Abstract
The recently emerged coronavirus designated as SARS-CoV-2 (also known as 2019 novel coronavirus (2019-nCoV) or Wuhan coronavirus) is a causative agent of coronavirus disease 2019 (COVID-19), which is rapidly spreading throughout the world now. More than 1.21 million cases of SARS-CoV-2 infection and more than 67,000 COVID-19-associated mortalities have been reported worldwide till the writing of this article, and these numbers are increasing every passing hour. The World Health Organization (WHO) has declared the SARS-CoV-2 spread as a global public health emergency and admitted COVID-19 as a pandemic now. Multiple sequence alignment data correlated with the already published reports on SARS-CoV-2 evolution indicated that this virus is closely related to the bat severe acute respiratory syndrome-like coronavirus (bat SARS-like CoV) and the well-studied human SARS coronavirus (SARS-CoV). The disordered regions in viral proteins are associated with the viral infectivity and pathogenicity. Therefore, in this study, we have exploited a set of complementary computational approaches to examine the dark proteomes of SARS-CoV-2, bat SARS-like, and human SARS CoVs by analysing the prevalence of intrinsic disorder in their proteins. According to our findings, SARS-CoV-2 proteome contains very significant levels of structural order. In fact, except for nucleocapsid, Nsp8, and ORF6, the vast majority of SARS-CoV-2 proteins are mostly ordered proteins containing less intrinsically disordered protein regions (IDPRs). However, IDPRs found in SARS-CoV-2 proteins are functionally important. For example, cleavage sites in its replicase 1ab polyprotein are found to be highly disordered, and almost all SARS-CoV-2 proteins contains molecular recognition features (MoRFs), which are intrinsic disorder-based protein–protein interaction sites that are commonly utilized by proteins for interaction with specific partners. The results of our extensive investigation of the dark side of SARS-CoV-2 proteome will have important implications in understanding the structural and non-structural biology of SARS or SARS-like coronaviruses.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03603-x) contains supplementary material, which is available to authorized users.
Keywords: SARS coronavirus, Intrinsically disordered proteins, Molecular recognition features, Nucleotide-binding regions, Coronavirus disease 2019
Introduction
The emerging coronavirus disease 2019 (COVID-19) is a recent pandemic which has been recently declared as a public health emergency by the World Health Organization (WHO). Since its first appearance in visitors of the Wuhan’s seafood and meat market, China, reported in December 2019, COVID-19 has now a large-scale socioeconomic impact [1]. According to WHO, till 6 April 2020, the infection has spread over to at least 170 countries and territories, where there have been more than 1.21 million confirmed cases, with more than 67,000 deaths due to COVID-19. One should also keep in mind that these data on the COVID-19 spread and related casualties are rapidly becoming outdated, almost with the speed of typing of these sentences [2].
According to the International Committee on Taxonomy of Viruses (ICTV), SARS-CoV-2 comes under the coronavirinae sub-family of coronaviridae family of order nidovirales. Viruses of the nidovirales order are enveloped, non-segmented positive-sense, single-stranded RNA viruses [3]. The family coronaviridae comprises of vertebrate infecting viruses that transmit horizontally, mainly through the oral/fecal route and cause gastrointestinal and respiratory problems to the host [4]. Sub-family coronavirinae consists of four genera, namely: alpha, beta, gamma, and delta coronaviruses based on the phylogenetic clustering of viruses [5, 6]. Coronavirinae having the largest genomes among the RNA viruses incorporate their ~ 30 kb genomes inside an enveloped capsid [7].
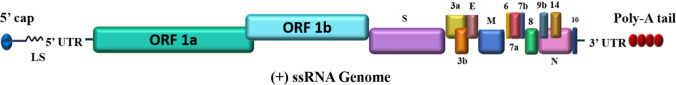
SARS-coronavirus genomic RNA includes a 5′ cap, leader sequence, UTR, a replicase gene, genes for structural and accessory proteins, 3′ UTR, and a poly-A tail (Fig. 1). Two-third of the genome codes for the replicase polyproteins (~ 20 kb) containing all non-structural viral proteins, while the remaining part of the genome (~ 10 kb) contains genes for accessory proteins interspersed between the genes responsible for coding structural proteins [7, 8]. The ~ 20 kb (replicase gene) ssRNA is translated first into two long polyproteins: replicase polyprotein 1a and 1ab inside host cells. The newly formed polyproteins, after cleavage by two viral proteases, result in 16 non-structural proteins (Nsps) that perform a wide range of functions for viruses inside the host cell [9, 10]. The genomic sequence of SARS-CoV-2 is reported to have 29,903 nucleotides with GenBank accession number NC_045512 [11].
Fig. 1.
Genome architecture of SARS-CoV-2. As a positive single-stranded RNA virus, SARS-CoV-2 contains a 5′ capped RNA which has a leader sequence (LS), poly-A tail at the 3′ end, and 5′ and 3′ UTR. It contains the following genes: ORF1a, ORF1b, spike (S), ORF3a, ORF3b, envelope (E), membrane (M), ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF14, nucleocapsid (N), and ORF10
In this study, we analysed the dark side of SARS-CoV-2 proteome (i.e. a part of a proteome that includes proteins or protein regions, which are not amenable to experimental structure determination by existing means and inaccessible to homology modeling), to better understand an interplay between the ordered and disordered components of the proteome. According to the “heretic” viewpoint of the “presence of functional intrinsic disorder in proteins”, a noticeable amount of biologically active proteins (of protein regions) fail to fold into the well-defined structures and instead remain disordered, existing as highly dynamic ensembles of rapidly interconverting conformations under the physiological conditions. These proteins and protein regions are known now as intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs), respectively. The propensity of being functional intrinsically disordered proteins (similar to the propensity of forming unique biologically active structures of ordered proteins) is determined by the amino acid sequences [12–14]. IDPs exhibit their biological functions in numerous biological processes commonly associated with cellular signalling, gene regulation, and control by interacting with their physiological partners [15–19]. These functions of IDPs and IDPRs are regulated by their protein–protein, protein–RNA, and protein–DNA interactions [20, 21]. Molecular recognition features (MoRFs) are the regions in IDPs implicated in the regulation of IDP function by protein–protein interactions and serve as the primary stage in molecular recognition.
It is known that the IDPs/IDPRs are present in all three kingdoms of life, and viral proteins often contain unstructured regions that have been strongly correlated with their virulence [22–25]. In this report, we have investigated the disordered side of SARS-CoV-2 proteome using a complementary set of computational approaches to check the prevalence of IDPRs in its proteins and to shed some light on their disorder-related functions. We also have comprehensively analysed IDPRs among the closely related viruses, human SARS and bat SARS-like CoVs. Furthermore, we have also identified protein functions related to protein–protein interactions, RNA binding, and DNA binding from all three viruses. Since these three viruses are closely related, our study provides an important means for a better understanding of the sequence and structural peculiarities of their evolution.
Materials and methods
Sequence retrieval and multiple sequence alignment
The protein sequences of bat CoV (SARS-like) and human SARS CoV were retrieved from UniProt (UniProt IDs for individual proteins are listed in Table 1). The translated sequences of SARS-CoV-2 proteins [GenBank database [26] (Accession ID: NC_045512.2)] were obtained from GenBank. We used these sequences for performing multiple sequence alignment (MSA) and predicting the IDPRs. We have used Clustal Omega [27] for protein sequence alignment and Esprit 3.0 [28] for constructing the aligned images.
Table 1.
Mean PPID scores of structural and accessory proteins from SARS-CoV-2, human SARS, and bat CoVs
| Proteins | SARS-CoV-2 | Human SARS-CoV | Bat CoV (SARS-like) | |||
|---|---|---|---|---|---|---|
| Length of protein (NCBI RefSeq accession ID) | Mean PPID | Length of protein (UniProt ID) | Mean PPID | Length of protein (UniProt ID) | Mean PPID | |
| Spike glycoprotein (S) |
1273 |
1.41 |
1255 (P59594) |
1.12 |
1242 (Q3LZX1) |
1.85 |
| Envelope (E) |
75 |
5.33 |
76 (P59637) |
6.58 |
76 (Q3LZW9) |
6.58 |
| Membrane (M) |
222 |
2.70 |
221 (P59596) |
1.36 |
221 (Q3LZX9) |
1.36 |
| Nucleocapsid (N) |
419 |
64.91 |
422 (P59595) |
71.09 |
421 (Q3LZX4) |
65.80 |
| ORF3a |
275 |
9.09 |
274 (P59632) |
8.76 |
274 (Q3LZX0) |
6.20 |
| ORF3b | 22* | 0 |
154 |
7.14 | 39* | 23.08 |
| ORF6 |
61 |
22.95 |
63 (P59634) |
20.63 |
63 (Q3LZX8) |
20.63 |
| ORF7a |
121 |
1.65 |
122 (P59635) |
0.82 |
122 (Q3LZX7) |
0.82 |
| ORF7b |
43 |
9.30 |
44 (Q7TFA1) |
4.55 |
44 (Q3LZX6) |
4.55 |
| ORF8 |
121 |
0.00 |
ORF8a–39 (Q7TFA0) |
2.56 |
121 (Q3LZX5) |
0.00 |
|
ORF8b–84 (Q80H93) |
2.38 | |||||
| ORF9b | 97* | 10.31 |
98 (P59636) |
26.53 |
97 (Q3LZX3) |
9.28 |
| ORF10# |
38 |
0.00 | – | – | – | – |
| ORF 14 | 73* | 0.00 |
70 (Q7TLC7) |
2.86 | 70* | 0.00 |
#No sequence similarity was found for this protein on performing BLAST
*These sequences are based on genome annotations conducted by Wu et al. [45]
Per-residue predictions of intrinsic disorder predisposition
For the prediction of the intrinsic disorder predisposition of CoV proteomes, we used multiple predictors, such as members of the PONDR® (Predictor of Natural Disordered Regions) family including PONDR®VLS2 [29], PONDR®VL3 [30], PONDR®FIT [31], and PONDR® VLXT [32], as well as the IUPred platform for predicting long (≥ 30 residues) and short IDPRs (< 30 residues) [33]. These computational tools predict residues/regions, which do not have the tendency to form an ordered structure. Residues with disorder scores exceeding the threshold value of 0.5 are considered as intrinsically disordered residues, whereas residues with the predicted disorder scores between 0.2 and 0.5 are considered flexible. Complete predicted percent of intrinsic disorder (PPID) in a query protein was calculated for every protein of all the three viruses from outputs of six predictors. The detailed methodology has been given in our previous reports [34, 35].
Combined CH–CDF analysis to predict disorder predisposition of proteins
The charge hydropathy plot [36] and PONDR® VLXT-based cumulative distribution function are two binary predictors of disorder (i.e. tool evaluating entire protein as mostly ordered or mostly disordered), which are available on the PONDR web server (https://www.pondr.com). Combining the result from these binary predictors helps to classify the proteins into different groups, depending on their global disorder [37].
Molecular recognition feature (MoRF) determination in CoV proteomes
The authentic online bioinformatics predictors that use a different set of algorithms for the prediction of MoRFs were used. These include MoRFchibi_web [38], ANCHOR [39, 40], MoRFPred [41], and DISOPRED3 [42]. The protein residues with ANCHOR, MoRFPred, and DISOPRED3 scores above the threshold value of 0.5 and MoRFchibi_web score above the threshold value of 0.725 are considered MoRF regions.
Identification of DNA- and RNA-binding regions in CoV proteomes
Often, IDPs and IDPRs facilitate interactions with RNAs and DNAs and regulate many cellular functions [43]. Thus, for predicting the DNA-binding residues in CoV proteins, we used two online servers: DRNAPred [20] and DisoRDPbind [43]. For RNA-binding residues, we used PPRInt (Prediction of Protein RNA- Interaction) [44] and DisoRDPbind servers [43].
Results and discussion
Comprehensive computational analysis of intrinsic disorder in structural and accessory proteins of SARS-CoV-2, human SARS and bat CoV (SARS-like)
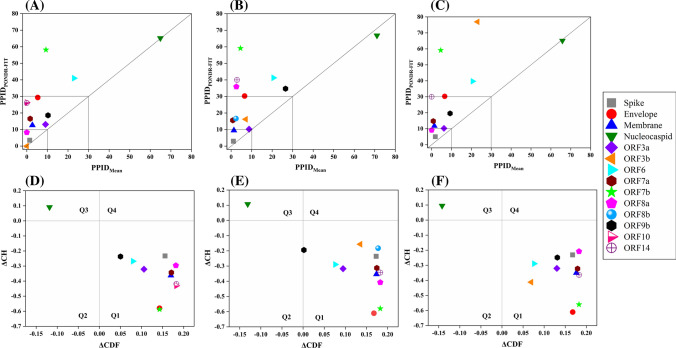
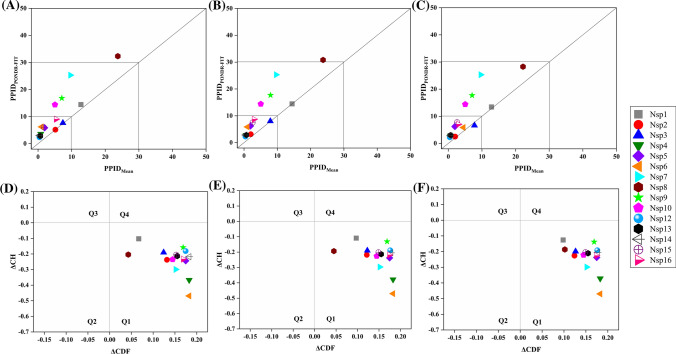
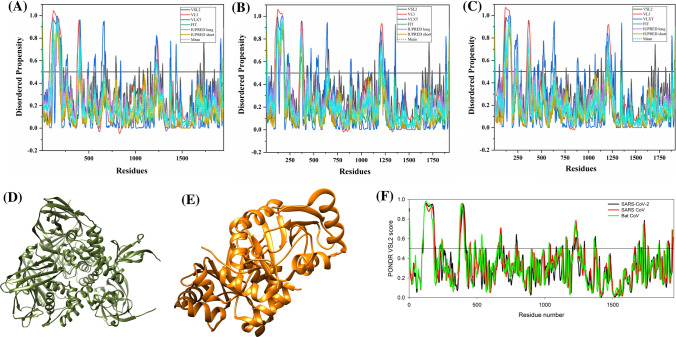
The mean values of the predicted percentage of intrinsic disorder scores (mean PPIDs) that were obtained by averaging the predicted disorder scores from six disorder predictors (Tables S1–S3) for structural and accessory proteins of SARS-CoV-2 as well as human SARS, and bat CoV are represented in Table 1.
Figure 2a–c are 2D-disordered plots generated for SARS-CoV-2, human SARS and bat CoV proteins, respectively, and represent the PPIDPONDR-FIT vs. PPIDMean plots. Based on their predicted levels of intrinsic disorder, proteins can be classified as highly ordered (PPID < 10%), moderately disordered (10% ≤ PPID < 30%) and highly disordered (PPID ≥ 30%) [46]. From the data in Table 1, Fig. 2a–c, as well as the PPID based classification, we conclude that the nucleocapsid protein from all three CoVs possess the highest percentage of disorder and, therefore, is classified as a highly disordered protein. The ORF3b protein in bat CoV, ORF6 protein of all three CoVs, and ORF9b proteins of SARS-CoV-2 and bat CoV belongs to the class of moderately disordered proteins. While the structured proteins, namely, spike glycoprotein (S), envelope protein (E) and membrane protein (M) as well as accessory proteins ORF3a, ORF7a, ORF8 (ORF8a and ORF8b in case of human SARS) of all three strains of CoVs are ordered proteins. ORF14 and ORF10 proteins are also ordered proteins.
