Abstract
All bacterial lipoproteins share a variably acylated N-terminal cysteine residue. Gram-negative bacterial lipoproteins are triacylated with a thioether-linked diacylglycerol moiety and an N-acyl chain. The latter is transferred from a membrane phospholipid donor to the α-amino terminus by the enzyme lipoprotein N-acyltransferase (Lnt), using an active-site cysteine thioester covalent intermediate. Many Gram-positive Firmicutes also have N-acylated lipoproteins, but the enzymes catalyzing N-acylation remain uncharacterized. The integral membrane protein Lit (lipoprotein intramolecular transacylase) from the opportunistic nosocomial pathogen Enterococcus faecalis synthesizes a specific lysoform lipoprotein (N-acyl S-monoacylglycerol) chemotype by an unknown mechanism that helps this bacterium evade immune recognition by the Toll-like receptor 2 family complex. Here, we used a deuterium-labeled lipoprotein substrate with reconstituted Lit to investigate intramolecular acyl chain transfer. We observed that Lit transfers the sn-2 ester-linked lipid from the diacylglycerol moiety to the α-amino terminus without forming a covalent thioester intermediate. Utilizing Mut-Seq to analyze an alanine scan library of Lit alleles, we identified two stretches of functionally important amino acid residues containing two conserved histidines. Topology maps based on reporter fusion assays and cysteine accessibility placed both histidines in the extracellular half of the cytoplasmic membrane. We propose a general acid base–promoted catalytic mechanism, invoking direct nucleophilic attack by the substrate α-amino group on the sn-2 ester to form a cyclic tetrahedral intermediate that then collapses to produce lyso-lipoprotein. Lit is a unique example of an intramolecular transacylase differentiated from that catalyzed by Lnt, and provides insight into the heterogeneity of bacterial lipoprotein biosynthetic systems.
Keywords: lipoproteins, Firmicutes, Toll-like receptor, acyl transfer, Enterococcus, Mut-Seq, immune evasion, virulence factor, membrane protein, intramolecular transferase, TLR2, lipoprotein, acyltransferase, Toll-like receptor (TLR), microbiology
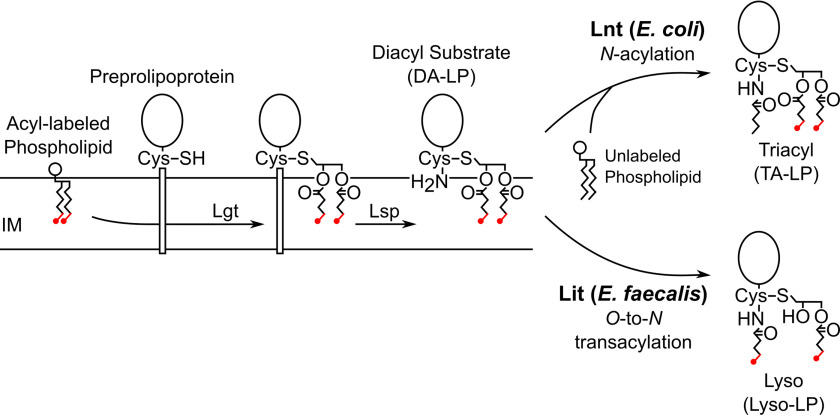
Lipoproteins are ubiquitous membrane-anchored proteins located on the extra-cytoplasmic surface of bacterial cell membranes (1–6). Lipoproteins perform a wide variety of functions at the membrane surface, including in the capture of nutrients, in adhesion, and as structural components of the cell envelope. Preprolipoproteins are exported from the cytoplasm and post-translationally modified by lipoprotein diacylglycerol transferase (Lgt) with a diacylglycerol moiety from a neighboring glycerophospholipid (Fig. 1) (7). Lipoprotein signal peptidase II (Lsp) then recognizes the lipobox and cleaves the leader peptide at the Cys + 1 position, liberating the cysteine α-amino group and forming a diacylated lipoprotein (DA-LP) (8). Although Lgt and Lsp are highly conserved in the lipoprotein biosynthetic pathways from diverse prokaryotes, further acyl tailoring modifications can occur. In the majority of Gram-negative bacteria and some high-GC Gram-positive species, lipoprotein N-acyltransferase (Lnt) attaches a single amide-linked acyl chain to the α-amino group of DA-LP using a neighboring phospholipid as the acyl donor to complete triacylated lipoprotein (TA-LP) (9). Despite lacking an lnt sequence ortholog, several low-GC Gram-positive Firmicutes also produce lipoproteins with N-terminal modifications (10).
Figure 1.
Lipoprotein biosynthetic pathways with Lnt and Lit N-terminal tailoring. The first two steps of the lipoprotein biosynthetic pathway are highly conserved in all prokaryotes. Lgt catalyzes thioether bond formation using a glycerophospholipid donor, followed by proteolytic cleavage of the signal peptide by Lsp to make DA-LP. In E. coli and other Gram-negative bacteria, Lnt then transfers the sn-1 lipid from a glycerophospholipid to the α-amino position of a lipoprotein (top). Lit from E. faecalis and other Firmicutes is proposed to function as an O-to-N intramolecular transacylase (bottom), in which case [d5]-DA-LP substrate with ω-labeled acyl chains (labels colored red) should be retained in the lys0-LP product. IM, inner membrane.
We previously identified an integral membrane protein in Enterococcus faecalis that forms lipoproteins with N-acyl-S-monoacyl (sn-1)-modified cysteine residues (11). This novel lipoprotein chemotype (lyso-LP) is directed by an enzyme (WMC_RS08810) that is narrowly distributed among enterococci, being confined to a subset of intestinal-associated strains that includes the opportunistic nosocomial pathogens Enterococcus faecalis and Enterococcus faecium (12). Lyso-LP formation may thus play a role in host adaptation, as lipoproteins are key ligands for Toll-like receptor 2 (TLR2) complexes of the innate immune system (13). E. faecalis expressing lyso-LP is recognized with over 100-fold less sensitivity by TLR2 compared with DA-LP expressing isogenic strains (14). Lyso-LP forming paralogs that are specifically induced by copper have been characterized on horizontally transmissible plasmid in select environmental isolates of Listeria monocytogenes (normally a DA-LP producer) (14). Copper, a widely used agricultural microbiocide and a natural defense mechanism in phagocytes (15, 16), was proposed to complex less tightly with N-acylated lyso-LP compared with the free α-amino nitrogen atom in DA-LP (14).
For the lyso-LP forming enzymes, there are no functionally characterized sequence orthologs, no conserved domains with annotation, and the catalytic mechanism is unknown. Two possible mechanisms for lyso-LP formation have been proposed: (i) N-acylation by an Lnt-like mechanism coupled with a O-deacylation sn-2 lipase activity; or (ii) transacylation through direct transfer of the sn-2 acyl chain from the diacylglycerol moiety to the N terminus (5). Because WMC_RS08810 is only ∼200 amino acids, we initially favored an intramolecular transfer model (Fig. 1, (ii)) and annotated WMC_RS08810 as lipoprotein intramolecular transferase (Lit) (11). Herein, a reconstituted Lit assay using isotopically labeled DA-LP substrate demonstrates the direct intramolecular migration of the sn-2 acyl chain to the α-amino cysteine group in the lyso-LP product. Alanine scanning mutagenesis of Lit using Mut-Seq (17, 18) has localized functionally important amino acid residues to two stretches containing two invariant histidine residues and no cysteine residues. Topology mapping experiments place both histidine residues in the outer leaflet of the membrane, so that a general acid-base catalytic model can be proposed. Unlike the well-characterized cysteine-based thioester mechanism of Lnt (19–22), we suggest the juxtaposition of the sn-2 acyl chain with the amino terminus promotes intramolecular cyclization and migration of the acyl chain from an ester (DA-LP) to amide (lyso-LP) bond.
Results
Isolation of deuterium-labeled LppK58A-strep tag DA-LP substrate
To test whether Lit functions as an inter- or intramolecular transacylase (Fig. 1), an E. coli strain was first engineered to produce substrate for in vitro reconstitution reactions with Lit (Fig. 2). DA-LP substrate with deuterium specifically incorporated into the diacylglycerol fatty acid ω termini could be used to ascertain N-acyl chain origin. Most Gram-negative bacteria, however, including E. coli, require N-acylation of lipoproteins for efficient trafficking from the cytoplasmic membrane to the outer membrane (23). Without N-acylation, outer membrane bound lipoproteins accumulate in the cytoplasmic membrane and some, notably Braun's lipoprotein (Lpp), form aberrant cross-links to peptidoglycan (24). The chromosomal lpp was therefore deleted and replaced with a streptomycin-tagged LppK58A allele to prevent covalent lysine-peptidoglycan cross-linking (23–26). LolCDE, the essential ABC (ABC) transporter that traffics lipoproteins from the inner to outer membrane in E. coli (27), was overexpressed to enhance transport of the nonoptimal DA-LP substrate. In this genetic background, lnt could be deleted without loss of cell viability as has been reported (23).
Figure 2.
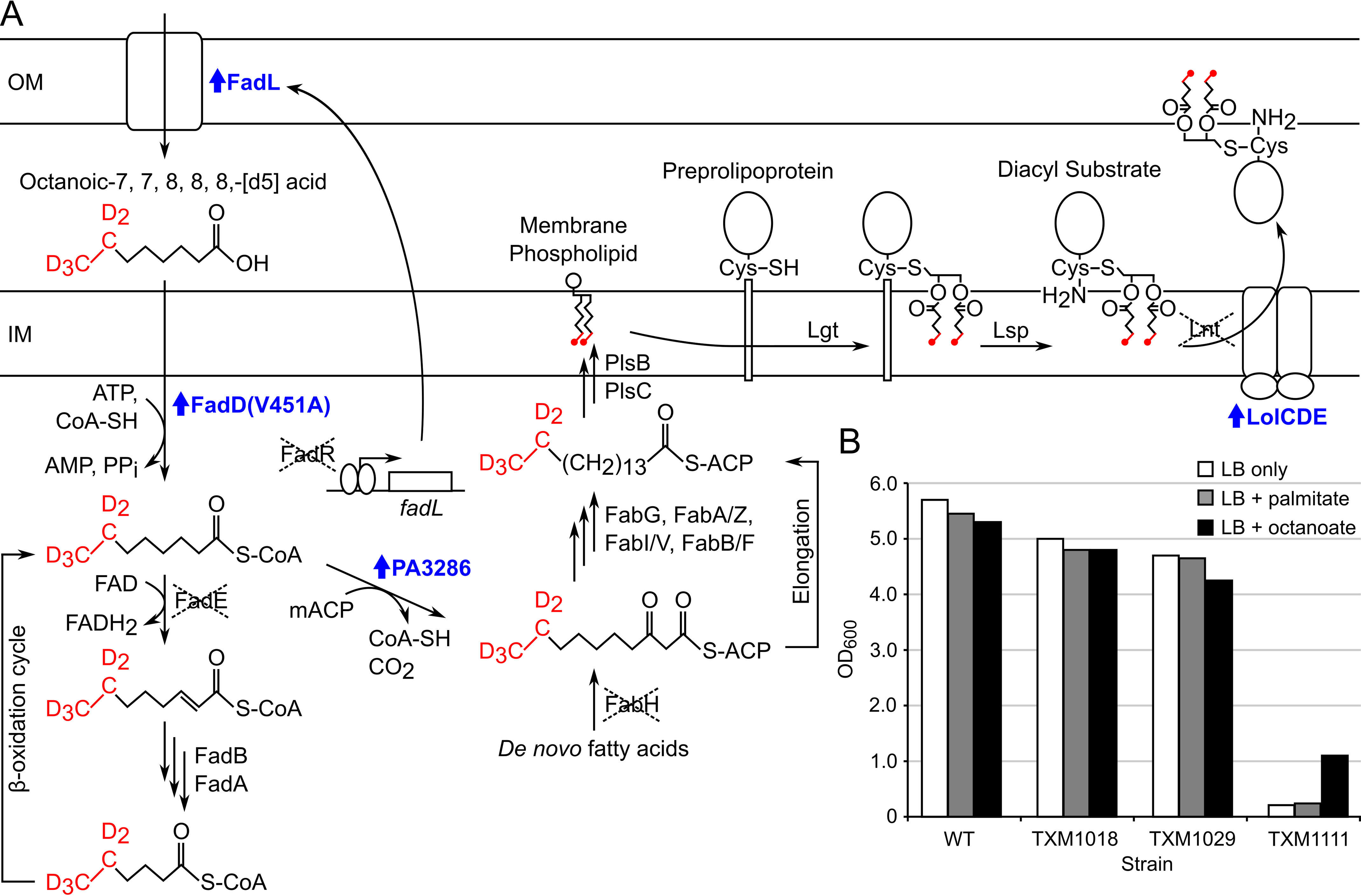
Production of acyl-labeled [d5]-DA-LP in strain TXM1111. A, exogenous octanoic-7,7,8,8,8-[d5] acid (labeled in red) is transported across the OM by constitutively expressed FadL upon deletion of FadR. Activation by esterification with CoA is enhanced by introduction of the FadD(V451A) allele. Entry of [d5]-octanoate substrate into the E. coli β-oxidation pathway is prevented by the deletion of FadE so that plasmid-encoded P. aeruginosa protein PA3286 can shunt the acyl-CoA ester intermediate into the FAS pathway by condensation with malonyl-ACP (mACP). Deletion of FabH precludes incorporation of unlabeled de novo fatty acids. Following cycles of elongation by FAS, [d5]-labeled acyl chains are incorporated into membrane phospholipids by PlsB and PlsC. Lipoprotein biosynthesis proceeds as normal, with the deuterium-labeled DA moiety transferred by Lgt from a phospholipid donor to the cysteine thiol of the preprolipoprotein. Lsp cleaves the signal peptide of the lipoprotein, exposing the α-amino group of the lipidated cysteine, to yield labeled [d5]-DA-LP. Overexpression of the ABC transporter LolCDE helps export the nonoptimal [d5]-DA-LP substrate to the OM and allows Lnt to be deleted. Up-regulated genes are labeled in blue. B, BW25113 WT (WT) and derivatives TXM1018 (fadR::Tmpr, lpp::FRT, fadE::Tetr), TXM1029 (fadR::Tmpr, lpp::FRT, fadE::Tetr + PA3286 + lolCDE), and TXM1111 (fadR::Tmpr, lpp::FRT, fadE::Tetr, fabH::FRT, lnt::Sptr + PA3286 + lolCDE + fadD(V451A)) were grown in LB only, or LB supplemented with 100 μg/ml of palmitate or octanoate. The final culture density (OD600) was measured after 19 h.
