Key Points
Question
How does a deep learning system for assessing prostate biopsy specimens compare with interpretations determined by specialists in urologic pathology and by general pathologists?
Findings
In a validation data set of 752 biopsy specimens obtained from 2 independent medical laboratories and a tertiary teaching hospital, this study found that rate of agreement with subspecialists was significantly higher for the deep learning system than it was for a cohort of general pathologists.
Meaning
The deep learning system warrants evaluation as an assistive tool for improving prostate cancer diagnosis and treatment decisions, especially where subspecialist expertise is unavailable.
Abstract
Importance
For prostate cancer, Gleason grading of the biopsy specimen plays a pivotal role in determining case management. However, Gleason grading is associated with substantial interobserver variability, resulting in a need for decision support tools to improve the reproducibility of Gleason grading in routine clinical practice.
Objective
To evaluate the ability of a deep learning system (DLS) to grade diagnostic prostate biopsy specimens.
Design, Setting, and Participants
The DLS was evaluated using 752 deidentified digitized images of formalin-fixed paraffin-embedded prostate needle core biopsy specimens obtained from 3 institutions in the United States, including 1 institution not used for DLS development. To obtain the Gleason grade group (GG), each specimen was first reviewed by 2 expert urologic subspecialists from a multi-institutional panel of 6 individuals (years of experience: mean, 25 years; range, 18-34 years). A third subspecialist reviewed discordant cases to arrive at a majority opinion. To reduce diagnostic uncertainty, all subspecialists had access to an immunohistochemical-stained section and 3 histologic sections for every biopsied specimen. Their review was conducted from December 2018 to June 2019.
Main Outcomes and Measures
The frequency of the exact agreement of the DLS with the majority opinion of the subspecialists in categorizing each tumor-containing specimen as 1 of 5 categories: nontumor, GG1, GG2, GG3, or GG4-5. For comparison, the rate of agreement of 19 general pathologists’ opinions with the subspecialists’ majority opinions was also evaluated.
Results
For grading tumor-containing biopsy specimens in the validation set (n = 498), the rate of agreement with subspecialists was significantly higher for the DLS (71.7%; 95% CI, 67.9%-75.3%) than for general pathologists (58.0%; 95% CI, 54.5%-61.4%) (P < .001). In subanalyses of biopsy specimens from an external validation set (n = 322), the Gleason grading performance of the DLS remained similar. For distinguishing nontumor from tumor-containing biopsy specimens (n = 752), the rate of agreement with subspecialists was 94.3% (95% CI, 92.4%-95.9%) for the DLS and similar at 94.7% (95% CI, 92.8%-96.3%) for general pathologists (P = .58).
Conclusions and Relevance
In this study, the DLS showed higher proficiency than general pathologists at Gleason grading prostate needle core biopsy specimens and generalized to an independent institution. Future research is necessary to evaluate the potential utility of using the DLS as a decision support tool in clinical workflows and to improve the quality of prostate cancer grading for therapy decisions.
To ascertain whether an assistive tool can improve prostate cancer diagnosis and treatment decisions, this study compares the ability of a deep learning system with those of general pathologists and urologic subspecialist pathologists to grade diagnostic prostate biopsy specimens.
Introduction
Prostate cancer is a leading cause of morbidity and mortality for men.1 Its treatment is determined based largely on the pathologic evaluation of a prostate biopsy,2 an imperfect diagnostic tool. The heterogeneous tumor growth patterns observed in a biopsy are characterized by the Gleason grading system in terms of their degree of differentiation (ranging from Gleason pattern 3, representing well-differentiated glands, to Gleason pattern 5, representing poorly differentiated cells). Ultimately, biopsy specimens are categorized into Gleason grade groups (GG) based on the proportions of the Gleason patterns present in a biopsy, with higher GG indicating greater clinical risk.
These GGs are inherently subjective by virtue of relying on the visual assessment of cell differentiation and Gleason pattern predominance. Consequently, it is common for different pathologists to assign a different GG to the same biopsy (30%-50% discordances).3,4,5,6,7,8 In general, pathologists with urologic subspeciality training show higher rates of interobserver agreement than general pathologists,9 and reviews by experts lead to more accurate risk stratification than reviews by less experienced pathologists.10,11 Because important treatment decisions rely on assessment of prostate biopsy specimens and there is limited availability of expert subspecialists, the development of an automated system for assessing prostate biopsy specimens with expert-level performance could help improve the clinical utility of the prostate biopsy.
We developed a deep learning system (DLS) for reading digitized prostate biopsy specimen sections with the intent of achieving performance comparable to expert subspecialists. We evaluated the rate of model agreement with the majority opinion of several experienced subspecialists and compared this performance to a panel of general pathologists who independently reviewed the same biopsy specimens.
Methods
Data Sets
Deidentified digitized images of formalin-fixed paraffin-embedded prostate needle core biopsy specimens were obtained from 4 sources, each with independent tissue processing and staining: 2 independent medical laboratories (ML1 and ML2), a tertiary teaching hospital, and a university hospital. The ML1, tertiary teaching hospital, and university hospital biopsy specimens were used for DLS development, and the ML1, ML2, and tertiary teaching hospital biopsy specimens were used for validation. Biopsy specimens from ML2 served as an external validation data set; these specimens were used for independent validation only and not used for DLS development (Table 1). Additional details are presented in the Slide Preparation and Image Digitization section of the eMethods in the Supplement. Ethics approval for the use of these deidentified slides in this study was granted by the Naval Medical Center San Diego Institutional Review Board, which also waived the need for obtaining informed patient consent because the data were deidentified. No patients received compensation or were offered any incentive for participating in this study.
