Significance
The human brain is organized into large networks. One important brain network is the Default network, which enables cognitive functions such as social thinking, memory, and reward. In group-averaged data, this network emerges as a unitary whole, despite its involvement in multiple cognitive functions. Here, we tested whether Default networks found in individual humans, rather than group-average networks, contain organized substructure. In individuals, we consistently found nine subnetworks within the Default network. These subnetworks matched brain activity patterns during cognitive tasks. Some subnetworks resembled brain circuits involved in specific Default functions. Others linked Default network to other large networks. In summary, this study describes a set of brain circuits within the Default networks of individual humans.
Keywords: fMRI, functional connectivity, individual variability, brain networks, Default network
Abstract
The human brain is organized into large-scale networks identifiable using resting-state functional connectivity (RSFC). These functional networks correspond with broad cognitive domains; for example, the Default-mode network (DMN) is engaged during internally oriented cognition. However, functional networks may contain hierarchical substructures corresponding with more specific cognitive functions. Here, we used individual-specific precision RSFC to test whether network substructures could be identified in 10 healthy human brains. Across all subjects and networks, individualized network subdivisions were more valid—more internally homogeneous and better matching spatial patterns of task activation—than canonical networks. These measures of validity were maximized at a hierarchical scale that contained ∼83 subnetworks across the brain. At this scale, nine DMN subnetworks exhibited topographical similarity across subjects, suggesting that this approach identifies homologous neurobiological circuits across individuals. Some DMN subnetworks matched known features of brain organization corresponding with cognitive functions. Other subnetworks represented separate streams by which DMN couples with other canonical large-scale networks, including language and control networks. Together, this work provides a detailed organizational framework for studying the DMN in individual humans.
The human brain is organized into spatially distributed networks that can be described in vivo using a functional MRI (fMRI)-based technique known as resting-state functional connectivity (RSFC). Findings from more than two decades of RSFC studies have converged on the existence of a set of large-scale brain networks that are identifiable and reproducible across populations, datasets, and analysis techniques (1–8). These include both sensory/motor networks and “higher-level cognitive” networks. The identification of these networks as an organizing principle of brain function has proven to be an indispensable framework for studies mapping cognition to brain function (6, 9).
One of the first to be identified and most studied brain networks is the Default-mode network (DMN) (10–12), which comprises regions in the bilateral medial parietal cortex, medial and superior prefrontal cortex, angular gyrus, medial and lateral temporal lobe, and cerebellum. While reliably identifiable as a single network using RSFC (13), this network paradoxically is associated with a variety of cognitive functions using task-based fMRI. Engagement of the DMN is most commonly associated with autobiographical memory and other types of internally oriented cognition (10, 14). However, various separable portions of the network have also been more strongly associated with processing contextual information (15), regulation of fear and anxiety (16), reward processing and reward-based decision making (17), social processing (18), and even task-oriented cognitive control (19–21).
Beyond task engagement, the DMN also exhibits substantial heterogeneity in other properties. For example, some DMN regions represent central, integrative elements of the overall system (22), while others represent connector hubs—pathways by which DMN processes communicate with other, non-DMN networks (23, 24). Further, signals within DMN exhibit heterogeneous temporal ordering, with some DMN regions consistently leading others by as much as 500 ms (25), suggesting differing functional roles within the network.
One explanation for the apparent functional heterogeneity of the DMN is that there may be discrete subnetwork structures within this network (26). This idea converges with work suggesting that brain network organization is fundamentally hierarchical and multiscale in nature (27). Specifically, while RSFC studies classically identify a common set of around 7 to 17 networks (5, 6, 8), other work suggests that these canonical networks can be divided into discrete, distributed subnetworks with differentiable structural connectivity, functional connectivity, engagement during task performance, and variability across subjects (22, 23, 28–32). Similarly, tract-tracing studies in animals identified subregions within the DMN that demonstrate overlapping but differentiable connectivity patterns (33–35). Thus, the canonical networks, while identifiable and consistent in most populations, are only one level of description of the brain’s complex, multiscale network structure. Describing DMN organization at other scales—and particularly the subnetwork structure present at finer scales—could improve our fundamental understanding of brain organization and its relationship to cognition.
Several previous works have used group-averaged RSFC data to analyze the subnetwork organization of the whole brain (36–38) or of the DMN in particular (22, 39–41). However, these works have not consistently identified DMN subdivisions that conform with patterns of activity evoked by cognitive functions. This may be because these examinations were fundamentally limited by their use of group-average data. RSFC-derived brain networks are spatially variable in their locations in individual human brains, even after optimized intersubject registration (42–45). This functional variability relative to anatomical features means that averaging RSFC data across individuals at each brain location impairs identification of detailed brain organizational features (46–48). This is particularly true for features that are physically small relative to the scale of interindividual spatial variation, as features with a small spatial extent will tend to overlap worse across subjects than features with a large spatial extent (49). Thus, group-average approaches that identify large-scale brain networks may be unable to accurately identify smaller substructures within those larger networks. Instead, the subnetwork organization of the brain is better served by examination at the level of the individual human (26).
Recent work has developed optimized approaches for studying individual human brains using RSFC (2, 50–53). Critically, this individualized approach has been extended to identify subnetworks within the DMN that are consistently present (although spatially variable) across individuals (26, 54, 55). The presence of DMN subnetworks that are consistent across individuals, but not detectable in group data, suggests the existence of important organizational principles at heretofore unexplored network scales. However, this recent work considered only two DMN subnetworks; these subnetworks dissociated episodic from social cognition, but they did not correspond to sets of brain regions that have been associated with other DMN cognitive functions (e.g., reward, anxiety processing) or to other types of DMN heterogeneity (cross-network hubs, heterogeneous temporal ordering). It is possible that more comprehensive network-function correspondence may require delineation of further subdivided individual-specific brain subnetworks, although the extent of subdivision required is unclear.
We used the individual-focused Midnight Scan Club (MSC) dataset (2) to explore and evaluate subnetworks of individual-specific brain networks at multiple scales. In evaluating these subnetworks, we reasoned that accurately described network structures should both reflect actual divisions within RSFC data (8, 56) and converge with patterns of coactivation during task performance (2, 6, 44, 55, 57). These two measures can serve as approximations of the internal and external validity of identified subnetworks, respectively. We employed these metrics to first determine whether examining brain subnetwork structure below the level of the canonical networks was warranted and then to understand whether there were specific hierarchical levels that most optimally described subnetwork structure.
While we delineated optimized subnetworks across the entire brain, in this paper, we focus on the DMN. We identified subnetworks within the canonical DMN that were consistently present across subjects. Just as large-scale networks are spatially variable but consistently identifiable across individuals based on their general spatial distributions (2, 43, 45, 54, 58), DMN subnetworks may exhibit some degree of spatial variability but should have broadly consistent spatial topographies across individuals. Further, these consistently present subnetworks should conform with previous focal localizations of cognitive function within the DMN. Finally, we determined whether specific DMN subnetworks might 1) serve as sets of connector hubs by linking to other canonical networks and 2) demonstrate heterogeneous temporal ordering, as evidenced by differential lead–lag relationships among blood oxygen level-dependent (BOLD) signals in different subnetworks.
Results
Individual-Specific Brain Networks Identified at Multiple Scales.
We described the network substructure of individual human brains using the data-driven Infomap community detection algorithm (59). In the classic Infomap approach, a matrix representing RSFC strength between every pair of brain locations (nodes) is thresholded to form a sparse graph containing a specific density of connections (edges), and networks are then identified using multiple density thresholds. In this approach, sparser density thresholds produce more subdivided networks. However, at very sparse edge densities, brain regions with weaker RSFC are disconnected, leaving large sections of the brain unassigned to any network (5) and limiting the potential depth of brain network subdivision.
We modified this approach to employ node-specific thresholding, allowing us to evaluate network structure at any edge density with no disconnections. Specifically, density thresholds were applied separately to the connections of each point in the brain, thus retaining (at least) the strongest X% of connections to each point. This allowed every point in the brain to have at least some connections and thus be assigned to its most connected brain network, no matter how sparsely the graph was thresholded (here, we used 0.01% ≤ X ≤ 5%). This location-specific thresholding approach shares the advantages of established “winner-take-all” methods of brain network detection (50, 52, 60–62) but without the need to impose previously identified priors on the data (which would be problematic, as subnetwork structures have not been characterized).
At the densest level tested (5% density), subjects averaged 7.7 ± 0.6 (mean ± SD) identified networks across the brain, and network structures (see example in SI Appendix, Fig. S1) broadly appeared similar to a widely accepted seven-network organization (8). At sparser levels (2 and 1% density), subjects averaged 13.1 ± 1.4 and 18.1 ± 1.6 networks, respectively, and network structures appeared similar to previous network characterizations containing 14 to 17 networks (5, 8). As densities became progressively sparser, numbers of networks increased beyond previous reports, ranging from 27.4 ± 2.2 networks at 0.5% density, to 82.5 ± 4.5 networks at 0.1% density, to 434.3 ± 20.0 networks at 0.01% density.
Internal and External Validity Metrics Are Maximized with ∼83 Subnetworks.
To determine whether these increasingly subdivided subnetworks represent true network substructure, we evaluated the internal and external validity of subnetworks identified at each density threshold, in each MSC subject. Internal validity was operationalized as subnetwork homogeneity (56). External validity was operationalized as the percentage of variance across vertices in a task-activation map that could be explained by subnetwork identities of those vertices (“task R2”), computed using a spatial ANOVA. This approach is similar in principle to the established technique of computing the similarity of two brain maps via spatial correlation. The ANOVA was calculated for each of three representative task-activation contrasts: 1) the “word stimuli > fixation” contrast in the semantic task; 2) the “hand > tongue” contrast in the motor task; and 3) the “scene > face” contrast in the implicit memory task.
