Summary
Large-scale population screening and in-home monitoring for patients with Parkinson's disease (PD) has so far been mainly carried out by traditional healthcare methods and systems. Development of mobile health may provide an independent, future method to detect PD. Current PD detection algorithms will benefit from better generalizability with data collected in real-world situations. In this paper, we report the top-performing smartphone-based method in the recent DREAM Parkinson's Disease Digital Biomarker Challenge for digital diagnosis of PD. Utilizing real-world accelerometer records, this approach differentiated PD from control subjects with an area under the receiver-operating characteristic curve of 0.87 by 3D augmentation of accelerometer records, a significant improvement over other state-of-the-art methods. This study paves the way for future at-home screening of PD and other neurodegenerative conditions affecting movement.
Keywords: deep learning, convolutional neural network, digital biomarker, signal processing, Parkinson's disease
Graphical Abstract

Highlights
-
•
Deep convolutional neural networks enable efficient screening of Parkinson's disease
-
•
Temporal and spatial data augmentation improves detection in in-home environments
-
•
Interpretable AI shows that deep learning identifies classical parkinsonian characteristics
-
•
The method paves a way for future population-level screening of motor-related disease
The Bigger Picture
In-home digital surveillance has been proposed as the future for chronic, neurodegenerative disease such as Parkinson's disease (PD), which can be monitored by wearable devices from its motor-related symptoms. However, the disparities between uncontrolled in-home environments have introduced obstacles to the population-level application of digital screening of PD. In this study, we developed the first-place solution in the recent DREAM Parkinson's Disease Digital Biomarker Challenge, which calls for optimal algorithms to extract digital biomarkers of PD from crowd-sourced movement records. To combat the spatial and temporal bias in different movement records, we applied a variety of data-augmentation methods, which significantly improves the performance of the deep-learning model. Besides PD, our method provides a path for large-scale population screening and in-home monitoring using wearable devices in other related neurodegenerative disorders.
The widespread use of wearable devices in our daily life has provided an ideal platform for the management of motor-related neurodegenerative diseases such as Parkinson's disease, while the application of such systems in the real-world situation has faced many challenges from in-home environments. In this study, we show how artificial intelligence could facilitate the efficient screening of Parkinson's disease in the general population and how interpretable artificial intelligence could provide useful information to understand the motor-related pathology of Parkinson's disease.
Introduction
Application of artificial intelligence to digital health monitoring opens the door to in-home disease screening and monitoring using widely available devices such as digital watches or smartphones. Parkinson's disease (PD) is a common neurodegenerative disease whose defining clinical features are movement dysfunction, namely bradykinesia (e.g., slowness of movement) associated with one or more other features of rest tremor, rigidity, or postural instability.1, 2, 3, 4, 5 Conventional clinical diagnosis depends on defined clinical criteria relying on human expert evaluation, which may be difficult to access and delays identification and treatment of PD.4,6, 7, 8, 9 Similarly, clinical management of PD and the great majority of clinical research on PD rely on face-to-face in-clinic evaluations, which may not capture sufficient or critical data. In-home evaluation, so far limited in large-scale clinical use, has the potential to significantly enhance accessibility to and reduce costs of clinical research via the currently widely available digital devices and generalizable algorithm design.10
Inertial sensors, such as accelerometers and gyroscopes, were originally introduced into smartphones to alert holders of sudden device falls and for maintaining display images upright.11 These sensors can provide useful instruments for detecting human motion and posture, and have been applied extensively in this context.12,13 A hindrance in utilizing smartphones for at-home study of PD is that the evaluation of PD movement usually needs to be corrected for specific tasks. For instance, when measuring the resting tremor, the clinician has to ensure that the patient’s hands are relaxed on the legs, and when measuring postural tremor, the patient’s arms must remain outstretched with fingers apart for several seconds. Another major challenge for in-home monitoring of PD is that movement recordings obtained by these mobile devices can be problematically influenced by the surrounding environments, i.e., the topographical orientation of the road, local gravities, and the positions and orientations of these devices when placed on the patient's body.14 These unrelated factors may create noise in the data used to extract PD-related pathological information from patients' walking records. This is a particular problem for multiple-parameter machine-learning models such as deep learning, which carry the risk of overfitting.15
A recent Dialog for Reverse Engineering Assessments and Methods (DREAM) PD Digital Biomarker (PDDB) Challenge called for methods to accurately identify PD in the general population using smartphone accelerometer and gyroscope records. Data science challenges such as DREAM provide participants with a training dataset and a testing dataset for a one-shot evaluation, helping to identify current state-of-the-art methods for specific problems without overfitting. We describe the top-scoring solution for this challenge, which performed significantly better than the prior state-of-the-art methods, such as Deep Learning by Convolutional Recurrent Attentive Neural Networks (CRANNs), as well as traditional machine-learning methods based on rigorously extracted features from Fourier transform and discrete wavelet transform.16,17 The key to improved performance was a generic methodology reflecting the physical properties of mobile device records. This method can be generalized to any accelerometer-based and/or gyroscope-based disease identification approach.
Results
Source and Descriptions of Smartphone-Collected Accelerometer and Gyroscope Data
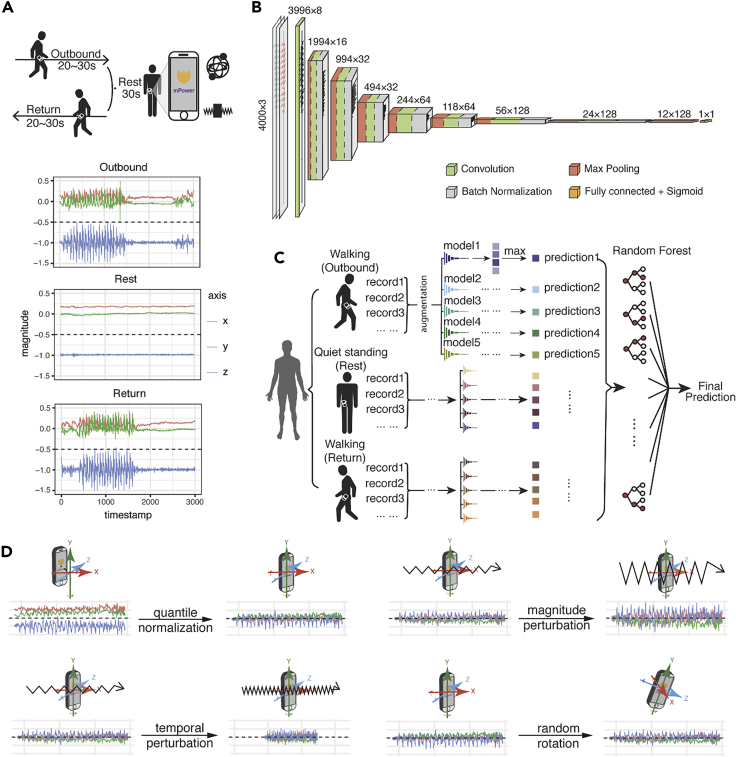
Our study was based on the latest released DREAM dataset (mPower) contributed by Sage Bionetworks, whereby a total of 2,804 subjects (656 self-reported patients and 2,148 healthy controls) participated in a simple walking evaluation and agreed to share their data with the broader research community.17,18 All participants contributed 34,632 walking records to the dataset. In the DREAM dataset (mPower), each participating individual may take multiple tests, termed “records.” The walking test consisted of two 30-step walking phases, here denoted as “Outbound” and “Return,” whereby the participant was asked to walk along a straight line. The two walking phases were interrupted by 30 s of quiet standing, here denoted as “Rest.” A representative walking record (outbound and return) will have an irregular cyclic pattern, and a representative quiet standing record (rest) will be relatively stable, with initial and ending noise periods (Figure 1A). In PD, tremors may occur during the quiet standing period, and oscillations may be observed.
Figure 1.
