Significance
Probabilistic bet hedging is a generalized diversification strategy to maximize fitness in unpredictable environments and has been proposed as an evolutionary basis for herpesvirus latency. However, the molecular mechanisms enabling probabilistic bet hedging have remained elusive. Here, we find that the human herpesvirus cytomegalovirus—a major cause of birth defects and transplant failures—utilizes stochastic variability in the abundance of a protein packaged into individual viral particles to enable probabilistic bet hedging between alternate viral states.
Keywords: stochastic variability, fate selection, herpesvirus, latency, tegument
Abstract
Probabilistic bet hedging, a strategy to maximize fitness in unpredictable environments by matching phenotypic variability to environmental variability, is theorized to account for the evolution of various fate-specification decisions, including viral latency. However, the molecular mechanisms underlying bet hedging remain unclear. Here, we report that large variability in protein abundance within individual herpesvirus virion particles enables probabilistic bet hedging between viral replication and latency. Superresolution imaging of individual virions of the human herpesvirus cytomegalovirus (CMV) showed that virion-to-virion levels of pp71 tegument protein—the major viral transactivator protein—exhibit extreme variability. This super-Poissonian tegument variability promoted alternate replicative strategies: high virion pp71 levels enhance viral replicative fitness but, strikingly, impede silencing, whereas low virion pp71 levels reduce fitness but promote silencing. Overall, the results indicate that stochastic tegument packaging provides a mechanism enabling probabilistic bet hedging between viral replication and latency.
Diverse biological systems share a common challenge to preserve reproductive fitness in unpredictable, changing environments. Faced with environmental variability, some organisms probabilistically generate a range of phenotypes to “hedge their bets” (1–7), in much the same way that financial houses diversify their assets to minimize risk against economic crashes. First proposed over 50 years ago (1) for desert annuals—where reproductive success is subject to unpredictable weather patterns—bet hedging theory noted that temporal variation in fitness could be minimized if husk thickness between seeds varied such that random chance biased the population’s germination potential. In this way, some seeds randomly enter dormancy irrespective of the environment, and a long-lived, desiccation-resistant subpopulation is always formed to avoid extinction during unforeseen droughts, but at the necessary expense of lowering germinative fitness. In the decades since, bet hedging has been studied in persistence phenotypes in bacteria (2, 5, 7, 8), yeasts (9, 10), and viruses (11–13). In herpesviruses, bet hedging theory has been proposed as a theoretical basis for the evolution of viral latency (11) and associated viral gene silencing. However, the molecular mechanisms that allow biological systems, such as viruses, to probabilistically generate the needed variability have remained unclear and an area of active study.
In the herpesviridae family, persistence within the host is mediated by establishment of a reversible latent state, wherein viral gene expression is largely silenced (14, 15). In human cytomegalovirus (CMV)—one of nine human herpesviruses and a leading cause of birth defects and transplant failure—lytic replication occurs in a variety of cell types, while latency and requisite silencing are established in myeloid-progenitor cells (14, 16, 17). The silent state is thought to enable evasion of host–immune responses and confer a selective advantage (11). To reactivate from a silenced state or initiate lytic replication, expression from CMV’s Major Immediate-Early Promoter (MIEP) is essential. During silencing, the MIEP is largely quiescent, but during lytic replication the MIEP drives expression of crucial viral genes including the 86-kDa Immediate-Early 2 (IE2) protein, a master regulator of lytic viral expression. Initiation of lytic replication requires that the MIEP be efficiently transactivated by proteins carried within the viral tegument (18), a proteinaceous region between the viral capsid and envelope. The pp150 (UL32) and pp71 (UL82) tegument proteins appear to be the chief tegument regulators of MIEP expression (19–21), with pp71 transactivating MIEP activity and pp150 repressing MIEP activity.
Building off previous quantitative analyses of tegument variability in other herpesviruses (22–24), we quantified CMV single-virion variability in pp71 and pp150 levels using superresolution fluorescence microscopy, and then examined how variability in particle-associated protein abundance affects CMV replicative strategies. We find that virion-associated pp71 levels are significantly more variant than pp150 levels or than expected by Poisson statistics. When virions with increased pp71 levels were generated, the population exhibited enhanced infectiousness and replicative fitness. This enhanced fitness conferred by higher pp71 raised the question of why selective pressures had not forced the pp71 distribution to a higher mean and lower variance, indicating that a putative counterbalancing selection pressure may exist to maintain the broad pp71 distribution. Indeed, we found that low pp71 levels promoted MIEP silencing, whereas high pp71 levels impeded silencing in undifferentiated cells. We propose a conceptual model for how heterogeneity in the levels of virion-packaged tegument transactivators may enable probabilistic bet hedging between replication and silencing in herpesviruses.
Results and Discussion
The HCMV Major Tegument Transactivator Protein pp71 Exhibits Super-Poissonian Variability in Virion-to-Virion Abundance.
To measure the variation in pp71 and pp150 abundance in individual virion particles, we utilized a superresolution imaging method (25) that enabled fluorescent imaging of pp71 or pp150 tegument proteins that were genetically tagged with yellow fluorescent protein (YFP) (Fig. 1A). To obtain the viral samples for imaging, recombinant virus was packaged in culture and viral preparations were gradient purified via ultracentrifugation to isolate infectious particles and exclude dense bodies and empty particles. Electron microscopy verified the purity of single, intact virions (SI Appendix, Fig. S1). For image quantification, we used an established superresolution segmentation algorithm (26) that ensures only singlet particles are analyzed, and scored and implemented segmentation based on the lipid envelope costaining to ensure that only envelope-associated particles were analyzed (SI Appendix, Fig. S2). Only virus-sized (∼200 nm diameter) singlet particles were included in the imaging analysis, and a minimum of 2,000 intact, singlet particles were quantified for each fusion virus. To control for contributions from instrument and photon shot noise, a “molecular ruler” (27), containing precisely 900 florescent-fused HSV-1 VP26 proteins per viral particle (24), was imaged in parallel and used for comparison.
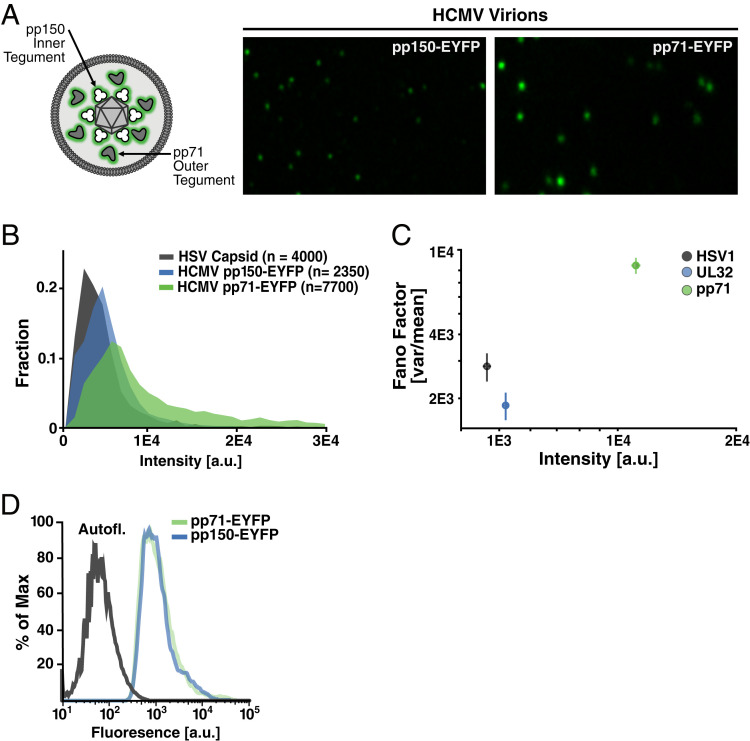
Fig. 1.