Due to the spatial autocorrelation of BOLD data, these homogeneity and task R2 measures necessarily increase as tested brain regions become more subdivided, regardless of the validity of the regions. Hence, these measures of validity must be contrasted against an appropriate null model. Following ref. 56, we employed a rotation-based null model, in which a subject’s subnetworks were iteratively rotated a random amount around the spherical expansion of the cortical surface, and both homogeneity and task R2 were recalculated for each random iteration. The individual specificity of the identified brain subnetworks was further evaluated using each MSC subject’s RSFC and task data to calculate the homogeneity and task R2 of the other subjects’ brain subnetworks.
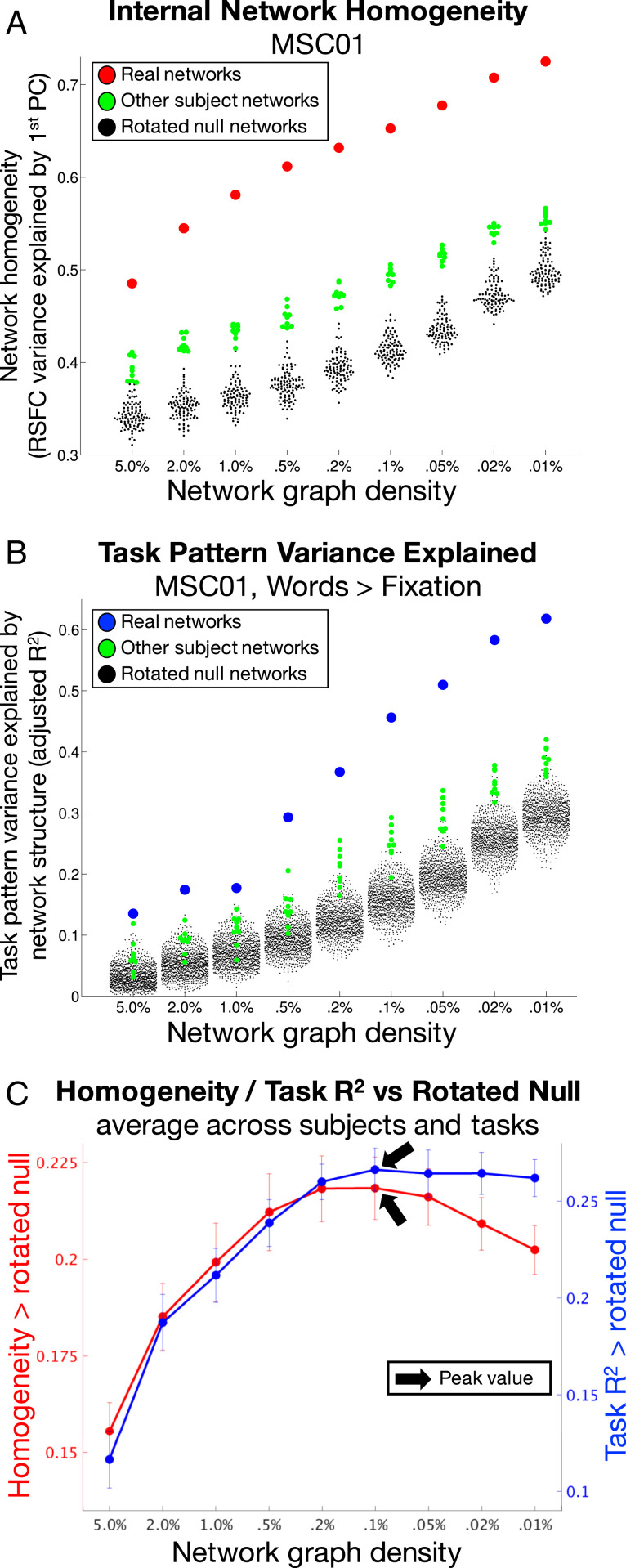
As expected, subnetwork homogeneity and task R2 increased in every subject as networks were increasingly subdivided. Fig. 1A (homogeneity, red dots) and 1B (task R2, blue dots) illustrate these effects for subject MSC01 for a single task contrast (Fig. 1B); see SI Appendix, Fig. S2 for all subjects and tasks. Importantly, in every subject and task, subnetworks at density levels of 0.1% and sparser exhibited greater homogeneity and task R2 than any rotated null iteration (Fig. 1 A and B, black dots) or any other subject’s subnetworks (green dots). This indicates that finely divided networks/subnetworks represent internally valid, externally valid, and individually specific descriptions of brain network organization.
Fig. 1.
Internal and external validity of subnetworks are maximized at 0.1% graph density. (A) RSFC homogeneity (y axis) within brain networks (red) was higher than within rotated networks (black) or other subjects’ networks (green), across all network densities (x axis). This is illustrated for subject MSC01. (B) The task-activation variance explained (y axis) by brain networks (blue) was consistently higher than by rotated networks (black) or other subjects’ networks (green), across all network densities (x axis). This is illustrated for the Words > Fixation task in subject MSC01. (C) When averaged across subjects, internal validity (homogeneity compared to null; red) and external validity (task variance explained compared to null, averaged across tasks; blue) peaked at 0.1% network density (black arrows). Error bars represent SEs across subjects. See SI Appendix, Fig. S2 for all subjects and all tasks.
We then compared homogeneity and task R2 from real subnetworks against the medians computed from rotated null descriptions. These differences represent null-corrected measures of validity. Across subjects, the average null-corrected homogeneity (Fig. 1C, red) was 0.16 at density levels of 5%, and increased with increasing network sparsity up to a maximum of 0.22 at 0.1% (black arrow). For networks sparser than 0.1%, homogeneity progressively decreased, down to 0.20 at 0.01%.
Similarly, we found that the average null-corrected task R2 across task contrasts and subjects (Fig. 1C, blue) was 0.12 at 5% density and increased with increasing network sparsity up to a maximum of 0.27 at 0.1% density (black arrow). For networks sparser than 0.1%, task R2 exhibited progressive decreases.
These results thus specify an optimal scale—a density of 0.1%, with an average of 82.5 network divisions—at which internal homogeneity and explanatory power of task activations are conjointly maximized.
DMN Subnetworks Exhibit Consistent Spatial Distributions across Subjects.
Neurobiologically meaningful subnetwork structures should exhibit some consistency in organization across individuals. Accordingly, we identified subnetworks within the large-scale DMN that could be matched across subjects based on spatial similarity. Since no priors were available, this identification was conducted similarly to procedures used by (54, 63), in which results from the automated network identification were visually compared across subjects for correspondence. The primary features used for correspondence included both the location of subnetwork regions on the anatomical structure of the brain (subnetwork topographies), as well as the location of regions relative to other subnetworks (subnetwork topologies). See Methods for details.
For each subnetwork that could be matched across subjects, we determined, for each subject, whether the subnetwork “existed” (i.e., it was identified at all), whether it was “complete” (i.e., was represented in all relevant brain structures), and whether it was “unitary” (i.e., existed as a single, spatially contiguous subnetwork, rather than being split into multiple pieces or merged with other subnetworks) (Table 1).
Table 1.
Percentage of DMN subnetworks across subjects that existed, were unitary (neither split nor merged with other subnetworks), and were complete (represented in all relevant brain structures)
| Exist | Unitary | Complete | Notes | |
| Parietal | 100% | 50% | 90% | Sometimes split into multiple subnetworks |
| Ventromedial | 100% | 70% | 70% | Merged with Pregenual in two subjects |
| Pregenual | 100% | 80% | 80% | Merged with Ventromedial in two subjects |
| Retrosplenial | 100% | 90% | 90% | |
| Posterior MTL | 100% | 100% | 90% | |
| Left Anterior Lateral | 100% | 80% | 90% | Merged with Right Anterior Lateral in one subject |
| Left Dorsal Lateral | 100% | 100% | 100% | |
| Right Anterior Lateral | 100% | 90% | 100% | Merged with Left Anterior Lateral in one subject |
| Right Dorsal Lateral | 100% | 100% | 100% |
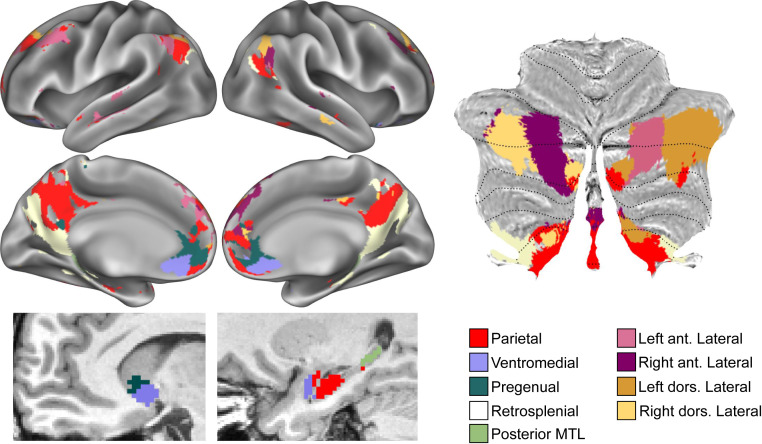
We observed nine separate subnetworks within the large-scale DMN that were consistent across subjects. See Fig. 2 for an example; see SI Appendix, Fig. S3 for all subjects.
Fig. 2.
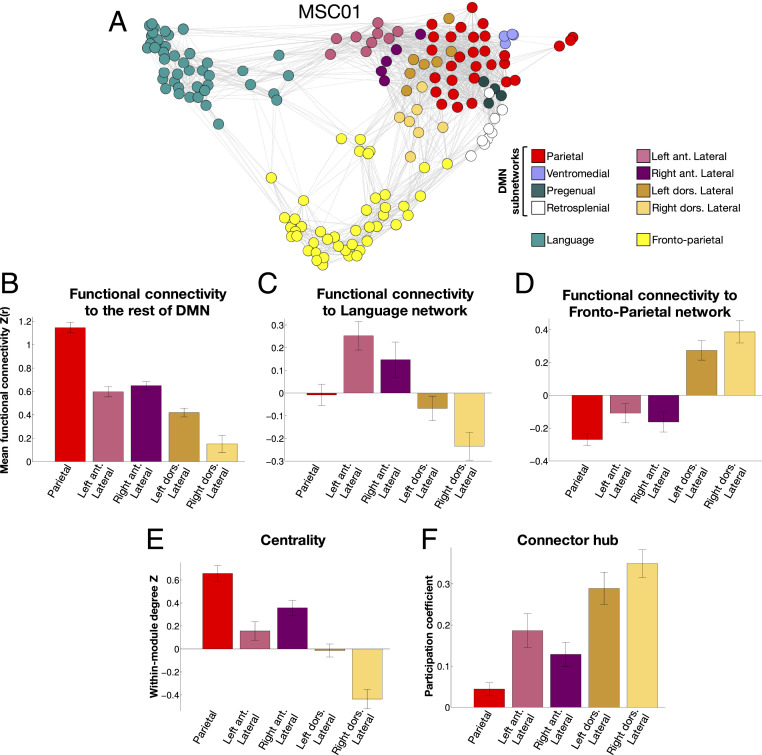
DMN subnetworks in subject MSC01. Different colors represent different subnetworks in cortex (Top Left), ventral caudate and MTL (Bottom Left), and cerebellum (Right). ant., anterior; dors., dorsal. See SI Appendix, Fig. S3 for all subjects.
