Abstract
Bacterial endosymbionts have enabled aphids to adapt to a range of stressors, but their effects in many aphid species remain to be established. The bird cherry‐oat aphid, Rhopalosiphum padi (Linnaeus), is an important pest of cereals worldwide and has been reported to form symbiotic associations with Serratia symbiotica and Sitobion miscanthi L‐type symbiont endobacteria, although the resulting aphid phenotype has not been described. This study presents the first report of R. padi infection with the facultative bacterial endosymbiont Hamiltonella defensa. Individuals of R. padi were sampled from populations in Eastern Scotland, UK, and shown to represent seven R. padi genotypes based on the size of polymorphic microsatellite markers; two of these genotypes harbored H. defensa. In parasitism assays, survival of H. defensa‐infected nymphs following attack by the parasitoid wasp Aphidius colemani (Viereck) was 5 fold higher than for uninfected nymphs. Aphid genotype was a major determinant of aphid performance on two Hordeum species, a modern cultivar of barley H. vulgare and a wild relative H. spontaneum, although aphids infected with H. defensa showed 16% lower nymph mass gain on the partially resistant wild relative compared with uninfected individuals. These findings suggest that deploying resistance traits in barley will favor the fittest R. padi genotypes, but symbiont‐infected individuals will be favored when parasitoids are abundant, although these aphids will not achieve optimal performance on a poor quality host plant.
Keywords: cereal aphid, Hamiltonella defensa, Hordeum spontaneum, Hordeum vulgare, symbiosis
Introduction
Aphids form a diverse range of associations with endosymbiotic bacteria, ranging from obligatory to facultative and beneficial to parasitic. The primary aphid endosymbiont, Buchnera aphidicola, provides nutritional supplementation to the aphid diet (Sasaki et al., 1991; Douglas & Prosser, 1992). Additional coobligatory symbioses with B. aphidiciola have been described in other aphid species, including cosymbiosis with Wolbachia sp. in the banana aphid, Pentalonia nigronervosa (Coquerel) (De Clerck et al., 2015), and Serratia symbiotica in Cinara species (Meseguer et al., 2017). The most frequently detected facultative endosymbionts of aphids are Hamiltonella defensa, Regiella insecticola, S. symbiotica, Rickettsia sp., Ricketsiella sp., Spiroplasma sp., the Pea Aphid X‐type Symbiont (PAXS) and Wolbachia sp. (Sandström et al., 2001; Oliver et al., 2003; Oliver et al., 2006; Degnan & Moran, 2008b; Guay et al., 2009; Oliver et al., 2010; Tsuchida et al., 2010; Łukasik et al., 2013b; De Clerck et al., 2014). A concise review of endosymbiont occurrence in aphid populations (Zytynska & Weisser, 2016) found that the facultative endosymbionts S. symbiotica and Wolbachia infected the highest proportion of the aphid species assessed (47% and 43%, respectively). Occasional associations have also been reported with Arsenophonus sp. (Jousselin et al., 2013; Wagner et al., 2015), infecting 7% of aphid species tested (Zytynska & Weisser, 2016), and two divergent Rickettsiacae species, known as SMLS (Sitobion miscanthis L‐type symbiont) and OLO (Orientia‐Like Organism) (Li et al., 2011, 2016). Variation in the frequency of aphid endosymbiont infection is thought to arise from a wide range of processes, including aphid utilization of different host plant species, compatibility between different aphid genotypes and symbiont strains, and aphid interactions with the biotic and abiotic environment (Zytynska &Weisser, 2016).
The consequences of endosymbiont infection for aphid fitness are not always clear, particularly for the most recently described taxa. A recent review by Guo et al. (2017) summarized the known effects of nine of these endosymbionts, although it is increasingly apparent that these effects are not always consistent between aphid species and endosymbiont strains. A well‐recognized fitness effect of endosymbiotic associations between facultative symbionts and aphid hosts is through their contribution to aphid resistance to parasitoid wasp species, particularly members of the Braconidae, which regulate aphid populations in natural and agricultural vegetation (Oliver et al., 2003; Oliver et al., 2010; Asplen et al., 2014; Cayetano & Vorburger, 2015). The primary mechanism of resistance against Braconid wasps in the pea aphid, Acyrthosiphon pisum (Harris), has been attributed to the Acyrthosiphon pisum Secondary Endosymbiont (APSE) bacteriophage that is frequently associated with H. defensa (Moran et al., 2005; Degnan & Moran, 2008a,b; Oliver et al., 2009). Phage‐derived factors have been reported to arrest the development of wasp embryos (Brandt et al., 2017). By contrast, resistance of the peach‐potato aphid, Myzus persicae (Sulzer), to Braconid wasps was associated with the facultative endosymbiont R. insecticola (von Burg et al., 2008; Vorburger et al., 2010). Experimental transfer of R. insecticola from M. persicae confirmed that this strain conferred resistance in Ac. pisum to the parasitoid Aphidius ervi (Haliday) (although a strain of R. insecticola derived from Ac. pisum was not protective) and this was attributed to a repertoire of pathogenicity factors in the virulent R. insecticola strain (Hansen et al., 2012).
The effect of many aphid endosymbionts on their host has been elucidated using Ac. pisum as a model. Alongside parasitoid defense, additional traits conferred to aphids by facultative endosymbionts include thermal tolerance (Russell & Moran, 2006) and adaptation to different host plant species (Tsuchida et al., 2004); however, in some cases, endosymbiont infection can lead to detrimental effects on aphid fitness as reported for the black bean aphid, Aphis fabae (Scopoli) (Vorburger & Gouskov, 2011) and Ac. pisum (Martinez et al., 2018). Research has also detected differences between aphid species in the effect of some facultative endosymbionts on aphid fitness. For example, the protective effect of H. defensa against parasitoid wasps is observed consistently for Ac. pisum (Oliver et al., 2003) and A. fabae (Schmid et al., 2012) but not for the English grain aphid, Sitobion avenae (Fabricius), or the potato aphid, Macrosiphum euphorbiae (Thomas) (Łukasik et al., 2013a; Clarke et al., 2017), which might be due to infection with nonprotective endosymbiont strains or infection with strains of endosymbionts that are ineffective against particular parasitoid species or genotypes (Vorburger & Rouchet, 2016; Dennis et al., 2017).
The bird cherry‐oat aphid, Rhopalosiphum padi (Linnaeus), is a worldwide agricultural pest of cereals (Leather et al., 1989) and a primary vector of economically damaging plant viruses, including Barley Yellow Dwarf Virus (BYDV) (Valenzuela & Hoffmann, 2015). Cereal yield losses due to BYDV infection can reach 35% (Perry et al., 2000) and might rise further as R. padi is anticipated to become a more persistent agricultural pest under a changing climate (Finlay & Luck, 2011). Despite the economic importance of R. padi, the endosymbionts associated with this aphid species, and their effects on aphid fitness, are not well described. Desneux et al. (2018) screened 18 R. padi lines and Henry et al. (2015) screened 11 lines of R. padi for the presence of endosymbionts, but neither study found evidence for the presence of facultative bacterial endosymbionts. By contrast, S. symbiotica was detected in populations of R. padi along the North Belgian coast by de la Peña et al. (2014), and Li et al. (2011) detected SMLS in R. padi collected from Jiangsu province in China. Functional characterization of these facultative endosymbionts in R. padi remains to be reported; although the role of SMLS in cereal aphids is not known, in other aphid species S. symbiotica has been reported to enhance aphid resistance against parasitoids, often in synergy with H. defensa (Oliver et al., 2003; Oliver et al., 2006). To address this knowledge gap, species‐specific research is needed to elucidate the role of these aphid endosymbionts, particularly in aphid species of agricultural and economic significance.
The primary strategy for controlling insect pests is via the application of insecticidal chemicals. However, due to their widespread environmental impacts (reviewed by Goulson, 2013), and the emergence of pesticide resistance (Field et al., 1988; Bass et al., 2014; Foster et al., 2014), the continued use of pesticides is considered unsustainable (Geiger et al., 2010). Alternative pest management solutions could include augmenting biocontrol using natural enemies (Ramsden et al., 2017), and plant‐mediated resistance. Mitchell et al. (2016) identified several resistance and tolerance traits that could be employed to increase plant resistance to arthropod pests, including physical barriers, chemical defenses, and reduced plant palatability. Indeed, resistance to cereal aphids has been identified in maize (Betsiashvili et al., 2015) and wheat (Girvin et al., 2017). A recent review by Jarosova et al. (2016) suggested that a strategy for tackling cereal aphid and BYDV control might lie in comparison of traits of susceptible modern crops with their wild relatives that display partial inherent resistance as a means to advise molecular breeding programmes. A comparative study of barley, Hordeum vulgare, and the wild relative, H. spontaneum, highlighted differential gene regulation in response to aphid infestation that might explain their differences in aphid susceptibility (Delp et al., 2009), although further work is needed to elucidate fully the underlying mechanism(s). Aphid endosymbionts have been reported to influence aphid fitness and adaptation to host plant species and plants that differ in quality (Tsuchida et al., 2004; Gauthier et al., 2015; Wagner et al., 2015). A better understanding of how endosymbionts modify the effects of plant resistance on aphid success might provide insights for improving the sustainability of insect pest management.