Substantial virion-to-virion heterogeneity in the level of the HCMV major tegument transactivator protein pp71. (A, Left) Schematic of HCMV virion showing relative locations of pp150 (UL32), which is capsid-associated, in the inner tegument, and pp71 (UL82), which is capsid-unassociated, in the outer tegument. (A, Right) Representative superresolution fluorescence micrographs of purified, infectious, recombinant HCMV virion particles where tegument proteins are genetically fused to EYFP: pp150-EYFP (Left), pp71-EYFP (Right) (pixel size; 40 nm). (B) Quantification of pp71 and pp150 abundance in individual virion particles relative to the HSV-1 VP26-GFP “molecular ruler” (900 copies per virion). (C) Normalized variance (Fano factor; σ2/μ) versus mean abundance (<intensity>) in pp71, pp150, and HSV-1 capsid in the virion populations. Error bars were estimated by bootstrapping the data with n = 1,000 particles per sample, 150 times. (D) Intracellular levels of tegument factors EYFP-pp71 and pp150-EYFP in infected human fibroblasts quantified by flow cytometry (Left).
Quantitative image analysis showed that the per-virion levels of the pp150 tegument protein, which is capsid-associated, fell within a relatively confined range, exhibiting two- to threefold particle-to-particle variation in intensity (Fig. 1B). This variation in pp150 was similar to the virion-to-virion variation in HSV-1 VP26 capsid levels (Fig. 1B), which are considered invariant (24). However, in striking contrast, pp71 tegument levels imaged under identical conditions and parameters varied >10-fold between the dimmest and brightest virion particles (Fig. 1B), with the normalized virion-to-virion variance (σ2/μ, a standardized measure of the histogram width referred to as the “Fano factor”) being fivefold greater for pp71 than pp150 (Fig. 1C). Importantly, an orthogonal confocal imaging method showed close agreement with the superresolution imaging measurements (SI Appendix, Fig. S2B). Moreover, these image-based quantifications of pp71 and pp150 are within the range of previously-reported ratios from mass spectrometry analyses (28).
To determine whether the large pp71 particle-to-particle variability resulted from a subpopulation of virus-producing cells that are “outliers” and express higher levels of pp71 but not pp150—or other extrinsic variability in the virus-producing cells—we analyzed pp71 and pp150 expression levels in virus-producing cells using flow cytometry. In contrast to virion-particle heterogeneity, pp71 and pp150 expression levels in virus-producing cells exhibited only minimal cell-to-cell variability that could not account for the relative amplification of pp71 heterogeneity in virion particles (Fig. 1D). While pp71 expression levels did not vary between cells, the distribution of pp71 puncta within virus-producing cells—assayed by superresolution imaging—showed high variability. Specifically, cytoplasmic pp71 foci that were virion-sized (i.e., smaller than 300 nm) showed a long-tailed distribution in intensity with a large normalized variance (σ2/μ = 16,782) (SI Appendix, Fig. S2C). While the distribution of pp150 puncta within virus-producing cells also exhibited similar variability, the capsid-associated nature of pp150 appears to effectively filter this heterogeneity, since overexpression of pp150 in cells did not increase pp150 levels in virions (SI Appendix, Fig. S2C), whereas pp71 overexpression in cells did increase pp71 in virions (18). These results argue that intrinsic heterogeneity in the spatial distribution of intracellular pp71, rather than extrinsic heterogeneity arising from expression-level differences between infected cells, contributes to virion-to-virion variability in pp71 levels.
High pp71 Offers an Evolutionary Advantage by Increasing Infectivity.
To test if this large intervirion pp71 variability carried a functional consequence, we generated CMV virion particles that packaged increased levels of pp71 protein (pp71HI) using a cell line expressing pp71, as previously described (29). Importantly, pp71 virion levels can be modulated without affecting packaging of other tegument factors (30). Superresolution image analysis verified that singlet pp71HI virions contained substantially more pp71, on average ∼2-fold higher than the pp71 levels in pp71WT virus produced from standard fibroblasts (Fig. 2A). In contrast, when virus was packaged on cells overexpressing pp150, no measurable increase in virion pp150 levels could be detected (SI Appendix, Fig. S2C), presumably because pp150 is capsid-associated and its levels are fixed by the invariable number of capsid proteins.
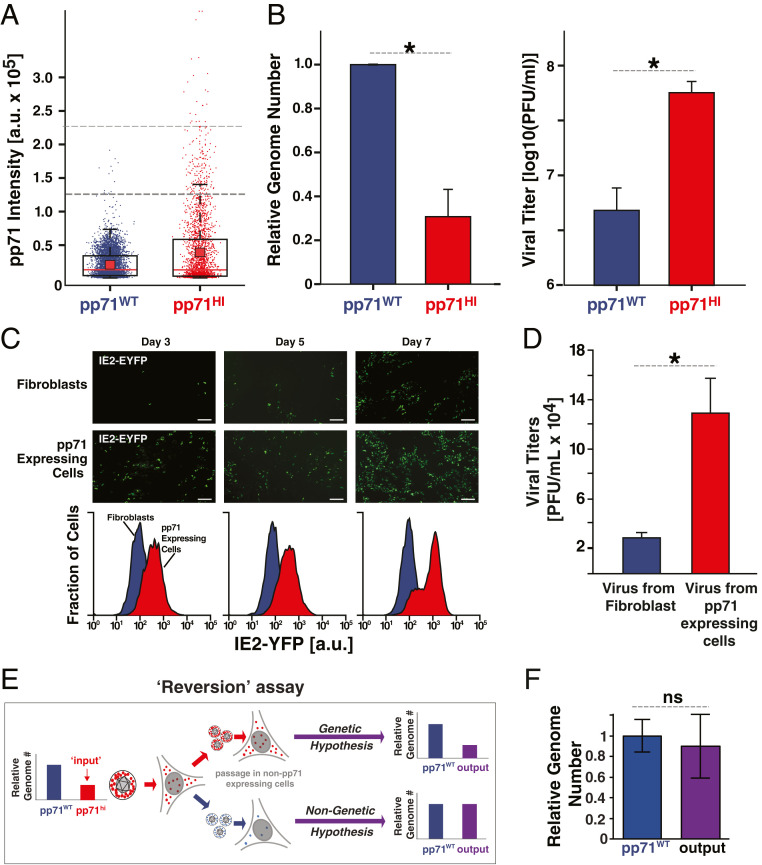
Fig. 2.
