Abstract
Asthma is a heterogeneous respiratory disease reflecting distinct pathobiologic mechanisms. These mechanisms are based, at least partly, on different genetic factors shared by many other conditions, such as allergic diseases and obesity. Investigating the shared genetic effects enables better understanding of the mechanisms of phenotypic correlations and is less subject to confounding by environmental factors. The increasing availability of large-scale genome-wide association study (GWAS) for asthma has enabled researchers to examine the genetic contributions to the epidemiologic associations between asthma subtypes and those between coexisting diseases and/or traits and asthma. Studies have found not only shared but also distinct genetic components between asthma subtypes, indicating that the heterogeneity is related to distinct genetics. This review summarizes a recently compiled analytic approach—genome-wide cross-trait analysis—to determine shared and distinct genetic architecture. The genome-wide cross-trait analysis features in several analytic aspects: genetic correlation, cross-trait meta-analysis, Mendelian randomization, polygenic risk score, and functional analysis. In this article, we discuss in detail the scientific goals that can be achieved by these analyses, their advantages, and their limitations. We also make recommendations for future directions: (1) ethnicity-specific asthma GWASs and (2) application of cross-trait methods to multiomics data to dissect the heritability found in GWASs. Finally, these analytic approaches are also applicable to complex and heterogeneous traits beyond asthma.
Key words: Asthma, heterogeneity, GWAS, shared genetics, cross-trait, allergic diseases, obesity
Abbreviations used: ASSET, Association analysis based on SubSETs; CAAPA, Consortium on Asthma among African-Ancestry Populations in the Americas; CPASSOC, Cross-phenotype association; ENCODE, Encyclopedia of DNA elements; eQTL, Expression quantitative trait locus; GTEx, Genotype-Tissue Expression Project; GWAS, Genome-wide association study; ImmGen, Immunological Genome Project; LD, Linkage disequilibrium; LDSC, Linkage disequilibrium score regression; SNP, Single-nucleotide polymorphism
Epidemiologic factors associated with asthma
Asthma is a common chronic respiratory disease that affects approximately 340 million individuals worldwide.1 Epidemiologic studies have identified many factors associated with asthma, including the following: genetic factors; demographics (eg, age and sex); personal and family history of allergic diseases and comorbid illnesses (eg, obesity and mental illnesses); and environmental (eg, air pollution), nutritional, and lifestyle (eg, physical activity) factors.2
Of the various factors associated with development of asthma, allergic diseases such as atopic dermatitis and allergic rhinitis play a pivotal role. The coexistence of allergic diseases with asthma in children and adolescences is high (eg, as high as 60% to 80% for allergic rhinitis).3 In both children and adults, allergic sensitization to allergens has been found to be an important risk factor for asthma and bronchial hyperresponsiveness.4, 5, 6, 7, 8 It is likely that the frequent coexistence of allergic diseases and asthma is due to their similar pathobiologic mechanisms.9
Another major factor associated with asthma is obesity in both children and adults.10 , 11 This relation is complex: “obese-asthma syndrome” consists of multiple subgroups (eg, de novo asthma, asthma modified by obesity, and obesity predisposed by asthma).10 Yet, studies have suggested the causal link from (anthropomorphically defined) overweight or obesity to asthma inception.10 , 12 , 13 Emerging evidence also suggests the role of adiposopathy—“sick fat” or adipose tissue dysfunction—in the pathogenesis of complex disease conditions, including asthma.10 Adiposopathy is characterized by impaired adipogenesis, altered lipid metabolism, and adipose and/or systemic inflammation (eg, upregulated IL-6, TH1 polarization, and TH17 pathways).10
Furthermore, studies have also reported that mental health disorders, such as attention-deficit/hyperactivity disorder,14 anxiety, and major depressive disorder, are a comorbidity of asthma.15 Yet, studies have suggested that such associations are potentially bidirectional.16 , 17 For example, anxiety can induce asthma symptoms,16 whereas living with an asthma condition (eg, poor asthma control and worse asthma-related quality of life) may have mental health implications.17 The exact mechanisms that underlie these mental health disorder–asthma associations remain uncertain.
Asthma heterogeneity and its related genetics
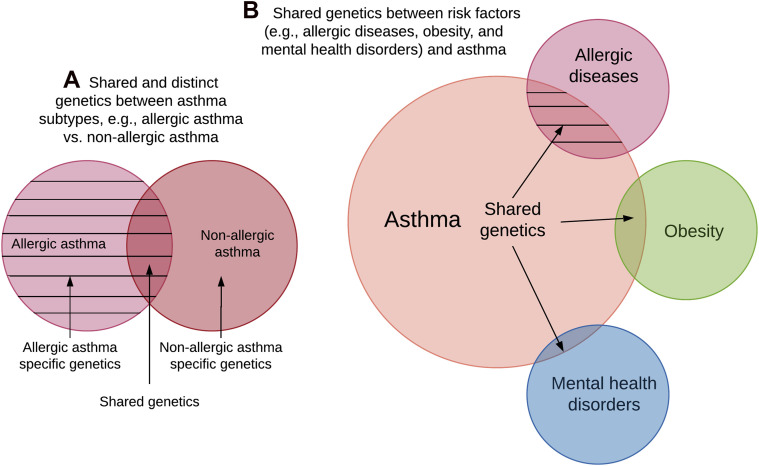
Understanding the exact pathobiology of asthma involves several major challenges—the identification of causal mechanisms, the effect of multiple environmental factors (eg, diet, physical activity, air pollution, and environmental microbiome), and the heterogeneity of asthma itself. Although asthma had been considered a single disease for decades, a growing body of literature has revealed that asthma comprises a range of heterogeneous subtypes differing in presentation and disease course18 and that the heterogeneity is based, at least partly, on different genetic factors for asthma subtypes (eg, childhood vs adult asthma, allergic vs nonallergic asthma).12 , 19 , 20 Accordingly, examinations of subtype-specific genetics in conjunction with shared genetic factors between coexistent diseases or traits (eg, allergic diseases and obesity) and asthma should inform research on the heterogeneity in asthma and provide insight into corresponding pathology (Fig 1 ).21 , 22
Fig 1.
Venn diagram of shared and distinct genetics between asthma subtypes and those between coexistent diseases or traits and asthma. A, Shared and distinct genetics between asthma subtypes (eg, allergic asthma vs nonallergic asthma). B, Shared genetics between coexistent diseases/traits (eg, allergic diseases, obesity, mental health disorders) and asthma. The overlapping size between 3 example coexistent diseases/traits and asthma are based on the order of their genetic similarities, with allergic diseases sharing the most genetic components with asthma, followed by obesity and mental health disorders. A and B, The area covered by horizontal cross-lines indicates similar underlying genetic components/mechanisms in allergic asthma and shared genetics of allergic diseases and asthma.
