Significance
The well-established up-regulation of genes coding for protective proteins during drying of desiccation-tolerant taxa has been central to previous efforts to understand (and manipulate) desiccation tolerance. Using comparative transcriptomics in closely related desiccation-tolerant (desert-derived) and intolerant (aquatic) green algae in the family Scenedesmaceae, here we show, however, that both desiccation-tolerant and intolerant algae up-regulated expression of common protective genes during desiccation. The feature distinguishing desert green algae from their aquatic relative was an extensive, controlled down-regulation of gene expression associated with diverse growth and energy-acquiring metabolic pathways during desiccation. These data suggest a new perspective on desiccation tolerance may be warranted, recognizing both up-regulation of genes coding for protective proteins and extensive down-regulation of diverse metabolic genes.
Keywords: aquatic green algae, desert-evolved green algae, extremophiles, microbiotic crusts, Scenedesmaceae
Abstract
Among green plants, desiccation tolerance is common in seeds and spores but rare in leaves and other vegetative green tissues. Over the last two decades, genes have been identified whose expression is induced by desiccation in diverse, desiccation-tolerant (DT) taxa, including, e.g., late embryogenesis abundant proteins (LEA) and reactive oxygen species scavengers. This up-regulation is observed in DT resurrection plants, mosses, and green algae most closely related to these Embryophytes. Here we test whether this same suite of protective genes is up-regulated during desiccation in even more distantly related DT green algae, and, importantly, whether that up-regulation is unique to DT algae or also occurs in a desiccation-intolerant relative. We used three closely related aquatic and desert-derived green microalgae in the family Scenedesmaceae and capitalized on extraordinary desiccation tolerance in two of the species, contrasting with desiccation intolerance in the third. We found that during desiccation, all three species increased expression of common protective genes. The feature distinguishing gene expression in DT algae, however, was extensive down-regulation of gene expression associated with diverse metabolic processes during the desiccation time course, suggesting a switch from active growth to energy-saving metabolism. This widespread downshift did not occur in the desiccation-intolerant taxon. These results show that desiccation-induced up-regulation of expression of protective genes may be necessary but is not sufficient to confer desiccation tolerance. The data also suggest that desiccation tolerance may require induced protective mechanisms operating in concert with massive down-regulation of gene expression controlling numerous other aspects of metabolism.
Colonization of land is one of the most important processes in evolutionary history, and with life on land comes the threat of desiccation. Most organisms die after drying to equilibrium even with moderately dry air. Water loss can cause extensive cellular damage, affecting membrane integrity, aggregating macromolecules, and altering lipid bodies (1). During desiccation, photosynthetic organisms face additional photooxidative stress, as light continues to be absorbed by the photosynthetic apparatus although carbon fixation is limited by drying. Desiccation tolerance is therefore rare in vegetative tissues of green plants (2, 3).
Still, there are more than 22 independently evolved lineages of green algae that have colonized terrestrial and even extreme desert microbiotic crust environments (4, 5), including the algal lineage ancestral to all Embryophytes (i.e., mosses, ferns, seed plants). Some species within these algal lineages can survive multiple consecutive cycles of desiccation and rehydration and, upon rehydration, regain photosynthetic activity within seconds that persists for hours (6, 7).
Among Embryophytes, desiccation tolerance is relatively frequent in bryophytes (e.g., mosses) but was lost during seed plant radiation (8). Desiccation tolerance in vegetative green tissues was secondarily acquired in (rare) “resurrection” seed plants by coopting metabolic pathways controlling seed development (9, 10). The similarity in physiological responses to desiccation among desiccation-tolerant (DT) cyanobacteria, green algae, and mosses (7, 11, 12) suggests there may be an even more ancestral origin of desiccation tolerance in photosynthetic tissues (13). Such an ancestral origin is also supported by the discovery of shared molecular responses to desiccation among DT Embryophytes and the terrestrial green algae most closely related to them (e.g., streptophyte algae Klebsormidium (14) and Zygnema (15)), but more distantly related DT green algae have not been examined. Shared molecular responses include up-regulation of transcripts coding for numerous late embryogenesis abundant (LEA) proteins, components of biosynthetic pathways for oligosaccharide osmolytes, and scavengers for reactive oxygen species (ROS), among others (14, 15).
Up-regulation in these protective gene groups has become central to the quest to understand and ultimately manipulate desiccation tolerance among green plants, including plants important to agriculture (16). To our knowledge, however, only very recently (17, 18), and only in the flowering plant genus Lindernia, has a critical question been asked: is drying-induced up-regulation of protective genes unique to DT taxa, or do close relatives that are intolerant of desiccation also exhibit the pattern? For DT and desiccation-intolerant Lindernia species, results were not straightforward. For example, comparative analysis of transcription of candidate protective genes showed that expression of a variety of LEA protein genes was up-regulated in both the desiccation-tolerant and intolerant Lindernia species during desiccation. Some LEA proteins were more strongly expressed in the DT taxon (∼30%), but a notable number (15%) were expressed more strongly in the desiccation-intolerant species (18).
