Summary
In completely insular microbial communities, evolution of community structure cannot be shaped by the immigration of new members. In addition, when those communities are run in steady state, the influence of environmental factors on their assembly is reduced. Therefore, one would expect similar community structures under steady‐state conditions. Yet, in parallel setups, variability does occur. To reveal ecological mechanisms behind this phenomenon, five parallel reactors were studied at the single‐cell level for about 100 generations and community structure variations were quantified by ecological measures. Whether community variability can be controlled was tested by implementing soft temperature stressors as potential synchronizers. The low slope of the lognormal rank‐order abundance curves indicated a predominance of neutral mechanisms, i.e., where species identity plays no role. Variations in abundance ranks of subcommunities and increase in inter‐community pairwise β‐diversity over time support this. Niche differentiation was also observed, as indicated by steeper geometric‐like rank‐order abundance curves and increased numbers of correlations between abiotic and biotic parameters during initial adaptation and after disturbances. Still, neutral forces dominated community assembly. Our findings suggest that complex microbial communities in insular steady‐state environments can be difficult to synchronize and maintained in their original or desired structure, as they are non‐equilibrium systems.
Introduction
Stochasticity represents a variation for which we do not have, or choose not to give, a mechanistic explanation. Demographic stochasticity, for example, represents the observation that individual organisms are affected by numerous processes that we cannot know in detail, but this variation is important for understanding the extinction risk (Grimm and Wissel, 2004). By contrast, for deterministic processes, such as the exponential growth of populations if resources are not limiting, random variation can often be ignored.
Both stochastic and deterministic processes determine community assembly, but to what degree it has long been debated in macroecology (Leibold et al., 2004; Chase, 2003, 2007, 2010). For tropical forests, which were at the origin of this debate, the current consensus is that the proportion of both mechanisms varies between communities and in time and space (Hubbell, 2001). For microbial communities, much less is known in this context, although they are the drivers of all biogeochemical cycles, support higher life in countless aspects (Nelson et al., 2016; Zhou and Ning, 2017) and are imperative for human health, nutrition and biotechnology (Schirmer et al., 2016; Johnson et al., 2017; Zeng et al., 2017; Byrd et al., 2018; Wackett, 2018). Microorganisms are ubiquitously distributed and assembled into loosely or strongly connected associations of individuals. Assembly characteristics, such as the type and number of microorganisms and stability of the assembly states, can influence a community's ability to either persist or be prone to extinction (Allison and Martiny, 2008). The extent to which deterministic or stochastic processes influence community assembly mechanisms must be understood if we are to rationally assess, exploit or even control microbial communities.
Deterministic mechanisms, such as niche differentiation, are found in defined localities where community assembly is shaped by selective abiotic parameters, e.g., by the conditions of a habitat, or by biotic interactions between species, e.g., by competition, cooperation and mutualism (Hibbing et al., 2010; Vellend, 2010; Faust and Raes, 2012; Nemergut et al., 2013). The effects of abiotic parameters that influence community assembly have been reported in both natural and bioengineering systems. For instance, bacterioplankton communities assemble according to turbidity, temperature and other water nutrient qualities, such as total phosphate and nitrogen (Peter et al., 2017). The composition of soil communities seems to depend, to a large degree, on the soil redox status, pH and carbon quality and quantity (Fierer, 2017). In full‐scale wastewater treatment plants, the fluctuation of the carbon load of the inflow has been found to select for specific phylotypes and even functions (Günther et al., 2012, 2016), and the core operating taxonomic units (OTUs) of communities from six different activated sludge basins from one region have been found to react in a synchronous manner to weather data (Griffin and Wells, 2017). While abiotic parameters constrain community assembly by limitations in space and resources, biotic parameters do so by species interactions. For example, a famous early study by Gause (1934) demonstrated competitive exclusion, whereby two different Paramecium species competing for the same resources led to the domination and survival of only one. Synthetic co‐cultures are well known to also mutually benefit from the cross‐feeding of organic acids and nutrients (e.g., McCully et al., 2017) and various individual‐based modelling approaches are developed to predict the individuals' competition for process control (Friedman et al., 2017; Daly et al., 2018). Additionally, predator–prey relationships contribute to microbial assembly dynamics, such as the bacterial predator Bdellovibrio spp., which feeds on gram‐negative bacterial species (Johnke et al., 2014), or host–parasite relationships in which genotype‐specific phages change community structures (Laanto et al., 2017). All these deterministic niche differentiation mechanisms ultimately lead to equilibria, corresponding to a steady‐state climax community (Morris and Blackwood, 2007; Nemergut et al., 2013).
By contrast, stochastic mechanisms disrupt strong interrelationships between organisms, and their biotic and abiotic circumstances and can, thereby, prevent deterministic processes from becoming dominant (Burns et al., 2016; Zhou and Ning, 2017). Neutral theory, for example, disregards the influence of species identities but declares all species essentially identical. Therefore, community assembly is random (Hubbell, 2001; Bell, 2000). Neutral forces encapsulate random birth, death, immigration and emigration events (Sloan et al., 2006; Ofiţeru et al., 2010). A typical characteristic of neutral forces is drifting in community assembly (Nemergut et al., 2013; Zhou and Ning, 2017) when niche differentiation mechanisms are weak (Ofiţeru et al., 2010). In addition, events such as random dispersal followed by birth can lead to different colonization patterns (Zhou et al., 2013; Evans et al., 2017). Initial assembly and colonization can also be strongly affected by the random death of microorganisms (Langenheder and Székely, 2011; Woodcock and Sloan, 2017), especially when cell densities are low (demographic stochasticity; Evans et al., 2017; Woodcock and Sloan, 2017). A similar effect can be caused by random cell loss from existent communities due to emigration or dilution of the medium (Nemergut et al., 2013).
A disturbance (a stochastic or an induced event) creates conditions for a new assembly of communities based on either the neutral behaviour of organisms or, and perhaps and, new niche differentiation processes. Under neutral conditions, a community can, after a disturbance, assume a vast number of likely variants of compositions (Ferrenberg et al., 2013). This high level of variation might be diminished by deterministic mechanisms, such as environmental filters or speciation effects, when specific microorganisms persist because of exclusive abilities. For instance, forest fire has been found to promote increasing abundances of spore‐forming Firmicutes in soil (Smith et al., 2008; Pérez‐Valera et al., 2017). Niche differentiation usually establishes dominant phylotypes, which, however, can be replaced after a disturbance by conditionally rare taxa that assemble into a new community with a different and, by chance, even higher diversity. According to the high functional redundancy in natural microbial communities (Allison and Martiny, 2008; De Vrieze et al., 2017; Tully et al., 2017), such disturbance events do not necessarily affect the functions of a community. Therefore, if a disturbance is of a stochastic nature and causes respective effects, it may also have the potential to reduce neutral forces, in which case it causes strong deterministic responses (Ferrenberg et al., 2013; Vanwonterghem et al., 2015; Choi et al., 2017).
The relative roles of niche differentiation and neutral mechanisms on microbial communities were estimated in this study in insular and steady‐state environments. Technical and operational parameters of such insular steady‐state reactors can be well controlled and are valuable for testing performances of biotechnologically relevant strains. Classical continuously operated bioreactors can grow a mono‐dominant strain under such conditions over infinite generations until disturbances, such as mutations or changes in substrate rates, may lead to extinction and, ultimately, to a loss of catalytic efficiency (Jannasch, 1974; Hoskisson and Hobbs, 2005). We have chosen to test species‐rich reactors because biodiversity is usually believed to increase a systems' stability (Hooper et al., 2005). Highly diverse microbial communities are assumed to be resilient catalysts with redundant and interconnected functions and are able to grow on waste substrates. A multitude of species can run identical metabolic pathways with similar efficiency, thus supporting the presence of neutral forces. By contrast, other species dispose of interdependent pathways, thus strengthening niche differentiation. From the ecological perspective, the question arises how highly diverse microbial communities assemble into insular and steady‐state bioreactors and whether they, concurrent with the abiotic conditions, also facilitate a steady‐state community structure. In addition, the engineering perspective requires available means to facilitate community structure control by, e.g., using environmental disturbances, as suggested by DeAngelis and Waterhouse (1987) and Wu and Loucks (1995) for macroecological systems and by Vanwonterghem et al. (2015) for anaerobic microbial systems. In our experimental set‐up, we used soft temperature stressors for this purpose.
Observations of the dynamics of species‐rich systems are challenging. Available studies have either used sequencing approaches (Faust et al., 2015; Shen et al., 2018) or flow cytometry (Props et al., 2016, 2017 and 2018; Koch and Müller, 2018), but the use of these technologies to measure dynamic community behaviour is still rather rare. Next‐generation sequencing platforms provided by, e.g., Illumina, PacBio and Nanopore (Goodwin et al., 2016) are currently the most up‐to‐date and commonly used methods to resolve microbial community composition and function. However, data generated with these methods are not quantitative, and their analysis and quantification are still costly and time‐consuming, rendering them impractical for rapid, cheap, on‐site enumeration of time series. By contrast, flow cytometry observes every cell in a community sample and allows the precise quantification of cells with similar properties. Recently, rapid data generation advanced flow cytometry to reveal the real‐time resilience behaviour in microbial communities (Liu et al., 2018).
In this study, a dense sampling strategy was used to assess whether only neutral mechanisms dominate community assembly. In this case, communities will drift in an unpredictable manner with no synchrony. The unpredictability of neutral mechanisms is problematic for biotechnologies, and there are already many indications that process control via niche differentiation is difficult to achieve (Zhou et al., 2013; Evans et al., 2017; Du et al., 2017). However, the principal aim of engineers in process control of bioreactors is to construct a niche with differentiating boundaries to strengthen and sustain an initially manufactured community. Due to the homogeneous environment of our reactor set‐ups, we expected community assembly to occur in synchrony at the outset due to adaptation processes; however, these synchronized assembly states should subsequently diverge under the chosen conditions. Disturbances may either add to this divergence due to their stochastic nature or shape community structures to re‐synchronize them if they possess niche‐differentiating power. We sought to determine how seemingly antagonistic paradigms are balanced in closed environments to irrevocably elucidate mechanisms for their control.
