Abstract
Study objective
The goal of this study is to create a predictive, interpretable model of early hospital respiratory failure among emergency department (ED) patients admitted with coronavirus disease 2019 (COVID-19).
Methods
This was an observational, retrospective, cohort study from a 9-ED health system of admitted adult patients with severe acute respiratory syndrome coronavirus 2 (COVID-19) and an oxygen requirement less than or equal to 6 L/min. We sought to predict respiratory failure within 24 hours of admission as defined by oxygen requirement of greater than 10 L/min by low-flow device, high-flow device, noninvasive or invasive ventilation, or death. Predictive models were compared with the Elixhauser Comorbidity Index, quick Sequential [Sepsis-related] Organ Failure Assessment, and the CURB-65 pneumonia severity score.
Results
During the study period, from March 1 to April 27, 2020, 1,792 patients were admitted with COVID-19, 620 (35%) of whom had respiratory failure in the ED. Of the remaining 1,172 admitted patients, 144 (12.3%) met the composite endpoint within the first 24 hours of hospitalization. On the independent test cohort, both a novel bedside scoring system, the quick COVID-19 Severity Index (area under receiver operating characteristic curve mean 0.81 [95% confidence interval {CI} 0.73 to 0.89]), and a machine-learning model, the COVID-19 Severity Index (mean 0.76 [95% CI 0.65 to 0.86]), outperformed the Elixhauser mortality index (mean 0.61 [95% CI 0.51 to 0.70]), CURB-65 (0.50 [95% CI 0.40 to 0.60]), and quick Sequential [Sepsis-related] Organ Failure Assessment (0.59 [95% CI 0.50 to 0.68]). A low quick COVID-19 Severity Index score was associated with a less than 5% risk of respiratory decompensation in the validation cohort.
Conclusion
A significant proportion of admitted COVID-19 patients progress to respiratory failure within 24 hours of admission. These events are accurately predicted with bedside respiratory examination findings within a simple scoring system.
Introduction
Background
Severe acute respiratory syndrome coronavirus 2 (coronavirus disease 2019 [COVID-19]) is a global pandemic with millions of cases and hundreds of thousands of deaths.1 , 2 Despite initial reports of patient characteristics and risk factors for critical illness, there is little evidence-based guidance available to aid provider decisionmaking in safely dispositioning patients with COVID-19.3 , 4 Inappropriate inpatient dispositions lead to increased provider contacts in the form of rapid response teams and the use of multiple care areas when hospital capacities are limited.5 , 6 More significantly, in other domains of emergency care, undertriage of patients is associated with worse morbidity and mortality than if patients are directly admitted to higher levels of care.7 , 8 Given the high morbidity and mortality associated with COVID-19 and uncertainty around the disease process and prognosis, there is great urgency in developing and validating effective clinical risk-stratification tools for COVID-19 patients.
Editor’s Capsule Summary.
What is already known on this topic
Patients with coronavirus disease 2019 (COVID-19) can experience respiratory deterioration after hospital admission.
What question this study addressed
Data were extracted retrospectively for 1,172 COVID-19 patients admitted to the hospital from 9 emergency departments to identify factors that may predict deterioration requiring oxygen greater than 10 L/min, noninvasive ventilation, or intubation; or that leads to death within 24 hours of hospital admission.
What this study adds to our knowledge
A model (quick COVID-19 Severity Index) with 3 variables (respiratory rate, pulse oximetry, and oxygen flow rate) outperformed other models, including the quick Sequential [Sepsis-related] Organ Failure Assessment and CURB-65, on an independent validation cohort.
How this might change clinical practice
The quick COVID-19 Severity Index model may be useful to assist level-of-care decisions for admitted COVID-19 patients. It is not known how well it performs relative to physician gestalt.
Importance
Expert-recommended admissions guidelines do not risk stratify among patients with severe COVID-19.9 International definitions of severe COVID-19 are evolving, but typically include respiratory rate less than or equal to 30 breaths/min, Spo 2 greater than or equal to 93%, Pao 2:Fio 2 less than or equal to 300 mm Hg, and infiltrates of greater than or equal to 50% of lungs.9 , 10 Critical COVID-19 exists on a spectrum with severe illness and involves organ failure, often leading to prolonged mechanical ventilation.9 In a large cohort of COVID-19 patients, severe and critical illness represented almost 20% of the studied population.10 In most institutions, dispositions for patients with critical respiratory failure (eg, those receiving ventilation or with nonrebreather masks) are largely apparent and determined by system protocols and capacity. Rapid progression from severe to critical illness, however, is a common problem and presents a prognostic challenge for ED providers determining admissions.
For this reason, we focus on patients for whom critical respiratory illness is not universally apparent in the ED; namely, those requiring nasal cannula with oxygen less than or equal to 6 L/min. In our health system, 6 L/min is typically the maximum flow rate delivered by nasal cannula. Greater than 90% of patients receiving oxygen at less than 6 L/min are admitted to the floors, but of those, greater than 10% were observed to have increased oxygen requirements within 24 hours. Conversely, among these patients admitted to higher levels of care, approximately 70% did not progress above nasal cannula oxygen at 6 L/min. These data suggest potential to improve our ability to risk stratify ED patients before admission.
Goals of This Investigation
The objective of this study was to derive a risk-stratification tool to predict 24-hour respiratory decompensation in admitted patients with COVID-19. Here, we expand on previous efforts describing the course of critical COVID-19 illness in 3 ways. First, we focused on ED prognostication by studying patient outcomes within 24 hours of admission, using data available during the first 4 hours of presentation.11 Although critical illness often occurs later in hospitalization, the relevance of these later events to ED providers is less clear. We emphasize oxygen requirements and mortality rather than ICU placement because we have observed the latter to have highly variable criteria, depending on total patient census.12 Second, to aid health care providers in assessing illness severity in COVID-19 patients, we presented predictive models of early respiratory failure during hospitalization and compare them with 3 benchmarks accessible with data in the electronic health record: the Elixhauser Comorbidity Index,13 the quick Sequential [Sepsis-related] Organ Failure Assessment (qSOFA),14 , 15 and the CURB-65 pneumonia severity score.16 Although many clinical risk models exist, these benefit from wide clinical acceptability and relative model parsimony because they require minimal input data for calculation. The Elixhauser Comorbidity Index was derived to enable prediction of hospital death with administrative data.13 The qSOFA score was included in SEPSIS-3 guidelines and can be scored at the bedside because it includes respiratory rate, mental status, and systolic blood pressure.14 The CURB-65 pneumonia severity score has been well validated for hospital disposition, but its utility in both critical illness and COVID-19 is unclear.16 , 17 Third, we made the quick COVID-19 Severity Index, a prognostic tool, available to the public through a Web interface.
