Abstract
Sepsis is the primary cause of burn-related mortality and morbidity. Traditional indicators of sepsis exhibit poor performance when used in this unique population due to their underlying hypermetabolic and inflammatory response following burn injury. To address this challenge, we developed the Machine Intelligence Learning Optimizer (MILO), an automated machine learning (ML) platform, to automatically produce ML models for predicting burn sepsis. We conducted a retrospective analysis of 211 adult patients (age ≥ 18 years) with severe burn injury (≥ 20% total body surface area) to generate training and test datasets for ML applications. The MILO approach was compared against an exhaustive “non-automated” ML approach as well as standard statistical methods. For this study, traditional multivariate logistic regression (LR) identified seven predictors of burn sepsis when controlled for age and burn size (OR 2.8, 95% CI 1.99–4.04, P = 0.032). The area under the ROC (ROC-AUC) when using these seven predictors was 0.88. Next, the non-automated ML approach produced an optimal model based on LR using 16 out of the 23 features from the study dataset. Model accuracy was 86% with ROC-AUC of 0.96. In contrast, MILO identified a k-nearest neighbor-based model using only five features to be the best performer with an accuracy of 90% and a ROC-AUC of 0.96. Machine learning augments burn sepsis prediction. MILO identified models more quickly, with less required features, and found to be analytically superior to traditional ML approaches. Future studies are needed to clinically validate the performance of MILO-derived ML models for sepsis prediction.
Subject terms: Infectious diseases, Trauma, Scientific data, Software
Introduction
Burn patients are at high risk for infections, with sepsis being the most common cause of morbidity and mortality1. Traditional indicators of sepsis defined previously by the Surviving Sepsis Campaign2 and other organizations exhibit poor performance when used in this unique population due to their underlying hypermetabolic and inflammatory response to burn injury. For example, the systemic inflammatory response syndrome2,3 lacks clinical sensitivity and specificity when applied to severely burned patients1, while the newer 2016 “Sepsis-3” criteria remain controversial in both burned and non-burned patients4–7. To this end, early and accurate recognition of sepsis represents a significant clinical knowledge gap in burn critical care.
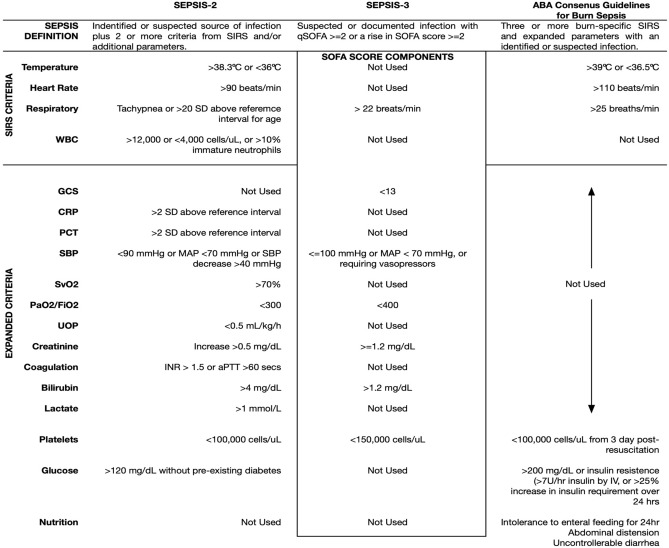
The American Burn Association (ABA) Consensus Guidelines published in 2007 was intended to better differentiate burn sepsis from the natural host-response to injury (Table 1)1. These guidelines recognized deficiencies of traditional indications of sepsis and removed less specific parameters such as white blood cell count (WBC). Fever was re-defined as temperatures > 39 °C to improve specificity and at the cost of sensitivity. Glycemic variability and thrombocytopenia were also included in the ABA Consensus Guidelines; however, measurement of glycemic variability is challenging without continuous glucose monitoring technology and platelet count aids burn sepsis recognition at later stages of severe infection.
Table 1.
Comparison of sepsis criteria.
ABA American Burn Association, aPTT activated partial thromboplastin time, CRP c-reactive protein, FiO2 fraction of inspired oxygen, GCS Glascow Coma Score, INR international normalized ratio, IV intravenous, NS not significant, PaO2 partial pressure of oxygen, PCT procalcitonin, PLT platelet count, SBP systolic blood pressure, SD standard deviation, SIRS systemic inflammatory response syndrome, SOFA sequential organ failure assessment, SvO2 saturation of venous oxygen, temp temperature, UOP urine output, WBC white blood cell count.
The emergence of artificial intelligence (AI) and machine learning (ML) provides an opportunity to improve burn sepsis recognition. Recent studies suggest AI/ML improves the diagnostic accuracy, and clinical sensitivity/specificity for predicting burn related sequalae such as acute kidney injury (AKI)8. However, widespread adoption of AI/ML in laboratory diagnostics is challenged by the lack of programming expertise in the medical community and accessibility of health data to develop clinically relevant models9. To this end, the development of automated ML platforms to facilitate pragmatic clinical studies are needed to fully realize the true capabilities of health AI. The objective of this study is to provide proof of concept clinical utility of ML for sepsis recognition in comparison to existing criteria in the high-risk burn population.