Walking Test Data Provided in the DREAM PDDB Challenge and Our Deep-Learning Model
(A) Example of the walking activities carried out by subjects and accelerometer records during the three activities. During the walking test, the velocity of participants is recorded by the two sets of sensors implemented in their phones—gyroscope and accelerometer—represented by a set of free-spinning wells and a rigid body attached to a spring.
(B) The architecture of the convolutional neural network in this study. The numbers at the top of the boxes indicate the size of the layers and the numbers of the filters.
(C) The model ensemble method in this study. We trained five models by reseeding the training and validation sets, for outbound walking, rest (quiet standing), and return walking, respectively. For our final prediction, we assemble the predictions of the 15 models of outbound/rest/return by random forest and create a final prediction for each individual.
(D) Data-augmentation strategies applied in this study. The original record is first normalized by quantile normalization and then applied to three data-augmentation operations, namely magnitude perturbation, temporal perturbation, and random rotation.
Two sets of independent data existed in the phone records to provide information on the participants' body movement as they were captured by different inertial sensors: acceleration, captured by an accelerometer (represented as a spring-attached rigid body), and rotationRate, captured by a gyroscope (represented as a spinning well with a fixed center) (Figure 1A). Both types of the signals could be represented as a 3Xn matrix, where n is the total sampled length of the record between ∼2,000 and ∼3,000 points, as the sampling frame is 100 per second for a total of 20–30 s. Our machine-learning PD diagnostic model would exploit PD pathological information from the two aforementioned independent accelerometer and gyroscope records.
Performance of Deep Convolutional Neural Network Model in Independent Cross-Validation and the DREAM PDDB Challenge
Prior studies of gait and gait abnormalities in neurodegenerative disorders used traditional signal-extraction techniques such as Fourier transform or wavelet analysis to identify extracted features, for example, walking pace, from accelerometer or gyroscope data.19, 20, 21, 22, 23, 24, 25 Classifiers such as Support Vector Machine or K-Nearest Neighbors were applied to extracted features to predict disease associations.26, 27, 28, 29, 30 These methodologies share the common limitation of feature extraction performed independently from the machine-learning step so that there is inevitably redundant information in the extracted features, diluting model power, while useful information is missed. Therefore, we implemented a deep convolutional neural network (DCNN), which can directly process the continuous accelerometer and gyroscope records (Figures 1B–1D).
Our model achieved an average area under the receiver-operating characteristic curve (AUROC) of 0.8558 (95% confidence interval [CI] 0.8529, 0.8588) in the 5-fold cross-validation using the same mPower dataset. The cross-validation results showed the consistently robust performance of our model. Our deep-learning prediction model also took first place in the DREAM PD prediction competition, in comparison with other teams that employed the aforementioned traditional machine-learning methods.31
Data Augmentation Significantly Improves Model Performance and Stability
Because convolutional neural networks (CNNs) have many parameters, deep-learning models are prone to overfitting. To combat overfitting, diverse data-augmentation techniques have been developed in the image and audio fields.32 In the past, three-dimensional (3D) information of movement in space has only been analyzed via intuitive approaches. For example, summed square, mean, or variance has been utilized to remove the differences in reference frames.33,34 To address the specific properties of an object moving in 3D space, we implemented data normalization and augmentation methods in the deep-learning framework (Figure 1D). We first quantile normalized the original 3D waveform signals by each axis. After normalization, we applied three types of data-augmentation strategy to the accelerometer and gyroscope recordings: timewise scaling, to mimic that the records are taken at a faster or a slower speed; magnitude scaling, to mimic the magnitude of the acceleration or rotation-rate disparities among records; and random rotation, to correct the disparity of phone orientation when a patient is taking the walking test (Figure 1D).
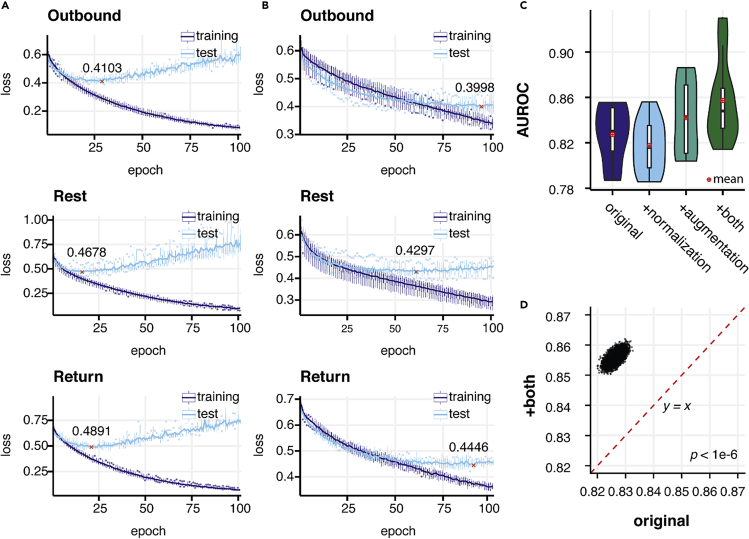
Without data augmentation the model quickly overfitted, as seen in the increase and bumpiness in the test error along a continuous drop in training error (Figure 2A). We found a significant improvement in performance when using augmentation, from 0.8261 (95% CI 0.8231, 0.8292) to 0.8496 (95% CI 0.8465, 0.8496) (p < 1 × 10−6) (Figures 2C, 2D, and S1C). On the other hand, both training and testing errors steadily dropped and were stabilized at a similar speed when we applied augmentations, indicating alleviation of overfitting (Figures 2B and S1B). We compared model performance with and without axis-wise normalization, and found substantial improvement by using normalization after augmentation was applied: 0.8558 (95% CI 0.8531, 0.8588) versus 0.8496 (95% CI 0.8465, 0.8496) (p < 1 × 10−6) (Figures 2C and S1E). This indicates that disparities in holding position leading to shifts in the axes constitute a confounding factor that is appropriately addressed by per-axis normalization.
Figure 2.
Normalization and Data Augmentation Improves the Performance and Stability of the Model
(A and B) The dynamics of training and testing loss during 100 epochs of training process for models applying no normalization and data augmentation (using raw signal) and both normalization and augmentation. Models that achieved the lowest test loss were used as final models to avoid underfitting or overfitting to the training set. The lowest test loss achieved during training is denoted and marked by red crosses. (A) Loss during training without augmentation and normalization. (B) Loss during training with both normalization and augmentation.
(C) Comparison of the performance of the model without both normalization and augmentation (original), with only normalization, with only augmentation, and with both operations.
(D). Pairwise AUROC comparison of the performance by models using original records and with both augmentation and normalization from 1,000,000 bootstrapping operations. The red dashed line denotes a baseline where the performances (AUROCs) of two operations are equal to each other.
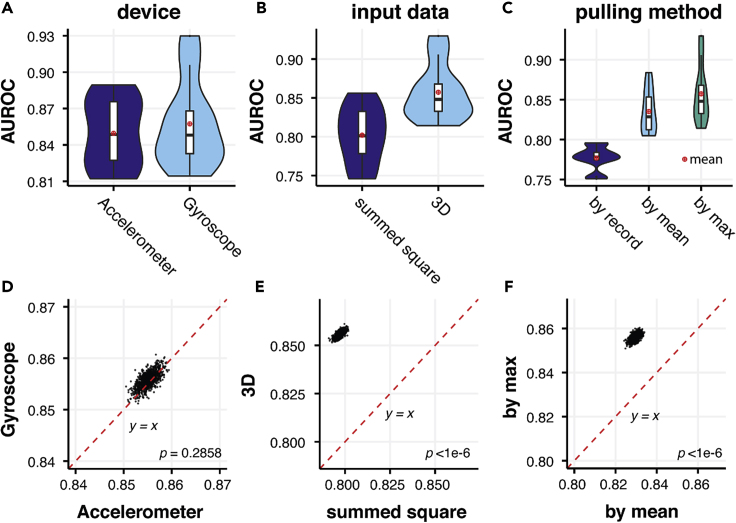
We also compared the performance of models using either accelerometer or gyroscope data as input, whereby performance was indistinguishable (Figures 3A and 3D). Furthermore, when we added accelerometer data to gyroscope data through a variety of network structures with six input channels, we did not observe an improvement in performance compared with either acceleration or rotationRate alone (Figure S2). We found that using summed squared values at each time point resulted in a substantial drop in performance (Figures 3B and 3E) (AUROC = 0.7971, 95% CI 0.7938, 0.8006) compared with using 3D coordinates with augmentation techniques discussed above (AUROC = 0.8558, 95% CI 0.8529,0.8588 (p < 1 × 10−6), likely due to the loss of information. We also found that taking the maximal prediction of each individual performed better than taking the mean prediction (Figures 3C and 3F), 0.8558 (95% CI 0.8531, 0.8588) versus 0.8294 (95% CI 0.8264,0.8325) (p < 1 × 10−6). This may imply that the symptoms of PD vary temporally in individuals and that the most severe records are more representative of the disease phenotype. Standard DCNN models, such as VGG-16 and its alternatives, were also tested in our experiments to search for the optimal deep-learning model (Figures S3–S5).35
Figure 3.
