Abstract
Diabetes mellitus (DM) is one of the deadliest diseases in the world, especially in developed nations. In recent years, it has become rampant in the developing nations such as Nigeria, posing more threats to individuals in the latter than those in the former. More than 415 million people were reported to suffer from DM worldwide as of 2015, with type 2 of the disease accounting for approximately 90% of the cases. The number of people with DM is expected to rise to 592 million by the year 2035. Therefore, DM is one of the growing public health concerns in Nigeria. In this study, the diagnostic dataset of DM type 2 was collected from the Murtala Mohammed Specialist Hospital, Kano, and used to develop predictive supervised machine learning models based on logistic regression, support vector machine, K-nearest neighbor, random forest, naive Bayes and gradient booting algorithms. The random forest predictive learning-based model appeared to be one of the best developed models with 88.76% in terms of accuracy; however, in terms of receiver operating characteristic curve, random forest and gradient booting predictive learning-based models were found to be the best predictive learning models with 86.28% predictive ability, respectively.
Keywords: Machine learning, Predictive model, Diabetes mellitus, Diabetes mellitus type 2, Random forest
Introduction
Machine learning (ML) is one of the sub-branches of artificial intelligence (AI) that deals with the ways in which machines learn from experience [1–3]. However, some of the computer scientists are of the opinion that the terms AI and ML are identical because of the possibility of learning from experience which is the main feature of the entity called intelligent system [4–6]. A detailed definition of the term ML was given by [7] as “a computer system is said to have learned from experience E with respect to some class of tasks T and performance measure P, if its performance at task in T, as measured by P, improves with experience E”. For many years, ML has solved many sophisticated and complex real world problems in the application areas such as marketing, business and retails applications, natural language processing, health care, autonomous vehicle system, intelligent robots, climate change, image processing, voice, gaming, among others. ML techniques had been used for the prediction and diagnosis of many diseases such as COVID-19 pandemic, malaria, typhoid, coronary artery diseases, diabetes mellitus, among others [8, 9]. ML algorithms are typically based on the trial and error approach which is quite opposite to conventional algorithms that follow the programming instruction based on like if-else decision statements [10].
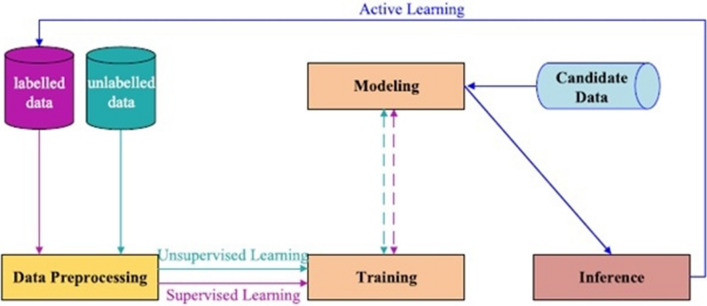
ML tasks are classified into four broad categories, namely supervised learning, unsupervised learning, active learning and reinforcement learning [11–13]. Supervised learning infers a function from the labeled training data, unsupervised learning infers a function from unlabeled training data, and active learning infers a function by choosing the most informative sample for labeling to train the model [12], while reinforcement learning interacts with a dynamic environment [10, 12]. The flowchart of the training process of ML tasks including supervised learning, unsupervised learning and active learning is shown in Fig. 1. As shown in Fig. 1, when training data are labeled, the training process is called supervised while otherwise it is called unsupervised training process. In contrast, when both labeled and unlabeled data are used for the training processing, this training process is called semi-supervised [10]. However, the task of supervised ML is the one most commonly used in real world applications, especially for the diagnosis and prediction of diseases. In this study, the supervised ML models were developed using supervised ML algorithms for the prediction of diabetes mellitus.
Fig. 1.