Here we use comparative transcriptomics to examine the full range of down-regulated as well as up-regulated genes differentially expressed during a desiccation/rehydration time course. We examine this gene expression in two extraordinarily desiccation-tolerant green algal species isolated from desert microbiotic crust (6) and a desiccation-intolerant aquatic relative. Such comparative analysis of broad gene expression using nonmodel organisms that are particularly well-suited to targeted scientific questions has only recently become possible with advances in bioinformatics and sequencing. In this case, the approach enables investigation of natural evolutionary innovations supporting desiccation tolerance among green plants. We study three closely related species in the green algal family Scenedesmaceae (SI Appendix, Fig. S1A): Acutodesmus deserticola and Flechtneria rotunda were isolated from desert microbiotic crust in the western United States and are extremely DT, surviving multiple consecutive cycles of desiccation and rehydration (6, 7) in the vegetative state (SI Appendix, Fig. S1 B and C). Each is descended from an independent ancestral leap from water to land (6, 19) (SI Appendix, Fig. S1A). The third taxon, aquatic Enallax costatus, is phylogenetically basal and closely related to F. rotunda and A. deserticola (respectively, 98.6% and 99.3% similarity in 18S ribosomic RNA (rRNA) gene sequence, SI Appendix, Fig. S1A) but is not desiccation-tolerant (6). We are using these three algal species because despite their close phylogenetic relationship (19) they harbor variation naturally developed in their strongly contrasting habitats, which should particularly facilitate investigation of determinants of desiccation tolerance. Further, because these organisms are more distant phylogenetically from Embryophytes, if recognized molecular responses to desiccation occur in these organisms, then the shared pattern strongly suggests that molecular circuitry supporting vegetative desiccation tolerance is indeed ancestral.
Results and Discussion
We used standard liquid culture (6) and slow-desiccation of 40-μL drops of algal culture (SI Appendix, Material and Methods and Fig. S2). Cell density was 3.2 ± 0.4 x107 algal cells mL−1 for A. deserticola, 4.9 ± 1.1 x107 algal cells mL−1 for F. rotunda, and 3.4 ± 1.2 x107 algal cells mL−1 for E. costatus. Full evaporation of culturing medium (the “Dry” stage) was achieved after 12 h (6), and samples were then rehydrated for 1 h (7). Because interpretation of desiccation-induced protective mechanisms can be confounded by light-induced ROS production during desiccation in the light, the desiccation time course was conducted in darkness (<1 µE). We compared gene expression changes in the three taxa, first examining commonalities in the broad metabolic responses and then focusing on the behavior of genes traditionally associated with desiccation tolerance in mosses and resurrection plants.
Changes in gene expression during the time course were identified using the samples collected in the initial “Hydrated” stage as the baseline. This “temporal” design (with samples compared to an initial stage), rather than a “parallel” experimental design (with simultaneous sampling of treated and untreated samples throughout the time course) is broadly used in time-series RNA-seq experiments in the field of desiccation tolerance (14, 18, 20–22). In a recent comparison (23), both experimental designs detected similar changes in gene expression during drought stress of two grasses. The temporal design allocates sequencing and labor effort to prioritizing replicates and (here) the number of species sampled. The temporal design cannot, however, separate effects of the treatment (here, desiccation) over time from the effects of any extraneous environmental factors that might affect the organisms during the sampling period. Particularly with multiple nonmodel species, consideration of differential response to extraneous factors is important. In this experiment, species-specific, differential respiratory drawdown of O2 concentration in the 40-μL drops in the dark might have contributed to extraneous environmental variation. We therefore conducted a companion control experiment that demonstrated that O2 diffusion even into full-volume algal dots was sufficient to counter respiratory drawdown of O2 by all species of algae in the dark, and during this control experiment the photosynthetic quantum yield of algae remained maximal for more than 12 h in the dark (see text in SI Appendix and SI Appendix, Fig. S3).
Differentially Expressed Genes (Dataset S1).
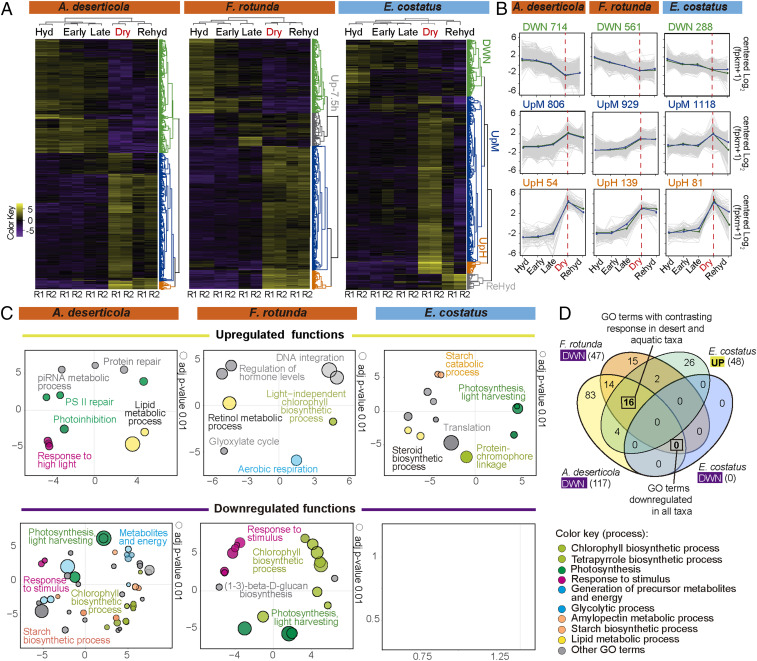
Over the desiccation–rehydration time course, 5–11% (∼1,500) of the genes in each reference transcriptome were identified as differentially expressed genes (DEGs). DEGs were identified using the criteria of ±4 fold change (FC) in expression over the time course at a false discovery rate <0.001 (Fig. 1A, and Dataset S1). Centralized Log2FC gene expression patterns are shown in heatmaps in Fig. 1A. Replicates grouped very closely in PCA plots (SI Appendix, Fig. S5A). Coexpressed DEGs largely fell into three main behaviors. Down-regulated DEGs clustered in group “DWN” (identified in green in the dendrograms to right of each heatmap). Up-regulated DEGs fell in groups “UpM” (moderately up-regulated, blue dendrogram) and “UpH” (highly up-regulated, orange dendrogram) (Fig. 1A). The number of DEGs, and their dynamics within each behavioral group, are shown in Fig. 1B. After 1 h of rehydration, expression in desert taxa returned toward the initial hydrated state (Fig. 1B). E. costatus, however, did not survive rehydration (6), and data from replicates of only that species diverged (SI Appendix, Fig. S5A).