The genetic effect of asthma is significant, with the heritability estimates ranging from 35% to 95%.23 Genome-wide association studies (GWASs) have been widely applied to complex diseases for more than 2 decades, with a greatly increased sample size. However, according to Schoettler et al, in the GWAS of asthma, a larger sample size with heterogeneous subtypes is not necessarily better than a smaller sample size for homogeneous subtypes to identify the relevant genetic variants because the genetic background between asthma subtypes may be different.24 Thus, investigating the shared genetic contribution to coexistent diseases or traits (eg, allergic disease, obesity) and specific asthma subtypes (eg, allergic asthma, obesity-associated asthma phenotype) would boost the power to detect subtype-specific variants that would have been masked by a traditional single-disease GWAS (Fig 1). A comprehensive characterization of these shared genetic architectures would improve understanding of the multiple dimensions of asthma pathobiology.
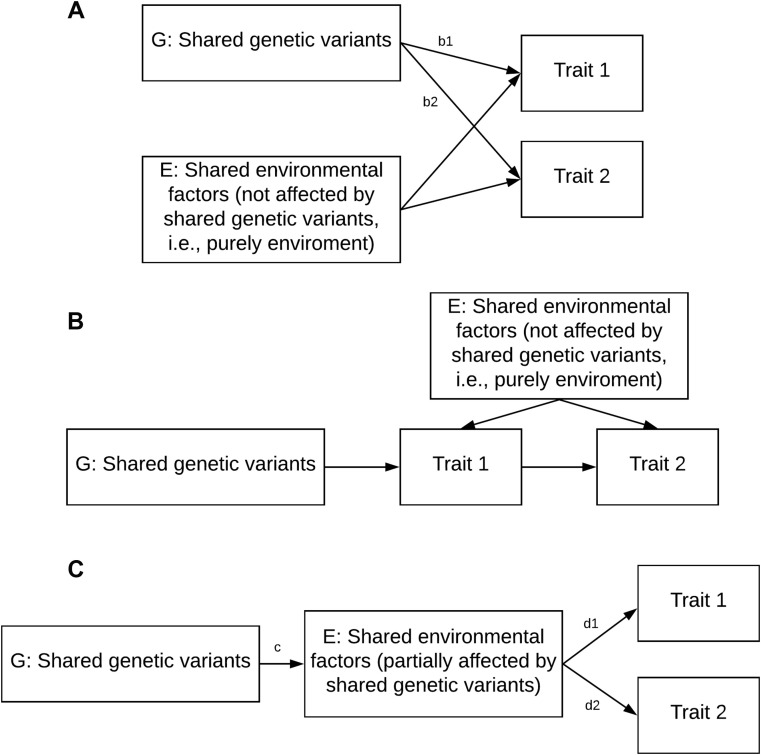
Traditionally, examining the phenotypic correlation or coexistence of other factors is a useful way to investigate the heterogeneity of asthma. However, this approach may have residual confounding and provide insufficient biologic insight as to which underlying mechanism(s) drive the association. A major advantage in going from phenotypic correlations to genetic correlations is improved understanding of the mechanism(s): shared genetic components can be identified at different levels from the whole genome to individual variants, providing insights into the reasons why asthma and coexistent diseases or traits are correlated. Furthermore, genetic correlations are less subject to confounding by environmental factors for several reasons. After adequate. control for population ancestry, genetic correlation would occur only if the germline genetic variant is causal or in linkage disequilibrium (LD) with the causal variant of both traits. A purely environmental confounding factor (eg, air pollution) would not lead to genetic correlation because it is not associated with any genetic variant (Fig 2 , A and B). In contrast, if an environmental factor is an intermediary step between the genetic variant and the trait, it is in the causal pathway and is not considered a confounder (ie, it does not create a false genetic correlation between the 2 traits) (Fig 2, C). Population stratification is arguably the only confounding factor in GWAS, but it can be effectively controlled by using principal components from genome-wide genetic markers.25 Once the genetic effect on diseases and traits has been robustly established, the genetic correlation between diseases and traits can be reliably estimated and replicated.26, 27, 28, 29 In the following sections, we will discuss a range of detailed analyses that can be used to compile a comprehensive investigation between asthma and other coexistent diseases or traits.
Fig 2.
Directed acyclic graphs of relationship between shared genetic or environmental factors with traits. A, Shared environment factors (not affected by shared genetic variants) will not bias genetic correlation. After appropriately control for population ancestry, the genetic effects b1 and b2 are unrelated to E, and therefore the genetic correlation—correlation b1 and b2—is not related to E. B, Another situation in which shared environment factors (not affected by shared genetic variants) will not bias genetic correlation. C, Shared environment factors (partially affected by shared genetic variants). In this case, E is not considered a confounder; rather, it is considered a mediator in the causal pathway of interest; c represents the effect of shared genetic variants on environmental factors, and d1 and d2 represent the effect of shared environmental factors on traits.
Genome-wide cross-trait analysis study design
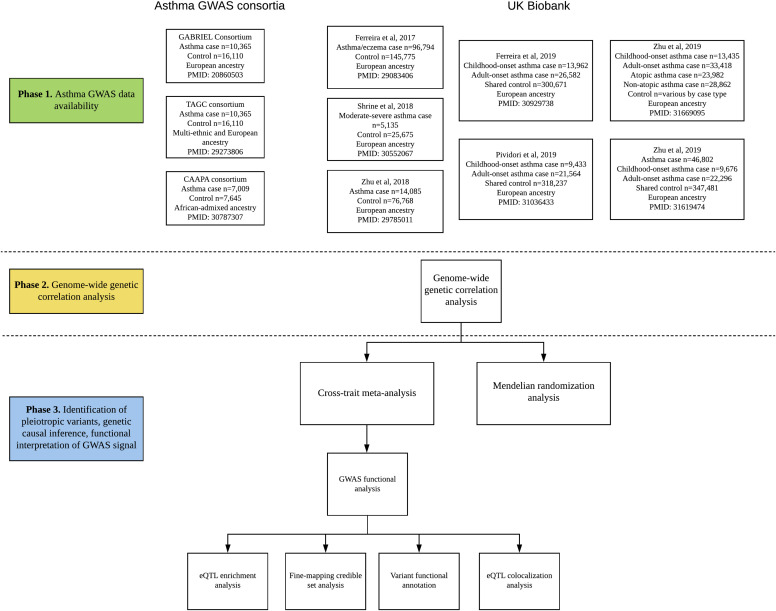
With the increasing availability of large-scale genetic data for asthma, such as the GABRIEL Consortium,30 the Trans-National Asthma Genetic Consortium,31 and the UK Biobank,12 , 21 , 22 , 26 , 32 as well as the advancement of genetic epidemiology and statistical genetics methods, researchers are now able to examine the genetic contribution to the epidemiologic associations between asthma subtypes and those between coexistent diseases or traits and asthma. For example, to understand the genetics of asthma heterogeneity, 2 recent studies examined the genetic overlap between asthma subtypes—childhood asthma and adult asthma—by using the UK Biobank and 23andMe data.19 , 20 They both found substantially shared (eg, IL1RL1, HLA–DQA1) but also distinct (eg, ORMDL3 specific for childhood asthma) genetic components between these 2 subtypes, supporting the idea that the heterogeneity is related to distinct genetics.19 , 20 These fundamental studies largely depend on single-trait analysis, and they can be further extended by our recently implemented study design called genome-wide cross-trait analysis, which is broadly applicable to asthma and many other diseases and/or traits. The design has been successfully applied to the UK Biobank and GWAS consortia data sets and has determined the shared genetic architectures between asthma and allergic diseases,22 obesity,12 and mental health disorders,21 which were reproducible in other studies.26, 27, 28, 29 A genome-wide cross-trait analysis features several analyses: genetic correlation, cross-trait meta-analysis, Mendelian randomization, polygenic risk score, and GWAS functional analysis. Each component is discussed in more detail in subsequent sections and depicted in Fig 3 . A glossary of the cross-trait GWAS terminology may be found in Table I . A summary of genome-wide cross-trait analysis methods may be found in Table II .
