Abstract
A large portion of the eukaryotic genome is packed into heterochromatin, a versatile platform that is essential to maintain genome stability. Often associated with a compact and transcriptionally repressed chromatin state, heterochromatin was earlier considered as a static and locked compartment. However, cumulative findings over the last 17 years have suggested that heterochromatin displays dynamics at different time and size scales. These dynamics are thought to be essential for the regulation of heterochromatin. This review illustrates how the key principles underlying heterochromatin structure and function have evolved along the years, and summarizes the discoveries that have led to the continuous revision of these principles. Using HP1-mediated heterochromatin as a context, we discuss a novel paradigm for heterochromatin organization based on two emerging concepts, phase separation and nucleosome structural plasticity. We also examine the broader implications of this paradigm for chromatin organization and regulation beyond heterochromatin.
Heterochromatin organization and function
Heterochromatin is a highly conserved, structurally poorly defined type of chromatin that is essential for the functional organization of eukaryotic genomes. It was first described by Emil Heitz in 1928 as a chromatin region that remained visible and condensed throughout the cell cycle [1]. Since its first observation, extensive research has identified many molecular features and mechanisms of action of heterochromatin. The two major types of heterochromatin are constitutive and facultative, defined by specific histone marks and chromatin-associated factors. Constitutive heterochromatin is a highly conserved type of heterochromatin present in many eukaryotes, and it is found in pericentromeric and telomeric regions of chromosomes where it forms specific chromosomal structures [2]. This type of heterochromatin is marked by methylation of lysine 9 of histone H3 (H3K9me), which is deposited by the Su(var)39 and G9a methyl-transferases, and recognized by the heterochromatin protein 1 (HP1) [2]. Facultative heterochromatin is marked by methylation of lysine 27 of histone H3 (H3K27me), which is deposited by the multi-subunit Polycomb complexes. Since this type of heterochromatin is found at genomic regions whose transcriptional state changes during development, it is often viewed as a less static structure than constitutive heterochromatin [3]. Constitutive and facultative heterochromatin share many common features, including their ability to propagate and spread across domains often causing heritable gene silencing.
Heterochromatin is essential for many aspects of genome function, as it (i) mediates gene silencing, (ii) protects genome integrity by repressing recombination of selfish elements, (iii) helps organize specialized structures such as centromeres and telomeres, and (iv) regulates nuclear morphology and rigidity [4,5]. Many of these functions have been proposed to arise from the ability of heterochromatin factors to occlude and condense the underlying DNA. For example, gene repression is thought to be achieved in part by the ability of heterochromatin to act as a barrier to transcriptional complexes. The concept of chromatin compaction as a means to make the underlying DNA inaccessible was initially proposed in 1976 by Aaron Klug and has since been supported by experimental observations based on staining, enzyme accessibility, and biochemical assays [6–8]. Consequently, earlier descriptions of heterochromatin imagined it as a locked and static structure.
While the many functions of heterochromatin seem to be tightly linked to its structure and reduced accessibility, the investigation of its three-dimensional organization has been a major challenge in the field. Models for how chromatin folds in the absence of heterochromatin proteins are also being constantly revised. Current textbook models describe DNA as packaged into chromatin via a set of hierarchically related structures [9]. The first level of packing in this model are nucleosomes, which consist of 147 bp of DNA wrapped around an octamer of histone proteins forming a particle with a ~10 nm diameter. An open and accessible chain of nucleosomes, often defined as a 10-nm fiber, has been considered to represent the architecture of the less dense and actively transcribed portions of chromatin, termed euchromatin. The next level of packing in the hierarchical model consists of the folding of the 10-nm fiber of nucleosomes into a higher-order structure that is specifically coiled to generate an ordered fiber of 30 nm diameter. High-resolution structures of biochemically reconstituted 30-nm fibers showed a structure with a regular periodicity generated by specific inter-nucleosome interactions [10–13]. Such structures were observed on chromatin preparations extracted from cells using defined extraction protocols [14]. Based on these observations, 30-nm fibers have, until recently, been argued to represent the underlying compact architecture of heterochromatin in cells.
