Significance
Cells avoid clashes between DNA repair machineries and the transcription apparatus by temporary halting gene expression in the vicinity of DNA double-strand breaks (DSBs), an emerging DNA damage response (DDR) that underlies genome integrity protection. In this study, we screened for novel activities that may be important in this DDR and have identified the DYRK1B kinase as a component of the mammalian DDR that specializes in fine-tuning DSB repair on actively transcribed chromatin. Moreover, global analysis of DYRK1B substrates has led to the identification of the histone methyltransferase EHMT2 as a DYRK1B target and effector. Our findings uncover the DYRK1B network as a DDR subpathway that preserves the integrity of active chromatin.
Keywords: DNA damage, transcription, DNA double-strand breaks, DNA repair, DYRK1B
Abstract
DNA double-strand breaks (DSBs) trigger transient pausing of nearby transcription, an emerging ATM-dependent response that suppresses chromosomal instability. We screened a chemical library designed to target the human kinome for new activities that mediate gene silencing on DSB-flanking chromatin, and have uncovered the DYRK1B kinase as an early respondent to DNA damage. We showed that DYRK1B is swiftly and transiently recruited to laser-microirradiated sites, and that genetic inactivation of DYRK1B or its kinase activity attenuated DSB-induced gene silencing and led to compromised DNA repair. Notably, global transcription shutdown alleviated DNA repair defects associated with DYRK1B loss, suggesting that DYRK1B is strictly required for DSB repair on active chromatin. We also found that DYRK1B mediates transcription silencing in part via phosphorylating and enforcing DSB accumulation of the histone methyltransferase EHMT2. Together, our findings unveil the DYRK1B signaling network as a key branch of mammalian DNA damage response circuitries, and establish the DYRK1B–EHMT2 axis as an effector that coordinates DSB repair on transcribed chromatin.
Repair of DNA double-strand breaks (DSBs) is accompanied by change in local chromatin architecture and its ongoing transactions, including transcription (1). Indeed, cells halt gene expression on DSB-flanking chromatin to prevent collision between the DNA repair machinery and the transcription apparatus, which in turn facilitates DNA repair and suppresses chromosomal instability (2). In this regard, the apical DNA damage kinase ATM targets a multitude of chromatin-modifying activities (3–5) and plays multifaceted roles to establish a DSB microenvironment to suppress local transcription (6). Interestingly, the DNA-dependent protein kinase (DNA-PK) complex has also been reported to arrest transcription (7) via proteasome-dependent eviction of RNA polymerase II (RNAPII) (8), although whether ATM and DNA-PK activities intersect remains undefined. Moreover, while transcription suppression on DSB-flanking chromatin also requires a growing number of chromatin remodelers and transcription regulators (9–13), it is currently enigmatic how DSB signals are propagated and translated to fine-tune local transcriptional activities to effect DNA repair processes.
DYRK1B (also known as MIRK) is a member of the evolutionarily conserved family of DYRK kinases, and encodes a dual-specificity serine/threonine (S/T) protein kinase with implicated roles in cell differentiation and survival (14, 15). DYRK1B is up-regulated in human cancers, and several lines of evidence have shown that DYRK1B maintains cancer cells in their quiescent state to confer chemoresistance (16, 17), and that pharmacological inhibition of its kinase activity may offer a means to sensitize cells to the cytotoxic effects of anticancer therapeutics (18). Aside from its putative oncogenic properties, DYRK1B is also involved in gene transcriptional control in growth and development (19), and genetic mutations are causally linked with an inherited form of metabolic syndrome (20). Notably, while DYRK1B inactivation may contribute to genome instability (21) and has been reported to reside in DSB repair macromolecular protein complexes in a proteomic study (22), a role of DYRK1B in DNA damage response control is not known.
Results
Chemical Screen Identifies DYRK1B as an Activity Required for Transcription Suppression on DSB-Flanking Chromatin.
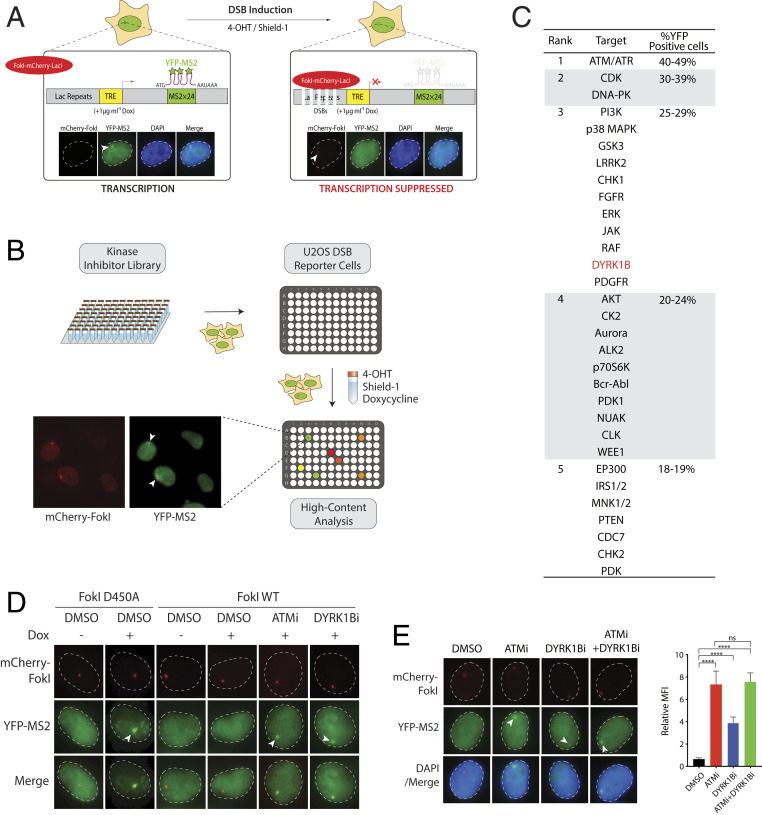
To isolate novel activities that orchestrate DSB repair on transcribed chromatin, a kinase inhibitor library consisting of 760 compounds was examined for its effect on DSB-induced transcription suppression. To this end, we took advantage of a previously established transcription reporter wherein nascent transcriptional activities can be monitored by local YFP-MS2 accumulation (23). Proximal DSBs can be induced by 4-hydroxytamoxifen (4-OHT) and Shield-1, which promote the docking of the estrogen receptor (ER)-fused FokI-mCherry-LacI nuclease onto LacO arrays (×256) located upstream of the transcriptional unit to ensure transcription suppression (Fig. 1A). Accordingly, DSBs were induced in reporter cells pretreated with individual kinase inhibitors, which were subjected to high-content microscopy and automated analyses of cells positive for YFP-MS2 foci (Fig. 1B). Consistent with a pivotal role of ATM in orchestrating DSB responses (24), including DSB-induced transcription silencing in cis (DISC) (6), chemical inhibitors that targeted the ATM kinase compromised DISC, as reflected by the high percentages of cells with YFP-MS2 foci (Fig. 1C, SI Appendix, Fig. S1, and Dataset S1). We filtered out kinase targets that reside in the cytoplasm (SI Appendix, Fig. S1A) and, among the list of nuclear kinase targets, we were drawn to DYRK1B (Fig. 1C and SI Appendix, Fig. S1B), as it was recently reported to interact with the DSB response factor RNF169 (22). We confirmed that chemical inhibition of DYRK1B, much like that of ATM, attenuated DSB-induced transcription suppression (Fig. 1D), and that nascent transcription can be inhibited by the transcription elongation inhibitor 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB) (SI Appendix, Fig. S2A). In support of the idea wherein DYRK1B may encode an ATM effector in mounting transcription suppression on damaged chromatin, we found that chemical inhibition of both ATM and DYRK1B compromised DISC to the same extent as that observed in ATM inhibitor-treated cells, suggesting that ATM and DYRK1B may be epistatic in this DNA damage response (DDR) (Fig. 1E).
Fig. 1.
Chemical screen identifies regulators of transcription at DSBs. (A) Schematic illustration of the DISC reporter in U2OS-DSB reporter cells. Doxycycline (Dox) induces nascent transcription and local YFP-MS2 accumulation at the transgene locus. Cell treatment with 4-OHT and Shield-1 leads to FokI-induced DSBs and suppression of proximal transcription. Arrowheads denote YFP-MS2 foci. (B) Workflow depicting the kinase inhibitor library screen using U2OS-DSB reporter cells. DSB-induced cells pretreated with each of the 760 kinase inhibitors were subjected to high-content imaging and automated analyses for YFP-MS2 focus. (C) Ranking of nuclear kinase inhibitor targets with putative roles in transcription silencing. (D) FokI-induced DSBs silence transcription in control but not in ATM- or DYRK1B-inhibited cells. Expression of FokI wild type (WT) but not its catalytically inactive mutant (D450A) suppressed doxycycline-induced transcription. Pretreatment with ATM inhibitor (ATMi; KU55933) or DYRK1B inhibitor (DYRK1Bi; AZ191) attenuated DSB-induced transcription silencing. U2OS-DSB reporter cells grown on coverslips were transfected with either FokI WT or D450A expression construct; 24 h posttransfection, cells were PFA-fixed and processed for fluorescence imaging. (E) ATM and DYRK1B act epistatically to mount DISC. U2OS-DSB reporter cells pretreated with DMSO or the indicated inhibitor(s) were induced with 4-OHT and Shield-1. Relative MFI of YFP-MS2 foci is shown and bars represent mean ± SEM; ****P < 0.0001; ns, not significant.
