Significance
Accurate gene expression relies on enzymes distinguishing correct from incorrect substrates that are chemically and structurally similar. During protein synthesis, correct base pairing between the tRNA anticodon and the mRNA codon is essential for accurate translation. Other tRNA elements outside of the anticodon also contribute to correct selection by the ribosome, but the structural basis for their contribution to selectivity is unknown. Here, we determine structures of the bacterial ribosome containing tRNAs with altered nucleotide pairings in the anticodon loop that are known to prevent the ribosome from distinguishing correct from incorrect tRNAs. Collectively, these structures reveal a previously unappreciated role for a ribosomal decoding site nucleotide in sensing the integrity of the tRNA that contributes to decoding fidelity.
Keywords: ribosome, tRNA, anticodon, miscoding, structural biology
Abstract
Bacterial transfer RNAs (tRNAs) contain evolutionarily conserved sequences and modifications that ensure uniform binding to the ribosome and optimal translational accuracy despite differences in their aminoacyl attachments and anticodon nucleotide sequences. In the tRNA anticodon stem−loop, the anticodon sequence is correlated with a base pair in the anticodon loop (nucleotides 32 and 38) to tune the binding of each tRNA to the decoding center in the ribosome. Disruption of this correlation renders the ribosome unable to distinguish correct from incorrect tRNAs. The molecular basis for how these two tRNA features combine to ensure accurate decoding is unclear. Here, we solved structures of the bacterial ribosome containing either wild-type or containing a reversed 32–38 pair on cognate and near-cognate codons. Structures of wild-type bound to the ribosome reveal 23S ribosomal RNA (rRNA) nucleotide A1913 positional changes that are dependent on whether the codon−anticodon interaction is cognate or near cognate. Further, the 32–38 pair is destabilized in the context of a near-cognate codon−anticodon pair. Reversal of the pairing in ablates A1913 movement regardless of whether the interaction is cognate or near cognate. These results demonstrate that disrupting 32–38 and anticodon sequences alters interactions with the ribosome that directly contribute to misreading.
The ribosome orchestrates the binding of messenger RNA (mRNA), protein translation factors, and transfer RNAs (tRNAs) in a sequential manner for the synthesis of all cellular proteins. This process is remarkably complicated and involves numerous steps that have been evolutionarily optimized to select correct tRNAs from a pool of structurally and chemically similar tRNAs. The tRNAs are the so-called “adaptor” molecules that decode the genetic code on mRNA and carry an aminoacyl group (1). As noted in a recent review (2), the term “adaptor” implies plasticity between the aminoacyl group and anticodon; however, other nucleotide sequences and modifications in each tRNA are also evolutionarily tuned to permit comparable binding affinities to both EF-Tu and the ribosome for optimal accuracy (3–5). This tuning occurs in all bacterial tRNAs (5), yet the structural basis for how optimized sequences contribute to efficient recognition on the ribosome is unclear.
All tRNAs adopt L-shaped structures that traverse both the small 30S and the large 50S subunits when bound to the ribosome. The aminoacyl group is attached to the 3′ end of the tRNA which is located ∼90 Å away from the anticodon (Fig. 1A). EF-Tu interacts extensively with the acceptor arm of tRNAs and surrounds the 3ʹ aminoacyl group. These aminoacyl groups represent a broad range of chemical diversity, yet all tRNAs bind to EF-Tu with the same relative affinities despite these intrinsic physical differences, due to compensatory binding contributions from the tRNA (2, 3, 6). Likewise, the anticodon regions of all tRNAs bind to their cognate mRNA codons on the ribosome with similar affinities, despite diverse codon−anticodon pairings that should exhibit differences in base pairing strengths (4). In both cases, the sequences of each tRNA have evolved to compensate for the chemical diversity of the aminoacyl group or the codon−anticodon strength to achieve similar rates of binding and optimal accuracy.
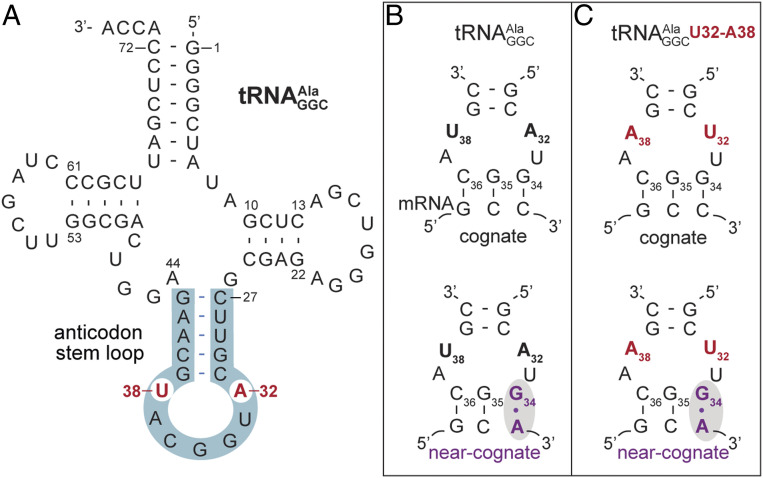
Fig. 1.
