Abstract
Summary
The kinetochore is a complex of proteins, broadly conserved from yeast to man, that resides at the centromere and is essential for chromosome segregation in dividing cells. There are no known functions of the core complex outside of the centromere. We now show that the proteins of the kinetochore have an essential post-mitotic function in neurodevelopment. At the embryonic neuromuscular junction of Drosophila melanogaster, mutation or knockdown of many kinetochore components cause neurites to overgrow and prevent formation of normal synaptic boutons. Kinetochore proteins were detected in synapses and axons in Drosophila. In post-mitotic cultured hippocampal neurons, knockdown of mis12 increased the filipodia-like protrusions in this region. We conclude that the proteins of the kinetochore are repurposed to sculpt developing synapses and dendrites and thereby contributes to the correct development of neuronal circuits in both invertebrates and mammals.
eTOC
The kinetochore has only been shown to function in chromosome segregation in dividing cells. Zhao et al. show that kinetochore proteins are repurposed during neuronal development and are required in the development of both fly and mammalian neurons for the correct formation of synapses and dendrites.
Keywords: Kinetochore, Drosophila, Neuromuscular Junction, Hippocampus, Centromere, Synapse
Introduction
The kinetochore complex mediates chromosome segregation in dividing cells and has been highly conserved in eukaryotic evolution. Kinetochores reside at the centromere of each chromosome and, by capturing spindle microtubules and attaching them to chromosomes, allow the spindle to pull chromosomes towards the poles (Cheeseman et al., 2006; Musacchio and Desai, 2017; Varma and Salmon, 2012). The proteins of the kinetochore form a load-bearing structure and enhance the stability of the bound microtubules while also providing a scaffold for key signaling events during cell division (Musacchio and Desai, 2017; Varma and Salmon, 2012). One subcomplex, the inner kinetochore, is bound to centromeric heterochromatin. Three other subcomplexes, named Mis12, Knl1, and Ndc80 after protein constituents, link the inner kinetochore to the microtubule-binding subunit ndc80 (Cheeseman et al., 2006; Musacchio and Desai, 2017; Przewloka et al., 2011; Varma and Salmon, 2012; Venkei et al., 2012). Heretofore, there are no known functions of this core kinetochore complex outside of cell division. However, in a forward genetic screen we identified mis12 as impacting Drosophila synaptic development. Mutations of many kinetochore proteins, including members of each kinetochore subcomplex, substantially altered the morphology of the Drosophila neuromuscular junction (NMJ), preventing proper formation of synaptic boutons. Kinetochore mutations also altered dendritic morphology in Drosophila sensory neurons and knockdown of mis12 altered the morphology of rat hippocampal dendrites, suggesting a conserved postmitotic function in neuronal development.
Results
Developmental defects in embryonic neuromuscular junctions of mis12 mutants
Synapse formation and function have been studied extensively at the neuromuscular junction of Drosophila (Budnik, 1996; Van Vactor and Sigrist, 2017). However, because most mutant screens have been conducted at the third-instar stage, essential synaptogenesis genes may have been missed due to their early lethality (Kurshan et al., 2009; Pack-Chung et al., 2007). We therefore screened for defects in embryonic synapse formation. The cell bodies of embryonic motor neurons in Drosophila arise from neuroblasts in the ventral nerve cord and, upon differentiation, extend axons into the periphery where they contact body wall muscles in a highly stereotyped fashion. Upon reaching the appropriate muscles, their growth cones are transformed into thin neurite branches and subsequently mature into rounded boutons that contain active zone proteins and are apposed to glutamate receptors on the muscle surface. To identify genes necessary for the process of forming synapses, we screened 185 chromosomal deficiencies from a collection previously identified as developing to late embryonic stages and uncovering approximately 50% of the Drosophila genome (Wright et al., 2010). For each deficiency, at least 5 homozygous embryos were dissected at 21 h after egg laying (AEL), a stage when motor neurons in wildtype embryos have contacted their appropriate target muscles and formed their stereotyped synapses. Embryos were stained with antibodies to horseradish peroxidase (HRP) to visualize neuronal membranes as well as to the presynaptic active zone marker Bruchpilot (Brp) and a postsynaptic glutamate receptor subunit (GluRIIC). Of the lines that survived to 21 h AEL, 6 showed distinct developmental abnormalities at the neuromuscular junctions on muscles 6,7,12, and 13 (Table S1). One of these, Df(3L)BSC27, was selected for further study because it had a particularly strong anatomical phenotype. The genetic locus was further refined by analysis of overlapping deficiencies in the same region, two of which, Df(3L)BSC224 and Df(3L)BSC374, showed the same phenotype (Fig. 1A). In wildtype neurons, by 21 h AEL, the growth cones of motor neurons have been transformed into a pair of short, thin branches at the boundary between muscles 6 and 7 and another pair on the surface of muscles 12 and 13. On each of these branches a few large, round synaptic boutons have formed (Fig. 1B). In contrast, in embryos homozygous for these deficiencies or in combination with one another, although the axons had reached the target muscles and branched in approximately the correct places, large boutons were seldom present (Fig. 1C). Moreover, the neurite branches on the muscles were abnormally long; in some embryos, these fine branches extended well beyond their normal range and encountered unrelated nearby nerves with which they appeared to fasciculate (Fig. 1C). The overgrowth phenotype was highly penetrant; it was apparent in each embryo and each segment examined.
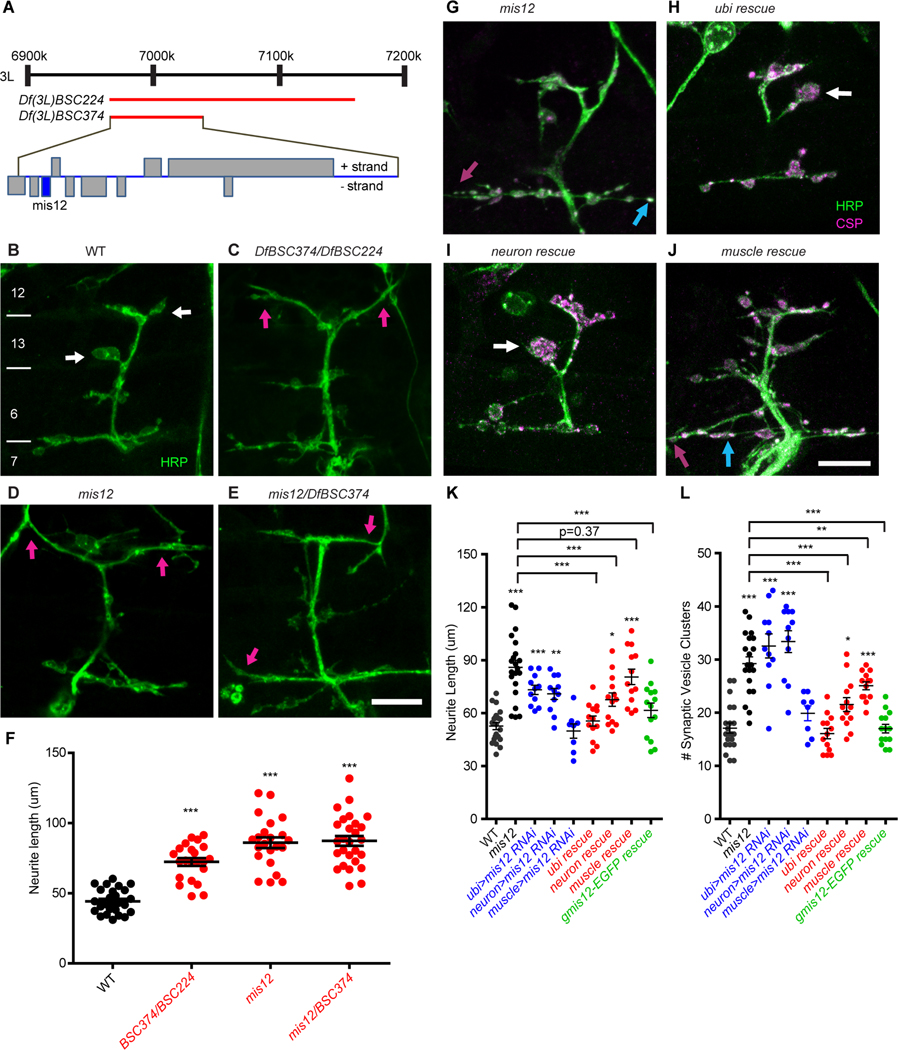
Figure 1. Neuronal Mis12 is required for embryonic development of the Drosophila neuromuscular junction.