The early model of a static and closed heterochromatin formed by repeated higher-order structures has been challenged at multiple levels over the years. Indeed, we have started to appreciate that heterochromatin is predominantly formed by less structurally ordered chromatin and that it is much more dynamic than previously thought. In this review we will discuss the findings that have contributed to the evolution and redefinition of models for heterochromatin.
Recent advances in methodologies have enabled studies of chromatin in cells with minimal sample disruption and demonstrate that the stereotypic 30-nm fiber architecture observed previously is not the predominant form of higher-order folding in cells. Several electron microscopy (EM)-based studies in yeast and mammalian cells showed that chromatin does not display any discrete and long-range repeating structures that resemble 30-nm fibers [15–19]. For example, a recent method that combines EM tomography with chemical labeling (ChromEMT) enabled visualization of chromatin organization in human cells and showed that chromatin consists of disordered chains with a diameter of 5–24 nm. Data based on quantitative super-resolution microscopy and Hi-C experiments that measure chromatin contacts and three-dimensional chromosome folding are also consistent with the absence of a predominant higher-order structure and suggest a more disorganized architecture [20,21]. Remarkably, euchromatin and heterochromatin appear to have the same underlying chromatin structure and differ mainly in the number of nucleosomes measured in a defined volume (i.e. density of fibers), with heterochromatin being denser [15].
These observations highlight the need for a revised model of heterochromatin that explains how its incredibly dense, yet apparently disordered architecture is formed, maintained, and dynamically used to create specific cellular programs. This review mainly focuses on recent discoveries made in the context of the heterochromatin protein 1 (HP1) that can help reconcile the lack of ordered 30-nm fibers with the presence of highly regulated cellular mechanisms.
HP1-heterochromatin as a model system for chromatin compaction
HP1 molecules are structural proteins that are at the core of constitutive heterochromatin organization and function [22]. Initially identified in Drosophila melanogaster, they are also highly conserved between yeast and humans. HP1 proteins consist of three conserved domains: the N-terminal chromodomain (CD), which binds the H3K9me3 tail; the hinge region (H), which is flexible and binds nucleic acid; and the chromoshadow domain (CSD), which mediates HP1 dimerization and interacts with other proteins (Fig. 1). Despite the high sequence conservation, differences in HP1 paralogs have been proposed to explain their distinct functions and cytological localization [23]. For example, humans possess three HP1 paralogs, HP1α, β and γ. HP1 α and β are mainly found at pericentric chromatin and are involved in gene silencing, while HP1γ is also found at transcriptionally active domains and is involved in transcriptional elongation and RNA processing [23]. Given their ability to self-oligomerize and bind chromatin marked by H3K9me, HP1 proteins are thought to compact heterochromatin by bridging across nucleosomes [22,24] (Fig. 2, panel a). More recently, human HP1α has been shown both in vitro and in vivo to form phase-separated liquid droplets that can include chromatin [25,26] (Fig. 2, panel b). Phase-separated liquid droplets are formed when a homogeneous solution demixes into separate phases with different chemical and physical properties. In biology, such physicochemical phenomena result in the formation of membraneless compartments that specifically partition and concentrate proteins and nucleic acids and whose function depends on their molecular composition [27]. These phase-separated compartments, named biomolecular condensates, are involved in many cellular processes including DNA damage responses and RNA metabolism [27]. Examples of biomolecular condensates are nucleoli and Cajal bodies in the nucleus, and stress and germ granules in the cytoplasm [28–30]. Weak intra- and intermolecular multivalent interactions have been shown to be a common organizing principle of these condensates [27,31]. Further, some or all of the component molecules display highly dynamic behavior within condensates and in some cases can exchange with the surrounding environment within seconds to minutes [26,27]. Insights from other biological phase separated condensates have greatly informed the significance of phase separation in the context of heterochromatin.
Figure 1: HP1 protein structure.