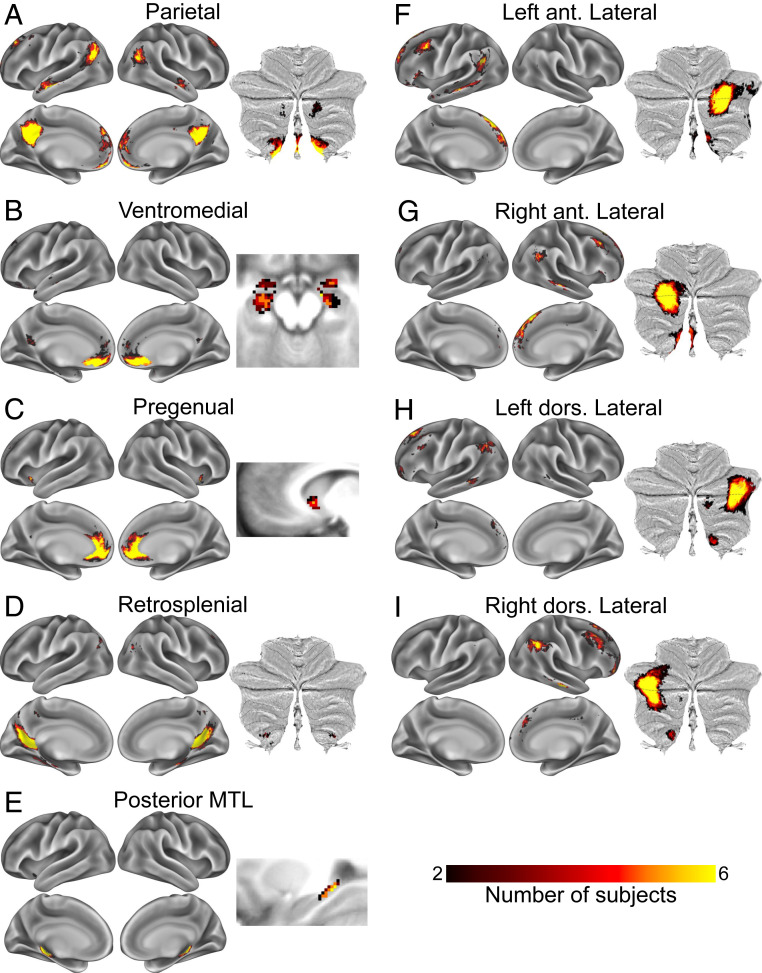
A Parietal subnetwork (Fig. 2, red; Fig. 3A) had relatively large representation in medial parietal cortex, moderately sized representation in bilateral middle angular gyrus, and small but consistent representation bilaterally in anterior ventromedial prefrontal cortex, anterior medial superior frontal gyrus, middle lateral superior frontal gyrus, anterior superior temporal sulcus, and ventral posterior cerebellar lobe and vermis. In some subjects, this subnetwork was split into multiple pieces, with the medial parietal node fragmenting. Note that for visual clarity, this fragmentation is not reflected in the figures.
Fig. 3.
(A–I) Spatial distributions of DMN subnetworks across subjects. Color indicates number of subjects with spatial overlap of matched subnetworks at each point in the brain. ant., anterior; dors., dorsal.
A Ventromedial subnetwork was observed in the ventromedial prefrontal cortex and amygdala and/or anterior hippocampus (Fig. 2, light blue; Fig. 3B). This subnetwork occasionally merged with the Pregenual network (below).
A Pregenual subnetwork was observed in the pregenual medial prefrontal cortex, anterior medial ventral caudate (usually), and ventral anterior insula (usually) (Fig. 2, dark teal; Fig. 3C). Note that while we include this as a subnetwork of the canonical DMN based on its anatomical location, and we believe that it represents the same object across individuals based on its anatomical consistency, in seven subjects, this subnetwork was a subdivision of the Salience network, not the DMN (as previously described in ref. 2).
A Retrosplenial subnetwork had representation in the retrosplenial cortex and the parietooccipital fissure, parahippocampal cortex, and occasionally posterior angular gyrus (Fig. 2, white; Fig. 3D). Note that in previous work, this network classically dissociates from DMN at ∼14 to 17 networks but not at 7 networks (5, 8).
A Posterior medial temporal lobe (MTL) subnetwork had representation in the bilateral posterior medial temporal cortex and caudal hippocampus (Fig. 2, light green; Fig. 3E).
Four lateralized subnetworks, termed the Left and Right Anterior and Posterior Lateral subnetworks, were observed in every subject. Overall, these four subnetworks were spatially variable in their cortical representation but had large representation straddling the horizontal fissure of the cerebellum and exhibited substantial topological consistency relative to each other and to the Parietal subnetwork. The first two (Fig. 2, magenta and purple; Fig. 3 F and G) were found in the left and right (respectively) anterior angular gyrus, middle frontal gyrus, anterior superior frontal gyrus, anterior middle temporal gyrus, and contralateral medial posterior cerebellum. These two lateralized subnetworks were merged in one subject. The other two Lateral subnetworks (Fig. 2, orange and yellow; Fig. 3 H and I) were represented more dorsally than the previous two subnetworks, being found in the left and right dorsal angular gyrus, posterior lateral superior frontal gyrus, middle frontal gyrus, and contralateral lateral posterior cerebellum.
Finally, to test the reliability and individual specificity of these subnetworks, we delineated the large-scale DMN (at 5% density) and DMN subnetworks (at 0.1% density) in separate split halves of each subject’s data (first five sessions vs. last five sessions). Within subjects, the DMN and DMN subnetworks followed similar spatial patterns between halves (SI Appendix, Fig. S4 A and B), as Dice coefficients of spatial overlap between split halves averaged 0.83 ± 0.07 for networks and 0.69 ± 0.10 for subnetworks. Dice overlaps were lower when comparing split-half networks/subnetworks across different subjects (whole DMN: Dice = 0.62 ± 0.02 [mean ± SD]; DMN subnetworks: Dice = 0.28 ± 0.02 [mean ± SD]). Notably, the differences between within-subject and cross-subject Dice overlaps were greater for subnetworks than for whole DMNs [paired t (9) = 8.19; P < 10−4], indicating that DMN subnetworks are more specific to each individual than the large-scale DMN.
DMN Subnetworks Exhibit Differential Task Engagement.
Fig. 1 indicates that subnetworks identified at 0.1% density better explain task activations than do networks identified using more typical, denser thresholds that yield canonical large-scale networks. A likely explanation for this phenomenon is that the tasks may differentially activate specific substructures within large-scale networks.
We tested this possibility by determining whether subnetworks identified at 0.1% density explained variance in task activation within the large-scale DMN identified at 5% density. See SI Appendix, Fig. S5 for relevant large-scale networks in each subject.
For each MSC subject, in each of the three task contrasts described above (word > fixation, hand > tongue, and scene > face), we selected all vertices within the 5% density-defined DMN. We then performed a spatial ANOVA to determine whether task activation was better explained by the subnetworks (0.1% density-defined) within that large-scale network than by rotated versions of the subnetworks. False discovery rate (FDR) correction for multiple comparisons across subjects and tasks was applied to a level of q < 0.05, which was found to be equivalent to P < 0.02.
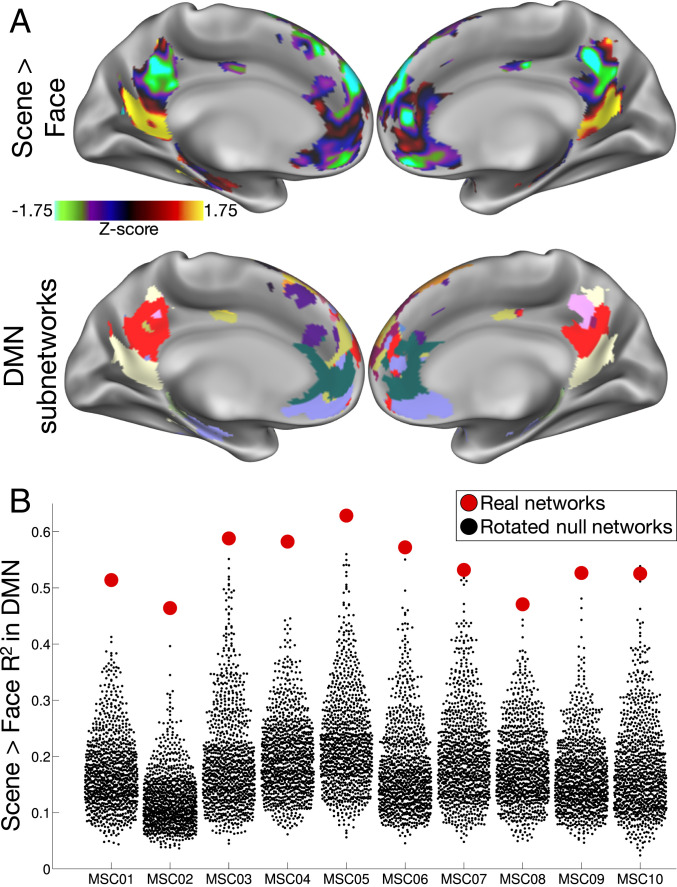
In every subject, we found that the scene > face contrast elicited strong positive activation in subnetworks within inferior medial parietal/retrosplenial and inferior temporal cortex, and strong negative activations in subnetworks in medial parietal and ventromedial prefrontal cortex (see Fig. 4A for an example subject). The spatial variance of these activation maps was significantly explained (i.e., to a greater extent than the null models) by DMN subnetworks in all subjects (mean ± SD R2 = 0.54 ± 0.05; all q values vs. rotated null networks: <0.05; P values ≤ 0.001; Fig. 4B). Activation driven by the words > fixation contrast similarly corresponded with DMN subnetworks in all subjects (mean ± SD R2 = 0.39 ± 0.05; all q values < 0.05), which was driven by strong deactivation within medial parietal/angular gyrus subnetworks and activation within lateral temporal and frontal subnetworks. However, activation patterns driven by the hand > tongue contrast did not vary within DMN subnetworks (all q values: not significant).
