Abstract
Fluorescence turn-on aptamers, in vitro evolved RNA molecules that bind conditional fluorophores and activate their fluorescence, have emerged as RNA counterparts of the fluorescent proteins. Turn-on aptamers have been selected to bind diverse fluorophores, and they achieve varying degrees of specificity and affinity. These RNA-fluorophore complexes, many of which exceed the brightness of green fluorescent protein (GFP) and their variants, can be used as tags for visualizing RNA localization and transport in live cells. Structure determination of several fluorescent RNAs revealed that they have diverse, unrelated overall architectures. As most of these RNAs activate the fluorescence of their ligands by restraining their photoexcited states into a planar conformation, their fluorophore binding sites have in common a planar arrangement of several nucleobases, most commonly a G-quartet. Nonetheless, each turn-on aptamer has developed idiosyncratic structural solutions to achieve specificity and efficient fluorescence turn-on. The combined structural diversity of fluorophores and turn-on RNA aptamers has already produced combinations that cover the visual spectrum. Further combined molecular evolution and structure-guided engineering is likely to produce fluorescent tags custom-tailored to specific applications.
Keywords: SELEX, RNA structure, X-ray crystallography, molecular engineering
1. Introduction
In cell biology, seeing is believing. Techniques that probe matter at atomic and near-atomic resolution provide insight into how molecular structure shapes life. Between the atomic and the macroscopic lies the mesoscopic organization of the cell. Light, as well as electron microscopy, provided observational insight into cell function, and fluorescent proteins (FPs) revolutionized cell biology by allowing real-time tracking of tagged proteins in live cells (Day; Rodriguez et al.). However, less than 3% of the human genome is translated into protein while most is transcribed to RNA (Pertea; Hangauer, Vaughn and McManus, 2013). Innovation in cellular RNA imaging technology is needed to uncover the dynamic and spatial properties of the transcriptome. To visualize RNA within cells, a number of fluorescent RNA tags have been developed. As there are no known naturally fluorescent RNAs, these tags are all products of in vitro and in vivo selection experiments as well as molecular engineering. Here, we review the current state of fluorescent RNA technology, highlighting the structural underpinnings for function of fluorogenic RNA aptamers, i.e., RNAs that bind and turn on the fluorescence of conditional small molecule fluorophores.
2. RNA imaging in cells
A popular approach for RNA imaging in live cells employs a fusion between a FP and a high-affinity RNA binding protein (RBP), such as the bacteriophage MS2 (Johansson et al.; Ni et al.) and PP7 (Chao et al.) coat proteins. Co-expression of the fusion protein with an RNA of interest engineered to contain the cognate binding site of the RNA-binding component produces a fluorescent tag (Bertrand et al.). This technique allows single RNA species to be imaged in live cells, but has several drawbacks, not the least of which is that the FP fusion needs considerable time to be translated and matured, requiring it to be present long before the RNA of interest can be examined. Because the FP-RBP fusion is intrinsically fluorescent, and the affinity of MS2 for its cognate RNA hairpin is finite, some fraction of the fusion protein will not be bound to the intended RNA, producing background fluorescence. To overcome this, the binding sites need to be multimerized, yielding a tag, expressed from a potentially recombinogenic locus, that can be several hundred kDa in molecular mass, possibly larger than the RNA of interest. These large fluorogenic tags have been demonstrated to adversely affect the metabolism of some tagged RNA (Garcia and Parker; Garcia and Parker). Several approaches have been taken to improve the signal-to-noise of the approach. Ever brighter FPs have been deployed to increase signal, but this does not reduce background fluorescence. Split green fluorescent protein (GFP) variants engineered to become fluorescent only upon colocalization on the RNA of interest have been reported (Valencia-Burton et al.; Wu, Chen and Singer) Nuclear export or import signals have also been used to reduce the background of unbound fusion protein, but may confound the cellular localization of the RNA of interest. More recently, catalytically inactive CRSPR-Cas9-FP fusions have been used to image RNA localization in live cells (Nelles et al.). This technique suffers from many of the same drawbacks of the GFP-MS2 fusion system, although the CRSPR guide RNA provides flexibility in targeting.
An optimal RNA tag would be small, genetically encodable, covalently attached to the RNA of interest, and exhibit bright fluorescence with tunable spectral properties. In vitro selection has been used to isolate RNA sequences that bind small molecules (Tuerk and Gold; Ellington and Szostak; Robertson and Joyce), and fluorescent RNA tags have been produced by selecting RNA sequences that bind to, and enhance the fluorescence of conditionally fluorescent small molecules (Troung and Ferré-D’Amaré; Trachman, Truong and Ferre-D’Amare, 2017; Ouellet, 2016; You and Jaffrey, 2015; Armitage, 2011; Dolgosheina and Unrau, 2016). Some of these fluorophores are cell-permeable and non-cytotoxic, and are minimally turned on by endogenous cellular components, therefore producing low background fluorescence. Expression of aptamer-tagged RNAs of interest in live cells perfused with the cognate fluorophores has allowed their visualization (e.g., (Autour et al.; Paige et al.; Braselmann et al.; Strack, Disney and Jaffrey). In addition to affinity-based SELEX, a number of other directed-evolution techniques such as fluorescence-based in vitro selection and live cell sorting-based selection, have been employed to generate or optimize fluorescence turn-on RNAs. The fluorophores themselves have seen improvements that alter their aptamer binding affinity and selectivity, as well as the brightness and spectral properties of their aptamer complexes. Numerous combinations of fluorophore-RNA complexes have been reported and are summarized in Table 1. By combining a variety of conditional fluorophores and structurally diverse RNAs, fluorescence turn-on tags are starting to rival the brightness and spectral range of the FPs (Figs 1, 2).
Table 1.
Fluorescence properties of select RNA-fluorophore complexes
| Aptamer | Fluorophore | Kd (nM) | λex (nm) | λem (nm) | ε (M−1 cm−1) | Φ | Brightness | Fl. E. | Length (nt) | PDB | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| eGFP | n.a. | 489 | 508 | 55,000 | 0.60 | 33,000 | n.a. | n.a. | 3EVP | 1,2 | |
| smURFP | biliverdin | 642 | 670 | 180,000 | 0.18 | n.a. | 6FZO | 3,4 | |||
| MGA | MG | 117 | 630 | 650 | 150,000 | 0.19 | 28,500 | 2360 | 38 | 1F1T | 5,6 |
| MGA | IMG | 666 | 642 | 673 | 0.32 | 2090 | 38 | - | 5 | ||
| MG-D3 | MG | 44 | N.D. | N.D. | 150,000 | N.D. | N.D. | 3660 | - | 7 | |
| Spinach | DFHBI | 537 | 469 | 501 | 24,300 | 0.72 | 17,500 | 1030 | 98 | 6B14 | 8,9 |
| Spinach2 | DFHBI | 430 | 454 | 498 | 26,100 | 0.70 | 18,300 | 1000 | 95 | - | 10 |
| Spinach2 | DFHBI-1T | 560 | 482 | 505 | 31,000 | 0.94 | 29,100 | 940 | 95 | - | 11 |
| Baby Spinach | DFHBI | N.D. | 466 | 503 | N.D. | N.D. | 16,625 | ND | 51 | - | 12 |
| iSpinach | DFHBI | 920 | 442 | 503 | 26,100 | 0.98 | 25,578 | 1505 | 69 | 5OB3 | 13,14 |
| Broccoli | DFHBI-1T | 360 | 472 | 507 | 29,600 | 0.94 | 27,800 | 940 | 49 | - | 15 |
| Orange Broccoli | DFHO | 230 | 513 | 562 | 34,000 | 0.28 | 9,520 | 793 | 49 | - | 16 |
| Red Broccoli | DFHO | 206 | 518 | 582 | 35,000 | 0.34 | 11,900 | 992 | 49 | - | 16 |
| Corn | DFHO | 70 | 505 | 545 | 29,000 | 0.25 | 7,250 | 604 | 36 (72) | 5BJO | 16 |
| Chili | DMHBI | 570 | 400 | 537 | 18,000 | 0.08 | 1,440 | 116 | 52 | - | 17 |
| Chili | DMHBI+ | 63 | 413 | 542 | 21,000 | 0.40 | 8,400 | 366 | 52 | - | 17 |
| Chili | DMHBI-Imi | 71 | 463 | 545,594 | 20,000 | 0.08 | 1,600 | 22 | 52 | - | 17 |
| Chili | DMHBO+ | 12 | 456 | 592 | 22,000 | 0.10 | 2,200 | 92 | 52 | - | 17 |
| Mango-I | TO1-Biotin | 3.6 | 510 | 535 | 77,500 | 0.14 | 10,850 | 1100 | 29 | 5V3F | 18,19 |
| Mango-I | TO3-Biotin | 8 | 635 | 660 | 130,000 | N.D. | N.D. | 35 | 29 | - | 18 |
| Mango-II | TO1-Biotin | 1 | 510 | 535 | 77,500 | 0.22 | 17,000 | 1300 | 30 | 6C63 | 20,21 |
| Mango-III | TO1-Biotin | 4 | 510 | 535 | 77,500 | 0.56 | 43,000 | 4360 | 31 | 6E8S | 20,22 |
| Mango-IV | TO1-Biotin | 11 | 510 | 535 | 77,500 | 0.41 | 32,000 | 3240 | 29 | - | 20 |
| Mango-II (A22U) | TO1-Biotin | 0.9 | 510 | 535 | 77,500 | 0.26 | 20,150 | 1543 | 30 | 6C65 | 21 |
| Mango-III (A10U) | TO1-Biotin | 1.7 | 510 | 534 | 77,500 | 0.66 | 51,150 | 5,185 | 31 | 6E8T | 22 |
| iMango-III | TO1-Biotin | 4.6 | 506 | 527 | 77,500 | 0.64 | 49,600 | 5,030 | 32 | 6E8U | 22 |
| DIR2s | DIR-Pro | 252 | 600 | 658 | 164,000 | 0.33 | 54,100 | ~80 | 57 | - | 23 |
| DIR2s | OTB-SO3 | 662 | 380 | 421 | 73,000 | 0.51 | 37,200 | 533 | 57 | 6DB8 | 23,24 |
| SRB-2 | SR-DN | 1400 | 579 | 596 | n.a. | 0.65 | n.a. | 8 | 54 | - | 25 |
| DNB | TMR-DN | 350 | 572 | 591 | 47,150 | 0.90 | 42,435 | 75 | 75 | - | 26 |
| RiboGlow (D) | Cbl-Cy5 | 3 | 646 | 666 | 271,000 | 0.25 | 67,750 | 2.7 | 130 | 4FRN | 27 |
(1)(Heim, Prasher and Tsien) (2)(Royant and Noirclerc-Savoye) (3)(Rodriguez et al., 2016) (4)(Fuenzalida-Werner et al.) (5)(Babendure, Adams and Tsien, 2003) (6)(Baugh, Grate and Wilson, 2000) (7)(Gotrik et al.) (8)(Paige, Wu and Jaffrey) (9)(Koirala et al., 2018) (10)(Strack, Disney and Jaffrey, 2013) (11)(Song et al., 2014) (12)(Warner et al., 2014) (13)(Autour, Westhof and Ryckelynck, 2016) (14)(Fernandez-Millan et al., 2017) (15)(Filonov et al., 2014) (16)(Song et al., 2017) (17)(Steinmetzger et al., 2019) (18)(Dolgosheina et al.) (19)(Trachman et al.) (20)(Autour et al., 2018) (21)(Trachman et al., 2018) (22)(Trachman III Robert J. Autour) (23)(Tan et al.) (24)(Shelke et al.) (25)(Sunbul and Jäschke) (26)(Arora, Sunbul and Jäschke) (27)(Braselmann et al.)
Fig. 1.
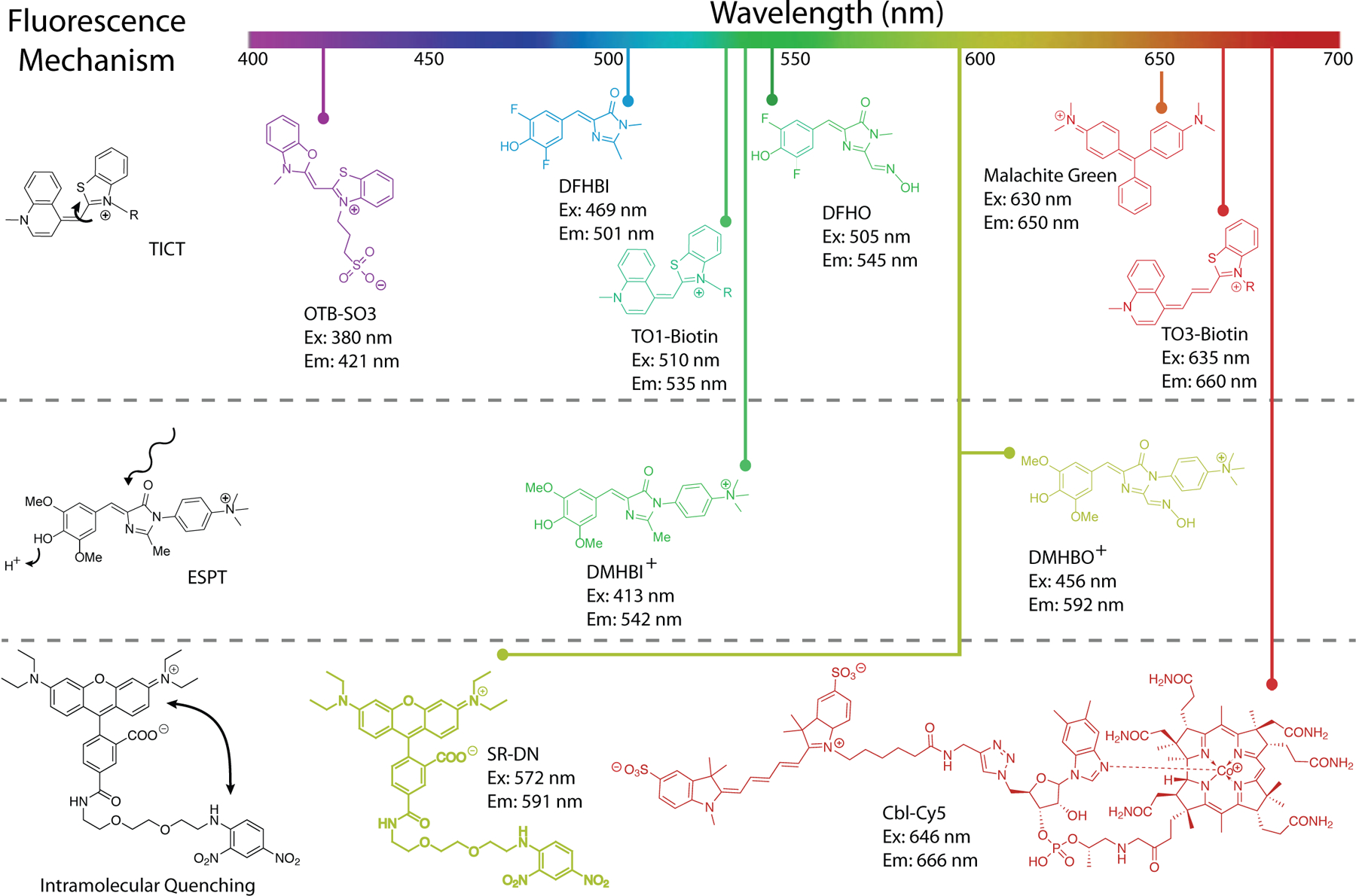
Overview of turn-on aptamer fluorophores and their mechanisms of activation. Reduction of rotation between conjugated heterocycles promoting twisted intramolecular charge transfer (TICT) is the most prevalent (top). Examples of TICT span almost the entire visible spectrum. Promoting excited state proton transfer (ESPT) has the benefit of producing large Stokes shifts (middle). The chromophores DMHBI+ and DMHBO+ are examples of ESPT, but function through a hybrid TICT/ESPT mechanism. Intramolecular quenching (IQ) in a fluorophore-quencher pair is relieved when an aptamer sequesters the quencher. Examples include SR-DN and Cbl-Cy5 (bottom), but numerous fluorophores can be used to expand the color spectrum of IQ aptamers.