DYRK1B Promotes DISC in a Kinase-Dependent Manner.
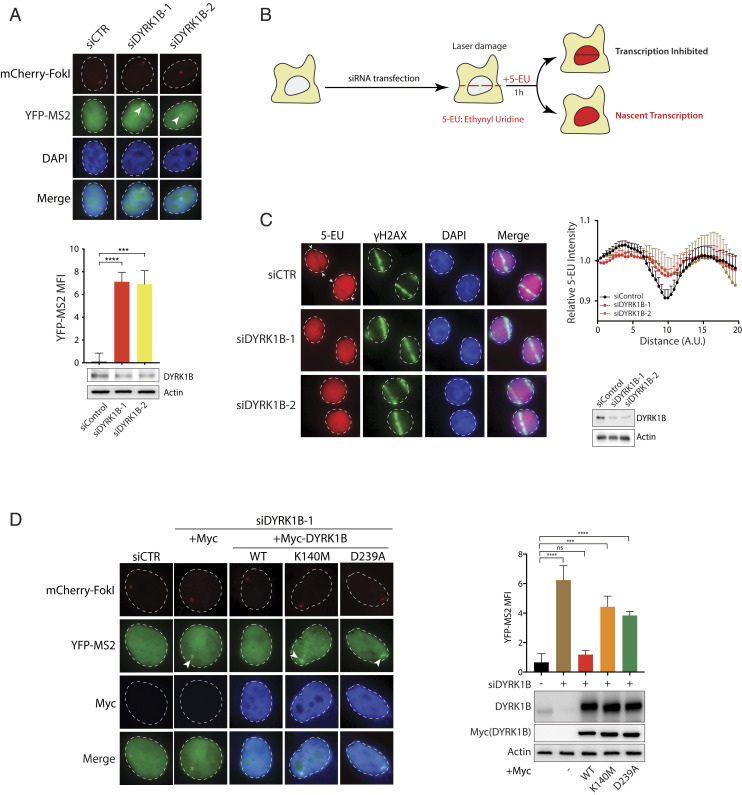
To corroborate that DYRK1B is important in transcription suppression on damaged chromatin, we depleted DYRK1B using two independent small interference RNAs (siRNAs) (SI Appendix, Fig. S2B) and found that DYRK1B silencing led to sustained transcription despite DSB induction in U2OS-DSB reporter cells (Fig. 2A). Similar observations were made in cells lentivirally transduced with two independent DYRK1B-targeting guide RNAs (gRNAs) (SI Appendix, Fig. S2 B and C). Moreover, in stark contrast to control cells, we found that DYRK1B-inactivated cells failed to suppress 5-ethynyl uridine (5-EU) incorporation at laser-microirradiated sites (Fig. 2 B and C and SI Appendix, Fig. S2D), indicating that DYRK1B is required to inhibit nascent transcription at DNA damage sites. Given that chemical inhibition of ATM or DYRK1B attenuated suppression of 5-EU incorporation at laser-induced DSBs (SI Appendix, Fig. S2E), we genetically examined whether DYRK1B kinase activity is important in DISC by reconstituting DYRK1B-depleted cells with wild-type DYRK1B or its kinase-inactive mutants K140M and D239A (25, 26). In line with the requirement of DYRK1B kinase activity in suppressing transcription on damaged chromatin, mutational inactivation of DYRK1B catalytic activity compromised DISC in reporter cells (Fig. 2D) as well as in laser-microirradiated cells (SI Appendix, Fig. S2F). Together, these data firmly establish the DYRK1B kinase as a transcription control factor at DSBs.
Fig. 2.
DYRK1B promotes DSB-induced transcription silencing. (A) U2OS-DSB reporter cells transfected with control (siCTR) or DYRK1B-specific siRNAs (siDYRK1B-1 and siDYRK1B-2) were induced with Dox, 4-OHT, and Shield-1 for 3 h. Cells were processed for fluorescence imaging to quantify MFI of YFP-MS2 or Western blotting experiments using the indicated antibodies. Arrowheads denote YFP-MS2 foci. Bars represent mean ± SEM; ***P < 0.001, ****P < 0.0001. (B) Schematic illustration of a 5-EU incorporation experiment to analyze nascent transcription. (C) HeLa cells transfected with control (siCTR) or DYRK1B-specific siRNAs (siDYRK1B-1 and siDYRK1B-2) were processed for laser microirradiation. Fixed cells were labeled for 5-EU and γH2AX. Nuclei were counterstained with DAPI. Representative images and quantification of relative 5-EU fluorescence intensity are shown. Arrowheads denote sites of laser microirradiation. Western blotting using the indicated antibodies was performed to evaluate RNAi depletion of DYRK1B. A.U., arbitrary units. (D) U2OS-DSB reporter cells pretreated with the indicated siRNAs were transfected with Myc vector or Myc-tagged DYRK1B WT and kinase mutants (K140M or D239A). Cells were processed 24 h posttransfection as in A and YFP-MS2 foci were analyzed and quantified. Western blotting experiments were performed to evaluate expression of Myc-DYRK1B alleles. Bars represent mean ± SEM; ***P < 0.001, ****P < 0.0001.
DYRK1B Promotes DISC-Associated Histone Ubiquitylation and RNAPII Dynamics.
Given that ATM mounts DISC by promoting local H2A ubiquitylation (6), we tested whether chemical inhibition of DYRK1B may similarly compromise DSB-associated histone ubiquitylation. Consistently, DYRK1B inactivation led to marked reduction in H2A ubiquitylation but did not noticeably affect levels of total ubiquitin conjugates (FK2) or K63-linked ubiquitin adducts at FokI-induced DSBs (SI Appendix, Fig. S3 A–C). Moreover, in line with a role in regulating RNAPII-dependent transcriptional activities on damaged chromatin, DYRK1B inhibition led to sustained accumulation and phosphorylation of RNAPII at FokI-induced DSBs in U2OS-DSB reporter cells (SI Appendix, Fig. S3 D–G). Together, these findings suggest that DYRK1B may regulate transcriptional activities at DSBs in an epistatic manner with ATM.
DYRK1B Is Recruited to DSBs.
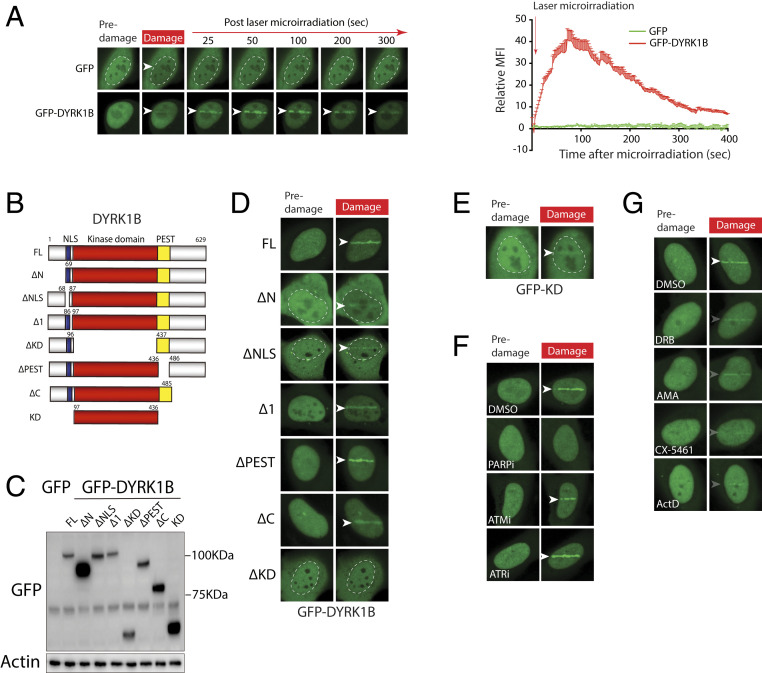
We next examined whether DYRK1B may play a direct role in DSB-flanking chromatin. To this end, we tested whether DYRK1B may be mobilized in response to DNA damage. Intriguingly, we found that laser microirradiation triggered swift and transient accumulation of green fluorescent protein (GFP)-tagged DYRK1B at DNA damage tracks (Fig. 3A). To decipher how DYRK1B docks at DSBs, we generated a panel of DYRK1B deletion mutants and analyzed their ability to accumulate at laser-induced DNA damage tracks (Fig. 3 B and C). Interestingly, we found a strict requirement of the DYRK1B kinase domain in supporting its accumulation at DSBs (Fig. 3D and SI Appendix, Fig. S4A), and that its kinase domain itself suffices in docking at laser-induced DNA damage tracks (Fig. 3 B and E). To examine if kinase activity is required for DYRK1B DSB accumulation, we monitored the dynamics of DYRK1B kinase-inactive mutants (i.e., K140M and D239A) at laser-induced DNA damage tracks. In contrast to the kinase domain deletion mutant, GFP-DYRK1B K140M and D239A displayed robust accumulation at laser-microirradiated sites (SI Appendix, Fig. S4B), suggesting that DYRK1B kinase activity per se is not a prerequisite for its recruitment to DSBs.