(A) Map of the genomic region containing mis12, adapted from Flybase, and showing the extent of two chromosome deficiencies (red bars) that were used in identifying the gene responsible for the synaptic phenotype. (B-E) Anti-HRP staining of the innervation of muscles 6, 7, 12, and 13 in 21-h embryos of the indicated genotypes. White bars in (B) mark the boundaries of the numbered muscles. Wildtype embryos (WT) have large boutons (white arrows) that are not present in the genotypes that disrupt mis12 which instead form overly long neurite branches (magenta arrows). Scale bar, 10 μm. (F) Quantification of neurite length for the genotypes shown in (B-E) ***p<0.001, one-way ANOVA with Dunnett’s multiple comparisons test. Error bars, mean ± s.e.m. (G-J) Embryonic neuromuscular junctions as in (B-E) stained with anti-HRP and anti-CSP. The NMJ phenotype of mis12f03756f03756 embryos (G) was rescued by ubiquitous (H) or neuronal (I), but not muscle (J) expression of Mis12. Scale bar, 10 μm. (K, L) Quantitation of the phenotypes in (G-J) and of neuromuscular junctions expressing mis12 RNAi (Fig. S2). Expression of RNAi either ubiquitously or selectively in neurons, but not in muscle, mimicked the mis12 phenotype. In addition, the mis12 phenotype was rescued by gmis12-EGFP, a transgene of genomic DNA in which EGFP-tagged mis12 is expressed under control of its endogenous promoter. Each genotype was compared to WT using one-way ANOVA with Dunnett’s multiple comparisons test, and, as indicated by brackets, rescue was compared to mis12 with an unpaired t test with Welch’s correction. ***p<0.001, **p<0.01, *p<0.05, Error bars, mean ± s.e.m.
At least 10 open reading frames are removed by Df(3L)BSC374, and so, to identify the gene responsible for the phenotype, we examined known alleles, P-element insertions, and RNAi lines for the genes in this region. A PBac-element insertion in mis12 (mis12f03756) (Schittenhelm et al., 2010; Schittenhelm et al., 2007; Venkei et al., 2011) was found to recapitulate the full mutant phenotype when either homozygous or placed over Df(3L)BSC374 (Fig. 1D-F). mis12f03756 contains a transposon insertion in the first intron of the mis12 locus and causes embryonic lethality with undetectable levels of Mis12 protein in late stage embryos (Venkei et al., 2011). Although the development of the synapse was defective in mis12 mutants, motor neurons had differentiated appropriately (Fig. S1 A-C) and the underlying muscles were grossly normal; in each hemisegment of the embryo, multinucleated muscles formed in the proper numbers and positions (Fig. S1D, E), and with at most a very slight (7%) decrease in the number of nuclei in each muscle 6 (Fig. S1 F-H, p=0.16).
Mis12 functions presynaptically in regulating NMJ growth
The role of mis12 was further characterized by expression of a mis12 transgene and staining of embryos for neuronal membranes and the synaptic vesicle marker Cysteine String Protein (CSP). The mis12f03756 phenotype could be rescued by ubiquitous (da::GAL4) or neuronal expression (elav::GAL4) of the mis12 transgene, but not by its expression in the muscle (C57::GAL4) (Fig. 1G-L), implying a presynaptic requirement for mis12. To further test the cell type responsible for the phenotype, a UAS::mis12RNAi transgene was expressed either broadly or exclusively in neurons or muscle. Knockdown of mis12, either ubiquitously or in neurons recapitulated the mis12f03756 phenotype but knockdown in muscle did not (Fig. 1 K, L and S2A-D). Immunolocalization of CSP revealed that the long neurite branches of the mutant synapses had multiple small clusters of vesicles present along their length, sometimes accompanied by small swellings of the axons, although large rounded boutons were seldom observed (Fig. 1G-L, Fig. S2A-D). These putative synaptic sites also contained the active zone protein Bruchpilot and were opposed by postsynaptic glutamate receptors (Fig. S2E, F). Thus, the mis12 mutant phenotype cannot be explained by developmental arrest at a stage prior to synapse maturation; rather the phenotype presents a selective defect in particular aspects of synapse formation that govern the length of branches and the formation of boutons.
Many kinetochore components are required for proper NMJ formation
The kinetochore is composed of a DNA-anchoring inner kinetochore that includes the histone Cenp-A (CID in Drosophila melanogaster) and Cenp-C, and three additional complexes of the outer kinetochore, the Mis12, Knl1, and Ndc80 complexes (Fig. 2G) (Cheeseman et al., 2006; Musacchio and Desai, 2017; Przewloka et al., 2011; Varma and Salmon, 2012; Venkei et al., 2012). Mis12 is an essential component of the kinetochore and forms a bridge between the inner kinetochore and the other complexes (Kline et al., 2006; Petrovic et al., 2016; Petrovic et al., 2010). Consequently, in misl203756 the microtubule-binding protein Ndc80 is not recruited to centromeres in late stage embryos (Venkei et al., 2011). Nevertheless, much of embryogenesis proceeds normally in these mutants due to the presence of maternally deposited mRNA and protein that can support the rapid cycles of mitosis in early embryos that generate most of the embryo (Venkei et al., 2011). The defects in synapse development in mis12 prompted us to examine other components of the kinetochore and thereby determine whether knockdown by RNAi or mutant alleles of those genes would produce similar phenotypes. We examined components of each of the kinetochore complexes and found for each complex at least one component that increased neurite length, increased the number of distinct vesicle clusters, and prevented large, normal boutons from forming (Fig. 2A-I and Fig. S3A-F). Thus, from the inner kinetochore Cenp-A (cid) gave a mis12-like phenotype as did nnf1a and 1b from the Mis12 complex, spc105R from the Knl1 complex, and ndc80 and spc25 from the Ndc80 complex. Not every genotype tested, however, differed from wildtype. Alleles of Cenp-C, nsl1, and nuf2, and RNAi against nuf2 had normal synaptic morphologies (Fig. 2H, I and Fig. S3C, D). The lack of a phenotype in these genotypes may indicate that the full complement of kinetochore proteins is not necessary for synapse formation, but it may also be due to poor efficacy of an RNAi line or indicate that, for those proteins, perdurance of the maternal contribution is sufficient to support synapse development. Cenp-C protein, for example, is still abundant in late-stage embryos even in cenp-Cprl41 homozygotes (Fig. S3G, H). The most unexpected finding was the NMJ phenotype of the histone Cenp-A (cid), as histones normally require binding to DNA for proper assembly. Drosophila Cenp-A, however, is unusual in that it can form a complex with Cenp-C during kinetochore formation prior to its association with DNA. This association requires the chaperone Cal1 to be present in the complex (Chen et al., 2014; Schittenhelm et al., 2010) and we therefore predicted that Cal1 would be required by the kinetochore proteins during synaptic development as well. As predicted, Cal1c03646embryos shared the defects in bouton formation and neurite overgrowth present in the other kinetochore mutants (Figures 2H, I and S3B).
