Abstract
Telomeres comprise specialized nucleic acid–protein complexes that help protect chromosome ends from DNA damage. Moreover, telomeres associate with subtelomeric regions through looping. This results in altered expression of subtelomeric genes. Recent observations further reveal telomere length–dependent gene regulation and epigenetic modifications at sites spread across the genome and distant from telomeres. This regulation is mediated through the telomere-binding protein telomeric repeat–binding factor 2 (TRF2). These observations suggest a role of telomeres in extra-telomeric functions. Most notably, telomeres have a broad impact on pluripotency and differentiation. For example, cardiomyocytes differentiate with higher efficacy from induced pluripotent stem cells having long telomeres, and differentiated cells obtained from human embryonic stem cells with relatively long telomeres have a longer lifespan. Here, we first highlight reports on these two seemingly distinct research areas: the extra-telomeric role of telomere-binding factors and the role of telomeres in pluripotency/stemness. On the basis of the observations reported in these studies, we draw attention to potential molecular connections between extra-telomeric biology and pluripotency. Finally, in the context of the nonlocal influence of telomeres on pluripotency and stemness, we discuss major opportunities for progress in molecular understanding of aging-related disorders and neurodegenerative diseases.
Keywords: stem cells, telomere, shelterin, gene regulation, neurodegenerative disease, de-differentiation, genome organization, chromosome end, pluripotency, extra-telomeric function
The ends of eukaryotic chromosomes have specialized nucleotide-protein complexes called telomeres. In mammalian cells, they are capped by a complex of six proteins, TRF1, TRF2, POT1, RAP1, TIN2, and TPP1, known as shelterin (1–3). The shelterin proteins have distinct roles. TRF1 and TRF2 bind to double-stranded telomeric DNA, whereas POT1 binds to single-stranded telomeric DNA. RAP1 associates with TRF2, whereas TPP1 and TIN2 primarily associate with POT1 (4) (Table 1). Together the shelterin proteins form subcomplexes that vary in ssDNA and dsDNA binding. These result in two broad functions: first, protection of telomeres to evade DNA damage repair at chromosome ends (which, when affected, results in chromosome end fusions and genomic instability (5, 6)) and, second, regulation of the recruitment of telomerase (the catalytic reverse transcriptase that synthesizes telomeres) to telomere ends to maintain the length of telomeres (7–9). The involvement of telomeres in cellular homeostasis, aging, and disease risk (10–12); initiation and progression of cancer (13–15); and variation of telomere length, during evolution and in different species (16–18), have been extensively reviewed (12).
Table 1.
Diverse functions of shelterin proteins
| Shelterins | Telomeric DNA binding | Chromatin organization | Gene regulation | Maintenance of the dedifferentiated state | Cancer stem cell | References |
|---|---|---|---|---|---|---|
| TRF1 | dsDNA | ✓ | ✗ | ✓ | ✗ | 4, 55–57, 86, 96, 97 |
| TRF2 | dsDNA | ✓ | ✓ | ✓ | ✓ | 4, 24, 58–62, 76, 77, 98–106 |
| RAP1 | ✗ | ✗ | ✓ | ✓ | ✓ | 4, 19, 46–52 |
| POT1 | ssDNA | ✗ | ✗ | ✓ | ✗ | 4, 109, 110 |
| TIN2 | ✗ | ✓ | ✓ | ✗ | ✗ | 4, 53, 54 |
| TPP1 | ✗ | ✗ | ✗ | ✓ | ✗ | 4, 107, 108 |
Relatively recent work shows association of shelterin proteins outside telomeres across the genome (19–21), suggesting functions that are extra-telomeric, or beyond telomeres. Extra-telomeric functions include how telomeres influence gene expression in the subtelomeric regions (∼10 Mb from telomeres (22, 23)), telomere length–dependent transcriptional activity, and epigenetic modifications at sites distant from telomeres (24). In addition, a large body of work suggests a role of telomeres, particularly telomere length, in self-renewal or pluripotency (25–28) (Table 1). Herein, we discuss literature that potentially bridges these two developing aspects, keeping in mind aging-related disorders that involve premature differentiation of stem cells (29).
Telomeres: Gene regulation, epigenetics, and genome organization
The role of telomeres in gene regulation first came to light in 1990. Gottschling et al. (30) noted heritable silencing of transgenes inserted within 4 kb from telomeric ends in yeast cells and reported this to be due to telomere position effect (TPE). Several years later, TPE was observed at chromosome 22 telomere in human lymphoblastoid cell lines (31). Extensive research followed to understand the TPE-related silencing of genes in subtelomeric regions of fungi and other organisms, such as Trypanosoma brucei, Plasmodium falciparum, Schizosaccharomyces pombe, Drosophila melanogaster, Pneumocystis carinii, and Candida glabrata (32). It was also observed that genes (e.g. ISG15, DSP, and C1S) positioned ∼10 Mb further from telomeres than found in TPE were down-regulated through physical association of telomeres. This was denoted as TPE-over long distance (TPE-OLD), which involves the long telomeres looping back to the chromatin, causing gene repression and shortening of telomeres, dissociating the loop leading to gene activation (Fig. 1) (22). Recent work shows telomerase reverse transcriptase gene hTERT is also regulated by TPE-OLD (33). TPE or TPE-OLD has been implicated in disorders such as idiopathic mental retardation, ring chromosome 17, and facio-scapulo-humeral dystrophy (32–34).
Figure 1.
TPE-OLD. Physical association of relatively long telomeres by looping to the subtelomeric regions results in transcriptional repression of genes located in the subtelomeres. In relatively short telomeres, the looping is lost, and genes become transcriptionally active.
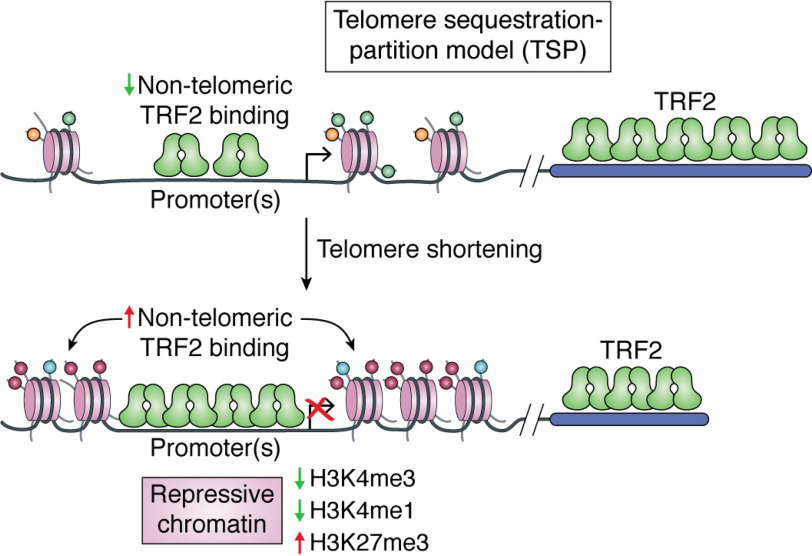
Recent findings show that telomere length influences transcription of genes as far as ∼60 Mb away from telomeres. It was demonstrated that this was because TRF2 binding across the genome (i.e. extra-telomeric sites) depended on telomere length—and TRF2 occupancy at promoters affected expression of target genes (24). TRF2 is known to bind to the G-rich TTAGGG motif present as repeats at the telomeres (35). Therefore, in human cells with elongated telomeres (i.e. increased number of TTAGGG repeats), telomeric TRF2 binding was enhanced as expected. On the other hand, extra-telomeric TRF2 binding was reduced relative to cells with shorter telomeres (with isogenic background) (24). TRF2 levels in the nucleus, however, remained unaltered in cells with long or short telomeres, consistent with a previous report showing relatively unchanged abundance of nuclear TRF2 in different types of cells with short/long telomeres (36). Based on this, it was postulated that redistribution of TRF2 binding between telomeric and extra-telomeric sites occurs as telomeres elongate (24). This is denoted as the telomere sequestration and partitioning (TSP) model, which describes altered extra-telomeric TRF2 binding in long/short telomeres resulting in differential expression of TRF2-target promoters (Fig. 2). Moreover, altered epigenetic state of the TRF2-target promoters (e.g. modification of histone activation (H3K4Me1 and H3K4Me3) and suppression (H3K27Me3) marks) was evident (24).