Fig. 4.
Subnetworks represent differentially task-activated divisions within large-scale networks. (A) Task heterogeneity within the large-scale DMN in example subject MSC01. Patterns of task activation (Top) driven by a Scene > Face contrast correspond very well with DMN subnetwork divisions (Bottom). (B) Variance in the pattern of Scene > Face task activation (y axis) explained by DMN subnetwork divisions (red) and by rotated DMN subnetworks (black), for each subject (x axis).
Supplemental analyses examined whether subnetworks similarly explained variance in task activation within other large-scale networks. The scene > face contrast was inconsistently explained by subnetwork divisions of the other large-scale networks, the most consistent being the Frontoparietal network (mean ± SD R2 = 0.32 ± 0.11; q values < 0.05 for six subjects). Activation driven by the hand > tongue contrast corresponded with subnetworks within the large-scale Somatomotor network in all subjects (mean ± SD R2 = 0.66 ± 0.04; all q values < 0.05; all P values ≤ 0.001; SI Appendix, Fig. S6 A and B) but not with subnetworks in any other large-scale network (no other network in which q < 0.05 for more than one subject). Activation driven by the words > fixation contrast corresponded in all subjects with subnetworks within the large-scale Frontoparietal network (mean ± SD R2 = 0.43 ± 0.06; all q values < 0.05; all P values ≤ 0.001; SI Appendix, Fig. S6 C and D) but corresponded with subnetworks within other large-scale networks only inconsistently, with q values < 0.05 for half of subjects or less.
Together, these results indicate that RSFC-derived subnetwork structures correspond well to task-activated subregions within DMN and other canonical large-scale networks.
Parietal Subnetwork Is Central within DMN, while Lateral Subnetworks Are Connector Hubs.
We next examined properties that are known to be heterogeneous within the larger DMN system to determine whether differences between subnetworks might explain this heterogeneity. For example, some DMN regions serve as central elements of the network (22). Others may represent pathways by which DMN processes interact with those of other networks, such as the Language and Frontoparietal networks, which are known to be partially connected to DMN regions (5, 23, 24).
For each subject, we first identified the canonical DMN, Frontoparietal, and Language networks, as defined at the densest edge density threshold at which it could be found. This density was usually 5%, but in some subjects, the Language network only separated from DMN at the 2% density level. See SI Appendix, Fig. S5 for networks in each subject. For each DMN subnetwork, we calculated the average strength of RSFC between that subnetwork and 1) the rest of the DMN, 2) the Language network, and 3) the Frontoparietal network. We note that in our previous work, the Language network identified here was referred to as the “Ventral Attention” network. We shift to a “Language” nomenclature based on recent findings by refs. 3 and 63.
Subnetwork-to-network relationships in individual subjects (example in Fig. 5A; see SI Appendix, Fig. S7 for all other subjects) suggested that the Parietal subnetwork was centrally connected to other DMN subnetworks, while the Anterior Lateral subnetworks connected to the Language network, and the Dorsal Lateral subnetworks connected to the Frontoparietal network.
Fig. 5.
DMN subnetworks are differentiated by functional connectivity to large-scale networks and by network role. (A) A spring-embedding plot illustrating relationships among DMN subnetworks and other networks in one example subject. Language and Frontoparietal networks are linked to specific DMN subnetworks. Nodes in this network are contiguous subnetwork regions. The Posterior MTL subnetwork is not shown here as it connects only to itself in this subject. (B) The Parietal subnetwork exhibited stronger RSFC to the rest of the DMN than any Lateral subnetwork. (C) The Anterior Lateral subnetworks both exhibited stronger connectivity to the Language network than any other subnetwork, except the Left Dorsal Lateral subnetwork. (D) The Dorsal Lateral subnetworks both exhibited stronger connectivity to the Frontoparietal network than any other subnetwork. (E) The Parietal subnetwork exhibited larger within-network degree Z scores than any Lateral subnetwork except the Right Anterior Lateral subnetwork. (F) The Parietal subnetwork exhibited smaller participation coefficients than any Lateral subnetwork. Error bars indicate SEs across subjects. ant., anterior; dors., dorsal.
Statistical testing revealed that the Parietal DMN subnetwork exhibited stronger RSFC to the rest of the DMN than any other subnetwork [all t (9) values > 5.3; all P values (corrected) < 0.05; Fig. 5B; see SI Appendix, Fig. S8A for connectivity strengths and comparisons in all nine DMN subnetworks]. By contrast, both of the Anterior Lateral subnetworks exhibited stronger RSFC to the Language network than any other subnetwork except each other [all t (9) values > 2.8; all P values (corrected) < 0.05; Fig. 5C and SI Appendix, Fig. S8B], with the exception of the comparison of the Right Anterior Lateral vs. the Left Dorsal Lateral subnetworks [t (9) = 2.58; P = 0.03 (uncorrected)]. Further, both of the Dorsal Lateral subnetworks exhibited stronger RSFC to the Frontoparietal network than any other subnetwork except each other [all t (9) values > 5.6; all P values (corrected) < 0.05; Fig. 5D and SI Appendix, Fig. S8C]. No other large-scale network exhibited positive RSFC to any of these subnetworks (SI Appendix, Fig. S9)
These findings further suggest that the Parietal subnetwork may play a primarily central, coordinating role within the broader DMN, while the Lateral networks may serve as sets of connector hubs that link DMN with other networks, as our previous work has suggested (24). Such functions can be quantified using graph theoretical measures: within-module degree quantifies the centrality of a node within its network, while participation coefficient quantifies how a node serves as a connector hub (64, 65). Here, we used contiguous subnetwork regions within the three large-scale networks as nodes to calculate these graph measures and calculated measures at 5% density.
When specifically examining DMN, Language, and Frontoparietal nodes, we found that the Parietal subnetwork exhibited significantly larger within-module degree than the Left Anterior [t (9) = 3.74; P (corrected) = 0.02), Left Dorsal [t (9) = 6.43; P (corrected) < 0.001], and Right Dorsal [t (9) = 7.88; P (corrected) < 0.001] Lateral subnetworks and numerically larger within-module degree (but not passing corrected significance thresholds) than the Right Anterior Lateral subnetwork [t (9) = 2.34; P (uncorrected) = 0.04] (Fig. 5E). By contrast, all four Lateral subnetworks exhibited larger participation coefficients than the Parietal subnetwork [all t (9) values > 3.89; all P values (corrected) < 0.015] (Fig. 5F). Follow-up analyses including all DMN subnetworks indicated that the Parietal subnetwork similarly exhibited higher within-module degree than any other subnetwork (SI Appendix, Fig. S8D), while the Lateral subnetworks exhibited higher participation coefficients than any other subnetwork (SI Appendix, Fig. S8E). These findings indicate that the Parietal, Anterior Lateral, and Dorsal Lateral subnetworks can be functionally differentiated based on their relationships within and between the DMN, Language, and Frontoparietal networks.
Lateral Subnetworks Systematically Lag Parietal Subnetworks.
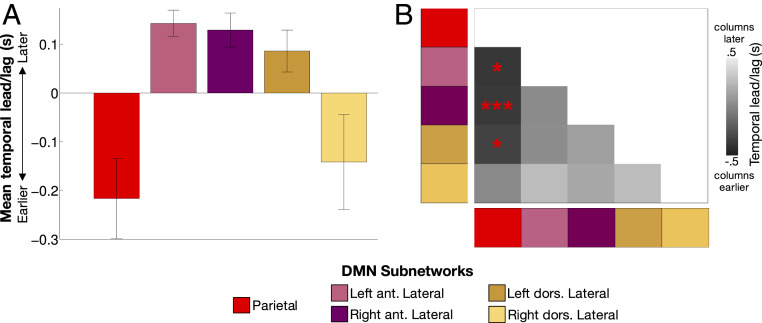
Signals within the DMN are known to exhibit particularly heterogeneous temporal ordering (25). To examine whether the subnetwork organization of the DMN contributes to this heterogeneity, we asked whether DMN subnetworks exhibit systematic temporal lags with respect to one another. For each subject and session, we first calculated the average time series of each Parietal and Lateral DMN subnetwork and then estimated the average pairwise latency between each subnetwork, forming a subnetwork-by-subnetwork time-delay (TD) matrix (66, 67).
We found that Parietal and Right Dorsal Lateral subnetwork signals occurred relatively early (mean time: −0.21 and −0.14 s earlier than other subnetworks), while the Right and Left Anterior and the Left Dorsal Lateral subnetwork signals occurred late (mean times: 0.14, 0.13, and 0.09 s later than other subnetworks, respectively; Fig. 6A). Examination of pairwise lags (Fig. 6B) revealed that in every subject, signals in the Parietal subnetwork preceded those in the Left Anterior Lateral [mean ± SD lag: 0.39 ± 0.28 s; range: 0.07 to 0.94 s; one-sample t (9) = 4.43, P (corrected) = 0.017] and Right Anterior Lateral subnetworks (mean ± SD lag: 0.38 ± 0.16 s; range: 0.12 to 0.63 s; t = 7.48; P [corrected] = 0.0004). Parietal signals similarly led Left Dorsal Lateral subnetwork signals in nine subjects (mean ± SD lag: 0.33 ± 0.28 s; range: −0.02 to 0.81 s; t = 3.75; P [corrected] = 0.046). No other pairwise comparisons were significant (all P values [corrected] > 0.2). Follow-up analyses indicated that this effect was consistent when cortical and cerebellar elements of the networks were examined separately (SI Appendix, Fig. S10 A and B) to account for the known lag in cerebellar signal (52).