The “advantage” of high pp71: high virion pp71 levels enhance HCMV replicative fitness. (A) Box-whisker plot of pp71 levels (by EYFP intensity) in purified virion particles quantified by superresolution microscopy: pp71WT EYFP virion particles (purple, n = 5,519); pp71HI EYFP virion particles (red, n = 2,463). Analysis was restricted to particles 100–300 nm in diameter. The red square depicts the mean μ, red line the median, the box encloses values between the first and third quantiles (25–75%), and the whiskers (error bars) show the minimum and maximum intensities. The horizontal-dashed lines represent intensity thresholds of 1.25 × 105 a.u. (dark gray) and 2.25 × 105 a.u. (light gray). For pp71WT, 0.25% of virions >1.25 × 105 a.u. and 0% of virions >2.25 × 105 a.u.; for pp71HI, 7.1% of virions >1.25 × 105 a.u. and 1% of virions >2.25 × 105 a.u. (these percentage are shown in Fig. 3F). (B, Left) qRT-PCR analysis of viral genome copy number in pp71HI and pp71WT stocks after stocks were normalized to have equivalent infectivity by infectious units (*P < 0.05, two-tailed t test). (B, Right) Infectivity of same pp71HI and pp71WT stocks, as measured by TCID50, after stocks were matched to have equivalent genome copy numbers (*P < 0.05, two-tailed t test). (C, Upper) Confocal micrographs of IE2-YFP reporter-virus infections (MOI 0.01) on human fibroblasts (HFFs) or a pp71-expressing HFF line (WF28). (Scale bars, 200 µm.) (C, Lower) Flow cytometry analysis of lytic IE2-YFP expression on indicated days. (D) Viral replication (titer) quantified by TCID50 7 d.p.i. of HFFs or pp71-expressing HFFs. (E) Schematic of “Reversion assay” to test if pp71HI virus harbors secondary mutations that influence its phenotype. If pp71HI phenotype has a genetic component, lower genome-copy number (i.e., higher infectious particle-to-genome ratio) relative to pp71WT will be retained and selected for due to its replicative advantage. In contrast, if the pp71HI phenotype is nongenetic, excess packaged pp71 will not be retained on low passage in naïve cells and phenotype will revert to pp71WT (i.e., genome-copy number and infectious particle-to-genome ratio equal to pp71WT). (F) qPCR analysis of virus output from the “reversion assay” (i.e., pp71HI virus after low passage in HFF). Titer of the resulting virus was measured by TCID50, matched to WT (as in Fig. 2B), and viral genomes in the MOI-matched isolate then quantified by qPCR (difference is not significant by t test). ns, not significant. (Magnification: 20× obj.)
We tested the functional consequence of increased pp71 by comparing the number of viral genomes present in pp71WT and pp71HI virus isolates at equivalent infectivity by qPCR. Surprisingly, three times as many viral genomes were required in a pp71WT virion isolate to generate the same infectivity as a pp71HI virion isolate (Fig. 2 B, Left). Conversely, when the viral isolates were then equalized for the number of viral genomes, the pp71HI virus was an order of magnitude more infectious than pp71WT virus (Fig. 2 B, Right).
Next, we tested how pp71 levels influence multiround viral replication kinetics by assaying expression of the lytic master regulator, the 86-kDa immediate early 2 (IE2) protein. We passaged an IE2-YFP expressing virus (31) on cells that conditionally overexpressed pp71 during late infection to produce virions carrying excess pp71 (32) (i.e., produce pp71HI virus). As predicted (31), increased pp71 generated increased single-round viral production as measured by IE2 expression (Fig. 2C), and passaging virus on conditionally pp71-overexpressing cells, to continually produce pp71HI virus, generated a >1-log increase in viral titer (Fig. 2D). The increased number of IE-expressing cells on day 5 (i.e., ∼24 h into the second round of infection) could not be accounted for by ectopic cellular pp71 in the overexpression cell line, since pp71 expression in this cell line is driven by a promoter activated late in CMV infection, and no increased IE expression was observed at 12 or 24 h postinfection (h.p.i.) (SI Appendix, Fig. S3), in agreement with previous reports (29).
To verify that the pp71HI phenotype was not a result of a secondary mutation in the virus, we performed a “reversion assay,” in which pp71HI virus was minimally passaged in fibroblasts that did not overexpress pp71 (Fig. 2E). We hypothesized that if a genetic mutation in the virus was responsible for the pp71HI phenotype, the infectivity of the resulting “output” virus would remain higher than pp71WT (i.e., the particle-to-genome ratio would remain three times higher than pp71WT). Furthermore, given the increased replicative fitness of pp71HI, any underlying mutation should be selected for, undergo “fixation,” and be maintained indefinitely. In contrast, if the phenotype was nongenetic and solely due to excess packaging of pp71, the excess pp71 would be diluted out during low passage, and the infectivity of the resulting “output” virus would revert to pp71WT. qPCR analysis indicated that the number of viral genomes present in multiplicity of infection (MOI)-matched pp71WT and “output” isolates was roughly equivalent (Fig. 2F), supporting a nongenetic basis for the pp71HI phenotype.
High pp71 Carries an Evolutionary Cost by Reducing Latency Establishment.
The enhanced infectiousness (Fig. 2B) and replicative fitness (Fig. 2C) conferred by higher pp71 levels presented an evolutionary conundrum given the broad virion-to-virion pp71 distribution (Fig. 1B). Specifically, Darwinian selection theory predicts that, absent a counterbalancing selection pressure, virus variants that package higher pp71 levels should be selected to promote replicative fitness. Thus, virus variants with high pp71 should sweep the population such that the virion-to-virion pp71 distribution would be narrow (low variability) with a high mean level, as opposed to the observed broad (high variability) pp71 distribution. To address this conundrum, we hypothesized that virions with lower pp71 levels may provide the putative counterbalancing selection pressure via an advantage in silencing lytic gene expression that is required to establish viral latency.
Previous studies demonstrated that during HCMV infection of undifferentiated cells, pp71 is excluded from the nucleus, promoting IE silencing and latency (33). Building off this finding, we hypothesized that pp71HI virus may overcome the putative threshold for nuclear exclusion and allow a fraction of pp71 to penetrate the nucleus, leading to desilencing and lytic expression. To test this, we infected embryonal undifferentiated NTera2 cells, an established latency model (34), with a newly developed two-color dual-reporter virus TB40E-IE-mCherry-EYFP (based on CMDR mutant described previously; ref. 18). In this reporter virus, mCherry-only fluorescence corresponds to IE1-only expression, whereas YFP fluorescence indicates that IE2 is expressed. TB40E-IE-mCherry-EYFP was packaged to generate either pp71WT or pp71HI virus, and lytic IE expression was assayed in undifferentiated NTera2 cells (Fig. 3A). No IE1 or IE2 expression was observed upon pp71WT infection, whereas pp71HI virus generated a significant percentage of dual-positive cells (Fig. 3 A and B and SI Appendix, Fig. S4A). Using confocal immunofluorescence microscopy, we then directly imaged the subcellular localization of pp71 and IE2 shortly after infection of undifferentiated NTera2 cells (Fig. 3C). The pp71HI virus generated a striking increase in nuclear pp71 levels compared to pp71WT virus, and nuclear IE2 expression levels in this latency model correlated with nuclear pp71 penetrance (Fig. 3D). A reversion assay in NTera2 cells verified that this pp71HI phenotype in NTera2 cells is nongenetic (SI Appendix, Fig. S5 A–C). In addition, IE expression generated by pp71HI could be phenocopied by supplying pp71 in trans (i.e., transient transfection of NTera2 cells with a pp71 expression vector) and infecting with pp71WT virus (SI Appendix, Fig. S5 D and E).