Flowchart for training process machine learning tasks [10]
Diabetes mellitus (DM), commonly referred to as diabetes, is a group of metabolic disorders of carbohydrate metabolism in which glucose is underutilized, producing hyperglycemia (increased glucose concentration in the blood) [14, 15]. The major symptoms of DM include frequent urination, increased thirst, and hunger. If left untreated, diabetes can cause many complications, such as diabetic ketoacidosis and the non-ketotic hyperosmolar coma [16–18]. The long-term complications caused by DM include cardiovascular diseases, strokes, chronic kidney failures, foot ulcers, and damages to the eyes and nerves. Diabetes occurs due to either the pancreas not producing enough insulin or the cells of the body not responding properly to the insulin produced [16]. DM is one of the deadliest diseases in the world, especially in developed nations. It is, however, becoming more rampant in the developing nations such as Nigeria, while posing more threats to individuals in the latter than those in the former [17, 18]. Over 415 million people were suffering from diabetes mellitus worldwide as of 2015, 8.3% being part of the adult population, with equal rates in both women and men. DM type 2 constitutes approximately 90% of the cases [18]. DM is estimated to have resulted in 1.5–5.0 million deaths each year in the period of 2012–2015 [14, 15, 19] and it doubles a person's risk of dying. The number of people with diabetes is expected to rise to 592 million by 2035 [15]. DM is one of the growing public health concerns in Nigeria. Four years ago, South Africa and Ethiopia had more cases of diabetes than Nigeria, but now Nigeria has the highest incidence of diabetes in the sub-Saharan Africa [18]. In this study, the diagnostic dataset of DM type 2 was collected from the Murtala Mohammed Hospital, Kano–Nigeria and used to develop the predictive supervised ML model based on logistic regression, support vector machine, K-nearest neighbor, random forest, naive Bayes, and gradient booting algorithms.
Related Work
Many works have been carried out using supervised ML to build predictive models in the health-care sector to complement and supplement the works of health workers in the course of diagnosing many diseases.
In the work of [10], COVID-19 prediction models that use supervised ML was developed. The model was developed based on liner regression (LR), support vector machine (SVM), least absolute shrinkage and selection (LASSO), and exponential smoothing (ES) algorithms. The study demonstrated the capability of the supervised ML algorithms to predict the number of upcoming COVID-19 patients that were affected. A supervised ML approach, which incorporated the generic algorithm and weighted K-nearest neighbor (WKNN) algorithms to predict and classify DM type 2 according to the presence or absence of coronary artery disease complications, was developed in the work of [20]. The supervised ML predictive model for acute ischemic stroke post intra-arterial therapy was developed in the work of [21]. The model showed a promising accuracy of the prediction and the study further proposed a robust learning model that can potentially optimize the selection process for medical treatment and endovascular activity in the management of acute strokes. In the work of [22], the predictive supervised ML model for the prediction of post-induction hypotension was developed. The result of the study showed that the success recorded in prediction post-induction hypotension demonstrates the ability of supervised ML models for predictive analytics in the field of anesthesiology. The predictive model for hospitalization due to heart disease was developed using supervised ML algorithms in the work of [23]. The dataset used for the development of the model was collected from an urban hospital in Boston and five models were developed using SVM, AdaBoost, LR, Naïve Bayes, and likelihood ratio test algorithms. A supervised ML model for rapid detection of heat rate fragmentation and cardiac arrhythmias was developed in the study of [24]. A random forest algorithm and a dataset of 300 instances of arrhythmic, non-arrhythmic coronary artery disease, and individuals without any medically significant cardiac conditions were used to develop a predictive model. The model was evaluated with 104 independent cases and proved to be very efficient. In the work of [25], supervised ML was used to generate a molecular signature that can classify metastatic hepatocellular carcinoma patients and identified genes that were relevant to metastatic and survival of the patients. In the work of [26], a review of supervised machine learning for population genetics was carried out and the review study found that there is a promising direction in the area. The study further found that supervised machine learning is an important and underutilized technique that has considerable potential for the evolutionary genomics. A supervised ML model for the identification of mosquitoes from the backscattered optical signal was developed in the study of [24]. The study showed that the optical sensor coupled with supervised ML can be a viable alternative means for monitoring the mosquito population. The predictive supervised ML approach for the estimation of the risk recurrence in early stages of oral tongue squamous cell carcinoma has been developed in the work of [27]. The result of the study showed the ability of supervised ML to predict locoregional recurrences. Supervised ML algorithms which include support vector machines, linear discriminant analysis, and K-nearest neighbor algorithms were used to identify dementia in the work of [28]. The result of the study showed that the algorithms are capable of predicting dementia.
Therefore, various related works so far reviewed in this section showed the potential ability of supervised ML algorithms to develop a model for the prediction of DM type 2.
Materials and Methods
Dataset
The diagnostic dataset for the DM type 2 patients was collected from the Murtala Mohammed Specialist Hospital, Kano State, in Nigeria. The dataset has nine attributes, including age, family history, glucose, cholesterol (CHOL), blood pressure (BP), HDL (high density lipoprotein), triglyceride, BMI (body mass index), and the diagnosis result. The dataset has 383 instances. Table 1 shows the description of units and ranges of risk attributes of the dataset.
Table 1.