Materials and Methods
Study Design and Setting
This was a retrospective observational cohort study to develop a prognostic model of early respiratory decompensation in patients admitted from the emergency department (ED) with COVID-19. The health care system is composed of a mix of suburban community (n=6), urban community (n=2), and urban academic (n=1) EDs. Data from 8 EDs were used in the derivation and cross validation of the predictive model, whereas data from the last urban community site was withheld for independent validation. We adhered to the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis checklist (Appendix E1, available online at http://www.annemergmed.com).18 This study was approved by our local institutional review board.
Data Collection and Processing
Patient demographics, summarized medical histories, vital signs, outpatient medications, chest radiograph reports, and laboratory results available during the ED encounter were extracted from our local Observational Medical Outcomes Partnership data repository and analyzed within our computational health platform.19 Data were collected into a research cohort with custom scripts in PySpark (version 2.4.5) that were reviewed by an independent analyst.
Nonphysiologic values likely related to data entry errors for vital signs were converted to missing values based on expert-guided rules (Appendix E1, available online at http://www.annemergmed.com [Table S1]). Laboratory values at minimum or maximum thresholds and encoded with “<“ or “>“ were converted to the numeric threshold value, and other nonnumeric values were dropped. Medical histories were generated by using diagnoses before the date of admission to exclude potential future information in modeling. Outpatient medications were mapped to the First DataBank Enhanced Therapeutic Classification System.20 Radiograph reports were manually reviewed by 2 physicians and categorized as “no opacity,” “unilateral opacity,” or “bilateral opacities.” One hundred radiograph reports were reviewed by both physicians to determine interrater agreement with weighted κ. Oxygen devices were similarly extracted from the Observational Medical Outcomes Partnership (Appendix E1, available online at http://www.annemergmed.com [Table S2]).
We defined critical respiratory illness in the setting of COVID-19 as any COVID-19 patient meeting one of the following criteria: oxygenation flow rate greater than or equal to 10 L/min, high-flow oxygenation, noninvasive ventilation, invasive ventilation, or death (Appendix E1, available online at http://www.annemergmed.com [Table S2]). We did not include ICU admission in our composite outcome because at the start of the COVID-19 pandemic, ICU admissions were protocolized to include even minimal oxygen requirements. A subset of outcomes was manually reviewed by physician members of the institutional computational health care team as part of a systemwide process to standardize outcomes for COVID-19–related research.
Data included visits from March 1, 2020, through April 27, 2020, because our institution’s first COVID-19 tests were ordered after March 1, 2020. This study included admitted COVID-19–positive patients as determined by test results ordered between 14 days before and up to 24 hours after hospital presentation. We included delayed testing because institutional guidelines initially restricted testing within the hospital to inpatient wards. Testing for COVID-19 was performed at local or reference laboratories by nucleic acid detection methods using oropharyngeal or nasopharyngeal swabs, or a combination oropharyngeal/nasopharyngeal swab. We excluded patients younger than 18 years and those who required oxygen at more than 6 L/min or otherwise met our critical illness criteria at any point within 4 hours of presentation. The latter was intended to exclude patients for whom critical illness was nearly immediately apparent to the medical provider and for whom a prediction would not be helpful. Patients who explicitly opted out of research were excluded from analysis (n<5). Data were extracted greater than 24 hours after the last included patient visit so that all outcomes could be extracted from the electronic health record.
We generated comparator models using the Elixhauser Comorbidity Index, qSOFA, and CURB-65 (Appendix E1, available online at http://www.annemergmed.com). International Statistical Classification of Diseases and Related Health Problems, 10th Revision codes from patient medical histories were mapped to Elixhauser comorbidities and indices with H-CUP Software and Tools (hcuppy package; version 0.0.7).21 , 22 qSOFA was calculated as the sum of the following findings, each of which was worth 1 point: Glasgow Coma Scale score less than 15, respiratory rate greater than or equal to 22 breaths/min, and systolic blood pressure less than or equal to 100 mm Hg. CURB-65 was calculated as the sum of the following findings, each of which was worth 1 point: Glasgow Coma Scale score less than 15, blood urea nitrogen level greater than 19 mg/dL, respiratory rate greater than or equal to 30 breaths/min, systolic blood pressure less than 90 mm Hg or diastolic blood pressure less than or equal to 60 mm Hg, and aged 65 years. Baseline models were evaluated on the training and internal validation cohort, using logistic regression on the calculated scores.
Samples from 8 hospitals were used in model generation and internal validation with the remaining large, urban community hospital serving as an independent site for validation. All models were fit on patient demographic and clinical data collected during the first 4 hours of patient presentation, and predictions are made with the most recently available data at the 4-hour point unless otherwise noted. We used an ensemble technique to identify and rank potentially important predictive variables based on their occurrence across multiple selection methods: univariate regression, random forest, logistic regression with LASSO, χ2 testing, gradient-boosting information gain, and gradient-boosting Shapley additive explanation (SHAP) interaction values (Appendix E1, available online at http://www.annemergmed.com).23, 24, 25 We counted the co-occurrences of the top 30, 40, and 50 variables of each of the methods before selecting features for a minimal scoring model (quick COVID-19 Severity Index) and machine-learning model (COVID-19 Severity Index) using gradient boosting. For the quick COVID-19 Severity Index, we used a point system guided by logistic regression (Appendix E1, available online at http://www.annemergmed.com). The gradient-boosting COVID-19 Severity Index model was fit with the XGBoost package and hyperparameters were set with Bayesian optimization with a tree-structured Parzen estimator (Appendix E1, available online at http://www.annemergmed.com).26 , 27 All analyses were performed in Python (version 3.8.2).