Methods
We developed and validated ML models for burn sepsis prediction using a retrospective dataset. The database was derived from a previous multicenter (5-site) randomized controlled trial (ClinicalTrials.gov#NCT01140269) evaluating the clinical impact of molecular pathogen detection in burn sepsis patients where vital signs and laboratory data was recorded daily for the duration of each patient’s intensive care unit (ICU) stay (Supplemental Data Fig. S1)10. Vital signs and laboratory data was consistently collected per study protocol. Contributing sites were ABA verified burn centers in the United States. Human subjects’ approval was obtained at each study site and through the United States Army Human Research Protection Office. From this study population, we used the data to help predict sepsis when analyzed using traditional statistics, as well as ML using traditional non-automated programming, and then compared against our novel automated ML approach. Study methods are described below.
Study population
The database consisted of 211 adult (age ≥ 18 years) patients with ≥ 20% total body surface area (TBSA) burns enrolled from across five United States academic hospitals. Patients with non-survivable injuries or lacking the ability to provide informed/surrogate consent were excluded. Relevant medical information including patient demographics, vital signs (i.e., heart rate, respiratory rate, systolic/diastolic blood pressure, mean arterial pressure, central venous pressure), laboratory results (i.e., blood gas indices, complete blood count, chemistry panels, coagulation status, and microbiology results), Glascow Coma Score (GCS), medical/surgical procedures (e.g., surgery, intravascular line placement/removal), mechanical ventilator settings, and prescribed antimicrobial medications were recorded daily over the course of their ICU stay. Outcome measures include sepsis status, and mortality were also recorded. Sepsis status was based on the 2007 ABA Consensus Guidelines1. The study population included patients with respiratory, urinary tract, soft tissue, and/or bloodstream infections. Recorded data variables are outlined in Table 2 and were used for curating the data to determine sepsis status, as well as for performing traditional statistical analyses, and ML model development and generalization.
Table 2.
Daily recorded variables for enrolled subjects.
| Vital sign | Blood gas | Chemistry | Heme/Coag | Microbiology | Calculated values | Clinical events |
|---|---|---|---|---|---|---|
| Heart rate | pH | Na+ | HGB | Blood culture | Anion gap | Surgery |
| Respiratory rate | pCO2 | K+ | HCT | Respiratory culture | BUN/creatinine ratio | Ventilatory status |
| SBP/DBP | pO2 | Cl− | WBC | Urine culture | MODS | Antibiotic therapy |
| CVP | HCO3− | TCO2 | Platelet count | Wound culture | SOFA | Dialysis status |
| MAP | FiO2 | Glucose | aPTT | PaO2/FiO2 | Survival status | |
| GCS | Creatinine | INR | ||||
| BUN | D-dimer | |||||
| Total bilirubin |
aPTT activated partial thromboplastin time, BUN blood urea nitrogen, CVP central venous pressure, DBP diastolic blood pressure, FiO2 fraction of inspired oxygen, HCT hematocrit, HGB hemoglobin, INR international normalized ratio, MAP mean arterial pressure, MODS multiple organ dysfunction score, PaO2 partial pressure of arterial oxygen, pCO2 partial pressure of CO2, pO2 partial pressure of oxygen, SBP systolic blood pressure, SOFA sequential organ failure assessment score, SO2 oxygen saturation, TCO2 total CO2, WBC white blood cell count.
Traditional machine learning method
Machine learning sepsis algorithms were first developed using our exhaustive “traditional” non-automated ML approach8,11. The process entailed manually selecting various feature set combinations aided with an unsupervised select percentile techniques such as ANOVA F-classification for feature selection from the original dataset followed by building a large number of models on various supervised algorithms. The five ML supervised algorithms employed for this task included: (a) logistic regression (LR), (b) k-nearest neighbor (k-NN), (c) random forest (RF), (d) support vector machine (SVM), and our multi-layer perceptron deep neural network (DNN). Scikit-Learn’s version 0.20.2 was used to construct models as in previous studies11. Cross validation and hyperparameter tuning studies were also performed for LR, RF, k-NN, SVM, and DNN methods using the Scikit-learn cross validation and grid search tools. This technique along with the grid search hyperparameter variations allowed us to develop and compare a large number (49,940) of unique models based on various feature set combinations (identified from our select percentile feature selection) within our five ML methods/algorithms. This approach enabled us to empirically assess and compare all models and to identify the best performing ML model for a given set of unique hyperparameters and feature set combinations.
