Abstract
Quantitative real-time PCR (qPCR) has become a widely used approach to analyze the expression level of selected genes. However, owing to variations in cell types and drug treatments, a suitable reference gene should be selected according to special experimental design. In this study, we investigated the expression level of ten candidate reference genes in hepatoma carcinoma cell (HepG2) and human hepatocyte cell line (L02) treated with ethanol (EtOH), hydrogen peroxide (H2O2), acetaminophen (APAP), and carbon tetrachloride (CCl4), respectively. To analyze raw cycle threshold values (Cp values) from qPCR run, three reference gene validation programs, including Bestkeeper, geNorm, and NormFinder, were used to evaluate the stability of ten candidate reference genes. The results showed that TATA-box binding protein (TBP) and tubulin beta 2a (TUBB2a) presented the highest stability for normalization under different treatments and were regarded as the most suitable reference genes of HepG2 and L02. In addition, this study not only identified the most stable reference genes of each treatment, but also suggested that β-actin (ACTB), glyceraldehade-3-phosphate dehydrogenase (GAPDH), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), and beta-2 microglobulin (B2M) were the least stable reference genes in HepG2 and L02. This work was the first report to systematically explore the stability of reference genes in injured models of HepG2 and L02.
1. Introduction
Quantitative real-time PCR (qPCR) is commonly used in analyzing gene expression levels owing to its credible precision and high-throughput competence [1, 2]. However, quantitative analysis of gene expressions is unavoidably affected by several factors such as sample amount, cell activity, RNA integrity, and cDNA quality [3–5]. Hence, in order to avoid quantitative errors and obtain a reliable experimental result, one or several reference genes should be applied as a suitable endogenous control for quantitative measurement of gene expression. Some literature indicated that at least three reference genes were needed to normalize the analysis of qPCR [6, 7]. In addition, numerous reports affirmed that the stability of reference genes might change based on various experimental designs and samples [8, 9]. Hence, a stable reference gene, which ensures the stability in various experimental conditions, should be identified.
Traditionally, GAPDH and ACTB are most frequently used for normalization; however, they have been demonstrated unsuitable for internal control because their stability varies in different experiments and samples [10–12]. For instance, in Li et al.'s [13] study, the mean Cp value of GAPDH was 23.88 in H2O2 treated human umbilical vein endothelial cells (HUVEC), while in cytokines treated HUVEC, the mean Cp values of GAPDH was distinctly below 20 [14]. Undoubtedly, the varied expressions of reference genes lead to the inaccuracy of results. Fortunately, an increasing number of researches have focused on selecting and identifying suitable reference genes of humans [15], plant [16], cell line [17], algae [18], animal [19], and bacteria [20]. However, a systematic research about the validation of suitable reference genes for liver cell (HepG2 and L02) injured models has not been reported.
HepG2 is an immortalized human hepatoma cell line, and L02 is an immortalized hepatocyte cell line [21, 22]. Additionally, HepG2 and L02 are widely accepted model systems for investigating hepatotoxicity, intracellular trafficking, and drug targeting in vitro [23–25]. Owing to the stability of reference genes varied with drug-treatments and differed in different cell lines [26]. Hence, in this study, we chose four liver cell injured models commonly used in pharmacology and toxicology: ethanol (EtOH) [27], hydrogen peroxide (H2O2) [28], acetaminophen (APAP) [29], and carbon tetrachloride (CCl4) [30], which represented alcoholic liver injury (EtOH), hepatic oxidative stress (H2O2), drug liver injury (APAP), and acute liver damage (CCl4), respectively, to find the most appropriate reference genes in different cell injured models of HepG2 and L02.
In this study, ten candidate reference genes, ACTB, B2M, GAPDH, TUBB2a, hypoxanthine phosphoribosyltransferase 1 (HPRT1), succinate dehydrogenase complex flavoprotein subunit A (SDHA), TBP, YWHAZ, cytochrome c isoform 1 (CYC1), and glucuronidase beta (GUSB), were selected to investigate the most stable reference genes for normalization in liver cell injured models. To evaluate the stability of candidate reference genes comprehensively, four types of experimental treatments (EtOH, H2O2, APAP, and CCl4) were investigated in two cell types (HepG2 and L02) in vitro. In addition, in order to analyze the correlation between different concentrations of drug treatment and expression levels of reference genes, we selected three groups of different concentrations (low dose group, middle dose group, and high dose group) for each treatment. All concentrations were chosen based on previous studies [31–36] which had performed a cell viability assay proving varying degrees cytotoxicity. To analyze the original data, three statistical algorithms named, geNorm [37], NormFinder [38], and Bestkeeper [39] were used based on the manufacturers' procedures. The calculation results of three kinds of software showed that TBP and TUBB2a were the most stable ones among all treatments. Moreover, geNorm was also used to calculate the optimal number of reference genes needed for normalization, and the results showed that it was sufficient for accuracy normalization to choose two reference genes in most groups. To our knowledge, this is the first study about the selection of the best reference genes in liver cell injured models, which would provide a proper choice of reference genes and guarantee a dependable result in liver cell injured model research.
2. Materials and Methods
2.1. Reagents
The ethanol (EtOH, 99.5% pure), hydrogen peroxide (H2O2, 30.0% pure), acetaminophen (APAP, 99.5% pure), and carbon tetrachloride (CCl4, 99.5% pure) were purchased from Aladdin Biochemical Technology Co., Ltd (Shanghai, China); Penicillin and streptomycin were obtained from Beyotime Institute of Biotechnology (Shanghai, China); Trypsin-EDTA Solution was purchased from Sangon Biotech (Shanghai, China).
2.2. Cell Culture and Treatment
The hepatoma carcinoma cells (HepG2) were obtained from the American Type Culture Collection (HB-8065), and the human hepatocyte cells were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Gibco), containing 100 U/ml penicillin-streptomycin and 10% fetal bovine serum (FBS, Bioind) under standard conditions (37°C and 5% CO2). The cells were grown to 80% confluence and then passaged using Trypsin-EDTA Solution (0.25% Trypsin, 0.02% EDTA). All cells were divided into four groups for treatments: (a) control group; (b) low dose group; (c) middle dose group; (d) high dose group. For HepG2, cells were treated with four different treatments, including ethanol (100 mM, 200 mM, 400 mM), H2O2 (200 μM, 400 μM, 800 μM), APAP (2.5 mM, 5 mM, 10 mM), and CCl4 (0.1%, 0.2%, 0.4%). For L02, cells were treated with four different treatments, including ethanol (100 mM, 200 mM, 400 mM), H2O2 (100 μM, 200 μM, 400 μM), APAP (2.5 mM, 5 mM, 10 mM), and CCl4 (0.05%, 0.1%, 0.2%). The CCl4 were dissolved into 0.25% DMSO and then were added to the serum-free DMEM; the ethanol, H2O2, and APAP were dissolved into serum-free DMEM directly. Cells were seeded in six-well plates before being subjected to treatments. For all groups, cells were incubated in the presence or absence of various treatments and different concentrations for 24 h.