We report summary statistics of model performance in predicting the composite outcome between 4 and 24 hours of hospital arrival. We used bootstrapped logistic regression with 10-fold cross validation to generate performance benchmarks for the Elixhauser, qSOFA, CURB-65, and quick COVID-19 Severity Index models and bootstrapped gradient boosting with 10-fold cross validation for the COVID-19 Severity Index model. Where necessary, data were imputed with training set median values of bootstraps. We report area under the receiver operating characteristic (ROC), accuracy, sensitivity and specificity at Youden’s index, area under the precision-recall curve,28 Brier score, F1 score, and average precision (Appendix E1, available online at http://www.annemergmed.com). Similarly, to evaluate model performance on the independent validation cohort, means and confidence intervals were calculated from bootstrap iterations of the test set, using sampling with replacement. We report 95% confidence intervals derived from the percentiles of the bootstrapped distribution or Welch’s 2-sample t test for statistical comparisons of model performance.29
Results
Characteristics of Study Subjects
Between March 1, 2020, and April 27, 2020, there were a total of 1,792 admissions for COVID-19 patients meeting our age criteria. Of these, 620 patients (35%) were excluded by meeting critical respiratory illness endpoints within 4 hours of presentation. Of the included patients, 144 (12.3%) had respiratory decompensation within the first 24 hours of hospitalization: 101 (8.6%) requiring oxygen flow at greater than 10 L/min, 112 (9.6%) with high-flow device support (Appendix E1, available online at http://www.annemergmed.com [Table S2]), 4 (0.3%) receiving noninvasive ventilation, 10 (0.8%) with invasive ventilation, and 1 (0.01%) who died. Fifty-nine patients (5%) were admitted to the ICU within the 4- to 24-hour period. Population characteristics including demographics and comorbidities for the study are shown in Table 1 . Study patient flow is shown in Figure 1 and patient characteristics for the development and validation populations are shown in Appendix E1 (available online at http://www.annemergmed.com [Tables S3 to S4]).
Table 1.
Characteristics of COVID-19–positive admitted patients stratified by primary outcome.
| Variable | Category | 24-Hour Critical Respiratory Illness |
||
|---|---|---|---|---|
| Missing | Negative, n=1,028 | Positive, n=144 | ||
| Age, mean (SD), y | 0 | 67.6 (16.9) | 64.8 (16.7) | |
| Age, y | 18–44 | 105 (10.2) | 19 (13.2) | |
| 45–64 | 340 (33.1) | 60 (41.7) | ||
| >65 | 583 (56.7) | 65 (45.1) | ||
| Sex | Women | 506 (49.2) | 61 (42.4) | |
| Men | 522 (50.8) | 83 (57.6) | ||
| Race | Black | 260 (25.3) | 40 (27.8) | |
| White | 517 (50.3) | 63 (43.8) | ||
| Other | 251 (24.4) | 41 (28.5) | ||
| Ethnicity | Hispanic or Latino | 233 (22.7) | 44 (30.6) | |
| Non-Hispanic | 776 (75.5) | 97 (67.4) | ||
| Unknown | 19 (1.8) | 3 (2.1) | ||
| Smoking status | Smoker | 39 (3.8) | 8 (5.6) | |
| Former smoker | 340 (33.1) | 45 (31.2) | ||
| Never smoker | 503 (48.9) | 66 (45.8) | ||
| Unknown | 185 (18.0) | 33 (22.9) | ||
| Insurance type | Commercial | 118 (11.5) | 21 (14.6) | |
| Medicaid | 136 (13.2) | 23 (16.0) | ||
| Medicare | 590 (57.4) | 68 (47.2) | ||
| Other | 92 (8.9) | 19 (13.2) | ||
| Self-pay | 92 (8.9) | 13 (9.0) | ||
| Comorbidities | None | 322 (31.3) | 47 (32.6) | |
| Fluid and electrolyte disorders | 378 (36.8) | 47 (32.6) | ||
| Other neurologic disorders | 320 (31.1) | 36 (25.0) | ||
| Deficiency anemias | 315 (30.6) | 48 (33.3) | ||
| Hypertension | 311 (30.3) | 47 (32.6) | ||
| Chronic pulmonary disease | 282 (27.4) | 32 (22.2) | ||
| Hypertension with complications | 264 (25.7) | 36 (25.0) | ||
| Diabetes with chronic complications | 263 (25.6) | 37 (25.7) | ||
| Obesity | 261 (25.4) | 40 (27.8) | ||
| Depression | 260 (25.3) | 31 (21.5) | ||
| Valvular disease | 235 (22.9) | 21 (14.6) | ||
| Peripheral vascular disease | 220 (21.4) | 31 (21.5) | ||
| Renal disease | 205 (19.9) | 30 (20.8) | ||
| Congestive heart failure | 203 (19.7) | 20 (13.9) | ||
| Hypothyroidism | 186 (18.1) | 22 (15.3) | ||
| Weight loss | 158 (15.4) | 18 (12.5) | ||
| Psychoses | 126 (12.3) | 16 (11.1) | ||
| Coagulation deficiency | 98 (9.5) | 10 (6.9) | ||
| Liver disease | 97 (9.4) | 15 (10.4) | ||
| Solid tumor without metastasis | 96 (9.3) | 10 (6.9) | ||
| Diabetes without chronic complications | 93 (9.0) | 19 (13.2) | ||
| Rheumatoid arthritis/collagen vascular | 74 (7.2) | 11 (7.6) | ||
| Paralysis | 71 (6.9) | 9 (6.2) | ||
| Anemia from blood loss | 68 (6.6) | 7 (4.9) | ||
| Metastatic disease | 66 (6.4) | 9 (6.2) | ||
| Pulmonary circulation disorders | 64 (6.2) | 6 (4.2) | ||
| Alcohol abuse | 63 (6.1) | 7 (4.9) | ||
| Drug abuse | 51 (5.0) | 12 (8.3) | ||
| Model variables, mean (SD) | Oxygen flow rate, L/min∗ | 9 | 1.8 (1.4) | 3.5 (1.5) |
| Respiratory rate, breaths/min∗ | 17 | 20.3 (4.2) | 22.3 (5.2) | |
| Minimum oxygen saturation (% oxygen) | 17 | 92.9 (3.2) | 89.9 (5.0) | |
| Aspartate aminotransferase | 323 | 53.8 (51.7) | 85.6 (227.1) | |
| Chloride | 45 | 100.1 (5.5) | 98.8 (5.1) | |
| Procalcitonin | 593 | 0.5 (2.3) | 0.8 (2.4) | |
| Minimum systolic blood pressure | 17 | 117.0 (17.8) | 113.7 (20.5) | |
| WBC count | 40 | 7.0 (3.7) | 7.6 (4.3) | |
| Blood urea nitrogen | 35 | 22.8 (18.6) | 28.3 (22.6) | |
| Creatinine | 35 | 1.4 (1.5) | 1.7 (2.0) | |
| Glucose | 34 | 142.8 (73.1) | 156.1 (82.4) | |
| C-reactive protein | 777 | 92.2 (70.0) | 153.9 (88.4) | |
| Ferritin | 781 | 812.4 (889.6) | 1,540.1 (3,342.2) | |
Unknown demographics are included under “other” or “unknown” where relevant.