Comparison of the Performance of Models Using Records from Different Devices, Processing Methods, and Pulling Methods
(A–C) Comparison of AUROCs between models using: (A) either accelerometer and gyroscope data; (B) the sum of the squared values of x, y, z axes and 3D data as input; (C) different pulling methods: on record level (by record), on individual level using average prediction across records of each individual (by mean), and on individual level using maximum prediction across all records of each individual (by max) in 5-fold cross-validations.
(D–F) Pairwise comparison between AUROCs of models mentioned in (A) to (C) in bootstrapping. No significant difference between accelerometer and gyroscope input data was observed, while more consistent improvement was observed by 3D input than summed square and by maximal prediction at the individual level than prediction at record level or the mean prediction at the individual level (p < 1 × 10−6).
Quiet Standing Records Predict PD More Accurately Than Outbound/Return and Were Paid Most Attention by DCNN Models
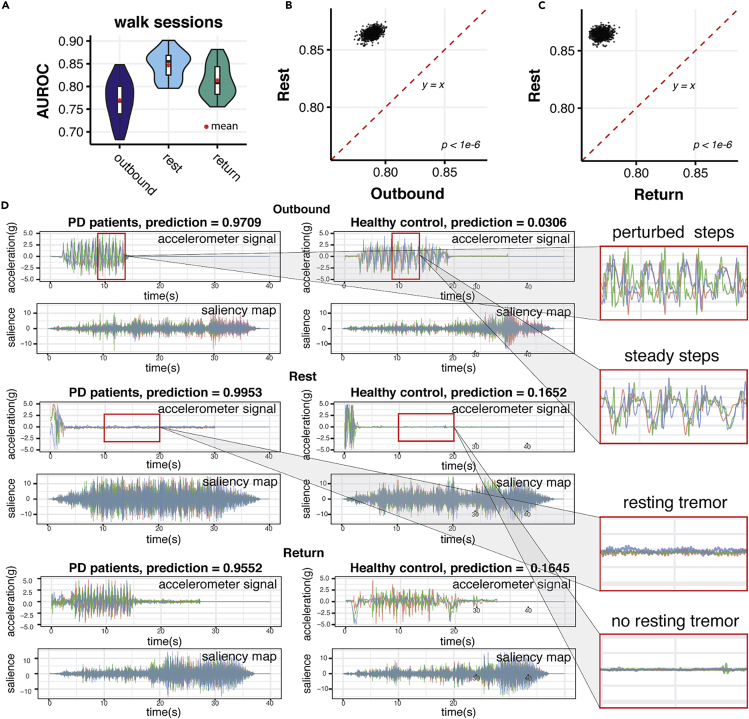
We compared the performance of the model using outbound walking, quiet standing, and return walking records as model inputs. Quiet standing (Rest) records performed substantially better than outbound and return walking records: 0.8548 (95% CI 0.8517, 0.8578) versus 0.7889 (95% CI 0.7802, 0.7971) and 0.7705 (95% HCI 0.7623, 0.7795) (Figures 4A–4C). Including outbound and return walking records did not improve the performance of the model (Figure S6).
Figure 4.
Visualization of Deep Convolutional Neural Networks Unveils the Importance of Quiet Standing Behaviors during Walking when Detecting Parkinson's Disease
(A) The comparisons of AUROCs performed by models on walking records during outbound walking (outbound), quiet standing (rest), and return walking (return).
(B) Pairwise comparison between AUROCs achieved by outbound walking (Outbound) and quiet standing (Return) models.
(C) Pairwise comparison between AUROCs achieved by return walking (Return) and quiet standing (Rest) models. Quiet standing consistently performed better than both outbound and return walking (p < 1 × 10−6).
(D) Saliency maps that the trained deep neural network extracted from walking records (after padding to 40 s) of both PD patients and healthy controls during outbound walking (Outbound), quiet standing (Rest), and return walking (Return). Ground-truth labels and predictions by the machine-learning models are denoted for each rotation-rate signal and corresponding saliency map extracted from each signal. On the right, PD characteristics of perturbed steps and tremors in comparison with controls are zoomed in to show them in detail.
To examine which part of the records supports PD predictions by the deep-learning model, we carried out a saliency map analysis.36 A saliency map is a visualized map signifying the first layer gradient of the neural network, where a higher value indicates the stronger signal in the original record that is captured by the CNN model.37 Saliency maps from both PD subjects and healthy controls revealed that our model extracted strongest PD prediction signals from quiet standing records and also from apparent quiet standing interruptions in both outbound and return walking records (Figures 4D and S7–S12). During quiet standing, all movement was expected to be involuntary, and the model was robust for detecting involuntary movements, such as tremors, as indicated by a waveform of low magnitude and high frequency (Figure 4D), a typical PD feature.38, 39, 40, 41 This provided an insight into the mechanism for the results above that quiet standing (Rest) records gave more accurate predictions than the other two types of movement records.
Meanwhile, our model also extracted relatively higher signals from the walking records with higher frequency and more disturbed waveforms, an obviously more perturbed walking pattern compared with healthy controls, as shown on the saliency maps of Outbound and Return records (Figures 4D and S7–12). Rather than the clear, synchronized waveform of the walking records of the healthy controls, the waveform of the walking records of PD patients is more curvy and disturbed, suggesting that the PD patients experienced struggles during walking and that their steps were unsteady. Moreover, the wavelength, or the distance between two peaks, of PD patients' walking records was much smaller than that of healthy controls, suggesting they were taking smaller steps and shorter strides, which was also a typical PD feature.3 The above visualization of the features extracted by the DCNN model suggests that our deep-learning model developed its own understanding of PD walking pathology by recognizing especially resting tremor and other typical PD gait characteristics.
Model Performance and Predictions in Groups of Different Demographic Status
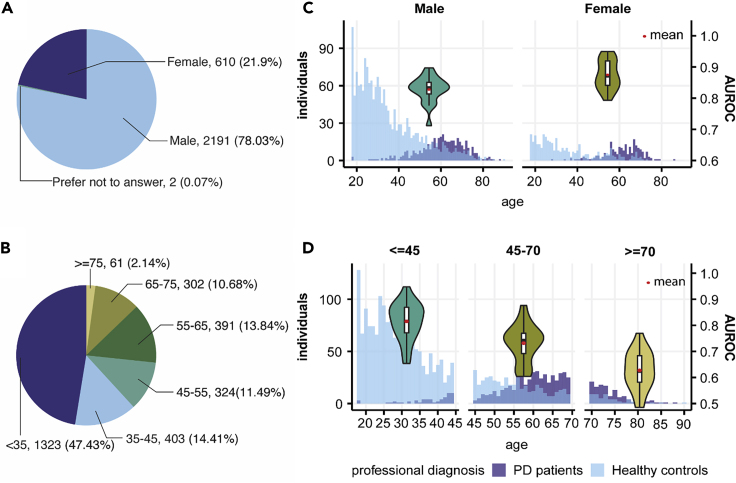
The mPower dataset also provided self-reported demographic information, year of the first PD diagnosis, and Unified Parkinson’s Disease Rating Scale (UPDRS) Part II score.18 We analyzed the model performance in different demographic subgroups of the mPower dataset (Figure 5). We found better performance of our model in female than in male subjects (Figure 5C). One potential reason for this is that women on average took more tests than male participants (19.08 versus 10.47 records per person). When participants were stratified by age, model performance decreased steadily with aging (Figure 5D).
