284, 21536–21544 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geng H. et al., Biochemical analysis of the human mismatch repair proteins hMutSα MSH2(G674A)-MSH6 and MSH2-MSH6(T1219D). J. Biol. Chem. 287, 9777–9791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivetti C., Guthold M., Bustamante C., Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J. 18, 4464–4475 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H., Tessmer I., Croteau D. L., Erie D. A., Van Houten B., Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Lett. 8, 1631–1637 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martik D., Baitinger C., Modrich P., Differential specificities and simultaneous occupancy of human MutSalpha nucleotide binding sites. J. Biol. Chem. 279, 28402–28410 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Josephs E. A., Zheng T., Marszalek P. E., Atomic force microscopy captures the initiation of methyl-directed DNA mismatch repair. DNA Repair (Amst.) 38, 71–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blackwell L. J., Wang S., Modrich P., DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. J. Biol. Chem. 276, 33233–33240 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Jeon Y. et al., Dynamic control of strand excision during human DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 113, 3281–3286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang Y., Marszalek P. E., Atomic force microscopy captures MutS tetramers initiating DNA mismatch repair. EMBO J. 30, 2881–2893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaur P., Longley M. J., Pan H., Wang H., Copeland W. C., Single-molecule DREEM imaging reveals DNA wrapping around human mitochondrial single-stranded DNA binding protein. Nucleic Acids Res. 46, 11287–11302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adkins N. L. et al., Nucleosome-like, single-stranded DNA (ssDNA)-histone octamer complexes and the implication for DNA double strand break repair. J. Biol. Chem. 292, 5271–5281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaur P. et al., Enhanced electrostatic force microscopy reveals higher-order DNA looping mediated by the telomeric protein TRF2. Sci. Rep. 6, 20513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu D. et al., Visualizing the path of DNA through proteins using DREEM imaging. Mol. Cell 61, 315–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henneman B., Heinsman J., Battjes J., Dame R. T., Quantitation of DNA-binding affinity using tethered particle motion. Methods Mol. Biol. 1837, 257–275 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Han L. et al., . “Calibration of tethered particle motion experiments” in Mathematics of DNA Structure, Function and Interactions. The IMA Volumes in Mathematics and its Applications, Benham C. J., Harvey S., Olson W. K., Sumners D. W., Swigon D., Eds. (Springer, New York, NY, 2009), Vol. 150, pp. 123–138. [Google Scholar]
- 85.Kovari D. T., Yan Y., Finzi L., Dunlap D., Tethered particle motion: An easy technique for probing DNA topology and interactions with transcription factors. Methods Mol. Biol. 1665, 317–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H. et al., DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. U.S.A. 100, 14822–14827 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hao P. et al., Recurrent mismatch binding by MutS mobile clamps on DNA localizes repair complexes nearby. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.1918517117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Räschle M., Dufner P., Marra G., Jiricny J., Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J. Biol. Chem. 277, 21810–21820 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Spampinato C., Modrich P., The MutL ATPase is required for mismatch repair. J. Biol. Chem. 275, 9863–9869 (2000). [DOI] [PubMed] [Google Scholar]
- 90.Hall M. C., Wang H., Erie D. A., Kunkel T. A., High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 312, 637–647 (2001). [DOI] [PubMed] [Google Scholar]
- 91.Hall M. C. et al., DNA binding by yeast Mlh1 and Pms1: Implications for DNA mismatch repair. Nucleic Acids Res. 31, 2025–2034 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mendillo M. L. et al., A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 106, 22223–22228 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kadyrova L. Y., Blanko E. R., Kadyrov F. A., CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 108, 2753–2758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li F., Tian L., Gu L., Li G. M., Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J. Biol. Chem. 284, 33056–33061 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schöpf B. et al., Interplay between mismatch repair and chromatin assembly. Proc. Natl. Acad. Sci. U.S.A. 109, 1895–1900 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodriges Blanko E., Kadyrova L. Y., Kadyrov F. A., DNA mismatch repair interacts with CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition. J. Biol. Chem. 291, 9203–9217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geng H. et al., In vitro studies of DNA mismatch repair proteins. Anal. Biochem. 413, 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Acharya S. et al., hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Varadhan G., Robinett W., Erie D., Taylor R. M. II, Fast simulation of atomic-force-microscope imaging of atomic and polygonal surfaces using graphics hardware. Proc. SPIE 4665, 116–124 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All primary data and computer codes for this paper are available in the Carolina Digital Repository (https://cdr.lib.unc.edu/) at https://doi.org/10.17615/wqe7-t470. The custom image analysis program ImageMetrics is freely available at http://imetrics.app.