Abstract
Objective
Measurement of the liver and spleen volumes has clinical implications. Although computed tomography (CT) volumetry is considered to be the most reliable noninvasive method for liver and spleen volume measurement, it has limited application in clinical practice due to its time-consuming segmentation process. We aimed to develop and validate a deep learning algorithm (DLA) for fully automated liver and spleen segmentation using portal venous phase CT images in various liver conditions.
Materials and Methods
A DLA for liver and spleen segmentation was trained using a development dataset of portal venous CT images from 813 patients. Performance of the DLA was evaluated in two separate test datasets: dataset-1 which included 150 CT examinations in patients with various liver conditions (i.e., healthy liver, fatty liver, chronic liver disease, cirrhosis, and post-hepatectomy) and dataset-2 which included 50 pairs of CT examinations performed at ours and other institutions. The performance of the DLA was evaluated using the dice similarity score (DSS) for segmentation and Bland-Altman 95% limits of agreement (LOA) for measurement of the volumetric indices, which was compared with that of ground truth manual segmentation.
Results
In test dataset-1, the DLA achieved a mean DSS of 0.973 and 0.974 for liver and spleen segmentation, respectively, with no significant difference in DSS across different liver conditions (p = 0.60 and 0.26 for the liver and spleen, respectively). For the measurement of volumetric indices, the Bland-Altman 95% LOA was −0.17 ± 3.07% for liver volume and −0.56 ± 3.78% for spleen volume. In test dataset-2, DLA performance using CT images obtained at outside institutions and our institution was comparable for liver (DSS, 0.982 vs. 0.983; p = 0.28) and spleen (DSS, 0.969 vs. 0.968; p = 0.41) segmentation.
Conclusion
The DLA enabled highly accurate segmentation and volume measurement of the liver and spleen using portal venous phase CT images of patients with various liver conditions.
Keywords: Deep learning, Artificial intelligence, Liver, Spleen, Segmentation, Volumetry
INTRODUCTION
Liver volume measurement is important for evaluating potential liver donors and patients who will undergo liver resection as remnant liver volume after hepatic resection is a major predictor of postoperative hepatic dysfunction and morbidity (1,2,3,4). Whole liver and spleen volume measurements also have clinical implications as the spleen volume and the liver to spleen volume ratio are associated with the severity of liver fibrosis and portal hypertension in patients with chronic liver disease (CLD) (5,6,7,8,9,10). Computed tomography (CT) volumetry is considered to be the most reliable noninvasive method for the volume measurement of abdominal organs. However, its time-consuming segmentation process limits its routine application in clinical practice.
Several automated methods based on image processing have been introduced to facilitate liver segmentation (11,12). However, these methods are not accurate enough for fully automated liver segmentation without user interaction (13,14,15,16). Recently, deep learning based on a convolutional neural network (CNN) has emerged as a method for image-based organ segmentation (12,17,18). Previous studies reported promising results of deep learning algorithms (DLAs)-based liver segmentation using CT images (7,19,20). However, these studies focused mainly on the technical feasibility of the algorithms without conducting full validation of the algorithms in diverse liver conditions. Variations in texture, morphology, and attenuation of the liver on CT images, depending on patients' liver conditions (i.e., presence of liver disease or previous liver surgery), may affect the segmentation performance of DLAs. Thus, for an algorithm to be applicable in clinical practice, it needs to be developed and validated with diverse liver conditions.
Therefore, the purpose of our study was to develop and validate a robust DLA for fully automated liver and spleen segmentation using portal venous phase CT images for use across various liver conditions.
MATERIALS AND METHODS
This study was approved by the Institutional Review Board of our institution. The requirement for informed consent was waived due to the retrospective nature of the study.
Development Dataset
The development dataset consisted of contrast agent-enhanced portal venous phase liver CT images from 813 patients. The dataset was derived from the study cohort of a previous study (19), in which a deep learning system for staging liver fibrosis was developed using retrospective CT data of 8352 adults between 2007 and 2016 at our institution and between 2014 and 2017 at two tertiary referral hospitals. To develop a robust algorithm that performs well with various liver conditions and using diverse CT techniques, the development dataset included CT data obtained using a variety of CT scanners (detailed later) from patients with a healthy liver; those with various liver diseases including fatty liver, non-cirrhotic CLD, and liver cirrhosis; and those who underwent a liver resection within the six months prior to CT. The characteristics of the development dataset are summarized in Table 1. Detailed information and a flow diagram of the development dataset are presented in Supplementary Materials and Supplementary Figure 1.
Table 1. Characteristics of Development and Test Datasets.