Results
Description of the ecological situation
Five insular reactors were inoculated with the identical natural community and continuously operated in parallel for 91 days at a dilution rate of D = 0.72 d−1, a working volume exchange rate of 33.3 h and, accordingly, a generation time ( ) of 23.1 h, allowing bacteria with slower growth rates to be sustained. Bacteria with
) of 23.1 h, allowing bacteria with slower growth rates to be sustained. Bacteria with  longer than 23.1 h were lost by continuous reactor operation and became extinct. The reactor steady‐state was reached by a fivefold working volume exchange after approximately 7 days (adaptation phase) and was maintained for 84 days (Supporting Information S1).
longer than 23.1 h were lost by continuous reactor operation and became extinct. The reactor steady‐state was reached by a fivefold working volume exchange after approximately 7 days (adaptation phase) and was maintained for 84 days (Supporting Information S1).
To precisely describe the conditions of our experiment, we are following Grimm and Wissel's (1997) recommendations (later applied to microbiological ecology studies by Liu et al., 2018) to specifying six features that define the background of a study. In this study, the features were as follows.
The level of description, which is a highly diverse natural microbial community originating from a wastewater treatment plant (Supporting Information S1). The individual microorganisms within this community were physiologically characterized using flow cytometry. A 3‐parameter descriptor of each of the 200 000 cells measured per sample (Supporting Information S4 and S5) was generated: forwards scatter (FSC) of light, which is related to cell size; side scatter (SSC) of light, which is related to cell density; and fluorescence related to chromosome information per cell, which is an extrinsic cell parameter. Chromosome information per cell is dependent on the type of cell (G/C content and genome size) and its state in the cell cycle (Müller, 2007). Therefore, every sample is characterized by 600 000 data points. In our study, 326 samples were measured. In addition, to verify the general trends visualized by the cytometric data 44 whole community and sorted 12 gates were analyzed by 16S rRNA gene amplicon sequencing and evaluated both on the class and genus level (Supporting Information S12, data repository https://osf.io/4tkcg/).
The second feature is the variable of interest, which is the change in the community structure over time. Here, a fingerprinting method (Koch et al., 2013, 2014) was used where groups of cells with similar cell parameters were declared as an entity type (Ovaskainen and Meerson, 2010). This is synonymous with a subcommunity, or technically, a gate (Supporting Information S5). Cells with similar parameter values belong to one gate, but they can also move between gates when their morphology, and hence their 3‐parameter descriptor changes while they proliferate. Thus, a unique subcommunity (gate) usually contains several distinct phylotypes but can also be mono‐dominant (Zimmermann et al., 2016). This has been verified by cell sorting combined with 16S rRNA gene amplicon sequencing in earlier studies (Lambrecht et al., 2017, van Gelder et al., 2018, Liu et al., 2018) and also in this study to reveal community and subcommunity compositions (Supporting Information S12). The used state variables were the numbers of gates (summed up to 68 gates, Supporting Information S5, Fig. S5.1 B), the position of gates in a fingerprint, and the numbers of cells per gate.
Reference dynamics, or space, must be defined via a set of states for which a community is considered to have preserved its key features. For bioreactors, this definition can be made normative, i.e., via desired features, which are unchanging community structures in our study. The rationale behind the approach lies in the ‘self‐identity’ of ecological systems (Jax et al., 1998), or its resilience: identity is maintained although essential state variables (community structures) change within certain boundaries. The boundary marking the reference space was determined by pairwise estimation of the deviation between parameter points of the state variables in a multidimensional space (e.g., by using a stability‐computation tool, URL: https://github.com/fcentler/EcologicalStabilityPropertiesComputation). Structural community variations around a reference point were allowed to fluctuate within a boundary of approximately 0.2 (Canberra distance, Liu et al., 2018). As long as a community changes within this boundary, it is defined as constant or as being essentially the same (Grimm and Wissel 1997). Each of the two control reactors, C1 and C2, showed small community structural changes only at the end of the processes, which were defined as reference space (Fig. 1a, C1 and C2). The radius r c was only slightly different between the two reactors, with r c values of 0.23 for reactor C1 and 0.24 for C2 respectively.
Figure 1.
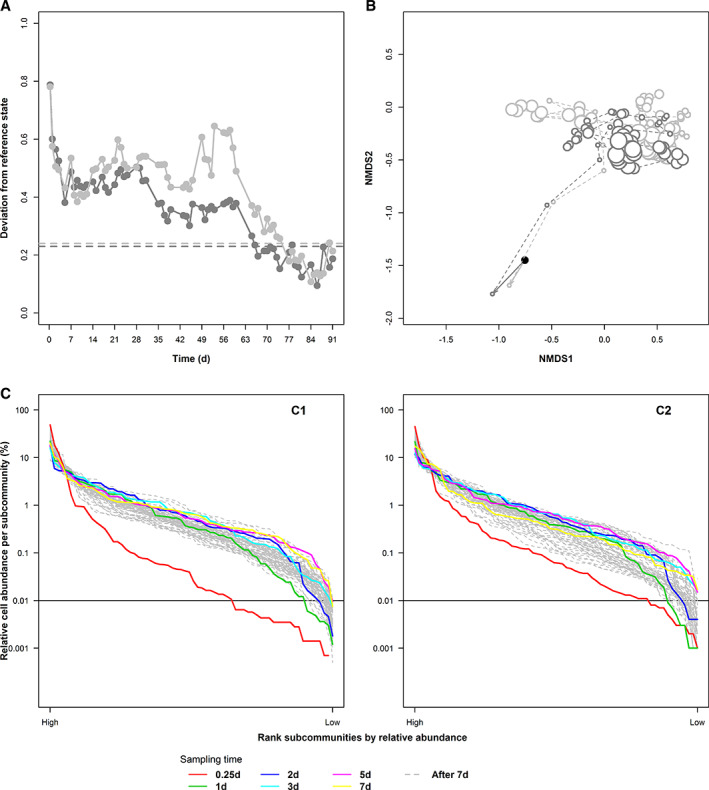
Community analysis based on all subcommunities in samples of control reactors C1 and C2. A. The boundary of the reference space was calculated using the last 10 time points (79–91 days). The reference state was determined by calculating the mean values per subcommunity of these 10 points. The pairwise deviations between the reference state and successive samples were estimated as Canberra distances (C1: dark grey, C2: light grey). The maximum value of deviation from the reference state was chosen as the boundary of reference space per reactor (dashed line). B. Dissimilarity analysis of community structures per reactor. Each point in the nMDS plot represented a community sample (inoculum: black, C1: dark grey, C2: light grey). The sampling time is represented by increasing point sizes. C. Rank‐order abundance curves of samples from C1 (left) and C2 (right); all 68 subcommunities per sample were ranked according to their relative cell abundances and displayed in decreasing order from left to right. The horizontal line marks the height of relative cell abundance at 0.01%. The slopes of samples at 0.25 day (red line) are C1: −0.055 and C2: −0.049; mean values of slopes after the adaptation (grey lines) are C1: −0.038 and C2: −0.041.
The next three features of the ecological study demand information on (iv) type and duration of disturbance, as well as the (v) spatial and (vi) temporal scales. The communities in three of the five reactors (D1, D2 and D3) were disturbed by a repeated and soft temperature stressor from 30 °C to 40 °C forwards and twice backwards, which was chosen to mimic common situations in wastewater treatment plants (Willers et al., 1998, Supporting Information S2). The spatial scale was set by the design of the continuous reactor operation while the temporal scale was bounded to 91 day (Supporting Information S1). Continuous reactor environments are obliged to avoid disturbances and guarantee steady‐state conditions. Pure cultures maintain invariable cell distributions under those conditions (Müller and Babel, 2003). In this study, significant variations in cell distributions could be identified, when, instead of a pure culture, a complex microbial community was cultivated (Movie 1). These variations were also reflected by the amplicon sequencing data (Supporting Information S12). The strength and possible causes of these variations are explored in the following two sections.
Neutral mechanisms in undisturbed communities
The insular reactor set‐up guaranteed, prior to the temperature disturbances, well‐mixed, homogeneous and constant environments, which would favour neutral mechanisms. They gave every organism with  shorter than 23.1 h identical chances to undergo growth and proliferation. In addition, continuous reactors are open to the environment by the outflow, which causes a random loss of organisms. To test for neutral behaviour, two control reactors, C1 and C2, were sampled and their community structures analyzed by flow cytometry. The community assembly processes were computed by using the cell data of C1 (dark grey) and C2 (light grey) and compared by dissimilarity analysis (Bray‐Curtis index, Fig. 1b). During nearly identical adaptation phases of the inoculum to the still unbalanced reactor conditions, a few subcommunities were dominant with extremely high cell abundance (greater than 45%, e.g., G36 of C1 at day 0.25, Supporting Information Table S7.1). This caused a more geometric‐like shape of the rank‐order abundance curves with a steep slope and fewer subcommunities with intermediate cell abundances (red line, Fig. 1c). Geometric‐like distributions and steeper slopes of rank‐order abundance curves are accepted indications for niche‐differentiating mechanisms (Tokeshi, 1993; Hubbell, 2001; Begon et al., 2006). After the adaptation phase, the microbial communities in C1 and C2 clearly evolved in different directions (Fig. 1b, confirmed by the ANOSIM procedure, Supporting Information S8, Table S8.1, R‐value of C1 × C2: 0.317). Here, an equal distribution of cells between all subcommunities (the average cell abundance per gate is 1.47%) of one reactor would support random community assembly due to the set‐up of the reactor favouring neutrality. Indeed, the S‐shape of the log‐normal rank‐order abundance curves indicated a high evenness for the majority of the subcommunities because only a small number of subcommunities with very high (above 10%) or very low (below 0.01%) cell abundancies were present in the assembly curves (Fig. 1c). These rank‐abundance distributions remained fairly constant in the studied time frame.
shorter than 23.1 h identical chances to undergo growth and proliferation. In addition, continuous reactors are open to the environment by the outflow, which causes a random loss of organisms. To test for neutral behaviour, two control reactors, C1 and C2, were sampled and their community structures analyzed by flow cytometry. The community assembly processes were computed by using the cell data of C1 (dark grey) and C2 (light grey) and compared by dissimilarity analysis (Bray‐Curtis index, Fig. 1b). During nearly identical adaptation phases of the inoculum to the still unbalanced reactor conditions, a few subcommunities were dominant with extremely high cell abundance (greater than 45%, e.g., G36 of C1 at day 0.25, Supporting Information Table S7.1). This caused a more geometric‐like shape of the rank‐order abundance curves with a steep slope and fewer subcommunities with intermediate cell abundances (red line, Fig. 1c). Geometric‐like distributions and steeper slopes of rank‐order abundance curves are accepted indications for niche‐differentiating mechanisms (Tokeshi, 1993; Hubbell, 2001; Begon et al., 2006). After the adaptation phase, the microbial communities in C1 and C2 clearly evolved in different directions (Fig. 1b, confirmed by the ANOSIM procedure, Supporting Information S8, Table S8.1, R‐value of C1 × C2: 0.317). Here, an equal distribution of cells between all subcommunities (the average cell abundance per gate is 1.47%) of one reactor would support random community assembly due to the set‐up of the reactor favouring neutrality. Indeed, the S‐shape of the log‐normal rank‐order abundance curves indicated a high evenness for the majority of the subcommunities because only a small number of subcommunities with very high (above 10%) or very low (below 0.01%) cell abundancies were present in the assembly curves (Fig. 1c). These rank‐abundance distributions remained fairly constant in the studied time frame.