Fig. 6.
BOLD signals in Lateral DMN subnetworks are delayed relative to those in the Parietal subnetwork. (A) Average temporal ordering of signals in each subnetwork relative to other subnetworks. Error bars indicate SEs across subjects. (B) TD matrix indicating relative lead/lag of each network pairing. Dark colors indicate the “column” subnetwork is earlier; light colors indicate the “row” subnetwork is earlier. *P < 0.05 corrected; ***P < 0.001 corrected. ant., anterior; dors., dorsal.
Further examination of temporal relationships across all DMN subnetworks did not indicate many other lead/lag relationships that were significant across subjects beyond the Parietal–Lateral effects described above. The exception was the Retrosplenial subnetwork, in which signals were observed to be very early (as in ref. 25), leading the Parietal subnetwork at P < 0.05 (corrected) and the Ventromedial and Left Dorsal Lateral subnetworks at P < 0.05 (uncorrected) (SI Appendix, Fig. S10 C and D). Thus, DMN subnetworks exhibit systematic temporal delays that contribute to the wide range of delays found at the whole-network level (25).
Discussion
The DMN has previously been divided into dorsal and ventral subsystems (41, 54, 68), with a Core linking the two (22, 37). The present work replicated these divisions and identified additional subdivisions apparent across individuals based on similarities in the location, shape, topology, and spatial distribution of the subnetwork.
Separate Anterior Lateral and Dorsal Lateral Subnetwork Streams Couple DMN to Control and Language Systems.
We observed a Parietal subnetwork matching that described by ref. 54 and similar to the Core described by refs. 22 and 37. We also found that more dorsal DMN structures were separable into two pairs of lateralized subnetworks in angular gyrus, lateral temporal cortex, and middle/superior frontal gyrus. These four subnetworks, while consistently exhibiting representation on the same gyri, were individually variable in their locations on the cortex but exhibited spatially consistent representations in cerebellum. This cerebellar representation of DMN substructures is generally consistent with previous group (60) and individual-specific (52) mappings of DMN in the cerebellum. However, while those studies suggested two mirrored DMN representations in the dorsal and ventral aspects of the posterior lobe, here, we demonstrate that this representation is not mirrored. Rather, the Parietal subnetwork was localized to bilateral inferior posterior lobe, while Lateral subnetworks were represented in contralateral (relative to cortex) superior posterior lobe straddling the horizontal fissure.
The analyses used here allow us to hypothesize potential differentiable roles for these Parietal and Lateral subnetworks. The Parietal subnetwork was strongly connected to other DMN subnetworks and exhibited a central role within the large-scale DMN, suggesting it may coordinate the activity of the other subnetworks. By contrast, the Anterior Lateral subnetworks uniquely exhibited positive connectivity to the Language network, while the Dorsal subnetworks uniquely exhibited positive connectivity to the Frontoparietal network. These appear to be specific circuits within the brain’s network structure serving as sets of connector hubs by enabling separate streams between the DMN and the Frontoparietal and Language networks.
These internetwork interactions may reflect separate inputs of control and language functions into the DMN. We found that BOLD signals within three of these four Lateral networks lagged the signals in the Parietal subnetwork by around 300 to 400 ms (Fig. 6), in addition to lagging the signals in most other subnetworks (SI Appendix, Fig. S10C). Previous work has described how infra-slow (<0.1 Hz) BOLD activity propagates in roughly reciprocal directions relative to faster delta-band activity (1 to 4 Hz), possibly helping to organize and regulate the fast delta-band information transfer (69, 70). While overall activity propagation between cortical regions is bidirectional, the delayed infra-slow BOLD activity observed in the Lateral subnetworks may represent a bias for higher-frequency activity to propagate more commonly from the Frontoparietal and Language large-scale networks to the Parietal subnetwork (via the Lateral subnetworks) than vice versa.
These two separate streams of activity propagation could represent very different functions. The Frontoparietal network is believed to exert top-down regulatory control over lower-level processing systems (71–73), including the DMN (74). As such, interactions between the Frontoparietal and DMN networks could represent the Frontoparietal network regulating the DMN via connector hubs (24, 75, 76), like the Dorsal Lateral subnetworks here. Interactions between the DMN and the Language network have also been previously reported (2, 5); we hypothesize that such interactions may represent linguistic input into the internally oriented functions of the DMN. Indeed, regions of the inferior parietal lobe and cerebellum corresponding with the Anterior Lateral DMN subnetwork are engaged by language, social, and self-referential processing (77–79), suggesting their common role in both linguistic and internally oriented processes.
Overall, the identification of separable, individual-specific coupling from the Frontoparietal network and Language network to the DMN suggests that the DMN does not operate in isolation but may be instantiated or regulated by inputs from control and Language networks. This conceptualization paves the way for testing hypotheses about these Frontoparietal network–DMN and Language–DMN interactions in disorders such as posttraumatic stress disorder (PTSD), in which lateral prefrontal-mediated top-down regulation of emotions is deficient (80), but verbal processing of memories can reduce intrusive symptoms (81). It also suggests that these network–network interactions may be specifically modifiable via treatments such as transcranial magnetic stimulation that can target individual-specific DMN subnetworks.
It is likely that temporally ordered interactions may also be present among other association networks such as the Dorsal Attention and Salience networks (82, 83). As such, future examination of subnetworks within these systems may reveal the individually specific pathways by which these interactions occur.
Subnetworks Represent Task Patterns More Specifically than Large-Scale Networks.
In comparison to classic RSFC-derived large-scale brain networks, the subnetwork divisions identified here likely represent a lower level of the brain’s hierarchically nested network structure, which allows them to demonstrate improved specificity in characterizing the brain’s functional organization in individuals. While large-scale brain networks are often used to help make sense of group averaged task-driven responses (6), in individual brains, these networks are often heterogeneously engaged by tasks, as measured by BOLD fMRI (55) or intracranial electroencephalogram (84, 85). This functional heterogeneity limits our ability to infer specific functions of these networks based on their task engagement. By contrast, subnetworks exhibit more homogenous and more individually specific task responses (Figs. 1 and 4). While large-scale networks are good representations of the systems level of brain organization (86) and thus may be appropriate units of analysis for many systems-level questions, the subnetworks delineated here better capture a level of organization corresponding with individual-specific brain regions engaged during task processing, especially in task contrasts where brain activation is regionally selective. Thus, achieving true individual-level specificity in quantifying task engagement may require the use of subnetworks.
Subnetworks also converge with known patterns of task engagement. Regions of the Parietal subnetwork previously have been associated with internally oriented (10) and socially oriented cognition (55). Pregenual subnetwork regions (pregenual medial prefrontal cortex, ventral anterior insula, and anterior striatum) have been associated with reward, expected value, and decision-related positive affect (87–92). Further, the ventral aspects of the DMN were divisible into separate Retrosplenial, Ventromedial, and Posterior MTL subnetworks, which each correspond with distinct, known functional processes. Retrosplenial cortex engages during processing of contextual and especially scene information (15), the ventromedial prefrontal cortex connects to the anterior MTL to regulate fear or anxiety (16, 89, 93), and the posterior MTL is critical for long-term memory (94–96), among other functions.
Individualized DMN Subnetworks May Be Critical for Use in Clinical Populations.
Identifying brain circuits relevant for fear and anxiety (Ventromedial), social cognition (Parietal), and reward processing (Pregenual) may be critical for the study of phobias, autism, and addiction, respectively. Similarly, the Lateral subnetwork links between DMN and Frontoparietal systems may be critical for understanding top-down control of emotion in PTSD and depression. However, even more important may be the ability to identify these subnetworks in individual patients. For example, within the ventromedial prefrontal cortex, the localizations of reward, social processing, and fear overlap in group-average studies, such that metaanalyses are needed to identify partial dissociations between functions (18, 89, 97, 98). By contrast, here, we are able to identify, in most individuals, three separate subnetworks that coincide with these functional dissociations (Pregenual, Parietal, and Ventromedial). Identification of such circuits in individual patients could enable dramatic advances in our ability to diagnose disorders, evaluate treatment efficacy and/or predict treatment response, and specify targets for deep-brain stimulation-based interventions.
Limits of Network Subdivision May Depend on Data Characteristics.
Subnetwork homogeneity and task-explanatory power was optimized at 0.1% graph density, and further network subdivision did not improve validity. This finding identifies a lower bound beyond which investigating network substructures using RSFC is no longer advantageous—at least with these data.
The nature of this apparent lower bound is not clear. One possibility is that, neurobiologically, brain network subdivisions do not exist below a certain scale, and so subdividing the networks present at that scale cannot produce additional information (beyond simply breaking those subnetworks into contiguous clusters, as in areal parcellation approaches; e.g., ref. 56). However, preliminary investigation into these sparser scales suggests this may not be the case. For example, SI Appendix, Fig. S11 illustrates how Frontoparietal subnetworks identified at the “optimal” 0.1% density in one subject exhibit inhomogenous activation patterns that appear better explained by the 0.05% density subnetworks, suggesting that in specific cases or specific subjects, more subdivided networks may contain additional information about brain organization.
A more likely explanation for this lower bound is that the validity measures calculated at sparse thresholds may be constrained by limitations on the effective spatial resolution of the present BOLD data—e.g., voxel size, spatial autocorrelation, and applied smoothing kernel—that reduce the observable validity of very small network structures. Thus, while the finding that brain networks are divisible is not in doubt, the limits of that divisibility should be further explored using higher-resolution single-subject data.
Improving and Expanding Subnetwork Delineation.