The -mRNA complexes used in this study. (A) The secondary structure of with the anticodon stem−loop highlighted in blue. (B) Complexes of wild-type (A32−U38 bolded) and (C) the variant containing U32−A38 (red) on either a cognate GCC or near-cognate GCA codons.
The evolution of tRNA sequences and modification patterns implicate previously unappreciated roles of specific tRNA regions in the process of translation, including decoding. During decoding, Watson–Crick base pairing between the three nucleotides of the mRNA codon and tRNA anticodon is monitored by the ribosome. Correct pairing causes conformational changes of the 30S subunit and GTPase activation of EF-Tu, leading to the release of the tRNA for elongation to proceed (7). Phylogenetic and biochemical analyses reveal that a universal feature of all bacterial tRNAs is a correlation between the nucleotide identities of the anticodon (nucleotides 34, 35, and 36) and nucleotides 32 and 38 located in the anticodon loop (5, 8–10) (Fig. 1A). Specifically, strong GC-rich codon−anticodon interactions are always balanced by a weaker 32–38 pairing and, conversely, a weak AU-rich codon−anticodon interaction is coupled with a stronger 32–38 pairing. This coordination of nucleotide identities ensures uniform binding affinities of all tRNAs to their cognate codons (4, 11). Further, when the nucleotide identity of the 32–38 pair is disrupted, the ribosome is unable to distinguish correct from incorrect tRNAs, establishing that this correlation is important for translation fidelity (9, 10).
Mutations of the A32−U38 pair in have provided significant insights into how 32–38 sequence changes alone disrupt the fidelity of translation (5, 8–10, 12). The wild-type A32−U38 pairing in is considered a weak interaction that counterbalances the GGC anticodon that binds tightly to the GCC alanine codon (9, 10). Without a weak A32−U38 pair, the GGC anticodon of binds very tightly to its own cognate codon but also binds tightly to other near-cognate codons (8), where near cognate is defined as a single mismatch pair between the codon and anticodon. When tRNAs bind too tightly to near-cognate codons, this can result in the incorrect tRNA outcompeting cognate tRNAs, resulting in the misincorporation of the tRNA or misreading. Changing the 32–38 pairing to UA, CG, or CA in causes efficient misreading of near-cognate codons, resulting in the misincorporation of alanine (8, 12). Specifically, the U32−A38 variant, which contains a reversed 32–38 pair, decodes the near-cognate GCA codon efficiently. While the initial binding affinity of the EF-Tu•GTP• U32−A38 ternary complex to the ribosome (either on cognate GCC or near-cognate GCA codons) is nominally increased as compared to that of wild-type , the GTP hydrolysis rates by EF-Tu and dipeptide formation are comparable to that of the wild-type tRNA decoding a cognate GCC codon (8). Therefore, the U32−A38 reversal circumvents proofreading mechanisms, allowing for the acceptance of the incorrect tRNA. In comparison, typically, all aspects of decoding are impaired including EF-Tu•GTP•tRNA binding, GTP hydrolysis, and dipeptide formation of wild-type tRNAs on near-cognate codons. In summary, these data strongly implicate bypassing of proofreading during decoding as the major outcome of dysregulation of the 32–38 pairing and anticodon sequence coordination (11). Further, while deletion of only incurs a minor growth defect (13), overexpression of certain variants, including the U32−A38 variant, is toxic in Escherichia coli (12). These data indicate that perturbing the 32–38 base pair in a single tRNA isoacceptor can overwhelm the canonical translation machinery, resulting in cell death.
Motivated by these compelling biochemical and in vivo assays, we solved four X-ray crystal structures of Thermus thermophilus ribosomes containing either wild-type or the U32−A38 variant in the aminoacyl (A) site bound to either a cognate GCC or near-cognate GCA codon (Fig. 1). We find that when wild-type decodes a near-cognate GCA codon (single mismatch at the third position of the codon−anticodon interaction), interactions between 23S ribosomal RNA (rRNA) nucleotide A1913 and the tRNA are ablated. However, in the case of the with the reversed 32–38 pair, A1913 maintains interactions with the tRNA regardless of whether the codon−anticodon interaction is cognate or near cognate, providing a molecular basis for how the ribosome recognizes such mismatches.
Results
Near-Cognate Interactions between and the GCA Codon Influence the Position of 23S rRNA A1913.