The primary aim of this study was to determine the presence and types of facultative endosymbionts associated with R. padi genotypes collected from U.K. populations and to ascertain the effects of any detected endosymbionts on aphid fitness. To achieve this, clonal lines were first established from R. padi individuals collected in Eastern Scotland and were characterized for aphid genotype and presence of facultative endosymbionts. Secondly, we tested the hypothesis that facultative endosymbionts influence aphid fitness by (i) examining variation in aphid susceptibility to parasitism by the common parasitoid wasp Aphidius colemani (Viereck) (Ronquim et al., 2004; McClure & Frank, 2015) and (ii) quantifying aphid performance on a susceptible modern cultivar of barley, H. vulgare cv. Concerto, and a barley wild relative, H. spontaneum 5 (HsP5), previously described as partially aphid‐resistant (Delp et al., 2009). We predicted that poor aphid performance relating to aphid genotype and/or endosymbiont status would be exacerbated on a partially resistant plant host.
Materials and methods
Plant material
Hordeum vulgare cv. Concerto and H. spontaneum 5 (HsP5) seeds were surface‐sterilized by rinsing in 2% (v/v) hypochlorite solution followed by three rinses in deionized water (ddH2O). Seeds were then kept moist in the dark. H. vulgare cv. Concerto seeds were stratified by incubating at room temperature for 48 h whereas HsP5 seeds were incubated at 4 °C for 14 d. Germinated seedlings were planted into a bulrush compost mix (Bulrush, Northern Ireland) under glasshouse conditions (16 : 8 h light and 20 : 15 °C day : night) until the first true leaf emerged (stage 1.2 on the Zadoks et al., 1974 decimal key) when they were used in insect assays.
Insect rearing
Individual apterous R. padi adults collected from cereal crops and grasses in Eastern Scotland, UK, in summer 2013 and summer 2016 were used to establish clonal lines. Cultures were reared on 1‐week‐old barley seedlings (H. vulgare cv. Optic; growth stage 1.1–1.2 on the Zadoks scale) contained in ventilated cups. These comprised two Perspex cups (50 mm width × 150 mm depth) placed one inside the other; barley seedling roots were placed into a c. 10 mm depth of water in the base of the outermost cup, with the stem inserted through a c. 5 mm circular hole in the base of the inner cup, and the cup surface was sealed with a mesh‐ventilated lid. A mixed population of the peach‐potato aphid, M. persicae (genotypes F and O; determined to be free from facultative endosymbiont infection by diagnostic PCR screening, as described below for R. padi), was reared in ventilated Perspex cages on young oilseed rape plants, Brassica napus cv. Mascot (growth stage 2.3–2.5 as determined using the Harper & Berkenkamp, 1975 staging key), produced in the growing medium and conditions described above. Plant material was replaced weekly.
Mummies of the Braconid wasp A. colemani, supplied by Fargro (West Sussex, UK), were transferred to plastic ventilated boxes supplied with a food source of 50% (v/v) honey, which is deemed suitable for rearing Hymenopteran parasitoids (Perera & Hemachandra, 2014), soaked into a cotton wool ball. A cohort of emerging wasps (5–7 d old) was transferred to M. persicae‐infested oilseed rape plants (growth stage 2.3–2.5, determined using the Harper and Berkenkamp key) enclosed in a fine mesh cage. After 12 d, aphid mummies were collected and transferred to a ventilated plastic box supplied with honey solution until the next generation of adult wasps had emerged. To ensure parasitoids had no prior experience of the experimental R. padi clones, wasps were reared through at least three generations on M. persicae before being used in bioassays. All insect cultures were maintained at 18 ± 2 °C and 16 h : 8 h (day : night).
Rhopalosiphum padi genotyping
DNA was extracted from frozen homogenized tissue of c. 20 aphids per clonal line, using the DNeasy Plant Mini Kit (Qiagen, UK) following the manufacturer's protocol. First, aphids were washed in 96%–100% ethanol (Sigma‐Aldrich, UK) for 5 min and rinsed three times with Gibco® distilled water (ThermoFischer Scientific, UK); samples were then flash‐frozen in liquid nitrogen and homogenized using a micropestle. Extracted DNA was quantified using a Nanodrop ND‐1000 (ThermoFischer Scientific, UK).
Asexual aphid lines were assigned to genotypes based on the length of six of the polymorphic microsatellite markers for R. padi identified by Simon et al. (2001) and an additional unpublished marker; two other microsatellite markers (R 1–35 and R 3–171; Simon et al., 2001) could not be amplified consistently across all asexual lines and were not used. Microsatellite primers are shown in Table S1. A ProFlex PCR System (Applied Biosystems, UK) was used to amplify the target microsatellites in 25 µL reactions, containing final reaction concentrations of 1.5 mmol/L MgCl2, 250 µmol/L of mixed deoxynucleotide triphosphate (dNTP), 1 µmol/L forward primer with a 6‐FAM fluorophore attached to the 5′ end, 1 µmol/L reverse primer, 1× Clear GoTaq® reaction buffer (Promega, UK), and 1.25 U GoTaq® DNA Polymerase (Promega, UK), with approximately 15 ng of DNA template. Thermocycling conditions consisted of 98 °C for 30 s, followed by 35 cycles of 98 °C for 30 s, an annealing step consisting of a temperature of either 52 °C or 60 °C for 30 s, and 72 °C for 45 s with a final extension step at 72 °C for 7 min; marker R 6‐3 was annealed at 52 °C, while all other markers were annealed at 60 °C.
Following successful amplification, which was determined by separating a 10 µL aliquot of the amplicons on 2% agarose gel stained with SYBR Safe®, PCR products were separated by capillary electrophoresis; first, the amplified products were diluted 1 : 10 with Gibco® distilled water, then 1 µL of the diluted sample was mixed with 0.16 µL of GeneScan™ 500 ROX™ dye size standard (ThermoFischer Scientific, UK) and suspended in 8.84 µL Hi‐Di™ Formamide (ThermoFischer, UK) in a nonskirted 96‐well plate and sealed with an adhesive film. PCR products were separated on an ABI 3730 DNA Analyser (Applied Biosystems, UK). Product size (bp) was assessed using Peak Scanner™ software v 1.0 (Applied Biosystems, UK), and aphid genotype was determined based on the pattern of PCR product sizes from the amplified alleles (Table S2).
Facultative endosymbiont detection
Diagnostic PCR screening
A diagnostic PCR screen was used targeting universal eubacterial 16S rDNA and the 16–23S rDNA (including the intergenic spacer), and the specific 16S rDNA target sequence of the seven most frequently detected aphid endosymbionts Regiella insecticola, Hamiltonella defensa, Serratia symbiotica, PAXS, Spiroplasma sp., Rickettsia sp., and Rickettsiella sp. Initially, extracted aphid DNA was pooled, using 5 µL of DNA from each R. padi asexual line, and screened for all diagnostic targets of aphid facultative endosymbionts (see Table S1 for primer details). The reactions were conducted using a G‐storm GS4822 thermocycler in a final reaction volume of 25 µL, with reaction concentrations of 1.5 mmol/L MgCl2, 250 µmol/L of mixed dNTP's, 1 µmol/L forward primer, 1 µmol/L reverse primer, 1× Green GoTaq® reaction buffer (Promega, UK) and 1.25 U GoTaq® DNA Polymerase (Promega, UK), and with approximately 15 ng of DNA template; thermocycling conditions are described in Table S3. An aliquot (10 µL) of the amplified product was separated and visualized on 1.5% agarose gel using SYBR Safe® DNA staining agent. In positive reactions, the residual 15 µL of amplified product was purified using the QIAquick PCR Purification Kit (Qiagen, UK) following the manufacturer's protocol. Purified products were quantified and analyzed for quality using a Nanodrop ND‐1000 (ThermoFischer Scientific, UK) and aliquots (250 ng template per 1.5 Kb product length) were prepared for sequencing using Sanger methodology. Sequencing reactions contained 1 µL primer (10 µmol/L), 2 µL of BigDye™ Terminator v3.1 mix (ThermoFisher Scientific, UK), and 1.0 µL of 5× BigDye™ dilution buffer (ThermoFischer Scientific, UK). Cycling was carried out on a Tetrad Cycler (Biorad, Hertfordshire, UK) using the following conditions: 96 °C for 20 s followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, 60 °C for 4 min. PCR products were purified by ethanol precipitation, air‐dried, and resuspended in 10 µL of Hi‐Di™ formamide (ThermoFisher Scientific, UK). Sequencing of products was carried out using a 36 cm capillary array on a 48 capillary ABI 3730 (ThermoFisher Scientific, UK).
Sequence data were subjected to a BLASTn search, using the NCBI online database, to check similarity to known aphid endosymbionts. The presence of detected endosymbionts in individual aphid lines was confirmed using diagnostic PCR of the appropriate 16S rDNA gene, and products from positive amplifications were purified and sequenced as described above.