Fig. 2.
Analysis of overall disorder status of proteins of SARS-CoV-2, human SARS, and bat CoV (SARS-like): 2D plots representing PPIDPONDR-FIT vs. PPIDMean for a SARS-CoV-2, b human SARS and c bat CoV. In the CH–CDF plot of the proteins of d SARS-CoV-2, e human SARS and f bat CoV, the Y coordinate of each protein spot signifies the distance of corresponding protein from the boundary in the CH plot and X coordinate value corresponds to the average distance of CDF curve for respective protein from the CDF boundary
To further investigate the nature of the disorder in proteins of all three CoVs, we utilized the combined CH–CDF tool that uses the outputs of two binary classifiers of disorder: charge hydropathy (CH) plot and cumulative distribution function (CDF) plot. This helped in retrieving more detailed characterization of the global disorder predisposition of query proteins and their classification according to the disorder “favors”. The CH plot is a linear classifier that differentiates between proteins that are predisposed to extended disordered conformations which includes random coils and pre-molten globules from proteins that have compact conformations (ordered proteins and molten globule-like proteins). The other binary predictor, CDF, is a nonlinear classifier that uses PONDR®VLXT scores to discriminate ordered globular proteins from all disordered conformations, which include native molten globules, pre-molten globules, and random coils. The CH–CDF plot can be divided into four quadrants: Q1 (bottom right quadrant) containing ordered proteins; Q2 (bottom left quadrant) includes proteins predicted to be disordered by CDF and compact by CH (i.e. native molten globules and hybrid proteins containing high levels of both ordered and disordered regions); Q3 (top left quadrant) contains proteins that are predicted to be disordered by both CH and CDF analysis (i.e. highly disordered proteins with the extended disorder); and Q4 (top right quadrant) possesses proteins disordered according to CH but ordered according to CDF analysis [34]. Figure 2d–f represent the CH–CDF analysis of proteins of SARS-CoV-2, human SARS, and bat CoV and shows that all the proteins are located within the two quadrants Q1 and Q2. The CH–CDF analysis leads to the conclusion that all proteins of all three CoVs are ordered except nucleocapsid protein, which is predicted to be disordered by CDF as well as CH and hence lies in Q3.
Molecular recognition features (MoRFs) are short interaction-prone disordered regions found within IDPs/IDPRs that commence a disorder-to-order transition upon binding to their partners [47, 48]. In this study, we have analysed and compared MoRFs (protein-binding regions) in SARS-CoV-2 with human SARS and bat CoVs. The results of this analysis are summarized in Table 2, which clearly shows that most of the SARS-CoV-2 proteins contain at least one MoRF. This is indicative of an important role played by disorder in functionality of these viral proteins. All of the SARS-CoV-2 proteins have been predicted to contain MoRFs except ORF7b and Nsp13 proteins. MoRFs in human SARS and bat CoV proteomes are listed in Tables S7 and S8. Similar to SARS-CoV-2 proteome, bat CoV proteins ORF7b, and Nsp13 are not predicted to have any MoRF by any of the servers used. In human SARS proteome, proteins ORF7b, Nsp13, Nsp2, and Nsp15 do not show the presence of any MoRF. Interestingly, the N protein from SARS-CoV-2, human SARS, and bat CoV shows high number of variable MoRFs, signifying its central role in virus pathogenesis.
Table 2.
Predicted MoRFs regions in SARS-CoV-2 proteins
| Proteins | MoRFchibi_web | ANCHOR | MoRFPred | DISOPRED3 |
|---|---|---|---|---|
| Spike glycoprotein (S) | 1265–1272 | – | 2–6, 819–823 | 1–10 |
| Envelope (E) | 46–75 | – | – | 26–30 |
| Membrane (M) | 187–221 | – | 214–222 | 117–132 |
| Nucleocapsid (N) | 1–20, 82–98, 102–118, 403–409 | 1–75, 104–113, 152–185, 201–236, 264–310, 347–361, 388–419 | 11–21, 48–58, 106–115, 218–228, 268–274, 354–358, 397–408 | 1–10,26–47,179–208,230–249,366–378,380–385,413–419 |
| ORF3a | 1–6, 261–268 | – | 7–12, 126, 230–233, 259–263 | 1–19 |
| ORF3b | NA | – | NA | – |
| ORF6 | 14–21, 26–61 | – | – | – |
| ORF7a | 29–34, 39–51, 77–89 | – | – | 1–10 |
| ORF7b | – | – | – | – |
| ORF8 | 1–5, 26–52, 69–91 | – | – | 1–10 |
| ORF9b | 3–29, 34–62, 79–84, | – | 89–97 | – |
| ORF10 | 3–7 | – | – | – |
| ORF14 | 1–45 | – | 6–12 | – |
| Nsp1 | 64–75, 93–97, 99–179 | 173–180 | 137–145, 172–179 | 1–6 |
| Nsp2 | 1–7 | – | – | – |
| Nsp3 | – | 105–171, 189–196 | 101–107, 184–194, 536–541, 980–984 | – |
| Nsp4 | – | – | 495–500 | 1–25 |
| Nsp5 | 3–8 | – | – | – |
| Nsp6 | – | – | – | 1–19 |
| Nsp7 | 1–30, 39–58, 65–83 | – | – | – |
| Nsp8 | 181–185 | – | 89–93 | – |
| Nsp9 | 40–44, 51–56, 62–77, 83–102 | – | – | – |
| Nsp10 | 25–32, 91–99, 133–138 | – | 11–18 | – |
| Nsp12 | 31–36 | – | 11–15 | 1–6 |
| Nsp13 | – | – | – | – |
| Nsp14 | – | – | 8–13, 441–445 | – |
| Nsp15 | – | – | 9–13 | – |
| Nsp16 | – | – | 151–156 | – |
Nucleotide-binding propensity in proteins of coronaviruses
In addition to protein–protein interactions/protein-binding functions, IDPs and IDRs also mediate functions by facilitating their interactions with nucleotides (DNA and RNA) [21, 49]. Therefore, we have used a combination of two different online servers for locating protein residues that show the propensity to bind with DNA as well as RNA. The nucleotide-binding residues in proteins of the three studied coronaviruses are listed in Tables S9–S11. Interestingly, all the viral proteins of SARS-CoV-2, human SARS, and bat CoV have shown the propensity to bind to nucleic acids. In particular, structural (S, M, and N proteins) and non-structural (Nsp 2, 3, 4, 5, 6, 12, 13, 14, 15, and 16) proteins of all three viruses display a large number of RNA-binding residues. However, ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, and ORF14 proteins show less RNA-binding and more DNA-binding residues.
Intrinsic disorder analysis of structural proteins of coronaviruses
Coronaviruses encode four structural proteins, namely, spike (S), envelope (E) glycoprotein, membrane (M), and nucleocapsid (N), which are translated from the last ~ 10 kb nucleotides and forms the outer cover of the CoVs, encapsulating their single-stranded genomic RNA.
Spike (S) glycoprotein
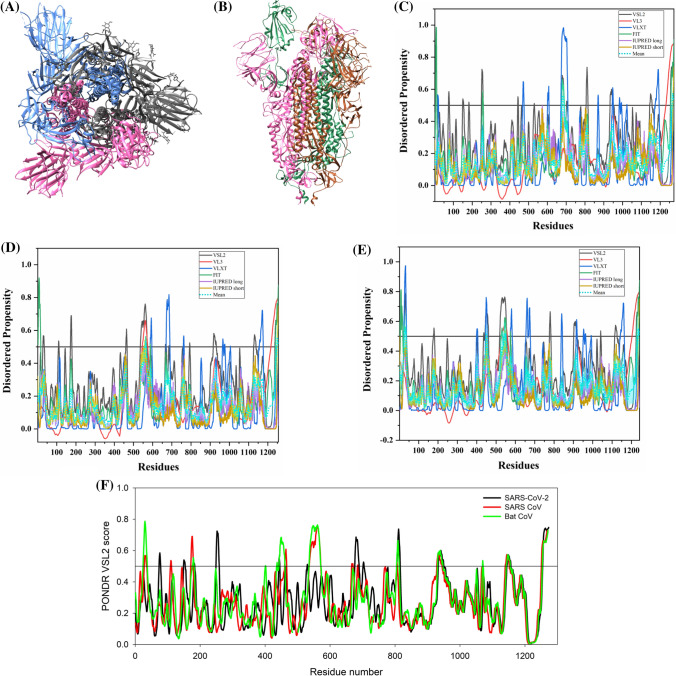
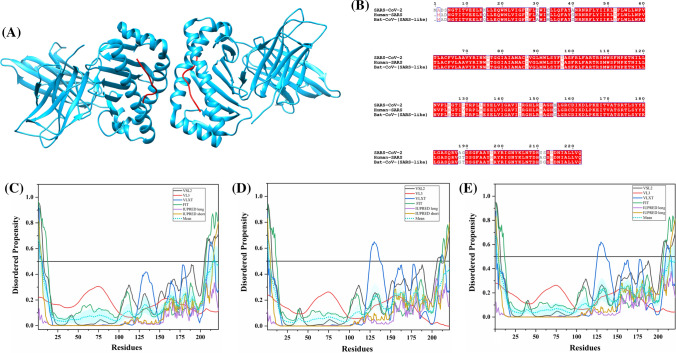
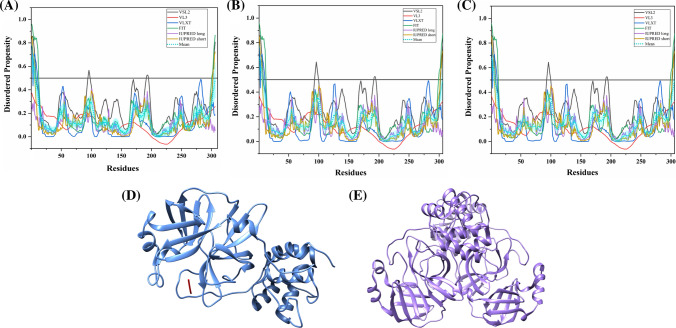
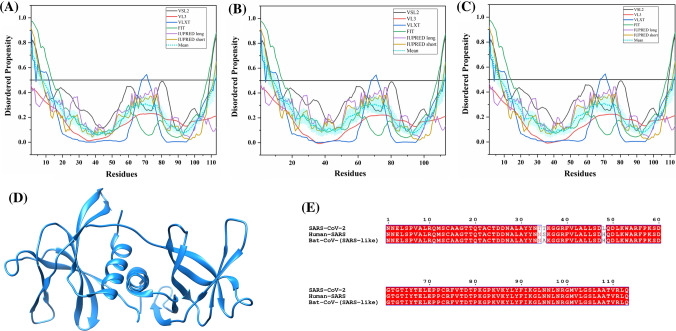
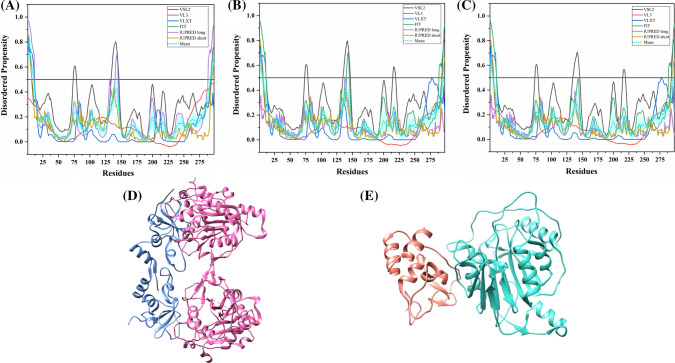
The S protein is a large multifunctional protein forming the exterior of the CoV particles [50, 51]. It forms surface homotrimers and contains two distinct ectodomain known as S1 and S2. Subunit S1 initiates viral infection by binding to the host cell receptors, while S2 acts as a class I viral fusion protein that mediates the fusion of the virion and cellular membranes and thereby promotes the viral entry into the host cells [52, 53]. It binds to specific surface receptor angiotensin-converting enzyme 2 (ACE2) on host cell plasma membrane through its N-terminal receptor-binding domain (RBD) [54].
S protein consists of an N-terminal signal peptide, a long extracellular domain, a single-pass transmembrane domain, and a short intracellular domain [55]. A 3.60 Å resolution structure (PDB ID: 6ACC) of human SARS S protein complexed with its host-binding partner ACE2 is obtained using cryo-electron microscopy (cryo-EM) (Fig. 3b). In this PDB structure, few residues (1–17, 240–243, 661–673, 812–831 and 1120–1203) are missing [56], suggesting their flexible nature. Also, the structure of S protein (3.5 Å) from SARS-CoV-2 has been recently deduced by Wrapp et al. using electron microscopy (PDB ID: 6VSB) [57] (Fig. 3a). In this structure, residues 1–26, 67–78, 96–98, 143–155, 177–186, 247–260, 329–334, 444–448, 455–490, 501–502, 621–639, 673–686, 812–814, 829–851, 1147–1288 are observed to be missing, again corresponding to the high conformational flexibility regions. Biophysical analysis of SARS-CoV-2 S protein has revealed a higher binding affinity with ACE2 receptor than S protein from human SARS [57].
Fig. 3.
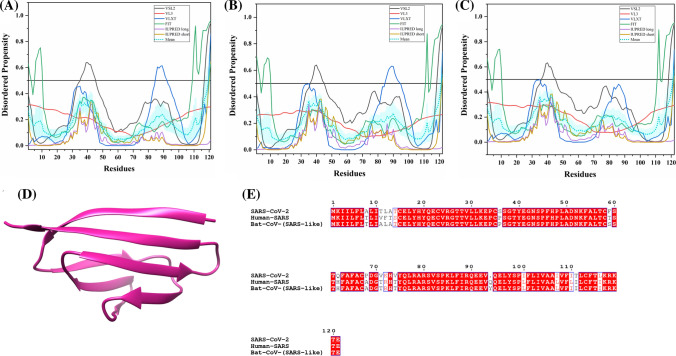
Structure and intrinsic disorder propensity of spike glycoprotein (S) from CoVs. a A 3.50 Å resolution structure (PDB ID: 6VSB) of SARS-CoV-2 S obtained through cryo-EM. This homotrimeric structure includes three chains, A (pink), B (dark grey), and C (turquoise). b A 3.6 Å resolution cryo-EM structure (PDB ID: 6ACC) of human SARS S protein complexed with its host-binding partner ACE2. In this structure, three chains are present: A (pink), B (green) and C (dark khaki). Evaluation of intrinsic disorder predisposition in S proteins of c SARS-CoV-2, d human SARS, and e bat CoVs. Graphs c–e depict the disorder profiles generated using six predictors: PONDR® VSL2 (black line), PONDR® VL3 (red line), PONDR® VLXT (blue line), PONDR® FIT (green line), IUPred long (purple line) and IUPred short (golden line). The mean disorder propensity calculated by averaging the disorder scores from all predictors is represented by a short dotted line (sky blue) in graphs. The light sky blue shadow region signifies the mean error distribution. f Aligned disorder profiles generated for all three S proteins is based on the outputs of the PONDR® VSL2
MSA analysis among all three coronaviruses demonstrates that the S protein of SARS-CoV-2 has a 77.71% sequence identity with bat CoV and 77.14% identity with human SARS (Fig. S1). As observed, there is a significant sequence variation in RBD located at the N-terminal region which might affect its virulence, receptor-mediated binding and entry into the host cell.