Figure 5.
Demographics of Data at the Individual Level and Model Performances in Different Demographic Groups
(A) Gender composition of participants in the mPower walking test.
(B) Age composition of participants in the mPower walking test.
(C) Age and PD patients/healthy controls distribution in male and female participants and comparison of AUROCs when our model is performed on either a male or female cohort in 5-fold cross-validation.
(D) Age and PD patients/healthy controls distribution in our dataset and comparison of AUROCs when our model is performed on participants aged under 45 (<45), aged between 45 and 70 (45–70), and aged over 70 (≥70) years in 5-fold cross-validation.
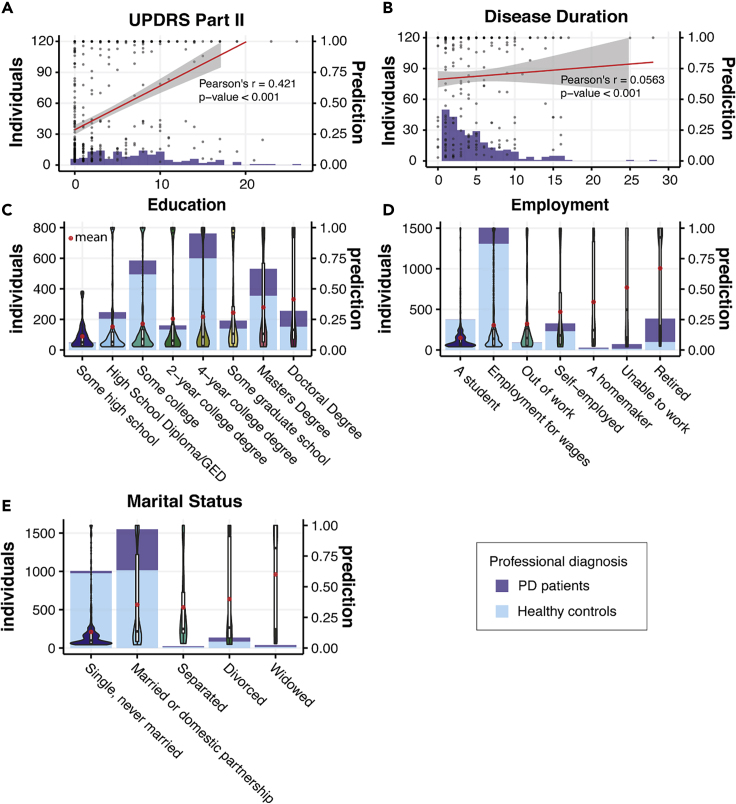
We also explored model predictions in other subgroups defined by demographic variables, such as participant education level, employment, and marital status. We found correlations between PD classification and higher educational levels as well as marital status and retirement status (Figures 6C–6E). Similar correlations were reported in some previous large-scale demographic screening studies, possibly reflecting the composition of study cohorts.42,43 While no substantial correlation was observed when we compared the PD classification with duration of disease as assessed by self-reported years of diagnosis, a strong correlation (Pearson's r = 0.421) was observed with self-reported UPDRS Part II score, which reflected the self-examined motor function in daily activities (Figures 6A and 6B).
Figure 6.
PD Prediction in Groups Separated by Demographic Status
(A) Composition of PD patients with different reported UPDRS Part II scores and PD predictions when applying our model to the patients. The red line denotes Pearson's correlation between the self-reported UPDRS Part II score and PD predictions.
(B) Composition of PD patients with different disease duration (years since when they were first diagnosed with PD) and PD predictions when applying our model to the patients. The red line denotes Pearson's correlation between the disease duration and PD predictions.
(C) Demographic groups divided by the highest education level the participants ever achieved.
(D) Demographic groups divided by employment status.
(E) Demographic groups divided by marital status.
Histograms in (C)–(E) denote the composition of PD patients and healthy controls in the demographic groups. Predictions of our model in each demographic group are presented as violin plots and box plots. The red dots denote the mean prediction of our model for each demographic group.
Training Set Size and Model Performance
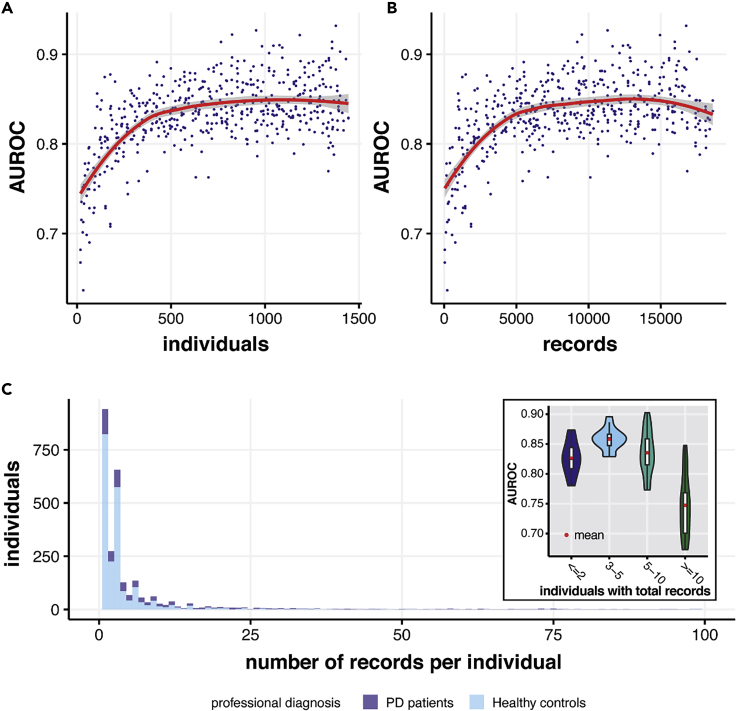
To test the relationship between the size of the training set and model performance, we tested a series of training set sizes and evaluated model performance (Figure 7). Substantial AUROC improvement was observed as training size increased from zero to 500 individuals or 5,000 records, with model performance plateauing subsequently. This result suggests that the dataset is adequate for training a generalizable model and that our model achieved excellent performance despite the variance and limitations of the training set. Also, the model predicts best in individuals with 3–5 records. Typically, with more records the predictions should be better. However, since only a few people in our dataset had more than five records, a decreasing model performance in individuals with more than five records is more likely to be the result of sampling bias.
Figure 7.
Relationship between Model Performance and Training Size/Records of Each Individual
(A) The AUROCs achieved by our models as the training size increases from 0 to 1,500 individuals. The model reached satisfactory performance at around 500 individuals.
(B) The AUROCs achieved by our models as the training size increases from 0 to 19,000 records. The red line in (A) and (B) shows the lowest fit between AUROC and individuals/records in the training set.
(C) The distribution of records per individual in our dataset (both PD patients and healthy controls) and comparison of AUROCs when our model performs on groups of individuals with ≤2, 3–5, 5–10, and >10 total walking records (a walking record here denotes a full round of outbound, quiet standing, and return activities). The maximum of the x axis was cut to 100 for better display.