Figure 2. Many kinetochore components are required for proper NMJ formation.
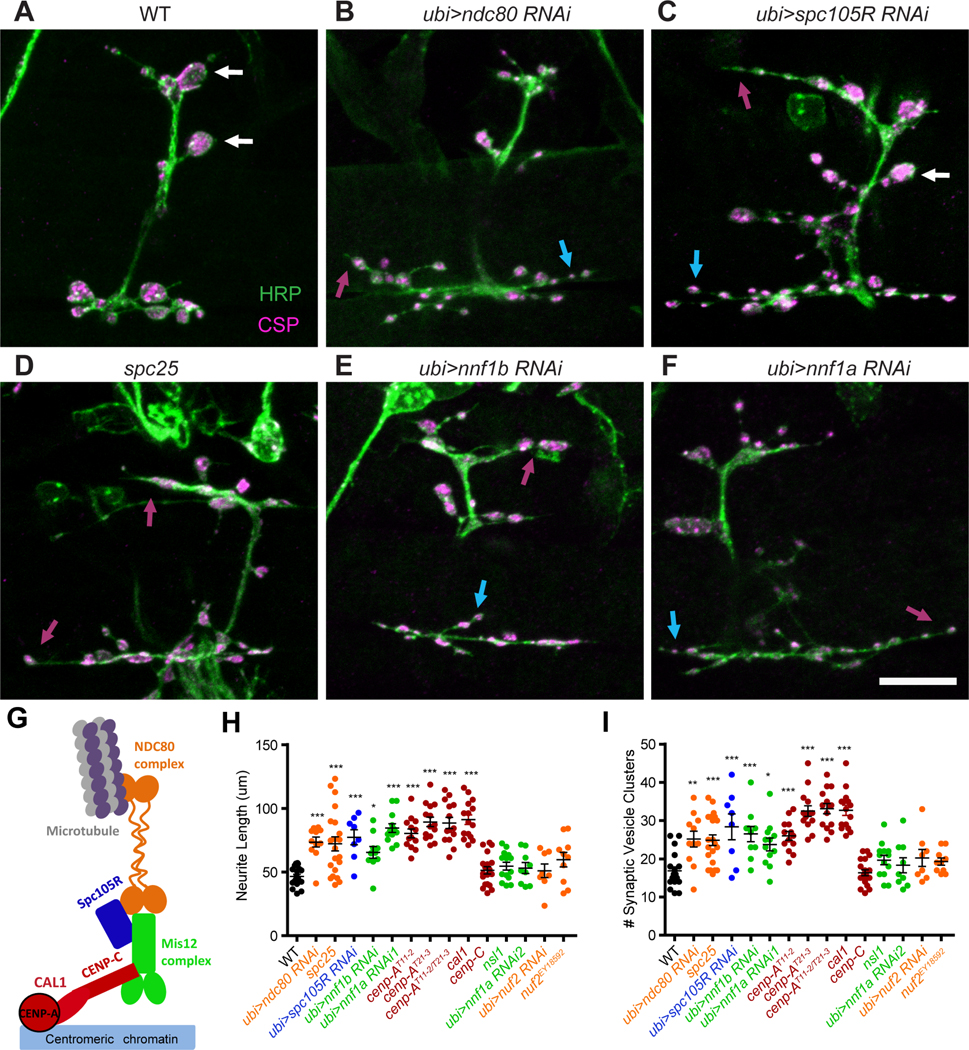
(A-F) For the indicated genotypes, loss of a kinetochore protein causes overly long neurites (magenta arrows) and multiple small vesicle clusters stained with anti-CSP (blue arrows), and few or no large boutons (white arrows). (G) Schematic of the subcomplexes that make up the kinetochore. (H, I) Quantification of the phenotypes for kinetochore mutants and RNAi knockdown, colored to correspond to the subcomplexes in (G). Each genotype was compared with WT using one-way ANOVA with Dunnett’s multiple comparisons test. Scale bars, 10 μm. ***p<0.001, **p<0.01, *p<0.05. Error bars, mean ± s.e.m.
Kinetochore proteins act broadly in neurodevelopment.
In the course of examining the embryos of kinetochore mutants, we noted that the neuropil of the ventral nerve cord was consistently abnormal. In Drosophila embryos, neuronal cell bodies form a cortex around the outside of the nerve cord and extend axons and dendrites into the two synapse-rich zones of neuropil that lie to either side of the midline. In embryos of kinetochore mutants, this neuropil is narrower, longer, and less densely stained with synaptic markers than that of control embryos (Fig. 3 AD). Although the complexity of the ventral nerve cord hinders a more detailed analysis of the underlying cellular defects, these phenotypes suggest a widespread requirement for the kinetochore proteins in the wiring of the central nervous system. To determine if kinetochore phenotypes extend to dendrite development we examined sensory neurons of the dorsal cluster because of their highly stereotyped branching (Corty et al., 2009; Grueber and Jan, 2004; Jan and Jan, 2010; Matthews et al., 2007). Because they are not contacted by presynaptic terminals, they afford an opportunity to look at dendritic development that is independent of contact with axon terminals. In late stage embryos the dendritic arbors of neurons in the dorsal cluster extend out from the somata but rarely reach or cross the segment boundaries. In mutations of mis12, cenp-A T11−2, cenp-A T21−3, and Call the arbors were larger and this was most apparent upon counting dendrite branches that crossed segment boundaries (Fig. S3I-N). The hyperextension of the dendrites resembles the overgrowth of axon endings at the NMJ and indicates that the neurodevelopmental function of kinetochore proteins is not restricted to the pre-synapse and indeed is likely to be widespread in the embryonic nervous system.
Figure 3. Kinetochore proteins localize to embryonic NMJs and synaptic neuropil.