Next, we rewired the fatty acid β-oxidation pathway of E. coli to uniformly label lipoproteins in the five terminal ω deuterium atoms in both acyl chains of DA-LppK58A-strep tags when fed octanoic-7,7, 8, 8, 8-[d5] acid (Fig. 2). E. coli will not readily take up exogenous medium and short chain fatty acids (28–30), however, because the cognate acyl-CoAs are poor ligands for the fatty acid metabolism regulatory protein FadR (31). Therefore we deleted FadR to de-repress expression of exogenous fatty acid catabolism genes, including the outer membrane transporter FadL (31, 32). The long chain fatty acid:acyl-CoA ligase FadD of E. coli has low intrinsic activity with octanoate substrate, so the FadD(V451A) allele was introduced to enhance conversion of octanoate to octyl-CoA (33, 34). To prevent further entry into the β-oxidation cycle, which would catabolize labeled octyl-CoA to acetyl-CoA and randomly distribute the deuterium label, the first enzyme in the β-oxidation cycle acyl-CoA dehydrogenase (FadE) was deleted as well (35). We have previously shown that the Pseudomonas aeruginosa enzyme PA3286 when expressed in E. coli specifically condenses octyl-CoA with malonyl-ACP (acyl carrier protein) to form the fatty acid synthesis (FAS) intermediate β-keto decanoyl-ACP (36). By expressing PA3286, terminally labeled β-keto decanoyl-ACP could now be made from fed octanoic-7,7,8,8,8-[d5] acid. Elongation to long chain acyl-ACP by FAS and incorporation into membrane glycerophospholipids by the acyltransferases PlsB and PlsC (37) thus provides labeled substrate for Lgt to form pro-DA-LP (7). Additionally, because the de novo fatty acid initiation enzyme FabH can be deleted in E. coli K-12 (38, 39), fabH was removed to favor incorporation of exogenously fed octanoate by shrinking the endogenous cellular fatty acid pool. Although the growth rate of the final DA-LP labeling strain (TXM1111) was significantly decreased in standard lyosogeny broth (LB) when compared with that of the parent WT and intermediate strains, octanoate supplementation of both solid (not shown) and liquid media improved growth (Fig. 2B). Long chain fatty acids such as palmitate, which are not shunted by PA3286 (36), did not improve growth and is consistent with a functional medium acyl-CoA substrate chain shunt rescuing defective de novo fatty acid initiation.
[d5]-DA-LppK58A-strep from strain TXM1111 is dually labeled and retains acyl chain asymmetry
Growth enhancement by octanoate, but not palmitate, suggested efficient incorporation of octanoate by the engineered PA3286 shunt (Fig. 2). MALDI-TOF MS analysis of trypsinized [d5]-DA-LppK58A-strep purified from TXM1111 grown in media supplemented with octanoic-7,7,8,8,8-[d5] acid confirmed a new series of peaks shifted by 10 u compared with the negative control fabH+ strain KA775 (Fig. 3, A versus D, Table 1). The most abundant peak (1167.8 u) can be assigned to the N-terminal lipopeptide of LppK58A with 10 deuterium atoms, consistent with two [d5]-labeled acyl chains (Fig. 3C). A low abundance peak corresponding to singly labeled lipopeptide can be observed at 1162.6 u. Notably absent is the unlabeled lipopeptide peak (1157.6 u), which is the most abundant ion in the spectrum of the control strain KA775. To further characterize both lipoprotein structure and extent of labeling, the 1167.8 u parent ion was fragmented by tandem MS/MS (Fig. 3B). The y series ions corresponding to the SSNAK peptide of Lpp were observed as expected, whereas fragment ions 575.6 u, 607.7 u, and 647.7 u confirm a free α-amino terminus (Fig. 3C). Ions 575.6 u and 607.7 u, respectively, represent the dehydroalanyl peptide and the thiolated congener, generated by the neutral loss of the diacylthioglycerol and diacylglycerol moieties. The 647.7 u ion is consistent with the loss of both labeled fatty acids, a characteristic DA-LP fragment peak (11, 40, 41). The MS/MS spectrum of the 1157.8 u parent ion of unlabeled Lpp from KA775 shared these nonacylated fragment ions, confirming peptide identity, free N-terminal structure, and supports specific isotope incorporation within the acyl chains in TXM1111 (Fig. 3, E and F). Distinct between the two spectra, additional ions at 909.1 u and 927.2 u of [d5]-DA-Lpp from TXM1111 correspond to the parent ion having lost a labeled [d5]-C16:1 fatty acid (C15D5H24COOH) or [d5]-C16:1 ketene (C14D5H22CH=C = O), respectively (Fig. 3B). The corresponding ions in the unlabeled sample are shifted 5 u lower (904.0 and 921.6 u) (Fig. 3, E and F). The acyl chain composition for the most abundant DA-LP species can thus be inferred as one monounsaturated (C16:1) and one saturated (C16:0) palmitate fatty acids.
Figure 3.
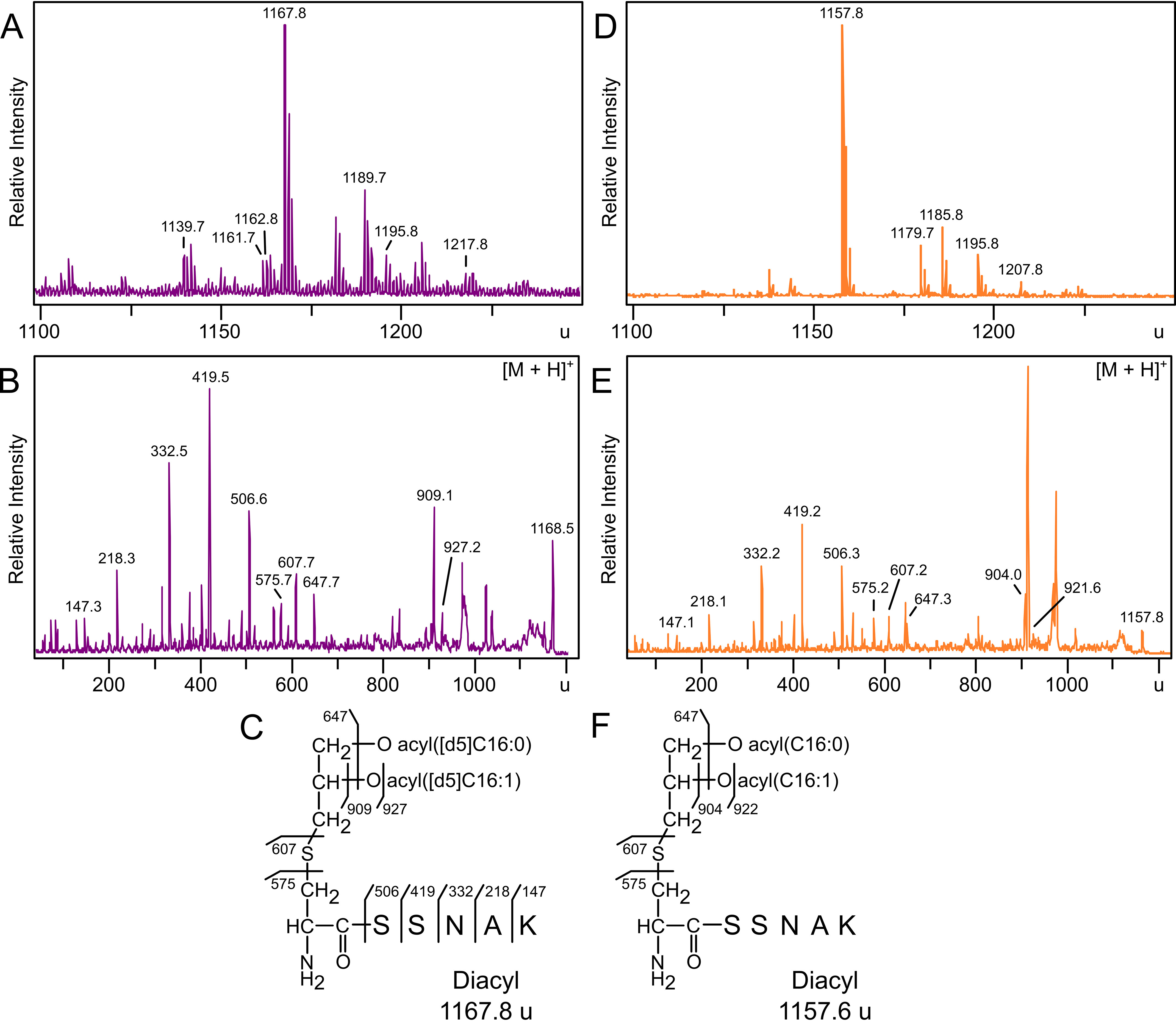
MALDI-TOF MS and MS/MS analysis of [d5]-DA-LppK58A-strep tag purified from strain TXM1111 and KA775. Trypsinized Lpp lipopeptides purified from strains grown with [d5]-octanoate were extracted from TXM1111 (A) or the control strain KA775 (D) and analyzed by MALDI-TOF MS. B, MS/MS spectra of the 1167.8 u parent ion was used to elucidate the N-terminal structure of Lpp as [d5]-DA-LppK58A with two labeled acyl chains (C). MS/MS spectra of the 1157.8 u parent ion (E) confirmed a free α-amino N terminus in DA-LppK58A (F). Calculated peak assignments are detailed in Table 1.
Table 1.
Calculated masses of [d5]-DA-LppK58A-strep
We next determined the position of the C16:1 fatty acid, either sn-1 or sn-2. Although WT E. coli preferentially incorporates unsaturated fatty acids at sn-2 (37, 42), the TXM1111 labeling strain has highly altered fatty acid metabolism and much slower growth kinetics (Fig. 2). Deletion of the fatty acid metabolic regulator FadR not only de-represses β-oxidation, but also decreases transcription of the unsaturated fatty acid biosynthetic enzymes fabAB (43). We utilized the sn-1 regiospecificity of lipoprotein lipase (LPL) from Pseudomonas spp., as reported by Asanuma et al. (44), to de-O-acylate [d5]-DA-LppK58A-strep. The MS spectra of digested [d5]-DA-LppK58A-strep contains a prominent peak at 924.6 u consistent in mass with the loss of the labeled saturated [d5]-C16:0 acyl chain, allowing regiospecific assignment of C16:1 to the sn-2 position in the [d5]-DA-LppK58A-strep substrate (Fig. S1). Together, these data indicate that the majority of [d5]-DA-LppK58A-strep population purified from strain TXM1111 is dually labeled with two C16 [d5]-acyl chains and has preferentially incorporated C16:1 in the sn-2 position.