Fig. 3.
DYRK1B is recruited to laser-induced DSBs. (A) Cells expressing GFP-DYRK1B or GFP alone were laser microirradiated and time-lapse images were captured to analyze protein accumulation at DNA damage tracks. Quantification of GFP-DYRK1B or GFP accumulation at DSBs was performed. Arrowheads denote sites of laser microirradiation. (B and C) Schematics and steady-state expression level of DYRK1B and its deletion mutant are shown. FL, full length; KD, kinase domain; NLS, nuclear localization signal; PEST, proline-, glutamic acid-, serine-, and threonine-rich domain. (D and E) U2OS cells expressing GFP-DYRK1B or mutants (B) were laser microirradiated. Representative images from predamaged and laser-damaged (100 s postmicroirradiation) cells are shown. (F and G) U2OS cells expressing GFP-DYRK1B were pretreated with the indicated inhibitors prior to laser microirradiation. Cells were processed as in A and representative images are shown as in D.
Given that PARP plays an early role in mobilizing early respondents to DSBs (27), we chemically inhibited PARylation using olaparib and examined whether DYRK1B accumulation at laser-induced DNA damage tracks requires PARP activity. We also pretreated cells with ATM- and ATR-specific inhibitors to explore whether these master regulators of DDRs are necessary for DYRK1B docking on damaged chromatin. Accordingly, while GFP-DYRK1B accumulation at laser-induced DSBs was only marginally attenuated in ATM/ATR-inhibited cells, PARP inhibition completely precluded DYRK1B from concentrating at DNA damage sites (Fig. 3F and SI Appendix, Fig. S4C). Inhibiting ATM and ATR, on the other hand, led to a substantial delay in GFP-53BP1 recruitment to laser-microirradiated sites (SI Appendix, Fig. S4D). Noting that DYRK1B is endowed with transcription regulatory roles on DSB-flanking chromatin, we also tested whether global transcription suppression may impact its migration to DSBs. Interestingly, chemical inhibition of transcription using a panel of small molecules that target different components of the host transcription apparatus led to quantitative attenuation of DYRK1B accumulation at DSBs (Fig. 3G and SI Appendix, Fig. S4E). These data led us to postulate that DYRK1B may preferentially target DSBs on transcribed chromatin.
DYRK1B Facilitates DSB Repair.
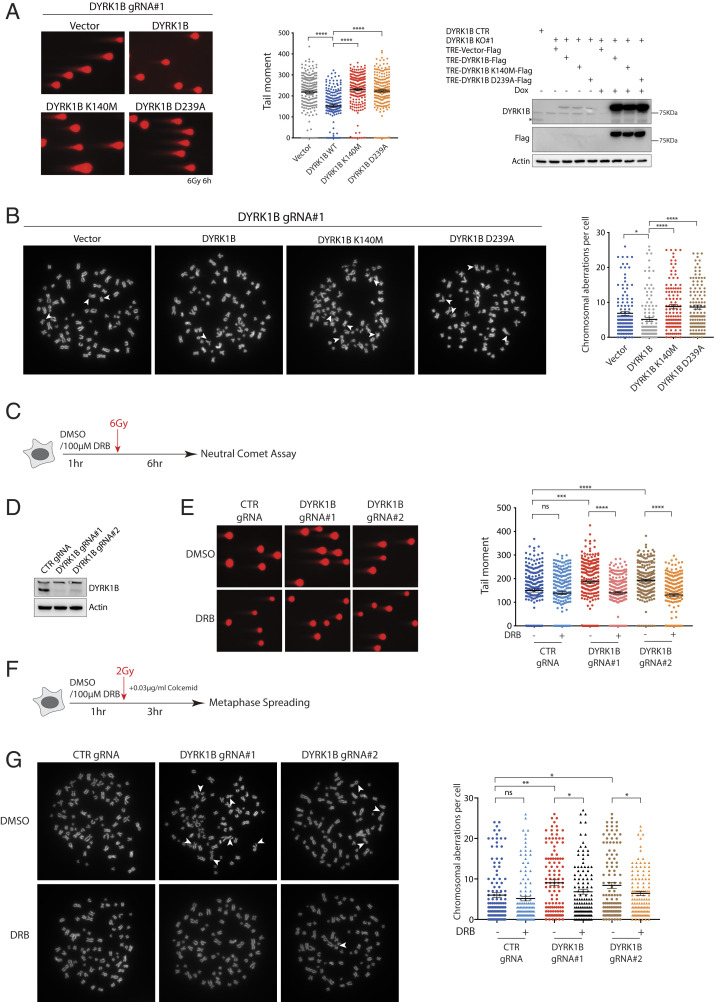
Our observations that DYRK1B promotes transcription silencing on DSB-flanking chromatin prompted us to test whether DYRK1B kinase may be required for efficient DNA repair. To this end, we measured DNA repair kinetics following cell exposure to ionizing radiation (IR) using the comet assay. In support of a role of DYRK1B in DSB repair, IR-induced DNA damage persisted in DYRK1B-inactivated cells (SI Appendix, Fig. S5A). Moreover, expression of wild-type DYRK1B but not its kinase-inactive mutants alleviated the DNA repair defect in DYRK1B-silenced cells (Fig. 4A), highlighting a role of its kinase activity in orchestrating DSB repair processes. To consolidate a role of DYRK1B in DSB repair, we further analyzed chromosome stability by scoring chromosome breaks in DYRK1B-deficient cells following IR treatment. Consistently, we found that DYRK1B promotes repair of IR-induced chromosome breaks (SI Appendix, Fig. S5B) and that its kinase activity is similarly required for recovery from IR treatment (Fig. 4B). Together, these data implicate DYRK1B-dependent transcription silencing as a key event that fine-tunes DSB repair.
Fig. 4.
DYRK1B facilitates repair of IR-induced DNA damage. (A) DYRK1B-silenced cells reconstituted with Dox-inducible expression vector (TRE-Vector-Flag) or those that harbor DYRK1B alleles were induced with doxycycline. Twenty-four hours posttreatment, cells were irradiated and allowed to recover. Cells were thereafter processed for the comet assay according to standard procedures. Relative tail moment of at least 200 cells from two independent experiments was quantified using ImageJ and results were plotted. Western blotting was performed to examine the expression of DYRK1B using the indicated antibodies. The asterisk denotes the protein band that corresponds to endogenous DYRK1B. Bars represent mean ± SEM; ****P < 0.0001. (B) Representative metaphase preparations from IR-treated HeLa derivatives are shown. HeLa cells lentivirally infected with DYRK1B gRNA (DYRK1B gRNA#1) were reconstituted with vector or DYRK1B alleles. Cells were thereafter irradiated and processed to determine the number of chromosomal breaks. At least 120 metaphases from two independent experiments were counted and results were plotted. Arrowheads denote chromosome breaks. Bars represent mean ± SEM; *P < 0.05, ****P < 0.0001. (C) Scheme depicting cell processing for the neutral comet assay following transient global inhibition of transcription using DRB. (D) Western blotting was performed to evaluate the expression of DYRK1B. (E) Representative images from single-cell electrophoresis to analyze relative tail moments. Quantification is shown and was derived from at least 200 cells from two independent experiments. Bars represent mean ± SEM; ***P < 0.001, ****P < 0.0001. (F) Scheme depicting cell processing for metaphase analyses following transient global inhibition of transcription using DRB. (G) Representative metaphase preparations from IR-treated HeLa derivatives are shown. The numbers of chromosomal breaks were quantified in HeLa cells processed as in B with or without pre-DRB treatment. Bars represent mean ± SEM; *P < 0.05, **P < 0.01.
Global Transcription Shutdown Alleviates DNA Repair Defects in DYRK1B-Inactivated Cells.
That DYRK1B may preferentially target DSBs on transcribing chromatin (Fig. 3G and SI Appendix, Fig. S4E) and is important for efficient DNA repair led us to speculate whether the kinase may facilitate repair of DSBs within transcriptionally active chromatin. We explored this possibility by performing both the comet assay (Fig. 4C) and by scoring chromosome breaks (Fig. 4F) with IR-challenged cells pretreated with the transcription inhibitor DRB. Notably, global transcription shutdown not only alleviated the DNA repair defect associated with DYRK1B loss (Fig. 4 D and E) but also suppressed chromosome breaks in otherwise DYRK1B-inactivated cells (Fig. 4G). Together, these data suggest that DYRK1B may facilitate repair of DSBs by orchestrating local transcriptional activities.
Phosphoanalysis of DYRK1B Substrates Identifies DISC Factors.