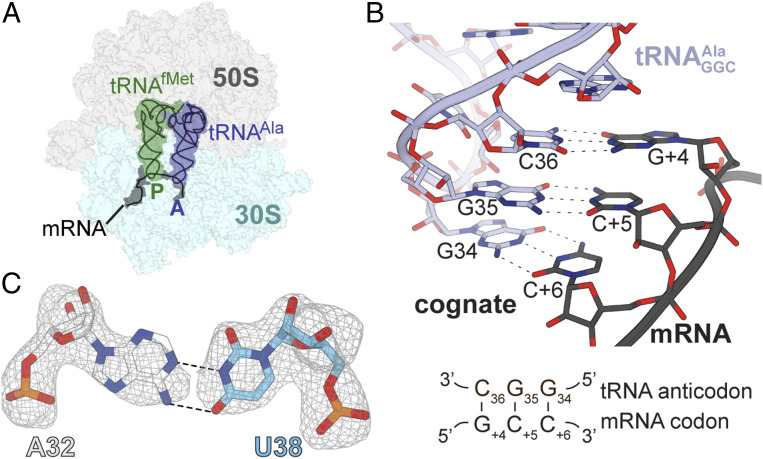
To determine the structural basis for how misreads codons upon reversal of its 32–38 pair, we first solved a structure of T. thermophilus 70S with wild-type bound to a cognate GCC codon in the A site (Figs. 1 A and B and 2A and Table 1). We formed ribosome complexes with mRNA containing an initiation AUG codon at the peptidyl (P) site and GCC alanine codon at the A site. E. coli tRNAfMet and were added to the P and the A sites, respectively, and crystallization trials were initiated according to standard procedures (14). In the structure solved to 3.2 Å, three Watson–Crick base pairs form between the codon−anticodon and are recognized by the A site as a cognate interaction by 16S rRNA nucleotides G530, A1492, A1493, and 23S rRNA A1913 (Fig. 2B and SI Appendix, Fig. S1) (the mRNA numbering refers to the first nucleotide position in the P site as +1, with A-site nucleotides denoted as +4, +5, and +6). These rRNA nucleotides surround the codon−anticodon pair and adopt “ON” positions whereby A1492 and A1493 interact directly with the first two positions of the codon−anticodon, G530 forms interactions with A1492 to lock the 30S in a closed conformation, and A1913 packs against the tRNA backbone of the anticodon stem (SI Appendix, Figs. S1 and S2) (15–17). Although the 32–38 nucleotides are commonly shown as unpaired in secondary structural representations, these two nucleotides often form interactions between their Watson–Crick faces (18) (SI Appendix, Fig. S3). In this structure, the A32−U38 pair in the anticodon loop adopts a canonical Watson–Crick pair with A1913 stacking against the phosphate backbone of U38 (Fig. 2C and SI Appendix, Fig. S1B).
Fig. 2.
Cognate interactions between and the GCC mRNA codon. (A) Overview of the bacterial 70S ribosome containing a P-site tRNAfMet and an A-site bound to a GCC codon. (B) Zoomed-in view of the anticodon stem−loop of showing three Watson–Crick pairings between the anticodon and the codon. The +1 mRNA numbering starts in the P site, with the A-site nucleotide positions labeled as +4, +5, and +6. (C) The A32−U38 pairing forms a well-ordered Watson–Crick base pair. The 2FO−FC electron density map (gray mesh) is contoured at 1.0σ.
Table 1.
Data collection and refinement statistics
| cognate codon | near-cognate codon | UA cognate codon | UA near-cognate codon | |
| PDB ID | 6OF6 | 6OJ2 | 6ORD | 6OPE |
| Data collection | ||||
| Wavelength (Å) | 1.0000 | 1.0000 | 0.9792 | 0.9791 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 211.3, 452.3, 626.5 | 211.0, 453.5, 625.4 | 209.4, 445.8, 616.0 | 209.4, 449.8, 620.0 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 43.9 to 3.2 (3.3 to 3.2) | 50 to 3.2 (3.3 to 3.2) | 49.9 to 3.1 (3.2 to 3.1) | 49.6 to 3.1 (3.2 to 3.1) |
| Rpim (%) | 7.4 (34.4) | 11.0 (74.0) | 12.7 (83.7) | 14.1 (77.0) |
| I/σI | 8.26 (2.04) | 4.88 (0.98) | 4.57 (0.71) | 6.29 (1.12) |
| CC1/2 | 0.993 (0.645) | 0.995 (0.329) | 0.991 (0.183) | 0.989 (0.387) |
| Completeness (%) | 91.15 (91.01) | 99.52 (99.92) | 95.19 (91.69) | 97.13 (99.13) |
| Redundancy | 6.6 (6.0) | 5.8 (5.6) | 3.0 (2.2) | 3.0 (3.0) |
| Refinement | ||||
| No. reflections | 889,696 | 971,076 | 981,305 | 1,016,419 |
| Rwork/Rfree (%) | 19.5/23.2 | 23.6/27.0 | 23.2/26.8 | 22.5/26.6 |
| No. atoms | ||||
| Macromolecules | 297,356 | 297,318 | 290,854 | 294,030 |
| Ligands | 1,319 | 1,053 | 2,965 | 951 |
| B-factors | ||||
| Macromolecules | 90.33 | 129.71 | 91.34 | 87.33 |
| Ligands | 56.13 | 130.73 | 60.81 | 59.98 |
| rms deviations | ||||
| Bond lengths (Å) | 0.005 | 0.006 | 0.008 | 0.012 |
| Bond angles (degrees) | 0.87 | 1.07 | 0.99 | 1.25 |
Values in parentheses are for highest-resolution shell.