Description of units and ranges of the dataset attributes
| SN | Attribute | Unit | Range |
|---|---|---|---|
| 1 | Age | Year | 1–150 |
| 2 | Family history | Yes (1), No (0) | 0, 1 |
| 3 | Glucose | mg/dL | 37–295 |
| 4 | Cholesterol (CHOL) | mg/dL | 128–575 |
| 5 | Blood pressure (BP) | mmHg | 90–190 |
| 6 | HDL | mg/dL | 10.6–73 |
| 7 | Triglyceride | mg/dL | 40–690 |
| 8 | BMI | kg/m2 | 20.28–40.25 |
| 9 | Diagnosis result | Positive (1), Negative (0) | 0, 1 |
Supervised Machine Learning Algorithms
Support Vector Machine
The support vector machine (SVM) is an elegant, powerful, and one of the most widely used supervised ML algorithms. SVM is used for both the regression and classification machine learning task problems due to its capability to non-linearly predict separable patterns by projecting the original feature into a hyperplane (higher-dimensional space) [27]. SVM is a non-parametric algorithm that recalls the training dataset for storing them all [28]. The SVM algorithm solves regression problems using linear functions, while in the case of non-linear regression problems, it maps the set of the input vector (a) to an n-dimensional space called a feature space (b) [13]. However, for multivariate training data (an) it is expressed in an N number of observations (bn), as a set of observed responses. The linear function can be shown as:
| 1 |
The objective is to make it as flat as possible: with as minimal norm values, and as such the problem fits in the minimization function;
| 2 |
Therefore with a special condition of the values of all the residual not more than ε, as in the equation below:
| 3 |
K-Nearest Neighbor
K-nearest neighbor is one of the simplest supervised ML algorithms that relies on the hypothesis “things that look alike” [28, 29]. The algorithm is a non-parametric and supervised classifier used for the regression and classification tasks [15]. In both tasks, the input features consists of K closest training examples or the dataset in the feature space, while the algorithm relies on labeled data for the learning process to produce appropriate outputs for unlabeled input features. The idea behind the KNN algorithm is that if a sample has k most similar neighbors in the feature space, m13ost of the samples belong to a certain category, then the sample also belongs to this category [4, 30, 31]. The voting method is generally used in the classification task, that is, the category label that appears frequently in the k sample is selected as the prediction result, while in the case of the regression task, the average method is used where the real value output labels of the k sample are used as the prediction result [5, 31].
Random Forest
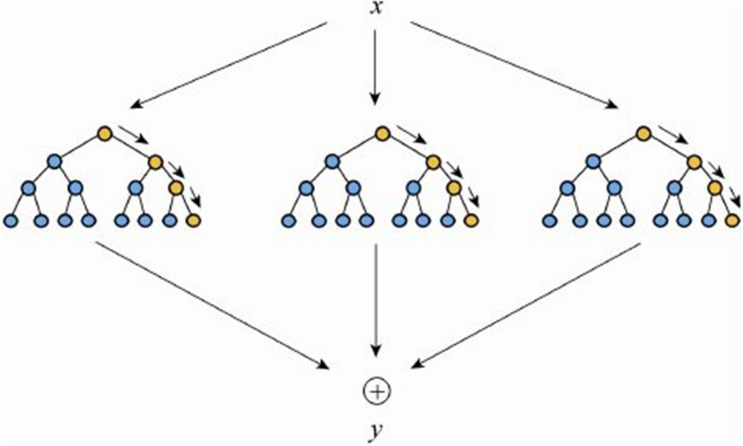
The random decision tree algorithm was proposed in 1995 by bell LABS Ho, which was advocated by aggregating many classifiers to improve the prediction accuracy. The idea behind the algorithm is to combine multiple decision tree classifiers, such as bagging and random space, to make predictions and get the final result by decision-making votes [4, 30]. Figure 2 shows the principle of filling the random forest.
Fig. 2.
Principle of filling random forest. The bootstrap resampling technique is firstly used where multiple samples are randomly selected from the original training dataset x to generate a new training dataset [32]. Then, multiple decision trees are built to form the random forest which then finally averages the output of each decision tree to determine the final filling result y [33].
Naive Bayes
Naïve Bayes is also a supervised ML algorithm based on the Bayes theorem. It learns by estimating the prior probability of each class using a training dataset [2]. The Bayes theorem is in the Eq. (4) below:
| 4 |
Gradient Boosting Algorithm
The gradient boosting algorithm combines a set of weaker learners to construct one strong learner. Unlike the bagging learning algorithm, where the models are made independently, gradient boosting makes its models sequentially by iteration to minimize the error of models learned earlier [34]. The gradient boosting algorithm learns a predictive model by combining M additive tree models (T0, T1, …, Tn) to predict the results as shown in the equation below:
| 5 |
The ensemble model can be optimized by reducing the expected generalization error L as shown in the equation below:
| 6 |
Logic Regression
The logic regression ML algorithm is an adaptive regression technique which constructs predicators as a Boolean combination of binary covariates [35]. The algorithm is used for a classification task with the aim of finding out a single Boolean expression that predicts a binary outcome. In the case of a regression task, many Boolean expressions can be investigated and simultaneously embedded into a liner regression model [36].