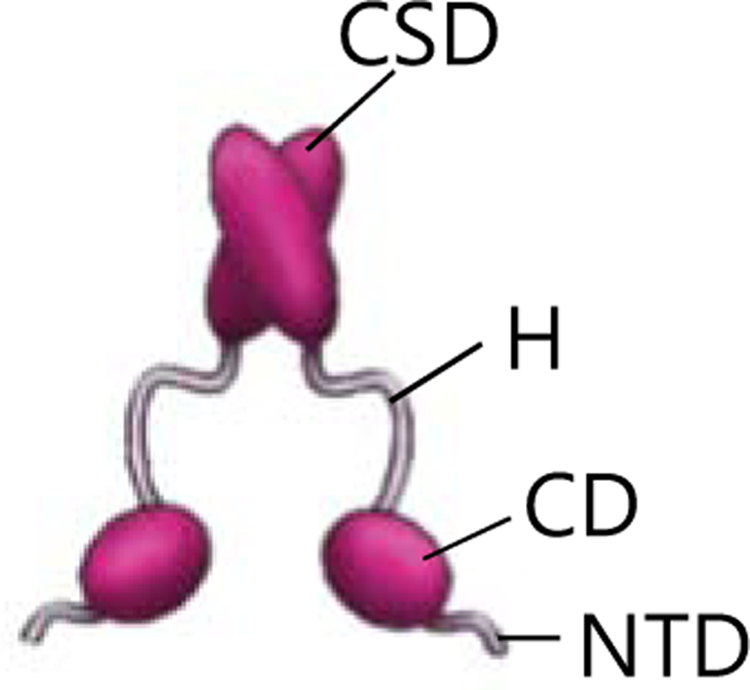
HP1 proteins contain three conserved domains: the chromodomain (CD) that binds H3K9me histone tails, the Hinge (H) that binds nucleic acids, the chromoshadow domain (CSD) that enables HP1 dimerization and interactions with other factors. The N-terminal domain (NTD) of HP1 is unstructured and has regulatory functions [23].
Figure 2: Model for HP1-heterochromatin.
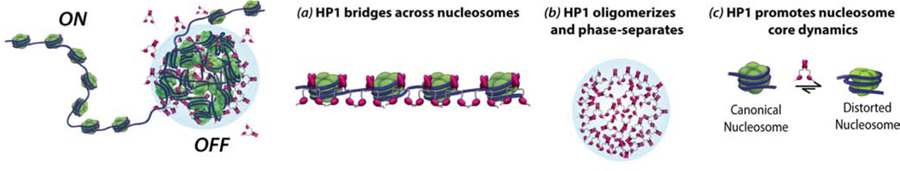
HP1 compacts chromatin into phase separated liquid droplets. This process relies on HP1 ability to: (a) bind and bridge across nucleosomes, (b) self-oligomerize, (c) and increase dynamics in the nucleosome core. Distorted nucleosomes expose buried regions of the histone core and enable inter-nucleosome interactions that drive chromatin phase separation. HP1 dimers are shown in pink.
A phase separation-based model for heterochromatin organization provides, at first glance, a simple and intuitive explanation of how heterochromatin can be condensed and organized without invoking higher-order structures beyond the 10-nm fibers. Within the HP1 condensates, HP1 could bring 10-nm fibers together in close proximity to create a chromatin domain with higher density than the surrounding phase. In this model, chromatin is condensed, yet dynamically accessible for regulation (Fig. 2).
Nucleosome conformational dynamics can regulate chromatin phase separation
An extra level of complexity has been added by recent work showing that Swi6, the major S. pombe HP1 protein, induces conformational dynamics within the core of individual nucleosomes to drive heterochromatin phase separation [32] (Fig. 2, panel c). Such dynamics transiently expose buried regions of the folded core of the histone proteins and promote phase separation of chromatin. It is proposed that such reshaping of individual nucleosomes helps drive weak and dynamic inter-nucleosome interactions thereby coupling phase-separation of chromatin to its condensation. This counterintuitive increase in accessibility at the level of individual nucleosomes results in the physical transformation of a chain of nucleosomes into a more compact and likely less accessible chromatin state within phase-separated droplets. Such a model is consistent with previous observations of lower histone turnover in S. pombe heterochromatin and lower DNA accessibility in Drosophila heterochromatin [33,34]. Hence, HP1 proteins regulate heterochromatin formation through multiple mechanisms: (i) nucleosome binding and bridging across nucleosomes, (ii) self-oligomerization, (iii) and induction of nucleosome structural dynamics that promote inter-nucleosome interactions (Fig. 2). Importantly, these molecular mechanisms are thought to be thermodynamically coupled to enable the formation of chromatin-HP1 phase-separated condensates. Together with previous data showing that in the context of heterochromatin in cells, HP1 molecules display “on-off” dynamics on time scales ranging from milliseconds to seconds, these discoveries indicate that heterochromatin is not a “locked-down” and static chromatin state [35–37]. Instead, heterochromatin behavior is consistent with a liquid-like compartment with molecular dynamics occurring at multiple size and time scales – ranging from atomic-scale conformational dynamics on the microsecond-millisecond timescale within the nucleosome core to meso-scale motions at the seconds timescale at the level of whole heterochromatin domains.