Figure 2.
TSP. The model implies partitioning of TRF2 between telomeric and extra-telomeric sites. Longer telomeres sequester more TRF2, thereby depleting TRF2 binding at extra-telomeric sites. Conversely, when telomeres shorten, an increase in TRF2 binding at promoters influences TRF2-mediated chromatin modifications and transcription.
Notable in this context are observations of extra-telomeric binding of TRF2 and/or TRF1 constituting 50 TRF2 (20) and 68 sites common to TRF1/TRF2, respectively (21). A majority of the extra-telomeric sites constituted telomere-like TTAGGG sequences and are therefore called interstitial telomeric sequence. Recent work, however, has revealed more extensive binding of TRF2 across the genome, with more than 20,000 sites from TRF2 ChIP-Seq in HT1080 fibrosarcoma cells. These interstitial TRF2 binding sites comprised G-rich repeat sequences, including interstitial telomeric sequence (37). In addition, ∼12,500 TRF2 peaks mapped within 20 kb of transcription start sites. Several of these promoters were tested and found to be epigenetically modified, and transcriptionally regulated, in a TRF2-dependent way (37).
Nucleosomes, the basic units of chromatin packaging in cells, comprise a complex of H2A, H2B, H3, and H4 histone proteins. Modifications (e.g. methylation or acetylation) of histone proteins are therefore closely related to how chromatin is packaged. These are known as epigenetic modifications that can impact gene regulation. Short telomeres, and consequent DNA damage, were noted to result in reduced histone biosynthesis (38, 39), affecting the state of chromatin.
Several studies further show shortened telomeres to be associated with genome-wide altered DNA methylation, nucleosome positioning, and histone modifications (reviewed in Ref. 40). Similar observations were also made during stem cell pluripotency, cell senescence, and cancer cell differentiation (41, 42).
Another line of investigation implicated the shelterin factor RAP1 more directly. On telomere shortening RAP1 was found to affect nucleosome occupancy, down-regulate histone genes, and increase expression of senescence-associated genes (40, 41, 43, 44). Together, these suggest a broader role of telomeres, particularly short telomeres, in the epigenetic state of the genome.
Functions of shelterin proteins independent of chromosome-end protection
In 2010, Martinez et al. (19) found extra-telomeric binding of RAP1 to the TTAGGGTTAGGG consensus motif in mice; 70% of RAP1 binding was intragenic or proximal to coding regions. Based on this, they described potential RAP1 target genes in the mouse genome. In RAP1-deficient mice, about one-third of the deregulated genes were noted to be RAP1 target genes, implicating RAP1-mediated gene regulation. In a later report, genome-wide RAP1 ChIP-Seq in telomerase-deficient mice showed altered RAP1 binding on telomere shortening (45). Further, 63 genes in the human genome were reported to have RAP1 occupancy (20).
It was reported that RAP1 interacted with the IκB kinase, a function not expected of shelterin. This resulted in activation of NF-κB, leading to up-regulation of NF-κB target genes (46). It was therefore postulated that expression of NF-κB targets might be telomere length–dependent (Fig. 3) (47) (Table 1).
Figure 3.
Extra-telomeric functions of shelterin proteins independent of telomeres. Examples of noncanonical function(s) of shelterin proteins are shown. Extra-telomeric binding of TRF2 to G-quadruplex–forming sequences across the genome induces epigenetic and transcriptional changes. NF-κB signaling is modulated through the shelterin factor RAP1: telomere-independent interaction of RAP1 with the IKK complex results in phosphorylation of the NF-κB p65 subunit, leading to activation of NF-κB target gene(s). Mitochondrial localization of TIN2, another shelterin protein, was reported to negatively regulate oxidative phosphorylation.
Extra-telomeric function of RAP1 was also noted in positive regulation of PPARα and PGC1α genes in mice, which affected cellular metabolism related to obesity (48, 49). RAP1 interaction with other co-factors was also reported in mesenchymal stem cell–based therapy for myocardial infarction and inflammation-dependent disorders (50, 51). Recently, Zhang et al. (52) observed another key function of RAP1 in epigenetic regulation of the RELN promoter in hematopoiesis (Table 1). Together, these show RAP1 functions that are clearly independent of its canonical role in the protection of telomeres.
Noncanonical extra-telomeric function was also observed for another shelterin protein, TIN2. A truncated isoform of TIN2, hTIN2S, was found to localize outside telomeres and affect heterochromatin organization (53). However, in cells with elongated telomeres, the dual localization was lost, such that hTIN2S redistributed from nontelomeric chromatin to telomeres exclusively. This is consistent with the TSP model described above (Fig. 2), where redistribution of a shelterin protein with change in telomere length would be expected. TIN2 was also shown to localize to mitochondria and regulate oxidative phosphorylation and glucose metabolism (Fig. 3) (54) (Table 1).
Noncanonical function of TRF1 was noted in phosphorylation of nontelomeric TRF1 by Aurora-A, which resulted in mitotic abnormality (55). This was also suggested by the role of TRF1 in chromosome segregation by positively regulating Aurora-B's centromeric function (56). Further, a crystallographic study showed that TRF1 interacts with TERB1, crucial for X-Y chromosome pairing, during meiosis (57) (Table 1).
A telomere-independent role(s) was also reported for TRF2 in the transcriptional activation of HS3ST4 (58) and PDGFRβ (59) and repression of the cell cycle–dependent kinase inhibitor CDKN1A (p21) (60). TRF2 was further found to associate with core histone proteins (61, 62), including the RE-1–silencing factor (REST) repressor complex (60). In addition to these, a role of telomere-independent TRF2 was reported in natural killer cell activation, angiogenesis, and intrinsic aspects like nucleosome formation and chromatin compaction (63) (Table 1).
As discussed above, recent work also showed TRF2-mediated regulation of genes spread across the genome. This involved interaction of TRF2 with DNA secondary structures called G-quadruplexes within promoters (37). It is important to mention here that sequences with potential to form G-quadruplex structures are enriched in regulatory regions throughout the genome across genera (64–69), and evidence suggests that G-quadruplexes might influence local epigenetic modifications (70–72).
Telomeres in 3D—correlation of telomere architecture and cell state
Telomeres are known to be organized within the nuclear matrix through interactions with lamin. Further telomeric association with distant parts of chromatin by looping are also known. Several studies show that these three-dimensional associations (i.e. nonlocal or extra-telomeric interactions) can impact cellular functions.
Early work from the Blackburn group (73) found telomeres to be motile with rapid motions of the telomere ends within the nucleus. Moreover, individual telomeres in a nucleus showed heterogeneity in motility. Relatively short uncapped telomeres in cancer cells had more motility than cells with long telomeres, possibly due to untethering of telomeres from the nuclear matrix (73–75). Furthermore, TRF2 association with lamin A/C proteins was observed to promote physical association of telomeres with interstitial chromatin through looping (76, 77) (Table 1).
Telomere-associated change in chromatin organization was noticed in other cell types also. A study in 2014 reported altered 3D telomeric architecture in buccal cells derived from Alzheimer's disease (AD) patients of mild, moderate, and extreme pathology using three-dimensional (3D) microscopy and quantitative fluorescence in situ hybridization. Telomere aggregates and overall numbers increased in mild to severe AD along with a decrease in telomere length (78). A follow-up study of the same parameters between AD and control group buccal cell samples found similar results (79).
Tichy et al. (80) reported altered telomere length and inconsistency in telomeric foci in the muscle stem cells of Duchenne muscular dystrophy patients when compared with the control group. On the other hand, muscle-stem cell telomere length remained unchanged between young and old healthy mice (80). This was similar to what was previously noted for humans and macaques (81), although mouse somatic tissues have longer telomeres and higher telomerase activity than humans and other primates.
