Abstract
Recent studies using mouse models suggest that interaction between the gut microbiome and IL-17/IL-22–producing cells plays a role in the development of metabolic diseases. We investigated this relationship in humans using data from the prediabetes study of the Integrated Human Microbiome Project (iHMP). Specifically, we addressed the hypothesis that early in the onset of metabolic diseases there is a decline in serum levels of IL-17/IL-22, with concomitant changes in the gut microbiome. Clustering iHMP study participants on the basis of longitudinal IL-17/IL-22 profiles identified discrete groups. Individuals distinguished by low levels of IL-17/IL-22 were linked to established markers of metabolic disease, including insulin sensitivity. These individuals also displayed gut microbiome dysbiosis, characterized by decreased diversity, and IL-17/IL-22–related declines in the phylum Firmicutes, class Clostridia, and order Clostridiales. This ancillary analysis of the iHMP data therefore supports a link between the gut microbiome, IL-17/IL-22, and the onset of metabolic diseases. This raises the possibility for novel, microbiome-related therapeutic targets that may effectively alleviate metabolic diseases in humans as they do in animal models.
Introduction
The human gut microbiome consists of trillions of microorganisms that are known to impact host physiology. Variation in composition of the gut microbiome has been linked to metabolic disorders such as hypertension (1,2), obesity (3,4), and insulin resistance (5), as well as to type 2 diabetes (T2D) (6–10).
While much remains to be learned about the functional mechanisms underpinning this relationship, growing evidence in mouse models points to an important role for the microbially mediated immune system (11,12). Deficiencies in TLR5 (13), RORγt (14), or IL-22 (15) are associated with a variety of metabolic disorders. Additionally, high-fat diet–induced obesity results in a reduction in IL-17–producing cells in the small intestine lamina propria (SILP) (14), while induction of Th17 cells (16), or treatment with gut-homing Th17 cells (17), low-dose IL-17 (18), or IL-22 (13,15), can ameliorate the obesity-associated metabolic phenotype. Collectively, these results suggest a protective role for IL-17/IL-22–producing cells during onset of diet-related metabolic disorders in mice. While Th17 cells are a major source of IL-17 and IL-22 (16,19), other cell types also produce these cytokines (20) and may be important in this process.
In humans, low IL-22 has been associated with impaired fasting glucose and T2D (21). However, the relationship between IL-17/IL-22 production, the microbiome, and metabolic diseases remains controversial (22–24) and comparatively understudied. Large-scale integrated omics studies provide an ideal opportunity to investigate this relationship. Here, we present an ancillary study of the recently released prediabetes arm of the Integrated Human Microbiome Project (iHMP) (25), in which our goal was to investigate the relationship between IL-17/IL-22 profiles, the gut microbiome, and aspects of the metabolic syndrome. Specifically, we aimed to test hypotheses in humans that have hitherto only been convincingly demonstrated in mice.
Research Design and Methods
This study is an ancillary analysis of data collected as part of the iHMP. An overview of the iHMP is provided below, along with a description of the collection of the specific data sets included in this analysis. However, full details of study design, recruitment, and sample collection can be found in our iHMP flagship article (26).
iHMP Overview
The iHMP consists of 103 human participants identified as at risk for developing T2D, who were followed over a 4-year period (Supplementary Figs. 1 and 2). During this time, detailed multiomic profiling was carried out at quarterly intervals and more frequently during periods of stress or upper respiratory infection. Participants were recruited following Stanford University Institutional Review Board Protocol no. 23062.
Blood and Insulin Sensitivity Measurements
Blood samples were collected from participants following an overnight fast (26) and used for lipid and metabolic panels, as well as fasting plasma glucose (FPG) and hemoglobin A1c (HbA1c) tests. Measurements of FPG ≥126 mg/dL were classified as diabetes, while measurements of FPG between 100 and 125 mg/dL were classified as prediabetes. HbA1c was assumed to be an indicator of 3-month average glucose levels, with measurements ≥6.5% (48mmol/mol) classified as diabetes and measurements between 5.7% and 6.5% (39–48 mmol/mol) classified as prediabetes. In addition to standard tests, a subset of study participants (n = 65) underwent a one-time measurement of steady-state plasma glucose (SSPG) levels via a modified insulin-suppression test (26). Briefly, individuals were infused with glucose (240 mg/m2/min), octreotide (0.27 µg/m2/min), and insulin (25 mU/m2/min) for 180 min after an overnight fast. Starting from 150 min, blood was drawn at 10-min intervals. Four plasma samples (from blood drawn at 150 min, 160 min, 170 min, and 180 min) were measured for glucose and insulin concentrations. SSPG was the mean of the four plasma glucose concentrations. At these time points, insulin concentrations were at a steady state and were similar in all subjects (65 μU/mL); thus, the SSPG provides a direct measure of the relative ability of insulin to dispose of a glucose load: the higher the SSPG concentration, the more insulin resistant the individual. Individuals with SSPG <150 mg/dL were classified as insulin sensitive, while individuals with SSPG ≥150 mg/dL were classified as insulin resistant.
Microbiome Measurements
Stool samples were collected and DNA was extracted according to the Human Microbiome Project standard protocol (no. 07-001. V12.0). Bacterial relative abundance was then determined by sequencing the V1–V3 region of the bacterial 16S rRNA gene on the MiSeq platform (Illumina, San Diego, CA).