Table 1. Characteristics of the Validation Sets.
| Source or diagnosis | Entire validation set, No.a | Total | ||
|---|---|---|---|---|
| ML1 | Tertiary teaching hospital | External validation set (ML2)b | ||
| Biopsy specimens from each source | 387 | 52 | 371 | 810 |
| Biopsy specimens excluded due to image quality, poor staining, or artifacts impeding diagnosis | 1 | 6 | 48 | 55 |
| Biopsy specimens excluded due to presence of ungradable variants | 2 | 0 | 1 | 3 |
| Cases included (1 biopsy specimen per case) | 384 | 46 | 322 | 752 |
| Nontumor | 94 | 13 | 147 | 254 |
| Tumor-containing | 290 | 33 | 175 | 498 |
| Grade group | ||||
| 1 | 147 | 24 | 76 | 247 |
| 2 | 72 | 6 | 44 | 122 |
| 3 | 46 | 2 | 22 | 70 |
| 4-5 | 25 | 1 | 33 | 59 |
The validation sets contain prostate core biopsy cases from 3 institutions: a large tertiary teaching hospital and 2 medical laboratories (ML1 and ML2) in the United States. A representative core specimen was selected from each case. Despite overlap in the data source for ML1 and the tertiary teaching hospital between the development and validation data sets, the cases and biopsy specimens did not overlap.
The deep learning system was developed using data from ML1 and the tertiary teaching hospital sources, but not from ML2. Thus ML2 represents an external validation data set.
Each specimen was randomly assigned to either the development or validation sets such that there was no overlap in slides between the development and validation sets. One specimen per case was selected for inclusion in the study, with selection of a tumor-containing specimen where available. Specimens with nongradable prostate cancer variants or with quality issues preventing diagnosis were excluded from the study. Additional details including the splitting of the development set for DLS training and tuning are presented in Table 1 and in eTable 1 in the Supplement.
Pathologic Examination of Prostate Biopsy Specimens
All pathologists participating in this study, including the general pathologists and urologic subspecialists (M.B.A., A.J.E., J.W.S., C.C., T.K., A.R.S., M.Z., R.A., and P.A.H.), were US board-certified or Canadian board-certified, and reviewed the pathology slides for each biopsy in a manner consistent with the International Society of Urological Pathology 2014 and College of American Pathologists guidelines with no time constraint.12,13 If the specimen did not contain Gleason-gradable adenocarcinoma, it was classified as nontumor. Otherwise, to assign the final GG, the pathologists provided the relative amount of tumor corresponding to each Gleason pattern, specifically, the percentage of each that was considered Gleason pattern 3, 4, or 5. Gleason patterns 1 and 2 are not used in contemporary Gleason grading. The corresponding GG (GG1, GG2, GG3, or GG4-5) was then derived from the relative proportions of the Gleason patterns (Box).13 Because of their low incidence and often similar treatment implications, GG4 and GG5 were collapsed into a single group.
Box. Simplified 5-Step Procedure of the Gleason Grading System.
Biopsy review involves the following 5 steps
Identify whether a tumor is present.
When a tumor is present, categorize regions of the tumor as 1 of 3 Gleason patterns: 3, 4, or 5.
Quantify the relative amounts of each pattern.
Sum the top 2 most prevalent patterns to determine the Gleason score. Under certain conditions, a third-most prevalent pattern is also used at this step.
Map the Gleason score to a grade group. Both the Gleason score and grade group are part of standard reporting. The grade group system was designed to facilitate mapping of Gleason scores into discrete prognostic groups.12
Biopsy Specimen Reviews
Reviews were collected for 2 purposes: first for DLS development (training and tuning) and second for assessment of the DLS system performance using a separate validation data set. Biopsy specimen reviews for DLS development are detailed in the eMethods in the Supplement. For DLS validation, 6 urologic subspecialists reviewed the validation set (eFigure 1A in the Supplement). The subspecialists (M.B.A., A.J.E., T.K., M.Z., R.A., and P.A.H.) represented 5 institutions and had 18 to 34 years of clinical experience after residency (mean, 25 years).
To reduce potential Gleason pattern ambiguity due to issues such as tangential cuts of the specimen, 2 adjacent sections (levels) of the specimens were provided to the subspecialists. These 3 levels were made available to the pathologists for establishing the reference standard, but not made available to the DLS, which interpreted only the middle section of each specimen. Furthermore, 1 additional section per specimen was stained with the PIN-4 immunohistochemistry cocktail (P504S plus p63 plus high molecular weight cytokeratin) to help the subspecialists identify cancer.
For each of the 752 biopsy specimens in the validation set, reviews were performed by 2 of the 6 aforementioned expert subspecialists. A third subspecialist reviewed the specimens when there were discordances between the first 2 subspecialists (176 specimens [23%]). For cases without a majority opinion after 3 independent reviews (13 cases [1.7%]), the median classification was used. We then evaluated the accuracy of the DLS compared with this majority opinion of the subspecialists for each biopsy.