Indicates the most recent documented value at 4 hours.
Figure 1.
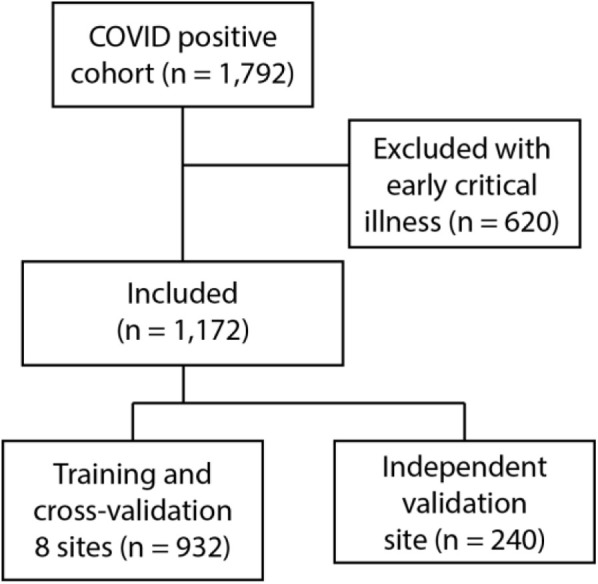
Model development strategy. Exclusions were for critical illness within 4 hours of ED presentation.
Our full data set included 713 patient variables available during the first 4 hours of the patient encounters (Appendix E1, available online at http://www.annemergmed.com [Table S5]). These included demographics, vital signs, laboratory values, comorbidities, chief complaints, outpatient medications, tobacco use histories, and radiographs. Radiologist-evaluated radiographs were classified into 3 categories, with strong interrater agreement (κ=0.81). Associations between radiographic findings and outcomes are shown in Appendix E1 (available online at http://www.annemergmed.com [Table S6]). We preferentially selected variables available at bedside for derivation of the quick COVID-19 Severity Index. Our ensemble approach identified 3 bedside variables as consistently important across the variable selection models: nasal cannula requirement, minimum recorded pulse oximetry, and respiratory rate (Appendix E1, available online at http://www.annemergmed.com [Figure S1]). These 3 features appeared in at least 5 of the 6 variable selection methods.
We divided each of these 3 clinical variables into value ranges according to clinical experience and used logistic regression to derive weights for the quick COVID-19 Severity Index scoring system (Table 2 ). Normal physiology was used as the baseline category, and the logistic regression odds ratios were offset to assign normal clinical parameters zero points in the quick COVID-19 Severity Index (Appendix E1, available online at http://www.annemergmed.com). The quick COVID-19 Severity Index score ranges from 0 to 12.
Table 3.
Performance characteristics for the COVID-19 Severity Index, quick COVID-19 Severity Index, and comparison models on independent validation.
| AU-ROC | Accuracy | Sensitivity | Specificity | AU-PRC | Brier Score | F1 Score | Average Precision | |
|---|---|---|---|---|---|---|---|---|
| CSI | 0.76 (0.65–0.86)∗ | 0.79 (0.72–0.86) | 0.73 (0.56–0.88) | 0.81 (0.72–0.89) | 0.38 (0.23–0.54) | 0.25 (0.25–0.25) | 0.47 (0.34–0.61) | 0.40 (0.25–0.56) |
| CURB-65 | 0.50 (0.40–0.60) | 0.64 (0.42–0.89) | 0.57 (0.03–0.97) | 0.52 (0.18–1.00) | 0.18 (0.09–0.30) | 0.12 (0.09–0.15) | 0.13 (0.00–0.27) | 0.16 (0.10–0.24) |
| Elixhauser | 0.61 (0.51–0.70) | 0.49 (0.26–0.74) | 0.82 (0.45–1.00) | 0.42 (0.15–0.78) | 0.19 (0.11–0.29) | 0.12 (0.09–0.15) | 0.28 (0.20–0.37) | 0.20 (0.13–0.30) |
| qCSI | 0.81 (0.73–0.89) | 0.82 (0.77–0.88) | 0.79 (0.63–0.93) | 0.79 (0.71–0.87) | 0.47 (0.30–0.64) | 0.10 (0.07–0.13) | 0.49 (0.36–0.62) | 0.44 (0.29–0.60) |
| qSOFA | 0.59 (0.50–0.68) | 0.83 (0.79–0.88) | 0.47 (0.06–0.66) | 0.72 (0.64–1.00) | 0.22 (0.11–0.35) | 0.12 (0.09–0.15) | 0.08 (0.00–0.23) | 0.20 (0.12–0.29) |
Table 2.
Quick COVID-19 Severity Index and COVID-19 Severity Index model variables.
| qCSI variable | Points | Additional CSI variables |
|---|---|---|
| Respiratory rate, breaths/min | Aspartate transaminase | |
| ≤22 | 0 | Alanine transaminase |
| 23–28 | 1 | Ferritin |
| >28 | 2 | Procalcitonin |
| Pulse oximetry, %∗ | Chloride | |
| >92 | 0 | C-reactive protein |
| 89–92 | 2 | Glucose |
| Oxygen flow rate, L/min | ||
| ≤88 | 5 | Urea nitrogen WBC count |
| ≤2 | 0 | Age |
| 3–4 | 4 | |
| 5–6 | 5 |
qCSI, Quick COVID-19 Severity Index; CSI, COVID-19 Severity Index.