| Characteristics | Development Dataset | Test Dataset | |
|---|---|---|---|
| Test Dataset-1 | Test Dataset-2 | ||
| No. of patients | 813 | 150 | 50 |
| Age (y)* | 50.0 ± 13.7 | 48.6 ± 14.2 | 56.0 ± 9.5 |
| No. of male patients | 460 (56.6) | 101 (67.3) | 39 (78.0) |
| Underlying liver disease | |||
| No† | 134 (16.5) | 55 (36.7) | 6 (12.0) |
| B viral hepatitis | 264 (32.5) | 71 (47.3) | 39 (78.0) |
| C viral hepatitis | 97 (11.9) | 3 (2.0) | 3 (6.0) |
| Alcoholic liver disease | 73 (9.0) | 6 (4.0) | 1 (2.0) |
| NAFLD | 32 (3.9) | 2 (1.3) | 0 (0.0) |
| Autoimmune‡ | 157 (19.3) | 7 (4.7) | 0 (0.0) |
| Others§ | 56 (6.9) | 6 (4.0) | 1 (2.0) |
| Pathologic liver fibrosis stage | |||
| F0 | 162 (19.9) | 59 (39.3) | 10 (20.0) |
| F1 | 82 (10.1) | 2 (1.3) | 1 (2.0) |
| F2 | 130 (16.0) | 23 (15.3) | 6 (12.0) |
| F3 | 117 (14.4) | 19 (12.7) | 9 (18.0) |
| F4 | 322 (39.6) | 47 (31.3) | 24 (48.0) |
| Moderate to sever fatty liver | 67 (8.2) | 29 (19.3) | 0 (0.0) |
| Focal hepatic lesion | |||
| No | 670 (82.4) | 112 (74.7) | 11 (22.0) |
| Hepatic cyst | 39 (4.8) | 3 (2.0) | 0 (0.0) |
| Benign tumor | 21 (2.6) | 0 (0.0) | 1 (2.0) |
| HCC | 70 (8.6) | 31 (20.7) | 35 (70.0) |
| Other malignancy | 13 (1.6) | 4 (2.7) | 3 (6.0) |
| Diameter of largest focal hepatic lesion* | 2.92 ± 1.53 (1.00–9.40) | 2.89 ± 1.32 (1.00–6.50) | 2.92 ± 0.96 (1.20–4.90) |
Unless otherwise indicated, data are expressed as number of participants; data in parentheses are percentages. *Data are expressed as mean ± standard deviation; data in parentheses are range, †Included healthy donor candidates for living donor liver transplant, ‡Included autoimmune hepatitis, autoimmune cholangitis, primary biliary cirrhosis, and primary sclerosing cholangitis, §Included viral hepatitis A, toxic hepatitis, Wilson disease, and liver disease due to unknown etiology. F0 = no fibrosis, F1 = portal fibrosis, F2 = periportal fibrosis, F3 = septal fibrosis, F4 = cirrhosis, HCC = hepatocellular carcinoma, NAFLD = nonalcoholic fatty liver disease
Test Dataset
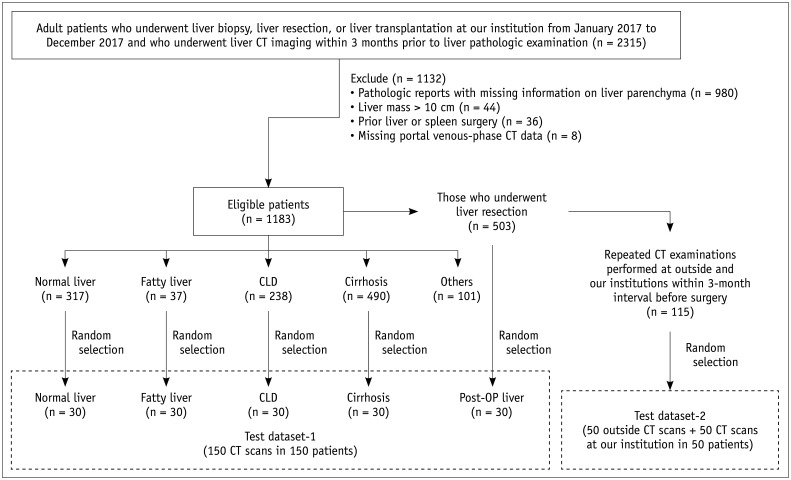
Two test datasets that included 250 portal venous phase CT image sets from 200 patients were used for the validation of the DLA (Fig. 1). The test datasets were retrospectively obtained from 1183 patients who underwent liver biopsy, resection, or transplantation at our institution in 2017 and who met the following eligibility criteria: 1) liver CT scan performed within three months of a pathologic examination of the liver, 2) pathologic reports including the findings of liver parenchyma, 3) no hepatic tumor larger than 10 cm in diameter, and 4) no previous liver or spleen surgery. Dataset-1 (150 CT examinations in 150 patients) was used to test the performance of the DLA in various liver conditions. We randomly selected 30 subjects who had one of the following liver conditions: 1) healthy liver, i.e., living liver donors with no clinical and pathologic evidence of liver disease; 2) fatty liver, i.e., pathologic macro-vesicular steatosis ≥ 33%; 3) non-cirrhotic CLD, i.e., any CLD with pathologically proven periportal or septal fibrosis; and 4) cirrhosis, i.e., pathologically proven cirrhosis from any cause. We additionally included post-hepatectomy CT images obtained within one month (n = 15) or from one to six months (n = 15) after liver resection in 30 randomly selected patients who had undergone liver resection. Dataset-2 was prepared for the intra-individual comparison of the segmentation performance of the DLA between the external and internal CT data and for the assessment of the reproducibility of automated volumetric measurements. Dataset-2 included 50 pairs of CT examinations performed on 50 patients at our institution and another institution. The data were preoperative CT data derived from 50 randomly selected patients among those who underwent liver resection at our institution and repeated CT examinations at an outside institution and our institution within a 3-month interval. The mean time interval between the two CT examinations was 31.8 ± 17.3 days (range: 5–90 days). A flow diagram of the test datasets is presented in Figure 1, and its characteristics are summarized in Table 1.
Fig. 1. Flow diagram of test datasets.
