While HSV antivirals reduce the severity and duration of clinical disease in some individuals, there is no vaccine or cure. Therefore, understanding the mechanisms regulating latency and reactivation as a potential to elucidate targets for better therapeutics is important. There are at least 20 confirmed HSV-1 miRNAs, yet the roles of individual miRNAs in the context of viral infection remain largely uncharacterized. The present study focuses on two of the miRNAs (miR-H1/H6) that are encoded within the latency-associated transcript (LAT) region, a portion of the genome that has been associated with efficient reactivation. Here, we demonstrate that the deletion of the seed sequences of these miRNAs results in a severe reduction in reactivation of HSV-1 in the mouse and rabbit models. These results suggest a linkage between these miRNAs and reactivation.
KEYWORDS: LAT, herpes simplex virus, latency, miRNA, reactivation
ABSTRACT
During all stages of infection, herpes simplex virus 1 (HSV-1) expresses viral microRNAs (miRNAs). There are at least 20 confirmed HSV-1 miRNAs, yet the roles of individual miRNAs in the context of viral infection remain largely uncharacterized. We constructed a recombinant virus lacking the sequences for miR-H1-5p and miR-H6-3p (17dmiR-H1/H6). The seed sequences for these miRNAs are antisense to each other and are transcribed from divergent noncoding RNAs in the latency-associated transcript (LAT) promoter region. Comparing phenotypes exhibited by the recombinant virus lacking these miRNAs to the wild type (17syn+), we found that during acute infection in cell culture, 17dmiR-H1/H6 exhibited a modest increase in viral yields. Analysis of pathogenesis in the mouse following footpad infection revealed a slight increase in virulence for 17dmiR-H1/H6 but no significant difference in the establishment or maintenance of latency. Strikingly, explant of latently infected dorsal root ganglia revealed a decreased and delayed reactivation phenotype. Further, 17dmiR-H1/H6 was severely impaired in epinephrine-induced reactivation in the rabbit ocular model. Finally, we demonstrated that deletion of miR-H1/H6 increased the accumulation of the LAT as well as several of the LAT region miRNAs. These results suggest that miR-H1/H6 plays an important role in facilitating efficient reactivation from latency.
IMPORTANCE While HSV antivirals reduce the severity and duration of clinical disease in some individuals, there is no vaccine or cure. Therefore, understanding the mechanisms regulating latency and reactivation as a potential to elucidate targets for better therapeutics is important. There are at least 20 confirmed HSV-1 miRNAs, yet the roles of individual miRNAs in the context of viral infection remain largely uncharacterized. The present study focuses on two of the miRNAs (miR-H1/H6) that are encoded within the latency-associated transcript (LAT) region, a portion of the genome that has been associated with efficient reactivation. Here, we demonstrate that the deletion of the seed sequences of these miRNAs results in a severe reduction in reactivation of HSV-1 in the mouse and rabbit models. These results suggest a linkage between these miRNAs and reactivation.
INTRODUCTION
The majority of the U.S. population (50 to 90%) is latently infected with herpes simplex virus 1 (HSV-1) by the time they are adults (1). HSV-1 causes the common cold sore and, less frequently, herpes stromal keratitis, the leading cause of infectious blindness in the United States (2). Primary infection typically occurs in the mucosal epithelium, and the virus spreads by retrograde axonal transport to the nuclei of peripheral neurons, the site of viral latency. During latency, viral gene expression is restricted to select noncoding RNAs (ncRNAs), which include the latency-associated transcripts (LATs) and viral microRNAs (miRNAs or miRs). Paradigm phenotypes of latency associated with the LAT locus include aiding in the establishment of latency, maintaining latency by protecting neurons from apoptosis, and facilitating efficient reactivation (3).
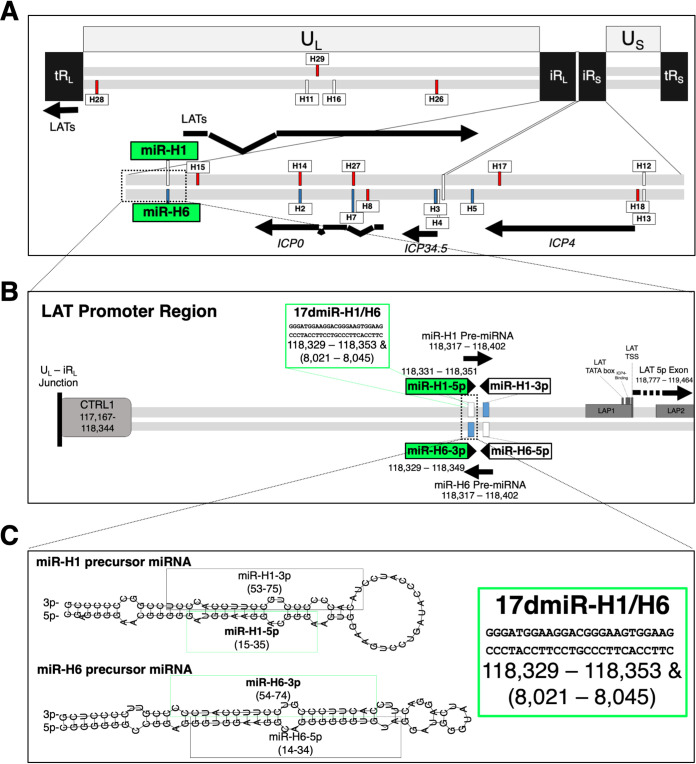
In 2004, miRNAs were first described in herpesviruses, and soon after, several HSV-1 miRNAs were predicted or shown to be transcribed from within the LAT region (4, 5). LAT phenotypes have largely been identified from the characterization of recombinant viruses with deletions in the LAT promoter, which, in retrospect, also reduced viral miRNA expression during latency (6). Since a hallmark feature of all herpesviruses is the ability to persist as lifelong infections, it is not surprising that of the known virally encoded miRNAs, 91% are found in herpesviruses (7). Thus, understanding the 10 known HSV-1 miRNAs in the LAT region (Fig. 1A), and separating their importance from the phenotypes ascribed to the LATs, may be key in understanding the dynamic nature of viral latency.
FIG 1.
Complexity of the latency-associated transcript (LAT) locus. (A) The HSV-1 genome (drawn to scale) is depicted. The unique long (UL) and short (US) segments are flanked by long (TRL) and short (TRs) terminal repeats that are inverted (IRL/IRs) on the inner repeat segments. The inner inverted repeats are enlarged to show transcripts in the LAT region. Viral miRNAs, colored by their upregulation during acute infection in white, latency in blue, and reactivation in red, as well as select viral transcripts relevant to the text, are labeled. (B) The LAT promoter region is enlarged to indicate the LAT promoter region elements: CTRL1, LAP1, LAP2, LAT TATA box, LAT transcriptional start site (TSS), and an ICP4-binding region. The miR-H1 and miR-H6 precursor miRNAs (as well as the 17dmiR-H1/H6 deletion) are also labeled. The nucleotide positions refer to the 17syn+ reference genome (NC_001806.2). (C) The predicted structures of miR-H1 and miR-H6 precursor miRNAs (RNAfold webserver, http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi) with the mature miRNA sequences highlighted and the deletion corresponding to 17dmiR-H1/H6.
Individual viral miRNAs in herpesviruses are currently being characterized in a systematic fashion by a number of investigators. For example, the biological significance of miR-H2 was quickly examined with multiple recombinant viruses due to the importance of its proposed target, ICP0 (8–11). A recombinant virus that disrupted miR-H2 function in the 17syn+ wild-type background resulted in increased replication in neuronal cells (8). In addition, another recombinant virus mutating the miR-H2 in the McKrae strain resulted in increased neurovirulence and increased reactivation upon explant in mice (9). However, an miR-H2 mutant in the KOS strain background exhibited no changes in pathogenesis or explant-induced reactivation in mice, suggesting that there are strain-specific differences in miR-H2 function (11). Studies have also examined the effects of a number of LAT promoter deletion viruses, used to prescribe paradigm phenotypes of the LAT, on the expression of viral miRNAs. For example, one study comparing two LAT promoter mutants in the KOS background, dlLAT1.8, which deletes a 1.8-kb region encompassing the LAT promoter and a portion of the primary LAT and has reduced reactivation (12), and KOSΔPstLAT, with a smaller deletion of 202 bases of the LAT promoter (13, 14), found no effect on viral miRNA expression during the acute infection in culture or in mice (6). However, during latent infection in mice, there was vastly reduced viral miRNA expression (6). These results suggest these viral miRNAs exert little or no influence on the acute infection or establishing latency but leave open the possibility that reduced viral miRNA expression during latency contributes to, or is responsible for, the reduced reactivation seen in LAT promoter mutants. Further, reduced spontaneous reactivation in the rabbit ocular infection model was observed in dLAT2903, a recombinant virus in the McKrae strain background with a 1.5-kb deletion encompassing the LAT promoter and the first 1.5 kb of the primary LAT (15), by stem-loop reverse transcription-quantitative PCR (RT-qPCR) that miR-H2, -H3, -H4, -H5, -H7, and -H8 in dLAT2903 were significantly reduced in expression relative to that of the wild-type McKrae parent (10). The viral miRNA expression of another LAT promoter deletion virus, 17ΔPst (16), was examined and found to have significantly reduced viral miRNA expression compared to its parental virus (10). To further examine the function of miR-H2, the authors mutated the miR-H2 sequence in the dLAT2903 virus and found increased neurovirulence and reactivation upon explant in mice (10). This finding was striking, since dLAT2903 was found to have reduced spontaneous reactivation, and the phenotype was reversed with mutation of just miR-H2, further highlighting the need to investigate the contributions of individual viral miRNAs to latency and reactivation.