Discussion
We describe a significant step toward population-level screening for PD. Our application of deep learning to smartphone-based gait data from a simple walking task demonstrated superior accuracy (AUROC = 0.86) when tested on an independent blind test set compared with the second-placed method, which was the prior state-of-the-art method for predicting PD using CRANNs (AUROC = 0.7), as well as traditional machine-learning and hand-crafted feature-extraction methods using wavelet or Fourier transform.16,17 Our model also achieved an average AUROC of 0.856 in 5-fold cross-validation on 34,632 walking records of 2,804 participating subjects, showing consistent generalizability and potential for future large-scale application.
The big-data era has produced opportunities for remote at-home monitoring of chronic diseases such as PD. The biggest challenge, however, in utilizing these datasets is how to extract the gold from the mud—in other words, how to extract the information that really matters in terms of pathophysiology from extensive noise in the coarse real-world data, and how to deal with the unavoidable missing values.
We applied two major strategies to address spatiotemporal bias within these real-world motion records to detect PD features. First, we identified the major spatial bias that could confound the walking record of each individual, such as the placement of the phone when recording the motion of participants. A novel augmentation method was applied to simulate the random reference frames of accelerometer records. Second, we noticed that the records taken by the same individual may not be equally reliable for PD identification, as they might be taken at time points reflecting different physical and medical conditions. This was solved by pulling out the most severe record of each individual, i.e., model ensemble from the maximum predictions. When incorporating these strategies into an appropriately tuned DCNN model, the performance is further boosted, as deep learning is suitable for handling complicated patterns in continuous time-series input, demonstrated by our first place in the DREAM PDDB Challenge.31
Our results suggest that resting tremor might be an important indicator of parkinsonian movement for efficient PD screening in the general population, as quiet standing records and the walking records with longer standing periods provided a more accurate prediction of PD. The importance of resting tremor in PD's digital detection was also observed and pinpointed in previous studies.44, 45, 46 Our model also paid the most attention to quiet standing periods as revealed by the model's saliency map (Figure 4). These results indicated that resting tremor was the most salient feature automatically identified by our DCNN model to distinguish between PD and non-PD participants. Although not all PD patients experience tremor, it is recognized as a common feature, present in 70%–100% PD patients,47,48 with resting tremor being a relatively specific feature.
We did not observe a significant correlation between our model's prediction and disease duration (Figure 7A). We did, however, observe a strong correlation between our model's prediction and self-reported UPDRS Part II scores (Figure 6A). The poor correlation between disease duration and PD prediction could be because of variation in PD progression, effects of treatments, or inaccurate recall of the year of first PD diagnosis. As tremor often develops early in PD patients,49 differences between early and advanced patients might not be very distinguishable by our model.
Our model provided a promising instrument for enhancing telemedicine and early screening of PD. Identification and accurate diagnosis of PD is often delayed, likely causing increased morbidity in the periods prior to diagnosis.50 Fang et al., for example, showed that head injury occurs more frequently in the year prior to diagnosis of PD.51 Therefore, earlier identification and large-scale screening of PD might significantly improve clinical outcomes in PD. Willis et al. also showed that a substantial fraction of PD patients in the United States do not obtain access to neurologist care.52 Accuracy of initial clinician-based diagnosis may be as low as 75%–80%, although it often improves markedly with serial patient follow-up.53, 54, 55 An algorithm with satisfactory performance in evaluating real-world in-home data might provide useful screening of potential PD patients and earlier referral for expert evaluations.
While our PD detection model achieved satisfactory performance with crowd-sourced data, it is important to note that there are limitations of our algorithm inherent to the nature of this dataset. It is possible that the mPower dataset used in this study overenrolled tremor-dominant PD subjects. It is also likely that many of the PD subjects enrolled in this dataset were treated, and since resting tremor does not always respond robustly to treatment,56 the relative sensitivity of tremor detection (versus detection of bradykinesia) for identifying PD might be enhanced. It is also plausible that smartphone monitoring and our model captured PD-specific resting tremor that cannot be observed by the naked eye. To carefully evaluate the specificity and sensitivity of this approach, future evaluations should include robust numbers of subjects with other forms of tremor and other forms of parkinsonism. The mPower dataset is imbalanced in terms of age, class, and gender (Figure 5). While we rebalanced the training set by oversampling the scarcer PD samples, mPower is under-represented in age-matched controls. The non-PD participants are mainly younger than 45 years. This might result in bias in our model given that our model performs better in the younger group (under age 45) than in the older group (over age 70) (Figure 5D). Furthermore, since the demographic information and clinical diagnosis of PD were self-reported by the participants through a smartphone App rather than by professional clinicians, we could inevitably have used inaccurate information in building the PD detection model. The lack of clinical data, such as UPDRS Part III subscores and total scores, evaluations in “ON” and “OFF” conditions related to levodopa intake, total levodopa equivalent daily doses, and Hoehn and Yahr scores, are limitations of this dataset. Extension of our model would benefit from involvement of neurologists and PD specialists in the data-collecting steps.
While our PD identification model is based on simple walking task data, smartphones can also collect multimodal data from subjects, raising the prospect of multimodal evaluations to identify PD. The mPower program also collected other PD-relevant performance data including voice, tapping, and memory.18 Integration of deep-learning models based on all data might generate more reliable assessments of PD. The techniques used in this study are generic, such as the augmentation by altering the reference frame of the accelerometer or gyroscope data, and can be used for monitoring other diseases with smartphones, watches, or professional medical devices57 in other neurologic disorders.58, 59, 60, 61 Sleep quality and breathing patterns associated with sleep disorders are also frequently monitored with accelerometers and gyroscopes.62, 63, 64 In-home monitoring of PD through wearable sensors may also elevate the effectiveness of therapy in real-world settings, as it can provide useful information for clinicians to evaluate medication regimens or monitor deep brain stimulation parameters.65 Wider application of these techniques may facilitate widespread use of smartphone gyroscope and accelerometer data in the digital health area.
Experimental Procedures
Resource Availability
Lead Contact
Yuanfang Guan is the lead contact of this study and can be reached through e-mail: gyuanfan@umich.edu.
Materials Availability
The machine-learning models generated in this study can be obtained via our public github repository: https://github.com/GuanLab/PDDB.
Data and Code Availability
All code associated with this paper can be freely accessed and downloaded via https://github.com/GuanLab/PDDB/. The mPower dataset used in this paper can be accessed via the mPower public research portal: https://www.synapse.org/#!Synapse:syn4993293/wiki/247859.
Collection of Mobile Phone Accelerometer and Gyroscope Data from the Participants
Motion-related data collected from smartphones consisted of the following six categories: timestamp, attitude, rotationRate, userAcceleration, acceleration, and gravity. The latter five categories were sampled every 0.01 s (100 frames/s). Following the mPower App protocol, participants signal the test start on their smartphone, pocket the phone, and perform the task, then take their phone out and stop the test. Each participant is able to make multiple records numbering from 1 to over 500, and at different time points, such as when they are feeling at their best (just after they have taken the medication) or at their worst (immediately before the medication) and other times. Demographic and clinical information were self-reported through the App.
The aforementioned six categories of data provide two independent sources of information. The first source of information is acceleration, taken from the accelerometer, which describes the change of speed of a mass point. This change is represented by projections along three orthogonal x, y, z axes in 3D space. userAcceleration is equal to the acceleration that the user imparts to the device plus local gravity.
The second source of information is the rotationRate taken from the gyroscopes, which describes the speed of the rotation around the reference frame: pitch for x, roll for y, and yaw for z.66 For rotationRate, the mobile phone is considered as a rigid body having 6° of freedom in motion (acceleration + rotation). Even if the center of the phone is still, or moving at a constant direction and speed, the rotationRate is not necessarily zero, because the phone can be rotated at its center point. The reference frame of the rotation is the phone. Both attitude and rotationRate recorded in the cell phones contain the same information about the rotation, but are expressed in different mathematical terms. rotationRate is expressed directly by x, y, z, which stand for pitch, roll, and yaw, respectively, while attitude is expressed as a unit vector (x′, y′, z′) and an angle (w) around this vector between 0° and 360°, according to Euler's rotation theorem.