Fig 3.
Data availability for GWAS of asthma and study design of genome-wide cross-trait analysis. Genome-wide genetic correlation analysis is used to examine the genetic correlation between a pair of traits by using genome-wide SNPs. Cross-trait meta-analysis is used to determine the shared genetic variants between multiple traits. Mendelian randomization analysis is used to examine the causal effect of the exposure trait on the other trait by using the genetic variants for exposure trait as the instrument variables. eQTL enrichment analysis is used to determine the enrichment of genetic variants associated with complex traits in eQTL. Fine mapping credible set analysis is used to examine whether there is a potential causal variant at each locus. Variant functional annotation is used to predict the functional effect of an individual SNP on a transcript. eQTL colocalization analysis is used to determine the shared causal variants between GWAS signals and eQTL signals. CAAPA, Consortium on Asthma among African-ancestry Populations in the Americas; TAGC, Trans-National Asthma Genetic Consortium.
Table I.
Glossary of terms related to genome-wide cross-trait analysis
| Term | Definition |
|---|---|
| Cross-trait meta-analysis | A meta-analysis testing the null hypothesis that none of the traits being examined is associated with the genetic variant. One genetic variant is tested at a time. |
| eQTLs | Genetic variants that are associated with the gene expression levels. |
| Genetic correlation | Assuming that all genetic variants have some effect on a trait and that their effect size follows a gaussian distribution (called the infinitesimal model), the genetic correlation between 2 traits (A and B) measures the Pearson correlation between the genetic variant effect on traits A and B. |
| GWAS | An analytic method that tests the association between each genetic variant and a specific phenotype (a disease status or a quantitative trait). One genetic variant is tested at a time. |
| HLA/MHC region | A genomic region of an approximately 3.6-Mb genome sequence located on the chromosome 6p21, which is mainly known for its pervasive pleiotropic effect and immune-related function. The extended MHC region is at 25 to 34 Mb on chromosome 6. |
| Horizontal pleiotropy | A genetic variant or gene having independent effects on multiple traits that do not have a causal effect on each other. |
| Instrumental variables | Variables that are associated with the modifiable exposure or risk factor of interest and affect the outcome only through the exposure or risk factor. |
| Mendelian randomization | An analytic approach that examines the causality of an observed association of a modifiable exposure or risk factor with an outcome of interest by using ≥1 genetic instrumental variables. |
| Polygenic risk score | A score based on a set of disease and/or trait-associated genetic variants, commonly defined as the weighted sum of their genotypes. Weights are chosen by their association effect on the disease and/or trait, directly from GWAS or further modified on the basis of a suitable statistical model incorporating all genetic variants on the genome. |
| Vertical pleiotropy (genetic causality) | A genetic variant or gene having an effect on a trait that has causal effect on another trait. |
Table II.
Summary of genome-wide cross-trait analysis methods
| Analysis method | Software | Advantages | Disadvantages | Examples of application in asthma or complex traits/PMID |
|---|---|---|---|---|
| Genetic correlation | LDSC/S-LDSC | Requires only GWAS summary statistics; computationally efficient; accounts for additive confounding in single-trait heritability (such as population stratification) and confounding in genetic correlation (such as overlapping samples); can allow relatively flexible heritability architecture in MAF, LD, and functional categories (the authors LDSC recommended S-LDSC) | Is sensitive to other genetic architectures not captured by the baseline LD model; requires that the reference panel LD and GWAS summary statistics be computed from the same population | Childhood asthma and adult asthma/30929738, allergic diseases and asthma/29785011, obesity and asthma/31669095, mental health disorders and asthma/31619474 |
| GCTA/GCTA-LDMS | Estimates genetic correlation with high accuracy; the LD and association effect are computed from the same genotype data; accounts for different genetic architectures by MAF and LD categories (the authors of GCTA recommended GCTA–LDMS-I) | Requires genotype data; computation is infeasible for an extremely large data set | Complex traits/ 21167468 | |
| SumHer/BLD-LDAK | Is similar to LDSC but assumes a specific parametric model for MAF/LD-dependent genetic architecture and multiplicative inflation bias due to population stratification or family relatedness; can allow the same baseline LD categories as in LDSC (the authors of LDAK recommended BLD-LDAK/BLD-LDAK-alpha) | Is sensitive to other genetic architecture deviated from the assumed parametric model; requires that the reference panel LD and GWAS summary statistics be computed from the same population | Complex traits/ 32203469 | |
| Cross-trait meta-analysis | ASSET | Accounts for overlapping samples | Is applicable only to binary traits | Allergic diseases and asthma/29785011, mental health disorders and asthma/31619474 |
| CPASSOC | Is applicable to both binary and continuous traits | Yields potential false positives due to overlapping samples | Obesity and asthma/31669095 | |
| MTAG | Accounts for possibly unknown sample overlap | Requires an assumption that all variants share the same genetic correlation across all traits (ie, no subset-specific effect is assumed) | Complex traits/29292387 | |
| Mendelian randomization | Inverse variance–weighted approach | Is applicable when the genetic variants’ pleiotropic effects (genetic variant–outcome direct effect) happen to cancel out | Accounts for only the designed scenario; requires independent variants | Asthma and cancer/32006205 |
| Egger regression | Is applicable when the genetic variant–exposure association is independent of the pleiotropic effect; appears to protect false positives in several simulation studies | Accounts for only the designed scenario; requires independent variants; when the outcome GWAS is low-power, its power to detect causal effect could be substantially smaller than that of other methods | Asthma and cancer/32006205 | |
| Weighted median estimator | <50% (counts or total weights) of the genetic variants are invalid instruments | Accounts for only the designed scenario | Asthma and cancer/32006205 | |
| Weighted mode-based estimate (weighted MBE) | Is applicable when the variants satisfying the exclusion restriction assumption give a causal effect estimate that is the majority among the effect estimates from all variants in the analysis; appears to protect false positives in several simulation studies | Accounts for only the designed scenario | Asthma and cancer/32006205 | |
| GSMR | Accounts for LD between variants; detects and accounts for outliers that could violate the exclusion restriction assumption | Requires sufficient numbers of GWAS significant variants; requires a genetic variant–exposure association that is independent of the pleiotropic effect | Obesity and asthma/31669095, mental health disorders and asthma/31619474 | |
| MR-PRESSO | Detects and accounts for outliers that could violate the exclusion restriction assumption | Requires independent and sufficient numbers of GWAS significant variants; requires a genetic variant–exposure association that is independent of the pleiotropic effect | Asthma and cancer/32006205 | |
| LCV | Distinguishes genetic correlation from genetic causality; uses all genome-wide variants | Does not estimate causal effect size, but does provide a scale parameter with higher magnitude indicating that it is closer to causality; requires that the LD reference match the study populations | Obesity and asthma/31669095 | |
| CAUSE | Allows genetic correlation and genetic causality for different variants; can estimate causal effect size; uses all genome-wide variants; has better power avoid false positive trade-off than other methods | Requires independent variants by pruning GWAS results; may have a higher than expected false-positive rate than Egger regression and MBE when a large fraction of variants affect the exposure and outcome through a strong shared factor; in addition, the power for both exposure and outcome GWAS is high | Complex traits/32451458 | |
| Polygenic risk score | LDpred | Accounts for LD between SNPs | Is computation-intensive when the number of SNPs is more than a couple of million | Asthma/32522462 |
| MTGBLUP | Simultaneously estimates genetic effect and genetic correlation for multiple traits | Assumes an infinitesimal genetic architecture. Is computation-intensive when the number of SNPs is more than a couple of million and becomes prohibitive for more than hundreds of thousands of samples | Complex traits/25640677 | |
| MTAG | Provides improved polygenic prediction thanks to a consistent estimator, and its effect estimates always have a lower genome-wide mean squared error than the corresponding single-trait GWAS estimates do | Requires an assumption that all variants share the same genetic correlation across all traits (ie, no subset-specific effect is assumed); power might be reduced when the genetic correlations between traits are not very high | Complex traits/29292387 | |
| CTPR | Optimizes the prediction accuracy for the primary trait of interest by taking advantage of shared genetic effects among multiple traits; secondary traits of GWAS can be individual-level data or summary statistics | Requires individual-level data for the primary trait of interest | Complex traits/30718517 |
BLD, Background linkage disequilibrium; CAUSE, causal analysis using summary effect estimates; CTPR, cross-trait penalized regression; GCTA, genome-wide complex trait analysis; GSMR, generalized summary data–based Mendelian randomization; LCV, latent casual variable; LDMS-I: linkage disequlibrim and minor allele frequency stratified-I; MAF, minor allele frequency; MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; MTAG, multitrait analysis of GWAS; MTGBLUP: multi-trait genomic best linear unbiased prediction; PMID, PubMed identifier; S-LDSC, stratified LDSC; SumHer, single-nucleotide polymorphism heritability.