2.3. Screening of Candidate Reference Genes and Primer Design
According to previous studies [9, 40], a total of ten candidate reference genes (ACTB, B2M, GAPDH, TUBB2a, HPRT1, SDHA, TBP, YWHAZ, CYC1, and GUSB) were selected to ascertain the best reference genes of HepG2 and L02 in liver cell injured conditions. The nucleotide sequences were downloaded, using Primer 5 to design primers. Full gene names and accession numbers, as well as primer length and intron-spanning primers, were listed in Table 1. The data of qPCR were repeated three times of biological and technical replicates.
Table 1.
Details of the ten candidate reference genes and primers used in the qPCR.
| Gene | Description | Primer: forward/reverse(5′-3′) | Length (bp) | Accession number |
|---|---|---|---|---|
| ACTB | β-Actin | F: AAGGCCAACCGCGAGAAGAT R: GCCAGAGGCGTACAGGGATA |
102 | NM_001101 |
| B2M | Beta-2 microglobulin | F: GTTTACTCACGTCATCCAGC R:AGACAAGTCTGAATGCTCCA |
141 | NM_004048 |
| GAPDH | Glyceraldehade-3-phosphate dehydrogenase | F: GCCTCCTGCACCACCAACTG R: CCATCACGCCACAGTTTCCC |
149 | NM_002046 |
| TUBB2a | Tubulin beta 2a | F: AACGCCACCCTCTCTGTCCA R: GCCGACACCAGGTGGTTGAG |
143 | NM_001069 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | F: ACTGAACGTCTTGCTCGAGA R: TGATGTAATCCAGCAGGTCA |
112 | NM_000194 |
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A | F: AAAGATCACGTCTACCTGCA R: CATGTTATAATGCACGGTGG |
150 | NM_004168 |
| TBP | TATA-box binding protein | F: GTTCAGCAGTCAACGTCCCA R: TCATGGGGGAGGGATACAGT |
127 | NM_003194 |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | F: CAGGCTGAGCGATATGATGA R: CCTACGGGCTCCTACAACAT |
126 | NM_003406 |
| CYC1 | Cytochrome c isoform 1 | F: CCAAAACCATACCCCAACAG R: AGTCCTCACCACCATGCCTA |
103 | NM_001916 |
| GUSB | Glucuronidase beta | F: GTTCCTTTTGCGAGAGAGAT R: ACACGCAGGTGGTATCAGTC |
124 | NM_000181 |
2.4. Total RNA, DNA Extraction and cDNA Synthesis
Total RNA was extracted from HepG2 and L02 and purified using the RNAiso Plus total RNA kit (TransGen Biotech, Dalian, China) according to the manufacturer's instruction. And then, DNase I (Takara, Dalian, China) was added to the sample to eliminate DNA contamination for RNA purity. The purity of the total RNA was assessed by measuring the absorbance ration at 260/280 nm of the samples. In addition, the quality of the RNA was confirmed by agarose gel electrophoresis. Purified RNA was reverse transcribed immediately after extraction. For qPCR experiments, HiScript® Q RT SuperMix for qPCR Kit (Vazyme, Nanjing, China) and a quantity of 1 μg total RNA were added into a 20 μl reaction volume to synthesize cDNA.
2.5. Quantitative Real-Time PCR
The sample reaction was run in 96-well plate. Real-time quantitative PCR with AceQ qPCR SYBR Green Master Mix (Vazyme, Nangjing, China) was performed at LightCycler 480 (Roche Molecular Biochemicals, Mannheim, Germany). Each reaction system was 20 μl, respectively. AceQ qPCR SYBR Green Master Mix 10 μl, forward and reverse primers were 0.4 μM each, template cDNA 2 μl, and added ddH2O to the final volume of 20 μl. Each sample was repeated 3 times. The optimizing reaction conditions of real-time quantitative PCR as follows: 1 cycle of 95°C for 5 min, 40 cycles of 95°C for 10 sec, and then 60°C for 30 sec.
2.6. Analysis of Reference Genes Stability
In order to evaluate the stability of ten selected reference genes, three reference gene validation programs (geNorm, NormFinder, and BestKeeper) were used under the manufacture's instruction. NormFinder was applied to calculate the stability value (M) for finding the steadiest candidate genes. For geNorm, the calculation could determine the optimal number of reference genes and, similar to geNorm, evaluate the stability of candidate genes. BestKeeper was based on the coefficient of variance (CV) and the standard deviation (SD) of the Cp values to assess the steadiness of reference genes. Three biological and technical repeats were used for different experimental conditions.
3. Results
3.1. Verification of the Primers Specificity
We used PCR to identify the specificity of the designed primers by agarose gel electrophoresis, as S2 and S3 Figs shows, the single band and peak of a melting curve indicated primers possessed the good specificity.
3.2. Evaluating Expression of the Reference Gene
The most suitable reference genes would have stable expression levels in various treatments and concentrations. And the Cp value of ten candidate reference genes underwent diver treatments were listed in Figure 1, ranging from 14.7 to 34.49 (HepG2 14.7 to 34.49, L02 14.85 to 34.48), suggesting that they have a noticeable variance in expression level. Particularly, most of the Cp values were in a range of 20 to 27. ACTB, B2M, GAPDH, HPRT1, and YWHAZ expressed lower Cp value around 20, while the rest of the genes showed that higher Cp value was greater than 25, especially the GUSB, which had the highest mean Cp values (HepG2 30.02 ± 1.49, L02 30.99 ± 1.66). Notably, ACTB showed the minimal change of Cp values from 18.44 to 24.90 under different treatments in HepG2, and meanwhile, the Cp values of HPRT1 from 21.46 to 27.02 showed the low variation in L02, suggesting that the two genes might have a constant expression under various treatments and could be a suitable reference gene. In short, Cp values, combined with box-plot, presented the expression of the reference genes, and as well provided us a general understanding of gene stability.
