Abstract
More than 100 trillion symbiotic microorganisms constitutively colonize throughout the human body, including the oral cavity, the skin, and the gastrointestinal tract. The oral cavity harbors one of the most diverse and abundant microbial communities within the human body, second to the community that resides in the gastrointestinal tract, and is composed of >770 bacterial species. Advances in sequencing technologies help define the precise microbial landscape in our bodies. Environmental and functional differences render the composition of resident microbiota largely distinct between the mouth and the gut and lead to the development of unique microbial ecosystems in the 2 mucosal sites. However, it is apparent that there may be a microbial connection between these 2 mucosal sites in the context of disease pathogenesis. Accumulating evidence indicates that resident oral bacteria can translocate to the gastrointestinal tract through hematogenous and enteral routes. The dissemination of oral microbes to the gut may exacerbate various gastrointestinal diseases, including irritable bowel syndrome, inflammatory bowel disease, and colorectal cancer. However, the precise role that oral microbes play in the extraoral organs, including the gut, remains elusive. Here, we review the recent findings on the dissemination of oral bacteria to the gastrointestinal tract and their possible contribution to the pathogenesis of gastrointestinal diseases. Although little is known about the mechanisms of ectopic colonization of the gut by oral bacteria, we also discuss the potential factors that allow the oral bacteria to colonize the gut.
Keywords: periodontal disease/periodontitis, bacteria, systemic health/disease, host-pathogen interactions, microbiology, mucosal immunity
Introduction
The human body is colonized by >100 trillion symbiotic microorganisms, almost equivalent to the number of human cells and collectively referred to as the human microbiota (Qin et al. 2010; Sender et al. 2016). Due to environmental differences, each site in the body is home to a distinct microbial ecosystem (Sender et al. 2016). Among these sites, the most diverse bacterial populations are found in the intestinal tract (Human Microbiome Project 2012; Blum 2017). The human gut microbiota contributes to host physiologic development and maintenance, including education of the host immune system, nutrient digestion, and defense against colonization by pathogenic microorganisms (Kamada, Seo, et al. 2013; Gilbert et al. 2018). Because of its enormous impact, disturbance of the gut microbiota, so-called gut dysbiosis, has been shown to underlie multiple intestinal pathologies, including irritable bowel syndrome (IBS), inflammatory bowel disease (IBD), and colorectal cancer (CRC). However, we lack a comprehensive understanding of which bacteria act as disease-associated pathobionts and how they contribute to the pathogenesis of disease. In this regard, it has been reported that patients with diseases of the gut exhibit an abnormal enrichment of typical oral bacteria in the luminal contents and the gut mucosal tissues (Gevers et al. 2014; Yachida et al. 2019). Thus, it is conceivable that the oral cavity serves as a reservoir of oral pathobionts whose ectopic gut colonization contributes to the pathogenesis of intestinal diseases.
The oral cavity is a primary gateway to the human body and has the second-largest and diverse microbiota after the gut, harboring >770 species of bacteria (Escapa et al. 2018). A variety of microbial habitats in the oral cavity (e.g., teeth, buccal mucosa, soft and hard palate, and tongue) makes the ecologic system complex and attracts diverse microorganisms, called oral microbiome, including bacteria, fungi, and viruses (Kilian 2018). Within oral microbiome, bacteria are the major components and form distinct microbial communities in each oral habitat; they primarily comprise members of the phyla Firmicutes, Fusobacteria, Proteobacteria, and Actinobacteria (Costalonga and Herzberg 2014). Diverse structural and nutritional difference creates a unique microbial ecosystem in each body site, benefiting human health. However, compelling evidence indicates that certain bacteria can be disseminated from one site to the others and cause systemic diseases. In this regard, numerous studies have elaborated that oral microbes can spread through the body and have been found in a variety of systemic diseases, such as cardiovascular diseases, adverse pregnancy outcomes, and rheumatoid arthritis (Hajishengallis 2015; Graves et al. 2019). Importantly, in addition to these sterile organs, oral microbes can be ingested and naturally translocate to the upper and lower digestive tract (i.e., esophagus, stomach, small and large intestine). Unlike sterile organs, the digestive tract harbors indigenous microbial communities that prevent the colonization of exogenous microbes that invade from the extraintestinal compartment (i.e., the mouth) via multiple means (Kamada, Chen, et al. 2013). However, under certain circumstances, oral microbes can ectopically colonize the upper and lower digestive tract. In this regard, increasing evidence suggests that ectopic colonization by oral microbes may be detrimental and cause diseases in the digestive tract. In this review, we summarize the current knowledge of the dissemination and its role of oral microorganisms in extraoral diseases, particularly diseases in the lower digestive tract. Among the oral microbiota, bacteria are the most well-studied microorganisms with respect to the possible involvement in extraoral diseases. Hence, in this review, we focus on the role that oral bacteria play in the pathogenesis of gastrointestinal diseases.
Oral Bacteria in Gut Pathology
Despite the environmental segregation of the mouth and gut, it has been reported that more than half of microbial species (e.g., Streptococcus and Veillonella) frequently detected in both sites show evidence of oral-gut translocation, even in healthy individuals (Schmidt et al. 2019). Gut colonization by oral bacteria such as Veillonella spp. is known to modulate host immunity (Geva-Zatorsky et al. 2017). Thus, ectopic colonization by oral bacteria in the healthy gut may in part contribute to the physiologic development and/or maintenance of gut immunity. However, ectopic gut colonization by specific oral bacteria and/or under certain conditions might be linked to the pathogenesis of diseases in the gastrointestinal tract. In Table 1, we define oral bacteria per the following criteria: 1) bacteria identified as a constituent of the oral microbiome by the Human Oral Microbiome Database (http://www.homd.org/) and 2) bacteria that have higher abundance in the oral cavity than in the gut samples of healthy individuals on the basis of the NIH Human Microbiome Project (HMP1; https://hmpdacc.org/hmp/). In addition, some bacteria that were previously reported as bacteria involved in oral pathology (e.g., some species/genera belonging to the Enterobacteriaceae family, Staphylococcus, Fusobacterium varium, and Porphyromonas gingivalis) are listed as possible oral bacteria even if they do not meet criterion 2 (see Appendix Table 1).