However, there was high variation in the order of subcommunities within the rank‐order abundance curves. During the adaptation phase, nearly identical subcommunities dominated each of the very high and very low‐abundant communities, but this trend changed dramatically afterwards. The dominant subcommunities, which are those subcommunities that contain cell numbers above the calculated mean cell abundance value per gate (55 unique subcommunities for both C1 and C2, Fig. 2a), were not only frequently changing their ranking but also receded to values below the mean cell abundance value (e.g., from C1: G16, G37‐G44; from C2: G13‐G14, G37‐G41) or, more seldom, re‐emerged (e.g., in C1: G1 and G12; in C2: G2 and G49). In addition, the community structures developed into different patterns and were not identical between C1 and C2 (Fig. 2a), although both reactors were performing in a steady‐state and started from identical situations.
Figure 2.
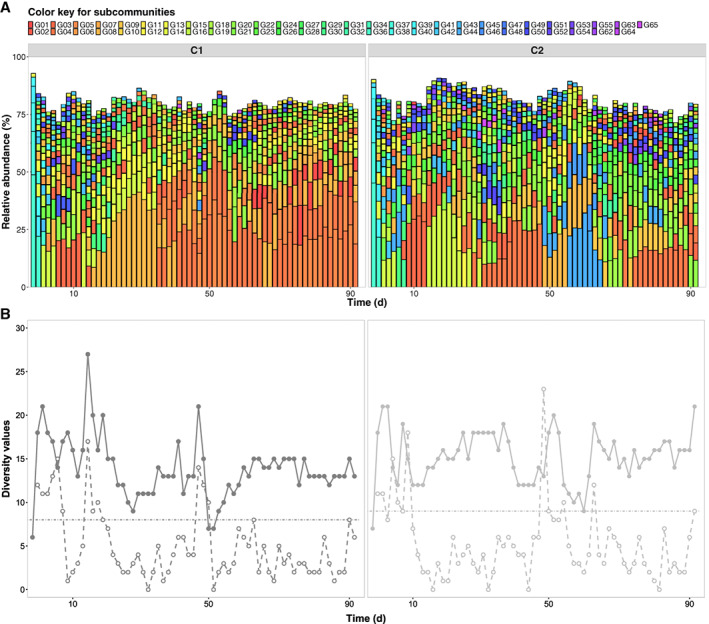
Community analysis based on dominant subcommunities in samples taken from control reactors C1 and C2. A. Variations of community structures through time were displayed as successive columns. Each column represents a community sample where dominant subcommunities are shown as squares filed with unique colours. The length and position of a square indicate the corresponding dominant subcommunities with its relative abundance value and the rank‐order of this abundance value (relative abundances of dominant subcommunities per sample displayed in decreasing order from bottom to top). B. Values of cytometric α‐diversity (solid points) and intra‐community β‐diversity (empty points) measured through the time. The threshold (dashed line) for recognizing drifts was defined regarding to the maximum of intra‐community β‐diversity values in reference spaces which were 8 for C1 and 9 for C2.
The observation that the rank‐abundance distributions were constant and yet the gates, occupying each particular rank, varied significantly is indicative of neutral dynamics, especially given that the environmental and initial conditions were the same.
Additional indications for community structure changes can be provided by cytometric α‐ and β‐diversity values. The cytometric α‐diversity value (Günther et al., 2016; Koch and Müller, 2018) uses the number of dominant subcommunities per sample to describe variations between samples in one reactor set‐up (Fig. 2b). The cytometric α‐diversity can also include evenness information, calculated using the Hill numbers Dq = 1,2 (based on all subcommunities per sample; Hill, 1973; Liu et al., 2018; or by bins, Props et al., 2016). The comparison between cytometric α‐diversity values for dominant subcommunities (Dq = 0) and those calculated for all subcommunities (Dq = 1,2) is provided in Supporting Information S9 (Fig. S9.1) and showed analogous trends. Therefore, only the Dq = 0 for dominant subcommunities was used for further analyses. The variations in α‐diversity values were calculated as 6 to 27 dominant subcommunities for C1 and 7 to 21 for C2, suggesting huge variations in the respective community structures during their steady‐state cultivations. In addition, high inter‐community β‐diversity values between the C1 and C2 reactors were determined (Supporting Information Table S9.2, C1 × C2, up to 27 at day 46), which highlights the diverse trajectories of the two independently grown communities. Finally, intra‐community differences were calculated by intra‐community β‐diversity values, which regard only those dominant subcommunities that are not present in the other sample when successive samples are compared pairwise (Fig. 2b). A threshold defined the highest value for intra‐community β‐diversity within the reference spaces (C1: 8; C2: 9). After the adaptation phase, the intra‐community β‐diversity values fluctuated frequently, but mostly below the threshold. However, some of the β‐diversity values surpassed this threshold. We found two time points in C1 (days 14 and 46) and three time points in C2 (days 9, 49 and 65) in which the community assemblies dramatically changed. These huge dissimilarities between community structures are also recognizable in Fig. 1b. The data suggest that fluctuations occurred in the steady‐state reactor environments. Such fluctuations can be regarded as drift, an event that reveals the random change in relative species abundance (Vellend, 2010; Nemergut et al., 2013; Zhou and Ning, 2017). In this study, we consider the term ‘drift’ as a result of the neutral background that stochastically changes, despite the steady‐state condition of the continuous reactor set‐up, the structure of a microbial community. In our set‐up, drift events were acknowledged only after adaptation. The data suggest that the microbial communities in reactors C1 and C2 changed their structures by those drift events in a very pronounced manner (Fig. 2b).
Niche differentiation in undisturbed communities
Niche differentiation comprises the influence from both the biotic and abiotic environment. In a continuous reactor environment, a niche differentiation process occurred in the adaptation phase until the steady‐state was reached. In addition, as all organisms with  longer than 23.1 h were washed out, this condition selected for organisms that could grow faster. The provided medium with carbon sources and nutrients as well as oxygen availability also promoted certain organisms over others (Supporting Information S3).
longer than 23.1 h were washed out, this condition selected for organisms that could grow faster. The provided medium with carbon sources and nutrients as well as oxygen availability also promoted certain organisms over others (Supporting Information S3).
Niche differentiation during adaptation was expected to result in increased interrelationships between organisms. Therefore, we expected fewer interrelationships after the adaptation phase and a rapid occupation of the reference space where interactions should be low. Steady‐states can be considered to favour neutral dynamics and promote drift events as random or neutral processes. However, after a drift event, the new communities must reorganize themselves to the steady‐state conditions, which are accomplished via new interconnections between microorganisms, which might then also influence their immediate environment (self‐amplification, Wu and Loucks, 1995). Therefore, testing for the strength of interconnections between microorganisms and their environment may reveal situations in which niche differentiation is or is not fulfilled. Recent studies have confirmed that the number of correlations between subcommunities and abiotic parameters can unravel these interconnections (Faust and Raes, 2012; Günther et al., 2016).
In this study, correlation analysis was performed with Spearman's rank‐order correlation coefficient (rho) (Koch et al., 2013; Gao et al., 2016). Both the cell abundance per subcommunity (Supporting Information S7, Table S7.1–5) and the respective abiotic parameters (Supporting Information S7, Table S7.6–10) were included in the correlation analysis (Supporting Information S8, SC vs. SC and SC vs. Abio). To count meaningful significant correlations, the data were subsampled into three phases, with the first phase representing adaptation, the second phase comprising the majority of the data and drift events and the third phase indicating the reference spaces. The number of significant correlations per defined phase was counted after testing the p value (< 0.05, Benjamini and Hochberg, 1995) and choosing a strong correlation coefficient (rho value >0.75 or < −0.75, Supporting Information S8).
The first phase mirrored the adaptation of the community to the new situation, where the number of correlations was the highest of all phases per reactor (summed up to C1: 92 and C2: 108, Table 1), and the niche differentiation mechanisms were superior. In the second phase, the numbers of correlations were lower, with C1 representing 85 and 86 and C2 61, 40 and 63 correlations respectively (Table 1), for comparable time intervals. While clearly only a minority of the subcommunity correlations depended on abiotic parameters (SC vs. Abio), the majority of correlations were found between microorganisms. These findings clearly indicate unbalanced community states. In the third phase, climax communities were expected with low numbers of correlations between microorganisms and their environment. Although, the communities in C1 and C2 were both considered to have entered reference spaces according to their structure, it seems that only the community in C1 approached a state near a climax definition according to the counted correlations (summed up to C1: 12, Table 1). By contrast, an unexpectedly high number of correlations was still found for C2 (summed up to C2: 77, Table 1), suggesting that even after 91 days of steady‐state cultivation, the interrelationships in C2 were still strong. It can be assumed that the enhanced interrelationships might occur in response to the three drift events in C2, which requires a new balance between the subcommunities and their environment (Fig. 1b, 2a). Therefore, apart from the adaptation processes, niche differentiation mechanisms were also found in the steady‐state environments, herein probably as response to changes in community structures due to drift events.
Table 1.