CLD = chronic liver disease, CT = computed tomography, OP = operative
CT Examination
CT scans were performed using various CT scanners and techniques (Supplementary Table 1). All CT images were obtained using multi-detector row CT systems, mostly using 16- or higher detector row systems (722 of 813 examinations [88.8%] in the development dataset and 248 [99.2%] of 250 examinations in the test datasets). Portal venous phase imaging was performed at 70–80 seconds after intravenous administration of a contrast agent, mostly with a tube voltage of 120 kVp (752 [92.5%] examinations in the development dataset and 114 [45.6%] in the test datasets) or 100 kVp (60 [7.4%] examinations in the development dataset and 132 [52.8%] in the test datasets). Most axial CT images were reconstructed at a section thickness of 5 mm (607 [74.7%] examinations in the development dataset and 241 [96.4%] in the test datasets) with no gaps.
Ground Truth
To obtain a large amount of labeled CT data in a time-efficient manner, CT imaging data in the development dataset (n = 813) were first processed by the prototype CNN algorithm for liver and spleen segmentation described previously (19). Briefly, the algorithm was developed on a three-dimensional U-net and trained using liver and spleen outlines drawn by an experienced radiologist on portal venous phase CT images of 50 patients. One radiology technician reviewed the liver and spleen marks generated by the CNN algorithm. Using an in-house software package (AsanJ; Asan Medical Center, Seoul, Korea) plugged into ImageJ software (http://rsb.info.nih.gov/ij/), the technician manually edited them to correctly outline the margins of the liver and spleen while excluding hepatic and splenic vessels and any focal hepatic lesions visible on the portal venous phase CT images. For the test dataset, including 250 CT scans from 200 patients, the radiology technician manually drew liver and spleen outlines while excluding vessels and focal hepatic lesions on the portal venous phase CT images using the software (AsanJ) without the assistance of the prototype CNN algorithm. To reconfirm the accuracy of both the development and test datasets, the liver and spleen segmentation results were re-evaluated by one of three radiologists (with 15 years of experience in liver imaging, with five years of experience in radiology, and with three years of experience in radiology). Any inaccuracies in the liver or spleen margins were corrected by the radiologists.
Development of the DLA
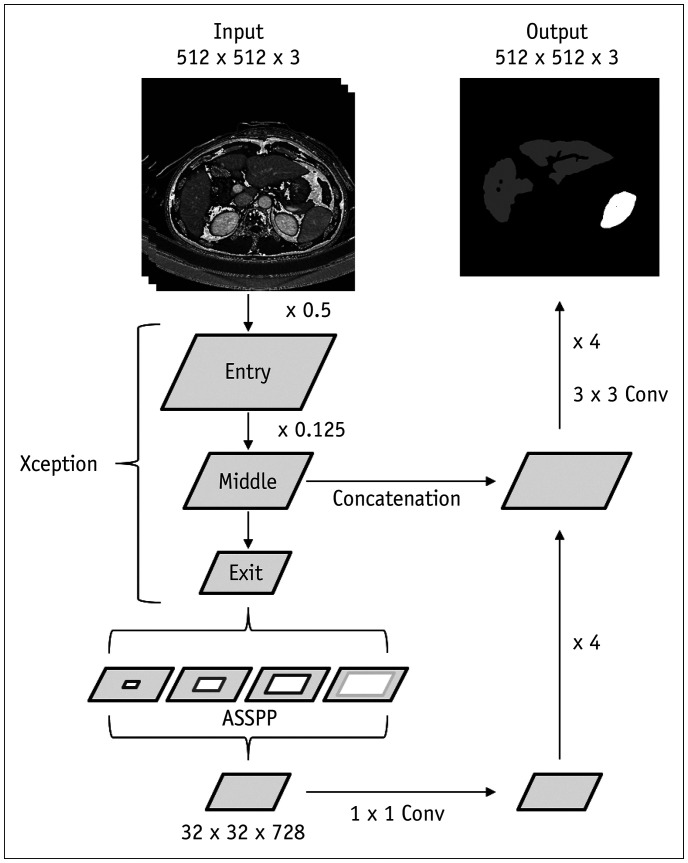
The development dataset was divided into the training and validation sets at an 8:2 ratio. The validation set was used to select the final optimized network architecture and hyperparameters by comparing the performances of different network architectures and various hyperparameters. The main objective of our DLA was to perform three-class automated segmentation (i.e., liver, spleen, and background) using three-dimensional spatial information on portal venous phase CT images. A CNN referred to as DeepLabV3+ was adapted to perform segmentation using CT images (21). We employed a 2.5-dimensional input set-up that imported three consecutive sections of CT images (i.e., a CT image of interest as well as CT image sections above and below the CT image of interest) as input data (22). This input set-up enabled the network to perform segmentation tasks on a given CT image using three-dimensional spatial information of three consecutive CT images. The final CNN architecture comprised an encoder and a decoder (Fig. 2). The encoder performs downsampling, depth-wise convolution, and global average pooling. The decoder includes bilinear upsampling layers with skip connections with the encoder layers. The network extracts multiple feature maps with progressively reduced resolution from input CT images and then upsamples them to provide a probability map with the same resolution as that of the input CT images. The final output is compared with that of the ground truth method using loss function, and the error is backpropagated to improve the accuracy of the algorithm by optimizing weights. Analysis of one slice of a CT image using the trained DLA took approximately 350 milliseconds, resulting in a computation time of approximately 25 seconds for a CT examination containing 70 image sections. The details of the DLA are described in Supplementary Materials and Supplementary Figure 2.
Fig. 2. Schematic diagram of deep learning algorithm for liver and spleen segmentation.