Reconstitution of lit:lipoprotein N-acyltransferase activity in vitro
With structurally defined [d5]-DA-LppK58A-strep substrate in hand, we next reconstituted Lit enzyme activity. We have previously shown that replacement of Lnt with Lit produces lyso-form LppK58A (11), indicating Lit is expressed and active in E. coli. For in vitro reaction conditions, we used conditions established by Hillmann et al. (45) with Lnt so activity could be directly compared in a substrate competition assay. Inner membranes were isolated from E. coli strains expressing Lnt, Lit, or both, and used as the enzyme source. Membranes were mixed with purified recombinant DA-LppK58A-strep lipoprotein substrate in the presence of detergent, and product formation was assayed by α-strep tag immunoblotting (Fig. 4A). When DA-LppK58A is incubated with E. coli Lnt, a higher molecular weight band is observed over time, consistent with conversion of DA-LP into TA-LP. DA-LP and lyso-LP cannot be separated by SDS-PAGE because they are the same mass (11). The persistence of a lower molecular weight band in reactions after 24 h with both Lnt and Lit (Lnt/Lit) is indirect evidence of Lit activity, however, as any DA-LppK58A converted to the lyso-LP by Lit cannot be converted back into the TA-LP chemotype by Lnt. This also supports an intra-molecular transfer, as a bifunctional N-acyltransferase (DA-LP to TA-LP) and de-O-acylation lipase (TA-LP to lyso-LP) Lit activity would be expected to convert the entire substrate population into lyso-LP (Fig. 1, bottom panel).
Figure 4.
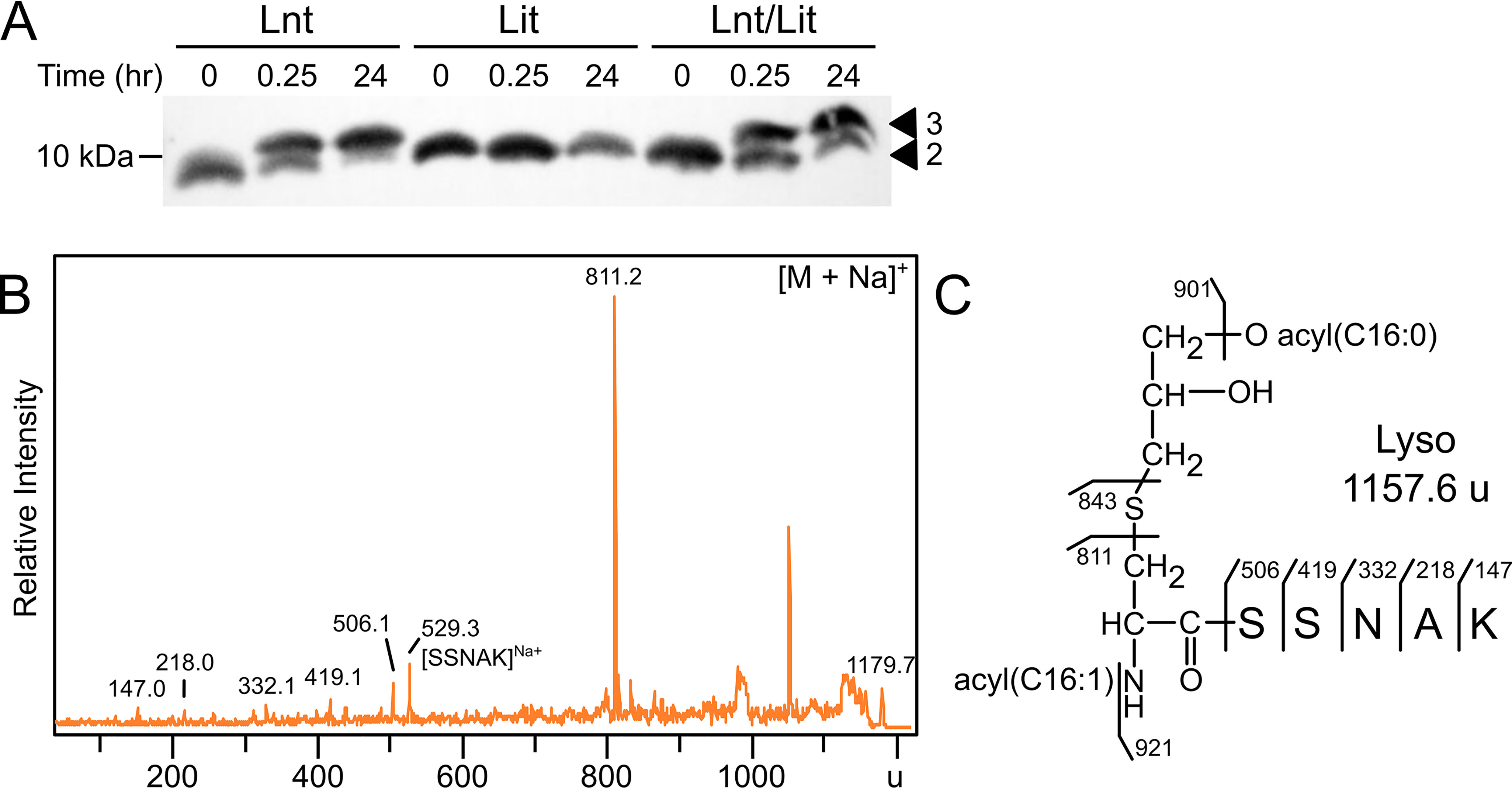
In vitro reconstitution of Lit. A, DA-LppK58A-strep tag was combined with inner membranes containing Lnt (from strain KA893), Lit (from KA801), or both Lnt/Lit (from KA736) for 24 h, with aliquots removed at 0, 0.25, and 24 h. Reactions containing Lnt show conversion to the slower migrating triacyl form (3), whereas Lit did not change migration. The reaction containing both Lnt and Lit shows persistence of LppK58A with two acyl chains (2), presumably due to lyso-LP formation. B, LppK58A was purified from the 24-h in vitro reaction with membranes containing Lit, digested with trypsin, and analyzed by MALDI-TOF MS. Fragmentation of the sodiated 1179.7 u parent ion produced the N-acylated dehydroalanyl ion at 811.2 u, indicating conversion of DA-LppK58A substrate to the lyso- form (C).
To confirm Lit-catalyzed lyso-LppK58A formation under our reconstitution conditions, Lit reactions were separated by SDS-PAGE and the product was analyzed by MS. The parent spectra obtained from membranes containing neither Lit/Lnt, nor only Lit, shared a prominent peak at 1157.7 u, consistent with diacylated Lpp lipopeptide (Fig. S2). The lyso-LP diagnostic N-acyl(C16:1) dehydroalanyl fragment ion at 811.2 u was only observed for reactions that included Lit (sodium adduct, 1179.7 u) (Fig. 4B). The preference for C16:1 incorporation at the N terminus is in full agreement with the in vivo acyltransferase activity of Lit (11).
Lit catalyzes an intramolecular transfer of the sn-2 acyl chain to the N terminus
To determine the origin of the amide-linked N terminus acyl chain in lyso-LP, Lit activity was reconstituted as above using deuterium-labeled d5-DA-LppK58A-strep substrate. If Lit functions intermolecularly, using a phospholipid acyl chain donor similar to that of Lnt (45), an unlabeled acyl chain from the phospholipid pool will be incorporated into the N-terminal lipopeptide. If Lit functions intramolecularly, the [d5]-C16:1–labeled acyl chain should migrate to the N terminus. Membranes were prepared from lnt+ E. coli cells and incubated with the [d5]-DA-LppK58A-strep tag substrate. Lipoprotein products were affinity purified with StrepTactin-coated magnetic beads, and analyzed by MS. Membranes with Lnt yielded products with masses consistent with TA*-LppK58A-strep (1406.3 u) (Fig. 5A). This mass is 10 u higher than the previously characterized unlabeled variant (11), consistent with two [d5]-labeled acyl chains and a single unlabeled acyl chain. Fragmentation produced the 814 u dehydroalanyl ion, indicating an unlabeled C16:0 N-acyl chain at the α-amino terminus (Fig. 5B). Additional peaks at 885.9 u, 1146.9 u, and 1164.9 u confirmed the retention of both [d5]-acyl chains on the diacylglycerol moiety and assignment of all acyl chains positions (Fig. 5C). The bulk phospholipid pool in the reconstituted system is thus the acyl donor used by Lnt in TA-LP synthesis, as previously reported (46).
Figure 5.
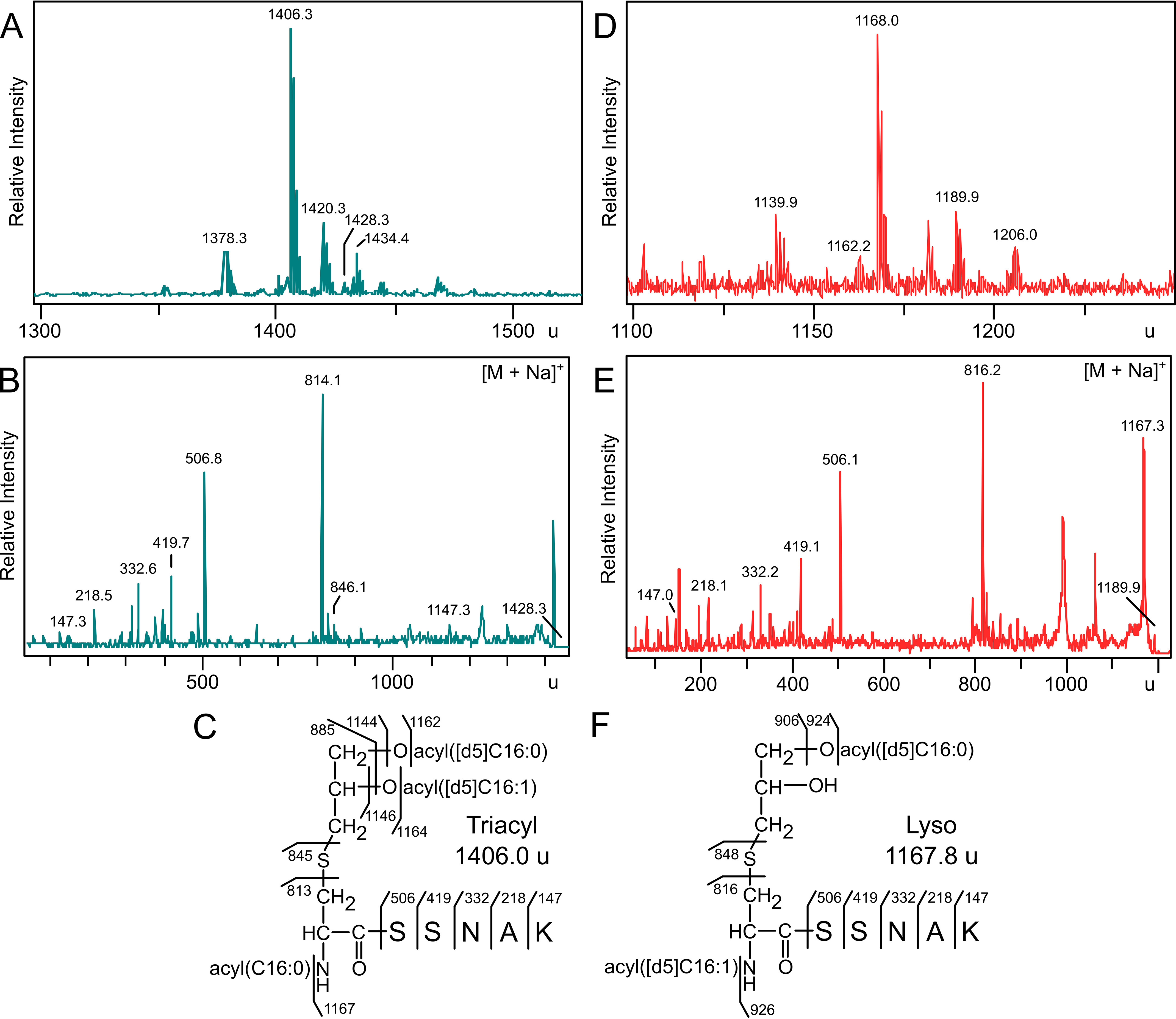
Reaction of [d5]-DA-LppK58A-strep with Lnt and Lit. [d5]-DA-LppK58A-strep purified from reactions with membranes containing Lnt (from strain KA893; panels A and B) or Lit (from strain KA801; panels D and E) were digested with trypsin and analyzed by MALDI-TOF MS. B, the MS/MS spectrum of the sodiated parent triacylated 1428.3 u ion yielded the dehydroalanyl ion at 814.1 u, indicating an unlabeled N-acyl chain on LppK58A-strep (C). E, MS/MS of the sodiated parent ion diacylated 1189.9 u ion from Lit generated a dehydroalanyl ion at 816.2 u, consistent with a lyso-LP chemotype containing a [d5]-labeled N-acyl chain (F).