Counts of significant correlations between dominant subcommunities and abiotic parameters for two control reactors (C1 and C2). Only strong correlations were involved using the Spearman's rank order correlation coefficient (rho > 0.75 or < −0.75; p value < 0.05, tested according to Benjamini and Hochberg, 1995). Correlations were tested for successive phases with equal sample numbers (10 time points). First phase: adaptation phase; second phase: correlations calculated starting at the height of drift events onwards (C1: days 14 and 46 and C2: days 9, 49 and 65); third phase: reference spaces. Significant correlations were visualized as networks in Supporting Information S8, Fig. S8.1.
| C1 | Phases | 1st phase | 2nd phase | 3rd phase | ||
| Drift 1 | Drift 2 | |||||
| SC vs. SC | 57 | 57 | 77 | 8 | ||
| SC vs. Abio | 35 | 28 | 9 | 4 | ||
| Sum | 92 | 85 | 86 | 12 | ||
| C2 | Phases | 1st phase | 2nd phase | 3rd phase | ||
| Drift 1 | Drift 2 | Drift 3 | ||||
| SC vs. SC | 75 | 40 | 29 | 35 | 45 | |
| SC vs. Abio | 33 | 21 | 11 | 28 | 32 | |
| Sum | 108 | 61 | 40 | 63 | 77 | |
Neutral and niche differentiation mechanisms in disturbed communities
Circumstantial evidence was provided for random drifts in community structure and, linked to these events, for high numbers of correlations between subcommunities in the control reactors. This begs the question posed by Ferrenberg et al. (2013) concerning whether an environmental disturbance could synchronize the community structure in isolated communities and drive them towards some new niche‐differentiated climax community. Here we introduced moderate changes in the temperature regime of three reactors [D1 (blue), D2 (red) and D3 (green)] running alongside the control reactors, with otherwise unchanged environments. Moderate temperature changes from 30 °C to 40 °C and the reverse were applied two times in succession (Supporting Information S2). The data were evaluated using the same workflow as for C1 and C2.
Using the state variables, the reference spaces for communities in reactors D1‐D3 were tested, but only D1 reached a similar value to the controls (r c value of 0.25). The other communities were not constant (D2: r c = 0.59, D3: r c = 0.47; Fig. 3a). The stressor clearly did not contribute to approaching the reference space synchronously and in time.
Figure 3.
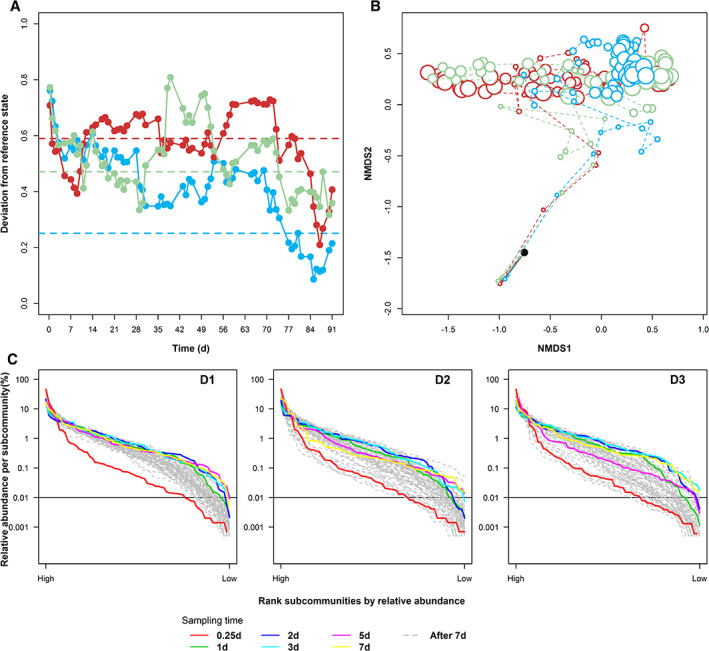
Community analysis based on all subcommunities in samples of disturbed reactors D1, D2 and D3. A. The boundary of the reference space was calculated using the last 10 time points (79–91 days). The reference state was determined by calculating the mean values per subcommunity of these 10 samples. The pairwise deviations between the reference state and successive samples were estimated as Canberra distances (D1: blue, D: red and D3: green). The maximum value of deviation from the reference state was chosen as the boundary of reference space per reactor (dashed line). B. Dissimilarity analysis of community structures per reactor. Each point on the nMDS plot represented a community sample (inoculum: black, D1: blue, D2: red and D3: green). The sampling time is represented increasing point sizes. c: Rank‐order abundance curves of samples from reactors D1 (left), D2 (middle) and D3 (right); all 68 subcommunities per sample were ranked according to their relative cell abundances and displayed in decreasing order from left to right. The horizontal line marks the height of relative cell abundance at 0.01%. The slopes of samples at 0.25 day (red line) are D1: −0.054, D2: −0.055 and D3: −0.056; mean values of slopes after the adaptation (grey lines) are D1: −0.049, D2: −0.049 and D3: −0.052.
Other ecological measures confirmed this trend. The dissimilarity analysis, for which the data for all five reactors were computed together, showed that the initial adaptation phase traversed nearly identically to the control reactors (Figs 1b and 3b). However, we subsequently found again diverging assembly processes (Fig. 3b), despite the soft disturbances. The cytometric community structure comparison (Fig. 3b) showed matching deviations between the samples of D2 and D3 in comparison to the control reactors, while D1 clustered separately from all of them (confirmed by ANOSIM analysis, see Supporting Information S8, Table S8.1, R‐values of D2 × D3: 0.029 in comparison to D1 × D2: 0.176, D1 × D3: 0.119). The numbers of cytometric α‐diversity values were comparable to those in C1 and C2, with 8 to 22 in D1, 3 to 23 in D2 and 5 to 24 in D3 (Fig. 4b) respectively. In D1, only gradual structural changes were found within the starting 18 days (Fig. 4b) and almost no further variations until the end of the experiment, in contrast to D2 and D3. When the temperature relapsed from 40 °C to 30 °C (Fig. 4b), only two identical subcommunities dominated both reactors for approximately 10 days, although at different times (G44 and G45, Fig. 4a). The Miseq data (Supporting Information Fig. S12.2) demonstrated that G44 and G45 revealed exclusively Alpha‐proteobacteria as the dominant phylotype. Respective mono‐dominant communities emerged in both reactors at days 81 (D2) and 49 (D3), which clearly confirmed a speciation event. Supported by the intra‐community β‐diversity values, only short‐lived and dissimilar stable states could be identified in D2 and D3 (Fig. 4b). The huge variations in α‐diversity and intra‐community β‐diversity values suggested unequal responses of the communities in D1, D2 and D3 to the identical temperature disturbance.
Figure 4.
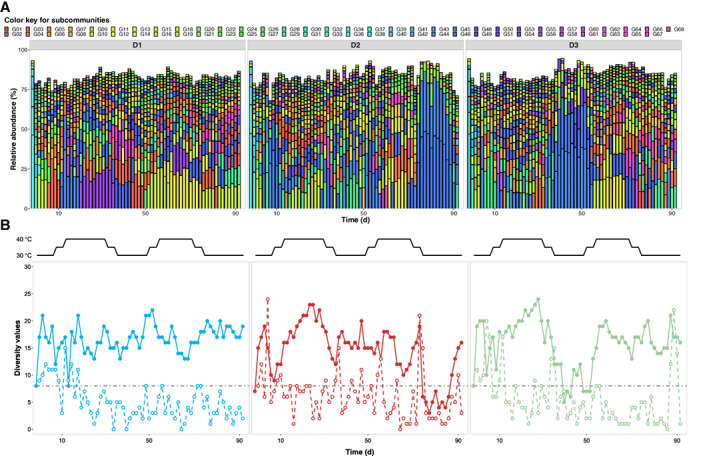
Community analysis based on dominant subcommunities in samples taken from disturbed reactors D1, D2 and D3, which were treated by identical temperature changes (black lines in the middle). A. Variations of community structures through time were displayed as successive columns. Each column represents a community sample where dominant subcommunities are shown as squares filed with unique colours. The length and position of a square indicate the corresponding dominant subcommunity with its relative abundance value and the rank‐order of this abundance value (relative abundances of dominant subcommunities per sample displayed in decreasing order from bottom to top). B. Values of cytometric α‐diversity (solid points) and intra‐community β‐diversity (empty points) measured through the time (D1: blue; D2: red and D3: green). The threshold (dashed line) for recognizing drifts was defined as for C1 (intra‐community β‐diversity = 8).
Furthermore, the rank‐order abundance curves of D2 and D3 were not as neutrally assembled as for the control reactors (C1 and C2) and D1 (Fig. 3c). After the adaptation phase, they showed steeper slopes (more geometric‐like curves, Supporting Information S8) and more subcommunities with low‐cell abundances (below 0.01%; C1: 986, C2: 1076, D1: 1239, D2: 1532 and D3: 1576 out of 4012 (59 samples × 68 gates per reactor)), which also indicated selection and dominance of fewer subcommunities (Fig. 3c). In addition, the abundance ranking of the dominant subcommunities did not differ between all three reactors as for the controls (Fig. 4a, 63 unique subcommunities for D1, D2 and D3). These findings suggested that temperature shifts transiently but asynchronously affected community structures by niche differentiation, at least in the disturbed reactors D2 and D3.
To further assess the niche differentiation forces, significant correlations were calculated for D1, D2 and D3 (Table 2), where the first (the adaptation phase) and the third phases (the reference space) were the same as previously defined. The second phase was subsampled according to the temperature stressors (T1‐T4, Supporting Information S8). The numbers of correlations in the first phase were high at D1: 98, D2: 98 and D3: 91 (Table 2) and comparable to those of the control reactors (Table 1). In the second phase, higher correlation numbers were found (D1max: 109, D2max: 138 and D3max: 125, Table 2) in comparison to those caused by drifts in controls. In addition, according to the correlation analysis, the parameter temperature (T) was related to the majority of dominant subcommunities for almost all situations in which temperature stress occurred (Supporting Information S8, Fig. S8.1). In the second phase, we found 38 edges for D1, 30 edges for D2 and 41 edges for D3 in comparison to the summarized 2 edges for all reactors in the first phase. The third phase was sustained with lower numbers of correlations (D1: 84, D2: 71 and D3: 62), although the value of C1 was clearly not reached (Table 1). Thus, the increased numbers of correlations and the presence of edges between temperature and subcommunities during the temperature pressure also suggested highly active niche differentiation mechanisms in the disturbed reactors.
Table 2.