Pulse oximetry represents the lowest value recorded during the first 4 hours of the patient encounter.
We identified an additional 12 features from the predictive factor analysis for use in a machine-learning model (COVID-19 Severity Index) with gradient boosting (Table 2 and Appendix E1, available online at http://www.annemergmed.com [Figure S1]). These variables were selected by balancing the goals of model parsimony, minimizing highly correlated features (ie, various summaries of vital signs), and predictive performance. We used SHAP methods to understand the importance of various clinical variables in the COVID-19 Severity Index (Figure 2 ).25 , 30, 31, 32 SHAP values are an extension of the game-theoretic Shapley values that seek to describe variable effects on model output, defined as the contribution of a specific variable to the prediction itself.30 The key advantage of the related SHAP values is that they add interpretability to complex models such as gradient boosting, which otherwise provide opaque outputs. SHAP values are dimensionless and represent the log odds of the marginal contribution a variable makes on a single prediction. In the case of our gradient-boosting COVID-19 Severity Index model, we used an isotonic regression step for model calibration, so the SHAP values reflect a relative weighting of contributions.33
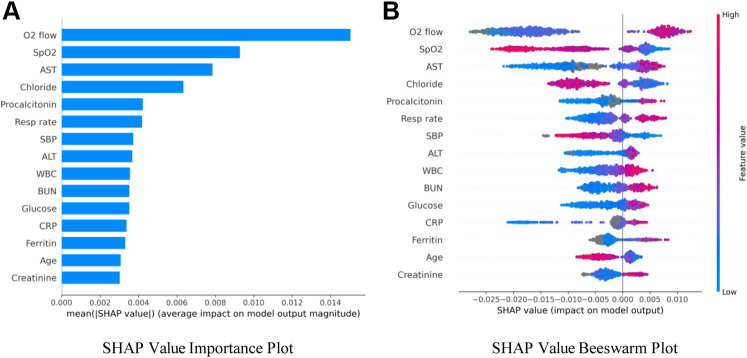
Figure 2.
SHAP variable importance and bee swarm plots. A, Mean absolute SHAP values suggest a rank order for variable importance in the COVID-19 Severity Index. B, In the bee swarm plot, each point corresponds to an individual person in the study. The points’ position on the x axis shows the effect that feature has on the model’s prediction for a given patient. Color corresponds to relative variable value.
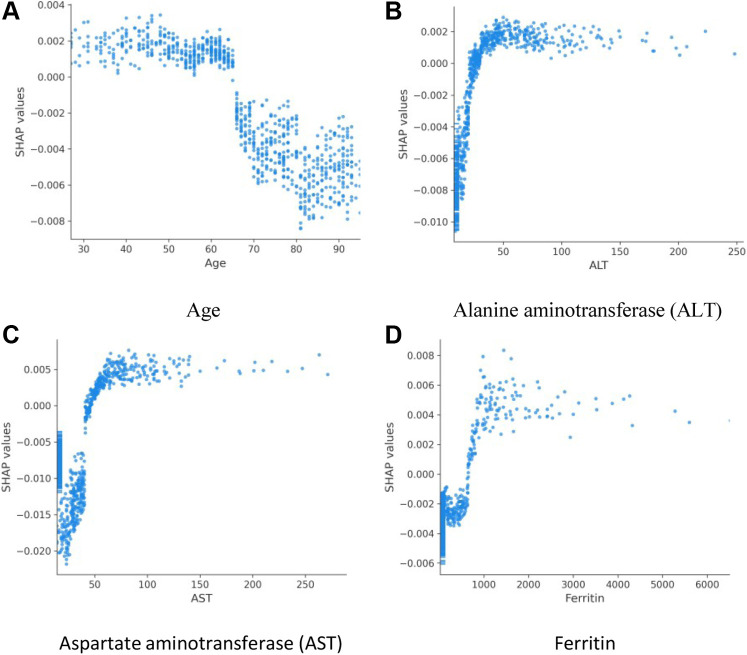
The rank order of average absolute SHAP values across all variables in a model suggests the most important variables in assigning modeled risk. For the COVID-19 Severity Index, these were flow rate by nasal cannula, followed by lowest documented pulse oximetry level and aspartate aminotransferase level (Figure 2 A). As did researchers in previous studies, we observed utility of inflammatory markers, ferritin, procalcitonin, and C-reactive protein. We then explored how ranges of individual feature values affected model output (Figure 2 B). For example, low oxygen flow rates (blue) are protective, as indicated by negative SHAP values, as are high pulse oximetry values (red). To better investigate clinical variable effects on predicted patient risk, we generated individual variable SHAP value plots (Figure 3 ). Age displayed a nearly binary risk distribution, with an inflection point between aged 60 and 70 years (Figure 3 A). Younger patients displayed a higher risk of 24-hour critical illness than did older patients. We also observed that elevated levels of aspartate aminotransferase, alanine aminotransferase, and ferritin were associated with elevated model risk, but the SHAP values reached their asymptotes well before the maximum value for each of these features (Figure 3 B to D). Aspartate aminotransferase and alanine aminotransferase SHAP values reached their maximum within normal or slightly elevated ranges for these laboratory tests. The inflection point in risk attributable to ferritin levels, however, was close to 1,000 ng/mL, above institutional normal range for this test (30 to 400 ng/mL).
Figure 3.
SHAP value plots for age (A), alanine aminotransferase (B), aspartate aminotransferase (C), and ferritin (D). Scatter plots show the effects of variable values (x axis) on the model predictions as captured by SHAP values (y axis).