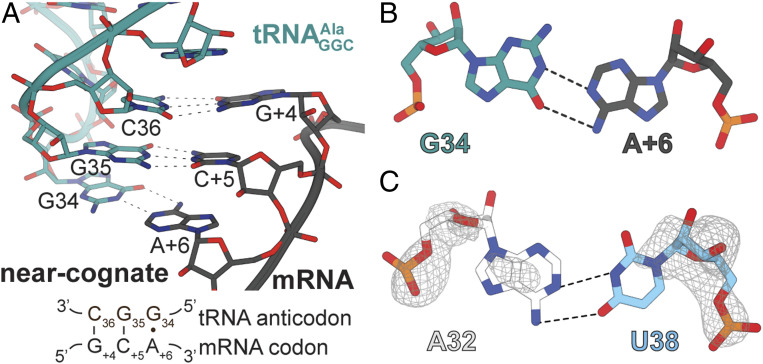
We next solved a 3.2-Å structure of the T. thermophilus 70S wild-type bound to a near-cognate GCA codon at the A site (Figs. 1B and 3A and Table 1). Two Watson–Crick base pairs form at the first two positions of the codon−anticodon, and a single A+6•G34 mismatch forms at the third, or wobble, position (Fig. 3B). Both A+6 and G34 adopt anti conformations containing two hydrogen bonds between the N6 amino of A+6 and the carbonyl O6 of G34, and between the N1 of A+6 and the N1 of G34 (Fig. 3B). The C1ʹ−C1ʹ distance between a Watson−Crick base pair is typically 10.5 Å, while G•A pairs have a distance of ∼12.6 Å (19); the C1ʹ−C1ʹ distance of the A+6• G34 pair in this study is ∼12.6 Å, consistent with previous studies. To our knowledge, a A+6• G34 mismatch has not been observed in the decoding center of the ribosome. At the wobble position, G•U pairs and chemically modified anticodon nucleotides that interact with mismatched codon nucleotides can be decoded as Watson–Crick-like base pairs by the ribosome, a mechanism known as wobble base decoding (16). By this definition, a A+6• G34 mismatch should be rejected by the ribosome, resulting in rapid dissociation of the ternary complex after initial selection.
Fig. 3.
A near-cognate codon−anticodon interaction in influences the stability of the A32−U38 pair. (A) Zoomed-in view of the interaction between the anticodon stem−loop of with a near-cognate GCA codon. G+4-C36 and C+5-G35 form Watson–Crick base pairs; a A+6•G34 mismatch forms at the third or wobble position. (B) Two hydrogen bonds form between the cis Watson–Crick A+6•G34 interaction. (C) The A32−U38 pair is destabilized in a structure of the 70S bound to in the A site decoding a near-cognate GCA codon. The 2FO−FC electron density map (gray mesh) is contoured at 1.0σ.
The near-cognate interaction between the codon−anticodon also causes the nucleobases of the A32−U38 base pair to become disordered, as indicated by the lack of electron density (Fig. 3C). This lack of electron density is notable because the cognate and the near-cognate, codon−anticodon-containing structures are at comparable resolutions (both 3.2 Å). The destabilization of the 32–38 pairing has also been seen when the tRNA−mRNA pairs are near cognate and cause mRNA frameshifts (20, 21). These data further emphasize the critical role of the 32–38 pair in the accurate decoding of cognate codons.