Biopsy Specimen Reviews by General Pathologists for Comparison
To measure the rate of agreement between the general pathologists and subspecialists, each biopsy specimen in the validation set was reviewed by several (median, 3, range, 1-6) US board-certified pathologists from the cohort of 19 participating in this study. The median number of biopsy specimens reviewed by each general pathologist was 84 (range, 41-312). To simulate routine clinical workflow, these pathologists had access to 3 sections per specimen, but not the immunohistochemistry-stained section.
Deep Learning System
The DLS operates in 2 stages, mimicking pathologists’ mental workflow by first characterizing individual regions into Gleason patterns, followed by assigning a GG to the entire biopsy specimen (eFigure 1B in the Supplement). To train the first stage of the DLS, we collected detailed region-level annotations from prostatectomy and biopsy specimens, which generated 114 million labeled image patches. The second stage of the DLS was trained using 580 biopsy specimen reviews (eTable 1 in the Supplement). Additional details, such as how the DLS neural network architecture was adapted for Gleason grading via Neural Architecture Search14 and refined from the system used in prior work15 as well as hyperparameter tuning16 using the development set, are available in the Deep Learning System section of the eMethods in the Supplement.
Statistical Analysis
Prostate biopsy specimen interpretation involves first determining the presence or absence of prostate cancer. To evaluate the performance of the DLS for tumor detection, we calculated the DLS agreement rate with the subspecialists’ majority opinion for tumor vs nontumor classification. For comparison, we also computed the agreement rate of the general pathologists with the subspecialists’ majority opinion for tumor vs nontumor classification. To represent each general pathologist equally, we calculated each individual general pathologist’s agreement rate with subspecialists separately and calculated a mean rate across the 19 general pathologists.
When a tumor is identified in the specimen, the next step of Gleason grading involves characterizing the Gleason pattern of each tumor region and estimating the proportion of each Gleason pattern present in the specimen (Box). To evaluate the ability of the DLS to quantitate Gleason patterns in the tumors, we computed the mean difference (mean absolute error) between the DLS-provided quantitation results and the mean of the subspecialist quantitation results for each Gleason pattern. For comparison, we also computed the mean absolute error between the general pathologists’ Gleason pattern quantitation results and the mean of the subspecialists’ quantitation results.
The final step of Gleason grading involves determining the top 2 most prevalent Gleason patterns in each specimen, which determines the GG (Box). For evaluating the DLS in determining the GG for prostate biopsy specimens, we calculated the exact rate of agreement of the DLS categorization with the majority opinion of the subspecialists in categorizing specimens as nontumor, GG1, GG2, GG3, or GG4-5. For comparison, we also calculated the general pathologists’ rate of agreement with the majority opinion of the subspecialists. Similar to the tumor vs nontumor evaluation, we calculated each individual general pathologist’s agreement rate with subspecialists separately and calculated the mean rate across the 19 general pathologists. We additionally performed several subanalyses, which are detailed in the Statistical Analysis section of the eMethods in the Supplement.
Finally, we conducted receiver operating characteristic curve analysis at 2 clinically meaningful decision thresholds: GG1 vs GG2-5 (representing the clinical threshold for potential eligibility for active surveillance vs prostatectomy or definitive treatment17,18) and GG1-2 vs GG3-5 (because some cases classified as GG2 with a low percentage of Gleason pattern 4 may still be managed with active surveillance17,18).
Confidence intervals for all evaluation metrics were computed using a bootstrap approach by sampling specimens with replacement, with 1000 iterations. All statistical tests were 2-sided (see Statistical Analysis in the eMethods of the Supplement), and P < .05 was considered statistically significant. No adjustment was made for multiple comparisons. These analyses were performed using Python, version 2.7.6, and the scikit-learn library, version 0.20.0.19
Results
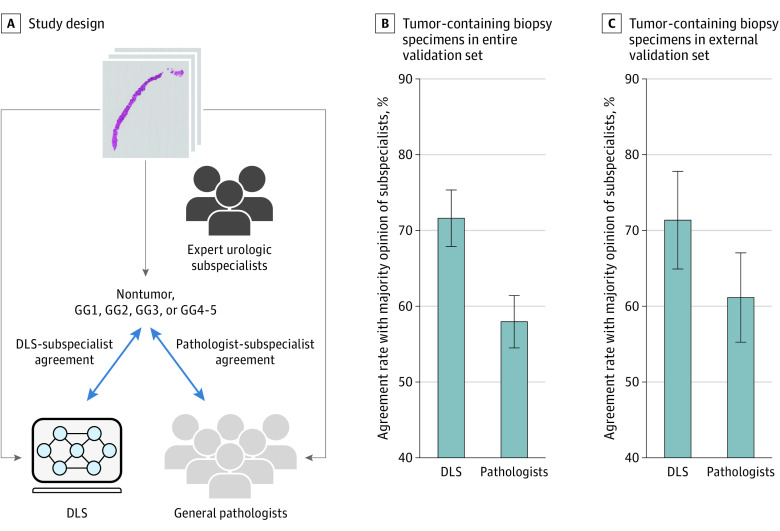
Evaluation was performed using an independent validation set from 3 institutions (752 biopsy specimens, 1 specimen per case) (Table 1), each reviewed by at least 2 expert subspecialists (3 subspecialists when there was discordance between the first 2). Using these data, we evaluated the performance of the DLS for tumor detection, Gleason pattern quantitation, and GG classification (Figure 1A).
Figure 1. Comparison of deep learning system (DLS) and Pathologist Agreement Rates With Subspecialists at Gleason Grading of Tumor-Containing Biopsy Specimens.