Similar observations were noted in other cases. For example, esophageal squamous cell carcinoma cells have altered 3D telomere organization compared with normal epithelial cells from the same patient with relatively long telomeres (82), and thyrospheres constituting stem cells from four subgroups of papillary thyroid carcinoma patients were found to have telomeric localization that was unique for each subgroup (83).
These data, describing studies of telomere-dependent gene expression and chromatin folding mediated through telomeres or telomere-associated factors, provide a clear view of how extra-telomeric functions may contribute to the role of telomeres in basic biological processes. Now we turn to the functions and implications of telomeres in integrated systems during health and disease.
Telomeres in cellular pluripotency and “stemness”—emerging observations
Induced pluripotent stem cells (iPSCs) have rapidly gained significance in basic and applied biological sciences (84) and serve as a facile model system for cellular differentiation and development. The important role of telomere elongation and homeostasis in formation/maintenance of iPSCs, including their implications in aging, is known (25, 85). In the following sections, we discuss the importance of telomeres in self-renewal and chromosome stability in iPSCs and consider emerging literature on how extra-telomeric function of telomere-associated factors might influence pluripotency.
Telomere elongation and pluripotency
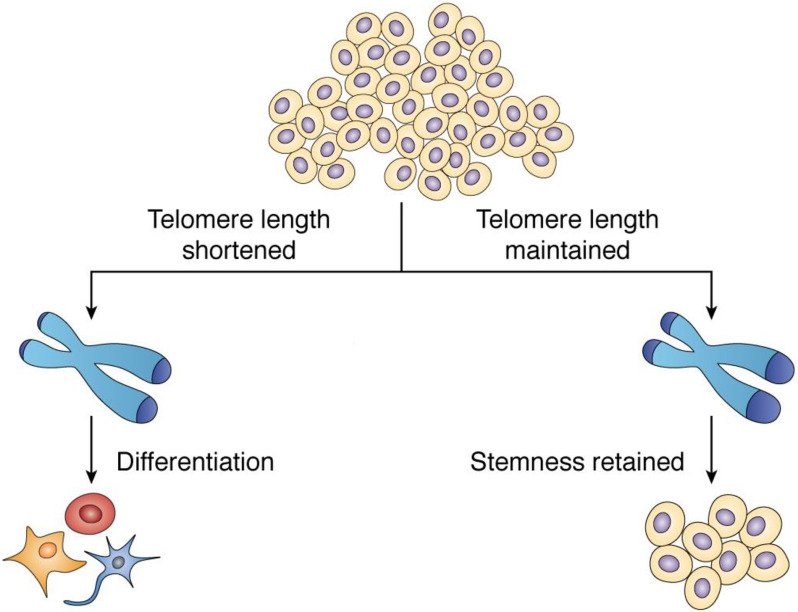
The presence of relatively long telomeres has been generally observed in pluripotent cells. For example, reprogramed iPSCs obtained from mice showed elongated telomeres in the pluripotent cells (86); iPSCs generated from dyskeratosis congenita patient samples had relatively long telomeres (87), and human fibroblast TIG-1 cells reprogrammed to iPSCs, telomeres increased from 6 to 8 Kb (88). Similarly, in many cancers like liposarcoma, hepatocellular carcinoma, and in pliocytic astrocytoma, elongated telomeres were noted in the dedifferentiated or pluripotent cells (89–91).
Consistent with this, shortening of telomeres was associated with differentiation. Telomere attrition was associated with loss of stemness markers in cardiac progenitor cells isolated from adult human heart failure cases (92); unstable differentiation was observed in ESCs with telomere dysfunction because of critically shorter telomeres (41); regenerative capacity of stem cells declined because of progressive telomere attrition in aging cells (93); and reduced proliferation and differentiation to osteoblasts was observed in mesenchymal stromal cells isolated from adults compared with those isolated from children, suggesting the impact of telomere attrition (94). On the other hand, longer life span was observed in cells that differentiated from human ESCs possessing relatively long telomeres (28) and cardiomyocytes differentiated with improved efficacy from iPSCs that had relatively long telomeres (26).
Furthermore, impaired differentiation due to poor telomere maintenance was observed in keratinocytes (27), and telomere elongation was found to be key for telomere length homeostasis in mouse embryonic stem cells (95). Together, these studies show the importance of telomere length maintenance in stem cells and how telomere shortening or attrition (with aging) impacts differentiation (Fig. 4).
Figure 4.
Role of telomeres in stem cell homeostasis. Importance of telomere length in maintaining stemness. Stem cells with relatively long telomeres are reported to retain stemness, whereas reduction of telomere length is generally observed during differentiation and/or in differentiated cells.
Role of telomere-associated factors in pluripotency or “stemness” and disease
The pluripotency factor Oct3/4 was reported to positively regulate TRF1 during the induction and maintenance of pluripotency (96). Consistent with this, TRF1 was observed to be up-regulated in ESCs and iPSCs (82, 92), and in the presence of the small molecule ETP-47037, which inhibits TRF1, reprograming efficiency in mice was reduced (86). Moreover, increase in TRF1 expression was observed during in vitro derivation of ESCs from the inner cell mass (97) (Table 1).
Several reports implicated TRF2 in pluripotency. Self-renewal and maintenance potential was perturbed when TRF2 was deleted in alveolar stem cells (98), and human mesenchymal stem cells showed increased sensitivity to irradiation when TRF2 was knocked down (99, 100). Further, in TRF2-null mice, terminal differentiation was triggered during skin carcinogenesis (101), and increase in TRF2 was implicated in aggressive proliferation of liver cancer stem cells (102).
In the above studies, it was not clear whether the function of TRF2 was involved as a telomeric and/or extra-telomeric factor. However, we noted further work suggesting extra-telomeric function of TRF2 in stemness. This includes nuclear interaction of TRF2 with REST, which was reported to be important in maintenance of the neural stem cell population (103, 104). Further, TRF2 depletion resulted in reduced proliferation and enhanced differentiation of glioblastoma stem cells due to both telomeric dysfunction and loss of REST-mediated repression (105), and silencing of TRF2 resulted in the reduction of the Yamanaka factors in oral cancer stem cells (106) (Table 1). In addition to these, computational modeling indicated high binding affinity of TRF2 to the stem-cell factor KLF4 (106).
TPP1-mediated recruitment of the reverse transcriptase telomerase (TERT) for telomere elongation was observed. Here, abrogation of TPP1 affected the reprograming of mouse embryonic fibroblasts (107). Later TPP1 was also shown to be important in maintaining the length of telomeres in human ESCs (108).
Depletion of POT1, on the other hand, triggered DNA damage response and thereby telomeric dysfunction, resulting in reduced survival of hematopoietic stem cells (HSCs) and bone marrow failure that mimicked the phenotypes of dyskeratosis congenita (109). Exogenous expression of POT1 induced self-renewal of human HSCs by inhibiting generation of reactive oxygen species (Table 1) (110). In addition, POT1-mediated metabolic control and transcriptional regulation in HSCs was shown (111). Together, these implicate extra-telomeric functions of POT1 in pluripotency.
Furthermore, mutations within shelterin genes were found to be associated with hematological malignancies due to telomere deprotection, showing the importance of the shelterin factors in hematopoiesis and self-renewal (111).
The role of the noncoding RNA transcribed from telomeres called telomeric repeat–containing RNA (TERRA) in pluripotency is also notable. TERRA was found to be overexpressed and contributed to the self-renewal of mesenchymal stem cells (112). In addition, decline in TERRA resulted in differentiation, and overexpression resulted in rescue of the self-renewal activity (112). TERRA foci formation (i.e. clustered presence of TERRA molecules as seen in microscopy) due to elevated expression and aggregation of TERRA was reported to occur in both developing cerebellar neural progenitors and medulloblastoma (113). A more recent study showed how TERRA through a TRF1-dependent mode regulates the transcriptional state of ESCs such that a naive state is maintained (114).