Cytokine Measurements
Cytokine data were generated from blood samples using a 63-plex Luminex antibody-conjugated bead capture assay (Affymetrix, Santa Clara, California). Raw cytokine data were normalized to median fluorescence intensity (MFI) to eliminate batch effects. Further details of approaches used to generate sequence and cytokine data can be found in our companion article (26). According to the manufacturer’s protocol, CHEX1–CHEX4 are different types of background control for Luminex MFI data. Based on preliminary examination of these data, any samples with substantial background noise (determined as >5 SD ± mean value [mean ± 5 * SD]) for one or more CHEX measurements were removed.
Diet Data
An assessment of the frequency of consumption of 25 food items was carried out during some, but not all, sample collection visits. Details of the food items monitored as well as the results of this questionnaire can be found in Supplementary Table 1. Full details of the questionnaire design and sample collection can be found in our companion article (27).
Statistical Analysis
A two-sided Student t test was used for significance testing when data were normally distributed; otherwise, a two-sided Wilcoxon signed rank test or Mann-Whitney U test was used. A χ2 test was used to determine whether the proportion of insulin-resistant individuals was different between high-activity (HA) and low-activity (LA) groups. Linear discriminant analysis based on effect size (LEfSe) (28) was performed to determine whether the microbial taxon abundances differed between HA and LA groups. All statistical tests were performed using R (version 3.5.0). Exploration of diet data was performed by principal components analysis using the prcomp command in R package stats. Diet scores were log transformed prior to analysis.
Data Modeling
Of the 103 iHMP study participants, not all had a sufficient number of repeated measurements for inclusion in this longitudinal study. An overview of the number of participants available for each analysis described below is provided in Supplementary Fig. 2. Key characteristics of the individuals included in the principal analyses are provided in Supplementary Table 2.
Mixture Model of Individuals Based on IL-17/IL-22
Participants with five or more longitudinal cytokine measurements (n = 68) were included in a general mixture model (GMM), built using the R package mclust (29). The longitudinal IL-17A, IL-17F, and IL-22 MFI data were summarized as mean value and SD for each individual and then scaled in R. For determination of the optimal number of Gaussian distributed clusters, models with 1–9 clusters were evaluated using the Bayesian information criterion, resulting in three clusters selected for further analyses (Supplementary Fig. 3). Cluster 1 comprised 25 individuals (LA group), cluster 2 comprised 32 (indeterminate-activity [IA] group), and cluster 3 comprised 11 (HA group), and the mixing probability of each cluster was 0.3634941, 0.4749886, and 0.1615172, respectively. Participants assigned to each cluster were associated with a “confidence of assignment” probability (0%–100%); those with <99% confidence (eight individuals in total) were removed from the subsequent analyses.
Linear Mixed Models
Linear mixed models were built using the R package lme4 (30). Models were built separately to test different hypotheses, as described below.
For testing of whether metabolic profiles were different among three groups, the equation used was as follows: target of interest = group + days + sex + BMI + Adj.age + (1/subject_ID). Fixed effects included group as the categorical variable derived from the GMM model, sex was a binary categorical variable, and BMI and adjusted age (Adj.age) were continuous variables. The term days indicates a numerical measurement of how many days after the overall study start date the sample was collected. Adjusted age described the average age of an individual during the study period. Random effects included a random intercept for each participant (1 / subject_ID).
For red blood cell distribution width (RDW), for adjustment of the previous known effect of mean corpuscular hemoglobin (MCH) and mean corpuscular volume (MCV) on the readout of RDW (31), the model was built as follows: RDW = group + days + MCH + MCV + sex + BMI + Adj.age + (1 / subject_ID). For analysis related to microbiome alpha diversity and Firmicutes-to-Bacteroidetes ratio, our goal was to understand the fixed effect, group, in this mixed model, so the model is built as follows: microbiome diversity (or Firmicutes-to-Bacteroidetes ratio) = group + sex + BMI + Adj.age + (1 / subject_ID).
Bayesian Mixed-Effects Model for Taxa and Cytokine Interactions Conditional on Cluster Assignment
Participants with five or more coinciding measurements for both cytokines and microbiome (n = 53) were included in a Bayesian negative binomial longitudinal mixed-effects model to evaluate the relationship between individual microbes and IL-17. To account for the zero-inflated nature of microbiome abundance, we used a Bayesian framework on a sparse matrix with a negative binomial distribution (32). Cytokine-related group (HA vs. LA) and cytokine (either IL-17A or IL-17F) were scored as the interaction term and fixed effect, respectively, to test the combined effect of cytokine-related group and cytokine on microbe abundance. IL-22 was excluded from this analysis because a large proportion of IL-22 measurements appeared to be lower than the accurate detection threshold of the Luminex assay (Supplementary Fig. 4). We included the cytokine-related group–cytokine interaction term with the aim of testing the hypothesis that significant microbe-cytokine associations may be detected in HA, but not LA, subjects or vice versa.
Each microbe was modeled as the response variable with a random intercept for each participant, and with fixed effects for time, and an interaction term for the cluster identity (defined by the GMM, described above) and the cytokine of interest, thereby evaluating whether the relationship between a microbe and cytokine pair differed depending on the identity of the cluster. This followed standard matrix notation:
where Mi is a vector of microbe relative abundances for each participant i, Xi is the matrix of fixed effects, Zi is the random effects vector of 1s denoting a random intercept, bi is a scalar for each participant, and εi is a zero-centered error term. The fixed-effects matrix Xi comprised the days post–study start Di and an interaction term for cluster (Ci) and cytokine (Yi).