A, Subspecialists review every biopsy to determine its grade group (GG) (see Box and Methods). Next, those GG determinations are compared with those of the DLS and the general pathologists. B, Agreement rates with subspecialists for the DLS and pathologists across all 498 tumor-containing biopsy specimens. C, Subanalysis considering 175 tumor-containing biopsy specimens from only the external validation set (medical laboratory 2). Because every pathologist reviewed only a subset of the cases, to represent every pathologist equally, the agreement rate shown for the general pathologists is the mean across all general pathologists. For the subanalysis presented in panel C, pathologists who conducted fewer than 20 reviews were excluded to avoid skewing the results (applied to 4 pathologists). Error bars represent 95% CIs.
Tumor Detection
In distinguishing 752 biopsy specimens containing tumor from those without tumor, the rate of agreement with subspecialists was similar for the DLS and for general pathologists (DLS, 94.3%; 95% CI, 92.4%-95.9% vs pathologists, 94.7%; 95% CI, 92.8%-96.3%; P = .58). The DLS detected tumors more often than general pathologists, at the cost of more false-positives (Table 2). Of the false-positives committed by the DLS, one-third were noted by subspecialists as precancerous: high-grade prostatic intraepithelial neoplasia (HGPIN) or atypical small acinar proliferation (ASAP). The remaining false-positives tended to occur on small artifact-containing tissue regions (median tissue area called as tumor in these cases, 1%).
Table 2. Agreement Rates of the DLS and General Pathologists With the Subspecialists’ Majority Opinion at 3 Clinically Important Decision Cutoffsa.
| Clinical task, evaluation metric | % (95% CI) | |
|---|---|---|
| DLS | General pathologist | |
| Nontumor vs tumor determination (n = 752) | ||
| Agreement with subspecialist majority opinion | 94.3 (92.4-95.9) | 94.7 (92.8-96.3)b |
| Sensitivity | 95.5 (93.7-96.8)b | 92.8 (90.0-95.1) |
| Specificity | 91.7 (88.2-94.6) | 97.0 (95.1-98.6)b |
| Grading of tumor-containing biopsy specimensc | ||
| Agreement with subspecialist majority opinion for GG1 vs GG2-5 (n = 498) | 86.1 (83.1-89.2)b | 80.6 (77.9-83.5) |
| Agreement with subspecialist majority opinion for GG1-2 vs GG3-5 (n = 498) | 92.8 (90.8-94.9)b | 86.0 (83.2-88.5) |
Abbreviations: DLS, deep learning system; GG, grade group.
Similar to Figure 1, the agreement rate of the general pathologists represents the mean rate across all general pathologists.
The higher value in the row.
Agreement on 2 Gleason grading thresholds.
Gleason Pattern Quantitation
The DLS Gleason pattern quantitation error was lower than that of general pathologists across all patterns (Table 3). In particular, on GG2 slides (n = 122), where small differences in pattern 4 can substantially alter patient prognosis and treatment,17 the DLS quantitation error rate was substantially lower than that of the general pathologists (DLS, 12.0%; 95% CI, 10.4%-13.6% vs pathologists, 22.0%; 95% CI, 19.6%-24.6%; P < .001).
Table 3. Mean Absolute Difference in Gleason Pattern Quantitation Relative to Subspecialistsa.
| Gleason pattern | No. | Subspecialist discordance, % (95% CI) | |
|---|---|---|---|
| Deep learning system | Pathologist | ||
| 3 (Tumor-containing specimens) | 498 | 9.2 (8.0-10.5)b | 14.0 (12.4-15.6) |
| 4 (Tumor-containing specimens) | 498 | 10.0 (8.6-11.2)b | 16.3 (14.6-18.1) |
| 5 (Tumor-containing specimens) | 498 | 1.5 (0.9-2.1)b | 3.2 (2.2-4.3) |
| 4 (Grade group 2 specimens only) | 122 | 12.0 (10.4-13.6)b | 22.0 (19.6-24.6) |
Gleason pattern quantitation reflects the proportion of tumor in each biopsy specimen that is characterized as each Gleason pattern. The mean absolute differences in Gleason pattern quantitation are measured against the mean of subspecialist quantitation results for all tumor-containing biopsy specimens (rows 1-3) or grade group 2 biopsy specimens only (row 4).
Lower absolute differences (higher agreement rate in Gleason pattern quantitation).
Grading Tumor-Containing Biopsy Specimens
For Gleason grading of tumor-containing biopsy specimens (n = 498), the rate of DLS agreement with the subspecialists (71.7%; 95% CI, 67.9%-75.3%) was significantly higher than the general pathologist agreement rate with subspecialists (58.0%; 95% CI, 54.5%-61.4%) (P < .001) (Figure 1B). The DLS outperformed 16 of the 19 general pathologists in this comparison (eTable 3 in the Supplement).
In a subanalysis of biopsy specimens from the external validation set (ML2, n = 175), the rate of DLS agreement with subspecialists remained significantly higher than the rate of general pathologist agreement with subspecialists (71.4%; 95% CI, 65.7%-77.7% vs 61.2%; 95% CI, 55.7%-67.0%; P = .01) (Figure 1C; eTables 4 and 5 in the Supplement; additional subanalyses and sensitivity analyses are provided in eFigures 5 and 6 and in eTables 7, 9, and 10 in the Supplement).