The reaction was repeated with membranes containing only Lit. Parent spectra of isolated lipoprotein products revealed a predominant peak at 1168.0 u, consistent in mass with LppK58A-strep containing two [d5]-acyl chains (Fig. 5D). As this ion could possibly be unreacted [d5]-DA-LppK58A-strep substrate, the corresponding sodium adduct 1189.9 u was subject to MS/MS (Fig. 5E). This ion preferentially fragmented to give the expected mass for the N-[d5]-acyl-dehydroalanyl ion (816.2 u) (Fig. 5F). These data confirms that Lit is an intramolecular sn-2 ester to N-terminal amide lipoprotein transacylase (Fig. 1).
Mut-seq analysis of important amino acids in lit
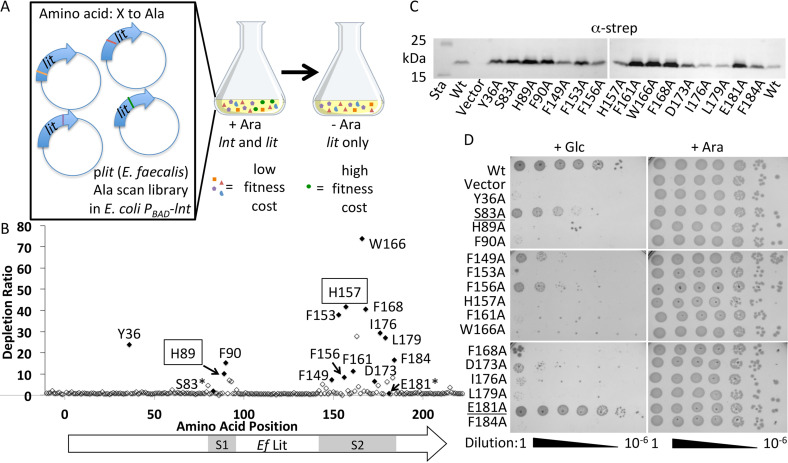
There are few examples of enzymes catalyzing analogous intramolecular ester to amide acyl chain migration reactions. We initially presumed one of the three cysteine residues (Cys-16, Cys-100, and Cys-187) in Lit formed an acyl thioester type intermediate, as has been characterized for Cys-387 in the active site of Lnt (19, 26, 45, 47). However, none of the cysteine residues in the chromosomal copy of Lit (11) appear to be conserved in the plasmid borne, copper-inducible Lit2 paralog (14). To better understand how Lit catalyzes transfer of the sn-2 acyl chain, we established a selection system to monitor growth rescue in E. coli by an alanine (Ala) scan library of E. faecalis Lit alleles (Fig. 6A). We have previously shown lit can functionally replace lnt in an E. coli Δlpp background through lyso-LP formation, with the N-acylation restoring lipoprotein trafficking to the outer membrane (11). A plasmid-based Ala scan allele library of the lit gene was systematically constructed to replace every coding amino acid with an Ala triplet (GCG) and introduced into an E. coli strain that conditionally expresses lnt (PBAD-lnt) provided arabinose inducer is present (Fig. 6A). Plasmids were isolated from the input library as well as from a culture serially passaged in the absence of arabinose inducer. Residues important for Lit function and phenotypic rescue were determined using Mut-seq analysis of next generation sequencing data to measure depletion ratios (17, 18). Every coding triplet except two nonsynonymous triplets of the 212 amino acids were detected above background in the input library (Fig. S3A). Deletion of any three Cys residues did not attenuate function (Fig. 6B), indicating the active site does not utilize a thioester intermediate and further differentiates Lit from Lnt. Except for L118A that modestly increased fitness (Fig. S3B), conversion to Ala at a single position either had no effect or led to a decrease in fitness when Lnt was depleted. Two prominent stretches of amino acids (S1 and S2) where Ala replacement led to marked depletion were identified (Fig. 6B), suggesting local regions of defined structure and the potential location of catalytic residues. Interestingly, many functionally important aromatic residues are located within both of these stretches (His-89, Phe-90 from S1 and Phe-149, Phe-153, Phe-156, His-157, Phe-161, Trp-166, Phe-168, Phe-184 from S2), in addition to a single N-terminal tyrosine residue (Tyr-36). We tested whether these residues were important due to changes in protein expression levels or protein stability by immunoblotting (Fig. 6C). Although the level of Lit varied between each Ala mutant, all constructs expressed protein at levels at least equal to that observed in WT Lit. All mutants displaying significant depletion ratios in the Mut-seq analysis when reconstructed and introduced into E. coli PBAD-lnt recapitulated the phenotype in a spot titer assay (Fig. 6D). A role for these aromatic residues in establishing tertiary structure is thus more likely. Aromatic residues, through π–π and other interactions, are critical determinants in the self-assembly of multipass α-helical transmembrane (TM) segments in integral membrane proteins (48–50). Phe in particular is one of the most common partners in interhelical contact pairs (51). Lit is predicted to encode multiple TM domains (see below), suggesting these key residues may be part of dynamic interhelical interactions involved in lipoprotein substrate gating and/or binding.
Figure 6.
Mut-seq analysis of E. faecalis Lit. A, a library of plasmids expressing lit from E. faecalis was systematically constructed to replace every amino acid (212 total) with an alanine residue. The plasmid library was pooled, transformed into E. coli Δlpp PBAD-lnt, and maintained in media with 0.2% arabinose (w/v) inducer so that lnt remained expressed. The library was then passaged in the absence of arabinose to make growth dependent on the activity of the respective lit X-to-Ala mutant allele. Plasmid DNA was isolated from the library before and after withdrawal of arabinose, the lit coding region was amplified with flanking primers, and the amplicon pools were subject to next generation sequencing. B, representation of each Ala codon (GCG) triplet substitution in the initial library was calculated as a percent of total reads containing Ala substitution, and compared with the composition after passaging to determine depletion ratios (Fig. S3). Residues rechecked by spot titer are shaded black, negative controls are indicated (*), and the putative catalytic His residues subjected to randomization (Fig. S4) are boxed. The Ef Lit coding region targeted for Ala scan mutagenesis is indicated along with the two functional stretches (S1 and S2). Flanking regions (10 in-frame triplets upstream of ATG start codon and eight in-frame triplets downstream) are shown for comparison to spontaneous mutations/sequencing error rate. C, the level of Lit expression for each Lit Ala point mutant was determined by immunoblotting with anti-strep tag. Sta, molecular weight standard; Vector, empty vector. D, strains of E. coli expressing each Lit variant were 10-fold serially diluted and spotted on LB agar supplemented with either glucose (+Glc) or arabinose (+Ara) to repress or induce expression of PBAD-lnt.
Residues deemed to be important by Mut-seq analysis would be expected to be highly conserved across Lit sequences from different organisms. Sequence alignment confirms that most depleted Ala substitution positions are indeed highly conserved residues across Lit from different bacterial genera (chromosomal Lit) as well as in plasmid-borne Lit2 paralogs located in a copper-resistance operon (Fig. 7). However, not all conserved residues are required for function. Both Ser-83 and Glu-181 are highly conserved, polar, and located within important stretches (S1 and S2), but are not necessary for catalysis (Fig. 6D). Overall, Mut-seq analysis implicated few polar residues as critical to function, which is surprising given the catalytic residues are likely polar. Presumably, the sn-2 position O-acyl chain shown to be transferred intramolecularly (Figs. 1, bottom panel, and 5D) is subject to nucleophilic attack by either: (i) an amino acid side chain to form an acylated covalent enzyme intermediate or (ii) through direct attack by the α-amino terminus of the of DA-LP substrate itself. Two prime candidates for general acid-base chemistry to activate the carbonyl are the functionally required (Fig. 6, B and D) and invariant (Fig. 7) His-89 and His-157 residues. We randomized both histidine positions with all 20 amino acids to determine whether any substitutions besides Ala could retain function and performed a second round of Mut-seq analysis. All amino acid replacements were rapidly depleted from the population (Fig. S4), confirming a key role of these His residues in Lit function.
Figure 7.
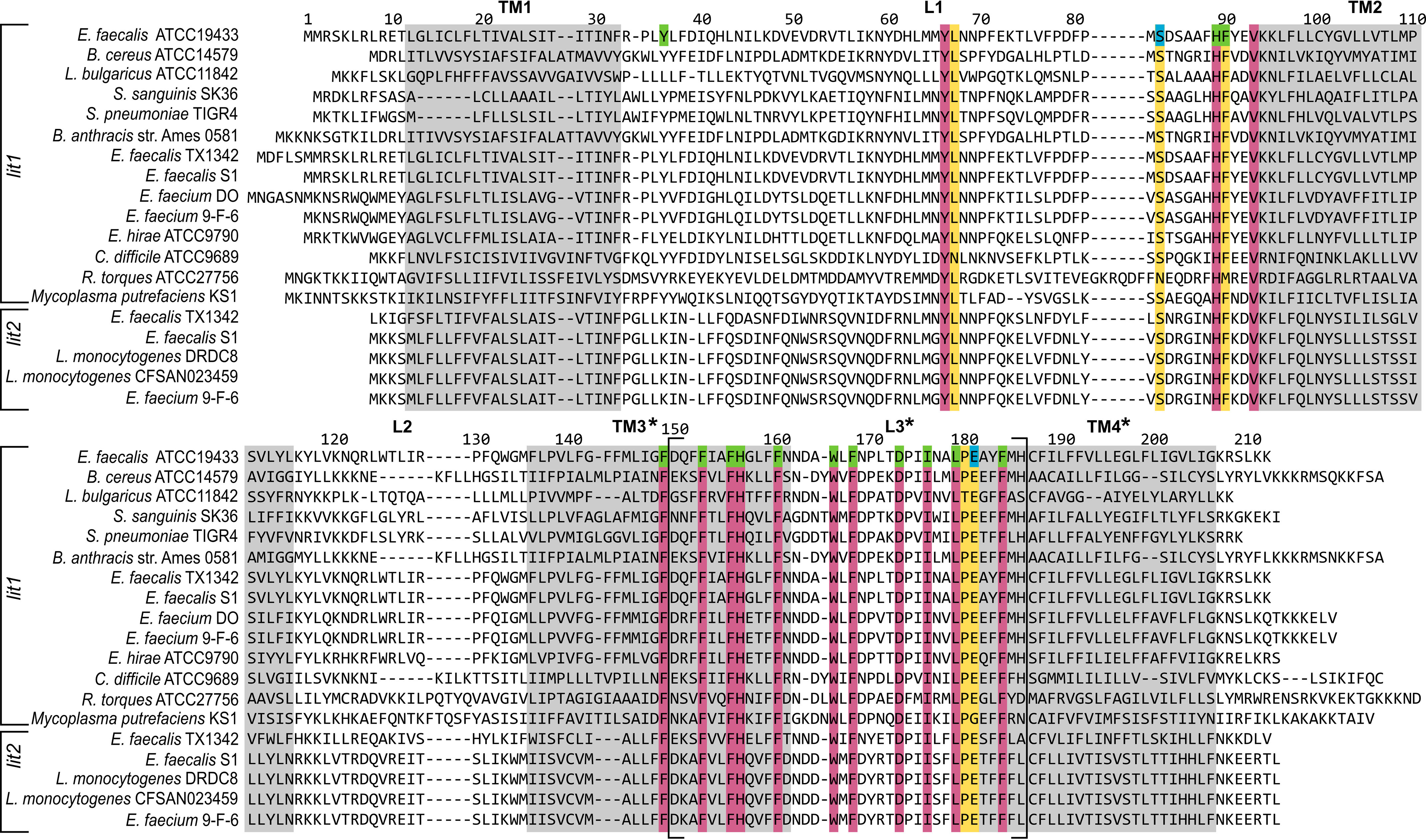
Primary amino acid sequence alignment of chromosomal Lit and accessory Lit2 proteins. Sequences of the chromosomal Lit (Lit1) and the copper-resistance associated accessory transposon/plasmid located (Lit2) paralogs from the indicated strains was generated by STRAP (36). Highly conserved amino acids are colored (purple for 100% and yellow for retention in all but one sequence). Residues deemed essential (green) or nonessential (blue) by Mut-seq analysis and confirmed for function (Fig. 6D) are indicated. Predicted TM segments (by Protter (26); see Fig. 8) are boxed in gray with uncertainty in the boundaries of loop 3 (L3) denoted by brackets and asterisks (Table S2).