Counts of significant correlations between dominant subcommunities and abiotic parameters for three reactors treated with temperature stressors (D1, D2 and D3). Only strong correlations were involved using the Spearman's rank order correlation coefficient (rho > 0.75 or < −0.75; p value < 0.05, tested according to Benjamini and Hochberg (1995). Correlations were tested for successive phases with equal sample numbers (10 time points). First phase: adaptation phase; second phase: correlations calculated starting at the inset of temperature stressors (T1‐T4); third phase: reference spaces.
| D1 | Phases | 1st phase | 2nd phase | 3rd phase | |||
| T1 | T2 | T3 | T4 | ||||
| SC vs. SC | 64 | 69 | 54 | 27 | 57 | 65 | |
| SC vs. Abio | 34 | 34 | 22 | 26 | 52 | 19 | |
| Sum | 98 | 103 | 78 | 53 | 109 | 84 | |
| D2 | Phases | 1st phase | 2nd phase | 3rd phase | |||
| T1 | T2 | T3 | T4 | ||||
| SC vs. SC | 79 | 59 | 46 | 80 | 113 | 46 | |
| SC vs. Abio | 19 | 25 | 17 | 32 | 25 | 25 | |
| Sum | 98 | 84 | 63 | 112 | 138 | 71 | |
| D3 | Phases | 1st phase | 2nd phase | 3rd phase | |||
| T1 | T2 | T3 | T4 | ||||
| SC vs. SC | 53 | 84 | 83 | 82 | 33 | 42 | |
| SC vs. Abio | 38 | 19 | 27 | 43 | 30 | 20 | |
| Sum | 91 | 103 | 110 | 125 | 63 | 62 | |
Stability properties of undisturbed and disturbed communities
Both neutral and niche differentiation mechanisms seemed to be linked under steady‐state cultivation conditions. Drift events were observed frequently, and soft disturbances increased niche differentiation effects. Calculating stability properties for all five community structures will reveal the degree to which communities resist drifts or disturbances and recover despite neutral and niche differentiation forces. To validate this situation, the stability of the communities was estimated by calculating deviations pairwise in a multidimensional space by using the state variables and Canberra distance (Lance and Williams, 1967; Kuczynski et al., 2010; Liu et al., 2018). The variations were quantified by the deviation d c, which is calculated on the basis of a reference point that was placed before the inset of a disturbance or drift event. The reference space was obtained from C1 (r c = 0.23, Fig. 1a). All communities were found to deviate from the reference point and pass through the boundary of the reference space (Supporting Information S10, Fig. S10.2). The highest community average deviation (accessed with d c. peak) was found for the reactor D3 with d c.peak.ave = 0.70 and the lowest for C1 with d c.peak.ave = 0.46. Consistently, resistance was lowest in the disturbed reactor D3 (average value RS = 0.29), while constancy was highest for C1 (average value RS = 0.54). The average values RS of the other three reactors did not vary remarkably (C2 = 0.39, D1 = 0.45 and D2 = 0.35). Recovery values were also calculated but found to be extremely low. None of the communities recovered to their original structure (Supporting Information Table S10.1).
These data revealed that soft temperature disturbances prompted low resistance, but also drift events in the controls caused similar high deviations. All five reactors failed to recover from drifts or temperature disturbances despite continuous steady‐state conditions. Both neutral and niche differentiation mechanisms led to unsteady communities and low‐stability properties.
Discussion
The aims of this study were (i) to unravel ecological principles of microbial community assembly in insular environments and (ii) to find concepts for maintaining an intended community structure over longer time periods. We found that both niche differentiation and neutral forces are responsible for different and intermittent degrees of fluctuations in dynamic community assembly.
The five communities could not be synchronized despite established steady‐state continuous conditions as can be done for pure cultures (Skarstad et al., 1985, Strässle et al., 1989); rather, they developed unique structures. The respective communities did not converge to a climax community even in individual reactors. Stability measures, such as constancy, resistance and recovery values, calculated from references points prior to drift and disturbances were low or not existent. Instead, the community assembly characteristics were different between all reactors and, remarkably, continuously changing with high frequency. We observed drifts occurring both in the undisturbed reactors and also superimposed on the temperature‐related variations in the disturbed environments. The dynamics clearly showed that the use of disturbance did not re‐synchronize the communities.
The relative roles of niche differentiation vs. neutral mechanisms
To clarify the relative roles of niche differentiation vs. neutral mechanisms, ecological measures were used. Our analyses support that the niche differentiating mechanism became only temporarily more dominant based on two major ecological measures.
First, niche differentiation effects are suggested by the geometric‐like rank order assembly curves with steeper slopes and by including more subcommunities with low cell abundances (below 0.01%). Such curves were found for communities during adaptation (in all five reactors) and under disturbance. In addition, in D2 and D3, not only a very low number of highly dominant subcommunities were prevalent temporarily, but these were also the same ones.
Second, niche differentiating mechanisms were disclosed by the numbers of detected significant correlations. An increased number of positive or negative correlations suggest both increasing interactions between organisms and between organisms and abiotic parameters (Seebacher and Franklin, 2012; Faust and Raes, 2012; Needham and Fuhrman, 2016; Günther et al., 2016). Adaptations to new conditions are known to cause not only changes in cell states, e.g. by cell growth and proliferation but also to alter community structures and, in response, lead to variations in abiotic parameters. Large numbers of correlations were detected in the adaptation phases of all reactors, but especially in response to drifts and the soft temperature stressor. While the high correlation numbers during the adaptation process were obviously caused by the different environmental background of the origin of the inoculum and the continuous set‐up of the reactors (also see Miseq analysis, Supporting Information Fig. S12.2), the random drifts and temperature disturbances initiated new interactions between microorganisms.
Neutral mechanisms were expected in our set‐up because the environment was maintained in a steady‐state. In addition, the set‐up provided no means to buffer species loss. In this context, both equal competitive abilities per cell and elimination by continuous dilution can be assumed to be vital features of our set‐up. We found three lines of evidence for the presence of neutral mechanisms.
First, the strongest evidence for neutral drifts is provided by the observation that, after adaptation, the shape of most rank‐order abundance curves of all reactors showed lognormal‐like distributions that were not steep. This phenomenon was predominantly observed in the controlled but also in the disturbed systems and is typical for a neutral assembly of communities (Hubbell, 2001; Matthews and Whittaker, 2014; Alroy, 2015). The S‐shape of lognormal assembly curves showed communities with relatively more subcommunities with a medium cell abundance (relative cell abundance between 0.01% and 10%), thus uncovering the weak niche selection of reactor conditions.
Second, according to Ofiţeru et al. (2010), neutral mechanisms can also be proven by variations in the rank order of the entities. Again, this was found predominantly in the control but also in the disturbed systems.
Third, diverging inter‐community β‐diversity across reactors suggests active neutral mechanisms (Chase, 2010). This hypothesis was proposed even for an open system (with regional exchange), in which stochasticity in assembly history can create multiple stable equilibria of community structures across local sites, leading to a high inter‐community β‐diversity (Chase, 2003; Pagaling et al., 2017). In our study, inter‐community β‐diversity showed a clear positive trend (gradient value = 0.12, R 2 = 0.413) for the control reactors (C1 × C2, Fig. 5a). By contrast, the reactors treated with the soft temperature stressor (D1, D2 and D3) showed lower gradients with D1 × D2 = 0.09 (R 2 = 0.153, blue line) and D2 × D3 = 0.08 (R 2 = 0.159, green line), and even a negative gradient for D1 × D3 = −0.01 (R 2 = 0.005, red line, Fig. 5b).
Figure 5.
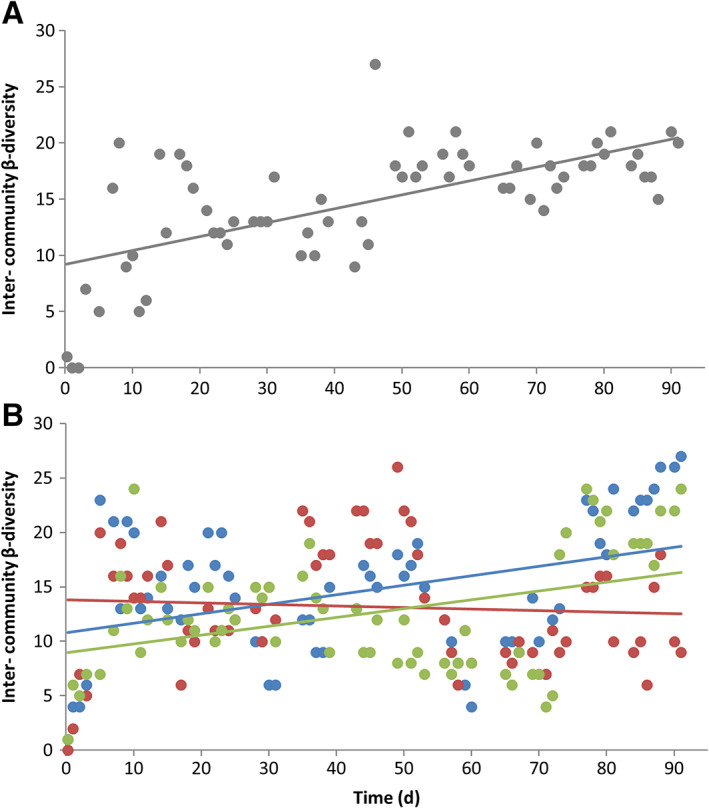
Inter‐community β‐diversity values (points) determined for samples compared pairwise for two reactors which were operated under the same conditions. Linear regression of inter‐community β‐diversity vs. time (lines) showed gradients of differences in diversities between communities. A. pairwise comparison of reactors C1 × C2 (grey). B. pairwise comparison of reactors treated by temperature stressors: D1 × D2 (blue), D1 × D3 (red) and D2 × D3 (green).
The results suggest that neutral forces were stronger in the control reactors but lower in the disturbed ones. Temperature stress, especially the second one, contributed only temporarily to more similar structures of compared communities, a phenomenon that was also demonstrated by Chase (2007) in pond communities. Nevertheless, the disturbed communities reorganized themselves within only a few generations into an altered higher cytometric α‐diversity (Fig. 4, Supporting Information Table S9.1) which is supported by data from sequencing analysis on the class level (D2: 81–90 days; D3: 49–70 days, Supporting Information Fig. S12.2) and the genus level (https://osf.io/4tkcg). Such results show that neutral mechanisms were also robust in the disturbed reactors, probably due to the upcoming conditionally rare taxa. We have previously suggested that the upcoming conditionally rare taxa might represent a response to stress situations (Günther et al., 2016).