Model receives three consecutive CT images as three-channel input using 2.5-dimensional input set-up and performs segmentation task on center section of CT images. Encoder part is based on modified Xception model, which contains series of downsampling layers and ASSPP unit. Output of encoder is feature maps, which are 32 × 32 × 728 in size. Decoder is series of bilinear upsampling layers with skip connections from encoder. Final output of model is three-channel (liver, spleen, and background) logit maps, which are same size as that of input CT image. ASSPP = Atrous Separable Spatial Pyramid Pooling, Conv = convolution
Performance Validation
Segmentation results of the deep learning system were compared with those of the ground truth (i.e., manually segmented liver and spleen excluding the vessels and focal hepatic lesions) in the test datasets. The performance was analyzed using the dice similarity score (DSS), defined as 2 × true positive pixels / (2 × true positive pixels + false negative pixels + false positive pixels). Liver and spleen volumes were calculated by summing the consecutive areas of the liver and spleen, respectively, multiplied by the slice thickness (23) and are expressed in cm3. The liver volume, spleen volume, and liver/spleen volume ratio calculated using deep learning segmentation were compared with those calculated using ground truth liver and spleen segmentation.
Statistical Analysis
In test dataset-1, the difference in the DSS values for the liver and spleen segmentations were compared among five subgroups of liver conditions (normal liver, fatty liver, non-cirrhotic CLD, cirrhosis, postoperative liver) using the Kruskal-Wallis test. The agreement between the liver volume, spleen volume, and liver/spleen volume ratio measured by the DLA and those measured by ground truth segmentation was evaluated using the Bland-Altman 95% limits of agreement (LOA) expressed as a percentage of the measured values. The Bland-Altman 95% LOA results are presented as the mean difference ± 1.96 × standard deviation (SD) of the difference, where the mean difference represents systematic bias, and 1.96 × SD of the difference represents the measurement error. The difference in DSS values for liver and spleen segmentations in test dataset-2 was compared between the internal and external CT data (i.e., data from our and other institutions, respectively) using the Wilcoxon signed-rank test. The agreement between volumetric indices measured by the DLA and those measured by ground truth segmentation was evaluated using the Bland-Altman 95% LOA for the internal and external CT data. Finally, the measurement reproducibility of volumetric indices over repeated CT examinations (i.e., at our institution after outside institutions) was assessed for the DLA and ground truth segmentation using the percentage reproducibility coefficient (RC), which is calculated as follows (24):
where %wCV is the within-subject coefficient of variation expressed as a percentage. Statistical analyses were performed using SPSS version 21.0 (IBM Corp., Armonk, NY, USA). A p value of less than 0.05 was considered to be statistically significant.
RESULTS
Segmentation Performance in Various Liver Conditions
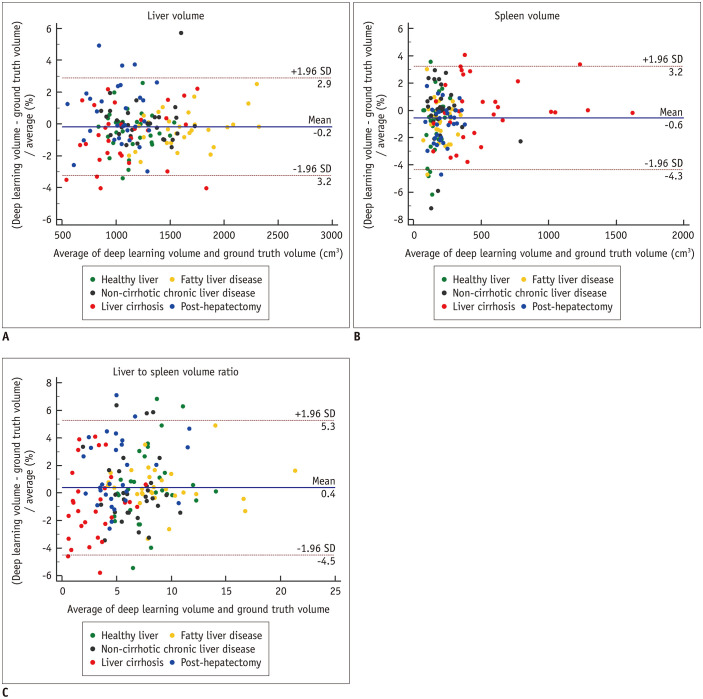
For test dataset-1, the mean DSS values representing the segmentation performance of the DLA were 0.973 for the liver and 0.974 for the spleen (Table 2). The DSS was not significantly different among the five subgroups of liver conditions for both the liver (p = 0.60) and spleen (p = 0.26). Figure 3 and Supplementary Figure 3 show representative segmentation results produced by the DLA compared with those of ground truth segmentation. For the measurement of volumetric indices, the Bland-Altman 95% LOAs between the DLA and ground truth were −0.2 ± 3.1% for liver volume, −0.6 ± 3.8% for spleen volume, and 0.4 ± 4.9% for the liver/spleen volume ratio (Table 3, Fig. 4). No statistically significant bias was noted for liver volume (p = 0.19) and liver/spleen volume ratio (p = 0.06) as measured by the DLA, while the DLA resulted in a slight underestimation of spleen volume in contrast to ground truth segmentation (mean bias, −0.56%; p < 0.001). In the five subgroups, the 1.96 × SD of the difference indicating the magnitude of measurement error ranged from 2.2% to 3.6% for liver volume, 2.8% to 4.3% for spleen volume, and 3.0% to 5.3% for the liver/spleen volume ratio.