Membrane topology of lit
As Lit is the sole member of a novel class of enzymes with no sequence similarity to characterized acyltransferases, we sought to map the membrane topology of Lit. According to several membrane topology mapping algorithms (Table S2), Lit is predicted to have four TM passes along with one intracellular and two extracellular loops, with both the N and C termini located in the cytoplasm (Fig. 8A). There is, however, poor consensus in boundaries for the second predicted extracellular loop (loop 3), which overlaps with S2 identified in Mut-seq analysis (Fig. 6B). To gain further insight into the membrane topology, a series of C-terminal reporter fusions using either β-gal (LacZ) or alkaline phosphatase (PhoA) were constructed within each putative loop. Localization of the reporter proteins to the cytoplasm (LacZ+) and the periplasm (PhoA+) was assessed using the respective colorimetric substrates (52). LacZ fusions within loop 2 (LitR129-LacZ) and at the C terminus (LitK212-LacZ) confirmed cytoplasmic locations, whereas loop 1 PhoA fusion (LitD52-PhoA) indicated an extracellular periplasmic location (Fig. 8, B and C). Loop 3 reporters for both intra- and extracellular (LitD173-LacZ and LitD173-PhoA, respectively) orientations were negative. Because loop 3 contains the functionally important S2 amino acid stretch, we tested additional fusion sites within loop 3 (after residues Leu-159, Ala-165, Leu-179, and Met-185) as well as an intervening linker to separate the reporter enzyme domain from Lit (18-residue linker VPDSYTQVASWTEPFPFC (53)). However, no reporter enzyme function was observed (data not shown).
Figure 8.
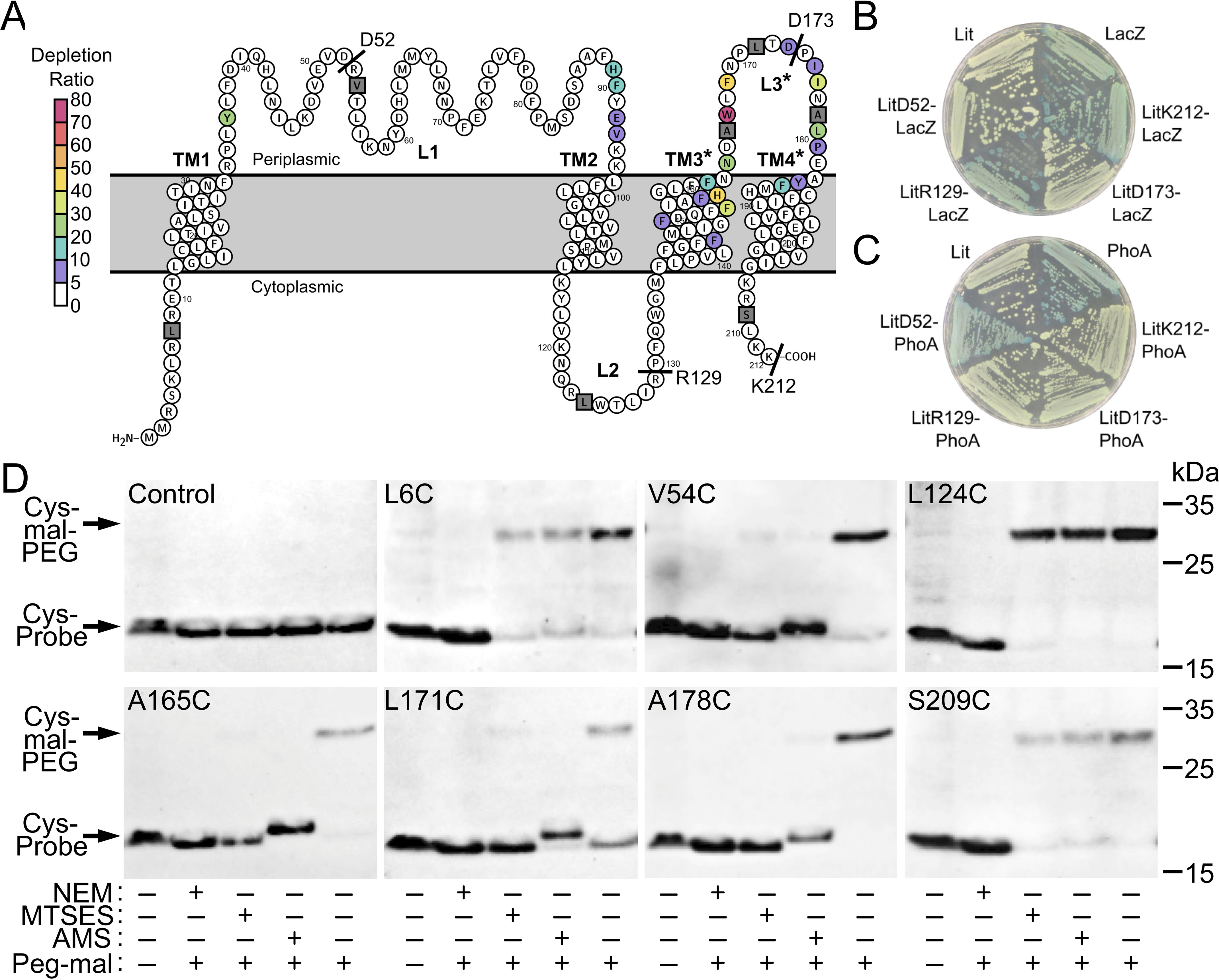
Membrane topology of Lit. A, the membrane topology of Lit as predicted by Protter (26). Uncertainty in membrane boundaries is denoted by an asterisk (*) on TM passes TM3 and TM4, and periplasmic loop L3 (see Table S2). Depletion ratios determined by Mut-seq analysis are indicated (see Fig. 6). The position of LacZ and PhoA C-terminal fusions are labeled with text, whereas positions probed using the SCAM are gray squares. B, strains expressing native Lit (SS851), native LacZ (SS904), and the fusions LitD52-LacZ (SS951), LitR129-LacZ (SS935), LitD173-LacZ (SS932), and LitK212-LacZ (SS915) were struck on solid media containing X-gal. Blue colonies (LacZ, LitR129-LacZ, or LitK212-LacZ) indicate active LacZ localized in the cytoplasm. C, strains expressing native Lit (SS851), native PhoA (SS964), and the fusions LitD52-PhoA (SS950), LitR129-PhoA (SS936), LitD173-PhoA (SS940), and LitK212-PhoA (SS916) were struck on solid media containing the alkaline phosphatase colorimetric indicator BCIP. Blue colonies (PhoA and LitD52-PhoA) indicate active PhoA localized in the periplasm. Neither of the predicted loop 3 fusions (LitD173-lacZ or LitD173-PhoA) demonstrated activity. D, SCAM was used to determine topology at the indicated positions (gray squares in panel A) in a strep-tagged Lit construct lacking all endogenous cysteine residues (C16A, C100A, C187A). Cells expressing Lit variants with a single cysteine residue were either unlabeled (−, first column) or labeled (+, second to fifth columns) with mal-PEG (average molecular mass 5,000 Da). Samples labeled with mal-PEG were pretreated with either a membrane permeable NEM or impermeable MTSES or AMS thiol reactive probe as indicated (+/−). Cells were lysed and protein extracts were separated by SDS-PAGE before detection of Lit with α-strep tag-specific antibody. Formation of Lit-mal-PEG adducts (higher molecular weight) occurs provided the cysteine residue is inaccessible to probe in whole cells (55, 56).
The failure to reconstitute activity for any fusion reporter construct in loop 3 suggests a highly structured local region closely associated with or even possibly embedded within the membrane. This would explain the difficulty in predicting a clear boundary consensus for TM3 (Table S2). As LacZ/PhoA fusion experimental analysis was likewise inconclusive, the substituted cysteine accessibility method (SCAM) (54) was used to interrogate topology at multiple positions within loop 3 (Fig. 8D). Cysteine exchanges were made within each loop and in both termini using a strep-tagged Lit construct lacking all endogenous cysteine residues (C16A, C100A, C187A). Methoxypolyethylene glycol maleimide (mal-PEG, average molecular mass 5,000 Da) was used to detect free cysteines after pretreatment with either membrane-permeable (N-ethylmaleimide (NEM)) or impermeable (2-sulfonatoethyl methanethiosulfonate (MTSES) or 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS)) thiol reactive probes (55, 56). Lit without any cysteine residues was unreactive with mal-PEG. Intracellular cysteine residues (N terminus L6C, intracellular loop 2 L124C, and C terminus S209C) were only protected by pretreatment with the membrane-permeable probe NEM, indicating a cytoplasmic location and suggested by the Lit-LacZ reporter construct activity assay (Fig. 8B). The loop 2 construct (V54C) was protected by all probes, consistent with the extracellular topology suggested by the Lit-PhoA reporter fusion data (Fig. 8C). Three positions were chosen within the putative loop 3 (A165C, L171C, and A178C), and the membrane-impermeable probes (MTSES and AMS) protected Lit from mal-PEG labeling in all three cases. The AMS (536.4 g/mol) membrane-impermeable probe, which is larger than MTSES (242.2 g/mol), induced an upward molecular mass shift in AMS-labeled samples in conjunction with decreased amounts of Lit-mal-PEG adduct. These positions within loop 3 of Lit thus appear to be at least somewhat exposed to small molecule polar probes on the extracellular membrane surface, although it remains unknown whether loop 3 is truly an extended disordered loop as shown in Fig. 8A or in a state that is more intimately associated with the membrane.
Discussion
Bacteria have evolved large and diverse enzyme families to catalyze acyl transfers (57). Lit, however, has no apparent sequence similarity to any of these enzyme families, reflecting the unique nature of an enzyme-catalyzed intramolecular acyl chain migration. From a thermodynamic perspective, the ester-to-amide acyl chain exchange would be expected to favor lyso-LP over DA-LP at equilibrium. Indeed, residual DA-LP is at the limit of detection by MALDI-TOF MS in WT E. faecalis (11). Unlike the unimolecular Lit reaction mechanism, Lnt in Gram-negative bacteria catalyzes a bimolecular acyl transfer reaction. The high effective phospholipid donor concentration, in concert with the N-acyl substrate selectivity of the Lol transport system (58), favors chemotype conversion of the lipoprotein population to TA-LP. Lol acts as a quality control mechanism to ensure only mature Lnt-processed lipoproteins are removed from the inner membrane, a particularly relevant factor in controlling the DA-LP/TA-LP ratio given that out of a an estimated 3-4 million total proteins in a typical E. coli cell (59), up to a million are outer membrane-bound lipoproteins (3, 60, 61). Because there is no Lol system to ensure N-acylation of the bulk of lipoproteins in Gram-positive bacteria, Lit must act rapidly as many lipoproteins are part of multicomponent protein complexes that will become inaccessible once assembled. For instance, if only ∼1% of the lipoprotein population does not get converted to lyso-LP, the TLR2 detection sensitivity will be doubled because DA-LP is detected with 100-fold more sensitivity (14). An intramolecular transfer mechanism may also help explain why both lgt2 and lit2 occur in tandem on a horizontally transmissible, divergently transcribed copper resistance operon (14). The Lit mechanism described here means Lgt/Lgt2 selects the Lit/Lit2 acyl chain substrate when condensing the preprolipoprotein with a particular diacylglycerophospholipid. Nonoptimal substrate acyl chains installed at sn-2 by Lgt/Lgt2 in DA-LP would retard the catalytic efficiency of Lit, limiting lyso-LP conversion ratios when acquired by hosts with mismatched chromosomal Lgt acyl chain specificities. Having both Lgt2-Lit2 would allow transmission between hosts with different acyl chain pools, as is the case with the Lgt2-Lit2 cassette, which has been passed between strains of Enterococcus sp. (even-chain fatty acids with unsaturation (62, 63)) and L. monocytogenes (odd-chain branched saturated fatty acid pool (64)).