Fig. 1.
Altered gene expression over the time course, in desiccation-tolerant A. deserticola and F. rotunda and intolerant E. costatus. (A) Heatmaps showing changes in transcription of all DEGs (Dataset S1) (purple down-regulated, gold up-regulated). Samples fell into hierarchical groups based on overall similarity (as illustrated above each heatmap). R1 and R2 are biological replicates. (B) Expression profiles in each behavioral group. Gray lines represent individual DEGs; colored lines mark the mean expression for the group, with replicate algal samples indicated in blue and green. (C) Biological processes (GO analysis, Dataset S2) enriched in up-regulated and down-regulated clusters of DEGs during desiccation. In the scatterplots, each bubble represents a significantly enriched term in a two-dimensional space derived by applying multidimensional scaling to a matrix of the GO terms’ semantic similarities (48). Bubble size is calculated as |Log10adj_p-value| (47). As reference, the bubble size for adj_p-value = 0.01 is provided for each plot. Related terms are indicated with the same color to facilitate comparison (see SI Appendix, Fig. S8). (D) Venn diagram showing overlap between GO terms down-regulated in tolerant taxa and up-regulated in the intolerant taxon. Venn diagrams showing overlap among down-regulated and among up-regulated GO terms are presented in SI Appendix, Fig. S9.
During the time course, down-regulation was gradual across the entire desiccation time frame (Fig. 1B, DWN group) and included the down-regulation of photosynthetic genes that has been previously observed during desiccation even in the light (3). In contrast, most gene expression within the UpM and UpH groups remained relatively constant in all three species from the initial “Hydrated” state through 7.5 h into the desiccation time course (the “Late” stage), although ∼60% of the original dot sample volume had been evaporated (SI Appendix, Fig. S2H). This stability was observed even in the aquatic species E. costatus (Fig. 1B) which we had anticipated would be more sensitive to water loss. At some time after the 7.5-h timepoint (but prior to full desiccation), all three species responded with a sharp increase in transcription of hundreds of genes (Fig. 1B and Dataset S1).
Gene Ontology Enrichment Analysis (Dataset S2).
In Fig. 1C, the leftmost and center upper panels show that during the desiccation time course, the desert taxa A. deserticola and F. rotunda up-regulated biological processes (grouped by gene entology [GO] analysis) that are traditionally associated with photosystem repair, high light stress (although these algae were desiccated in the dark), protective pigment production, hormone levels, and neutral lipids and triglyceride metabolism. The rightmost panel in Fig. 1C shows that, in contrast, the aquatic species E. costatus up-regulated aspects of photosynthesis (energetic metabolism), translation, catabolism of starch and glucan, and generation of precursor metabolites (Fig. 1 C, Upper Right). Remarkably, more than half of the 32 processes down-regulated during desiccation by both desert taxa (Fig. 1C, A. deserticola and F. rotunda lower panels, and Fig. 1D) were up-regulated in the aquatic species (Fig. 1C, E. costatus upper panel, and Fig. 1D). The extensive down-regulation of gene expression in desert taxa suggests a metabolic slowdown affecting processes such as photosynthesis, starch metabolism, and generation of precursor metabolites and energy (Fig. 1 C, Lower). No biological processes identified by GO analyis were down-regulated in the aquatic species E. costatus (Fig. 1C).
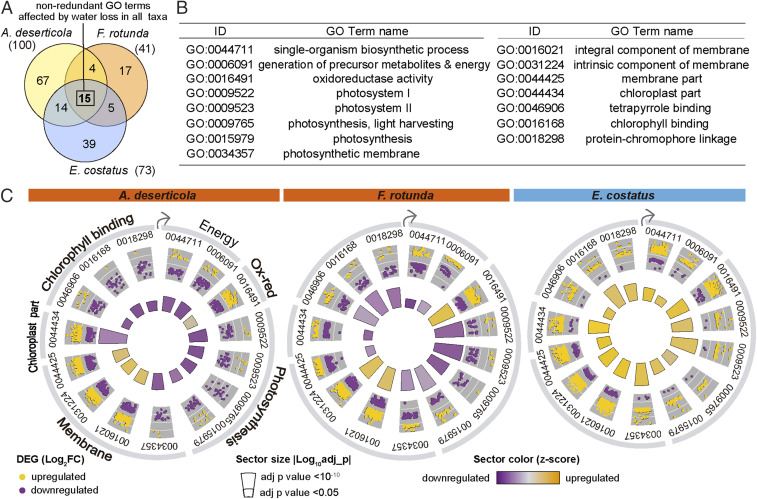
Up- and Down-Regulation of Shared GO Terms.
This contrasting genetic response during the desiccation time course is further analyzed in Fig. 2. To facilitate comparison among species, we used GO term enrichment analysis to identify those functions that were impacted (positively or negatively) by desiccation across all taxa (Fig. 2 A and B). Those responses are plotted in polar graphs (Fig. 2C) where each sector represents one GO term in each species. Changes in expression as Log2FC of each DEG annotated within each GO term are shown as positive (gold dots) or negative (purple dots) (Fig. 2C, outer circle). To capture the overall tendency toward up- or down-regulation of each term, a z score was calculated () and is displayed coded by color in the inner circle of Fig. 2C. Both DT taxa up-regulate oxidoreductase activity and membrane-related properties (gold z-score blocks) and down-regulate energy production and photosynthesis (purple blocks). Desiccation-intolerant E. costatus up-regulates every function analyzed in this cross-species comparative framework (Fig. 2C).