Table 2. Performance of Deep Learning Algorithm in Liver and Spleen Segmentation in Test Dataset-1.
| Dice Similarity Score | ||
|---|---|---|
| Liver Segmentation | Spleen Segmentation | |
| Total | 0.973 ± 0.019 (0.907–0.999) | 0.974 ± 0.018 (0.940–0.999) |
| Subgroups | ||
| Healthy liver | 0.975 ± 0.017 (0.974–0.999) | 0.971 ± 0.017 (0.948–0.998) |
| Fatty liver disease | 0.976 ± 0.019 (0.952–0.999) | 0.974 ± 0.018 (0.947–0.999) |
| Non-cirrhotic chronic liver disease | 0.974 ± 0.019 (0.945–0.999) | 0.974 ± 0.018 (0.944–0.999) |
| Liver cirrhosis | 0.970 ± 0.016 (0.926–0.997) | 0.978 ± 0.018 (0.940–0.999) |
| Post-hepatectomy | 0.968 ± 0.030 (0.907–0.999) | 0.972 ± 0.020 (0.947–0.999) |
| p value* | 0.6 | 0.26 |
Unless otherwise indicated, data are expressed as mean ± standard deviation; data in parentheses are range. *p values for comparison of dice similarity score among five subgroups using Kruskal-Wallis test.
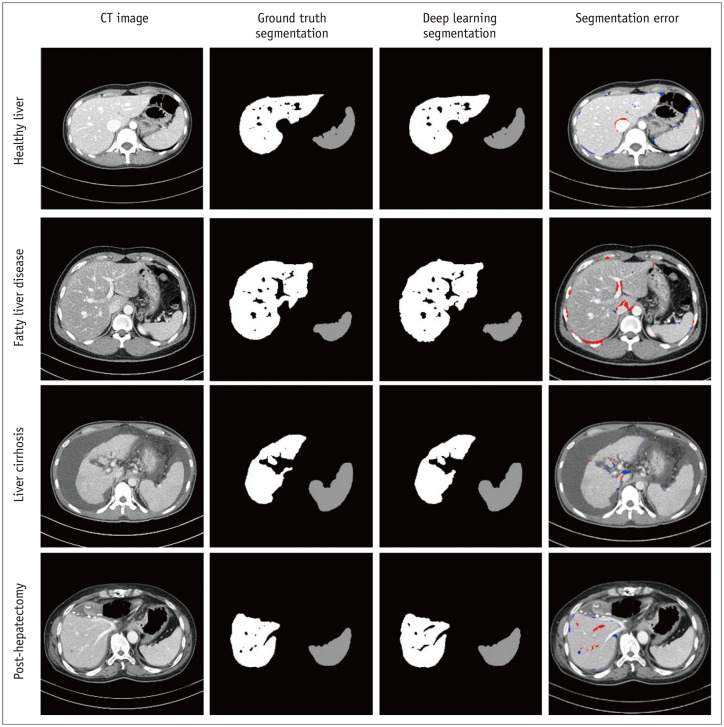
Fig. 3. Representative images showing deep learning-based liver and spleen segmentation results in various liver conditions.
Each row demonstrates original CT image, image of ground truth segmentation, image of deep learning segmentation, and image of segmentation error overlaid on CT image (red mask = false-positive segmentation; blue mask = false-negative segmentation). Images were obtained from healthy liver (first row), fatty liver disease (second row), liver cirrhosis (third row), and post-hepatectomy (fourth row) subgroups in test dataset-1.
Table 3. Agreement of Volumetric Indices between Deep Learning Segmentation and Ground Truth Segmentation in Test Dataset-1.
| Liver Volume | Spleen Volume | Liver/Spleen Volume Ratio | ||||
|---|---|---|---|---|---|---|
| 95% LOA* | P† | 95% LOA* | P† | 95% LOA* | P† | |
| Total | -0.17 ± 3.07 | 0.19 | -0.56 ± 3.78 | 0.001 | 0.39 ± 4.89 | 0.06 |
| Subgroups | ||||||
| Healthy liver | -0.42 ± 2.49 | 0.08 | -0.97 ± 4.15 | 0.02 | 0.55 ± 5.30 | 0.28 |
| Fatty liver disease | -0.33 ± 2.23 | 0.12 | -0.77 ± 2.84 | 0.007 | 0.44 ± 3.04 | 0.13 |
| Non-cirrhotic chronic liver disease | 0.12 ± 2.63 | 0.64 | -0.14 ± 4.32 | 0.74 | 0.25 ± 4.95 | 0.59 |
| Liver cirrhosis | -0.75 ± 3.58 | 0.03 | 0.06 ± 4.21 | 0.88 | -0.81 ± 5.15 | 0.10 |
| Post-hepatectomy | 0.55 ± 3.64 | 0.11 | -0.99 ± 2.79 | 0.001 | 1.54 ± 4.82 | 0.002 |
*Data are Bland-Altman 95% LOA expressed in percentages as mean difference ± 1.96 × standard deviation of difference, †p values for statistically significant difference in mean difference from zero. LOA = limits of agreement
Fig. 4. Bland-Altman plots for agreement between the liver volume (A), spleen volume (B), and liver/spleen volume ratio (C) measured using deep learning segmentation and those by ground truth segmentation.
Solid lines indicate mean differences and dashed lines indicate upper and lower limits of 95% limits of agreement. SD = standard deviation
Segmentation Performance in the External CT Data
For test dataset-2, the segmentation performance of the DLA in the external CT data was not significantly different from the performance of the DLA in the internal CT data for both the liver (DSS, 0.982 vs. 0.983; p = 0.28) and spleen (DSS, 0.969 vs. 0.968, respectively; p = 0.41) (Table 4). The 95% Bland-Altman LOA of the volumetric indices between the DLA and ground truth were also similar in the external and internal CT data (Table 4), with 1.96 × SD of the difference ranging from 2.7% to 5.2% for the external CT data and 3.5% to 4.0% for the internal CT data.