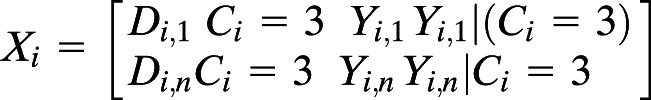 |
Sampling was performed with four chains with 5,000 iterations per sample and a burn-in of 1,000 iterations. Samples were drawn using the No-U-Turn Sampler implemented in the brms package (33–35). Chain convergence was confirmed by visual inspection of iteration plots and posterior predictive distributions.
Data and Resource Availability
Microbial sequence and cytokine data included in this study can be downloaded from the iHMP data depository website (https://www.hmpdacc.org/ihmp/). Diet data are included in Supplementary Table 1.
Results
Overview of the iHMP Prediabetes Cohort
We used the recently released iHMP data set as the basis for a detailed longitudinal analysis of the relationship between the gut microbiome and serum levels of IL-17A, IL-17F, and IL-22. The 103 individuals in the iHMP cohort were well characterized with respect to glucose-related measures, including fasting glucose, HbA1c, and insulin resistance (SSPG). Notably, however, these measures showed little concordance, suggesting they do not provide a consistent representation of the progression to T2D, which has been shown to be highly variable between individuals in this cohort (36).
Based on HbA1c measurements alone, 4 individuals in the iHMP cohort had diabetes at their first time of measurement (HbA1c ≥6.5% [48mmol/mol]), while 37 had prediabetes (5.7% ≤ HbA1c < 6.5% [39 ≤ HbA1c <48 mmol/mol]). Additionally, a further four individuals came to have diabetes at one or more points during the course of the study; however, their HbA1c measurements did not stay within the diabetes range (26,36). For the majority of study participants, HbA1c measurements did not increase across the course of the study (Supplementary Fig. 1).
Given the complexity of T2D diagnosis, we chose to focus on SSPG, which was measured once for the majority of participants and is a robust measure of insulin resistance (37). This focus was in keeping with our central hypothesis that microbiome-mediated changes in IL-17/IL-22 may affect insulin sensitivity.
Individuals Show Discrete IL-17/IL-22 Profiles Associated With Insulin Sensitivity
From the complete iHMP cohort, we selected 68 individuals who had five or more longitudinal cytokine measurements. Using Gaussian mixture modeling based on the mean cytokine level and longitudinal variance in each individual, we observed that 60 of these individuals could be optimally separated into three groups (Supplementary Figs. 2 and 3). Individuals at one extreme were characterized by consistently low cytokine levels and variance, while individuals at the other extreme were characterized by high levels or variance of at least one cytokine (Fig. 1A and Supplementary Fig. 3). Henceforth, we refer to these three groups as LA (n = 20), IA (n = 30), and HA (n = 10) to represent differences in their temporally integrated levels of serum IL-17 and IL-22 activity (Fig. 1A). Importantly, identifying the three groups in this manner required estimating intraindividual variation (Supplementary Fig. 5), which is not available from cross-sectional data, highlighting the advantage of the longitudinal design. Further investigation indicated that the longitudinal IL-17/IL-22 profiles characterizing each group were not significantly impacted by periods of stress or upper respiratory infection reported during iHMP visits (see additional analyses in Supplementary Materials and Supplementary Figs. 6–8). Baseline characteristics for these 60 individuals, including blood and insulin sensitivity measurements used in subsequent analyses, are provided in Supplementary Table 2.
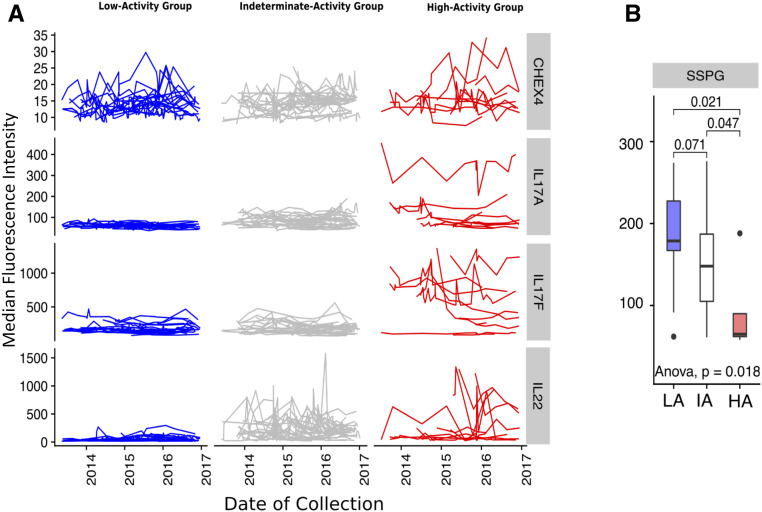
Figure 1.
Participants grouped according to IL-17/IL-22 cytokines. A: Gaussian mixture modeling of cytokine mean abundance and variance separates study participants into three discrete groups (columns). Lines within each panel represent repeated measurements of serum cytokine abundance for one individual over the study period. Rows represent serum cytokines (IL-17A, IL-17F, IL-22). CHEX4 is a measurement of background fluorescence intensity and can be treated as a negative control. (Note: different scales on y-axis for each row.) B: SSPG (mg/dL) measurement by group. P values for pairwise Wilcoxon test are labeled above the bar plot, and the P value for a one-way ANOVA test is labeled under the bar plot. The analysis in A was based on 297, 371, and 112 repeated measurements for HA, IA, and LA subjects, respectively.