Fig. 2.
Contrasting response of functions identified by GO analysis, during desiccation. (A) Venn diagram showing the overlap of GO terms (after filtering for redundancy) that were enriched in DEGs identified during Late and Dry stages in desert-evolved (A. deserticola and F. rotunda) and aquatic taxa (E. costatus). The number between parentheses is the total number of enriched GO terms. (B) Identity of the 15 GO terms enriched in DEGs in all species, during desiccation. (C) Polar graphs indicating the direction of response of enriched GO terms in each species. The outer ring shows the Log2FC (calculated with DESeq2 (46)) of each DEG annotated within the GO category (up-regulated colored gold, down-regulated colored purple, Datasets S3–S5). In the inner ring, the size of each sector is proportional to the statistical significance of the term (quantified as the adjusted P value) from the GO enrichment analysis in GOseq (47). The color indicates the overall behavior of the GO term based on the value of its z score, calculated using GOplot (49). Up-regulated terms are indicated in gold, down-regulated in purple.
Emerging from this analysis is a picture of DT desert taxa up-regulating genes coding for protective functions while extensively down-regulating genes associated with metabolism as desiccation stress mounts. Notably, E. costatus also up-regulates genes coding for aspects of protection (see next section); in contrast to the desert taxa, however, it broadly activates genes necessary for energetic metabolism and exhibits very limited down-regulation during desiccation (Fig. 1 C and D).
Expression of Specific Photosynthetic, Protective, and Regulatory Genes.
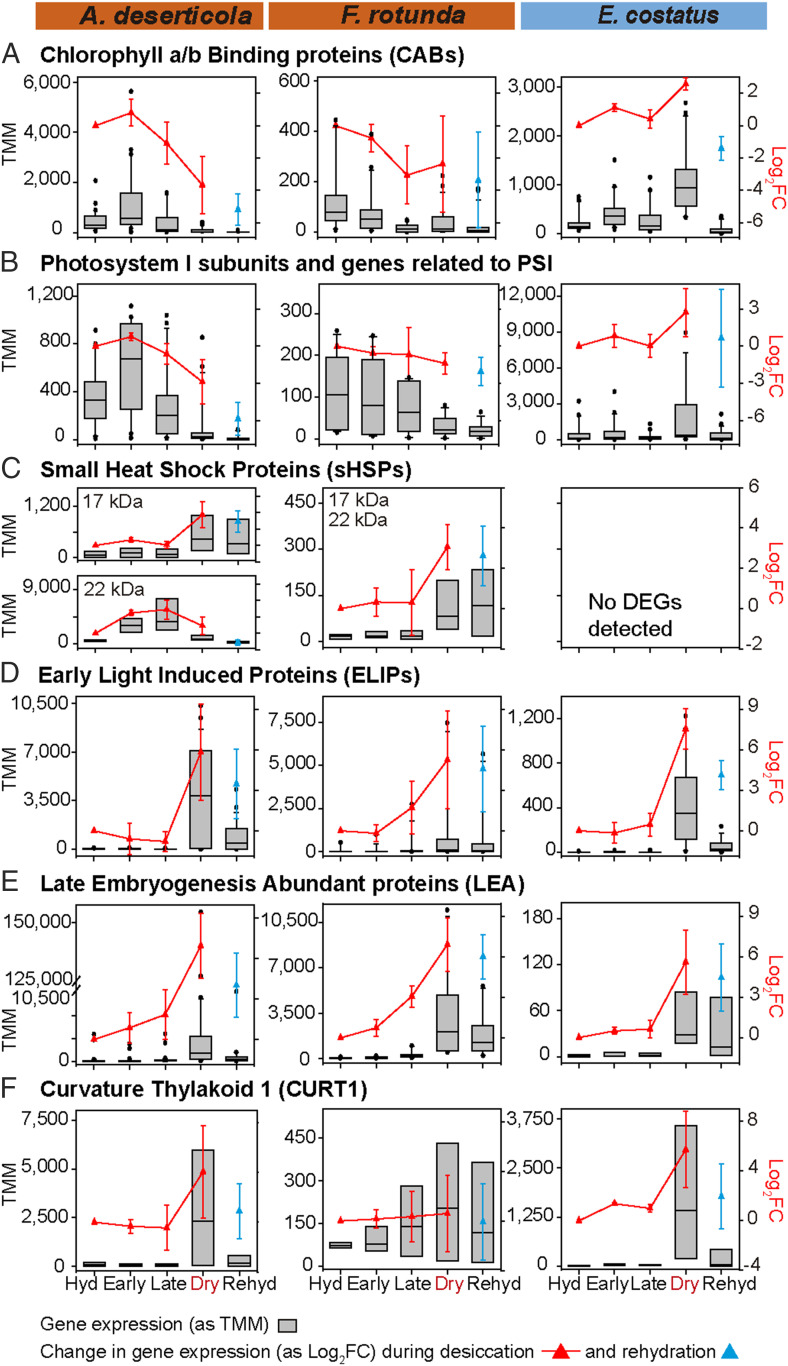
This overall difference in behavior of desert and aquatic taxa was also observed among specific genes commonly analyzed in the desiccation tolerance literature (Fig. 3, see Dataset S7 for additional details). During the desiccation time course, desert taxa reduced the gene expression of chlorophyll a-b binding proteins (CABs) (Fig. 3A) and of highly conserved genes associated with photosystem I (PSI) (Fig. 3B), likely minimizing photodamage. This is a common response in the desiccation tolerance literature (2, 21, 22, 24), even when desiccation occurs in the presence of light (9, 14, 15, 18). In contrast, E. costatus up-regulated expression of both CABs and PSI genes (Fig. 3 A and B). Desert taxa up-regulated expression of potentially protective 17.9-kDa and 22-kDa small heat shock proteins (sHSPs, Fig. 3C) during desiccation. No differentially expressed transcripts encoding sHSPs were detected for E. costatus (Fig. 3C). (Although sHSP transcripts were annotated in the reference transcriptome of E. costatus, their expression levels were very low, see Dataset S7). sHSPs serve as ATP-dependent chaperones in green plants (20, 25), preventing protein aggregation during stress (2, 3, 26).