Fig. 2.
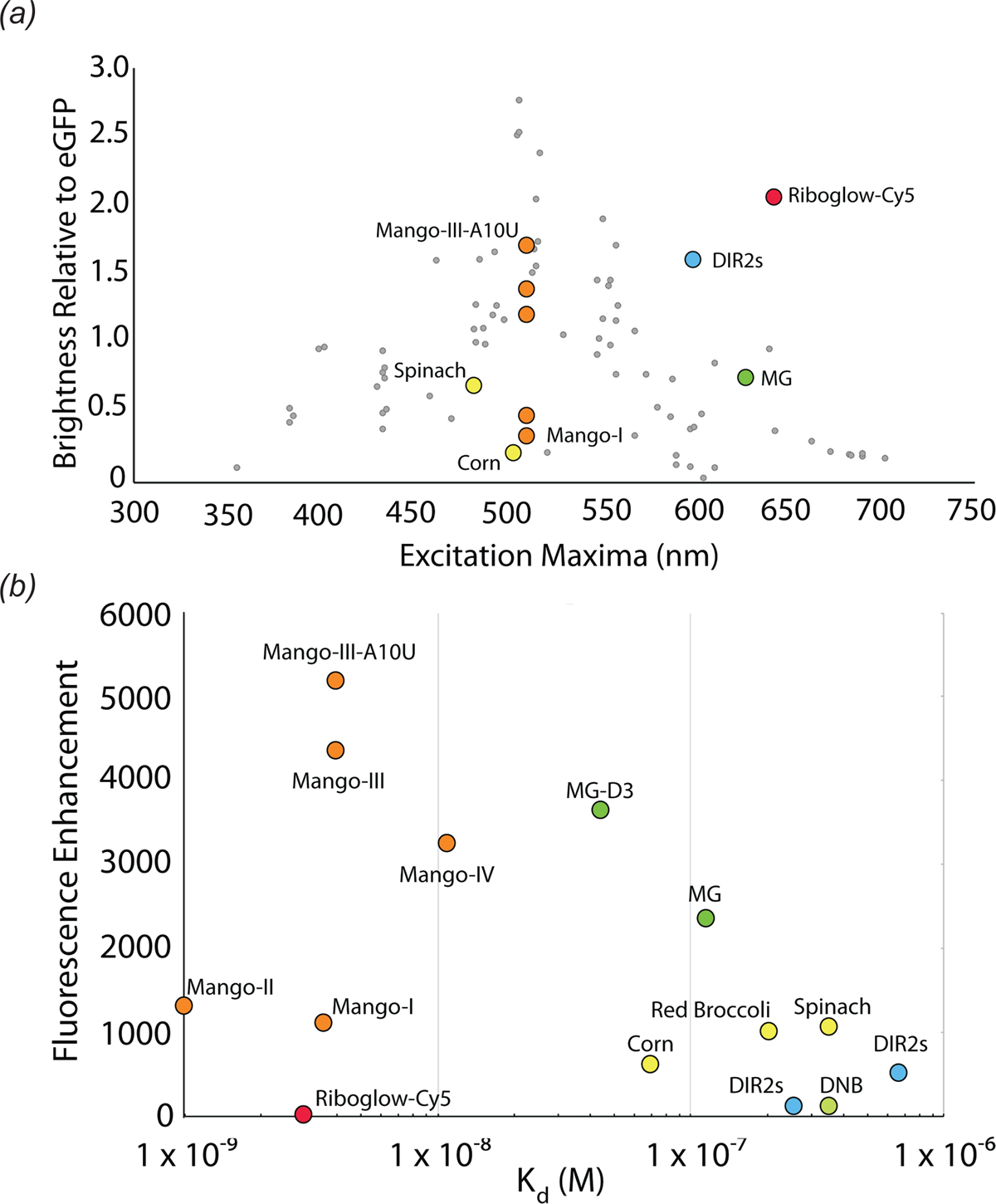
Photophysical comparison of fluorescent aptamers. (a) Excitation maxima and brightness relative to eGFP for select fluorescent aptamers. Aptamer complexes are colored according to sequence or fluorophore lineage (Fig. 1). DIR-2 (light blue), DNB (dark blue), Chili (purple), HBI derivatives (yellow), Riboglow (red), Malachite Green (green), Mango RNAs (orange). Grey points denote fluorescent proteins, adapted from Rodriquez et al. 2017 (b) Scatter plot of aptamer-fluorophore dissociation constants (Kd) and fluorescence enhancements (FE), depicting diverse fluorescence efficiencies (Kd * FE).
3. Mechanisms of fluorescence activation
Selection or engineering of a fluorescence turn-on aptamer starts with choice of a conditional fluorophore (Klymchenko). Fluorophores of interest are small molecules that exhibit minimal fluorescence when free in solution, but become fluorescent upon changing local environment (in particular, binding to a cognate RNA). There are three principal mechanisms of fluorescence turn-on that could be exploited by turn-on aptamers: twisted intramolecular charge transfer (TICT), excited state proton transfer (ESPT), and unquenching of fluorophore-quencher conjugates (Fig. 1). Fluorophores exhibiting TICT have increased vibrational modes in solution and readily undergo non-radiative decay when the conjugated network of the excited fluorophore is disrupted by bond rotation. The majority of fluorescent aptamers rigidify TICT fluorophores to promote fluorescence enhancement (Fig. 1). A relatively recent addition to the fluorescent aptamer tool box, ESPT exploits proton exchange in the excited state by incorporating cationic groups on the fluorophore. These fluorophores mimic the FP fluorophores of LSSmOrange (Shcherbakova et al.) and LSSmKate (Piatkevich et al.; Piatkevich et al.) to produce large Stokes shifts. Fluorophore-quencher conjugates suppress fluorescence of an otherwise constitutively fluorescent molecule through non-covalent interactions with a covalently linked quencher. Aptamers sequester the quencher moiety, allowing fluorescence.
Regardless of fluorescence turn-on mechanism, several properties of small molecule fluorophores are important for imaging. Ideal fluorescent tags for live-cell imaging are bright and redshifted. Brightness is the product of the extinction coefficient (ε) and the quantum yield (Φ). For TICT fluorophores, Φ is a function of rigidity and conformation of the fluorophore in the photoexcited state, and can in principle be improved with stringent aptamer selection. However, ε is almost exclusively dictated by intrinsic properties of the fluorophore. Fluorophores with large ε (i.e., > 50,000) and small Φ (i.e., < 0.001) when free in solution are ideal. Fluorescent imaging in cells is subject to large background signal from autofluorescence of the cytoplasm and nucleus. Background is mainly in the blue-green range of the visible spectrum (Monici), so fluorophores that emit in the near-infrared are optimal. Fluorescence-activating proteins (FAPs) derived from bacterial phytochromes bind in trans to biliverdin and turn on fluorescence (Fig. 3). Like FAPs, fluorescence turn-on aptamers exhibit a finite affinity for their fluorophores. Sufficient binding affinity is required to image low copy RNAs and the fluorescent signal produced by free ligand must be accounted for. The fluorescence efficiency of an RNA has been defined as a function of both binding affinity (Kd) and fluorescence enhancement (generally approximated to Φbound / Φunbound). Fluorescent aptamers with low Kd (< 10 nM) and large fluorescence enhancements (>1,000) are desirable (Fig. 2). Not least, selectivity of the fluorophore-RNA interaction is important. If the fluorophore is turned on nonspecifically by cellular nucleic acids (or other endogenous molecules), high background will ensue.
Fig. 3.
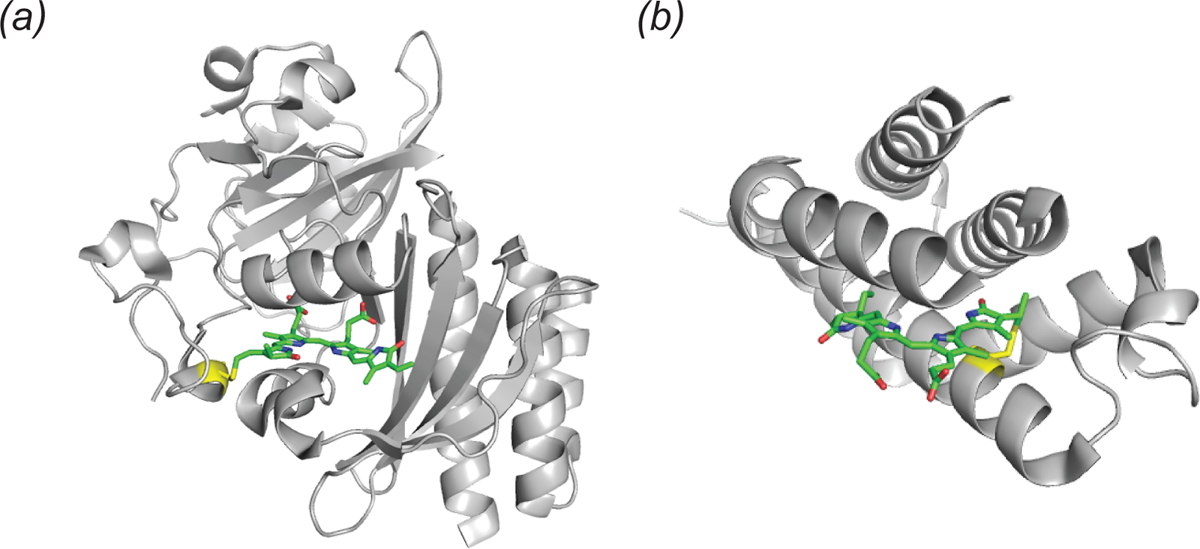
Structure of two fluorescence activating proteins bound to biliverdin. (a) Cartoon representation of the infrared fluorescent protein IFP 2.0 (PDB ID: 4CQH). The biliverdin chromophore (green sticks) is shown with the covalent linkage to cystine 24 (yellow). (b) Cartoon representation of smURFP (PDB ID: 6FZN). The biliverdin chromophore (green sticks) is shown with the covalent linkage to cystine 52 (yellow sticks).
4. The malachite green aptamer
The malachite green (MG) aptamer was the first turn-on aptamer for which a three-dimensional structure was determined. It was originally selected for use in chromophore-assisted laser inactivation (CALI) (Jay and Keshishian; Grate and Wilson). Using microinjected Fab-MG conjugates, CALI sought to employ the specificity of antibodies coupled to the hydroxyradical production by photoexcited MG to specifically knock out proteins of interest. The MG aptamer was engineered to perform the same function on RNA species using a genetically encodable aptamer.
The MG aptamer was found to have higher affinity for the planar analogue tetramethylrosamine (TMR; Kd = 40 nM) despite being selected to bind malachite green (Kd = 117 nM). The crystal structure of the MG aptamer-TMR complex demonstrated that the larger ring system of the fluorophore stacks between a base quadruple and a base pair (Baugh, Grate and Wilson, 2000), suggesting that the RNA would stabilize a planar conformation of MG (Fig. 4). Subsequent solution NMR structures of the aptamer bound to TMR and MG showed that the RNA structure can accommodate either ligand with only minor perturbations, although MG is bound in a slightly twisted conformation (Flinders et al., 2004; Nguyen et al., 2002; Brackett and Dieckmann, 2006).
Fig. 4.
Structure of the malachite green aptamer. (a) Cartoon representation of the malachite green aptamer bound to TMR. The TMR ligand is shown as green ball-and-stick with flanking A-form helices and unstructured nucleotides (grey). Tetrad (yellow) and binding pocket nucleotides (marine and light blue) are colored. (b) Side view of the Spinach binding pocket (ball-and-stick) colored according to (a). (c) Top view of the nalachite green ligand-binding pocket.
MG and other triphenylmethane dyes fluoresce weakly in solution due to vibrational de-excitation. As with other TICT fluorophores, intramolecular rotation results in non-radiative decay in aqueous solution. Fluorescence can be enhanced by increasing the solution viscosity (Baptista and Indig). Based on the crystal structure of the MG aptamer-TMR complex, it was hypothesized that the aptamer may be able to enhance fluorescence of the otherwise dark MG. Remarkably, the aptamer was shown to increase quantum yield of MG by four orders of magnitude, producing a fluorescence enhancement of 2,360 (Table 1) (Babendure, Adams and Tsien). MG and the fluorescent aptamer SRB (Holeman et al.; Grate and Wilson) were the first aptamers demonstrated to turn on the fluorescence of small molecules. Their utility to image RNA in live cells, however, is limited by promiscuous binding of MG, toxicity upon illumination, and the low aptamer-MG affinity (and therefore low fluorescence efficiency (Table 1). A recent reselection sought to improve the fluorescence efficiency of the MG aptamer (Gotrik et al.). By reducing the concentration of fluorophore during selection while increasing stringency of the fluorescence sorting gate, both high affinity and bright aptamers were selected against MG. The best aptamer exhibited a Kd of 44 nM and a fluorescence enhancement of 3,660.
