Abstract
The essence of shotgun lipidomics is to maintain consistency of the chemical environment of lipid samples during mass spectrometry acquisition. This strategy is suitable for large-scale quantitative analysis. This strategy also allows sufficient time to collect data to improve the signal-to-noise ratio. The initial approach of shotgun lipidomics was the electrospray ionization (ESI)-based direct infusion mass spectrometry strategy. With development of mass spectrometry for small molecules, shotgun lipidomics methods have been extended to matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) and ambient mass spectrometry, including MS imaging methods. Furthermore, the object of analysis has extended from organ and body fluid levels to tissue and cell levels with technological developments. In this article, we summarize the status and technical challenges of shotgun lipidomics at different resolution of measurements from the mass spectrometry perspective.
Keywords: High resolution mass spectrometry, Lipidomics, Mass spectrometry imaging, Single-cell analysis, Shotgun lipidomics
1. Introduction
The complete set of lipids in tissues or cells is referred to as the lipidome [1]. According to the classification of the LIPID Metabolites and Pathway Strategy (LIPID MAPS) project, lipids can be divided into eight main categories: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides [2, 3]. Each category can be further classified into different lipid classes and subclasses, based on their polarities, charge(s), sizes, linkages to the backbone, and distinct functional groups. The LIPID MAPS structure database currently records 43,616 unique lipid structures (as of 9/13/2019) [4]. Lipid functions are highly related to their structure, concentration, and spatial and temporal distribution. The composition, distribution, and dynamics of the lipidome reflect the status of cellular metabolism [5]. Dysregulation of lipid metabolism is associated in the onset and progression of many diseases, including cardiovascular diseases, neurodegeneration, diabetes, and cancer [5, 6].
Lipidomics is the study of lipids based on qualitative and quantitative analysis of lipidomes in a systematic fashion [7, 8]. Lipidomics analysis is facilitated by decades of biological mass spectrometry advancements [9–14]. Due to the huge difference in lipid properties from water-soluble metabolites, lipidomics is very different from conventional metabolomics in terms of sample preparation, mass spectrometric analysis, and data processing. Due to the chemical nature of lipid molecules, metabolomics and proteomics techniques developed based on aqueous systems are not always suitable for lipidomics analysis. Lipidomic analysis requires non-aqueous phase extraction and ionization, and it considers lipid aggregation under certain conditions, which are unique features of lipid analysis [15].
Shotgun lipidomics analysis occurs under specific experimental conditions with a constant lipid solution concentration [12, 16–18]. In most cases, there is no separation process conducted prior to the mass spectrometric analysis. Thus, shotgun lipidomics can provide sample signals from a uniform chemical environment. Since the chemical environment of the solution do not change during analysis, differences in signal response of homologues are minimized. The signal-to-concentration relationship can be calibrated readily. Accurate absolute quantification after normalization to a common denominator can be achieved with a limited number of internal standards [15, 19]. As a result, high-accuracy and large-scale absolute quantitative analysis is easier to achieve by shotgun lipidomics than by separation-based lipidomics.
According to the essential features of shotgun lipidomics, the scope of shotgun lipidomics has been extended to cover analyses after both direct infusion and desorption (including imaging analysis) [20]. The direct infusion-based electrospray ionization is the earliest ionization strategy used in shotgun lipidomics. This is mainly because, in the early stages of shotgun lipidomics, it was the best approach for direct analysis of lipid extraction in homogeneous solutions. This approach is still the most widely used in quantitative shotgun lipidomics [16, 21]. With the development of MALDI matrices that are specific to small molecules, technical approaches in shotgun lipidomics have been expanded [22–26]. Recently, with the development of ambient ion sources (such as desorption electrospray ionization (DESI), laser ablation electrospray ionization (LAESI), and liquid extraction surface analysis-mass spectrometry (LESA-MS)) and mass spectrometry imaging (MSI) technologies, the scope of shotgun lipidomics has been expanded into direct desorption analysis [27–31]. Currently, analytical methods based on direct desorption or imaging analysis are becoming more important in lipidomics analysis because they provide valuable information on the spatial distribution of lipids. The combined use of these approaches should be very effective to understand lipidomes on different resolution. Figure 1 shows a number of methodological approaches to expand the scope of shotgun lipidomic analysis. Table 1 summarized the significance and purpose of shotgun lipidomics analysis at different levels of measurements for biomedical research.
Figure 1.

Approaches to expand the scope of shotgun lipidomic analysis.
Table 1.
Different measurement levels of shotgun lipidomics for the scenarios and objectives of biomedical research.
| Macro/Organ Level | Micro/Tissue Level | Single-cell Level | |
|---|---|---|---|
| Representative samples | Whole organs (e.g., liver, heart)/Body Fluids (e.g., serum, plasma) | Tissue section/Local part of organs (e.g., islet, dorsal root ganglion)/Collection of organelles (e.g., mitochondrial) | Cultured cells/Tissue section |
| Application scenario | Functional research. The role of lipidome in the development of the disease is understood from the perspective of the overall physiological function of the organs or the whole body.[135, 136] | Corresponds to research at the resolution of histological level or above. It is of great significance for the study of drug functions and release, as well as the mechanism or the development of diseases. [29, 122, 137, 138] | Corresponds to studies at the cellular and molecular levels. It is the cornerstone of basic research.[85, 92, 97] |
| Technical maturity of biomedical applications | High maturity. There is a need to increase the automation and standardization of quantification. The analysis of the fine structure of lipid molecules is at the forefront of methodological research. | Generally, technical mature in the qualitative analysis. The quantification analysis is not mature enough. Increased sensitivity is the key to solving the problem. | Technically immature. The current research is focused on methodological study. In addition to improving sensitivity, micrometer/nanometer operations and control are also technical bottlenecks. |
The ultimate goal of lipidomic analysis is to achieve a full coverage of precise structural analysis, accurate quantification, and a comprehensive understanding of lipid dynamics at all levels. This goal is far from the existing analytical capabilities. We believe this requires a combination of lipidomics based on both efficient separation and direct analysis. Shotgun lipidomics offers unique advantages in both quantitative and spatial distribution analysis.
In this review, we overview the progress in and challenges of shotgun lipidomics analysis at different resolution of measurements, with an emphasis on instrumental analysis and sample processing. We discuss current technical characteristics and bottlenecks, to provide insights that inspire new ideas and drive novel technological advancement.
2. Shotgun lipidomics at spatial levels
2.1. Analysis at macro/organ level
This level is a macroscopic explanation of changes in the whole body or organs. For solid tissue, this level of analysis is for the representative part of the organ or entire organ. Its resolution is usually in the order of a few millimeters or tens of millimeters. For body fluid samples, its analytical difficulty depends on the type of body fluid. Lipid concentrations vary widely among different body fluids. For serum or plasma, approximately lipid content in one hundred microliters corresponds to lipids contents in 10 milligrams of tissue. For sample with a very low lipid content such as cerebrospinal fluid, several times or tens of times of the volume of plasma or serum are required. Analysis of serum and plasma is relatively easier to achieve than tissue samples. The main reason is that the quantity of samples is sufficient, the samples are homogenate, and the lipid content is usually abundant. However, for samples of cerebrospinal fluid, the total amount of samples that can be obtained is limited, and it is hard to quantify low abundance lipid species. At the level of organs and body fluids, quantitative shotgun lipidomics is widely used in mechanism studies of diseases associated with lipid metabolism or in biomarker discovery studies [20, 32–34]. The most commonly measured lipid classes are sphingolipids (SL), glycerophospholipid (GPL), glycerolipids (GL), and non-esterified fatty acid (NEFA). They are involved deeply in the construction of cellular membrane, energy metabolism, transportation, and signal transduction [6]. More than 45 classes of lipids are quantifiable by shotgun lipidomic analysis [5]. In this macrolevel analysis, tens of milligrams of tissue samples are typically required for quantitative analysis. The mass spectrometry strategies or modes that can be used are very diverse, including precursor-ion scan, neutral loss scan, high-resolution mass spectrometry, and multi-dimensional mass spectrometry strategy (MDMS) [11, 35–39]. Absolute quantitative analysis has been easily implemented in a homogeneous direct infusion system. In the case of quantitative analysis using the MDMS method, it is possible to achieve at least one internal standard for each lipid class. The choice of the internal standard needs to avoid interference with endogenous substances, and the spiking content is not less than 10% of the high abundance component. In the calculation, the correction of the abundance of 13C isotope with molecular weight changes should be considered. The in-depth review by Wang et al. [15] should be helpful for considering the selection of internal standards. In this level, the overall quantitative accuracy for lipid species that present in modest-to-high abundance can be greater than 90% [40]. For a brief introduction to the basic technical concepts of lipidomics, please refer to a tutorial written by Wang et al. [41].
From the perspective of instrumental analysis, there is no practical difficulty in the quantitative accuracy of the macro/organ level. However, if the sample collection and quality control does not attract enough attention, it will affect the accuracy of the quantification [42]. At the macro/organ level, the minimal sample size for analysis is relatively easy to guarantee. Since human to small animal samples are used, there are still certain requirements for sample consistency to avoid data variation. The difficulty of sample collection is varied among different organs. Samples from the liver and muscle are relatively homogenous and easily collected. However, if only a partial part of the organ is collected, the distribution of fat and blood vessels needs to be considered. For organs with complex and non-homogeneous morphological structures, such as the brain and kidneys, partial excision and collection is more likely to cause data variations. Further, caution should be taken when collecting from small animals, such as mice. The accuracy of the anatomical position and degree of exfoliation of the adherent tissue have a great influence on the sample consistency.