Here, we identified DMN subnetworks by conducting community detection separately at multiple scales. While the Infomap algorithm employed here is relatively stable across scales (2, 5), other approaches can conduct community detection in a truly multiscale fashion, by considering all densities/network scales at once (29–31, 99, 100). Future work may explore whether a multiscale approach could improve the robustness of detected subnetworks and allow better comparison across scales.
While we focused on identifying and characterizing subnetwork structures of DMN, we believe that such subnetwork structure is likely a general feature of brain networks. Indeed, previous work suggests that subnetwork structure exists within Frontoparietal (23, 37, 101) and Visual (37, 102–104) networks, while the data presented here suggest that the Frontoparietal and Somatomotor networks also exhibit finer task-relevant subnetwork structure than has previously been described (SI Appendix, Fig. S6). Future work elucidating such structures and their roles within their large-scale network will allow a more detailed mapping between network structures and cognitive functions, which will in turn enable a more detailed understanding of the healthy and pathological network organization of individual human brains.
Methods
Subjects.
Data were collected from 10 healthy, right-handed, young adult subjects (5 females; age: 24 to 34 y). Two of the subjects were authors (N.U.F.D. and S.M.N.), and the remaining subjects were recruited from the Washington University community. Informed consent was obtained from all participants. The study was approved by the Washington University School of Medicine Human Studies Committee and Institutional Review Board. For details of the acquisition parameters, task design and analysis, and processing procedures, refer to SI Appendix, Supplemental Methods. Other findings using these participants and processing procedures have been previously reported (2, 50, 52, 53, 105).
Mapping Multiscale Network Structure.
We first calculated the cross-correlation matrix of time courses from all brain vertices (on the cortex) and voxels (in subcortex), concatenated across sessions. Correlations between vertices/voxels within 30 mm (within-hemisphere connections: geodesic distance; subcortical–cortical connections: Euclidean distance) of each other were set to zero in order to avoid effects of spatial smoothing. Connections between subcortical structures were disallowed, as strong correlations within nearly the entire basal ganglia prevented cortical–subcortical network structures from emerging at sparse graph densities. Interhemispheric connections between cortical surfaces were retained.
We observed that correlations within regions with low BOLD signal due to susceptibility artifact dropout were effectively random. Thus, we calculated a set of low-signal regions as the vertices in which the average mode-1,000 normalized BOLD signal across subjects and time points was less than 750 (56). All connections to low-signal vertices were set to zero.
This matrix was thresholded at multiple levels in such a way to retain at least the strongest X% of connections to each vertex and voxel. Because we did not enforce a requirement that exactly X% of connections survive, this did not result in a homogenous degree distribution. Some voxels/vertices with only relatively weak connections had close to X% of their connections survive thresholding. Other voxels/vertices had much more than X% of their connections survive, because they retained not only their own top X% connections but also many other connections that were in the top X% of connections of other vertices. Unlike previous approaches (5), this approach did not result in a graph with an a priori density, but rather in a fully connected graph in which every node was connected to at least X% of the other nodes. The values of X employed varied by approximate powers of two (rounded to base-10 values) and were as follows: 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, and 5%. For a visualization of how these procedures affected the connectivity matrix, see SI Appendix, Fig. S12.
These thresholded matrices were used as inputs for the Infomap algorithm (59), which calculated community assignments (representing brain network structures) separately for each threshold. Small networks with 10 or fewer vertices/voxels were considered unassigned and removed from consideration. This analysis was conducted in each subject.
Calculating Network Internal Validity.
For each separate brain network at each density in each subject, we measured the homogeneity of the functional connectivity patterns within the network, following ref. 56. For all vertices within the network, we computed a functional connectivity pattern as the Fisher-transformed correlation between the vertex’s time course and the time courses of every other point in the brain. The connectivity patterns of all vertices within the network were then entered into a principal components analysis. Homogeneity was calculated as the percent variance in connectivity patterns explained by the first (largest) principal component (i.e., the most common connectivity pattern). To assess the total homogeneity of all networks delineated at a given density, we averaged homogeneities across networks, weighting each value by the number of vertices within its network. Calculations were restricted to cortical surface vertices, as subcortical voxels could not be included in a rotation-based null model (below).
Importantly, the spatial autocorrelation of smoothed BOLD data means that small networks are more likely to contain a single connectivity pattern than large networks. As such, homogeneity-based evaluations of a network should consider whether the network is more homogenous than a randomly placed network of the same size and shape. Thus, we calculated homogeneities of matched null networks consisting of randomly placed networks with the same size, shape, and relative position to each other.
To create such matched random networks, we rotated each hemisphere of the original networks a random amount around the x, y, and z axes on the spherical expansion of the cortical surface. This procedure randomly relocated each network while maintaining networks’ size, shape, and relative positions to each other. Random rotation was repeated 100 times to generate distributions of homogeneities calculated from random networks. Vertices rotated into the medial wall or into low signal-to-noise ratio (SNR) regions were not included in the homogeneity calculation.
The null-corrected internal validity of a set of networks was calculated as the size-weighted average homogeneity of all networks (as above) minus the median of the size-weighted average homogeneities calculated from rotated networks.
Calculating Network External Validity.
For each density in each subject, we measured how well identified network structures explained spatial patterns of three task contrasts: the (left hand + right hand) > (tongue) contrast from the motor task; the (all scenes) > (all faces) contrast from the implicit memory task; and the (semantic judgment) > (fixation) contrast from the mixed design task. Z-transformed contrast t values from (high-SNR) cortical vertices were entered as the dependent variable in a one-way ANOVA, and vertex network identities were entered as the independent factor. The primary outcome of interest from this ANOVA was the R2 value, representing variance in the task-activation pattern explained by the networks. Here, we used adjusted R2, which accounted for number of levels (i.e., network numbers) within the ANOVA factor varying across density thresholds. As above, calculations were restricted to cortical vertices.
As with homogeneity, this R2 measure must account for network size. Thus, we compared R2 measures calculated from real networks against null R2 measures calculated from 1,000 randomly rotated networks, using the same rotation techniques as above.
For a given task contrast, the null-corrected external validity of a set of networks was calculated as the R2 value of the real networks explaining that task contrast minus the median of the R2 values from all rotated networks applied to that task contrast.
Determining Individual-Specific Large-Scale Network Identities.
For each subject, we identified canonical DMN, Frontoparietal, and Language networks at the 5% density level following the approach described in ref. 2. Briefly, all 5% density Infomap-derived communities within an individual were compared (using spatial overlap, quantified with the Jaccard index) to each network in turn from an independent set of group networks (described more comprehensively in ref. 2; see SI Appendix, Fig. S13 for these independent group networks). Note that, at the 5% level, some group networks (e.g., somatomotor hand, mouth, and foot) were typically combined together. The best-matching community was assigned that network identity. Matches lower than Jaccard = 0.1 were not considered (to avoid matching based on only a few vertices). Notably, in some subjects, the Language network was integrated into the DMN at the 5% level; in these cases, we reidentified networks at 2% density (SI Appendix, Fig. S5).
Identifying DMN Subnetworks in Individuals.
For each subject, we visually examined the cortical and subcortical topographies of each subnetwork, as well as their topological arrangement relative to each other. Subnetworks were identified based on the following heuristic rules:
The Parietal subnetwork was the subnetwork most closely following the classic distribution of DMN regions. We searched for a subnetwork with strong bilateral medial parietal and angular gyrus representation, as well as some presence in superior frontal gyrus, anterior medial prefrontal cortex, and anterior lateral temporal cortex.
The Ventromedial subnetwork was present bilaterally in a swath running from subgenual cingulate to ventral anterior ventromedial prefrontal cortex.
The Pregenual subnetwork was represented in bilateral pregenual cingulate and had a characteristic shape wherein it extended dorsally along the anterior cingulate and forward into anterior medial prefrontal cortex.
The Retrosplenial subnetwork was present in bilateral retrosplenial cortex extending dorsally and posteriorly into the precuneus, with representation in the inferior temporal cortex and in posterior angular gyrus, abutting the angular gyrus Parietal subnetwork cluster.
The Posterior MTL subnetwork was present in the bilateral posterior medial temporal cortex and hippocampus, with little representation elsewhere.
The Dorsal Lateral subnetworks had lateralized representation in the angular gyrus, superior frontal cortex, and posterior cerebellum, but little representation in the medial parietal cortex. The angular gyrus cluster was required to be dorsal to the Parietal subnetwork cluster in the angular gyrus.
The Anterior Lateral subnetworks had lateralized representation in the angular gyrus, superior frontal cortex, and posterior cerebellum, but little representation in the medial parietal cortex. The angular gyrus cluster was anterior to the angular gyrus Parietal subnetwork cluster, and the cerebellar cluster was medial to the cerebellar cluster of the Dorsal Lateral subnetwork.
Calculating Connectivity to Other Networks.
For each DMN subnetwork matched across subjects, we calculated the average time course across subnetwork voxels/vertices. We then calculated the average time courses across voxels/vertices of 1) the DMN, 2) the Language network, and 3) the Frontoparietal network. Voxels/vertices that overlapped with that subnetwork were excluded from these averages. We then calculated the functional connectivity between each DMN subnetwork and each of the three large-scale networks as the Fisher-transformed correlation of the two time courses.
We compared subnetworks’ network-specific connectivities to each other using paired t tests (subnetwork vs. subnetwork) and applying FDR correction for multiple comparisons.
Subnetwork–Network Relationships.
Visualization of subnetwork–network relationships in individual subjects was conducted using spring-embedded plots (5), as implemented in Gephi (https://gephi.org/). In each subject, nodes were defined as congruent clusters of subnetworks larger than 20 mm2. Nodes were selected from the nine matched DMN subnetworks, as well as from other subnetworks within the large-scale Frontoparietal and Language networks (as defined at the 5% density level). Pairwise connectivity between nodes was calculated as the Z-transformed correlation of their mean time courses. For visualization purposes, graphs were constructed by thresholding node-to-node connectivity matrices at 10% density (conclusions did not change at 5% density).
Calculation of Graph-Theoretical Measures.