Table 1.
Oral Bacteria Found in the Gut of Patients with Gut Pathology.
| Percentage Abundance in Healthy Individuals | |||||
|---|---|---|---|---|---|
| Gut Pathology: Oral Bacterial Species Detected in the Guta | Sample Type | Saliva | Gingiva | Buccal | Stool |
| Irritable bowel syndrome | |||||
| Streptococcus (genus) | Stool | 13.94 | 10.04 | 51.49 | 0.04 |
| Streptococcus thermophilus | Stool | 2.63b | 0.10b | 1.92b | 0.01b |
| Veillonella (genus) | Stool | 11.64 | 3.91 | 3.35 | 0.08 |
| Haemophilus (genus) | Stool | 12.65 | 4.06 | 15.12 | 0.07 |
| Prevotella (genus) | Stool | 13.01 | 8.79 | 2.27 | 3.4 |
| Fusobacterium (genus) | Stool | 6.42 | 13.75 | 2.54 | 0.06 |
| Dialister invisus | Stool | 0.17 | 0.38 | 0.04 | 0.67 |
| Gammaproteobacteria (class) | Stool | 13.7 | 6.13 | 15.89 | 0.24 |
| Enterobacteriaceae (family) | Stool | 0.01 | <0.005 | <0.005 | 0.01 |
| Inflammatory bowel disease | |||||
| Veillonellaceae (family) | Tissue | 12.7 | 4.5 | 3.47 | 0.94 |
| Pasteurellaceae (family) | Tissue | 13.61 | 5.83 | 15.83 | 0.07 |
| Neisseriaceae (family) | Tissue | 6.57 | 6.07 | 3.34 | <0.005 |
| Peptostreptococcaceae (family) | Tissue | 0.51 | 0.52 | 0.08 | 0.11 |
| Atopobium parvulum | Tissue | 0.42 | 0.05 | 0.06 | <0.005 |
| Fusobacteriaceae (family) | Stool, tissue | 6.42 | 13.75 | 2.54 | 0.06 |
| Fusobacterium varium | Tissue | <0.005 | ND | ND | 0.02 |
| Campylobacter (genus) | Tissue | 1.93 | 0.99 | 0.2 | <0.005 |
| Campylobacter concisus | Stool, tissue | 1.66 | 0.1 | 0.01 | <0.005b |
| Aggregatibacter segnis | Stool | 0.11 | 0.27 | 0.02 | <0.005 |
| Streptococcus (genus) | Stool, tissue | 13.94 | 10.04 | 51.49 | 0.04 |
| Streptococcus anginosus | Stool | <0.005b | 0.02b | 0.05b | <0.005b |
| Gemellaceae (family) | Stool, tissue | 0.87 | 0.53 | 6.32 | <0.005 |
| Enterobacteriaceae (family) | Stool, tissue | 0.01 | <0.005 | <0.005 | 0.01 |
| Escherichia coli | Stool, tissue | 0.01b | <0.005 | <0.005b | <0.005 |
| Colorectal cancer | |||||
| Porphyromonas (genus) | Stool, rectal swab | 4.67 | 3.6 | 2.66 | <0.005 |
| Porphyromonas gingivalis | Stool | <0.005 | 0.01 | <0.005 | <0.005 |
| Porphyromonas uenonis | Stool | ND | ND | ND | ND |
| Fusobacterium (genus) | Tissue, rectal swab | 6.42 | 13.75 | 2.54 | 0.06 |
| Fusobacterium nucleatum | Stool | 0.78 | 8.45 | 0.38 | <0.005 |
| Streptococcus (genus) | Stool | 13.94 | 10.04 | 51.49 | 0.04 |
| Peptostreptococcaceae (family) | Stool, rectal swab | 0.51 | 0.52 | 0.08 | 0.11 |
| Peptostreptococcus stomatis | Stool | 0.3 | 0.08 | 0.05 | <0.005 |
| Peptostreptococcus anaerobius | Stool | <0.005 | <0.005b | <0.005b | <0.005 |
| Prevotella (genus) | Stool | 13.01 | 8.79 | 2.27 | 3.4 |
| Prevotella intermedia | Stool | 0.13 | 0.66 | 0.04 | ND |
| Gemella morbillorum | Stool, rectal swab | 0.87b | 0.53b | 6.32b | <0.005b |
| Solobacterium moorei | Stool | 0.01 | <0.005 | <0.005 | <0.005 |
| Atopobium parvulum | Stool | 0.42 | 0.05 | 0.06 | <0.005 |
| Actinomyces odontolyticus | Stool | 1.43 | 0.16 | 0.49 | <0.005 |
| Parvimonas micra | Stool | 0.16 | 0.51 | 0.05 | <0.005 |
| Escherichia coli | Stool, tissue | 0.01b | <0.005 | <0.005b | <0.005 |
| Klebsiella (genus) | Stool | <0.005 | <0.005 | <0.005 | <0.005 |
| Helicobacter pylori | Tissue | <0.005 | <0.005 | <0.005 | <0.005 |
| Mogibacterium | Stool | 0.15 | 0.03 | 0.02 | <0.005 |
| Dialister pneumosintes | Tissue | 0.04 | 0.07 | 0.01 | <0.005 |
| Celiac disease | |||||
| Staphylococcus (genus) | Stool | 0.01 | 0.01 | 0.01 | 0.19 |
| Staphylococcus epidermidis | Tissue | 0.01b | 0.01b | 0.01b | 0.19b |
| Enterobacteriaceae (family) | Tissue | 0.01 | <0.005 | <0.005 | 0.01 |
| Klebsiella oxytoca | Tissue | <0.005b | <0.005b | <0.005b | <0.005b |
ND, not detected.