Automated ML (auto-ML) platform
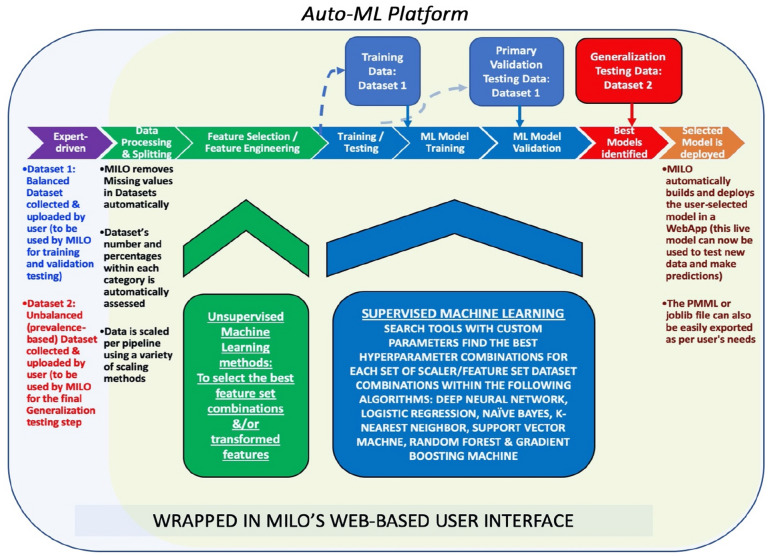
In addition to the above manual ML approach, we developed the Machine Intelligence Learning Optimizer (MILO) platform to perform a similar task in a fully automated fashion (Fig. 1)12. The MILO infrastructure includes an automated data processor, a data feature selector (ANOVA F select percentile feature selector and RF Feature Importances Selector) and data transformer (e.g., principal component analysis), followed by custom supervised ML model building using our custom hyperparameter ranges used with search tools (i.e., grid search and random search tools) to help identify the optimal hyperparameter combinations for DNN, LR, naïve Bayes (NB), k-NN, SVM, RF, and XGBoost gradient boosting machine (GBM) techniques. Additionally, MILO enables addition of other algorithms and hyperparameter combinations—allowing us to easily add in NB and GBM for analysis.
Figure 1.
Machine intelligence learning optimizer: the MILO auto-machine learning (ML) infrastructure consists of begins with two datasets: (a) balanced data (Data Set 1) set used for training and validation, and (b) an unbalanced dataset (Data Set 2) for generalization. MILO removes missing values, assessed and scaled by the software. Unsupervised ML is then used for feature selection and engineering. The generated models are trained and then tested with the Data Set 1 during the supervised ML stage. Primary validation is then performed using Data Set 1 and followed by generalization using Data Set 2. Selected models can then be deployed thereafter as predictive model markup language (PMML) files.
Following the training and validation of models, MILO executes an automated performance assessment with results exported for user viewing. In the end, MILO employs a combination of unsupervised and supervised ML platforms from a large set of algorithms, scalers and feature selectors/transformers to create greater than 1,000 unique pipelines (i.e., set of automated machine learning steps)—ultimately generating > 100,000 models that are then statistically assessed to identify optimal algorithms for use (Supplemental Data Table S1).
For this study, we imported the trial data into MILO using sepsis status as the outcome measure for analysis. The following functions are then performed automatically by MILO. First, rows with any missing values are removed (e.g., laboratory results that were not performed for a given day). Next the information is assessed to ensure model training and the initial validation step is based on a balanced dataset. A balanced dataset is used for training because the system was built to work with small amounts of training data and since accuracy was a scoring discriminator, the measured accuracy can then be better assessed against a lower null accuracy baseline which ultimately would minimize overfitting. This balanced dataset is then split into training and validation test sets in an 80–20 split, respectively. Since many algorithms benefit from scaling, in addition to using the unscaled data, the training dataset also underwent two possible scaling transformations (i.e., standard scaler and minmax scaler). To evaluate the effect of various features within the datasets, a combination of various statistically significant feature was then selected to build new datasets with less or transformed features. The features selected in this step are derived from several well-established unsupervised ML techniques including ANOVA F-statistic value select percentile, RF Feature Importances or transformed using our principle component analysis approach9. A large number of ML models are then built from these datasets with optimal parameters on large number of pipelines which include a combination of various algorithms (i.e., DNN, SVM, NB, LR, k-NN, RF, and GBM), scalers, hyper-parameters, and feature sets. All pipelines generated by MILO undergo generalization assessment no matter their performance. For model validation, MILO creates and assesses hundreds of thousands of models. All models for each category are then identified and passed onto the next phase of the software pipeline for generalization assessment. Machine learning model performance data is then tabulated by MILO and reported as clinical sensitivity, specificity, accuracy, F1 score, receiver operator characteristic (ROC) curves, and reliability curves. Finally, to evaluate if ABA Consensus Guidelines1 and Sepsis-3 criteria4 are compatible with ML applications, MILO algorithms were generated using their respective parameters (ABA Consensus Guidelines: body temperature, respiratory rate, and heart rate, platelet count (PLT), and glucose; Sepsis-3: respiratory rate, PaO2/FiO2, GCS, PLT, MAP, and total bilirubin) and compared against models evaluating optimized features from the dataset. American Burn Association Consensus Guideline1 criteria such as insulin rates/resistance, intolerance to enteral feedings, abdominal distension, and uncontrollable diarrhea were not available for the study dataset.