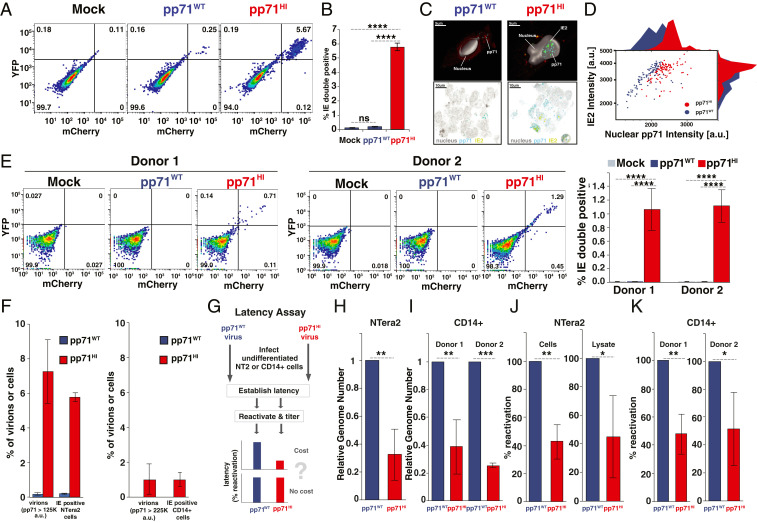
Fig. 3.
The “cost” of high pp71: high virion pp71 levels overcome nuclear exclusion in undifferentiated cells and impede establishment of viral silencing. (A) Flow cytometry analysis of undifferentiated NTera2 cells infected with dual-reporter TB40E-IE-mCherry-EYFP. Cells were either mock infected (Left), infected with pp71WT virus (Center), or with pp71HI virus (Right) at MOI = 3. (B) Quantification of % IE double-positive population for three biological replicates from the flow cytometry data shown in Fig. 3A (P values from Student’s t test: ****<0.0001). (C) Representative confocal immunofluorescence micrographs of infected NTera2 cells (MOI = 3) assayed 6 h.p.i. with either pp71WT or pp71HI virus expressing IE2-YFP. Two-dimensional images along with a single confocal plane from a three-dimensional (3D) z-stack are shown along with a 3D reconstruction of the cell nucleus (solid). pp71 (stained via α-pp71 antibody) is visible as fluorescent puncta (teal) proximal to IE2-YFP puncta (green). Extranuclear fluorescence (red) is due to cytoplasmic autofluorescence. (D) Image-based quantification of pp71 and IE2 intensity levels in cells exhibiting both IE2 expression and nuclear pp71 levels for cells infected with pp71WT (purple; 105 cells) or pp71HI (red; 109 cells). Histograms on axes are derived from projecting the dot-plot data onto respective axis. (E, Left) Flow cytometry analysis of donor-derived human CD14+ primary monocytes infected with dual-reporter TB40E-IE-mCherry-EYFP (MOI = 2), 6 h.p.i., for two donors. Cells were either mock infected, infected with pp71WT virus or pp71HI virus. (E, Right) Quantification of %IE double-positive CD14(+) monocytes per donor, as assayed by flow cytometry 6 h.p.i. (P values as in B). (F) Comparative analysis of percentage of virions with high pp71 levels (from Fig. 2A) vs. cells with IE expression (Fig. 3 C and E). (G) Schematic of the latency assay for pp71WT and pp71HI virus. Briefly, undifferentiated NTera2 or CD14+ cells are infected with either pp71WT or pp71HI virus, latency established over 4–10 d, and virus then reactivated from latency. Latency is quantified by qPCR for viral genomes as well as titering on HFF after reactivation. (H) qPCR quantification of latent CMV genomes 4 d after TB40E infection of NTera2 cells. (I) qPCR quantification of latent CMV genomes 10 d after TB40E infection of CD14+ monocytes. (J) Analysis of latent reactivation in NTera2 cells (initial infection with pp71WT or pp71HI TB40E-IE-mCherry-YFP virus at MOI = 3). Four days postinfection, NTera2 cells were treated with TSA for 24 h, washed three times, and serially diluted and cocultured with HFFs for 10 d and plaque-forming units/mL then calculated by TCID50. NTera2 cell lysate was titered in parallel for 10 d. Average of three biological replicates shown with pp71WT titers normalized to 100% reactivation. (K) Analysis of latent reactivation in human primary CD14+ monocytes (initial infection with pp71WT or pp71HI TB40E-IE-mCherry-YFP virus at MOI = 2). Ten days postinfection, CD14+ cells were cocultured with HFFs (10-fold serial dilution) supplemented with reactivation media (IL3, IL6, G-CSF, GM-CSF) for 15 d and viral titers then analyzed by TCID50. The experiment was performed for two donors with three biological replicates and shown with pp71WT titers normalized to 100% reactivation. (P value <0.05 was considered statistically significant: *<0.05, **<0.01, ***<0.001, ****<0.0001, two-tailed t test).
To determine if high levels of pp71 also impeded silencing in human primary CD14+ monocytes—which serve as a reservoir for harboring latent HCMV (17, 35, 36)—we infected donor-derived primary human CD14+ monocytes with pp71WT or pp71HI dual-reporter TB40E-IE-mCherry-EYFP virus at MOI = 2, and assayed for IE expression by flow cytometry (Fig. 3E and SI Appendix, Fig. S4B). In agreement with the NTera2 data, in primary monocytes from two separate donors, pp71HI infection generated a significant number in IE dual-positive cells, whereas pp71WT virus generated no detectable IE expression (Fig. 3E). Immunofluorescence imaging verified that pp71 was only present in the nucleus in CD14+ monocytes infected with pp71HI virus and correlated with IE expression (SI Appendix, Fig. S5G).
To determine if the increased pp71 in pp71HI virions could explain the frequency of IE-positive undifferentiated cells, we analyzed the percentage of virions with the highest pp71 levels in both the pp71HI and pp71WT isolates, compared to the percentage of IE-expressing cells generated. The virion imaging data (Fig. 2A) show that ∼0.25% of pp71WT virions lay above a 1.25 × 105 a.u. intensity threshold, whereas ∼7.1% pp71HI virions surpassed this threshold. In comparison, pp71WT generated IE expression in 0.25% of NTera2 cells, whereas pp71HI generated IE expression in 5.7% of NTera2 cells (the difference between 5.7% and 7.1% was not statistically significant) (Fig. 3F). Similarly, ∼1% of pp71HI virions surpassed a 2.25 × 105 a.u. threshold, and ∼1% of CD14+ monocytes expressed IE after pp71HI infection, whereas 0% of pp71WT virions surpassed 2.25 × 105 a.u., and 0% of monocytes expressed IE after pp71WT infection (Fig. 3F). These data indicate that virions with high pp71 abundance generate IE expression in undifferentiated cells and likely do so by overcoming the cytoplasmic pp71 restriction.