Fig. 3.
Expression patterns of genes traditionally associated with desiccation tolerance during desiccation and rehydration. (A–F) Each panel presents normalized expression data (as standardized trimmed mean of M-values, TMM) for all DEGs annotated using nr NCBI and/or Uniprot90-SwissProt databases as members of the given gene family (Dataset S7). Box plots summarize the expression of the category in each stage of the desiccation rehydration time course. In the graphs, the boundaries of the boxes indicate the lower and upper quartiles, a black line within each box marks the median, and the whiskers above and below the box indicate the 5th and 95th percentiles. Black points above and below the whiskers indicate outliers outside that range. Average changes in expression of all DEGs within the category during the desiccation time course are indicated with a red line (as Log2FC ± SD). Changes during rehydration are indicated in blue (as Log2FC ± SD). For each gene family, the right-hand y axis Log2FC scale is identical across all species.
Still, expression of several well-known genes for protective proteins was induced during the desiccation time course in all three green algal species, suggesting their induction may be necessary, but not sufficient, to confer desiccation tolerance. (Whether increased transcription of these many protective genes was coupled with enhanced translation in the desiccation-intolerant taxon remains unknown.) All three taxa increased expression of genes coding for Early Light-Induced Proteins (ELIPs, Fig. 3D) and LEA proteins (Fig. 3E). LEA proteins are well known to accumulate in response to stress including water deficit and during late-stage seed maturation (2, 3), stabilizing enzymes, preventing protein aggregation, and scavenging ROS (27). ELIPs are closely related to CABs and PSBS, and they are known to prevent photodamage during diverse abiotic stresses by binding antenna complex chlorophyll (28). For the desert taxa studied here, we have shown previously (6) that protection clearly occurs during desiccation because within seconds of rehydration, light can stimulate notable photosynthetic activity. This photosynthetic capacity is evident, although transcription of CABs and PSI genes does not recover even after 1 h of rehydration in the desert taxa (Fig. 3 A and B). Though not previously linked to desiccation tolerance, expression of a variant of Curvature Thylakoid 1A (CURT1) (29) strongly increased during desiccation in all three species (Fig. 3F). This pattern is intriguing because CURT1 affects thylakoid structure and compartmentalization, influencing PSII repair in Arabidopsis (30) and response to heat (31) and oxidative stress in Chlamydomonas reinhardtii. A homolog of CURT1 is also important during osmotic stress in the cyanobacterium Synechocystis (32).
The extensive metabolic reprogramming evidenced in Figs. 1 and 2 is likely orchestrated by transcription factors (TFs). To date, no cis-elements unique to desiccation tolerance have been identified in the literature, suggesting tolerance may be acquired through rewiring of conserved, preexisting regulatory networks that control ontogeny and mediate green plant responses to abiotic stress (2, 18). Here, five stress-responsive families were differentially expressed exclusively in DT taxa: MYB, B3, TRAF, GNAT, and CSD (Dataset S6). Among them, expression of a MYB98-like TF (homologous to protein Cre03.g197100) was increased (>3 Log2FC). MYBs are involved in cellular cycle control, development, ABA-mediated responses, abiotic stress, and tolerance to drought and desiccation (16), making this MYB98-like TF a particularly interesting candidate to modify cellular responses in DT taxa during water loss.
Conclusion
Overall, our results extend the green plant phylogenetic space over which common molecular pathways supporting desiccation tolerance are known to operate, suggesting strongly that they may be ancestral in the green plant clade. Further, the critical comparison of closely related desert and aquatic green algae revealed that up-regulation of expression of genes with long-recognized protective function (e.g., ELIPs, LEA proteins) is shared across taxa and may be necessary but is clearly insufficient to confer desiccation tolerance. Rather, the evolution of desiccation tolerance may have required both harnessing the capacity for up-regulation of such protective functions and rewiring regulatory mechanisms to reduce gene expression for more generalized cellular metabolism, thereby orchestrating an ordered, metabolic slowdown during water stress.
Materials and Methods
Algal Isolates.
Acutodesmus deserticola (isolate BCP-SNI-2), Flechtneria rotunda (isolate BCP-SEV3-VF49), and Enallax costatus (isolate CCAP276-31) are all in the green algal family Scenedesmaceae (Chlorophyta) and diverge from one another diverge less than 2% in 18S rRNA gene sequence (SI Appendix, Fig. S1A). A. deserticola and F. rotunda are independently evolved, desert-dwelling, unicellular microalgae (19), extremely tolerant to multiple cycles of desiccation and rehydration (6). E. costatus is coenobial, aquatic, and desiccation-intolerant (SI Appendix, Fig. S1 B and C). A full description of each alga can be found in SI Appendix, Material and Methods.