Recent studies have also revealed that chromatin has an intrinsic ability to phase separate under physiological conditions [32,38]. Further, not only chromatin-binding factors, but also post-translational modifications (PTMs) on histone tails and nucleosome conformational dynamics can fine-tune this process. For example, histone tail PTMs that are known to prevent chromatin compaction such as acetylation and disulphide bonds that reduce histone core dynamics inhibit chromatin’s intrinsic ability to phase separate [32,38]. Conversely, the presence of the linker histone H1, which is known to compact chromatin, promotes the ability of chromatin to phase separate [38].
A liquid-like model for heterochromatin
Overall, these recent data reinforce the concept of a highly dynamic heterochromatin and provide the following new conceptualizations. First, chromatin is a fluid polymer whose compaction is coupled to its phase separation. Second, chromatin phase separation relies on inter-nucleosomal interactions that can be regulated by chromatin-binding factors such as HP1, and by chromatin properties, such as histone tail PTMs and nucleosome conformational dynamics. Third, these inter-nucleosomal interactions are transient and weak in order to enable the formation of liquid-like condensates, and they do not require the formation of a higher-ordered and static structure such as a 30-nm fiber (Fig. 3). Importantly, this new model proposes the critical role of transient exposure of buried nucleosome core regions to drive inter-nucleosomal interactions, in addition to the previously described interactions between histone tails, nucleosomal DNA and the acidic patch surface of the nucleosome [10].
Figure 3: Model for genome packaging.
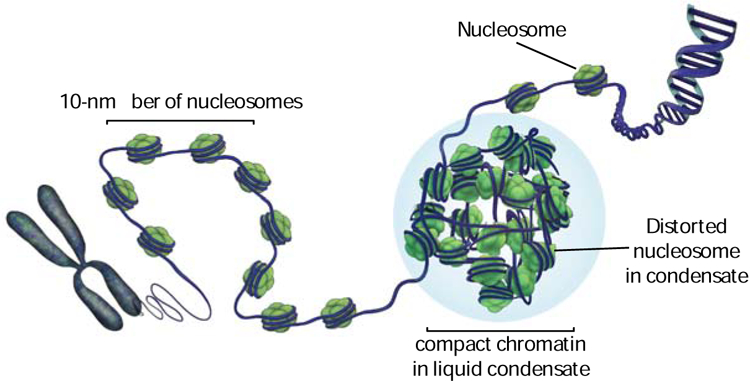
DNA is wrapped around nucleosomes, the fundamental units of chromatin. The 10-nm fiber of nucleosomes is a fluid polymer that organizes through weak and multivalent interactions. Nucleosome conformational dynamics, histone PTMs and chromatin-binding proteins fine-tune and regulate such interactions. Proteins such as HP1 can promote condensation of chromatin into phase-separated condensates. Phase separation and nucleosome dynamics might more generally regulate chromatin compaction, for example, in the context of mitotic chromosomes.
The liquid-like model introduces two novel fundamental principles of chromatin organization that have been missing from previous models: the role of nucleosome conformational dynamics as a new layer of chromatin regulation and chromatin phase separation as a chromatin self-organizing property. Below we will discuss the implications of these emerging principles.
Implications of nucleosome conformational dynamics
HP1-mediated nucleosome conformational dynamics on the atomic-scale appear to be tightly coupled with the three-dimensional folding of chromatin and compaction into liquid droplets [32]. In this framework, we can imagine that changes in nucleosome structural flexibility will directly impact chromatin organization. Nucleosomes have been shown to adopt conformations distinct from the one seen in the crystal structure and recent work suggests that nucleosomes in mitotic chromatin may adopt non-canonical conformations [16,39–41]. It is therefore possible that nucleosome conformational flexibility might be an additional layer of chromatin regulation that could broadly tune chromatin compaction and genome organization. For example, the formation of chromosomal domains or highly compacted chromatin states such as mitotically condensed chromatin may also rely on the deformation of the histone core (Fig. 3).