16–23S rDNA sequencing for screening of endosymbionts not targeted by diagnostic PCR
Briefly, the 16–23S rDNA region of a pooled R. padi DNA sample was amplified using the thermocycling conditions described above. Amplified products were purified using the QIAquick PCR Purification Kit (Qiagen), following the manufacturer's protocol. To the purified DNA template, a ‐CACC‐ tag was cloned into the 5′ region using the altered 16–23S rDNA F primer: 5′‐CACC AGTTTGATCATGGCTCAGATTG‐3′; this cloning procedure was carried out in a G‐storm GS4822 thermocycler in a final volume of 25 µL containing Phusion® High‐Fidelity DNA Polymerase (0.02 U/µL), 1× High‐Fidelity Buffer (ThermoFisher Scientific, UK), 200 µmol/L of each dNTP, and 0.5 µmol/L of each primer, under the following thermocycling conditions: 98 °C for 3 min, followed by 35 cycles of 98 °C for 30 s, 67 °C for 45 s, and 72 °C for 45 s, with a final elongation step of 72 °C for 10 min. The amplified 5′‐tagged 16–23S rDNA region was purified using the QIAquick PCR Purification Kit (Qiagen) following the manufacturer's instructions. The purified product was cloned into a pENTR™/D‐TOPO® vector with kanamycin resistance and transformed into One Shot® Chemically Competent E. coli Cells, following the instructions in the pENTR™ Directional TOPO® Cloning Kit manual (ThermoFisher Scientific, UK).
Transformed E. coli were incubated on Luria–Bertani (LB) plates supplemented with 50 µL/mL kanamycin for 16 h at 37 °C; this step was repeated twice to isolate individual colonies. Five individual colonies were selected from each of 25 plates and individually grown in 5 mL of LB broth, supplemented with 50 µL/mL kanamycin, for 16 h at 300 r/min and 37 °C. Plasmids were extracted from 4 mL of the resulting LB culture using the QIAprep Miniprep Kit (Qiagen, UK) following the manufacturer's protocol. Extracted plasmid DNA was quantified and checked for quality using a Nanodrop ND‐1000 (ThermoFischer Scientific, UK), and aliquots (at least 250 ng of template per 5 Kb vector) were subject to Sanger sequencing. The reaction mix comprised 1 µL M13F primer 5′‐GTAAAACGACGGCCAG‐3′ (10 µmol/L), 2 µL of BigDye™ Terminator v3.1 mix (ThermoFisher Scientific, UK) and 1 µL of 5× BigDye™ dilution buffer (ThermoFischer Scientific, UK). The reaction was carried out using a Tetrad Cycler (Biorad, Hertfordshire, UK) with the following conditions: 96 °C for 20 s, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, 60 °C for 4 min. Sequencing products were purified by ethanol precipitation, then air‐dried and resuspended in 10 µL of Hi‐Di™ formamide (ThermoFisher Scientific). Sequencing was carried out using a 360 mm capillary array on a 48 capillary ABI 3730 (ThermoFisher Scientific, UK). Sequences were compared with known prokaryotic sequences held on the NCBI BLAST server.
Detection of the APSE bacteriophage
Aphid lines harboring H. defensa were subjected to additional diagnostic PCR screening for the detection of the lysogenic bacteriophage, APSE, using three APSE genomic markers (P3, P35, and P51) using the thermocycling conditions described in Table S3. Amplified PCR products were purified, visualized on 1.5% agarose gel, and sequenced as described above. The best BLASTn hits of the sequences are shown in Table S4.
Aphid parasitism assay
Parasitism assays were conducted on four clonal lines of a single aphid genotype (genotype E), two lines harboring H. defensa (DL 16/04, DL 16/05) and two lines free from H. defensa infection (DL 16/06, DL 16/13), with a total of seven assays per aphid line. Assay arenas were constructed using four leaves of barley (H. vulgare cv. Optic, growth stage 1.1–1.2 on the Zadoks scale) fixed adaxial side up into 1% (w/v) agarose in Petri dishes of 120 mm diameter. Ten R. padi nymphs (1st–3rd instar) were transferred into each arena and a single A. colemani female (5–7 d old), presumed mated, was introduced. After each wasp oviposition event, the attacked nymph was transferred to fresh leaves of H. vulgare cv. Optic in a ventilated cup. Attacked nymphs were examined daily for 12 d postparasitism. Mummies were carefully removed using fine forceps and placed in ventilated plastic boxes until eclosion. Rhopalosiphum padi mortality was measured as the proportion of nymphs mummified out of the ten nymphs that had been attacked, and sex determination of the emerged wasps was scored based on the presence of an ovipositor.
Aphid performance assays
Three separate aphid performance experiments were carried out, using two experimental methods, on plants initially at the first true leaf stage (1.2 on the Zadoks decimal growth scale; Zadoks et al., 1974), which were conducted as follows.
Experiments one and two each consisted of 12 replicates for each experimental treatment, and used Perspex clip cages (MacGillivray & Anderson, 1957) to contain the aphids onto the experimental plants; these two experiments were conducted under glasshouse conditions (16 h : 8 h L : D and 20 : 14 °C) and assessed the performance of the same four genotype E aphid lines with differential H. defensa infection that were used in the parasitism assays (see above and Table 1). Plants were infested with a single apterous aphid, which was allowed to reproduce overnight. A total of three nymphs were retained on each plant and mean nymph mass was recorded after 48 h and 144 h, after which a single nymph, selected at random, was returned to the plant; for this focal nymph, data was collected on the prereproductive period (d) and the intrinsic rate of population increase (rm).
Table 1.
Presence of Hamiltonella defensa and APSE in 16 asexual lines of R. padi
| Endosymbiont marker | APSE marker | |||||||
|---|---|---|---|---|---|---|---|---|
| R. padi asexual line | Genotype | 16S rDNA (+ve for B. aphidicola) | 16–23S rDNA (+ve for facultative symbiont presence) | H. defensa 16S rDNA | Accession number of sequenced H. defensa 16S rDNA | APSE P51 | APSE P35 | APSE P3 |
| AK 13/33 ‡ | A | + | – | – | ||||
| AK 13/34 ‡ | B | + | – | – | ||||
| DL 16/14 | C | + | + | + | MG595523 | – | + | – |
| DL 16/12 | D | + | – | – | ||||
| DL 16/02 | E | + | – | – | ||||
| DL 16/03 | E | + | + | + | MG595518 | + | + | + |
| DL 16/04 †, ‡ | E | + | + | + | MG595519 | + | + | + |
| DL 16/05 †, ‡ | E | + | + | + | MG595520 | + | + | + |
| DL 16/06 †, ‡ | E | + | – | – | ||||
| DL 16/07 | E | + | + | + | MG595521 | + | + | + |
| DL 16/08 | E | + | + | + | MG595522 | + | + | + |
| DL 16/10 | E | + | – | – | ||||
| DL 16/13 †, ‡ | E | + | – | – | ||||
| DL 16/15 | F | + | – | – | ||||
| DL 16/16 | F | + | – | – | ||||
| JB | G | + | − | − | ||||
†Indicates aphid lines used in parasitism experiments.
‡Indicates aphid lines used in performance experiments.
Experiment three assessed the performance of aphid lines representing aphid genotypes A, B, and E, consisted of 10 replicates per experimental treatment and was conducted in a Sanyo controlled environment cabinet (PAR 150 µmol/m−2 s−1, 16 : 8 h L : D and 20 °C ± 2 °C); the same four genotype E aphid lines used in experiments one and two were used, along with genotype A (aphid line AK 13/33) and genotype B (AK 13/34), and aphids were contained on plants using microperforated bags. Plants were infested with a single apterous aphid, which was allowed to reproduce overnight. The entire progeny of the aphid was retained on the plant for 48 h, at which point all nymphs were removed and the mass of a single nymph, selected at random, was recorded and returned to the plant for further monitoring of nymph mass at 144 h, prereproductive period and rm. Aphid survival was also recorded in experiment 3 until aphids were 21 d old.
For all three experiments, nymph mass gain was calculated as the change in mass between 144 and 48 h. Aphid rm was calculated using the equation of Wyatt and White (1977), where d is the time period between aphid birth and production of first progeny, and Fd is the total progeny over a time period equal to d:
Statistical analysis
All statistical analyses were carried out using R Studio Desktop version 1.0.143 running R version 3.4.0 (R Core Team, 2014), with additional packages broom (v. 0.4.2) (Robinson, 2017), car (v. 2.1–4) (Fox & Weisberg, 2011), coxme (v. 2.2–7) (Therneau, 2018), ggplot2 (v. 2.2.1) (Wickham, 2009), ggpubr (v. 0.1.2) (Kassambara, 2017), lme4 (v. 1.1–13) (Bates et al., 2015), lmerTest (v. 2.0–33), pkbrtest (v.0.4–7) (Halekoh & Højsgaard, 2014), pastecs (v. 1.3–18) (Grosjean & Ibanez, 2014), survival (v. 2.41–3) (Therneau & Grambsch, 2000), and survminer (v. 0.3.1) (Kassambara & Kosinski, 2017).