Table 4. Comparison of Segmentation Performance and Volumetric Measurement Results of Deep Learning Algorithm between Internal CT Data and External CT Data in Test Dataset-2.
| Performance Statistics | External CT Data | Internal CT Data | P* |
|---|---|---|---|
| Dice similarity score† | |||
| Liver | 0.982 ± 0.011 (0.932–0.999) | 0.983 ± 0.007 (0.954–0.998) | 0.28 |
| Spleen | 0.969 ± 0.011 (0.930–0.994) | 0.968 ± 0.010 (0.936–0.993) | 0.41 |
| 95% LOA of volumetric indices‡ | |||
| Liver volume | -0.34 ± 2.67 | -0.03 ± 3.47 | NA |
| Spleen volume | -0.63 ± 4.34 | -0.33 ± 3.70 | NA |
| Liver/spleen volume ratio | 0.30 ± 5.22 | 0.50 ± 4.02 | NA |
Data are expressed as mean ± standard deviation; data in parentheses are range. *p values for comparison of dice similarity score between internal CT data and external CT data using Wilcoxon test, †Data are expressed as mean ± standard deviation; data in parentheses are range, ‡Data are Bland-Altman 95% LOA expressed in percentage as mean difference ± 1.96 × standard deviation of difference. NA = not applicable
Measurement Reproducibility of the Liver and Spleen Volumetric Indices
For test dataset-2, the measurement reproducibility of the volumetric indices was evaluated over two CT examinations performed at different institutions (i.e., ours and another institution). The percentage RCs for the automated measurement of the volumetric indices using the DLA were 16.7% (95% confidence interval [CI], 13.1–19.8%) for the liver volume, 19.9% (95% CI, 16.7–24.7%) for the spleen volume, and 22.5% (95% CI, 18.8–27.9%) for the liver/spleen volume ratio. These results were similar to that for ground truth segmentation, which were 18.1% (95% CI, 15.1–22.4%) for the liver volume, 18.8% (95% CI, 15.7–23.4%) for the spleen volume, and 21.5% (18.0–26.7%) for the liver/spleen volume ratio.
DISCUSSION
In our study, we developed and evaluated a DLA for the fully automated segmentation of the liver and spleen using portal venous phase CT images. Our study demonstrated that the DLA, which was trained using a large amount of labeled CT data, allowed for highly accurate segmentation and volume measurements of the liver and spleen in a fully automated manner. In the two test datasets, the segmentation performance of the DLA represented by the DSS was higher than 0.97. Volumetric indices obtained by deep learning segmentation showed close agreement with those measured by the radiologist manually, with a small bias (i.e., −1% to 0.6% of the measured volumetric indices for all indices) and measurement error (i.e., < 5.2% of the measured volumetric indices for all indices). Of note, there was a small but statistically significant systematic bias in the volumetric indices measured with the DLA compared with the ground truth, mostly toward an underestimation of the volumetric indices with the DLA (range of mean bias, −1–0.6%). However, we considered that this small bias would not cause a real problem in clinical practice.
A variety of image processing methods have been proposed for the automated segmentation of the liver using CT images, including statistical shape models (25), atlas-based models, and three-dimensional deformable models (25). However, these methods may not fully account for variations in liver shape and may fail in pathologic or postoperative livers (11). In our study, we evaluated our DLA in test datasets, including CT data from patients with a healthy liver, fatty liver, CLD, liver cirrhosis, and post-hepatectomy status and CT data from our institution and outside institutions. We found that our algorithm was robust across various liver conditions without a significant difference in the segmentation performance and showed comparable performance between internal and external CT data, indicating the generalizability of our DLA to various clinical settings.
There may be multiple potential applications of our DLA in clinical practice. Our algorithm allows for the accurate and automated measurements of liver and spleen volumes and may facilitate clinical applications of liver and spleen volumetry, such as liver volume measurements in potential living liver donors. Our DLA may be used for monitoring liver regeneration after liver resection (2). In patients with CLD, spleen volume and the liver/spleen volume ratio may be used as quantitative imaging biomarkers to assess the severity of CLD and portal hypertension, as suggested by previous studies (5,6,7,8,9,10). The actual utility and clinical implications of our DLA should be evaluated in future studies.
In our study, the measurement reproducibility of volumetric indices was similar between the DLA (percentage RC of 16.7–22.5%) and the radiologist's manual segmentation (percentage RC of 18.1% to 21.5%). The percentage RC values represent the range of variability of volumetric indices (i.e., measurement error) measured on repeated CT examinations which are performed in a reproducibility condition involving different institutions, scanners, or observers on the same subject, assuming that there is no true change in organ volume (26). Our results indicate that a difference up to 22% in volumetric indices on follow-up CT may be due to measurement error and, thus, are not considered true changes. Although not fully understood, multiple factors may have contributed to the variability of volumetric indices over repeated CT examinations in our study, including the different scanners and imaging parameters used for CT examinations. Although the time interval between two CT examinations was relatively short (i.e., less than three months), we do not completely exclude a possibility of true changes in the liver or spleen volumes between the two CT examinations in exceptional cases. Furthermore, compared with volume measurements using three-dimensional isometric volume CT data, volume measurement using two-dimensional CT images was reported to be less accurate, leading to an underestimation of volume in proportion to slice thickness of the CT images (27,28). Thus, our two-dimensional volume measurement approach may have added some degree of measurement error. Despite this inherent limitation, we consider the two-dimensional volume measurement to be a practical method since processing three-dimensional volume data requires more time, higher computational capacity, and larger data storage capacity and may potentially increase the need for operators' correction in cases of inaccurate segmentation results in contrast to our approach. Thus, most of the previous studies involving volume measurement of the liver and spleen have utilized two-dimensional CT images (18,29,30,31,32).