5. Spinach and related aptamers
In early studies seeking to characterize the intrinsic fluorophore of the Aequora victoria GFP, p-HBI was synthesized (Niwa et al., 1996). This soluble form of the GFP fluorophore was found to exhibit minimal fluorescence in solution, also consistent with the loss of fluorescence of the FP upon denaturation (Ward and Bokman, 1982). In vitro selection experiments in which RNAs were selected to bind to various derivatives of p-HBI produced several turn-on aptamer-fluorophore combinations (Paige et al, 2011). The brightest of these, termed Spinach, in complex with DFHBI, exhibited fluorescence ~50% as bright as that of enhanced GFP (EGFP), and was shown to be useful to visualize tagged RNAs in live cells. Further optimization of the aptamer to improve affinity and fluorescence enhancement yielded Spinach2, which in complex with a variant fluorophore, DFHBI-1T, achieves fluorescence 88% as bright as that of EGFP (Strack, Disney and Jaffrey).
Crystallographic structure determination of the 98 nucleotide (nt) Spinach in complex with DFHBI revealed the fluorophore to be constrained in a planar conformation (consistent with its high Φ of 0.74) by sandwiching between a G-quartet and a base triple, while also making lateral interactions with a ribose 2’-OH, an unpaired guanine base, and well-ordered solvent (Huang et al.; Warner et al.). Overall, the aptamer folds as a single coaxial stack of two antiparallel A-form duplexes flanking a core G-quadruplex of mixed parallel and antiparallel connectivity (Fig. 5). The RNA chain needs to traverse the continuous coaxial stack and change polarity as well as strand number while maintaining base stacking; the resulting connectivity of the Spinach fold is of unprecedented complexity for a G-quadruplex nucleic acid. Mutational as well as solution NMR analyses confirmed the importance of the G-quadruplex structure for fluorescence turn-on. Small-angle X-ray scattering analyses and a fluorophore-free crystal structure indicate that Spinach is pre-folded, and an unpaired nucleotide occupies the DFHBI binding pocket in the unliganded state. The outlying A-form duplex elements of Spinach could be truncated to produce the 51-nt Baby Spinach, which retained the G-quadruplex-base triple core and exhibited the full brightness of the parent aptamer.
Fig. 5.
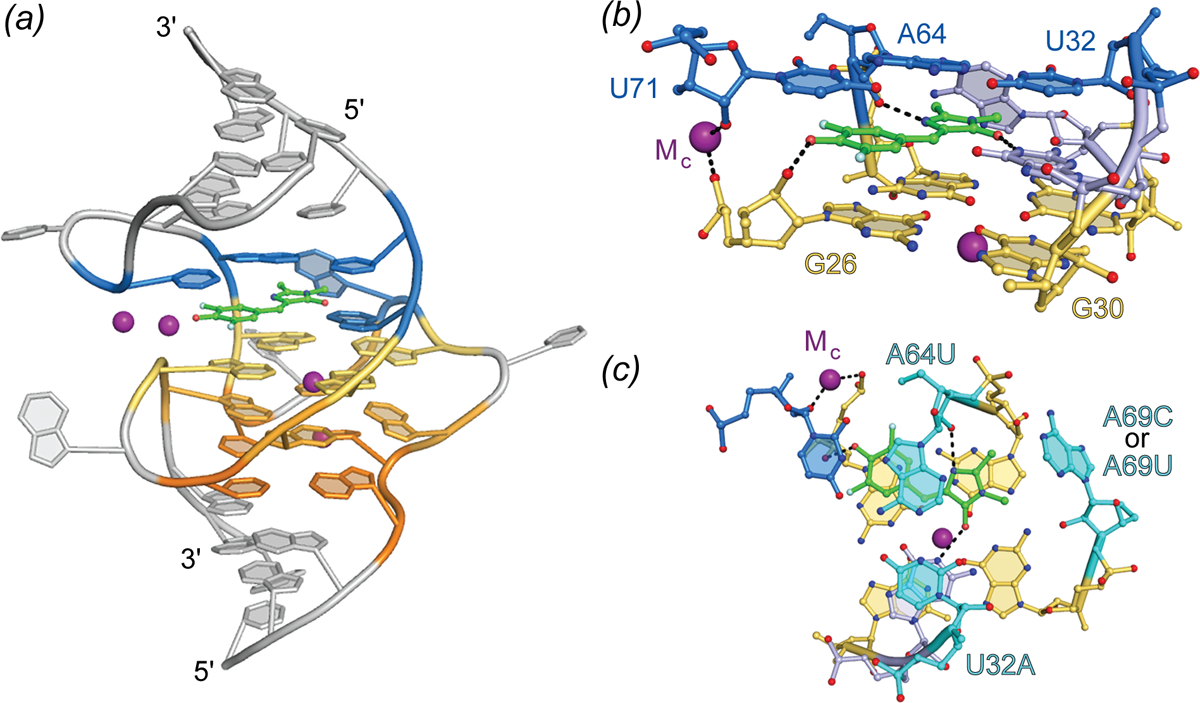
Structure of Spinach aptamer. (a) Cartoon representation of the Spinach aptamer core (PBD ID: 4TSO). The DFHBI fluorescent ligand is shown as green ball-and-stick with flanking A-form helices and unstructured nucleotides (grey), G-quadruplex nucleotides (warm colors), triplex and flanking binding pocket nucleotides (blue), shown in cartoon representation. Bound K+ ions represented as purple spheres. (b) Side view of the Spinach binding pocket (ball-and-stick) colored according to (a). Hydrogen bonds and metal coordination are represented as black dashed lines. (c) Top view of the Spinach ligand-binding pocket. Residues demonstrated to alter spectral properties in HBI-derivative binding aptamers are colored cyan and labeled with mutation details.
A variety of approaches have been employed to tune and improve properties of Spinach. Mutational redesign of the base triple stacking on DFHBI resulted in hypsochromic shifts in excitation and emission maxima of 45 nm and 11 nm, respectively (Fig. 5) (Warner et al.). Directed evolution by fluorescence-activated cell sorting (FACS) produced the 49-nt aptamer Broccoli with improved brightness (Filonov et al.). Further selection to bind and activate the extended-conjugation fluorophore DFHO yielded the red shifted aptamers Orange Broccoli and Red Broccoli, that differ from each other in the identity of a single nucleotide, while preserving the overall Spinach sequence. A microfluidic, droplet-based screening method was employed to reduce the folding dependence of the aptamer on K+ and select for brighter DFHBI binding aptamers. This selection produced iSpinach, a 69-nt aptamer that binds and enhances fluorescence of DFHBI to levels almost 50% brighter than Spinach2 (Autour, Westhof and Ryckelynck). This selection was carried out in high concentrations of NaCl resulting in an aptamer with vastly improved activity in Na+. The crystal structure of iSpinach revealed overall and binding pocket structures almost identical to those of Spinach; both RNA and water interactions with DFHBI are conserved between the two aptamers (Fernandez-Millan et al.)
6. Quasisymmetry in the Corn aptamer
While Spinach and its derivatives demonstrated that a turn-on aptamer could achieve brightness comparable to that of many FPs, and could be used as a fluorescent tag for imaging RNAs in vivo, these aptamers have properties that limit their imaging applications. In addition to poor folding efficiency both in vitro and in vivo (possibly a result of their relatively large size and complex fold) and modest fluorophore binding affinity (Kd ~ 0.5 μM), fluorophore complexes of Spinach and related aptamers (including the various Broccoli RNAs and iSpinach) were found to lose fluorescence rapidly upon illumination (Paige, Wu and Jaffrey; Wang et al.; Han et al.) Interestingly, Spinach was found to regain fluorescence in the dark in the presence of excess DFHBI, suggesting that the photodamaged or photoisomerized fluorophore is bound only loosely and can readily exchange with fresh fluorophore. Together with the crystal structures, which showed only two direct hydrogen bonds between the RNA and DFHBI, this suggested that a fluorophore with additional polar groups and an RNA pocket that bound these may improve photostability. To test this hypothesis, DFHO, a variant fluorophore that extends the conjugation of DFHBI through addition of an oxime moiety was synthesized. It was reasoned that this additional moiety would not only allow for more RNA-fluorophore interactions, but also red-shift the fluorescence.
Selection from a randomized library for an RNA to bind and enhance fluorescence of DFHO produced a yellow fluorescence turn-on aptamer, Corn (Song et al.). The small size of Corn (36 nt) was exploited to successfully image activity of RNA polymerase III transcriptional units in live cells. Crystallographic structure determination revealed that Corn exists as a homodimer that binds one molecule of DFHO at its interprotomer interface, and biochemical analyses confirmed this 2:1 RNA:DFHO stoichiometry. DFHO binds in a planar conformation, with a plane of mirror symmetry. Since Corn RNA is chiral, it cannot match the symmetry of the bound fluorophore and breaks symmetry locally at the binding site. Overall, however, the dimer is symmetric, the two protomers being identical away from the interface. The dimerization interface of Corn is unique among known dimeric RNAs not only in being quasisymmetric (Jones and Ferré-D’Amaré) but because it lacks any base-pairing interactions between protomers. Each Corn RNA comprises an A-form duplex that stacks on a 4-tiered quadruplex, with an antiparallel connectivity much simpler than that of Spinach. Two of the tiers are canonical G-quartets, and one G-quartet from each protomer sandwiches the bound fluorophore. In addition, three unpaired adenosines from each RNA surround the bound fluorophore, and because of the quasisymmetry, adenines from the two protomers adopt different conformations. Consistent with the greatly improved photostability of Corn over Spinach (by over a factor of 1000), one of the interfacial adenines makes a bidentate interaction through its Watson-Crick face with the oxime of DFHO.
The quasisymmetry of the DFHO-bound Corn homodimer hinted that it might be possible to engineer the aptamer into variants that would function as obligate heterodimers. Such variants may be employed as RNA analogs of split GFP (and other split protein reporter systems), to monitor co-expression, co-localization and dimerization of cellular RNAs. Mutagenesis of the interfacial adenines of Corn confirmed the importance of these residues for fluorescence turn-on, as most mutants were inactive, and even the most active was only a quarter as bright as the parental aptamer. However, when point mutants of Corn were combined pairwise, three combinations produced fluorescence approaching 50% of the wild-type, suggesting this may be a promising approach to generate ‘split Corn’.
To gain further insight into how the quasisymmetric Corn interface arises, its crystal structures in the absence of fluorophore, and also in the presence of the non-specific but G-quadruplex-preferent fluorophores thioflavin-T (ThT) and thiazole orange (TO) were determined (Sjekloca & Ferré-D’Amaré, 2019). The ligand-free Corn crystallized as an exactly symmetric homodimer, but at least in the crystalline form of the RNA, the ligand binding pocket had collapsed, such that the G-quadruplexes of the two protomers stacked directly on each other. Numerous studies have shown that ThT and TO bind non-specifically to a variety of nucleic acids and undergo variable levels of fluorescence turn-on (Guan et al.; Turaev et al.; Nygren, Svanvik and Kubista; Jarikote et al.). More recently, it was found that G-quadruplex containing DNAs and RNAs strongly activate ThT fluorescence (Xu et al.; Mohanty et al.; Renaud et al.). This is the case for Corn, which achieves Φ of 0.41 and 0.53 for ThT and TO, respectively, considerably higher than for its cognate fluorophore DFHO (Φ = 0.25). The Corn co-crystal structures with these non-specific fluorophores revealed a third type of Corn homodimer, in this case strictly symmetric but with a binding pocket at the interprotomer interface. ThT and TO bind to Corn with 2:2 and 2:1 (RNA:fluorophore, respectively) stoichiometry, but symmetrically (or in two equivalent conformations in the case of TO). Neither fluorophore makes interactions with nucleobases or riboses; both ThT and TO appear to bind exclusively by stacking between the G-quartets of the two Corn protomers. Overall, these results suggest that Corn, whose dimerization Kd is less than 1 nM, exists in solution as a mixture of open and closed symmetric forms, and that fluorophore binding either maintains symmetry (for ligands such as ThT and TO that do not engage in hydrogen bonding with RNA functional groups) or breaks symmetry, for ligands such as DFHO that make specific interactions with interfacial nucleotides.
7. Excited-state proton transfer in Chili
DFHBI and DFHO, the Spinach and Corn fluorophores, are thought to function by canonical TICT; the turn-on aptamers stabilize the planar conformation of their bound fluorophores, thereby dramatically increasing Φ. One of the structurally unrelated fluorescence turn-on aptamers isolated in the Spinach selection, termed 13–2min, in conjunction with the variant fluorophore DMHBI, exhibited an unusually large Stokes shift of 137 nm (Paige et al 2011), which suggests that it may also incorporate ESPT in its fluorescence turn-on mechanism, as this underlies the large Stokes shifts of FPs such as LSSmOrange (Shcherbakova et al.) and LSSmKate (Piatkevich et al.) The sequence of the ~60-nt 13–2min aptamer suggests a structure different from that of Spinach, and 1D solution NMR indicates that while this aptamer also incorporates a G-quadruplex at or near its fluorophore binding site, it is different from the Spinach G-quadruplex (Warner et al.). No atomistic structure of 13–2min has been reported. In an effort to improve properties of this fluorescent tag system, several fluorophore variants of DFHBI containing the cationic moiety N-trimethylaniline were synthesized (Steinmetzger et al, 2019). It was found that in conjunction with the same aptamer, rechristened Chili, a variety of colors could be obtained. For one of the new fluorophores, DMHBO+, the large Stokes shift (136 nm) was maintained, and the affinity for the RNA improved nearly 50-fold to a Kd of 12 nM. Its large Stokes shift, and its emission maximum at 592 nM, makes the Chili-DMHBO+ complex attractive for studies in which Förster resonant energy transfer (FRET), for instance to the rhodamine dye Atto 590, is sought, even though its modest Φ of 0.1 limits applications where sensitivity is important. Another trimethylanilinium-containing fluorophore, DMHBI+, in complex with Chili, produced a Stokes shift of 127 nM, and while the affinity of the complex was weaker (Kd = 63 nM), Φ was 0.4. Thus, further improvement of the Chili aptamer and its complexes with cationic p-HBI derivatives hold promise.