We next considered the possibility that discrete IL-17/IL-22 cytokine profiles may reflect different stages of metabolic disease progression. Previous studies in mice demonstrated that HFD-induced onset of metabolic disease is associated with loss of CD4+ IL-17–producing cells in the SILP (14), while studies in humans have shown a negative correlation between serum IL-22 levels and physiological indicators of T2D (21). We therefore hypothesized that individuals with an LA profile would show a more severe metabolic phenotype than individuals with an HA profile. Individuals with an IA profile were excluded from this and subsequent analyses because we reasoned that, while they may reflect progression from HA to LA, they may also reflect a healthy state (i.e., pre-HA) prior to the onset of the chronic inflammation that is a characteristic of the metabolic syndrome (38).
Within the two-thirds (40 of 60) of study participants for whom an SSPG measurement was available (Supplementary Table 2), we observed that SSPG levels were significantly lower in HA subjects compared with LA subjects (two-sided Wilcoxon test, W = 64.5, P = 0.021 [Fig. 1B]). Accordingly, individuals in the LA group were more frequently insulin resistant (Supplementary Fig. 9A); however, mean FPG levels, age, and BMI did not vary significantly across groups (Supplementary Fig. 9A).
IL-17/IL-22 Inactivity Is Associated With a More Severe Metabolic Phenotype
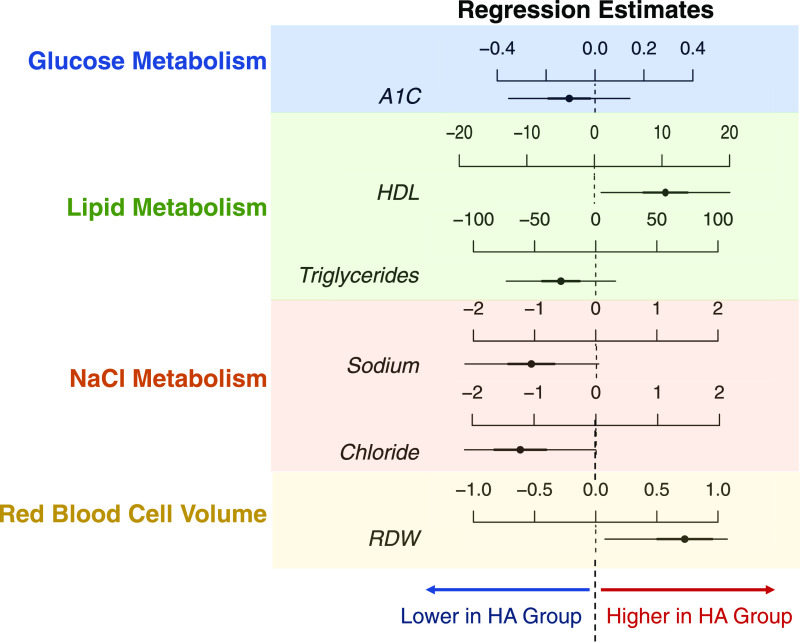
Longitudinal modeling of clinical data collected across the study period also revealed multiple markers that showed significant differences between LA compared with HA subjects (Fig. 2 and Supplementary Fig. 10). Participants classified as HA showed higher plasma HDL and lower plasma triglycerides. The RDW was higher in HA subjects, consistent with previous findings of high RDW as being associated with high HDL and low triglycerides (39). Additionally, serum sodium and chloride levels were lower in the HA group, and high insulin level has been associated with increased sodium retention in T2D (40). Established markers of T2D, including HbA1c and serum glucose, did not vary significantly between groups (Fig. 2 and Supplementary Figure 10). This was not unexpected, given the limited concordance between glucose-related measurements and that the fact that the majority of participants did not develop T2D during the course of this study (36) (Supplementary Fig. 1).
Figure 2.
Linear mixed model estimates on fixed effects introduced by LA and HA group. Results for full linear mixed models are shown in Supplementary Fig. 9. The comparisons of active vs. inactive groups are presented here. Dashed lines represent the LA group, while regression estimates for the HA group are displayed as horizontal lines. The center of each horizontal line is the β-coefficient of regression, while thick lines represent 50% credible intervals or ±1 SD and thin lines represent 95% credible intervals or ±2 SD. A1C, hemoglobin A1c. This analysis is based on 88 and 229 repeated measurements for HA and LA subjects, respectively.
IL-17/IL-22 Activity Is Associated With Variation in the Composition of the Gut Microbiome but Not Diet
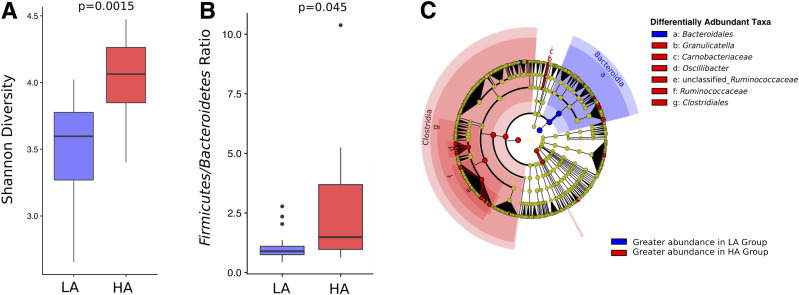
The close association between IL-17/IL-22–producing cells and gut microbiota prompted us to next ask whether individuals distinguished by cytokine activity levels differed in the composition of their gut microbiome. As an individual’s gut microbiome remained relatively stable throughout the course of this study (26) (Supplementary Fig. 5), we began by comparing mean microbiome abundance for each participant between HA and LA groups. We found LA subjects had a significantly lower alpha diversity (two-sided Wilcoxon test, W = 34.0, P = 0.003 [Fig. 3A and Supplementary Fig. 11]) and a lower Firmicutes-to-Bacteroidetes ratio (two-sided Wilcoxon test, W = 54.0, P = 0.044 [Fig. 3B and Supplementary Fig. 12]) compared with HA subjects. LefSE revealed differences between LA and HA groups at multiple taxonomic levels. Most notably, the classes Bacteroidia and Clostridia were more abundant in the LA and HA groups, respectively (Fig. 3C), indicating that members of these taxa were likely responsible for observed differences in the Firmicutes-to-Bacteroidetes ratio.