Evaluation Metrics
In this study, we used two evaluation techniques to determine the performance of each developed predictive learning model based on various supervised ML algorithms. These techniques include the following:
Accuracy is used to evaluate the supervised ML predictive models. The accuracy has the following definition:
| 7 |
For binary predictive models, accuracy can be calculated in terms of positive and negative as follows:
| 8 |
where tp is the true positive, tn is the true negative, and fp is the false positive, while fn is the false negative.
The receiver operating characteristic curve (ROC) is used to determine the diagnostic or predictive ability of the ML model as its discrimination threshold is varied. The curve is created by plotting the rate of true positive against the rate of false positive at various threshold settings. Figure 3 shows a typical ROC curve.
Fig. 3.

Typical ROC curve
Predictive Supervised Machine Learning Models
In this study, the predictive supervised ML model for diabetes type 2 was developed using a diagnostic dataset for DM type 2 patients. The dataset was collected at the Murtala Mohammed General Hospital Kano State in Nigeria and was used to develop predictive learning models. Table 2 shows the sample of the dataset. Figure 4 shows the work flow and how the models were developed.
Table 2.
Sample of the dataset
| Age (years) | Family history | Glucose (mg/dL) | CHOL (mg/dL) | BP (mmHg) | HDL (mg/dL) | Triglyceride (mg/dL) | BMI (kg/m2) | Diagnosis result |
|---|---|---|---|---|---|---|---|---|
| 62 | 1 | 281 | 135 | 312 | 56 | 234 | 56 | 1 |
| 42 | 1 | 201 | 171 | 391 | 71 | 98 | 43 | 1 |
| 39 | 0 | 281 | 140 | 309 | 45 | 62 | 45 | 0 |
| 62 | 1 | 136 | 140 | 129 | 32 | 201 | 32 | 0 |
| 60 | 1 | 149 | 120 | 134 | 60 | 119 | 44 | 1 |
| 57 | 1 | 120 | 135 | 178 | 11 | 300 | 14 | 0 |
| 59 | 0 | 130 | 180 | 341 | 67 | 65 | 15 | 1 |
| 63 | 1 | 199 | 130 | 198 | 15 | 123 | 15 | 1 |
| 74 | 0 | 178 | 118 | 169 | 32 | 56 | 32 | 1 |
| 61 | 1 | 201 | 176 | 190 | 21 | 319 | 32 | 1 |
| 34 | 0 | 123 | 130 | 231 | 17 | 21 | 17 | 0 |
Fig. 4.
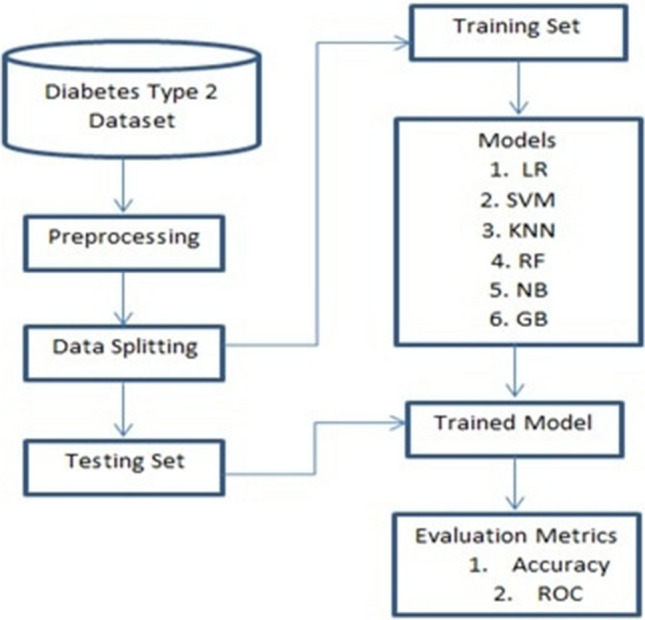
Workflow of the predictive models
The dataset collected was preprocessed and prepared into a comma-separated values file (CSV) format. CSV uses a comma to separate values, where each line of the file is a data record called data instance. Each data instance consists of one or more fields called columns, separated by a comma. A field separator using commas is the source of the name of the CSV file format. The dataset has 363 instances (records) without a missing value. The dataset has two demographic attributes, which include age and family history, as well as clinical attributes including BP, glucose, CHOL, triglycerides, HDL, BMI, and the diagnostic result of the doctor which can be either positive or negative. Table 3 shows the data type of the dataset attribute.