Building Augmentation Methods for Accelerometer and Gyroscope Records
A data-augmentation strategy is vital for overfitting prone machine-learning algorithms. Before being fed into the DCNN, we transformed the raw data by quantile normalization and three data-augmentation operations (Figure 1D and Supplemental Information).We carried two loss-included data-augmentation strategies, by timewise rescaling between 0.8- and 1.2-fold to mimic that the records are taken at a faster or a slower speed, and magnitude rescaling between 0.8- and 1.2-fold to mimic the magnitude of the acceleration or rotation-rate disparities among records.
Also, the placement of the phone on the subject's body during movement can have a fundamental impact on the measurement by inertial sensors. For example, for a phone lying flat on a table, its z-axis acceleration is around 9.8 m2/s while the acceleration along the x and y axes is zero. If we keep the phone still but hold it vertically, its y axis acceleration becomes 9.8 m2/s while the acceleration along the other two axes is zero. Normalization removes such disparities. Thus, we used a transformation of the reference frame to create loss-free augmentations of the same record (see Supplemental Information). After a record is transformed by augmentation, it is fed into the deep learning network (Figures 1B and 1C). A single mobile record can be augmented to generate multiple records randomly on-the-fly. This means every epoch of the same records in the training set can be randomly transformed into a new record. For example, if the training process stopped early, say, converged at the 43rd epoch, 43 new records would be generated from the same training record.
Applying DCNN to Continuous Walking Records
Deep learning is now the preferred algorithm for datasets with spatial (e.g., image) and time (e.g., sound) continuity.67 Feature extraction in deep learning is obtained by convolution operations that extract local information and progressively summarize local information into a global prediction (Figure 1B). Because network parameters are trained directly to fit prediction targets, deep learning is a more direct prediction approach compared with traditional feature extraction, which is followed by classification or regression. In many of the machine-learning analyses of data with spatial and time continuity, performance of the deep-learning models reaches or surpasses human performance (i.e., human visual interpretation, hand labeling, or diagnoses by clinicians).68,69
Convolutional neural networks (CNNs) learn information derived from time- and space-continuous data and have been widely used to address various biomedical problems.70,71 Mobile accelerometer and gyroscope records exhibit time and space continuity, and deep CNNs are a natural route to build models for such data. We constructed a one-dimensional CNN using Theano and Lasagne (v.1.0) Python libraries, where the input channels were the x, y, z values of acceleration or rotation rates. The DCNN model consists of nine building blocks, each consisting of a maximum pooling layer, convolution layer, and batch normalization layer (Figure 1B). The features extracted by convolution layers then are put into a dense layer for finalized output of PD prediction.
On each occasion when a record was fed into the CNN, the three augmentations mentioned in the previous section were executed. This allowed the network to assess more different examples. Models for quiet standing and walking periods of records were trained independently.
Assembling the Multiple Models' and Records' Predictions of the Individuals
For each individual, we took the maximal value across all outbound walking, quiet standing, and return walking records, respectively. The outputs of these three types of models were stacked together in a random forest learner for the final prediction (Figure 1C). For each record, which contained a section of walking and a section of quiet standing, we calculated the mean values of the five models for walking and quiet standing, respectively. According to the mPower App, participants were instructed to carry out the walking task including two sessions of walking (Outbound and Return) interrupted by one session of quiet standing (Rest). Some participants might stop after the first walking session, leading to missing quiet standing and return walking data in a record. If quiet standing data were missing, we first searched for other quiet standing records from the same individual and used the mean prediction of those records to replace the missing data. If such a replacement was not found, we replaced its predicted score of its outbound walking data if available. If an individual had more than one record, we took the maximum or the average value of all records for outbound/return walking and quiet standing separately. The final prediction score was combined by a random forest learner from prediction scores of the walking and quiet standing records of the individual.
Visualization of Features Extracted by DCNN Models via Saliency Maps
To better interpret the information extracted by our PD prediction deep-learning model, we drew saliency maps of the DCNN model corresponding to the walking records of patients and healthy individuals by computing the gradient of the last layer corresponding to the input layer. Both negative and positive saliency were computed and visualized (see Supplemental Information). The saliency maps show significantly stronger signals during quiet standing periods, as the example in Figure 4D shows. More examples can be found in Figures S7–S12.
Acknowledgments
This study was supported by NSF-US14-PAF07599 Career Award #1452656: On-line Service for Predicting Protein Phosphorylation Dynamics Under Unseen Perturbations, NIH Project 1R35-GM133346-01: Machine Learning for Drug Response Prediction, American Parkinson's Disease Association AWD007950: Digital Biomarkers in Voices for Parkinson's Disease, and Michael J. Fox Foundation Project #17373: Interpretation of Accelerometer Data with deep learning, to Y.G. This study was also supported by NIH Project P50NS091856: Cholinergic Mechanisms of Attentional-Motor Integration and Gait Dysfunction in Parkinson's Disease, NIH Project P30AG053760: Core A: Administrative Core, R21NS114749, and Parkinson’s Foundation Research Center of Excellence to R.L.A.; and by the American Heart Association and Amazon Web Services Data Grant Portfolio 3.0: Artificial Intelligence and Machine Learning Training Grants award 19AMTG34850176 to H.L. We thank the GPU donation from Nvidia and the AWS donation from Amazon.
Author Contributions
Conceptualization, Y.G.; Methodology, Y.G.; Formal Analysis, Y.G. and H.Z.; Investigation, Y.G. and H.Z.; Writing – Original Draft, Y.G.; Writing – Review & Editing, H.Z., K.D., H.L., and R.L.A.; Visualization, H.Z.; Funding Acquisition, Y.G., H.L., and R.L.A.; Resources, Y.G.; Supervision, Y.G.
Declaration of Interests
Y.G. serves as scientific advisor and receives personal payment from Genentech, Inc., Eli Lilly & Co., and F. Hoffmann-La Roche AG. Y.G. serves as chief scientist and holds equity shares at Ann Arbor Algorithms Inc. Y.G. receives research grants and/or hardware support from Merck KGaA, Amazon, and Nvidia.
Published: May 28, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2020.100042.