Of the LAT region miRNAs, HSV-1 miR-H1 and miR-H6 (termed miR-H1/H6) are of particular interest for several reasons. HSV-1 miR-H1/H6 is composed of two miRNAs, miR-H1-5p and miR-H6-3p, that are antisense to each other and transcribed from within the LAT promoter region (Fig. 1). Mutating the seed sequence of one impacts both seed sequences, complicating analyses. As stated above, many of the paradigm phenotypes associated with the LATs have been revealed using promoter deletion mutants, which also inadvertently affect miRNA expression (6). One of the central goals of this current study was to determine if miR-H1/H6 phenotypes contributed to paradigm phenotypes associated with the LATs. miR-H6 was one of the first HSV-1 miRNAs identified and was the first to be shown to target ICP4, an essential immediate-early lytic viral gene (5). In this seminal study, Umbach et al. showed that miR-H6 targeted ICP4 using transient expression data from cell culture experiments (5). Here, the authors found a decrease in ICP4 protein by Western blotting but not in ICP4 mRNA by qPCR (5). Further, Duan et al. showed, using miR-H6 mimics and HSV-1 infection of human corneal epithelial cells, that miR-H6 expression corresponded with a decrease in viral replication and reduced interleukin-6 (IL-6) signaling (17). Aside from targeting ICP4, there is strong evidence that suggests that miR-H6 is expressed during all phases of infection. Northern blot analysis confirmed miR-H6 precursor miRNA (6, 18, 19) and mature miRNA (18, 19) following acute infection in cell culture. In addition, miR-H6 accumulation during acute infection in cell culture has been analyzed by stem-loop RT-qPCR (5, 6, 20, 21), deep sequencing of RNA (18, 22), and RNA immunoprecipitation sequencing (RIP-seq) (8). During latency in vivo, miR-H6 expression has been detected in mouse ganglia by stem-loop RT-qPCR (6, 21) and deep RNA sequencing (18). It has also been validated in human postmortem trigeminal ganglia (TG) samples following stem-loop RT-qPCR (23) and deep RNA sequencing (20).
There is also strong evidence that miR-H1 is expressed during all temporal phases of lytic infection, except during reactivation (21). Recently, miR-H1 was shown to target host transcripts Sort1 (24) and Ubr1 (25) by transient expression; however, these studies were based on transfections and not done in the context of viral infection. An additional study that highlights the importance of examining these miRNAs in the context of infection to validate transient expression assays was the proposed targeting of ATRX by miR-H1. In this particular study, an miR-H1 mimic was shown to target the mRNA of the host intrinsic effector, ATRX, which is a component of ND10 bodies (26). However, a recombinant virus lacking miR-H1/H6 in the HSV-1 strain KOS background had no effect on ATRX depletion by Western blotting (26). Nonetheless, if the functions of miRs H1 and H6 are biologically significant in the context of latency and reactivation, rigorous in vivo characterization of a recombinant virus lacking miRs H1 and H6 would effectively show whether or not these interactions are biologically significant.
The locations of miR-H1 and miR-H6 within the LAT locus, as well as their temporal expression during all phases of infection, provide a strong scientific premise that these miRNAs play a significant role in regulating HSV-1 latency and reactivation. We hypothesized that if miR-H6 functions to downregulate ICP4, a recombinant virus lacking miRs H1 and H6 would have increased pathogenesis compared to that of wild-type virus due to an increase in the activation of the lytic cascade. We further hypothesized that this fine-tuning of gene regulation by HSV-1 miRNAs contributes to the dynamic state of viral latency.
RESULTS
An HSV-1 recombinant with deletions in miRNAs H1 and H6 displayed modest increases in viral yields in vitro.
HSV-1 miRNAs miR-H1 and miR-H6 are both expressed during acute infection in vitro (21). While a recombinant virus containing a deletion in miR-H1/H6 had previously been constructed in HSV-1 strain KOS, this deletion had little effect on pathogenesis (26). Because strain KOS is relatively avirulent in the mouse (27) and does not reactivate in the rabbit eye model (28), we assessed the contributions of these two miRNAs to pathogenesis and reactivation utilizing the more pathogenic strain 17syn+ (abbreviated as 17+ in the figures). Therefore, we constructed a recombinant virus, 17dmiR-H1/H6, which contained a deletion of the antisense mature miRNAs miR-H1-5p and miR-H6-3p (Fig. 1C). The 25-bp deletion corresponding to the mature miRNA duplex of miR-H1-5p and miR-H6-3p was confirmed by Southern blotting, PCR analysis, and Sanger sequencing of the fragment containing the sequences flanking the deletion (data not shown). The deletion and genetic integrity of the entire genome was confirmed by Illumina sequencing (see the supplemental material). Sixty-two putative SNPs, including 10 nonsynonymous substitutions, were observed, but none resulted in nonsense, no-start, no-stop, or frameshift mutations. Therefore, this mutant virus, 17dmiR-H1/H6 (abbreviated as dH1/H6 in the figures), was compared directly to its parent wild-type virus, 17syn+.
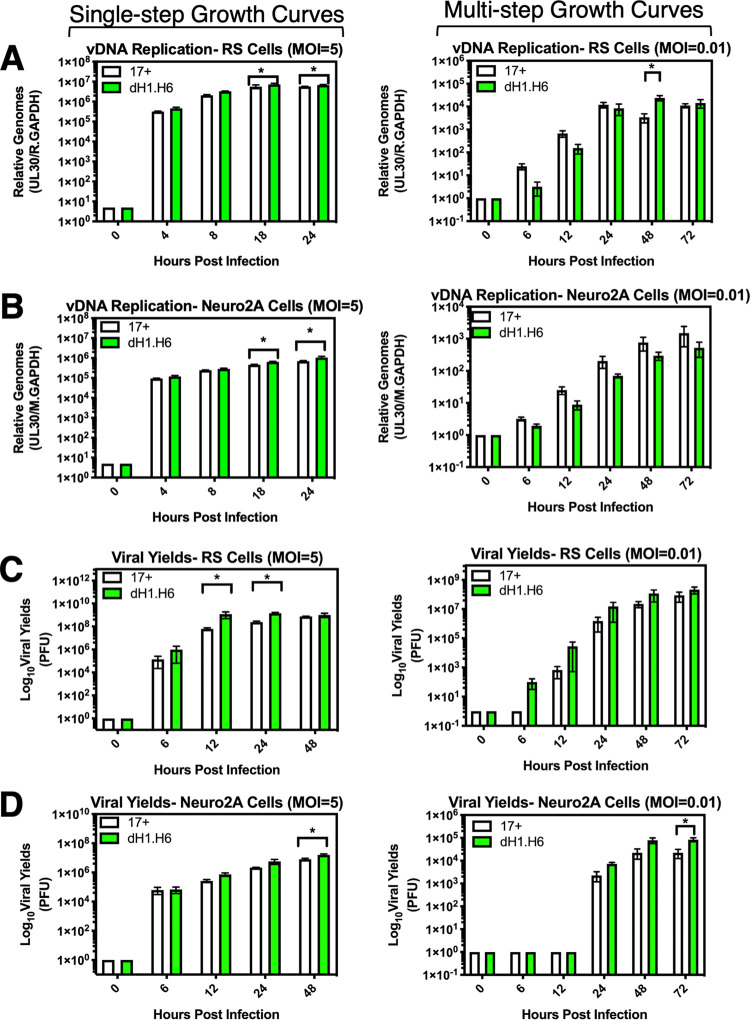
Replication kinetics of 17dmiR-H1/H6 was assessed in cell lines from species relevant to the animal models of HSV-1 infection, including rabbit skin (RS) and mouse neuroblastoma (Neuro2A) cells. At a high multiplicity of infection (MOI) (5 PFU/cell), viral DNA (vDNA) accumulation for 17dmiR-H1/H6 showed a slight, though significant, increase at 18 and 24 h postinfection (hpi) in RS and Neuro2A cells (*, P < 0.05 by ordinary two-way analysis of variance [ANOVA] with Sidak’s multiple-comparison test) (Fig. 2). At a high MOI, viral yields were also significantly increased at 12 and 24 hpi in the absence of miR-H1/H6 in RS cells (Fig. 2C) and at 48 hpi in Neuro2A cells (Fig. 2D). At low MOI (0.01 PFU/cell), there were no consistently significant differences in viral yields or vDNA replication.
FIG 2.
HSV-1 recombinant with deletions in miRNAs H1 and H6 displays modest increases in viral yields in vitro. (A and B) Viral DNA (vDNA) replication in rabbit skin (RS) cells (A) and mouse neuroblastoma (Neuro2A) cells (B). In a 24-well plate format, cells were infected with wild-type virus 17syn+ or 17dmiR-H1/H6 at a multiplicity of infection (MOI) of 5 or 0.01, as indicated, and harvested at the indicated hours postinfection (hpi). gDNA was extracted and subjected to qPCR probing for UL30, the viral polymerase gene, and rabbit (A) or mouse (B) GAPDH (M.GAPDH). RS (C) or Neuro2A (D) cells were infected with wild-type virus 17syn+ or 17dmiR-H1/H6 at an MOI of 5 or 0.01 as indicated. Cells and supernatant were harvested at the given time point, lysed by one freeze/thaw cycle, and subjected to limited dilution plaque assays. These data represent three independent experiments, and significance was determined by ordinary two-way ANOVAs with Sidak’s multiple-comparison test (*, P < 0.05).
Deletion of the HSV-1 miR-H1/H6 resulted in a slight increase in virulence in the mouse footpad infection model.
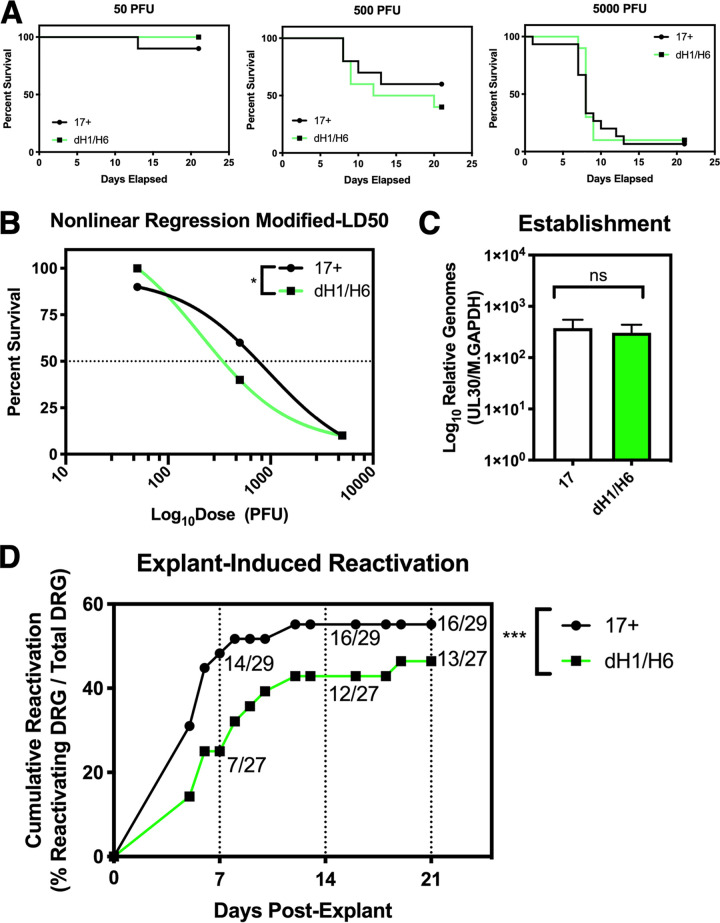
To measure the effects of deleting miR-H1 and miR-H6 on pathogenesis, we utilized the murine footpad infection model because it sensitively assesses the ability of HSV-1 to replicate in the footpad epithelium and invade the peripheral to central nervous systems (29). Thus, these experiments were conducted to provide a sensitive measure of potential differences in neurovirulence and neuroinvasiveness due to deletions of miR-H1 and miR-H6. It is important to note that we utilized the 17syn+ strain as the backbone for making the miR-H1 and miR-H6 deletions in an effort to better detect even subtle effects of the deletion due to its higher pathogenicity, which is not observed in the KOS background (26), where 17syn+ is virulent following footpad infection in the mouse (50% lethal dose [LD50] of 500 PFU) compared to LD50s of >2,000,000 PFU for KOS (27). Mice were infected with either 17dmiR-H1/H6 or 17syn+ at 50, 500, and 5,000 PFU/mouse on both rear footpads and monitored daily in a masked fashion. When the mice reached the defined clinical endpoint where they would likely succumb to HSV-1 infection (see Materials and Methods), they were humanely euthanized. The resulting survival curves are shown in Fig. 3A. The percent survival was plotted relative to dose using nonlinear regression models. We found the modified LD50 of 17syn+ to be 618 PFU and the modified LD50 of 17dmiR-H1/H6 to be 210.5 PFU (Fig. 3B). Significant differences in survival between mice infected with 17syn+ and 17dmiR-H1/H6 at 500 PFU were observed (P = 0.017 by log-rank Mantel-Cox test). Therefore, these results indicated that deletion of miR-H1/H6 from HSV-1 resulted in a significant increase in virulence relative to that of wild-type virus.