In addition, different biological models should be considered. Usually, sample deviation among patients are very large. If it is a model organism, the wild type should be stable. However, due to the introduction of additional interventions, the level of gene expression is not usually consistent among samples in the experimental group, thus variations in lipid quantification may increase. Therefore, in addition to careful handling of the sampling operation, background information of the sample should be considered to determine the appropriate number of samples for statistical analysis.
There are primarily two analytical challenges at this level: accurate quantification of low-to-modest abundance lipids and determination of fine structural information. The concentration range of lipids in a biological sample varies from amol to nmol/mg of protein (estimated from LIPID MAPS). Ion suppression by high abundant species during ionization or trapping capacity limits the dynamic range of a mass spectrometer. However, the quantitative dynamic range of shotgun lipidomics is only approximately four orders of magnitude. Therefore, using the MDMS strategy, selective ionization, and pre-separation of the lipid extraction by liquid-liquid partitioning or liquid-solid partitioning should be very useful to significantly increase the dynamic range [38, 43]. For example, triglycerides are the predominant component in adipose tissue. Pre-separation of triglycerides from other polar lipids is necessary for shotgun lipidomics analysis of the majority of lipid classes [8]. This challenge is not only due to the full range of lipid concentration distribution, but also because the lipids self are prone to aggregation. To avoid or reduce the adverse effects of aggregation, the concentration of each lipid for infusion should be less than 10 pmol/μl.
Derivatization strategies can be used to significantly improve signal quality in multiple ways [44]. First, derivatization can lead to increases in ionization sensitivity by introducing a functional group that is prone to ionization. Second, derivatization can improve the linear dynamic range of lipid analysis by selectively improving ionization efficiency of low abundance lipid classes. For example, Trimethylsilyl (TMS) derivatization of phosphatidylinositol phosphate [45] and N-(4-aminomethylphenyl)pyridinium (AMPP) derivatization of NEFA [46] greatly enhanced the quantification range. Moreover, derivatization can enhance analytical specificity with tagged moieties. For example, phosphatidylglycerol (PG) and bis(monoacylglycero)phosphate (BMP) are isomers. They can be distinguished by TMS derivatization due to differences in the characteristics of derivatized fragments [47]. In addition to expanding the derivatization coverage of lipid classes, simplification of the derivatization process and methods that do not require purification are important development directions, especially for smaller size analyses. For a more detailed understanding of derivatization methods, please refer to recent review articles written by Hu et al. and Ryan et al. [44, 48].
Comprehensive coverage of fine structural information of lipid molecules is an important research area at this level. Positional determination of unsaturated bonds in lipid molecules and differentiation of cis and trans isomers are currently critical challenges [49]. In diseases such as cancer, the content of isomers differs from the normal state [50]. Therefore, determination of fine structure and quantitative or ratio analysis are required. Current studies on the localization and quantification of carbon-carbon double bonds employ oxidation reactions on double bonds, including ozone induced dissociation (OzID) [51], photochemical method such as Paternò–Büchi reaction [52], and ultraviolet photodissociation (UVPD) [53]. A recently published review article [54] provides detailed information about this topic.
In the high vacuum environment of common mass spectrometry, we do not consider ion collisions because their mean free path is greater than the dimensions of the vacuum cavity. However, after the introduction of collision gas, the mean free path of ions and the average kinetic energy change. These changes are related to the collision cross section of ions. The collision cross section is determined by their molecular structures. Ion mobility mass spectrometry (IMS-MS) is an effective method to distinguish three-dimension conformation such as cis-trans isomers [55]. There are many variants of ion mobility mass spectrometry. These methods combine different collision gas conditions with different electric field modes to produce different separation fields. The ions are separately detected based on differences in their collision cross sections and mass-to-charge ratios. High field asymmetric waveform ion mobility spectrometry (FAIMS) [56], drift tube ion mobility spectrometry (DTIMS) [57], and traveling wave ion mobility spectrometry (TWIMS) [58] are major IMS-MS technologies that have been used for lipidomic analysis. Due to similarities in the common structure of a specific lipid class and small differences in the collision cross section of each lipid species, the separation power of complex mixtures is limited. Nevertheless, the combination of ion mobility mass spectrometry with MALDI, DESI, or other ambient ionization methods for lipid extraction could accurately distinguish high abundance components [59, 60]. A recently published review article [61] can be consulted to learn new methods and ideas for lipid separation and structural elucidation by ion mobility mass spectrometry.
At the macro level, direct infusion analysis is still a dominant approach. Table 2 summarizes the comparison between related technologies and challenges.
Table 2.
Technical characteristics of shotgun lipidomics at different level of measurements.
| Scope of Analysis | MS | Identification | Quantification | Derivatization | Fine structure characterization | Major technology trends |
|---|---|---|---|---|---|---|
| Macro/Organ (Of or above 1 mm) | ESI MALDI Ambient MS | >1000 species; > 45 lipid classes | Absolute quantification | No limitation | ***** Limited quantitative methods |
Automated high-throughput process |
| Micro/Tissue(20–200 μm) | ESI MALDI Ambient MS | <1000 species; Major species of SL, GPL, GL and NEFA | Relative quantification; Limited Absolute quantification | Limited choices | ** | Efficient extraction and derivatization methods |
| Single-cell/Subcellular(<10 μm;Below mean cell size) | ESI MALDI SIMS LAESI LESA | <100 species; High abundant species of SL, GPL, GL and NEFA | Relative quantification | Rare | * | Improvement of ionization efficiency |
2.2. Analysis at micro/tissue level
In this review, we refer to analysis of a sample size of less than 2 mg or a spatial resolution for imaging from 20 to 200 μm as micro- or tissue-level analysis. There are three scenarios for direct infusion analysis at this level. First, some changes occur only in local areas of the organ, and if analyzed at the organ level, this change is likely to be masked in the background. Therefore, the number of available samples for anatomical sampling of these small areas is very limited. Examples are the dorsal root ganglion and islets. For this type of bulk sample, homogenization before lipid extraction is usually performed, followed by a direct infusion strategy.
Second, samples are obtained from isolated primary cells or organelles. For isolated primary cells, considering the loss during isolation and purification, only up to hundreds of thousands of cells can be obtained. For organelle analysis of cultured cells, in theory, this amount of sample is sufficient to ensure quantification; however, the purity of the sample will affect the accuracy and stability of the results. For example, the separation of subcellular organelles is usually carried out by ultracentrifugation, density gradient centrifugation, size exclusion, or immunoaffinity capture. Such methods provide inadequate purity or lipidome integrity for exosome analysis [62]. Recently, the application of novel separation methods, such as flow field-flow fractionation (FIFFF), provided better purity and showed improvements in lipidomic analysis [63]. For cell or organelle samples, the sample can be extracted after sufficient dispersion. Direct infusion analysis is a commonly used strategy.
Third, tissue-level samples are usually obtained from pathological sections or tissue biopsies. For tissue section samples that do not require imaging analysis, the weight of the tissue section is usually around 1 mg, and the strategy of homogenization-extraction and direct infusion analysis is often used; however, analysis from the tissue section requires tissue transfer from a glass slide. This differs from bulk samples in that protein-based quantification becomes more difficult. This is because the tissue must be stripped from the glass slide to quantify protein content. Collection of tissue section samples can be effectively achieved using laser microdissection [64]. In addition to quantification based on protein content, other normalizers (e.g., DNA content, sample weight, or sample volume) can also be used for quantification.
The strategy for direct infusion analysis is characterized by maximum utilization of multiple MS acquisition modes. The major advantages of this approach are high accuracy of targeted or untargeted quantitative analysis. It should be noted that due to the very small amount of sample, such as < 1 mg, conventional extraction methods can cause a large amount of transfer loss. It is possible that global analysis of broad lipid classes is not achievable. Without a simplified extraction strategy, it is possible that there is no signal detectable due to severe sample loss. In this case, a compromise strategy is unavoidable. With a simplified extraction strategy, it is possible to selectively retain some lipid classes for effective detection with a small sample size [64]. For example, a simplified liquid extraction procedure of conventional biphasic lipid extraction such as methyl tert-butyl ether-methanol (MTBE-MeOH) method and butanol-MeOH (BUME) method, or a solid phase extraction (SPE) extraction strategy, can be employed [65–69]. If direct infusion analysis is coupled with monophasic liquid extraction methods (such as MeOH, ethanol, and isopropanol), a signal drop of major lipids caused by the loss of samples during transferring can be reduced [70–73]. Thus, an orthogonal extraction strategy that combines multiple simplified extraction methods, each of which consumes a portion of the sample, and the final combined data can effectively increase the data amount for lipidomic analysis. A recent review provided a good guide for solvent selection in lipid extraction [74]. In addition, the available methods of chemical derivatization in small volume samples are limited. Multiple sample manipulation steps will result in transferring losses, such that selection and development of one-pot derivatization methods, such as Fmoc derivatization of phosphatidylethanolamine (PE) [75] and 2-picolylamine derivatization of fatty acids [76], which do not require separation and purification are necessary.
Microfluidic devices may have the potential to enhance micro level lipid analysis [77]. In this study by Sun et al., the authors integrated solid-phase microextraction materials into microfluidic tubing. The advanced point of this idea is the selective separation of lipid from samples to reduce the suppression effects of other non-lipid molecules. However, current microfluidic devices are mostly developed for operation in an aqueous phase system. The materials used in microfluidic devices are usually not compatible with solvents required for comprehensive lipid extraction, because chloroform is required. Although the fully implemented example has not yet appeared, we believe that development of a slipchip microfluidic device designed with all-glass materials can be used to achieve comprehensive extraction of lipids via microlevel biphasic separation such as Bligh-Dyer extraction [78, 79].