The taxonomic rank is provided in parentheses only if the species information is not defined in the reference. Oral bacteria are defined per the following criteria: 1) bacteria identified as a constituent of the oral microbiome by the Human Oral Microbiome Database (Escapa et al. 2018) and 2) bacteria having higher abundance in the oral tissues than in the gut samples. The percentage abundance in the saliva, gingiva, buccal mucosa, and stool in healthy individuals, based on the NIH Human Microbiome Project (HMP1), is shown. In addition, some bacteria that were previously reported as bacteria involved in oral pathology are listed as possible oral bacteria even if they do not meet criterion 2. References are provided in Appendix Table 1.
Amplicon sequence variants also match some other taxa (likely in the same genus).
Irritable Bowel Syndrome
IBS is the most common functional gastrointestinal disorder, characterized by recurrent episodes of abdominal pain (Simren and Tack 2018). The global prevalence of IBS is estimated to be 11.2%, with geographic variations ranging from 7% to 21% (Lovell and Ford 2012; Canavan et al. 2014). A study revealed the approximately 7-fold increased risk of IBS development after microorganism-driven gastroenteritis (Halvorson et al. 2006). Given the evidence that gut dysbiosis may lead to the activation of gut immune systems and subsequent low-grade inflammation of the gut (Ohman and Simren 2010), it is likely that microbes residing in the gut play a role in the pathogenesis of IBS. In this context, notwithstanding the huge variations in the gut microbial composition of patients with IBS (Ohman et al. 2015; Chong et al. 2019; Hugerth et al. 2020), there are some common features in IBS, including an increase in the families Enterobacteriaceae and Lactobacillaceae and a decrease in the genera Clostridium, Faecalibacterium, and Bifidobacterium, as compared with controls (Pittayanon et al. 2019). Interestingly, alterations in the microbial composition of patients with IBS include enrichment of certain types of typical oral bacteria in the gut. For example, Streptococcus spp. have repeatedly been reported to be enriched in the gut of patients with IBS (Wyatt et al. 1988; Kassinen et al. 2007; Rajilic-Stojanovic et al. 2011; Vich Vila et al. 2018). Likewise, an increased abundance of the family Veillonellaceae in the gut is often observed in IBS (Table 1). In this regard, overweight patients with IBS who have significantly higher IVP (induced visceral pain) scores exhibit a higher abundance of Veillonellaceae than normal-weight patients with IBS. Also, infants with colic—characterized by gastrointestinal discomfort caused by the accumulation of lactate, hydrogen (H2), or hydrogen sulfide (H2S)—have an increased level of Veillonellaceae in the gut (Pham et al. 2017). The ability of Veillonella spp. to produce a robust quantity of H2 suggests that Veillonella spp. may play a role in determining the pathogenesis of IBS.
Inflammatory Bowel Disease
IBD is an idiopathic disorder that causes chronic inflammation of the digestive tract, comprising Crohn’s disease (CD) and ulcerative colitis. Imbalance of the gut microbiota appears to be an essential factor in the pathogenesis of IBD (Sartor 2008). Multiple studies have shown the difference in the gut microbial composition between patients with and without IBD (i.e., healthy individuals; Lloyd-Price et al. 2019). Gut dysbiosis in IBD is characterized by a decrease in the bacterial diversity and species richness of the microbiota. In this context, one large multicenter microbiome study involved the collection of >400 treatment-naïve pediatric CD samples from multiple concurrent gastrointestinal sites (e.g., stool, rectal, ileum). The results clearly demonstrated a significant correlation between microbial alterations in rectal and ileal mucosa and disease status, with an increased abundance of Veillonellaceae, Pasteurellaceae, Enterobacteriaceae, Nisseriaceae, Gemellaceae, and Fusobacteriaceae and a decreased abundance of Bacteroidales, Erysipelotrichales, and Clostridiales (Gevers et al. 2014). Notably, most of the bacteria enriched in the gut of these pediatric patients with CD were resident oral bacteria rather than typical resident bacteria in the gut, implying the contribution of oral bacteria to the pathogenesis of CD. Enterobacteriaceae are generally considered gut bacteria but not typical oral bacteria. However, a recent study showed that Enterobacteriaceae that reside in the saliva can elicit pathogenic immune responses when they ectopically colonize the gut (Atarashi et al. 2017). In this study, members of the family Enterobacteriaceae, in particular Klebsiella spp. (K. pneumoniae and K. aeromobilis [also known as K. aerogenes]), were isolated from gnotobiotic animals colonized by salivary microbiota derived from patients with IBD and identified as potent Th1 inducers in the gut. These Klebsiella strains are capable of eliciting severe gut inflammation when colonized in hosts genetically susceptible to IBD. Although Enterobacteriaceae, including Klebsiella spp., are only a minor constituent of oral microbiota, multiple studies reported that Enterobacteriaceae, including Klebsiella and E. coli, reside in the oral cavity in humans (Souto 2006; Baek et al. 2018; Zawadzki et al. 2016). Thus, at least a part of Enterobacteriaceae enriched in the gut of patients with CD could also be originated from the oral cavity (Table 1).