Aphid mortality (mummification) and the proportion of female : male wasps emerging from the mummified aphids were each modeled using a generalized linear mixed effects model fitted with binomial distribution with wasp generation and batch incorporated as random factors and aphid asexual line as a random factor nested within endosymbiont association. Model simplification in both cases was carried out through manual backward stepwise model selection. Throughout both model simplification processes, analysis of deviance using a Type II Wald χ 2 test and observing changes in AIC ensured that model simplification was justified and that the data fitted the model parameters. Fitted‐residual plots of the final models were assessed for model suitability.
Aphid performance data were split into two subDatasets. One, labeled “Genotype,” assessed insect performance in relation to aphid genotype, host plant (cv. Concerto or HsP5) and genotype × host plant interaction in aphid lines uninfected with H. defensa, namely AK 13/33, AK 13/34, DL 16/06, and DL 16/13 belonging to genotypes A, B, E, and E, respectively. Another subDataset, labeled “Endosymbiont,” assessed aphid performance in relation to host plant, facultative endosymbiont association with H. defensa and the plant × endosymbiont interaction in four aphid lines from genotype E with differential H. defensa infection: DL 16/04 (Hd +), DL 16/05 (Hd +), DL 16/06 (Hd–), and DL 16/13 (Hd–). In each subDataset, nymph mass gain, prereproductive period and rm were assessed in separate linear mixed effects models, incorporating experiment number and experimental block as random factors. To minimize the influence of multiple aphid lines representing genotype E on the model outcome, aphid line was incorporated into the models as a nested random factor and was nested within aphid genotype in the “Genotype” subDataset and within Endosymbiont association in the “Endosymbiont” subDataset. All models were simplified using manual backward stepwise model selection to reach the final models with Type II Wald χ 2 analysis of deviance and observing changes in AIC to ensure that model simplification was justified and that the data fitted the model parameters. Calculation of the differences of Least Squares Means was used as a post hoc test on the final models to identify which levels in each factor were significantly different.
For survival analysis, the two subDatasets were modeled separately by fitting a Cox proportional hazards regression model with experimental block incorporated as a random factor and aphid line incorporated as a nested factor within aphid genotype or endosymbiont association. Model simplification was carried out using manual backward stepwise model selection.
Results
Hamiltonella defensa associates with Rhopalosiphum padi
Based on the banding patterns of the six microsatellite markers used, the R. padi asexual lines were grouped into one of seven genotypes (labeled A–G; Table S2). Hamiltonella defensa was detected in six asexual lines within genotypes C and E only (Table 1). These six aphid lines were also positive for the APSE P35 genomic marker, with five lines positive for all three APSE genomic markers (P3, P35, P51; Table 1). Additional sequencing of pooled 16–23S rDNA extracted from all aphid lines did not detect the presence of additional eubacterial endosymbionts. Differential presence of H. defensa and APSE was detected in genotype E only (Table 1); comparative assays to detect the effect(s) of H. defensa infection on aphid performance focused on four aphid lines from genotype E, two with and two without H. defensa infection (DL 16/04, DL 16/05, DL 16/06, and DL 16/13). Insect fitness was compared between multiple aphid genotypes using H. defensa‐free clonal lines of genotypes A (AK 13/33), B (AK 13/34), and E (DL 16/06, DL 16/13).
Hamiltonella defensa confers protection against A. colemani in R. padi
Aphid mortality after parasitoid attack was significantly lower for aphid lines harboring the facultative endosymbiont H. defensa (χ 2 1,24 = 92.07, P < 0.001; Fig. 1; Table 2). The ratio of female : male wasps emerging from mummified aphids can be used as an indicator of aphid host suitability, with a higher proportion of female progeny indicative of a good quality host (King, 1987; Pandey & Singh, 1999); the observed female : male ratio was consistent across all treatments (50 : 50, on average) indicating that aphid genotype and presence of H. defensa did not alter aphid quality as a host for A. colemani (Table 2).
Figure 1.
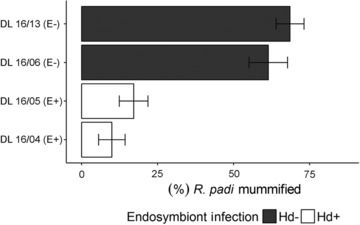
The effect of facultative endosymbiont presence (+/–) in aphid lines of genotype E on aphid mortality after attack by A. colemani. Values are means ± SE. Number of observations in model = 28.
Table 2.
Summary of statistical modeling outputs for aphid parasitism assays. For each performance parameter, the treatment factor, model basis, error distribution, analysis method, and statistical outputs are shown. Type II Wald χ 2 analysis of deviance and observation of fitted‐residual plots was conducted throughout the modeling simplification process to ensure data fitted the model parameters and that model simplification was justified
| Response variable | Treatmentfactor | Model basis | Error distribution | Modelanalysis | Test statistic | Degrees of freedom (residuals) | P value |
|---|---|---|---|---|---|---|---|
| Susceptibility to parasitism (number of aphid mummies) | Endosymbiont infection | Generalized linear mixed effects model | Binomial | Type II Wald χ 2 analysis of deviance (χ 2 Test) | χ 2 = 92.07 | 1 (24) | <0.001 |
| Sex of wasp progeny | Endosymbiont infection | Generalized linear mixed effects model | Binomial | χ 2 = 0.07 | 1 (100) | 0.787 |
Aphid genotype and host plant identity are dominant factors influencing aphid performance
Aphid prereproductive period and survival probability were unaffected by host plant identity, aphid genotype, and endosymbiont infection (Tables 3 and 4). However, assessment of the “Genotype” subDataset indicated that host plant identity significantly affected nymph mass gain (F 1,130 = 6.49, P = 0.012; Fig. 2A; Table 3) and aphid rm (F 1,104 = 11.94, P < 0.001; Fig. 2B; Table 3), with lowest values on HsP5 compared with cv. Concerto. This effect was also detected in the “Endosymbiont” subDataset (F 1,170 = 31.77, P < 0.001; Fig. 2A; Table 4). In addition, nymph mass gain varied significantly between genotypes (F 2,131 = 4.48, P = 0.013; Fig. 2A; Table 3) with significantly lower values in genotype E compared with genotype A (Table 5).
Table 3.
Summary of statistical modeling outputs for aphid performance parameters using the Genotype subDataset. For each performance parameter, the treatment factor, model basis, error distribution or statistical method applied, analysis method, and the statistical outputs are shown. Type II Wald χ 2 analysis of deviance and observation of fitted‐residual plots was conducted throughout the modeling simplification process to ensure data fitted the model parameters and that model simplification was justified
| Response variable | Treatment factor | Model basis | Error distributionor statisticalmethod | Model analysis | Test statistic | Degrees of freedom (Residuals) | P value |
|---|---|---|---|---|---|---|---|
| Nymph mass gain | Genotype | Linear mixed effects model | Maximum Likelihood (ML) | Type III analysis of variance with satterthwaite approximation for degrees of freedom (Type III ANOVA) | F = 4.48 | 2 (131) | 0.013 |
| Plant | F = 6.49 | 1 (130) | 0.012 | ||||
| Genotype × Plant | F = 1.01 | 2 (131) | 0.366 | ||||
| Prereproductive Period | Genotype | Linear mixed effects model | ML | Type III ANOVA | F = 0.24 | 2 (109) | 0.784 |
| Plant | F = 1.61 | 1 (94) | 0.208 | ||||
| Genotype × Plant | F = 0.15 | 1 (150) | 0.861 | ||||
| rm | Genotype | Linear mixed effects model | ML | Type III ANOVA | F = 0.92 | 2 (104) | 0.454 |
| Plant | F = 11.94 | 1 (104) | < 0.001 | ||||
| Genotype × Plant | F = 0.78 | 2 (103) | 0.462 | ||||
| Aphid survival | Genotype | Cox proportional hazards regression | N/A | Type II Wald χ 2 analysis of deviance (χ 2 Test) | χ 2 = 0.86 | 2 | 0.649 |
| Plant | χ 2 = 0.92 | 1 | 0.919 |
Table 4.
Summary of statistical modeling outputs for aphid performance experiments using the Endosymbiont subDataset. For each performance parameter, the treatment factor, model basis, error distribution or statistical method applied, analysis method, and the statistical outputs are shown. Type II Wald χ 2 analysis of deviance and observation of fitted‐residual plots was carried out throughout the modeling simplification process to ensure data fitted the model parameters and that model simplification was justified
| Response variable | Treatment factor | Model basis | Error distributionor statisticalmethod | Model analysis | Test statistic | Degrees of freedom (residuals) | P value |
|---|---|---|---|---|---|---|---|
| Nymph mass gain | Endosymbiont | Linear mixed effects model | Maximum Likelihood (ML) | Type III analysis of variance with Satterthwaite approximation for degrees of freedom (Type III ANOVA) | F = 4.69 | 1 (203) | 0.031 |
| Plant | F = 24.76 | 1 (203) | < 0.001 | ||||
| Endosymbiont × Plant | F = 5.03 | 1 (203) | 0.026 | ||||
| Prereproductive period | Endosymbiont | Linear mixed effects model | ML | Type III ANOVA | F = 2.61 | 1 (201) | 0.464 |
| Plant | F = 2.67 | 1 (202) | 0.104 | ||||
| Endosymbiont × Plant | F = 0.46 | 1 (202) | 0.497 | ||||
| rm | Endosymbiont | Linear mixed effects model | ML | Type III ANOVA | F = 3.10 | 1 (170) | 0.807 |
| Plant | F = 31.77 | 1 (170) | < 0.001 | ||||
| Endosymbiont × Plant | F = 1.08 | 1 (170) | 0.300 | ||||
| Aphid survival | Endosymbiont | Cox proportional hazards regression | N/A | Type II Wald χ 2 analysis of deviance (χ 2 Test) | χ 2 = 0.14 | 1 | 0.713 |
| Plant | χ 2 = 2.19 | 1 | 0.139 |
Figure 2.