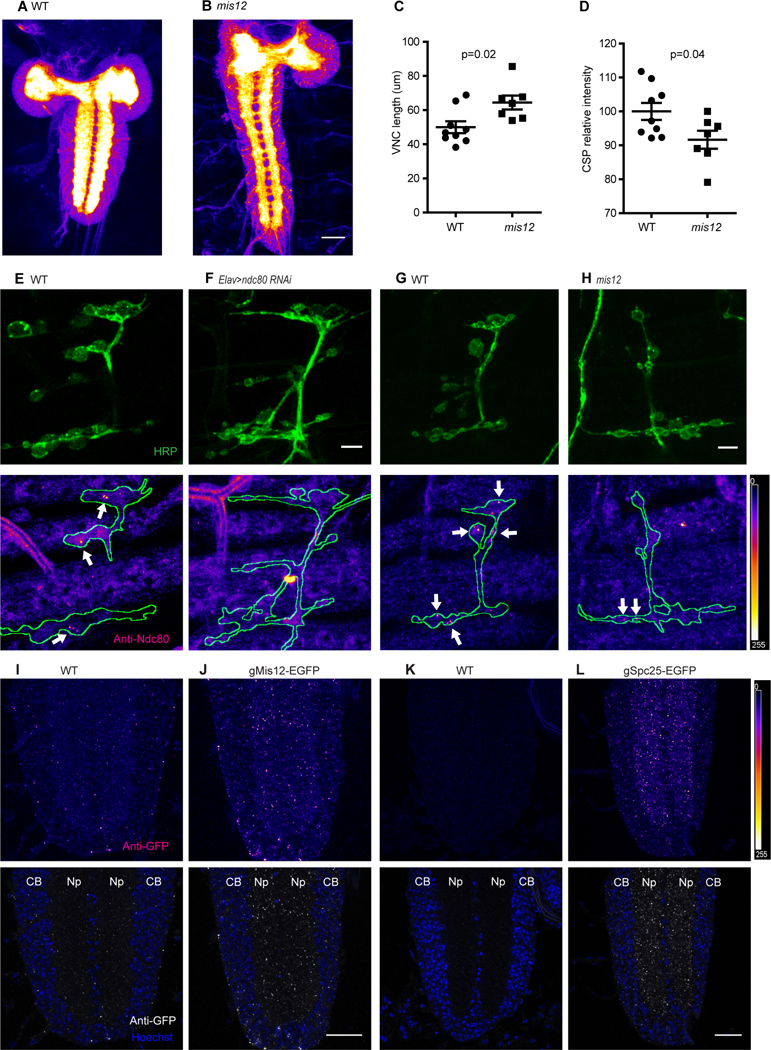
(A, B) Anti-CSP staining of WT and mis12 mutant nervous systems. The synaptic neuropil, forming the center of the brain lobes and parallel stripes in the ventral nerve cord (VNC) is intensely stained in contrast to the surrounding cortices of cell bodies. (C, D) Quantification of VNC length (C) and anti-CSP signal intensity (D) for the genotypes in (A, B). (E-H) ndc80 antibody staining of NMJs in wildtype embryos (E, G), and in embryos expressing ndc80 RNAi in neurons (F) or in mis12f03756f03756 embryos (H). Nerve endings were identified by anti-HRP staining (top panels) and then outlined (below) in images of anti-ndc80 immunoreactivity. Sparse puncta of ndc80 (arrows) were present in boutons of wildtype embryos, but largely absent when ndc80 was knocked down by RNAi and also fewer and fainter in mis12 mutants. (I-L) Anti-GFP staining for EGFP fusion proteins of Mis12 and Spc25, in genomic constructs that place their expression under control of the endogenous promoters. In the neuropil of the embryonic ventral nerve cord Mis12-EGFP (J) and Spc25-EGFP (L) formed bright puncta of GFP immunoreactivity that were absent from embryos lacking the transgenes (I and K). CB, cell body; Np, neuropil. Scale bar, 5 μm (E-H) and 20 μm (A-D and I-L).
Kinetochore proteins localize to synaptic neuropil and axons
Although core kinetochore proteins have only been observed at centromeres, the neurodevelopmental phenotypes suggested extra-nuclear functions. We therefore tested available antibodies to components of the Drosophila kinetochore complex and found that antibodies to Ndc80 (Venkei et al., 2011) were the most effective for immunostaining of late-stage embryos. Ndc80 immunoreactivity was present in sparse but bright puncta within boutons of the embryonic NMJ. Typically, only 1 or 2 puncta could be seen in any bouton (Fig. 3E). The specificity of the staining was confirmed by expression of ndc80 RNAi (Fig. 3F). Moreover, the ndc80 puncta were largely absent in mis12 mutant embryos, suggesting that their presence was dependent on the formation of kinetochore-like complex (Fig. 3G, H).
Because kinetochore mutations also altered the development of the central neuropil, and because the high density of synapses in this region could be expected to enhance the ability to detect sparse complexes, we also examined the neuropil for the presence of kinetochore proteins. Anti-Ndc80 stained the synaptic neuropil, a region devoid of nuclei (Fig.S4A) in which, consequently, the presence of the protein cannot be attributed to its conventional centromeric function. Moreover, Ndc80 immunoreactivity in the neuropil was diminished in both a mis12 and a Cenp-A/cid mutant background (Fig. S4B-D).
To determine if mis12 was also present in the neuropil, we took advantage of a previously reported construct that expresses GFP-tagged Mis12 under control of its endogenous promotor and that had been shown appropriately to label centromeres and to rescue the chromosome segregation phenotype of mis12 (Schittenhelm et al., 2007). The GFP-tagged construct is thus functional and likely to reflect the expression pattern and localization of the native protein. We found that this genomic GFP-mis12 construct could also rescue the embryonic NMJ phenotype of mis12 (Fig. 1K, L). GFP-Mis12, detected with anti-GFP, formed discrete puncta in the neuropil regions of the VNC (Fig. 3I, J). We also examined the segmental nerves of third-instar larvae and observed discrete puncta of mis12-GFP to be present there as well (Fig. S4 E, F). In each tissue, the specificity of the anti-GFP immunostaining was clearly distinct from background immunofluorescence in the absence of the tagged transgene (Fig 3I, S4E). To determine whether additional kinetochore components are present in synaptic and axonal regions, we examined additional genomic transgenes bearing GFP-tagged kinetochore proteins whose expression is driven by their endogenous promoters and were previously shown to rescue mitotic defects (Schittenhelm et al. 2007). EGFP-Nuf2, like Mis12-GFP, formed sparse puncta within the axons of segmental nerves of third instar larvae (Fig. S4G). Spc25-GFP, like Ndc80, gave rise to puncta in the central neuropil of the embryonic ventral nerve cord (Fig. 3K, L). In a mis12 mutant background, spc25-EGFP is more abundant and much of this Spc25-GFP signal was redistributed to the cell body region (Fig. S4H, I). In aggregate, the presence of multiple kinetochore components in axonal and synaptic regions devoid of nuclei is consistent with a post-mitotic role in neurons that is distinct from their known chromosomal function.
Mis12 functions postmitotically in hippocampal dendritic development.