Table 3.
Data type of the dataset attributes
| SN | Attribute | Data type |
|---|---|---|
| 1 | Age | int64 |
| 2 | Family history | int64 |
| 3 | Glucose | int64 |
| 4 | Cholesterol | int64 |
| 5 | Blood pressure | int64 |
| 6 | High density lipoprotein | int64 |
| 7 | Triglyceride | int64 |
| 8 | Body mass index | int64 |
| 9 | Diagnosis result | int64 |
The Python programming language is used for the development of the learning predictive models. The Python programming language is an open source language and generally one of the most powerful and well-known dynamic programming languages used for development of predictive learning models and other data analysis tasks. In this study, the Python programming language has been used to develop the predictive supervised ML models for DM type 2 with a diagnostic dataset for DM type 2 patients. Figure 5 shows the description of the values of each attribute of the dataset, which includes the number of non-null observations, mean, standard deviation, the minimum value, 25% values, 50% values, 75% values, and the maximum value for each variable of the dataset.
Fig. 5.
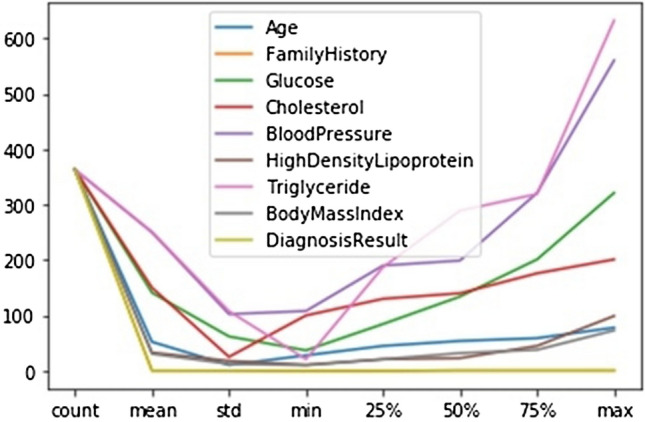
Description of the values of the dataset attributes
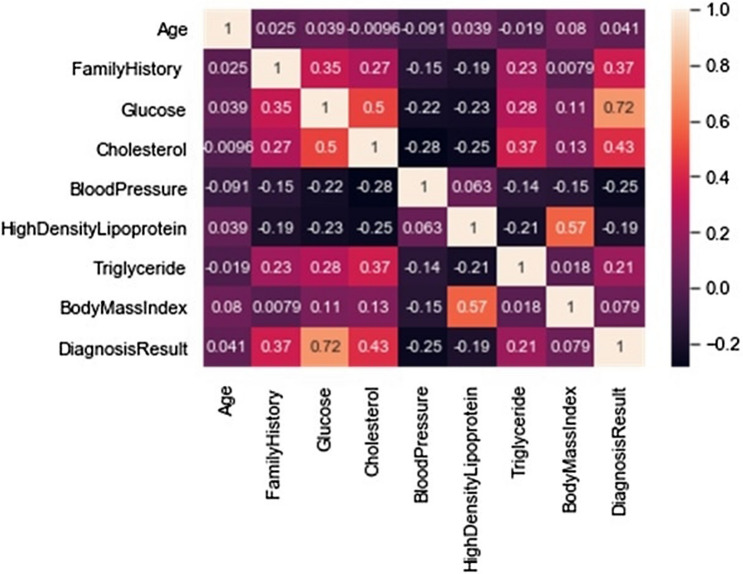
In this study, the correlation coefficient analysis of the dataset attributes was also carried out. The correlation coefficient r is used to measure or determine the strength and direction of the linear relationship between two features or variables of the dataset on a scatterplot [37]. Figure 6 shows the scatterplot correlation coefficient of the attributes of the dataset, while Fig. 7 shows the correlation matrix which depicts the correlation coefficients between sets of variables.
Fig. 6.
Scatterplot correlation coefficient of the dataset attributes
Fig. 7.
The correlation matrix of the dataset attributes
In the correlation coefficient analysis, the value r is always a finite number between − 1 to + 1. As for the regression analysis, the correlation coefficient is used for modeling the association between the dependent variable and the independent variable. Table 4 shows the r value and the correlation coefficient status between the dependent variables and independent variables of the dataset used in this study.