Interactions of niche differentiation and neutral mechanisms during community assembly can be assumed using neutral models as the null hypothesis (Leibold et al., 2004; Dumbrell et al., 2010; Faust and Raes, 2012; Zhou et al., 2013). Computational neutral models were already well used in macroecology (Fisher et al., 1943; Preston, 1948; MacArthur, 1960) until Hubbell developed the unified neutral theory of biodiversity and biogeography (UNTB, 2001), which has since been modified for microorganisms (Sloan et al., 2006; Woodcock et al., 2007; Ofiţeru et al., 2010). When the immigration rate is equal to zero, a steady loss of richness can be expected, and the climax community will be random mono‐species‐dominant (Hubbell, 2001; Sloan et al., 2006). The precondition for such a development is a disturbance‐free environment. We did not find indications for developments to mono‐species‐dominant communities, but we argue that the dilution rate itself acted as an intervention in two directions: it provided new resources continuously, thereby lowering competition between species, and it created sufficient space for competitive inferiors to co‐exist because the slow growth rate allowed them to stay in the reactor with high probability (Chesson and Huntly, 1997). Although, the niche differentiation mechanisms in our set were less strong, we found, like others, that they and neutral processes are not mutually exclusive (Gravel et al., 2006; Adler et al., 2007; Ofiţeru et al., 2010). For instance, the decrease in the ammonium concentration in the medium of the control reactors was accompanied by a decrease in pH and electrical conductivity (Supporting Information S3). Thereby, the activity of the ammonium oxidizing bacteria caused, in the undisturbed reactors, a change in reactor conditions, which indicated niche differentiation forces in otherwise neutral environments. This exemplary relationship shows the interconnection between neutral and niche differentiation mechanisms and that the two mechanisms do not appear independently.
The Intermediate Disturbances Hypothesis (IDH) backs high α‐diversity values
In the disturbed reactors, some α‐diversity values were temporarily very low but subsequently recovered to the starting values. The α‐diversity medians were even slightly higher (Fig. 6, D1: 17; D2:15; D3:17) in comparison to the control reactors (Fig. 6, C1: 13; C2: 15). These findings can be observed in the frame of the IDH, which states that disturbance regimes that are intermediate in terms of frequency and/or intensity increases diversity, while for low frequencies diversity is low due to the dominance of a few species and for high frequencies, overall extinction rates are too high for most species to persist (Connell, 1978; Flöder and Sommer, 1999; Buckling et al., 2000; Wu et al., 2002; Griffiths and Philippot, 2013). For intermediate disturbance rates or intensities, many species can co‐exist because enough space is created for the weaker competitors without compromising their persistence.
Figure 6.
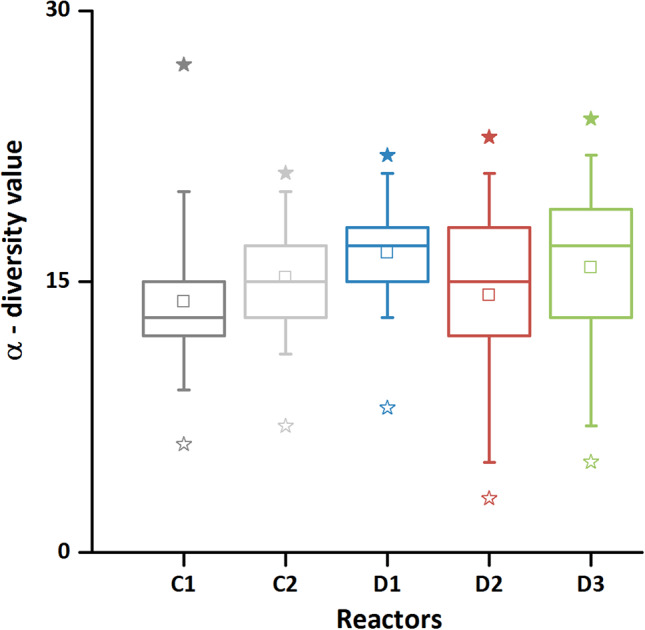
Box plot analysis of cytometric α‐diversity per each reactor (C1: dark grey; C2: light grey; D1: blue; D2: red and D3: green). α‐diversity values of 65 samples per each reactor were ranked in decreasing order from top to bottom, where whiskers mark the rank positions of 5% and 95% and stars mark the max (solid) and min (empty) values respectively. Box areas cover the rank range from 25% to 75% and the middle line is the position for medians.
Indeed, we temporarily determined low α‐diversity values for D2 and D3 and concurrent decreases in cell numbers from 6 × 106 cells mL−1 to approximately 6 × 104 ml−1 (Supporting Information Fig. S3.1). However, both diversity values and cell numbers were ultimately recovering, indicating disturbances that act at intermediate levels and allow the persistence of communities in their niches. Nevertheless, it needs to be stressed that persistence in cell number and α‐diversity values did not accompany persistence in community structures because all the communities developed disparate structures.
Slow competitive displacement causes non‐equilibrium community states
The purpose of stable community management is to maintain the community in a desired equilibrium with its environments. Constant environments are expected to contribute to the establishment of synchronized or at least multiple equilibria (DeAngelis and Waterhouse, 1987; Chase, 2003). However, no synchronization and no such equilibria were found in our reactors, as community structures varied disparately and over time. Under steady‐state performance, temperature disturbances caused unique niche differentiation processes. We showed that, while being reduced to low values intermittently, a high and unequal α‐diversity was finally maintained, although approximately 100 generations must have been produced. In non‐equilibrium theories, slow‐competitive displacement (Chesson and Case, 1986) was considered as one causal mechanism. An intermediate disturbance may suggest no or slow competitive displacement of organisms; therefore, the non‐equilibria in the disturbed reactors may have been supported by mechanisms of the IDH. In addition, Chesson and Case (1986) described the non‐equilibrium to be connected to the competitive equality of organisms in communities, which can go extinct only by random drift, a situation established predominantly in the control reactor set‐up. Therefore, competitive equality amongst organisms may always lead to non‐equilibrium in insular steady‐state systems that do not allow species inflow.
In summary, our findings describe the relative roles of neutral and niche differentiation mechanisms of microbial community assembly in steady‐state insular environments. The neutral mechanisms were indicated by (i) the absence of steep lognormal‐like abundance curves, (ii) varied order of ranked subcommunities per sample and (iii) increased inter‐community β‐diversity by pairwise community comparisons over time. The niche differentiation mechanisms were indicated by (i) geometric‐like rank‐order abundance curves with steeper slopes, which represented strong selections for specific dominant subcommunities and (ii) active interactions between abiotic and biotic parameters, as represented by the number of significant correlations. The neutral and niche differentiation mechanisms led to communities with low stability properties (resistance and recovery). Neutral mechanisms (especially drifts) were found to influence the assembly of microbial communities with superior strength in our system. Thus, disparate assembly trajectories of communities were found, therefore qualifying the communities in our reactors as non‐equilibrium systems. Furthermore, the tested periodic temperature disturbances were not an effective means to synchronize communities, and IDH suggested that the applied disturbances fell into intermediate disturbance pressure frequencies that caused competition in which species can co‐exist. These findings suggest that complex microbial communities in insular steady‐state environments probably cannot be synchronized or maintained in the original or intended assembly.
Experimental procedures
Origin and cultivation of the microbial communities
A natural wastewater sample was obtained from a full‐scale wastewater treatment plant and cultivated in parallel in five insular continuous steady‐state reactors. Specific conditions and details are provided in Supporting Information S1, and the set‐up of disturbances is shown in Supporting Information S2.
Flow cytometric measurements
Cells were directly stabilized after sampling and stored until measurement (Supporting Information S4). Cells were stained for DNA and measured by flow cytometry as described previously (Liu et al., 2018, Supporting Information S4). Samples were analyzed with a BD Influx v7 Sorter (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). The details of the instrumental and analysis set‐up are provided in Supporting Information S5. In addition to the optical calibration, 0.5 μm and 1 μm beads were amended into every sample as internal standards to monitor the instrument stability and allow for a correct comparison of cytometric data. All raw data can be accessed at the FlowRepository (URL: https://flowrepository.org/) under accession number: FR‐FCM‐ZYWX. Quality control of the cytometric measurements was performed with triplicate measures (Supporting Information S6). The abundance dynamics of cells per single subcommunity are shown in Supporting Information S11.
Analytical methods
Parameters were measured according to the German DIN guidelines as well as standard procedures. The details of the analytic procedures and values are provided in Supporting Information S3.
Analysis of ecological metrics and statistics
If not stated otherwise, all calculations were performed in RStudio (V1.0.143, Boston, MA, USA) with R (V3.3.3, R Core Team, 2017), and graphs were generated with the package ggplot2 (Wickham and Chang, 2017) and R‐script (URL https://github.com/LiuZishu/MCFlowCytoAnalysis). The R packages vegan (Oksanen et al., 2017) and flowCyBar (Koch et al., 2013, http://bioconductor.org/packages/flowCyBar/) were used for dissimilarity analysis, and the packages Hmisc (Harrell, 2017), psych (Revelle, 2017), and qgraph (Epskamp et al., 2017) were used for correlation analysis. For the stability calculation, an R‐script from Liu et al. (2018) was used. The data in Supporting Information S6, S9 and S10 were visualized with the software OriginPro (V9.0, OriginLab, Northampton, MA, USA).
Analysis of community composition by MiSeq
16S rRNA gene (V3‐V4 region) amplicon sequencing was performed using Illumina Miseq (San Diego, CA, USA) for exemplary whole community samples and sorted subcommunities. The procedure for DNA extraction, library preparation and sequencing data evaluation are documented in the supplementary information (Supporting Information S12). The sequencing data supported the conclusions drawn from the flow cytometry data as they verified general trends such as community adaptation to reactor conditions as well as the deterministic influence of the temperature disturbance on community composition and its recovery after disturbance. The samples were resolved on the class (Supporting Information S12) and genus level (Supporting Information S12, data repository https://osf.io/4tkcg/). One mock community (MBARC26, Singer et al., 2016) was added to the sequencing project to ensure the quality of the sequencing run as well as the sequencing analysis. All raw data are available under the BioProject (URL: https://www.ncbi.nlm.nih.gov/bioproject/) accession number: PRJNA437592.
Supporting information
Appendix S1 Supporting Information.