According to our intrinsic disorder propensity analysis, the S protein from all three CoVs are found to be highly structured (Table 1). The mean PPID scores of SARS-CoV-2, human SARS, and bat CoVs are calculated to be 1.41%, 1.12%, and 1.85%, respectively. Figure 3c–e represent the intrinsic disorder profiles of S proteins from SARS-CoV-2, human SARS and bat CoV obtained from six disorder predictors. Finally, Fig. 3f shows aligned disorder profiles of S proteins from these CoVs and illustrates remarkable similarity in their disorder propensity, especially in the C-terminal region.
It is of interest to map known functional regions of S proteins to their corresponding disorder profiles. The maturation of S protein requires specific post-translational modifications (PTM), proteolytic cleavage that happens at two stages. First, host cell furin or another cellular protease nicks the S precursor to generate S1 and S2 proteins, whereas the second cleavage takes place after the viral attachment to host cell receptors which leads to the release of a fusion peptide generating the S2′ subunit. In human SARS, the first and second cleavage site are located at residues R667 and R797, respectively, whereas in bat CoV, the corresponding cleavage sites are residues R654 and R784. As it follows from Fig. 3, these cleavage sites are located within the IDPRs. In human SARS S protein, fusion peptide (residues 770–788) located within a flexible region is characterized by a mean disorder score of 0.232 ± 0.053. Similarly, in bat CoV S protein, fusion peptide (residues 757–775) has a mean disorder score of 0.320 ± 0.046. It contains two heptad repeat regions that form coiled-coil structure during viral and target cell membrane fusion, assuming a trimer-of-hairpins structure needed for the functional positioning of fusion peptide. In human SARS, heptad repeat regions are formed by residues 902–952 and 1145–1184, which have mean disorder scores of 0.458 ± 0.067 and 0.353 ± 0.062, respectively. The analogous situation is observed for S protein of bat CoV, where these heptads repeat regions are positioned at residues 889–939 (0.44 ± 0.11) and 1132–1171 (0.353 ± 0.062). Another functional region found in S proteins is the RBD (residues 306–527 and 310–514 in human SARS and bat CoVs, respectively) containing a receptor-binding motif responsible for interaction with human ACE2. In S protein of human SARS, this motif (residues 424–494) is not only characterized by structural flexibility, possessing a mean disorder score of 0.30 ± 0.16, but also contains a disordered region (residues 461–466). Since S protein is known as spike glycoprotein, it contains numerous glycosylation sites. Due to rather close similarity of disorder profiles of S proteins analysed here, we can assume that all the aforementioned indications of the functional importance of disorder and flexible regions in S proteins from human SARS and bat CoVs are also applicable to SARS-CoV-2 S protein. Finally, Table 2 shows that S protein from SARS-CoV-2 contains a MoRF region at its C-terminal (residues 1265–1272) as predicted by MoRFchibi_web, two MoRF regions ((residues 2–6, 819–823) by MoRFPred, and one MoRF region at the N-terminal (residues 1–10) by DISOPRED3. These results indicate that intrinsic disorder is important for its interaction with binding partners. Strikingly, the N-terminal region of S protein (residues 1–10) from all three viruses are predicted to be a MoRF by two servers (MoRFPred and DISOPRED3). This displays its role in viral interaction with host receptor, while the C-terminal MoRF is engaged in interaction with M protein for assembly of viral particles [58]. Moreover, MoRF regions lying in the N- and C-terminal regions suggest their possible role during cleavage as well. In addition to protein-binding regions, S protein also shows many nucleotide-binding residues. Tables S9–S11 shows numerous RNA-binding residues predicted by PPRInt in all three viruses. Further, DRNApred and DisoRDPbind predicted the presence of many DNA binding residues in all three S proteins. These results signify the molecular recognition (protein–protein interaction, RNA binding, and DNA binding) and interactions with host cell membrane and further viral infection. Therefore, IDPs/IDPRs and residues/regions in S proteins that are crucial for molecular recognition can be targeted for disorder-based drug discovery.
Envelope (E) small membrane protein
Envelope (E) protein is a small, multifunctional membrane protein that plays an important role in the assembly and morphogenesis of virions in the cell [59–61]. It consists of two ectodomains associated with N- and C-terminal regions, and a middle transmembrane domain. It homo-oligomerizes into a pentameric membrane destabilizing transmembrane hairpins to form a pore necessary for its ion channel activity [62]. Figure 4a shows the NMR-structure (PDB ID: 2MM4) of human SARS envelope glycoprotein of 8–65 residues [63].
Fig. 4.
Analysis of structural features and intrinsic disorder predisposition of envelope glycoprotein (E). a NMR solution structure (PDB ID: 2MM4) of human SARS E protein (residues 8–65). b Multiple sequence alignment (MSA) profile of all three E proteins. Graphs c–e represent the intrinsic disorder profiles of E proteins of c SARS-CoV-2, d human SARS, and e bat CoVs. Color schemes are similar to given in Fig. 3
MSA results illustrate (Fig. 4b) that this protein is highly conserved, with only three amino acid substitutions in E protein of SARS-CoV-2 conferring its 96% sequence similarity with human SARS and bat CoV. Also, bat CoV shares 100% sequence identity with human SARS. Mean PPID calculated for SARS-CoV-2, human SARS, and bat CoV E proteins are 5.33%, 6.58%, and 6.58%, respectively (Table 1). The E protein is found to have a reasonably well-predicted structure; however, residues of N- and C-terminals display a higher tendency for the disorder (disorder profiles in Fig. 4c–e). Evidences show that the last 18 hydrophilic residues (residues 59–76) adopt a random-coil conformation with and without the addition of lipid membranes [64]. Further, the last four amino acids of the C-terminal region containing a PZD-binding motif are involved in protein–protein interactions with a tight junction protein PALS1. PALS1 is involved in maintaining the polarity of epithelial cells in mammals [65]. Our results support the existing literature, as we identified a long N-terminal region of ~ 30 residues as a MoRF region in all three viruses (see Tables 2, S7, S8). We speculate that the disordered regions may facilitate interactions with other proteins as well. In agreement with this hypothesis, the C-terminal domain of SARS-CoV-2 E protein serves as a protein-binding region. We also found that residues from 45–75 is a long MoRF in E proteins of all three viruses as predicted by MoRFchibi_web. As aforementioned, these randomly coiled binding residues at the C-terminus may gain structure while assisting the protein–protein interaction mediated by E protein. One more MoRF region (residues 26–30) in the transmembrane domain was observed by DISOPRED3 in all three E proteins. Since these residues are part of the ion channel, they may be involved in guiding the specific function of ion channel activity. Few nucleotide-binding residues are predicted for all three E proteins (Tables S9–S11).
Membrane (M) glycoprotein
It plays an important role in virion assembly by interacting with the nucleocapsid (N) and E proteins [66–68]. Protein M interacts specifically with coronavirus RNA containing a short viral packaging signal in the absence of N protein, highlighting an important nucleocapsid-independent viral RNA packaging mechanism inside the host cells [69]. Cryo-EM and tomography data reveal its two distinct conformations, a compact structure having high flexibility and low spike density, and an elongated M protein having a rigid structure and narrow range of membrane curvature [70]. Although no structural information is available for full-length M protein, a short peptide of the membrane glycoprotein (residues 88–96) from human SARS is co-crystallized with a complex of A-2 α chain of the HLA class I histocompatibility antigen and β2-microglobulin (PDB ID:3I6G) [71]. Figure 5a shows the extended conformation of M protein.
Fig. 5.
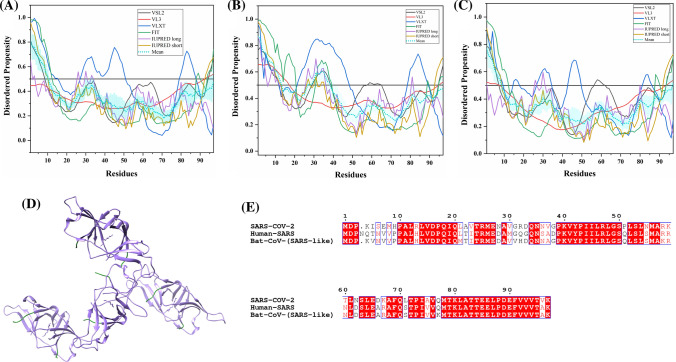
Analysis of intrinsic disorder propensity of membrane glycoprotein (M). a A 2.20 Å resolution crystal structure (PDB ID: 3I6G) of human SARS M protein (residues 88–96) in complex with A-2 α chain of HLA class I histocompatibility antigen and β2-microglobulin. Chains in this dimer corresponding to M are shown in red, while A-2 α chain and β2-microglobulin complex are shown using ice blue colour. b MSA profile of all three M proteins. Graphs c–e represent intrinsic disorder profiles of M protein of c SARS-CoV-2, d human SARS, and e bat CoV. Color schemes are similar to those given in Fig. 3
The M protein of SARS-CoV-2 has a sequence similarity of 90.1% with bat CoV and 89.6% with human SARS M proteins (Fig. 5b). Disorder profiles in Fig. 5c–e show a relatively low level of disorder in M proteins of SARS-COV-2 (2.70%), human SARS CoV (1.36%,), and bat CoV (1.36%). This is consistent with a previous publication by Goh et al. on human SARS HKU4, where they found the mean PPID of 4% using additional predictors such as TopIDP and FoldIndex along with the predictors used in our study [72]. The last 20 residues of MERS-CoV M protein are important for intracellular trafficking and contain a determinant that localizes it into the Golgi network [73]. MoRF analysis revealed that the disordered C-tail of M protein contains a MoRF region which can serve as a binding site for its partner required during localization inside the host cell. A long MoRF region (residues 186–220) at the C-terminal of M protein in all three viruses is located by MoRFchibi_web. Two MoRF regions [one at N-terminus (residues 1–16) and one at the C-terminus (residues 205–221)] are predicted by DISOPRED3 in human SARS and bat CoV. However, only a single MoRF (residues 117–132) is observed in SARS-CoV-2 (by DISOPRED3) (Tables 2, S7, S8). Furthermore, the M protein from all three viruses displays strong tendency to bind with RNA (as predicted by PPRInt and DisoRDPbind) and DNA (as predicted by DRNApred and DisoRDPbind) (see Tables S9–S11). Our understanding of M protein of CoVs (IDPs and MoRF at C-terminus) elucidates its critical role in interaction with the N and E proteins for viral assembly.
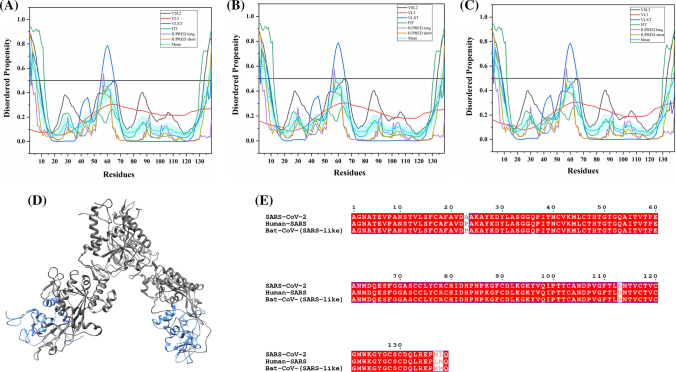
Nucleocapsid (N) protein
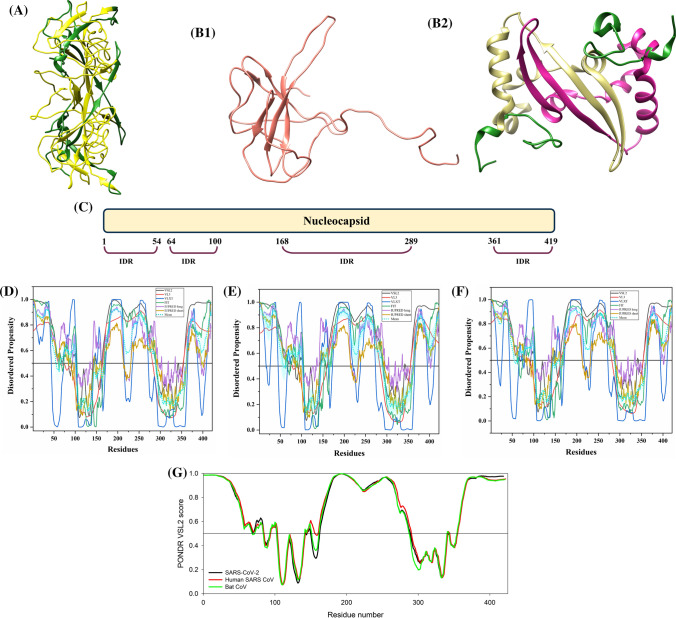
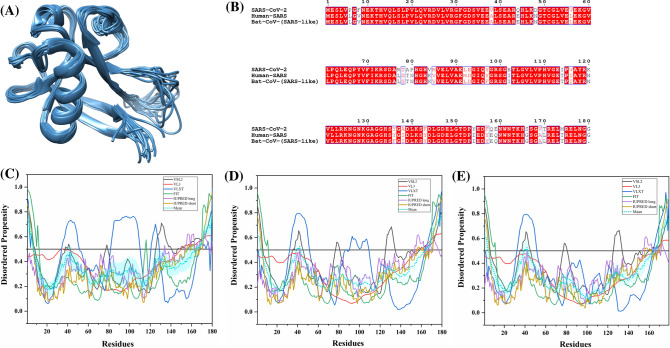
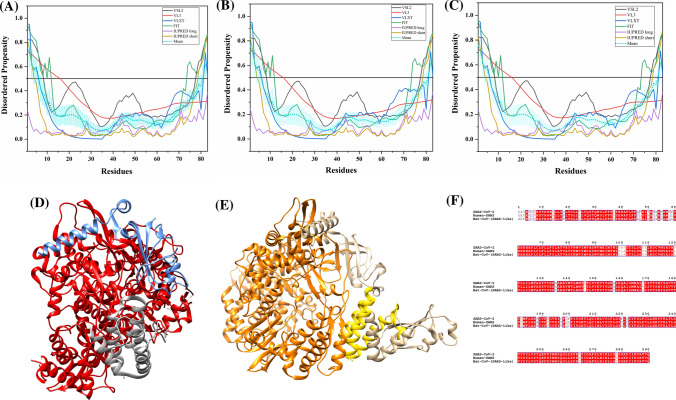
It is one of the major viral proteins playing an essential role during transcription, and virion assembly of CoVs [74]. It binds to viral genomic RNA forming a ribonucleoprotein core required for RNA encapsidation during viral particle assembly [75]. It consists of two structural domains, the N-terminal RNA-binding domain (NTD: 45–181 residues) and the C-terminal dimerization domain (CTD: 248–365 residues) with a disordered patch between these domains. It is demonstrated to bind with viral RNA using both NTD and CTD [76]. Recently, residues 50–173 of the N protein of SARS-CoV-2 has been crystallized (PDB ID: 6VYO) (Fig. 6a). Figure 6b1 displays the NMR solution structure of NTD (45–181 residues) of human SARS N protein (PDB ID: 1SSK) [77]. Figure 6b2 shows an X-ray crystal structure of CTD of human SARS N protein (270–366 residues) (PDB ID: 2GIB) [78]. A model of domain organization of N-protein from SARS-CoV-2 is shown in Fig. 6c.
Fig. 6.