Traditional statistical analysis
JMP software (SAS Institute, Cary, NC) was used for statistical analysis. Descriptive statistics were calculated for patient demographics. Data was also assessed for normality using the Ryan-Joiner Test. Continuous parametric variables were analyzed using the 2-sample t-test, while discrete variables were compared using the non-parametric Chi-square test. As appropriate, continuous non-parametric variables, the Mann–Whitney U Test was used. Multivariate LR was used to determine predictors of sepsis with age and burn size serving as covariates with 95% confidence intervals (CI) reported. Repeated measures ANOVA was used for time series data. A P-value < 0.05 was considered statistically significant with ROC analysis also performed to compare sepsis biomarker performance. Bootstrapping (minimum of 2,500 bootstrap samples) via JMP software was employed to calculate 95% confidence intervals (CI) for the area under the ROC curves.
Results
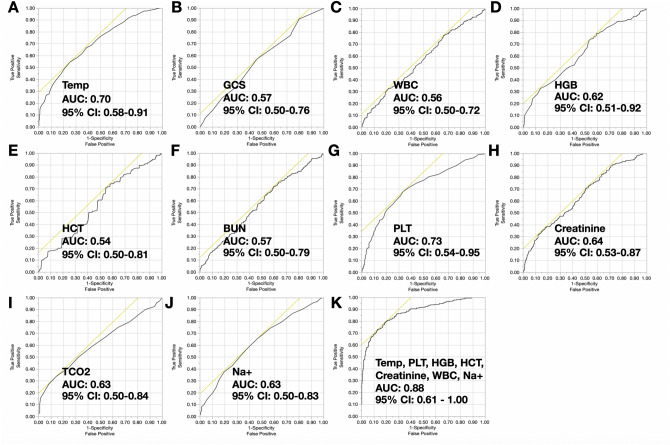
The study dataset was compromised of data from 211 patients with 704 incidents requiring collection of cultures in suspicion of sepsis. Table 3 highlights the demographics for septic versus non-septic burn patients. Briefly, septic patients significantly differed in terms of vital signs and laboratory parameters compared to their non-septic counterparts. Multiple organ dysfunction score (MODS) (4.42 [2.8] vs. 3.87 [2.9], P = 0.006) and sequential organ failure assessment (SOFA) scores (4.10 [2.7] vs. 3.17 [2.3], P < 0.001) were also found to be significantly higher in sepsis versus non-sepsis patients. Burn sepsis patients also exhibited greater disease severity scores (maximum MODS: 17 vs. 13 and maximum SOFA: 14 vs. 10). Figure 2 compares ROC curves for these statistically significant variables. Multivariate LR identified body temperature, WBC, HGB, HCT, Na+, and PLT as predictors of sepsis when controlled for age and burn size (OR 2.8, 95% CI 1.99–4.04, P = 0.032). The area under the ROC curve when using these seven predictors was 0.88 (95% CI 0.61–1.00).
Table 3.
Comparison of septic versus non-septic burn patients.
| Variable | Septic (n = 92) | Non-septic (n = 119) | P-value |
|---|---|---|---|
| A. Demographics | |||
| Mean (SD) age (years) | 44.5 (18.1) | 38.6 (15.7) | NS |
| Mean (SD) TBSA (%) | 38.9 (16.9) | 21.7 (10.8) | 0.033 |
| Gender (M/F) | 59/33 | 80/39 | NS |
| Inhalation injury (%) | 14.1% | 13.9% | NS |
| ICU LOS (days) | 58.7 (25.6) | 40.2 (26.2) | 0.021 |
| Median (IQR) # infections per patient | |||
| Bloodstream | 5.5 (6.0) | N/A | N/A |
| Pneumonia | 7.2 (5.5) | N/A | N/A |
| Urinary tract | 3.6 (2.7) | N/A | N/A |
| Wound | 4.4 (3.6) | N/A | N/A |
| Mortality (%) | 31.5 | 11.8 | 0.001 |
| Mean GCS (SD) | 10.3 (2.8) | 11.4 (2.8) | 0.005 |
| Mean (SD) MODS | 4.4 (2.8) | 3.9 (2.7) | 0.006 |
| Mean (SD) SOFA | 4.1 (2.7) | 3.2 (2.3) | < 0.001 |
| B. Laboratory data | |||
| Median Temperature (IQR) (ºC) | 39.2 (4.0) | 38.0 (3.5) | 0.010 |
| Median HCT (IQR) (%) | 24.0 (7.0) | 25.5 (8.6) | 0.003 |
| Median HGB (IQR) (g/dL) | 7.8 (2.2) | 8.5 (3.0) | < 0.001 |
| Median WBC (IQR) (cells/µL) | 13.1 (10.7) | 12.1 (9.0) | 0.001 |
| Median creatinine (IQR) (mg/dL) | 0.80 (0.91) | 0.71 (0.38) | < 0.001 |
| Median BUN (IQR) (mg/dL) | 21 (20.8) | 13 (9.8) | < 0.001 |
| Mean Na+ (SD) (mmol/L) | 139.2 (5.28) | 136.8 (4.53) | < 0.001 |
| Mean TCO2 (SD) (mmol/L) | 26.8 (4.9) | 24.8 (3.68) | 0.001 |
| Median PLT (IQR) (cells/µL) | 289 (300.5) | 352.5 (344.8) | < 0.001 |
BUN blood urea nitrogen, HCT hematocrit, HGB hemoglobin, ICU intensive care unit, F female, GCS Glascow Coma Score, IQR interquartile range, LOS length-of-stay, M male, MODS multiple organ dysfunction score, N/A not applicable, Na+ ionized sodium, NS not significant, PLT platelet count, SD standard deviation, SOFA sequential organ failure assessment score, TBSA total body surface area, TCO2 total CO2, and WBC white blood cell count.