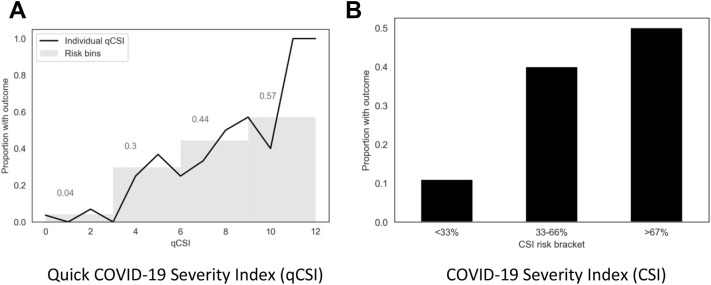
Across the cohort, 72% of patients did not have a Glasgow Coma Scale score documented. On cross validation, the quick COVID-19 Severity Index had an area under the ROC of 0.89 (0.84, 0.95), COVID-19 Severity Index score 0.92 (0.86, 0.97), qSOFA score 0.76 (0.69 to 0.85), Elixhauser score 0.70 (0.62 to 0.80), and CURB-65 score 0.66 (0.58 to 0.77) (Appendix E1, available online at http://www.annemergmed.com [Table S7]) (P<.05). On the independent validation cohort, the area under the ROCs of the quick COVID-19 Severity Index and COVID-19 Severity Index were 0.81 (0.73, 0.89) and 0.76 (0.65 to 0.86), respectively.3 We tested the calibration of the quick COVID-19 Severity Index and COVID-19 Severity Index scores by assigning all patients in the independent validation cohort each of the scores and comparing them with known outcomes (Figure 4 and Appendix E1, available online at http://www.annemergmed.com [Figures S2 to 3]).34 These calibration curves suggest that outcome rates increased with quick COVID-19 Severity Index and COVID-19 Severity Index scores. A quick COVID-19 Severity Index score of less than or equal to 3 has a sensitivity of 0.79 (0.65 to 0.93), specificity 0.78 (0.72 to 0.83), PPV 0.36 (0.25 to 0.47), NPV 0.96 (0.93 to 0.99), LR+ 3.55 (3.51 to 3.59), and LR- 0.27 (0.26 to 0.28).
Figure 4.
Calibration of quick COVID-19 Severity Index and COVID-19 Severity Index on the independent validation data set. A, Each patient in the validation cohort was assigned a score by quick COVID-19 Severity Index, and the percentage who had a critical respiratory illness outcome were plotted with a line plot. Patients were then grouped into risk bins by quick COVID-19 Severity Index intervals (0 to 3, 4 to 6, 7 to 9, and 10 to 12); the percentage of patients in each group with the outcome is indicated in the bar plot. B, Each patient in the validation cohort was assigned a COVID-19 Severity Index score, a percentage risk from 0% to 100% using gradient boosting and isotonic regression. The percentage of patients with COVID-19 Severity Index scores of 0% to 33%, 33% to 66%, and 66% to 100% who experienced critical respiratory illness at 24 hours is shown.
The quick COVID-19 Severity Index is available at https://covidseverityindex.org. The quick COVID-19 Severity Index calculator includes selection boxes for each of the 3 variables, which are summed to generate a score and prediction as estimated with the independent validation cohort.
Limitations
The data in this study were observational and provided from a single health system, and so they may not be generalizable according to local testing and admissions practices. Our data were extracted from an electronic health record, which is associated with known limitations, including propagation of old or incomplete data. There are important markers of oxygenation that were out of the scope of our study, including alveolar-arterial gradients. Because of data availability, no signs or symptoms or provider notes were included as candidate predictor variables.
Retrospective observational studies lack control of variables, so prospective studies will be required to assess validity of the presented models and the specificity of the features we identify as important to COVID-19 progression. Because of the retrospective nature of this study and the use of electronic health records, data imputation and assumptions about missingness were required, which introduced biases into our results. We assumed a Glasgow Coma Scale score of 15 unless documented otherwise, which may underestimate severity in qSOFA and CURB-65. Likewise, comorbidities were populated from previous in-system diagnoses; patients without system visits are likely to have lower Elixhauser indices than those whose care was integrated within the health system. In the quick COVID-19 Severity Index calculations, nasal cannula flow rate was imputed if nasal cannula was documented without a flow rate. In the COVID-19 Severity Index, no specific imputations were required because gradient boosting natively handles missing values. Chest radiograph interpretation was conducted manually with radiology reports, but without reviewing the radiography, which introduces subjectivity as reflected in the interrater agreement metric.
There are limitations in model performance, with confidence intervals reflective of moderate study size. We additionally did not compare the models with unstructured provider judgment, and thus one cannot make conclusions about whether this tool has utility beyond clinical gestalt. Most significant, however, is that management of COVID-19 is evolving, so future clinical decisions may not match those standards used in the reported clinical settings.
Discussion
Consistent with clinical observations, we noted a significant rate of progression to critical respiratory illness within the first 24 hours of hospitalization in COVID-19 patients. We used 6 parallel approaches to identify a subset of variables for the final quick COVID-19 Severity Index and COVID-19 Severity Index models. The quick COVID-19 Severity Index ultimately requires only 3 variables, all of which are accessible at the bedside.
We propose that a quick COVID-19 Severity Index score of 3 or less be considered low likelihood for 24-hour respiratory critical illness, with a mean outcome rate of 4% in the independent validation cohort (Figure 4) and a LR- of 0.27 (0.26 to 0.28). This score is achievable under the following patient conditions: respiratory rate less than or equal to 28 breaths/min, minimum pulse oximetry reading of greater than or equal to 89%, and oxygen flow rate of less than or equal to 2 L. In the validation cohort, a quick COVID-19 Severity Index cutoff greater than 3 had a sensitivity of 0.79 (0.65 to 0.93) in predicting progression of respiratory failure. However, few patients in the validation cohort had a quick COVID-19 Severity Index score of 3 (Spo 2 of 89% to 92% and respiratory rates of 23 to 28 breaths/min with oxygen requirement ≤2 L/min) (Appendix E1, available online at http://www.annemergmed.com [Figure S2]). In the validation cohort, patients with a quick COVID-19 Severity Index score of 4 to 6 had a 30% rate of decompensation, whereas the group with a score of 7 to 9 had a 44% rate and the group with a score of 10 to 12 had a 57% rate. A quick COVID-19 Severity Index score of greater than 9 had a specificity of 0.99 in predicting respiratory failure, with a LR of 8.36 (7.98 to 8.76). Taken together, the quick COVID-19 Severity Index provides an objective tool for planning hospital dispositions. Patients with low quick COVID-19 Severity Index scores are unlikely to have respiratory decompensation, whereas those with high scores may benefit from higher levels of care.