Colorectal Cancer
The colon is exposed to an infinite number of microorganisms, corresponding to about 70% of the estimated human microbiome (Sekirov et al. 2010). Given that most of the known colon cancer risks (e.g., age, inflammation, obesity) are closely associated with gut dysbiosis, it is conceivable that certain gut microbes contribute to tumor cell generation, by directly or indirectly shaping a microenvironment in the gut that is more favorable to tumor development. Many studies have shown that patients with CRC have a distinct gut microbial composition as compared with healthy individuals (Table 1). Of note, many of the bacteria enriched in colonic adenomas and carcinomas are related to the typical resident oral bacteria, including the families Streptococcaceae and Neisseriaceae and the genera Staphylococcus, Porphyromonas, Veillonella, and Fusobacterium (Kostic et al. 2013; Geng et al. 2014). This observation received validation from 3 recent large cohort studies demonstrating reproducible CRC-associated gut microbial signatures (Thomas et al. 2019; Wirbel et al. 2019; Yachida et al. 2019). These studies showed that patients with CRC have an enrichment of members of the oral microbes, including Fusobacterium, Atopobium, Actinomyces, Parvimonas, Peptostreptococcus, Porphyromonas, and Solobacterium, in the gut (Table 1). Furthermore, patients diagnosed with CRC had higher transmission rates of bacteria from the mouth to the gut when compared with healthy individuals. In particular, the transmission of Fusobacterium nucleatum, Parvimonas micra, and Peptostreptococcus stomatis was increased in patients with CRC. These results likewise support a potential link between the oral and gut microbiome in the context of CRC (Schmidt et al. 2019).
Mechanistic Insights into the Role of Oral Bacteria in Gut Pathology
Despite increasing knowledge of oral bacterial dissemination to the gut, the functional role of oral bacteria in the development of intestinal pathology remains unexplored. Published reports demonstrate plausible molecular mechanisms by which oral bacteria affect the host responses to enhance diseases of the gut. The mechanistic role of 3 bacteria are discussed in turn and summarized in Table 2.
Table 2.
Possible Mechanisms of Oral Bacteria in the Gut Pathogenesis.
| Pathways in Host Cells | ||||
|---|---|---|---|---|
| Oral Bacteria: Target Cells | Effector | Receptor | Related Signals | Pathologic Functions |
| Fusobacterium nucleatum | ||||
| Epithelial cells | Fap2 | Gal-GalNAc | Tumor binding and enrichment | |
| Epithelial cells | Metalloproteinase collagenase | Cellular migration and invasive properties | ||
| Epithelial cells | FadA | Ecad | Wnt/β-catenin | Tumor cell proliferation |
| NK cells, T cells | Fap2 | TIGIT | Immune evasion | |
| Epithelial cells | LPS | TLR4 | miR-4802, miR-18a* | Chemoresistance (autophagy activation) |
| Epithelial cells | LPS | TLR4 | Myd88, miR-21 | Tumor cell proliferation |
| Recruitment of tumor-infiltrating immune cells (MDSC, TAM, regDC) | ||||
| Fusobacterium varium epithelial cells | Adhesion and invasion, IL-8 and TNF-a production | |||
| Porphyromonas gingivalis | ||||
| Epithelial cells | Jak1/Akt/Stat3, PI3K/Akt | Cell survival (antiapoptotic) | ||
| Epithelial cells | Cyclin D and E, PI3K | Cell proliferation | ||
| Epithelial cells | Gingipain | β-catenin destruction, complex degradation | β-catenin | Cell proliferation |
| Epithelial cells | Immune evasion (B7-H1 and B7-DC upregulation) | |||
| Epithelial cells | Gingipain | PAR | NF-kB, ERK1/2, p38 | Tumor invasiveness (MMPs expression↑) |
| Epithelial cells and others | Epithelial disruption, proinflammatory cytokine induction, gut dysbiosis | |||
| Epithelial cells and others | Epithelial disruption, immune activation, gut dysbiosis | |||
| Neutrophils | TLR1-TLR2 | Myd88 | Impaired antimicrobial response, Impaired killing activity | |
| Mø and DC | Fimbrial proteins (FimA and Mfa1) | CR3 or DC-SIGN | MMP and C1q | Hijack and direct host immune cells (distant tissue destruction) |
| Klebsiella pneumoniae, K. aerogenes (K. aeromobilis) epithelial cells | TLR | IL18 and Myd88 | Th1 cell generation | |
| Atopobium parvulum: unknown | H2S | Mitochondrial dysfunction in host with impaired H2S detoxification | ||
| Campylobacter concisus epithelial cells | Epithelial disruption | |||
| Staphylococcus aureus epithelial cells, T cells | Enterotoxins | Epithelial disruption, immune activation | ||
Fusobacterium nucleatum
F. nucleatum is enriched in the colonic mucosa of patients with IBD and CRC (Table 1). A recent study showed that identical strains of F. nucleatum are detected in both the saliva and colonic tumors of patients with CRC, indicating that F. nucleatum colonized in the colonic tumors originates in the oral microbiota (Komiya et al. 2019). Unlike other oral bacteria, the mechanistic role of F. nucleatum has been relatively well explored. F. nucleatum is highly adhesive to the gut epithelium through Fap2 adhesin–mediated binding to Gal-GalNAc (galactose N-acetyl-d-galactosamine; Abed et al. 2016). Reports that F. nucleatum promotes the proliferation of tumor cells in vitro and in vivo may be explained by the F. nucleatum FadA adhesin–mediated activation of the Wnt/β-catenin pathway (Rubinstein et al. 2013). Furthermore, F. nucleatum plays a pivotal role in controlling CRC chemoresistance in response to chemotherapy drugs (e.g., oxaliplatin) by selectively targeting miRNAs and activating the autophagy pathway (Yu et al. 2017). Beyond a direct interaction with epithelial cells, F. nucleatum can shape the tumor microenvironment by altering the cytotoxic functions of tumor-infiltrating lymphocytes and natural killer cells. This action is mediated by the interaction between the inhibitory immunoreceptor TIGIT on these immune cells and Fap2 (Gur et al. 2015). Given that the overrepresentation of Fusobacterium in CRC positively correlates with lymph node metastasis, Fusobacterium spp. may have further malignant potential that needs to be clarified. Notably, the ectopic gut colonization by fusobacteria can be a biomarker for the detection of CRC. A recent study reported that the fecal abundance of fusobacteria in combination with that of the potentially beneficial populations (e.g., Bifidobacterium) might serve as a biomarker for early CRC (Guo et al. 2018). Also, measuring anti-Fn-IgA level with the conventional biomarkers, such as carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen, may increase the sensitivity for the detection of early CRC (Wang et al. 2016).