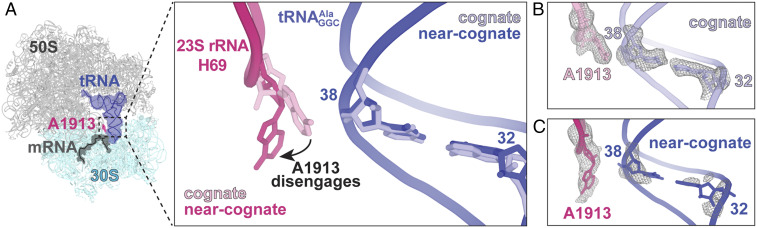
The ribosome closely monitors the codon−anticodon interaction (SI Appendix, Figs. S1A and S2), but the rest of the anticodon stem−loop (ASL) is minimally inspected in the A site, providing a conundrum in understanding how the correlation between the anticodon and the 32–38 pairing could tune tRNA binding and acceptance by the ribosome (8–10). The closest ribosomal nucleotide or protein to anticodon stem nucleotides 32–38 is 23S rRNA nucleotide A1913 (Fig. 4A and SI Appendix, Fig. S2). A1913 is located in the loop of Helix 69 (H69) which is a universally conserved helix that forms an intersubunit bridge, contacts 16S rRNA nucleotide A1493 during decoding (22), and is also important for release factor recognition of stop codons (22, 23). A1913 typically packs against nucleotide 38 of the A-site tRNA and forms a hydrogen bond with the 2ʹ-OH of nucleotide 37 (SI Appendix, Fig. S1B). In the case of wild-type decoding a near-cognate GCA codon that causes disordering of A32−U38, we find that the nucleobase of A1913 shifts ∼7 Å from the tRNA as compared to when binds to a cognate codon (as measured between N1 atoms of the nucleobase) (Fig. 4 B and C and SI Appendix, Fig. S1C).
Fig. 4.
Interaction of 23S rRNA A1913 with tRNA and its ablation when the 32–38 pair is destabilized. (A) The 23S rRNA A1913 (light pink) in helix 69 packs against the backbone of U38 of the A-site tRNA (purple) in the context of a cognate tRNA−mRNA pair. When a near-cognate tRNA−mRNA pair is present at the A site, A1913 (dark pink) moves away from the tRNA. (B) Interactions between A1913 showing its nucleobase is proximal to the U38−A32 pair in tRNA when decodes a cognate GCC codon. A1913 also adopts this conformation in the structures where the U32−A38 variant is a cognate GCC or near-cognate GCA codon. (C) When decodes a near-cognate GCA codon, the 32–38 pair becomes disordered and A1913 moves away from the A-site tRNA. The 2FO−FC electron density maps are contoured at 1.0σ.
Disrupting the 32–38 Pair of Renders the Ribosome Unable to Distinguish Cognate from Near-Cognate Codon−Anticodon Pairs.
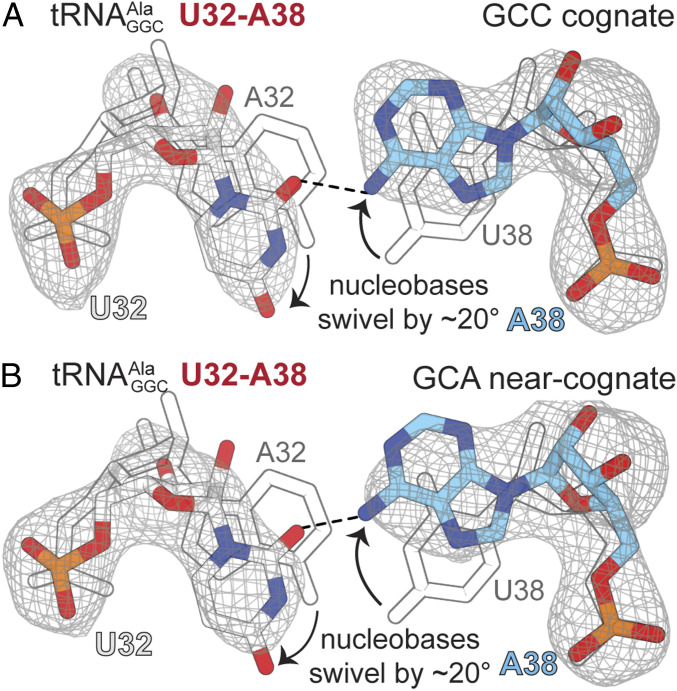
To understand the structural basis for how reversing the 32–38 pairing in tRNAs leads to miscoding, we solved a structure of the ribosome bound to an A-site U32−A38 variant decoding either a cognate GCC or a near-cognate GCA codon (SI Appendix, Fig. S4 and Table 1). Three Watson–Crick base pairs form between the U32−A38 variant and a GCC cognate codon similar to the wild-type −GCC codon interaction (Fig. 2B and SI Appendix, Fig. S4A). Similarly, the near-cognate interaction formed between U32−A38 and the GCA codon is identical to how wild-type interacts with the near-cognate codon (Fig. 3A and SI Appendix, Fig. S4B). However, in contrast to the disordering of the A32−U38 nucleobases observed in wild-type decoding a near-cognate GCA codon (Fig. 3C), all regions of the ASL of the U32−A38 variant are well ordered whether bound to a cognate GCC or near-cognate GCA codon (SI Appendix, Fig. S5). The only notable difference in the structures of the U32−A38 variant is the position of the mutated U32−A38 nucleobases; U32−A38 swivel ∼20° away from each other as compared to wild-type (Fig. 5). The N6 amino of A38 now forms a single hydrogen bond with the carbonyl O2 of U32, in contrast to a typical Watson–Crick base pair that forms with wild-type (Figs. 2C and 3C). In both structures of the U32−A38 variant decoding either a cognate or a near-cognate codon when the ribosome is unable to distinguish between correct and incorrect tRNAs (8), A1913 remains packed against the A-site tRNA variant (SI Appendix, Fig. S6). These results indicate reversing the 32–38 pair results in changes in their pairing that, in turn, influence how A1913 interacts with the ASL.