8. The Mango aptamers
8.1. Mango-I fluorescence is limited by a non-planar fluorophore
Although TO interacts nonspecifically with DNAs and RNAs yielding variable fluorescence turn-on, addition of a substituent to its benzothiazole heterocycle greatly reduces this intercalation-mediated binding. The TO derivative TO1-Biotin (in which a PEG linker connects the fluorophore to a biotin moiety useful for immobilization of the fluorophore) was employed to search for high-affinity turn-on aptamers of moderate size. An aptamer called Mango-I (originally ‘RNA Mango’), with a 29-nt conserved core sequence resulted from the in vitro selection experiment performed so as to increase the on-rate of the aptamer while reducing the off-rate, and exhibited high affinity for TO1-Biotin (Kd = 3.6 nM). Despite its modest Φ (0.14), the substantial fluorescence enhancement achieved by the aptamer (~1100 fold) and the high ε of the TO moiety allowed imaging of Mango-I-TO1-Biotin in microinjected live Caenorhabditis elegans.
The 1.7 Å-resolution co-crystal structure of the Mango-I-TO1-Biotin complex (Trachman et al.) revealed an RNA of relatively simple structure in which a 3-tiered G-quadruplex is linked flexibly to an A-form helix through an interrupted GNRA tetraloop-like junction (Fig. 7A). The TO moiety of the fluorophore binds to one of the exposed planar faces of the G-quadruplex and is held in place by the nucleobases of two unpaired adenines from propeller loops of the mostly parallel G-quadruplex. Unexpectedly, the PEG linker and biotin of the fluorophore also pack against the G-quadruplex. When bound to Mango-I, TO1-Biotin adopts a circular conformation in which the biotin and methylquinoline heterocycles are closely juxtaposed. This conformation of the fluorophore appears to maximize the solvent-inaccessible interface between the small molecule and the RNA, probably contributing to the high affinity of the complex. The crystal structure shows that aptamer binding imposes a 45° angle between the heterocycles of TO (Fig. 7B). This conformation, suboptimal for TICT, may be responsible for the modest quantum yield of the complex. To improve fluorophore planarity, the biotin of TO1-Biotin was replaced with desthiobiotin. Presumably by relief of steric crowding, this resulted in 50% increase in fluorescence enhancement.
Fig. 7.
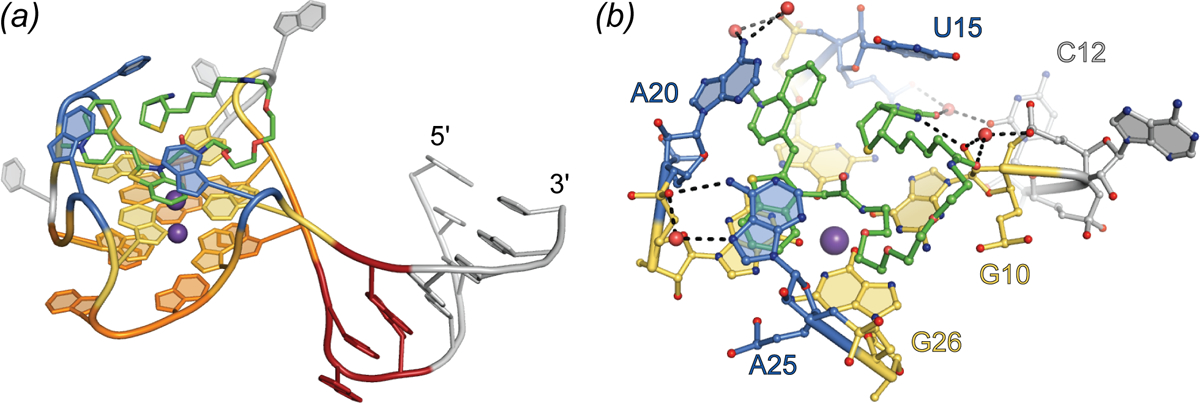
Structure of the Mango-I aptamer. (a) Cartoon representation of the Mango-I aptamer (PDB ID: 5V3F) bound to TO1-Biotin (green sticks). Variable helical element and disordered nucleotides shown in grey. GAAÂ tetraloop-like junction shown in red. Binding pocket nucleotides are shown in warm, orange-yellow colors for the quadruplex and blue for nucleotide flaps. (b) Side view of the Mango-I binding pocket. Nucleotides and TO1-Biotin in ball-and-stick representation
8.2. Mango-II and Mango-IV have improved brightness
Following the success in isolating the compact, high-affinity turn-on aptamer Mango-I, it was hypothesized that brighter, similarly high affinity, compact aptamers may exist in the sequence pool from which that RNA was selected. To search for such molecules, the final pool from the Mango-I selection was subjected to a microfluidics-based, in vitro compartmentalization selection (Autour et al.) RNA transcripts were expressed in aqueous droplets immersed in oil and incubated with TO1-Biotin. Droplets were sorted based on brightness relative to Mango-I. Sequential rounds of fluorescence sorting were performed with increasing concentration of small molecule competitors to select for high-affinity aptamers. This functional re-selection yielded three new aptamer sequences of 24 nt to 29 nt core length, named Mango-II, Mango-III, and Mango-IV. All three new aptamers were brighter than Mango-I, with Mango-III being the brightest (Table 1). Dissociation constants for TO1-Biotin ranged from 1 nM to 11 nM (Table 1). Both Mango-II and Mango-IV have high sequence similarity to Mango-I; Mango-III is more divergent. The new RNAs were tested for fluorescence activity in live and fixed mammalian cells by grafting them into 5S, U6, and box C/D RNAs. Subcellular localization of the tagged RNAs was consistent with localization of the native RNAs, demonstrating the usefulness of these aptamers for imaging both nuclear and cytoplasmic RNAs. Despite being the second brightest and lowest affinity TO1-Biotin aptamer in vitro, Mango-IV was the brightest of the fluorescent aptamers in live and formaldehyde-fixed cells.
Crystallographic structure determination of Mango-II revealed functionally significant differences between it and Mango-I, despite their overall similar folds (the structure of Mango-IV has not been reported). As in Mango-I, the fluorophore binds on the top face of the three-tiered G-quadruplex of Mango-II, but its thiazole orange headgroup adopts a planar conformation, and is held in place by adenine flaps in a considerably more open binding pocket. Neither the biotin nor most of the PEG linker bind to Mango-II (Fig. 8). The crystallographic asymmetric unit (A.S.U.) comprised three independent aptamers with overall similar structure. In all three molecules, the TO1 headgroup was coplanar with the G-quadruplex, but in one, it was rotated by 90° around the G-quadruplex 4-fold axis, suggesting limited stereoselectivity of the fluorophore-binding pocket (Fig. 8). TO3-Biotin, a derivative of TO1-Biotin with extended conjugation from which the aptamer elicits red-shifted fluorescence and for which it has comparable affinity, was also co-crystalized with Mango-II. In this structure, the three RNAs in the A.S.U are near-identical, but only two are bound to TO3-Biotin (Fig. 8B). The conformation of the unpaired nucleobase of A22 (in one of the propeller loops) differs between the Mango-II complexes with the two fluorophores. In the TO1-Biotin complexes, A22 extends upward and away from the fluorophore while in the TO3-Biotin complex, the purine base packs against the dimethyne linker of the headgroup (Fig. 8B). Site-directed mutagenesis produced the more selective A22U mutant, which exhibited 18% improved fluorescence enhancement of TO1-Biotin and reduced affinity and fluorescence enhancement of TO3-Biotin. The crystal structure of Mango-II(A22U)-TO1-Biotin showed all three complexes in the A.S.U. adopting similar conformations (Fig. 8C), suggesting improved stereoselectivity. Biophysical experiments with analogs of TO1-Biotin imply that residue 22 modulates selectivity of Mango-II by interacting with the PEG linker of the fluorophore.
Fig. 8.
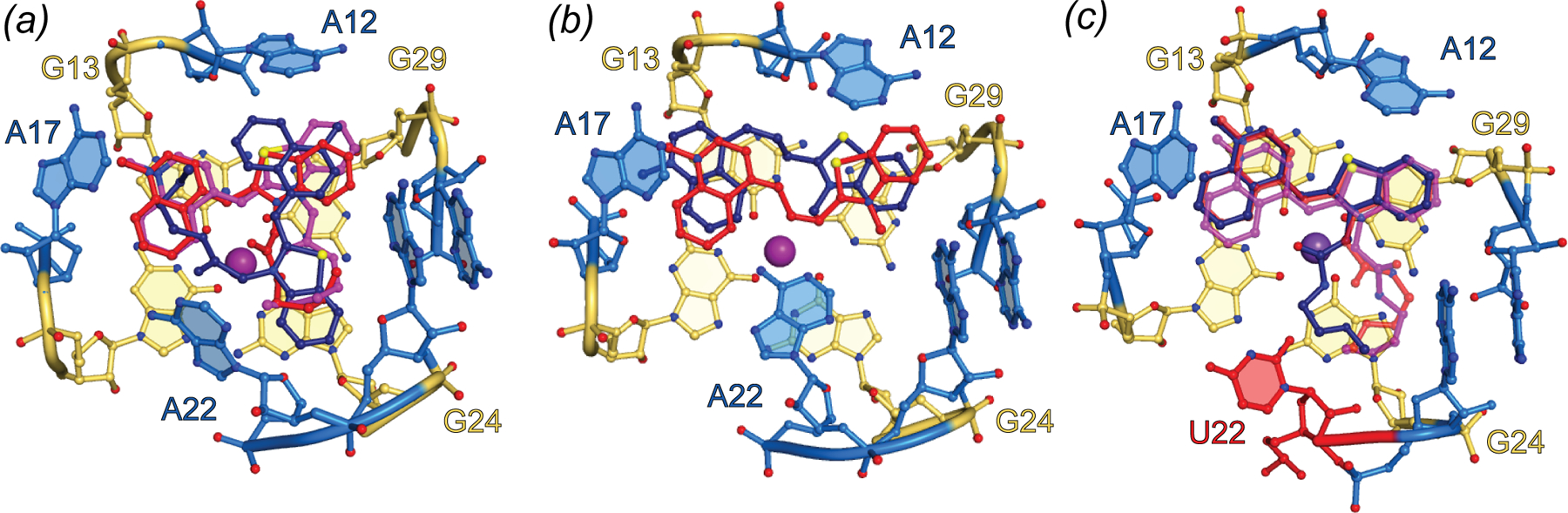
Binding Pocket of Mango-II. (a) Ball-and-stick representation of the Mango-II-TO1-Biotin binding pocket. TO1-Biotin structures from three protomers in the asymmetric unit (red, purple and dark blue) are superimposed. (b) Ball-and-stick representation of the Mango-II-TO3-Biotin binding pocket. TO3-Biotin structures from two protomers in the asymmetric unit (red, purple and dark blue) are superimposed. (c) Ball-and-stick representation of the structure guided Mango-II(A22U) mutant bound to TO1-Biotin. TO1-Biotin colored according to (a) with mutated residue, 22, colored red.
8.3. Mango-III is an aptamer of unusual structural complexity
Crystallographic structure determination of the brightest and most sequence-divergent Mango aptamer in complex with TO1-Biotin revealed an unusually complex 3D-structure for a small RNA. Mango-III folds around a 2-tiered G-quadruplex that stacks co-axially on a partially water-mediated base triple, which in turn stacks on an A-form duplex. Completely unexpected, one of the propeller loops of the G-quadruplex base pairs with a segment 3’ to the quadruplex to form a second duplex consisting of three non-canonical pairs. For two of these, the backbone runs in the parallel direction, and all three are trans pairs (canonically, nucleic acid duplexes are comprised of cis, anti-parallel base pairs). This helix is unprecedented not only because of its local structure; globally, it introduces a connectivity to Mango-III that is analogous to that of a canonical (H-type) pseudoknot (Pleij), that would be formed by base pairing of a loop at the apex of a conventional stem-loop with an unpaired segment distal to it. In the case of Mango-III, the G-quadruplex takes the place of the stem of the conventional stem-loop. Finally, two nucleotides, one each from two propeller loops diagonally across the G-quadruplex, form a trans A10•U17 Watson-Crick base pair that closes over the G-quadruplex. The overall complexity of Mango-III compared to Mango-I becomes apparent when the number of nucleotides that participate in hydrogen bonding is considered. Of the core 28 nucleobases of Mango-III, 26 participate in hydrogen bonding, while for the 22-nucleitide core of Mango-I only 16 do (92% vs. 72%).
The co-crystal structure shows that the headgroup of TO1-Biotin binds to Mango-III in a planar conformation (consistent with its quantum yield of 0.55), sandwiched between the G-quadruplex and the trans Watson-Crick pair of the aptamer (Fig. 9). During in vitro selection, it was noted that a transition mutation of nucleotide 12 (crystal structure numbering) from cytosine to uridine improved fluorescence of the Mango-III pool by ~ 40% (Autour et al.) The crystal structure shows the nucleobase of U12 in an unusual conformation, projecting into the binding pocket perpendicular to the planes of the G-quadruplex and trans Watson-Crick base pair, packing against the PEG linker of TO1-Biotin. Thus, it appears that while the G-quadruplex and trans Watson-Crick base pair prevent rotation of the photoexcited fluorophore heterocycles, U12 packs against the PEG linker of TO1-Biotin prevent sliding of the TO1-Biotin (Fig. 9C), functioning in a manner analogous to that of residue 22 of Mango-II.
Fig. 9.