FIG 3.
Deletion of HSV-1 miR-H1/H6 results in a slight increase in virulence in the mouse footpad infection model and a significant reduction in reactivation from latency in explanted mouse ganglia. Female 4- to 6-week-old ND4 Swiss Webster mice were infected with 17syn+ or 17dmiR-H1/H6 using the mouse footpad infection model. (A) Infection doses of 50, 500, and 5,000 PFU (n = 15 mice/dose/virus) were used to determine dose-dependent differences in viral replication and spread in vivo over 21 days. (B) Survival for each dose is shown and combined to show the percent survival plotted against the log dose. The combined data were fit using a nonlinear regression model to calculate the modified LD50 (17syn+ = 618, 17dmiR-H1/H6 = 210.6). These curves are significantly different (*, P < 0.05 by Mantel-Cox log-rank test). (C) Twenty-eight days postinfection, the dorsal root ganglia (DRG) were dissected and gDNA was extracted (6 ganglia from each mouse were pooled for 1 gDNA sample and 5 mice per virus were analyzed; n = 5 mice/virus) and subjected to qPCR probing for UL30 and mouse GAPDH (M.GAPDH). Unpaired t tests were used to determine significance (*, P < 0.05; ns, not significant). (D) Twenty-eight days postinfection, individual DRG were dissected and seeded onto monolayers of rabbit skin cells (6 individual ganglia per mouse and 5 mice per virus; n = 29 total DRG for 17syn+ and 27 total DRG for 17dmiR-H1/H6 [some ganglia were excluded due to contamination]). Cytopathic effects were monitored daily and scored for evidence of virus reactivation over 21 total days postexplant. Cumulative reactivation refers to explant DRG on a rabbit skin cell monolayer positive for cytopathic effects relative to the total number of explant DRG. Reactivation was not tracked on a per-mouse basis. Significance was determined by Mann-Whitney test (***, P < 0.0001).
17dmiR-H1/H6 established latency in the murine dorsal root ganglia equivalently to 17syn+ following footpad infection.
Mice were infected at 500 PFU with either 17dmiR-H1/H6 or 17syn+ on both rear footpads, as described in Materials and Methods. At 28 days postinfection (dpi), the dorsal root ganglia (DRG) from surviving mice were dissected, DNA was extracted, and the relative load of latent HSV-1 genomes was determined by qPCR (P = 0.75 by unpaired t test) (Fig. 3C). These analyses detected no significant differences in the establishment of latency between 17dmiR-H1/H6 and 17syn+.
Deletion of HSV-1 miR-H1/H6 resulted in a significant reduction in reactivation from latency in explanted mouse ganglia.
We next utilized the murine explant cocultivation model to determine if 17dmiR-H1/H6 displayed any alteration in its ability to reactivate from latency. Mice were infected at 500 PFU with either 17dmiR-H1/H6 or 17syn+ on both rear footpads, as described in Materials and Methods. At 28 dpi, the DRG from surviving mice were dissected (L4, L5, and L6), and individual ganglia were placed onto monolayers of RS cells and monitored for reactivation by observing cytopathic effects (scoring as positive or negative for reactivation) over 21 days (30). At 5 days postexplant, reactivation was observed for both viruses. At 7 days postexplant, 14/29 ganglia for 17syn+ had reactivated, while only 7/27 ganglia for 17dmiR-H1/H6 had reactivated. Overall, reactivation of 17syn+ peaked by day 12 postexplant, and for 17dmiR-H1/H6, reactivation occurred at a much lower rate, peaking at 19 days postexplant (P < 0.001 by Mann-Whitney test) (Fig. 3D). By 21 days postexplant, we found that 17dmiR-H1/H6 reactivated from 12/27 ganglia (44%), compared to 16/29 ganglia (55%) for 17syn+. Given that there was no significant reduction in the ability of 17dmiR-H1/H6 to establish latency, these results indicate that the deletion of miR-H1/H6 results in a decreased ability of 17dmiR-H1/H6 to reactivate from latency in the murine explant model.
In the rabbit ocular infection model, the absence of miR-H1/H6 dramatically reduced adrenergically induced reactivation.
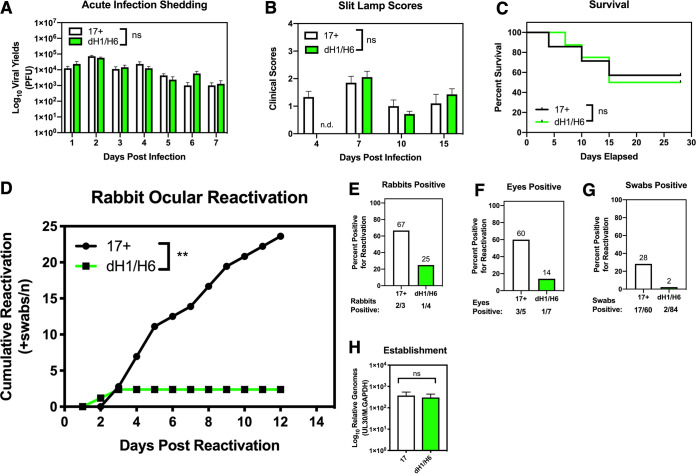
Given the decreased reactivation of 17dmiR-H1/H6 that was observed in the explanted mouse ganglia, we sought to determine the ability of 17dmiR-H1/H6 to reactivate in the rabbit ocular model. The rabbit ocular model parallels a number of aspects of ocular HSV-1 infection in humans more closely than in the mouse (31, 32), including the ability to observe and quantify corneal lesions during the acute infection period as well as the ability to very efficiently and synchronously induce reactivation in latently infected rabbits following the administration of adrenergic agents, such as epinephrine (33). Rabbits were infected with 200,000 PFU/eye of either 17dmiR-H1/H6 or 17syn+ and monitored for survival, virus shedding, and corneal pathology, as described in Materials and Methods. During the acute infection, no significant differences were observed in the amount of virus present in tear swabs through 7 days postinfection (*, P < 0.05 by ordinary two-way ANOVA with Sidak’s multiple-comparison test; ns, not significant) (Fig. 4A), and we did not monitor spontaneous reactivation between 7 and 30 days postinfection. There were no significant differences in slit lamp clinical scores grading the severity of the corneal lesions (Fig. 4B) or in survival (*, P < 0.05 by Mantel-Cox log-rank test) (Fig. 4C). Therefore, 17dmiR-H1/H6 was phenotypically indistinguishable from 17syn+ during the acute phase of infection in the rabbit eye.
FIG 4.
In the rabbit ocular infection model, the absence of miR-H1/H6 dramatically reduces adrenergically induced reactivation. Male and female New Zealand White rabbits (2 to 3 kg) were infected in each cornea with either 17syn+ (7 rabbits) or 17dmiR-H1/H6 (8 rabbits) at 200,000 PFU and monitored over the course of 41 days. During acute infection (1 to 15 days postinfection), the course of infection was monitored by shedding of virus by plaque assay (*, P < 0.05 ordinary two-way ANOVA with Sidak’s multiple-comparison test) (A), clinical scores (see Materials and Methods) assessed by slit lamp (ND, not determined; 17dmiR-H1/H6 eyes at 4 dpi were not scored) (B), and survival (*, P < 0.05 by Mantel-Cox log-rank test) (C). (D) Thirty days postinfection, reactivation was induced by iontophoresis of epinephrine. Each eye was treated as an individual biological replicate (17syn+, n = 5 eyes from 3 rabbits; 17dmiR-H1/H6, n = 7 eyes from 4 rabbits), and eyes with evidence of corneal scarring were excluded from the reactivation experiments. Swabs were taken daily, and reactivation was assessed by plaque assay. Data are shown as cumulative reactivation (swabs positive for infectious virus) as a function of time (**, P < 0.005 by Mann-Whitney test). Proportions of reactivation are represented for each rabbit (n = 3 for 17syn+ and n = 4 for 17dmiR-H1/H6) (E), eye (n = 5 for 17syn+ and n = 7 for 17dmiR-H1/H6) (F), and swab (n = 60 for 17syn+ and n = 84 for 17dmiR-H1/H6) (G). After the reactivation experiment, the infection returned to latency (determined by the absence of infections virus), and trigeminal ganglia (n = 8 TG per virus) were extracted to assess relative viral genomes (H) (*, P < 0.05 by unpaired t test).
At 30 days postinfection, reactivation was induced by iontophoresis of epinephrine, as previously described (34). The eyes were swabbed daily, and the presence of infectious virus was determined by plaque assay. Compared to 17syn+, 17dmiR-H1/H6 exhibited dramatically reduced cumulative reactivation when assessed by the cumulative number of positive swabs (P = 0.0024 by Mann-Whitney test) (Fig. 4D), total percentage of rabbits that reactivated (Fig. 4E), total percentage of rabbit eyes that reactivated (Fig. 4F), or percentage of swabs that were positive for reactivation (Fig. 4G). Strikingly, 17dmiR-H1/H6 reactivated in only 14% of the eyes compared to 60% of eyes latently infected with 17syn+. These data demonstrate that 17dmiR-H1/H6 is significantly impaired in epinephrine-induced reactivation in the rabbit ocular model and that the degree of impairment approached that of the LAT promoter deletion 17ΔPst (16).