Figure 1.

Expression levels of ten reference genes (ACTB, B2M, GAPDH, TUBB2a, HPRT1, SDHA, TBP, YWHAZ, CYC1, GUSB) in HepG2 (a) and L02 (b). Squares in the middle of the box represent the mean values, horizontal lines in the box represent the median, and whiskers represent the highest and lowest values.
3.3. Expression Stability of Candidate Reference Genes
The data obtained from different treatments (wild-type APAP, CCl4, ethanol, and H2O2) and each reference gene were analyzed with three Excel-based programs (geNorm, NormFinder, and BestKeeper) for further evaluation on the stability of putative reference genes.
3.4. geNorm Analysis
To ascertain the stability of candidate reference genes, geNorm was applied to evaluate the expression stability measurement (M) value by Cp values of each gene in groups. According to the analysis of geNorm, genes with the highest M values were considered as the least stable ones and the lowest the most. As shown in Figure 2 and S1 Figure, different reference genes had different M values in different treatments. For instance, in the L02 groups, TBP with the M value of 0.51 in 400 μM H2O2 treatment would be the steadiest reference genes, while the GUSB was more than twice TBP, with M value was 1.37 in the same treatment. More interestingly, even the same reference gene had different expressions in different treatments. In the HepG2 groups, the gene with the lowest M values in 10 mM APAP treatment was HPRT1, which owned the highest M values in 200 μM H2O2 treatment, meaning that HPRT1 was the most stable genes in 10 mM APAP treatment and the least stable ones in 200 μM H2O2 treatment.
Figure 2.
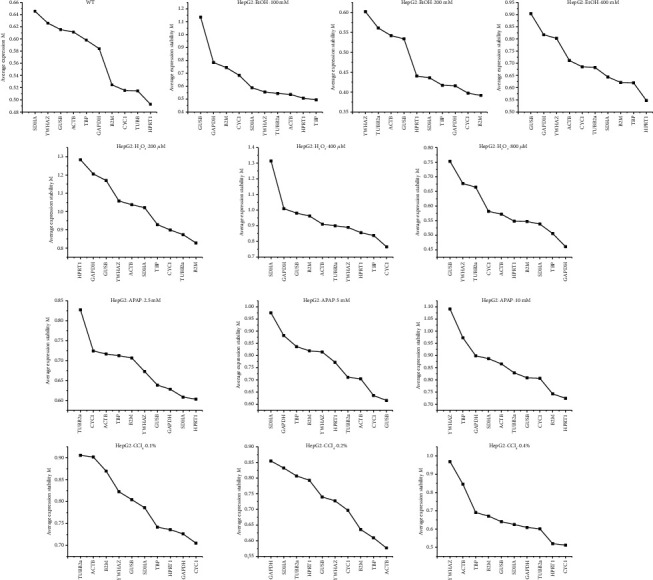
Expression stability of the reference genes in HepG2 evaluated by geNorm M values represents the average expression stability. From left to right, the value of M decreased in turn, indicating the stability gradually increased. Smaller M value means higher stability. The control group, ethanol, hydrogen peroxide, acetaminophen, and carbon tetrachloride were abbreviated to WT, EtOH, H2O2, APAP, and CCl4, respectively.
3.5. NormFinder Analysis
NormFinder was used to evaluate the optimal gene for normalization in each experiment. The raw Cp values obtained from qPCR were firstly log-transformed and used as the input value for the NormFinder, and then used to analyze the expression stability according to the similarity of the expression profiles of candidate genes. Genes with lower values that were close to zero were regarded as the best candidate ones. As shown in Table 2 and S1 Table, the rank of M values was increasing from top to bottom of the table, whereas genes on the top of the table were the most stable reference genes. Therefore, the most stable candidate genes could be easily found from the table. In the HepG2 group, CYC1 (7 times to be the top 3 candidate genes in 13 treatments) and HPRT1 (8 times to be the top 3 in 13 treatments) were considered as the most stable reference genes; the results were similar to that of geNorm analysis. Nevertheless, in the L02 group, the results of geNorm and NormFinder analysis seemed to be different. For instance, in the NormFinder analysis, TUBB2a (9 times to be the top 3 in all treatments) was the steadiest ones, while GAPDH, the most stable genes in the geNorm analysis, appeared only once to be the top 3 of all treatments. Hence, the third analysis method should be used.
Table 2.
Expression stability values of ten candidate reference genes in HepG2 analyzed by NormFinder.