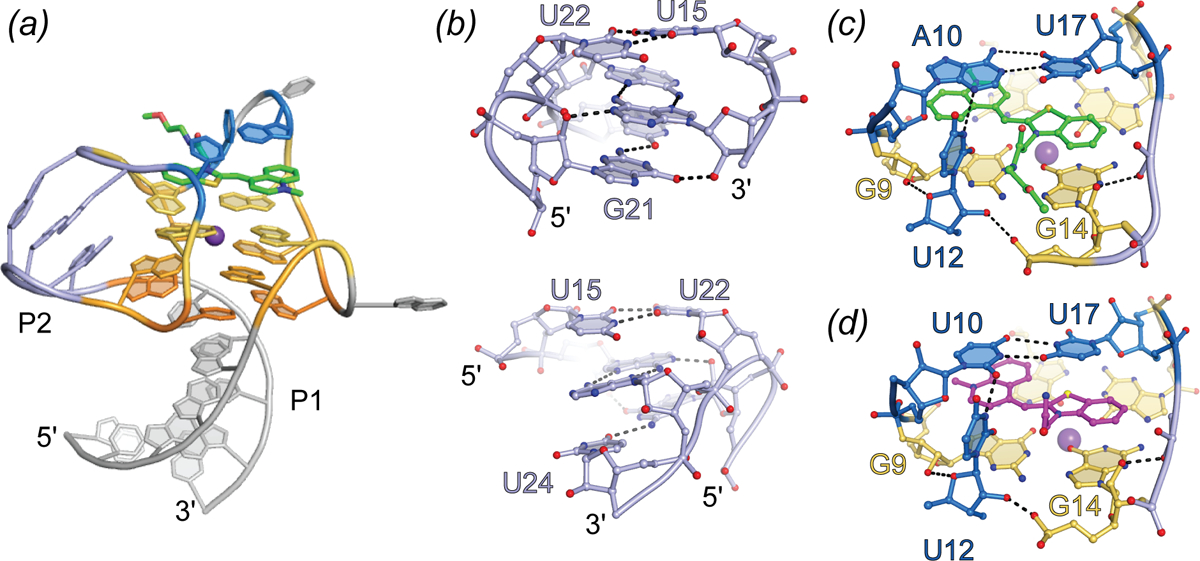
Structure of the Mango-III aptamer. (a) Cartoon representation of the Mango-III aptamer in complex with TO1-Biotin (green sticks), highlighting the triplex and quadruplex nucleotides (warm orange-yellow colors), P2 helix (light blue), and binding pocket nucleotides (marine). (b) Front (top) and back (bottom) view of the P2 helical element. (c) Ligand binding pocket of the Mango-III-TO1-Biotin complex. Hydrogen bonds are represented with black dashes. (d) Ligand binding pocket of the Mango-III (A10U) mutant in complex with (Z)-TO1-Biotin.
8.4. Structure-guided engineering and reselection of Mango-III
Electronic coupling between RNA and fluorophore would be expected to impact fluorescence properties. Mutagenesis of the trans Watson-Crick pair to all sixteen possible variants revealed that a U10•U17 pair improved fluorescence enhancement by 16% (Trachman et al. 2019). The co-crystal structure of the Mango-III(A10U)-TO1-Biotin complex contained four complexes in the A.S.U. The aptamers are highly similar among themselves, and also to the parental RNA. The mutated residue A10U forms a trans Watson-Crick U•U base pair, and the fluorophore binds in the same manner as in Mango-III. Notably, while in two of the complexes in the A.S.U. the TO1-Biotin is bound in the (E) conformation (as in all previously determined Mango aptamer structures), in two of the Mango-III(A10U) complexes the fluorophore is in the (Z) conformation (Fig. 9D). Thus, it appears that while interaction of the TO1-Biotin with the homopyrimidine base pair improves brightness, its reduced steric confinement leads to less stereoselectivity.
To search for additional variants of Mango-III with improved properties, a pool of variants was prepared by mutagenizing residues surrounding the fluorophore as well as those in the co-axial junction between the duplex and the quadruplex. Functional, microfluidic-based re-selection of this pool yielded four sequence variants. The most abundant improved variant (58% of the final pool), termed iMango-III, exhibited 13% greater fluorescence enhancement of TO1-Biotin than Mango-III. The 1.5 Å-resolution co-crystal structure of the iMango-III-TO1-Biotin complex revealed several differences from the parental Mango-III (Fig. 10). First, the trans Watson-Crick pair is formed by two uridines, confirming this is the optimal solution. Second, the sequence of the “pseudoknot” duplex has changed, and this duplex is shortened by one residue. Third, an insertion has allowed rearrangement of the base triple at the co-axial junction, which is now a canonical (non-water-mediated) base triple. Examination of the photophysical properties of this new aptamer indicated that its excitation and emission wavelengths are blue-shifted by 4–7 nm relative to Mango-III and Mango-III(A10U) (the latter two have exactly overlapping spectra). Moreover, the fluorescence lifetime of the iMango-III complex was also consistently shorter than those of the other two aptamers. Reversion of the trans Watson-Crick pair of iMango-III [i.e., iMango-III (U10A)] did not lengthen the fluorescence lifetime or eliminate the hypsochromic shift. Thus, it appears that iMango-III residues that are not in direct contact with the fluorophore (either at the coaxial junction or in the non-canonical “pseudoknot” helix) are responsible for these changed photophysical properties of the improved aptamer resulting from this structure-guided re-selection experiment.
Fig. 10.
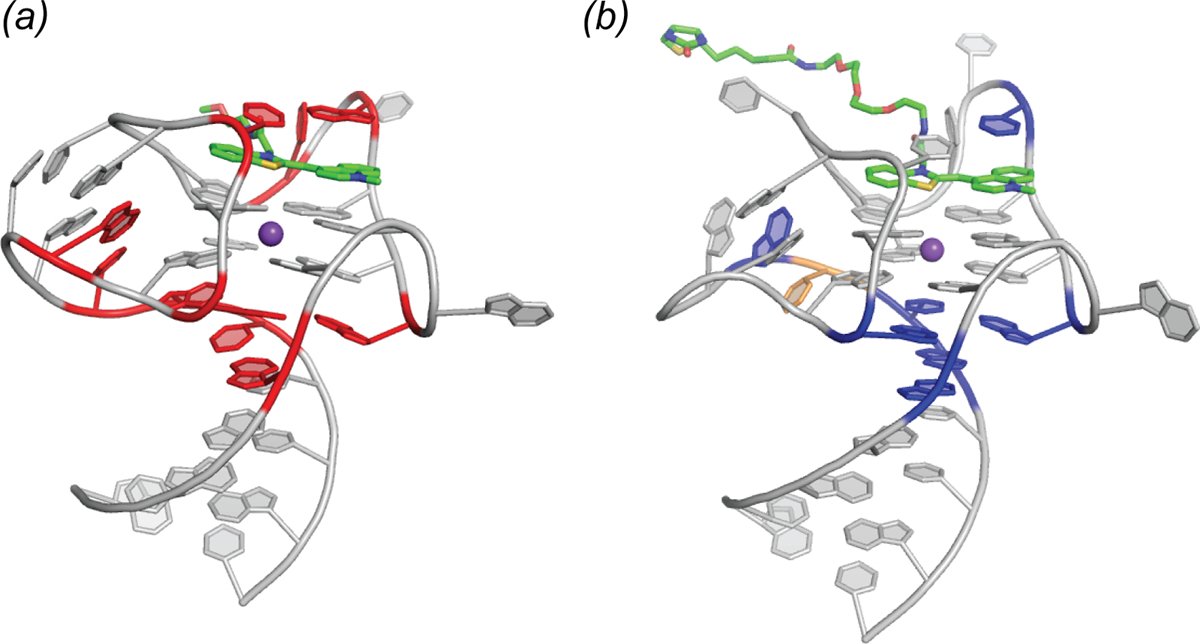
Structure guided engineering of the Mango-III aptamer. (a) Structure of the Mango-III aptamer with 10 randomized nucleotide positions (red). Two rounds of SELEX followed by four rounds of microfluidic fluorescence sorting were carried out. (b) Locations of nucleotides that varied from the original sequence (blue) mapped onto the iMango-III structure. Yellow nucleotide represents the place at which an insertion was introduced and selected.
9. The dimethyl indole red aptamers
Cyanine dyes, such as thiazole orange (TO), generally have large extinction coefficients, making them attractive conditional fluorophores for turn-on aptamer development. However, the hydrophobic and cationic characteristics inherent to these fluorophores promotes non-specific binding to nucleic acids and membranes. The asymmetrical cyanine dye dimethyl indole red (DIR) was designed and synthesized in order to suppress nonspecific binding to nucleic acids (Fig. 1) (Constantin et al.) The dimethyl substituent was intended to have reduced the ability the indole dye to intercalate into nucleic acids while the anionic propylsulfonate moiety yields a zwitterion with lessened non-specific electrostatic binding to nucleic acids. A propyne conjugated linker separates the two conjugated heterocylces to promote red emission and a large extinction coefficient (ε = 134,000 M−1 cm −1). Aptamers to DIR were generated through SELEX (Constantin et al.). Three aptamers were identified that enhance fluorescence of DIR by greater than 20-fold. As this modest fluorescence enhancement is not sufficient for in-cell imaging a second selection was performed against DIR using a starting pool enriched with a stem-loop structure (Tan et al.). This selection yielded an RNA, DIR2s, that promiscuously binds cyanine dyes with submicromolar dissociation constants. In complex with the conditional fluorophore DIR-Pro (ε = 164,000 M−1 cm−1; Kd = 252 nM), DIR2s fluoresced at 658 nm. To test function in live cells, DIR2s was fused to an EGFR aptamer (Li et al.) and internalization of the EGFR-DIR2s-DIR chimeric complex was monitored with microscopy with a red channel filter (Tan et al.).
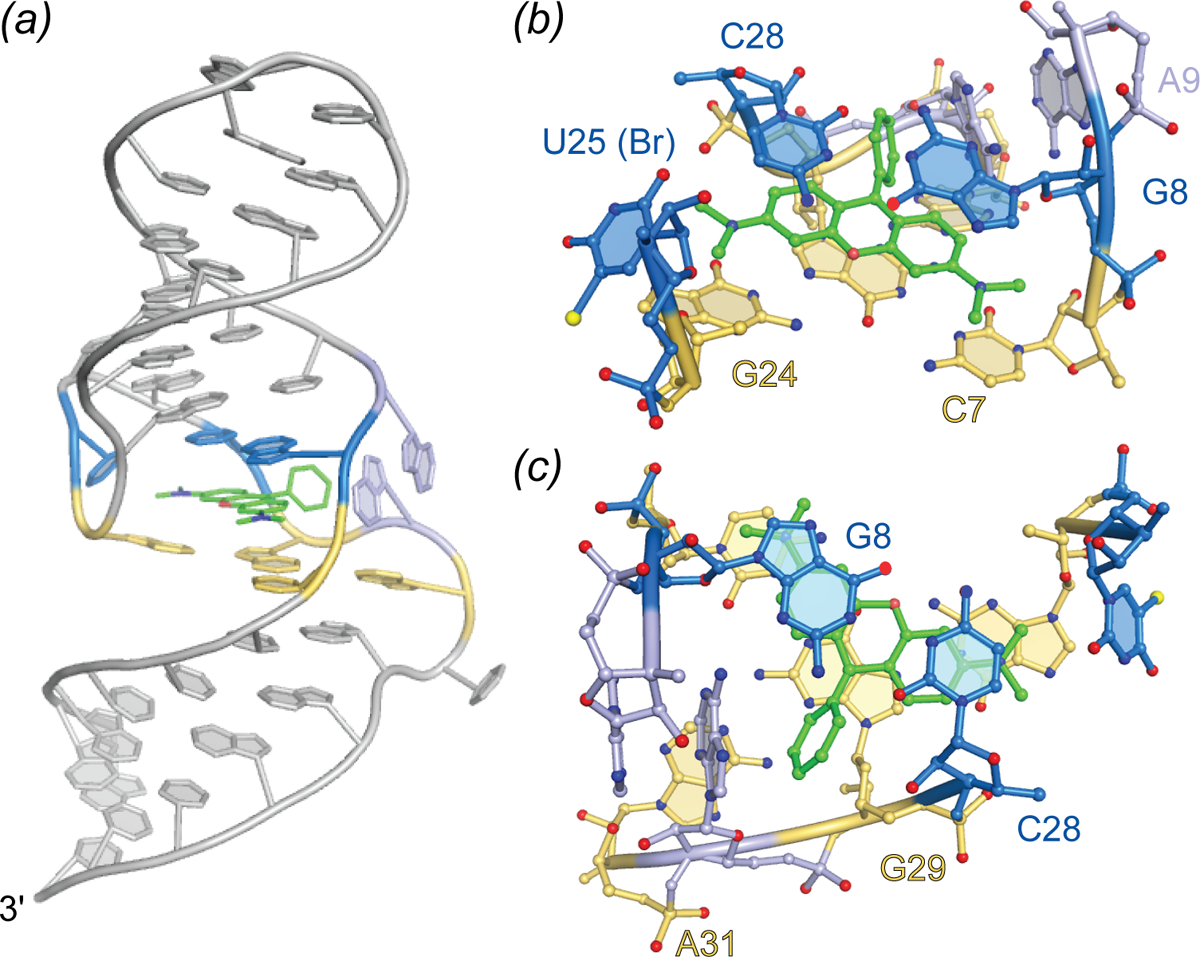
Crystal structures of the DIR2s aptamer, unliganded and bound to the oxazole thiazole blue (OTB-SO3) fluorophore have been reported (Fig. 11) (Shelke et al.). These locate the ligand at a kissing-loop-type junction, in which the fluorophore headgroup stacks between a mixed-sequence base tetrad and a single adenine “flap” (Fig. 11B). Similar to the malachite green aptamer, DIR2s lacks a G-quadruplex motif, yet employ a base tetrad to promote planarity of the fluorophore heterocycles. In addition to the stacking interaction, two hydrogen bonds form between the Watson-Crick face of a guanine and the sulfate OTB-SO3 (Fig. 11B). The only substantial change to the structure induced by fluorophore binding appears to be the adenine flap swinging over the ligand (Fig. 11C). This turn-on aptamer system has the potential for bright fluorescence, and the available structural information should drive further improvements.
Fig. 11.