Analysis of the structural properties and intrinsic disorder propensity of the nucleocapsid (N) protein. a 1.70 Å resolution structure (PDB ID: 6VYO) of RNA-binding domain of SARS-CoV-2 N obtained using X-ray diffraction. Residues 64–100 are found to be disordered which are represented with forest green colour. b1 NMR solution structure (PDB ID: 1SSK) of the NTD (residues 45–181) of human SARS N. b2 X-ray diffraction-based crystal structure (PDB ID: 2GIB) of CTD (residues 270–366) of human SARS N. The structure is a homodimer of chains A (violet-red) and B (dark khaki). Residues 270–289 and 362–366 showing disorder propensity are represented using forest green colour. c Representation of predicted disordered regions in SARS-CoV-2 N protein. Graphs d–f shows the intrinsic disorder profiles of N protein of d SARS-CoV-2, e human SARS, and f bat CoV. g Aligned disorder profiles generated for all three N proteins is based on the outputs of PONDR® VSL2. Color schemes are similar to given in Fig. 3
The 419 amino acid-long N protein of SARS-CoV-2 shows a percentage identity of 88.76% and 89.74% with N proteins of bat and human SARS CoVs (Fig. S2). Our analysis revealed the highest levels of intrinsic disorder in N proteins of all three CoVs (graphs in Fig. 6d–f), which is in accordance with the previously evaluated intrinsic disorder predisposition [72]. In fact, N proteins from SARS-CoV-2, human SARS, and bat CoVs are characterized by the mean PPIDs of 64.91%, 71.09%, and 65.80%, respectively. This is further supported by Fig. 6g, where PONDR® VSL2-generated disorder profiles of these three proteins are overlapped to show almost complete coincidence of their major disorder-related features. In particular, SARS-CoV-2 N protein residues 1–57, 64–102, 145–162, 166–289, and 362–422 are found to be disordered (Fig. 6d). Many of these residues lie within the NTD and CTD regions, which due to their structural plasticity does not get crystallized in human SARS N protein crystal structure. Overall, all three N proteins are found to be highly disordered.
Tables 2, S7 and S8 show that the N protein is heavily decorated with MoRFs, suggesting that this protein is a promiscuous binder. Long disorder-based protein bonding regions at the N- and C- terminus of the N protein of all three viruses are observed by all four predictors. The N protein from human SARS has one phosphorylation site (residue S177) and several regions with compositional biases, such as Ser-rich (residues 181–213), Poly-Leu, Poly-Gln, and Poly-Lys (residues 220–225, 240–245, and 370–376), all predicted to be disordered. Similarly, the N protein of bat CoV, S176 is phosphorylated and has Ser-rich, Poly-Leu, and Poly-Lys regions (residues 176–206, 219–224, and 369–375, respectively), all of which are disordered. It has been reported to interact using the central disordered region with M protein, hnRNP A1, and self-N–N interaction [79–81]. The middle flexible region is also responsible for its RNA-binding activity [82]. Deletion of 184–196 residues, 169–308 residues, and 161–210 residues of N abolishes its multimerization, RNA-binding capacity, and hnRNP A1 interactions, respectively. The MoRFs present in the aforementioned regions may mediate these interactions of N proteins. Figure 6b2 represents another important disorder-related functional feature of N protein. CTD homodimer shown is characterized by highly intertwined morphology, which is typically a result of binding-induced folding [83–85], indicating that a very significant part of CTD gains structure during dimerization. We identified numerous RNA-binding residues in all three viruses using PPRInt server. This finding supports the function of N protein as it interacts with genomic RNA for a ribonucleoprotein core formation, which is a crucial step for RNA encapsidation. Additionally, DRNApred and DisoRDPbind predict multiple DNA-binding residues in N protein of all the studied CoVs. The flexible (IDPRs) regions at the N- and C-terminus of SARS-CoV-2 have long protein-binding as well as nucleotide-binding regions that may play a vital role in its interaction with viral RNA. These flexible regions can be targeted to inhibit the interaction of N protein with viral genomic RNA.
Intrinsic disorder analysis of accessory proteins of coronaviruses
Literature suggests that some viral proteins are translated from the genes interspersed in between the genes of structural proteins. These proteins are known as accessory proteins, and many of them are proposed to be involved in viral pathogenesis [86].
Proteins ORF3a and ORF3b
ORF3a is a multifunctional protein (of molecular weight ~ 31 kDa) that performs a major function during virion assembly by co-localizing with E, M, and S viral proteins [87–91]. The homo-tetrameric complex of ORF3a has been demonstrated to form a potassium-ion channel on the host cell plasma membrane [92]. ORF3b protein can be found in the cytoplasm, nucleolus, and outer membrane of mitochondria of the host cells [93, 94]. In Huh 7 cells, its over-expression has been linked with the activation of AP-1 via the ERK and JNK pathways [95].
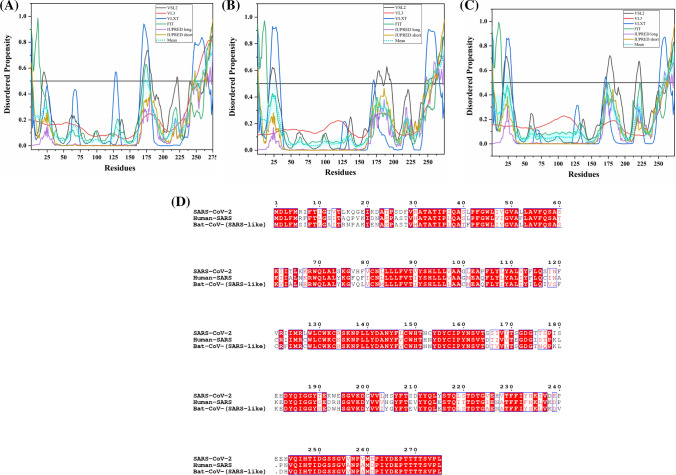
On performing MSA (Fig. 7d), we found that ORF3a protein of SARS-CoV-2 is almost equally closer to ORF3a proteins of bat (73.36%) and human SARS CoV (72.99%). The graphs in Fig. 7a–c depict the propensity for disorder in ORF3a proteins of novel SARS-CoV-2, human SARS, and bat CoVs, respectively (mean PPIDs are listed in Table 1). SARS-CoV-2 ORF3a shows protein-binding regions at its N-terminus (by MoRFchibi_web (residues 1–6), MoRFPred [residues 7–12), and DISOPRED3 (residues 1–19)] and at the C-terminus (by MoRFchibi_web [residues 261–268) and MoRFPred (residues 259–263)] (Table 2). Similarly, ORF3a of human SARS and bat CoV also have MoRFs at the N- and C-terminus as predicted by MoRFchibi_web and MoRFPred (Tables S7, S8). These protein-binding regions in ORF3a may have a role in its co-localization with E, M, and S viral proteins. In conjunction with MoRFs, ORF3a proteins have a maximum number of nucleotide-binding residues among all accessory proteins.
Fig. 7.
Analysis of intrinsic disorder propensity of ORF3a protein. Graphs a–c represent intrinsic disorder profiles of ORF3a protein of a SARS-CoV-2, b human SARS, and c bat CoV. d MSA profile of all three ORF3a proteins. Color schemes are similar to those given in Fig. 3
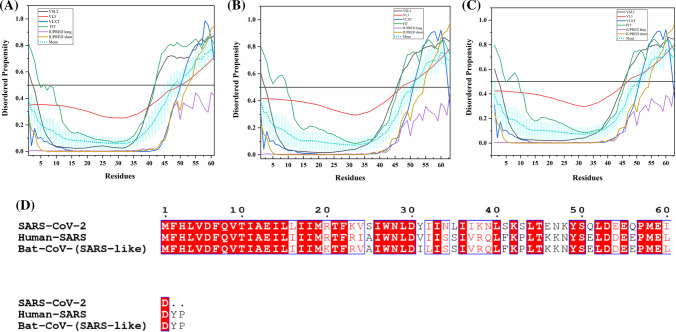
Mean PPID values of ORF3b proteins of SARS-CoV-2, human SARS, and bat CoV are 0%, 7.1%, and 23.1% respectively, as represented in Fig. 8a–c. MSA results (Fig. 8d) demonstrate that ORF3b of SARS-CoV-2 is little evolutionarily closer to ORF3b proteins of human SARS and bat CoV, having a sequence similarity of only 54.6% and 59.1%, respectively. As we can see in Table 2, there is not a single MoRF located in SARS-CoV-2 ORF3b. However, for human SARS, MoRFchibi_web server has identified three MoRFs (residues 32–37, 41–70, and 125–153), whereas, for bat CoV, a single MoRF at N-terminus is observed (residues 1–38).
Fig. 8.
Analysis of intrinsic disorder propensity of ORF3b protein. Graphs a–c represent intrinsic disorder profiles of ORF3b protein of a SARS-CoV-2, b human SARS, and c bat CoV. d MSA profile of all three ORF3b proteins. Color schemes are similar to those given in Fig. 3
Protein ORF6
Also known as P6, this membrane-associated protein serves as an interferon (IFN) antagonist [96]. Using its C-terminal residues, ORF6 disrupts karyopherin import complex in cytosol and, therefore, hampers the movement of transcription factors like STAT1 into the nucleus resulting in downregulation of the IFN pathway [96, 97]. It contains a YSEL motif near its C-terminal region that functions in protein internalization from the plasma membrane into the endosomal vesicles [98].
MSA results demonstrate that (Fig. 9d), SARS-CoV-2 ORF6 is closer to human SARS ORF6, having a sequence similarity of 68.85% than to bat CoV ORF6 (67.21%). Novel SARS-CoV-2 ORF6 is predicted to be the second most disordered structural protein with a PPID of 22.95%, containing a disordered C-terminal region.
Fig. 9.
Analysis of intrinsic disorder propensity of ORF6 protein. Graphs a–c represent the intrinsic disorder profiles of ORF6 protein of a SARS-CoV-2, b human SARS, and c bat CoV. d MSA profile of all three ORF6 proteins. Colour schemes are similar to those given in Fig. 3
The mean PPID of the other two ORF6 proteins are listed in Table 1. Graphs in Fig. 9a–c illustrate that all three ORF6 proteins are moderately disordered with the presence of high disorder near C-terminal residues. As aforementioned, this hydrophilic region contains lysosomal targeting motif (YSEL) and diacidic motif (DDEE) responsible for its binding and recognition during translocation [98], this region is important for the biological activities of ORF6. Moreover, the N-terminus does not contain any prominent disorder. First, 38 amino acids of human SARS ORF6 are described to form an α-helical structure spanning the membrane [99]. A long MoRF region [(residues 26–61 in SARS-CoV-2), (residues 31–63 in Human SARS), and (residues 30–60 in bat Cov)] is also present near the C-terminus. It represents very few RNA- and DNA-binding residues.
ORF7a and ORF7b proteins
ORF7a is a type I transmembrane protein [100, 101]. It contributes to viral pathogenesis by activating the release of pro-inflammatory cytokines and chemokines, such as IL-8 and RANTES [102, 103]. The presence of a KRKTE motif near the C-terminal region is needed for its import from ER to Golgi apparatus [100, 101]. On the other hand, ORF7b is an integral membrane protein that has been shown to localize in the Golgi complex [104, 105]. These reports also confirm the role of ORF7b as an accessory as well as a structural protein in SARS-CoV virion [104, 105].
Figure 10d represents the 1.8 Å X-ray crystal structure of the 14–96 fragment of the ORF7a from human SARS (PDB ID: 1XAK) and demonstrates the compact seven β-stranded topology of this protein similar to the Ig-superfamily members [106]. Importantly, in this crystal structure, residues 82–96 constitute the region with missing electron density, indicating the highly dynamic nature of this segment. In line with this hypothesis, NMR solution structure of the 16–99 fragment of ORF7a of human SARS (PDB ID: 1YO4) showed that residues 81–99 are highly disordered [107].
Fig. 10.
Analysis of intrinsic disorder propensity of ORF7a protein. Graphs a–c represent the intrinsic disorder profiles of ORF7a protein of a SARS-CoV-2, b human SARS, and c bat CoV. d A 1.8 Å resolution X-ray diffraction-based structure (PDB ID:1XAK) of human SARS ORF7a protein (residues 14–96) is illustrated using pink colour. e MSA profile of all three ORF7a proteins. Color schemes are similar to those given in Fig. 3
We found that 121 residues-long ORF7a protein of SARS-CoV-2 shares 89.26% and 85.95% sequence identity with ORF7a proteins of bat CoV and human SARS, respectively (Fig. 10e). In contrast, SARS-CoV-2 ORF7b is found to be closer to human SARS ORF7b (81.40%) than bat CoV ORF7b (79.07%) (see Fig. S3D).
As observed from Table 1 and Fig. 10a–c, our disorder predisposition analyses resulted in the overall PPID values for ORF7a proteins—1.65% (SARS-CoV-2), 0.82% (bat CoV), and 0.82% (Human SARS). The mean PPIDs estimated for ORF7b proteins are 9.30% for SARS-CoV-2, 4.55% for bat CoV and 4.55% human SARS. Table 2 shows the presence of several MoRFs in ORF7a, indicating its potential involvement in disorder-dependent protein–protein interactions. At the N-terminus, one MoRF region (residues 1–10) is predicted by DISOPRED3 in all three ORF7a proteins. In addition to protein-binding regions, ORF7a also contains several RNA- and DNA-binding residues. Analysis also reveals the low disorder content in all three ORF7b proteins (Fig. S3A–C), and subsequently no MoRFs. Although ORF7b does not contain protein-binding regions, it has many nucleotide (both RNA and DNA)-binding residues. Figures S3A, 3B, and 3C depict the residues predisposed for disorder in ORF7b proteins of SARS-CoV-2, human SARS CoV, and bat CoV, respectively. In particular, both proteins in all three studied viruses have ordered structures.
Proteins ORF8a and ORF8b
In isolates from early human infections, the ORF8 gene codes for a single ORF8 protein. However, in late infections, more specifically, at middle and late stages, a 29 nucleotide deletion in the ORF8 gene led to the formation of two distinct proteins, ORF8a and ORF8b containing 39 and 84 residues, respectively [108, 109]. Both proteins have conformations different from that of the longer ORF8 protein and interacts with different structural proteins [110]. The disorder-based protein-binding regions of this protein identified in this study may have an important role in interaction with other proteins.
ORF8 protein found in early SARS-CoV-2 isolates having 121 residues shares a 90.05% sequence identity with bat CoV ORF8 (Fig. S4C). Furthermore, Figs. S4A and S4B illustrates the absence of intrinsic disorder in both ORF8 proteins. Therefore, these two proteins are predicted to be completely structured (mean PPID of 0.00%). In ORF8a and ORF8b proteins of the human SARS, the predicted disorder is estimated to be 2.56% and 2.38%, respectively (Table 1). Graphs in Figs. S5A and 5B illustrate the presence of some disorder near the N- and C-terminals of ORF8a and ORF8b proteins. Table 2 shows three MoRF regions (residues 1–5, 26–52, and 69–91) by MoRFchibi_web and one MoRF region (residues 1–10) by DISOPRED3 in SARS-CoV-2 ORF8. Bat CoV has four protein-binding regions (residues 26–53, 70–91, 98–104, and 113–130) identified by MoRFchibi_web server (Table S8). Furthermore, in human SARS, the N-terminus of both ORF8a (residues 1–39) and ORF8b (residues 1–83) is predicted to be MoRF by the MoRFchibi_web server (Table S7). In addition with protein-binding regions, ORF8, ORF8a and ORF8b proteins contain many nucleotide-binding residues (Tables S9–S11).