| Rank | WT | EtOH 100 mM |
EtOH 200 mM |
EtOH 400 mM |
H2O2 200 μM |
H2O2 400 μM |
H2O2 800 μM |
APAP 2.5 mM |
APAP 5 mM |
APAP 10 mM |
CCl4 0.1% |
CCl4 0.2% |
CCl4 0.4% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HPRT1 0.011 |
HPRT1 0.004 |
CYC1 0.007 |
HPRT1 0.008 |
B2M 0.018 |
CYC1 0.013 |
TBP 0.011 |
HPRT1 0.013 |
GUSB 0.004 |
HPRT1 0.012 |
TBP 0.015 |
TBP 0.011 |
HPRT1 0.005 |
| 2 | TUBB2a 0.012 |
TBP 0.005 |
TBP 0.009 |
TUBB2a 0.013 |
TUBB2a 0.019 |
TUBB2a 0.017 |
GAPDH 0.011 |
GUSB 0.015 |
CYC1 0.010 |
B2M 0.018 |
CYC1 0.017 |
ACTB 0.013 |
CYC1 0.008 |
| 3 | CYC1 0.013 |
TUBB2a 0.005 |
SDHA 0.011 |
B2M 0.014 |
CYC1 0.020 |
TBP 0.019 |
SDHA 0.012 |
SDHA 0.018 |
TUBB2a 0.014 |
SDHA 0.019 |
SDHA 0.020 |
B2M 0.021 |
GUSB 0.009 |
| 4 | B2M 0.017 |
YWHAZ 0.012 |
B2M 0.011 |
TBP 0.016 |
TBP 0.027 |
HPRT1 0.024 |
HPRT1 0.015 |
YWHAZ 0.022 |
ACTB 0.020 |
TUBB2a 0.020 |
HPRT1 0.021 |
TUBB2a 0.021 |
TUBB2a 0.016 |
| 5 | TBP 0.018 |
SDHA 0.013 |
HPRT1 0.013 |
SDHA 0.018 |
GUSB 0.030 |
GUSB 0.025 |
B2M 0.017 |
CYC1 0.023 |
HPRT1 0.021 |
CYC1 0.022 |
TUBB2a 0.024 |
CYC1 0.021 |
SDHA 0.019 |
| 6 | SDHA 0.019 |
ACTB 0.014 |
GAPDH 0.015 |
CYC1 0.020 |
SDHA 0.030 |
YWHAZ 0.027 |
TUBB2a 0.017 |
TBP 0.024 |
TBP 0.022 |
ACTB 0.024 |
GUSB 0.024 |
GUSB 0.022 |
TBP 0.024 |
| 7 | ACTB 0.020 |
CYC1 0.022 |
GUSB 0.015 |
ACTB 0.025 |
YWHAZ 0.037 |
ACTB 0.031 |
CYC1 0.017 |
ACTB 0.028 |
YWHAZ 0.025 |
GUSB 0.025 |
YWHAZ 0.028 |
YWHAZ 0.022 |
B2M 0.029 |
| 8 | GUSB 0.021 |
B2M 0.032 |
TUBB2a 0.016 |
GUSB 0.027 |
ACTB 0.039 |
B2M 0.037 |
ACTB 0.019 |
GAPDH 0.029 |
B2M 0.029 |
TBP 0.029 |
GAPDH 0.032 |
SDHA 0.024 |
GAPDH 0.031 |
| 9 | YWHAZ 0.023 |
GAPDH 0.041 |
ACTB 0.020 |
YWHAZ 0.028 |
HPRT1 0.046 |
SDHA 0.043 |
GUSB 0.022 |
TUBB2a 0.030 |
SDHA 0.031 |
GAPDH 0.035 |
B2M 0.034 |
HPRT1 0.029 |
ACTB 0.037 |
| 10 | GAPDH 0.029 |
GUSB 0.042 |
YWHAZ 0.023 |
GAPDH 0.041 |
GAPDH 0.061 |
GAPDH 0.048 |
YWHAZ 0.024 |
B2M 0.032 |
GAPDH 0.039 |
YWHAZ 0.037 |
ACTB 0.037 |
GAPDH 0.044 |
YWHAZ 0.039 |
3.6. BestKeeper Analysis
BestKeeper was an Excel-based tool used to analyze the expression stability of the candidate reference gene. The standard deviation (SD) and coefficient of variation (CV) were calculated by BestKeeper to assess the stability of candidate reference genes in each group. Genes with the lowest SD and CV would be the most stable reference ones. As shown in Table 3 and S2 Table, the (CV ± SD) values of ten candidate reference genes progressively increased from top to bottom of tables, showing their decreasingly stability. As an example, TBP was listed on the top of Table 3, with a lower (CV ± SD) value of (0.85 ± 0.24), representing the most stable genes in 800 μM H2O2 induced oxidative stress in HepG2, and meanwhile, GUSB, having a (CV ± SD) value of (5.42 ± 1.62), was listed at the bottom of the table. In HepG2 groups, some reference genes, namely, TBP, CYC1, and TUBB2a, might be the best suitable genes for the reason that they were listed on top 3 of the rank in majority treatments. Similarly, CYC1 and TBP occupied most of the top 3 in the table, suggesting that the two candidate genes would be the steadiest genes in L02 treatments.
Table 3.
Expression stability values of the reference genes calculated by BestKeeper in HepG2.
| Rank | WT | EtOH 100 mM |
EtOH 200 mM |
EtOH 400 mM |
H2O2 200 μM |
H2O2 400 μM |
H2O2 800 μM |
APAP 2.5 mM |
APAP 5 mM |
APAP 10 mM |
CCl4 0.1% |
CCl4 0.2% |
CCl4 0.4% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GUSB 1.57 ± 0.45 |
HPRT1 1.10 ± 0.24 |
SDHA 0.74 ± 0.20 |
B2M 1.57 ± 0.29 |
CYC1 1.37 ± 0.33 |
CYC1 1.49 ± 0.36 |
TBP 0.85 ± 0.24 |
SDHA 1.78 ± 0.44 |
CYC1 0.95 ± 0.25 |
GUSB 1.75 ± 0.59 |
GUSB 0.90 ± 0.26 |
HPRT1 1.67 ± 0.38 |
TBP 1.15 ± 0.30 |
| 2 | ACTB 1.89 ± 0.39 |
CYC1 1.24 ± 0.3 |
TBP 1.00 ± 0.26 |
TBP 1.67 ± 0.44 |
TUBB2a 1.68 ± 0.48 |
YWHAZ 1.51 ± 0.35 |
CYC1 0.95 ± 0.24 |
TBP 2.50 ± 0.63 |
GUSB 1.47 ± 0.46 |
CYC1 2.60 ± 0.75 |
CYC1 1.10 ± 0.27 |
GUSB 1.68 ± 0.51 |
SDHA 1.53 ± 0.38 |
| 3 | TUBB2a 2.22 ± 0.63 |
TBP 1.28 ± 0.33 |
CYC1 1.08 ± 0.27 |
SDHA 1.70 ± 0.46 |
SDHA 1.90 ± 0.51 |
GUSB 1.54 ± 0.47 |
B2M 1.08 ± 0.21 |
TUBB2a 2.83 ± 0.72 |
TUBB2a 1.59 ± 0.45 |
TUBB2a 3.25 ± 0.94 |