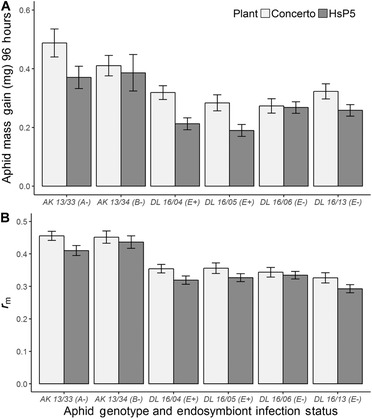
Effects of plant identity, aphid genotype, and H. defensa infection (+/−) on (A) nymph mass gain (mg) over a 96‐h period and (B) aphid rm. Values are means ± SE. Number of observations in model = 280. [Correction added on 21 February 2019, after first online publication: Figure 2's image has been replaced.]
Table 5.
Post hoc test of the least squares means for observed differences in aphid nymph mass gain between aphid genotypes, showing pairwise comparisons (t and P values) for each set of aphid genotypes
| Aphid genotype | Aphid genotype | Fitness parameter | subDataset | t value | P value | |
|---|---|---|---|---|---|---|
| A | vs. | B | Nymph mass gain | Genotype | 0.88 | 0.380 |
| A | vs. | E | 2.84 | 0.005 | ||
| B | vs. | E | 1.81 | 0.072 |
Interactive effects of plant identity and H. defensa presence on R. padi nymph mass gain
Analysis of the “Endosymbiont” subDataset highlighted an endosymbiont × host plant interaction for nymph mass gain, which was due to significantly lower mass gain in H. defensa‐infected genotype E nymphs feeding on HsP5 (F 1,203 = 5.03, P = 0.026; Fig. 2A; Table 4)
Discussion
This study reports on the presence of the facultative bacterial endosymbiont, Hamiltonella defensa, in the bird cherry‐oat aphid, R. padi, and assesses the effect of this endosymbiont on aphid fitness. Novel data are presented on the association between R. padi genotypes and H. defensa sampled from U.K. populations. Intraspecific variation in R. padi performance was detected in relation to aphid genotype and H. defensa infection and led to differential outcomes for aphid interactions with two host plant species and a natural enemy, which are summarized in Figure 3.
Figure 3.
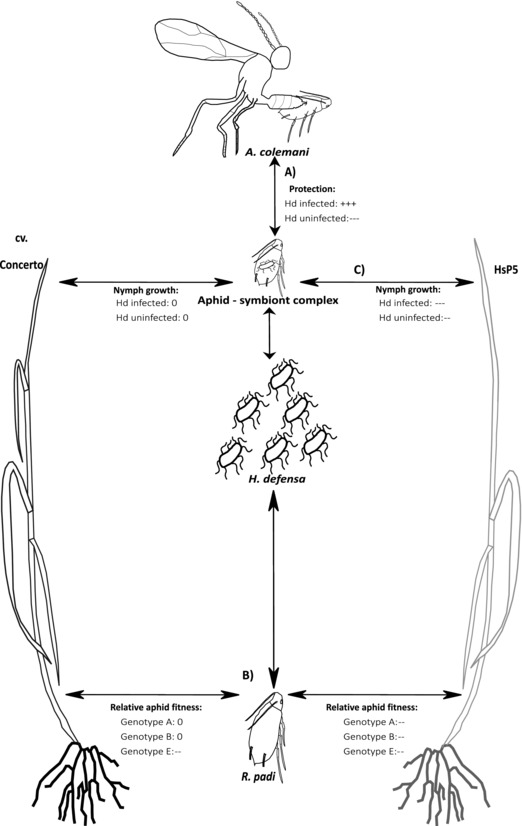
Summary of interactions between aphid genotype, H. defensa and other trophic groups. Arrows indicate trophic interactions that had positive (+), neutral (0), or negative (−) effects on aphid fitness; the relative magnitude of effect is shown by the number of symbols. (A) H. defensa conferred protection against the parasitoid wasp, A. colemani. (B) Aphid genotype was the main determinant of aphid fitness on H. vulgare cv. Concerto compared with H. spontaneum (HsP5). (C) Association with H. defensa was detrimental to juvenile aphid growth when feeding on unfavorable host plants.
The outcome of a symbiont–aphid–parasitoid relationship is species‐specific
This study provides evidence that R. padi forms associations with the facultative endosymbiont H. defensa, which was detected in c. 38% of the aphid lines assessed and in two out of seven aphid genotypes, although not in all representative lines of these genotypes. Polymorphic associations with H. defensa were only detected in genotype E. Previous studies of R. padi have not detected H. defensa but have reported the presence of two other facultative endosymbionts, S. symbiotica (de la Peña et al., 2014), and SMLS (Li et al., 2011), although their effects on aphid fitness remain to be established. The number of aphid lines infected with H. defensa in this study (6 out of a total of 16 lines) is representative of the intermediate infection frequencies detected for heritable symbionts in aphid populations (Russell et al., 2013).
To date, the most frequent effect on aphid fitness attributed to H. defensa is resistance to Hymenopterous parasitoids, primarily Braconid wasps (Oliver et al., 2010) such as A. ervi attacking Ac. pisum (Oliver et al., 2003) and Lysiphlebus fabarum (Marshall) attacking Ap. fabae (Schmid et al., 2012). Consistent with these studies, our findings show that the H. defensa–APSE complex can provide protection to R. padi against the parasitoid wasp A. colemani (Fig. 3A) and reinforces the defensive role attributed to this symbiont.
However, endosymbiont‐conferred protection is not necessarily observed consistently against all potential parasitoids of an aphid species. McLean and Godfray (2015) assessed the efficacy of endosymbiont‐mediated resistance to Braconid and Aphelinid wasps in relation to H. defensa strains selected from Ac. pisum biotypes adapted to different host plants. The authors detected differences in parasitism susceptibility due to H. defensa strain, with one strain able to confer protection against the Aphelinid wasp Aphelinus abdominalis (Dalman), but unable to provide resistance to the Braconid wasp A. ervi. Differences in parasitoid wasp susceptibility were also attributed to aphid biotype, indicating that aphid adaptation to host plant species could influence the efficacy of endosymbiont‐mediated resistance. A hypothesis was put forward by Hopper et al. (2018) to explain why H. defensa does not confer widespread protection against Aphelinid wasps, and relates to the anhydropic chlorinated eggs produced by the Aphelinidae, which are thought to be less susceptible to secreted APSE toxins.
Additionally, the cowpea aphid, Aphis craccivora (Koch), is attacked by a number of Braconid wasps, including Binodoxys communis (Gahan), B. koreanus (Stary), L. orientalis (Stary & Rakhshani), and A. colemani; H. defensa infection did not protect aphids against L. orientalis or A. colemani, but did provide resistance to B. communis and B. koreanus (Asplen et al., 2014). The authors hypothesized that differential protection conferred by endosymbionts against particular parasitoid species might be linked to particular H. defensa–APSE combinations, and recent work provides evidence for specificity of symbiont defense in relation to APSE strain and aphid and parasitoid genotype (Dennis et al., 2017; Käch et al., 2018; Martinez et al., 2018). These factors could explain why H. defensa provided protection to R. padi against A. colemani (this study) and to Ap. fabae against A. colemani (Cayetano & Vorburger, 2015), but did not protect Ap. craccivora against A. colemani (Asplen et al., 2014).
The APSE genome has been reported to undergo rapid recombination resulting in strain‐dependent variation in the identity of toxins and their protective effects (Degnan & Moran, 2008a,b; Dennis et al., 2017), which might also explain differences between wasp genera in their susceptibility to symbiont–APSE‐mediated protection (Käch et al., 2018). Other studies have highlighted the existence of aphid‐encoded resistance to parasitism irrespective of endosymbiont presence in M. euphorbiae and Ac. pisum (Martinez et al., 2014; Clarke et al., 2017), indicating that aphid‐encoded traits could be another factor influencing the specificity of parasitoid resistance in aphids. Experimental manipulation of symbiont infection in different aphid genotypes would be a useful next step to identify the contribution of these factors to the defensive phenotype in R. padi.