Supplemental Information
References
- 1.Rogers M.W. Disorders of posture, balance, and gait in Parkinson’s disease. Clin. Geriatr. Med. 1996;12:825–845. [PubMed] [Google Scholar]
- 2.Boonstra T.A., van der Kooij H., Munneke M., Bloem B.R. Gait disorders and balance disturbances in Parkinson’s disease: clinical update and pathophysiology. Curr. Opin. Neurol. 2008;21:461–471. doi: 10.1097/WCO.0b013e328305bdaf. [DOI] [PubMed] [Google Scholar]
- 3.Hausdorff J.M. Gait dynamics in Parkinson’s disease: common and distinct behavior among stride length, gait variability, and fractal-like scaling. Interdiscip. J. Nonlin. Sci. 2009;19:026113. doi: 10.1063/1.3147408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postuma R.B., Berg D., Stern M., Poewe W., Warren Olanow C., Oertel W., Obeso J., Marek K., Litvan I., Lang A.E. MDS clinical diagnostic criteria for Parkinson’s disease. Movement Disord. 2015;30:1591–1601. doi: 10.1002/mds.26424. [DOI] [PubMed] [Google Scholar]
- 5.Poewe W., Seppi K., Tanner C.M., Halliday G.M., Brundin P., Volkmann J., Schrag A.-E., Lang A.E. Parkinson disease. Nat. Rev. Dis. Primers. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 6.Patel S., Park H., Bonato P., Chan L., Rodgers M. A review of wearable sensors and systems with application in rehabilitation. J. Neuroeng. Rehabil. 2012;9:21. doi: 10.1186/1743-0003-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pahwa R., Lyons K.E. Early diagnosis of Parkinson’s disease: recommendations from diagnostic clinical guidelines. Am. J. Manag. Care. 2010;16 Suppl:S94–S99. [PubMed] [Google Scholar]
- 8.DeKosky S.T., Marek K. Looking backward to move forward: early detection of neurodegenerative disorders. Science. 2003;302:830–834. doi: 10.1126/science.1090349. [DOI] [PubMed] [Google Scholar]
- 9.Brooks D.J. The early diagnosis of Parkinson’s disease. Ann. Neurol. 1998;44:S10–S18. doi: 10.1002/ana.410440704. [DOI] [PubMed] [Google Scholar]
- 10.Espay A.J., Bonato P., Nahab F.B., Maetzler W., Dean J.M., Klucken J., Eskofier B.M., Merola A., Horak F., Lang A.E. Technology in Parkinson’s disease: challenges and opportunities. Mov. Disord. 2016;31:1272–1282. doi: 10.1002/mds.26642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King A.D. Inertial navigation-forty years of evolution. GEC Rev. 1998;13:140–149. [Google Scholar]
- 12.Luinge H.J., Veltink P.H. Measuring orientation of human body segments using miniature gyroscopes and accelerometers. Med. Biol. Eng. Comput. 2005;43:273–282. doi: 10.1007/BF02345966. [DOI] [PubMed] [Google Scholar]
- 13.Wong W.Y., Wong M.S., Lo K.H. Clinical applications of sensors for human posture and movement analysis: a review. Prosthet. Orthot. Int. 2007;31:62–75. doi: 10.1080/03093640600983949. [DOI] [PubMed] [Google Scholar]
- 14.Ozer E., Feng M.Q. Direction-sensitive smart monitoring of structures using heterogeneous smartphone sensor data and coordinate system transformation. Smart Mater. Struct. 2017;26:045026. [Google Scholar]
- 15.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015. Going deeper with convolutions.https://arxiv.org/abs/1409.4842v1 [Google Scholar]
- 16.Schwab P., Karlen W. PhoneMD: learning to diagnose Parkinson’s disease from smartphone data. arXiv. 2018 http://arxiv.org/abs/1810.01485 [Google Scholar]
- 17.Sage Bionetworks i. Aboensis III Parkinson DREAM challenge submissions (subchallenge 1) 2020. https://www.synapse.org/#!Synapse:syn10849965/wiki/470064
- 18.Bot B.M., Suver C., Neto E.C., Kellen M., Klein A., Bare C., Doerr M., Pratap A., Wilbanks J., Dorsey E.R. The mPower study, Parkinson disease mobile data collected using ResearchKit. Sci. Data. 2016;3:160011. doi: 10.1038/sdata.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Najafi B., Aminian K., Paraschiv-Ionescu A., Loew F., Büla C.J., Robert P. Ambulatory system for human motion analysis using a kinematic sensor: monitoring of daily physical activity in the elderly. IEEE Trans. Biomed. Eng. 2003;50:711–723. doi: 10.1109/TBME.2003.812189. [DOI] [PubMed] [Google Scholar]
- 20.Figo D., Diniz P.C., Ferreira D.R., Cardoso J.M.P. Preprocessing techniques for context recognition from accelerometer data. Pers. Ubiquit. Comput. 2010;14:645–662. [Google Scholar]
- 21.Thang H.M., Viet V.Q., Thuc N.D., Choi D. 2012 International Conference on Control, Automation and Information Sciences (ICCAIS) 2012. Gait identification using accelerometer on mobile phone. [DOI] [Google Scholar]
- 22.Brajdic A., Harle R. Proceedings of the 2013 ACM International Joint Conference on Pervasive and Ubiquitous Computing (UbiComp ’13) 2013. Walk detection and step counting on unconstrained smartphones. [DOI] [Google Scholar]
- 23.Abdulhay E., Arunkumar N., Narasimhan K., Vellaiappan E., Venkatraman V. Gait and tremor investigation using machine learning techniques for the diagnosis of Parkinson disease. Future Generation Comput. Syst. 2018;83:366–373. [Google Scholar]
- 24.Wu Y., Krishnan S. Statistical analysis of gait rhythm in patients with Parkinson’s disease. IEEE Trans. Neural Syst. Rehabil. Eng. 2010;18:150–158. doi: 10.1109/TNSRE.2009.2033062. [DOI] [PubMed] [Google Scholar]
- 25.Zheng H., Yang M., Wang H., McClean S. Machine learning and statistical approaches to support the discrimination of neuro-degenerative diseases based on gait analysis. In: McClean S., Millard P., El-Darzi E., Nugent C., editors. Intelligent Patient Management. Springer Berlin Heidelberg; 2009. pp. 57–70. [Google Scholar]
- 26.Siirtola P., Röning J. Recognizing human activities user-independently on smartphones based on accelerometer data. Int. J. Interact. Multimed. Artif. Intell. 2012;1:38. [Google Scholar]
- 27.Muniz A.M.S., Liu H., Lyons K.E., Pahwa R., Liu W., Nobre F.F., Nadal J. Comparison among probabilistic neural network, support vector machine and logistic regression for evaluating the effect of subthalamic stimulation in Parkinson disease on ground reaction force during gait. J. Biomech. 2010;43:720–726. doi: 10.1016/j.jbiomech.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Shetty S., Rao Y.S. 2016 International Conference on Inventive Computation Technologies (ICICT) 2016. SVM based machine learning approach to identify Parkinson’s disease using gait analysis. [DOI] [Google Scholar]
- 29.Kim H., Lee H.J., Lee W., Kwon S., Kim S.K., Jeon H.S., Park H., Shin C.W., Yi W.J., Jeon B.S. Unconstrained detection of freezing of Gait in Parkinson’s disease patients using smartphone. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2015;2015:3751–3754. doi: 10.1109/EMBC.2015.7319209. [DOI] [PubMed] [Google Scholar]
- 30.Wahid F., Begg R.K., Hass C.J., Halgamuge S., Ackland D.C. Classification of Parkinson’s disease gait using spatial-temporal gait features. IEEE J. Biomed. Health Inform. 2015;19:1794–1802. doi: 10.1109/JBHI.2015.2450232. [DOI] [PubMed] [Google Scholar]
- 31.Businesswire.com Sage Bionetworks in collaboration with the Michael J. Fox Foundation announce winners in the DREAM Parkinson’s disease digital biomarker challenge. 2020. https://www.businesswire.com/news/home/20180117006187/en
- 32.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017;60:84–90. [Google Scholar]
- 33.Albert M.V., Toledo S., Shapiro M., Kording K. Using mobile phones for activity recognition in Parkinson’s patients. Front. Neurol. 2012;3 doi: 10.3389/neur.2012.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth J., Klucken J., Kugler P., Kammerer T., Steidl R., Winkler J., Hornegger J., Eskofier B. Biometric and mobile gait analysis for early diagnosis and therapy monitoring in Parkinson’s disease. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2011;2011:868–871. doi: 10.1109/IEMBS.2011.6090226. [DOI] [PubMed] [Google Scholar]
- 35.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2014 https://arxiv.org/abs/1409.1556 [Google Scholar]
- 36.Itti L., Koch C., Niebur E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998;20:1254–1259. [Google Scholar]