HPRT1 1.43 ± 0.32 |
TBP 1.72 ± 0.45 |
CYC1 1.72 ± 0.43 |
| 4 | CYC1 2.41 ± 0.57 |
SDHA 1.79 ± 0.47 |
GUSB 1.12 ± 0.34 |
YWHAZ 2.08 ± 0.47 |
TBP 2.28 ± 0.61 |
HPRT1 1.77 ± 0.40 |
SDHA 1.16 ± 0.32 |
GUSB 2.97 ± 0.86 |
HPRT1 1.64 ± 0.41 |
ACTB 3.27 ± 0.77 |
B2M 1.99 ± 0.37 |
CYC1 1.84 ± 0.47 |
TUBB2a 1.97 ± 0.54 |
| 5 | B2M 2.65 ± 0.48 |
TUBB2a 1.92 ± 0.54 |
ACTB 1.16 ± 0.25 |
HPRT1 2.12 ± 0.49 |
ACTB 2.58 ± 0.53 |
TUBB2a 1.80 ± 0.52 |
HPRT1 1.22 ± 0.29 |
YWHAZ 3.25 ± 0.71 |
B2M 1.74 ± 0.35 |
TBP 3.38 ± 0.98 |
TBP 2.19 ± 0.57 |
YWHAZ 2.40 ± 0.54 |
HPRT1 2.31 ± 0.53 |
| 6 | TBP 2.78 ± 0.72 |
ACTB 2.01 ± 0.41 |
B2M 1.19 ± 0.22 |
CYC1 2.16 ± 0.53 |
B2M 2.84 ± 0.51 |
TBP 1.84 ± 0.49 |
GAPDH 1.39 ± 0.25 |
HPRT1 3.28 ± 0.76 |
YWHAZ 2.17 ± 0.53 |
B2M 3.90 ± 0.88 |
SDHA 2.42 ± 0.61 |
TUBB2a 2.58 ± 0.73 |
GUSB 2.72 ± 0.81 |
| 7 | HPRT1 3.10 ± 0.70 |
YWHAZ 2.13 ± 0.47 |
TUBB2a 1.28 ± 0.37 |
TUBB2a 2.20 ± 0.62 |
GUSB 2.96 ± 0.88 |
ACTB 2.78 ± 0.57 |
GUSB 1.59 ± 0.50 |
CYC1 3.31 ± 0.82 |
ACTB 2.20 ± 0.50 |
HPRT1 4.04 ± 1.11 |
YWHAZ 2.45 ± 0.54 |
ACTB 2.84 ± 0.57 |
YWHAZ 2.97 ± 0.67 |
| 8 | SDHA 3.41 ± 0.91 |
GUSB 2.18 ± 0.65 |
GAPDH 1.45 ± 0.25 |
GUSB 2.58 ± 0.78 |
YWHAZ 3.48 ± 0.78 |
B2M 3.14 ± 0.59 |
TUBB2a 1.62 ± 0.47 |
ACTB 3.39 ± 0.67 |
TBP 2.59 ± 0.72 |
SDHA 4.41 ± 1.30 |
TUBB2a 2.63 ± 0.74 |
B2M 2.92 ± 0.55 |
ACTB 3.92 ± 0.75 |
| 9 | YWHAZ 3.63 ± 0.82 |
B2M 2.82 ± 0.51 |
HPRT1 1.55 ± 0.35 |
ACTB 3.37 ± 0.71 |
HPRT1 4.86 ± 1.10 |
SDHA 3.84 ± 1.05 |
ACTB 1.65 ± 0.36 |
B2M 4.86 ± 0.92 |
SDHA 2.68 ± 0.73 |
GAPDH 4.81 ± 0.97 |
ACTB 2.95 ± 0.60 |
SDHA 3.32 ± 0.84 |
B2M 4.41 ± 0.83 |
| 10 | GAPDH 3.99 ± 0.66 |
GAPDH 3.30 ± 0.55 |
YWHAZ 2.32 ± 0.54 |
GAPDH 4.03 ± 0.71 |
GAPDH 5.26 ± 0.89 |
GAPDH 4.21 ± 0.71 |
YWHAZ 2.24 ± 0.53 |
GAPDH 4.95 ± 0.78 |
GAPDH 3.43 ± 0.63 |
YWHAZ 6.38 ± 1.64 |
GAPDH 3.42 ± 0.57 |
GAPDH 4.38 ± 0.73 |
GAPDH 4.46 ± 0.73 |
3.7. Optimal Numbers of Reference Genes for Normalization
The minimal numbers of reference genes for accurate normalization could also be determined by geNorm, according to the calculation of pairwise variation (variation coefficient, V) between the normalization factors (NF) in various treatment sets using Vn/n + 1 < 0.15 as a criterion cut-off value [37]. Based on this rule, the calculation was listed in Figure 3. As we can see, there were enough to choose two or three reference genes in most treatments of HepG2 and L02 for normalization. Moreover, 10 mM APAP treatment in HepG2, 200 mM EtOH, and 400 mM EtOH in L02, respectively, required four, five, and nine reference genes for normalization.
Figure 3.
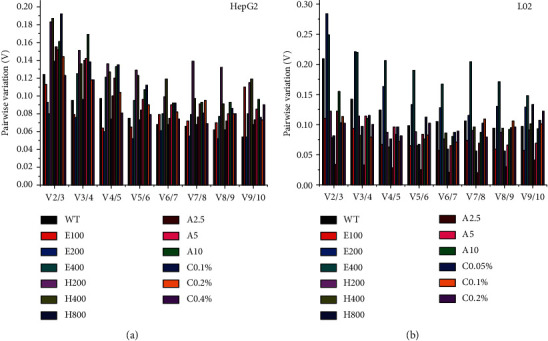
Calculation of the optimal number of reference genes for quantitative analysis using geNorm. Pairwise variation (Vn/n + 1) of reference genes analyzed in different treatments was listed in (a, b) set the cut-off threshold value 0.15, calculating the optimal number of reference genes for precise quantitative in this work. WT, E100 (E200, E400), H100 (H200, H400, H800), A2.5 (A5, A10), and C0.05% (C0.1%, C0.2%, C0.4%), respectively, were the abbreviation for the control group, EtOH-100 mM (200 mM, 400 mM), H2O2-100 μM (200 μM, 400 μM, 800 μM), APAP-2.5 mM (5 mM, 10 mM, 20 mM), CCl4-0.05% (0.1%, 0.2%, 0.4%).
4. Discussion
Quantitative real-time PCR is one of the most accurate and commonly used techniques for analysis of gene transcript levels. Selection of suitable reference gene is indispensable to guarantee the accuracy and consistency of the data and minimize the experimental errors. To confirm the precise expression analysis of putative genes, numerous steady reference genes have been verified in different experimental designs [41, 42]. Hence, the purpose of this study was to investigate the expression stability of ten candidate reference genes (ACTB, B2M, GAPDH, TUBB2a, HPRT1, SDHA, TBP, YWHAZ, CYC1, GUSB) in two in-vitro cell types, namely, HepG2 cells and L02 cells, and determine the optimal candidate genes under the treatment of alcoholic liver injury (EtOH), hepatic oxidative stress (H2O2), drug liver injury (APAP), and acute liver damage (CCl4). After that, raw data was input and calculated in three Excel-based programs: geNorm, NormFinder, and BestKeeper.