Additionally, nucleosome conformational dynamics can provide solutions to some longstanding questions in chromatin regulation. For example, several histone modifications are being discovered on buried histone regions, but their functions remain unknown [42]. Similarly, while different histone variants have distinct biological effects, their crystal structures within nucleosomes often appear similar, limiting our understanding of the structural basis for their different functions. This new model can help address these questions because some histone modifications and histone variants may function by directly affecting the conformational plasticity of the histone octamer and their interaction with proteins like HP1, thereby altering inter-nucleosome interactions. For example, it has been shown that in vitro, the interaction of HP1α with nucleosomes is enhanced by the histone variant H2A.Z as is the ability of HP1α to compact chromatin [43,44]. Therefore, it is possible that changes in the dynamics of nucleosomes containing H2A.Z might contribute to the increased compaction by HP1. In another example, the H3 histone variant CENP-A has been reported to contribute to the increase in elasticity and accessibility of CENP-A-containing nucleosomes [45]. Part of this effect might arise from the specific nucleosome dynamics enabled by CENP-A. Alterations in nucleosome dynamics might also be one mechanism by which onco-histone mutations, especially in the globular domains, misregulate chromatin function in cancers [46]. The application of solution methods such as NMR and hydrogen-deuterium exchange experiments will be essential in the future to better characterize and understand the extent and function of nucleosome conformational plasticity.
The model also suggests new mechanisms for how HP1 ligands and chromatin properties can regulate phase separation of heterochromatin. For example, the presence of histone marks could promote chromatin phase separation by tuning the extent of octamer deformation mediated by HP1. This is consistent with data showing that deformation of the octamer core by Swi6 is energetically coupled to recognition of H3K9me3 [32]. In contrast, specific ligands of Swi6 could compete or weaken the interaction with chromatin thereby inhibiting phase separation.
The significant increase in nucleosome breathing mediated by HP1 also challenges the current paradigm of DNA inaccessibility in heterochromatin. By inducing conformational changes in nucleosomes, HP1 could in principle increase DNA breathing and accessibility within the heterochromatin phase. In this framework, the increased nucleosome breathing, could be an opportunity for fine-tuning molecular interactions in specific contexts within heterochromatin. For example, this phenomenon could explain how HP1 proteins promote open chromatin and transcriptional activation at specific genomic sites, such as in the case of chromosome 4 in Drosophila [47–49].
Implication of phase separation by chromatin
Phase separation has not only been observed in the context of HP1-mediated heterochromatin. It has also been reported that phase separation of the Polycomb complex PRC1 is important for Polycomb mediated chromatin compaction and gene regulation [50]. Furthermore, phase separation of many other chromatin-binding factors such as BRD4 has been linked to their functions on chromatin [51–56]. The discovery of an increasing number of chromatin factors bearing phase-separation ability, together with the intrinsic ability of chromatin to phase separate, suggests a more general role for phase separation in genome compartmentalization and regulation. In this context, the potentially rapid and fine-tunable aspects of phase separation processes might enable the high plasticity of genome regulatory programs.
Viewing heterochromatin organization through the lens of phase separation provides a starting point to discover novel regulatory mechanisms relevant to health and disease. For example, given the link between neurodegenerative disease and the formation of defective glass-like condensates or aggregates in the cytoplasm, we can speculate that similar defects within heterochromatin condensates might be linked to premature aging and other diseases [57].
Challenges for the future
Despite having identified some of the key interactions that drive formation of HP1-mediated heterochromatin condensates, it remains unclear how HP1 and chromatin are structurally organized within droplets and how heterogeneous and diverse any such organization may be. For example, HP1 can phase separate upon phosphorylation, in presence of DNA, or upon interaction with both un-methylated and H3K9me chromatin [32,58]. In these different contexts, the critical concentration required for phase separation is different and even the molecular interactions involved are different. At a superficial level these condensates display similar morphologies but whether and how their material properties and internal structural organizations differ remains a mystery [59]. It is also unclear whether molecules within these different condensates are homogeneously distributed and whether or not they display similar diffusion behaviors and similar conformational states. New tools are thus needed to allow a more in-depth study of the structure and physicochemical properties of condensates, and their functions.