MS imaging analysis at the tissue level is readily available. Desorption electrospray ionization (DESI) and MALDI mass spectrometry imaging are the most common methods. At the modest-to-high spatial resolution level, DESI imaging achieves resolutions between 100 μm and 200 μm. Although the resolution of the DESI method is not high, this method can realize the analysis of the native state of the tissue. This method has been widely used for clinical sample biomarker discovery and interoperation tissue boundary recognition of cancer [27, 28, 80, 81]. If the need for rapid diagnosis is not taken into account, the use of higher resolution methods such as MALDI imaging can provide more detailed information for pathological analysis, making it easier to match the results of histochemistry. In the field of application, the resolution achieved by MALDI imaging is between 20 μm and 100 μm. For example, in a model study of colorectal cancer liver metastasis, the authors screened a matrix that effectively enhanced glycerophospholipid signals in liver tissue, revealing fingerprinting lipid species for tumor metastasis at different spatial resolutions [82]. In the high-resolution mode, small tumor foci at the early-stage of metastasis of the mouse liver were revealed. There are many excellent reviews on mass spectrometry imaging methods that are worth reading [80, 83–86]. Here, we mainly discuss some technical aspects for lipidomic analysis.
The advantage of the DESI-based imaging method is its convenience to obtain essential information needed for pathological diagnosis without additional sample processing [27–29]. This method keeps the sample in its natural state. Inspired by the MDMS strategy [38, 39], which utilizes multiple MS modes and selective ionization, lipid classes can also be selectively ionized by changing the solution composition in DESI. A major drawback of this type of method is the inability to conduct absolute quantification because it lacks a precise way to add internal standards.
The MALDI imaging method enables a higher resolution analysis than DESI. A large number of small molecule matrices are available for lipidomic analysis [87]. In imaging analysis, lipids cannot be physically separated. Therefore, selective ionization is an optimal approach to improve the dynamic range and lipid coverage. Although many matrices have been developed for lipid analysis, there are not sufficient to construct a complete selection list for selective ionization analysis. The current matrix combinations for selective ionization do not compare to the flexibility of direct infusion analysis [22, 24]. For MALDI analysis, discovery of a novel matrix may also promote improvements of ionization efficiency. However, discovery of these matrices, which can significantly improve sensitivity for lipid analysis, is difficult to predict. Nanomaterial matrices are a type of matrix that is expected. However, although the nanomaterial matrix exhibits excellent low background interference properties, their sensitivity to analyzing lipids cannot be compared to organic matrices. Currently, nanomaterial matrices are more advantageous for the analysis of small molecular weight metabolites, but not lipids because the organic matrix (even those specific for metabolites analysis) still has strong or unpredictable interference peaks in the range of molecular weights less than 500 Da.
At modest-to-high resolution levels, MALDI imaging can provide absolute quantitative analysis [88]. An internal standard addition method based on a spotter or inkjet printer can accurately provide a given amount of internal standard at a modest resolution level [89]. As far as we know, there are no example that using a pneumatic spray method for accurate addition of internal standards for high-resolution imaging. However, in the case of correction with optic images, a precisely-controlled pneumatic spray method allows the accurate addition of internal standard. It should be noted that both the internal standard and matrix require a uniform distribution and co-crystallization with lipids in these sections. Therefore, the thickness of the tissue section and deposition time of the matrix need to be optimized. A thickness less than 10 μm is more conducive to quantitative analysis.
At the tissue level of analysis, sampling and extraction of small volume samples, as well as accurate and absolute quantification in mass spectrometry imaging, are currently major challenges. The acquisition of depth information, such as quantification of low abundance lipids and interpretation of structural information, becomes more difficult due to sensitivity or sample size limitations [90, 91]. Therefore, future developments may come by improving ionization efficiency. This is discussed in the following sections. Table 2 provides some comparisons and technical features of related technologies.
2.3. Single-cell level analysis
Single-cell lipidomic analysis is very important for revealing fundamental biological processes [92]. For example, cell division is a highly dynamic process that is rapidly regulated in the cell cycle [93]. This process involves dramatic changes in cell membrane structure and composition. It is known that hundreds of proteins are involved in regulation, synthesis, decomposition, modification, and transport of lipids at complex spatial and temporal levels [6]. However, people understanding of protein and gene expression regulation in this process is far beyond our understanding of lipid changes. In addition, the plasma membrane is the basis for maintaining cellular homeostasis. Analysis of its changes is necessary to reveal the disease mechanism.
Lipids are not synthesized under the guidance of the DNA sequence. Lipid synthesis can be directly derived from conversion of exogenous uptake or occur ab initio [94]. Analysis of the genome at the single-cell level can be performed using amplification and labeling methods to achieve highly sensitive analysis [95]. Proteomics can use the antibody or labeling technology to achieve high-resolution analysis with the laser ablation inductively coupled plasma mass spectrometry imaging [96]. However, the challenge of lipidomics in high-resolution analysis is even greater. There are no methods to amplify, and a labeling method that can be used for signal enhancement is lacking. In addition, due to its chemical properties, many aqueous phase analysis methods are not suitable for lipid analysis. The most important choice to overcome these mismatches and achieve a technological breakthrough is the advancement of mass spectrometry.
Analysis at the single-cell level is divided into two categories: imaging analysis at single-cell or subcellular resolution and sensitivity, which is subject to the sampling area size, mainly through MALDI or secondary ion mass spectrometry (SIMS). This method is characterized by providing important spatial information. As resolution increases, sub-cellular level imaging analysis can be achieved. The other analysis strategy is to sample and analyze single cells directly using infusion ionization.
The cytosol lipids of cells can be extracted by capillary micro sampling [97]. The advantage of this method is that they can be directly sampled from the tissue surface or cultured adherent cells without cell isolation. The disadvantage of this method is that it does not reflect the actual or even major lipid components of a cell. Because the cytoplasm is not uniform, the extracted cytosol is not complete, and the cell membrane components cannot be analyzed. However, this is an effective method when it is necessary to exclude interference from the cell membrane. There have been many studies on single-cell metabolomics and lipidomics methods for extracting cytosol from cells. These methods include different ionization methods [98–100], optimization of sampling and direct infusion volumes [101–103], and use of SPE for selective extraction [104, 105].
Analysis of intact cells can provide more comprehensive information about the cellular lipidome [106, 107]. However, application of this analytical method requires isolation of adherently-grown cells or analysis of suspended cells. It should be noted that the method of analyzing single cells based on droplets and microfluidic systems has matured in single-cell manipulation. However, online direct infusion mass spectrometry is not optimized for lipid analysis because the design of these microfluidic methods is based on aqueous phase analysis. Currently a large number of compromise strategies are the separation of cells using a microfluidic system followed by offline detection using MALDI MS [108, 109].
Currently, relative quantitative analysis of direct infusion single-cell lipidomics is based on a comparison of normalized mass spectral signal intensities [103, 107–111]. This relative comparison cannot be consistent due to the ionization environment, and the precision and reliability of the quantification are affected to some extent. Achieving absolute quantification of single-cell lipidome analysis or quantification via a stable reference is an important direction that needs to be addressed in the future.
MS imaging analysis is an important approach for lipidomic analysis at the single-cell and subcellular levels. For high-resolution MALDI imaging, there are two configurations to perform laser adsorption and ionization: the transmission (Figure 2A) and reflection (Figure 2B, C) modes. Due to the characteristics of the MALDI instrument, the distance from the exit objective lens to the sample is usually a few centimeters long. In the ordinary optical design, it is difficult to focus below 10 μm in the reflective mode. To improve resolution (to decrease the laser spot size), researchers have proposed an improved configuration based on the microscope design, which uses a transmission mode to allow the laser to be incident from the back of the sample to focus on the surface of the sample section [112]. This is a brilliant and simple method that draws directly from proven technologies. This design allows the lens to be placed very close to the sample for single-cell level imaging. This transmission-mode geometry optical design makes it easier to achieve subcellular resolution. However, the transmission mode has high requirements for sample uniformity. Due to the texture of the sample and influences of crystallization of the matrix, the effective intensity of the laser reaching the surface after passing through the sample varies. This inhomogeneity of light intensity affects the ionization efficiency [112].
Figure 2.