The Lit enzymes have a narrow phylogenetic distribution and thus limited amino acid sequence divergence (Fig. 7). None the less, residues identified by Mut-seq analysis as important for function are highly conserved (Fig. 6). Aside from Tyr-36, key amino acid residues cluster within two stretches (S1 and S2). A number of aromatic residues, particularly Phe, within S1 and S2 proved critical for growth rescue. The Phe to Ala point mutations did not impact Lit protein levels, and most were located within putative TM passes (Fig. 8). Aromatic residues are important determinants for proper insertion and stability of TM passes, and their overrepresentation at the membrane-water boundary in integral membrane proteins has been described as the aromatic belt (65). However, Phe has no polar heteroatom functionality for H-bonding and is the one aromatic residue not typically enriched at the membrane interface. In the case of Lit, Phe may rather play a role in directing and stabilizing inter helical contacts through π–π or other interactions within the membrane interior. Few conserved polar residues and possible active site candidates were deemed essential by Mut-seq. Given the absence of any obvious candidates for an acyl thioester (Cys) or analogous ester (Ser/Thr) or mixed anhydride (Asp/Glu) type intermediate, there are no mechanistic parallels with Lnt. This underscores the independent origin of lipoprotein N-terminal tailoring reactions in what is otherwise a highly conserved pathway.
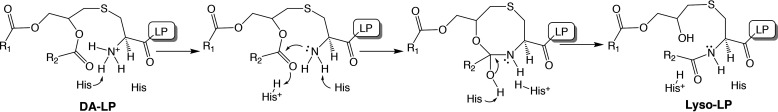
Two polar residues with high depletion ratios, located within stretches of functional importance, and absolutely required for activity were His-89 and His-157 (Fig. 6). In addition to being conserved (Fig. 7), both His residues were irreplaceable by any other amino acid (Fig. S4). Topology mapping places both residues in an extracellular or membrane interface having access to DA-LP substrate (Fig. 8). In the case of His-157, precise mapping of TM3 and the putative loop 3 boundary encompassing S2 was complicated. Computational (Table S2) and experimental colorimetric fusion reporter assays could not verify topology (Fig. 8, B and C). SCAM labeling with membrane-impermeable thiol-reactive probes did support a degree of extracellular exposure of the putative loop 3 (Fig. 8D). It is possible that this TM3-loop3 boundary region is partially embedded or parallel with the membrane, defining a cavity that directly interacts with DA-LP. Histidine residues are common catalytic residue in many acyltransferases (57), and in the case of Lit, presents two possible catalytic mechanisms. In a general acid-base mechanism (Fig. 9), Lit positions the free α-amino terminus of DA-LP substrate in proximity to the sn-2 carbonyl for nucleophilic attack. An 8-member cyclic tetrahedral intermediate then forms before collapsing to produce N-acylated lyso-LP product. This mechanism is simplest, requiring protonation/deprotonation events and polarization of the ester carbonyl. The candidate His residues could fulfill these catalytic roles. A second plausible mechanism invokes direct nucleophilic attack by His on the sn-2 carbonyl to form a covalent acyl-Lit enzyme intermediate that then transfers the acyl chain to the N terminus. Although our data cannot discriminate between the two scenarios, we currently favor the first mechanism. Examples of histidines in acyltransferases acting as general acid-base catalysts are common, whereas covalent acylhistidine intermediates are rare (57).
Figure 9.
Putative catalytic mechanism of Lit. The direct attack mechanism using general acid-base catalysis to activate the sn-2 carbonyl of DA-LP substrate. Nucleophilic attack by the α-amino group forms an 8-member cyclic intermediate that collapses to yield N-acyl containing lyso-LP product. His-89 and His-157 are candidate residues for directing protonation and deprotonation events in the catalytic cycle.
Lit represents a novel class of acyltransferases catalyzing lipoprotein N-acylation in select Firmicutes. Lit shares no primary sequence or mechanistic similarity with the only other characterized lipoprotein N-acylating enzyme Lnt from Gram-negative bacteria. There are other options for lipoprotein N-acylating systems besides Lit and Lnt as well, including the recently identified two-component LnsAB system in Staphylococcus aureus, which bears no resemblance to either Lit or Lnt.3 In the context of the lyso-LP formation by Lit, what is the advantage of intramolecular transacylation over other known mechanisms of N-acylation? Extracellular proteins in Firmicutes have evolved to exclude cysteine as many lack machinery to reduce aberrant disulfide bonds formed under oxidative stress (38). Lit does not utilze an active site cysteine (Fig. 6), a relevant detail given Lit2 is specifically induced by copper, which readily oxidizes thiols. Formation of TA-LP by Lnt type activity also generates a lysophospholipid byproduct from the glycerophospholipid acyl donor (46). Physically similar to detergents, lysophospholipids are cytolytic and perturb membrane integrity (66). In E. coli, lysophospholipids are reacylated by the phospholipid repair system LplT/Aas (67–70), a system largely absent in Gram-positive bacteria. In light of this, N-acylation by intramolecular transacylation avoids forming lysophospholipids and may be advantageous. As more is learned regarding the various lipoprotein N-acylating systems in different bacteria, insights into why each system has been acquired in a particular host will become more clear.
Experimental procedures
Bacterial strains and growth conditions
E. coli strains used in this study, listed in Table 2, are derivatives of either K-12 (BW25113) or BL21(DE3). All strains were grown in lysogeny broth-Miller medium (LB) at 37 °C with agitation. Antibiotic markers were selected with carbenicillin (100 μg/ml), apramycin (100 μg/ml), spectinomycin (50 μg/ml), tetracycline (5 μg/ml), kanamycin (15 or 30 μg/ml), and trimethoprim (50 μg/ml). Where appropriate, fatty acids were supplemented to 100 μg/ml.
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/phenotype1 | Reference |
|---|---|---|
| Escherichia coli | ||
| BL21(DE3) strains | ||
| KA729 | pET22b(+)-Eflit (from E. faecalis ATCC 19433) | This study |
| KA736 | lpp::Cmr + pET22b(+)-Eflit | This study |
| KA801 | lpp::Cmr lnt::Specr chiQ::Aprr + pET22b(+)-Eflit | This study |
| KA818 | lpp::Cmr lnt::Specr chiQ::Aprr + pET22b(+)-Eflit + pKA810 | This study |
| KA893 | lpp::Cmr | This study |
| E. coli | ||
| BW25113 strains | ||
| TXM327 | lpp::Cmr | 8 |
| KA349 | lpp::Cmr ybeX-(Kanr-rrnB TT-araC-PBAD)-lnt | 11 |
| TXM541 | gut::Kanr-rrnB TT-araC-PBAd-lnt, lnt::Specr, chiQ::Aprr | 8 |
| KA775 | TXM327 lnt::Specr + pCT763 (spontaneous suppressor mutant) | Laboratory stock |
| KA827 | phoA::Kanr | Keio collection |
| KA845 | E. coli BW25113 fadR::Trimr | This study |
| SS851 | KA827 + pET22b(+)-Eflit; Carbr | This study |
| SS904 | KA827 + pET22b(+)-lacZ; Carbr | This study |
| SS915 | KA827 + pET22b(+)-EflitK212-(+9)lacZ fusion; Carbr | This study |
| SS916 | KA827 + pET22b(+)-EflitK212-(+13)phoA fusion; Carbr | This study |
| SS932 | KA827 + pET22b(+)-EflitD173-(+9)lacZ fusion; Carbr | This study |
| SS935 | KA827 + pET22b(+)-EflitR129-(+9)lacZ fusion; Carbr | This study |
| SS936 | KA827 + pET22b(+)-EflitR129-(+13)phoA fusion; Carbr | This study |
| SS940 | KA827 + pET22b(+)-EflitD173-(+13)phoA fusion; Carbr | This study |
| SS950 | KA827 + pET22b(+)-EflitD52-(+13)phoA fusion; Carbr | This study |
| SS951 | KA827 + pET22b(+)-EflitD52-(+9)lacZ fusion; Carbr | This study |
| SS964 | KA827 + pET22b(+)-phoA; Carbr | This study |
| TXM1014 | KA845 lpp::Kanr flanked by FRT sites | This study |
| TXM1015 | TXM1014 lpp::FRT | This study |
| TXM1018 | TXM1015 fadE::Tetr | This study |
| TXM1O29 | TXM1018 + pTXM1O26 | This study |
| TXM1036 | TXM1O29 lnt::Sptr | This study |
| TXM1058 | TXM1036 fabH::Cmr flanked by FRT sites | This study |
| TXM1067 | TXM1058 fabH::FRT | This study |
| TXM1111 | TXM1067 + pTM1100 | This study |
| Plasmids | ||
| pCL25 | Cloning vector with RepA origin | 39 |
| pSEVA434 | General cloning vector with pBBR1 origin | 71 |
| pKA522 | pCL25(ori) lpp(K58A)-Strep-tag Kanr | 8 |
| pET22b(+) | Expression vector with T7 promoter lacI; Carbr | Novagen |
| pET22b(+)-Eflit | pET22b(+) Eflit (from E. faecalis ATCC 19433), 212 amino acid ORF | This study |
| pET22b(+)-Eflit strep | pET22b(+) Eflit strep tag (from E. faecalis ATCC 19433) | This study |
| pET22b(+)-Eflit strep | pET22b(+) Eflit strep tag (from E. faecalis ATCC 19433) lacking all cysteine residues | This study |
| C16A C100A C187A | ||
| pLI50-Ppen Gfpmut2 | Mid-copy plasmid number with Ppen promoter controlling insert expression; Carbr | 72 |
| pLI50-Eflit strep | pLI50-Ppen lit lacking all endogenous cysteine residues | This study |
| C16A C100A C187A | ||
| pCT763 | pKA522 PA3286 (from Pseudomonas aeruginosa) | This study |
| pKA810 | pCL25(ori) Tmpr-lpp(K58A)-Strep-tag | This study |
| pTXM1O26 | pBBR1(ori) PKan-lolCDE-PA3286 Kanr | This study |
| pTM1100 | pCL25(ori) lpp(K58A)-Strep-tag-fadD(V451A) Cmr | This study |
1Resistance phenotypes: Aprr, apramycin; Carbr, carbenicillin; Cmr, chloramphenicol; Kanr, kanamycin; Sptr, spectinomycin; Tetr, tetracycline; and Tmpr, trimethoprim.
Construction of deletion strains and plasmids
Gene deletions in E. coli were constructed using the Red-recombinase method and transduced into recipient strains by P1vir (73). Plasmids were assembled using the In-Fusion HD cloning kit (Takara Bio). Plasmids with point mutations were constructed by inverse PCR. Primers used in this study are listed in Table S1.
Deuterium labeling and affinity purification of lpp(K58A) strep tag
Strain TXM1111 was constructed and used to produce deuterium labeled [d5]-labeled *DA-LppK58A-strep tag. For labeling, octanoic-7,7,8,8,8-[d5] acid (CDN Isotopes) was added to LB solid media at 100 μg/ml. After overnight incubation of strain TXM1111 at 37 °C, colonies were collected from the agar surface by scraping and washed with PBS (PBS, pH 7.4, 5.6 mm Na2HPO4, 1.06 mm KH2PO4, 154 mm NaCl). Labeled DA-LppK58A-strep tag was purified from the resulting biomass using affinity chromatography as described previously (11), except additional purification was performed to isolate the DA-LppK58A-strep tag from any phospholipid contaminants. After purification using Strep-Tactin®XT column (Iba Life Sciences), protein was further purified using a 1-ml HiTrap Q HP anion exchange column (GE Healthcare) in buffer containing 100 mm Tris-HCl, pH 7.4, 50 mm NaCl, and 0.5 mm n-dodecyl-β-d-maltoside (DDM). The Q column flow through containing DA-LppK58A-strep tag was then loaded onto Superdex 10/300 sizing column (GE Healthcare) equilibrated with 10 mm Hepes, pH 7.4, and 0.5 mm DDM buffer. Fractions containing purified protein were combined and flash frozen. Labeled DA-LppK58A-strep tag was characterized by MALDI-TOF MS and lipase treatment (described below). Purified protein was determined to contain two [d5]-labeled acyl chains on the α-amino terminus (referred to as [d5]-DA-LppK58A-strep tag). Unlabeled DA-LppK58A-strep tag preparations were likewise isolated from E. coli TXM1111 but grown in LB agar containing unlabeled sodium octanoate (Sigma).
Growth assay with fatty acids
Single colonies of strains WT BW25113, TXM1018, TXM1029, and TXM1111 were pregrown in LB media. For TXM1111, pregrowth media contained sodium octanoate. Once cultures had reached exponential growth, cells were washed once with LB, inoculated into fresh LB to an OD600 of 0.0001 (∼5 × 104 colony forming units/ml), and supplemented with either 100 μg/ml of sodium palmitate (Sigma) or sodium octanoate. Cultures were grown at 37 °C for 19 h before measuring the final OD600.