Figure 3.
Differences in the gut microbiome of IL-17/IL-22 LA and HA subjects. A: Shannon diversity estimates for the HA and LA. Mean value of diversity for each participant across the study period is used to generate this plot. The P value from a Wilcoxon test is labeled above the plot. B: Firmicutes-to-Bacteroidetes ratio of HA and LA. Mean value of Firmicutes-to-Bacteroidetes ratio for each participant across the study period is used to generate this plot. The P value from a Wilcoxon test is labeled above the plot. C: Cladogram representing the LEfSe results for comparing taxa abundance between HA and LA groups. Circles on the cladogram represent the phylogenetic relationship of taxa that are tested, with phylum at the center and operational taxonomic unit (OTU) on the edges. Each point represents a taxonomic unit. Red color covering a dot/region indicates the taxa that are more abundant in the HA group, and blue color covering a dot/area indicates the taxa are more abundant in the LA group.
As diet profoundly influences the composition of the gut microbiome (41,42), and a high-fat diet results in a loss of IL-17–producing cells in mice (14), we next considered whether IL-17/IL-22 activity was associated with dietary habits recorded as part of the iHMP. Diet composition appeared stable and distinct between individuals (Supplementary Fig. 13A), which was consistent with trends observed in the microbiome (Supplementary Fig. 5A). However, there was no evidence that diet varied significantly between LA and HA groups (Supplementary Fig. 13B).
Finally, we took advantage of intraindividual variation in microbe and cytokine abundance (Supplementary Fig. 5) by performing longitudinal modeling to look at pairwise relationships between individual bacterial genera and cytokine abundances within HA versus LA subjects. Previous studies identified significant associations between cytokines and gut microbes but only revealed interindividual variation (43,44) due to their cross-sectional design. The repeated and longitudinal measurements of the iHMP study allowed us to test the possibility that intraindividual variation in cytokine/bacteria abundance may be used to identify additional host-microbe associations of biological interest. To accommodate within-individual correlation, we used a mixed-effects model with random effects by individual.
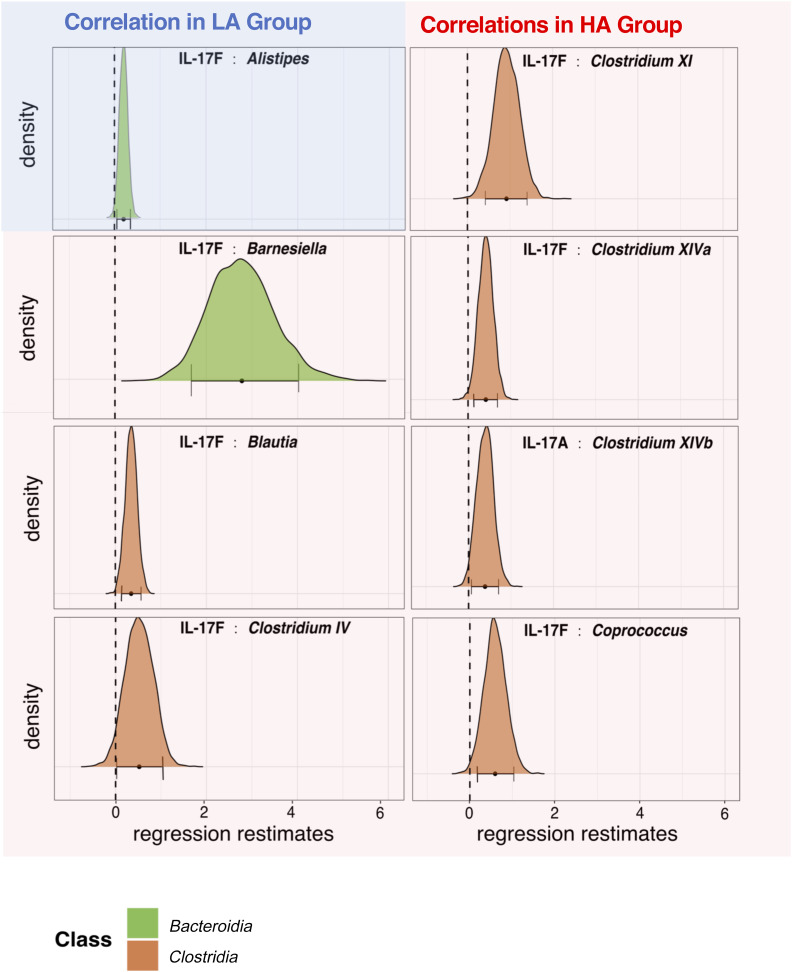
We first observed the abundance of Alistipes was positively correlated with changes in serum IL-17F levels (Fig. 4). As models were designed to compare cytokine versus microbe interactions in the context of cytokine activity (encoded as HA vs. LA), this result indicates Alistipes was significantly associated with IL-17F in LA subjects. In contrast, seven bacterial taxa were significant for the activity group–cytokine interaction term (Fig. 4), indicating their relative abundance was significantly associated with IL-17F or IL-17A levels in HA subjects. Notably, six of these seven significant relationships involved taxa belonging to the class Clostridia, and in all eight associations, the cytokine abundance was positively correlated with taxon relative abundance. In conclusion, analyses of the taxonomic abundance of the gut microbiome both between individuals (Fig. 3) and within individuals (Fig. 4) provide evidence that members of the class Clostridia are positively associated with increased levels of IL-17 activity.