- 37.Itti L., Koch C. Computational modelling of visual attention. Nat. Rev. Neurosci. 2001;2:194–203. doi: 10.1038/35058500. [DOI] [PubMed] [Google Scholar]
- 38.Chen W., Hopfner F., Becktepe J.S., Deuschl G. Rest tremor revisited: Parkinson’s disease and other disorders. Transl. Neurodegener. 2017;6 doi: 10.1186/s40035-017-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jankovic J. Parkinson’s disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 40.Fahn S. Description of Parkinson’s disease as a clinical syndrome. Ann. N. Y. Acad. Sci. 2003;991:1–14. doi: 10.1111/j.1749-6632.2003.tb07458.x. [DOI] [PubMed] [Google Scholar]
- 41.Duval C. Rest and postural tremors in patients with Parkinson’s disease. Brain Res. Bull. 2006;70:44–48. doi: 10.1016/j.brainresbull.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Frigerio R., Elbaz A., Sanft K.R., Peterson B.J., Bower J.H., Ahlskog J.E., Grossardt B.R., de Andrade M., Maraganore D.M., Rocca W.A. Education and occupations preceding Parkinson disease: a population-based case-control study. Neurology. 2005;65:1575–1583. doi: 10.1212/01.wnl.0000184520.21744.a2. [DOI] [PubMed] [Google Scholar]
- 43.Blume J., Rothenfusser E., Schlaier J., Bogdahn U., Lange M. Educational attainment and motor burden in advanced Parkinson’s disease—the emerging role of education in motor reserve. J. Neurol. Sci. 2017;381:141–143. doi: 10.1016/j.jns.2017.08.3241. [DOI] [PubMed] [Google Scholar]
- 44.Arora S., Baig F., Lo C., Barber T.R., Lawton M.A., Zhan A., Rolinski M., Ruffmann C., Klein J.C., Rumbold J. Smartphone motor testing to distinguish idiopathic REM sleep behavior disorder, controls, and PD. Neurology. 2018;91:e1528–e1538. doi: 10.1212/WNL.0000000000006366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lonini L., Dai A., Shawen N., Simuni T., Poon C., Shimanovich L., Daeschler M., Ghaffari R., Rogers J.A., Jayaraman A. Wearable sensors for Parkinson’s disease: which data are worth collecting for training symptom detection models. NPJ Digit Med. 2018;1:64. doi: 10.1038/s41746-018-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hssayeni M.D., Jimenez-Shahed J., Burack M.A., Ghoraani B. Wearable sensors for estimation of parkinsonian tremor severity during free body movements. Sensors. 2019;19 doi: 10.3390/s19194215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumann C.R. Epidemiology, diagnosis and differential diagnosis in Parkinson’s disease tremor. Parkinsonism Relat. Disord. 2012;18(Suppl 1):S90–S92. doi: 10.1016/S1353-8020(11)70029-3. [DOI] [PubMed] [Google Scholar]
- 48.Calne D.B., Snow B.J., Lee C. Criteria for diagnosing Parkinson’s disease. Ann. Neurol. 1992;32(Suppl):S125–S127. doi: 10.1002/ana.410320721. [DOI] [PubMed] [Google Scholar]
- 49.Koller W.C., Vetere-Overfield B., Barter R. Tremors in early parkinsonʼs disease. Clin. Neuropharmacology. 1989;12:293–297. doi: 10.1097/00002826-198908000-00006. [DOI] [PubMed] [Google Scholar]
- 50.Breen D.P., Evans J.R., Farrell K., Brayne C., Barker R.A. Determinants of delayed diagnosis in Parkinson’s disease. J. Neurol. 2013;260:1978–1981. doi: 10.1007/s00415-013-6905-3. [DOI] [PubMed] [Google Scholar]
- 51.Fang F., Chen H., Feldman A.L., Kamel F., Ye W., Wirdefeldt K. Head injury and Parkinson’s disease: a population-based study. Mov. Disord. 2012;27:1632–1635. doi: 10.1002/mds.25143. [DOI] [PubMed] [Google Scholar]
- 52.Willis A.W., Schootman M., Evanoff B.A., Perlmutter J.S., Racette B.A. Neurologist care in Parkinson disease: a utilization, outcomes, and survival study. Neurology. 2011;77:851–857. doi: 10.1212/WNL.0b013e31822c9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes A.J., Daniel S.E., Lees A.J. Improved accuracy of clinical diagnosis of Lewy body Parkinson’s disease. Neurology. 2001;57:1497–1499. doi: 10.1212/wnl.57.8.1497. [DOI] [PubMed] [Google Scholar]
- 54.Dickson D.W. Neuropathology of Parkinson disease. Parkinsonism Relat. Disord. 2018;46(Suppl 1):S30–S33. doi: 10.1016/j.parkreldis.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marsili L., Rizzo G., Colosimo C. Diagnostic criteria for Parkinson’s disease: from james Parkinson to the concept of prodromal disease. Front. Neurol. 2018;9:156. doi: 10.3389/fneur.2018.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yahr M.D., Duvoisin R.C. Drug therapy of parkinsonism. N. Engl. J. Med. 1972;287:20–24. doi: 10.1056/NEJM197207062870105. [DOI] [PubMed] [Google Scholar]
- 57.Wu K., Wu X. 2007 1st International Conference on Bioinformatics and Biomedical Engineering. 2007. A wireless mobile monitoring system for home healthcare and community medical services. [DOI] [Google Scholar]
- 58.Nejati H., Pomponiu V., Do T.-T., Zhou Y., Iravani S., Cheung N.-M. Smartphone and mobile image processing for assisted living: health-monitoring apps powered by advanced mobile imaging algorithms. IEEE Signal. Process. Mag. 2016;33:30–48. [Google Scholar]
- 59.Srivastava S., Soman S., Rai A., Srivastava P.K. Deep learning for health informatics: recent trends and future directions. International Conference on Advances in Computing, Communications and Informatics (ICACCI); 2017; 2017. [DOI] [Google Scholar]
- 60.Zhou C., Yao J., Motani M. Optimizing autoencoders for learning deep representations from health data. IEEE J. Biomed. Health Inform. 2018;23:103–111. doi: 10.1109/JBHI.2018.2856820. [DOI] [PubMed] [Google Scholar]
- 61.Shin J., Shin D., Shin D., Her S., Kim S., Lee M. Human movement detection algorithm using 3-axis accelerometer sensor based on low-power management scheme for mobile health care system. Adv. Grid Pervasive Comput. 2010:81–90. [Google Scholar]
- 62.Sathyanarayana A., Joty S., Fernandez-Luque L., Ofli F., Srivastava J., Elmagarmid A., Arora T., Taheri S. Sleep quality prediction from wearable data using deep learning. JMIR Mhealth Uhealth. 2016;4:e125. doi: 10.2196/mhealth.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krejcar O., Jirka J., Janckulik D. Use of mobile phones as intelligent sensors for sound input analysis and sleep state detection. Sensors. 2011;11:6037–6055. doi: 10.3390/s110606037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alqassim S., Ganesh M., Khoja S., Zaidi M., Aloul F., Sagahyroon A. 2012 IEEE 14th International Conference on E-Health Networking, Applications and Services (Healthcom) 2012. Sleep apnea monitoring using mobile phones. [DOI] [Google Scholar]
- 65.Chirra M., Marsili L., Wattley L., Sokol L.L., Keeling E., Maule S., Sobrero G., Artusi C.A., Romagnolo A., Zibetti M. Telemedicine in neurological disorders: opportunities and challenges. Telemed. J. E. Health. 2019;25:541–550. doi: 10.1089/tmj.2018.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scarborough J.B. The gyroscope. theory and applications. Math. Gaz. 1959;43:304–305. [Google Scholar]
- 67.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 68.He K., Zhang X., Ren S., Sun J. 2015 IEEE International Conference on Computer Vision (ICCV) 2015. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. [DOI] [Google Scholar]
- 69.Xiong W., Droppo J., Huang X., Seide F., Seltzer M.L., Stolcke A., Yu D., Zweig G. Toward human parity in conversational speech recognition. IEEE/ACM Trans. Audio, Speech, Lang. Process. 2017;25:2410–2423. [Google Scholar]
- 70.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A.W.M., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 71.Jiang Y.Q., Xiong J.H., Li H.Y., Yang X.H., Yu W.T., Gao M., Zhao X., Ma Y.P., Zhang W., Guan Y.F. Recognizing basal cell carcinoma on smartphone-captured digital histopathology images with a deep neural network. Br. J. Dermatol. 2019;182:754–762. doi: 10.1111/bjd.18026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code associated with this paper can be freely accessed and downloaded via https://github.com/GuanLab/PDDB/. The mPower dataset used in this paper can be accessed via the mPower public research portal: https://www.synapse.org/#!Synapse:syn4993293/wiki/247859.