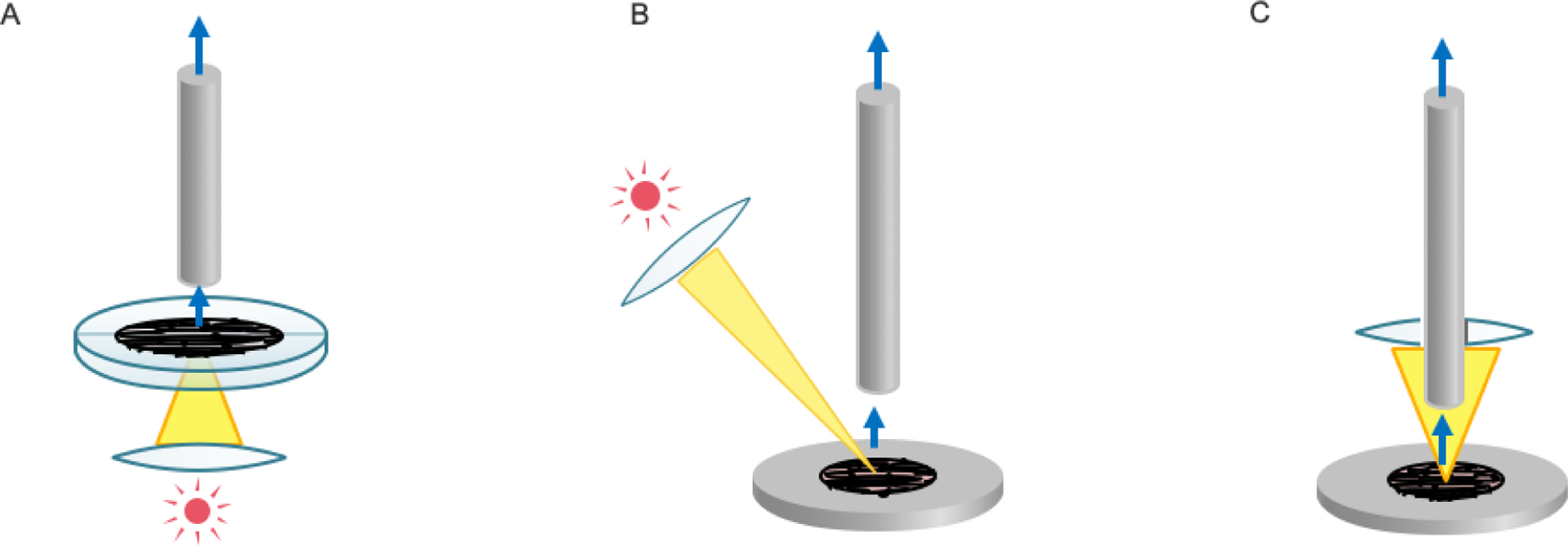
Schematic diagram of MALDI MS imaging configuration. A, Transmission mode. B, Reflection mode. C, SMALDI.
Most high-resolution MS imaging systems use the reflection mode. In commercially available products, the resolution for tissue imaging has reached 10 μm. In theory, it reaches the threshold for single-cell resolution. In fact, although the diameter of cells in tissue is mostly in the range of 10–20 μm, an MSI resolution below 5 μm is similar to a single-cell resolution level due to the heterogeneity of cell morphology. To improve the resolution in the reflection mode, the resolution of the 5 μm level can be achieved by expanding the laser spot diameter before focusing, improving the spot shape of the light source, and optimizing the lens-to-sample distance [113]. Due to inherent characteristics of optical systems, efforts to further reduce the spot in the reflective mode require more complex optical designs and are very difficult. It is well known that reducing the distance from the lens to the sample surface can easily reduce the size of the laser spot. To enable the lens to be placed close to the sample for high-resolution imaging while avoiding disadvantages of the transmission mode, an improved reflection mode was created by generating a hollow lens (Figure 2C). In this way, the sample can fly along the lens axil through a hole in the reflection mode, and the laser can focus on the sample surface through the peripheral annular area of the lens. This method is called scanning microprobe matrix-assisted laser desorption ionization (SMALDI) [114, 115]. Recently, the best resolution of mass spectrometry imaging at 1.4 μm using this mode has been reached [116]. At this resolution power, changes in lipids during mitosis can be observed. The autofocus system is an integral part of the system because of the depth of field decreases dramatically. The lens of SMALDI can be very close to the sample to achieve high-resolution imaging; however, the splashed sample can also cause contamination of the lens, affecting the stability for long-term acquisition. In addition, since the central portion of the lens with the best optical performance cannot be utilized, and the shape of the spot (laser energy profile) is not optimized. Yet, overall, this design can well balance the effects of various constraints. By further improving the design of the lens system, it is expected that the resolution will be enhanced to less than 1 μm in the near future.
For the reflection mode, it may be difficult to reduce the spatial resolution to below 400–500 nm for lipid MS imaging. At the 400–500 nm resolution level, the amount of sample that can be utilized is only 1/10 of the current best resolution. To improve the sensitivity, mainly by increasing the ionization efficiency, it would be effective to use a laser with a higher energy density, a shorter pulse, or a method of introducing post-ionization. We are optimistic about achieving resolutions at the near 400–500 nm level, as the major technical obstacles have been resolved in recent years. This is because focusing on this level can be achieved by expanding the laser beam diameter before objective lens focusing [113] and shortening the distance from the objective lens to the sample. The matrix sublimation deposition technique also has no obstacles at this level. The morphology of the tissue section was also stable at atmospheric pressure, and the deformation of the sample was also minimized [117]. The main problem with MALDI at atmospheric pressure is a decrease in sensitivity. The integration of other technologies, such as laser-induced post-ionization technique, can hopefully achieve imaging analysis at the 400–500 nm resolution. Therefore, the SMALDI configuration is likely to be the most accurate MS imaging technology that is able to match optical microscope imaging. It is highly possible that for quantitative lipid imaging, the internal standards can be quantitatively deposited using a pneumatic spray or electrospray-assisted pneumatic spray, and the resolution level around1 μm can be achieved.
In theory, the optical resolution can be greatly improved in the transmission mode, especially under an ultraviolet excitation source, and the Abbe limit can be as low as about 150 nm. However, considering the problems mentioned above, the non-uniformity of the transmitted laser beam must be solved to ensure the stability and reliability of the signal. The technical complexity of achieving the requirements can be very high because crystallization of the matrix can affect laser focusing. In addition, a sophisticated and complex adaptive optics system is required. It is necessary to equip with a more accurate autofocus system, automatic energy compensation system, and short pulse laser. In addition, matrix deposition at a nanometer scale is also a technical problem. Although the matrix sublimation method achieved the best-known minimum crystal size, there are no effective approaches to achieve quantitative sublimation of internal standards. Therefore, it is difficult to perform accurate quantitative MS imaging.
Sensitivity is a central constraint factor for accessing rich information from each pixel. As spatial resolution increases, the amount of limitedly available sample in each pixel leads to insufficient sensitivity. In addition to the quantification challenge, it is increasingly difficult to perform identification by MS/MS in situ.
In the MALDI process, the ionization efficiency is less than 1 out of 1000 desorbed molecules [118]. The sensitivity can be greatly improved by increasing the ionization efficiency. In addition to the development of new matrices and increased the sensitivity of a detector, increasing the energy profile of the laser is a direct way, such as changing the tunable wavelength and improved spot energy distribution. Derivatization is another way to increase the sensitivity. This method works well for targeted analysis. However, derivatization at the single-cell resolution level is much more difficult to implement than the tissue level.
From the perspective of the instrumentation, universal improvement of the ionization efficiency by post-ionization is a fundamental way for single-cell level analysis. An improved post-ionization method (called MALDI-2) based on MALDI imaging was designed by Soltwisch and co-workers (Figure 3A) [119]. A sample plume produced by the first MALDI process was post-ionized by a second laser beam to generate secondary MALDI-like ionization processes in the gas phase. At a 5-μm resolution, the signal increased by two orders of magnitude. A large number of neutral molecules in the gas plume are ionized. The ionization efficiency of low-abundance species, which are suppressed by high-abundance species, is greatly enhanced. This increase in ionization efficiency also increases the dynamic range. The application of laser post-ionization technology in MALDI imaging has proven that this idea is one of the most potential and scalable technologies for single-cell level MS imaging. We expect the combination of SMALDI and MALDI-2 to archive accurate mass spectrometry imaging of lipids at subcellular resolution.
Figure 3.
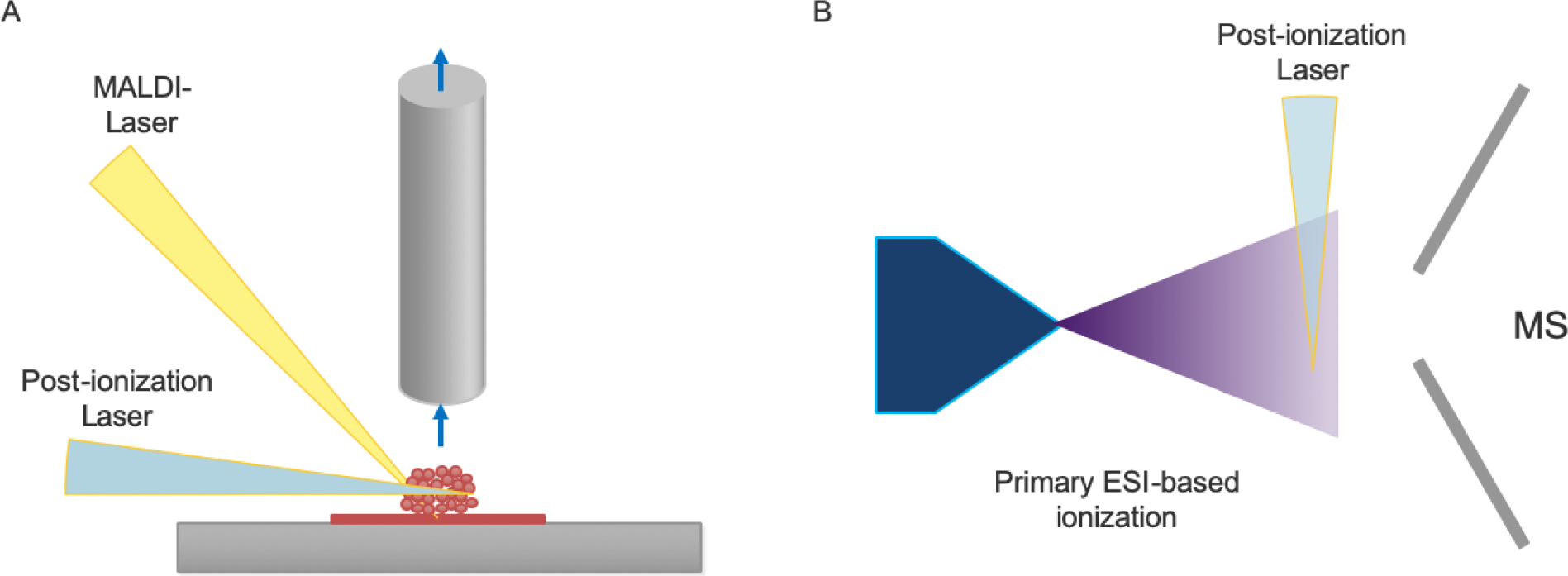
Schematic diagram of laser-induced post-ionization. A, MALDI MS imaging with laser-induced post-ionization. B, ESI-based direct infusion MS with laser-induced post-ionization.