In each subject, the graph used was defined using a 5% density threshold. This relatively dense threshold was used because we observed that at the 0.1% density level, nodes rarely had connections outside their subnetworks. This caused the graph to be mostly connected only in subnetwork clumps, with no overall network structure evident, and resulted in graph measures being mostly meaningless. Module identities for each node were input as the node’s large-scale network identity (DMN, Frontoparietal, or Language).
Node centrality was calculated as within-module degree Z score, while connector hubness was calculated as participation coefficient; both were computed using the Brain Connectivity Toolbox (106). For each subject, within-module degree Z scores and participation coefficients were averaged across nodes separately for each subnetwork.
We compared subnetworks’ within-module degree Z scores and participation coefficients to each other using paired t tests (subnetwork vs. subnetwork) and applying FDR correction for multiple comparisons across both measures and all pairwise subnetwork comparisons.
Calculating Subnetwork Time Delays.
We computed TD estimates using a previously published method (66). Briefly, we averaged BOLD signals across all voxels/vertices within each subnetwork. Then, for each session, we computed a lagged cross-covariance function (CCF) between each pair of subnetwork time courses. These lags used the same temporal resolution as the acquired data (volume time of repetition [TR] = 2,200 ms). To account for censored frames, we computed CCFs over blocks of contiguous frames and averaged these CCFs, weighted by block duration, to obtain a single-session CCF. We excluded TDs greater than 4 s as, in our experience, these tend to reflect sampling error or artifact. Thus, CCFs were computed over three TR shifts in the positive and negative directions, making the minimum block duration [3 (TR shifts) + 1 (zero-lag)] × TR = 8.8 s. Lags were then more precisely determined by estimating the cross-covariance extremum of the session CCF using three-point parabolic interpolation. The resulting lags were assembled into an antisymmetric matrix capturing all pairwise TDs (TD matrix) for each session, which was averaged across sessions to yield subject-level TD matrices.
Each subject’s TD matrix was averaged across rows to summarize the average time shift from one subnetwork to all other subnetworks. Statistics were performed on TD matrices as one-sample t tests testing whether the TD for each pair of subnetworks was different from zero across subjects, Bonferroni correcting for multiple comparisons.
Supplemental analyses repeated the above procedures after segregating cortical from cerebellar aspects of the network.
Data and Software Availability.
Data from the MSC Dataset have been deposited in the Openneuro data repository (https://openneuro.org/datasets/ds000224/versions/1.0.1) under the label “Midnight Scan Club.” Code to perform all preprocessing and analysis is available at GitHub (https://github.com/MidnightScanClub/Gordon_2020_PNAS).
Supplementary Material
Acknowledgments
We acknowledge Dr. Marc Raichle for his helpful comments and suggestions. This work was supported by US Department of Veterans Affairs Clinical Sciences Research and Development Service Grant 1IK2CX001680 (to E.M.G.); by NIH Grants F31NS110332 (to D.J.N.), NS088590 (to N.U.F.D.), TR000448 (to N.U.F.D.), MH1000872 (to T.O.L.), 1R25MH112473 (to T.O.L.), 5T32 MH100019-02 (to S.M.), and MH104592 (to D.J.G.), 1P30NS098577 (to the Neuroimaging Informatics and Analysis Center); the Kiwanis Neuroscience Research Foundation (N.U.F.D. and B.L.S.); Jacobs Foundation Grant 2016121703 (to N.U.F.D.); the Child Neurology Foundation (N.U.F.D.); the McDonnell Center for Systems Neuroscience (N.U.F.D. and B.L.S.); Mallinckrodt Institute of Radiology Grant 14-011 (to N.U.F.D.); the Hope Center for Neurological Disorders (N.U.F.D., B.L.S., and S.E.P.). The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the US government.
Footnotes
Competing interest statement: N.U.F.D. is cofounder of Nous Imaging.
This article is a PNAS Direct Submission.
Data deposition: Code to perform all preprocessing and analysis is available at GitHub (https://github.com/MidnightScanClub/Gordon_2020_PNAS).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005238117/-/DCSupplemental.
References
- 1.Damoiseaux J. S. et al., Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U.S.A. 103, 13848–13853 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon E. M. et al., Precision functional mapping of individual human brains. Neuron 95, 791–807.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji J. L. et al., Mapping the human brain’s cortical-subcortical functional network organization. Neuroimage 185, 35–57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marek S. et al., Identifying reproducible individual differences in childhood functional brain networks: An ABCD study. Dev. Cogn. Neurosci. 40, 100706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Power J. D. et al., Functional network organization of the human brain. Neuron 72, 665–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith S. M. et al., Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U.S.A. 106, 13040–13045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith S. M. et al.; WU-Minn HCP Consortium , Resting-state fMRI in the human connectome project. Neuroimage 80, 144–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeo B. T. T. et al., The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen S. E., Sporns O., Brain networks and cognitive architectures. Neuron 88, 207–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckner R. L., Andrews-Hanna J. R., Schacter D. L., The brain’s default network: Anatomy, function, and relevance to disease. Ann. N. Y. Acad. Sci. 1124, 1–38 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Raichle M. E., Snyder A. Z., A default mode of brain function: A brief history of an evolving idea. Neuroimage 37, 1083–1090, NaN–1099 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Raichle M. E. et al., A default mode of brain function. Proc. Natl. Acad. Sci. U.S.A. 98, 676–682 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greicius M. D., Krasnow B., Reiss A. L., Menon V., Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. U.S.A. 100, 253–258 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews-Hanna J. R., Reidler J. S., Huang C., Buckner R. L., Evidence for the default network’s role in spontaneous cognition. J. Neurophysiol. 104, 322–335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar M., Aminoff E., Cortical analysis of visual context. Neuron 38, 347–358 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Phelps E. A., Delgado M. R., Nearing K. I., LeDoux J. E., Extinction learning in humans: Role of the amygdala and vmPFC. Neuron 43, 897–905 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Sescousse G., Caldú X., Segura B., Dreher J.-C., Processing of primary and secondary rewards: A quantitative meta-analysis and review of human functional neuroimaging studies. Neurosci. Biobehav. Rev. 37, 681–696 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Lieberman M. D., Straccia M. A., Meyer M. L., Du M., Tan K. M., Social, self, (situational), and affective processes in medial prefrontal cortex (MPFC): Causal, multivariate, and reverse inference evidence. Neurosci. Biobehav. Rev. 99, 311–328 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Leech R., Kamourieh S., Beckmann C. F., Sharp D. J., Fractionating the default mode network: Distinct contributions of the ventral and dorsal posterior cingulate cortex to cognitive control. J. Neurosci. 31, 3217–3224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spreng R. N., Schacter D. L., Default network modulation and large-scale network interactivity in healthy young and old adults. Cereb. Cortex 22, 2610–2621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spreng R. N., Stevens W. D., Chamberlain J. P., Gilmore A. W., Schacter D. L., Default network activity, coupled with the frontoparietal control network, supports goal-directed cognition. Neuroimage 53, 303–317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews-Hanna J. R., Reidler J. S., Sepulcre J., Poulin R., Buckner R. L., Functional-anatomic fractionation of the brain’s default network. Neuron 65, 550–562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon M. L., et al. , Heterogeneity within the frontoparietal control network and its relationship to the default and dorsal attention networks. Proc. Natl. Acad. Sci. U.S.A. 115, E1598–E1607 (2018). Correction in: Proc. Natl. Acad. Sci. U.S.A.115, E3068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon E. M. et al., Three distinct sets of connector hubs integrate human brain function. Cell Rep. 24, 1687–1695.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raut R. V. et al., Organization of propagated intrinsic brain activity in individual humans. Cereb. Cortex 30, 1716–1734 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckner R. L., DiNicola L. M., The brain’s default network: Updated anatomy, physiology and evolving insights. Nat. Rev. Neurosci. 20, 593–608 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Betzel R. F., Bassett D. S., Multi-scale brain networks. Neuroimage 160, 73–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Oort E. S. B., van Cappellen van Walsum A. M., Norris D. G., An investigation into the functional and structural connectivity of the Default Mode Network. Neuroimage 90, 381–389 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Ashourvan A., Telesford Q. K., Verstynen T., Vettel J. M., Bassett D. S., Multi-scale detection of hierarchical community architecture in structural and functional brain networks. PLoS One 14, e0215520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betzel R. F. et al., Multi-scale community organization of the human structural connectome and its relationship with resting-state functional connectivity. Netw. Sci. 1, 353–373 (2013). [Google Scholar]
- 31.Jeub L. G. S., Sporns O., Fortunato S., Multiresolution consensus clustering in networks. Sci. Rep. 8, 3259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betzel R. F. et al., The community structure of functional brain networks exhibits scale-specific patterns of inter- and intra-subject variability. Neuroimage 202, 115990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmichael S. T., Price J. L., Connectional networks within the orbital and medial prefrontal cortex of macaque monkeys. J. Comp. Neurol. 371, 179–207 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Ongür D., Price J. L., The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb. Cortex 10, 206–219 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Parvizi J., Van Hoesen G. W., Buckwalter J., Damasio A., Neural connections of the posteromedial cortex in the macaque. Proc. Natl. Acad. Sci. U.S.A. 103, 1563–1568 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abou Elseoud A. et al., Group-ICA model order highlights patterns of functional brain connectivity. Front. Syst. Neurosci. 5, 37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doucet G. et al., Brain activity at rest: A multiscale hierarchical functional organization. J. Neurophysiol. 105, 2753–2763 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Meunier D., Lambiotte R., Fornito A., Ersche K. D., Bullmore E. T., Hierarchical modularity in human brain functional networks. Front. Neuroinform. 3, 37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahn I., Andrews-Hanna J. R., Vincent J. L., Snyder A. Z., Buckner R. L., Distinct cortical anatomy linked to subregions of the medial temporal lobe revealed by intrinsic functional connectivity. J. Neurophysiol. 100, 129–139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kernbach J. M. et al., Subspecialization within default mode nodes characterized in 10,000 UK Biobank participants. Proc. Natl. Acad. Sci. U.S.A. 115, 12295–12300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uddin L. Q., Kelly A. M., Biswal B. B., Castellanos F. X., Milham M. P., Functional connectivity of default mode network components: Correlation, anticorrelation, and causality. Hum. Brain Mapp. 30, 625–637 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon E. M., Laumann T. O., Adeyemo B., Petersen S. E., Individual variability of the system-level organization of the human brain. Cereb. Cortex 27, 386–399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison S. J. et al., Large-scale probabilistic functional modes from resting state fMRI. Neuroimage 109, 217–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M. et al., Performing group-level functional image analyses based on homologous functional regions mapped in individuals. PLoS Biol. 17, e2007032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D. et al., Parcellating cortical functional networks in individuals. Nat. Neurosci. 18, 1853–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bijsterbosch J. D. et al., The relationship between spatial configuration and functional connectivity of brain regions. eLife 7, e32992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feilong M., Nastase S. A., Guntupalli J. S., Haxby J. V., Reliable individual differences in fine-grained cortical functional architecture. Neuroimage 183, 375–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon E. M. et al., Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage 146, 918–939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison S. J., et al. , Modelling subject variability in the spatial and temporal characteristics of functional modes. bioRxiv:10.1101/544817 (8 February 2019). [DOI] [PMC free article] [PubMed]