We further examined several clinically important thresholds on 498 tumor-containing cases (Table 2; eFigure 3 in the Supplement). The rate of agreement with subspecialists was higher for the DLS than for the general pathologists at distinguishing GG1 vs GG2-5 cases, a threshold with important implications for active surveillance vs definitive treatment (DLS: 86.1%; 95% CI, 83.1%-89.2% vs general pathologists: 80.6%, 95% CI, 77.9%-83.5%; P < .001). Results were similar for distinguishing GG1-2 vs GG3-5 cases (Table 2). The receiver operating characteristic curve analysis at these GG thresholds is shown in eFigure 3 in the Supplement.
The contingency tables comparing GG classification by the DLS and by the general pathologists relative to the subspecialist majority opinion are provided in eTable 2 in the Supplement. Most of the improvement in GG accuracy by the DLS was due to reduced overgrading. On tumor-containing cases, pathologists had a 25.7% frequency of overgrading vs 8.9% overgrading by the DLS. By contrast, the DLS was slightly more likely to undergrade tumor-containing cases relative to specialists (frequency of undergrading: by pathologists, 14.7% vs by the DLS, 19.6%).
DLS Grading Examples
Figure 2 and eFigure 2 in the Supplement contain example visualizations of the DLS’s precise, interpretable glandular-level Gleason grading. They illustrate the potential of the DLS to be helpful in assisting pathologists in tumor detection, grading, and Gleason pattern quantitation.
Figure 2. Illustrative Concept of How Deep Learning System (DLS) Results May Be Presented to a Pathologist.
These cases were graded by both the DLS and subspecialists as grade groups 1 (A), 2 (B), and 3 (C). The DLS provides both a glandular-level Gleason pattern categorization and a biopsy-level Gleason score and grade group. Left column represents low-power magnification view of the Gleason pattern categorization; middle column, 10 × magnification of the indicated area from the left column; right column, the DLS-generated Gleason score and Gleason pattern quantitation. In the left column, green represents DLS-categorized Gleason pattern 3; yellow, DLS-categorized Gleason pattern 4.
Discussion
We have presented a system for Gleason grading prostate biopsy specimens with a rigorous evaluation involving numerous experienced urologic subspecialists from diverse backgrounds, having a mean of 25 years of experience, with access to several histologic sections and immunohistochemical-stained sections for every specimen. First, the DLS showed similar overall tumor detection rates compared with general pathologists, by catching more cases of tumor than general pathologists at the cost of some false-positives. This trade-off suggests that the DLS could help alert pathologists to tumors that may otherwise be missed20,21 while relying on pathologist judgment to overrule false-positive categorizations on small tissue regions. Second, the DLS showed better agreement rates with subspecialists than pathologists did for Gleason pattern quantitation, which is an important prognostic signal and independent predictor of biochemical recurrence22,23 and part of recommended reporting by the College of American Pathologists, International Society of Urological Pathology, World Health Organization, and European Association of Urology guidelines.12,13,24,25 Third, in summarizing the overall GG for the biopsy specimens (which is derived from the proportions of Gleason patterns present in the specimen and ultimately used in risk stratification with the National Comprehensive Cancer Network guidelines), the DLS showed significantly greater agreement rates with subspecialists than general pathologists did. Finally, the rate of agreement of the DLS with subspecialists on an external validation set remained similar, suggesting DLS robustness to interlaboratory and patient cohort differences.
Over the years, prostate cancer treatment has evolved such that the role of conservative management has been recognized in men with low-risk disease. In particular, several trials have shown the safety of active surveillance compared with radical prostatectomy or radiation therapy in carefully selected patients with localized prostate cancer.26,27,28 In this decision-making process, guidelines endorsed by the American Society of Clinical Oncology recommend consideration of both the GG and relative amount of Gleason pattern 4.17,18 Owing to the recognized interobserver variability in Gleason grading, intradepartmental consults have been recommended to improve consistency and quality of care.29,30 In this regard, the DLS could function as a valuable decision support tool when deciding between GGs for patients with localized disease, with important downstream implications on treatment.
A DLS such as this could therefore create efficiencies for health care systems by improving consistency of grading, reducing the consultation-associated costs and turnaround delays, and potentially decreasing treatment-related morbidity for men with low-risk disease. In particular, the DLS was substantially less likely to overgrade (especially at the clinically important GG1 vs GG2 distinction) while being slightly more likely to undergrade cases than general pathologists, especially at higher GGs (eTable 2 in the Supplement). These findings suggest that DLS assistance could be particularly helpful in accurately identifying low-risk cases that are eligible for more conservative management. The exact implementation and benefit of using such a tool remains to be determined but must be guided by prospective validation studies that examine the influence on diagnostic reporting and patient outcomes.