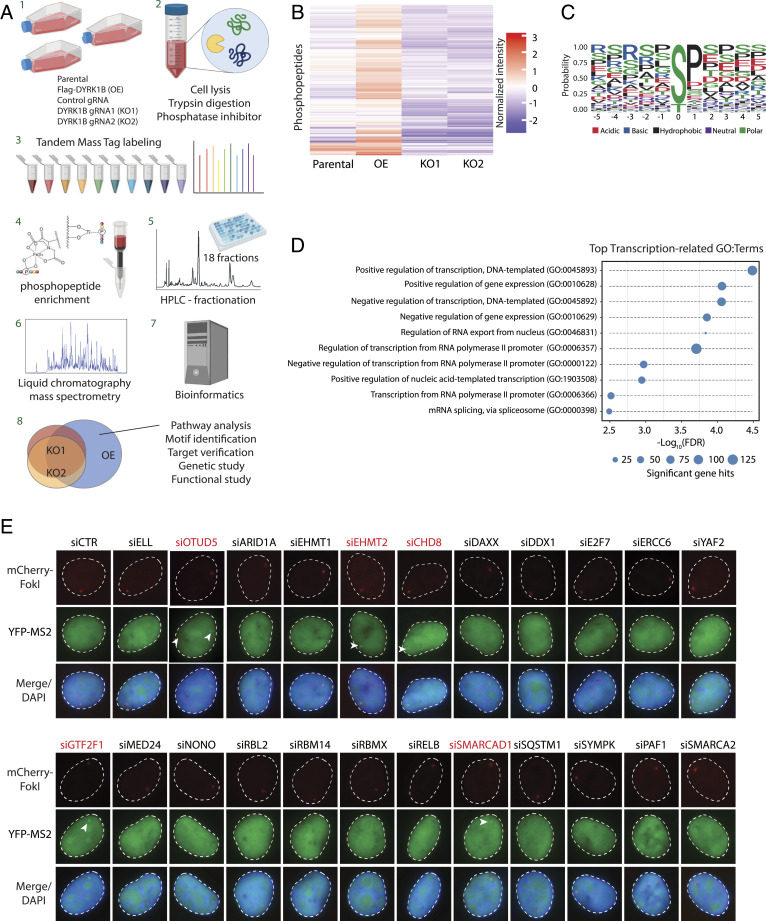
Given the requirement of DYRK1B kinase activity in orchestrating DSB-induced transcription silencing and in DNA repair, we performed a global profiling of DYRK1B targets with the aim to delineate how DYRK1B mediates these DSB responses (Fig. 5A). We sampled and enriched phosphopeptides obtained from parental RPE-1 cells as well as DYRK1B-overexpressing and DYRK1B-inactivated counterparts. Accordingly, bioinformatic analyses of phosphopeptides revealed a high degree of overlap between parental RPE-1 cells and those that overexpress DYRK1B, as well as those between the two isogenic DYRK1B gRNA-targeted cell lines (SI Appendix, Fig. S6A). Notably, analysis of phosphopeptides that are significantly enriched in cells with ectopic expression of DYRK1B overexpression over its knockout counterparts (KO1+KO2) (Fig. 5B and Dataset S2) uncovered that DYRK1B preferentially targets substrates on the serine/threonine-proline (S/T-P) motif (Fig. 5C). Furthermore, Gene Ontology (GO) enrichment analysis of putative DYRK1B targets indicated that DYRK1B may be involved in a plethora of biochemical pathways, including those that regulate transcription (Fig. 5D, SI Appendix, Fig. S6B, and Dataset S3).
Fig. 5.
Global phosphoproteomic identification of DYRK1B substrates. (A) Schematic diagram of phosphoproteomic analysis using high-resolution mass spectrometry. (B) Heatmap expressing the different relative abundances of phosphopeptides between parental cells, DYRK1B overexpression (OE) cells, and DYRK1B knockout (KO) cells. (C) DYRK1B phosphorylation motif determined from the significantly enriched phosphopeptides in OE vs. KO samples. (D) Top biological process GO: terms related to transcription within the total top 50 terms as determined by EnrichR comparing OE with KO cell lines. The size of the data points signifies the number of significant proteins within each pathway. Data represent four biological replicates. FDR, false discovery rate. (E) RNAi-based validation screen for DYRK1B targets in DISC. U2OS-DSB reporter cells pretreated with the indicated siRNA pools were induced with Dox, 4-OHT, and Shield-1. Arrowheads denote YFP-MS2 foci. Cells were thereafter processed to visualize mCherry-FokI and YFP-MS2 foci.
With a focus to decipher how DYRK1B mediates DISC, we selected a number of candidates from transcription-related pathways (Fig. 5D, SI Appendix, Fig. S6C, and Dataset S3) and performed an RNA interference (RNAi)-based validation screen to isolate novel players that are important in transcription suppression on damaged chromatin (Fig. 5E). Among those that are required for DISC, we were drawn to EHMT2 (also known as G9a), as it has established roles in gene repression (28) and represents a promising drug target for cancer (29). More recently, EHMT2 has also been implicated in cell responses to DNA damage (30–32). Accordingly, we confirmed the DYRK1B–EHMT2 interaction using a coimmunoprecipitation approach (SI Appendix, Fig. S7A) and that DYRK1B promotes EHMT2 phosphorylation in a kinase-dependent manner (SI Appendix, Fig. S7B).
EHMT2 Promotes Transcription Silencing on Damaged Chromatin.
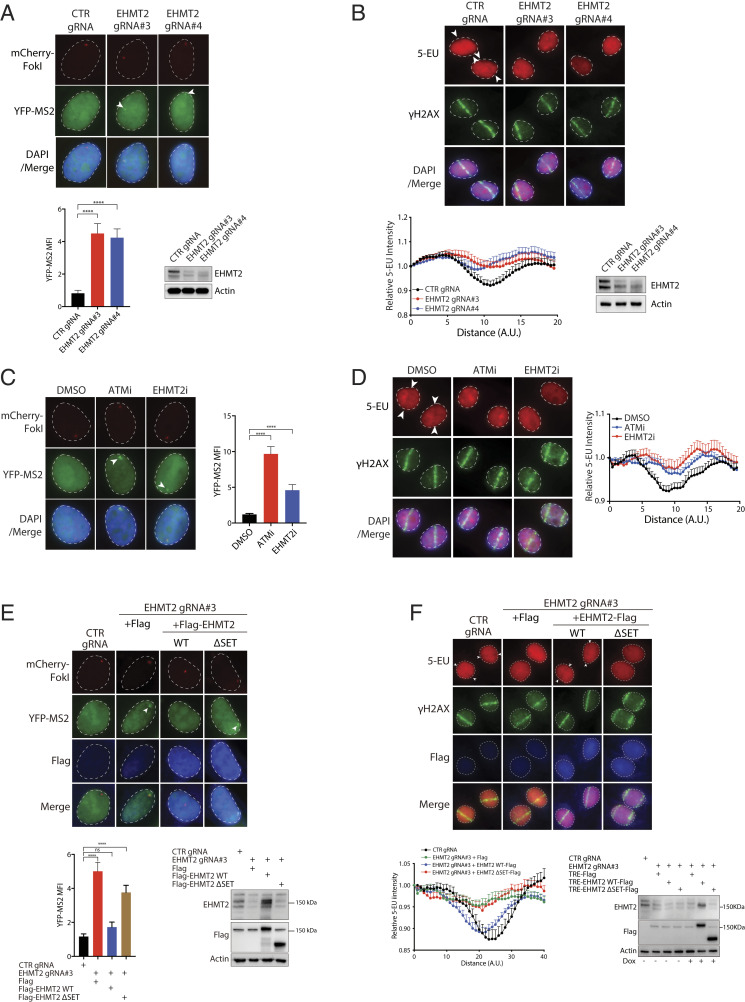
In line with a requirement of EHMT2 in pausing transcriptional activities proximal to FokI-induced DSBs in U2OS-DSB reporter cells (Figs. 5E and 6A), genetic inactivation of EHMT2 also led to sustained 5-EU incorporation at laser-induced DNA damage tracks (Fig. 6B). Given that EHMT2 is endowed with histone lysine methyltransferase activity (28), we chemically inhibited EHMT2 using UNC0638 to examine if its catalytic activity may be required for transcription suppression on DSB-flanking chromatin (33, 34). Indeed, chemical inhibition of EHMT2 led to sustained nascent transcription at FokI-induced DSBs as well as at laser-induced DNA damage tracks (Fig. 6 C and D). To further corroborate a role of EHMT2 activity in DISC, we reconstituted EHMT2-inactivated cells with either wild-type EHMT2 or its catalytic mutant (ΔSET) and assayed if its methyltransferase activity is required for transcription silencing at FokI- and laser-induced DSBs. In line with a requirement of EHMT2 activity in DISC, reexpression of wild-type EHMT2 but not its ΔSET mutant in EHMT2-inactivated cells restored transcription suppression following DSB induction (Fig. 6 E and F). Together, these data establish the histone methyltransferase EHMT2 as an important activity in mounting DISC.
Fig. 6.