Culturing Conditions and Slow Desiccation.
For each species, two independent algal cultures were grown for 6 wk in 250-mL Erlenmeyer flasks containing 150 mL of autoclaved 1:1 mix of Bold’s Basal medium and Woods Hole medium (SI Appendix, Fig. S2A). Each flask was inoculated with algal cells from stocks on slants and bubbled with room air. Cultures were unialgal but not axenic. All cultures were grown at 25 °C in a Conviron PGW36DE growth chamber (Conviron, Winnipeg, Canada) with a 12:12 h Light/Dark cycle and 40 µE from mixed metal halide and sodium lamps.
To generate manipulable experimental units, referred to from here on as algal dots, 40 μL of concentrated algal cultures were pipetted onto round coverslips (8-mm-diameter German Glass, Electron Microscopy Sciences) (SI Appendix, Fig. S2C). All cultures were adjusted to a density of 3–5 × 107 algal cells mL−1 prior to pipetting.
Coverslips were then placed on microscopy slides and introduced into the desiccation chamber of a custom-made drying apparatus (6) (SI Appendix, Fig. S2 D–F). Humidified air flowed through the desiccation chamber, slowly desiccating the algal dots. A low evaporation rate (<5 μL h−1) was achieved by maintaining a high relative humidity in the desiccation chamber (SI Appendix, Fig. S2F). All desiccation experiments were carried out in near darkness (<1 µE) to avoid light stress (and associated production of reactive oxygen species) during desiccation. Desiccation was monitored by recording the diminishing size of the algal dots using a very sensitive astronomy camera (Acton PI 1 kb Versarray Cooled Camera, Princeton Instruments, with WinView/32 software). Full evaporation of culturing medium (the “Dry” stage) was achieved after 12–13 h (SI Appendix, Fig. S2 E and G). Dry algal dots remained in the drying chamber 11 additional hours until being rehydrated with 40 μL of sterile distilled water (∼24 h after the beginning of the experiment)
Time-Course Sampling, RNA Extraction, Library Prep, and Sequencing.
For each species and replicate, five timepoints of the desiccation and rehydration time course were used in RNA-seq experiments (SI Appendix, Fig. S2H). “Early” and “Late” desiccation stages corresponded to a volume loss of 25% and 60%, respectively, and were collected 2.5 and 7.5 h into the desiccation time course. The Dry stage corresponded to fully flattened dots (SI Appendix, Fig. S2E). Dry algal dots remained in the drying chamber 11 additional hours until collection (∼24 h after the beginning of the experiment). Finally, the “Rehydrated” stage corresponded to algal dots collected 1 h after rehydration. These dots retained at least 90% of the initial rehydration volume (SI Appendix, Fig. S2G).
Total RNA was extracted from ground algal dots using ZR Plant RNA MiniPrep (Zymo) followed by RNA Clean & Concentrator-5 (Zymo). Quantity, purity, and integrity of each sample were determined using Qubit RNA BR Assay Kit for Qubit 2 (ThermoFisher Scientific), NanoDrop microvolume spectrophotometer (ThermoFisher Scientific), and Agilent RNA 6000 Pico Kit in an Agilent 2100 bioanalyzer (Agilent Technologies). RNA samples had A260/A280 ratios ∼2 and RNA integrity number (RIN) in excess of 8.
Strand-specific libraries were prepared from total RNA using the Ovation Arabidopsis RNA-seq Systems approach (NuGEN) following manufacturer instructions. This approach enriches for messenger RNA in the sample without using polyA selection that could affect the recovery of organellar mRNA (33). Nuclear, chloroplast, mitochondrial, and bacterial rRNA were removed during the InDA-C adaptor cleavage step using a combination of commercially available oligos targeting prokaryote and green plant rRNA genes (Arabidopsis kit; NuGEN S02070, S02076, R01758, F01278) supplemented with 105 custom-made oligos designed from rRNA sequence data from these algae within the Scenedesmaceae including GenBank sequences AY510465.1, KJ680140.1, KC145438.1, and HQ246446.1. Six libraries per lane were pooled in equimolar concentrations for paired-end multiplexed sequencing (2 × 150 nt) using Illumina Nextseq 500 (Illumina) at the W. M. Keck Ecological and Evolutionary Genetics Facility (Marine Biological Laboratory).
De Novo Transcriptome Assembly and Annotation.
For each species, pooled filtered reads were used for de novo assembly of transcriptomes using Trinity v2.1 (34) (Jaccard clip option, strand-specific, k-mer length of 25). Assemblies were refined (see SI Appendix, SI Material and Methods for specifics), and reference transcriptomes were annotated using BLAST homologies captured against Uniprot (35) and nr NCBI (36) databases (evalue <1e−5). Protein sequences were predicted with TransDecoder v. 2.0.1 (transdecoder.github.io/). PFAM domains were identified with HMMER (37), signal peptides with SignalP v. 4.1 (38), transmembrane regions with TMHMM v. 2.0 (39). All annotations were integrated following Trinotate v 3.0.1 pipeline (github.com/Trinotate/Trinotate.github.io/wiki). Functional annotations included KEGG (40), GO (41), eggnog (42), and InterPro (43).
Differential Expression Analysis.