The GG plays a pivotal role in patient treatment,26,27,28 and grading among subspecialists is substantially more concordant than grading among general pathologists, both in our study (eFigure 4 and eTables 3 and 6 in the Supplement) and in the literature.6,31 However, discordance remains even among subspecialists due to the inherent subjectivity and difficulty of Gleason grading. The subspecialists participating in the present study had at least a decade of urologic pathology experience and access to 3 levels and immunohistochemistry of each biopsy specimen in the validation set. These discordances highlight the need to further improve risk stratification for prostate cancer. One possibility is to develop systems to directly predict clinical risk with more precision than is possible by human graders. Such machine learning models could identify novel histoprognostic signals that are undiscovered or not evident to the human eye,32,33 and may help stratify patient risk in a manner similar to existing molecular tests.34,35
Other works have applied deep learning to Gleason grading.36,37,38,39,40 Ström et al39 trained and validated a DLS using biopsy specimens graded by the same urologic subspecialist (validation data set sizes: 1631 biopsy specimens from 246 men, and 330 biopsy specimens from 73 men) and additionally compared grading with 23 subspecialists on a smaller set of 87 biopsy specimens. Bulten et al40 validated a DLS on 550 biopsy specimens from 210 randomly selected patients from the same institution used for development, using consensus grades from 3 experienced subspecialists at 2 institutions, and further compared with 15 pathologists or trainees on a smaller set of 100 biopsy specimens. Our study improved on these efforts via substantial subspecialist-reviewed glandular annotations to enable gland-level Gleason grading for assistive visualizations and explainability (Figure 2); via a rigorous review process involving several subspecialists from different institutions as well as 3 specimen levels and immunohistochemistry samples for every case; through the use of a sizable, independent clinical data set for validation; and finally by assessment of Gleason pattern quantitation in addition to Gleason grading of biopsy specimens.
Limitations
This study has limitations. First, we used 1 biopsy specimen per case although each clinical case typically involves 12 to 18 biopsy specimens. Second, this study did not evaluate the correlation of the DLS Gleason grading with clinical outcomes, which would be less subjective than to subspecialist review. However, unlike a previous analysis on radical prostatectomies,15 such an analysis for biopsy specimens would be challenging due to confounding factors such as divergent treatment pathways based on the original diagnosis, tissue sampling variability inherent to small biopsy specimens, other clinical variables, and patient preferences. Third, the effect of rescanning the specimens on model performance will need to be evaluated in future work. Fourth, additional aspects such as nonadenocarcinoma prostate cancer variants or precancerous findings were not evaluated in this study.
Conclusions
To conclude, we have presented a DLS for Gleason grading of prostate biopsy specimens that is highly concordant with subspecialists and that maintained its performance on an external validation set. Future work will need to assess the diagnostic and clinical effect of the use of a DLS for increasing the accuracy and consistency of Gleason grading to improve patient care.
eMethods
eFigure 1. Overview of the Reviews Provided by Expert Subspecialists for the Validation Set and an Overview of the Deep Learning System
eFigure 2. Example Overcalls by the DLS on Biopsies Without Tumor
eFigure 3. Validation Set Performance for Gleason Grading and Tumor Detection
eFigure 4. Evaluation of Inter-Subspecialist Agreement
eFigure 5. Comparison of the Deep Learning System (DLS) With Individual Subspecialists on Tumor-Containing Slides
eFigure 6. Sensitivity of the DLS’s Gleason Grade Group Classification Accuracy to Differences in Gleason Pattern (GP) Thresholds for GP 3 vs 4, 4 vs 5, and 3 vs 5
eTable 1. Characteristics of the Development Set
eTable 2. Breakdown of Discordances (A) Between Subspecialists, (B) Between the Majority Opinion of Subspecialists and the Deep Learning System, DLS, and (C) Between the Majority Opinion of Subspecialists and General Pathologists
eTable 3. Individual Pathologist Gleason Scoring Agreement With Subspecialist Majority Opinion for the 19 Pathologists
eTable 4. DLS Performance For Gleason Grading Compared to the Majority Opinion of Subspecialists
eTable 5. Confusion Matrix (5 × 5 Contingency Table) Showing the Breakdown of Classifications for the DLS on Biopsies From ML2
eTable 6. Interpathologist Agreement on Tumor-Containing Slides
eTable 7. Sensitivity Analysis When Separating GG4 and GG5 into Separate Classification Categories
eTable 8. Hyperparameters for the Deep Learning System
eTable 9. Sensitivity of the DLS-Subspecialist Agreement in Tumor-Containing Cases to the Availability of Annotations From Each Individual Subspecialist
eTable 10. Sensitivity of the DLS-Subspecialist Agreement in Tumor-Containing Cases to the Availability of Annotations From Each Individual Subspecialist
eReferences
References
- 1.Litwin MS, Tan H-J. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317(24):2532-2542. doi: 10.1001/jama.2017.7248 [DOI] [PubMed] [Google Scholar]