EHMT2 promotes transcription silencing on damaged chromatin. (A) EHMT2 inactivation compromised DSB-induced transcription silencing. U2OS-DSB reporter cells lentivirally transduced with the indicated gRNAs were incubated with Dox, 4-OHT, and Shield-1 to induce DSBs proximal to the transcription unit. Thereafter, cells were processed to visualize YFP-MS2 and mCherry-FokI foci. Nuclei were counterstained with DAPI. Arrowheads denote YFP-MS2 foci. MFI of YFP-MS2 was quantified. Bars represent mean ± SEM; ****P < 0.0001. Western blotting was performed to evaluate expression of EHMT2. (B) EHMT2 silencing led to sustained nascent transcription at laser-induced DSBs. HeLa cells transduced with the indicated gRNAs were laser microirradiated. Cells were processed 1 h after to evaluate 5-EU incorporation at laser-induced DNA damage tracks. Arrowheads denote sites of laser microirradiation. Quantification of 5-EU incorporation at laser-induced γH2AX-marked DSBs was performed. Data represent mean ± SEM from three independent experiments. Western blotting was performed to evaluate expression of EHMT2. (C and D) Chemical inhibition of EHMT2 attenuated DISC. Cells pretreated with either ATM inhibitor (KU55933) or EHMT2 inhibitor (UNC0638) were processed for visualization of mCherry-FokI and YFP-MS2 (C) or 5-EU incorporation assay (D). Representative images are shown. Arrowheads denote YFP-MS2 focus or anti–5-EU stripes. Data represent mean ± SEM from three independent experiments. (E and F) Genetic inactivation of EHMT2 methyltransferase activity compromised transcription silencing proximal to FokI- (E) and laser-induced (F) DSBs. Control gRNA- or EHMT2 gRNA-targeted cells reconstituted with vector control, WT EHMT2, or its SET deletion mutant (ΔSET) were subjected to either the U2OS-DSB reporter assay as in A or the 5-EU incorporation assay as in B. Quantification and Western blotting analyses were performed as in A and B. Note that the EHMT2 antibodies were raised against a synthetic peptide corresponding to the carboxyl terminus of the protein, and do not recognize the EHMT2 SET deletion mutant.
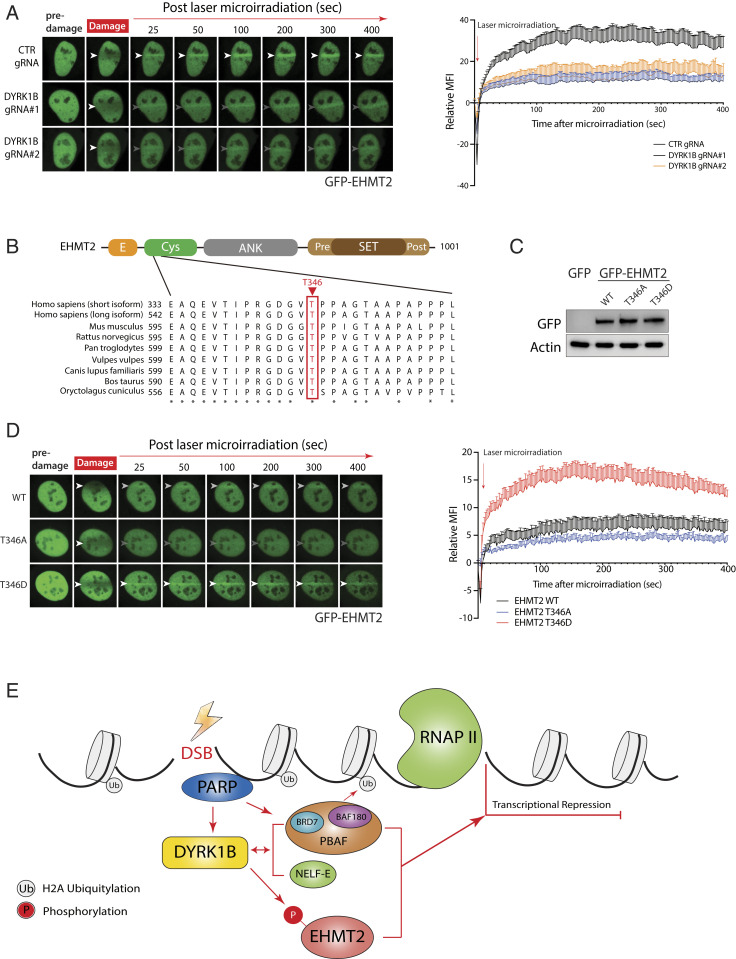
DYRK1B Promotes EHMT2 Accumulation at DSBs.
To further explore how DYRK1B may effect DISC via EHMT2 phosphorylation, we first examined if EHMT2 recruitment to DSBs may depend on DYRK1B. Time-lapse imaging of GFP-EHMT2 following laser microirradiation indicated that DYRK1B enforces EHMT2 accumulation at DSBs, as GFP-EHMT2 recruitment was quantitatively attenuated in DYRK1B-inactivated cells (Fig. 7A) as well as in DYRK1B-inhibited cells (SI Appendix, Fig. S7C), although DYRK1B deficiency did not noticeably alter EHMT2 protein expression level (SI Appendix, Fig. S7D). Moreover, similar to those that underlie DYRK1B recruitment to DSBs, EHMT2 deposition at DSBs required PARP and transcriptional activities (SI Appendix, Fig. S7C). These findings prompted us to investigate whether DYRK1B may promote EHMT2 deposition at DSBs via its phosphorylation. To this end, we generated EHMT2 phosphomutants (i.e., T346A and T346D) according to the DYRK1B-enriched EHMT2 phosphopeptide (Fig. 7 B and C and Dataset S3C), and examined their migration kinetics to laser-induced DSBs. Notably, while the nonphosphorylatable T346A mutation partially hampered EHMT2 recruitment to DSBs, the phosphomimicking T346D mutant accumulated with much more robust kinetics when compared with wild-type EHMT2 (Fig. 7D). These data suggest that DYRK1B facilitates EHMT2 docking at DSBs, at least in part, via targeted T346 phosphorylation.
Fig. 7.
DYRK1B facilitates EHMT2 recruitment to DSBs. (A) Control (CTR gRNA) or cells transduced with DYRK1B gRNAs (DYRK1B gRNA#1 and DYRK1B gRNA#2) were transiently transfected with the GFP-EHMT2 expression construct. Twenty-four hours posttransfection, cells were microirradiated and time-lapse images were captured to monitor GFP-EHMT2 migration to laser-induced DNA damage tracks. Arrowheads denote sites of laser microirradiation. Quantification is shown and is derived from two independent experiments of at least 10 cells each. (B) Schematic illustration of EHMT2 protein domains and the DYRK1B-enriched EHMT2 phosphopeptide surrounding T346. Note that T346 corresponds to T555 in the long isoform of EHMT2. (C and D) GFP-tagged WT EHMT2 or its phosphomutants (T346A and T346D) were expressed in U2OS cells prior to laser microirradiation and time-lapse imaging experiments as done in A. (E) Working model depicting DYRK1B in orchestrating transcription suppression on DSB-flanking chromatin. DYRK1B accumulates at DSBs in a PARP-dependent manner, and promotes EHMT2 T346 phosphorylation and concentration at DSBs to effect DISC.
Discussion
In this study, we have uncovered the DYRK1B network as a branch of mammalian DDR pathways that orchestrates transcriptional activities on damaged chromatin for effective DNA repair (Fig. 7E). By conducting a kinase inhibitor library screen, we identified DYRK1B as a S/T-P-targeting kinase that accumulates at DSBs to promote transcription suppression on DSB-flanking chromatin. Phosphoprofiling of DYRK1B substrates further unveiled that DYRK1B-dependent DISC is, at least in part, effected via EHMT2 phosphorylation and its docking at DSBs. Our findings thus highlight the DYRK1B kinase as a specialized mediator of ATM-dependent DDRs that serves to maintain genome stability of transcribed chromatin.
Our appreciation for the interplay between DSB metabolism and local transcriptional activities has grown since the introduction of elegant platforms in which DSBs can be induced at specific genomic loci (6, 7, 35–37), thus permitting the study of DSB microenvironment (e.g., histone marks) and its impact on local chromatin transactions (38, 39). Notably, it has now become evident that DSB repair can be dynamic (40, 41) and that DSB signal output is limited by local chromatin architecture and activities (36, 42). In particular, the molecular events that underlie transcriptional control on damaged chromatin have garnered much interest (11, 36, 42, 43), in part due to their emerging impact on genome stability and its relevance to human diseases. In this connection, seeing that DYRK1B has been associated with a metabolic syndrome (20), we subcloned the clinically derived DYRK1B mutations (i.e., H90P and R102C) and examined their impact on DYRK1B-dependent DDRs (SI Appendix, Fig. S8). Intriguingly, while both DYRK1B mutations compromised DSB repair in the comet assay (SI Appendix, Fig. S8A), R102C exhibited perturbed DSB recruitment kinetics (SI Appendix, Fig. S8B) and failed to suppress transcription following FokI- and laser-induced DSBs (SI Appendix, Fig. S8 C and D). Given the emerging links between DSB repair and metabolic homeostasis (44), it would be of significant interest to explore how defective DISC may impact cell differentiation and tissue homeostasis.