Pleiotropy and causality
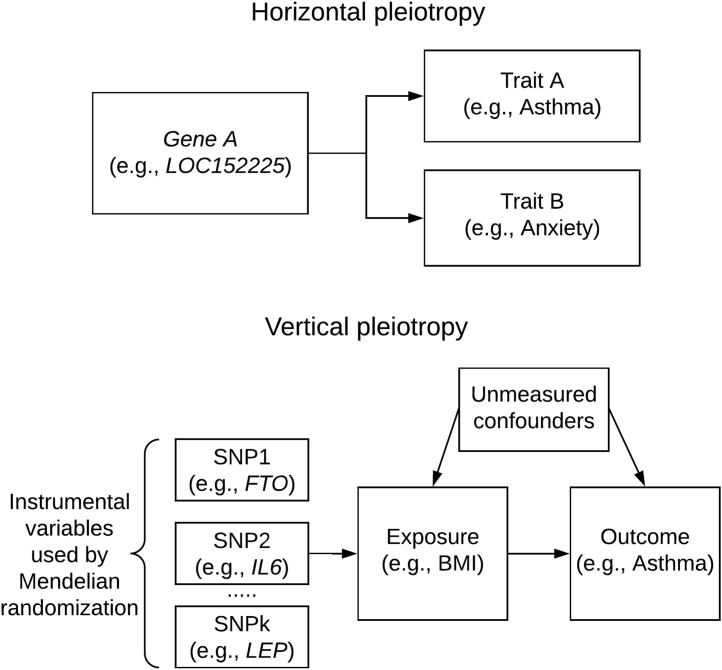
Genetic pleiotropy refers to the same gene simultaneously influencing multiple traits.33 A gene can be associated with more than 1 trait in 2 ways: horizontal pleiotropy and vertical pleiotropy (Fig 4 ). Horizontal pleiotropy, which is often simplified as pleiotropy, is defined as 1 genetic variant having independent effects on multiple traits. Vertical pleiotropy, which is often called genetic causality, is defined as a genetic variant having an effect on a trait via its genetic effect on an intermediate trait. Identification of pleiotropy may improve the understanding and utility of disease-gene biology in multiple ways. First, there is a potential to detect the broad biologic impact of a gene, such as through phenome-wide association studies.34 Second, if a pharmacologic genetic target could affect multiple traits or diseases, it might allow a drug developed for 1 disease to be repurposed for other diseases. For example, 3-hydroxy-3-methyl-glutaryl–coenzyme A reductase inhibitors, commonly known as statins, have also been found to have several molecular actions beyond cholesterol reduction,35 such as reducing the development of type 2 diabetes.36 Third, knowing the genetic causality can help develop disease prevention strategies via intervention in relation to nongenetic modifiable factors. For example, because the obesity-asthma genetic association suggests obesity-to-asthma effects, reduction of body mass index in patients with obesity might counteract the genetic effect, thereby potentially preventing the development of asthma. Therefore, distinguishing horizontal pleiotropy from vertical pleiotropy in cases in which both contribute to genetic correlations is important and can be challenging. We discuss methods for these analyses in the following sections on cross-trait meta-analysis and Mendelian randomization.
Fig 4.
Diagram of horizontal pleiotropy and vertical pleiotropy with examples for asthma. BMI, Body mass index.
Genetic correlation
The genetic correlation between 2 traits (eg, A and B) measures the Pearson correlation between the genetic variant effect on traits A and B.37 , 38 It could be the result of horizontal pleiotropic action of genes on trait A and B or a causal link between A and B. Genetic correlation indicates intrinsic correlations between 2 traits unaffected by environmental confounders, which are common and often unavoidable in conventional epidemiologic studies. Several methods have been developed to estimate genetic correlations, such as linkage disequilibrium score regression (LDSC/stratified-LDSC),37 genome-wide complex trait analysis (GCTA/GCTA-linkage disequilibrium and minor allele frequency stratified-I),38 and SumHer/BLD-LDAK from Linkage-Disequilibrium Adjusted Kinships (LDAK).39 Among these, LDSC becomes one of the most commonly used methods for estimating genetic correlations because it uses GWAS summary statistics, which largely reduces computational burden and accounts for confounding in single-trait heritability (such as population stratification) and confounding in genetic correlation (such as shared study subjects).22 , 40 The genetic correlation estimate (the Rg value) ranges from –1 to 1, where –1 indicates a perfect negative genetic correlation and 1 indicates a perfect positive genetic correlation. For example, Ferreira et al recently reported that the genetic correlation between childhood and adult asthma is 0.67 using same set of control samples, indicating both shared and distinct genetics between 2 asthma subtypes at the genome-wide level.19 A recent study also conducted sensitivity analyses to examine the potential bias in LDSC due to overlapping subjects; the study used the following 4 scenarios while maintaining the same sample size: (1) no case overlap or control overlap, (2) case overlap and no control overlap, (3) no case overlap and control overlap, and (4) case overlap and control overlap. The sensitivity analyses showed that the Rg estimate from LDSC is unbiased to overlapping cases and/or controls.22
Cross-trait meta-analysis
Genetic correlation depicts the genome-wide average sharing of genetic effect between traits. To identify genetic variants with pleiotropic effects, cross-trait GWASs have used a range of conventional and recently developed meta-analysis methods. Typically, summary statistics of distinct but potentially related traits are combined in a meta-analysis framework to detect specific loci with shared associations. Such univariate approaches do not require access to individual-level genotype data and thus are readily applicable to existing GWAS results. Combining results across studies of different traits also improves the statistical power of detecting modest cross-trait genetic effects that may not have reached genome-wide significance for any single trait.