Finally, 12 days after the induction of reactivation and no further virus shedding was observed, the rabbits were euthanized, and TG were dissected to quantify latent viral genomes. DNA was extracted from the TG, and relative quantities of HSV-1 genomes were determined by qPCR. Although there is a potential for virus to replicate and reestablish latency following reactivation, these analyses revealed no significant differences in genome copies between wild-type and 17dmiR-H1/H6 viruses (Fig. 4H). Therefore, the decreased reactivation in the rabbit ocular model exhibited by 17dmiR-H1/H6 was due to the deletion of miR-H1/H6 and not decreased levels of establishment of latency.
Deletion of the viral miRs H1 and H6 results in increased LAT 5p exon and viral miRNA accumulation during acute infection in cell culture.
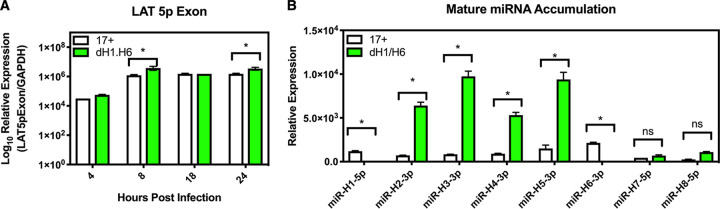
Due to the location of miR-H1 and miR-H6 within the LAT region (Fig. 1A), we speculated that these miRNAs could exert a regulatory influence on the LAT itself. Therefore, we next examined the impact of the dH1/H6 deletion on the accumulation of latency-associated viral transcripts (Fig. 1A). During acute infection, significant increases in the accumulation of the LAT 5p exon (Fig. 5A) were found at 8 and 24 hpi compared to levels of wild-type infection (*, P < 0.05 by ordinary two-way ANOVAs with Sidak’s multiple-comparison test). This increase of LAT 5p exon expression following infection with 17dmiR-H1/H6 suggested that miR-H1 and/or miR-H6 negatively regulate LAT accumulation. Due to the complexity of this region, we hypothesized that increased latency-associated transcription could also impact viral miRNA accumulation, since miRs H1 through H8 reside in the LAT locus (Fig. 1A).
FIG 5.
Viral miRs H1 and H6 negatively regulate the LAT 5p exon and viral miRNA accumulation during acute infection in cell culture. (A) Rabbit skin cells were infected (MOI of 5) and harvested at several time points to analyze 5p exon expression (*, P < 0.05 by ordinary two-way ANOVAs with Sidak’s multiple-comparison test). (B) Rabbit skin cells were infected with 17syn+ or 17dmiR-H1/H6 (MOI of 5) and harvested 12 h later to analyze viral miRNA (miR-H1 through miR-H8) accumulation using stem-loop primers (*, P < 0.05 by unpaired t-tests). These data represent three independent experiments.
To investigate whether the modulation of LAT expression by miR-H1/H6 resulted in downstream effects on the accumulation of other miRNAs in the LAT region, we examined the relative accumulation of viral miRNAs (miRs H1 through H8) in RS cells during acute infection at 12 hpi (MOI of 5) by stem-loop RT-qPCR (Fig. 5B). As expected, we did not detect mature miR-H1 or miR-H6 transcripts from the 17dmiR-H1/H6 virus. We did, however, find significant increases (*, P < 0.05 by unpaired t tests) in the expression of other viral miRNAs in this region. These findings once again make it difficult to separate the effects of the deletion of miR-H1/H6 and those corresponding to increases in LAT and the expression of the other viral miRNAs, which both appear to increase approximately 2-fold in 17dmiR-H1/H6. For this reason, we examined the role of Drosha, a component of the canonical miRNA biogenesis pathway that processes the primary hairpin miRNA into the precursor miRNA, which is then exported into the cytoplasm by Exportin-5 (35), in the accumulation of these different species of HSV-1 ncRNAs.
Latency-associated transcript accumulation was Drosha dependent.
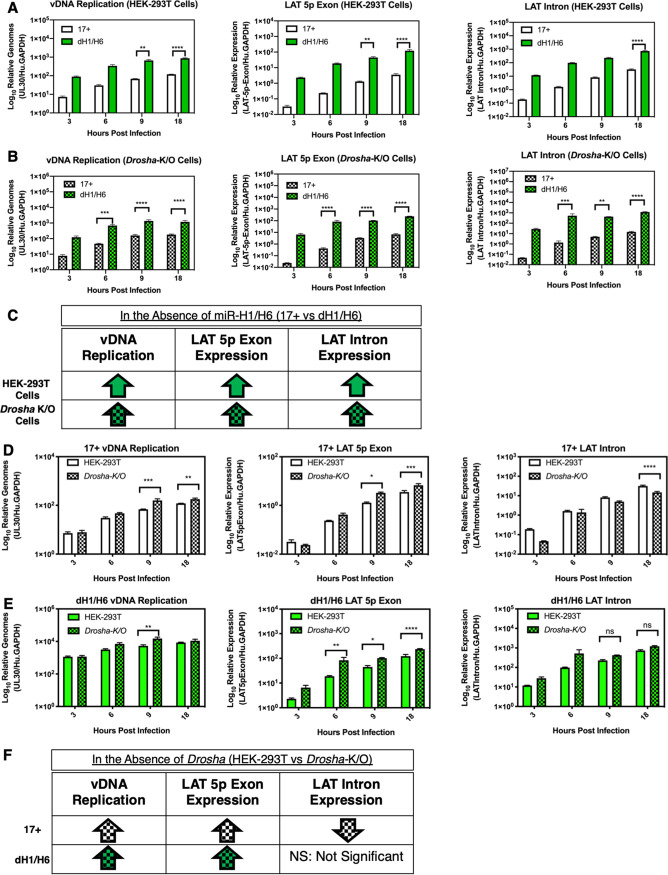
The LAT primary transcript is 8.3 kb and is spliced to form a stable 2-kb intron. The 5p and 3p exons are degraded rapidly, yet the mechanism for this rapid degradation of the spliced transcript remains unknown. Following the discovery of the viral miRNAs in this region, it was hypothesized that the spliced product acts as an lncRNA precursor for the viral miRNAs in this region, which includes miR-H7, miR-H8, and miR-H14, but this hypothesis has yet to be experimentally validated (5). In an attempt to address this question and determine whether Drosha-mediated miRNA processing is involved in LAT accumulation, we utilized a human embryonic kidney (HEK-293T) CRISPR Drosha−/− knockout (K/O) cell line (36) and compared the results to those for the wild-type HEK-293T cell line in the context of 17syn+ or 17dmiR-H1/H6 infection (Fig. 6). In theory, miR-H1/H6 would not be processed for 17syn+ in the Drosha-K/O cells, allowing a possible rescue of the miR-H1/H6 phenotypes. However, in designing this experiment, we also appreciated that the Drosha-K/O cells would ablate the processing of all other host and viral miRNAs in the canonical miRNA biogenesis pathway, resulting in the dysregulation of many biological processes mediated downstream of both host and viral miRNA regulation.
FIG 6.
Latency-associated transcript accumulation is Drosha dependent. Differences in human embryonic kidney cells (HEK-293T) and a CRISPR knockout of Drosha (HEK-293t-Drosha-K/O) cell lines were examined following infection with 17syn+ or 17dmiR-H1/H6 at an MOI of 5. The effects of the miR-H1/H6 deletion on vDNA replication and LAT expression were assessed in HEK-293T (A) and Drosha-K/O (B) cells. Relative genomes were determined by gDNA qPCR probing for UL30, the viral polymerase, normalized to human GAPDH. RT-qPCR of the LAT 5p exon and LAT intron was normalized to human GAPDH. (C) Summary of the findings comparing the two viruses. Comparison of results in HEK-293T and HEK-293t-Drosha-K/O cells for 17syn+ (D) and 17dmiR-H1/H6 (E). (F) Summary of the findings comparing the two cell lines. These data represent three independent experiments, and significance was determined by ordinary one-way ANOVA with Sidak’s multiple-comparison test (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; ****, P < 0.00001).
First, we assessed the effect of deleting miR-H1/H6 by comparing the results between 17syn+ and 17dmiR-H1/H6 during acute infection in HEK-293T cells (Fig. 6A). Consistent with our findings in rabbit skin cells (Fig. 2A and 5A), we found that the deletion of miR-H1/H6 resulted in increased vDNA replication and increased LAT expression. These results indicate that 17dmiR-H1/H6 infection of HEK-293T cells is similar to that observed for rabbit skin cells and suggests that functional miR-H1/H6 is responsible in part for repressing vDNA replication and LAT expression. In the Drosha-K/O HEK-293T cells, we also found increases in vDNA replication and LAT expression for both viruses (Fig. 6B), suggesting miR-H1/H6 (or their deletion) are not solely responsible for regulating vDNA replication and LAT expression. There are likely other biological processes downstream of global miRNA regulation, apart from those attributed to miR-H1/H6, contributing to this phenotype in the Drosha-K/O cells. Alternatively, the results for the miR-H1/H6 deletion in the absence of Drosha may be alluding to a function for miR-H1/H6, such as antisense regulation, cis-regulation, or noncanonical miRNA biogenesis (see Discussion), which may be amplified by the loss of global Drosha-mediated miRNA regulation.
We next assessed the impact of the absence of Drosha, comparing the results between the HEK-293T cells and the Drosha-K/O cells. We found that for 17syn+ in the Drosha K/O line, vDNA replication significantly increased by 9 hpi (Fig. 6D), which was expected, since it is known that host miRNAs can be antiviral and are perturbed during HSV-1 infection (19, 37). We also examined the effects of Drosha on the accumulation of the LAT 5p exon and the LAT intron by RT-qPCR. The LAT intron values are higher than those of the LAT 5p or 3p exon, since, following splicing, the intron forms a highly stable lariat structure (38). We found that in the absence of Drosha, the relative accumulation of the LAT 5p exon increased and LAT intron levels decreased during wild-type infection. To our knowledge, this is the first experimental evidence to show that Drosha, presumably through the processing of miRNAs, is responsible for degradation of the LAT primary transcript. This suggests functional miRNAs processed by Drosha, or regulated by Drosha-mediated miRNAs, are responsible for repressing vDNA replication and LAT 5p exon levels but facilitate LAT intron accumulation.