COVID-19 Severity Index performance on the validation cohort was not superior to that of the quick COVID-19 Severity Index. We hypothesize that this may be related to cohort differences or COVID-19 Severity Index overfitting on the development cohort. The COVID-19 Severity Index offers opportunities to examine further potential COVID-19 prognostic factors. We used gradient-boosting models rather than logistic regression because gradient boosting allowed us to better capture nonlinear relationships, such as those observed in the liver chemistries, and natively handles missing values without imputation. Lower age had higher SHAP values, suggesting potential bias in the admitted patient cohort; young admitted patients may be more acutely ill than older ones. In alignment with current hypotheses about COVID-19 severity, multiple variable selection techniques identified inflammatory markers, including C-reactive protein and ferritin, as potentially important predictors. More striking, however, was the importance of aspartate aminotransferase and alanine aminotransferase in COVID-19 Severity Index predictions as calculated with SHAP values.35 , 36 The transition point at which the SHAP value analysis identified model risk associated with liver chemistries was at the high end of normal, consistent with previous observations that noted normal to mild liver dysfunction among COVID-19 patients. We hypothesize that the asymptotic quality of the investigated variables with respect to COVID-19 Severity Index risk contributions reflects our moderate study size. We expect that scaling COVID-19 Severity Index training to larger cohorts will further elucidate the effects of more extreme laboratory values. Although our data set included host risk factors, including smoking history, obesity, and body mass index, these did not appear to play a prominent role in predicting acute deterioration. Here, we recognize 2 important considerations: first, that predictive factors may not be mechanistic or causative factors in disease, and second, that these factors may be related to disease severity without providing predictive value for 24-hour decompensation.
We included radiographs for 1,170 visits in this cohort. Radiographs are of significant clinical interest because previous studies have shown high rates of ground-glass opacity and consolidation.37 Chest computed tomography may have superior utility for COVID-19 investigation, but the procedure is not being widely performed at our institutions as part of risk stratification or prognostic evaluation.38 , 39 Radiograph reports were classified according to containing bilateral, unilateral, or no opacities or consolidations. We found high interrater agreement in this coding, but radiographs were not consistently identified by our variable selection models. A majority of patients were coded as having bilateral consolidations, limiting the specificity of the findings. Further studies using natural language processing of radiology reports or direct analysis of radiographs with tools such as convolutional neural networks will provide more evidence regarding utility of these studies in COVID-19 prognostication.40 Furthermore, we do not consider other applications of radiographs including the identification of other pulmonary findings like diagnosis of bacterial pneumonia.
The Elixhauser Comorbidity Index, qSOFA, and CURB-65 baseline models provided the opportunity to test well-known risk-stratification and prognostication tools with a COVID-19 cohort. These tools were selected, in part, for their familiarity within the medical community, and because each has been proposed as having potential utility within the COVID-19 epidemic. These metrics have relatively limited predictive performance, and there were limitations in electronic health records; none were designed to address the clinical question addressed here. We observed both a high rate of missing mental status documentation and a significant proportion of the population without documented medical histories. In particular, we hypothesize that the CURB-65 pneumonia severity score may still have utility in determining patient disposition with respect to discharge or hospitalization.
Future studies will be required to expand on this work in a number of ways. First, external validation is needed, as is comparison with physician judgment. Second, future studies may evaluate prospective robustness and utility of this scoring metric. Third, we expect related models to be extended to patient admission decisions as well as continuous hospital monitoring.41, 42, 43 Fourth, we anticipate potential applications in stratifying patients for therapeutic interventions. Early proof-of-concept studies for the viral ribonucleic acid polymerase inhibitor remdesivir included patients with severe COVID-19 as defined by pulse oximetry level of less than or equal to 94% on ambient air or with any oxygen requirement.44 , 45 Given ongoing drug scarcity, improved pragmatic, prognostic tools such as the quick COVID-19 Severity Index may offer a route to expanded inclusion criteria for ongoing trials or for early identification of patients who might benefit from therapeutics.
Taken together, these data show that the quick COVID-19 Severity Index provides easily accessed risk stratification relevant to ED providers.
Acknowledgments
Drs. Haimovich and Ravindra served as co-first authors and contributed equally to the work.
Footnotes
Please see page 443 for the Editor’s Capsule Summary of this article.
Supervising editor: Gregory J. Moran, MD. Specific detailed information about possible conflict of interest for individual editors is available at https://www.annemergmed.com/editors.
Author contributions: AH, NGR, and RAT designed the project. NGR, HPY, WLS, and RAT extracted and processed the data. AH, NGR, FPW, DvD, and RAT created the models. SS designed the Web interface. All authors contributed substantially to article revisions. RAT takes responsibility for the paper as a whole.
All authors attest to meeting the four ICMJE.org authorship criteria: (1) Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; AND (2) Drafting the work or revising it critically for important intellectual content; AND (3) Final approval of the version to be published; AND (4) Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding and support: By Annals policy, all authors are required to disclose any and all commercial, financial, and other relationships in any way related to the subject of this article as per ICMJE conflict of interest guidelines (see www.icmje.org). Dr. Wilson acknowledges funding from the National Institutes of HealthR01DK113191 and P30DK079310. Dr. Schulz was an investigator for a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; is a technical consultant to HugoHealth, a personal health information platform; is cofounder of Refactor Health, an artificial intelligence–augmented data mapping platform for health care; and is a consultant for Interpace Diagnostics Group, a molecular diagnostics company.
Trial registration number: XXXXXXXX
Funding sources had no involvement in the design of the study. Researchers are independent from funders.
Readers: click on the link to go directly to a survey in which you can provide feedback to Annals on this particular article.