Decrease in telomere length with age and associated telomere dysfunction contributes to initiation and progress of cancer (10, 115). It is also widely known that in more than 90% of human cancers, telomerase (hTERT)—the enzyme necessary for telomere synthesis—is reactivated, and as a result telomeres are maintained, unlike in normal adult somatic cells (12). However, despite reactivation, most cancer cells and cancer stem cells have shorter telomeres than surrounding normal cells (12, 116, 117).
Expression of hTERT was shown to involve TPE-OLD (33). In normal cells, the chromosome 5p telomere folds backs and associates with the hTERT loci ∼1.3 Mb away. Kim et al. (33) concluded that through this interaction telomeric TRF2 associates with the hTERT promoter. Further, the TRF2 interaction was lost in cells with relatively short telomeres where the telomeric loop was unable to form (33). Loss of TRF2 from the hTERT promoter correlated with increased hTERT expression. However, it was not clear whether this involved TRF2-mediated regulation or was a result of telomere-induced gene silencing as noted for several genes in earlier studies (22, 30).
More recent work, on the other hand, suggested that hTERT regulation is under direct transcriptional control of TRF2 (118). Here, TRF2 presence on the hTERT promoter was independent of telomeres (i.e. there was involvement of extra-telomeric TRF2). This was also clear from TRF2 occupancy at the exogenously inserted hTERT promoter ∼46 Mb away from telomeres (118)—where looping due to physical proximity like the 5p telomere end was unlikely. Together, these leads suggest the involvement of the TSP model discussed above in hTERT regulation, where the presence of extra-telomeric TRF2 on the hTERT promoter is of interest and depends on how much TRF2 is free or sequestered at the telomere ends.
How might aspects of extra-telomeric biology impact stem cells? Stem cells, that replenish “worn out” cells, undergo telomere shortening, as reviewed earlier (112, 113), suggesting that many of the mechanisms described above could be in play. It must be mentioned here that although the literature suggests a potential role of extra-telomeric function(s) in pluripotency/stemness, evidence supporting direct causal links remains to be established to the best of our knowledge.
The platelet-derived growth factor receptor (PDGFR) was found to be significantly abrogated in the myocardium of people with increasing age, suggesting the role of PDGFR signaling in cardiomyocyte regeneration and proliferation (119). This was consistent with the telomere length–dependent differentiation of cardiomyocytes observed frequently (reviewed in Ref. 26). As mentioned above, PDGFR-β is a transcriptional target of extra-telomeric TRF2 (59). Furthermore, it was demonstrated that PDGFR-β is regulated epigenetically by TRF2 in a telomere length-dependent fashion (24). Therefore, it is likely that regulation of PDGFR-β by extra-telomeric TRF2, which depends on telomere length (24) (described above as TSP), plays a more direct role in telomere-dependent cardiomyocyte differentiation described above.
A recent study demonstrated the increased expression of genes related to neurogenesis and neuronal maturation in sporadic Alzheimer's disease, suggesting a potential link between neuronal differentiation and this debilitating neurodegenerative disease (29). Telomere shortening, a hallmark of aging, is also widely observed in neurodegenerative diseases like AD (119, 120, 122). Like the other cell types discussed above, short telomeres affect the proliferative capacity of neural stem cells and reduce the self-renewal potential of progenitors required for normal adult neurogenesis (123). Could a telomere function, and particularly an extra-telomeric function, serve as a molecular trigger underlying the pathophysiology of the disease (124)?
The recent study on pathophysiology in AD also showed that accelerated differentiation of neural stem cells in AD was associated with deregulated levels of REST (29). Notably, earlier work had reported that extra-telomeric TRF2-mediated stabilization of REST was critical for neuronal differentiation (125). Furthermore, recent findings showed that REST binding to nontelomeric chromatin was also dependent on extra-telomeric TRF2 (60). As described above, in TSP (Fig. 2), the presence of extra-telomeric TRF2 depends on telomere length. Therefore, these findings suggest a direct causal link between telomere length, extra-telomeric TRF2, and REST in neural stem cells, which might be key to neuronal differentiation. It will be fascinating to determine whether cellular renewal, rather than the more canonical aggregation hypothesis, might play a driving role in this and other degenerative diseases.
Conclusions and future perspectives
The notion that telomeres influence function beyond chromosome ends is relatively recent. Findings from many research groups, including ours, reveal this to be through two primary modes: (a) physical looping of telomeres (mostly within subtelomeric regions) (22) or (b) extra-telomeric function of shelterin factor(s) (37, 47, 54), which in the case of TRF2 depends on telomere length, as described above in the TSP model (Fig. 2) (24). The role of telomeres, particularly telomere length, has been observed closely during both pluripotency and stem cell differentiation. However, the underlying molecular processes that link telomeres to pluripotency are only beginning to emerge.
Notably, recent work has shown that TRF2 binding throughout the genome results in epigenetic modifications. Further, this depends on telomere length (24). Together, these findings contribute to a new understanding of telomeric factors. Moreover, these data suggest that a novel set of protein-protein interactions are possibly induced instead of the canonical shelterin complex at telomeres. It will be interesting to explore how these interactions with TRF2 and other telomeric factors are regulated (e.g. with distinct post-translational modifications that direct nontelomeric binding).
Based on RAP1 and NF-κB interactions (46), association of telomeric factors with proteins independent of DNA binding is another molecular aspect that might be worthwhile to study. Contextually, whether other telomeric factors associate with nuclear or cytoplasmic factors—and how such interactions are affected as telomere length changes—would be of interest.
Telomerase—the only protein that synthesizes telomeres—is overexpressed in most cancers. Recent findings suggest that telomeres exert control over telomerase through telomeric or extra-telomeric mechanisms (33, 118). Teasing out molecular details of these controls, including whether and how the TSP model might be involved in telomere-dependent control of telomerase, remains to be studied in further detail, considering its broad and significant implications.
Taken together, these new aspects of extra-telomeric biology—dependent on telomere length (and thereby aging)—may reveal a novel understanding of the molecular processes underlying pluripotency. Moreover, whether and how, particularly in what context, premature differentiation is linked to aging through telomeres would be of interest in improving our understanding of diseases associated with aging.
Acknowledgments
We acknowledge all members of the Chowdhury group for fruitful discussions on many aspects of the review and Dr. Munia Ganguli for assisting with editing of the manuscript.
Funding and additional information—This work was supported by Wellcome Trust/DBT India Alliance Fellowship IA/S/18/2/504021 (to S. C.). S. V. and A. G. acknowledge CSIR and DBT (GAP171), respectively, for research fellowships.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TPE
- telomere position effect
- TPE-OLD
- TPE-over long distance
- TSP
- telomere sequestration and partitioning
- AD
- Alzheimer's disease
- 3D
- three-dimensional
- iPSC
- induced pluripotent stem cell
- ESC
- embryonic stem cell
- TERT
- telomerase reverse transcriptase
- HSC
- hematopoietic stem cell
- TERRA
- telomeric repeat–containing RNA
- PDGFR
- platelet-derived growth factor receptor
- REST
- RE-1–silencing factor.