The data from qPCR run of ten candidate genes were listed in Figure 1, where the expression level and mean Cp values of the candidate genes ranging from 14.7 to 34.49 (HepG2 14.7 to 34.49, L02 14.85 to 34.48) could be easily seen. However, the scope of the Cp values of some selected genes inconsistent with the previous study [43, 44] might be a result of liver damage treatments. Based on the fact that the genes with the highest expression levels owned minimal Cp values, GAPDH with the lowest mean Cp values of 17.47 in HepG2 and 17.25 in L02 means that the GAPDH was abundantly distributed in the two cell types. Considering that a wide distribution range tends to be low stability and moreover, Cp values with low variation would be more suitable for reference gene selection. The variation of Cp values suggested that ACTB and HPRT1 were the best reference genes in HepG2 and L02, while HPRT1 and YWHAZ were the least ones. The verification above was a little different from the calculation of the three Excel-based programs. For instance, the Cp values of ACTB and HPRT1 might not fluctuate significantly, but in the calculations of the three kinds of software, the two genes appeared at the bottom of ranking frequently, which reflected more instability. Accordingly, more calculation results needed to be combined.
Furthermore, some literature reported that the expression level of reference genes would change under different concentration treatments [26, 45]. Hence, we investigated the stability of the reference genes in the same treatment of different concentrations with the results of three software calculations. Since the results of three software varied based on different algorithms, we selected the top five (six in some groups) reference genes for each calculation to evaluate them comprehensively. We identified the top five genes under three concentrations, and then found that genes were common in geNorm, NormFinder, and Bestkeeper. For instance, in CCl4 of three concentrations treated L02 cells, GUSB, TUBB2a, and TBP commonly appeared to be the top five of Bestkeeper calculation results, while SDHA, GAPDH, and TUBB2a of geNorm and SDHA, TBP, and TUBB2a of NormFinder, respectively. Apparently, TUBB2a appeared to be one of the top five of each calculation. Hence, we recommended TUBB2a as the most stable reference genes in CCl4 treated L02 cells. Likewise, in L02 cells, TBP was considered as the most stable genes in EtOH, H2O2, and APAP treatments, and TUBB2a was the steadiest in CCl4 treatments. In HepG2 cells, we suggested TBP being the most stable reference genes in EtOH and H2O2 treatments, and GUSB and CYC1 in APAP and CCl4 treatments, respectively. In the same way, we evaluated the least stable reference genes in each group. On the whole, ACTB, GAPDH, YWHAZ, and B2M always ranked the last, meaning that they were considered as the least stable. Hence, we did not recommend ACTB, GAPDH, YWHAZ, or B2M as internal control for normalization. However, in Bridget's study [44], they identified GAPDH as the most stable reference genes in APAP treated HepG2, opposite to our research, which was mainly because they evaluated the stability only using geNorm, which might lead to inaccurate results.
Based on the analysis above, we found the best suitable reference genes in different treatments. Nevertheless, how many reference genes were required to be chosen for optimal data normalization required further investigation. Hence, we chose the geNorm software, which could calculate the optimal number of reference genes in a qPCR experiment to solve these problems. According to the handbook [37], the V score of 0.15 as a criterion value was recommended, and an additional gene was included until (Vn/Vn + 1) was lower than 0.15. In this study, the results showed that the majority of pairwise V values were lower than 0.15 after a total of 26 treated groups. and the calculation was shown in Figure 3, among which 23 out of 26 groups with a low V < 0.15, signifying the inclusion of additional reference genes was unnecessary in those 23 groups. Hence, two or three reference genes would suffice for reliable normalization in these groups above. Unfortunately, not all groups had a suitable V value of lower than 0.15. For instance, in the 400 mM EtOH treated L02 group, all of pairwise V values were greater than 0.15. Vandesompele et al. recommended that it was a waste of resources to quantify more genes than necessary, and hence, the V value (V6/7 and V9/10 values were close to 0.15) indicated that six reference genes should be a good choice for normalization in 400 mM treated L02 group.
5. Conclusion
In this study, 10 candidate genes were selected and evaluated in two types of liver cells (HepG2 and L02) for four types of liver cell injured treatments using the three different algorithms, namely, BestKeeper, geNorm, and NormFinder. To the best of our knowledge, this was the first systematic selection of reference genes in the liver cell injured model and laid the basis for further research in HepG2 and L02. Based on the analysis, we identified the best reference genes of HepG2 and L02 under the treatments of EtOH, H2O2, APAP, and CCl4. The results of gene expression revealed that TBP and TUBB2a were the most stable reference genes for normalization in different treatments. On one hand, in the HepG2, the most stable reference genes of EtOH and H2O2 treatments were TBP, while GUSB and CYC1 were, respectively, the most suitable reference genes of APAP and CCl4 treatments. In the L02, TBP was identified as the most stable reference genes of EtOH, H2O2, and APAP treatments, while TUBB2a was the steadiest reference genes of CCl4 treatment. On the other hand, ACTB, GAPDH, YWHAZ, and B2M were the least stable reference genes in EtOH, H2O2, APAP, and CCl4 treated HepG2 and L02. In short, our study provided a credible selection of reference gene in HepG2 and L02 injured models.
Acknowledgments
This work was funded by the Practical Training Program for Young Teachers in Higher Vocational Colleges in Jiangsu Province (2019QYSJPX171) and Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJD430005). This project was also funded by the Qinglan project of excellent teaching team in Jiangsu and teaching and research project of Jiangsu Health Vocational College (JKKYTD201701, JKA201812).
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors did not report any conflict of interest.
Authors' Contributions
Jiyu Chen, Zhenzhen Bao, and Yanli Huang contributed equally to this work.
Supplementary Materials
S1 Figure: expression stability of the housekeeping genes in L02 evaluated by geNorm M values represents the average expression stability. From left to right, the value of M decreased in turn, indicating the stability gradually increased. Smaller M value means higher stability. The control group, ethanol, hydrogen peroxide, acetaminophen, and carbon tetrachloride were abbreviated to WT, EtOH, H2O2, APAP, and CCl4, respectively. S2 Figure: agarose gel (1%) electrophoresis of the ten candidate housekeeping genes. 1-10 represent ACTB, B2M, GAPDH, TUBB2a, HPRT1, SDHA, TBP, YWHAZ, CYC1, and GUSB, respectively. S3 Figure: melt curves of the ten candidate housekeeping genes. S1 Table: expression stability values of ten candidate housekeeping genes in L02 analyzed by NormFinder. S2 Table: expression stability values of the housekeeping genes calculated by BestKeeper in L02.