Porphyromonas gingivalis
P. gingivalis accumulates in the gut of patients with CRC. Although the precise role of P. gingivalis in the pathogenesis of CRC remains unknown, some pathologic functions ofP. gingivalis implicate a pathogenic role for this bacterium in CRC (Table 2). For instance, oral administration of P. gingivalis to mice is reported to disrupt the gut epithelial integrity (e.g., reduced expression of tight junction proteins; Arimatsu et al. 2014; Nakajima et al. 2015). P. gingivalis also inhibits epithelial apoptosis through multiple mechanisms, including activation of the JAK1/STAT3 and PI3K/Akt signaling pathways (Yilmaz et al. 2004; Mao et al. 2007), inactivation of caspase 3 and 9 (Mao et al. 2007; Yao et al. 2010), and prevention of P2X7-mediated apoptosis (Yilmaz et al. 2008). Similar to F. nucleatum, P. gingivalis is capable of potentiating epithelial cell proliferation through activation of the Wnt/β-catenin pathway (Zhou et al. 2015), as well as by controlling the activity of PI3K, p53, and cyclins (Kuboniwa et al. 2008; Pan et al. 2014). Furthermore, P. gingivalis contributes to the invasive properties of tumors through the activation of matrix metalloproteinases (MMPs), including MMP-1, MMP-9, MMP-10, and MMP-13 (Inaba et al. 2014; Ha et al. 2015; Inaba et al. 2015). Also, P. gingivalis is known to invade macrophages and dendritic cells through the interaction between its fimbrial proteins and complement receptor 3 (or DC-SIGN) on the immune cell surface. After hijacking the immune cells, P. gingivalis instigates the production of proteins that destroy tissue, such as MMP-9, from the infected cells. The role that MMPs play in controlling the invasive properties of tumors suggests that oral bacteria residing in colonic tumors may contribute to the metastatic potential of these tumors (Hajishengallis 2015).
Klebsiella Species
A recent study showed that the colonization of germ-free mice with salivary microbiota isolated from patients with CD resulted in potent Th1 cell differentiation in the gut (Atarashi et al. 2017). In this study, the authors determined that certain Klebsiella spp. (e.g., K. pneumoniae, K. aerogenes/aeromobilis) residing in the salivary microbiota are responsible for the induction of Th1 cells. Interestingly, the expansion of Th1 cells due to the ectopic gut colonization by oral Klebsiella spp. does not lead to the development of spontaneous gut inflammation. However, these oral Klebsiella spp. are capable of inducing the development of Th1-skewed IBD-like colitis in mice lacking the immunosuppressive cytokine IL-10. Given that impaired IL-10 signaling is associated with the risk for very-early-onset IBD (Moran et al. 2013), oral bacteria such as Klebsiella spp. may contribute to the pathogenesis of certain subsets of IBD.
Other Oral Bacteria
Members of the oral microbiota found in the gut are known to produce carcinogenic substances. Similar to F. nucleatum and P. gingivalis, certain types of oral bacteria (e.g., Atopobium spp., Veillonella spp., Prevotella spp., Streptococcus spp., and Aggregatibacter spp.) are known to liberate H2S, a genotoxic and inflammatory substance, from sulfur-containing amino acids. Also, many species of indigenous oral bacteria, such as Streptococcus spp. and Neisseria spp., have been reported to produce acetaldehyde by catabolizing ethanol and glucose (Tagaino et al. 2019). Given the high genotoxic capacity of these bacterial metabolites, even at low concentrations, it is conceivable that ectopic colonization of the gut by these oral bacteria could induce genomic instability or mutations, leading to colonic tumor development.
Possible Pathways of Ectopic Gut Colonization by Oral Bacteria
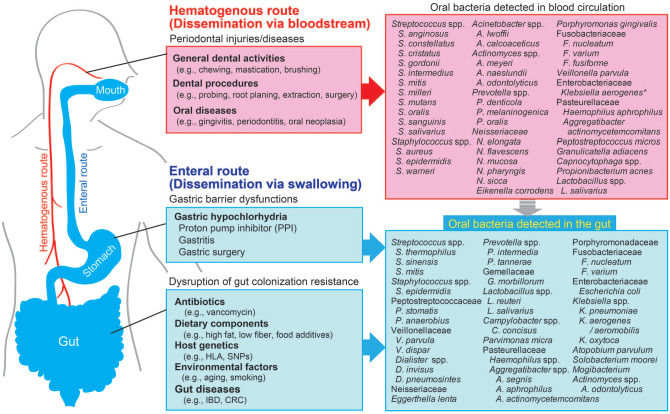
Although the mode of the relocation of oral bacteria from the oral cavity to the gut mucosa is uncertain, 2 routes have been proposed: hematogenous and enteral.
