Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by the novel coronavirus named severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), has led to an unprecedented medical crisis. Current lack of data from experimental animals makes it difficult to understand the pathophysiological mechanisms of COVID-19. A clinical study from COVID-19–positive patients in Hubei, China, revealed that 46% presented with gastrointestinal (GI) problems.1 Also, SAR-CoV-2 viral RNA has been detected in the stools of COVID-19 patients. These data suggest the underestimated importance of intestinal infection in SARS-CoV-2-induced severe respiratory response.2
Angiotensin-converting enzyme 2 (ACE2), the receptor for SARS-CoV-2,3 is best known for its role in blood pressure regulation. Additionally, ACE2 has another important function of curbing intestinal inflammation. Ace2−/− mice with a reshaped gut microbiota were susceptible to intestinal inflammation.4 Transfer of the Ace2−/− gut microbiota to germ-free (GF) mice worsened colitis pathogenesis,4 suggesting gut microbiota-mediated protective effects of ACE2. These GI benefits of ACE2 may be masked during coronavirus infection, where its expression in the intestine represents a possible avenue for viral entry. We speculated that gut microbiota, which is highly variable between individuals, could be an additional factor modulating colonic Ace2 expression and thereby influencing COVID-19 infectivity.
Methods
Animals
7 weeks old male GF Sprague Dawley (SD) rats (N=5) and GF rats co-housed with conventional SD rats for 10 days (GFC) (N=6) were obtained from Taconic Biosciences (https://www.taconic.com/). They were immediately sacrificed with excess of isoflurane anesthesia upon arrival at the University of Toledo. Fecal samples were collected from the colon for analysis.
16S rRNA gene sequencing
Fecal DNA was extracted from fecal pellet. PCR library preparation, 16S rRNA gene sequencing and analysis were performed as previously described.5
Real-time PCR
A total of 1000ng of RNA from each sample was used to synthesize cDNA using High Capacity cDNA Reverse Transcription Kit (#15596026, Thermo Fisher Scientific, Waltham, MA). The primers used are: Ace2 forward ACCCTTCTTACATCAGCCCTACTG, reverse TGTCCAAAACCTACCCCACATAT, and Gapdh forward ACCACAGTCCATGCCATCAC, reverse TCCACCACCCTGTTGCTGTA.
Serum Lcn2 measurement
Serum Lipocalin 2 was measured by DuoSet enzyme-linked immunosorbent assay kits from R&D Systems (#DY3508, Minneapolis, MN) according to the manufacturer’s protocol.
Flow cytometry
Single-cell suspension was stained with APC Mouse Anti-Rat CD11b (#562102, BD Bioscience, Franklin Lakes, NJ) and PE Mouse Anti-Rat Granulocytes (#550002, BD Bioscience, Franklin Lakes, NJ) prior to flow cytometer analysis (Accuri c6; BD Biosciences, Franklin Lakes, NJ). Results were presented as the percentage of CD11b+ Granulocytes+ population from the cells gated in the forward vs. side scatter plot.
Correlation analysis
The Fragments Per Kilobase of transcript per Million mapped reads (FPKM) data from WKY and SHR colonic epithelium were obtained from the previous study.6 The Spearman r correlation analysis was performed using Graphpad 8.3.0 (538) (Graphpad Software, San Diego, CA) on the combined data from both WKY and SHR rats.
Results and Discussion
To test the hypothesis that microbiota influences Ace2 expression, we compared two groups of concomitantly raised gnotobiotic (germ-free,GF) rats, with one group acquiring gut microbiota through co-housing for 10 days with conventional specific pathogen-free rats. 16S rRNA sequencing analysis of fecal samples confirmed successful colonization of 9 bacterial phyla in the conventionalized GF (GFC) rats (Figure A). Reconstitution of the gut microbiota for 10 days markedly decreased the colonic Ace2 expression in GFC rats, compared with GF rats (Figure B). Since the only variable between the two groups was the presence or absence of the microbiota, our observation indicates that the gut microbiota colonized in the GFC rats caused the decrease in colonic Ace2 expression. Whether this is directly or indirectly occurring via microbial metabolites remains unknown. The conventionalization also resulted in increasing systemic inflammatory tone in GFC compared with GF rats. This was demonstrated by significantly higher levels of lipocalin 2 (Lcn2) (Figure C) and neutrophilia determined by an elevated population of CD11b+Granulocyte+ cells (Figure D). In support of altered ACE2 expression impacting metabolism in the gut,4 a metabolomic analysis showed that compared to GF, GFC rats had higher levels of tryptophan metabolites, kynurenic acid and hydroxy kynurenine (Figure E). Considering that individuals with pre-existing conditions, including hypertension, are at higher risk for SARS-CoV-2 infection, we next examined whether colonic Ace2 expression was altered in a well-validated rat model of hypertension. A Spearman correlation analysis using the colonic gene expression profile from Wistar Kyoto (WKY) rats and spontaneously hypertensive rats (SHR)6 found that colonic Ace2 levels were inversely correlated with both Lcn2 and NOD-like receptor family CARD domain containing 5 (Nlrc5), which encodes a protein involved in anti-viral responses (Figure F).