Figure 2.
Receiver operator characteristic curves for statistically significant burn sepsis biomarkers: (A–J) represent receiver operator characteristic (ROC) curves and the area under the curve (AUC) analysis (in fractions) with 95% confidence intervals (CI) for statistically significant predictors of burn sepsis. (K) is the ROC curve for the multivariate model that best predicts sepsis using logistic regression. The tangent line for each ROC curve identifies the point where sensitivity and specificity are optimized. BUN blood urea nitrogen, GCS Glascow coma score, HCT hematocrit, HGB hemoglobin, Na+ sodium, PLT platelet, TCO2 total carbon dioxide, TCO2 total carbon dioxide.
Generalization performance of ML algorithms produced by traditional ML versus the MILO approach
Traditional ML approach required 400 h to train and test various models and utilizing manual interventions and discussions, while MILO automatically completed the same programming tasks plus additional comparisons in 20 h.
Table 4 compares the generalization performance for the top five performing ML models on each given algorithm produced by our traditional ML programming approach versus MILO’s Auto-ML approach which included DNN, LR, NB, k-NN, SVM, RF, and GBM methods. For ML model development using both traditional and MILO techniques, our heuristic approach employed a balanced training dataset of 250 culturing events that resulted in sepsis and 250 that did not result in sepsis pulled from the 704 events derived from the 211 patient dataset. The remaining 204 culturing events which represented the study population’s sepsis prevalence (48 sepsis and 156 non-sepsis), were then used for the final validation/generalization of each of the ML models.
Table 4.
Machine learning algorithm performance for the top 5 models identified by traditional programming versus MILO.
| Method | Accuracy (95% CI) | AUROC (95% CI)* | Sensitivity (95% CI) | Specificity (95% CI) | Features |
|---|---|---|---|---|---|
| A. Traditional programming | |||||
| Logistic regression | 86 (80–90) | 0.96 (0.88–1.00) | 98 (89–100) | 82 (75–88) | 16a |
| Deep neural network | 81 (75–86) | 0.96 (0.85–1.00) | 94 (83–99) | 77 (70–83) | 10b |
| k-nearest neighbor | 81 (75–86) | 0.92 (0.84–1.00) | 98 (89–100) | 76 (68–82) | 10b |
| Support vector machine | 85 (79–89) | 0.97 (0.86–1.00) | 98 (89–100) | 81 (74–87) | 14c |
| Random forest | 79 (73–85) | 0.92 (0.84–1.00) | 94 (83–99) | 75 (67–82) | 10b |
| B. MILO | |||||
| k-nearest neighbor | 90 (85–94) | 0.96 (0.85–1.00) | 96 (86–99) | 88 (82–93) | 5e |
| Logistic regression | 87 (81–91) | 0.95 (0.83–1.00) | 98 (89–100) | 83 (77–89) | 23f |
| Naïve bayes | 89 (84–93) | 0.95 (0.84–1.00) | 94 (83–99) | 87 (81–92) | 11d |
| Random forest | 84 (79–89) | 0.94 (0.84–1.00) | 96 (86–99) | 81 (74–87) | 23f |
| Deep neural network | 84 (79–89) | 0.95 (0.85–1.00) | 100 (93–100) | 80 (72–86) | 17 g |
| Support vector machine | 86 (80–90) | 0.97 (0.87–1.00) | 98 (89–100) | 82 (75–88) | 11d |
| Gradient boosting machine | 81 (75–86) | 0.94 (0.88–1.00) | 96 (86–99) | 76 (69–83) | 5e |
BUN blood urea nitrogen, CI confidence interval, CVP central venous pressure, DBP diastolic blood pressure, GCS Glascow Coma Score, HCT hematocrit, HGB hemoglobin, HR heart rate, MAP mean arterial pressure, MODS multiple organ dysfunction score, PLT platelet count, RR respiratory rate, SBP systolic blood pressure, SO2 oxygen saturation, TCO2 total CO2, and WBC white blood cell count.