References
- 1.Bustin S. A., Benes V., Garson J. A., et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Zhang D., Li W., Chen J., Peng Y., Cao W. A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Research. 2003;31(20):123e–1123. doi: 10.1093/nar/gng123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alemayehu S., Feghali K. C., Cowden J., Komisar J., Ockenhouse C. F., Kamau E. Comparative evaluation of published real-time PCR assays for the detection of malaria following MIQE guidelines. Malaria Journal. 2013;12(1) doi: 10.1186/1475-2875-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsgren E., Locke B., Semberg E., Laugen A. T., Miranda J. R. . Sample preservation, transport and processing strategies for honeybee RNA extraction: Influence on RNA yield, quality, target quantification and data normalization. Journal of Virological Methods. 2017;246:81–89. doi: 10.1016/j.jviromet.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Huang X., Baumann M., Nikitina L., et al. RNA degradation differentially affects quantitative mRNA measurements of endogenous reference genes in human placenta. Placenta. 2013;34(7):544–547. doi: 10.1016/j.placenta.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Kong Q., Yuan J., Gao L., et al. Evaluation of appropriate reference genes for gene expression normalization during watermelon fruit development. Plos One. 2015;10(6, article e0130865) doi: 10.1371/journal.pone.0130865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicot N., Hausman J. F., Hoffmann L., Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56(421):2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- 8.Heckmann L. H., Connon R., Hutchinson T. H., Maund S. J., Sibly R. M., Callaghan A. Expression of target and reference genes in Daphnia magna exposed to ibuprofen. BMC Genomics. 2006;7(1) doi: 10.1186/1471-2164-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Ding L., Sandford A. J. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Molecular Biology. 2005;6(1) doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori R., Wang Q., Danenberg K. D., Pinski J. K., Danenberg P. V. Both beta-actin and GAPDH are useful reference genes for normalization of quantitative RT-PCR in human FFPE tissue samples of prostate cancer. Prostate. 2008;68(14):1555–1560. doi: 10.1002/pros.20815. [DOI] [PubMed] [Google Scholar]
- 11.Murthi P., Fitzpatrick E., Borg A. J., Donath S., Brennecke S. P., Kalionis B. GAPDH, 18S rRNA and YWHAZ are Suitable Endogenous Reference Genes for Relative Gene Expression Studies in Placental Tissues from Human Idiopathic Fetal Growth Restriction. Placenta. 2008;29(9):798–801. doi: 10.1016/j.placenta.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Pérez-Rico A., Crespo F., Sanmartín M. L., De Santiago A., Vega-Pla J. L. Determining ACTB, ATP5B and RPL32 as optimal reference genes for quantitative RT-PCR studies of cryopreserved stallion semen. Animal Reproduction Science. 2014;149(3-4):204–211. doi: 10.1016/j.anireprosci.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Li T., Diao H., Zhao L., et al. Identification of suitable reference genes for real-time quantitative PCR analysis of hydrogen peroxide-treated human umbilical vein endothelial cells. BMC Molecular Biology. 2017;18(1) doi: 10.1186/s12867-017-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Vallejo J. J., Van het Hof B., Robben J., et al. Approach for defining endogenous reference genes in gene expression experiments. Analytical Biochemistry. 2004;329(2):293–299. doi: 10.1016/j.ab.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 15.Ohl F., Jung M., Radonić A., Sachs M., Loening S. A., Jung K. Identification and validation of suitable endogenous reference genes for gene expression studies of human bladder cancer. Journal of Urology. 2006;175(5):1915–1920. doi: 10.1016/S0022-5347(05)00919-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y., Luo J., Xu S., et al. Selection of Reference Genes for Gene Expression Normalization in Peucedanum praeruptorum Dunn under Abiotic Stresses, Hormone Treatments and Different Tissues. Plos One. 2016;11(3, article e0152356) doi: 10.1371/journal.pone.0152356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceelen L., De Spiegelaere W., David M., et al. Critical selection of reliable reference genes for gene expression study in the HepaRG cell line. Biochemical Pharmacology. 2011;81(10):1255–1261. doi: 10.1016/j.bcp.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Dong M., Zhang X., Chi X., et al. The validity of a reference gene is highly dependent on the experimental conditions in green alga Ulva linza. Current Genetics. 2012;58(1):13–20. doi: 10.1007/s00294-011-0361-3. [DOI] [PubMed] [Google Scholar]
- 19.Yuanyuan S., Qin S., Rongrong X., et al. Reference gene selection for real-time quantitative PCR analysis on ovarian cryopreservation by vitrification in mice. Journal of Assisted Reproduction and Genetics. 2015;32(8):1277–1284. doi: 10.1007/s10815-015-0503-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiter L., Kolstø A.-B., Piehler A. P. Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. Journal of Microbiological Methods. 2011;86(2):210–217. doi: 10.1016/j.mimet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Roe A. L., Snawder J. E., Benson R. W., Roberts D. W., Casciano D. A. HepG2 Cells: An in Vitro Model for P450-Dependent Metabolism of Acetaminophen. Biochemical and Biophysical Research Communications. 1993;190(1):15–19. doi: 10.1006/bbrc.1993.1003. [DOI] [PubMed] [Google Scholar]
- 22.de Diesbach P., N'Kuli F., Berens C., et al. Receptor-mediated endocytosis of phosphodiester oligonucleotides in the HepG2 cell line: evidence for non-conventional intracellular trafficking. Nucleic Acids Research. 2002;30(7):1512–1521. doi: 10.1093/nar/30.7.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaeschke H., McGill M. R., Ramachandran A. Pathophysiological Relevance of Proteomics Investigations of Drug-Induced Hepatotoxicity in HepG2 Cells. Toxicological Sciences. 2011;121(2):428–430. doi: 10.1093/toxsci/kfr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lihua Y., Shi Z., Liu H., Yang X., Wang K. Gadolinium induced apoptosis of human embryo liver L02 cell line by ROS- mediated AIF pathway. Journal of Rare Earths. 2011;29(2):178–184. doi: 10.1016/S1002-0721(10)60427-9. [DOI] [Google Scholar]