References
- 1. Palm W., and de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- 2. Xin H., Liu D., and Songyang Z. (2008) The telosome/shelterin complex and its functions. Genome Biol. 9, 232 10.1186/gb-2008-9-9-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Červenák F., Juríková K., Sepšiová R., Neboháčová M., Nosek J., and Tomáška L. (2017) Double-stranded telomeric DNA binding proteins: diversity matters. Cell Cycle 16, 1568–1577 10.1080/15384101.2017.1356511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmutz I., and De Lange T. (2016) Shelterin. Curr. Biol. 26, R397–R399 10.1016/j.cub.2016.01.056 [DOI] [PubMed] [Google Scholar]
- 5. Timashev L. A., Babcock H., Zhuang X., and de Lange T. (2017) The DDR at telomeres lacking intact shelterin does not require substantial chromatin decompaction. Genes Dev. 31, 578–589 10.1101/gad.294108.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Lange T. (2018) Shelterin-mediated telomere protection. Annu. Rev. Genet. 52, 223–247 10.1146/annurev-genet-032918-021921 [DOI] [PubMed] [Google Scholar]
- 7. Lim C. J., Zaug A. J., Kim H. J., and Cech T. R. (2017) Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat. Commun. 8, 1075 10.1038/s41467-017-01313-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pike A. M., Strong M. A., Ouyang J. P. T., and Greider C. W. (2019) TIN2 functions with TPP1/POT1 to stimulate telomerase processivity. Mol. Cell Biol. 39, e00593–18 10.1128/mcb.00593-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nandakumar J., and Cech T. R. (2013) Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14, 69–82 10.1038/nrm3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martínez P., and Blasco M. A. (2015) Replicating through telomeres: a means to an end. Trends Biochem. Sci. 40, 504–515 10.1016/j.tibs.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 11. Blackburn E. H., Epel E. S., and Lin J. (2015) Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198 10.1126/science.aab3389 [DOI] [PubMed] [Google Scholar]
- 12. Shay J. W., and Wright W. E. (2019) Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 20, 299–309 10.1038/s41576-019-0099-1 [DOI] [PubMed] [Google Scholar]
- 13. Shay J. W. (2016) Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 10.1158/2159-8290.CD-16-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maciejowski J., and De Lange T. (2017) Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 18, 175–186 10.1038/nrm.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okamoto K., and Seimiya H. (2019) Revisiting telomere shortening in cancer. Cells 8, 107 10.3390/cells8020107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baird D. M. (2018) Telomeres and genomic evolution. Philos. Trans. R. Soc. B Biol. Sci. 373, 20160437 10.1098/rstb.2016.0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Monaghan P., and Ozanne S. E. (2018) Somatic growth and telomere dynamics in vertebrates: relationships, mechanisms and consequences. Philos. Trans. R. Soc. B Biol. Sci. 373, 20160446 10.1098/rstb.2016.0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian X., Doerig K., Park R., Can Ran Qin A., Hwang C., Neary A., Gilbert M., Seluanov A., and Gorbunova V. (2018) Evolution of telomere maintenance and tumour suppressor mechanisms across mammals. Philos. Trans. R. Soc. B Biol. Sci. 373, 20160443 10.1098/rstb.2016.0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez P., Thanasoula M., Carlos A. R., Gómez-López G., Tejera A. M., Schoeftner S., Dominguez O., Pisano D. G., Tarsounas M., and Blasco M. A. (2010) Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 12, 768–780 10.1038/ncb2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang D., Xiong Y., Kim H., He Q., Li Y., Chen R., and Songyang Z. (2011) Human telomeric proteins occupy selective interstitial sites. Cell Res. 21, 1013–1027 10.1038/cr.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simonet T., Zaragosi L.-E., Philippe C., Lebrigand K., Schouteden C., Augereau A., Bauwens S., Ye J., Santagostino M., Giulotto E., Magdinier F., Horard B., Barbry P., Waldmann R., and Gilson E. (2011) The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 21, 1028–1038 10.1038/cr.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robin J. D., Ludlow A. T., Batten K., Magdinier F., Stadler G., Wagner K. R., Shay J. W., and Wright W. E. (2014) Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 28, 2464–2476 10.1101/gad.251041.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robin J. D., Ludlow A. T., Batten K., Gaillard M.-C., Stadler G., Magdinier F., Wright W. E., and Shay J. W. (2015) SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 25, 1781–1790 10.1101/gr.190660.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mukherjee A. K., Sharma S., Sengupta S., Saha D., Kumar P., Hussain T., Srivastava V., Roy S. D., Shay J. W., and Chowdhury S. (2018) Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 14, e1007782 10.1371/journal.pgen.1007782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrington L. (2004) Does the reservoir for self-renewal stem from the ends? Oncogene 23, 7283–7289 10.1038/sj.onc.1207948 [DOI] [PubMed] [Google Scholar]
- 26. Aguado T., Gutiérrez F. J., Aix E., Schneider R. P., Giovinazzo G., Blasco M. A., and Flores I. (2017) Telomere length defines the cardiomyocyte differentiation potency of mouse induced pluripotent stem cells. Stem Cells 35, 362–373 10.1002/stem.2497 [DOI] [PubMed] [Google Scholar]
- 27. Martínez P., Ferrara-Romeo I., Flores J. M., and Blasco M. A. (2014) Essential role for the TRF2 telomere protein in adult skin homeostasis. Aging Cell 13, 656–668 10.1111/acel.12221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zou Y., Tong H. J., Li M., Tan K. S., and Cao T. (2017) Telomere length is regulated by FGF-2 in human embryonic stem cells and affects the life span of its differentiated progenies. Biogerontology. 18, 69–84 10.1007/s10522-016-9662-8 [DOI] [PubMed] [Google Scholar]
- 29. Meyer K., Feldman H. M., Lu T., Drake D., Lim E. T., Ling K.-H., Bishop N. A., Pan Y., Seo J., Lin Y.-T., Su S. C., Church G. M., Tsai L.-H., and Yankner B. A. (2019) REST and neural gene network dysregulation in iPSC models of Alzheimer's disease. Cell Rep. 26, 1112–1127.e9 10.1016/j.celrep.2019.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gottschling D. E., Aparicio O. M., Billington B. L., and Zakian V. A. (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63, 751–762 10.1016/0092-8674(90)90141-Z [DOI] [PubMed] [Google Scholar]
- 31. Baur J. A., Zou Y., Shay J. W., and Wright W. E. (2001) Telomere position effect in human cells. Science 292, 2075–2077 10.1126/science.1062329 [DOI] [PubMed] [Google Scholar]
- 32. Ottaviani A., Gilson E., and Magdinier F. (2008) Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 90, 93–107 10.1016/j.biochi.2007.07.022 [DOI] [PubMed] [Google Scholar]
- 33. Kim W., Ludlow A. T., Min J., Robin J. D., Stadler G., Mender I., Lai T.-P., Zhang N., Wright W. E., and Shay J. W. (2016) Regulation of the human telomerase gene TERT by telomere position effect—over long distances (TPE-OLD): implications for aging and cancer. PLoS Biol. 14, e2000016 10.1371/journal.pbio.2000016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim W., and Shay J. W. (2018) Long-range telomere regulation of gene expression: telomere looping and telomere position effect over long distances (TPE-OLD). Differentiation 99, 1–9 10.1016/j.diff.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M. R., Schnapp G., and de Lange T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol. 20, 1659–1668 10.1128/mcb.20.5.1659-1668.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takai K. K., Hooper S., Blackwood S., Gandhi R., and de Lange T. (2010) In vivo stoichiometry of shelterin components. J. Biol. Chem. 285, 1457–1467 10.1074/jbc.M109.038026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukherjee A. K., Sharma S., Bagri S., Kutum R., Kumar P., Hussain A., Singh P., Saha D., Kar A., Dash D., and Chowdhury S. (2019) Telomere repeat-binding factor 2 binds extensively to extra-telomeric G-quadruplexes and regulates the epigenetic status of several gene promoters. J. Biol. Chem. 294, 17709–17722 10.1074/jbc.RA119.008687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Sullivan R. J., Kubicek S., Schreiber S. L., and Karlseder J. (2010) Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 17, 1218–1225 10.1038/nsmb.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hauer M. H., Seeber A., Singh V., Thierry R., Sack R., Amitai A., Kryzhanovska M., Eglinger J., Holcman D., Owen-Hughes T., and Gasser S. M. (2017) Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 24, 99–107 10.1038/nsmb.3347 [DOI] [PubMed] [Google Scholar]