Figure.
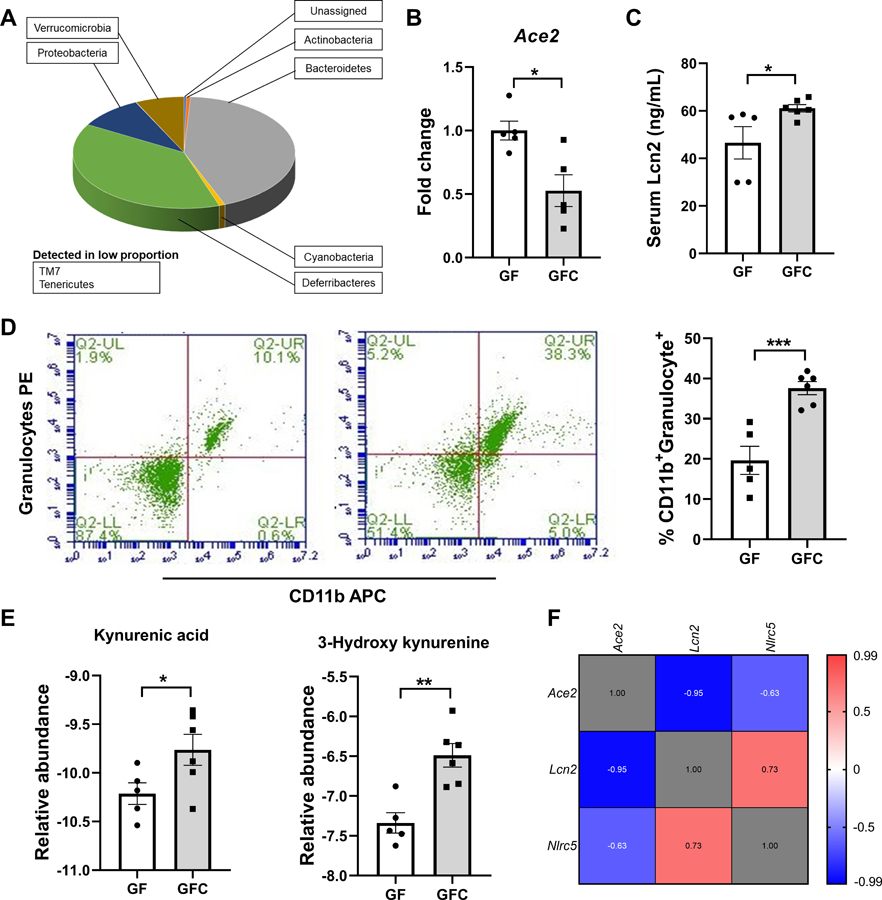
A. Gut microbial composition at the phylum level in GFC rats. Gut microbial compositions were analyzed by 16S rRNA gene sequencing and taxonomy assignments were based on Greengenes as reference. No phyla were detected in GF rats. B. Decreased Ace2 expression in GFC rats. The colonic expression of Ace2 was determined by real-time PCR and normalized to Gapdh. * p<0.05, unpaired t-test. C. Increased level of Lcn2 in serum from GFC rats. Hemolysis-free sera were obtained by centrifugation at 10,000 rpm, 10 min, 4°C. Lcn2 level in serum was determined by ELISA. * p<0.05; unpaired t-test. D. Increased proportion of CD11b+Granulocyte+ cells in peripheral blood of GFC rats. Whole blood was lysed with red blood cell lysis buffer and stained for CD11b+Granulocyte+ cells. *** p<0.001; unpaired t-test. E. Increased kynurenic acid and 3-hydroxy kynurenine in serum of GFC rats. Targeted metabolomic analyses were performed by HPLC-MS to measure the amount of kynurenic acid and 3-hydroxy kynurenine. The data were normalized with internal standards and log2-transformed per-sample basis. * and ** indicate p<0.05 and <0.01, respectively; unpaired t-test. F. Negative correlative expression of Ace2 with Lcn2 and Nlrc5 in the rats. The correlation analysis was performed on combined data from WKY and SHR rats. The expression levels of Ace2, Lcn2 and Nlrc5 in the WKY and SHR were evaluated by RNA-seq and presented as FPKM6.