- 25.Faibish D., Suzuki M., Bartlett J. D. Appropriate real-time PCR reference genes for fluoride treatment studies performed in vitro or in vivo. Archives of Oral Biology. 2016;62:33–42. doi: 10.1016/j.archoralbio.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szuster-Ciesielska A., Kandefer-Szerszen M. Protective effects of betulin and betulinic acid against ethanol-induced cytotoxicity in HepG2 cells. Pharmacological Reports. 2005;57(5):588–595. [PubMed] [Google Scholar]
- 27.Sohn S. H., Kim S. K., Kim Y. O., et al. A comparison of antioxidant activity of Korean White and Red Ginsengs on H2O2-induced oxidative stress in HepG2 hepatoma cells. Journal of Ginseng Research. 2013;37(4):442–450. doi: 10.5142/jgr.2013.37.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y., Liu C., Xia X., Liu L. Conformal microcapsules encapsulating microcarrier-L02 cell complexes for treatment of acetaminophen-induced liver injury in rats. Journal of Materials Chemistry B. 2017;5(10):1962–1970. doi: 10.1039/C6TB03033E. [DOI] [PubMed] [Google Scholar]
- 29.Ou Y., Zheng S., Lin L., Jiang Q., Yang X. Protective effect of C-phycocyanin against carbon tetrachloride-induced hepatocyte damage in vitro and in vivo. Chemico-Biological Interactions. 2010;185(2):94–100. doi: 10.1016/j.cbi.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Yarnpakdee S., Benjakul S., Kristinsson H. G., Bakken H. E. Preventive effect of Nile tilapia hydrolysate against oxidative damage of HepG2 cells and DNA mediated by H2O2 and AAPH. Journal of Food Science and Technology. 2015;52(10):6194–6205. doi: 10.1007/s13197-014-1672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozerkan D., Ozsoy N., Yilmaz E. Vitamin D and melatonin protect the cell’s viability and ameliorate the CCl4 induced cytotoxicity in HepG2 and Hep3B hepatoma cell lines. Cytotechnology. 2015;67(6):995–1002. doi: 10.1007/s10616-014-9738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Q., Wang L. H., Jiang J. G. Hepatoprotective effect of flavonoids from Cirsium japonicum DC on hepatotoxicity in comparison with silymarin. Food & Function. 2016;7(5):2179–2184. doi: 10.1039/C6FO00068A. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., Huang H., Lin J., Zhang Q., Tan J., Ren J. Suppressive effects of genomic imprinted gene PEG10 on hydrogen peroxide-induced apoptosis in L02 cells. Journal of Huazhong University of Science and Technology [Medical Sciences] 2009;29(6):705–709. doi: 10.1007/s11596-009-0606-2. [DOI] [PubMed] [Google Scholar]
- 34.Hu C. F., Sun L. P., Yang Q. H., Lu D. X., Luo S. Ginsenosides from stems and leaves of ginseng prevent ethanol-induced lipid accumulation in human L02 hepatocytes. Chinese Journal of Integrative Medicine. 2017;23(6):438–444. doi: 10.1007/s11655-016-2617-8. [DOI] [PubMed] [Google Scholar]
- 35.Kumar K. J. S., Liao J.-W., Xiao J.-H., Vani M. G., Wang S.-Y. Hepatoprotective effect of lucidone against alcohol-induced oxidative stress in human hepatic HepG2 cells through the up-regulation of HO-1/Nrf-2 antioxidant genes. Toxicology in Vitro. 2012;26(5):700–708. doi: 10.1016/j.tiv.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Vandesompele J., De Preter K., Pattyn F., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen C. L., Jensen J. L., Orntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26(6):509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 39.Chen L., Zhong H. Y., Kuang J. F., Li J. G., Lu W. J., Chen J. Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta. 2011;234(2):377–390. doi: 10.1007/s00425-011-1410-3. [DOI] [PubMed] [Google Scholar]
- 40.Jung M., Ramankulov A., Roigas J., et al. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Molecular Biology. 2007;8(1) doi: 10.1186/1471-2199-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun W., Jin Y., He L., Lu W. C., Li M. Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. Journal Of Insect Science. 2010;10(208):1–12. doi: 10.1673/031.010.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chua S. L., Too W. C. S., Khoo B. Y., Few L. L. UBC and YWHAZ as suitable reference genes for accurate normalisation of gene expression using MCF7, HCT116 and HepG2 cell lines. Cytotechnology. 2011;63(6):645–654. doi: 10.1007/s10616-011-9383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cicinnati V. R., Shen Q., Sotiropoulos G. C., Radtke A., Gerken G., Beckebaum S. Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC Cancer. 2008;8(1) doi: 10.1186/1471-2407-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox B. C., Devonshire A. S., Schutte M. E., et al. Validation of reference gene stability for APAP hepatotoxicity studies in different in vitro systems and identification of novel potential toxicity biomarkers. Toxicology in Vitro. 2010;24(7):1962–1970. doi: 10.1016/j.tiv.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Gebeh A. K., Marczylo E. L., Amoako A. A., Willets J. M., Konje J. C. Variation in stability of endogenous reference genes in fallopian tubes and endometrium from healthy and ectopic pregnant women. International Journal of Molecular Sciences. 2012;13(3):2810–2826. doi: 10.3390/ijms13032810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 Figure: expression stability of the housekeeping genes in L02 evaluated by geNorm M values represents the average expression stability. From left to right, the value of M decreased in turn, indicating the stability gradually increased. Smaller M value means higher stability. The control group, ethanol, hydrogen peroxide, acetaminophen, and carbon tetrachloride were abbreviated to WT, EtOH, H2O2, APAP, and CCl4, respectively. S2 Figure: agarose gel (1%) electrophoresis of the ten candidate housekeeping genes. 1-10 represent ACTB, B2M, GAPDH, TUBB2a, HPRT1, SDHA, TBP, YWHAZ, CYC1, and GUSB, respectively. S3 Figure: melt curves of the ten candidate housekeeping genes. S1 Table: expression stability values of ten candidate housekeeping genes in L02 analyzed by NormFinder. S2 Table: expression stability values of the housekeeping genes calculated by BestKeeper in L02.
Data Availability Statement
The data used to support the findings of this study are included within the article.