Many cross-trait meta-analysis methods are available.41 Two specific methods, association analysis based on SubSETs (ASSET)42 for binary traits and cross phenotype association (CPASSOC)43 for continuous or binary traits, have been shown to outperform a range of alternatives for detecting pleiotropy effects shared in all traits or a subset of traits.41 ASSET combines an all-subsets fixed-effects GWAS meta-analysis with a bayesian method to evaluate the best-fit configuration of genotype-phenotype associations.42 Similar to the ASSET, CPASSOC assumes that effects may exist only within a subset of traits.43 This feature is not only useful for detecting overall pleiotropy but is also important for detecting subset-specific effect (eg, variants shared with only some but not all asthma subtypes because of heterogeneity). Unlike ASSET, CPASSOC identifies the subset of studies with effects by sequentially adding a trait by an incremental order of their association significance. Among the sequentially examined subsets, the one with the highest meta-statistics is selected. Both methods are designed to correct for inflation due to overlapping subjects. However, our previous simulation analysis showed that caution is advised for CPASSOC when almost all controls were shared between studies.41 Another caveat is that it is sometimes difficult to retrieve the exact number of overlapping subjects between 2 studies, which is required for ASSET. A recently developed method called multitrait analysis of GWAS44 describes a promising strategy to address the problem by using LDSC.37 However, this method assumes that all single-nucleotide polymorphisms (SNPs) share the same variance-covariance matrix among all traits, which could be violated when some SNPs are associated only with a subset of traits.44
In a previous cross-trait analysis of allergic diseases and asthma,22 ASSET was used to determine 38 shared loci, some of which (such as variations in EVI5, NRROS, and C11orf30) are newly discovered from the cross-trait meta-analysis. In the cross-trait analysis of mental health disorders and asthma,21 the use of ASSET identified 7 loci that are jointly associated with attention-deficit/hyperactivity disorder and asthma, 1 locus that is jointly associated with anxiety disorder and asthma, and 10 loci that are jointly associated with major depressive disorder and asthma. Of note, the HLA region (chromosome 6, 25-34 Mb) was found to be shared in the cross-trait meta-analysis of allergic disease and asthma and in that between major depressive disorder and asthma. The HLA region was commonly reported to have important pleotropic effects.45 However, because of its high gene density and extensive LD, GWAS signals within the HLA region are difficult to map in fine resolution. Thus, a considerable amount of work, which was recently developed in the sequencing of the entire HLA region,46 has established its central role in the biology of the immune system and in predisposition to a large number of inflammatory diseases. However, the extent to which these diseases share the same causal risk variants or genes remains unclear. Finally, in the cross-trait analysis of obesity and asthma, CPASSOC has been used to determine potential shared loci and variations in MYL6 and ACOXL and replicated them in a mouse model.12
Many cross-trait effects are not surprising. For example, variants in the C11orf30, IL1R1, and FLG genes, as well as in the HLA region, are associated with allergic diseases and asthma,22 as was found in previous studies with individual traits.26 Others are perhaps less intuitive and can shed light on hitherto unknown connections between traits. For example, variants in the POLI gene have been shown to affect risk of both major depressive disorder and asthma. These seemingly unrelated diseases share pathways involved in immune processes.21 These results demonstrated a key benefit of cross-trait meta-analysis, namely, discovery of shared loci that have not been reported as having genome-wide significance (P < 5 × 10–8) from the individual-trait GWAS. As another example, whereas the RERE gene was found to be associated only with asthma in the single-trait GWAS,22 the top variant, rs301817, within RERE gene was found to be shared by major depressive disorder and asthma,21 although the biologic role of RERE between asthma and major depression needs further exploration.
Mendelian randomization
When 2 traits are known to be correlated (phenotypically or genetically), identification of causality between the traits is imperative. Here, GWAS data for these traits provide a unique opportunity to make robust causal inference by using GWAS summary statistics without requiring individual-level data for both traits in the same set of subjects. Mendelian randomization refers to an analytic approach that uses the genetic variants for 1 trait (exposure [eg, body mass index]) as the instrument variables to examine the causal effect of the exposure trait on the other trait (outcome [eg, asthma]). This method is based on the theory that the germline alleles of these variants are randomly allocated, which can be seen analogously to the randomized treatment assignment in a randomized controlled trial resulting in an unconfounded exposure-outcome relationship.47 It provides a valuable tool—especially when randomized controlled trials are not feasible and observational studies provide a biased estimate for causal effects because of unmeasured confounding, model misspecification, or reverse causation. Of note, the validity of Mendelian randomization depends on 3 key assumptions: (1) the relevance assumption, (2) the independence assumption, and (3) the exclusion restriction, which have been thoroughly described in a previous review.47 Briefly, relevance assumption refers to the genetic variants that are strongly associated with the exposure of interest; independence assumption refers to the effect of genetic variants on the outcome of interest that is unconfounded; and exclusion restriction refers to the genetic variants that affect the outcome only through the exposure of interest.
With the availability of large-scale GWASs, it has become easier to find genetic variants that are strongly associated with the exposure trait (satisfying the relevance assumption). Although population stratification is arguably the only potential confounder between the genetic variants and the outcome, it can be effectively accounted for in standard GWAS analysis. Accordingly, the independence assumption is not difficult to satisfy. The exclusion restriction assumption is the nonexistence of other pathway(s) from the genetic variant to the outcome except through the exposure. This assumption is difficult to verify and hence has led to the development of various Mendelian randomization methods that relax this assumption in 3 ways: (1) by using less stringent assumptions, such as an inverse variance weighted approach,48 Mendelian randomization–Egger regression,49 a weighted median estimator,50 and a mode-based estimate51; (2) by removing genetic variants that violate the assumption, such as generalized summary data–based Mendelian randomization52 and Mendelian randomization pleiotropy residual sum and outlier53; and (3) by estimating the likelihood from pure horizontal pleiotropy to pure causality, such as the latent causal variable54 and causal analysis using summary effect estimates.55 The major features of each method can be found in Table II. Users can consider the likely pathobiology of the exposure and outcome to choose the most appropriate assumption and pick the most appropriate method. Often, however, which assumptions have been met is not clear. In such situations, we suggest conducting the Mendelian randomization by using multiple methods as a sensitivity analysis.56 If multiple methods arrive at a similar conclusion, then we would consider the results robust. However, when the conclusions reached by using these methods do not agree, we recommend close comparisons of their results, including for consistency of the direction for causal effect estimates, the magnitude of the effect, and the statistical significance (related to the power and sample size). Later methods (eg, latent causal variable, causal analysis using summary effect estimates) might address some of the drawbacks of the methods developed earlier (eg, the inverse variance–weighted method). Thus, their disagreement would become more interpretable. Accordingly, we recommend cautious interpretation of the results when different methods yield discordant results.