Deletion of miR-H1/H6 in the Drosha-K/O cells (Fig. 6E) also resulted in increased vDNA replication and LAT 5p exon levels, suggesting that functional miRNAs other than miR-H1/H6 are responsible, at least in part, for repressing vDNA replication and LAT 5p exon levels. However, the miR-H1/H6 deletion in the Drosha-K/O cells had no effect on restoring LAT intron levels, suggesting wild-type LAT intron accumulation is dependent on functional miR-H1/H6 by an unknown mechanism independent of global Drosha-mediated miRNAs and their downstream targets.
The increase in LAT 5p exon accumulation in Drosha-K/O cells was also observed in the 17dmiR-H1/H6 infections, but the LAT intron accumulation did not significantly decrease, as was observed in the wild-type infection, suggesting the dH1/H6 deletion alters the splicing or stability of the LAT intron by a non-miRNA-mediated mechanism. Nonetheless, these findings clearly demonstrate that (i) the deletion of miR-H1/H6 results in an overall increase in LAT accumulation, consistent with the increase in LAT region miRNAs seen in Fig. 5, and (ii) these effects are Drosha dependent, suggesting that the processing of the miRNAs from the LAT plays a significant role in the regulation of LAT accumulation.
DISCUSSION
Much effort in the field of HSV-1 latency has been focused on the characterization of viral noncoding RNAs (ncRNAs) that are transcribed during latency, with particular focus on the LAT. Hallmark phenotypes attributed to the LAT include facilitating establishment and maintenance of latency (39, 40) and also facilitating the ability of the virus to reactivate (12, 16, 41, 42). These hallmark phenotypes were assigned to the LAT by comparing recombinant viruses with various deletions in the LAT promoter (for a comprehensive review, see Phelan et al. [3]). A limitation of these studies is that by deleting the LAT promoter, the expression of other ncRNAs within the LAT region, namely, a number of miRNAs, was also impacted (Fig. 5 and 6). Notably, several of the LAT promoter deletions also deleted or reduced the expression of miR-H1 and miR-H6 (reviewed in reference 3). Therefore, we have examined the phenotypes attributed to miR-H1-5p and miR-H6-3p, utilizing a recombinant virus in the 17syn+ wild-type background in which the mature miRNAs for miR-H1/H6 (25 bp) are deleted. It should be noted that this deletion is at least 70 bp from the nearest known LAT promoter element. Using this recombinant with the 25-bp deletion (17dmiR-H1/H6), we found slight increases in viral DNA replication and viral yields in rabbit and mouse cell lines relative to wild-type 17syn+ (Fig. 2), along with increases in LAT 5p exon and LAT region miRNA expression during acute infection in cell culture (Fig. 5). In the mouse footpad infection model, 17dmiR-H1/H6 was slightly more virulent than 17syn+ (Fig. 3). However, by far the most significant phenotype we observed for 17dmiR-H1/H6 was decreased reactivation from latency in the mouse explant and rabbit ocular models (Fig. 3 and 4). It is noteworthy that the degree of reduced reactivation efficiency was similar to that observed previously for LAT promoter mutants. Significantly, in both the mouse and rabbit models, the decrease in reactivation was present in the absence of any detectable decrease in the establishment of latency. This is consistent with a recent study utilizing an LAT promoter deletion virus (17ΔPst) with significantly reduced viral miRNA expression, which also had no impact on the establishment of latency in mice (6). The results presented here represent the first robust phenotype described for a recombinant of HSV-1 deleted specifically in miR-H1/H6 and indicates that one or both of these miRNAs function to enhance the ability of HSV-1 to reactivate from latency.
Previous studies with miR-H1/H6 recombinant viruses.
Previous studies examining HSV-1 miR-H1/H6 with recombinant viruses have not detected any phenotypes. An analysis of the growth kinetics of a recombinant virus lacking miR-H1/H6, in the KOS wild-type background, found no significant differences during acute infection in cell culture (26). As stated earlier, we utilized the 17syn+ wild-type strain as the backbone for our miRNA mutant due to the higher pathogenicity, an LD50 of 500 PFU in the mouse footpad model, and the efficient reactivation phenotype in the rabbit ocular model. These robust phenotypes for 17syn+ improve the sensitivity necessary for observing even subtle differences. A lack of virulence and reactivation phenotypes for miR-H1/H6 in the KOS background would be consistent with decreased virulence in the footpad and limited reactivation in rabbit ocular models (26–28). Overall, differences in virulent or avirulent wild-type and infection models (murine ocular, murine footpad, or rabbit ocular) may explain the lack of phenotypes for miR-H1/H6 in the literature and the phenotypes described here.
miR-H6 regulation of ICP4.
The possible mechanisms of miR-H1/H6 are likely layered in complexity. Since miR-H6 may target ICP4, the regulation of ICP4 and its role in repressing LAT expression must be discussed. ICP4 is an important transactivator of other viral genes, and in high abundance it can bind its own promoter and repress its own expression later during infection, and it has been shown that ICP4 can bind the promoter of LAT and limit LAT expression (43). Therefore, it is possible that miR-H6 expression flattens the curve of ICP4 expression by reducing its peak levels before it eventually represses its own expression and inhibits LAT expression. Therefore, the effect of miR-H6 during the acute infection is predicted to be subtle. Indeed, in the case of herpes simplex virus 2 (HSV-2), a recombinant virus lacking the HSV-2 ortholog of miR-H6 exhibited no significant differences in absolute ICP4 levels during acute infection in culture and no difference in the guinea pig reactivation model (44). In addition, an HSV-2 recombinant virus in which the ICP4-binding sequence in the LAT promoter was deleted (Fig. 1B) exhibited no change in replication kinetics, virulence, viral miRNA expression, or reactivation (45), providing further evidence that an ICP4-mediated repression of LAT expression mechanism for miR-H6 is unlikely.
Antisense regulation by miR-H1/H6.
Since, in the case of HSV-1, we are not able to specifically delete miR-H6 separately from miR-H1 due to their overlap, it is intriguing to speculate that miR-H1 plays a dominant role in the reactivation phenotype that we identify here by deleting both miR-H1 and miR-H6. A recent study examined miR-H6-5p and miR-H1-3p, which are considered the star strands of the miR-H1/H6 miRNA duplex (Fig. 1C), meaning they are presumably degraded quickly (46), as opposed to the miRNAs examined here, miR-H1-5p/H6-3p. These authors found miR-H6-5p accumulated more rapidly than its antisense partner, miR-H1-3p, which accumulated about 6 h later (46). To examine the functions of miR-H6-5p, miR-H6-5p was inhibited with the transfection of a plasmid expressing miR-H1-3p or an miRNA sponge for miR-H6-5p transiently, resulting in reduced vDNA replication, viral yields, and viral protein levels of the F wild-type HSV-1 strain in culture. To examine the effects of inhibiting miR-H6-5p, a recombinant virus was constructed expressing an miR-H6-5p sponge between the viral genes UL3 and UL4 and resulted in reduced viral yields and viral protein levels during acute infection in culture and no differences in viral gene expression upon reactivation in the mouse trigeminal ganglia explant-induced reactivation model compared to that of wild-type virus (46). Overall, these results suggest that the antisense partners miR-H1 and H6 play a minor role in reducing lytic gene expression during acute infection, consistent with our results, but we are not convinced by the negative results in vivo, since the miRNA sponge was encoded between the lytic genes UL3 and UL4, which likely would not be expressed during latency, since this part of the genome is heterochromatic. This highlights the limitations of an ectopic expression approach for examining miRNA effects during HSV-1 latency.
Consideration of the complexity of the LAT locus.
Further resolution of miR-H1/H6 mechanisms will require specific knockdown of the individual precursor miRNAs, possibly by ribozymes (47), and this work is underway. However, this approach is not straightforward, since there are multiple possible primary precursor lncRNAs for miR-H1 and -H6, including AL (48) or TAL (49) for H6 and UOL (50) or ATAL for H1 (49), and the expression of these transcripts may be precisely regulated for an efficient latent infection. Further, an alternatively spliced transcript, named pri-miR-H6/UDG, derived from UL1 and UL2, a viral uracil-DNA glycosylase (UDG) gene, was found to be expressed in KOS (51). Pri-miR-H6/UDG expression was found to be dependent on ICP27 (51), implicating differential regulation of a spliced miR-H6 precursor during acute infection and a Drosha-processed miR-H6 precursor in the absence of ICP27 during the establishment of latency. Importantly, this suggests a possible cis-regulation mechanism of the biallelic LAT miRNAs that would be regulated differently if they were next to UL1 and UL2 (tRL) or in the iRL, where insulator elements were recently shown to influence epigenetic repression during HSV-1 latency (52). In addition, these multiple cis elements in the LAT promoter region make ectopic expression approaches likely to influence the epigenetic repression of an ectopically expressed miR-H1 or miR-H6 in another part of the virus. For these reasons, we did not pursue an ectopic expression approach where miR-H1/H6 would be expressed in a different part of the HSV-1 genome, and we are designing ribozymes to specifically target the miR-H1/H6 precursors.
Previous studies with LAT promoter mutants and ectopic expression of miR-H1/H6.
The ectopic expression of miR-H1/H6 was examined retrospectively. A recombinant virus, dLAT2903, in the McKrae background lacks the LAT promoter and the first 1.5 kb of LAT and exhibits a reduced reactivation phenotype in the rabbit ocular model (53). With another recombinant virus, LAT15a, the LAT promoter (including miR-H1/H6) and the first 1.5 kb of LAT were deleted (as in dLAT2903), and this segment was inserted between UL37 and UL38 to examine the ectopic expression of LAT from its native promoter (15). These authors found that the spontaneous reactivation phenotype of LAT was rescued (15). These results support our findings here, that miR-H1/H6 are involved in HSV-1 latency and reactivation, because miR-H1/H6 also were ectopically expressed from the LAT promoter region included in LAT15a. However, a follow-up study examining the ectopic expression of the LAT promoter and a smaller segment of the LAT 5p exon resulted in only a partial rescue of the spontaneous reactivation phenotype (54). This raises a question regarding whether or not ectopic expression of the LAT promoter (including miR-H1/H6) and partial 5p exon sequences fully or partially restore spontaneous reactivation.
Different models to study HSV-1 reactivation.