- 40. Song S., and Johnson F. (2018) Epigenetic mechanisms impacting aging: a focus on histone levels and telomeres. Genes (Basel) 9, 201 10.3390/genes9040201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pucci F., Gardano L., and Harrington L. (2013) Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell 12, 479–486 10.1016/j.stem.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harrington L., and Pucci F. (2018) In medio stat virtus: unanticipated consequences of telomere dysequilibrium. Philos. Trans. R. Soc. B Biol. Sci. 373, 20160444 10.1098/rstb.2016.0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Platt J. M., Ryvkin P., Wanat J. J., Donahue G., Ricketts M. D., Barrett S. P., Waters H. J., Song S., Chavez A., Abdallah K. O., Master S. R., Wang L. S., and Johnson F. B. (2013) Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev. 27, 1406–1420 10.1101/gad.218776.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song S., Perez J. V., Svitko W., Ricketts M. D., Dean E., Schultz D., Marmorstein R., and Johnson F. B. (2020) Rap1‐mediated nucleosome displacement can regulate gene expression in senescent cells without impacting the pace of senescence. Aging Cell 19, e13061 10.1111/acel.13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martínez P., Gómez-López G., Pisano D. G., Flores J. M., and Blasco M. A. (2016) A genetic interaction between RAP1 and telomerase reveals an unanticipated role for RAP1 in telomere maintenance. Aging Cell 15, 1113–1125 10.1111/acel.12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teo H., Ghosh S., Luesch H., Ghosh A., Wong E. T., Malik N., Orth A., de Jesus P., Perry A. S., Oliver J. D., Tran N. L., Speiser L. J., Wong M., Saez E., Schultz P., et al. (2010) Telomere-independent Rap1 is an IKK adaptor and regulates NF-κB-dependent gene expression. Nat. Cell Biol. 12, 758–767 10.1038/ncb2080 [DOI] [PubMed] [Google Scholar]
- 47. Crabbe L., and Karlseder J. (2010) Mammalian Rap1 widens its impact. Nat. Cell Biol. 12, 733–735 10.1038/ncb2088 [DOI] [PubMed] [Google Scholar]
- 48. Martínez P., Gómez-López G., García F., Mercken E., Mitchell S., Flores J. M., de Cabo R., and Blasco M. A. (2013) RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep. 3, 2059–2074 10.1016/j.celrep.2013.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yeung F., Ramírez C. M., Mateos-Gomez P. A., Pinzaru A., Ceccarini G., Kabir S., Fernández-Hernando C., and Sfeir A. (2013) Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep. 3, 1847–1856 10.1016/j.celrep.2013.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cai Y., Kandula V., Kosuru R., Ye X., Irwin M. G., and Xia Z. (2017) Decoding telomere protein Rap1: its telomeric and nontelomeric functions and potential implications in diabetic cardiomyopathy. Cell Cycle 16, 1765–1773 10.1080/15384101.2017.1371886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding Y., Liang X., Zhang Y., Yi L., Shum H. C., Chen Q., Chan B. P., Fan H., Liu Z., Tergaonkar V., Qi Z., Tse H., and Lian Q. (2018) Rap1 deficiency-provoked paracrine dysfunction impairs immunosuppressive potency of mesenchymal stem cells in allograft rejection of heart transplantation. Cell Death Dis. 9, 386 10.1038/s41419-018-0414-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang X., Liu Z., Liu X., Wang S., Zhang Y., He X., Sun S., Ma S., Shyh-Chang N., Liu F., Wang Q., Wang X., Liu L., Zhang W., Song M., et al. (2019) Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell 10, 649–667 10.1007/s13238-019-0610-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaminker P. G., Kim S.-H., Desprez P.-Y., and Campisi J. (2009) A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell Cycle 8, 931–939 10.4161/cc.8.6.7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen L.-Y., Zhang Y., Zhang Q., Li H., Luo Z., Fang H., Kim S. H., Qin L., Yotnda P., Xu J., Tu B. P., Bai Y., and Songyang Z. (2012) Mitochondrial localization of telomeric protein TIN2 links telomere regulation to metabolic control. Mol. Cell 47, 839–850 10.1016/j.molcel.2012.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohishi T., Hirota T., Tsuruo T., and Seimiya H. (2010) TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res. 70, 2041–2052 10.1158/0008-5472.CAN-09-2008 [DOI] [PubMed] [Google Scholar]
- 56. Ohishi T., Muramatsu Y., Yoshida H., and Seimiya H. (2014) TRF1 ensures the centromeric function of Aurora-B and proper chromosome segregation. Mol. Cell Biol. 34, 2464–2478 10.1128/MCB.00161-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Long J., Huang C., Chen Y., Zhang Y., Shi S., Wu L., Liu Y., Liu C., Wu J., and Lei M. (2017) Telomeric TERB1-TRF1 interaction is crucial for male meiosis. Nat. Struct. Mol. Biol. 24, 1073–1080 10.1038/nsmb.3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Biroccio A., Cherfils-Vicini J., Augereau A., Pinte S., Bauwens S., Ye J., Simonet T., Horard B., Jamet K., Cervera L., Mendez-Bermudez A., Poncet D., Grataroli R., de Rodenbeeke C. T., Salvati E., et al. (2013) TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 15, 818–828 10.1038/ncb2774 [DOI] [PubMed] [Google Scholar]
- 59. El Maï M., Wagner K.-D., Michiels J.-F., Ambrosetti D., Borderie A., Destree S., Renault V., Djerbi N., Giraud-Panis M.-J., Gilson E., and Wagner N. (2014) The telomeric protein TRF2 regulates angiogenesis by binding and activating the PDGFRβ promoter. Cell Rep. 9, 1047–1060 10.1016/j.celrep.2014.09.038 [DOI] [PubMed] [Google Scholar]
- 60. Hussain T., Saha D., Purohit G., Kar A., Kishore Mukherjee A., Sharma S., Sengupta S., Dhapola P., Maji B., Vedagopuram S., Horikoshi N. T., Horikoshi N., Pandita R. K., Bhattacharya S., Bajaj A., et al. (2017) Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci. Rep. 7, 11541 10.1038/s41598-017-11177-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Su C.-H., Cheng C., Tzeng T.-Y., Lin I.-H., and Hsu M.-T. (2016) An H2A histone isotype, H2ac, associates with telomere and maintains telomere integrity. PLoS ONE 11, e0156378 10.1371/journal.pone.0156378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konishi A., Izumi T., and Shimizu S. (2016) TRF2 protein interacts with core histones to stabilize chromosome ends. J. Biol. Chem. 291, 20798–20810 10.1074/jbc.M116.719021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaur P., Wu D., Lin J., Countryman P., Bradford K. C., Erie D. A., Riehn R., Opresko P. L., and Wang H. (2016) Enhanced electrostatic force microscopy reveals higher-order DNA looping mediated by the telomeric protein TRF2. Sci. Rep. 6, 20513 10.1038/srep20513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rawal P., Kummarasetti V. B. R., Ravindran J., Kumar N., Halder K., Sharma R., Mukerji M., Das S. K., and Chowdhury S. (2006) Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 16, 644–655 10.1101/gr.4508806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huppert J. L., and Balasubramanian S. (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413 10.1093/nar/gkl1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Verma A., Halder K., Halder R., Yadav V. K., Rawal P., Thakur R. K., Mohd F., Sharma A., and Chowdhury S. (2008) Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 51, 5641–5649 10.1021/jm800448a [DOI] [PubMed] [Google Scholar]