ORF9b protein
This protein is expressed from an alternative ORF within the N gene through a leaky ribosome-binding process [111]. This protein is shown to interact with a nuclear export protein receptor Exportin 1 (Crm1), using which it's translocated out of the nucleus [112]. Our MoRFs analysis shows the presence of disorder-based protein-binding regions in ORF9b protein which may have a role in its interaction with Crm1 for translocation outside the nucleus. A 2.8 Å resolution crystal structure of ORF9b protein from human SARS CoV (PDB ID: 2CME) shows the presence of a dimeric tent-like β-structure along with the central hydrophobic amino acids (Fig. 11d) [113].
Fig. 11.
Analysis of intrinsic disorder propensity of ORF9b protein. Graphs a–c represent intrinsic disorder profiles of ORF9b protein of a SARS-CoV-2, b Human SARS, and c bat CoV. d A 2.8 Å resolution crystal structure (PDB ID: 2CME) of human SARS ORF9b protein. The structure includes four ORF9b homodimers where chains A–H are shown in purple colour and disordered residues (1–10) are depicted in green. e MSA profile of all three ORF9b proteins. Color schemes are similar to those given in Fig. 3
Based on the sequence availability (accession ID NC_045512.2), translated protein sequence of ORF9b is not reported for SARS-CoV-2. However, based on a report by Wu and colleagues [45], corresponding annotated sequence is used for intrinsic disorder analysis. According to the MSA (results shown in Fig. 11e), ORF9b protein from SARS-CoV-2 shares 73.2% identity with human SARS and 74.23% identity with bat CoV.
Our IDP analysis (Table 1) exposed moderate disorder content in ORF9b of human SARS having a mean PPID of 26.53%. As depicted in Fig. 11a–c, disorder in human SARS ORF9b protein mainly lies near the N-terminal end (residues 1–10) and near the central region (residues 28–40) with a well-ordered inner core. The X-ray crystal structure of ORF9b has a missing electron density of first 8 residues and 26–37 residues near the central region. This indicates that the corresponding regions are disordered, which are difficult to crystallize due to their highly dynamic structural organization. SARS-CoV-2 ORF9b with a mean PPID of 10.31% also has an N-terminal (1–10 residues) disordered segment. ORF9b of bat CoV is shown to have an intrinsic disorder content of 9.28%, comparatively lower than the other two ORF9b proteins. MoRFs lies in the N-terminal region of ORF9b proteins of all three viruses (Tables 2, S7, S8). In the absence of other viral proteins, its first 41 residues are demonstrated to induce membranous structures similar to DMVs [99]. The available crystal structure also has a missing electron density in the N-terminal region suggesting that these flexible amino acids are likely to interact with host lipids. The 3–29 amino acid segment of SARS-CoV-2 is identified as disorder-based protein-binding region that may mediate its interaction with host lipids for the formation of DMVs.
ORF10 protein
The newly emerged SARS-CoV-2 has an ORF10 protein of 38 amino acids. ORF10 of SARS-CoV-2 has a 100% sequence similarity with ORF10 of bat CoV strain bat-SL-CoVZC45 [11]. However, we did not conduct the disorder analysis for ORF10 from the bat-SL-CoVZC45 strain, since all our studies reported here are related to a different strain of bat CoV (reviewed strain HKU3-1). Therefore, we have only reported the results of disorder analysis for the ORF10 protein from SARS-CoV-2, according to which this protein has a mean PPID of 0.00% (see also Fig. S6 for disorder profile of ORF10). This protein contains a MoRF from three to seven residues at its N-terminus as predicted by MoRFchibi_web. Further, we predicted its binding tendency to nucleotides and found the presence of few RNA-binding sites; however, it does not contain DNA-binding residues.
Protein ORF14
This is a 70 amino acid long uncharacterized protein of unknown function. According to the MSA, ORF14 of SARS-CoV-2 has 77.1% identity with human-SARS and 72.9% identity with bat CoV as represented in Fig. S7D. Figure S7A–C shows the resulting disorder profiles of all three ORF14 proteins (mean PPIDs are listed in Table 1). Further, these proteins have calculated mean PPID values of 0.00%, 2.86%, and 0.00%, respectively. These proteins have flexible N- and C-terminal regions. It can use intrinsic disorder or structural flexibility for protein–protein interactions since it possesses MoRFs. It mainly contains MoRFs at the N- and C-terminal regions (Tables 2, S7, S8) and several RNA- and DNA-binding residues (Tables S9–S11). These regions indicate its vital role in protein function related to protein–RNA and protein–DNA interaction.
Intrinsic disorder analysis of non-structural proteins of coronaviruses
In coronaviruses, due to ribosomal leakage during translation, two-third of the RNA genome is processed into two polyproteins: (i) replicase polyprotein 1a and (ii) replicase polyprotein 1ab. Both contains non-structural proteins (Nsp1-10) in addition to different proteins required for viral replication and pathogenesis. Replicase polyprotein 1a contains an additional Nsp11 protein of 13 amino acids, the function of which has not been investigated yet. The longer replicase polyprotein 1ab of 7073 amino acids accommodates five other non-structural proteins (Nsp12-16) [114, 115].
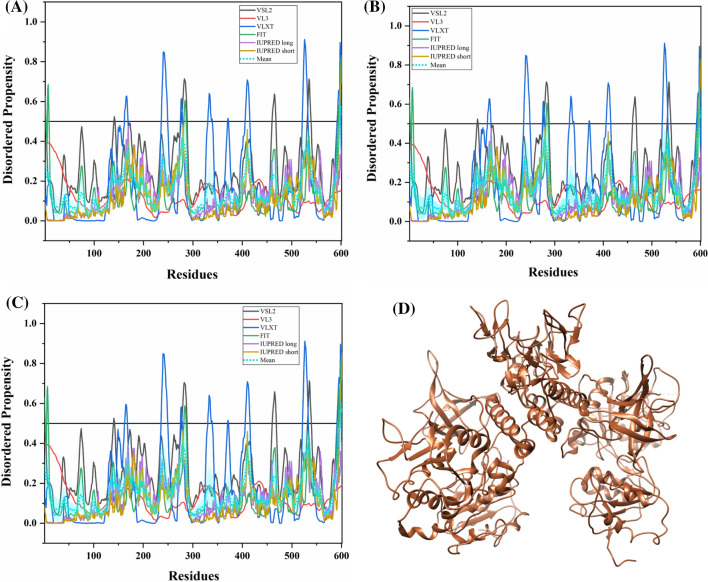
Global analysis of intrinsic disorder in the replicase polyprotein 1ab
Table 3 represents the mean PPID scores of 15 Nsps derived from the replicase polyprotein 1ab in SARS-CoV-2, human SARS, and bat CoV. These values were obtained by combining the results from six disorder predictors (see Tables S4–S6). Figure 12a–c represents 2D-disordered plots of the Nsps coded by ORF1ab in SARS-CoV-2, Human SARS, and bat CoV, respectively. Based on the mean PPID scores in Table 3, Fig. 12a–c, and taking into PPID based classification [46], we conclude that none of the Nsps in SARS-CoV-2, human SARS, and bat CoV are highly disordered. Only Nsp1 and Nsp8 proteins are found to be moderately disordered (10% ≤ PPID ≤ 30%). We also observed that Nsp2, Nsp3, Nsp5, Nsp6, Nsp7, Nsp9, Nsp10, Nsp15, and Nsp16 have less than 10% disordered residues and hence, belong to the category of mostly ordered proteins. Other non-structural proteins, namely, Nsp4, Nsp12, Nsp13, and Nsp14 have negligible levels of disorder (PPID < 1%) and are concluded to be highly structured.
Table 3.
Evaluation of the mean predicted percentage disorder in non-structural proteins of novel SARS-CoV-2, human SARS, and bat CoV
| Proteins | SARS-CoV-2 | Human SARS (UniProt ID: P0C6X7) | Bat CoV (SARS-like) (UniProt ID: P0C6W2) | |||
|---|---|---|---|---|---|---|
| Length of protein (residues) (NCBI RefSeq accession ID) | Mean PPID | Length of protein (residues) | Mean PPID | Length of protein (residues) | Mean PPID | |
| Nsp1 (host translation inhibitor) |
180 (1–180) |
12.78 |
180 (1–180) |
14.44 |
179 (1–179) |
12.85 |
| Nsp2 |
638 (181–818) |
5.17 |
638 (181–818) |
2.04 |
639 (180–818) |
2.03 |
| Nsp3 (papain-like proteases) |
1945 (819–2763) |
7.40 |
1922 (819–2740) |
7.91 |
1916 (819–2734) |
7.78 |
| Nsp4 |
500 (2764–3263) |
0.80 |
500 (2741–3240) |
0.60 |
500 (2735–3234) |
0.60 |
| Nsp5 (3CL-Pro) |
306 (3264–3569) |
1.96 |
306 (3241–3546) |
1.96 |
306 (3235–3540) |
1.96 |
| Nsp6 |
290 (3570–3859) |
1.03 |
290 (3547–3836) |
1.03 |
290 (3541–3830) |
1.03 |
| Nsp7 (multimeric RNA polymerase) |
83 (3860–3942) |
9.64 |
83 (3837–3919) |
9.64 |
83 (3831–3913) |
9.64 |
| Nsp8 (multimeric RNA polymerase) |
198 (3943–4140) |
23.74 |
198 (3920–4117) |
23.74 |
198 (3914–4111) |
22.22 |
| Nsp9 |
113 (4141–4253) |
7.08 |
113 (4118–4230) |
7.96 |
113 (4112–4224) |
7.08 |
| Nsp10 |
139 (4254–4392) |
5.04 |
139 (4231–4369) |
5.04 |
139 (4225–4363) |
5.04 |
| Nsp12 (RNA-directed RNA polymerase, RDRP) |
932 (4393–5324) |
0.43 |
932 (4370–5301) |
0.43 |
932 (4364–5295) |
0.43 |
| Nsp13 (viral helicase) |
601 (5325–5925) |
0.67 |
601 (5302–5902) |
0.67 |
601 (5296–5896) |
0.67 |
| Nsp14 (guanine-N7 methyltransferase/ExoN-nsp14) |
527 (5926–6452) |
0.38 |
527 (5903–6429) |
0.38 |
527 (5897–6423) |
0.57 |
| Nsp15 (uridylate-specific endoribonuclease/NendoU) |
346 (6453–6798) |
1.73 |
346 (6430–6775) |
2.60 |
346 (6424–6769) |
2.60 |
| Nsp16 (2′-O-methyltransferase) |
298 (6799–7096) |
5.37 |
298 (6776–7073) |
3.02 |
298 (6770–7067) |
3.02 |
Fig. 12.
Analysis of overall intrinsic disorder status of non-structural proteins (Nsps): 2D plot representing PPIDPONDR-FIT vs PPIDMean in a SARS-CoV-2 b human SARS and c bat CoV. In CH–CDF plot of the proteins of d SARS-CoV-2 e human SARS and f bat CoV, the Y coordinate of each protein spot signifies the distance of the corresponding protein from the boundary in the CH plot and the X coordinate value corresponds to the average distance of the CDF curve for the respective protein from the CDF boundary
The CH–CDF analysis of Nsps from SARS-CoV-2, human SARS and bat CoV is depicted in Fig. 12d–f respectively. It was observed that all Nsps of the three CoVs are located within the quadrant Q1 of the CH–CDF phase space, which is indicative of their ordered structure.
Replicase polyprotein 1ab
The longer replicase polyprotein contains 15 Nsps listed in Table 3. Nsp1, Nsp2, and Nsp3 are cleaved using a viral papain-like proteinase (Nsp3/PL-Pro), while the rest of the Nsps are cleaved by another viral 3C-like proteinase, Nsp5/3CL-Pro.We mapped the cleavage sites of the replicase 1ab polyprotein from human SARS CoV to the disorder profile of this polyprotein. Figure 13 represents the results of this analysis by showing zoomed-in regions surrounding all the cleavage sites with few residues spanning at both terminals. Interestingly, we observed that all the cleavage sites are largely disordered, suggesting that intrinsic disorder may have a crucial role in the maturation of individual non-structural proteins. As Nsps of human SARS are evolutionarily closer to Nsps of SARS-CoV-2, we hypothesize that cleavage sites in the SARS-CoV-2 replicase 1ab polyprotein are also intrinsically disordered or flexible. To shed more light on other implications of IDPRs, the structural and functional properties of Nsps and their predicted IDPRs are thoroughly described below.
Fig. 13.
Intrinsic disorder at the cleavage sites of the replicase 1ab polyprotein of human SARS. Plots a–n denotes the cleavage sites (magenta coloured bar for PL-Pro protease and grey coloured bar for 3CL-Pro protease) in relation to disordered regions present between the individual proteins (Nsp1-16) of replicase 1ab polyprotein of human SARS. All proteins are represented by different colored horizontal bars
Non-structural protein 1 (Nsp1)
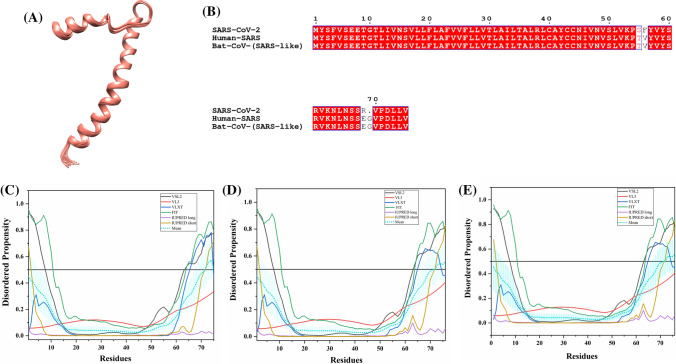
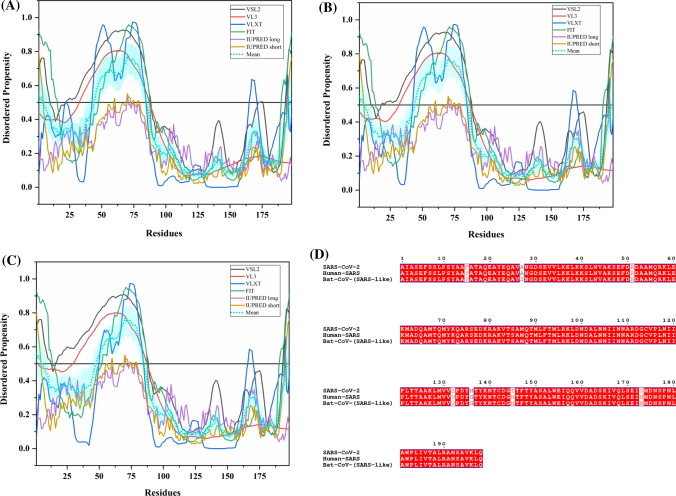
This protein acts as a host translation inhibitor as it binds to the 40S subunit of ribosome and blocks the translation of cap-dependent mRNAs as well as mRNAs that uses the internal ribosome entry site (IRES) [116]. Figure 14a shows the NMR solution structure (PDB ID: 2GDT) of human SARS Nsp1 protein (13–128 residues) [117].
Fig. 14.