We did not measure spontaneous reactivation for 17dmiR-H1/H6 in rabbits in this study and instead utilized epinephrine-induced reactivation of latently infected (>30 dpi) rabbits. These conflicting results warrant a discussion regarding spontaneous versus induced reactivation in the rabbit ocular infection model. Following ocular infection of rabbits with HSV-1, there is an initial period of sustained acute shedding in the eyes that can be assessed by swabbing of the eyes and assaying for infections virus. This shedding peaks at days 3 to 4 and generally decreases to undetectable by days 8 to 10, while viral lytic transcripts can still be detected in the TG until days 16 to 20. Spontaneous reactivation analysis makes use of the observation that one can detect frequent episodes of positive ocular swabs between days 20 and 30. After 30 days, ocular shedding is infrequent. It has been argued that spontaneous reactivation actually is an incomplete resolution of the acute infection period as the virus establishes stable latency, perhaps through more complete heterochromatinization of the viral genomes. Nonetheless, this spontaneous reactivation phenomenon mirrors the more frequent appearance of lesions that occurs in humans immediately after primary infection. In contrast, adrenergically induced reactivation is performed on rabbits that are in a more stable state of latency and involves a synchronous induction of reactivation by iontophoresis of epinephrine, an adrenergic agent. This model closely parallels the periodic reactivation episodes that occur throughout HSV patients’ lives.
Over the years, differences have been observed between spontaneous reactivation and induced reactivation phenotypes for different HSV-1 virus strains and mutants. With recombinant virus X10-13 (55), which had a deletion of the LAT promoter and a portion of the 5′ LAT and was an HSV-1/2 chimera (HSV2-HG52 sequence inserted close to UL29), explant-induced reactivation (55) and spontaneous reactivation was observed (56). However, induced reactivation yielded a reduced reactivation phenotype compared to that of a rescue virus (56). However, for the most part, phenotypes for LAT mutant viruses are consistent between models (10, 28, 57–61).
Impact of miR-H1/H6 deletion on ncRNA expression from the LAT locus.
In this study, we further examined the roles of miR-H1/H6 on the expression of other ncRNAs and transcripts in the LAT region and found that LAT region expression may be downstream of Drosha-mediated processing of miR-H1/H6 (Fig. 6). We experimentally showed that Drosha is responsible for the rapid degradation of the spliced LAT primary transcript, and the effects of Drosha on the accumulation of LAT differ between 17dmiR-H1/H6 and 17syn+. In the absence of Drosha, there were significant decreases in the LAT intron and significant increases in the LAT 5p exon (Fig. 6). Since canonical miRNA biogenesis is Drosha mediated, in the Drosha-K/O cells, miR-H1/H6 most likely would not be processed and functional in 17syn+-infected Drosha-K/O cells. If the only impact of the miR-H1/H6 deletion was the loss of a functional miR-H1/H6, one hypothesis is that 17dmiR-H1/H6 would not have a phenotype similar to that of 17syn+ in the Drosha-K/O cells. The increased vDNA replication and LAT expression for 17dmiR-H1/H6 compared to that of 17syn+ found in HEK-293T cells was also found in the Drosha-K/O cells. These results suggest the differences in vDNA replication and LAT expression for 17dmiR-H1/H6 are independent of Drosha. A broader conclusion would be that the phenotypes found here cannot be attributed to miR-H1/H6. However, since Drosha mediates canonical miRNA biogenesis, the majority of host and viral miRNAs also would not be functional, and miRNA regulation can have many effects downstream from any given target. Therefore, we favor the most obvious interpretation that, in a wild-type infection setting, miR-H1/H6 is responsible for the phenotypes presented here. It is possible that the phenotype for 17dmiR-H1/H6 compared to 17syn+ in Drosha-K/O cells is attributed to the miR-H1/H6 deletion itself, and the effects may be amplified by the absence of global miRNA regulation. The phenotypes found for 17dmiR-H1/H6 compared to 17syn+ in the Drosha-K/O cells are also suggestive of a possible compensation mechanism, where the observed increased in LAT and viral miRNA accumulation (Fig. 5) may have been in response to the miR-H1/H6 deletion. The phenotypes observed following the deletion of miR-H1/H6 and the combined dysregulation of ncRNAs in this region highlight the complexity of the LAT locus and the importance of its precise regulation. The export of precursor miRNAs in the canonical miRNA biogenesis pathway has been shown to be inhibited by HSV-1 during acute infection in a vDNA replication- and ICP27-dependent manner, resulting in a high precursor-to-mature miRNA ratio (19). More investigation is required to determine if HSV-1 miRNAs, including miR-H1/H6, are regulated in both a canonical Drosha-dependent manner and possibly a Drosha-independent mirtron mechanism arising from the alternative splicing of pri-miR-H6/UDG.
Possible functions of miR-H1/H6.
Based on the data presented in this study, we propose that there are multiple functions for miR-H1/H6. During wild-type infection, miR-H1/H6 negatively regulate lytic transcription, resulting in an acute infection with moderate levels of pathogenesis. This facilitates the establishment of dynamic latency, whereby lytic genes are epigenetically repressed to evade the immune system, and this repression occurs in a way that retains the capacity to reactivate efficiently. In the absence of miR-H1/H6, there is a modest increase in pathogenesis, which results in a latent infection with a reduced ability to reactivate. Further, the expression of other viral miRNAs in the LAT region is increased. This could be very significant, given the evidence that miR-H2, miR-H3, and miR-H4 limit viral replication and pathogenesis by targeting and downregulating important viral genes, including ICP0 (miR-H2) and ICP34.5 (miRs H3 and H4) (8–11). Thus, when miRs H1 and H6 are absent, either by deletion or by reduced expression, the virus may possess an alternate program to regulate lytic gene expression by an increase in the expression of the other viral miRNAs in this region. We hypothesize that the reduced ability of 17dmiR-H1/H6 to reactivate ultimately may be due to qualitative differences in the establishment of latency resulting from the altered acute infection program. This may be manifested in the ratios of heterochromatin marks on lytic genes that are constitutive (H3K9me3) or facultative (H3K27me3), similar to the differences found for other LAT promoter mutants (13, 62, 63).
Finally, it is possible and likely that miR-H1/H6 target multiple host and viral transcripts, some of which may be cell type or species specific. For instance, in human corneal epithelium, the transfection of miR-H6 mimics was found to result in decreased IL-6 secretion and decreased viral replication (17), suggesting miR-H6 has multiple roles in balancing lytic replication relative to host immune responses. Therefore, it is possible that miR-H1 and miR-H6 will be found to target other viral and cellular transcripts, including host restriction factors, since it is advantageous for a virus with limited coding capacity to globally modulate its lytic and latent cycles. Future analyses using modified cross-linking and sequencing of hybrids (qCLASH) (64) and miRNA mimic and sponge approaches will be needed to further elucidate the importance and global nature of individual HSV-1 miRNAs in the regulation of lytic and latent infections.
In conclusion, our studies demonstrate that HSV-1 miRNAs miR-H1 and miR-H6 are likely important regulators of HSV-1 reactivation. Herpesviruses have evolved complex regulation mechanisms to limit pathogenesis in order to increase fitness and transmission. Evolutionarily, viruses with uncontrolled lytic transcription paired with frequent and severe reactivation episodes would be selected against in favor of viruses with stealthy shedding of virus, sensed and regulated by both host and viral factors, which set the stage for efficient and dynamic HSV-1 latency.
MATERIALS AND METHODS
Cell culture, viruses, and infections.
Low-passage immortalized rabbit skin (RS) cells (gift from B. Roizman) were cultured (passages 5 to 30) in minimum essential medium (MEM; 11095-098; Gibco) complete with 5% bovine serum (26170-043; Gibco) and 1% PSG (100 U penicillin, 100 mg/ml streptomycin, 0.292 mg/ml l-glutamine; SV30082.01; HyClone). Neuro2A cells were obtained from the ATCC (CCL-13) and were cultured in MEM plus 10% fetal bovine serum (FBS; 16140-0630; Gibco) with 1% nonessential amino acids (NEAA; SH30238.010; HyClone) and 1% PSG. HEK-293T (CRL-11268; ATCC) and HEK293T-Drosha-K/O (gift from S. Gu) cells were cultured in Dulbecco’s modified Eagle medium (DMEM; MT100114CV; ThermoFisher) with 10% FBS and 1% PSG. Low-passage-number wild-type virus 17syn+ was obtained from J. Stevens and used between passages 5 and 7. Cells were seeded at 50% density in 24-well plates and infected at the indicated multiplicity of infection (MOI) for 1 h in a volume of 250 μl, with gentle rocking every 15 min. Following the 1-h inoculation, the cells were washed with phosphate-buffered saline (PBS; SH30256.01; HyClone) and overlaid with complete medium. For plaque assays, cells and supernatants were harvested at the corresponding time points and lysed by 1 freeze (−80°C)/thaw cycle. Limiting dilution series (1:10) were used to determine viral yields. For plaque assays, the overlay medium (complete) was supplemented with 0.3 mg/ml human IgG serum (18640-100MG; Sigma), and 48 h later, cells were fixed and stained with crystal violet solution (0.01% g/ml crystal violet [C0775-100G; Sigma] with 22% 190-proof ethanol [2801G; Decon Labs] and deionized water). Plaques were counted and averaged across three technical and three biological replicates.
Construction of recombinant virus 17dmiR-H1/H6.
17dmiR-H1/H6 includes a biallelic deletion of 25 nucleotides from the TRL/IRL segments of the genome (Fig. 1). The In-Fusion advantage PCR cloning kit (639649; TaKaRa) was used to subclone ∼150-bp recombination arms and the deletion corresponding to the seed sequences of miR-H1-5p and miR-H6-3p into a pBluescript plasmid by following the manufacturer’s protocol. Primers used to construct the recombination plasmid were the following: BS-H1-5f 5p, TCCCCCGGGCTGCAGGAATTCCCAGTCTCCTCGCCTTCTC; D-H1-5r 5p, ATCAGGACCGTTCCCCTCGGTTGTTC; D-H1-3f 5p, GGGAACGGTCCTGATACCCATCCTACACCC; and BS-H1-3r 5p, GATAAGCTTGATATCGAATTCCTGCCTCTGCCGCTTGTG. The wild-type strain 17syn+ HSV-1 DNA was transfected along with the recombination plasmid by calcium phosphate precipitation at a ratio of 10:1 as described previously (65). Three independent isolates were screened by dot blot TMAC hybridization using oligonucleotide probes and plaque purified at least 5 times as described previously (65). The biallelic deletion was confirmed by PCR, Sanger sequencing, and Southern blotting (data not shown), and no off-target mutations were found by Illumina sequencing. The sequencing data for the 17dmiR-H1/H6 virus are readily available (SRA accession number SRR9121361).