To directly test the central tenet of bet hedging theory—i.e., that the phenotype with a fitness advantage under one condition carries a “cost” under another condition—we used an established approach (17, 34, 36) to analyze the percentage of NTera2 cells and primary CD14+ monocytes that establish and reactivate latency (Fig. 3G). In this assay, cells are infected with either pp71HI or pp71WT virus and then incubated until IE expression silences—i.e., 4 d for NTera2 and 10 d for CD14+ cells (SI Appendix, Fig. S6 A and B)—latency establishment is quantified by the number of genome copies, and latency reversal is quantified on permissive cells. If pp71HI virus carries a “cost,” latency establishment and reactivation are predicted to be diminished in comparison to pp71WT virus. In agreement with bet hedging theory, pp71HI virus exhibited a significant reduction in both latency establishment (Fig. 3 H and I) and reactivation (Fig. 3 J and K) in both NTera2 cells and CD14+ monocytes. Collectively, these data indicate that increasing virion-associated pp71 abundance promotes a lytic-expression program in undifferentiated cells leading to reduced latency establishment.
Super-Poissonian Variability in Virion Tegument Abundance Enables Bet Hedging.
Overall, our results show that heterogeneity in tegument protein abundance in CMV viral particles creates a phenotypically diverse virion population, providing a molecular mechanism for probabilistic bet hedging between lytic and latent infection (Fig. 4). Interestingly, a positive autoregulatory circuit mediated by the 76-kDa immediate early 1 (IE1) protein provides a mechanism for further amplification of pp71 phenotypic diversity. Positive feedback loops are known to amplify variability and generate bimodality in other viral and nonviral systems (37, 38). We speculate that variable pp71 levels in infecting virions provide an initial “kernel” level of variability, which is then processed by IE1 positive-feedback circuitry. If the incoming pp71 level in the infecting virion is below the threshold for activation of the IE1 positive-feedback circuit, it will undergo proteolysis within its characteristic 8-h half-life without sufficiently activating the immediate-early viral expression program, and the infected cell will enter a quiescent state of expression. In contrast, if the incoming pp71 level in the infecting virion is above the threshold, the immediate-early viral expression program will be activated and the cell will proceed to lytic infection. Stochastic variability in packaged tegument levels in virions may explain the differing data on early IE expression in undifferentiated cells (36, 39, 40).
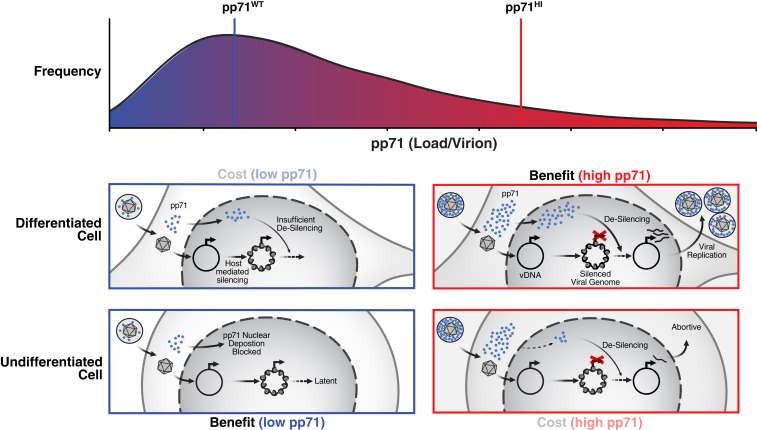
Fig. 4.
Summary model: super-Poissonian variability in tegument abundance enables bet hedging between replication and latency. In differentiated cells (Middle), pp71HI virions (Right) have an “advantage” over virions with low pp71: pp71HI enables desilencing and enhanced replicative fitness compared to pp71WT virions that are more likely to generate a silenced infection. In undifferentiated cells (Bottom), pp71HI virions (Right) have a “cost” as they impede viral silencing and reduce establishment of latency. In undifferentiated cells, virions with low pp71 levels promote establishment of viral silencing and latency.
Stochastic tegument packaging has both similarities and differences with other proposed bet hedging mechanisms. In a sense, the stochastic tegument packaging mechanism can be considered a phenomenological analog of early bet hedging theories of variability in seed-husk thickness (1). In influenza virus, variations in virus composition, morphology, and sequence also promote phenotypic diversity (6, 41), whereas in the lentiviridae, probabilistic establishment of proviral latency and persistence is regulated by stochastic fluctuations in viral transcription (37, 42, 43), with the HIV promoter generating very large stochastic fluctuations in gene expression. Surprisingly, the variability in virion pp71 levels (Fig. 1) is extreme, even in comparison to the variability generated by HIV (44, 45), with σ/μ and σ2/μ2 values being ∼5-fold greater. As such, stochastic tegument packaging may represent a mechanism for further amplifying phenotypic variability downstream of gene expression fluctuations. Tegument protein loads also appear variable in other herpesviruses (22–24), suggesting that stochastic packaging may be a generalized mechanism for probabilistically maximizing fitness in variant host environments and immune-stimulation conditions. Overall, approaches to modulate tegument expression levels may provide an avenue for limiting viral persistence or, alternatively, reversing viral silencing and persistence.
Materials and Methods
Virus Cloning and Purification.
Viral recombinants TB40E IE2-YFP, HCMV AD169 pp71-EYFP, pp150-EYFP, and dual-tagged TB40E-IE-mCherry-EYFP have previously been described (18, 30, 31, 46, 47)—the two-color TB40E-IE-mCherry-EYFP dual-reporter virus was cloned as an intermediate of the previously described CMDR virus (18). Viruses were propagated in MRC5 human foreskin fibroblast (HFF) cells (American Type Culture Collection; ATCC) and upon infection reaching ∼90% viral cytopathic effect or ∼90% GFP, the culture supernatant was collected and filtered through a 0.2-μm filter. HCMV pp71HI virus was generated by expanding either HCMV AD169 pp71-EYFP virus (30) or IE2-YFP virus (31) on life-extended HFFs (21) stably transduced with either a pp71-EYFP or a pp71 expression vector, respectively, as described (30).