Figure 4.
Bacterial genera whose abundance correlates with serum IL-17. Significant correlations between serum IL-17 and bacterial genus abundance are shown for HA subjects (red panels) and LA subjects (blue panel). Distributions show estimated effect sizes from Bayesian Markov chain Monte Carlo draws after parameter convergence. Panels show bacteria for which the estimated effect is significantly greater or less than zero (95% credible interval does not include zero). This analysis was based on 264 and 100 repeated measurements for HA and LA subjects, respectively.
Discussion
In this ancillary analysis of the iHMP, we present evidence that individuals at risk for developing T2D display distinct, longitudinal IL-17/IL-22 cytokine profiles, which can be associated with altered severity in a number of established markers for metabolic disorders. By subsequently providing evidence for a link between IL-17/IL-22 and the composition of the gut microbiome, we validate previous findings in mouse models and thus provide further support for the hypothesis that microbe–immune system interactions are relevant to human metabolic homeostasis.
Sustained loss of IL-17/IL-22 activity in iHMP study participants was associated with increased insulin resistance, as well as variation in metabolic markers that included lower HDL and increased triglycerides. A trend for higher HbA1c in LA subjects was not statistically significant. This is consistent with the previous observation that SSPG and HbA1c measures provide different perspectives on insulin resistance and glucose metabolism (36). It may also reflect the fact that few individuals were classified as having diabetes at any point during the course of this 4-year study. In spite of such inconsistencies between measures, our observations are in line with evidence that RORγt−/− and IL-22−/− mouse models show reduced insulin sensitivity on a chow diet. Furthermore, low-dose administration of IL-17 (18) or IL-22 (15) suppresses the metabolic phenotype induced by a high-fat diet. Taken together, these murine studies suggest that circulating levels of IL-17/IL-22 can be protective. Our work suggests that similar mechanisms may also apply to humans.
One explanation for the protective effects of IL-17/IL-22 is that these cytokines are directly, or indirectly, involved in regulating the composition of the gut microbiome, e.g., by regulating the production of antimicrobial peptides that limit the abundance of potentially pathogenic taxa (45–47). Another possibility is that IL-17/IL-22 may influence tight junction function, meaning their deficiency could result in a leaky gut (47–51). This could in turn contribute to translocation of gut bacteria to the blood, which has been associated with T2D (52). Alternatively, IL-22 may directly influence β-cell function (53), in which case changes in the gut microbiome may be correlative, rather than directly contributing to the phenotype reported here.
While IL-17/IL-22 may influence the gut microbiome, the composition of the gut microbiome may reciprocally affect metabolic diseases via the ability of certain taxa to directly, or indirectly, influence IL-17/IL-22 production. In a cross-sectional study of the influence of the human gut microbiome on cytokine production, Schirmer et al. (44) previously demonstrated that IL-17 production from peripheral blood mononuclear cells exposed to Staphylococcus aureus was positively correlated with the relative abundance of Clostridium in the host gut microbiome. This is consistent with our observations that 1) IL-17/IL-22 HA subjects had greater mean relative abundance of Clostridium in gut microbiome across the course of this study and that 2) in longitudinal analysis, three members of the class Clostridia (Clostridium IV, Clostridium XI, Clostridium XIVa) were positively correlated with IL-17F production and one (Clostridium XIVb) with IL-17A production. Human isolates from three of these Clostridium clusters (XIVa, XIVb, IV) were previously found to induce Th17 cells (a major producer of IL-17 and IL-22 [54,55] in mice [56]). Notably, germ-free mice do not carry Th17 cells in the SILP (57), but inoculation with certain bacteria, including segmented filamentous bacteria, can induce Th17 cell development (58,59). While segmented filamentous bacteria remain an ambiguous clade, they may be related to the family Clostridiales (60). In conclusion, while our study does not characterize sources of IL-17/IL-22 production, the trends we report are consistent with previous evidence that members of the class Clostridia can induce development of IL-17/IL-22–producing cells. This in turn represents one plausible way in which the gut microbiome could influence human cytokine profiles and thereby influence metabolic diseases.
Establishing the causative relationship of IL-17/IL-22, the gut microbiome, and metabolic diseases is beyond the scope of these data and will benefit from a deeper understanding of the mechanisms that underpin this interaction. The role of diet in shaping both the immune environment and composition of the gut microbiome is likely to be of particular interest, given that loss of IL-17–producing cells has been linked to a high-fat diet (14). No significant variation was observed between the diets of subjects with IL-17/IL-22 activity versus inactivity in this study, which may suggest that important differences in the composition of the microbiome are influenced by factors other than diet. However, it may also be that the available diet information was not sufficient to capture differences relevant to this study.
Regardless of whether differences in the microbiome are due to diet, or other environmental and host genetic factors, the molecular mechanisms underpinning interaction between members of the Clostridia and the host immune system warrant further investigation. One possibility is that gut microbiome–derived aryl hydrocarbon receptor signaling is critical for maintenance of Th17 cells (61,62). Bacterial-produced aryl hydrocarbon receptor ligand is reduced after mice are switched to a high-fat diet (63). Alternatively, Qin et al. (9) established a correlational association between the loss of members of the Clostridia in T2D patients and reduced short-chain fatty acid (SCFA) production. A later study confirmed this observation and demonstrated that supplementing SCFA-producing bacteria strains to T2D patients can improve their clinical outcomes (64), possibly via the ability of SCFA to influence Th17 production (65).