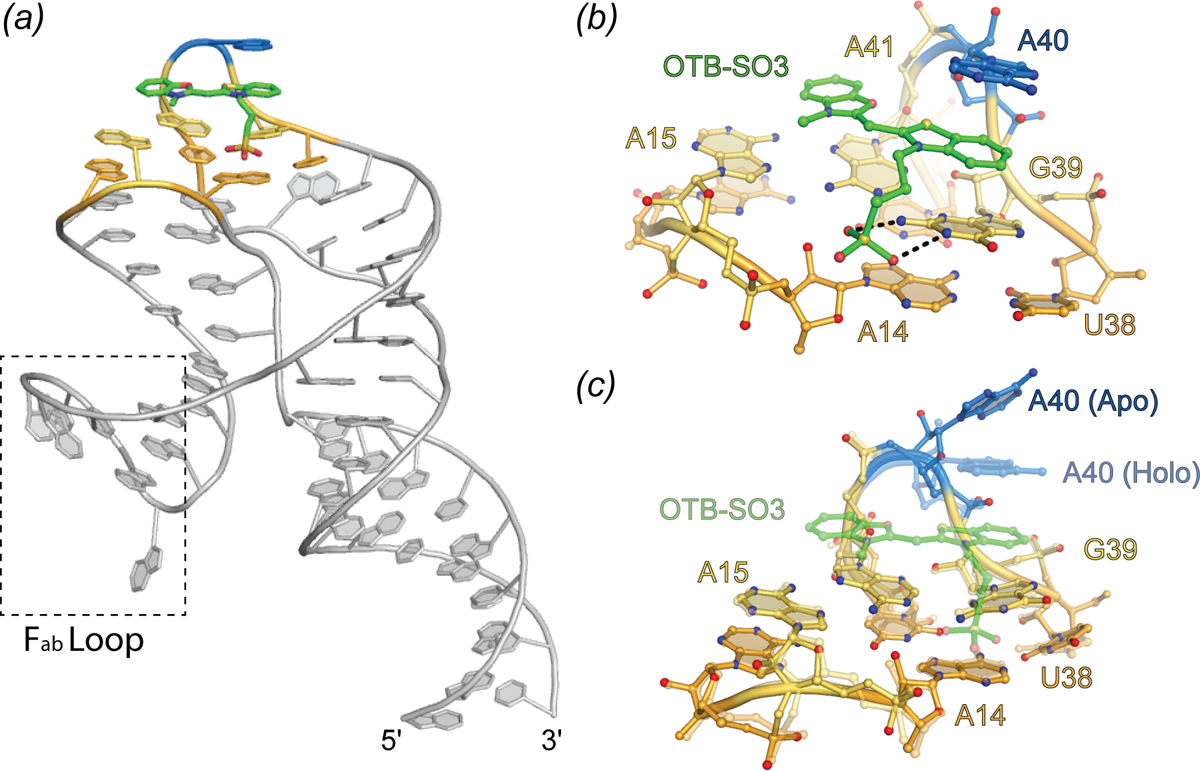
Structure of the DIR2s aptamer. Cartoon representation of DIR2s aptamer bound to OTB-SO3 (green sticks). Nucleotides composing the ligand platform are colored orange and yellow and the adenine lid (A40) is colored blue. Dashed box denotes the Fab binding loop used to facilitate crystallization. (b) Ligand binding pocket of the DIR2s aptamer (ball-and-stick) bound to OTB-SO3 (green ball-and-stick). (c) Overlay of the Apo-DIR2s structure with DIR2s-OTB-SO3 (transparent).
10. Aptamers to fluorophore-quencher conjugates
All the fluorescence turn-on aptamers discussed above bind to a conditional fluorophore to promote fluorescence by TICT (as well as ESPT for Chili). Aptamers have also been designed to induce fluorescence by reducing intramolecular quenching. By covalently attaching a contact quencher through a flexible linker to a fluorophore, intrinsic fluorescence is dramatically reduced. The first report of selection of a fluorescence turn-on aptamer employing a fluorophore-quencher conjugate was of an RNA selected to bind the quencher dimethylaniline (Sparano and Koide; Sparano and Koide). Binding of dimethylaniline by the DMA aptamer enhanced fluorescence of the conjugated fluorescein, but applications of this RNA were limited by its micromolar dissociation constant for dimethylaniline. Subsequently, an effort to repurpose the sulforhodamine blue (SRB) aptamer was undertaken by ligating various quenchers to SRB (Sunbul and Jäschke). Fluorescence enhancement was promoted by eliminating contact quenching by binding the fluorophore SRB. The SRB aptamer was still limited by micromolar affinity to its fluorophore. A combination of the SRB aptamer and an aptamer selected to bind dinitroaniline, an effective contact quencher, was used as proof of principle for two-color imaging of RNA (Arora, Sunbul and Jäschke). More recently, Riboglow was developed using the aptamer domain of the bacterial cobalamin riboswitch. Cobalamin effectively quenches fluorescence of fluorescent molecules through contact quenching. The high affinity of the cobalamin riboswitch (Kd ~ 4 nM), coupled with the folding efficiency of the biologically evolved RNA provided dramatic improvement over previously developed fluorophore-quencher conjugates (Braselmann et al.). Riboglow was able to outperform the MS2 cassette system to image stress granules in live cells. Overall, aptamers that induce fluorescence of fluorophore-quencher conjugates hold promise for their ability to accommodate spectrally diverse fluorophores with minimal RNA engineering. However, these systems suffer from high background fluorescence, inherent to contact quenching in solution.
11. Discussion
11.1. Optimization of fluorophores and aptamer selectivity
Rapid development of fluorescence turn-on aptamers has been facilitated by the use of established fluorophores, which can be modified to improve molecular characteristics (Lavis; Lavis and Raines). Limits to the ultimate photophysical and imaging capabilities of aptamer-fluorophore complexes are largely set by the fluorophores. For example, cyanine dyes and triphenylmethane dyes often bind with high affinity to fluorescence turn-on aptamers and can yield high brightness, as a result of their large absorption cross-sections (ε). However, hydrophobic, and sometimes cationic properties inherent to these fluorophores can result in non-specific interactions with membranes and nucleic acids, and the concomitant fluorescence turn-on can raise the background fluorescence. As noted above, specificity of the Mango aptamers for TO1-Biotin is in part mediated by interactions with the PEG linker (as well as the biotin itself, in the case of Mango-I), and the linker also reduces non-specific nucleic acid binding and turn-on. Thus, substituting fluorophores with hydrophilic tags provides a plausible approach to promoting specificity and photostability (Shank et al.; Shank et al.; Lavis; Grimm et al.).
Fluorophores derived from FPs, such as DFHBI and DFHO, have smaller ε than those of cyanine dyes, limiting their ultimate brightness. However, being more polar and often anionic, they are less prone to non-specific interactions. In addition, extensive studies of fluorescent protein fluorophores facilitates rational design of synthetic FP-derived conditional fluorophores with consistently improving properties (see, e.g., Paige 2011 and Song 2017). Bright cyanine dye-contact quencher conjugates provide easy access to red-shifted emission spectra, and can comprise a variety of quenchers, including polar or anionic quenchers that may reduce non-specific cellular interactions, for which aptamers can be selected. Fluorophore-quencher conjugates often suffer from high background fluorescence and potential non-specific interactions. This is not to say that these systems cannot be utilized or further improved. At present, turn-on aptamers that bind their fluorophores with the highest affinity and produce the largest fluorescence enhancements employ cyanine dye derivatives (Table 1), and it has been demonstrated that this type of fluorophore, diffused into cells at low concentrations, can result in bright aptamer fluorescence with low background (Autour, Westhof and Ryckelynck; Braselmann et al.).
To be effective imaging tools in vivo, it is desirable that turn-on aptamers exhibit high specificity for their cognate fluorophores, and discriminate strongly against other fluorophores. Such discrimination is essential if multiple turn-on aptamers are to be deployed simultaneously for studying different RNAs concurrently, as FRET pairs, or possibly as orthogonal FRET pairs (Jepsen et al.). Structural and biochemical studies of RNA-small molecule interactions indicate that molecular specificity arises primarily from polar interactions, while van der Waals and stacking interactions provide the driving force for association through the hydrophobic effect. Thus, shape complementarity between the ligand and the RNA pocket may be sufficient for binding and fluorescence turn-on, but may not provide specificity. For instance, the Corn aptamer was found to promote bright fluorescence of two general nucleic acid dyes (ThT and TO), but crystal structures of these complexes revealed disorder in the binding pockets (Sjekloca and Ferré-D’Amaré). DFHO, the cognate fluorophore of Corn, was designed by adding a hydrogen bonding “handle” (an oxime function) onto DFHBI, that the aptamer could use to increase specificity. Indeed, the Corn-DFHO co-crystal structure demonstrates that the aptamer makes two hydrogen bonds with this functional group. This recognition scheme is consistent with Corn-DFHO being 400-times brighter than the non-cognate Corn-DFHBI complex. Spinach, in contrast, is only 20 times brighter bound to its cognate fluorophore DFHBI, than in complex with the Corn fluorophore DFHO (Warner 2017). The Spinach fluorophore binding pocket appears to have generally limited selectivity, even being able to turn on fluorescence of TO1-Biotin. Indeed, the Spinach-TO1-Biotin complex is nearly five times brighter than the cognate TO1-Biotin complex of Mango-I (Jeng et al.). Because turn-on aptamers all employ planar arrangements of nucleobases to enhance fluorescence by TICT, some level of cross-reactivity is to be expected. Nonetheless, a combination of strict shape and hydrogen-bonding group complementarity, as seen for instance in some riboswitches (Peselis and Serganov; Wakeman, Winkler and Dann; Edwards, Klein and Ferré-D’Amaré), may allow a higher degree of aptamer-fluorophore specificity.
11.2. The diversity of turn-on aptamer structures
Most of the fluorescence turn-on aptamers whose structures have been determined to date employ G-quadruplexes to organize their cores, and to bind to their cognate fluorophores. This is unexpected because G-quadruplexes are unknown among structures of other small-molecule binding RNA aptamers and ribozymes (catalytic RNAs) evolved by SELEX, and absent as well in the known structures of naturally evolved metabolite-responsive RNAs (riboswitches). The 3D-architectures of small-molecule binding aptamers, ribozymes, and riboswitches are typically organized around multi-helical junctions or around pseudoknots. Some of these RNAs incorporate both structural elements. However, there is no known example of a natural small-molecule binding RNA with a G-quadruplex, and the only known examples of in vitro evolved RNAs other than turn-on aptamers that incorporate a G-quadruplexes are protein-binding aptamers, such as the fragile-X mental retardation protein aptamer (Vasilyev et al.; Phan et al.). Because G-quadruplexes are common among in vitro evolved DNAs (Tucker et al.), their absence from small-molecule binding RNAs (other than turn-on aptamers) likely is not a result of negative selection during the in vitro evolution process (e.g., by blocking reverse-transcription or causing aggregation). Thus, the prevalence of G-quadruplexes among turn-on RNA aptamers suggests that this nucleic acid structural motif is uniquely suited to activate conditional fluorophores.
The maximally fluorescent conformation of typical TICT fluorophores is one in which their conjugated heterocycles and linkers are coplanar. G-quadruplexes may be frequent in the ligand binding sites of turn-on RNA aptamers because by virtue of their cyclic symmetry and stabilization by the centrally bound cation (canonically the abundant intracellular monovalent ion, K+), their nucleobases are typically more coplanar than the bases of base pairs or triples, which often twist and buckle. Thus, G-quadruplexes can more easily restrain a stacked fluorophore into a conformation with a high quantum yield. In addition, the planar faces of G-quadruplexes are large enough to accommodate, not only the TICT fluorophore, but also additional RNA elements that can make lateral interactions with the fluorophore, thus contributing to affinity and selectivity (and possibly modulation of photophysical properties). Not all turn-on aptamers comprise G-quadruplexes. Those that do not, such as the DIR2s and MG aptamers, nonetheless employ base triples and quadruples to stabilize their fluorophores in a planar conformation.
G-quadruplexes typically display great thermodynamic stability under physiological conditions. An important design goal of fluorescence turn-on aptamers is small size, so as to allow for efficient transcription and to be minimally disruptive of the biological properties of a fluorescently tagged cellular RNA. The structural cores of most turn-on aptamers are only ~ 30 nucleotides long, yet these RNAs can make high-affinity, multidentate interactions with their cognate ligands that endow them with specificity, and also restrain their photoexcited states so as to induce fluorescence with high quantum yields. The Mango-III aptamer, whose structure results from functionalizing a G-quadruplex with two duplexes, a lone tertiary base pair, and a triplex interhelical junction, illustrates the unprecedented structural complexity that is possible in a small RNA. As structural complexity (and high information (Szostak)) is a prerequisite for functional complexity, G-quadruplexes may also be common in turn-on aptamers because they enable stable folding of even very small RNAs.
11.3. Applications of turn-on aptamers
Although the initial impetus for the development of fluorescence turn-on aptamer RNA aptamers was to serve as genetically-encoded tags for cellular RNAs to track their expression, localization, transport and turnover of transcripts, they have been deployed for other applications. Similar to split GFP, split aptamers have been engineered to characterize quaternary interactions (Rogers et al.; Alam et al.; Chandler et al.) making it possible to visualize colocalization of RNA in living cells. Split aptamers benefit from diffusion limited maturation as opposed to FP maturation that can take minutes to hours (Bevis and Glick; Subach et al.). Additionally, concentrations of small molecules have been determined in live cells through fluorescent aptamer-riboswitch chimeras (Paige et al.; Kellenberger et al.). This is not to say that biological sciences is the only field to benefit from this new technology. It has recently been shown that a robust in vitro transcription assay using a fluorescent aptamer read-out can assay toxin levels in polluted water (Alam et al.). These examples illustrate the diversity of applications possible for fluorescent RNA technologies and hint at the potential to expand on these in creative ways.
11.4. Perspectives
Fluorescence turn-on RNA aptamers hold great promise for unlocking the spatial and dynamic aspects the transcriptome in live cells. There are still improvements to be made in regard to design and engineering of optimal tags. The bimolecular nature of fluorescent aptamers enables tuning and selection of both the fluorophore and RNA component to achieve optimal properties. Recent developments in imaging dye technology (Grimm et al.) can be applied so as to expand upon the current set of fluorophores used for RNA imaging. Fluorophore design and synthesis is likely to focus on generating conditional fluorophores with a large ε, as well as sufficient polar functionalization to promote solubility and specificity. Ideally, new fluorophores will have excitation maxima corresponding to popular diode laser wavelengths to facilitate use with widely available instrumentation. Additionally, an avenue of research suggested by FAPs, the use endogenous conditional fluorophores such as biliverdin, is promising.
Although the workflow, SELEX → functional selection → structure guided engineering and re-selection has produced the various complexes of Mango-III variants and TO1-Biotin, fluorescent tags brighter than EGFP, continued progress in methods for generating bright and stable fluorescent aptamers remains important. In vitro selection experiments can only partially mimic conditions encountered within a cell, and in vivo aptamer selection is still under active development. In terms of structure-guided design, all extant turn-on aptamer crystal structures were solved using X-ray diffraction data collected over seconds to minutes in ambient light. The resulting structures likely represent the dark states of the aptamer-fluorophore complexes. In order to understand in predictive detail how RNA structure contributes to fluorescence turn-on, and how to rationally engineer fluorescence properties, high-resolution structures of photoexcited states are needed. Serial femtosecond crystallography, enabled by X-ray free electron lasers (Chapman), holds promise for determining excited-state structures, in principle with high spatial and temporal resolution. Such structures would open the way to detailed quantum mechanical analyses of the photophysics of these fluorescent complexes.