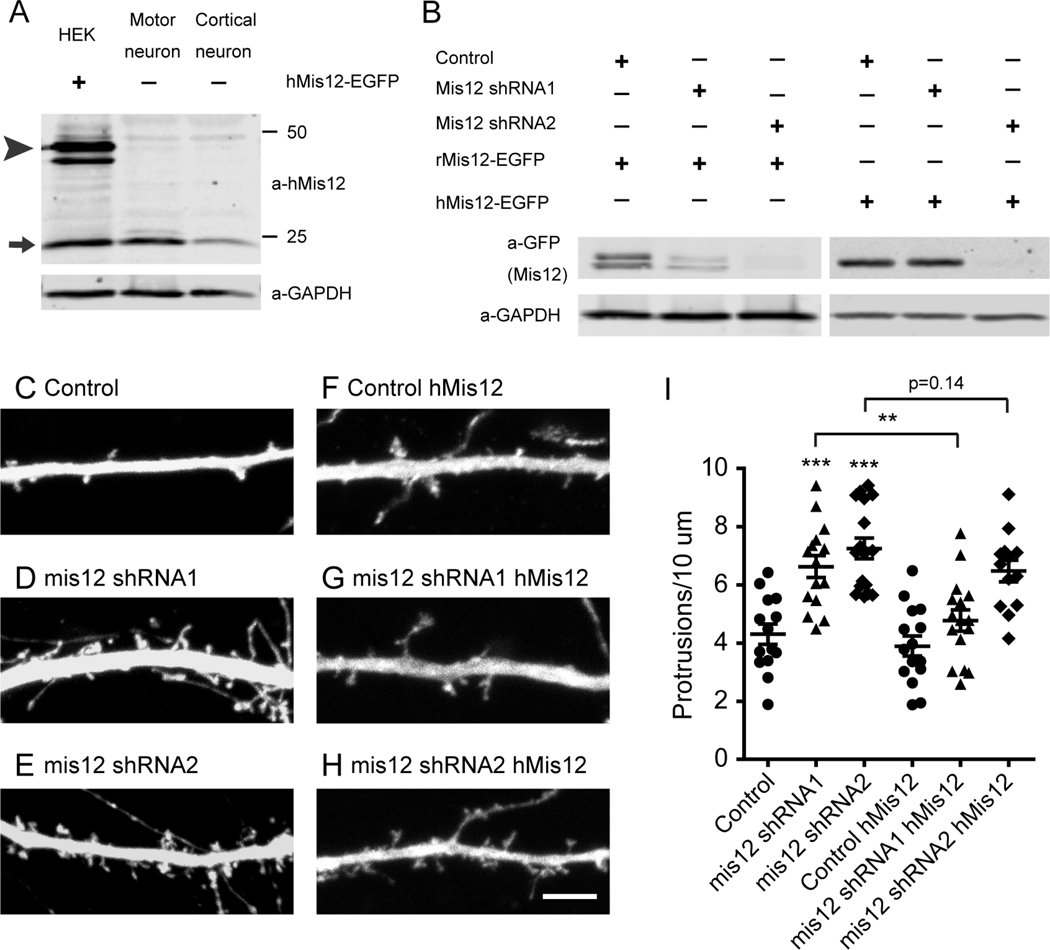
To determine if this unexpected post-mitotic role of kinetochore proteins was also relevant to mammalian neuronal development we undertook a preliminary examination of mis12 in post-mitotic mammalian neurons. We obtained antibodies to human mis12 (Petrovic et al., 2016) and verified that they detected both endogenous human Mis12 and a GFP-tagged human Mis12 transgene on immunoblots of HEK293T cells. To detect neuronal expression and avoid the confounding presence of dividing non-neuronal cells, we used cultures of iPS-derived human motor neurons and cortical neurons that were postmitotic and cultured in the absence of any non-neuronal supporting cells. Anti-hMis12 detected a protein of the appropriate size in both motor neuron and cortical neuron lysates that comigrated with that of Mis12 from HEK293T cells (Fig. 4A). To assess the functional role of mis12 in postmitotic neurons, we tested two shRNA constructs for efficacy in disrupting expression of both GFP-tagged rat and human Mis12 upon expression in HEK293T cells. shRNA1 was effective in knockdown of rat, but not human, Mis12 while shRNA2 was effective in knockdown of both (Fig. 4B). We then transfected primary cultures of rat hippocampal neurons with either of the two mis12 shRNA constructs. At DIV 15, a stage at which extensive synapse formation is underway, knockdown of Mis12 by either shRNA construct, but not a control construct, caused a clear alteration in the number of protrusions formed along dendrites (Fig. 4C-E). This effect was quantified by determining the density of these filipodia-like projections along a specified stretch of dendrite 20 μm from the cell body. The average density of the protrusions was increased by 50% upon knockdown of Mis12 (Fig. 4I). To verify the specificity of the shRNA, we cotransfected human mis12 and, as predicted by the specificity profile of the shRNA (Fig. 4B), expression of the human mis12 cDNA could rescue the dendritic phenotype of shRNA1, but only poorly rescue that of shRNA2 (Fig. 4F-I). Thus, disruption of mis12 expression in postmitotic neurons alters dendritic development.
Figure 4. Mis12 knockdown phenotype in rat hippocampal dendrites.
(A) Immunoblot of HEK293T cells transfected with EGFP-tagged human Mis12, and of motor neurons and cortical neurons differentiated from human iPSCs and cultured in the absence of non-neuronal supporting cells. In HEK293T cells, anti-hMis12 identified both the EGFP-tagged Mis12 (arrowhead) and endogenous Mis12 (arrow). In neurons anti-hMis12 labeled the endogenous Mis12 band. (B) Efficacy of mis12 shRNA. The indicated shRNA were cotransfected into HEK293T cells with EGFP-tagged rat or human Mis12 cDNAs. shRNA1 knocked down expression of rat Mis12 but not human Mis12, whereas shRNA2 was effective on both. (C-H) Dendrites of rat hippocampal neurons were cotransfected with cytoplasmic Dsred (to visualize dendritic morphology), the indicated shRNA (from constructs that also expressed GFP), and, in (F-H), human mis12 DNA (res). Scale bar, 5 μm. (I) Quantification of the density of dendritic protrusions in each condition revealed that knockdown of Mis12 increased protrusions and this effect was reversed by the rescue of the shRNA1 upon expression of the resistant human clone. Each genotype was compared to control using one-way ANOVA with Dunnett’s multiple comparisons test, and, as indicated by brackets, rescue by hMis12 expression was analyzed with an unpaired t test with Welch’s correction. ***p<0.001, **p<0.01, *p<0.05. Error bars, mean ± s.e.m.
Discussion
Although there is no precedent for core kinetochore proteins functioning outside of chromosome mechanics, several lines of evidence argue that the observed Drosophila phenotypes in neurons are not secondary to chromosome segregation defects. 1. Errors of kinetochore assembly are lethal during cell division, arresting at the spindle assembly checkpoint, and this would have prevented motor neurons from forming; we saw no loss of motor neurons in mis12 mutants (Fig. S1A-C). 2. Were aneuploid or polyploid cells sometimes to escape that lethality, they would do so rarely, randomly, and with heterogeneity in the chromosomes lost. We, however, detect synaptic phenotypes consistently at the NMJs of all mutant embryos examined. 3. The selective nature of the NMJ defect is difficult to reconcile with chromosomal aberrations: muscles are correctly patterned and have the correct complement; motor neurons are born and target consistently and appropriately to their muscles; and synaptic specializations form with appropriate components, despite the failure to form large boutons. 4. Ndc80 immunoreactivity is present at the embryonic NMJ and elsewhere in the nervous system; tagged kinetochore proteins, expressed under control of their normal promoters, were detected outside of nuclei in synaptic and axonal regions. In mammalian neurons, although only mis12 has been examined so far, the function of mis12 is clearly postmitotic. It was detected by Western blot in postmitotic neurons and the knockdown of mis12 in postmitotic hippocampal neurons altered the morphology of hippocampal dendrites. A similar post-mitotic requirement for the proteins of the kinetochore has also been demonstrated in C. elegans, where the degradation of kinetochore proteins selectively in postmitotic and differentiated sensory neurons disrupts their morphogenesis (Cheerambathur and Desai, accompanying submission). Interestingly, there is precedent for the repurposing of other mitotic proteins, such as those of the anaphase promoting complex, for post-mitotic functions in synaptogenesis (Juo and Kaplan, 2004; van Roessel et al., 2004; Yang et al., 2009).