Polygenic risk score
A genetic risk score aggregates genetic variants aiming to predict disease risk or trait level, and it can thereby help guide early prevention, targeted intervention, and characterization of subtypes for complex diseases (including asthma).57 With the arrival of large-scale publicly available GWAS summary statistics, a polygenic risk score integrating genome-wide variants regardless of statistical significance is a promising approach to realize its clinical utility and potential public health benefit.58 , 59 Among the many available polygenic risk models, those that are widely used include LD pruning followed by P value thresholding (probability + threshold) and LDpred, a bayesian framework that estimates posterior mean causal effect sizes from GWAS summary statistics by assuming a prior for the genetic architecture and LD information from a reference panel.60 This method was recently applied to investigate the association between the asthma polygenic risk scores and COVID-19.61 More recent methods have further improved prediction accuracy by combining effects across multiple genetically related traits; these methods include multi-trait genomic best linear unbiased prediction,62 multitrait analysis of GWAS,44 and cross-trait penalized regression (which attempts to optimize the prediction accuracy for the primary trait of interest by taking advantage of shared genetic effects among multiple traits through a multivariate penalized least-squares method). These methods can use either GWAS summary statistics or individual-level genotype data and can thus provide an opportunity to integrate many GWAS results.63 The cross-trait design has demonstrated an advantage over a single-trait design in terms of prediction accuracy and model calibration.63 One limitation of most of the current polygenic risk score methods is the generalizability of the algorithms to non-European racial/ethnic groups, which may exacerbate health disparities.64 Large-scale application of polygenic risk scores in asthma studies is still ongoing and remains to be seen in future publications, especially in studies involving non-European racial/ethnic groups.
Cross-trait gwas functional analysis
Cross-trait meta-analysis methods are helpful to identify novel genetic loci that are associated with multiple diseases and traits. However, the functional impact of the shared loci on disease risks or traits remain to be clarified. In the past decade, there has been development of large-scale studies for genomic functions annotation, including the Encyclopedia of DNA Elements65 and National Institute of Health Roadmap Epigenomics Project,66 which can be readily used to categorize the functions for GWAS-discovered loci. Another useful source of functional annotation is use of expression quantitative trait loci (eQTLs), namely, the discovery of genetic variants that explain the variations in gene expression levels, providing annotation to the regulatory regions outside coding sequence.
As of now, several eQTL databases are publicly available for GWAS functional analysis. The databases that are especially useful for asthma and other inflammatory diseases include the Genotype-Tissue Expression (GTEx) project67 and Immunological Genome Project (ImmGen).68 , 69 The GTEx project is a resource database for studying the relationship between genetic variation and gene expression in different human tissues.67 GTEx version 8 contains 54 types of human tissues and 17,382 RNA-sequencing samples from 948 donors. Among these, 49 tissues have eQTL data. The application of GTEx eQTL in asthma can be useful. For example, in a cross-trait analysis, the use of GTEx revealed that shared genes between allergic diseases and asthma (eg, FLG) are most significantly enriched in epithelial tissues, such as skin.22 Traditionally, researchers look to eQTL databases to identify gene expression associated with disease variants. However, a recent GTEx study has also pointed out that nearly all common variations show some associations with expression of at least 1 gene in at least 1 tissue and that care is required when using their eQTLs as demonstration of a causal gene.67 Thus, we also recommend colocalization analysis70 to determine the shared causal variants between GWAS signals and eQTL signals. Colocalization analysis70 has adapted a bayesian model to compute the posterior probabilities for 4 scenarios that the 2 traits sharing causal variants at a specific locus: (1) no causal variant exists for either trait, (2) causal variants exist for only 1 trait, (3) causal variants exist for both traits, but they are different variants, and (4) causal variants exist for both traits, and they are the same variants. For example, in the cross-trait meta-analysis of allergic disease and asthma,22 the variant rs34290285 in the 2q14 region was related to both GAL3ST2 and D2HGDH genes, with both genes showing eQTL signals in multiple tissues in GTEx. However, the colocalization analysis has shown that asthma might share causal variants at the rs34290285 locus with GAL3ST2 and in multiple tissues of GTEx but not with D2HGDH.
In addition to GTEx, other publicly available eQTL resources focus on a specific area, such as the immune-related eQTL database ImmGen.68 The ImmGen contains gene expression data of 292 specific immune cell types from mice. It has established the baseline measurements of variations in the hematopoietic transcriptome that allow for many eQTL analyses. For example, Finucane et al recently developed a method based on LDSC to investigate heritability enrichment in specific eQTL data,71 including ImmGen. They found that both eczema and asthma exhibited enrichment in TH and natural killer T cells but not in other immune cell types. In addition to eQTL, quantitative trait locus mapping for other functional data such as methylation69 would also provide important functional insight into the shared genetic variants discovered.
In addition to being applicable in eQTL analyses, genome-wide cross-trait analysis is also applicable to other types of functional investigation. For example, in addition to identifying shared causal variants between disease GWAS signals and functional data as already mentioned, colocalization analysis is also applicable to cross-disease analysis. For example, a recent study showed that 33 loci share causal variants (scenario 4) between allergic diseases and asthma.22 These shared loci can then be prioritized for the further functional investigation and pathway analysis72 to identify shared biologic pathways. For analysis involving more than 2 intermediate traits, it is possible to examine the mediating role of these intermediate traits between exposure and disease by using GWAS summary statistics based on 2-step Mendelian randomization.73
Discussion and future directions
Genome-wide cross-trait analysis offers opportunities to investigate shared genetics among complex traits by using large-scale publicly available GWAS data. In this review, we have summarized the current status of cross-trait genetics studies for the shared and distinct genetic effects between asthma subtypes and their shared genetics with coexistent diseases or traits.12 , 19, 20, 21, 22 , 26, 27, 28, 29 , 31 We have surveyed a broad range of major analytic methods at each phase of the genome-wide cross-trait analysis, namely, genetic correlation, cross-trait meta-analysis, Mendelian randomization, polygenic risk score, and functional analysis. We have also discussed scientific goals for each phase, as well as the advantages and limitations for these methods. Major challenges to future genome-wide cross-trait studies involve the availability of comprehensive and consistent phenotype data, combining data from different GWAS imputation panels, cross-ethnic genetic analysis, and integration of multiomics data sets. We discuss these challenges and new opportunities in the following paragraph.