This laser-induced post-ionization strategy can be used not only for MALDI imaging, but also for ESI-based single-cell analysis (Figure 3B) [99]. If the ionization efficiency is high enough, it can improve sensitivity and reduce the ion suppression effect, which is more conducive for quantitative analysis.
Unlike MALDI and DESI, we believe that the SIMS method is very suitable for providing surface information because there is no extraction process. The gas cluster ion beam (GCIB) technology using the super heavy ion beam in the SIMS can achieve minimal molecular fragments, while the sensitivity is also greatly reduced [120]. If the ion beam energy is increased, the resolution can be increased accordingly; however, the fragmentation becomes severe. So although the idealized resolution can be higher, the current practical resolution is 2 μm [121]. The main development direction of this method is to generate ion beams with a larger cluster size. If a significant breakthrough is made in the development of ion beams, this approach may provide a wealth of meaningful information for lipidomic surface imaging, even including three-dimensional imaging. The advantage of nanoSIMS is that it can achieve a 200-nm resolution [122]; however, valid qualitative information is limited. As such, it is difficult to expect that this technique will have a wide range of applications in lipidomics.
In lipidomic analysis at the single-cell level, direct infusion-based analysis and imaging-based analysis complement each other. At this level of analysis, no matter which method is used, the sensitivity becomes the most important constraint factor. For both electrospray and MALDI ionization processes, there are 2–3 orders of magnitude of signal improvement in ionization efficiency to be achieved. As the performance of ion transmission and detectors increases, detection sensitivity will increase further. Table 2 provides some comparisons and summaries of related technologies and challenges.
3. Perspectives on shotgun lipidomics at temporal levels
The difficulty of analyzing dynamic changes of lipids on a time resolution is far greater than analysis on spatial resolution. In circadian physiology, such as the effects of circadian rhythm changes on metabolism or in stress response studies, such as anesthesia resuscitation or inflammatory response, there have been some studies on lipid changes at different time points, and many interesting discoveries have been revealed [123–128]. These studies are all in hours or days as a time unit. By reducing the sampling interval, temporal resolution can be increased, but the temporal resolution that can be improved is very limited. Stable isotope labeling methods are often used for comparative/quantitative analysis. For cultured cell samples, stable isotope labeling can be used to track the flux of lipid metabolism. Some representative methods, such as X13CMS or lipidome isotope labeling of yeast (LILY) [129, 130], have demonstrated that lipid labeling can be efficiently quantified. The stable isotope labeling method can also be used for labeling in small animals. There is little research in applying isotopic labeling for high temporal resolution lipidomics. We believe that lipidomic analysis with higher temporal resolution can be obtained by selecting multiple isotopic labels and applying appropriate time or transient labeling.
Due to the fluidity of the membrane, even if the time interval of sample collection is shortened or stable isotope labeling is used, dynamic information of membrane lipids may change due to a long duration of removal from a physiological environment. Ultra-fast and low-temperature freezing can reduce this effect. The SIMS technology is a surface analysis method that is compatible with cryo-freeze during sample preparation [131]. Therefore, it is possible to study dynamic changes of the cellular lipidome by this means, and simultaneously, high-resolution imaging can be achieved. It is known that PE is usually enriched on the inner leaflet of the cell membrane, whereas in mitotic cytokinesis, PE is distributed on the outer leaflet in the cleave furrow [132]. Similar to such dynamic changes of PE during mitosis, the analysis of time snapshots obtained by SIMS cell surface scanning may reveal many fundamental discoveries that are yet unknown.
Since mass spectrometry requires substantial ‘extraction’ of molecules from the sample, the challenges of using mass spectrometry to analyze dynamic lipidome changes at high temporal resolution far exceeds the challenge in spatial resolution. Optical-based analysis methods have significant advantages in high time-resolved dynamic analysis and real-time analysis in vivo. Fluorescent probe labeling is commonly used in optical imaging methods[133]. The advantage of this type of approach is high sensitivity. The disadvantage is that the structure of the fluorescent probe is inconsistent with the natural structure of the lipid molecule. Since the lipid molecules themselves are relatively small, this type of probe may have a greater negative effect on the results than the protein probe. Therefore, real-time imaging methods based on Raman spectroscopy have been developed in recent years[134]. This method overcomes the shortcomings of fluorescent probes. The insufficiency of Raman imaging is that the sensitivity needs to be improved. Currently, it is usually used to observe specific cells type rich in lipids. Although optical-based methods can achieve high temporal resolution, the specificity is limited. The result often displays the overall functioning of a class of lipids. It is difficult to distinguish specific lipid species.
4. Conclusion
Mass spectrometric analysis of lipids has evolved for nearly 40 years. Shotgun lipidomics has also exceeded 15 years of research history. During the development process, the scope and methods of shotgun lipidomics have undergone profound changes. The scope of shotgun lipidomics has also expanded from massive tissue sample level to micro- or even single-cell levels. The analysis ranges from qualitative and quantitative determinations of lipid molecular species expended to position determinations of unsaturated bonds and isomer characterization and quantification. Data dimensions of the analysis extend from mixtures to spatial distribution of lipid molecules and dynamic change characterization. The expansion of these research areas has been driven by advances in a range of instruments and analytical methods. Quantitative analysis is a major advantage of shotgun lipidomics, including its increased sensitivity as a prerequisite. Increasing ionization efficiency or selective ionization improves sensitivity and the dynamic detection range, which is crucial for shotgun lipidomics.
Acknowledgments
This work was partially supported by NIH/NIA (RF1 AG061872), the institutional research funds from the University of Texas Health Science Center at San Antonio (UT Health SA), the Mass Spectrometry Core Facility at UT Health SA, and the Methodist Hospital Foundation.
References
- [1].Kishimoto K, Urade R, Ogawa T, Moriyama T, Nondestructive quantification of neutral lipids by thin-layer chromatography and laser-fluorescent scanning: suitable methods for “lipidome” analysis, Biochem Biophys Res Commun, 281 (2001) 657–662. [DOI] [PubMed] [Google Scholar]
- [2].Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA, Update of the LIPID MAPS comprehensive classification system for lipids, J Lipid Res, 50 Suppl (2009) S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH Jr., Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA, A comprehensive classification system for lipids, J Lipid Res, 46 (2005) 839–861. [DOI] [PubMed] [Google Scholar]
- [4].Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH Jr., Murphy RC, Raetz CR, Russell DW, Subramaniam S, LMSD: LIPID MAPS structure database, Nucleic Acids Res, 35 (2007) D527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Han X, Lipidomics for studying metabolism, Nat Rev Endocrinol, 12 (2016) 668–679. [DOI] [PubMed] [Google Scholar]
- [6].van Meer G, Voelker DR, Feigenson GW, Membrane lipids: where they are and how they behave, Nat Rev Mol Cell Bio, 9 (2008) 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shevchenko A, Simons K, Lipidomics: coming to grips with lipid diversity, Nat Rev Mol Cell Bio, 11 (2010) 593–598. [DOI] [PubMed] [Google Scholar]
- [8].Gross RW, Han X, Lipidomics at the interface of structure and function in systems biology, Chem Biol, 18 (2011) 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Han X, Gross RW, Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids, Proc Natl Acad Sci USA, 91 (1994) 10635–10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kerwin JL, Tuininga AR, Ericsson LH, Identification of Molecular-Species of Glycerophospholipids and Sphingomyelin Using Electrospray Mass-Spectrometry, Journal of Lipid Research, 35 (1994) 1102–1114. [PubMed] [Google Scholar]
- [11].Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD, Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry, Proc Natl Acad Sci USA, 94 (1997) 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Han X, Gross RW, Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics, J Lipid Res, 44 (2003) 1071–1079. [DOI] [PubMed] [Google Scholar]