To purify particles for single-virion imaging, viral recombinants (HCMV AD169 pp71-EYFP or pp150-EYFP, and HSV-1 Strain F VP26-EGFP) were used to synchronously infect 10 confluent 15-cm culture dishes containing either MRC5 cells (HCMV AD169) or Vero cells (HSV-1 Strain F). Supernatants were collected from infected tissue-culture plates after a single round of replication, corresponding to ∼30 h.p.i. for HSV-1, 96 h.p.i. for HCMV AD169 pp71-EYFP, and 168 h.p.i. for HCMV AD169 pp150-EYFP. Supernatants were then clarified by low-speed spin at 3,000 rpm for 10 min, and the resulting debris pellet was discarded. For HCMV purifications, the clarified supernatant was then layered over a 20% sorbitol gradient supplemented with 100 μg/mL Bacitracin. For HSV-1 purifications, supernatant was layered over a 30% sucrose gradient supplemented with 100 μg/mL Bacitracin. All overlays were centrifuged in a Beckman Coulter Optima L-80 XP Ultracentrifuge in an SW-28 rotor at 20,000 rpm and 18 °C for 1.5 h. Pellets were resuspended in 1 mL of Tris-sodium chloride buffer and overlaid on top of a glycerol tartrate gradient (48). This gradient was then centrifuged in a Beckman Coulter Optima L-80 XP Ultracentrifuge in an SW-41 rotor at 28,000 rpm and 4 °C for ∼15 min. Lower bands corresponding to the infectious virion population were harvested by sequentially removing upper layers of the gradient and isolating the appropriate band. 1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate was then added to isolated bands such that the final concentration was 10% vol/vol and allowed to incubate for 2 h protected from light at room temperature (RT). After incubation, the samples were fixed in 2% (vol/vol) formaldehyde (Tousimis Research Corporation, catalog no. 1008A diluted 1:10 in phosphate-buffer saline [PBS]).
Superresolution Imaging of Viral Particles.
All viral purifications were mounted on 3 inch × 1 inch × 1 mm microscope slides (Fisherfinest, 12-544-1) with plasma-treated 22 mm × 22 mm #1.5 coverslips (Fisherbrand, 12-541-B). A Plasma Etch PE50 unit was used to clean coverslips at 100 W for 5 min prior to mounting. Three microliters of each purification was dotted onto the microscope slide and overlaid with a plasma-treated #1.5 coverslip. Coverslips were sealed with fast-drying clear nail polish. All mounted slides were stored at 4 °C and protected from light until imaging. Viral particle purifications were imaged at super resolution using a Zeiss LSM880 Airyscan microscope using a 63× 1.4 N.A. oil-immersion DIC M27 objective (Apochromat) and laser lines for 488 nm and 561 nm. Each x-y position was sampled at 10 z planes (1.665-μm intervals) to identify the z plane where virions particles were at maximal concentration and this z was chosen for image analysis. For confocal imaging of the viral particles, a Zeiss Observer Z1 Yokagawa Spinning Disk Confocal Microscope equipped with a Yokogawa spinning disk, a CoolSNAP HQ2 14-bit camera (PhotoMetrics), and laser lines for 488 nm and 561 nm was used with a 100× 1.45 N.A. oil-immersion objective (Apocromat). Zeiss Immersol 518F immersion oil was used during imaging of all purifications (Carl Zeiss Microscopy, 444970-9000-000). Viral particle preparations were imaged under a 488-nm laser excitation (3.5-s exposure) at 20% laser power and a 531-nm laser for 300 ms at 11.5% laser power.
Superresolution Image Analysis.
Superresolution images of purified particles were analyzed using an established superresolution algorithm (26) that restricted analysis to particles 100–300 nm in diameter and fit each particle’s intensity distribution to a single Gaussian distribution to exclude doublets. Mean intensity of each particle was calculated from the fitted single-Gaussian distribution. Point-spread functions were calculated, and images were also size-standardized against fluorescently labeled size-calibration microspheres (TetraSpeck ThermoFisher) imaged under the same conditions. For parallel confocal imaging of purified viral particles, images were analyzed using a custom MATLAB script. Briefly, the algorithm is designed to first locate and separate potential virion particles from background pixels in a micrograph. Once thresholded, each of the pixels in a segmented particle was normalized to the maximum pixel intensity within that same segmented particle. An additional normalized threshold for pixel intensity of 50% was introduced in order to isolate and characterize the peak of the point spread function associated with each particle (akin to full width at half maximum [FWHM]). Once segmented and normalized, several quality control steps were implemented in order to remove objects unlikely to be single viral particles. First, a strict pixel-area size threshold of 4–11 pixels was introduced for segmented objects based upon the measured camera pixel scaling of 0.129 μm per pixel. These pixel area values were verified against size-standardized fluorescently labeled microspheres imaged under the same conditions. Following size exclusion, a series of exclusion steps based on the composition and shape of segmented particles was used. The final quality control step was achieved by identifying objects with at least 1-pixel intersections between the green and red channels. These colocalized particles were then quantified by summing pixel intensities within a colocalized viral particle to produce a cumulative sum of fluorescence intensity for an individual virion. Quantifications of viral particles from each set of micrographs were then pooled together and placed into corresponding histograms. Virion particles with fluorescence intensities ±1 SD from their respective means were excluded to remove any possible contribution from viral dense bodies or twinned viral particles.
Cell-Culture Conditions and Drug Perturbations.
HFFs, MRC5 fibroblasts, and telomerase life-extended HFFs (21) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 50 U/mL penicillin-streptomycin at 37 °C and 5% CO2 in a humidified incubator. NTera2 cells were obtained from ATCC and maintained in DMEM supplemented with 10% FBS and 50 U/mL penicillin-streptomycin at 37 °C and 5% CO2 in a humidified incubator.
Negative Staining and Electron Microscopy.
Infectious virion was purified using density gradient centrifugation as mentioned above and negative staining of virion was performed. Briefly, virion was diluted 1:100 in PBS buffer, 2 μL of virion was added on holey film TEM grid (EM Resolutions), washed three times with distilled water, and stained with 10 μL of 2% phosphotungstic acid for 1 min. Grids were subjected to electron microscopy using JEOL JEM-1230 transmission electron microscope (JEOL USA).
Viral Titering and qPCR Analysis of Viral Genome Copy Numbers.
pp71WT and pp71HI viruses were generated by expanding TB40E IE2-YFP or TB40E-IE-mCherry-EYFP virus on HFF or WF28 HFF cells that were stably transduced with a pp71 expression cassette (30), respectively. Viral preps were titered by 50% tissue culture infective dose (TCID50). For analysis of viral genome copy numbers, viral DNA was extracted from equal amounts of virus using a Nucleospin virus kit (MACHEREY-NAGEL, 740983.10). To account for potential differences in efficiency of DNA extraction, 5 ng of a reference plasmid (pNL4-3, Addgene) was spiked into each DNA preparation and used for qPCR normalization (SI Appendix, Table S1). The number of viral genomes was normalized to the reference carrier DNA level in each sample. The relative difference in genomes between the two viral preparations was then used to match the viral stocks by viral genome copy number.
Confocal Imaging and Flow Cytometry.
Confocal imaging of IE2-YFP, IE-mCherry-EYFP, and α-pp71–stained NTera2 cells was performed on an Axiovert inverted fluorescence microscope (Carl Zeiss), equipped with a Yokogawa spinning disk, a CoolSNAP HQ2 14-bit camera (PhotoMetrics), and laser lines for 488 nm and 561 nm. Flow cytometry analysis of IE2-YFP and IE-mCherry-EYFP expression for HFF, NTera2, and human primary CD14+ monocytes were performed with on an LSRII flow cytometer (BD Biosciences).
Donor-Derived Human Peripheral Blood Monocyte Isolation.