Supplementary Data
References
- 1.World Health Organization Novel coronavirus (2019-nCoV) situation reports; 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ Available at:
- 2.CDC U Coronavirus disease 2019 (COVID-19) cases in US; 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html Available at:
- 3.Singer A.J., Morley E.J., Meyers K. Cohort of 4404 persons under investigation for COVID-19 in a NY hospital and predictors of ICU care and ventilation. Ann Emerg Med. 2020 doi: 10.1016/j.annemergmed.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haimovich A., Warner F., Young H.P. Patient factors associated with SARS-CoV-2 in an admitted emergency department population. https://onlinelibrary.wiley.com/doi/abs/ 10.1002/emp2.12145 Available at: [DOI] [PMC free article] [PubMed]
- 5.Chan P.S., Jain R., Nallmothu B.K. Rapid response teams: a systematic review and meta- analysis. Arch Intern Med. 2010;170:18–26. doi: 10.1001/archinternmed.2009.424. [DOI] [PubMed] [Google Scholar]
- 6.Badawi O., Liu X., Berman I. Impact of COVID-19 pandemic on severity of illness and resources required during intensive care in the greater New York City area. https://www.medrxiv.org/content/early/2020/04/14/2020.04.08.20058180 Available at:
- 7.Kennedy M., Joyce N., Howell M.D. Identifying infected emergency department patients admitted to the hospital ward at risk of clinical deterioration and intensive care unit transfer. Acad Emerg Med. 2010;17:1080–1085. doi: 10.1111/j.1553-2712.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simchen E., Sprung C.L., Galai N. Survival of critically ill patients hospitalized in and out of intensive care. Crit Care Med. 2007;35:449–457. doi: 10.1097/01.CCM.0000253407.89594.15. [DOI] [PubMed] [Google Scholar]
- 9.Berlin DA, Gulick RM, Martinez FJ. Severe Covid-19. N Engl J Med. 10.1056/NEJMcp2009575. [DOI] [PubMed]
- 10.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 11.Horwitz L.I., Green J., Bradley E.H. US emergency department performance on wait time and length of visit. Ann Emerg Med. 2010;55:133–141. doi: 10.1016/j.annemergmed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maves R.C., Downar J., Dichter J.R. Triage of scarce critical care resources in COVID-19: an implementation guide for regional allocation: an expert panel report of the Task Force for Mass Critical Care and the American College of Chest Physicians. Chest. 2020 doi: 10.1016/j.chest.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Walraven C., Austin P.C., Jennings A. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Med Care. 2009:626–633. doi: 10.1097/MLR.0b013e31819432e5. [DOI] [PubMed] [Google Scholar]
- 14.Singer M., Deutschman C.S., Seymour C.W. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira M., Blin T., Collercandy N. Critically ill SARS-CoV-2–infected patients are not stratified as sepsis by the qSOFA. Ann Intensive Care. 2020;10:1–3. doi: 10.1186/s13613-020-00664-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim W.S., Van der Eerden M., Laing R. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–382. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilg A., Moskowitz A., Konanki V. Performance of the CURB-65 score in predicting critical care interventions in patients admitted with community-acquired pneumonia. Ann Emerg Med. 2019;74:60–68. doi: 10.1016/j.annemergmed.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moons K.G., Altman D.G., Reitsma J.B. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 19.McPadden J., Durant T.J., Bunch D.R. Health care and precision medicine research: analysis of a scalable data science platform. J Med Internet Res. 2019;21 doi: 10.2196/13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.First DataBank First DataBank enhanced therapeutic classification system (ETC) http://www.firstdatabank.com/Products/therapeutic-classification-system-nddf.aspx Available at:
- 21.Elixhauser A., Steiner C., Harris D.R. Comorbidity measures for use with administrative data. Med Care. 1998:8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Agency for Healthcare Research and Quality . Agency for Healthcare Research & Quality; Rockville, MD: 2020. HCUP Tools and Software. Healthcare Cost and Utilization Project (HCUP)http://www.hcup-us.ahrq.gov/tools_software.jsp Available at: [Google Scholar]
- 23.Cohen SB, Ruppin E, Dror G. Feature Selection Based on the Shapley Value. In: IJCAI. vol. 5; 2005. p. 665-670.
- 24.Guyon I., Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 25.Lundberg S.M., Erion G., Chen H. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2:2522–5839. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen T, Guestrin C. Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining; 2016:785-794.
- 27.Bergstra J, Yamins D, Cox DD. Making a science of model search: hyperparameter optimization in hundreds of dimensions for vision architectures. In: Proceedings of the 30th International Conference on International Conference on Machine Learning, Volume 28. ICML ‘13. 2013:I-115-I-123.
- 28.Saito T., Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efron B., Tibshirani R.J. Chapman & Hall/CRC; Boca Raton, FL: 1993. An Introduction to the Bootstrap. No. 57 in Monographs on Statistics and Applied Probability. [Google Scholar]
- 30.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Advances in neural information processing systems; 2017:4765-4774.
- 31.Lundberg S.M., Nair B., Vavilala M.S. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2:749. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Artzi N.S., Shilo S., Hadar E. Prediction of gestational diabetes based on nationwide electronic health records. Nat Med. 2020;26:71–76. doi: 10.1038/s41591-019-0724-8. [DOI] [PubMed] [Google Scholar]
- 33.Niculescu-Mizil A, Caruana R. Predicting good probabilities with supervised learning. In: Proceedings of the 22nd International Conference on Machine Learning; 2005:625-632.
- 34.Backus B., Six A., Kelder J. A prospective validation of the HEART score for chest pain patients at the emergency department. Int J Cardiol. 2013;168:2153–2158. doi: 10.1016/j.ijcard.2013.01.255. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C., Shi L., Wang F.S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai Q., Huang D., Yu H. Characteristics of liver tests in COVID-19 patients. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong H.Y.F., Lam H.Y.S., Fong A.H.T. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020:201160. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung M., Bernheim A., Mei X. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements and prognosis of COVID-19 pneumonia using computed tomography. Cell. [DOI] [PMC free article] [PubMed]
- 40.Rajpurkar P., Irvin J., Zhu K. CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. http://arxiv.org/abs/1711.05225 Available at:
- 41.Henry K.E., Hager D.N., Pronovost P.J. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med. 2015;7:299ra122. doi: 10.1126/scitranslmed.aab3719. [DOI] [PubMed] [Google Scholar]
- 42.Simonov M., Ugwuowo U., Moreira E. A simple real-time model for predicting acute kidney injury in hospitalized patients in the US: a descriptive modeling study. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomasev N., Glorot X., Rae J.W. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572:116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grein J., Ohmagari N., Shin D. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Zhang D., Du G. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janssen A., Pauls T. How do bootstrap and permutation tests work? Ann Stat. 2003;31:768–806. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.