Hematogenous Route
One possible route of oral bacteria dissemination is by hematogenous spreading from the oral cavity (Fig.). Studies have shown that oral mechanical injuries caused by daily dental activity (e.g., hard mastication, brushing) and dental procedures (e.g., orthodontics, extraction) enable oral bacteria to spread into the systemic circulation (Lockhart et al. 2008; Parahitiyawa et al. 2009). Patients with periodontal diseases such as periodontitis and oral cancer have an elevated level of oral bacteria in their blood. Moreover, a ligature-induced murine periodontitis model showed that periodontal inflammation triggers oral bacterial dissemination to the liver and spleen, indicating a key role for oral inflammation in the systemic dissemination of oral bacteria through the bloodstream (Tsukasaki et al. 2018). This role was supported by another study that identified periodontal pathogens such as P. gingivalis in the bloodstream of patients with periodontitis (Horliana et al. 2014). Furthermore, as described in Table 2, oral bacteria is known to invade and survive inside immune cells, such as dendritic cells and macrophages, indicating that oral bacteria may hijack host immune cells to serve as Trojan horses for dissemination from the oral mucosa to the gut mucosa (Hajishengallis 2015).
Figure.
Possible routes of oral bacteria transmigration from the mouth to the gut. The diagram depicts possible routes of oral bacteria transmigration from the oral cavity to the gut and potential factors contributing to the ectopic colonization of oral bacteria in the gut. Laboratory and clinical studies discussed in this review reveal 2 possible routes: hematogenous (red) and enteral (blue). *Enterobacter aerogenes was renamed Klebsiella aerogenes in 2017.
Enteral Route
Another possible route of oral bacteria dissemination is by enteral spreading. People swallow about 600 times a day, and ~1.5 L of saliva contains numerous resident oral bacteria (Humphrey and Williamson 2001; Pedersen et al. 2002). However, ingested oral bacteria seldom reach and colonize the healthy gut because of the barrier functions along the gastrointestinal tract. The colonization resistance by the gut resident microbiota is considered the major barrier that prevents the ectopic colonization by swallowed oral bacteria. In other words, the disruption of the healthy gut microbiota results in the increased gut colonization by oral bacteria (Fig). For instance, antibiotics (e.g., vancomycin) used to treat bacterial infections are known to perturb gut microbial composition and to generate niches for translocated oral bacteria to colonize and expand in the gut. Th1 cell–inducing Klebsiella spp. that reside in the saliva of patients with IBD possess resistance to multiple antibiotics, including ampicillin (Atarashi et al. 2017). Hence, ampicillin treatment can result in the gut colonization by oral Klebsiella spp. and subsequent pathogenic Th1 cell expansion in the gut, suggesting that inadequate use of antibiotics may increase the risk for oral bacteria–driven gut pathology (Atarashi et al. 2017). Other than the aforementioned gut dysbiosis–inducing factors, multiple factors that elicit gut dysbiosis (e.g., gut inflammation, diets, artificial sweeteners) may increase the opportunistic gut colonization by oral bacteria. Considering all these factors, gut dysbiosis may be a prerequisite for the ectopic colonization of oral pathobionts.
Also, gastric acidity is an important bottleneck for oral bacteria. Since the majority of oral resident bacteria are sensitive to the gastric acid, ingested oral bacteria might be significantly reduced while they are passing the stomach. Consistent with this notion, patients who have gastric dysfunction related to achlorhydria caused by the long-term use of proton pump inhibitors exhibit a significant increase in gut colonization by oral bacteria (e.g., Streptococcus spp., Veillonella spp., Haemophilus spp.; Fig.). Another example of reduced exposure of ingested bacteria to gastric juice may occur in individuals who have gastritis and gastric surgery (e.g., gastric bypass and removal; Castaner et al. 2018; Paganelli et al. 2019). These individuals have an altered gut microbial composition, accompanied by a significant increase in the level of resident oral bacteria (e.g., Streptococcus spp., Veillonella spp., and Enterobacteriaceae) in the gut. Interestingly, certain types of oral bacteria, such as P. gingivalis, are able to tolerate the harsh acidic environment in the stomach and consequently may pass through the stomach barrier (Walker et al. 2018). Thus, gastric acidity can prevent the enteral transmission of oral bacteria but might be less effective for bacteria that are tolerated to the acidic environment.
Other Possible Factors Associated with the Ectopic Colonization of Oral Bacteria
In addition to the aforementioned mechanisms, other factors may be responsible for the ectopic colonization of oral bacteria. For example, immune-compromised individuals (e.g., patients infected with HIV) have gut dysbiosis accompanied by the accumulation of oral bacteria, such as the Prevotellaceae, Erysipelotrichaceae, and Veillonellaceae families and the Proteobacteria phylum (Crakes and Jiang 2019). Given the importance of host immunity in shaping gut microbiota composition and its colonization resistance, it is likely that immune depression by multiple factors (e.g., aging, drugs, virus infection) may promote the ectopic gut colonization by oral bacteria. Furthermore, overgrowth of oral pathogenic bacteria in the diseased oral cavity may increase the supply of oral bacteria, resulting in increased oral bacterial colonization in the gut. For instance, periodontal pathogens such as F. nucleatum and P. gingivalis expand in the oral cavity of patients with periodontitis (Socransky et al. 1998). Notably, a recent large cohort study (n = 77,443) revealed that women with fewer teeth and presumably moderate or severe periodontal inflammation have up to a 48% increased risk for developing CRC (Momen-Heravi et al. 2017). Given the high prevalence of certain periodontal pathogens in the gut of individuals with CRC (Table 1), it is conceivable that poor oral health, accompanied by the expansion of certain oral bacteria, may cause an oversupply of the oral bacteria to the gut, increasing the chance of oral bacterial colonization in the gut. However, only a few studies have focused on the link between periodontal disease and gut pathology. Further large cohort studies are needed to clarify the clinical relevance of periodontal disease in the development of gut pathology.