Reverse transcription and qPCR.
Genomic DNA (gDNA) and RNA were extracted using DNA and RNA miniprep kits (D7001; Zymo Research) by following the manufacturer’s instructions. RNA was DNase treated using TURBO DNase (AM2239; ThermoFisher) and reverse transcribed using random decamers (5722G; ThermoFisher/Invitrogen) and the Omniscript reverse transcription kit (205113; Qiagen). gDNA or cDNA was subjected to qPCR (StepOnePlus real-time PCR system; 4376600; ThermoFisher/Applied Biosystems) with fast qPCR master mix (4352042; ThermoFisher/Applied Biosystems) and custom primer probes (Table 1). Relative genomes (probing for UL30, the viral polymerase gene) or relative expression (relative to human [assay Hs02786624_g1; ThermoFisher/Applied Biosystems], mouse [assay Mm99999915_g1], or rabbit [assay AI09030] glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were determined based on standard curves from 1:10 dilutions of amplicon PCR products. For miRNA accumulation assays, small RNAs were extracted using the Zymo Quick-RNA miniprep kits (R1055) by following the manufacturer’s protocol for isolating small RNAs (<200 nucleotides [nt]) and TURBO DNase treated. Stem-loop reverse transcription primers (miR-H1-5p assay 464923_mat, miR-H2-3p assay 005632, miR-H3-3p assay 97242_mat, miR-H4-3p assay 197191_mat, miR-H5-3p assay 197213_mat, miR-H6-3p assay 197219_mat, miR-H7-5p assay 462602_mat, and miR-H8-5p assay 241862_mat) were used for reverse transcription (Table 1). For extracting DNA from animal tissue, murine dorsal root ganglia or rabbit trigeminal ganglia were dissected, and cells were suspended and snap-frozen in RNA lysis buffer (D7001; Zymo Research). The ganglia then were sonicated (Bioruptor-Twin; UCD-400 TO; Diagenode) on the high setting for 30 s on and 30 s off for 3 to 5 cycles, followed by gDNA extraction by following the manufacturer’s protocol.
TABLE 1.
Custom TaqMan primer/probe sequences
| Transcript | Forward primera | Reverse primer | Probe |
|---|---|---|---|
| LAT 5p exon | GGCTCCATCGCCTTTCCT | AAGGGAGGGAGGAGGGTACTG | TCTCGCTTCTCCCC |
| LAT intron | CGCCCCAGAGGCTAAGG | GGGCTGGTGTGCTGTAACA | CCACGCCACTCGCG |
| UL30 | AGAGGGACATCCAGGACTTTGT | CAGGCGCTTGTTGGTGTAC | ACCGCCGAACTGAGCA |
The primer and probe sequences are in the 5p-to-3p orientation.
Murine footpad infections.
Four- to 6-week-old female ND-40 Swiss-Webster mice were pretreated with saline injections in both rear footpads 2 h prior to infection. The mice were anesthetized and the cornified epithelium layer abraded with an emery board, and 10 to 25 μl of diluted virus stock was applied to each footpad and allowed to absorb for 1 h until the mice regained consciousness. Mice were monitored twice daily during the acute infection (5 to 15 days postinfection) and humanely euthanized prior to succumbing to infection based on the University of Florida’s Institutional Animal Care and Use Committee-approved defined endpoints. Mice that survived the acute infection had no obvious signs of infection, and latency was established. Twenty-eight days postinfection, mice were euthanized and the L4 to L6 DRG were dissected and analyzed by qPCR as described above (65). For explant-induced reactivation assays, individual DRG were dissected 28 dpi and seeded onto RS cell monolayers in a 24-well format. Each well was monitored for cytopathic effects daily and scored positive or negative for reactivation over 21 days as described previously (30).
Rabbit ocular infection model.
Male and female New Zealand White rabbits (2 to 3 kg) were infected in each cornea at doses of 200,000 PFU, as described previously (47). Briefly, eyes were scarified and inoculated with 25 μl virus in MEM plus 5% FBS and massaged for 30 s. Eyes were examined by slit lamp microscopy (SLE) with 0.1% fluorescein 3, 5, 7, 10, and 14 dpi and assessed for the presence of ocular lesions. Lesion scores were determined by the surface area of cornea the lesion occupied, as previously described. Rabbits were considered latent when no corneal lesions were observed. Ocular swabs were done to confirm that no infectious virus could be detected 30 dpi. Thirty-one days postinfection, rabbits were subjected to transcorneal iontophoresis of 0.01% epinephrine (0.8 mA for 8 min) daily for 3 days for reactivation, as previously described. Ocular swabs were taken from each eye daily for 12 days postreactivation, and the presence of infectious virus was determined by plaque assay as a measure of reactivation. By 12 days postreactivation, infectious virus was no longer detected in the ocular swabs, and trigeminal ganglia were dissected and subjected to qPCR as described above.
HSV-1 genome sequencing.
A total of 500 ng of nucleocapsid viral genomic DNA was purified (66) and sonicated in a Covaris Model S2 (intensity, 5; duty cycle, 10; cycle per burst, 200; treatment time, 180 s). Libraries were made using SPRIworks System I for the Illumina Genome Analyzer (A88267; Beckman Coulter) and Illumina DNA barcodes. Samples were pooled at equal nanomolar concentrations and sequenced on MiSeq with 2× 75-nt paired-end reads. MiSeq reads were trimmed using Trimmomatic, quality filtered using FASTQC, and mapped to the HSV-1 17syn+ genome (GenBank accession no. NC_001806.2) using SeqMan NGen (version 17.0.2.2). Out of 6.0 million paired reads, template coverage was 99.897% with median coverage of 6,797×. Variants were analyzed using SeqMan Ultra (version 17.0.2 [1]). The intended 25-bp deletion of mature miR-H1-5p/miR-H6-3p was observed at NC_001806.2 nt 8021 to 8045 and 118329 to 118353, representing the tRL and iRL segments, respectively. Ten nonsynonymous substitutions were observed but did not result in nonsense, no-start, no-stop, or frameshift mutations.
Statistical methods.
All statistics were done using GraphPad/Prism (version 8.3.0). Data from cell culture experiments represent at least three independent experiments, acute mouse infections were repeated independently three times, and the reactivation experiments in the mouse and rabbit models were not repeated, although the similar results in the two different reactivation models strongly suggest these results were cross-validated. Significance for experiments with one time point was determined by unpaired t tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Significance for time course infection experiments in cell culture was determined by ordinary two-way ANOVAs with Sidak’s multiple-comparison test. In the mouse modified LD50 experiment, the percent survival relative to dose was fit using a nonlinear regression model to calculate the modified LD50s. Survival was found to be significantly different at 500 PFU (Mantel-Cox log rank test). Significance for cumulative reactivation in the mouse explant and adrenergically induced rabbit ocular reactivation models was determined by Mann-Whitney tests.
Data availability.
The sequencing data for the 17dmiR-H1/H6 virus are available in the Sequence Read Archive (SRA) under accession number SRR9121361.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants R01-AI097376 (D.C.B.), R01-AI048633 (D.C.B.), and F31-AI140713 (E.R.B.) from the NIH. E.R.B. was supported by a Florida Education Fund McKnight Doctoral Fellowship, a UF Graduate School Fellowship, a UF Top-Up Fellowship, and the Laura & Kenneth Berns Award from the UF Genetics Institute. S.N. was supported by a Japan Herpesvirus Infections Forum Scholarship Award in Herpesvirus Infection Research. D.M.N. was supported by NIH grant P30-EY016665, granted to the University of Wisconsin, and an Unrestricted Grant from Research to Prevent Blindness, granted to the Department of Ophthalmology and Visual Sciences at the University of Wisconsin.