*Area under the ROC curves are reported in fractions.
aMAP, RR, body temperature, GCS, WBC, HGB, HCT, PLT, Na+ , K+ , BUN, creatinine, BUN/creatinine, glucose, TCO2, and MODS.
bBody temperature, WBC, HGB, HCT, Na+ , K+ , BUN, creatinine, BUN/creatinine, and TCO2.
cRR, body temperature, GCS, WBC, HGB, HCT, PLT, Na+ , K+ , BUN, creatinine, BUN/creatinine, TCO2, and MODS.
dSBP, MAP, HR, TEMP, HCT, Na+ , K+ , BUN, BUN/creatinine, anion gap, and TCO2.
eHR, body temperature, HGB, BUN, and TCO2.
fSBP, DBP, MAP, CVP, RR, HR, body temperature, GCS, SO2, WBC, HGB, HCT, PLT, Na+ , K+ , Cl−, anion gap, BUN, creatinine, BUN/creatinine, glucose, TCO2, and MODS.
gMAP, HR, RR, TEMP, WBC, HGB, HCT, PLT, Na+ , K+ , BUN, creatinine, BUN/creatinine, glucose, anion gap, TCO2, and MODS.
First, using our traditional ML approach, comparisons (i.e., 4,540 models times 11 categories) were made for the generated 49,940 ML models. Within this approach, the best performing ML model used LR with 16 features (i.e., mean arterial pressure, respiratory rate, body temperature, GCS, WBC, HGB, HCT, PLT, Na+ , K+ , BUN, plasma creatinine, glucose, TCO2, and MODS) with an accuracy of 86% and an area under the ROC curve of 0.96.
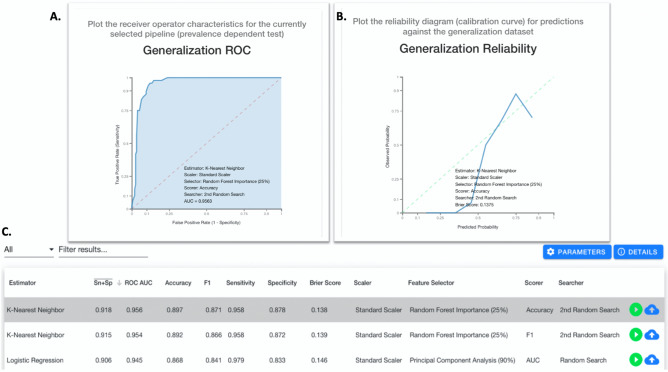
For MILO, 345,330 model comparisons were automatically produced for the ML models using the same training-generalization split for the data (Supplemental Data Table S2). Based on the MILO approach, a k-NN model using heart rate, body temperature, HGB, BUN, and TCO2 as features was found to be the best performer. The MILO k-NN accuracy was found to be 89.7% with an area under the ROC curve of 0.96 (95% CI 0.85–1.0). Clinical sensitivity and specificity were 95.8% and 87.8% respectively (Fig. 3). When using only ABA Consensus Guidelines consisting of body temperature, respiratory rate, heart rate, PLT, and glucose, the optimal model produced by MILO achieved an accuracy of 67.1% with an area under the ROC curve of 0.76 (95% CI 0.68–0.82) using the RF approach. Clinical sensitivity and specificity were 75.0% and 65.7% respectively. In contrast, applying Sepsis-3 criteria with ML exhibited an optimized accuracy of 74.5% using NB with an area under the ROC curve of 0.55 (95% CI 0.50–0.81), and sensitivity of 61.2% and specificity of 55.1%.
Figure 3.
MILO ROC for optimal ML model: screenshot of the optimal machine learning (ML) model generated by MILO based on logistic regression. (A) is the MILO read out for the receiver operator characteristic (ROC) curve using the selected ML model (i.e., logistic regression). (B) is the generalization/reliability plot for the selected ML model. (C) is the filtered list of ML models displaying other parameter such as average sensitivity and specificity (Sn + Sp “bar”), area under the curve (AUC) for the ROC analysis, F1 score, binary sensitivity and specificity, Brier Score, scaler used, feature selector used, scorer used, and searcher used.
Discussion
Burn sepsis exhibits high mortality attributed, in part, to delayed recognition of infection. Studies have indicated traditional sepsis criteria are not suitable for this unique population1. Recent investigations suggest ML may be able to identify unique pathologic patterns not recognized by the “human eye” and enhance the performance of traditional biomarkers for certain diseases (e.g., acute kidney injury)8,11. In this article, we report the use of ML for predicting sepsis in this unique high-risk burn population. Study data was based on a unique ABA sponsored multicenter randomized controlled trial that ensured complete and high-quality data for the entirety of each patient’s ICU stay.