Future fluorescent RNA tag development will likely recapitulate in part the trajectory of fluorescent protein development. Engineering photoactivatable, photoconvertible, and photoswitchable fluorescent RNA aptamer-fluorophore complexes will improve coordinated tracking and super-resolution imaging of RNA complexes. Similar to FAPs, autocatalysis can be employed to covalently fuse the RNA-fluorophore complex and alter photophysical properties (Yu et al.; Rodriguez et al.). However, despite performing a similar function, fluorescent RNA aptamers must function under a different set of principles than FAPs. The hydrophobic core of a protein, marked by a low dielectric constant, is vastly different from the large, negative, electrostatic field generated by RNA (Misra and Draper; Draper). Inherent differences in polymer makeup such as this may result in new and interesting fluorescence activation mechanisms for RNA.
Fig. 6.
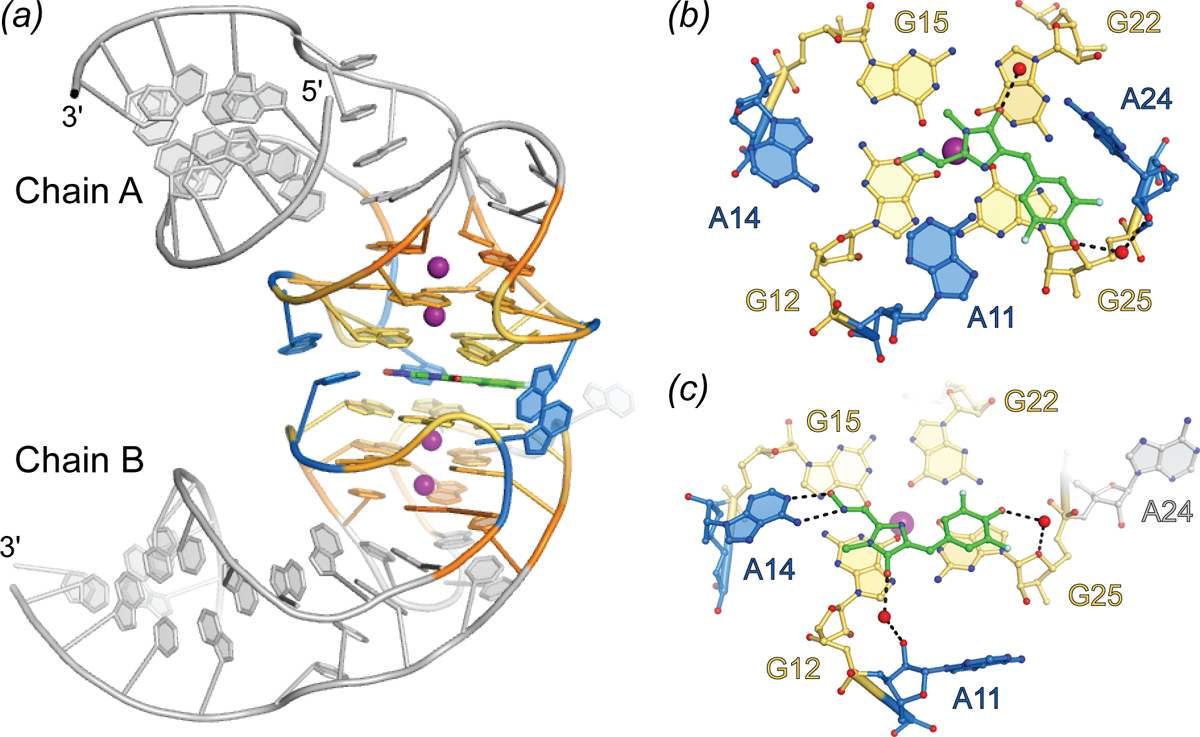
Structure of the Corn aptamer. (a) Cartoon representation of the Corn aptamer-fluorophore complex (PDB ID 5BJO). Quadruplex nucleotides are shown in warm colors and adenine flaps, at dimer interface, shown in blue. DFHO ligand is represented as green sticks. (b) Ball-and-stick representation of DFHO interactions with Chain A at the dimer interface. Nucleotides and DFHO are colored according to (a) with water molecules shown as red spheres. Hydrogen bonds are represented as black dashed lines. (c) Ball-and-stick representation of DFHO interactions with Chain B at the dimer interface.
References
- Alam KK, Jung JK, Verosloff MS, Clauer PR, Lee JW, Capdevila DA, Pastén PA, Giedroc DP, Collins JJ and Lucks JB (2019) ‘Rapid, Low-Cost Detection of Water Contaminants Using Regulated In Vitro Transcription’, bioRxiv, pp. 619296. [Google Scholar]
- Alam KK, Tawiah KD, Lichte MF, Porciani D and Burke DH (2017) ‘A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly In Vivo’, ACS Synth Biol, 6(9), pp. 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage BA (2011) ‘Imaging of RNA in live cells’, Curr Opin Chem Biol, 15(6), pp. 806–12. [DOI] [PubMed] [Google Scholar]
- Arora A, Sunbul M and Jäschke A (2015) ‘Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers’, Nucleic Acids Res, 43(21), pp. e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autour A, C Y Jeng S, D Cawte A, Abdolahzadeh A, Galli A, Panchapakesan SSS, Rueda D, Ryckelynck M and Unrau PJ (2018) ‘Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells’, Nat Commun, 9(1), pp. 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autour A, Westhof E and Ryckelynck M (2016) ‘iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications’, Nucleic Acids Res, 44(6), pp. 2491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babendure JR, Adams SR and Tsien RY (2003) ‘Aptamers switch on fluorescence of triphenylmethane dyes’, Journal of the American Chemical Society, 125(48), pp. 14716–14717. [DOI] [PubMed] [Google Scholar]
- Baptista M and Indig G (1998) ‘Effect of BSA binding on photophysical and photochemical properties of triarylmethane dyes’, Journal of Physical Chemistry B, 102(23), pp. 4678–4688. [Google Scholar]
- Baugh C, Grate D and Wilson C (2000) ‘2.8 angstrom crystal structure of the malachite green aptamer’, Journal of Molecular Biology, 301(1), pp. 117–128. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH and Long RM (1998) ‘Localization of ASH1 mRNA particles in living yeast’, Molecular Cell, 2(4), pp. 437–445. [DOI] [PubMed] [Google Scholar]
- Bevis BJ and Glick BS (2002) ‘Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed)’, Nat Biotechnol, 20(1), pp. 83–7. [DOI] [PubMed] [Google Scholar]
- Brackett DM and Dieckmann T (2006) ‘Aptamer to ribozyme: the intrinsic catalytic potential of a small RNA’, Chembiochem, 7(5), pp. 839–43. [DOI] [PubMed] [Google Scholar]
- Braselmann E, Wierzba AJ, Polaski JT, Chromiński M, Holmes ZE, Hung ST, Batan D, Wheeler JR, Parker R, Jimenez R, Gryko D, Batey RT and Palmer AE (2018) ‘A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells’, Nat Chem Biol, 14(10), pp. 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M, Lyalina T, Halman J, Rackley L, Lee L, Dang D, Ke W, Sajja S, Woods S, Acharya S, Baumgarten E, Christopher J, Elshalia E, Hrebien G, Kublank K, Saleh S, Stallings B, Tafere M, Striplin C and Afonin KA (2018) ‘Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology’, Molecules, 23(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Almo SC and Singer RH (2008) ‘Structural basis for the coevolution of a viral RNA-protein complex’, Nature Structural & Molecular Biology, 15(1), pp. 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HN (2019) ‘X-Ray Free-Electron Lasers for the Structure and Dynamics of Macromolecules’, Annu Rev Biochem. [DOI] [PubMed] [Google Scholar]
- Chen MC, Tippana R, Demeshkina NA, Murat P, Balasubramanian S, Myong S and Ferré-D’Amaré AR (2018) ‘Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36’, Nature, 558(7710), pp. 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin TP, Silva GL, Robertson KL, Hamilton TP, Fague K, Waggoner AS and Armitage BA (2008) ‘Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules’, Organic Letters, 10(8), pp. 1561–1564. [DOI] [PubMed] [Google Scholar]
- Day RND, M. W (eds.) (2014) The Fluorescent Protein Revolution. Boca Raton, Florida, USA: CRC Press. [Google Scholar]
- Dolgosheina EV, Jeng SC, Panchapakesan SS, Cojocaru R, Chen PS, Wilson PD, Hawkins N, Wiggins PA and Unrau PJ (2014) ‘RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking’, ACS Chem Biol, 9(10), pp. 2412–20. [DOI] [PubMed] [Google Scholar]
- Dolgosheina EV and Unrau PJ (2016) ‘Fluorophore-binding RNA aptamers and their applications’, Wiley Interdisciplinary Reviews-Rna, 7(6), pp. 843–851. [DOI] [PubMed] [Google Scholar]
- Draper DE (2004) ‘A guide to ions and RNA structure’, Rna-a Publication of the Rna Society, 10(3), pp. 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Klein DJ and Ferré-D’Amaré AR (2007) ‘Riboswitches: small-molecule recognition by gene regulatory RNAs’, Curr Opin Struct Biol, 17(3), pp. 273–9. [DOI] [PubMed] [Google Scholar]
- Ellington AD and Szostak JW (1990) ‘In vitro selection of RNA molecules that bind specific ligands’, Nature, 346(6287), pp. 818–22. [DOI] [PubMed] [Google Scholar]
- Fernandez-Millan P, Autour A, Ennifar E, Westhof E and Ryckelynck M (2017) ‘Crystal structure and fluorescence properties of the iSpinach aptamer in complex with DFHBI’, RNA, 23(12), pp. 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Moon JD, Svensen N and Jaffrey SR (2014) ‘Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution’, J Am Chem Soc, 136(46), pp. 16299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinders J, DeFina SC, Brackett DM, Baugh C, Wilson C and Dieckmann T (2004) ‘Recognition of planar and nonplanar ligands in the malachite green-RNA aptamer complex’, Chembiochem, 5(1), pp. 62–72. [DOI] [PubMed] [Google Scholar]
- Fuenzalida-Werner JP, Janowski R, Mishra K, Weidenfeld I, Niessing D, Ntziachristos V and Stiel AC (2018) ‘Crystal structure of a biliverdin-bound phycobiliprotein: Interdependence of oligomerization and chromophorylation’, J Struct Biol, 204(3), pp. 519–522. [DOI] [PubMed] [Google Scholar]
- Garcia JF and Parker R (2015) ‘MS2 coat proteins bound to yeast mRNAs block 5’ to 3’ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system’, RNA, 21(8), pp. 1393–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF and Parker R (2016) ‘Ubiquitous accumulation of 3 ‘ mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays’, RNA, 22(5), pp. 657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotrik M, Sekhon G, Saurabh S, Nakamoto M, Eisenstein M and Soh HT (2018) ‘Direct Selection of Fluorescence-Enhancing RNA Aptamers’, J Am Chem Soc, 140(10), pp. 3583–3591. [DOI] [PubMed] [Google Scholar]
- Grate D and Wilson C (1999) ‘Laser-mediated, site-specific inactivation of RNA transcripts’, Proceedings of the National Academy of Sciences of the United States of America, 96(11), pp. 6131–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grate D and Wilson C (2001) ‘Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex’, Bioorg Med Chem, 9(10), pp. 2565–70. [DOI] [PubMed] [Google Scholar]
- Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N and Lavis LD (2017) ‘A general method to fine-tune fluorophores for live-cell and in vivo imaging’, Nat Methods, 14(10), pp. 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan AJ, Zhang XF, Sun X, Li Q, Xiang JF, Wang LX, Lan L, Yang FM, Xu SJ, Guo XM and Tang YL (2018) ‘Ethyl-substitutive Thioflavin T as a highly-specific fluorescence probe for detecting G-quadruplex structure’, Sci Rep, 8(1), pp. 2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU and Bartel DP (2016) ‘RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria’, Science, 353(6306). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KY, Leslie BJ, Fei JY, Zhang JC and Ha T (2013) ‘Understanding the Photophysics of the Spinach-DFHBI RNA Aptamer-Fluorogen Complex To Improve Live-Cell RNA Imaging’, Journal of the American Chemical Society, 135(50), pp. 19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW and McManus MT (2013) ‘Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs’, PLoS Genet, 9(6), pp. e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington C, Lan Y and Akman SA (1997) ‘The identification and characterization of a G4-DNA resolvase activity’, J Biol Chem, 272(39), pp. 24631–6. [DOI] [PubMed] [Google Scholar]
- Heim R, Prasher DC and Tsien RY (1994) ‘Wavelength mutations and posttranslational autoxidation of green fluorescent protein’, Proceedings of the National Academy of Sciences of the United States of America, 91(26), pp. 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeman LA, Robinson SL, Szostak JW and Wilson C (1998) ‘Isolation and characterization of fluorophore-binding RNA aptamers’, Folding & Design, 3(6), pp. 423–431. [DOI] [PubMed] [Google Scholar]
- Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA and Piccirilli JA (2014) ‘A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore’, Nature Chemical Biology, 10(8), pp. 686–U128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarikote DV, Krebs N, Tannert S, Röder B and Seitz O (2007) ‘Exploring base-pair-specific optical properties of the DNA stain thiazole orange’, Chemistry, 13(1), pp. 300–10. [DOI] [PubMed] [Google Scholar]
- Jay DG and Keshishian H (1990) ‘Laser inactivation of fasciclin I disrupts axon adhesion of grasshopper pioneer neurons’, Nature, 348(6301), pp. 548–50. [DOI] [PubMed] [Google Scholar]
- Jepsen MDE, Sparvath SM, Nielsen TB, Langvad AH, Grossi G, Gothelf KV and Andersen ES (2018) ‘Development of a genetically encodable FRET system using fluorescent RNA aptamers’, Nat Commun, 9(1), pp. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng SCY, Chan HHY, Booy EP, McKenna SA and Unrau PJ (2016) ‘Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers’, RNA, 22(12), pp. 1884–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson HE, Dertinger D, LeCuyer KA, Behlen LS, Greef CH and Uhlenbeck OC (1998) ‘A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein’, Proceedings of the National Academy of Sciences of the United States of America, 95(16), pp. 9244–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP and Ferré-D’Amaré AR (2015) ‘RNA quaternary structure and global symmetry’, Trends Biochem Sci, 40(4), pp. 211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Chen C, Whiteley AT, Portnoy DA and Hammond MC (2015) ‘RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP’, J Am Chem Soc, 137(20), pp. 6432–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymchenko AS (2017) ‘Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications’, Acc Chem Res, 50(2), pp. 366–375. [DOI] [PubMed] [Google Scholar]
- Koirala D, Shelke SA, Dupont M, Ruiz S, DasGupta S, Bailey LJ, Benner SA and Piccirilli JA (2018) ‘Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography’, Nucleic Acids Res, 46(5), pp. 2624–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Bugaut A, Huppert JL and Balasubramanian S (2007) ‘An RNA G-quadruplex in the 5’ UTR of the NRAS proto-oncogene modulates translation’, Nat Chem Biol, 3(4), pp. 218–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD (2017a) ‘Chemistry Is Dead. Long Live Chemistry!’, Biochemistry, 56(39), pp. 5165–5170. [DOI] [PubMed] [Google Scholar]
- Lavis LD (2017b) ‘Teaching Old Dyes New Tricks: Biological Probes Built from Fluoresceins and Rhodamines’, Annu Rev Biochem, 86, pp. 825–843. [DOI] [PubMed] [Google Scholar]