In light of the current insufficient animal data on COVID-19, our results, although introductory, are timely and significant for 4 reasons: (1) This is the first demonstration of direct modulation of colonic ACE2 by the gut microbiota in the rat, which is closer in gut microbial composition and function to human than mouse.7 (2) This work lays the scientific foundation for examining the variability in gut microbial composition as a factor impacting susceptibility to COVID-19. (3) There is a close, strong relationship between hypertension and the gut microbiota.8,9 Therefore, administering broad-spectrum antibiotics to combat bacterial infection in COVID-19 may be given a second thought as this may increase Ace2 expression in patients. (4) Our data demonstrating an inverse correlation of colonic Ace2 with Lcn2 and Nlrc5, coupled with the previous finding that oral administration of Angiotensin-converting enzyme inhibitor (ACEi) significantly re-shaped gut microbiota10 indicates that elevated colonic ACE2 in hypertensive patients on ACEi11 may also present with a weakened intestinal immune response depending on the extent of gut dysbiosis.
In summary, this study clearly showed that gut microbiota represents a critical factor for the regulation of colonic Ace2 expression and associated colonic and systemic factors that likely contribute to the pathology of the gut-lung axis during COVID-19. Therefore, further studies are necessary to examine the gut microbial composition and its role in ACE2 expression in the COVID-19 susceptible and resistant populations, which would importantly inform on better clinical management of COVID-19.
Sources of Funding
This work was supported by the National Institutes of Health R01HL143082 (B. Joe), R00GM118885 (C.F. Wenceslau), R01CA219144 (M. Vijay-Kumar), Biocodex Microbiota Foundation USA (T. Yang), DK083890 and DK099071 (A.T Gewirtz), and the American Heart Association 18POST34060003 (C.G. McCarthy). The metabolomics core at Baylor College of Medicine was supported by the Cancer Prevention and Research Institute of Texas Core Facility Support Award RP170005 “Proteomic and Metabolomic Core Facility,” National Cancer Institute Support Grant P30CA125123, and intramural funds from the Dan L. Duncan Cancer Center.
Footnotes
Disclosures
None
References
- 1.Pan L, Mu M, Yang P, et al. Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am J Gastroenterol April 2020. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell April 2020;181(2):271–280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature July 2012;487(7408):477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galla S, Chakraborty S, Cheng X, et al. Disparate effects of antibiotics on hypertension. Physiol Genomics 10 2018;50(10):837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang T, Li H, Oliveira AC, et al. Transcriptomic signature of gut microbiome-contacting cells in colon of spontaneously hypertensive rats. Physiol Genomics March 2020;52(3):121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan H, Guo R, Zhu J, et al. A gene catalogue of the Sprague-Dawley rat gut metagenome. Gigascience 05 2018;7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mell B, Jala VR, Mathew AV, et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics June 2015;47(6):187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang T, Santisteban MM, Rodriguez V, et al. Gut Dysbiosis Is Linked to Hypertension. Hypertension June 2015;65(6):1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T, Aquino V, Lobaton GO, et al. Sustained Captopril-Induced Reduction in Blood Pressure Is Associated With Alterations in Gut-Brain Axis in the Spontaneously Hypertensive Rat. J Am Heart Assoc February 2019;8(4):e010721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vuille-dit-Bille RN, Camargo SM, Emmenegger L, et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids April 2015;47(4):693–705. [DOI] [PubMed] [Google Scholar]