Table 4.
r value and correlation coefficient
| SN | Dependent variable | Independent variable | r value | Correlation coefficient relationship |
|---|---|---|---|---|
| 1 | Age | Diagnosis result | 0.041 | A weak positive correlation coefficient relationship |
| 2 | Family history | Diagnosis result | 0.37 | A moderate positive correlation coefficient relationship |
| 3 | Glucose | Diagnosis result | 0.72 | A strong positive correlation coefficient relationship |
| 4 | Cholesterol | Diagnosis result | 0.43 | A moderate positive correlation coefficient relationship |
| 5 | Blood pressure | Diagnosis result | − 0.25 | A weak negative correlation coefficient relationship |
| 6 | High density lipoprotein | Diagnosis result | − 0.19 | A weak negative correlation coefficient relationship |
| 7 | Triglyceride | Diagnosis result | 0.21 | A weak positive correlation coefficient relationship |
| 8 | Body mass index | Diagnosis result | 0.079 | A weak positive correlation coefficient relationship |
The predictive supervised ML models for DM type 2 based on logistic regression, support vector machine, K-nearest neighbor, random forest, naive Bayes, and gradient booting algorithms were developed. The algorithms were directly applied on the dataset with the help of Python programming and its built-in libraries to develop the models. The accuracy and ROC performance evaluation of the predictive supervised ML models for DM type 2 was carried out. Table 5 shows the result of the performance evaluation of the models.
Table 5.
Performance evaluation result of the model
| S/N | Supervised machine learning model | Accuracy (%) | ROC (%) |
|---|---|---|---|
| 1 | Logistic regression | 80.88 | 80.73 |
| 2 | Support vector machine | 85.29 | 84.74 |
| 3 | K-nearest neighbor | 82.35 | 81.94 |
| 4 | Random forest | 88.76 | 86.28 |
| 5 | Naive Bayes | 77.94 | 77.43 |
| 6 | Gradient booting | 86.76 | 86.28 |
Results and Discussion
In this study, predictive learning models for DM type 2 based on logistic regression, support vector machine, K-nearest neighbor, random forest, naive Bayes, and gradient booting algorithms were developed. The performance evaluation in terms of accuracy and receiver operating characteristic curve (ROC) of each developed model was determined. In terms of accuracy, the random forest predictive learning-based model happened to be the best model with an accuracy of 88.76%, followed by the gradient booting-based model with an accuracy of 86.76%, the support vector machine-based model with an accuracy of 85.29%, the K-nearest neighbor-based model with an accuracy of 82.35%, the logistic regression-based model with an accuracy of 80.88%, and lastly the naive Bayes-based model with an accuracy of 77.94%. However, in terms of the receiver operating characteristic curve, the random forest and gradient booting happened to be the best predictive models with an 86.28% predictive ability, respectively, followed by the support vector machine-based predictive model with 84.74%, the K-nearest neighbor-based predictive model with 81.94%, the logistic regression-based predictive model with 80.73%, and the naive Bayes-based predictive model with 77.73%. Figure 8 shows the results of the performance evaluation in terms of accuracy and ROC in each predictive model. Figure 9 shows the visualization of the random tree predictive model, which happened to be the best models in terms of accuracy and one of the best in terms of ROC.
Fig. 8.

Performance evaluation result of the predictive models
Fig. 9.
Visualization of the random tree predictive model
The predictive model in the figure shows that the glucose dataset attribute appeared to be the first splitting attribute and that the attribute is the most important for the diagnosis of DM type 2 in patients. This also corroborates the strong positive correlational coefficient relationship found between glucose attributes against the diagnosis result attribute of the dataset in the correlation coefficient analysis carried out earlier in the study. A set of rules that can be used for the diagnosis and prediction of DM type 2 can be extracted from the figure, which shows the visualization of the random tree predictive model. Below are the samples of the rules extracted from the predictive learning model:
-
(i)
If the glucose level of the patient is greater than 125 mg/dL and his/her blood pressure is between 129 and 225 mmHg, then the patient is diabetic.
-
(ii)
If the glucose level of the patient is less or equal to 125 mg/dL, his/her HDL is less than 53 mg/dL and his/her triglyceride is greater than 45 mg/dL, then the patient is diabetic.
-
(iii)
If the glucose level of the patient is less or equal to 125 mg/dL, his/her HDL is less than 53 mg/dL and his/her triglyceride is less than 45 mg/dL, and his blood pressure is less than 247 mmHg, then the patient is not diabetic.
Conclusion
In the present study, predictive learning models for DM type 2 based on logistic regression, support vector machine, K-nearest neighbor, random forest, naive Bayes, and gradient booting algorithms were developed. However, the random forest predictive learning-based model was found to be the best model among the developed models with 88.76% in terms of accuracy, while in terms of the receiver operating characteristic curve, random forest and gradient booting appeared to be the best predictive learning models with 86.28%, respectively. The model will help health workers and medical personnel when diagnosing and predicting DM type 2 among those patients suspected to have diabetes mellitus.