A close link between IL-17/IL-22 and members of the Clostridia supports previous assertions that manipulating the relationship between cytokines and the gut microbiome presents novel therapeutic opportunities. In humans, transferring the gut microbiome from lean donors to patients with metabolic syndrome has been shown to increase the insulin sensitivity of the recipients. Notably, among 16 bacteria strains that increased in gut microbiome of recipients postprocedure, 12 strains belonged to the class Clostridia (66). In mice, metabolic disorders accompanied by a lack of Th17 cells and IL-22 could be rescued, either by induction or adaptive transfer of Th17 cells (16,17), low-dose IL-17 (18), and IL-22 (15) or by supplementing the gut microbiome with symbiotics that increase Th17 cell abundance (14). Our demonstration of a close relationship between IL-17/IL-22 and members of the Clostridia therefore provides valuable insight into the biological processes that underpin the efficacy of these approaches.
In conclusion, our analysis of the newly released iHMP data set suggests novel avenues of research and raises the possibility of therapeutic targets related to IL-17/IL-22 that may effectively alleviate metabolic diseases in humans as they do in animal models.
Article Information
Acknowledgments. The authors thank Monika Avina and The Human Immune Monitoring Center for performing cytokine assays.
Funding. This work was supported by the National Institutes of Health Common Fund Human Microbiome Project (HMP) (1U54DE02378901).
Duality of Interest. M.S. is a cofounder of Personalis, Q bio, SensOmics, January, Filtricine, and Akna and an advisor for GenapSys. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. G.M.W. and M.S. designed the study. W.Z., Y.Z., E.S., G.M.W., and M.S. oversaw data collection, curation, and storage. X.Z., J.S.J., and D.S. analyzed data. X.Z., J.S.J., D.S., and G.M.W. wrote manuscript. J.S.J. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
X.Z. and J.S.J. equally contributed to the article.
This article contains supplementary material online at https://doi.org/10.2337/figshare.12213893.
References
- 1.Al Khodor S, Reichert B, Shatat IF. The microbiome and blood pressure: can microbes regulate our blood pressure? Front Pediatr 2017;5:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan Q, Gu Y, Li X, et al. . Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol 2017;7:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiao N, Baker SS, Nugent CA, et al. . Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: a meta-analysis. Physiol Genomics 2018;50:244–254 [DOI] [PubMed] [Google Scholar]
- 4.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome, and gastrointestinal disease. Clin Transl Gastroenterol 2015;6:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caricilli AM, Saad MJ. The role of gut microbiota on insulin resistance. Nutrients 2013;5:829–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forslund K, Hildebrand F, Nielsen T, et al.; MetaHIT consortium . Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlsson FH, Tremaroli V, Nookaew I, et al. . Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103 [DOI] [PubMed] [Google Scholar]
- 8.Larsen N, Vogensen FK, van den Berg FW, et al. . Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin J, Li Y, Cai Z, et al. . A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60 [DOI] [PubMed] [Google Scholar]
- 10.Wu H, Esteve E, Tremaroli V, et al. . Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017;23:850–858 [DOI] [PubMed] [Google Scholar]
- 11.Grossmann V, Schmitt VH, Zeller T, et al. . Profile of the immune and inflammatory response in individuals with prediabetes and type 2 diabetes. Diabetes Care 2015;38:1356–1364 [DOI] [PubMed] [Google Scholar]
- 12.Shu CJ, Benoist C, Mathis D. The immune system’s involvement in obesity-driven type 2 diabetes. Semin Immunol 2012;24:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. . Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010;328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garidou L, Pomié C, Klopp P, et al. . The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab 2015;22:100–112 [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Ota N, Manzanillo P, et al. . Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature 2014;514:237–241 [DOI] [PubMed] [Google Scholar]
- 16.Martins LMS, Perez MM, Pereira CA, et al. . Interleukin-23 promotes intestinal T helper type17 immunity and ameliorates obesity-associated metabolic syndrome in a murine high-fat diet model. Immunology 2018;154:624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong CP, Park A, Yang BG, et al. . Gut-specific delivery of T-helper 17 cells reduces obesity and insulin resistance in mice. Gastroenterology 2017;152:1998–2010 [DOI] [PubMed] [Google Scholar]
- 18.Mohamed R, Jayakumar C, Chen F, et al. . Low-dose IL-17 therapy prevents and reverses diabetic nephropathy, metabolic syndrome, and associated organ fibrosis. J Am Soc Nephrol 2016;27:745–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol 2010;10:479–489 [DOI] [PubMed] [Google Scholar]
- 20.Valeri M, Raffatellu M. Cytokines IL-17 and IL-22 in the host response to infection. Pathog Dis 2016;74:74:ftw111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Fang Y, Zhu H, Ge W. Plasma interleukin-22 levels are associated with prediabetes and type 2 diabetes in the Han Chinese population. J Diabetes Investig 2018;9:33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdel-Moneim A, Bakery HH, Allam G. The potential pathogenic role of IL-17/Th17 cells in both type 1 and type 2 diabetes mellitus. Biomed Pharmacother 2018;101:287–292 [DOI] [PubMed] [Google Scholar]
- 23.Galvan DL, Danesh FR. Paradoxical role of IL-17 in progression of diabetic nephropathy. J Am Soc Nephrol 2016;27:657–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yousefidaredor H, Zare-Bidaki M, Hakimi H, Assar S, Bagheri V, Arababadi MK. IL-17A plays an important role in induction of type 2 diabetes and its complications. Asian Pac J Trop Dis 2014;4:412–415 [Google Scholar]
- 25.Integrative HMP (iHMP) Research Network Consortium The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 2014;16:276–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W, Sailani MR, Contrepois K, et al. . Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 2019;569:663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahadi S, Zhou W, Schüssler-Fiorenza Rose SM, et al. . Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat Med 2020;26:83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segata N, Izard J, Waldron L, et al. . Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scrucca L, Fop M, Murphy TB, Raftery AE. Mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J 2016;8:289–317 [PMC free article] [PubMed] [Google Scholar]
- 30.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. 23 June 2014 [preprint]. arXiv:14065823.