Fig. 5.
Reversing the 32–38 pair in causes both nucleotides to swivel ∼20° in opposite directions, disrupting canonical Watson–Crick base pairing. (A) The structure of 70S U32−A38 interacting with a cognate GCC codon. (B) The 70S U32-A38 variant interacting with a near-cognate GCA codon. In both structures, the nucleotides swivel away from each other, as compared to the position of the A32−U38 base pair in wild-type tRNA bound to the cognate codon (shown in gray outline). The 2FO−FC electron density maps in gray are contoured at 1.0σ.
Discussion
To maintain efficient and accurate protein synthesis, tRNAs acquired diverse sequences and chemical modifications that enable their specific recognition by specific aminoacyl-tRNA synthetases and the decoding of cognate mRNA codons (24, 25). In addition, these tRNA elements are evolutionarily optimized to ensure that all tRNAs have similar binding affinities to both EF-Tu and the ribosome, thus preventing potential thermodynamic differences from contributing to the decoding process. As part of the tuning of tRNAs to bind uniformly to the ribosome, the nucleotide identities and strength of the 32–38 base pair and the anticodon nucleotides are correlated (9, 10). Since the ribosome closely monitors the codon−anticodon interaction but the rest of the ASL is minimally inspected in the A site, it was unclear how disrupting this correlation would influence the overall tRNA structure and whether this dysregulation affects how the ribosome interacts with the A-site tRNA. Here, we solved X-ray crystal structures of ribosome complexes containing or a U32−A38 variant known to cause high levels of miscoding (8). The U32−A38 variant binds tighter to both cognate and near-cognate codons, resulting in increased misreading as assessed by biochemical assays and cell death in vivo upon overexpression (8, 12). The reversal of U32−A38 does not affect other aspects of tRNA selection, including GTP hydrolysis by EF-Tu or dipeptide formation kinetics (8), establishing as an ideal system to study these miscoding events.
In this study, we report two observations that may explain the misreading propensity when the identity of the 32–38 pair and the anticodon nucleotides are dysregulated: In the context of the wild-type decoding a near-cognate codon, the 32–38 pairing becomes disordered, and 23S rRNA A1913 moves away from the backbone of the ASL of the tRNA (Figs. 3C and 4C). Normally, when wild-type decodes a cognate GCC codon, nucleotides 32 and 38 form a stable Watson–Crick base pair with well-defined electron density (Fig. 2C). However, when the same tRNA is bound to a near-cognate codon, resulting in a A+6•G36 mismatch at the wobble position, the electron density of the 32–38 base pair is weakened, signifying that these nucleotides are more mobile (Fig. 3C). These data provide hints to how noncanonical interactions between the codon and the anticodon may be sensed by other regions of the tRNA. A similar disruption of the 32–38 pairing was previously observed in the context of mutant tRNAs that cause mRNA frameshifts in the +1 direction (20, 21). Notably, in these structures, there is a complete lack of density for nucleotide 32 of the anticodon loop and 5ʹ of the anticodon stem (nucleotides 28 to 31), suggesting this disorder and/or instability may be a common feature of tRNA−mRNA pairs that cause miscoding.