How specificity of partitioning into the droplets is achieved and how such specificity determines which proteins or DNA can be included or excluded from a phase is also an intriguing aspect to be addressed in the future. By addressing how these emerging principles contribute to heterochromatin organization, we will be able to better understand the functional compartmentalization of the genome and how to eventually restore it when defects arise.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Heitz E Das Heterochromatin der Moose. Jahrb Wiss Botanik 1928;69:762–818. [Google Scholar]
- 2.Grewal SIS, Jia S: Heterochromatin revisited. Nat. Rev. Genet 2007, 8:35–46. [DOI] [PubMed] [Google Scholar]
- 3.Margueron R, Reinberg D: The Polycomb complex PRC2 and its mark in life. Nature 2011, 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon N: Heterochromatin structure and function. Biol. Cell 2004, 96:631–637. [DOI] [PubMed] [Google Scholar]
- 5.Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, Marko JF: Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 2018, 29:220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finch JT, Klug A: Solenoidal model for superstructure in chromatin. PNAS 1976, 73:1897–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert N, Allan J: Distinctive higher-order chromatin structure at mammalian centromeres. PNAS 2001, 98:11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallrath LL, Elgin SC: Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 1995, 9:1263–1277. [DOI] [PubMed] [Google Scholar]
- 9.Widom J: Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct 1998, 27:285–327. [DOI] [PubMed] [Google Scholar]
- 10.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ: Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 2004, 306:1571–1573. [DOI] [PubMed] [Google Scholar]
- 11.Widom J, Finch JT, Thomas JO: Higher-order structure of long repeat chromatin. EMBO J 1985, 4:3189–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schalch T, Duda S, Sargent DF, Richmond TJ: X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 2005, 436:138–141. [DOI] [PubMed] [Google Scholar]
- 13.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D: EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. PNAS 2006, 103:6506–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA: Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 2004, 118:555–566. [DOI] [PubMed] [Google Scholar]
- 15.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC: ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357:eaag0025.(*) By developing a method that combines EM tomography with chemical labeling, this study investigates chromatin organization in human cells. The authors show that chromatin forms flexible chains with diameters between 5 and 24 nm packed together at different densities.
- 16.Cai S, Böck D, Pilhofer M, Gan L: The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin. Mol. Biol. Cell 2018, 29:2450–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDowall AW, Smith JM, Dubochet J: Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J 1986, 5:1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eltsov M, MacLellan KM, Maeshima K, Frangakis AS, Dubochet J: Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. PNAS 2008, 105:19732–19737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Lim HH, Shi J, Tamura S, Maeshima K, Surana U, Gan L: Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo. Mol. Biol. Cell 2016, 27:3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP: Chromatin Fibers Are Formed by Heterogeneous Groups of Nucleosomes In Vivo. Cell 2015, 160:1145–1158. [DOI] [PubMed] [Google Scholar]
- 21.Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. : Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A 2015, 112:E6456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maison C, Almouzni G: HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol 2004, 5:296–304. [DOI] [PubMed] [Google Scholar]
- 23.Canzio D, Larson A, Narlikar GJ: Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol 2014, 24:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B: Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 2011, 41:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ: Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547:236–240.(**) This is one of the two initial studies proposing that heterochromatic domains are organized into phase-separated liquid compartments. This work uses biochemical assays to characterize human HP1a phase separation.
- 26.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH: Phase separation drives heterochromatin domain formation. Nature 2017, 547:241–245.(**) This is one of the two initial studies proposing that heterochromatic domains are organized into phase-separated liquid compartments. This work shows that Drosophila HP1a exhibits liquid demixing in vitro and in vivo.