- 31.Pilling LC, Atkins JL, Kuchel GA, Ferrucci L, Melzer D. Red cell distribution width and common disease onsets in 240,477 healthy volunteers followed for up to 9 years. PLoS One 2018;13:e0203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bürkner P-C. Advanced Bayesian multilevel modeling with the R package brms. 31 May 2017 [preprint]. arXiv:170511123.
- 34.Bürkner P-C. brms: an R package for Bayesian multilevel models using Stan. J Stat Softw 2017;80:1–28 [Google Scholar]
- 35.Hoffman MD, Gelman A. The No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. J Mach Learn Res 2014;15:1593–1623 [Google Scholar]
- 36.Schüssler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ, et al. . A longitudinal big data approach for precision health. Nat Med 2019;25:792–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pei D, Jones CNO, Bhargava R, Chen YDI, Reaven GM. Evaluation of octreotide to assess insulin-mediated glucose disposal by the insulin suppression test. Diabetologia 1994;37:843–845 [DOI] [PubMed] [Google Scholar]
- 38.Sell H, Habich C, Eckel J. Adaptive immunity in obesity and insulin resistance. Nat Rev Endocrinol 2012;8:709–716 [DOI] [PubMed] [Google Scholar]
- 39.Pilling LC, Atkins JL, Duff MO, et al. . Red blood cell distribution width: genetic evidence for aging pathways in 116,666 volunteers. PLoS One 2017;12:e0185083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brands MW, Manhiani MM. Sodium-retaining effect of insulin in diabetes. Am J Physiol Regul Integr Comp Physiol 2012;303:R1101–R1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmody RN, Gerber GK, Luevano JM Jr, et al. . Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015;17:72–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David LA, Maurice CF, Carmody RN, et al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Oosting M, Deelen P, et al. . Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat Med 2016;22:952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schirmer M, Smeekens SP, Vlamakis H, et al. . Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016;167:1125–1136.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abusleme L, Moutsopoulos NM. IL-17: overview and role in oral immunity and microbiome. Oral Dis 2017;23:854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon BR, Radin JN, Piazuelo MB, Contreras DC, Algood HM. IL-17a and IL-22 induce expression of antimicrobials in gastrointestinal epithelial cells and may contribute to epithelial cell defense against Helicobacter pylori. PLoS One 2016;11:e0148514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenewicz LA, Yin X, Wang G, et al. . IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 2013;190:5306–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol 2015;33:747–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JS, Tato CM, Joyce-Shaikh B, et al. . Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 2015;43:727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maxwell JR, Zhang Y, Brown WA, et al. . Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity 2015;43:739–750 [DOI] [PubMed] [Google Scholar]
- 51.O’Connor W Jr, Kamanaka M, Booth CJ, et al. . A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol 2009;10:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato J, Kanazawa A, Ikeda F, et al. . Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care 2014;37:2343–2350 [DOI] [PubMed] [Google Scholar]
- 53.Hasnain SZ, Borg DJ, Harcourt BE, et al. . Glycemic control in diabetes is restored by therapeutic manipulation of cytokines that regulate beta cell stress. Nat Med 2014;20:1417–1426 [DOI] [PubMed] [Google Scholar]
- 54.Liang SC, Tan XY, Luxenberg DP, et al. . Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 2006;203:2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov 2012;11:763–776 [DOI] [PubMed] [Google Scholar]
- 56.Atarashi K, Tanoue T, Ando M, et al. . Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015;163:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanov II, Frutos RdeL, Manel N, et al. . Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008;4:337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivanov II, Atarashi K, Manel N, et al. . Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan TG, Sefik E, Geva-Zatorsky N, et al. . Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA 2016;113:E8141–E8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin Y, Wang Y, Zhu L, et al. . Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J 2013;7:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chewning JH, Weaver CT. Development and survival of Th17 cells within the intestines: the influence of microbiome- and diet-derived signals. J Immunol 2014;193:4769–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue J, Nguyen DT, Habtezion A. Aryl hydrocarbon receptor regulates pancreatic IL-22 production and protects mice from acute pancreatitis. Gastroenterology 2012;143:1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Natividad JM, Agus A, Planchais J, et al. . Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab 2018;28:737–749.e4 [DOI] [PubMed] [Google Scholar]
- 64.Zhao L, Zhang F, Ding X, et al. . Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018;359:1151–1156 [DOI] [PubMed] [Google Scholar]
- 65.Asarat M, Apostolopoulos V, Vasiljevic T, Donkor O. Short-chain fatty acids regulate cytokines and Th17/Treg cells in human peripheral blood mononuclear cells in vitro. Immunol Invest 2016;45:205–222 [DOI] [PubMed] [Google Scholar]
- 66.Vrieze A, Van Nood E, Holleman F, et al. . Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–916.e7 [DOI] [PubMed] [Google Scholar]