Libraries were individually aligned to their respective reference transcriptomes using Bowtie 2 v.2.2 (44). Transcript abundance was estimated using RSEM v.1.2.28 (45). Differential expression analysis was performed on count data generated with RSEM (45) using DESeq2 (46). A gene was classified as differentially expressed if the change in its expression over the time course exceeded fourfold (Log2 2-FC) in pairwise comparisons with a significance level <0.001 (adjusted P value using Benjamini–Hochberg (BH) correction for multiple testing (46). DEGs were organized by similarity from Euclidean distance matrices and grouped with hclust (complete-linkage method). An automatic partitioning of similarity of dendrograms (60% maximum height) was used to identify clusters of coexpressed DEGs along the desiccation/rehydration time course. For each cluster, we plotted the expression data of all individual genes within the cluster as centered to the mean Log2 (fpkm +1) (fragments per kilobase million). The complete list of the DEGs in each group is provided in Dataset S1. Expression data of genes of interest are presented in box plots (Fig. 3) generated with SigmaPlot10 (Systat Software).
GO and Transcription Factors.
GO term assignments were extracted from the annotated reference transcriptome for each species. GO term enrichment tests were performed using GOseq (47). A GO term was considered enriched if its significance level was <0.05 (adjusted P value using BH correction for multiple testing (47).
We conducted two complementary GO term enrichment analyses. First, we explored the response to desiccation by identifying those functions enriched in DEGs with overall up-regulation (UpM and UpH groups in Fig. 1A) and down-regulation (DWN group) during the time course. Second, we compared the specific response functions that were identified in common across all three taxa as affected by desiccation. Results were visualized in scatterplots generated with REVIGO (48) (Fig. 1C). For each species, we conducted GO term enrichment tests using GOseq (47) on total DEGs identified in Late and Dry stages. GO term lists were reduced with REVIGO (48). Overlapping terms across species were identified using Venn diagrams. We used the GOplot v.1.0.1 (49) package run in R v.3.1 to visualize the overall direction of response of each GO term, quantified as its z score. A negative z score indicates a down-regulated function, and a positive z score indicates up-regulation (49). Results are presented in polar graphs generated using the circ command in GOplot (49). Transcription factors, transcription regulators, and protein kinases were identified using the software package iTAK v.1.7 (50) (database 17.09, 167 genomes including multiple Chlorophyte algae) (Dataset S6).
Data Availability.
Algal isolates used in this study are deposited at the George Safford Torrey Herbarium, University of Connecticut. (A. deserticola, CONN00226458; F. rotunda, CONN00181061) or at the Culture Collection of Algae and Protozoa, Argyll, Scotland (E. costatus, isolate CCAP276-31). Culturing techniques, methods, analysis strategies, and results are discussed in detail in SI Appendix and Datasets S1–S7. Custom InDA-C primers are available from the corresponding author upon request. Raw sequencing data are available at the NCBI’s SRA database, de novo transcriptome assemblies at the Transcriptome Shotgun Assembly (TSA) database, and expression data at GEO database under the SuperSeries record GSE133354. A. deserticola, PRJNA529464. SRA accessions SRR8794168–SRR8794177, GEO (GSE133353), TSA (GHRQ00000000). F. rotunda, PRJNA529457. SRASRR8793708–SRR8793717, GEO (GSE133352), TSA (GHRR00000000). E. costatus, PRJNA529437. SRA accessions SRR8793529–SRR8793538, GEO (GSE133350), TSA (GHUV00000000).
Supplementary Material
Acknowledgments
Dr. Louise Lewis (University of Connecticut) provided F. rotunda and A. deserticola. Suzanne Thomas and Jordan Stark provided expert technical assistance. This work was supported by the NSF, Division of Integrative Organismal Systems (1355085 to Z.G.C.), and an anonymous donor (to Z.G.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data are available at the National Center for Biotechnology’s (NCBI) Sequence Read Archive (SRA) database, de novo transcriptome assemblies at the Transcriptome Shotgun Assembly (TSA) database, and expression data at Gene Expression Omnibus (GEO) database under the SuperSeries record GSE133354. Acutodesmus deserticola, PRJNA529464. SRA accessions SRR8794168–SRR8794177, GEO (GSE133353), TSA (GHRQ00000000). Flechtneria rotunda, PRJNA529457. SRASRR8793708–SRR8793717, GEO (GSE133352), TSA (GHRR00000000). Enallax costatus, PRJNA529437. SRA accessions SRR8793529–SRR8793538, GEO (GSE133350), TSA (GHUV00000000).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906904117/-/DCSupplemental.