Our study had several limitations. First, our algorithm was developed and validated using portal venous phase CT images. The application of our algorithm to other CT images may require further training of the algorithm using additional training data through transfer learning (18,26). Second, our algorithm only provides whole liver segmentation. For the measurement of lobar or segmental liver volumes, user interaction is required to divide the segmented liver. Third, we did not compare our DLA with other methods of automated or semi-automated organ segmentation, which could have helped demonstrate the clinical usefulness of our DLA. Lastly, our study only validated the performance of the DLA in the segmentation and volume measurement of the liver and spleen. The actual clinical impact of our algorithm on patient care should be evaluated in future research. Although we tried to validate our DLA with diverse liver conditions and CT techniques, the generalizability of our DLA has not been thoroughly evaluated and thus needs to be further evaluated using a large amount of external CT data.
In conclusion, we developed and validated a DLA for the fully automated segmentation and volume measurement of the liver and spleen using portal venous-phase CT data in patients with various liver conditions. As the DLA enables highly accurate segmentation and volume measurement, we expect that our algorithm can be used for CT-based liver and spleen volumetry in clinical practice and research.
Acknowledgments
Seung Soo Lee was responsible for the clinical study, and Heung-Il Suk was responsible for the development of the deep learning algorithm. Both authors contributed equally to this study and assume the role of corresponding authors.
Footnotes
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2020R1F1A1048826) and the Bio and Medical Technology Development Program of the NRF, which is funded by the Ministry of Science and ICT (NRF-2016M3A9A7918706).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Supplementary Materials
The Data Supplement is available with this article at https://doi.org/10.3348/kjr.2020.0237.
Flow diagram of development dataset.
CT Imaging Techniques Used for Development and Test Datasets
Diagrams showing detailed architecture of deep learning algorithm.
Representative images showing examples of segmentation errors generated by deep learning algorithm in test dataset-1.
References
- 1.Lim MC, Tan CH, Cai J, Zheng J, Kow AW. CT volumetry of the liver: where does it stand in clinical practice? Clin Radiol. 2014;69:887–895. doi: 10.1016/j.crad.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Schindl MJ, Redhead DN, Fearon KC, Garden OJ, Wigmore SJ Edinburgh Liver Surgery and Transplantation Experimental Research Group (eLISTER) The value of residual liver volume as a predictor of hepatic dysfunction and infection after major liver resection. Gut. 2005;54:289–296. doi: 10.1136/gut.2004.046524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogasawara K, Une Y, Nakajima Y, Uchino J. The significance of measuring liver volume using computed tomographic images before and after hepatectomy. Surgery Today. 1995;25:43–48. doi: 10.1007/BF00309384. [DOI] [PubMed] [Google Scholar]
- 4.Prodeau M, Drumez E, Duhamel A, Vibert E, Farges O, Lassailly G, et al. An ordinal model to predict the risk of symptomatic liver failure in patients with cirrhosis undergoing hepatectomy. J Hepatol. 2019;71:920–929. doi: 10.1016/j.jhep.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Huang B, Kan T, Yang B, Yuan M, Wang J. Liver-to-spleen ratio as an index of chronic liver diseases and safety of hepatectomy: a pilot study. World J Surg. 2014;38:3186–3192. doi: 10.1007/s00268-014-2717-6. [DOI] [PubMed] [Google Scholar]
- 6.Berzigotti A, Seijo S, Arena U, Abraldes JG, Vizzutti F, García-Pagán JC, et al. Elastography, spleen size, and platelet count identify portal hypertension in patients with compensated cirrhosis. Gastroenterology. 2013;144:102–111.e1. doi: 10.1053/j.gastro.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Iranmanesh P, Vazquez O, Terraz S, Majno P, Spahr L, Poncet A, et al. Accurate computed tomography-based portal pressure assessment in patients with hepatocellular carcinoma. J Hepatol. 2014;60:969–974. doi: 10.1016/j.jhep.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 8.Murata Y, Abe M, Hiasa Y, Azemoto N, Kumagi T, Furukawa S, et al. Liver/spleen volume ratio as a predictor of prognosis in primary biliary cirrhosis. J Gastroenterol. 2008;43:632–636. doi: 10.1007/s00535-008-2202-9. [DOI] [PubMed] [Google Scholar]
- 9.Pickhardt PJ, Malecki K, Hunt OF, Beaumont C, Kloke J, Ziemlewicz TJ, et al. Hepatosplenic volumetric assessment at MDCT for staging liver fibrosis. Eur Radiol. 2017;27:3060–3068. doi: 10.1007/s00330-016-4648-0. [DOI] [PubMed] [Google Scholar]
- 10.Son JH, Lee SS, Lee Y, Kang BK, Sung YS, Jo S, et al. Assessment of liver fibrosis severity using computed tomography-based liver and spleen volumetric indices in patients with chronic liver disease. Eur Radiol. 2020;30:3486–3496. doi: 10.1007/s00330-020-06665-4. [DOI] [PubMed] [Google Scholar]
- 11.Gotra A, Sivakumaran L, Chartrand G, Vu KN, Vandenbroucke-Menu F, Kauffmann C, et al. Liver segmentation: indications, techniques and future directions. Insights Imaging. 2017;8:377–392. doi: 10.1007/s13244-017-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61:8676–8698. doi: 10.1088/1361-6560/61/24/8676. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama Y, Li Q, Katsuragawa S, Ikeda R, Hiai Y, Awai K, et al. Automated hepatic volumetry for living related liver transplantation at multisection CT. Radiology. 2006;240:743–748. doi: 10.1148/radiol.2403050850. [DOI] [PubMed] [Google Scholar]