Analysis of intrinsic disorder propensity of non-structural protein 1 (Nsp1). a NMR solution structure (PDB ID: 2GDT) of 13–128 residue fragment of human SARS Nsp1. b MSA profile of all three Nsp1 proteins. Graphs c–e represent the intrinsic disorder profiles of Nsp1 protein of c SARS-CoV-2, d Human SARS, and e bat CoV. Color schemes are similar to those given in Fig. 3
SARS-CoV-2 Nsp1 shares 84.44% and 83.80% sequence identity with Nsp1s of human SARS and bat CoV, respectively (Fig. 14b). The respective mean PPIDs of Nsp1s from SARS-CoV-2, Human SARS, and bat CoV are 12.78%, 14.44%, and 12.85% (disorder profiles in Fig. 14c–e). In particular, the following regions are predicted to be disordered: SARS-CoV-2 (residues 1–7 and 165–180), human SARS (residues 1–5 and 165–180), and bat CoV (residues 1–5 and 165–179). NMR solution structure of Nsp1 from human SARS revealed the presence of two unstructured segments near the N-terminal (1–12 residues) and C-terminal (129–179 residues) regions [117]. The disordered region (128–180 residues) at the C-terminus is already mapped important for its expression [118]. Based on sequence homology with human SARS Nsp1, the predicted disordered C-terminal region of SARS-CoV-2 Nsp1 may play a critical role in its expression. Alanine mutants at K164 and H165 near the C-terminal region are reported to abolish its binding with the 40S subunit of the host ribosome [119]. In conjunction with this data, several MoRFs are present in the unstructured segments of Nsp1 proteins. These regions are shown in Tables 2, S7 and S8.
Non-structural protein 2 (Nsp2)
This protein functions by disrupting the host survival pathway via interaction with the host proteins prohibitin-1 and prohibitin-2 [120]. Reverse genetic deletion in the coding sequence of Nsp2 of SARS virus attenuated little viral growth as well as replication and allowed the recovery of mutant virulent viruses [121].
The sequence identity of Nsp2 protein of SARS-CoV-2 with Nsp2s of human SARS and bat CoV amounts to 68.34% and 68.97%, respectively (Fig. S8). We have estimated the mean PPIDs of Nsp2s of SARS-CoV-2, human SARS, and batbat CoV to be 5.17%, 2.04%, and 2.03% respectively (see Table 3) (per-residue predisposition of intrinsic disorder is depicted in Fig. S9A–C). According to the results, residues 570–595 (SARS-CoV-2), residues 110–115 (Human SARS), and residues 112–116 (bat CoV) are predicted to be disordered. As listed in Tables 2, S7 and S8, human SARS does not contain MoRF, while SARS-CoV-2 and bat CoV have a N-terminally located MoRF region predicted by MoRFchibi_web.
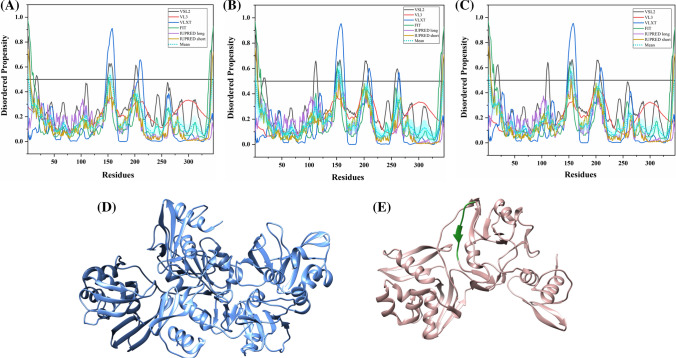
Non-structural protein 3 (Nsp3)
Nsp3 is a viral papain-like protease (PLP) that affects the phosphorylation and activation of IRF3 and, therefore, antagonizes the IFN pathway [122]. It's also reported to stabilize NF-κβ inhibitor which further blocks the NF-κβ pathway [122]. Figure 15d represents the 1.85 Å resolution X-ray crystal structure of the catalytic core of Nsp3 protein from human SARS CoV (PDB ID: 2FE8) [123]. The structure consisting of residues 723–1036 revealed folds similar to a deubiquitinating enzyme in vitro, the deubiquitinating activity of which was found to be efficiently high [123]. A 1.45 Å resolution structure (PDB ID: 6W6Y) of SARS-CoV-2 Nsp3 homodimer (chains A and B from 207–374 residues) is recently generated using X-ray diffraction (Fig. 15e) [124].
Fig. 15.
Analysis of intrinsic disorder propensity of Nsp3. Graphs a–c represent the intrinsic disorder profiles of Nsp3 protein of a SARS-CoV-2, b human SARS, and c bat CoV. d A 1.85 Å resolution crystal structure (PDB ID: 2FE8) of residues 723–1036 of Nsp3 of human SARS CoV. e A 1.45 Å resolution crystal structure (PDB ID: 6W6Y) of ADP ribose phosphatase of Nsp3 [residues 207–374 (orange colour)] of SARS CoV-2. f Aligned disorder profiles generated for all three Nsp3 is based on the outputs of the PONDR® VSL2. Colour schemes are similar to those given in Fig. 3
Nsp3 protein of SARS-CoV-2 contains several substituted residues throughout the protein. It is equally close to both Nsp3 proteins of human SARS and bat CoV, sharing 76.69% and 76.31% identity respectively (Fig. S10). According to our results, the mean PPIDs of Nsp3 proteins of SARS-CoV-2, human SARS, and bat CoV are 7.40%, 7.91%, and 7.78% respectively (Table 3). Disorder profiles in Fig. 15a–c shows that all three Nsp3 proteins are highly structured. This is further supported by Fig. 15f, where PONDR® VSL2-generated disorder profiles of these three proteins are overlapped to show almost complete coincidence of their major disorder-related features. According to the mean disorder analysis (see Fig. 15a–c), Nsp3 proteins are predicted to have the following IDPRs: SARS-CoV-2 (1–5, 105–199, 1221–1238), human SARS (102–189, 355–384, 1195–1223) and bat CoV (107–182, 352–376, 1191–1217). The first 112 residues represent a ubiquitin-like globular fold, while 113–183 residues form the flexible acidic domain rich in glutamic acid. It is thought to bind and ubiquitinate viral E protein using the N-terminal acidic domain [125, 126]. This unstructured segment has many MoRFs predicted by ANCHOR and MoRFPred servers which may facilitate the protein–protein interaction (Table 2). Interestingly, Nsp3 of all three viruses is found to have the highest number of RNA-binding residues (Tables S9–S11).
Non-structural protein 4 (Nsp4)
Nsp4 is reported to induce the formation of DMVs for optimal replication inside host cells [127–129]. Although no crystal or NMR solution structure is reported, Nsp4 is demonstrated to contain a tetra-spanning transmembrane region with its N- and C-terminals present in cytosol [130].
Nsp4 protein of SARS-CoV-2 has multiple substitutions near the N-terminal region and has a quite conserved C-terminus (Fig. S11). It is found to be closer to Nsp4 of bat CoV (81.40% identity) than to human SARS Nsp4 (80%). The low level of intrinsic disorder illustrated in Fig. S12A–C and mean PPIDs of Nsp4 proteins (Table 3) classify it as a highly structured protein which, however, contains some flexible regions. Likewise, only N- and C-terminal MoRFs which possibly assist in its cleavage from long polyproteins 1a and 1ab are shown in Table 2.
Non-structural protein 5 (Nsp5)
Also referred to as 3CL-pro, it works as a protease and cleaves the replicase polyproteins (1a and 1ab) at 11 major sites [131, 132]. Recently, the X-ray diffraction-based crystal structure of SARS-CoV-2 Nsp5 in complex with an inhibitor N3 has been solved (PDB ID:6LU7) (Fig. 16d) [133]. An X-ray crystal structure (PDB ID: 5C5O) obtained for human SARS CoV Nsp5 is shown in Fig. 16e. Here, 3CL-protease is bound to a phenyl-beta-alanyl (S, R)-N-decalin type inhibitor [134].
Fig. 16.
Analysis of intrinsic disorder propensity of Nsp5. Graphs a–c represent intrinsic disorder profiles of Nsp5 protein of a SARS-CoV-2, b human SARS, and c bat CoV. Colour schemes are similar to those given in Fig. 3. d A 2.16 Å X-ray diffraction-based crystal structure (PDB ID: 6LU7) of SARS-CoV-2 Nsp5 in complex with its inhibitor N3. e A 1.50 Å crystal structure (PDB ID: 5C5O) of Nsp5 of human SARS CoV
Nsp5 protein is found to be highly conserved in all three studied CoVs. SARS-CoV-2 Nsp5 shares a 96.08% sequence identity with human SARS Nsp5 and 95.42% with bat CoV Nsp5 (Fig. S13). Therefore, it is not surprising that our analysis demonstrated the identical mean PPID values of 1.96% for all three Nsp5s (Table 3). As the graphs (Fig. 16a–c) depict, Nsp5s have several flexible regions and N-terminally IDPR of six residues. Due to the low flexibility of this protein, a single MoRF predicted by MoRFchibi_web is present in the N-terminal region (residues 3–8) in all Nsp5s (Tables 2, S7, S8). Further, the identified nucleotide-binding residues in Nsp5 proteins are tabulated in Tables S9–S11.
Non-structural protein 6 (Nsp6)
Nsp6 protein is involved in blocking ER-induced autophagosome/autolysosome vesicle formation that functions in restricting viral production inside host cells. It induces autophagy by activating the omegasome pathway, which is normally utilized by cells in response to starvation [135].
Nsp6 of SARS-CoV-2 is equally close to Nsp6s from both human SARS and bat CoV, having a sequence identity of 87.24% (Fig. S14D). Similarly, mean PPIDs for all three Nsp6 proteins is calculated to be 1.03%. The graphs in Fig. S14A–C further illustrates its highly structured nature. As Nsp6 is a membrane protein, all three proteins are predicted to have a single MoRF near the N-terminal region (residues 1–19 in SARS-CoV-2, residues 1–22 in human SARS, and residues 1–21 in bat CoV) by the DISOPRED3 server. The role of this protein-binding region for the induction of autophagy needs to be elucidated.
Non-structural proteins 7 and 8 (Nsp7 and 8)
The ~ 10 kDa Nsp7 helps in primase-independent de novo initiation of viral RNA replication by forming a hexadecameric ring-like structure with Nsp8 protein [136, 137]. Both Nsp 7 and 8 contribute 8 molecules to the ring-structured multimeric viral RNA polymerase (Nsp12) [136]. Figure 17d depicts the 2.90 Å resolution structure (PDB ID: 6M71) of SARS-CoV-2 Nsp12 with its cofactors Nsp7 and Nsp8 [138]. Another 3.1 Å resolution electron microscopy-based structure (PDB ID: 6NUR) of human SARS Nsp12–Nsp8–Nsp7 complex is shown in Fig. 17e [139].
Fig. 17.
Analysis of intrinsic disorder propensity of Nsp7. Graphs a–c represent intrinsic disorder profiles of Nsp7 protein of a SARS-CoV-2, b human SARS, and c bat CoV. d A 2.90 Å resolution structure (PDB ID: 6M71) of SARS-CoV-2 Nsp12 with its cofactors Nsp7 and Nsp8. Chain A represents Nsp12 of residues 31–50, 69–102, 112–895, 906–919 (red colour), chain C represents Nsp7 of residues 2–71 (blue colour), and chains B and D represent Nsp8 from residues 84–122 and 129–132 (dark grey colour). e A 3.10 Å resolution cryo-EM structure (PDB ID: 6NUR) of Nsp12–Nsp8–Nsp7 complex. Chain C includes 2–71 residues of Nsp7 (gold colour), chains B and D (dark khaki) represent 77–191 residues of Nsp8 and chain A signifies residues 117–896 and 907–920 of Nsp12 (RNA-directed RNA polymerase) (orange colour) from human SARS CoV. f MSA profile of all three Nsp7 proteins. Colour schemes are similar to those given in Fig. 3
In this study, we found that Nsp7 of SARS-CoV-2 shares 100% sequence identity with the other two Nsp7 proteins (Fig. 17f), while SARS-CoV-2 Nsp8 is slightly closer to Nsp8 of human SARS (97.47%) than to other Nsp8 protein (96.46%) (Fig. 18d).
Fig. 18.
Analysis of intrinsic disorder propensity of Nsp8. Graphs a–c represent intrinsic disorder profiles of Nsp8 protein of a SARS-CoV-2, b human SARS, and c bat CoV. d MSA profile of all three Nsp8 proteins. Colour schemes are similar to those given in Fig. 3
Due to the similar sequence identities, mean PPIDs of all Nsp7s proteins are 9.64%, indicating their ordered structure (disorder profiles in Fig. 17a–c). Both SARS-CoV-2 and human SARS Nsp8 proteins have a mean PPID of 23.74%, while Nsp8 of bat CoV has a PPID of 22.22% (disorder profiles in Fig. 18a–c). As moderately disordered proteins, Nsp8s are predicted to have a long IDPR (residues 44–84) in both SARS-CoV-2 and human SARS, and a bit shorter IDPR in bat CoV (residues 48–84). Furthermore, SARS-CoV Nsp7 using its N-terminus residues (V11, C13, V17, and V21) forms a hydrophobic core with Nsp8 residues (M92, M95, L96, M99, and L103). Additionally, H-bonding takes place between Nsp7 Q24 and Nsp8 T89 residues [137]. These amino acids are the part of MoRFs predicted in these proteins. The results are tabulated in Tables 2, S7 and S8. Three protein-binding regions in Nsp7 of SARS-CoV-2 (residues 1–30, 39–58, and 65–83), human SARS (residues 1–30, 44–58, and 64–83), and bat CoV (residues 1–30, 39–58, and 65–83) are identified by MoRFchibi_web server. Nsp7 shows the presence of very few nucleotide-binding regions while Nsp8 contains several DNA- as well as RNA-binding residues (see Tables S9–S11).
Non-structural protein 9 (Nsp9)
Nsp9 protein is a single-stranded RNA-binding protein [140]. It might protect RNA from nucleases by binding and stabilizing viral nucleic acids during replication or transcription [140]. Our results on nucleotide-binding tendency of Nsp9 shows the presence of several RNA-binding and few DNA-binding residues in Nsp9 of SARS-CoV-2, Human SARS, and bat CoV (Tables S9–S11). Presumed to evolve from a protease, Nsp9 forms a dimer using its GXXXG motif [141, 142]. Figure 19d shows a 2.7 Å crystal structure of human SARS Nsp9 homodimer (PDB ID: 1QZ8) that identified a unique and previously unreported oligosaccharide/oligonucleotide fold-like fold [140]. Here, each monomer contains a cone-shaped β-barrel and a C-terminal α-helix arranged into a compact domain [140].
Fig. 19.
Analysis of intrinsic disorder propensity of Nsp9. Graphs a–c represent the intrinsic disorder profiles of Nsp9 protein of a SARS-CoV-2, b human SARS, and c bat CoV. d A 2.70 Å crystal structure (PDB ID: 1QZ8) of residues 3–113 of human SARS Nsp9. e MSA profile of all three Nsp9 proteins. Colour schemes are similar to those given in Fig. 3
Nsp9 of SARS-CoV-2 is equally similar to other two Nsp9 proteins (with a percentage identity of 97.35%). The difference in the three amino acids at 34, 35 and 48 positions accounts for its similarity (Fig. 19e). Mean PPIDs of all Nsp9s are listed in Table 3. Graphs in Fig. 19a–c show that all three Nsp9s are rather structured, but contain flexible regions. It contains conserved residues (R10, K52, Y53, R55, R74, F75, K86, Y87, F90, K92, R99, and R111) of positively charged side chains suitable for binding with the negatively charged phosphate backbone of RNA and aromatic side-chain amino acids providing stacking interactions [140]. These residues are a part of multiple disorder-based binding sites predicted by MoRFchibi_webserver (Tables 2, S7, S8).