Movie 1: Cytometric community dynamics of two control and three disturbed reactors. Temperature changes for are shown at the top‐right corner.
Acknowledgements
We acknowledge the support of the Central Innovation Programme for SMEs (ZIM) of the Federal Ministry of Economic Affairs and Energy (BMWi) (INAR‐ABOS,16KN043222), the China Scholarship Council (CSC), the European Regional Development Funds (EFRE—Europe Funds Saxony, grant 100192205) and the Helmholtz Association within RP Renewable Energies.
References
- Adler, P. B. , HilleRisLambers, J. , and Levine, J. M. (2007) A niche for neutrality. Ecol Lett 10: 95–104. [DOI] [PubMed] [Google Scholar]
- Allison, S. D. , and Martiny, J. B. (2008) Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105: 11512–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy, J. (2015) The shape of terrestrial abundance distributions. Sci Adv 1: e1500082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon, M. , Townsend, C. R. , and Harper, J. L. (2006) The nature of the community: patterns in space and time In Ecology: From Individuals to Ecosystems, 4th ed. Malden, MA, USA: Blackwell Publishing. [Google Scholar]
- Benjamini, Y. , and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Bell, G. (2000) The distribution of abundance in neutral communities. Am Nat 155: 606–617. [DOI] [PubMed] [Google Scholar]
- Buckling, A. , Kassen, R. , Bell, G. , and Rainey, P. B. (2000) Disturbance and diversity in experimental microcosms. Nature 408: 961–964. [DOI] [PubMed] [Google Scholar]
- Burns, A. R. , Stephens, W. Z. , Stagaman, K. , Wong, S. , Rawls, J. F. , Guillemin, K. , and Bohannan, B. J. (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, A. L. , Belkaid, Y. , and Segre, J. A. (2018) The human skin microbiome. Nat Rev Microbiol 16: 143–155. [DOI] [PubMed] [Google Scholar]
- Chase, J. M. (2003) Community assembly: when should history matter? Oecologia 136: 489–498. [DOI] [PubMed] [Google Scholar]
- Chase, J. M. (2007) Drought mediates the importance of stochastic community assembly. Proc Natl Acad Sci U S A 104: 17430–17434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, J. M. (2010) Stochastic community assembly causes higher biodiversity in more productive environments. Science 328: 1388–1391. [DOI] [PubMed] [Google Scholar]
- Chesson, P. L. , and Case, T. J. (1986) Overview: nonequilibrium community theories: chance, variability, history In Community Ecology, Diamond J., and Case T. J. (eds). New York, USA: Harper and Row Publishers. [Google Scholar]
- Chesson, P. , and Huntly, N. (1997) The roles of harsh and fluctuating conditions in the dynamics of ecological communities. Am Nat 150: 519–553. [DOI] [PubMed] [Google Scholar]
- Choi, S. , Song, H. , Tripathi, B. M. , Kerfahi, D. , Kim, H. , and Adams, J. M. (2017) Effect of experimental soil disturbance and recovery on structure and function of soil community: a metagenomic and metagenetic approach. Sci Rep 7: 2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell, J. H. (1978) Diversity in tropical rain forests and coral reefs. Science 199: 1302–1310. [DOI] [PubMed] [Google Scholar]
- Daly, A. J. , Baetens, J. M. , Vandermaesen, J. , Boon, N. , Springael, D. , and De Baets, B. (2018) Individual‐based modelling of invasion in bioaugmented sand filter communities. Processes 6: 2. [Google Scholar]
- De Vrieze, J. , Christiaens, M. E. , Walraedt, D. , Devooght, A. , Ijaz, U. Z. , and Boon, N. (2017) Microbial community redundancy in anaerobic digestion drives process recovery after salinity exposure. Water Res 111: 109–117. [DOI] [PubMed] [Google Scholar]
- DeAngelis, D. L. , and Waterhouse, J. C. (1987) Equilibrium and nonequilibrium concepts in ecological models. Ecol Monogr 57: 1–21. [Google Scholar]
- Du, C. , Cui, C. , Qiu, S. , Xu, S. , Shi, S. , Sangeetha, T. , and Ma, F. (2017) Microbial community shift in a suspended stuffing biological reactor with pre‐attached aerobic denitrifier. World J Microbiol Biotechnol 33: 148. [DOI] [PubMed] [Google Scholar]
- Dumbrell, A. J. , Nelson, M. , Helgason, T. , Dytham, C. , and Fitter, A. H. (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4: 337–345. [DOI] [PubMed] [Google Scholar]
- Evans, S. , Martiny, J. B. , and Allison, S. D. (2017) Effects of dispersal and selection on stochastic assembly in microbial communities. ISME J 11: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epskamp, S. , Costantini, G. , Haslbeck, J. , Cramer, A. O. J. , Waldorp, L. J. , Schmittmann, V. D. , and Borsboom, D. (2017) qgraph: Graph Plotting Methods, Psychometric Data Visualization and Graphical Model Estimation. R Package Version 1.4.3, [WWW document]. URL https://cran.r-project.org/web/packages/qgraph
- Faust, K. , Lahti, L. , Gonze, D. , de Vos, W. M. , and Raes, J. (2015) Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol 25: 56–66. [DOI] [PubMed] [Google Scholar]
- Faust, K. , and Raes, J. (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10: 538–550. [DOI] [PubMed] [Google Scholar]
- Ferrenberg, S. , O'neill, S. P. , Knelman, J. E. , Todd, B. , Duggan, S. , Bradley, D. , et al (2013) Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J 7: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer, N. (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15: 579–590. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A. , Corbet, A. S. , and Williams, C. B. (1943) The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol 12: 42–58. [Google Scholar]
- Flöder, S. , and Sommer, U. (1999) Diversity in planktonic communities: an experimental test of the intermediate disturbance hypothesis. Limnol Oceanogr 44: 1114–1119. [Google Scholar]
- Friedman, J. , Higgins, L. M. , and Gore, J. (2017) Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol 1: 0109. [DOI] [PubMed] [Google Scholar]
- Gao, P. , Xu, W. , Sontag, P. , Li, X. , Xue, G. , Liu, T. , and Sun, W. (2016) Correlating microbial community compositions with environmental factors in activated sludge from four full‐scale municipal wastewater treatment plants in Shanghai, China. Appl Microbiol Biotechnol 100: 4663–4673. [DOI] [PubMed] [Google Scholar]
- Gause, G. F. (1934) Experimental analysis of Vito Volterra's mathematical theory of the struggle for existence. Science 79: 16–17. [DOI] [PubMed] [Google Scholar]
- Goodwin, S. , McPherson, J. D. , and McCombie, W. R. (2016) Coming of age: ten years of next‐generation sequencing technologies. Nat Rev Genet 17: 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel, D. , Canham, C. D. , Beaudet, M. , and Messier, C. (2006) Reconciling niche and neutrality: the continuum hypothesis. Ecol Lett 9: 399–409. [DOI] [PubMed] [Google Scholar]
- Griffin, J. S. , and Wells, G. F. (2017) Regional synchrony in full‐scale activated sludge bioreactors due to deterministic microbial community assembly. ISME J 11: 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, B. S. , and Philippot, L. (2013) Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37: 112–129. [DOI] [PubMed] [Google Scholar]
- Grimm, V. , and Wissel, C. (1997) Babel, or the ecological stability discussions: an inventory and analysis of terminology and a guide for avoiding confusion. Oecologia 109: 323–334. [DOI] [PubMed] [Google Scholar]
- Grimm, V. , and Wissel, C. (2004) The intrinsic mean time to extinction: a unifying approach to analysing persistence and viability of populations. Oikos 105: 501–511. [Google Scholar]
- Günther, S. , Faust, K. , Schumann, J. , Harms, H. , Raes, J. , and Müller, S. (2016) Species‐sorting and mass‐transfer paradigms control managed natural metacommunities. Environ Microbiol 18: 4862–4877. [DOI] [PubMed] [Google Scholar]
- Günther, S. , Koch, C. , Hubschmann, T. , Roske, I. , Müller, R. A. , Bley, T. , et al (2012) Correlation of community dynamics and process parameters as a tool for the prediction of the stability of wastewater treatment. Environ Sci Technol 46: 84–92. [DOI] [PubMed] [Google Scholar]
- Harrell, F. E. (2017) Hmisc: Harrell Miscellaneous. R Package Version 4.0–3, [WWW document]. URL https://cran.r-project.org/web/packages/Hmisc
- Hill, M. O. (1973) Diversity and evenness: a unifying notation and its consequences. Ecology 54: 427–432. [Google Scholar]
- Hibbing, M. E. , Fuqua, C. , Parsek, M. R. , and Peterson, S. B. (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, D. U. , Chapin, F. S. , Ewel, J. J. , Hector, A. , Inchausti, P. , Lavorel, S. , et al (2005) Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75: 3–35. [Google Scholar]
- Hubbell, S. P. (2001) The Unified Neutral Theory of Biodiversity and Biogeography. Princeton, NJ, USA: Princeton University Press. [Google Scholar]
- Hoskisson, P. A. , and Hobbs, G. (2005) Continuous culture–making a comeback? Microbiology 151: 3153–3159. [DOI] [PubMed] [Google Scholar]
- Jannasch, H. W. (1974) Steady state and the chemostat in ecology. Limnol Oceanogr 19: 716–720. [Google Scholar]
- Jax, K. , Jones, C. G. , and Pickett, S. T. A. (1998) The self‐identity of ecological units. Oikos 82: 253–264. [Google Scholar]
- Johnson, E. L. , Heaver, S. L. , Walters, W. A. , and Ley, R. E. (2017) Microbiome and metabolic disease: revisiting the bacterial phylum bacteroidetes. J Mol Med 95: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnke, J. , Cohen, Y. , de Leeuw, M. , Kushmaro, A. , Jurkevitch, E. , and Chatzinotas, A. (2014) Multiple micro‐predators controlling bacterial communities in the environment. Curr Opin Biotechnol 27: 185–190. [DOI] [PubMed] [Google Scholar]
- Koch, C. , Günther, S. , Desta, A. F. , Hübschmann, T. , and Müller, S. (2013) Cytometric fingerprinting for analyzing microbial intracommunity structure variation and identifying subcommunity function. Nat Protoc 8: 190–202. [DOI] [PubMed] [Google Scholar]
- Koch, C. , Harnisch, F. , Schröder, U. , and Müller, S. (2014) Cytometric fingerprints: evaluation of new tools for analyzing microbial community dynamics. Front Microbiol 5: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, C. , and Müller, S. (2018) Personalized microbiome dynamics–cytometric fingerprints for routine diagnostics. Mol Aspects Med 59: 123–134. [DOI] [PubMed] [Google Scholar]