Human peripheral PBMCs were isolated from deidentified blood samples obtained from the University Medical Center at the University of Arizona, in accordance with the Institutional Review Board. Briefly, 240 mL of blood samples was centrifuged through a Ficoll Histopaque 1077 gradient (Sigma-Aldrich) at 200 × g for 30 min at RT. Mononuclear cells were collected and washed six times with sterile 0.9% sodium chloride saline solution (Baxter) to remove platelets at 200 × g for 10 min at RT. Monocytes were then layered on top of a 45% and 52.5% isosmotic Percoll gradient and centrifuged for 30 min at 400 × g at RT yielding an average of 90% monocyte population. Cells were washed three times with saline at 200 × g for 10 min at RT to remove residual Percoll and suspended in RPMI medium 1640 (Cellgro) supplemented with 5% human serum (Sigma-Aldrich), unless otherwise stated. University of Arizona Institutional Review Board and Health Insurance Portability and Accountability Act guidelines for the use of human subjects were followed for all experimental protocols in our study.
Virus Reactivation Assay.
Virus reactivation assay was performed for two different cell types. Virus reactivation assay in Ntera2 cells was performed as described previously (34) with minor modifications. Briefly, NTera2 cells were infected with pp71WT or pp71HI dual-reporter TB40E-IE-mCherry-EYFP virus at an MOI = 3, and at 4 dpi, cells were treated with trichostatin A (TSA) (100 ng/mL) (Sigma-Aldrich) and incubated for 24 h followed by washing cells three times in PBS and coculturing at 10-fold dilution with HFFs in a 96-well plate or performing TCID50 with the cell lysate. Cells were scored for YFP and mCherry at 15 d after serial dilution. To make sure there was no reactivation of virus prior to TSA treatment, cells were lysed at 4 dpi, and the lysate was added to HFFs and monitored for IE expression for 15 d (SI Appendix, Fig. S7A). Virus reactivation assay in CD14+ cells was performed as described previously with minor modifications (17). Briefly, for CD14+ cells, cells from two donors were infected with pp71WT or pp71HI dual-reporter TB40E-IE-mCherry-EYFP virus at an MOI = 2, at 10 dpi when the virus had established latency, cells were twofold serially diluted and cocultured with fibroblast in a 96-well plate with reactivation media (RPMI supplemented with 20% FBS, 100 u/mL penicillin, 100 mg/mL streptomycin, 20 ng/mL each IL3 IL6, G-CSF, GM-CSF; Sigma-Aldrich). Fibroblasts were monitored for YFP, mCherry expression for 15 d. Equal number of cells were lysed, serially diluted, and plated on HFFs as a control (SI Appendix, Fig. S7B). The frequency of reactivation was calculated by TCID50 assay and %reactivation for pp71HI was reported relative to %reactivation for pp71WT virus, which was normalized to 100%. To quantify viral genome copy number total DNA was extracted from cells and subjected to qPCR on 7900HT Fast Real-Time PCR System (catalog no: 4329003, Thermo-Fisher Scientific) using Fast SYBR Green Master Mix (catalog no: 4385612, Applied Biosystems) and normalized with β-actin using sequence specific primers (SI Appendix, Table S1).
Immunofluorescent Microscopy, Transient Transfection, and Western Blots.
For immunofluorescent microscopy, NTera2 cells were grown on glass coverslips and infected with either pp71WT or pp71HI dual-reporter TB40E-IE-mCherry-EYFP virus (MOI = 3) for 6 h, coverslips were harvested and washed with PBS (1×) (Sigma-Aldrich) three times, and fixed, permeabilized, and immunostained as described below. CD14+ cells (grown in suspension) were infected with either pp71WT or pp71HI dual-reporter TB40E-IE-mCherry, EYFP (MOI = 2). At 6 h.p.i., cells were harvested by centrifuging at 900 rpm, washed three times in PBS, fixed, permeabilized, and immunostained as mentioned below. Briefly, cells on glass coverslips (NTera2) or in suspension (CD14+) were fixed using 4% paraformaldehyde (pH 7.4) (Electron Microscopy Sciences) for 10 min on ice, washed three times with 500 mL of PBS, permeabilized using 0.1% Triton X-100 in PBS for 10 min, and immunostained. Cells were incubated with primary antibodies (pp71 antibody kindly provided by Thomas Shenk, dilution 1:100 in 0.1% BSA; Daxx antibody [MABE1911, Sigma-Aldrich], dilution 1:50 in 0.1% BSA) for 3 h at RT with gentle shaking followed by three PBS washes. Cells were incubated with secondary antibody (goat anti-mouse Alexa Fluor 405, catalog no: A-31553 [Invitrogen, Thermo Fisher Scientific], dilution 1:500 or donkey anti-mouse Alexa Fluor 647, catalog no: A-31571 [Invitrogen, Thermo Fisher Scientific], dilution: 1:500 in PBS supplemented with 0.1% BSA) for 1 h at RT, and washed three times with PBS. For samples stained for DAPI, cells were stained with DAPI (0.1 mg/mL in PBS) at RT for 10 min and imaged. Transient transfection of pp150 in HFF was performed by transfecting pp150-expressing plasmid (kindly provided by Edward Mocarski) using Amaxa Nucleofection kit V (Lonza Inc.). The pp71-expressing plasmid (pCGN-pp71) was transfected into NTera2 cells using lipofectamine-3000 transfection reagent (Invitrogen) following the manufacturer’s instructions. For Western blot analysis, infectious virus was resuspended in 20 μL of PBS buffer and analyzed by Western blot. Briefly, 5 μL of purified virion sample was added to 1× loading buffer (100 mM Tris⋅HCl [pH 6.8], 200 mM dithiothreitol, 4% sodium dodecyl sulfate [SDS], 0.1% Bromophenol blue, 20% glycerol) and boiled for 10 min at 95 °C. Samples were loaded on 12% SDS-Polyacrylamide gel (Bio-Rad), and ran at 90 V for 2 h in Tris-glycine running buffer (25 mM Tris, 250 mM glycine and 0.1% SDS). Gel was blotted on a polyvinylidene difluoride membrane (Bio-Rad) using Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad) at 20 V for 45 min, and was blocked with 10 mL of Li-Cor odyssey blocking buffer (Li-Cor Biosciences) for 1 h at RT with gentle shaking. Membrane was treated with primary antibody, anti-pp150 (1:200 dilution, mouse MAb 36–10, gift from William Britt, University of Alabama, Birmingham, AL), for 2 h at RT with gentle shaking. The membrane was then washed three times with wash buffer (1× PBS + 0.01% Tween-20), treated with secondary Li-Cor detection antibody (Li-Cor Biosciences) (1:20,000 dilution, goat anti-mouse 800CW), and incubated in dark for 1 h at RT. The membrane was washed three times with wash buffer and scanned on Li-Cor Odyssey system (Li-Cor Biosciences).
Data Availability.
All of the data are made available in the manuscript and in SI Appendix.