References
- 1.Alpert P., Constraints of tolerance: Why are desiccation-tolerant organisms so small or rare? J. Exp. Biol. 209, 1575–1584 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Giarola V., Hou Q., Bartels D., Angiosperm plant desiccation tolerance: Hints from transcriptomics and genome sequencing. Trends Plant Sci. 22, 705–717 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Farrant J. M., Moore J. P., Programming desiccation-tolerance: From plants to seeds to resurrection plants. Curr. Opin. Plant Biol. 14, 340–345 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Lewis L. A., McCourt R. M., Green algae and the origin of land plants. Am. J. Bot. 91, 1535–1556 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Cardon Z. G., Gray D. W., Lewis L. A., The green algal underground: Evolutionary secrets of desert cells. Bioscience 58, 114–122 (2008). [Google Scholar]
- 6.Cardon Z. G., Peredo E. L., Dohnalkova A. C., Gershone H. L., Bezanilla M., A model suite of green algae within the Scenedesmaceae for investigating contrasting desiccation tolerance and morphology. J. Cell Sci. 131, jcs212233 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Gray D. W., Lewis L. A., Cardon Z. G., Photosynthetic recovery following desiccation of desert green algae (Chlorophyta) and their aquatic relatives. Plant Cell Environ. 30, 1240–1255 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Oliver M. J., Tuba Z., Mishler B. D., The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 151, 85–100 (2000). [Google Scholar]
- 9.VanBuren R. et al., Seed desiccation mechanisms co-opted for vegetative desiccation in the resurrection grass Oropetium thomaeum. Plant Cell Environ. 40, 2292–2306 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Proctor M. C. F. et al., Desiccation-tolerance in bryophytes : A review. Bryologist 110, 595–621 (2007). [Google Scholar]
- 11.Proctor M. C. F., Smirnoff N., Rapid recovery of photosystems on rewetting desiccation-tolerant mosses: Chlorophyll fluorescence and inhibitor experiments. J. Exp. Bot. 51, 1695–1704 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Rajeev L. et al., Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J. 7, 2178–2191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaff D. F., Oliver M., The evolution of desiccation tolerance in angiosperm plants: A rare yet common phenomenon. Funct. Plant Biol. 40, 315–328 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Holzinger A. et al., Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS One 9, e110630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rippin M., Becker B., Holzinger A., Enhanced desiccation tolerance in mature cultures of the streptophytic green alga Zygnema circumcarinatum revealed by transcriptomics. Plant Cell Physiol. 58, 2067–2084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore J. P., Le N. T., Brandt W. F., Driouich A., Farrant J. M., Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci. 14, 110–117 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Giarola V., Jung N. U., Singh A., Satpathy P., Bartels D., Analysis of pcC13-62 promoters predicts a link between cis-element variations and desiccation tolerance in Linderniaceae. J. Exp. Bot. 69, 3773–3784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanBuren R. et al., Desiccation tolerance evolved through gene duplication and network rewiring in Lindernia. Plant Cell 30, 2943–2958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis L. A., Flechtner V. R., Cryptic species of Scenedesmus (Chlorophyta) from desert soil communities of western North America. J. Phycol. 40, 1127–1137 (2004). [Google Scholar]
- 20.Gechev T. S. et al., Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cell. Mol. Life Sci. 70, 689–709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez M. C. S. et al., Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 63, 212–228 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Xu Z. et al., Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Mol. Plant 11, 983–994 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Qiu F., Bachle S., Nippert J. B., Ungerer M. C., Comparing control options for time-series RNA sequencing experiments in nonmodel organisms: An example from grasses. Mol. Ecol. Resour. 20, 681–691 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Yobi A. et al., Sporobolus stapfianus: Insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol. 17, 67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alamillo J., Almoguera C., Bartels D., Jordano J., Constitutive expression of small heat shock proteins in vegetative tissues of the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 29, 1093–1099 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Oliver M. J., Dowd S. E., Zaragoza J., Mauget S. A., Payton P. R., The rehydration transcriptome of the desiccation-tolerant bryophyte Tortula ruralis: Transcript classification and analysis. BMC Genomics 5, 89 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campos F., Cuevas-Velazquez C., Fares M. A., Reyes J. L., Covarrubias A. A., Group 1 LEA proteins, an ancestral plant protein group, are also present in other eukaryotes, and in the archeae and bacteria domains. Mol. Genet. Genomics 288, 503–517 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Alamillo J. M., Bartels D., Effects of desiccation on photosynthesis pigments and the ELIP-like dsp 22 protein complexes in the resurrection plant Craterostigma plantagineum. Plant Sci. 160, 1161–1170 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Armbruster U. et al., Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25, 2661–2678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puthiyaveetil S. et al., Compartmentalization of the protein repair machinery in photosynthetic membranes. Proc. Natl. Acad. Sci. U.S.A. 111, 15839–15844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemme D. et al., Systems-wide analysis of acclimation responses to long-term heat stress and recovery in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26, 4270–4297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S. et al., Thylakoid membrane architecture in Synechocystis depends on curT, a homolog of the granal CURVATURE THYLAKOID1 proteins. Plant Cell 28, 2238–2260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone J. D., Storchova H., The application of RNA-seq to the comprehensive analysis of plant mitochondrial transcriptomes. Mol. Genet. Genomics 290, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Haas B. J. et al., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The UniProt Consortium , UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W., GenBank. Nucleic Acids Res. 44, D67–D72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn R. D., Clements J., Eddy S. R., HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen T. N., Brunak S., von Heijne G., Nielsen H., SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M., KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashburner M. et al.; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell S. et al., eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter S. et al., InterPro: The integrative protein signature database. Nucleic Acids Res. 37, D211–D215 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young M. D., Wakefield M. J., Smyth G. K., goseq: Gene Ontology testing for RNA-seq datasets reading data. Genome Bio. 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter W., Sánchez-Cabo F., Ricote M., GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y. et al., iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Algal isolates used in this study are deposited at the George Safford Torrey Herbarium, University of Connecticut. (A. deserticola, CONN00226458; F. rotunda, CONN00181061) or at the Culture Collection of Algae and Protozoa, Argyll, Scotland (E. costatus, isolate CCAP276-31). Culturing techniques, methods, analysis strategies, and results are discussed in detail in SI Appendix and Datasets S1–S7. Custom InDA-C primers are available from the corresponding author upon request. Raw sequencing data are available at the NCBI’s SRA database, de novo transcriptome assemblies at the Transcriptome Shotgun Assembly (TSA) database, and expression data at GEO database under the SuperSeries record GSE133354. A. deserticola, PRJNA529464. SRA accessions SRR8794168–SRR8794177, GEO (GSE133353), TSA (GHRQ00000000). F. rotunda, PRJNA529457. SRASRR8793708–SRR8793717, GEO (GSE133352), TSA (GHRR00000000). E. costatus, PRJNA529437. SRA accessions SRR8793529–SRR8793538, GEO (GSE133350), TSA (GHUV00000000).