Burn sepsis recognition is challenged by the underlying hypermetabolic and inflammatory response that persists for days or months following injury1. Using a combination of heart rate, body temperature, HGB, BUN, and TCO2, k-NN was able to predict burn sepsis with high accuracy, sensitivity, and specificity—exceeding the performance of both the ABA Consensus Guidelines and Sepsis-3 criteria reported in literature. These findings are clinically significant, since this is the first time burn sepsis has been detected with this level of accuracy, sensitivity, and specificity. Interestingly, PLT, which is often included in burn sepsis criteria was excluded in our optimal ML algorithm. This observation may be due to PLT representing a late-stage indicator of burn sepsis, while inclusion of HGB and TCO2 may be attributed to hematological derangements and acid–base/electrolyte disturbances encountered during sepsis13–15. Interestingly, when MILO was limited to the ABA Consensus Guidelines or Sepsis-3 criteria, the optimal model performed marginally better than traditional statistics. These findings highlight the capacity for ML, and especially MILO’s ability to run an exhaustive search of models and across all collected variables to identify clinically significant patterns not observed by the human eye or by traditional statistics.
In the end, the ABA Consensus Guidelines provides a framework to improve burn sepsis recognition, however its clinical performance remains limited with an area under the ROC curve of 0.62 based on current literature16. Studies by Mann-Salinas et al. proposed novel predictors that could outperform the ABA Consensus Guidelines with an area under the ROC curve of 0.78. We were able to improve predictive performance using traditional statistics in our study and achieve an area under the ROC curve of 0.88. Sepsis-3 has also been investigated, however its heavy reliance on the SOFA score remains controversial in multiple populations including in burned patients. It must be noted that a new study recently suggested Sepsis-3 may be superior to ABA Consensus Guidelines5, however, performance appears to still lag behind AI/ML models produced in this study.
Despite the promising performance of various ML algorithms reported in literature for in vitro diagnostic applications9, the accessibility of ML in healthcare remains limited to facilities employing experienced programmers and data science experts17. Unique to this study, we employed an automated ML application, called MILO. MILO eliminates these limitations and improves the accessibility and feasibility of ML-based data sciences, and more importantly, helps identify the optimal ML models in an unbiased and transparent way. No assumptions are made on a given dataset using MILO and programming expertise is not necessary. The benefits of using an automated ML platform not only accelerates development of new predictive models (400 vs. 20 h), but also identifies the best performing algorithm after running a very large feature and ML model combinations. Ultimately, MILO serves to limit selection and method bias to make ML more accessible to the general public.
Limitations of this study include its retrospective nature and the use of protocolized clinical trial data from another study. In particular, clinical trial data is more controlled compared to what is encountered on a routine basis, however, the data does offer the benefit of having been vetted for accuracy and completeness. Reproducibility of AI is also a concern due to differences in populations, test methodology, differences in test practices across institutions/disciplines, and ML methods9,18. Thus, the generalizability of the data is limited to this retrospective dataset, therefore additional studies are needed to further verify performance and assess how these ML algorithms perform under real-world conditions prior to any clinical implementation.
Conclusions
Sepsis contributes to high mortality in the severely burned patient population. Existing sepsis criteria are not well suited to differentiate between the host response to infection versus burn injury. Machine learning offers unique opportunities to exploit and enable sophisticated computer-based pattern recognition tools to predict sepsis in the burn patient population. Traditional methods for creating ML is both time consuming and relies on experienced programmers to work through a large permutation of features across a range of models. The deployment of MILO not only accelerates the development of ML models, but quickly helps identify optimal features and algorithms for burn sepsis prediction. Future multicenter studies are needed to refine the performance and confirm generalizability of ML models. Next steps include deploying the MILO burn sepsis algorithm at our institution to observationally test its accuracy when using prospective data.
Supplementary information
Acknowledgements
Study data was provided by a United States Army Medical Research and Material Command (USAMRMC) Combat Casualty Care Grant (Award Number W81XWH-09-2-0194) grant (Study PI: Tran). We thank the American Burn Association, UC Davis Data Coordination Center, Mr. Keith Brock, and Dr. Daniel Mecozzi for their support. MILO development was independent of the USAMRMC grant and is the sole property of the Regents of the University of California (Inventors: Rashidi, Albahra, and Tran).