Funding
No funding sources.
Compliance with Ethical Standards
Conflict of Interest
The authors have declared that no conflict of interest exists.
Footnotes
This article is part of the topical collection “Advances in Computational Approaches for Artificial Intelligence, Image Processing, IoT and Cloud Applications” guest edited by Bhanu Prakash K N and M. Shivakumar”.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
L. J. Muhammad, Email: lawan.jibril@fukashere.edu.ng
Ebrahem A. Algehyne, Email: e.algehyne@ut.edu.sa
Sani Sharif Usman, Email: ssu992@fukashere.edu.ng.
References
- 1.Muhammad LJ, Usman SS. Power of artificial intelligence to diagnose and prevent further COVID-19 outbreak: a short communication. 2020. arXiv:2004.12463 [cs.CY]
- 2.Muhammad LJ, Islam MM, Usman SS, et al. Predictive data mining models for novel coronavirus (COVID-19) infected patients’ recovery. Springer Nat Comput Sci. 2020 doi: 10.1007/s42979-020-00216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh P. Supervised machine learning. In: Learn PySpark. Apress, Berkeley. 2019.
- 4.Muhammad LJ, et al. Performance evaluation of classification data mining algorithms on coronary artery disease dataset. In: IEEE 9th international conference on computer and knowledge engineering (ICCKE 2019), Ferdowsi University of Mashhad. 2019.
- 5.Muhammad LJ, et al. Performance evaluation of classification data mining algorithms on coronary artery disease dataset. In: IEEE 9th international conference on computer and knowledge engineering (ICCKE 2019), Ferdowsi University of Mashhad. IEEE. 2019.
- 6.Kavakiotis I, et al. Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J. 2017;15:104–116. doi: 10.1016/j.csbj.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell T. Machine learning. New York: McGraw Hill; 1997. [Google Scholar]
- 8.Haruna AA, Muhammad LJ, Yahaya BZ, et al. An improved C4.5 data mining driven algorithm for the diagnosis of coronary artery disease. In: International conference on digitization (ICD), Sharjah, United Arab Emirates, 2019. p. 48–52.
- 9.Muhammad LJ, Garba EJ, Oye ND, et al. On the problems of knowledge acquisition and representation of expert system for diagnosis of coronary artery disease (CAD) Int J u- and e-Serv Sci Technol. 2018;11(3):50–59. [Google Scholar]
- 10.Rustam F, et al. COVID-19 future forecasting using supervised machine learning models. IEEE Access. 2020 doi: 10.1109/ACCESS.2020.2997311. [DOI] [Google Scholar]
- 11.Muhammad LJ, et al. Using decision tree data mining algorithm to predict causes of road traffic accidents, its prone locations and time along Kano –Wudil highway. Int J Database Theory Appl. 2017;10(11):197–208. doi: 10.14257/ijdta.2017.10.1.18. [DOI] [Google Scholar]
- 12.Gong Z, Zhong P, Hu W. Diversity in machine learning. IEEE Access. 2019;7:64323–64350. doi: 10.1109/ACCESS.2019.2917620. [DOI] [Google Scholar]
- 13.Sadiq H, Muhammad LJ, Yakubu A. Mining social media and DBpedia data using Gephi and R. J Appl Comput Sci Math. 2018;12(1):14–20. doi: 10.4316/JACSM.201801002. [DOI] [Google Scholar]
- 14.Ishaq FS, Muhammad LJ, Yahaya BZ, et al. Fuzzy based expert system for diagnosis of diabetes mellitus. Int J Adv Sci Technol. 2020;136:39–50. [Google Scholar]
- 15.Ishaq FS, Muhammad LJ, Yahaya BZ, et al. Data mining driven models for diagnosis of diabetes mellitus: a survey. Indian J Sci Technol. 2018;11:42. doi: 10.17485/ijst/2018/v11i42/132665. [DOI] [Google Scholar]
- 16.Garcia MA. ESDIABETES (an expert system in diabetes) Eur J Sci Res. 2001;50(3):166–175. [Google Scholar]
- 17.American Diabetes Association Type 2 diabetes in children and adolescents. Pediatrics. 2000;105(36):71–680. doi: 10.1542/peds.105.3.671. [DOI] [PubMed] [Google Scholar]
- 18.Ajikobe D. Does Nigeria have the most people with diabetes in sub-Saharan Africa? Africa Check Sorting fact from fiction. https://africacheck.org/reports/nigeria-people-diabetes-sub-saharan-africa. Accessed 25 Apr 2020.