We were fortunate to find the synaptogenic role of mis12 in our screen: had the maternal contribution been less, the motor neurons might not have formed and had the contribution persisted into late stage embryos, it might have been sufficient fully to support synaptogenesis. The observation that at least eight kinetochore components, including representatives of each of the kinetochore subcomplexes, give rise to the same phenotype at the NMJ suggests that they are functioning in a complex very much like that at the centromere. This supposition is further strengthened by our observations that, in several cases we examined, the proper localization of one component was altered in a genetic background mutant for another component (Figs. 3 and S4). Although some alleles and RNAi lines did not have a phenotype, their role may have been obscured by the persistence of sufficient maternal contribution, as in the case of Cenp-C (Fig. S3G, H), or poor efficacy of the RNAi line. This is particularly true for Nuf2 which was detectable in larval axons (Fig. S4G) although its RNAi line lacked a phenotype (Fig. 2H, I, and Fig. S3D). It remains to be determined if the complete complement of kinetochore proteins or only mis12 function in the development of hippocampal dendrites.
In light of the knockdown phenotype of ndc80, the microtubule-binding subunit (Powers et al., 2009), at the embryonic NMJ and its presence in sparse puncta at that synapse, one parsimonious hypothesis is that the kinetochore proteins interact with neuronal microtubules akin to their function at the centromere. Microtubule dynamics are crucial to the formation of both synapses and dendrites, and this is consistent with our observation of significant overextension of both synaptic branches and sensory dendrites in Drosophila (Fig. S3I-N) and alterations in dendritic morphology in hippocampal neurons (Fig. 4C-I). At the embryonic NMJ and in larval nerves, where individual puncta of kinetochore proteins could be resolved, the puncta were sparse, with just one or two per bouton. The axonal puncta in larval nerves were not as bright as those in nearby glial nuclei. Whereas multiple kinetochores and microtubule + ends are present at each centromere, the dim axonal puncta may represent individual kinetochore-like complexes at individual + ends and this would account for the difficulty of imaging the proteins outside of the densely synaptic neuropil of the ventral nerve cord. In axons, the + ends of axonal microtubules are oriented towards growth cones and synapses and, while microtubules are splayed in growth cones, they are replaced during synaptogenesis by more stable bundles (Van Vactor and Sigrist, 2017). When microtubules are not appropriately stabilized, synapse formation is perturbed, giving rise to abnormal extensions of axons and improper bouton formation (Borgen et al., 2017; Bourgeois et al., 2015; Graf et al., 2011; Pawson et al., 2008). The phenotypes now reported suggest that this developmental transition requires a kinetochore-like complex. Hippocampal dendrites contain microtubules of both polarities (Baas and Lin, 2011) and the increased filipoidia-like protrusions that appear upon knockdown of mis12 may arise from misorganization or overgrowth of dendritic microtubules. Future studies will need to clarify how the kinetochore influences microtubule organization. Analysis of kinetochore mutations has thus uncovered a previously unknown mechanism that appears to coopt the fundamental mitotic functions of the ancient kinetochore complex for non-mitotic functions in both invertebrate and vertebrate neurodevelopment. A deeper understanding of these synaptogenic functions should therefore illuminate a process central to the accurate wiring of the brain.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas Schwarz (Thomas.Schwarz@childrens.harvard.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Drosophila stocks
The P-element insertion mutant of mis12 (mis12f03756), cenp-A mutant (cidT11−2 and cidT21−3), cenp-C mutant (cenp-Cprl41), cal1c03646 and spc25 (spc25A341) were examined as homozygotes unless otherwise noted. These lines, the elav-GAL4, OK6-Gal4, MHC-GAL4, Da-GAL4 and actin-GAL4 drivers, UAS-mCD8::GFP, and all other stocks were obtained from the Bloomington Drosophila Stock Center unless otherwise noted. Flies carrying the GFP-tagged kinetochore components, gEGFP-nuf2, gSpc25-EGFP, and gMis12-EGFP were generously provided by Dr. Christian Lehner (University of Zurich, Switzerland). All the RNAi Drosophila stocks (table S1) were from the Vienna Drosophila Resource Center (VDRC). Wild type refers to w1118. All the fly stocks were maintained on standard cornmeal food at 25°C. The mixed sex of flies were used in this study.
HEK293 cells and culture condition
HEK293T Cells from ATCC were cultured in DMEM (ThermoFisher Scientific) supplemented with L-glutamine, penicillin/streptomycin (ThermoFisher Scientific), and 10% FBS (Atlanta Biologicals).
iPSC-derived human neurons and culture conditions
For induction of cortical neurons from iPSCs, the iPSC line, LAM78, was derived from peripheral blood mononuclear cells of a normal individual by episomal reprogramming with 7 factors (OCT4, SOX2, NANOG, LIN28, L-MYC, KLF4, SV40LT). iPSCs were maintained in mTeSR-1 media (STEMCELL Technologies, #85850) on Geltrex (ThermoFisher Scientific, #A1413301), and passaged about once a week with Gentle Cell Dissociation Reagent (STEMCELL Technologies, #07174).
Cortical neurons were differentiated according to a protocol published by (Zhang et al., 2013) with minor modifications as described in the detailed method. Neurons were cultured in human astrocyte-conditioned media after day 6 and collected for western blot analysis after day 12.
Human motor neurons were derived from WA01 (WiCell) stem cells based on a modified protocol for adherent cells (Hill et al., 2016). Stem cells were plated in Matrigel-coated plates and maintained in StemFlex media (ThermoFisher Scientific), and dissociated to single cells by Accutase before differentiation.
hippocampal neuron culture
Hippocampal neurons were cultured as in (Teodoro et al., 2013). Specifically, hippocampal neurons were obtained from E18 mixed sex rat embryonic brains, plated on 24 well glass bottom plates (Cellvis, cat# P24–1.5H-N) coated with 20 μg/mL poly-L-Lysine (Sigma-Aldrich) and 3.5 μg/mL laminin (ThermoFisher Scientific). The neurons were maintained in Neurobasal medium (ThermoFisher Scientific) supplemented with B27 (ThermoFisher Scientific), L-glutamine, and penicillin/streptomycin.