Isolation of inner membranes for enzyme reconstitution assays
As a source of Lnt, Lit, and Lnt/Lit, inner membranes of KA893, KA801, and KA736, respectively, were harvested as previously described, with some modifications (11). One liter of cells were grown to an OD600 of 1.0, harvested by centrifugation, washed once with PBS, and stored frozen until use. Cell pellets were resuspended in ∼35 ml of 50 mm Tris, pH 7.8 (adjusted with HCl), containing 1 mm EDTA and disrupted with 3 passes through a French pressure cell at 14,000 p.s.i. Phenylmethylsulfonyl fluoride (PMSF) was then added to 1 mm and the lysates centrifuged at 3,200 × g for 10 min at 4 °C to remove unbroken cells. Total membranes were collected by ultracentrifugation at 110,000 × g for 90 min at 4 °C, then homogenized in 3 ml of 10 mm Hepes, pH 7.4 (adjusted with NaOH), with a 26-gauge needle. The membrane suspension was layered on top of a discontinuous sucrose gradient as described previously and centrifuged at 80,000 × g for 19 h at 4 °C. The top volume of sucrose was discarded and the inner membrane layer (∼4-6 ml) transferred to a new ultracentrifuge tube and then brought up to ∼40 ml total with Hepes buffer. The solution was centrifuged again at 110,000 × g for 90 min at 4 °C. The final inner membrane pellet was resuspended in Hepes buffer supplemented with 10% glycerol and the protein concentration normalized using Bradford regent. Aliquots were stored frozen at −80 °C.
Small scale reaction with unlabeled DA-LppK58A-strep tag
The reaction buffer, composed of 50 mm Tris, pH 7.2, 150 mm NaCl, and 0.1% Triton X-100, was optimized previously by Hillmann et al. (45) in activity assays with purified Lnt enzyme. To the reaction, inner membrane fractions containing Lnt, Lit, or Lnt/Lit were added to a final concentration of 0.75 mg/ml of total protein and sonicated for 1 min in a water sonicator bath. Purified DA-LppK58A was then added to 0.004 mg/ml and mixed by further sonication before incubation at 37 °C. At the indicated time intervals, 10-μl samples were removed and the reaction halted with SDS-PAGE loading buffer and flash frozen. Samples were separated by SDS-PAGE over a 16.5% Tris-Tricine gel and immunoblotted for LppK58A-strep tag using Precision Protein StrepTactin-horseradish peroxidase (strep-HRP) conjugate (Bio-Rad) as described previously (11).
Large scale lit acylation reaction and product purification
To 500 μl in the aforementioned reaction buffer supplemented with 1 mm PMSF and 0.75 mg/ml of total inner membrane protein fractions containing Lnt or Lit, 3 μg total of purified deuterium-labeled [d5]-DA-LppK58A-strep tag substrate was added. Reactions were incubated at 37 °C overnight with constant shaking and intermittent sonication (∼30 s in sonicator bath). MagStrep “type 3” XT beads (IBA Life Sciences) were used to purify LppK58A from the reaction following the manufacturer's recommendations with minor modifications. The reaction was first clarified by centrifugation at 4,000 × g for 2 min, then combined with an excess of 3 μl of magnetic beads. The reaction was inverted for 1.5 h at 4 °C and the beads washed as recommended. LppK58A-strep tag was eluted from the beads under denaturing conditions by boiling in 25 μl of SDS loading buffer for 10 min. Samples were taken from the start and end points of the initial reaction, the pelleted fraction, the unbound fraction, and the final elution fraction and visualized by Western blotting to assess for recovery, with an estimated ∼40% recovery of total strep-tagged protein in the eluent. The entire elution fraction was loaded into a single lane on an SDS-PAGE gel for immunoblotting and further analysis by MS (see below).
Lipoprotein lipase treatment [d5]-DA-LppK58A-strep tag
Lyophilized LPL from Pseudomonas sp. (Sigma) was reconstituted in water to 1 mg/ml, then added to a final concentration of 80 ng/μl in a 15-μl reaction with 1.8 μg total of [d5]-DA-LppK58A-strep tag. The reaction was incubated at 37 °C for 19 h and halted by the addition of SDS loading buffer and boiling for 10 min. The reaction volume was loaded into a single lane on an SDS-PAGE gel for immunoblotting and further analysis by MS (see below).
MALDI-TOF MS analysis
For MS analysis of lipoprotein substrates, preparations were transferred from a 16.5% Tris-Tricine SDS-PAGE gel to a nitrocellulose membrane. The band corresponding to Lpp was excised, digested in a trypsin solution overnight, then sequentially washed with 0.1% TFA, 10% acetonitrile, and finally 20% acetonitrile before eluting the N-terminal lipopeptides in 10 mg/ml of α-cyano-4-hydroxycinnamic acid in 2:1 chloroform-methanol, as described previously (11, 40). MS and MS/MS spectra were acquired on an Ultraflextreme (Bruker Daltonics) MALDI-TOF mass spectrometer in positive anion mode.
Alanine scanning mutagenesis and Mut-seq analysis of E. faecalis lit
The lit gene was amplified from E. faecalis genomic DNA (primers TM1990 and SS1314) and cloned into pET22b(+) using the NdeI/XhoI restriction sites. This resulting plasmid (pET22b(+)-Eflit) was then used as template for inverse PCR using primer pairs that introduced an alanine codon (GCG) at each position. Primers were 25 bp long, with 6 bp on the 5'-side and 16 bp on the 3'-side retaining homology to DNA flanking the targeted codon. The forward and reverse primers shared 15 bp of complimentary homology on the 5' ends to facilitate assembly by re-circularization. Separate PCR (212 total for each Lit amino acid encoding triplet) were set up in 96-well microplates with pET22b(+)-Eflit as template (1 ng/20-μl reaction). After PCR, 5-μl aliquots were resolved on 1% Tris acetate-EDTA (TAE)-agarose gels in sets of 12. Each set of PCR products was pooled according to relative individual intensities to normalize respective concentrations and gel purified as a batch. Purified PCR product sets were circularized using In-Fusion HD cloning (Takara Bio) and transformed into E. coli Stellar chemically competent cells (Takara Bio). Bacterial transformant lawns for each set were collected by scraping, plasmid pools isolated by mini-prep, and all sets of plasmid DNA were then mixed together in equal quantities to generate the final plasmid library to balance representation of each Ala mutant construct. Aliquots were then transformed into the conditional arabinose-inducible lnt-depletion strain E. coli KA349 (PBAD-lnt) and maintained on 0.2% arabinose for growth. Transformant lawns were scraped, pooled, and resuspended in LB, 15% glycerol for storage at −80 °C.
Library aliquots were pre-grown in LB, 0.2% arabinose at 37 °C to return to exponential growth, washed three times with LB to remove inducer, and an aliquot set aside for plasmid isolation (input library). Fresh cultures were then inoculated (OD600 of 0.0001 (∼5 × 104 colony forming units/ml)) into pre-warmed LB media without arabinose and outgrown for a total of 24 h, with samples being rediluted every 8 h for a total of three rounds of outgrowth. Plasmids were isolated from each round, and used as template for limited PCR amplification (primers TM1924 and TM1925, 15 cycles) of the lit encoding region. Amplicons were sequenced by next generation sequencing (142-bp post trim read length with 15,000- to 20,000-fold coverage depth at each triplet) and aligned to the reference WT lit sequence. For each triplet targeted for conversion to Ala (GCG), representation within the library was calculated by summing the number of G, C, and G reads at positions one, two, and three of the in-frame triplet, respectively. This sum was then divided by the average coverage at the corresponding three base positions. To correct for synonymous changes involving bases held in common with GCG, this raw value was adjusted by dividing by 3 for HDH, by 2 for GDH/HCH/HDG, and 1 for HCG/GDG/GCH targeted codons to generate GCG enrichment values for each in-frame triplet in the library. Depletion ratios were then calculated by dividing the enrichment values for each Lit codon in the input library by the lnt-depleted library after three passages.
Phenotypic confirmation of select lit Ala mutants
Select mutants were re-made by inverse PCR and transformed into KA349 using the method described above for the Ala library. Transformants were grown to mid-logarithmic growth in LB supplemented with 0.2% arabinose, washed, and serially diluted in PBS before spot tittering on LB agar plates containing either 0.2% glucose or 0.2% arabinose to compare complementation efficiency. Plates were incubated overnight at 37 °C before imaging. To measure Lit protein expression levels, the corresponding pET22b(+)-Eflit plasmids were converted into C-terminal epitope pET22b(+)-Eflit strep-tagged variants by inverse PCR with primers encoding for the strep tag (TM1987 and TM1988). The expression level of pET22b(+)-Eflit strep Ala mutants was measured by immunoblotting (StrepMAB-Classic HRP conjugate, IBA Life Sciences) using total protein extracts obtained from exponentially growing cells at identical optical densities.
His-89 and His-157 randomization and Mut-seq analysis
Primers targeting His-89 (TM2182 and TM2183) and His-157 (TM2184 and TM2185) were designed to introduce the degenerate NNK/MNN triplet and create a library of 32 unique triplets, encompassing 31 amino acids and a stop codon. All 20 amino acids are represented by at least one codon. Plasmids construction, selection post lnt-depletion, and next generation sequencing was performed as described above for the alanine scan library. Reads from each passage were aligned to the reference WT lit sequence (10,000- to 25,000-fold coverage depth at His-89 and His-157 to generate at minimal 100 reads per randomized codon). Representation of each NNK codon was calculated as a percentage of the entire library in the input and passaged libraries.
LacZ and PhoA lit fusion activity assays
A series of pET22b(+)-based constructs were made by fusing lit to either β-gal (lacZ) or alkaline phosphatase (phoA) reporters lacking their endogenous N-terminal signal peptides (from amino acid +13 for PhoA and +9 for LacZ) (26). Fusions were made after each predicted TM pass in every loop of Lit. Constructs encoding native lacZ and phoA genes were used as positive controls. For colorimetric activity assays on solid media, the indicated strains were struck on LB agar containing 100 μg/ml of carbenicillin and 1 mm isopropyl β-d-1-thiogalactopyranoside. To assay for LacZ activity, media also contained 40 μg/ml of 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-gal), and for PhoA activity, 40 μg/ml of 5-bromo-4-chloro-3'-indoly phosphate (BCIP).
SCAM for lit topology mapping
Native cysteine residues were replaced by three rounds of inverse PCR using the corresponding alanine scan library primers to construct pET22b(+)-Eflit strep C16A, C100A, C187A plasmid. The cysteine-less lit gene was amplified by PCR (primers TM2149 and TM2150) and cloned into the XbaI/AscI sites of pLI50-Ppen for low level constitutive expression. Inverse PCR using the corresponding primers (Table S1) was used to reintroduce Cys in targeted positions. SCAM experiments were conducted as described (55, 56), except probe labeling was conducted with 2.5 mm thiol probe NEM (Acros Organics), MTSES, (Biotium), or AMS (Invitrogen) for 20 min before quenching with addition of 100 mm cysteine. Cells were then washed three times with PBS before proceeding to lysis and labeling with 5 mm mal-PEG (average molecular mass 5,000 Da, Sigma). Lysed protein extracts were resolved on a 12% SDS-PAGE Tris glycine gel, transferred to nitrocellulose membranes, and immunoblotted with StrepMAB-Classic HRP conjugate (IBA Life Sciences).
Data availability
All data described herein are contained within this manuscript.
Supplementary Material
Acknowledgments
We thank Tania Laremore (Proteomics and Mass Spectrometry Core Facility, Penn State University) for technical assistance.
This article contains supporting information.
Author contributions—K. M. A., G. K., and T. C. M. conceptualization; K. M. A., G. K., and T. C. M. formal analysis; K. M. A., G. K., and T. C. M. investigation; K. M. A., G. K., and T. C. M. methodology; K. M. A., G. K., and T. C. M. writing-original draft; K. M. A., G. K., and T. C. M. writing-review and editing; T. C. M. resources; T. C. M. supervision; T. C. M. funding acquisition; T. C. M. project administration.
Funding and additional information—This work was supported by the National Institutes of Health Grant NIGMS R01GM127482 (to T. C. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest.