Aphid genotype is a key determinant of aphid fitness
Intraspecific variation in R. padi mass gain in the present study was attributed mainly to aphid genotype and not to H. defensa infection (Fig. 3B), with individuals belonging to genotype E generally performing poorly compared with genotypes A and B. Aphid genotype is often identified as a key determinant of aphid performance, for example in S. avenae (Figueroa et al., 2004) and M. euphorbiae (Karley et al., 2017). A recent study reported that M. euphorbiae genotypes capable of forming endosymbiotic associations with H. defensa had higher fitness than those genotypes which did not support H. defensa infection, at least when feeding on a susceptible host plant species (Clarke et al., 2017), which contrasts with the findings for R. padi lines assessed in the current study. Differential effects of aphid genotype on fitness might also depend, however, on plant suitability for aphids (Karley et al., 2017). In general, all R. padi genotypes examined in the present study performed poorly on the wild species HsP5 compared with the commercial barley cultivar Concerto, indicating that HsP5 is partially resistant to aphids irrespective of aphid genetic variation. To understand the implications of these findings in the context of pest control, further work is needed to assess whether the frequency of R. padi genotypes detected in this study are representative of field populations.
Endosymbiont infection exacerbates the effects of poor plant quality
Infection with H. defensa has previously been shown to decrease the longevity of Ap. fabae (Vorburger & Gouskov, 2011). Although we did not identify any negative effects of H. defensa infection on aphid longevity, a key finding of this study was that symbiont‐infected individuals exhibited reduced growth during their juvenile stages compared with symbiont‐free individuals, but only on the partially resistant plant, HsP5 (Fig. 3C), in line with our original prediction. The mechanism of aphid resistance in HsP5 has not been fully characterized, but partial resistance to aphids in the wild relative of wheat, Triticum monococccum, is thought to be phloem‐mediated, linked to increased secondary metabolite concentrations (Greenslade et al., 2016). Whatever the causal mechanism of resistance, it is possible that the decrease in nymph growth rate on HsP5 in aphid lines harboring H. defensa resulted from resource demand by the endosymbiont, which intensified the negative effects of feeding on a poor quality plant host. Indeed a similar observation was made by Chandler et al. (2008), where growth of Ap. fabae differed between two host plants—a favorable host (Vicia faba) and an unfavorable host (Lamium purpureum). The negative effect of L. purpureum on aphid fitness was exacerbated by the presence of the facultative endosymbionts Regiella insecticola and H. defensa. This observation was thought to relate to low phloem concentrations of amino acids in L. purpureum, which disrupted the ability of the aphid to regulate facultative endosymbiont titres in aphid tissues, leading to greater symbiont resource demand and decreased insect growth (Chandler et al., 2008). The possibility that the symbiont‐associated decrease in nymph growth of R. padi on HsP5 is linked to symbiont resource demand and poor quality phloem sap is an interesting avenue for further research, especially if it reveals a trade‐off with aphid resistance to parasitoid wasps.
Conclusions
This study highlights both large and small magnitude effects of a facultative endosymbiont on aphid fitness that could influence aphid ecology and population dynamics by modifying the outcome of aphid interactions with host plants and natural enemies. Our findings show that infection with the defensive endosymbiont H. defensa provides protection to R. padi against a common parasitoid wasp through a 5 fold increase in aphid survival after parasitoid attack (Fig. 3A). However, this benefit could be partly mitigated by the 16% reduction, on average, in growth of symbiont‐infected nymphs observed on a partially resistant host plant (Fig. 3C), although this might be a relatively small price to pay for parasitoid protection. Finally, while most genotypes exhibited reduced fitness on the partially resistant host (Fig. 3B), the fittest genotypes still performed better on this host than the least fit genotypes. In summary, these findings suggest that plant resistance factors will favor the fittest R. padi genotypes, but symbiont‐infected individuals will be favored when parasitoids are abundant (Käch et al., 2018), although these aphids might not achieve optimal performance on a poor quality host plant. While the consequences of symbiont‐conferred parasitoid resistance for aphid biocontrol are increasingly recognized (Vorburger, 2018), symbiont‐mediated fitness trade‐offs that interact with plant defensive traits have received relatively little attention until recently (e.g., Frago et al., 2017; Karley et al., 2017) and should be taken into account when deploying crop resistance and natural enemies for integrated management of crop pests.
Disclosure
The authors declare no conflicts of interest.
Data archiving and deposition
16S rDNA sequences of the sequenced H. defensa strains from the R. padi lines assessed have been deposited into GenBank within the National Centre for Biotechnology Information database, with accession numbers MG595518–MG595523.
Supporting information
Table S1 Primer names, targets, 5′–3′ sequence, use, and source for all primers used in this study for genotyping R. padi asexual lines using microsatellite markers and for facultative endosymbiont screens.
Table S2 R. padi genotyping results showing aphid asexual line, collection site, original collection host plant, assigned genotype, and the allele sizes (in bp) for seven microsatellite loci.
Table S3 Thermocycling conditions for diagnostic PCR of aphid facultative endosymbionts and APSE amplification.
Table S4 Best BLASTn hits for APSE endosymbiont marker sequences amplified from R. padi lines infected with H. defensa. Sequences were analyzed for BLASTn similarity against sequences held on the NCBI database.
Acknowledgments
DJL was funded by the James Hutton Institute and the Universities of Aberdeen and Dundee through a Scottish Food Security Alliance (Crops) PhD studentship. AJK and TAV were funded through the strategic research programme funded by the Scottish Government's Rural and Environment Science and Analytical Services Division. JIBB was funded through a Royal Society of Edinburgh Personal Fellowship and an ERC starting grant (APHIDHOST). We thank Gaynor Malloch (James Hutton Institute) and Brian Fenton (Scotland's Rural College) for supplying aphid line “JB,” with additional thanks to Gaynor Malloch for advice on microsatellite scoring. We also thank Stephen Hubbard (University of Dundee) for statistical advice, Rob Hancock (James Hutton Institute) for helpful comments on the manuscript, and three anonymous reviewers for their comments and advice to improve the manuscript.
[The copyright line for this article was changed on 5 September 2019 after original online publication.]
References
- Asplen, M.K. , Bano, N. , Brady, C.M. , Desneux, N. , Hopper, K.R. , Malouines, C . et al (2014) Specialisation of bacterial endosymbionts that protect aphids from parasitoids. Ecological Entomology, 39, 736–739. [Google Scholar]
- Bass, C. , Puinean, A.M. , Zimmer, C.T. , Denholm, I. , Field, L.M. , Foster, S.P . et al (2014) The evolution of insecticide resistance in the peach potato aphid, Myzus persicae . Insect Biochemistry and Molecular Biology, 51, 41–51. [DOI] [PubMed] [Google Scholar]
- Bates, D. , Maechler, M. , Bolker, B. and Walker, S. (2015) Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Betsiashvili, M. , Ahern, K.R. and Jander, G. (2015) Additive effects of two quantitative trait loci that confer Rhopalosiphum maidis (corn leaf aphid) resistance in maize inbred line Mo17. Journal of Experimental Botany, 66, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, J.W. , Chevignon, G. , Oliver, K.M. and Strand, M.R. (2017) Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proceedings of the Royal Society B: Biological Sciences, 284, 10.1098/rspb.2017.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayetano, L. and Vorburger, C. (2015) Symbiont‐conferred protection against Hymenopteran parasitoids in aphids: how general is it? Ecological Entomology, 40, 85–93. [Google Scholar]
- Chandler, S.M. , Wilkinson, T.L. and Douglas, A.E. (2008) Impact of plant nutrients on the relationship between a herbivorous insect and its symbiotic bacteria. Proceedings of the Royal Society B: Biological Sciences, 275, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, H.V. , Cullen, D. , Hubbard, S.F. and Karley, A.J. (2017) Susceptibility of Macrosiphum euphorbiae to the parasitoid Aphidius ervi: larval development depends on host aphid genotype. Entomologia Experimentalis et Applicata, 162, 148–158. [Google Scholar]