METHOD DETAILS
Immunocytochemistry
21 h Drosophila embryos were dechorionated in 50% bleach for 4 minutes and dissected on Sylgard slides with GLUture Topical Adhesive (Abbot Laboratories) in phosphate-buffered saline, fixed in 4% paraformaldehyde (vol/vol) for 25 min (for CSP and HRP antibodies) or Bouin’s fixative for 5 min (for Brp and GluRIIC antibodies). Fixed embryos were washed five times in PBS + 0.1% Triton-X100 (PBST) before blocking with 5% normal goat serum (NGS, Sigma, NS02L), 1% Bovine Serum Albumin (BSA, Sigma, A9418) in PBST for 30 minutes. All washes were 10–15 minutes. Embryos were incubated with primary antibodies in 5% normal goat serum, 1% Bovine Serum Albumin overnight at 4°C. After 5 washes with PBST, incubate with secondary antibodies in 5% normal goat serum, 1% Bovine Serum Albumin for 3 hours at room temperature. After 5 washes with PBST, embryos were cleared in 70% glycerol +PBS overnight at 4°C. Finally, embryos were mounted in Vectashield (Vector Laboratories, H-1000).
We used mouse anti-Brp (1:50; Developmental Studies Hybridoma Bank, DSHB), mouse anti-CSP (6D6, 1:400; DSHB), mouse anti-GFP (3E6, 1:500; ThermoFisher Scientific), , rabbit anti-GluRIIC (1:2,000; A. DiAntonio, Washington University), Rabbit anti-Ndc80 (1:500, Marcin R. Przewloka, University of Southampton, UK), rabbit anti-cenp-C (1:1000, Marcin R. Przewloka, University of Southampton, UK), and Cy5-, Cy3- or FITC-conjugated antibodies to HRP (1:100; Jackson ImmunoResearch) as primary antibodies, and Alexa Fluor-488, Alexa Fluor-568 or Alexa Fluor-647-conjugated antibodies (1:400; ThermoFisher Scientific) as secondary antibodies. Embryos were imaged with an LSM700 or LSM710 laser-scanning confocal microscope (Zeiss) and a 63× 1.4 NA objective using separate channels and processed using the Fiji (ImageJ) or Adobe illustrator. To visualize GFP-tagged kinetochore proteins (Fig. S4), embryos or third instar filets were stained in parallel with anti-GFP and images were collected at the same gain settings.
Biochemistry in HEK293 cells
Transfections for HEK293 cells were done at 90% confluence using TransIT®-LT1 Transfection Reagent (MIR2300) 24 hours after plating, and cells were harvested 72 h after the transfection. The plasmids for GFP-tagged Mis12 and mis12 shRNA were used at a ratio of 1:10. For Western blots, the primary antibodies were used at the following dilutions: mouse anti-GFP (ThermoFisher Scientific, A11120, 3E6) at 1:2500; mouse anti-a-tubulin (Sigma-Aldrich, catalog no. T6199) at 1:10,000; mouse anti-human Mis12 (Andrea Musacchio, Max Planck Institute of Molecular Physiology, Germany) at 1:1000 (Petrovic et al., 2016); rabbit anti-GAPDH (Santa Cruz Biotechnology, (FL-335, SC-25778 ) at 1:4000.
iPSC-derived human neurons and biochemistry
For induction of cortical neurons from iPSCs, the iPSC, LAM78, line was derived from peripheral blood mononuclear cells of a normal individual by episomal reprogramming with 7 factors (OCT4, SOX2, NANOG, LIN28, L-MYC, KLF4, SV40LT). This iPSC line has a normal karyotype and expresses pluripotency markers. iPSCs were maintained in mTeSR-1 media (STEMCELL Technologies, #85850) on Geltrex (ThermoFisher Scientific, #A1413301), and passaged about once a week with Gentle Cell Dissociation Reagent (STEMCELL Technologies, #07174).
Cortical neurons were differentiated according to a protocol published by (Zhang et al., 2013) with minor modifications described below. iPSCs were treated with Accutase (Innovative Cell Technologies, #AT104) and plated at 90,000 cells/cm2 in mTeSR-1 media supplemented with 10uM ROCK inhibitor, Y-27632 (Cayman Chemical, #10005583) on Geltrex-coated 12-well plates. On day 2, no mouse glia cells were added and we used Ara-C (Sigma-Aldrich, #C1768) at a final concentration of 2uM. Neurons were cultured in human-astrocyte-conditioned media after day 6 and collected for western blot analysis after day 12.
Human motor neurons were derived from WA01 (WiCell) stem cells based on a modified protocol for adherent cells (Hill et al., 2016) Stem cells were plated in Matrigel-coated plates and maintained in StemFlex media (ThermoFisher Scientific), and dissociated to single cells by Accutase before differentiation. Then cells were changed to the DMEM/F12:Neurobasal media, supplemented with N2, B27, non-essential-amino-acids and glutamax. For the first 6 days of culture, the neuronal differentiation media was supplemented with SB431542 (10uM), LDN (0.1uM), retinoic acid (1uM), SAG (1uM), and then switched to retinoic acid (1uM), SAG (1uM), SU5402 (4uM), and DAPT (5uM) for the next 8 days. Then cells were dissociated by accutase, and enriched through magnetic sorting using anti-NCAM antibody prior to lysis for analysis on immunoblots as described above for HEK cells biochemistry.
Analysis of hippocampal neurons
DIV 10 hippocampal neurons were transfected with the indicated constructs using Lipofectamine 2000 (Life Technologies, Carlsbad, CA), and at DIV 15 fixed for 10 min with 4% paraformaldehyde, 4% sucrose in phosphate buffered saline at room temperature. Fixed cells were immunostained in Superblock blocking buffer (ThermoFisher, 37515) overnight at 4°C with mouse anti-GFP (ThermoFisher Scientific, A11120, 3E6) at 1: 500, and subsequently with goat anti-mouse Alexa-488 (1:500 in Superblock) for 3 hours at room temperature. Immunostained samples were mounted with cover slips using Vectashield antifade mounting medium (VECTOR Laboratories). Z-stack images were acquired for quantification of dendritic protrusions using a Zeiss LSM700 confocal microscope (Zeiss, Thornwood, NY). All transfected neurons with a pyramidal morphology that expressed both GFP (which marked the shRNA vector) and Dsred were imaged in an unbiased manner. To quantify dendritic protrusions, neurons were selected based on DsRed staining at a low magnification, at which protrusions were not visible, so as not to bias the selection. Dendritic segments that were 20–30 μm from the cell body were then examined at higher magnification. For each phenotype, at least 14 neurons from two independent experiments were examined. Optical sections of dendrites were acquired with a 63X, 1.4 NA oil immersion objective with 0.5 μm intervals and 1024×1024 pixel resolution and averaged twice. Dendritic protrusions were then analyzed manually by using Fiji ImageJ software (available at https://fiji.sc/).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of Synaptic Phenotypes
For neurite length measurements, the length of each branch on muscles 6, 7, 12, and 13 was summed for each hemisegment examined. To quantify the number of synaptic vesicle clusters, puncta positive for CSP immunoreactivity were counted that fell within the anti-HRP-labelled neurite branches on those muscles. For dendrites of sensory neuron cross the segment, we count the total number of dendrites which is HRP positive cross the segments boundary.
Quantification of the morphology of the VNC neuropil
21h embryos were stained with anti-CSP antibody. Z-stack images were acquired using the same setting for control and mis12 mutants. The VNC length was measured along a line from the posterior tip to the middle of the brain. For the CSP staining intensity, an arbitrary threshold was set and used for all relevant images, the mean gray value was analyzed.