- [13].Jung HR, Sylvanne T, Koistinen KM, Tarasov K, Kauhanen D, Ekroos K, High throughput quantitative molecular lipidomics, Biochim Biophys Acta, 1811 (2011) 925–934. [DOI] [PubMed] [Google Scholar]
- [14].Hsu FF, Turk J, Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: mechanisms of fragmentation and structural characterization, J Chromatogr B, 877 (2009) 2673–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang M, Wang C, Han X, Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why?, Mass Spectrom Rev, 36 (2017) 693–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hsu FF, Mass spectrometry-based shotgun lipidomics - a critical review from the technical point of view, Anal Bioanal Chem, 410 (2018) 6387–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A, Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry, Proc Natl Acad Sci USA, 106 (2009) 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Han X, Gross RW, Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples, Mass Spectrom Rev, 24 (2005) 367–412. [DOI] [PubMed] [Google Scholar]
- [19].Yang K, Han X, Accurate quantification of lipid species by electrospray ionization mass spectrometry - Meet a key challenge in lipidomics, Metabolites, 1 (2011) 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang K, Han X, Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences, Trends Biochem Sci, 41 (2016) 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sustarsic EG, Ma T, Lynes MD, Larsen M, Karavaeva I, Havelund JF, Nielsen CH, Jedrychowski MP, Moreno-Torres M, Lundh M, Plucinska K, Jespersen NZ, Grevengoed TJ, Kramar B, Peics J, Hansen JB, Shamsi F, Forss I, Neess D, Keipert S, Wang J, Stohlmann K, Brandslund I, Christensen C, Jorgensen ME, Linneberg A, Pedersen O, Kiebish MA, Qvortrup K, Han X, Pedersen BK, Jastroch M, Mandrup S, Kjaer A, Gygi SP, Hansen T, Gillum MP, Grarup N, Emanuelli B, Nielsen S, Scheele C, Tseng YH, Faergeman NJ, Gerhart-Hines Z, Cardiolipin Synthesis in Brown and Beige Fat Mitochondria Is Essential for Systemic Energy Homeostasis, Cell Metab, 28 (2018) 159–174 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schroter J, Fulop A, Hopf C, Schiller J, The combination of 2,5-dihydroxybenzoic acid and 2,5-dihydroxyacetophenone matrices for unequivocal assignment of phosphatidylethanolamine species in complex mixtures, Anal Bioanal Chem, 410 (2018) 2437–2447. [DOI] [PubMed] [Google Scholar]
- [23].Shanta SR, Zhou LH, Park YS, Kim YH, Kim Y, Kim KP, Binary Matrix for MALDI Imaging Mass Spectrometry of Phospholipids in Both Ion Modes, Anal Chem, 83 (2011) 1252–1259. [DOI] [PubMed] [Google Scholar]
- [24].Wang J, Wang C, Han X, Enhanced coverage of lipid analysis and imaging by matrix-assisted laser desorption/ionization mass spectrometry via a strategy with an optimized mixture of matrices, Anal Chim Acta, 1000 (2018) 155–162. [DOI] [PubMed] [Google Scholar]
- [25].Cheng H, Sun G, Yang K, Gross RW, Han X, Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix, J Lipid Res, 51 (2010) 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ibrahim H, Jurcic K, Wang JS, Whitehead SN, Yeung KK, 1,6-Diphenyl-1,3,5-hexatriene (DPH) as a Novel Matrix for MALDI MS Imaging of Fatty Acids, Phospholipids, and Sulfatides in Brain Tissues, Anal Chem, 89 (2017) 12828–12836. [DOI] [PubMed] [Google Scholar]
- [27].Zhang J, Yu W, Ryu SW, Lin J, Buentello G, Tibshirani R, Suliburk J, Eberlin LS, Cardiolipins Are Biomarkers of Mitochondria-Rich Thyroid Oncocytic Tumors, Cancer Res, 76 (2016) 6588–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sans M, Gharpure K, Tibshirani R, Zhang J, Liang L, Liu J, Young JH, Dood RL, Sood AK, Eberlin LS, Metabolic Markers and Statistical Prediction of Serous Ovarian Cancer Aggressiveness by Ambient Ionization Mass Spectrometry Imaging, Cancer Res, 77 (2017) 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Abbassi-Ghadi N, Golf O, Kumar S, Antonowicz S, McKenzie JS, Huang J, Strittmatter N, Kudo H, Jones EA, Veselkov K, Goldin R, Takats Z, Hanna GB, Imaging of Esophageal Lymph Node Metastases by Desorption Electrospray Ionization Mass Spectrometry, Cancer Res, 76 (2016) 5647–5656. [DOI] [PubMed] [Google Scholar]
- [30].Ellis SR, Brown SH, Het Panhuis M, Blanksby SJ, Mitchell TW, Surface analysis of lipids by mass spectrometry: more than just imaging, Prog Lipid Res, 52 (2013) 329–353. [DOI] [PubMed] [Google Scholar]
- [31].Hall Z, Chu Y, Griffin JL, Liquid Extraction Surface Analysis Mass Spectrometry Method for Identifying the Presence and Severity of Nonalcoholic Fatty Liver Disease, Anal Chem, 89 (2017) 5161–5170. [DOI] [PubMed] [Google Scholar]
- [32].Ha SJ, Bailey L, Ferreira C, Wang HY, Bailey B, Ding JX, Saadatzadeh MR, Pollok KE, Clase K, Shotgun lipidomics analysis of temozolomide-treated glioblastoma, Cancer Research, 77 (2017). [Google Scholar]
- [33].Perez O, Margolis M, Santander AM, Martinez M, Bhattacharya S, Torroella-Kouri M, Breast cancer and obesity impact the lipid composition of breast adipose tissue: a preliminary study using shotgun lipidomics, Cancer Research, 74 (2014). [Google Scholar]
- [34].Lynes MD, Shamsi F, Sustarsic EG, Leiria LO, Wang CH, Su SC, Huang TL, Gao F, Narain NR, Chen EY, Cypess AM, Schulz TJ, Gerhart-Hines Z, Kiebish MA, Tseng YH, Cold-Activated Lipid Dynamics in Adipose Tissue Highlights a Role for Cardiolipin in Thermogenic Metabolism, Cell Rep, 24 (2018) 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A, Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ Orbitrap mass spectrometers, Anal Chem, 83 (2011) 5480–5487. [DOI] [PubMed] [Google Scholar]
- [36].Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A, Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra, Anal Chem, 79 (2007) 4083–4093. [DOI] [PubMed] [Google Scholar]
- [37].Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A, Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition, Anal Chem, 78 (2006) 585–595. [DOI] [PubMed] [Google Scholar]
- [38].Yang K, Cheng H, Gross RW, Han X, Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics, Anal Chem, 81 (2009) 4356–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Han X, Gross RW, Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes, Expert Rev Proteomic, 2 (2005) 253–264. [DOI] [PubMed] [Google Scholar]
- [40].Wang M, Wang C, Han RH, Han X, Novel advances in shotgun lipidomics for biology and medicine, Prog Lipid Res, 61 (2016) 83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang J, Wang C, Han X, Tutorial on lipidomics, Anal Chim Acta, 1061 (2019) 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Han Xianlin, Lipidomics : comprehensive mass spectrometry of lipids, Wiley, Hoboken, New Jersey, 2016. [Google Scholar]
- [43].Han X, Yang K, Gross RW, Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics, Rapid Commun Mass Spectrom, 22 (2008) 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ryan E, Reid GE, Chemical Derivatization and Ultrahigh Resolution and Accurate Mass Spectrometry Strategies for “Shotgun” Lipidome Analysis, Acc Chem Res, 49 (2016) 1596–1604. [DOI] [PubMed] [Google Scholar]
- [45].Wang C, Palavicini JP, Wang M, Chen L, Yang K, Crawford PA, Han X, Comprehensive and Quantitative Analysis of Polyphosphoinositide Species by Shotgun Lipidomics Revealed Their Alterations in db/db Mouse Brain, Anal Chem, 88 (2016) 12137–12144. [DOI] [PubMed] [Google Scholar]
- [46].Wang M, Han RH, Han X, Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach, Anal Chem, 85 (2013) 9312–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang M, Palavicini JP, Cseresznye A, Han X, Strategy for Quantitative Analysis of Isomeric Bis(monoacylglycero)phosphate and Phosphatidylglycerol Species by Shotgun Lipidomics after One-Step Methylation, Anal Chem, 89 (2017) 8490–8495. [DOI] [PubMed] [Google Scholar]
- [48].Hu C, Wang C, He L, Han X, Novel strategies for enhancing shotgun lipidomics for comprehensive analysis of cellular lipidomes, Trends Anal Chem, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rustam YH, Reid GE, Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics, Anal Chem, 90 (2018) 374–397. [DOI] [PubMed] [Google Scholar]
- [50].Chatgilialoglu C, Ferreri C, Melchiorre M, Sansone A, Torreggiani A, Lipid geometrical isomerism: from chemistry to biology and diagnostics, Chem Rev, 114 (2014) 255–284. [DOI] [PubMed] [Google Scholar]
- [51].Vu N, Brown J, Giles K, Zhang Q, Ozone-induced dissociation on a traveling wave high-resolution mass spectrometer for determination of double-bond position in lipids, Rapid Commun Mass Spectrom, 31 (2017) 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ma X, Chong L, Tian R, Shi R, Hu TY, Ouyang Z, Xia Y, Identification and quantitation of lipid C=C location isomers: A shotgun lipidomics approach enabled by photochemical reaction, Proc Natl Acad Sci U S A, 113 (2016) 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Williams PE, Klein DR, Greer SM, Brodbelt JS, Pinpointing Double Bond and sn-Positions in Glycerophospholipids via Hybrid 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry, J Am Chem Soc, 139 (2017) 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hancock SE, Poad BL, Batarseh A, Abbott SK, Mitchell TW, Advances and unresolved challenges in the structural characterization of isomeric lipids, Anal Biochem, 524 (2017) 45–55. [DOI] [PubMed] [Google Scholar]