- 14.Fananapazir G, Bashir MR, Marin D, Boll DT. Computer-aided liver volumetry: performance of a fully-automated, prototype post-processing solution for whole-organ and lobar segmentation based on MDCT imaging. Abdom Imaging. 2015;40:1203–1212. doi: 10.1007/s00261-014-0276-9. [DOI] [PubMed] [Google Scholar]
- 15.Huynh HT, Karademir I, Oto A, Suzuki K. Computerized liver volumetry on MRI by using 3D geodesic active contour segmentation. AJR Am J Roentgenol. 2014;202:152–159. doi: 10.2214/AJR.13.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grieser C, Denecke T, Rothe JH, Geisel D, Stelter L, Cannon Walter T, et al. Gd-EOB enhanced MRI T1-weighted 3D-GRE with and without elevated flip angle modulation for threshold-based liver volume segmentation. Acta Radiol. 2015;56:1419–1427. doi: 10.1177/0284185114558975. [DOI] [PubMed] [Google Scholar]
- 17.Huo Y, Terry JG, Wang J, Nair S, Lasko TA, Freedman BI, et al. Fully automatic liver attenuation estimation combing CNN segmentation and morphological operations. Med Phys. 2019;46:3508–3519. doi: 10.1002/mp.13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Mamidipalli A, Retson T, Bahrami N, Hasenstab K, Blansit K, et al. Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol Artif Intell. 2019;1:180022. doi: 10.1148/ryai.2019180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, et al. Development and validation of a deep learning system for staging liver fibrosis by using contrast agent-enhanced CT images in the Liver. Radiology. 2018;289:688–697. doi: 10.1148/radiol.2018180763. [DOI] [PubMed] [Google Scholar]
- 20.Dou Q, Yu L, Chen H, Jin Y, Yang X, Qin J, et al. 3D deeply supervised network for automated segmentation of volumetric medical images. Med Image Anal. 2017;41:40–54. doi: 10.1016/j.media.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoderdecoder with atrous separable convolution for semantic image segmentation; The European conference on computer vision (ECCV); 2018 September 8–14; Munich, Germany. [Google Scholar]
- 22.Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S, et al. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. Med Image Comput Comput Assist Interv. 2014;17:520–527. doi: 10.1007/978-3-319-10404-1_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto T, Sugawara Y, Tamura S, Hasegawa K, Kishi Y, Kokudo N, et al. Estimation of standard liver volume in Japanese living liver donors. J Gastroenterol Hepatol. 2006;21:1710–1713. doi: 10.1111/j.1440-1746.2006.04433.x. [DOI] [PubMed] [Google Scholar]
- 24.Serai SD, Obuchowski NA, Venkatesh SK, Sirlin CB, Miller FH, Ashton E, et al. Repeatability of MR elastography of liver: a meta-analysis. Radiology. 2017;285:92–100. doi: 10.1148/radiol.2017161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito A, Yamamoto S, Nawano S, Shimizu A. Automated liver segmentation from a postmortem CT scan based on a statistical shape model. Int J Comput Assist Radiol Surg. 2017;12:205–221. doi: 10.1007/s11548-016-1481-5. [DOI] [PubMed] [Google Scholar]
- 26.Barnhart HX, Barboriak DP. Applications of the repeatability of quantitative imaging biomarkers: a review of statistical analysis of repeat data sets. Transl Oncol. 2009;2:231–235. doi: 10.1593/tlo.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prionas ND, Ray S, Boone JM. Volume assessment accuracy in computed tomography: a phantom study. J Appl Clin Med Phys. 2010;11:3037. doi: 10.1120/jacmp.v11i2.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hori M, Suzuki K, Epstein ML, Baron RL. Computed tomography liver volumetry using 3-dimensional image data in living donor liver transplantation: effects of the slice thickness on the volume calculation. Liver Transpl. 2011;17:1427–1436. doi: 10.1002/lt.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotra A, Chartrand G, Vu KN, Vandenbroucke-Menu F, Massicotte-Tisluck K, de Guise JA, et al. Comparison of MRI- and CT-based semiautomated liver segmentation: a validation study. Abdom Radiol (NY) 2017;42:478–489. doi: 10.1007/s00261-016-0912-7. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki K, Kohlbrenner R, Epstein ML, Obajuluwa AM, Xu J, Hori M. Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms. Med Phys. 2010;37:2159–2166. doi: 10.1118/1.3395579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris A, Kamishima T, Hao HY, Kato F, Omatsu T, Onodera Y, et al. Splenic volume measurements on computed tomography utilizing automatically contouring software and its relationship with age, gender, and anthropometric parameters. Eur J Radiol. 2010;75:e97–e101. doi: 10.1016/j.ejrad.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Dello SA, Stoot JH, van Stiphout RS, Bloemen JG, Wigmore SJ, Dejong CH, et al. Prospective volumetric assessment of the liver on a personal computer by nonradiologists prior to partial hepatectomy. World J Surg. 2011;35:386–392. doi: 10.1007/s00268-010-0877-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow diagram of development dataset.
CT Imaging Techniques Used for Development and Test Datasets
Diagrams showing detailed architecture of deep learning algorithm.
Representative images showing examples of segmentation errors generated by deep learning algorithm in test dataset-1.