First, although we listed several publicly available asthma GWAS data in Fig 3, other uncommon phenotypes may not be available in large-scale GWASs, such as food allergy, which may also contains subtypes.74 Additionally, consistency between the same phenotype across different studies is a challenge. For example, the definition of childhood and adult asthma in the study by Ferreira et al19 is different from that in the study by Pividori et al.20 Such inconsistency in phenotype definition may lead to different interpretations of cross-trait results. Second, there are challenges in combining data from different GWAS platforms and imputation panels used. We note that this is a major limitation for many cross-trait meta-analysis methods. For example, conducting cross-trait meta-analysis between a GWAS imputation based on 1000 Genome and HapMap panels will usually lead to use of only common SNPs between the 2 panels, which may greatly reduce the chance of identifying cross-trait signals. Thus, standardization and imputation of GWASs is strongly recommended before cross-trait analysis. Third, most asthma GWASs have been performed primarily in European populations despite the widely recognized racial/ethnic disparities in asthma.75 It is important to extend asthma GWASs to non-European racial/ethnic groups, thereby providing opportunities to gradually map the causal variants for the diseases and traits by leveraging population specific LD, develop a more accurate polygenic risk prediction model, and provide population-matched GWAS summary statistics for 2-sample Mendelian randomization. Several asthma GWASs in other non-European populations are ongoing. For example, one of the largest non-European asthma GWASs was recently conducted by the Consortium on Asthma among African-Ancestry Populations in the Americas, which identified 2 loci that are potentially specific to asthma risk in the population with African ancestry.76 Yet the sample size and diversity in population ethnicity are still far from sufficient. Fourth, GWASs have successfully uncovered genetic variants related to asthma in the past decades.12 , 19, 20, 21, 22, 23 , 26 , 31 , 32 However, these variants alone account for a limited proportion of the phenotypic variance in asthma.77 The total additive heritability provides an upper bound estimate for how much asthma risk can be explained by the genetics. This has generated interest in alternative sources of phenotypic variance (eg, host-microbiome-environment interrelationships).78 Integrating GWAS with other omics data, such as epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics data, will provide a potential for defining the pathobiology of asthma and its subtypes. Additionally, we also note that to develop novel prevention and treatment strategies, it is imperative to apply these approaches integrated with causal inference methods to multiomics data. Examples of these causal inference methods are cross-trait methods (including Mendelian randomization, as presented in this review), causal mediation methods,79 and causal structure learning.80
In summary, this review has illustrated the utility of large-scale genetic data coupled with advanced statistical genetic tools to understand the shared and distinct components between asthma subtypes as well as between coexistent diseases or traits and asthma. The success of current cross-trait studies in asthma also suggests useful applications to other complex and heterogeneous traits beyond asthma, including diseases of major interest to the allergy and immunology community, such as atopic dermatitis, food allergy, and allergic rhinitis.
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.Gobal Initiative for Asthma. Global strategy for asthma management and prevention. 2018. Available at: www.ginasthma.org. Accessed December 19, 2019.
- 2.Subbarao P., Mandhane P.J., Sears M.R. Asthma: epidemiology, etiology and risk factors. CMAJ. 2009;181:E181–E190. doi: 10.1503/cmaj.080612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Groot E.P., Duiverman E.J., Brand P.L. Comorbidities of asthma during childhood: possibly important, yet poorly studied. Eur Respir J. 2010;36:671–678. doi: 10.1183/09031936.00185709. [DOI] [PubMed] [Google Scholar]
- 4.Priftis K.N., Mermiri D., Papadopoulou A., Papadopoulos M., Fretzayas A., Lagona E. Asthma symptoms and bronchial reactivity in school children sensitized to food allergens in infancy. J Asthma. 2008;45:590–595. doi: 10.1080/02770900802032941. [DOI] [PubMed] [Google Scholar]
- 5.Plaschke P., Janson C., Norrman E., Bjornsson E., Ellbjar S., Jarvholm B. Association between atopic sensitization and asthma and bronchial hyperresponsiveness in Swedish adults: pets, and not mites, are the most important allergens. J Allergy Clin Immunol. 1999;104:58–65. doi: 10.1016/s0091-6749(99)70114-4. [DOI] [PubMed] [Google Scholar]
- 6.Vink N.M., Postma D.S., Schouten J.P., Rosmalen J.G., Boezen H.M. Gender differences in asthma development and remission during transition through puberty: the tracking adolescents' individual lives survey (trails) study. J Allergy Clin Immunol. 2010;126:498–504.e1-6. doi: 10.1016/j.jaci.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 7.Macsali F., Real F.G., Plana E., Sunyer J., Anto J., Dratva J., et al. Early age at menarche, lung function, and adult asthma. Am J Respir Crit Care Med. 2011;183:8–14. doi: 10.1164/rccm.200912-1886OC. [DOI] [PubMed] [Google Scholar]
- 8.Larsen G.L. Differences between adult and childhood asthma. J Allergy Clin Immunol. 2000;106:S153–S157. doi: 10.1067/mai.2000.109421. [DOI] [PubMed] [Google Scholar]
- 9.Simons F.E.R. What's in a name? The allergic rhinitis–asthma connection. 2003;3:9–17. [Google Scholar]
- 10.Peters U., Dixon A.E., Forno E. Obesity and asthma. J Allergy Clin Immunol. 2018;141:1169–1179. doi: 10.1016/j.jaci.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camargo C.A., Jr., Weiss S.T., Zhang S., Willett W.C., Speizer F.E. Prospective study of body mass index, weight change, and risk of adult-onset asthma in women. Arch Intern Med. 1999;159:2582–2588. doi: 10.1001/archinte.159.21.2582. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z., Guo Y., Shi H., Liu C.L., Panganiban R.A., Chung W., et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK biobank. J Allergy Clin Immunol. 2020;145:537–549. doi: 10.1016/j.jaci.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granell R., Henderson A.J., Evans D.M., Smith G.D., Ness A.R., Lewis S., et al. Effects of BMI, fat mass, and lean mass on asthma in childhood: a Mendelian randomization study. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortese S., Sun S., Zhang J., Sharma E., Chang Z., Kuja-Halkola R., et al. Association between attention deficit hyperactivity disorder and asthma: a systematic review and meta-analysis and a swedish population-based study. Lancet Psychiatry. 2018;5:717–726. doi: 10.1016/S2215-0366(18)30224-4. [DOI] [PubMed] [Google Scholar]
- 15.Scott K.M., Von Korff M., Ormel J., Zhang M.Y., Bruffaerts R., Alonso J., et al. Mental disorders among adults with asthma: results from the world mental health survey. Gen Hosp Psychiatry. 2007;29:123–133. doi: 10.1016/j.genhosppsych.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen E., Miller G.E. Stress and inflammation in exacerbations of asthma. Brain Behav Immun. 2007;21:993–999. doi: 10.1016/j.bbi.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavoie K.L., Cartier A., Labrecque M., Bacon S.L., Lemiere C., Malo J.L., et al. Are psychiatric disorders associated with worse asthma control and quality of life in asthma patients? Respir Med. 2005;99:1249–1257. doi: 10.1016/j.rmed.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Borish L., Culp J.A. Asthma: a syndrome composed of heterogeneous diseases. Ann Allergy, Asthma Immunol. 2008;101:1–9. doi: 10.1016/S1081-1206(10)60826-5. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira M.A.R., Mathur R., Vonk J.M., Szwajda A., Brumpton B., Granell R., et al. Genetic architectures of childhood- and adult-onset asthma are partly distinct. Am J Hum Genet. 2019;104:665–684. doi: 10.1016/j.ajhg.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pividori M., Schoettler N., Nicolae D.L., Ober C., Im H.K. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med. 2019;7:509–522. doi: 10.1016/S2213-2600(19)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z., Zhu X., Liu C.L., Shi H., Shen S., Yang Y., et al. Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur Respir J. 2019;54 doi: 10.1183/13993003.01507-2019. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Z., Lee P.H., Chaffin M.D., Chung W., Loh P.R., Lu Q., et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. 2018;50:857–864. doi: 10.1038/s41588-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ober C., Yao T.C. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoettler N., Rodriguez E., Weidinger S., Ober C. Advances in asthma and allergic disease genetics - is bigger always better? J Allergy Clin Immunol. 2019 doi: 10.1016/j.jaci.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price A.L., Zaitlen N.A., Reich D., Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira M.A., Vonk J.M., Baurecht H., Marenholz I., Tian C., Hoffman J.D., et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49:1752–1757. doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickrell J.K., Berisa T., Liu J.Z., Segurel L., Tung J.Y., Hinds D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melen E., Himes B.E., Brehm J.M., Boutaoui N., Klanderman B.J., Sylvia J.S., et al. Analyses of shared genetic factors between asthma and obesity in children. J Allergy Clin Immunol. 2010;126:631–637.e1-8. doi: 10.1016/j.jaci.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehto K., Pedersen N.L., Almqvist C., Lu Y., Brew B.K. Asthma and affective traits in adults: a genetically informative study. Eur Respir J. 2019;53 doi: 10.1183/13993003.02142-2018. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demenais F., Margaritte-Jeannin P., Barnes K.C., Cookson W.O.C., Altmuller J., Ang W., et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrine N., Portelli M.A., John C., Soler Artigas M., Bennett N., Hall R., et al. Moderate-to-severe asthma in individuals of european ancestry: a genome-wide association study. Lancet Respir Med. 2019;7:20–34. doi: 10.1016/S2213-2600(18)30389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solovieff N., Cotsapas C., Lee P.H., Purcell S.M., Smoller J.W. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny J.C., Bastarache L., Roden D.M. Phenome-wide association studies as a tool to advance precision medicine. Annu Rev Genomics Hum Genet. 2016;17:353–373. doi: 10.1146/annurev-genom-090314-024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takemoto M., Liao J.K. Pleiotropic effects of 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors. Arterioscler Thromb Vasc Biol. 2001;21:1712–1719. doi: 10.1161/hq1101.098486. [DOI] [PubMed] [Google Scholar]
- 36.Tonolo G., Melis M.G., Formato M., Angius M.F., Carboni A., Brizzi P., et al. Additive effects of simvastatin beyond its effects on LDL cholesterol in hypertensive type 2 diabetic patients. Eur J Clin Invest. 2000;30:980–987. doi: 10.1046/j.1365-2362.2000.00735.x. [DOI] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speed D., Holmes J., Balding D.J. Evaluating and improving heritability models using summary statistics. Nat Genet. 2020;52:458–462. doi: 10.1038/s41588-020-0600-y. [DOI] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J. Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Z., Anttila V., Smoller J.W., Lee P.H. Statistical power and utility of meta-analysis methods for cross-phenotype genome-wide association studies. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharjee S., Rajaraman P., Jacobs K.B., Wheeler W.A., Melin B.S., Hartge P., et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–835. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X., Feng T., Tayo B.O., Liang J., Young J.H., Franceschini N., et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am J Hum Genet. 2015;96:21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivakumaran S., Agakov F., Theodoratou E., Prendergast J.G., Zgaga L., Manolio T., et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89:607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 47.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess S., Scott R.A., Timpson N.J., Davey Smith G., Thompson S.G. EPIC-Interact Consortium. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z., Zheng Z., Zhang F., Wu Y., Trzaskowski M., Maier R., et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Connor L.J., Price A.L. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet. 2018;50:1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrison J., Knoblauch N., Marcus J.H., Stephens M., He X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat Genet. 2020 doi: 10.1038/s41588-020-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang X., Dimou N.L., Zhu Z., Bonilla C., Lewis S.J., Lindstrom S., et al. Allergy, asthma, and the risk of breast and prostate cancer: a Mendelian randomization study. Cancer Causes Control. 2020;31:273–282. doi: 10.1007/s10552-020-01271-7. [DOI] [PubMed] [Google Scholar]
- 57.Belsky D.W., Sears M.R., Hancox R.J., Harrington H., Houts R., Moffitt T.E., et al. Polygenic risk and the development and course of asthma: an analysis of data from a four-decade longitudinal study. Lancet Respir Med. 2013;1:453–461. doi: 10.1016/S2213-2600(13)70101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugrue L.P., Desikan R.S. What are polygenic scores and why are they important? JAMA. 2019;321:1820–1821. doi: 10.1001/jama.2019.3893. [DOI] [PubMed] [Google Scholar]
- 60.Vilhjalmsson B.J., Yang J., Finucane H.K., Gusev A., Lindstrom S., Ripke S., et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu Z., Hasegawa K., Ma B., Fujiogi M., Camargo C.A., Jr., Liang L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J Allergy Clin Immunol. 2020 doi: 10.1016/j.jaci.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maier R., Moser G., Chen G.B., Ripke S. Cross-Disorder Working Group of the Psychiatric Genomics C, Coryell W, et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am J Hum Genet. 2015;96:283–294. doi: 10.1016/j.ajhg.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung W., Chen J., Turman C., Lindstrom S., Zhu Z., Loh P.R., et al. Efficient cross-trait penalized regression increases prediction accuracy in large cohorts using secondary phenotypes. Nat Commun. 2019;10:569. doi: 10.1038/s41467-019-08535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.GTEx Consortium The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heng T.S., Painter M.W., Immunological Genome Project Consortium The immunological genome project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 69.Huan T., Joehanes R., Song C., Peng F., Guo Y., Mendelson M., et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun. 2019;10:4267. doi: 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallace C. Statistical testing of shared genetic control for potentially related traits. Genet Epidemiol. 2013;37:802–813. doi: 10.1002/gepi.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khatri P., Sirota M., Butte A.J. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Relton C.L., Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rona R.J., Keil T., Summers C., Gislason D., Zuidmeer L., Sodergren E., et al. The prevalence of food allergy: a meta-analysis. J Allergy Clin Immunol. 2007;120:638–646. doi: 10.1016/j.jaci.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 75.Oraka E., Iqbal S., Flanders W.D., Brinker K., Garbe P. Racial and ethnic disparities in current asthma and emergency department visits: findings from the national health interview survey, 2001-2010. J Asthma. 2013;50:488–496. doi: 10.3109/02770903.2013.790417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daya M., Rafaels N., Brunetti T.M., Chavan S., Levin A.M., Shetty A., et al. Association study in African-admixed populations across the americas recapitulates asthma risk loci in non-African populations. Nat Commun. 2019;10:880. doi: 10.1038/s41467-019-08469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeVries A., Vercelli D. Epigenetic mechanisms in asthma. Ann Am Thorac Soc. 2016;13(Suppl 1):S48–S50. doi: 10.1513/AnnalsATS.201507-420MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.VanderWeele T.J. Mediation analysis: a practitioner's guide. Annu Rev Public Health. 2016;37:17–32. doi: 10.1146/annurev-publhealth-032315-021402. [DOI] [PubMed] [Google Scholar]
- 80.Heinze-Deml C., Maathuis M.H., Meinshausen N. Causal structure learning. 2018;5:371–391. [Google Scholar]