Non-structural protein 10 (Nsp10)
Nsp10 forms a complex with Nsp14 for hydrolysing dsRNA in 3′–5′ direction [143]. In addition to activating the exonuclease activity of Nsp14, it also stimulates its methyltransferase (MTase) activity required during RNA-cap formation after replication [144]. Figure 20d represents the X-ray crystal structure of the Nsp10/Nsp14 complex (PDB ID: 5C8T) [145]. In agreement with the results of previous biochemical experimental studies, the structure identified important interactions with the ExoN (exonuclease domain) of Nsp14 without affecting its N7-MTase activity [143, 144].
Fig. 20.
Analysis of intrinsic disorder propensity of Nsp10. Graphs a–c represent the intrinsic disorder profiles of Nsp10 protein of a SARS-CoV-2, b human SARS, and c bat CoV. d A 3.20 Å crystal structure (PDB ID: 5C8T) of SARS CoV Nsp10/Nsp14 complex. In this structure, A and C chains (cornflower blue colour) signifies 1–131 residues of Nsp10, while B and D chains corresponds to residues 1–453 and 465–525 of Nsp14 (dim grey colour). e MSA profile of all three Nsp10 proteins. Colour schemes are similar to those given in Fig. 3
SARS-CoV-2 Nsp10 protein is quite conserved having a 97.12% sequence identity with Nsp10 of human SARS and 97.84% with Nsp10 of bat CoV (Fig. 20e). Mean PPIDs of all three studied Nsp10 proteins are found to be 5.04%. Figure 20a–c represents the disorder profiles of Nsp10s and signifies the lack of long IDPRs. Furthermore, Tables 2, S7 and S8 shows that all three Nps10 proteins have multiple MoRFs. For SARS-CoV-2, three MoRFs (residues 25–32, 91–99, and 133–138) were identified by MoRFchibi_web server and one MoRF (residues 11–18) was predicted by MoRFPred server. Interestingly, the SARS-CoV Nsp10 residues F16, F19, and V21 form van der Waals interactions with many of the Nsp14 amino acids [145] out of which one residue (F16) is located in the MoRF region identified in this study. Furthermore, many nucleotide-binding residues which are found in all three Nsp10s are listed in Tables S9–S11.
Non-structural protein 12 (Nsp12)
In coronaviruses, Nsp12 acts an RNA-dependent RNA polymerase (RDRP). It accomplishes both primer-independent and primer-dependent synthesis of viral RNA with Mn2+ as its metallic co-factor and viral Nsp7 and 8 as protein co-factors [146]. As aforementioned, a 3.1 Å resolution structure of human SARS Nsp12 in association with Nsp7 and Nsp8 proteins (PDB ID: 6NUR) has been reported using electron microscopy (Fig. 17e). Nsp12 has a polymerase domain similar to “right hand” containing finger subdomain (398–581, 628–687 residues), palm subdomain (582–627, 688–815 residues) and a thumb subdomain (816–919) [139].
SARS-CoV-2 Nsp12 protein has a highly conserved C-terminal region (Fig. S16). It is found to share a 96.35% sequence identity with human SARS Nsp12 and 95.60% with bat CoV Nsp12. Mean PPID values for all three Nsp12s are estimated to be 0.43% (Table 3). Graphs in Fig. S15A–C show that although Nsp12s are mostly ordered, they have multiple flexible regions. As RDRP protein is observed to be mostly structured, significant MoRFs in disordered regions are not found (Tables 2, S7, S8).
Non-structural protein 13 (Nsp13)
Nsp13 functions as a viral helicase and unwinds dsDNA/dsRNA in 5′–3′ direction [147]. Recombinant viral helicase expressed in E.coli Rosetta 2 strain was reported to unwind ~ 280 bp per second [147]. Figure 21d represents 2.8 Å crystal structure of human SARS Nsp13 (PDB ID: 6JYT) [148]. This helicase contains a β19–β20 loop on 1A domain, which is primarily responsible for its unwinding activity. Furthermore, the study revealed an important interaction of Nsp12 with Nsp13 which further enhances its helicase activity [148].
Fig. 21.
Analysis of intrinsic disorder propensity of Nsp13. Graphs a–c represent intrinsic disorder profiles of Nsp13 protein of a SARS-CoV-2, b human SARS, and c bat CoV. Colour schemes are similar to those given in Fig. 3. d A 2.80 Å crystal structure (PDB ID: 6JYT) of human SARS Nsp13 (residues 1–596)
Nsp13 of SARS-CoV-2 is found to be almost conserved as it shares 99.83% with Nsp13 of human SARS and 98.84% with Nsp13 of bat CoV (Fig. S17). Accordingly, mean PPIDs of all three Nsp13 proteins are estimated to be 0.67%. Graphs in Fig. 21a–c show that Nsp13s contain multiple flexible regions but does not possess significant disorder. As expected, being ordered proteins, Nsp13s does not contain any MoRF (Tables 2, S7, S8), but has several nucleotide-binding residues (Tables S9–S11).
Non-structural protein 14 (Nsp14)
Nsp14 is a multifunctional viral protein that acts as an exoribonuclease (ExoN) and methyltransferase (N7-MTase) in SARS coronaviruses. Its 3′–5′ exonuclease activity lies in conserved DEDD residues related to the exonuclease superfamily [149]. Its guanine-N7 methyltransferase activity depends upon the S-adenosyl-l-methionine (AdoMet) as a cofactor [144]. As mentioned previously, Nsp14 requires Nsp10 for activating its ExoN and N7-MTase activity inside host cells. Figure 20d depicts the 3.2 Å crystal structure of human SARS nsp10/nsp14 complex (PDB ID: 5C8T), where amino acids 1–287 form the ExoN domain and 288–527 residues form the N7-MTase domain of nsp14. A loop (residues 288–301) is essential for its N7-MTase activity [145].
SARS-CoV-2 Nsp14 protein shares a 95.07% identity with human SARS Nsp14 and 94.69% with bat CoV Nsp14 (Fig. S18). Low mean PPID values for all three Nsp14s (Table 3) and disorder profiles depicted in Fig. S19A–C shows its highly structured nature. Likewise, all three Nsp14 proteins contains two protein binding regions (residues 8–13 and 441–445) predicted by the MoRFPred.
Non-structural protein 15 (Nsp15)
Nsp15 is a uridylate-specific RNA endonuclease (NendoU) which creates a 2′–3′ cyclic phosphates after cleavage. Its endonuclease activity depends upon Mn2+ ions as co-factors. Conserved in Nidoviruses, it acts as an important genetic marker due to its absence in other RNA viruses [150]. A crystal structure of SARS-CoV-2 Nsp15 (207–374 residues) has been resolved using X-ray diffraction [151] (depicted in Fig. 22d). Figure 22e represents a 2.6 Å crystal structure of human SARS Nsp15 (PDB ID: 2H85) deduced by Bruno and colleagues [152].
Fig. 22.
Analysis of intrinsic disorder propensity of Nsp15. Graphs a–c represent intrinsic disorder profiles of Nsp15 protein of a SARS-CoV-2, b human SARS, and c bat CoV. Colour schemes are similar to given in Fig. 3. d A 1.9 Å resolution structure (PDB ID: 6W01) of Nsp15 of SARS CoV-2 consisting of 207–374 residues is represented in cornflower blue colour. e A 2.60 Å crystal structure (PDB ID: 2H85) of Nsp15 from human SARS CoV (rosy brown colour) where residues 151–157 predicted to be disordered are represented in forest green colour
SARS-CoV-2 Nsp15 shares 88.73% sequence identity with human SARS and 88.15% with bat CoV (Fig. S20). The calculated mean PPIDs of Nsp15s from SARS-CoV-2, human SARS, and bat CoV are 1.73%, 2.60%, and 2.60%, respectively. Similar to many other Nsps, all three Nsp15 proteins are predicted to possess multiple flexible regions but contain virtually no IDPRs (see Fig. 22a–c). Also, no significant disorder-binding regions are predicted in Nsp15 proteins (Table 2). SARS-CoV-2 and bat CoV Nsp15s possesses very short binding regions, while human SARS Nsp15 does not contain any MoRF (Tables S7, S8). Tables S9–S11 depict the presence of many RNA-binding residues and few DNA-binding residues in Nsp15 of all three viruses.
Non-structural protein 16 (Nsp16)
Nsp16 protein is another MTase domain-containing protein. As methylation of CoV mRNAs occurs in steps, three proteins Nsp10, Nsp14, and Nsp16 act one after another. First event requires the initiation trigger from Nsp10 protein, after which Nsp14 methylates capped mRNAs forming cap-0 (7Me) GpppA-RNAs. Nsp16 protein, along with its co-activator protein Nsp10, acts on cap-0 (7Me) GpppA-RNAs to give rise to final cap-1 (7Me)GpppA(2′OMe)-RNAs [144, 153]. The crystal structure (PDB ID: 6W75) of Nsp10–Nsp16 complex of SARS-CoV-2 is generated using X-ray diffraction (Fig. 23d). A 2 Å X-ray crystal structure of human SARS Nsp10–Nsp16 complex is depicted in Fig. 23e (PDB ID: 3R24) [154]. The structure consists of a characteristic fold present in class I MTase family comprising α-helices and loops surrounding a seven-stranded β-sheet [154].
Fig. 23.
Analysis of intrinsic disorder propensity of Nsp16. Graphs a–c represent the intrinsic disorder profiles of Nsp9 protein of a SARS-CoV-2, b human SARS, and c bat CoV. Colour schemes are similar to those given in Fig. 3. d A 1.95 Å resolution crystal structure (PDB ID: 6W75) of the Nsp10–Nsp16 complex of SARS-CoV-2. Nsp16 of residues 2–298 is represented using pink colour, while Nsp10 of residues 18–139 is shown in cornflower blue colour. e A 2.60 Å crystal structure (PDB ID: 3R24) of human SARS Nsp10–Nsp16 complex. Chain A shown in turquoise colour corresponds to residues 3–294 of Nsp16
Nsp16 of SARS-CoV-2 is found to be identical with other two Nsp16 proteins (93.29%) (Fig. S21). Mean PPIDs for Nsp16s from SARS-CoV-2, human SARS, and bat CoV are 5.37%, 3.02%, and 3.02%, respectively. In line with these PPID values, graphs in Fig. 23a–c show that these proteins are mostly ordered having several flexible regions. Correspondingly, only a single MoRF (residues 151–156) is present in all three Nsp16s. Further, several RNA-binding and few DNA-binding residues are also identified (Tables S9–S11).
Replicase polyprotein 1a
Since replicase polyprotein 1a contains non-structural proteins 1–10 identical to those found in replicase polyprotein 1ab, we did not perform their disorder analysis separately. However, replicase polyprotein 1a has one additional non-structural protein designated as Nsp11.
Non-structural protein 11 (Nsp11)
Nsp11 is a small uncharacterized protein with unknown function and requires extensive experimental insights to reveal its structural indentity. The intrinsic disorder-predicting software used in this study requires amino acid sequences which are at least 30-residue long. Therefore, because of their short sequences (just 13 residues), Nsp11s from all three studied coronaviruses are not checked for the intrinsic disorder, disorder-based protein-binding regions, and nucleotide-binding residues. Based on the MSA outputs, Nsp11 from SARS-CoV-2 is found to have a sequence identity of 84.62% with Nsp11s from human SARS and bat CoV (Fig. S22).
Concluding remarks
Emergence of new viruses and associated deaths around the globe represent one of the major concerns of modern times. Despite its pandemic nature, there is very little information available in the public domain regarding the structures and functions of SARS-CoV-2 proteins. Based on its similarity with human SARS CoV and bat CoV, the published reports have suggested the functions of SARS-CoV-2 proteins. In this study, we utilized information available on SARS-CoV-2 genome as well as translated proteome from GenBank, and carried out a comprehensive computational analysis of the prevalence of intrinsic disorder in SARS-CoV-2 proteins. Additionally, a comparison is also made with proteins from close relatives of SARS-CoV-2 from the same group of beta coronaviruses, human SARS CoV and bat CoV. Our analysis revealed that in these three CoVs, the N proteins are highly disordered, possessing the PPID values of more than 60%. These viruses also have several moderately disordered proteins, such as Nsp8, ORF6, and ORF9b. Although other proteins have shown lower disorder content, almost all of them contain at least one IDPR. Importantly, our study provides novel information on the presence of intrinsic disorder at the cleavage sites of replicase polyprotein 1ab of SARS CoVs. This observation confirms the crucial role of IDPRs in maturation of individual proteins. We also established that many of these proteins contain disorder-based binding motifs. Since IDPs/IDPRs might undergo structural transition upon association with their physiological partners, our study generates important grounds for better understanding of the functionality of these proteins, their interactions with other viral proteins, as well as interaction with host proteins in different physiological conditions.
Future perspective
The periodical outbreaks of pathogens worldwide always pinpoint the lack of suitable drugs or vaccines for proper cure or treatment. In 2003, nearly 750 deaths were reported due to the SARS outbreak in more than 24 countries. But this time, the outbreak of Wuhan’s novel coronavirus (SARS-CoV-2) has quickly surpassed this number, indicating more casualities soon. The lack of accurate information and ignorance of primary symptoms are major reasons, which cause many infection cases. Although efficient transmission from human to human has been confirmed, the actual reasons for fast SARS-CoV-2 spread are still unknown, but some assumptions are made by researchers and the Chinese authorities. The fast spread of SARS-CoV-2, COVID-19 pandemic, and associated introduction of quarantine also have made major impacts on economy and education worldwide due to several restrictions, such as limited transportation, restrained or frozen travel, halted attendance of mass events and the introduction of distant teaching and learning. Due to advancements in sequencing techniques, the full genomic sequence of SARS-CoV-2 was made available in a few days of the first infection report from Wuhan, China. However, massive subsequent research needs to be done to identify the actual cause of SARS-CoV-2 infectivity and to design suitable treatment in the future. Certain possibilities can be explored with the available information. The mutational pressure study on this virus will be very interesting to see if this virus transforms from bat SARS to human SARS to SARS-CoV-2. More in-depth experimental studies using molecular and cell biology techniques to establish structure–function relationships are required for a better understanding of the functioning of SARS-CoV-2 proteins. Additionally, based on the sequence homology and information on protein–protein interactions, the associated viral and host proteins should be explored, for finding means suitable for limiting replication, maturation, and ultimately pathogenesis of this virus. Although structural biology techniques (so-called rational drug design) can be used in drug development utilizing high-throughput screening of compounds virtually or experimentally, the applicability of these techniques is limited by the presence of intrinsic disorder in target proteins. Therefore, the thorough disorder analysis of three coronaviruses conducted in this study will help structural biologists to rationally design experiments keeping this information in mind.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
All the authors would like to thank IIT Mandi for providing facilities. MS and BRG were supported with funding from MHRD. KG was supported by the Department of Biotechnology (DBT), India (BT/PR16871/NER/95/329/2015). PK was supported by IIT Mandi-IIT Ropar-PGI Chandigarh, BioX consortium grant (IITM/INT/RG/18). TB is grateful to the Department of Science and Technology for her INSPIRE fellowship.