- De Clerck, C. , Fujiwara, A. , Joncour, P. , Léonard, S. , Félix, M.L. , Francis, F . et al (2015) A metagenomic approach from aphid's hemolymph sheds light on the potential roles of co‐existing endosymbionts. Microbiome, 3, 10.1186/s40168-015-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clerck, C. , Tsuchida, T. , Massart, S. , Lepoivre, P. , Francis, F. and Jijakli, M.H. (2014) Combination of genomic and proteomic approaches to characterize the symbiotic population of the banana aphid (Hemiptera: Aphididae). Environmental Entomology, 43, 29–36. [DOI] [PubMed] [Google Scholar]
- de la Peña, E. , Vandomme, V. and Frago, E. (2014) Facultative endosymbionts of aphid populations from coastal dunes of the North Sea. Belgian Journal of Zoology, 144, 41–50. [Google Scholar]
- Degnan, P.H. and Moran, N.A. (2008a) Diverse phage‐encoded toxins in a protective insect endosymbiont. Applied and Environmental Microbiology, 74, 6782–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan, P.H. and Moran, N.A. (2008b) Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin‐encoding bacteriophage. Molecular Ecology, 17, 916–929. [DOI] [PubMed] [Google Scholar]
- Delp, G. , Gradin, T. , Åhman, I. and Jonsson, L.M. (2009) Microarray analysis of the interaction between the aphid Rhopalosiphum padi and host plants reveals both differences and similarities between susceptible and partially resistant barley lines. Molecular Genetics and Genomics, 281, 233–248. [DOI] [PubMed] [Google Scholar]
- Dennis, A.B. , Patel, V. , Oliver, K.M. and Vorburger, C. (2017) Parasitoid gene expression changes after adaptation to symbiont‐protected hosts. Evolution, 71, 2599–2617. [DOI] [PubMed] [Google Scholar]
- Desneux, N. , Asplen, M.K. , Brady, C.M. , Heimpel, G.E. , Hopper, K.R. , Luo, C . et al (2018) Intraspecific variation in facultative symbiont infection among native and exotic pest populations: potential implications for biological control. Biological Control, 116, 27–35. [Google Scholar]
- Douglas, A.E. and Prosser, W.A. (1992) Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. Journal of Insect Physiology, 38, 565–568. [Google Scholar]
- Field, L.M. , Devonshire, A.L. and Forde, B.G. (1988) Molecular evidence that insecticide resistance in peach‐potato aphids Myzus persicae (Sulz.) results from amplification of an esterase gene. Biochemical Journal, 251, 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, C.C. , Simon, J.C. , Le Gallic, J.F. , Prunier‐Leterme, N. , Briones, L.M. , Dedryver, C.A . et al (2004) Effect of host defense chemicals on clonal distribution and performance of different genotypes of the cereal aphid Sitobion avenae . Journal of Chemical Ecology, 30, 2515–2525. [DOI] [PubMed] [Google Scholar]
- Finlay, K.J. and Luck, J.E. (2011) Response of the bird cherry‐oat aphid (Rhopalosiphum padi) to climate change in relation to its pest status, vectoring potential and function in a crop–vector–virus pathosystem. Agriculture Ecosystems & Environment, 144, 405–421. [Google Scholar]
- Foster, S.P. , Paul, V.L. , Slater, R. , Warren, A. , Denholm, I. , Field, L.M . et al (2014) A mutation (L1014F) in the voltage‐gated sodium channel of the grain aphid, Sitobion avenae, is associated with resistance to pyrethroid insecticides. Pest Management Science, 70, 1249–1253. [DOI] [PubMed] [Google Scholar]
- Fox, J. and Weisberg, S. (2011) An {R} Companion to Applied Regression. Sage, Thousand Oaks, CA. [Google Scholar]
- Frago, E. , Mala, M. , Weldegergis, B.T. , Yang, C. , McLean, A. , Godfray, H.C.J . et al (2017) Symbionts protect aphids from parasitic wasps by attenuating herbivore‐induced plant volatiles. Nature Communications, 8, 1860 10.1038/s41467-017-01935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, J.P. , Outreman, Y. , Mieuzet, L. and Simon, J.C. (2015) Bacterial communities associated with host‐adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS ONE, 10, http://doi.org/10.1371%2Fjournal.pone.0120664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, F. , Bengtsson, J. , Berendse, F. , Weisser, W.W. , Emmerson, M. , Morales, M.B . et al (2010) Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic and Applied Ecology, 11, 97–105. [Google Scholar]
- Girvin, J. , Whitworth, R.J. , Rojas, L.M.A. and Smith, C.M. (2017) Resistance of select winter wheat (Triticum aestivum) cultivars to Rhopalosiphum padi (Hemiptera: Aphididae). Journal of Economic Entomology, 110, 1886–1889. [DOI] [PubMed] [Google Scholar]
- Goulson, D. (2013) Review: an overview of the environmental risks posed by neonicotinoid insecticides. Journal of Applied Ecology, 50, 977–987. [Google Scholar]
- Greenslade, A.F.C. , Ward, J.L. , Martin, J.L. , Corol, D.I. , Clark, S.J. , Smart, L.E . et al (2016) Triticum monococcum lines with distinct metabolic phenotypes and phloem‐based partial resistance to the bird cherry–oat aphid Rhopalosiphum padi . Annals of Applied Biology, 168, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, P. and Ibanez, F. (2014) pastecs: package for analysis of space‐time ecological series. CRAN Repository. https://github.com/phgrosjean/pastecs. [Google Scholar]
- Guay, J.F. , Boudreault, S. , Michaud, D. and Cloutier, C. (2009) Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. Journal of Insect Physiology, 55, 919–926. [DOI] [PubMed] [Google Scholar]
- Guo, J.Q. , Hatt, S. , He, K. , Chen, J. , Francis, F. and Wang, Z.Y. (2017) Nine facultative endosymbionts in aphids: a review. Journal of Asia‐Pacific Entomology, 20, 794–801. [Google Scholar]
- Halekoh, U. and Højsgaard, S. (2014) A Kenward‐Roger approximation and parametric bootstrap methods for tests in linear mixed models—the R Package pbkrtest. Journal of Statistical Software, 59, 1–32.26917999 [Google Scholar]
- Hansen, A.K. , Vorburger, C. and Moran, N.A. (2012) Genomic basis of endosymbiont‐conferred protection against an insect parasitoid. Genome Research, 22, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, F. and Berkenkamp, B. (1975) Revised growth‐stage key for Brassica campestris and B. napus . Canadian Journal of Plant Science, 55, 657–658. [Google Scholar]
- Henry, L.M. , Maiden, M.C.J. , Ferrari, J. and Godfray, H.C.J. (2015) Insect life history and the evolution of bacterial mutualism. Ecology Letters, 18, 516–525. [DOI] [PubMed] [Google Scholar]
- Hopper, K.R. , Kuhn, K.L. , Lanier, K. , Rhoades, J.H. , Oliver, K.M. , White, J.A . et al (2018) The defensive aphid symbiont Hamiltonella defensa affects host quality differently for Aphelinus glycinis versus Aphelinus atriplicis . Biological Control, 116, 3–9. [Google Scholar]
- Jarosova, J. , Beoni, E. and Kundu, J.K. (2016) Barley yellow dwarf virus resistance in cereals: approaches, strategies and prospects. Field Crops Research, 198, 200–214. [Google Scholar]
- Jousselin, E. , Cœur d'Acier, A. , Vanlerberghe‐Masutti, F. and Duron, O. (2013) Evolution and diversity of Arsenophonus endosymbionts in aphids. Molecular Ecology, 22, 260–270. [DOI] [PubMed] [Google Scholar]
- Käch, H. , Mathé‐Hubert, H. , Dennis, A.B. and Vorburger, C. (2018) Rapid evolution of symbiont‐mediated resistance compromises biological control of aphids by parasitoids. Evolutionary Applications, 11, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karley, A.J. , Emslie‐Smith, M. and Bennett, A.E. (2017) Potato aphid Macrosiphum euphorbiae performance is determined by aphid genotype and not mycorrhizal fungi or water availability. Insect Science, 24, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Kassambara, A. (2017) ggpubr: “ggplot2” based publication ready plots. CRAN Repository. http://www.sthda.com/english/rpkgs/ggpubr. [Google Scholar]
- Kassambara, A. and Kosinski, M. (2017) survminer: drawing survival curves using “ggplot.” CRAN Repository. http://www.sthda.com/english/rpkgs/survminer/. [Google Scholar]
- King, B.H. (1987) Offspring sex ratios in parasitoid wasps. The Quarterly Review of Biology, 62, 367–396. [Google Scholar]
- Leather, S. , Walters, K.A. and Dixon, A.G. (1989) Factors determining the pest status of the bird cherry‐oat aphid, Rhopalosiphum padi (L.) (Hemiptera: Aphididae), in Europe: a study and review. Bulletin of Entomological Research, 79, 345–360. [Google Scholar]
- Li, T. , Wu, X.J. , Jiang, Y.L. , Zhang, L. , Duan, Y. , Miao, J . et al (2016) The genetic diversity of SMLS (Sitobion miscanthi L type symbiont) and its effect on the fitness, mitochondrial DNA diversity and Buchnera aphidicola dynamic of wheat aphid, Sitobion miscanthi (Hemiptera: Aphididae). Molecular Ecology, 25, 3142–3151. [DOI] [PubMed] [Google Scholar]
- Li, T. , Xiao, J.H. , Xu, Z.H. , Murphy, R.W. and Huang, D.W. (2011) Cellular tropism, population dynamics, host range and taxonomic status of an aphid secondary symbiont, SMLS (Sitobion miscanthi L type symbiont). PLoS ONE, 6, 10.1371/journal.pone.0021944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łukasik, P. , Dawid, M.A. , Ferrari, J. and Godfray, H.C.J. (2013a) The diversity and fitness effects of infection with facultative endosymbionts in the grain aphid, Sitobion avenae . Oecologia, 173, 985–996. [DOI] [PubMed] [Google Scholar]