- 67. Yadav V. K., Abraham J. K., Mani P., Kulshrestha R., and Chowdhury S. (2008) QuadBase: genome-wide database of G4 DNA–occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 36, D381–5 10.1093/nar/gkm781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dhapola P., and Chowdhury S. (2016) QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 44, W277–W283 10.1093/nar/gkw425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yadav V., Hemansi, Kim N., Tuteja N., and Yadav P. (2017) G quadruplex in plants: a ubiquitous regulatory element and its biological relevance. Front. Plant Sci. 8, 1163 10.3389/fpls.2017.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Saha D., Singh A., Hussain T., Srivastava V., Sengupta S., Kar A., Dhapola P., Dhople V., Ummanni R., and Chowdhury S. (2017) Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J. Biol. Chem. 292, 15205–15215 10.1074/jbc.M117.792077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guilbaud G., Murat P., Recolin B., Campbell B. C., Maiter A., Sale J. E., and Balasubramanian S. (2017) Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 9, 1110–1117 10.1038/nchem.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mukherjee A. K., Sharma S., and Chowdhury S. (2019) Non-duplex G-quadruplex structures emerge as mediators of epigenetic modifications. Trends Genet. 35, 129–144 10.1016/j.tig.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang X., Kam Z., Carlton P. M., Xu L., Sedat J. W., and Blackburn E. H. (2008) Rapid telomere motions in live human cells analyzed by highly time-resolved microscopy. Epigenetics Chromatin 1, 4 10.1186/1756-8935-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Burla R., La Torre M., and Saggio I. (2016) Mammalian telomeres and their partnership with lamins. Nucleus 7, 187–202 10.1080/19491034.2016.1179409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gonzalo S., and Eissenberg J. C. (2016) Tying up loose ends: telomeres, genomic instability and lamins. Curr. Opin. Genet. Dev. 37, 109–118 10.1016/j.gde.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wood A. M., Danielsen J. M. R., Lucas C. A., Rice E. L., Scalzo D., Shimi T., Goldman R. D., Smith E. D., Le Beau M. M., and Kosak S. T. (2014) TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 5, 5467 10.1038/ncomms6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith E. D., Garza-Gongora A. G., MacQuarrie K. L., and Kosak S. T. (2018) Interstitial telomeric loops and implications of the interaction between TRF2 and lamin A/C. Differentiation 102, 19–26 10.1016/j.diff.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 78. Mathur S., Glogowska A., McAvoy E., Righolt C., Rutherford J., Willing C., Banik U., Ruthirakuhan M., Mai S., and Garcia A. (2014) Three-dimensional quantitative imaging of telomeres in buccal cells identifies mild, moderate, and severe Alzheimer's disease patients. J. Alzheimers Dis. 39, 35–48 10.3233/JAD-130866 [DOI] [PubMed] [Google Scholar]
- 79. Garcia A., Mathur S., Kalaw M. C., McAvoy E., Anderson J., Luedke A., Itorralba J., and Mai S. (2017) Quantitative 3D telomeric imaging of buccal cells reveals Alzheimer's disease-specific signatures. J. Alzheimers Dis. 58, 139–145 10.3233/JAD-161169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tichy E. D., Sidibe D. K., Tierney M. T., Stec M. J., Sharifi-Sanjani M., Hosalkar H., Mubarak S., Johnson F. B., Sacco A., and Mourkioti F. (2017) Single stem cell imaging and analysis reveals telomere length differences in diseased human and mouse skeletal muscles. Stem Cell Reports 9, 1328–1341 10.1016/j.stemcr.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gardner J. P., Kimura M., Chai W., Durrani J. F., Tchakmakjian L., Cao X., Lu X., Li G., Peppas A. P., Skurnick J., Wright W. E., Shay J. W., and Aviv A. (2007) Telomere dynamics in macaques and humans. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 62, 367–374 10.1093/gerona/62.4.367 [DOI] [PubMed] [Google Scholar]
- 82. Sunpaweravong S., Sunpaweravong P., Sathitruangsak C., and Mai S. (2016) Three-dimensional telomere architecture of esophageal squamous cell carcinoma: comparison of tumor and normal epithelial cells. Dis. Esophagus 29, 307–313 10.1111/dote.12317 [DOI] [PubMed] [Google Scholar]
- 83. Caria P., Dettori T., Frau D. V., Lichtenzstejn D., Pani F., Vanni R., and Mai S. (2019) Characterizing the three-dimensional organization of telomeres in papillary thyroid carcinoma cells. J. Cell. Physiol. 234, 5175–5185 10.1002/jcp.27321 [DOI] [PubMed] [Google Scholar]
- 84. Takahashi K., and Yamanaka S. (2013) Induced pluripotent stem cells in medicine and biology. Development 140, 2457–2461 10.1242/dev.092551 [DOI] [PubMed] [Google Scholar]
- 85. Liu L. (2017) Linking telomere regulation to stem cell pluripotency. Trends Genet. 33, 16–33 10.1016/j.tig.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 86. Marión R. M., López de Silanes I., Mosteiro L., Gamache B., Abad M., Guerra C., Megías D., Serrano M., and Blasco M. A. (2017) Common telomere changes during in vivo reprogramming and early stages of tumorigenesis. Stem Cell Reports 8, 460–475 10.1016/j.stemcr.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Agarwal S., Loh Y.-H., McLoughlin E. M., Huang J., Park I.-H., Miller J. D., Huo H., Okuka M., dos Reis R. M., Loewer S., Ng H.-H., Keefe D. L., Goldman F. D., Klingelhutz A. J., Liu L., et al. (2010) Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature 464, 292–296 10.1038/nature08792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kamada M., Mitsui Y., Matsuo T., and Takahashi T. (2016) Reversible transformation and de-differentiation of human cells derived from induced pluripotent stem cell teratomas. Hum. Cell 29, 1–9 10.1007/s13577-015-0119-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wood L. D., Heaphy C. M., Daniel H. D.-J., Naini B. V., Lassman C. R., Arroyo M. R., Kamel I. R., Cosgrove D. P., Boitnott J. K., Meeker A. K., and Torbenson M. S. (2013) Chromophobe hepatocellular carcinoma with abrupt anaplasia: a proposal for a new subtype of hepatocellular carcinoma with unique morphological and molecular features. Mod. Pathol. 26, 1586–1593 10.1038/modpathol.2013.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lee J.-C., Jeng Y.-M., Liau J.-Y., Tsai J.-H., Hsu H.-H., and Yang C.-Y. (2015) Alternative lengthening of telomeres and loss of ATRX are frequent events in pleomorphic and dedifferentiated liposarcomas. Mod. Pathol. 28, 1064–1073 10.1038/modpathol.2015.67 [DOI] [PubMed] [Google Scholar]
- 91. Rodriguez F. J., Brosnan-Cashman J. A., Allen S. J., Vizcaino M. A., Giannini C., Camelo-Piragua S., Webb M., Matsushita M., Wadhwani N., Tabbarah A., Hamideh D., Jiang L., Chen L., Arvanitis L. D., Alnajar H. H., et al. (2019) Alternative lengthening of telomeres, ATRX loss and H3-K27M mutations in histologically defined pilocytic astrocytoma with anaplasia. Brain Pathol. 29, 126–140 10.1111/bpa.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hariharan N., Quijada P., Mohsin S., Joyo A., Samse K., Monsanto M., De La Torre A., Avitabile D., Ormachea L., McGregor M. J., Tsai E. J., and Sussman M. A. (2015) Nucleostemin rejuvenates cardiac progenitor cells and antagonizes myocardial aging. J. Am. Coll. Cardiol. 65, 133–147 10.1016/j.jacc.2014.09.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Diao D., Wang H., Li T., Shi Z., Jin X., Sperka T., Zhu X., Zhang M., Yang F., Cong Y., Shen L., Zhan Q., Yan J., Song Z., and Ju Z. (2018) Telomeric epigenetic response mediated by Gadd45a regulates stem cell aging and lifespan. EMBO Rep. 19, e45494 10.15252/embr.201745494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Choumerianou D. M., Martimianaki G., Stiakaki E., Kalmanti L., Kalmanti M., and Dimitriou H. (2010) Comparative study of stemness characteristics of mesenchymal cells from bone marrow of children and adults. Cytotherapy 12, 881–887 10.3109/14653249.2010.501790 [DOI] [PubMed] [Google Scholar]