Global phosphoprofiling of DYRK1B targets led to the uncovering of the histone lysine methyltransferase EHMT2 as one of the candidate downstream effectors that promotes DSB-induced transcription silencing (Fig. 5). Indeed, that EHMT2 plays established roles in gene repression (28) suggested that the DYRK1B–EHMT2 axis may represent a novel branch of DDRs with specialized roles in preserving transcriptionally active chromatin. In line with this notion, we found that DYRK1B targets EHMT2 at a highly conserved S/T-P motif (Fig. 7B) and that EHMT2 carrying the phosphomimetic T346D mutation much more readily accumulates at laser-induced DSBs (Fig. 7D), suggesting that DYRK1B may promote EHMT2 docking and activity at DSBs. While the mechanistic details that underlie EHMT2-dependent DISC remain to be defined, that both chemical inhibition and genetic inactivation of EHMT2 methyltransferase activity led to sustained nascent transcription at damaged chromatin (Fig. 6 C–F) suggests that EHMT2 may exert gene repression by contributing to the local DSB microenvironment, potentially by targeting histone molecules.
Our observation that a number of putative DYRK1B targets may be important in transcription suppression on DSB-flanking chromatin (Fig. 5E) suggests that DYRK1B may orchestrate DSB repair on transcriptionally active chromatin via multipartite mechanisms. To further explore this possibility, we examined whether DYRK1B may support DSB recruitment of chromatin-remodeling complexes and chromatin architectural factors recently implicated in DISC (3, 10). Notably, we found that DYRK1B deficiency impinged on, to varying extents, migration of PBAF (i.e., BRD7 and BAF180) and cohesin (i.e., STAG2) subunits to laser-induced DNA damage tracks but not that of the Polycomb repressive complex components BMI1 and EZH2 (SI Appendix, Fig. S9). On the other hand, DYRK1B appears to have minimal impact on DSB accumulation of the negative elongation factors NELF-A/E (SI Appendix, Fig. S10) and did not detectably affect the metabolism of R loops (SI Appendix, Fig. S11). Together, while DYRK1B likely effects transcription suppression via multiple targets on DSB-flanking chromatin, the fact that DYRK1B-associated DSB repair defects can be alleviated by global transcription inhibition (Fig. 4 C–G) establishes DYRK1B and its network as key signaling intermediates that promote DISC, a dedicated ATM-dependent DSB response that has evolved to suppress chromosomal instability and maintain gene expression programs important in cell proliferation and organismal development. We await further work to decipher how DYRK1B coordinates local transcription during DSB metabolism and how it translates to DSB repair and maintenance of chromosome stability.
Materials and Methods
Cell Lines, Cell Culture, Plasmids, and Chemicals.
HeLa, U2OS, hTERT RPE-1, and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco, Thermo Fisher Scientific) at 37 °C in 5% CO2. Details of chemicals, plasmids, and antibodies are in SI Appendix, Tables S1–S3, respectively.
RNA Interference.
Cells were transfected twice at 24-h intervals with either nontargeting control or gene-specific siRNAs (Dharmacon) using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. Sequences of siRNAs are listed in SI Appendix, Table S4.
Lentiviral Particle Packaging and Transduction.
HEK293T cells were transiently cotransfected with lentiviral-based expression plasmids psPAX2 and pMD2.G at a ratio of 4:3:1 using polyethyleneimine to produce lentiviral particles. Forty-eight hours after transfection, supernatants containing lentiviruses were filtered with an Acrodisc 25-mm syringe filter with a 0.45-µm membrane (PALL Life Sciences) and were used for cell transduction in the presence of 8 µg⋅mL−1 polybrene (Sigma-Aldrich).
Genome Editing Using the CRISPR-Cas9 Method.
Gene-targeting gRNAs were subcloned into the LentiCRISPR v2 vector (Addgene; 52961) following standard procedures. Viral particles containing gRNAs were applied to recipient cells twice at 24-h intervals in the presence of 8 µg⋅mL−1 polybrene (Sigma-Aldrich). Transduced cells were pooled and selected with DMEM supplemented with 1 µg⋅mL−1 puromycin (Sigma-Aldrich) for 1 wk. Gene-edited cells were validated by Western blotting. Sequences of gRNAs are listed in SI Appendix, Table S5.
Western Blotting and Coimmunoprecipitation.
Cells were harvested and lysed with NETN buffer (20 mM Tris·HCl, pH 8.0, 100 mM NaCl, 0.5% Nonidet P-40, and 1 mM ethylenediaminetetraacetic acid [EDTA]) supplemented with benzonase nuclease (ChemCruz) for 30 min on ice. Whole-cell lysates were boiled in sodium dodecyl sulfate (SDS) loading buffer, resolved on polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene fluoride (PVDF) membranes, and immunoblotted with the indicated antibodies. For coimmunoprecipitation, cells were lysed with NETN buffer for 30 min on ice. After centrifugation at 15,000 rpm for 15 min at 4 °C, supernatants were incubated with 200 µL streptavidin-conjugated beads (GE Healthcare, Sigma-Aldrich) for 4 h at 4 °C with gentle rotation. Protein-bound beads were washed with ice-cold NETN buffer three times and subjected to immunoblotting.
Denaturing Immunoprecipitation.
HEK293T cells transiently cotransfected with S protein-Flag-streptavidin–binding peptide (SFB)-tagged EHMT2 mutants with myc epitope-tagged vector or myc epitope-tagged DYRK1B were lysed with denaturing buffer (20 mM Tris·HCl, pH 8.0, 50 mM NaCl, 0.5% Nonidet P-40, 0.5% SDS, 0.5% deoxycholate, and 1 mM EDTA) on ice for 15 min and subsequently boiled at 95 °C for 5 min. The cell lysates were cooled down on ice for 5 min and incubated with Anti-Flag Affinity Gel (Bimake.com) for 3 h at 4 °C with gentle rotation. Protein-bound beads were washed with ice-cold denaturing buffer four times and boiled with SDS/PAGE sample-loading buffer before being subjected to immunoblotting.
Immunofluorescence.
Cells cultured on coverslips were processed and at the indicated time points washed with ice-cold 1× phosphate-buffered saline (PBS) twice and thereafter fixed with 3% paraformaldehyde (PFA) for 30 min at room temperature. Cells were subsequently permeabilized with 0.5% Triton X-100 for 30 s after two PBS washes. Coverslips were blocked with 5% milk before incubating with primary antibodies for 1 h at room temperature. Cells were then washed twice with PBS and incubated with secondary antibodies for 40 min. Nuclei were counterstained with DAPI for 10 s before coverslips were mounted with fluorescence mounting medium (Dako, Agilent) onto glass slides. Images were acquired by an Olympus BX51 fluorescence microscope (UPlanSApo 40×/0.95 objective).
Laser Microirradiation and Live-Cell Imaging.
Laser microirradiation was carried out on an inverted two-photon microscope (LSM780; Carl Zeiss) equipped with an inverted Axio Observer.Z1 stand, motorized scanning stage, and integrated laser microbeam system, with total UV laser output set to 750 nm (8%). Cells cultured on glass-bottomed confocal dishes (SPL Life Sciences) were subjected to laser microirradiation in a temperature-controlled (37 °C) environmental chamber supplied with 5% CO2 at 24 h after transient transfection with GFP-tagged indicated plasmids. Time-lapse images were acquired by ZEN 2012 (Carl Zeiss) software with a Plan Apochromat 40×/1.4 oil differential interference contrast (DIC) M27 objective and further processed by ImageJ software to analyze mean fluorescence intensity (MFI) across the laser-microirradiated regions. MFI was quantified as the difference between the average fluorescence intensity in the laser-microirradiated regions versus the average fluorescence intensity from adjacent undamaged regions of the same size in the same nucleus.
5-Ethynyl Uridine Incorporation Assay.
Nascent transcription at laser-microirradiated sites was detected by a Click-iT RNA Alexa Fluor 594 Imaging Kit (C10330; Thermo Fisher Scientific). Briefly, cells grown on glass-bottomed confocal dishes (SPL Life Sciences) for 24 h were subjected to laser microirradiation by a live Carl Zeiss LSM780 inverted confocal microscope (10× objective) with a 750-nm laser (8% output). Subsequently, cells were cultured with complete media containing 1 mM 5-EU for 1 h after laser microirradiation. 5-EU labeling was performed following manufacturer instructions. Cells were immunostained for γH2AX and nuclei were counterstained with DAPI before mounting. Images were captured by Olympus BX51 fluorescence microscope (PlanApo N 60×/1.42 oil-immersion objective). ImageJ was used to analyze relative fluorescence intensity across the laser-microirradiated stripes. An analyzing line drawn by a line tool was perpendicular to the damaged stripes, centered at the stripes with two ends at undamaged regions, and the fluorescence intensity was multiplotted. Relative fluorescence intensity was normalized to each end from the same cell.