A1913 adopts two different conformations that appear to be dependent on whether the codon−anticodon interaction is cognate or near cognate (Fig. 4 and SI Appendix, Fig. S1). In the case of a cognate codon−anticodon interaction, A1913 packs against the backbone of tRNA nucleotide 38 in a position that is observed in most ribosome structures solved to date (Fig. 4B and SI Appendix, Fig. S3). In contrast, in the case of a near-cognate, codon−anticodon interaction, A1913 moves ∼7 Å away from the tRNA (Fig. 4C). We propose that A1913 is part of the response of the ribosome to monitor the structural integrity of the A-site tRNA [previously termed “ON” (17)]. When the 32–38 nucleotides are reversed in , the position of A1913 is always “ON,” packed against the tRNA backbone, regardless of whether the codon−anticodon interaction is cognate or near cognate (SI Appendix, Fig. S6). These data strongly suggest that, in the case of the U32−A38 variant, the ribosome recognizes both the cognate and the near-cognate interaction as correct (or “ON”), consistent with previous biochemical analyses demonstrating that U32-A38 bypasses decoding checkpoints to misread near-cognate codons (8, 9).
To our knowledge, the position of A1913 has never been seen to move away from the tRNA backbone in the context of a mismatched codon−anticodon interaction in the A site (Fig. 4). In structures containing single C•A, A•C, A•A, U•G, and G•U mismatches at the first (16, 26) or second (16, 17, 27) position of the codon−anticodon interaction, A1913 packs against the tRNA; in one structure with a U•G mismatch at the second A-site position, A1913 is conformationally dynamic and unresolvable in the electron potential map (17). In these cases, the mismatched interaction was formed by systematically changing the sequence of the mRNA codon using four standard tRNAs in the absence of prior biochemical knowledge of how these pairs impact the decoding process (SI Appendix, Fig. S7). It is now well appreciated that certain codon−anticodon pairs undergo high levels of misreading in vivo, while other pairings do not (28). Therefore, perhaps the movement of A1913 wasn’t previously identified because the mismatched complexes affect a different stage of initial tRNA selection than what was captured in the structure, or the mismatched codon−anticodon pair does not cause high levels of miscoding. Future studies are required to understand this previously unappreciated role of A1913 in maintaining the fidelity of the decoding process.
Materials and Methods
In Vitro Transcription of .
Two DNA oligos spanning the A32−U38 variant were annealed, PCR amplified, and subcloned into a linearized pUC18 plasmid. The tRNA sequence was flanked by a T7 RNA polymerase promoter and a BsaI restriction digest site. E. coli DH5α were transformed with pUC18-Ala and grown overnight in super broth (3.5% tryptone, 2.0% yeast extract, 0.5% NaCl, 5 mM NaOH) supplemented with 100 µg/mL ampicillin at 37 °C. The cell pellet was harvested by centrifugation, plasmid DNA was purified and digested using BsaI, run-off transcription was performed, and the RNA was purified as previously described (29).
Crystallization, X-ray Data Collection, and Structural Determination.
The 70S ribosomes were purified from T. thermophilus using previously established protocols (14). The ribosome complex was formed by incubating 4.4 µM 70S with 8.8 µM mRNA (IDT) in buffer (5 mM Hepes-KOH, pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(CH3COO)2, 6 mM β-mercaptoethanol (β-Me)) at 55 °C for 5 min. Then 11 µM tRNAfMet (Chemical Block) and 22 µM tRNAAla were sequentially incubated at 55 °C for 15 min. The reaction was cooled to 37 °C, and 0.1 mM paromomycin was added and incubated at 37 °C. After equilibrating at 20 °C, a final concentration of 2.8 µM deoxy BigCHAP (Hampton Research) was added to the complex. Crystals grew from either a polyethylene glycol (PEG) condition (0.1 M Tris-HOAc pH 7.0, 0.2 M KSCN, 4–4.5% [weight/vol] PEG 20K, 4.5 to 5.5% [vol/vol] PEG 550MME, 10 mM Mg(CH3COO)2) or a 2-methyl-2,4-pentanediol (MPD) condition (0.1 M l-arginine HCl, 0.1 M Tris⋅HCl pH 7.5, 3% PEG 20K, 10 to 16.5% MPD, 1 mM β-Me). Data collection was performed at the SER-CAT 22-ID and NE-CAT 24ID-C beamlines at the Advanced Photon Source. Data were integrated and scaled using XDS (30), molecular replacement performed in PHENIX (31) using coordinates from Protein Data Bank (PDB) structure 4Y4O (32). Initial refinement was done using rigid-body restraints in PHENIX, followed by jelly-body refinement in REFMAC5 (33) in the CCP4i2 suite (34), and further iterative rounds of crystallographic refinements were performed in PHENIX. Model building was performed in Coot (35), and figures were generated using PyMol (36).
Data Availability.
Crystallography, atomic coordinates, and structure factors have been deposited in the PDB, https://www.wwpdb.org/ (PDB ID codes 6OF6, 6OJ2, 6OPE, 6ORD).