- 19.Ajmalahamed A, Nandhini KM, Anand SK. Designing a rule based fuzzy expert controller for early detection and diagnosis of diabetes. ARPN J Eng Appl Sci. 2014;9(5):21–32. [Google Scholar]
- 20.Giardina M, Azuaje F, McCullagh P, et al. Supervised learning approach to predicting coronary heart disease complications in type 2 diabetes mellitus patients. In: 6th IEEE symposium on bioinformatics and bioengineering (BIBE'06), Arlington, 2006. p. 325–33.
- 21.Asadi H, Dowling R, Yan B, Mitchell P, et al. Machine learning for outcome prediction of acute ischemic stroke post intra-arterial therapy. PLoS ONE. 2014;9:2. doi: 10.1371/journal.pone.0088225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samir K, Prathamesh K, Andrew DR, et al. Supervised machine-learning predictive analytics for prediction of postinduction hypotension. Anesthesiology. 2018;129:675–688. doi: 10.1097/ALN.0000000000002374. [DOI] [PubMed] [Google Scholar]
- 23.Dai W, Brisimia TS, Adams WG, Mela T, Saligrama V, Ioannis Ch. Paschalidisa. Int J Med Inform. 2015;84–3:189–197. doi: 10.1016/j.ijmedinf.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopalan A, Vollmer M. Rapid detection of heart rate fragmentation and cardiac arrhythmias: cycle-by-cycle rr analysis, supervised machine learning model and novel insights. In: Riaño D, Wilk S, ten Teije A, editors. Artificial intelligence in medicine. AIME 2019. Lecture notes in computer science. Springer, Cham. 2019. p. 11526.
- 25.Ye Q, Qin L, Forgues M, et al. Predicting hepatitis B virus–positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 26.Daniel R, Schrider A, Kern D. Supervised machine learning for population genetics: a new paradigm. Trend Genet. 2018;34–4:301–312. doi: 10.1016/j.tig.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasheed OA, Mohammed E, Iris S, et al. Comparison of supervised machine learning classification techniques in prediction of locoregional recurrences in early oral tongue cancer. Int J Med Inform. 2020;136:104068. doi: 10.1016/j.ijmedinf.2019.104068. [DOI] [PubMed] [Google Scholar]
- 28.Mathkunti NM, Rangaswamy S. Machine learning techniques to identify dementia. SN Comput Sci. 2020;1:118. doi: 10.1007/s42979-020-0099-4. [DOI] [Google Scholar]
- 29.Hussain S, et al. Performance evaluation of various data mining algorithms on road traffic accident dataset. In: Satapathy S, Joshi A, editors. Information and communication technology for intelligent systems. Smart Innovation, Systems and Technologies. 2019. p. 106.
- 30.Lan H, Pan Y. A crowdsourcing quality prediction model based on random forests. In: 2019 IEEE/ACIS 18th international conference on computer and information science (ICIS), Beijing, China. 2019. p. 315–319. 10.1109/ICIS46139.2019.8940306.
- 31.Zhang W, Chen X, Liu Y. A distributed storage and computation k-nearest neighbor algorithm based cloud-edge computing for cyber-physical-social systems. IEEE Access. 2020;8:50118–50130. doi: 10.1109/ACCESS.2020.2974764. [DOI] [Google Scholar]
- 32.Deng W, Guo Y, Liu J, et al. A missing power data filling method based on improved random forest algorithm. Chin J Electr Eng. 2019;5(4):33–39. doi: 10.23919/CJEE.2019.000025. [DOI] [Google Scholar]
- 33.Breiman L. Random forests. Mach Learn. 2001;45:1. [Google Scholar]
- 34.Xia Y. A novel reject inference model using outlier detection and gradient boosting technique in peer-to-peer lending. IEEE Access. 2019;7:92893–92907. doi: 10.1109/ACCESS.2019.2927602. [DOI] [Google Scholar]
- 35.Charles K, Ingo R, Michael LL, Li H. Sequence analysis using logic regression. Genet Epidemiol. 2001;21:S626–S631. doi: 10.1002/gepi.2001.21.s1.s626. [DOI] [PubMed] [Google Scholar]
- 36.Schwender H, Ruczinski I. Logic regression and its extensions. Adv Genet. 2010;72:25–45. doi: 10.1016/B978-0-12-380862-2.00002-3. [DOI] [PubMed] [Google Scholar]
- 37.Deborah JR. How to interpret a correlation coefficient r. Dummies. https://www.dummies.com/education/math/statistics/how-to-interpret-a-correlation-coefficient-r/. Accessed 12 June 2020.