- 27.Banani SF, Lee HO, Hyman AA, Rosen MK: Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 2017, 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA: Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- 29.Brangwynne CP, Mitchison TJ, Hyman AA: Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A 2011, 108:4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin Y, Chang Y-C, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP: Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 2018, 175:1481–1491.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. : Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ: HP1 reshapes nucleosome core to promote heterochromatin phase separation. Nature 2019, 29:220.(**) This study uses biophysical method to show that the S.pombe HP1 protein, Swi6, increases structural dynamics of the nucleosome core to compact chromatin in phase separated condensates. It also shows that nucleosome dynamics regulate chromatin compaction in condensates.
- 33.Aygün O, Mehta S, Grewal SIS: HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol 2013, 20:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elgin SCR, Reuter G: Position-Effect Variegation, Heterochromatin Formation, and Gene Silencing in Drosophila. Cold Spring Harb Perspect Biol 2013, 5:a017780–a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T: Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 2003, 299:721–725. [DOI] [PubMed] [Google Scholar]
- 36.Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D: Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science 2003, 299:719–721. [DOI] [PubMed] [Google Scholar]
- 37.Hinde E, Cardarelli F, Gratton E: Spatiotemporal regulation of Heterochromatin Protein 1-alpha oligomerization and dynamics in live cells. Sci Rep 2015, 5:12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK: Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179:470–484.e21.(**) This study reports that chromatin can undergo liquid–liquid phase separation under physiological conditions. It further proposes that histone post-translational modifications and chromatin-binding factors regulate chromatin phase separation.
- 39.Bilokapic S, Strauss M, Halic M: Cryo-EM of nucleosome core particle interactions in trans. Sci Rep 2018, doi: 10.1038/s41598-018-25429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bilokapic S, Strauss M, Halic M: Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol 2017, doi: 10.1038/s41594-017-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha KK, Gross JD, Narlikar GJ: Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science 2017, 355:eaaa3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman GD, Poirier MG: Post-Translational Modifications of Histones That Influence Nucleosome Dynamics. Chem. Rev 2014, 115:2274–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan JY, Rangasamy D, Luger K, Tremethick DJ: H2A.Z Alters the Nucleosome Surface to Promote HP1α-Mediated Chromatin Fiber Folding. Mol. Cell 2004, 16:655–661. [DOI] [PubMed] [Google Scholar]
- 44.Ryan DP, Tremethick DJ: The interplay between H2A.Z and H3K9 methylation in regulating HP1α binding to linker histone-containing chromatin. Nucleic Acids Res 2018, 46:9353–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melters DP, Pitman M, Rakshit T, Dimitriadis EK, Bui M, Papoian GA, Dalal Y: Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners. PNAS 2019, doi: 10.1073/pnas.1911880116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, Kundra R, Schultz N, Muir TW, Allis CD: The expanding landscape of “oncohistone” mutations in human cancers. Nature 2019, 567:473–478.(*) The authors report the first complete dataset of histone mutations associated with cancers. They list mutations occurring in all four core histones, in both the N-terminal tails and globular folded domains.
- 47.Cryderman DE, Vitalini MW, Wallrath LL: Heterochromatin protein 1a is required for an open chromatin structure. Transcription 2011, 2:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon SH, Workman JL: The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays 2011, 33:280–289. [DOI] [PubMed] [Google Scholar]
- 49.Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, et al. : Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 2011, 21:147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, Kingston RE: Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev 2019, 33:799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA: A Phase Separation Model for Transcriptional Control. Cell 2017, 169:13–23.(*) This is the first paper suggesting a possible role of phase separation in transcriptional control.
- 52.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. : Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175:1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Bertulat B, Tencer AH, Ren X, Wright GM, Black J, Cardoso MC, Kutateladze TG: MORC3 Forms Nuclear Condensates through Phase Separation. iScience 2019, 17:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II: Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, Wang S, Suter T, Alshareedah I, Gamliel A, et al. : Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol 2019, 26:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, et al. : RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol 2018, 25:833–840. [DOI] [PubMed] [Google Scholar]
- 57.Alberti S, Dormann D: Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet 2019, 53:annurev–genet–112618–043527. [DOI] [PubMed] [Google Scholar]
- 58.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ: Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alberti S, Gladfelter A, Mittag T: Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176:419–434.The authors discuss guidelines for rigorous experimental characterization of LLPS processes in vitro and in cells, the caveats of common experimental approaches, and point out experimental and theoretical gaps in the field.