Author contributions
N.T. serves as study PI and including providing fund for the overall study. Performed statistical analysis of the study data as well as supporting the ML algorithm development with Dr. Rashidi. Significantly contributed to the writing, review, and editing of the manuscript. Generated Figures for the manuscript. S.A. is co-investigator/inventor for the MILO component. He also helped develop MILO and significantly contributed to the development of this paper. T.P. is site-PI for one of the study sites that contributed significantly to subject enrollment. Significantly contributed to the development of this paper. J.H. is site-PI for one of the study sites that contributed significantly to subject enrollment. Significantly contributed to the development of this paper. D.G. is Chief of the Burn Division at the core study site where patients were enrolled. Significantly contributed to the parent grant, as well as burn sepsis components of the manuscript. T.L.P. provided burn/trauma clinical expertise. Significantly contributed to the writing, review, and editing of the manuscript. J.W. provided clinical informatics expertise. Significantly contributed to the writing, review, and editing of the manuscript. H.R. developed and validated the ML algorithms used in the manuscript. Significantly contributed to the writing, review, and editing of the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nam K. Tran, Email: nktran@ucdavis.edu
Hooman H. Rashidi, Email: hrashidi@ucdavis.edu
Supplementary information
is available for this paper at 10.1038/s41598-020-69433-w.
References
- 1.Greenhalgh DG, Saffle JR, Holmes JH, 4th, Gamelli RL, Palmieri TL, Horton JW, et al. American Burn Association consensus conference to define sepsis and infection in burns. J. Burn Care Res. 2007;28(6):776–790. doi: 10.1097/BCR.0b013e3181599bc9. [DOI] [PubMed] [Google Scholar]
- 2.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. Definitions for sepsis and organ failure and guidelines for use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 3.Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving sepsis champaign: International guidelines for management of severe sepsis and septic shock, 2012. Intens. Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shankar-Hari M, Phillips GS, Levy ML, Seymour CW, Liu VX, Deuschman CS, et al. Developing a new definition and assessing new clinical criteria for septic shock: For the third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:775–787. doi: 10.1001/jama.2016.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan J, Hill WF, Rehou S, Pinto R, Shahrokhi JMG. Sepsis criteria versus clinical diagnosis of sepsis in burn patients: A validation of current sepsis scores. Surgery. 2018;164:1241–1245. doi: 10.1016/j.surg.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon J, Kym D, Hur J, Kim Y, Yang HT, Yim H, et al. Comparative usefulness of Sepsis-3, burn sepsis, and conventional sepsis criteria in patients with major burns. Crit. Care Med. 2018;46:e656–e662. doi: 10.1097/CCM.0000000000003144. [DOI] [PubMed] [Google Scholar]
- 7.Sartelli M, Kluger Y, Ansaloni L, Hardcastle TC, Rello J, Watkins RR, et al. Raising concerns about the Sepsis-3 definitions. World J. Emerg. Surg. 2018;13:6. doi: 10.1186/s13017-018-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran NK, Sen S, Palmieri TL, Lima K, Falwell S, Wajda J, et al. Artificial intelligence and machine learning for predicting acute kidney injury in severely burned patients: A proof of concept. Burns. 2019;45:1350–1358. doi: 10.1016/j.burns.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 9.Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial intelligence and machine learning in pathology: The present landscape of supervised learning methods. Acad. Pathol. 2019 doi: 10.1177/2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran, N. K., Pham, T. N., Holmes, I. V. J., Greenahlgh, D. G., Palmieri, T. L. Near-patient quantitative molecular detection of Staphylococcus aureus in severely burned adult patients. A pragmatic randomized study (2019) [submitted].
- 11.Rashidi HH, Sen S, Palmieri TL, Blackmon T, Wajda J, Tran NK. Early recognition of burn- and trauma-related acute kidney injury: A pilot comparison of machine learning techniques. Sci. Rep. 2020;10:205. doi: 10.1038/s41598-019-57083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rashidi, H. H., Albahra, S., Tran, N. K. Regents of the University of California. United States Provisional Patent Application, November 20, 2019.
- 13.Lawrence C, Atac B. Hematological changes in massive burn injury. Crit. Care Med. 1992;20:1284–1288. doi: 10.1097/00003246-199209000-00015. [DOI] [PubMed] [Google Scholar]
- 14.Sen S, Tran NK, Chan B, Palmieri TL, Greenhalgh DG, Cho K. Sodium variability is associated with increased mortality in severe burn injury. Burns Trauma. 2017;5:34. doi: 10.1186/s41038-017-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen S, Hsei L, Tran N, Romanowski K, Palmieri TL, Greenhalgh DG, et al. Early clinical complete blood count changes in sever burn injuries. Burns. 2019;45:97–102. doi: 10.1016/j.burns.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Mann-Salinas EA, Baun MM, Meininger JC, Murray CK, Arden JK, Wolf SE, et al. Novel predictors of sepsis outperform the American Burn Association sepsis criteria in the burn intensive care unit patient. J. Burn Care Res. 2013;34(1):31–43. doi: 10.1097/BCR.0b013e31826450b5. [DOI] [PubMed] [Google Scholar]
- 17.Health Management website. https://healthmanagement.org/c/it/news/ai-skills-shortage-stunting-healthcare. Accessed 6 Jan 2020.
- 18.Beam AL, Manrai AJ, Ghassemi M. Challenges to the reproducibility of machine learning models in health care. JAMA. 2020;323:305. doi: 10.1001/jama.2019.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.