We thank S. Gu for the HEK-293T Drosha K/O cell line. Finally, we acknowledge T. Edwards, D. Phelan, T. Grams, J. Singh, W. Canty, and A. Adolphson for discussions and comments in preparing the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bradley H, Markowitz LE, Gibson T, McQuillan GM. 2014. Seroprevalence of herpes simplex virus types 1 and 2—United States, 1999–2010. J Infect Dis 209:325–333. doi: 10.1093/infdis/jit458. [DOI] [PubMed] [Google Scholar]
- 2.Liesegang TJ. 1989. Epidemiology of ocular herpes simplex: natural history in Rochester, Minn, 1950 through 1982. Arch Ophthalmol 107:1160–1165. doi: 10.1001/archopht.1989.01070020226030. [DOI] [PubMed] [Google Scholar]
- 3.Phelan D, Barrozo ER, Bloom DC. 2017. HSV1 latent transcription and non-coding RNA: a critical retrospective. J Neuroimmunol 308:65–101. doi: 10.1016/j.jneuroim.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 5.Umbach J, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM. 2011. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology 417:239–247. doi: 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. 2013. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol 87:6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, Brown D, Osorio N, Hsiang C, Li L, Chan L, BenMohamed L, Wechsler SL. 2015. A herpes simplex virus type 1 mutant disrupted for microRNA H2 with increased neurovirulence and rate of reactivation. J Neurovirol 21:199–209. doi: 10.1007/s13365-015-0319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, Brown D, Osorio N, Hsiang C, BenMohamed L, Wechsler SL. 2016. Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2. J Neurovirol 22:38–49. doi: 10.1007/s13365-015-0362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan D, Pesola JM, Li G, McCarron S, Coen DM. 2017. Mutations inactivating herpes simplex virus-1 miR-H2 do not detectably increase ICP0 gene expression in infected cultured cells or mouse trigeminal ganglia. J Virol 91:e02001-16. doi: 10.1128/JVI.02001-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, Knipe DM, Schaffer PA. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol 63:2893–2900. doi: 10.1128/JVI.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobson AT, Sederati F, Devi-Rao G, Flanagan WM, Farrell MJ, Stevens JG, Wagner EK, Feldman LT. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J Virol 63:3844–3851. doi: 10.1128/JVI.63.9.3844-3851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perng GC, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol 70:976–984. doi: 10.1128/JVI.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom DC, Devi-Rao GB, Hill JM, Stevens JG, Wagner EK. 1994. Molecular analysis of herpes simplex virus type 1 during epinephrine-induced reactivation of latently infected rabbits in vivo. J Virol 68:1283–1292. doi: 10.1128/JVI.68.3.1283-1292.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan F, Liao J, Huang Q, Nie Y, Wu K. 2012. HSV-1 miR-H6 inhibits HSV-1 replication and IL-6 expression in human corneal epithelial cells in vitro. Clin Dev Immunol 2012:192791–192798. doi: 10.1155/2012/192791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol 84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan D, Li G, Morris-Love J, Qi S, Feng L, Mertens ME, Jurak I, Knipe DM, Coen DM. 2019. Herpes simplex virus 1 lytic infection blocks microRNA (miRNA) biogenesis at the stage of nuclear export of pre-miRNAs. mBio 10:e02856-18. doi: 10.1128/mBio.02856-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du T, Han Z, Zhou G, Zhou G, Roizman B. 2015. Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation. Proc Natl Acad Sci U S A 112:E49–E55. doi: 10.1073/pnas.1422657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munson DJ, Burch AD. 2012. A novel miRNA produced during lytic HSV-1 infection is important for efficient replication in tissue culture. Arch Virol 157:1677–1688. doi: 10.1007/s00705-012-1345-4. [DOI] [PubMed] [Google Scholar]
- 23.Held K, Junker A, Dornmair K, Meinl E, Sinicina I, Brandt T, Theil D, Derfuss T. 2011. Expression of herpes simplex virus 1-encoded microRNAs in human trigeminal ganglia and their relation to local T-cell infiltrates. J Virol 85:9680–9685. doi: 10.1128/JVI.00874-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naqvi AR, Shango J, Seal A, Shukla D, Nares S. 2018. Viral miRNAs alter host cell miRNA profiles and modulate innate immune responses. Front Immunol 9:433. doi: 10.3389/fimmu.2018.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng K, Liu Q, Wang S, Ren Z, Kitazato K, Yang D, Wang Y. 2018. HSV-1-encoded microRNA miR-H1 targets Ubr1 to promote accumulation of neurodegeneration-associated protein. Virus Genes 54:343–350. doi: 10.1007/s11262-018-1551-6. [DOI] [PubMed] [Google Scholar]
- 26.Jurak I, Silverstein LB, Sharma M, Coen DM. 2012. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J Virol 86:10093–10102. doi: 10.1128/JVI.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javier RT, Sedarati F, Stevens JG. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234:746–748. doi: 10.1126/science.3022376. [DOI] [PubMed] [Google Scholar]
- 28.Hill JM, Field MAR, Haruta Y. 1987. Strain specificity of spontaneous and adrenergically induced HSV-1 ocular reactivation in latently infected rabbits. Curr Eye Res 6:91–97. doi: 10.3109/02713688709020074. [DOI] [PubMed] [Google Scholar]
- 29.Bloom DC, Stevens JG. 1994. Neuron-specific restriction of a herpes simplex virus recombinant maps to the UL5 gene. J Virol 68:3761–3772. doi: 10.1128/JVI.68.6.3761-3772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devi-Rao GB, Bloom DC, Stevens JG, Wagner EK. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J Virol 68:1271–1282. doi: 10.1128/JVI.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webre JM, Hill JM, Nolan NM, Clement C, McFerrin HE, Bhattacharjee PS, Hsia V, Neumann DM, Foster TP, Lukiw WJ, Thompson HW. 2012. Rabbit and mouse models of HSV-1 latency, reactivation, and recurrent eye diseases. J Biomed Biotechnol 2012:612316. doi: 10.1155/2012/612316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner EK, Bloom DC. 1997. Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev 10:419–443. doi: 10.1128/CMR.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon BS, Gangarosa LP, Burch KD, deBack J, Hill JM. 1981. Induction of ocular herpes simplex virus shedding by iontophoresis of epinephrine into rabbit cornea. Investig Ophthalmol Vis Sci 21:442–449. [PubMed] [Google Scholar]
- 34.Hill JM, Dudley JB, Shimomura Y, Kaufman HE. 1986. Quantitation and kinetics of induced HSV-1 ocular shedding. Curr Eye Res 5:241–246. doi: 10.3109/02713688609020049. [DOI] [PubMed] [Google Scholar]
- 35.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 36.Dai L, Chen K, Youngren B, Kulina J, Yang A, Guo Z, Li J, Yu P, Gu S. 2016. Cytoplasmic Drosha activity generated by alternative splicing. Nucleic Acids Res 44:10454–10466. doi: 10.1093/nar/gkw668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brdovčak M, Zubković A, Jurak I. 2018. Herpes simplex virus 1 deregulation of host microRNAs. ncRNA 4:36. doi: 10.3390/ncrna4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukerjee R, Kang W, Suri V, Fraser NW. 2004. A non-consensus branch point plays an important role in determining the stability of the 2-kb LAT intron during acute and latent infections of herpes simplex virus type-1. Virology 324:340–349. doi: 10.1016/j.virol.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 39.Thompson RL, Sawtell NM. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol 71:5432–5440. doi: 10.1128/JVI.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 41.Leib DA, Nadeau KC, Rundle SA, Schaffer PA. 1991. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc Natl Acad Sci U S A 88:48–52. doi: 10.1073/pnas.88.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloom DC, Hill JM, Devi-Rao G, Wagner EK, Feldman LT, Stevens JG. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol 70:2449–2459. doi: 10.1128/JVI.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivera-Gonzalez R, Imbalzano AN, Gu B, Deluca NA. 1994. The role of ICP4 repressor activity in temporal expression of the IE-3 and latency-associated transcript promoters during HSV-1 infection. Virology 202:550–564. doi: 10.1006/viro.1994.1377. [DOI] [PubMed] [Google Scholar]
- 44.Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. 2011. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J Virol 85:4501–4509. doi: 10.1128/JVI.01997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawamura Y, Bosch-Marce M, Tang S, Patel A, Krause PR. 2018. Herpes simplex virus 2 latency-associated transcript (LAT) region mutations do not identify a role for LAT-associated microRNAs in viral reactivation in guinea pig genital models. J Virol 92:18. doi: 10.1128/JVI.00642-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang R, Zhou X, Ren S, Liu X, Han Z, Zhou G. 2019. Effect of loss-of-function of the herpes simplex virus-1 microRNA H6-5p on virus replication. Virol Sin 34:386–311. doi: 10.1007/s12250-019-00111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson ZL, Washington SD, Phelan DM, Lewin AS, Tuli SS, Schultz GS, Neumann DM, Bloom DC. 2018. In vivo knock-down of the HSV-1 latency-associated transcript reduces reactivation from latency. J Virol 92:e00812-18. doi: 10.1128/JVI.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perng GC, Maguen B, Jin L, Mott KR, Kurylo J, BenMohamed L, Yukht A, Osorio N, Nesburn AB, Henderson G, Inman M, Jones C, Wechsler SL. 2002. A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5’ end of the latency-associated transcript produces a protein in infected rabbits. J Virol 76:8003–8010. doi: 10.1128/jvi.76.16.8003-8010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan D, Cohen J, Naito J, Mott KR. 2006. A mutant deleted for most of the herpes simplex virus type 1 (HSV-1) UOL gene does not affect the spontaneous reactivation phenotype in rabbits. J Neurovirol 12:5–16. doi: 10.1080/13550280500516401. [DOI] [PubMed] [Google Scholar]
- 51.Tang S, Patel A, Krause PR. 2019. Hidden regulation of herpes simplex virus 1 pre-mRNA splicing and polyadenylation by virally encoded immediate early gene ICP27. PLoS Pathog 15:e1007884. doi: 10.1371/journal.ppat.1007884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Washington SD, Singh P, Johns RN, Edwards TG, Mariani M, Frietze S, Bloom DC, Neumann DM. 2019. The CCCTC binding factor, CTRL2, modulates heterochromatin deposition and the establishment of HSV-1 latency in vivo. J Virol 93:e00415-19. doi: 10.1128/JVI.00415-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perng GC, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68:8045–8055. doi: 10.1128/JVI.68.12.8045-8055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drolet BS, Perng GC, Villosis RJ, Slanina SM, Nesburn AB, Wechsler SL. 1999. Expression of the first 811 nucleotides of the herpes simplex virus type 1 latency-associated transcript (LAT) partially restores wild-type spontaneous reactivation to a LAT-null mutant. Virology 253:96–106. doi: 10.1006/viro.1998.9492. [DOI] [PubMed] [Google Scholar]
- 55.Javier RT, Stevens JG, Dissette VB, Wagner EK. 1988. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology 166:254–257. doi: 10.1016/0042-6822(88)90169-9. [DOI] [PubMed] [Google Scholar]
- 56.Hill JM, Sedarati F, Javier RT, Wagner EK, Stevens JG. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 57.Steiner I, Spivack J, Lirette R, Brown S, MacLean A, Subak-Sharpe J, Fraser N. 1989. Herpes simplex virus type 1 latency‐associated transcripts are evidently not essential for latent infection. EMBO J 8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Junejo F, Brown SM. 1995. Latent phenotype analysis of three deletion variants of herpes simplex virus type 1 (HSV-1) in mouse model. J Pakistan Med Assoc 45:99–99. [PubMed] [Google Scholar]
- 59.Trousdale M, Steiner I, Spivack J, Deshmane S, Brown S, MacLean A, Subak-Sharpe JH, Fraser N. 1991. In vivo and in vitro reactivation impairment of an HSV-1 latency-associated transcript variant in a rabbit eye model. J Virol 65:6989–6993. doi: 10.1128/JVI.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cook S, Hill J, Lynas C, Maitland N. 1991. Latency-associated transcripts in corneas and ganglia of HSV-1 infected rabbits. Br J Ophthalmol 75:644–648. doi: 10.1136/bjo.75.11.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.BenMohamed L, Osorio N, Srivastava R, Khan AA, Simpson JL, Wechsler SL. 2015. Decreased reactivation of a herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) mutant using the in vivo mouse UV-B model of induced reactivation. J Neurovirol 21:508–517. doi: 10.1007/s13365-015-0348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giordani NV, Neumann DM, Kwiatkowski DL, Bhattacharjee PS, McAnany PK, Hill JM, Bloom DC. 2008. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J Virol 82:6056–6060. doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gay LA, Sethuraman S, Thomas M, Turner PC, Renne R. 2018. Modified cross-linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi’s sarcoma-associated herpesvirus microRNA targets in endothelial cells. J Virol 92:17. doi: 10.1128/JVI.02138-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bloom DC. 1998. HSV vectors for gene therapy. Methods Mol Med 10:369–386. doi: 10.1385/0-89603-347-3:369. [DOI] [PubMed] [Google Scholar]
- 66.Szpara M. 2014. Isolation of herpes simplex virus nucleocapsid DNA, p 31–41. In Diefenbach R, Fraefel C (ed), Herpes simplex virus. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for the 17dmiR-H1/H6 virus are available in the Sequence Read Archive (SRA) under accession number SRR9121361.