- Łukasik, P. , van Asch, M. , Guo, H. , Ferrari, J. and Godfray, C.J. (2013b) Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecology Letters, 16, 214–218. [DOI] [PubMed] [Google Scholar]
- MacGillivray, M. and Anderson, G. (1957) Three useful insect cages. The Canadian Entomologist, 89, 43–46. [Google Scholar]
- Martinez, A.J. , Doremus, M.R. , Kraft, L.J. , Kim, K.L. , Oliver, K.M. and Fenton, A. (2018) Multi‐modal defences in aphids offer redundant protection and increased costs likely impeding a protective mutualism. Journal of Animal Ecology, 87, 464–477. [DOI] [PubMed] [Google Scholar]
- Martinez, A.J. , Ritter, S.G. , Doremus, M.R. , Russell, J.A. and Oliver, K.M. (2014) Aphid‐encoded variability in susceptibility to a parasitoid. BMC Evolutionary Biology, 14, 10.1186/1471-2148-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, T. and Frank, S. (2015) Grain diversity effects on banker plant growth and parasitism by Aphidius colemani . Insects, 6, 772–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, A.H.C. and Godfray, H.C.J. (2015) Evidence for specificity in symbiont‐conferred protection against parasitoids. Proceedings of the Royal Society B: Biological Sciences, 282, 10.1098/rspb.2015.0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseguer, A.S. , Manzano‐Marín, A. , d'Acier, A.C. , Clamens, A.L. , Godefroid, M. and Jousselin, E. (2017) Buchnera has changed flatmate but the repeated replacement of co‐obligate symbionts is not associated with the ecological expansions of their aphid hosts. Molecular Ecology, 26, 2363–2378. [DOI] [PubMed] [Google Scholar]
- Mitchell, C. , Brennan, R.M. , Graham, J. and Karley, A.J. (2016) Plant defense against herbivorous pests: exploiting resistance and tolerance traits for sustainable crop protection. Frontiers in Plant Science, 7, 10.3389/fpls.2016.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N.A. , Degnan, P.H. , Santos, S.R. , Dunbar, H.E. and Ochman, H. (2005) The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proceedings of the National Academy of Sciences USA, 102, 16919–16926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M. , Degnan, P.H. , Burke, G.R. and Moran, N.A. (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annual Review of Entomology, 55, 247–266. [DOI] [PubMed] [Google Scholar]
- Oliver, K.M. , Degnan, P.H. , Hunter, M.S. and Moran, N.A. (2009) Bacteriophages encode factors required for protection in a symbiotic mutualism. Science, 325, 992–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M. , Moran, N.A. and Hunter, M.S. (2006) Costs and benefits of a superinfection of facultative symbionts in aphids. Proceedings of the Royal Society B: Biological Sciences, 273, 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M. , Russell, J.A. , Moran, N.A. and Hunter, M.S. (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proceedings of the National Academy of Sciences USA, 100, 1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S. and Singh, R. (1999) Host size induced variation in progeny sex ratio of an aphid parasitoid Lysiphlebia mirzai . Entomologia Experimentalis et Applicata, 90, 61–67. [Google Scholar]
- Perera, M. and Hemachandra, K. (2014) Study of longevity, fecundity and oviposition of Trichogrammatoidea bactrae Nagaraja (Hymenoptera: Trichogrammatidae) to facilitate mass rearing. Tropical Agricultural Research, 25, 602–609. [Google Scholar]
- Perry, K.L. , Kolb, F.L. , Sammons, B. , Lawson, C. , Cisar, G. and Ohm, H. (2000) Yield effects of Barley yellow dwarf virus in soft red winter wheat. Phytopathology, 90, 1043–1048. [DOI] [PubMed] [Google Scholar]
- R‐Core (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ramsden, M. , Menendez, R. , Leather, S. and Wäckers, F. (2017) Do natural enemies really make a difference? Field scale impacts of parasitoid wasps and hoverfly larvae on cereal aphid populations. Agricultural and Forest Entomology, 19, 139–145. [Google Scholar]
- Robinson, D. (2017) Broom: convert statistical analysis objects into tidy data frames. CRAN Repository. http://github.com/tidyverse/broom. [Google Scholar]
- Ronquim, J.C. , Pacheco, J.M. and Ronquim, C.C. (2004) Occurrence and parasitism of aphids (Hemiptera: Aphididae) on cultivars of irrigated oat (Avena spp.) in São Carlos, Brazil. Brazilian Archives of Biology and Technology, 47, 163–169. [Google Scholar]
- Russell, J.A. and Moran, N.A. (2006) Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proceedings of the Royal Society B: Biological Sciences, 273, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J.A. , Weldon, S. , Smith, A.H. , Kim, K.L. , Hu, Y. , Lukasik, P . et al (2013) Uncovering symbiont‐driven genetic diversity across North American pea aphids. Molecular Ecology, 22, 2045–2059. [DOI] [PubMed] [Google Scholar]
- Sandström, J.P. , Russell, J.A. , White, J.P. and Moran, N.A. (2001) Independent origins and horizontal transfer of bacterial symbionts of aphids. Molecular Ecology, 10, 217–228. [DOI] [PubMed] [Google Scholar]
- Sasaki, T. , Hayashi, H. and Ishikawa, H. (1991) Growth and reproduction of the symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum maintained on artificial diets. Journal of Insect Physiology, 37, 749–756. [Google Scholar]
- Schmid, M. , Sieber, R. , Zimmermann, Y.S. and Vorburger, C. (2012) Development, specificity and sublethal effects of symbiont‐conferred resistance to parasitoids in aphids. Functional Ecology, 26, 207–215. [Google Scholar]
- Simon, J.C. , Leterme, N. , Delmotte, F. , Martin, O. and Estoup, A. (2001) Isolation and characterization of microsatellite loci in the aphid species, Rhopalosiphum padi . Molecular Ecology Notes, 1, 4–5. [Google Scholar]
- Therneau, T. (2018) coxme: mixed effects Cox models. CRAN Repository. https://cran.r‐project.org/web/packages/coxme/index.html [Google Scholar]
- Therneau, T.M. and Grambsch, P.M. (2000) Modeling Survival Data: Extending the Cox Model. Springer, New York. [Google Scholar]
- Tsuchida, T. , Koga, R. and Fukatsu, T. (2004) Host plant specialization governed by facultative symbiont. Science, 303, 1989–1989. [DOI] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Horikawa, M. , Tsunoda, T. , Maoka, T. , Matsumoto, S . et al (2010) Symbiotic bacterium modifies aphid body color. Science, 330, 1102–1104. [DOI] [PubMed] [Google Scholar]
- Valenzuela, I. and Hoffmann, A.A. (2015) Effects of aphid feeding and associated virus injury on grain crops in Australia. Austral Entomology, 54, 292–305. [Google Scholar]
- von Burg, S. , Ferrari, J. , Müller, C.B. and Vorburger, C. (2008) Genetic variation and covariation of susceptibility to parasitoids in the aphid Myzus persicae: no evidence for trade‐offs. Proceedings of the Royal Society B: Biological Sciences, 275, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger, C. (2018) Symbiont‐conferred resistance to parasitoids in aphids—challenges for biological control. Biological Control, 116, 17–26. [Google Scholar]
- Vorburger, C. , Gehrer, L. and Rodriguez, P. (2010) A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biology Letters, 6, 109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger, C. and Gouskov, A. (2011) Only helpful when required: a longevity cost of harbouring defensive symbionts. Journal of Evolutionary Biology, 24, 1611–1617. [DOI] [PubMed] [Google Scholar]
- Vorburger, C. and Rouchet, R. (2016) Are aphid parasitoids locally adapted to the prevalence of defensive symbionts in their hosts? BMC Evolutionary Biology, 16, 271 10.1186/s12862-016-0811-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, S.M. , Martinez, A.J. , Ruan, Y.M. , Kim, K.L. , Lenhart, P.A. , Dehnel, A.C . et al (2015) Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Functional Ecology, 29, 1402–1410. [Google Scholar]
- Wickham, H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer‐Verlag, New York. [Google Scholar]
- Wyatt, I. and White, P. (1977) Simple estimation of intrinsic increase rates for aphids and tetranychid mites. Journal of Applied Ecology, 14, 757–766. [Google Scholar]
- Zadoks, J.C. , Chang, T.T. and Konzak, C.F. (1974) A decimal code for the growth stages of cereals. Weed Research, 14, 415–421. [Google Scholar]
- Zytynska, S.E. and Weisser, W.W. (2016) The natural occurrence of secondary bacterial symbionts in aphids. Ecological Entomology, 41, 13–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primer names, targets, 5′–3′ sequence, use, and source for all primers used in this study for genotyping R. padi asexual lines using microsatellite markers and for facultative endosymbiont screens.
Table S2 R. padi genotyping results showing aphid asexual line, collection site, original collection host plant, assigned genotype, and the allele sizes (in bp) for seven microsatellite loci.
Table S3 Thermocycling conditions for diagnostic PCR of aphid facultative endosymbionts and APSE amplification.
Table S4 Best BLASTn hits for APSE endosymbiont marker sequences amplified from R. padi lines infected with H. defensa. Sequences were analyzed for BLASTn similarity against sequences held on the NCBI database.