- 2.Mohler JL, Antonarakis ES, Armstrong AJ, et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17(5):479-505. doi: 10.6004/jnccn.2019.0023 [DOI] [PubMed] [Google Scholar]
- 3.Veloso SG, Lima MF, Salles PG, Berenstein CK, Scalon JD, Bambirra EA. Interobserver agreement of Gleason score and modified Gleason score in needle biopsy and in surgical specimen of prostate cancer. Int Braz J Urol. 2007;33(5):639-646. doi: 10.1590/S1677-55382007000500005 [DOI] [PubMed] [Google Scholar]
- 4.Özdamar ŞO, Sarikaya S, Yildiz L, Atilla MK, Kandemir B, Yildiz S. Intraobserver and interobserver reproducibility of WHO and Gleason histologic grading systems in prostatic adenocarcinomas. Int Urol Nephrol. 1996;28(1):73-77. doi: 10.1007/BF02550141 [DOI] [PubMed] [Google Scholar]
- 5.Egevad L, Ahmad AS, Algaba F, et al. Standardization of Gleason grading among 337 European pathologists. Histopathology. 2013;62(2):247-256. doi: 10.1111/his.12008 [DOI] [PubMed] [Google Scholar]
- 6.Allsbrook WC Jr, Mangold KA, Johnson MH, et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: urologic pathologists. Hum Pathol. 2001;32(1):74-80. doi: 10.1053/hupa.2001.21134 [DOI] [PubMed] [Google Scholar]
- 7.Melia J, Moseley R, Ball RY, et al. A UK-based investigation of inter- and intra-observer reproducibility of Gleason grading of prostatic biopsies. Histopathology. 2006;48(6):644-654. doi: 10.1111/j.1365-2559.2006.02393.x [DOI] [PubMed] [Google Scholar]
- 8.Abdollahi A, Meysamie A, Sheikhbahaei S, et al. Inter/intra-observer reproducibility of Gleason scoring in prostate adenocarcinoma in Iranian pathologists. Urol J. 2012;9(2):486-490. [PubMed] [Google Scholar]
- 9.Kvåle R, Møller B, Wahlqvist R, et al. Concordance between Gleason scores of needle biopsies and radical prostatectomy specimens: a population-based study. BJU Int. 2009;103(12):1647-1654. doi: 10.1111/j.1464-410X.2008.08255.x [DOI] [PubMed] [Google Scholar]
- 10.Bottke D, Golz R, Störkel S, et al. Phase 3 study of adjuvant radiotherapy versus wait and see in pT3 prostate cancer: impact of pathology review on analysis. Eur Urol. 2013;64(2):193-198. doi: 10.1016/j.eururo.2013.03.029 [DOI] [PubMed] [Google Scholar]
- 11.van der Kwast TH, Collette L, Van Poppel H, et al. ; European Organisation for Research and Treatment of Cancer Radiotherapy and Genito-Urinary Cancer Groups . Impact of pathology review of stage and margin status of radical prostatectomy specimens (EORTC trial 22911). Virchows Arch. 2006;449(4):428-434. doi: 10.1007/s00428-006-0254-x [DOI] [PubMed] [Google Scholar]
- 12.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA; Grading Committee . The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40(2):244-252. [DOI] [PubMed] [Google Scholar]
- 13.Paner GP, Srigley JR, Zhou M, et al. Protocol for the examination of specimens from patients with carcinoma of the prostate gland. Protocol Posting Date June 2017. Accessed June 19, 2020. https://documents.cap.org/protocols/cp-malegenital-prostate-18protocol-4030.pdf
- 14.Bender G, Liu H, Chen B, et al. Can weight sharing outperform random architecture search? an investigation with TuNAS. IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Accessed June 19, 2020. http://openaccess.thecvf.com/content_CVPR_2020/papers/Bender_Can_Weight_Sharing_Outperform_Random_Architecture_Search_An_Investigation_With_CVPR_2020_paper.pdf
- 15.Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med. 2019;2:48. doi: 10.1038/s41746-019-0112-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Chen PC, Krause J, Peng L. How to read articles that use machine learning: users’ guides to the medical literature. JAMA. 2019;322(18):1806-1816. doi: 10.1001/jama.2019.16489 [DOI] [PubMed] [Google Scholar]
- 17.Chen RC, Rumble RB, Jain S. Active surveillance for the management of localized prostate cancer (Cancer Care Ontario Guideline): American Society of Clinical Oncology Clinical practice guideline endorsement summary. J Oncol Pract. 2016;12(3):267-269. doi: 10.1200/JOP.2015.010017 [DOI] [PubMed] [Google Scholar]
- 18.Morash C, Tey R, Agbassi C, et al. Active surveillance for the management of localized prostate cancer: Guideline recommendations. Can Urol Assoc J. 2015;9(5-6):171-178. doi: 10.5489/cuaj.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825-2830. [Google Scholar]
- 20.Liu Y, Kohlberger T, Norouzi M, et al. Artificial intelligence-based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Med. 2019;143(7):859-868. doi: 10.5858/arpa.2018-0147-OA [DOI] [PubMed] [Google Scholar]
- 21.Steiner DF, MacDonald R, Liu Y, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol. 2018;42(12):1636-1646. doi: 10.1097/PAS.0000000000001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauter G, Steurer S, Clauditz TS, et al. Clinical utility of quantitative Gleason grading in prostate biopsies and prostatectomy specimens. Eur Urol. 2016;69(4):592-598. doi: 10.1016/j.eururo.2015.10.029 [DOI] [PubMed] [Google Scholar]
- 23.Cole AI, Morgan TM, Spratt DE, et al. Prognostic value of percent Gleason grade 4 at prostate biopsy in predicting prostatectomy pathology and recurrence. J Urol. 2016;196(2):405-411. doi: 10.1016/j.juro.2016.01.120 [DOI] [PubMed] [Google Scholar]
- 24.Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO classification of tumours of the urinary system and male genital organs—part b: prostate and bladder tumours. Eur Urol. 2016;70(1):106-119. doi: 10.1016/j.eururo.2016.02.028 [DOI] [PubMed] [Google Scholar]