- Kuczynski, J. , Liu, Z. , Lozupone, C. , McDonald, D. , Fierer, N. , and Knight, R. (2010) Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods 7: 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanto, E. , Hoikkala, V. , Ravantti, J. , and Sundberg, L. R. (2017) Long‐term genomic coevolution of host‐parasite interaction in the natural environment. Nat Commun 8: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance, G. N. , and Williams, W. T. (1967) Mixed‐data classificatory programs. I agglomerative systems. Austral Comput J 1: 15–20. [Google Scholar]
- Langenheder, S. , and Székely, A. J. (2011) Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht, J. , Cichocki, N. , Hübschmann, T. , Koch, C. , Harms, H. , and Müller, S. (2017) Flow cytometric quantification, sorting and sequencing of methanogenic archaea based on F420 autofluorescence. Microb Cell Fact 16: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold, M. A. , Holyoak, M. , Mouquet, N. , Amarasekare, P. , Chase, J. M. , Hoopes, M. F. , et al (2004) The metacommunity concept: a framework for multi‐scale community ecology. Ecol Lett 7: 601–613. [Google Scholar]
- Liu, Z. , Cichocki, N. , Bonk, F. , Günther, S. , Schattenberg, F. , Harms, H. , et al (2018) Ecological stability properties of microbial communities assessed by flow cytometry. mSphere 3: e00564–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur, R. (1960) On the relative abundance of species. Am Nat 94: 25–36. [Google Scholar]
- Matthews, T. J. , and Whittaker, R. J. (2014) Neutral theory and the species abundance distribution: recent developments and prospects for unifying niche and neutral perspectives. Ecol Evol 4: 2263–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully, A. L. , LaSarre, B. , and McKinlay, J. B. (2017) Growth‐independent cross‐feeding modifies boundaries for coexistence in a bacterial mutualism. Environ Microbiol 19: 3538–3550. [DOI] [PubMed] [Google Scholar]
- Morris, S. J. , and Blackwood, C. B. (2007) The ecology of soil organisms In Soil Microbiology, Ecology and Biochemistry, 3rd ed. Cambridge, MA, USA: Academic Press, pp. 195–229. [Google Scholar]
- Müller, S. (2007) Modes of cytometric bacterial DNA pattern: a tool for pursuing growth. Cell Prolif 40: 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S. , and Babel, W. (2003) Analysis of bacterial DNA patterns—an approach for controlling biotechnological processes. J Microbiol Methods 55: 851–858. [DOI] [PubMed] [Google Scholar]
- Nelson, M. B. , Martiny, A. C. , and Martiny, J. B. (2016) Global biogeography of microbial nitrogen‐cycling traits in soil. Proc Natl Acad Sci U S A 113: 8033–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut, D. R. , Schmidt, S. K. , Fukami, T. , O'Neill, S. P. , Bilinski, T. M. , Stanish, L. F. , et al (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham, D. M. , and Fuhrman, J. A. (2016) Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1: 16005. [DOI] [PubMed] [Google Scholar]
- Ofiţeru, I. D. , Lunn, M. , Curtis, T. P. , Wells, G. F. , Criddle, C. S. , Francis, C. A. , and Sloan, W. T. (2010) Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci U S A 107: 15345–15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , et al (2017) vegan: Community Ecology Package. R Package Version 2.4–3, [WWW document]. URL https://cran.r-project.org/web/packages/vegan
- Ovaskainen, O. , and Meerson, B. (2010) Stochastic models of population extinction. Trends Ecol Evol 25: 643–652. [DOI] [PubMed] [Google Scholar]
- Pagaling, E. , Vassileva, K. , Mills, C. G. , Bush, T. , Blythe, R. A. , Schwarz‐Linek, J. , et al (2017) Assembly of microbial communities in replicate nutrient‐cycling model ecosystems follows divergent trajectories, leading to alternate stable states. Environ microbial 19: 3374–3386. [DOI] [PubMed] [Google Scholar]
- Peter, H. , Jeppesen, E. , De Meester, L. , and Sommaruga, R. (2017) Changes in bacterioplankton community structure during early lake ontogeny resulting from the retreat of the Greenland ice sheet. ISME J 12: 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Valera, E. , Goberna, M. , Faust, K. , Raes, J. , García, C. , and Verdú, M. (2017) Fire modifies the phylogenetic structure of soil bacterial co‐occurrence networks. Environ Microbiol 19: 317–327. [DOI] [PubMed] [Google Scholar]
- Props, R. , Kerckhof, F. M. , Rubbens, P. , De Vrieze, J. , Sanabria, E. H. , Waegeman, W. , et al (2017) Absolute quantification of microbial taxon abundances. ISME J 11: 584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Props, R. , Monsieurs, P. , Mysara, M. , Clement, L. , and Boon, N. (2016) Measuring the biodiversity of microbial communities by flow cytometry. Methods Ecol Evol 7: 1376–1385. [Google Scholar]
- Props, R. , Schmidt, M. L. , Heyse, J. , Vanderploeg, H. A. , Boon, N. , and Denef, V. J. (2018) Flow cytometric monitoring of bacterioplankton phenotypic diversity predicts high population‐specific feeding rates by invasive dreissenid mussels. Environ Microbiol 20: 521–534. [DOI] [PubMed] [Google Scholar]
- Preston, F. W. (1948) The commonness, and rarity, of species. Ecology 29: 254–283. [Google Scholar]
- Revelle, W. (2017) psych: Procedures for Psychological, Psychometric, and Personality Research. R Package Version 1.7.5, [WWW document]. URL https://cran.r-project.org/web/packages/psych
- Schirmer, M. , Smeekens, S. P. , Vlamakis, H. , Jaeger, M. , Oosting, M. , Franzosa, E. A. , et al (2016) Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebacher, F. , and Franklin, C. E. (2012) Determining environmental causes of biological effects: the need for a mechanistic physiological dimension in conservation biology. Philos Trans R Soc B‐Biol Sci 367: 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, D. , Jürgens, K. , and Beier, S. (2018) Experimental insights into the importance of ecologically dissimilar bacteria to community assembly along a salinity gradient. Environ Microbiol 20: 1170–1184. [DOI] [PubMed] [Google Scholar]
- Singer, E. , Andreopoulos, B. , Bowers, R. M. , Lee, J. , Deshpande, S. , Chiniquy, J. , et al (2016) Next generation sequencing data of a defined microbial mock community. Sci Data 3: 160081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad, K. , Steen, H. B. , and Boye, E. (1985) Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol 163: 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, W. T. , Lunn, M. , Woodcock, S. , Head, I. M. , Nee, S. , and Curtis, T. P. (2006) Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8: 732–740. [DOI] [PubMed] [Google Scholar]
- Smith, N. R. , Kishchuk, B. E. , and Mohn, W. W. (2008) Effects of wildfire and harvest disturbances on forest soil bacterial communities. Appl Environ Microbiol 74: 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässle, C. , Sonnleitner, B. , and Fiechter, A. (1989) A predictive model for the spontaneous synchronization of Saccharomyces cerevisiae grown in continuous culture. II. Experimental verification. J Biotechnol 9: 191–208. [Google Scholar]
- Tokeshi, M. (1993) Species abundance patterns and community structure In Advances in Ecological Research, Begon M., and Fitter A. H. (eds). Cambridge, MA, USA: Academic Press, 24: 111–186. [Google Scholar]
- Tully, B. J. , Wheat, C. G. , Glazer, B. T. , and Huber, J. A. (2017) A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J 12: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwonterghem, I. , Jensen, P. D. , Rabaey, K. , and Tyson, G. W. (2015) Temperature and solids retention time control microbial population dynamics and volatile fatty acid production in replicated anaerobic digesters. Sci Rep 5: 8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellend, M. (2010) Conceptual synthesis in community ecology. Q Rev Biol 85: 183–206. [DOI] [PubMed] [Google Scholar]
- Wackett, L. P. (2018) Microbial acid fermentation products. Microbial Biotechnol 11: 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willers, H. C. , Derikx, P. J. L. , ten Have, P. J. W. , and Vijn, T. K. (1998) Nitrification limitation in animal slurries at high temperatures. Bioresour Technol 64: 47–54. [Google Scholar]
- Wickham, H. , and Chang, W. (2017) ggplot2: Create elegant data visualisations using the grammar of graphics. R package version 2.2.1, [WWW document]. URL https://cran.r-project.org/web/packages/ggplot2
- Woodcock, S. , and Sloan, W. T. (2017) Biofilm community succession: a neutral perspective. Microbiology 163: 664–668. [DOI] [PubMed] [Google Scholar]
- Woodcock, S. , van der Gast, C. J. , Bell, T. , Lunn, M. , Curtis, T. P. , Head, I. M. , and Sloan, W. T. (2007) Neutral assembly of bacterial communities. FEMS Microbiol Ecol 62: 171–180. [DOI] [PubMed] [Google Scholar]
- Wu, X. L. , Chin, K. J. , and Conrad, R. (2002) Effect of temperature stress on structure and function of the methanogenic archaeal community in a rice field soil. FEMS Microbiol Ecol 39: 211–218. [DOI] [PubMed] [Google Scholar]
- Wu, J. , and Loucks, O. L. (1995) From balance of nature to hierarchical patch dynamics: a paradigm shift in ecology. Q Rev Biol 70: 439–466. [Google Scholar]
- Zeng, M. Y. , Inohara, N. , and Nuñez, G. (2017) Mechanisms of inflammation‐driven bacterial dysbiosis in the gut. Mucosal Immunol 10: 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Liu, W. , Deng, Y. , Jiang, Y. H. , Xue, K. , He, Z. , et al (2013) Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 4: e00584–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , and Ning, D. (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81: e00002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, J. , Hübschmann, T. , Schattenberg, F. , Schumann, J. , Durek, P. , Riedel, R. , et al (2016) High‐resolution microbiota flow cytometry reveals dynamic colitis‐associated changes in fecal bacterial composition. Eur J Immunol 46: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information.
Movie 1: Cytometric community dynamics of two control and three disturbed reactors. Temperature changes for are shown at the top‐right corner.