Abbreviations
- CoVs
Coronaviruses
- ACE2
Angiotensin-converting enzyme 2
- CDF
Cumulative distribution function
- CH
Charge hydropathy
- COVID-19
Coronavirus disease 2019
- CTD
C-terminal domain
- DMVs
Double-membrane vesicles
- ICTV
International Committee on Taxonomy of Viruses
- IDP
Intrinsically disordered proteins
- IDPRs
Intrinsically disordered protein regions
- IFN
Interferon
- MoRFs
Molecular recognition features
- MSA
Multiple sequence alignment
- Nsps
Non-structural proteins
- NTD
N-terminal domain
- PONDR
Predictor of natural disordered regions
- PPID
Predicted percentage of intrinsic disorder
- PPRInt
Prediction of protein RNA-interaction
- RBD
Receptor-binding domain
- SARS
Severe acute respiratory syndrome
- TRS
Transcriptional regulatory sequences
- ER
Endoplasmic reticulum
- WHO
World Health Organization
- Cryo-EM
Cryo-electron microscopy
- SF
Supplementary figures
- ST
Supplementary tables
Author contributions
RG: conception and design, interpretation of data, writing, and review of the manuscript, and study supervision. VNU and CJO: conception and design, acquisition and interpretation of data, writing, and review of the manuscript. TB, MS, PK, BRG, and KG: acquisition and interpretation of data, writing of the manuscript.
Compliance with ethical standards
Conflict of interest
All authors declare that there is no financial conflict of interest.
Footnotes
The infection caused by a novel coronavirus (SARS-CoV-2) that causes severe respiratory disease with pneumonia-like symptoms in humans is responsible for the current COVID-19 pandemic. No in-depth information on the structures and functions of SARS-CoV-2 proteins is currently available in public domain, and no effective anti-viral drugs and/or vaccines have been designed for the treatment of this infection. Our study provides the first comparative analysis of the order- and disorder-based features of SARS-CoV-2 proteome relative to human SARS and bat CoV that may be useful for structure-based drug discovery.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease 2019. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 29 Feb 2020
- 3.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Lienau J, Witzenrath M. Coronaviruses as the cause of respiratory infections. Internist. 2019;60:1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo PCY, Lau SKP, Lam CSF, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/jvi.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotten M, Lam TT, Watson SJ, et al. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis. 2013;19:736–742. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain S, Pan J, Chen Y, et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288–5295. doi: 10.1128/jvi.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/jvi.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/jvi.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 14.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Cortese MS, Romero P, et al. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 16.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/S0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 17.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Perumal NB, Oldfield CJ, et al. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, Kurgan L. DRNApred, fast sequence-based method that accurately predicts and discriminates DNA- and RNA-binding residues. Nucleic Acids Res. 2017;45:e84. doi: 10.1093/nar/gkx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng ZKL. High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder. Nucleic Acids Res. 2015;43:e121. doi: 10.1093/nar/gkv585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giri R, Kumar D, Sharma N, Uversky VN. Intrinsically disordered side of the Zika virus proteome. Front Cell Infect Microbiol. 2016;6:144. doi: 10.3389/fcimb.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue B, Williams RW, Oldfield CJ, et al. Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept Lett. 2010;17:932–951. doi: 10.2174/092986610791498984. [DOI] [PubMed] [Google Scholar]
- 24.Singh A, Kumar A, Yadav R, et al. Deciphering the dark proteome of Chikungunya virus. Sci Rep. 2018;8:5822. doi: 10.1038/s41598-018-23969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward JJ, Sodhi JS, McGuffin LJ, et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Clark K, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng K, Radivojac P, Vucetic S, et al. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng K, Vucetic S, Radivojac P, et al. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/S0219720005000886. [DOI] [PubMed] [Google Scholar]
- 31.Xue B, Dunbrack RL, Williams RW, et al. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero P, Obradovic Z, Li X, et al. Sequence complexity of disordered protein. Proteins Struct Funct Genet. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Mészáros B, Erdos G, Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gadhave K, Gehi BR, Kumar P, et al. The dark side of Alzheimer’s disease: unstructured biology of proteins from the amyloid cascade signaling pathway. Cell Mol Life Sci. 2020 doi: 10.1007/s00018-019-03414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garg N, Kumar P, Gadhave K, Giri R. The dark proteome of cancer: intrinsic disorderedness and functionality of HIF-1α along with its interacting proteins. Prog Mol Biol Transl Sci. 2019;166:371–403. doi: 10.1016/bs.pmbts.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Genet. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Huang F, Oldfield C, Meng J et al (2012) Subclassifying disordered proteins by the CH-CDF plot method. In: Pacific symposium on Biocomputing, pp 128–139 [PubMed]
- 38.Malhis N, Wong ETC, Nassar R, Gsponer J. Computational identification of MoRFs in protein sequences using hierarchical application of Bayes rule. PLoS ONE. 2015;10:e0141603. doi: 10.1371/journal.pone.0141603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mészáros B, Simon I, Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dosztányi Z, Mészáros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Disfani FM, Hsu W-L, Mizianty MJ, et al. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012;28:i75–83. doi: 10.1093/bioinformatics/bts209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng Z, Wang C, Uversky VN, Kurgan L. Prediction of disordered RNA, DNA, and protein binding regions using DisoRDPbind. Methods Mol Biol. 2017;1484:187–203. doi: 10.1007/978-1-4939-6406-2_14. [DOI] [PubMed] [Google Scholar]
- 44.Kumar M, Gromiha MM, Raghava GPS. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins. 2008;71:189–194. doi: 10.1002/prot.21677. [DOI] [PubMed] [Google Scholar]
- 45.Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajagopalan K, Mooney SM, Parekh N, et al. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem. 2011;112:3256–3267. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra PM, Uversky VN, Giri R. Molecular recognition features in Zika virus proteome. J Mol Biol. 2018;430:2372–2388. doi: 10.1016/j.jmb.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Gypas F, Tsaousis GN, Hamodrakas SJ. mpMoRFsDB: a database of molecular recognition features in membrane proteins. Bioinformatics. 2013;29:2517–2518. doi: 10.1093/bioinformatics/btt427. [DOI] [PubMed] [Google Scholar]
- 49.Oldfield CJ, Peng Z, Kurgan L. Disordered RNA-binding region prediction with DisoRDPbind. Methods Mol Biol. 2020;2106:225–239. doi: 10.1007/978-1-0716-0231-7_14. [DOI] [PubMed] [Google Scholar]
- 50.Cavanagh D, Davis PJ. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J Gen Virol. 1986;67(Pt 7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- 51.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. doi: 10.1128/jvi.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Haan CAM, te Lintelo E, Li Z, et al. Cooperative involvement of the s1 and s2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J Virol. 2006;80:10909–10918. doi: 10.1128/jvi.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 55.Broer R, Boson B, Spaan W, et al. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J Virol. 2006;80:1302–1310. doi: 10.1128/jvi.80.3.1302-1310.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song W, Gui M, Wang X, Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBride CE, Li J, Machamer CE. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J Virol. 2007;81:2418–2428. doi: 10.1128/jvi.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ujike M, Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeDiego ML, Alvarez E, Almazan F, et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81:1701–1713. doi: 10.1128/jvi.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torres J, Wang J, Parthasarathy K, Liu DX. The transmembrane oligomers of coronavirus protein E. Biophys J. 2005;88:1283–1290. doi: 10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Surya W, Claudine S, Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem. 2014;289:12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surya W, Samsó M, Torres J (2013) Structural and functional aspects of viroporins in human respiratory viruses: respiratory syncytial virus and coronaviruses. In: Mahboub BH (ed) Respiratory disease and infection - a new insight. IntechOpen. 10.5772/53957
- 65.Teoh KT, Siu YL, Chan WL, et al. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tseng Y-T, Chang C-H, Wang S-M, et al. Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS ONE. 2013;8:e64013. doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tseng Y-T, Wang S-M, Huang K-J, et al. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J Biol Chem. 2010;285:12862–12872. doi: 10.1074/jbc.M109.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corse E, Machamer CE. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312:25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Narayanan K, Chen C-J, Maeda J, Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol. 2003;77:2922–2927. doi: 10.1128/jvi.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J, Sun Y, Qi J, et al. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J Infect Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goh GK-M, Dunker AK, Uversky V. Prediction of intrinsic disorder in MERS-CoV/HCoV-EMC supports a high oral-fecal transmission. PLoS Curr. 2013 doi: 10.1371/currents.outbreaks.22254b58675cdebc256dbe3c5aa6498b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perrier A, Bonnin A, Desmarets L, et al. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 2019;294:14406–14421. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saikatendu KS, Joseph JS, Subramanian V, et al. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang C-K, Hsu Y-L, Chang Y-H, et al. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J Virol. 2009;83:2255–2264. doi: 10.1128/jvi.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang Q, Yu L, Petros AM, et al. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 78.Yu I-M, Oldham ML, Zhang J, Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J Biol Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He R, Leeson A, Ballantine M, et al. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo H, Chen Q, Chen J, et al. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–2628. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He R, Dobie F, Ballantine M, et al. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nelson GW, Stohlman SA. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J Gen Virol. 1993;74(Pt 9):1975–1979. doi: 10.1099/0022-1317-74-9-1975. [DOI] [PubMed] [Google Scholar]
- 83.Gunasekaran K, Tsai C-J, Nussinov R. Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J Mol Biol. 2004;341:1327–1341. doi: 10.1016/j.jmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 84.Oldfield CJ, Meng J, Yang JY, et al. Flexible nets: disorder and induced fit in the associations of p53 and 14–3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu Z, Hu G, Yang J, et al. In various protein complexes, disordered protomers have large per-residue surface areas and area of protein-, DNA- and RNA-binding interfaces. FEBS Lett. 2015;589:2561–2569. doi: 10.1016/j.febslet.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 86.Narayanan K, Huang C, Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan Y-J. The severe acute respiratory syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein. Virol J. 2005;2:5. doi: 10.1186/1743-422X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McBride R, Fielding BC. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu C-J, Chen Y-C, Hsiao C-H, et al. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan X, Li J, Shan Y, et al. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan Y-J, Teng E, Shen S, et al. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol. 2004;78:6723–6734. doi: 10.1128/jvi.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu W, Zheng B-J, Xu K, et al. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan X, Shan Y, Yao Z, et al. Mitochondrial location of severe acute respiratory syndrome coronavirus 3b protein. Mol Cells. 2006;21:186–191. [PubMed] [Google Scholar]
- 94.Yuan X, Yao Z, Shan Y, et al. Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res. 2005;114:70–79. doi: 10.1016/j.virusres.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Varshney B, Lal SK. SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry. 2011;50:5419–5425. doi: 10.1021/bi200303r. [DOI] [PubMed] [Google Scholar]
- 96.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, et al. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/jvi.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frieman M, Yount B, Heise M, et al. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgi membrane. J Virol. 2007;81:9812–9824. doi: 10.1128/jvi.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Netland J, Ferraro D, Pewe L, et al. Enhancement of murine coronavirus replication by severe acute respiratory syndrome coronavirus protein 6 requires the N-terminal hydrophobic region but not C-terminal sorting motifs. J Virol. 2007;81:11520–11525. doi: 10.1128/jvi.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou H, Ferraro D, Zhao J, et al. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J Virol. 2010;84:3542–3551. doi: 10.1128/jvi.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fielding BC, Tan Y-J, Shuo S, et al. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J Virol. 2004;78:7311–7318. doi: 10.1128/jvi.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang C, Ito N, Tseng C-TK, Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J Virol. 2006;80:7287–7294. doi: 10.1128/jvi.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanzawa N, Nishigaki K, Hayashi T, et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Law HKW, Cheung CY, Ng HY, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol. 2007;81:718–731. doi: 10.1128/jvi.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kopecky-Bromberg SA, Martinez-Sobrido L, Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J Virol. 2006;80:785–793. doi: 10.1128/jvi.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nelson CA, Pekosz A, Lee CA, et al. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hänel K, Stangler T, Stoldt M, Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J Biomed Sci. 2006;13:281–293. doi: 10.1007/s11373-005-9043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oostra M, de Haan CAM, Rottier PJM. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J Virol. 2007;81:13876–13888. doi: 10.1128/jvi.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 110.Keng C-T, Choi Y-W, Welkers MRA, et al. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu K, Zheng B-J, Zeng R, et al. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sharma K, Åkerström S, Sharma AK, et al. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS ONE. 2011;6:e19436. doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meier C, Aricescu AR, Assenberg R, et al. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thiel V, Ivanov KA, Putics Á, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 115.Fan K, Wei P, Feng Q, et al. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lokugamage KG, Narayanan K, Huang C, Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J Virol. 2012;86:13598–13608. doi: 10.1128/jvi.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Almeida MS, Johnson MA, Herrmann T, et al. Novel-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J Virol. 2007;81:3151–3161. doi: 10.1128/jvi.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jauregui AR, Savalia D, Lowry VK, et al. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS ONE. 2013;8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cornillez-Ty CT, Liao L, Yates JR, et al. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009;83:10314–10318. doi: 10.1128/jvi.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Graham RL, Sims AC, Brockway SM, et al. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol. 2005;79:13399–13411. doi: 10.1128/jvi.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Frieman M, Ratia K, Johnston RE, et al. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-B signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/jvi.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ratia K, Saikatendu KS, Santarsiero BD, et al. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Michalska K. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase (ADRP) from the apo form to ligand complexes. biorxiv. 2020 doi: 10.1101/2020.05.14.096081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Serrano P, Johnson MA, Almeida MS, et al. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J Virol. 2007;81:12049–12060. doi: 10.1128/jvi.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alvarez E, DeDiego ML, Nieto-Torres JL, et al. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402:281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013 doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hagemeijer MC, Ulasli M, Vonk AM, et al. Mobility and interactions of coronavirus nonstructural protein 4. J Virol. 2011;85:4572–4577. doi: 10.1128/jvi.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sakai Y, Kawachi K, Terada Y, et al. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oostra M, te Lintelo EG, Deijs M, et al. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol. 2007;81:12323–12336. doi: 10.1128/jvi.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tomar S, Johnston ML, St John SE, et al. Ligand-induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro): implications for nsp5 regulation and the development of antivirals. J Biol Chem. 2015;290:19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sparks JS, Donaldson EF, Lu X, et al. A novel mutation in murine hepatitis virus nsp5, the viral 3c-like proteinase, causes temperature-sensitive defects in viral growth and protein processing. J Virol. 2008;82:5999–6008. doi: 10.1128/jvi.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jin Z, Du X, Xu Y, et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 134.Anand K, Palm GJ, Mesters JR, et al. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.te Velthuis AJW, van den Worm SHE, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhai Y, Sun F, Li X, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gao Y, Yan L, Huang Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(80):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Egloff M-P, Ferron F, Campanacci V, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ponnusamy R, Moll R, Weimar T, et al. Variable oligomerization modes in coronavirus non-structural protein 9. J Mol Biol. 2008;383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Miknis ZJ, Donaldson EF, Umland TC, et al. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol. 2009;83:3007–3018. doi: 10.1128/jvi.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bouvet M, Imbert I, Subissi L, et al. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bouvet M, Debarnot C, Imbert I, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ahn D-G, Choi J-K, Taylor DR, Oh J-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Adedeji AO, Marchand B, Te Velthuis AJW, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jia Z, Yan L, Ren Z, et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Minskaia E, Hertzig T, Gorbalenya AE, et al. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ivanov KA, Hertzig T, Rozanov M, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ricagno S, Egloff M-P, Ulferts R, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Decroly E, Imbert I, Coutard B, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2’O)-methyltransferase activity. J Virol. 2008;82:8071–8084. doi: 10.1128/jvi.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen Y, Su C, Ke M, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2’-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.