- 50.Greene D. J. et al., Integrative and network-specific connectivity of the basal ganglia and thalamus defined in individuals. Neuron 105, 742.e6-758.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laumann T. O. et al., Functional system and areal organization of a highly sampled individual human brain. Neuron 87, 657–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marek S. et al., Spatial and temporal organization of the individual human cerebellum. Neuron 100, 977–993.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sylvester C. M. et al., Individual-specific functional connectivity of the amygdala: A substrate for precision psychiatry. Proc. Natl. Acad. Sci. U.S.A. 117, 3808–3818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braga R. M., Buckner R. L., Parallel interdigitated distributed networks within the individual estimated by intrinsic functional connectivity. Neuron 95, 457–471.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiNicola L. M., Braga R. M., Buckner R. L., Parallel distributed networks dissociate episodic and social functions within the individual. J. Neurophysiol. 123, 1144–1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon E. M. et al., Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26, 288–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavor I. et al., Task-free MRI predicts individual differences in brain activity during task performance. Science 352, 216–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong R. et al., Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb. Cortex 29, 2533–2551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosvall M., Bergstrom C. T., Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. U.S.A. 105, 1118–1123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buckner R. L., Krienen F. M., Castellanos A., Diaz J. C., Yeo B. T. T., The organization of the human cerebellum estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 2322–2345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi E. Y., Yeo B. T. T., Buckner R. L., The organization of the human striatum estimated by intrinsic functional connectivity. J. Neurophysiol. 108, 2242–2263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greene D. J. et al., Developmental changes in the organization of functional connections between the basal ganglia and cerebral cortex. J. Neurosci. 34, 5842–5854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braga R. M., DiNicola L. M., Buckner R. L., Situating the left-lateralized language network in the broader organization of multiple specialized large-scale distributed networks. bioRxiv:10.1101/2019.12.11.873174 (12 December 2019). [DOI] [PMC free article] [PubMed]
- 64.Guimerà R., Amaral L. A. N., Cartography of complex networks: Modules and universal roles. J. Stat. Mech. 2005, a35573 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Power J. D., Schlaggar B. L., Lessov-Schlaggar C. N., Petersen S. E., Evidence for hubs in human functional brain networks. Neuron 79, 798–813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitra A., Snyder A. Z., Hacker C. D., Raichle M. E., Lag structure in resting-state fMRI. J. Neurophysiol. 111, 2374–2391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitra A., Snyder A. Z., Blazey T., Raichle M. E., Lag threads organize the brain’s intrinsic activity. Proc. Natl. Acad. Sci. U.S.A. 112, E2235–E2244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Braga R. M., Van Dijk K. R. A., Polimeni J. R., Eldaief M. C., Buckner R. L., Parallel distributed networks resolved at high resolution reveal close juxtaposition of distinct regions. J. Neurophysiol. 121, 1513–1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitra A. et al., Human cortical-hippocampal dialogue in wake and slow-wave sleep. Proc. Natl. Acad. Sci. U.S.A. 113, E6868–E6876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitra A. et al., Spontaneous infra-slow brain activity has unique spatiotemporal dynamics and laminar structure. Neuron 98, 297–305.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dosenbach N. U. F. et al., Distinct brain networks for adaptive and stable task control in humans. Proc. Natl. Acad. Sci. U.S.A. 104, 11073–11078 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dosenbach N. U. F., Fair D. A., Cohen A. L., Schlaggar B. L., Petersen S. E., A dual-networks architecture of top-down control. Trends Cogn. Sci. 12, 99–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marek S., Dosenbach N. U. F., The frontoparietal network: Function, electrophysiology, and importance of individual precision mapping. Dialogues Clin. Neurosci. 20, 133–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen A. C. et al., Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proc. Natl. Acad. Sci. U.S.A. 110, 19944–19949 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole M. W. et al., Multi-task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 16, 1348–1355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gratton C., Sun H., Petersen S. E., Control networks and hubs. Psychophysiology 55, e13032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bzdok D. et al., Left inferior parietal lobe engagement in social cognition and language. Neurosci. Biobehav. Rev. 68, 319–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guell X., Gabrieli J. D. E., Schmahmann J. D., Triple representation of language, working memory, social and emotion processing in the cerebellum: Convergent evidence from task and seed-based resting-state fMRI analyses in a single large cohort. Neuroimage 172, 437–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morin A., Michaud J., Self-awareness and the left inferior frontal gyrus: Inner speech use during self-related processing. Brain Res. Bull. 74, 387–396 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Fitzgerald J. M., DiGangi J. A., Phan K. L., Functional neuroanatomy of emotion and its regulation in PTSD. Harv. Rev. Psychiatry 26, 116–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luo P. et al., Effects of different forms of verbal processing on the formation of intrusions. J. Trauma. Stress 26, 288–294 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Raccah O., Daitch A. L., Kucyi A., Parvizi J., Direct cortical recordings suggest temporal order of task-evoked responses in human dorsal attention and default networks. J. Neurosci. 38, 10305–10313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shine J. M. et al., Distinct patterns of temporal and directional connectivity among intrinsic networks in the human brain. J. Neurosci. 37, 9667–9674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daitch A. L., Parvizi J., Spatial and temporal heterogeneity of neural responses in human posteromedial cortex. Proc. Natl. Acad. Sci. U.S.A. 115, 4785–4790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Foster B. L., Dastjerdi M., Parvizi J., Neural populations in human posteromedial cortex display opposing responses during memory and numerical processing. Proc. Natl. Acad. Sci. U.S.A. 109, 15514–15519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sejnowski T. J., Churchland P. S., “Brain and cognition” in Foundations of Cognitive Science, Posner M. I., Ed. (MIT Press, Cambridge, MA, 1989), p. 888. [Google Scholar]
- 87.Clithero J. A., Rangel A., Informatic parcellation of the network involved in the computation of subjective value. Soc. Cogn. Affect. Neurosci. 9, 1289–1302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grabenhorst F., Rolls E. T., Value, pleasure and choice in the ventral prefrontal cortex. Trends Cogn. Sci. 15, 56–67 (2011). [DOI] [PubMed] [Google Scholar]
- 89.Hiser J., Koenigs M., The multifaceted role of the ventromedial prefrontal cortex in emotion, decision making, social cognition, and psychopathology. Biol. Psychiatry 83, 638–647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shenhav A., Buckner R. L., Neural correlates of dueling affective reactions to win-win choices. Proc. Natl. Acad. Sci. U.S.A. 111, 10978–10983 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shenhav A., Karmarkar U. R., Dissociable components of the reward circuit are involved in appraisal versus choice. Sci. Rep. 9, 1958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith D. V. et al., Distinct value signals in anterior and posterior ventromedial prefrontal cortex. J. Neurosci. 30, 2490–2495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kalisch R. et al., Context-dependent human extinction memory is mediated by a ventromedial prefrontal and hippocampal network. J. Neurosci. 26, 9503–9511 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ranganath C., Ritchey M., Two cortical systems for memory-guided behaviour. Nat. Rev. Neurosci. 13, 713–726 (2012). [DOI] [PubMed] [Google Scholar]
- 95.Squire L. R., Stark C. E. L., Clark R. E., The medial temporal lobe. Annu. Rev. Neurosci. 27, 279–306 (2004). [DOI] [PubMed] [Google Scholar]
- 96.Wig G. S. et al., Medial temporal lobe BOLD activity at rest predicts individual differences in memory ability in healthy young adults. Proc. Natl. Acad. Sci. U.S.A. 105, 18555–18560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Delgado M. R. et al., Viewpoints: Dialogues on the functional role of the ventromedial prefrontal cortex. Nat. Neurosci. 19, 1545–1552 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Schneider B., Koenigs M., Human lesion studies of ventromedial prefrontal cortex. Neuropsychologia 107, 84–93 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Mucha P. J., Richardson T., Macon K., Porter M. A., Onnela J.-P., Community structure in time-dependent, multiscale, and multiplex networks. Science 328, 876–878 (2010). [DOI] [PubMed] [Google Scholar]
- 100.Rosvall M., Bergstrom C. T., Multilevel compression of random walks on networks reveals hierarchical organization in large integrated systems. PLoS One 6, e18209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tobyne S. M., Osher D. E., Michalka S. W., Somers D. C., Sensory-biased attention networks in human lateral frontal cortex revealed by intrinsic functional connectivity. Neuroimage 162, 362–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dawson D. A. et al., Partial correlation-based retinotopically organized resting-state functional connectivity within and between areas of the visual cortex reflects more than cortical distance. Brain Connect. 6, 57–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Felleman D. J., Van Essen D. C., Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47 (1991). [DOI] [PubMed] [Google Scholar]
- 104.Genç E., Schölvinck M. L., Bergmann J., Singer W., Kohler A., Functional connectivity patterns of visual cortex reflect its anatomical organization. Cereb. Cortex 26, 3719–3731 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Gratton C. et al., Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron 98, 439–452.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rubinov M., Sporns O., Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.