Statistical analysis
Statistical analysis of data was done using GraphPad Prism 5.0 software. P values were derived using a student’s t-test with Welch-correction. P values lower than 0.05 were considered statistically significant. Data graphs were plotted in GraphPad Prism using the scatter dot plot functions showing the mean and standard error of the mean (SEM). p values for each experiment are denoted in the corresponding figure legend.
Supplementary Material
Supplemental table1: Screened deficiency list, related to Figure 1.
Highlights.
Kinetochore proteins act locally in development of postmitotic Drosophila neurons.
Mutations of kinetochore proteins alter synaptic morphology and neurite length
Knockdown of mis12 alters morphology of rat hippocampal dendrites
Acknowledgements
We thank Drs. Christian Lehner (University of Zurich), David Glover (University of Cambridge), Marcin R Przewloka (University of Southampton), Andrea Musacchio (Max Planck Institute of Molecular Physiology), James B. Skeath (Washington University) and Iain Cheeseman (MIT), and the Bloomington Stock Center (NIH P40OD018537), Vienna Drosophila RNAi Center, and Developmental Studies Hybridoma Bank, University of Iowa, for fly stocks and antibodies. We thank Dr. Mustafa Sahin (Boston Children’s Hospital) for providing the iPSC line, Dr. Thomas Sudhof (Stanford University) for the lentiviral plasmids, and the Human Neuron Core at Boston Children’s Hospital for differentiating neurons. We thank Dr.Xuan Huang and Dr. Clifford Woolf (Boston Children’ Hospital) for providing iPSC-derived motor neurons. We thank Drs. Arshad Desai (University of California, San Diego), Aaron DiAntonio (Washington University), Wesley Grueber (Columbia University), Liqun Luo (Stanford University) Matthew Pecot (Harvard Medical School), and Bing Ye (University of Michigan) for their comments and suggestions, and Dr. Ann Goldstein, Dr. Amos Gutnick, Himanish Basu and members of the Schwarz laboratory for helping with image processing, and for discussion and support. This work was supported by National Institute of Health grant R01 NS041062 to TLS and the IDDRC Cellular Imaging, Translational Neuroscience, and Molecular Genetics Cores (NIH P30 HD018655).
Footnotes
Author contributions
A.O., T.L.S. and G.Z. conceived the project. A.O. conducted the deficiency screen, Y.Y. analyzed mammailan Mis12 expression, and G.Z. performed all other experiments. T.L.S and G.Z. wrote the manuscript with feedback from all authors.
Declaration of interests
The authors have no competing interests. While working on this project, Dr. Asli Oztan Matos was affiliated with Boston Children’s Hospital as indicated on the title page. She is currently employed at Foundation Medicine, but has no competing interest in that capacity.
The deficiencies screened in this study are listed with their stock numbers from the Bloomington Drosophila Stock Center.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Baas PW, and Lin S (2011). Hooks and comets: The story of microtubule polarity orientation in the neuron. Dev Neurobiol 71, 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgen MA, Wang D, and Grill B (2017). RPM-1 regulates axon termination by affecting growth cone collapse and microtubule stability. Development 144, 4658–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois F, Messeant J, Kordeli E, Petit JM, Delers P, Bahi-Buisson N, Bernard V, Sigoillot SM, Gitiaux C, Stouffer M, et al. (2015). A critical and previously unsuspected role for doublecortin at the neuromuscular junction in mouse and human. Neuromuscul Disord 25, 461–473. [DOI] [PubMed] [Google Scholar]
- Budnik V (1996). Synapse maturation and structural plasticity at Drosophila neuromuscular junctions. Curr Opin Neurobiol 6, 858–867. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, and Desai A (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997. [DOI] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, and Mellone BG (2014). CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204, 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, and Grueber WB (2009). Molecules and mechanisms of dendrite development in Drosophila. Development 136, 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Heerssen HM, Wright CM, Davis GW, and DiAntonio A (2011). Stathmin is required for stability of the Drosophila neuromuscular junction. J Neurosci 31, 15026–15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, and Jan YN (2004). Dendritic development: lessons from Drosophila and related branches. Curr Opin Neurobiol 14, 74–82. [DOI] [PubMed] [Google Scholar]
- Hill SJ, Mordes DA, Cameron LA, Neuberg DS, Landini S, Eggan K, and Livingston DM (2016). Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci U S A 113, E7701–E7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, and Jan LY (2010). Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci 11, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, and Kaplan JM (2004). The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol 14, 2057–2062. [DOI] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, and Desai A (2006). The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshan PT, Oztan A, and Schwarz TL (2009). Presynaptic alpha2delta-3 is required for synaptic morphogenesis independent of its Ca2+-channel functions. Nat Neurosci 12, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, and Grueber WB (2007). Dendrite self-avoidance is controlled by Dscam. Cell 129, 593–604. [DOI] [PubMed] [Google Scholar]
- Musacchio A, and Desai A (2017). A Molecular View of Kinetochore Assembly and Function. Biology (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung E, Kurshan PT, Dickman DK, and Schwarz TL (2007). A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat Neurosci 10, 980–989. [DOI] [PubMed] [Google Scholar]
- Pawson C, Eaton BA, and Davis GW (2008). Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. J Neurosci 28, 11111–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Keller J, Liu Y, Overlack K, John J, Dimitrova YN, Jenni S, van Gerwen S, Stege P, Wohlgemuth S, et al. (2016). Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell 167, 1028–1040 e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, et al. (2010). The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol 190, 835–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, and Asbury CL (2009). The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, and Glover DM (2011). CENP-C is a structural platform for kinetochore assembly. Curr Biol 21, 399–405. [DOI] [PubMed] [Google Scholar]
- Schittenhelm RB, Althoff F, Heidmann S, and Lehner CF (2010). Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J Cell Sci 123, 3768–3779. [DOI] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, and Lehner CF (2007). Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116, 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro RO, Pekkurnaz G, Nasser A, Higashi-Kovtun ME, Balakireva M, McLachlan IG, Camonis J, and Schwarz TL (2013). Ral mediates activity-dependent growth of postsynaptic membranes via recruitment of the exocyst. EMBO J 32, 2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, and Brand AH (2004). Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119, 707–718. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, and Sigrist SJ (2017). Presynaptic morphogenesis, active zone organization and structural plasticity in Drosophila. Curr Opin Neurobiol 43, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, and Salmon ED (2012). The KMN protein network-chief conductors of the kinetochore orchestra. J Cell Sci 125, 5927–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkei Z, Przewloka MR, and Glover DM (2011). Drosophila Mis12 complex acts as a single functional unit essential for anaphase chromosome movement and a robust spindle assembly checkpoint. Genetics 187, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkei Z, Przewloka MR, Ladak Y, Albadri S, Sossick A, Juhasz G, Novak B, and Glover DM (2012). Spatiotemporal dynamics of Spc105 regulates the assembly of the Drosophila kinetochore. Open Biol 2, 110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AP, Fox AN, Johnson KG, and Zinn K (2010). Systematic screening of Drosophila deficiency mutations for embryonic phenotypes and orphan receptor ligands. PLoS One 5, e12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, and Bonni A (2009). A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science 326, 575–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table1: Screened deficiency list, related to Figure 1.