- [55].Paglia G, Kliman M, Claude E, Geromanos S, Astarita G, Applications of ion-mobility mass spectrometry for lipid analysis, Anal Bioanal Chem, 407 (2015) 4995–5007. [DOI] [PubMed] [Google Scholar]
- [56].Bowman AP, Abzalimov RR, Shvartsburg AA, Broad Separation of Isomeric Lipids by High-Resolution Differential Ion Mobility Spectrometry with Tandem Mass Spectrometry, J Am Soc Mass Spectrom, 28 (2017) 1552–1561. [DOI] [PubMed] [Google Scholar]
- [57].Groessl M, Graf S, Knochenmuss R, High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids, Analyst, 140 (2015) 6904–6911. [DOI] [PubMed] [Google Scholar]
- [58].Paglia G, Astarita G, Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry, Nat Protoc, 12 (2017) 797–813. [DOI] [PubMed] [Google Scholar]
- [59].Zhang X, Quinn K, Cruickshank-Quinn C, Reisdorph R, Reisdorph N, The application of ion mobility mass spectrometry to metabolomics, Curr Opin Chem Biol, 42 (2018) 60–66. [DOI] [PubMed] [Google Scholar]
- [60].Hinz C, Liggi S, Griffin JL, The potential of Ion Mobility Mass Spectrometry for high-throughput and high-resolution lipidomics, Curr Opin Chem Biol, 42 (2018) 42–50. [DOI] [PubMed] [Google Scholar]
- [61].Zheng XY, Smith RD, Baker ES, Recent advances in lipid separations and structural elucidation using mass spectrometry combined with ion mobility spectrometry, ion-molecule reactions and fragmentation approaches, Current Opinion in Chemical Biology, 42 (2018) 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li P, Kaslan M, Lee SH, Yao J, Gao Z, Progress in Exosome Isolation Techniques, Theranostics, 7 (2017) 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yang JS, Lee JC, Byeon SK, Rha KH, Moon MH, Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry, Anal Chem, 89 (2017) 2488–2496. [DOI] [PubMed] [Google Scholar]
- [64].Knittelfelder O, Traikov S, Vvedenskaya O, Schuhmann A, Segeletz S, Shevchenko A, Shevchenko A, Shotgun Lipidomics Combined with Laser Capture Microdissection: A Tool To Analyze Histological Zones in Cryosections of Tissues, Anal Chem, 90 (2018) 9868–9878. [DOI] [PubMed] [Google Scholar]
- [65].Gil A, Zhang W, Wolters JC, Permentier H, Boer T, Horvatovich P, Heiner-Fokkema MR, Reijngoud DJ, Bischoff R, One- vs two-phase extraction: re-evaluation of sample preparation procedures for untargeted lipidomics in plasma samples, Anal Bioanal Chem, 410 (2018) 5859–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Abbott SK, Jenner AM, Mitchell TW, Brown SH, Halliday GM, Garner B, An improved high-throughput lipid extraction method for the analysis of human brain lipids, Lipids, 48 (2013) 307–318. [DOI] [PubMed] [Google Scholar]
- [67].Birjandi AP, Bojko B, Ning Z, Figeys D, Pawliszyn J, High throughput solid phase microextraction: A new alternative for analysis of cellular lipidome?, J Chromatogr B Analyt Technol Biomed Life Sci, 1043 (2017) 12–19. [DOI] [PubMed] [Google Scholar]
- [68].Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D, Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics, J Lipid Res, 49 (2008) 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lofgren L, Stahlman M, Forsberg GB, Saarinen S, Nilsson R, Hansson GI, The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma, Journal of Lipid Research, 53 (2012) 1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lydic TA, Busik JV, Reid GE, A monophasic extraction strategy for the simultaneous lipidome analysis of polar and nonpolar retina lipids, J Lipid Res, 55 (2014) 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Shiva S, Enninful R, Roth MR, Tamura P, Jagadish K, Welti R, An efficient modified method for plant leaf lipid extraction results in improved recovery of phosphatidic acid, Plant Methods, 14 (2018) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li L, Yang SH, Lemr K, Havlicek V, Schug KA, Continuous flow-extractive desorption electrospray ionization: analysis from “non-electrospray ionization-friendly” solvents and related mechanism, Anal Chim Acta, 769 (2013) 84–90. [DOI] [PubMed] [Google Scholar]
- [73].Basu SS, Randall EC, Regan MS, Lopez BGC, Clark AR, Schmitt ND, Agar JN, Dillon DA, Agar NYR, In Vitro Liquid Extraction Surface Analysis Mass Spectrometry (ivLESA-MS) for Direct Metabolic Analysis of Adherent Cells in Culture, Anal Chem, 90 (2018) 4987–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cajka T, Fiehn O, Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics, Anal Chem, 88 (2016) 524–545. [DOI] [PubMed] [Google Scholar]
- [75].Han X, Yang K, Cheng H, Fikes KN, Gross RW, Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization, J Lipid Res, 46 (2005) 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wu Q, Comi TJ, Li B, Rubakhin SS, Sweedler JV, On-Tissue Derivatization via Electrospray Deposition for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging of Endogenous Fatty Acids in Rat Brain Tissues, Anal Chem, 88 (2016) 5988–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sun T, Pawlowski S, Johnson ME, Highly efficient microscale purification of glycerophospholipids by microfluidic cell lysis and lipid extraction for lipidomics profiling, Anal Chem, 83 (2011) 6628–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Du W, Li L, Nichols KP, Ismagilov RF, SlipChip, Lab Chip, 9 (2009) 2286–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang S, Chen S, Wang J, Xu P, Luo Y, Nie Z, Du W, Interface solution isoelectric focusing with in situ MALDI-TOF mass spectrometry, Electrophoresis, 35 (2014) 2528–2533. [DOI] [PubMed] [Google Scholar]
- [80].Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR, Mass spectrometry imaging under ambient conditions, Mass Spectrom Rev, 32 (2013) 218–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sans M, Zhang J, Lin JQ, Feider CL, Giese N, Breen MT, Sebastian K, Liu J, Sood AK, Eberlin LS, Performance of the MasSpec Pen for Rapid Diagnosis of Ovarian Cancer, Clin Chem, 65 (2019) 674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang J, Qiu S, Chen S, Xiong C, Liu H, Wang J, Zhang N, Hou J, He Q, Nie Z, MALDI-TOF MS imaging of metabolites with a N-(1-naphthyl) ethylenediamine dihydrochloride matrix and its application to colorectal cancer liver metastasis, Anal Chem, 87 (2015) 422–430. [DOI] [PubMed] [Google Scholar]
- [83].Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC, MALDI imaging of lipid biochemistry in tissues by mass spectrometry, Chem Rev, 111 (2011) 6491–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Norris JL, Caprioli RM, Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research, Chem Rev, 113 (2013) 2309–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Gilmore IS, Heiles S, Pieterse CL, Metabolic Imaging at the Single-Cell Scale: Recent Advances in Mass Spectrometry Imaging, Annu Rev Anal Chem (Palo Alto Calif), 12 (2019) null. [DOI] [PubMed] [Google Scholar]
- [86].Buchberger AR, DeLaney K, Johnson J, Li L, Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights, Anal Chem, 90 (2018) 240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Calvano CD, Monopoli A, Cataldi TRI, Palmisano F, MALDI matrices for low molecular weight compounds: an endless story?, Anal Bioanal Chem, 410 (2018) 4015–4038. [DOI] [PubMed] [Google Scholar]
- [88].Chumbley CW, Reyzer ML, Allen JL, Marriner GA, Via LE, Barry CE 3rd, Caprioli RM, Absolute Quantitative MALDI Imaging Mass Spectrometry: A Case of Rifampicin in Liver Tissues, Anal Chem, 88 (2016) 2392–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Luo Z, He J, He J, Huang L, Song X, Li X, Abliz Z, Quantitative analysis of drug distribution by ambient mass spectrometry imaging method with signal extinction normalization strategy and inkjet-printing technology, Talanta, 179 (2018) 230–237. [DOI] [PubMed] [Google Scholar]
- [90].Paine MRL, Poad BLJ, Eijkel GB, Marshall DL, Blanksby SJ, Heeren RMA, Ellis SR, Mass Spectrometry Imaging with Isomeric Resolution Enabled by Ozone-Induced Dissociation, Angew Chem Int Edit, 57 (2018) 10530–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bednarik A, Bolsker S, Soltwisch J, Dreisewerd K, An On-Tissue Paterno-Buchi Reaction for Localization of Carbon-Carbon Double Bonds in Phospholipids and Glycolipids by Matrix-Assisted Laser-Desorption-Ionization Mass-Spectrometry Imaging, Angew Chem Int Edit, 57 (2018) 12092–12096. [DOI] [PubMed] [Google Scholar]
- [92].Borland LM, Kottegoda S, Phillips KS, Allbritton NL, Chemical analysis of single cells, Annu Rev Anal Chem (Palo Alto Calif), 1 (2008) 191–227. [DOI] [PubMed] [Google Scholar]
- [93].Storck EM, Ozbalci C, Eggert US, Lipid Cell Biology: A Focus on Lipids in Cell Division, Annu Rev Biochem, 87 (2018) 839–869. [DOI] [PubMed] [Google Scholar]
- [94].Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr., Cellular fatty acid metabolism and cancer, Cell Metab, 18 (2013) 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gawad C, Koh W, Quake SR, Single-cell genome sequencing: current state of the science, Nat Rev Genet, 17 (2016) 175–188. [DOI] [PubMed] [Google Scholar]
- [96].Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B, Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry, Nat Methods, 11 (2014) 417–422. [DOI] [PubMed] [Google Scholar]