Conclusions and Perspective
Considering the results of many investigations, the mouth-to-gut transmission may be an important process in bacteria-driven pathologies in the gastrointestinal tract. However, as mentioned, most of the studies demonstrating the pathologic link between oral bacteria and extraoral diseases are observational and still at the stage of association. There is a need for more studies to elucidate the transmigration mechanisms of oral bacteria to extraoral sites and to understand the precise role of oral pathobionts in the pathogenesis of diseases at extraoral sites, including the gastrointestinal tract. In parallel and also essential are further epidemiologic cohort studies to clarify the clinical relevance of oral pathology, accompanied by the expansion of oral pathobionts, in the development of gut pathology. These efforts will pave the way for the future development of novel diagnostic and therapeutic interventions to target oral bacteria.
Author Contributions
S. Kitamoto, H. Nagao-Kitamoto, contributed to conception and design, drafted the manuscript; R. Hein, T.M. Schmidt, contributed to conception, design, and data analysis, drafted the manuscript; N. Kamada, contributed to conception and design, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520924633 for The Bacterial Connection between the Oral Cavity and the Gut Diseases by S. Kitamoto, H. Nagao-Kitamoto, R. Hein, T.M. Schmidt and N. Kamada in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported by National Institutes of Health grants DK119219 and AI142047 (N.K.), Crohn’s and Colitis Foundation (H.N.-K.), Clinical and Translational Science Awards Program, University of Michigan (S.K.), and Prevent Cancer Foundation (S.K.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iDs: S. Kitamoto  https://orcid.org/0000-0003-3597-6647
https://orcid.org/0000-0003-3597-6647
N. Kamada  https://orcid.org/0000-0002-1980-4178
https://orcid.org/0000-0002-1980-4178
References
- Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. 2016. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 20(2):215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, Gotoh K, Motooka D, Nakamura S, Iida T, et al. 2014. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. 4:4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. 2017. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 358(6361):359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K, Ji S, Choi Y. 2018. Complex intratissue microbiota forms biofilms in periodontal lesions. J Dent Res. 97(2):192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum HE. 2017. The human microbiome. Adv Med Sci. 62(2):414–420. [DOI] [PubMed] [Google Scholar]
- Canavan C, West J, Card T. 2014. The epidemiology of irritable bowel syndrome. Clin Epidemiol. 6:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow STE, Schroder H. 2018. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PP, Chin VK, Looi CY, Wong WF, Madhavan P, Yong VC. 2019. The microbiome and irritable bowel syndrome—a review on the pathophysiology, current research and future therapy. Front Microbiol. 10:1136. Erratum in: Front Microbiol. 2019. August 13;10:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 162(2 Pt A):22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crakes KR, Jiang G. 2019. Gut microbiome alterations during hiv/siv infection: implications for HIV cure; Front Microbiol. 10:1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems. 3(6):e00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Song Q, Tang X, Liang X, Fan H, Peng H, Guo Q, Zhang Z. 2014. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. 2017. Mining the human gut microbiota for immunomodulatory organisms. Cell. 168(5):928–943, e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. 2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 15(3):382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nat Med. 24(4):392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves DT, Correa JD, Silva TA. 2019. The oral microbiota is modified by systemic diseases. J Dent Res. 98(2):148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, et al. 2018. A simple and novel fecal biomarker for colorectal cancer: ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin Chem. 64(9):1327–1337. [DOI] [PubMed] [Google Scholar]
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. 2015. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor tigit protects tumors from immune cell attack. Immunity. 42(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha NH, Woo BH, Kim DJ, Ha ES, Choi JI, Kim SJ, Park BS, Lee JH, Park HR. 2015. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. 36(12):9947–9960. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 15(1):30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorson HA, Schlett CD, Riddle MS. 2006. Postinfectious irritable bowel syndrome—a meta-analysis. Am J Gastroenterol. 101(8):1894–1899. [DOI] [PubMed] [Google Scholar]
- Horliana AC, Chambrone L, Foz AM, Artese HP, Rabelo Mde S, Pannuti CM, Romito GA. 2014. Dissemination of periodontal pathogens in the bloodstream after periodontal procedures: a systematic review. PLoS One. 9(5):e98271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugerth LW, Andreasson A, Talley NJ, Forsberg AM, Kjellström L, Schmidt PT, Agreus L, Engstrand L. 2020. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 69:1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature. 486(7402):215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SP, Williamson RT. 2001. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 85(2):162–169. [DOI] [PubMed] [Google Scholar]
- Inaba H, Amano A, Lamont RJ, Murakami Y. 2015. Involvement of protease-activated receptor 4 in over-expression of matrix metalloproteinase 9 induced by Porphyromonas gingivalis. Med Microbiol Immunol. 204(5):605–612. [DOI] [PubMed] [Google Scholar]
- Inaba H, Sugita H, Kuboniwa M, Iwai S, Hamada M, Noda T, Morisaki I, Lamont RJ, Amano A. 2014. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 16(1):131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 14(7):685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Nunez G. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 13(5):321–335. [DOI] [PubMed] [Google Scholar]
- Kassinen A, Krogius-Kurikka L, Makivuokko H, Rinttila T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. 2007. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 133(1):24–23. [DOI] [PubMed] [Google Scholar]
- Kilian M. 2018. The oral microbiome—friend or foe? Eur J Oral Sci. 126 Suppl 1:5–12. [DOI] [PubMed] [Google Scholar]
- Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A. 2019. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 68(7):1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 14(2):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz O. 2008. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 10(2):122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 569(7758):655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. 2008. Bacteremia associated with toothbrushing and dental extraction. Circulation. 117(24):3118–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell RM, Ford AC. 2012. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol. 10(7):712–721, e714. [DOI] [PubMed] [Google Scholar]
- Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz O, Lamont RJ. 2007. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 9(8):1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F, Babic A, Tworoger SS, Zhang L, Wu K, Smith-Warner SA, Ogino S, Chan AT, Meyerhardt J, Giovannucci E, et al. 2017. Periodontal disease, tooth loss and colorectal cancer risk: results from the Nurses’ Health Study. Int J Cancer. 140(3):646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, Turner D, Wolters VM, Bandsma RH, Mouzaki M, Zachos M, et al. 2013. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 19(1):115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Arimatsu K, Kato T, Matsuda Y, Minagawa T, Takahashi N, Ohno H, Yamazaki K. 2015. Oral administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of enterobacteria to the liver. PLoS One. 10(7):e0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman L, Simren M. 2010. Pathogenesis of ibs: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol. 7(3):163–173. [DOI] [PubMed] [Google Scholar]
- Ohman L, Tornblom H, Simren M. 2015. Crosstalk at the mucosal border: importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol. 12(1):36–49. [DOI] [PubMed] [Google Scholar]
- Paganelli FL, Luyer M, Hazelbag CM, Uh HW, Rogers MRC, Adriaans D, Berbers RM, Hendrickx APA, Viveen MC, Groot JA, et al. 2019. Roux-Y gastric bypass and sleeve gastrectomy directly change gut microbiota composition independent of surgery type. Sci Rep. 9(1):10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Xu X, Tan L, Lin L, Pan Y. 2014. The effects of Porphyromonas gingivalis on the cell cycle progression of human gingival epithelial cells. Oral Dis. 20(1):100–108. [DOI] [PubMed] [Google Scholar]
- Parahitiyawa NB, Jin LJ, Leung WK, Yam WC, Samaranayake LP. 2009. Microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev. 22(1):46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AM, Bardow A, Jensen SB, Nauntofte B. 2002. Saliva and gastrointestinal functions of taste, mastication, swallowing and digestion. Oral Dis. 8(3):117–129. [DOI] [PubMed] [Google Scholar]
- Pham VT, Lacroix C, Braegger CP, Chassard C. 2017. Lactate-utilizing community is associated with gut microbiota dysbiosis in colicky infants. Sci Rep. 7(1):11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, Moayyedi P. 2019. Gut microbiota in patients with irritable bowel syndrome—a systematic review. Gastroenterology. 157(1):97–108. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 464(7285):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. 2011. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 141(5):1792–1801. [DOI] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/beta-catenin signaling via its fadA adhesin. Cell Host Microbe. 14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology. 134(2):577–594. [DOI] [PubMed] [Google Scholar]
- Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, et al. 2019. Extensive transmission of microbes along the gastrointestinal tract. Elife. 8:e42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol Rev. 90(3):859–904. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simren M, Tack J. 2018. New treatments and therapeutic targets for IBS and other functional bowel disorders. Nat Rev Gastroenterol Hepatol. 15(10):589–605. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Souto RA, de Andrade AFB, Uzeda M, Colombo A. 2006. Prevalence of “non-oral” pathogenic bacteria in subgingival biofilm of subjects with chronic periodontitis. Braz J Microbiol. 37(3):208–215. [Google Scholar]
- Tagaino R, Washio J, Abiko Y, Tanda N, Sasaki K, Takahashi N. 2019. Metabolic property of acetaldehyde production from ethanol and glucose by oral streptococcus and neisseria. Sci Rep. 9(1):10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, et al. 2019. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 25(4):667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukasaki M, Komatsu N, Nagashima K, Nitta T, Pluemsakunthai W, Shukunami C, Iwakura Y, Nakashima T, Okamoto K, Takayanagi H. 2018. Host defense against oral microbiota by bone-damaging T cells. Nat Commun. 9(1):701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, et al. 2018. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 10(472):eaap8914. [DOI] [PubMed] [Google Scholar]
- Walker MY, Pratap S, Southerland JH, Farmer-Dixon CM, Lakshmyya K, Gangula PR. 2018. Role of oral and gut microbiome in nitric oxide-mediated colon motility. Nitric Oxide. 73:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G. 2016. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 6:33440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. 2019. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 25(4):679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt GM, Bayliss CE, Lakey AF, Bradley HK, Hunter JO, Jones VA. 1988. The faecal flora of two patients with food-related irritable bowel syndrome during challenge with symptom-provoking foods. J Med Microbiol. 26(4):295–299. [DOI] [PubMed] [Google Scholar]
- Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. 2019. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 25(6):968–976. [DOI] [PubMed] [Google Scholar]
- Yao L, Jermanus C, Barbetta B, Choi C, Verbeke P, Ojcius DM, Yilmaz O. 2010. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Mol Oral Microbiol. 25(2):89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Jungas T, Verbeke P, Ojcius DM. 2004. Activation of the phosphatidylinositol 3-kinase/akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 72(7):3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, Lamont RJ, Ojcius DM. 2008. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 10(4):863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 170(3):548–563e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki PJ, Perkowski K, Starosciak B, Baltaza W, Padzik M, Pionkowski K, Chomicz L. 2016. Identification of infectious microbiota from oral cavity environment of various population group patients as a preventive approach to human health risk factors. Ann Agric Environ Med. 23(4):566–569. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Sztukowska M, Wang Q, Inaba H, Potempa J, Scott DA, Wang H, Lamont RJ. 2015. Noncanonical activation of beta-catenin by Porphyromonas gingivalis. Infect Immun. 83(8):3195–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520924633 for The Bacterial Connection between the Oral Cavity and the Gut Diseases by S. Kitamoto, H. Nagao-Kitamoto, R. Hein, T.M. Schmidt and N. Kamada in Journal of Dental Research