Chromosomal Aberration Analysis.
Chromosomal aberrations were analyzed by chromosome metaphase spreading. Cells were cultured in media containing 1 µg⋅mL−1 colcemid (KaryoMAX Colcemid Solution in Hanks' balanced salt solution; Thermo Fisher Scientific) for 3 h after 2-Gy irradiation and suspended in 0.8% sodium citrate for 15 min at 37 °C. Subsequently, freshly prepared fixative solution (methanol:acetic acid 3:1; vol/vol) was added and incubated for 5 min at 37 °C. After washing three times by the fixative solution, cells resuspended in a small volume of fixative solution were dropped onto alcohol-cleaned slides and air dried. Cells were stained with DAPI before mounting. Images were captured by a Nikon Ti2-E widefield confocal microscope (100× oil-immersion objective) using MetaMorph microscopy and image analysis software (Molecular Devices).
Neutral Comet Assay.
Cells were harvested and resuspended at 5 × 105 cells per milliliter in ice-cold Ca2+/Mg2+-free PBS. Diluted cells were fixed with 37 °C molten low-melting agarose (LMAgarose; Trevigen) at a ratio of 1:10 (vol/vol) and cell suspensions (60 µL) were transferred onto prewarmed comet slides (CometSlide; Trevigen). Slides were kept at 4 °C in the dark for 30 min and immersed in prechilled lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris⋅HCl, 1% N-lauroylsarcosine sodium, and 1% Triton X-100) for 1 h at 4 °C followed by further immersion in freshly prepared alkaline buffer for 30 min. Slides were washed with 1× TBE buffer (90 mM Tris, 90 mM boric acid, and 3 mM EDTA) twice and were thereafter subjected to TBE electrophoresis at 60 V for 5 min. Slides were then fixed in 100% ethanol for 5 min, air dried, and stained with 1 µg⋅mL−1 propidium iodide (Sigma-Aldrich) at room temperature in the dark for 20 min. Images were captured by an Olympus BX51 fluorescence microscope (20× objective). Tail moments of comets were quantified by ImageJ software with the OpenComet plugin.
Kinase Inhibitor Library Screen.
A kinase inhibitor library was purchased from Selleck Chemicals. U2OS-DSB reporter cells, a kind gift from Roger Greenberg (University of Pennsylvania, Philadelphia, PA) were used to assay transcription repression following DSB induction (23). DSBs were induced by cell pretreatment with Shield-1 and 4-OHT, which promote the nuclear translocation of the FokI-mCherry-LacI nuclease and its docking at the LacO transgene array. Nascent transcription at the reporter gene can be monitored by local accumulation of YFP-MS2 upon the addition of 1 µg⋅mL−1 doxycycline. U2OS-DSB reporter cells in 96-well microplates (PerkinElmer) were individually incubated with 10 µM kinase inhibitor for 3 h. Cells were fixed and subjected to a high-content imaging system (IN Cell Analyzer 6500HS). Dimethyl sulfoxide (DMSO) and ATM inhibitor (KU55933) were used as negative and positive controls, respectively. Data were processed by IN Carta analysis software.
Customized siRNA Screen.
Customized siRNAs (three siRNAs per gene target) were purchased from GenePharma to silence selected putative DYRK1B substrates for a role in DSB-induced transcription repression. U2OS-DSB reporter cells were transfected twice at 24-h intervals with either nontargeting control or gene-specific siRNAs, and local transcription was monitored by accumulation of YFP-MS2 at the LacO transgene array.
Cell Culture and Induction of DYRK1B for Phosphoproteomic Analysis.
Parental RPE-1, doxycycline-inducible DYRK1B-overexpressing RPE-1 tetracycline response element (TRE)-DYRK1B, RPE-1-DYRK1B-control, CRISPR-Cas9–mediated RPE-1 DYRK1B-KO#1, and RPE-1 DYRK1B-KO#2 were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. RPE-1 TRE-DYRK1B-SFB cells were induced with 1 µg⋅mL−1 doxycycline for 36 h prior to harvesting. Cells were harvested by trypsin, washed with PBS, flash frozen, and stored at −80 °C until needed.
Chloroform/Methanol Extraction and Generation of Tandem Mass Tag (TMT)-Labeled Tryptic Peptides.
Frozen cell pellets were thawed on ice and resuspended in lysis buffer (2% SDS and 100 mM Tris⋅HCl, pH 7.6) supplemented with fresh protease and phosphatase inhibitors (Pierce) and incubated on ice for 20 min. Viscosity of the samples was reduced using QIAshredders (Qiagen). Briefly, the lysed samples were applied to a QIAshredder spin column and centrifuged at 16,000 × g for 5 min. Protein concentration within the flowthrough was then quantified using a BCA protein assay (Pierce). Volumes equivalent to 300 µg protein were aliquoted into a fresh 1.5-mL tube and the volume was adjusted to 100 µL using lysis buffer. Proteins were reduced by addition of Tris(2-carboxyethyl)phosphine (TCEP) to a final concentration of 5 mM for 30 min at 37 °C. The free cysteine residues were alkylated by incubating with 10 mM iodoacetamide for 30 min protected from light. Proteins were then extracted by chloroform/methanol extraction with modification (45). Briefly, 400 µL of methanol was added and the sample was vortexed, followed by 100 µL of chloroform and vortexing; 300 µL of H2O was added and vortexed. The samples were centrifuged for 1 min at 14,000 × g. The aqueous layer was removed and 400 µL of methanol was added followed by vortexing. The samples were centrifuged at 20,000 × g for 5 min and as much supernatant as possible was removed. The samples were dried in a SpeedVac. The samples were then resuspended in 300 µL of 100 mM triethylammonium bicarbonate (TEAB) and sequencing-grade trypsin (Promega) in a protein:trypsin ratio of 50:1. Samples were tryptically digested overnight at 37 °C. The following day the samples were acidified with 0.1% formic acid. Acidified samples were desalted using a Waters C18 Sep-Pak and dried using a SpeedVac. Dried peptide samples were resuspended in 100 µL of 100 mM TEAB; 120 µg of peptide for each sample was used for labeling and 10 µL of each sample was combined to create a pooled sample for normalization between batches. Eleven-plex TMT labels were equilibrated to room temperature and centrifuged prior to resuspension in 60 µL of acetonitrile. TMT label (30 µL) was added to a unique sample and incubated at room temperature for 1 h. The labeling reaction was quenched by addition of 8 µL 5% hydroxylamine and incubation for 15 min at room temperature. Five microliters of each labeled sample was combined and analyzed by mass spectrometry (MS) to check for proper mixing. Mixing was adjusted according to the results and the combined samples were desalted by a Waters C18 Sep-Pak and dried by SpeedVac.
Phosphopeptide Enrichment and basic-pH High-Performance Liquid Chromatography (bHPLC) Off-Line Fractionation of TMT-Labeled Peptides.
Pooled TMT-labeled, tryptic peptides previously dried by SpeedVac were resuspended in phosphopeptide binding/wash buffer from the High-Select TiO2 Phosphopeptide Enrichment Kit (Pierce) and processed for enrichment. The flowthrough from the TiO2 column was applied to a second phosphopeptide enrichment kit (the High-Select Fe-NTA Phosphopeptide Enrichment Kit; Pierce), and the eluates of both kits were dried immediately following elution to prevent loss of phosphopeptides due to the high pH. Eluates from both kits were resuspended in basic buffer A (10 mM ammonium hydroxide, pH 10) and were separated into 36 fractions on a 100 × 1.0-mm Acquity BEH C18 column (Waters) using an UltiMate 3000 ultra high performance liquid chromatography (UHPLC) system (Thermo) with a 40-min gradient from 99:1 to 60:40 basic buffer A:B ratio (buffer B: 10 mM ammonium hydroxide, 99.9% acetonitrile, pH 10), and then consolidated into 18 superfractions.
Multinotch MS3 Analysis of TMT-Labeled Phosphopeptides.
Analysis of TMT-labeled samples was carried out as previously reported (46). In-line reverse-phase fractionation with Jupiter Proteo resin (Phenomenex) was employed to further reduce the complexity of each superfraction generated by off-line bHPLC. The resin was packed into a 200 × 0.075-mm column and HPLC was carried out on a Waters nanoAcquity ultra performance liquid chromatography (UPLC). A 95-min gradient was used to elute samples from the reverse-phase resin. The gradient consisted of 97:3 to 67:33 buffer A (0.1% formic acid and 0.5% acetonitrile) to buffer B (0.1% formic acid and 99.9% acetonitrile). Electrospray ionization at 2.5 kV was used to ionize eluted peptides into an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo). Multinotch MS3 reporter ion mass spectra was used to collect peptide sequence information and TMT reporter ion quantities. The Orbitrap was used to collect profile MS data over 375 to 1,500 m/z at 120,000 resolution. Collision-induced dissociation at 35.0 normalized collision energy fragmented ions for MS/MS. Centroided MS2 fragment ion mass spectra data were collected in the ion trap for ions between 400 and 2,000 m/z. Up to 10 MS2 ions were selected by synchronous precursor selection and fragmented by high-energy C-trap dissociation at 65.0 normalized collision energy to separate the TMT reporter ions from the precursors. Profile MS3 data from 100 to 500 m/z were collected at 50,000 within the Orbitrap for quantification of TMT reporters.