- [97].Zhang L, Vertes A, Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity, Angew Chem Int Edit, 57 (2018) 4466–4477. [DOI] [PubMed] [Google Scholar]
- [98].Wei Z, Xiong X, Guo C, Si X, Zhao Y, He M, Yang C, Xu W, Tang F, Fang X, Zhang S, Zhang X, Pulsed Direct Current Electrospray: Enabling Systematic Analysis of Small Volume Sample by Boosting Sample Economy, Anal Chem, 87 (2015) 11242–11248. [DOI] [PubMed] [Google Scholar]
- [99].Lee JK, Jansson ET, Nam HG, Zare RN, High-Resolution Live-Cell Imaging and Analysis by Laser Desorption/Ionization Droplet Delivery Mass Spectrometry, Anal Chem, 88 (2016) 5453–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jacobson RS, Thurston RL, Shrestha B, Vertes A, In Situ Analysis of Small Populations of Adherent Mammalian Cells Using Laser Ablation Electrospray Ionization Mass Spectrometry in Transmission Geometry, Anal Chem, 87 (2015) 12130–12136. [DOI] [PubMed] [Google Scholar]
- [101].Zhang L, Vertes A, Energy Charge, Redox State, and Metabolite Turnover in Single Human Hepatocytes Revealed by Capillary Microsampling Mass Spectrometry, Anal Chem, 87 (2015) 10397–10405. [DOI] [PubMed] [Google Scholar]
- [102].Zhang L, Sevinsky CJ, Davis BM, Vertes A, Single-Cell Mass Spectrometry of Subpopulations Selected by Fluorescence Microscopy, Anal Chem, 90 (2018) 4626–4634. [DOI] [PubMed] [Google Scholar]
- [103].Sun M, Yang Z, Metabolomic Studies of Live Single Cancer Stem Cells Using Mass Spectrometry, Anal Chem, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Piri-Moghadam H, Ahmadi F, Gomez-Rios GA, Boyaci E, Reyes-Garces N, Aghakhani A, Bojko B, Pawliszyn J, Fast Quantitation of Target Analytes in Small Volumes of Complex Samples by Matrix-Compatible Solid-Phase Microextraction Devices, Angew Chem Int Edit, 55 (2016) 7510–7514. [DOI] [PubMed] [Google Scholar]
- [105].Deng J, Yang Y, Xu M, Wang X, Lin L, Yao ZP, Luan T, Surface-coated probe nanoelectrospray ionization mass spectrometry for analysis of target compounds in individual small organisms, Anal Chem, 87 (2015) 9923–9930. [DOI] [PubMed] [Google Scholar]
- [106].Zhang XC, Zang Q, Zhao H, Ma X, Pan X, Feng J, Zhang S, Zhang R, Abliz Z, Zhang X, Combination of Droplet Extraction and Pico-ESI-MS Allows the Identification of Metabolites from Single Cancer Cells, Anal Chem, 90 (2018) 9897–9903. [DOI] [PubMed] [Google Scholar]
- [107].Chen F, Lin L, Zhang J, He Z, Uchiyama K, Lin JM, Single-Cell Analysis Using Drop-on-Demand Inkjet Printing and Probe Electrospray Ionization Mass Spectrometry, Anal Chem, 88 (2016) 4354–4360. [DOI] [PubMed] [Google Scholar]
- [108].Neumann EK, Comi TJ, Rubakhin SS, Sweedler J, Lipid heterogeneity between astrocytes and neurons revealed with single cell MALDI MS supervised by immunocytochemical classification, Angew Chem Int Edit, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Xie W, Gao D, Jin F, Jiang Y, Liu H, Study of Phospholipids in Single Cells Using an Integrated Microfluidic Device Combined with Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry, Anal Chem, 87 (2015) 7052–7059. [DOI] [PubMed] [Google Scholar]
- [110].Hua X, Li HW, Long YT, Investigation of Silver Nanoparticle Induced Lipids Changes on a Single Cell Surface by Time-of-Flight Secondary Ion Mass Spectrometry, Anal Chem, 90 (2018) 1072–1076. [DOI] [PubMed] [Google Scholar]
- [111].Do TD, Comi TJ, Dunham SJ, Rubakhin SS, Sweedler JV, Single Cell Profiling Using Ionic Liquid Matrix-Enhanced Secondary Ion Mass Spectrometry for Neuronal Cell Type Differentiation, Anal Chem, 89 (2017) 3078–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM, Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS, J Mass Spectrom, 47 (2012) 1473–1481. [DOI] [PubMed] [Google Scholar]
- [113].Feenstra AD, Duenas ME, Lee YJ, Five Micron High Resolution MALDI Mass Spectrometry Imaging with Simple, Interchangeable, Multi-Resolution Optical System, J Am Soc Mass Spectrom, 28 (2017) 434–442. [DOI] [PubMed] [Google Scholar]
- [114].Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B, A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer, Rapid Commun Mass Spectrom, 22 (2008) 3275–3285. [DOI] [PubMed] [Google Scholar]
- [115].Spengler B, Hubert M, Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis, J Am Soc Mass Spectrom, 13 (2002) 735–748. [DOI] [PubMed] [Google Scholar]
- [116].Kompauer M, Heiles S, Spengler B, Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-mum lateral resolution, Nat Methods, 14 (2017) 90–96. [DOI] [PubMed] [Google Scholar]
- [117].Kettling H, Vens-Cappell S, Soltwisch J, Pirkl A, Haier J, Muthing J, Dreisewerd K, MALDI mass spectrometry imaging of bioactive lipids in mouse brain with a Synapt G2-S mass spectrometer operated at elevated pressure: improving the analytical sensitivity and the lateral resolution to ten micrometers, Anal Chem, 86 (2014) 7798–7805. [DOI] [PubMed] [Google Scholar]
- [118].Knochenmuss R, Zhigilei LV, What determines MALDI ion yields? A molecular dynamics study of ion loss mechanisms, Anal Bioanal Chem, 402 (2012) 2511–2519. [DOI] [PubMed] [Google Scholar]
- [119].Soltwisch J, Kettling H, Vens-Cappell S, Wiegelmann M, Muthing J, Dreisewerd K, Mass spectrometry imaging with laser-induced postionization, Science, 348 (2015) 211–215. [DOI] [PubMed] [Google Scholar]
- [120].Tian H, Sparvero LJ, Amoscato AA, Bloom A, Bayir H, Kagan VE, Winograd N, Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry High-Resolution Imaging of Cardiolipin Speciation in the Brain: Identification of Molecular Losses after Traumatic Injury, Anal Chem, 89 (2017) 4611–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Passarelli MK, Pirkl A, Moellers R, Grinfeld D, Kollmer F, Havelund R, Newman CF, Marshall PS, Arlinghaus H, Alexander MR, West A, Horning S, Niehuis E, Makarov A, Dollery CT, Gilmore IS, The 3D OrbiSIMS-label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power, Nat Methods, 14 (2017) 1175–1183. [DOI] [PubMed] [Google Scholar]
- [122].Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP, Multiplexed ion beam imaging of human breast tumors, Nat Med, 20 (2014) 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, Asher G, Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides, Cell Metab, 19 (2014) 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Anitei M, Stange C, Czupalla C, Niehage C, Schuhmann K, Sala P, Czogalla A, Pursche T, Coskun U, Shevchenko A, Hoflack B, Spatiotemporal Control of Lipid Conversion, Actin-Based Mechanical Forces, and Curvature Sensors during Clathrin/AP-1-Coated Vesicle Biogenesis, Cell Rep, 20 (2017) 2087–2099. [DOI] [PubMed] [Google Scholar]
- [125].Aviram R, Manella G, Kopelman N, Neufeld-Cohen A, Zwighaft Z, Elimelech M, Adamovich Y, Golik M, Wang C, Han X, Asher G, Lipidomics Analyses Reveal Temporal and Spatial Lipid Organization and Uncover Daily Oscillations in Intracellular Organelles, Mol Cell, 62 (2016) 636–648. [DOI] [PubMed] [Google Scholar]
- [126].Man K, Loudon A, Chawla A, Immunity around the clock, Science, 354 (2016) 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Musiek ES, Holtzman DM, Mechanisms linking circadian clocks, sleep, and neurodegeneration, Science, 354 (2016) 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Panda S, Circadian physiology of metabolism, Science, 354 (2016) 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Rampler E, Coman C, Hermann G, Sickmann A, Ahrends R, Koellensperger G, LILY-lipidome isotope labeling of yeast: in vivo synthesis of (13)C labeled reference lipids for quantification by mass spectrometry, Analyst, 142 (2017) 1891–1899. [DOI] [PubMed] [Google Scholar]
- [130].Huang X, Chen YJ, Cho K, Nikolskiy I, Crawford PA, Patti GJ, X13CMS: global tracking of isotopic labels in untargeted metabolomics, Anal Chem, 86 (2014) 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Vickerman JC, Molecular imaging and depth profiling by mass spectrometry--SIMS, MALDI or DESI?, Analyst, 136 (2011) 2199–2217. [DOI] [PubMed] [Google Scholar]
- [132].Emoto K, Umeda M, An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine, J Cell Biol, 149 (2000) 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Fam TK, Klymchenko AS, Collot M, Recent Advances in Fluorescent Probes for Lipid Droplets, Materials (Basel), 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Fu D, Lu FK, Zhang X, Freudiger C, Pernik DR, Holtom G, Xie XS, Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy, J Am Chem Soc, 134 (2012) 3623–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Laaksonen R, Identifying new Risk Markers and Potential Targets for Coronary Artery Disease: The Value of the Lipidome and Metabolome, Cardiovasc Drugs Ther, 30 (2016) 19–32. [DOI] [PubMed] [Google Scholar]
- [136].Bhattacharya SK, Recent advances in shotgun lipidomics and their implication for vision research and ophthalmology, Curr Eye Res, 38 (2013) 417–427. [DOI] [PubMed] [Google Scholar]
- [137].Dehairs J, Derua R, Rueda-Rincon N, Swinnen JV, Lipidomics in drug development, Drug Discov Today Technol, 13 (2015) 33–38. [DOI] [PubMed] [Google Scholar]
- [138].Wood PL, Barnette BL, Kaye JA, Quinn JF, Woltjer RL, Non-targeted lipidomics of CSF and frontal cortex grey and white matter in control, mild cognitive impairment, and Alzheimer’s disease subjects, Acta Neuropsychiatr, 27 (2015) 270–278. [DOI] [PubMed] [Google Scholar]