Supplementary Material
Acknowledgments
We thank Edward Mocarski for generously providing us with pp150 expression plasmid and Thomas Stamminger for the generous contribution of the pp71-YFP reporter virus. We thank Elena Ingerman, Melanie Ott, JJ Miranda, Marielle Cavrois, Nandhini Raman, Elizabeth Tanner, Nadia Roan, Jason Neidleman and the L.S.W. laboratory for discussions and suggestions; Melissa Teng and Jinny Wong for technical support; and Kathryn Claiborn for reviewing the manuscript. We acknowledge the Gladstone Flow Cytometry Core, funded through NIH Grant P30 AI027763. S.C. was supported in part by generous donations to the C.B.-F. Memorial Fund. We acknowledge M. Ghassemian at the University of California at San Diego for technical support. L.S.W. acknowledges support from the Bowes Distinguished Professorship, the Alfred P. Sloan Research Fellowship, and NIH Director’s New Innovator Award OD006677 and Pioneer Award OD17181 programs.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914430117/-/DCSupplemental.
References
- 1.Cohen D., Optimizing reproduction in a randomly varying environment. J. Theor. Biol. 12, 119–129 (1966). [DOI] [PubMed] [Google Scholar]
- 2.Balaban N. Q., Persistence: Mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Golding I., Infection by bacteriophage lambda: An evolving paradigm for cellular individuality. Curr. Opin. Microbiol. 43, 9–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symmons O., Raj A., What’s luck got to do with it: Single cells, multiple fates, and biological nondeterminism. Mol. Cell 62, 788–802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S., Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Vahey M. D., Fletcher D. A., Low-fidelity assembly of influenza A virus promotes escape from host cells. Cell 176, 678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaumont H. J., Gallie J., Kost C., Ferguson G. C., Rainey P. B., Experimental evolution of bet hedging. Nature 462, 90–93 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Kussell E., Leibler S., Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Jarosz D. F. et al., Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 158, 1083–1093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarosz D. F., Lancaster A. K., Brown J. C. S., Lindquist S., An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 158, 1072–1082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stumpf M. P., Laidlaw Z., Jansen V. A., Herpes viruses hedge their bets. Proc. Natl. Acad. Sci. U.S.A. 99, 15234–15237 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin E. V., Wolf Y. I., Evolution of microbes and viruses: A paradigm shift in evolutionary biology? Front. Cell. Infect. Microbiol. 2, 119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouzine I. M., Weinberger A. D., Weinberger L. S., An evolutionary role for HIV latency in enhancing viral transmission. Cell 160, 1002–1012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mocarski E. S., Shenk T., Pass R. F., Fields’ Virology: Cytomegaloviruses, (Lippincott Villiams & Wilkins, Philadelphia, 2006). [Google Scholar]
- 15.Stevens J. G., Human herpesviruses: A consideration of the latent state. Microbiol. Rev. 53, 318–332 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrum F., Reeves M., Sinclair J., High K., Shenk T., Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110, 937–945 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hargett D., Shenk T. E., Experimental human cytomegalovirus latency in CD14+ monocytes. Proc. Natl. Acad. Sci. U.S.A. 107, 20039–20044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vardi N., Chaturvedi S., Weinberger L. S., Feedback-mediated signal conversion promotes viral fitness. Proc. Natl. Acad. Sci. U.S.A. 115, E8803–E8810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogdanow B. et al., Human cytomegalovirus tegument protein pp150 acts as a cyclin A2-CDK-dependent sensor of the host cell cycle and differentiation state. Proc. Natl. Acad. Sci. U.S.A. 110, 17510–17515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B., Stinski M. F., Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66, 4434–4444 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bresnahan W. A., Shenk T. E., UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 97, 14506–14511 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Rio T., Ch’ng T. H., Flood E. A., Gross S. P., Enquist L. W., Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J. Virol. 79, 3903–3919 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohannon K. P., Jun Y., Gross S. P., Smith G. A., Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture. Proc. Natl. Acad. Sci. U.S.A. 110, E1613–E1620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke R. W. et al., Two-color fluorescence analysis of individual virions determines the distribution of the copy number of proteins in herpes simplex virus particles. Biophys. J. 93, 1329–1337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard C. J., Mehta S. B., Heintzmann R., Superresolution by image scanning microscopy using pixel reassignment. Opt. Lett. 38, 2889–2892 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Bates M., Huang B., Dempsey G. T., Zhuang X., Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffman V. C., Wu J. Q., Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 37, 499–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varnum S. M. et al., Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J. Virol. 78, 10960–10966 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bresnahan W. A., Hultman G. E., Shenk T., Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74, 10816–10818 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavalai N., Kraiger M., Kaiser N., Stamminger T., Insertion of an EYFP-pp71 (UL82) coding sequence into the human cytomegalovirus genome results in a recombinant virus with enhanced viral growth. J. Virol. 82, 10543–10555 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teng M. W. et al., An endogenous accelerator for viral gene expression confers a fitness advantage. Cell 151, 1569–1580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn J. H., Brignole E. J. 3rd, Hayward G. S., Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18, 4899–4913 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saffert R. T., Penkert R. R., Kalejta R. F., Cellular and viral control over the initial events of human cytomegalovirus experimental latency in CD34+ cells. J. Virol. 84, 5594–5604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier J. L., Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: Role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75, 1581–1593 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrier R. D., Nelson J. A., Oldstone M. B., Detection of human cytomegalovirus in peripheral blood lymphocytes in a natural infection. Science 230, 1048–1051 (1985). [DOI] [PubMed] [Google Scholar]
- 36.Rossetto C. C., Tarrant-Elorza M., Pari G. S., Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 9, e1003366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razooky B. S., Pai A., Aull K., Rouzine I. M., Weinberger L. S., A hardwired HIV latency program. Cell 160, 990–1001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrell J. E., Jr., Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140–148 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Ibanez C. E., Schrier R., Ghazal P., Wiley C., Nelson J. A., Human cytomegalovirus productively infects primary differentiated macrophages. J. Virol. 65, 6581–6588 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins-McMillen D. et al., Alternative promoters drive human cytomegalovirus reactivation from latency. Proc. Natl. Acad. Sci. U.S.A. 116, 17492–17497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell A. B., Trapnell C., Bloom J. D., Extreme heterogeneity of influenza virus infection in single cells. eLife 7, e32303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahgoub M. et al., Sporadic on/off switching of HTLV-1 Tax expression is crucial to maintain the whole population of virus-induced leukemic cells. Proc. Natl. Acad. Sci. U.S.A. 115, E1269–E1278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen M. M. K. et al., A post-transcriptional feedback mechanism for noise suppression and fate stabilization. Cell 173, 1609–1621.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pai A., Weinberger L. S., Fate-regulating circuits in viruses: From discovery to new therapy targets. Annu. Rev. Virol. 4, 469–490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen M. M. K., Desai R. V., Simpson M. L., Weinberger L. S., Cytoplasmic amplification of transcriptional noise generates substantial cell-to-cell variability. Cell Syst. 7, 384–397.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moorman N. J. et al., Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3, 253–262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saykally V. R. et al., A bioreactor method to generate high-titer, genetically stable, clinical-isolate human cytomegalovirus. Bio Protoc. 7, e2589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talbot P., Almeida J. D., Human cytomegalovirus: Purification of enveloped virions and dense bodies. J. Gen. Virol. 36, 345–349 (1977). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are made available in the manuscript and in SI Appendix.