- 25.Habchi H, Mottet N. Management of prostate cancer: EAU guidelines on screening, diagnosis and local primary treatment. In: Bolla M, van Poppel H, eds. Management of Prostate Cancer: A Multidisciplinary Approach. Springer; 2017:399-411. doi: 10.1007/978-3-319-42769-0_26 [DOI] [Google Scholar]
- 26.Lane JA, Donovan JL, Davis M, et al. ; Protect study group . Active monitoring, radical prostatectomy, or radiotherapy for localised prostate cancer: study design and diagnostic and baseline results of the Protect randomised phase 3 trial. Lancet Oncol. 2014;15(10):1109-1118. doi: 10.1016/S1470-2045(14)70361-4 [DOI] [PubMed] [Google Scholar]
- 27.Wilt TJ. The Prostate Cancer Intervention Versus Observation Trial: VA/NCI/AHRQ Cooperative Studies Program #407 (PIVOT): design and baseline results of a randomized controlled trial comparing radical prostatectomy with watchful waiting for men with clinically localized prostate cancer. J Natl Cancer Inst Monogr. 2012;2012(45):184-190. doi: 10.1093/jncimonographs/lgs041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson E, Steineck G, Holmberg L, et al. ; SPCG-4 Investigators . Long-term quality-of-life outcomes after radical prostatectomy or watchful waiting: the Scandinavian Prostate Cancer Group-4 randomised trial. Lancet Oncol. 2011;12(9):891-899. doi: 10.1016/S1470-2045(11)70162-0 [DOI] [PubMed] [Google Scholar]
- 29.Chen RC, Rumble RB, Loblaw DA, et al. Active surveillance for the management of localized prostate cancer (Cancer Care Ontario Guideline): American Society of Clinical Oncology clinical practice guideline endorsement. J Clin Oncol. 2016;34(18):2182-2190. doi: 10.1200/JCO.2015.65.7759 [DOI] [PubMed] [Google Scholar]
- 30.Brimo F, Schultz L, Epstein JI. The value of mandatory second opinion pathology review of prostate needle biopsy interpretation before radical prostatectomy. J Urol. 2010;184(1):126-130. doi: 10.1016/j.juro.2010.03.021 [DOI] [PubMed] [Google Scholar]
- 31.Allsbrook WC Jr, Mangold KA, Johnson MH, Lane RB, Lane CG, Epstein JI. Interobserver reproducibility of Gleason grading of prostatic carcinoma: general pathologist. Hum Pathol. 2001;32(1):81-88. doi: 10.1053/hupa.2001.21135 [DOI] [PubMed] [Google Scholar]
- 32.Courtiol P, Maussion C, Moarii M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019;25(10):1519-1525. doi: 10.1038/s41591-019-0583-3 [DOI] [PubMed] [Google Scholar]
- 33.Wulczyn E, Steiner DF, Xu Z, et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE. 2020;15(6):e0233678. doi: 10.1371/journal.pone.0233678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marrone M, Potosky AL, Penson D, Freedman ANA. A 22 gene-expression assay, Decipher® (GenomeDx Biosciences) to predict five-year risk of metastatic prostate cancer in men treated with radical prostatectomy. PLoS Curr. 2015;7:7. doi: 10.1371/currents.eogt.761b81608129ed61b0b48d42c04f92a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay—a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. doi: 10.1186/1471-2164-14-690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucas M, Jansen I, Savci-Heijink CD, et al. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019;475(1):77-83. doi: 10.1007/s00428-019-02577-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301-1309. doi: 10.1038/s41591-019-0508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arvaniti E, Fricker KS, Moret M, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8(1):12054. doi: 10.1038/s41598-018-30535-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ström P, Kartasalo K, Olsson H, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 2020;21(2):222-232. doi: 10.1016/S1470-2045(19)30738-7 [DOI] [PubMed] [Google Scholar]
- 40.Bulten W, Pinckaers H, van Boven H, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 2020;21(2):233-241. doi: 10.1016/S1470-2045(19)30739-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eFigure 1. Overview of the Reviews Provided by Expert Subspecialists for the Validation Set and an Overview of the Deep Learning System
eFigure 2. Example Overcalls by the DLS on Biopsies Without Tumor
eFigure 3. Validation Set Performance for Gleason Grading and Tumor Detection
eFigure 4. Evaluation of Inter-Subspecialist Agreement
eFigure 5. Comparison of the Deep Learning System (DLS) With Individual Subspecialists on Tumor-Containing Slides
eFigure 6. Sensitivity of the DLS’s Gleason Grade Group Classification Accuracy to Differences in Gleason Pattern (GP) Thresholds for GP 3 vs 4, 4 vs 5, and 3 vs 5
eTable 1. Characteristics of the Development Set
eTable 2. Breakdown of Discordances (A) Between Subspecialists, (B) Between the Majority Opinion of Subspecialists and the Deep Learning System, DLS, and (C) Between the Majority Opinion of Subspecialists and General Pathologists
eTable 3. Individual Pathologist Gleason Scoring Agreement With Subspecialist Majority Opinion for the 19 Pathologists
eTable 4. DLS Performance For Gleason Grading Compared to the Majority Opinion of Subspecialists
eTable 5. Confusion Matrix (5 × 5 Contingency Table) Showing the Breakdown of Classifications for the DLS on Biopsies From ML2
eTable 6. Interpathologist Agreement on Tumor-Containing Slides
eTable 7. Sensitivity Analysis When Separating GG4 and GG5 into Separate Classification Categories
eTable 8. Hyperparameters for the Deep Learning System
eTable 9. Sensitivity of the DLS-Subspecialist Agreement in Tumor-Containing Cases to the Availability of Annotations From Each Individual Subspecialist
eTable 10. Sensitivity of the DLS-Subspecialist Agreement in Tumor-Containing Cases to the Availability of Annotations From Each Individual Subspecialist
eReferences