- 95. Dan J., Rousseau P., Hardikar S., Veland N., Wong J., Autexier C., and Chen T. (2017) Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep. 20, 1936–1949 10.1016/j.celrep.2017.07.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Schneider R. P., Garrobo I., Foronda M., Palacios J. A., Marión R. M., Flores I., Ortega S., and Blasco M. A. (2013) TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat. Commun. 4, 1946 10.1038/ncomms2946 [DOI] [PubMed] [Google Scholar]
- 97. Varela E., Schneider R. P., Ortega S., and Blasco M. A. (2011) Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc. Natl. Acad. Sci. U.S.A. 108, 15207–15212 10.1073/pnas.1105414108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Alder J. K., Barkauskas C. E., Limjunyawong N., Stanley S. E., Kembou F., Tuder R. M., Hogan B. L. M., Mitzner W., and Armanios M. (2015) Telomere dysfunction causes alveolar stem cell failure. Proc. Natl. Acad. Sci. U.S.A. 112, 5099–5104 10.1073/pnas.1504780112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Orun O., Tiber P. M., and Serakinci N. (2016) Partial knockdown of TRF2 increase radiosensitivity of human mesenchymal stem cells. Int. J. Biol. Macromol. 90, 53–58 10.1016/j.ijbiomac.2015.10.072 [DOI] [PubMed] [Google Scholar]
- 100. Serakinci N., Mega Tiber P., and Orun O. (2018) Chromatin modifications of hTERT gene in hTERT-immortalized human mesenchymal stem cells upon exposure to radiation. Eur. J. Med. Genet. 61, 288–293 10.1016/j.ejmg.2017.12.014 [DOI] [PubMed] [Google Scholar]
- 101. Lagunas A. M., Wu J., and Crowe D. L. (2017) Telomere DNA damage signaling regulates cancer stem cell evolution, epithelial mesenchymal transition, and metastasis. Oncotarget 8, 80139–80155 10.18632/oncotarget.20960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wu M., Lin Z., Li X., Xin X., An J., Zheng Q., Yang Y., and Lu D. (2016) HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci. Rep. 6, 36045 10.1038/srep36045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang P., Pazin M. J., Schwartz C. M., Becker K. G., Wersto R. P., Dilley C. M., and Mattson M. P. (2008) Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Curr. Biol. 18, 1489–1494 10.1016/j.cub.2008.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang P., Casaday-Potts R., Precht P., Jiang H., Liu Y., Pazin M. J., and Mattson M. P. (2011) Nontelomeric splice variant of telomere repeat-binding factor 2 maintains neuronal traits by sequestering repressor element 1-silencing transcription factor. Proc. Natl. Acad. Sci. U.S.A. 108, 16434–16439 10.1073/pnas.1106906108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bai Y., Lathia J. D., Zhang P., Flavahan W., Rich J. N., and Mattson M. P. (2014) Molecular targeting of TRF2 suppresses the growth and tumorigenesis of glioblastoma stem cells. Glia 62, 1687–1698 10.1002/glia.22708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Saha A., Roy S., Kar M., Roy S., Thakur S., Padhi S., Akhter Y., and Banerjee B. (2018) Role of telomeric TRF2 in orosphere formation and CSC phenotype maintenance through efficient DNA repair pathway and its correlation with recurrence in OSCC. Stem Cell Rev. Rep. 14, 871–887 10.1007/s12015-018-9823-z [DOI] [PubMed] [Google Scholar]
- 107. Tejera A. M., Stagno d'Alcontres M., Thanasoula M., Marion R. M., Martinez P., Liao C., Flores J. M., Tarsounas M., and Blasco M. A. (2010) TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev. Cell 18, 775–789 10.1016/j.devcel.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sexton A. N., Regalado S. G., Lai C. S., Cost G. J., O'Neil C. M., Urnov F. D., Gregory P. D., Jaenisch R., Collins K., and Hockemeyer D. (2014) Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 28, 1885–1899 10.1101/gad.246819.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. He H., Wang Y., Guo X., Ramchandani S., Ma J., Shen M.-F., Garcia D. A., Deng Y., Multani A. S., You M. J., and Chang S. (2009) Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol. Cell Biol. 29, 229–240 10.1128/MCB.01400-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hosokawa K., MacArthur B. D., Ikushima Y. M., Toyama H., Masuhiro Y., Hanazawa S., Suda T., and Arai F. (2017) The telomere binding protein Pot1 maintains haematopoietic stem cell activity with age. Nat. Commun. 8, 804 10.1038/s41467-017-00935-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hosokawa K., and Arai F. (2018) The role of telomere binding molecules for normal and abnormal hematopoiesis. Int. J. Hematol. 107, 646–655 10.1007/s12185-018-2432-4 [DOI] [PubMed] [Google Scholar]
- 112. Xu X., Guo M., Zhang N., and Ye S. (2018) Telomeric noncoding RNA promotes mouse embryonic stem cell self-renewal through inhibition of TCF3 activity. Am. J. Physiol. Cell Physiol. 314, C712–C720 10.1152/ajpcell.00292.2017 [DOI] [PubMed] [Google Scholar]
- 113. Deng Z., Wang Z., Xiang C., Molczan A., Baubet V., Conejo-Garcia J., Xu X., Lieberman P. M., and Dahmane N. (2012) Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J. Cell Sci. 125, 4383–4394 10.1242/jcs.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Marión R. M., Montero J. J., López de Silanes I., Graña-Castro O., Martínez P., Schoeftner S., Palacios-Fábrega J. A., and Blasco M. A. (2019) TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. Elife 8, e44656 10.7554/elife.44656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gunes C., Avila A. I., and Rudolph K. L. (2018) Telomeres in cancer. Differentiation 99, 41–50 10.1016/j.diff.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 116. Shay J. W., and Wright W. E. (2010) Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 584, 3819–3825 10.1016/j.febslet.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Barthel F. P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S. B., Akdemir K. C., Seth S., Song X., Wang Q., Lichtenberg T., Hu J., Zhang J., Zheng S., and Verhaak R. G. W. (2017) Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 49, 349–357 10.1038/ng.3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sharma S., Mukherjee A. K., Roy S. S., Bagri S., Lier S., Verma M., Sengupta A., Kumar M., Nesse G., Pandey D. P., and Chowdhury S. (2020) Human Telomerase Expression is under Direct Transcriptional Control of the Telomere-binding-factor TRF2. bioRxiv 10.1101/2020.01.15.907626 10.1101/2020.01.15.907626 [DOI]
- 119. Yue Z., Chen J., Lian H., Pei J., Li Y., Chen X., Song S., Xia J., Zhou B., Feng J., Zhang X., Hu S., and Nie Y. (2019) PDGFR-β signaling regulates cardiomyocyte proliferation and myocardial regeneration. Cell Rep. 28, 966–978.e4 10.1016/j.celrep.2019.06.065 [DOI] [PubMed] [Google Scholar]
- 120. Hiyama E., and Hiyama K. (2007) Telomere and telomerase in stem cells. Br. J. Cancer 96, 1020–1024 10.1038/sj.bjc.6603671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Blasco M. A. (2007) Telomere length, stem cells and aging. Nat. Chem. Biol. 3, 640–649 10.1038/nchembio.2007.38 [DOI] [PubMed] [Google Scholar]
- 122. Hou Y., Dan X., Babbar M., Wei Y., Hasselbalch S. G., Croteau D. L., and Bohr V. A. (2019) Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 10.1038/s41582-019-0244-7 [DOI] [PubMed] [Google Scholar]
- 123. Ferrón S., Mira H., Franco S., Cano-Jimenez M., Bellmunt E., Ramírez C., Fariñas I., and Blasco M. A. (2004) Telomere shortening and chromosomal instability abrogates proliferation of adult but not embryonic neural stem cells. Development 131, 4059–4070 [DOI] [PubMed] [Google Scholar]
- 124. 2019 Alzheimer's disease facts and figures (2019) Alzheimers Dement. 15, 321–387 10.1016/j.jalz.2019.01.010 [DOI] [Google Scholar]
- 125. Ovando-Roche P., Yu J. S. L., Testori S., Ho C., and Cui W. (2014) TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells 32, 2111–2122 10.1002/stem.1725 [DOI] [PubMed] [Google Scholar]