Supplementary Material
Acknowledgments
We thank Dunham laboratory member Dr. Eric Hoffer for technical assistance and helpful scientific insights, Dr. Graeme L. Conn and other members of the Dunham laboratory for critical reading of the manuscript, and staff members of the Northeast Regional Collaborative Access Team (NE-CAT) and Southeast Regional Collaborative Access Team (SER-CAT) beamlines for assistance during data collection. This work was supported by NIH Grant R01 GM093278. X-ray crystallography datasets were collected at the NE-CAT beamlines (funded by National Institute of General Medical Sciences (NIGMS) Grant P30 GM124165), using a Pilatus detector (RR029205) and an Eiger detector (OD021527), and at the SER-CAT beamlines (funded by its member institutions and NIH Equipment Grants S10_RR25528 and S10_RR028976). This research used resources of the Advanced Photon Source (APS), a US Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contracts DE-AC02-06CH11357 (NE-CAT) and W-31-109-Eng-38 (SER-CAT).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank (PDB ID codes 6OF6, 6OJ2, 6OPE, and 6ORD).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004170117/-/DCSupplemental.
References
- 1.Crick F. H., On protein synthesis. Symp. Soc. Exp. Biol. 12, 138–163 (1958). [PubMed] [Google Scholar]
- 2.Uhlenbeck O. C., Schrader J. M., Evolutionary tuning impacts the design of bacterial tRNAs for the incorporation of unnatural amino acids by ribosomes. Curr. Opin. Chem. Biol. 46, 138–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaRiviere F. J., Wolfson A. D., Uhlenbeck O. C., Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294, 165–168 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Fahlman R. P., Dale T., Uhlenbeck O. C., Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16, 799–805 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Ledoux S., Uhlenbeck O. C., Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell 31, 114–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louie A., Ribeiro N. S., Reid B. R., Jurnak F., Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 259, 5010–5016 (1984). [PubMed] [Google Scholar]
- 7.Schuette J. C. et al., GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 28, 755–765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledoux S., Olejniczak M., Uhlenbeck O. C., A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat. Struct. Mol. Biol. 16, 359–364 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olejniczak M., Dale T., Fahlman R. P., Uhlenbeck O. C., Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 12, 788–793 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Olejniczak M., Uhlenbeck O. C., tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88, 943–950 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Gromadski K. B., Daviter T., Rodnina M. V., A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol. Cell 21, 369–377 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Murakami H., Ohta A., Suga H., Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat. Struct. Mol. Biol. 16, 353–358 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Gabriel K., Schneider J., McClain W. H., Functional evidence for indirect recognition of G.U in tRNA(Ala) by alanyl-tRNA synthetase. Science 271, 195–197 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Selmer M. et al., Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Ogle J. M., Murphy F. V., Tarry M. J., Ramakrishnan V., Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G., A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Loveland A. B., Demo G., Grigorieff N., Korostelev A. A., Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auffinger P., Westhof E., Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 292, 467–483 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Westhof E., Fritsch V., RNA folding: Beyond Watson-Crick pairs. Structure 8, R55–R65 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Maehigashi T., Dunkle J. A., Miles S. J., Dunham C. M., Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc. Natl. Acad. Sci. U.S.A. 111, 12740–12745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagan C. E., Maehigashi T., Dunkle J. A., Miles S. J., Dunham C. M., Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA 20, 1944–1954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz-Meoz R. F., Green R., Helix 69 is key for uniformity during substrate selection on the ribosome. J. Biol. Chem. 286, 25604–25610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali I. K., Lancaster L., Feinberg J., Joseph S., Noller H. F., Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell 23, 865–874 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Agris P. F., Narendran A., Sarachan K., Väre V. Y. P., Eruysal E., The importance of being modified: The role of RNA modifications in translational fidelity. Enzymes 41, 1–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Väre V. Y., Eruysal E. R., Narendran A., Sarachan K. L., Agris P. F., Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules 7, 29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozov A., Westhof E., Yusupov M., Yusupova G., The ribosome prohibits the G•U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res. 44, 6434–6441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozov A. et al., Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 7, 10457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manickam N., Nag N., Abbasi A., Patel K., Farabaugh P. J., Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 20, 9–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linpinsel J. L., Conn G. L., General protocols for preparation of plasmid DNA template, RNA in vitro transcription, and RNA purification by denaturing PAGE. Methods Mol. Biol. 941, 43–58 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polikanov Y. S., Melnikov S. V., Söll D., Steitz T. A., Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov G. N. et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potterton L. et al., CCP4i2: The new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 74, 68–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Schrödinger LLC, The PyMOL Molecular Graphics System, Version 2.3.1, (Schrödinger LLC, New York, 2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallography, atomic coordinates, and structure factors have been deposited in the PDB, https://www.wwpdb.org/ (PDB ID codes 6OF6, 6OJ2, 6OPE, 6ORD).