Analysis of Phosphopeptides.
Phosphopeptides were identified and reporter ions were quantified using MaxQuant (Max Planck Institute) with a parent ion tolerance of 3 parts per million, a fragment ion tolerance of 0.5 Da, and a reporter ion tolerance of 0.001 Da. Oxidation of methionine and STY phosphorylation were searched as variable modifications, and carbamidomethylation of cysteine residues and TMT-10plex labeling of lysine residues and N termini of peptides were set as fixed modifications. The data were searched against the human UniProt database UP000005640 (74,458 proteins and added known contaminants). MaxQuant data were further analyzed using R and the packages Limma (47), ggplot2 (48), and heatmaply (49). A significance criterion of Limma adjusted P value < 0.05 and a log2 fold change of ≥1 were employed to produce a “significantly enriched phosphopeptide” list for each groupwise comparison. Pathway analysis was performed on the enriched phosphopeptides for each group using the webtool Enrichr (50) and GO term complexity was reduced using REVIGO (51).
Statistics and Reproducibility.
Quantitative data represent mean ± SEM from at least three independent experiments unless otherwise noted. Two-tailed Student’s t test was used for statistical analysis by GraphPad Prism 8. Statistical differences were considered significant at P < 0.05.
Data Availability Statement.
The phosphopeptide data generated in this study have been uploaded to ProteomeXchange under accession no. PXD019102.
Supplementary Material
Acknowledgments
We thank Dr. Roger Greenberg for the U2OS-DSB reporter cell line, Dr. Jing Guo and the Faculty Core Facility (The University of Hong Kong) for help with high-content screening, Dr. Wen Deng for advice on preparation of metaphases, Dr. Nabieh Ayoub for the GFP-NELF-A/E constructs, Dr. Jessica Downs for pEGFP-STAG2, Drs. William Hahn and David Root for pDONR223-DYRK1B, Dr. Kyle Miller for GFP-BRD7 and GFP-PBRM1, Dr. Thomas Tuschl for the RNaseH1 expression construct, Drs. John Doench and David Root for DYRK1B gRNA constructs, Dr. Ricky Edmondson and the Proteomics Core (University of Arkansas for Medical Sciences) for access to a Thermo Orbitrap Lumos Tribrid mass spectrometer, Dr. Feng Zhang for lentiviral plasmids, and Dr. Simon Bekker-Jensen for GFP-53BP1. The work is sponsored by funding from the Research Grants Council (Projects C7007-17GF and 17104618) (to M.S.Y.H.) and NIH (K22CA204354 and P20GM121293) and Arkansas Breast Cancer Research Program (AWD00053730) (to J.W.C.L.). X.Y.T. is a recipient of a Hong Kong PhD Fellowship, and J.L. is supported by a Lee Shau Kee Postgraduate Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The phosphopeptide data generated in this study have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository (PXD019102).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002193117/-/DCSupplemental.
References
- 1.D’Alessandro G., d’Adda di Fagagna F., Transcription and DNA damage: Holding hands or crossing swords? J. Mol. Biol. 429, 3215–3229 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Marnef A., Cohen S., Legube G., Transcription-coupled DNA double-strand break repair: Active genes need special care. J. Mol. Biol. 429, 1277–1288 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Kakarougkas A. et al., Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 55, 723–732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ui A., Nagaura Y., Yasui A., Transcriptional elongation factor ENL phosphorylated by ATM recruits Polycomb and switches off transcription for DSB repair. Mol. Cell 58, 468–482 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Ji J. H. et al., De novo phosphorylation of H2AX by WSTF regulates transcription-coupled homologous recombination repair. Nucleic Acids Res. 47, 6299–6314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A., ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pankotai T., Bonhomme C., Chen D., Soutoglou E., DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 19, 276–282 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Caron P. et al., WWP2 ubiquitylates RNA polymerase II for DNA-PK-dependent transcription arrest and repair at DNA breaks. Genes Dev. 33, 684–704 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awwad S. W., Abu-Zhayia E. R., Guttmann-Raviv N., Ayoub N., NELF-E is recruited to DNA double-strand break sites to promote transcriptional repression and repair. EMBO Rep. 18, 745–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meisenberg C. et al., Repression of transcription at DNA breaks requires cohesin throughout interphase and prevents genome instability. Mol. Cell 73, 212–223.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong F. et al., Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29, 197–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vivo A. et al., The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 47, 729–746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez A. et al., BMI1-UBR5 axis regulates transcriptional repression at damaged chromatin. Proc. Natl. Acad. Sci. U.S.A. 113, 11243–11248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker W., Emerging role of DYRK family protein kinases as regulators of protein stability in cell cycle control. Cell Cycle 11, 3389–3394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer S. E., Friedman E., Mirk/Dyrk1B: A multifunctional dual-specificity kinase involved in growth arrest, differentiation, and cell survival. Cell Biochem. Biophys. 45, 303–315 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Friedman E., Mirk/dyrk1B kinase in ovarian cancer. Int. J. Mol. Sci. 14, 5560–5575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman E., The kinase Mirk/dyrk1B: A possible therapeutic target in pancreatic cancer. Cancers 2, 1492–1512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker W., A wake-up call to quiescent cancer cells—Potential use of DYRK1B inhibitors in cancer therapy. FEBS J. 285, 1203–1211 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian G. et al., The zebrafish dyrk1b gene is important for endoderm formation. Genesis 48, 20–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keramati A. R. et al., A form of the metabolic syndrome associated with mutations in DYRK1B. N. Engl. J. Med. 370, 1909–1919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coelho M. C., Pinto R. M., Murray A. W., Heterozygous mutations cause genetic instability in a yeast model of cancer evolution. Nature 566, 275–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An L. et al., Dual-utility NLS drives RNF169-dependent DNA damage responses. Proc. Natl. Acad. Sci. U.S.A. 114, E2872–E2881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J. et al., Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiloh Y., Ziv Y., The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 (2013). [PubMed] [Google Scholar]
- 25.Lee K., Deng X., Friedman E., Mirk protein kinase is a mitogen-activated protein kinase substrate that mediates survival of colon cancer cells. Cancer Res. 60, 3631–3637 (2000). [PubMed] [Google Scholar]
- 26.Ashford A. L. et al., Identification of DYRK1B as a substrate of ERK1/2 and characterisation of the kinase activity of DYRK1B mutants from cancer and metabolic syndrome. Cell. Mol. Life Sci. 73, 883–900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posavec Marjanović M., Crawford K., Ahel I., PARP, transcription and chromatin modeling. Semin. Cell Dev. Biol. 63, 102–113 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Shinkai Y., Tachibana M., H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 25, 781–788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner T., Jung M., New lysine methyltransferase drug targets in cancer. Nat. Biotechnol. 30, 622–623 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Yang Q. et al., G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc. Natl. Acad. Sci. U.S.A. 114, E6054–E6063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginjala V. et al., Protein-lysine methyltransferases G9a and GLP1 promote responses to DNA damage. Sci. Rep. 7, 16613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe S. et al., MDC1 methylation mediated by lysine methyltransferases EHMT1 and EHMT2 regulates active ATM accumulation flanking DNA damage sites. Sci. Rep. 8, 10888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubicek S. et al., Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Vedadi M. et al., A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 7, 566–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitor A. C. et al., Single-molecule imaging of transcription at damaged chromatin. Sci. Adv. 5, eaau1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacovoni J. S. et al., High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purman C. E. et al., Regional gene repression by DNA double-strand breaks in G1 phase cells. Mol. Cell. Biol. 39, e00181-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clouaire T., Legube G., A snapshot on the cis chromatin response to DNA double-strand breaks. Trends Genet. 35, 330–345 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Tan X. Y., Huen M. S. Y., Perfecting DNA double-strand break repair on transcribed chromatin. Essays Biochem., 10.1042/EBC20190094 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Aymard F. et al., Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 24, 353–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemaître C. et al., Nuclear position dictates DNA repair pathway choice. Genes Dev. 28, 2450–2463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aymard F. et al., Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iannelli F. et al., A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks. Nat. Commun. 8, 15656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu I., Yoshida Y., Suda M., Minamino T., DNA damage response and metabolic disease. Cell Metab. 20, 967–977 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Wessel D., Flügge U. I., A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 46.Zybailov B. L. et al., Metaproteomics reveals potential mechanisms by which dietary resistant starch supplementation attenuates chronic kidney disease progression in rats. PLoS One 14, e0199274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie M. E. et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham H., Ggplot2: Elegant Graphics for Data Analysis. Use R!, (Springer, New York, 2009). [Google Scholar]
- 49.Galili T., O’Callaghan A., Sidi J., Sievert C., heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 34, 1600–1602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen E. Y. et al., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phosphopeptide data generated in this study have been uploaded to ProteomeXchange under accession no. PXD019102.