- Lavis LD and Raines RT (2008) ‘Bright ideas for chemical biology’, ACS Chem Biol, 3(3), pp. 142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Larson T, Nguyen HH, Sokolov KV and Ellington AD (2010) ‘Directed evolution of gold nanoparticle delivery to cells’, Chem Commun (Camb), 46(3), pp. 392–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra VK and Draper DE (2002) ‘The linkage between magnesium binding and RNA folding’, Journal of Molecular Biology, 317(4), pp. 507–521. [DOI] [PubMed] [Google Scholar]
- Mohanty J, Barooah N, Dhamodharan V, Harikrishna S, Pradeepkumar PI and Bhasikuttan AC (2013) ‘Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA’ J Am Chem Soc, 135(1) 367–76 [DOI] [PubMed] [Google Scholar]
- Monici M (2005) ‘Cell and tissue autofluorescence research and diagnostic applications’, Biotechnol Annu Rev, 11, pp. 227–56. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA and Yeo GW (2016) ‘Programmable RNA Tracking in Live Cells with CRISPR/Cas9’, Cell, 165(2), pp. 488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, DeFina SC, Fink WH and Dieckmann T (2002) ‘Binding to an RNA aptamer changes the charge distribution and conformation of malachite green’, Journal of the American Chemical Society, 124(50), pp. 15081–15084. [DOI] [PubMed] [Google Scholar]
- Ni CZ, Syed R, Kodandapani R, Wickersham J, Peabody DS and Ely KR (1995) ‘Crystal structure of the MS2 coat protein dimer: implications for RNA binding and virus assembly’, Structure, 3(3), pp. 255–63. [DOI] [PubMed] [Google Scholar]
- Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M and Tsuji FI (1996) ‘Chemical nature of the light emitter of the Aequorea green fluorescent protein’, Proceedings of the National Academy of Sciences of the United States of America, 93(24), pp. 13617–13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren J, Svanvik N and Kubista M (1998) ‘The interactions between the fluorescent dye thiazole orange and DNA’, Biopolymers, 46(1), pp. 39–51. [DOI] [PubMed] [Google Scholar]
- Ouellet J (2016) ‘RNA Fluorescence with Light-Up Aptamers’, Frontiers in Chemistry, 4, pp. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Nguyen-Duc T, Song WJ and Jaffrey SR (2012) ‘Fluorescence Imaging of Cellular Metabolites with RNA’, Science, 335(6073), pp. 1194–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Wu KY and Jaffrey SR (2011a) ‘RNA Mimics of Green Fluorescent Protein’, Science, 333(6042), pp. 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Wu KY and Jaffrey SR (2011b) ‘RNA mimics of green fluorescent protein’, Science, 333(6042), pp. 642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M (2012) ‘The human transcriptome: an unfinished story’, Genes (Basel), 3(3), pp. 344–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis A and Serganov A (2014) ‘Themes and variations in riboswitch structure and function’, Biochim Biophys Acta, 1839(10), pp. 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB and Patel DJ (2011) ‘Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction’, Nat Struct Mol Biol, 18(7), pp. 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich KD, Hulit J, Subach OM, Wu B, Abdulla A, Segall JE and Verkhusha VV (2010a) ‘Monomeric red fluorescent proteins with a large Stokes shift’, Proc Natl Acad Sci U S A, 107(12), pp. 5369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich KD, Malashkevich VN, Almo SC and Verkhusha VV (2010b) ‘Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift’, J Am Chem Soc, 132(31), pp. 10762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij CW, Rietveld K and Bosch L (1985) ‘A new principle of RNA folding based on pseudoknotting’ Nucleic Acids Res, 13(5) 1717–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud de la Faverie A, Guédin A, Bedrat A, Yatsunyk LA and Mergny JL (2014) ‘Thioflavin T as a fluorescence light-up probe for G4 formation’ Nucleic Acids Res, 42(8) e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL and Joyce GF (1990) ‘Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA’, Nature, 344(6265), pp. 467–468. [DOI] [PubMed] [Google Scholar]
- Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu XK, Zhang J and Tsien RY (2017) ‘The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins’, Trends in Biochemical Sciences, 42(2), pp. 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez EA, Tran GN, Gross LA, Crisp JL, Shu XK, Lin JY and Tsien RY (2016) ‘A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein’, Nature Methods, 13(9), pp. 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TA, Andrews GE, Jaeger L and Grabow WW (2015) ‘Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer’, ACS Synth Biol, 4(2), pp. 162–6. [DOI] [PubMed] [Google Scholar]
- Royant A and Noirclerc-Savoye M (2011) ‘Stabilizing role of glutamic acid 222 in the structure of Enhanced Green Fluorescent Protein’, J Struct Biol, 174(2), pp. 385–90. [DOI] [PubMed] [Google Scholar]
- Shank NI, Pham HH, Waggoner AS and Armitage BA (2013) ‘Twisted cyanines: a non-planar fluorogenic dye with superior photostability and its use in a protein-based fluoromodule’, J Am Chem Soc, 135(1), pp. 242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank NI, Zanotti KJ, Lanni F, Berget PB and Armitage BA (2009) ‘Enhanced photostability of genetically encodable fluoromodules based on fluorogenic cyanine dyes and a promiscuous protein partner’, J Am Chem Soc, 131(36), pp. 12960–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Hink MA, Joosen L, Gadella TW and Verkhusha VV (2012) ‘An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging’, J Am Chem Soc, 134(18), pp. 7913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelke SA, Shao Y, Laski A, Koirala D, Weissman BP, Fuller JR, Tan X, Constantin TP, Waggoner AS, Bruchez MP, Armitage BA and Piccirilli JA (2018) ‘Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif’, Nat Commun, 9(1), pp. 4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjekloca L and Ferré-D’Amaré AR (2019) ‘Binding between the G-quadruplexes at the homodimer interface of the Corn RNA aptamer strongly activates Thioflavin T fluorescence’. [DOI] [PMC free article] [PubMed]
- Song W, Filonov GS, Kim H, Hirsch M, Li X, Moon JD and Jaffrey SR (2017) ‘Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex’, Nat Chem Biol, 13(11), pp. 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Strack RL, Svensen N and Jaffrey SR (2014) ‘Plug-and-Play Fluorophores Extend the Spectral Properties of Spinach’, Journal of the American Chemical Society, 136(4), pp. 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparano BA and Koide K (2005) ‘A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA’, J Am Chem Soc, 127(43), pp. 14954–5. [DOI] [PubMed] [Google Scholar]
- Sparano BA and Koide K (2007) ‘Fluorescent sensors for specific RNA: a general paradigm using chemistry and combinatorial biology’, J Am Chem Soc, 129(15), pp. 4785–94. [DOI] [PubMed] [Google Scholar]
- Steinmetzger C, Palanisamy N, Gore KR and Höbartner C (2019) ‘A Multicolor Large Stokes Shift Fluorogen-Activating RNA Aptamer with Cationic Chromophores’, Chemistry, 25(8), pp. 1931–1935. [DOI] [PubMed] [Google Scholar]
- Strack RL, Disney MD and Jaffrey SR (2013) ‘A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA’, Nature Methods, 10(12), pp. 1219–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach F, Subach O, Gundorov I, Morozova K, Piatkevich K, Cuervo A and Verkhusha V (2009) ‘Monomeric fluorescent timers that change color from blue to red report on cellular trafficking’, Nature Chemical Biology, 5(2), pp. 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunbul M and Jäschke A (2013) ‘Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer’, Angew Chem Int Ed Engl, 52(50), pp. 13401–4. [DOI] [PubMed] [Google Scholar]
- Szostak JW (2003) ‘Functional information: Molecular messages’, Nature, 423(6941), pp. 689. [DOI] [PubMed] [Google Scholar]
- Tan X, Constantin TP, Sloane KL, Waggoner AS, Bruchez MP and Armitage BA (2017a) ‘Fluoromodules Consisting of a Promiscuous RNA Aptamer and Red or Blue Fluorogenic Cyanine Dyes: Selection, Characterization, and Bioimaging’, J Am Chem Soc, 139(26), pp. 9001–9009. [DOI] [PubMed] [Google Scholar]
- Tan XH, Constantin TP, Sloane KL, Waggoner AS, Bruchez MP and Armitage BA (2017b) ‘Fluoromodules Consisting of a Promiscuous RNA Aptamer and Red or Blue Fluorogenic Cyanine Dyes: Selection, Characterization, and Bioimaging’, Journal of the American Chemical Society, 139(26), pp. 9001–9009. [DOI] [PubMed] [Google Scholar]
- Trachman Robert J. III Autour AJ, Sunny CY. Abdolahzadeh Amir Andreoni Alessio Cojocaru Razvan Garipov Ramil Dolgosheina Elena V. Knutson Jay R. Ryckelynck Michael Unrau Peter J. Ferré-D’Amaré Adrian R. (2019) ‘Structure and functional reselection of the Mango-III fluorogenic RNA aptamer’, Nature Chemical Biology, Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachman RJ, Abdolahzadeh A, Andreoni A, Cojocaru R, Knutson JR, Ryckelynck M, Unrau PJ and Ferré-D’Amaré AR (2018) ‘Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness’, Biochemistry, 57(26), pp. 3544–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachman RJ, Demeshkina NA, Lau MWL, Panchapakesan SSS, Jeng SCY, Unrau PJ and Ferré-D’Amaré AR (2017) ‘Structural basis for high-affinity fluorophore binding and activation by RNA Mango’, Nat Chem Biol, 13(7), pp. 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachman RJ, Truong L and Ferre-D’Amare AR (2017) ‘Structural Principles of Fluorescent RNA Aptamers’, Trends in Pharmacological Sciences, 38(10), pp. 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troung LA and Ferré-D’Amaré AR (2019) From fluorescent proteins to fluorogenic RNAs: tools for imaging cellular macromolecules Protein Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker WO, Shum KT and Tanner JA (2012) ‘G-quadruplex DNA aptamers and their ligands: structure, function and application’, Curr Pharm Des, 18(14) 2014–26 [DOI] [PubMed] [Google Scholar]
- Tuerk C and Gold L (1990) ‘Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA-polymerase’, Science, 249(4968), pp. 505–510. [DOI] [PubMed] [Google Scholar]
- Turaev AV, Tsvetkov VB, Tankevich MV, Smirnov IP, Aralov AV, Pozmogova GE and Varizhuk AM (2019) ‘Benzothiazole-based cyanines as fluorescent “light-up” probes for duplex and quadruplex DNA’, Biochimie, 162, pp. 216–228. [DOI] [PubMed] [Google Scholar]
- Valencia-Burton M, McCullough RM, Cantor CR and Broude NE (2007) ‘RNA visualization in live bacterial cells using fluorescent protein complementation’, Nat Methods, 4(5), pp. 421–7. [DOI] [PubMed] [Google Scholar]
- Vasilyev N, Polonskaia A, Darnell JC, Darnell RB, Patel DJ and Serganov A (2015) ‘Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP’, Proc Natl Acad Sci U S A, 112(39), pp. E5391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman CA, Winkler WC and Dann CE (2007) ‘Structural features of metabolite-sensing riboswitches’, Trends Biochem Sci, 32(9), pp. 415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PC, Querard J, Maurin S, Nath SS, Le Saux T, Gautier A and Jullien L (2013) ‘Photochemical properties of Spinach and its use in selective imaging’, Chemical Science, 4(7), pp. 2865–2873. [Google Scholar]
- Ward WW and Bokman SH (1982) ‘Reversible denaturation of aquoria green-fluorescent protein: physical seperation and characterization of the renatured protein’, Biochemistry, 21(19), pp. 4535–4540. [DOI] [PubMed] [Google Scholar]
- Warner KD, Chen MC, Song WJ, Strack RL, Thorn A, Jaffrey SR and Ferre-D’Amare AR (2014) ‘Structural basis for activity of highly efficient RNA mimics of green fluorescent protein’, Nat. Struct. Mol. Biol, 21(8), pp. 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chen J and Singer RH (2014) ‘Background free imaging of single mRNAs in live cells using split fluorescent proteins’, Sci Rep, 4, pp. 3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li Q, Xiang J, Yang Q, Sun H, Guan A, Wang L, Liu Y, Yu L, Shi Y, Chen H and Tang Y (2016) Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes’ Sci Rep, 6:24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You MX and Jaffrey SR (2015) ‘Structure and Mechanism of RNA Mimics of Green Fluorescent Protein’, in Dill KA (ed.) Annu. Rev. Biophys Annual Review of Biophysics Palo Alto: Annual Reviews, pp. 187–206. [DOI] [PubMed] [Google Scholar]
- Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge WP, Thayer DA, Huang H, Kornberg TB, Royant A, Jan LY, Jan YN, Weiss WA and Shu XK (2014) ‘An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging’, Nat. Comm, 5, pp. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]