J. Gardiner, 4th, G. Komazin, M. Matsuo, K. Cole, F. Gotz, and T. C. Meredith, unpublished data.
- Lgt
- lipoprotein diacylglycerol transferase
- Lsp
- lipoprotein signal peptidase II
- Lit
- lipoprotein intramolecular transferase
- ABC
- ATP-binding cassette
- ACP
- acyl carrier protein
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- BCIP
- 5-bromo-4-chloro-3'-indoly phosphate
- CoA
- coenzyme A
- DA-LP
- diacylated lipoprotein
- DDM
- n-dodecyl-β-d-maltoside
- HRP
- horseradish peroxidase
- LacZ
- β-galactosidase
- LB
- lysogeny broth-Miller medium
- LPL
- lipoprotein lipase
- Lyso-LP
- lyso-form lipoprotein
- mal-PEG
- methoxypolyethylene glycol maleimide
- MTSES
- 2-sulfonatoethyl methanethiosulfonate
- NEM
- N-ethylmaleimide
- PhoA
- alkaline phosphatase
- PMSF
- phenylmethylsulfonyl fluoride
- SCAM
- substituted cysteine accessibility method
- TAE
- Tris acetate-EDTA
- TA-LP
- triacylated lipoprotein
- TM
- transmembrane
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-galactopyranoside
- mACP
- malonyl-ACP
- FAS
- fatty acid synthesis
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Lpp
- lipoprotein.
References
- 1. Braun V., and Hantke K. (2019) Lipoproteins: structure, function, biosynthesis. Subcell. Biochem. 92, 39–77 10.1007/978-3-030-18768-2_3 [DOI] [PubMed] [Google Scholar]
- 2. Zuckert W. R. (2014) Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim. Biophys. Acta 1843, 1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Narita S., and Tokuda H. (2010) Biogenesis and membrane targeting of lipoproteins. EcoSal Plus 4, 10.1128/ecosalplus.4.3.7 10.1128/ecosalplus.4.3.7 [DOI] [PubMed] [Google Scholar]
- 4. Nguyen M. T., and Gotz F. (2016) Lipoproteins of Gram-positive bacteria: key players in the immune response and virulence. Microbiol. Mol. Biol. Rev. 80, 891–903 10.1128/MMBR.00028-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buddelmeijer N. (2015) The molecular mechanism of bacterial lipoprotein modification: how, when and why?. FEMS Microbiol. Rev. 39, 246–261 10.1093/femsre/fuu006 [DOI] [PubMed] [Google Scholar]
- 6. Narita S. I., and Tokuda H. (2017) Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim. Biophys. Acta 1862, 1414–1423 10.1016/j.bbalip.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 7. Sankaran K., and Wu H. C. (1994) Lipid modification of bacterial prolipoprotein: transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706 [PubMed] [Google Scholar]
- 8. Hussain M., Ichihara S., and Mizushima S. (1982) Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257, 5177–5182 [PubMed] [Google Scholar]
- 9. Gupta S. D., and Wu H. C. (1991) Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli. FEMS Microbiol. Lett. 62, 37–41 10.1111/j.1574-6968.1991.tb04413.x [DOI] [PubMed] [Google Scholar]
- 10. Nakayama H., Kurokawa K., and Lee B. L. (2012) Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J. 279, 4247–4268 10.1111/febs.12041 [DOI] [PubMed] [Google Scholar]
- 11. Armbruster K. M., and Meredith T. C. (2017) Identification of the lyso-form N-acyl intramolecular transferase in low-GC firmicutes. J. Bacteriol. 199, e00099 10.1128/JB.00099-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fiore E., Van Tyne D., and Gilmore M. S. (2019) Pathogenicity of enterococci. Microbiol. Spectr. 7, GPP3-0053-2018 10.1128/microbiolspec.GPP3-0053-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oliveira-Nascimento L., Massari P., and Wetzler L. M. (2012) The role of TLR2 in infection and immunity. Front. Immunol. 3, 79 10.3389/fimmu.2012.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Armbruster K. M., Komazin G., and Meredith T. C. (2019) Copper-induced expression of a transmissible lipoprotein intramolecular transacylase alters lipoprotein acylation and the Toll-like receptor 2 response to Listeria monocytogenes. J. Bacteriol. 201, e00195 10.1128/JB.00195-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaturvedi K. S., and Henderson J. P. (2014) Pathogenic adaptations to host-derived antibacterial copper. Front. Cell. Infect. Microbiol 4, 3 10.3389/fcimb.2014.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dalecki A. G., Crawford C. L., and Wolschendorf F. (2017) Copper and antibiotics: discovery, modes of action, and opportunities for medicinal applications. Adv. Microb. Physiol. 70, 193–260 10.1016/bs.ampbs.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 17. Meeske A. J., Riley E. P., Robins W. P., Uehara T., Mekalanos J. J., Kahne D., Walker S., Kruse A. C., Bernhardt T. G., and Rudner D. Z. (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 10.1038/nature19331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robins W. P., Faruque S. M., and Mekalanos J. J. (2013) Coupling mutagenesis and parallel deep sequencing to probe essential residues in a genome or gene. Proc. Natl. Acad. Sci. U.S.A. 110, E848–857 10.1073/pnas.1222538110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buddelmeijer N., and Young R. (2010) The essential Escherichia coli apolipoprotein N-acyltransferase (Lnt) exists as an extracytoplasmic thioester acyl-enzyme intermediate. Biochemistry 49, 341–346 10.1021/bi9020346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gelis-Jeanvoine S., Lory S., Oberto J., and Buddelmeijer N. (2015) Residues located on membrane-embedded flexible loops are essential for the second step of the apolipoprotein N-acyltransferase reaction. Mol. Microbiol. 95, 692–705 10.1111/mmi.12897 [DOI] [PubMed] [Google Scholar]
- 21. Wiktor M., Weichert D., Howe N., Huang C. Y., Olieric V., Boland C., Bailey J., Vogeley L., Stansfeld P. J., Buddelmeijer N., Wang M., and Caffrey M. (2017) Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis. Nat. Commun. 8, 15952 10.1038/ncomms15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiseman B., and Hogbom M. (2020) Conformational changes in apolipoprotein N-acyltransferase (Lnt). Sci. Rep. 10, 639 10.1038/s41598-020-57419-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Narita S., and Tokuda H. (2011) Overexpression of LolCDE allows deletion of the Escherichia coli gene encoding apolipoprotein N-acyltransferase. J. Bacteriol. 193, 4832–4840 10.1128/JB.05013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yakushi T., Tajima T., Matsuyama S., and Tokuda H. (1997) Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J. Bacteriol. 179, 2857–2862 10.1128/jb.179.9.2857-2862.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grabowicz M., and Silhavy T. J. (2017) Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 114, 4769–4774 10.1073/pnas.1702248114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robichon C., Vidal-Ingigliardi D., and Pugsley A. P. (2005) Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J. Biol. Chem. 280, 974–983 10.1074/jbc.M411059200 [DOI] [PubMed] [Google Scholar]
- 27. Yakushi T., Masuda K., Narita S., Matsuyama S., and Tokuda H. (2000) A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218 10.1038/35008635 [DOI] [PubMed] [Google Scholar]
- 28. Klein K., Steinberg R., Fiethen B., and Overath P. (1971) Fatty acid degradation in Escherichia coli: an inducible system for the uptake of fatty acids and further characterization of old mutants. Eur. J. Biochem. 19, 442–450 10.1111/j.1432-1033.1971.tb01334.x [DOI] [PubMed] [Google Scholar]
- 29. Nunn W. D., Giffin K., Clark D., and Cronan J. E. Jr. (1983) Role for fadR in unsaturated fatty acid biosynthesis in Escherichia coli. J. Bacteriol. 154, 554–560 10.1128/JB.154.2.554-560.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weeks G., Shapiro M., Burns R. O., and Wakil S. J. (1969) Control of fatty acid metabolism. I. Induction of the enzymes of fatty acid oxidation in Escherichia coli. J. Bacteriol. 97, 827–836 10.1128/JB.97.2.827-836.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DiRusso C. C., Metzger A. K., and Heimert T. L. (1993) Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Mol. Microbiol. 7, 311–322 10.1111/j.1365-2958.1993.tb01122.x [DOI] [PubMed] [Google Scholar]
- 32. Black P. N. (1991) Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J. Bacteriol. 173, 435–442 10.1128/jb.173.2.435-442.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Black P. N., DiRusso C. C., Metzger A. K., and Heimert T. L. (1992) Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J. Biol. Chem. 267, 25513–25520 [PubMed] [Google Scholar]
- 34. Ford T. J., and Way J. C. (2015) Enhancement of E. coli acyl-CoA synthetase FadD activity on medium chain fatty acids. Peer J. 3, e1040 10.7717/peerj.1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campbell J. W., and Cronan J. E. Jr. (2002) The enigmatic Escherichia coli fadE gene is yafH. J. Bacteriol. 184, 3759–3764 10.1128/jb.184.13.3759-3764.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan Y., Leeds J. A., and Meredith T. C. (2012) Pseudomonas aeruginosa directly shunts β-oxidation degradation intermediates into de novo fatty acid biosynthesis. J. Bacteriol. 194, 5185–5196 10.1128/JB.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y. M., and Rock C. O. (2008) Thematic review series: glycerolipids: acyltransferases in bacterial glycerophospholipid synthesis. J. Lipid Res. 49, 1867–1874 10.1194/jlr.R800005-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao Z., Davis R. M., Kishony R., Kahne D., and Ruiz N. (2012) Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, E2561–2568 10.1073/pnas.1209742109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan Y., Sachdeva M., Leeds J. A., and Meredith T. C. (2012) Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by the FabY class of β-ketoacyl acyl carrier protein synthases. J. Bacteriol. 194, 5171–5184 10.1128/JB.00792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Armbruster K. M., and Meredith T. C. (2018) Enrichment of bacterial lipoproteins and preparation of N-terminal lipopeptides for structural determination by mass spectrometry. J. Vis. Exp. 135, 56842 10.3791/56842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kurokawa K., Ryu K. H., Ichikawa R., Masuda A., Kim M. S., Lee H., Chae J. H., Shimizu T., Saitoh T., Kuwano K., Akira S., Dohmae N., Nakayama H., and Lee B. L. (2012) Novel bacterial lipoprotein structures conserved in low-GC content Gram-positive bacteria are recognized by Toll-like receptor 2. J. Biol. Chem. 287, 13170–13181 10.1074/jbc.M111.292235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rock C. O., Goelz S. E., and Cronan J. E. Jr. (1981) Phospholipid synthesis in Escherichia coli: characteristics of fatty acid transfer from acyl-acyl carrier protein to sn-glycerol 3-phosphate. J. Biol. Chem. 256, 736–742 [PubMed] [Google Scholar]
- 43. Cronan J. E. Jr., and Subrahmanyam S. (1998) FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 29, 937–943 10.1046/j.1365-2958.1998.00917.x [DOI] [PubMed] [Google Scholar]
- 44. Asanuma M., Kurokawa K., Ichikawa R., Ryu K. H., Chae J. H., Dohmae N., Lee B. L., and Nakayama H. (2011) Structural evidence of α-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J. 278, 716–728 10.1111/j.1742-4658.2010.07990.x [DOI] [PubMed] [Google Scholar]
- 45. Hillmann F., Argentini M., and Buddelmeijer N. (2011) Kinetics and phospholipid specificity of apolipoprotein N-acyltransferase. J. Biol. Chem. 286, 27936–27946 10.1074/jbc.M111.243519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jackowski S., and Rock C. O. (1986) Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J. Biol. Chem. 261, 11328–11333 [PubMed] [Google Scholar]
- 47. Vidal-Ingigliardi D., Lewenza S., and Buddelmeijer N. (2007) Identification of essential residues in apolipoprotein N-acyl transferase, a member of the CN hydrolase family. J. Bacteriol. 189, 4456–4464 10.1128/JB.00099-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ridder A., Skupjen P., Unterreitmeier S., and Langosch D. (2005) Tryptophan supports interaction of transmembrane helices. J. Mol. Biol. 354, 894–902 10.1016/j.jmb.2005.09.084 [DOI] [PubMed] [Google Scholar]
- 49. Sal-Man N., Gerber D., Bloch I., and Shai Y. (2007) Specificity in transmembrane helix-helix interactions mediated by aromatic residues. J. Biol. Chem. 282, 19753–19761 10.1074/jbc.M610368200 [DOI] [PubMed] [Google Scholar]
- 50. Unterreitmeier S., Fuchs A., Schäffler T., Heym R. G., Frishman D., and Langosch D. (2007) Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J. Mol. Biol. 374, 705–718 10.1016/j.jmb.2007.09.056 [DOI] [PubMed] [Google Scholar]
- 51. Adamian L., and Liang J. (2001) Helix-helix packing and interfacial pairwise interactions of residues in membrane proteins. J. Mol. Biol. 311, 891–907 10.1006/jmbi.2001.4908 [DOI] [PubMed] [Google Scholar]
- 52. Manoil C. (1991) Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Methods Cell Biol. 34, 61–75 10.1016/s0091-679x(08)61676-3 [DOI] [PubMed] [Google Scholar]
- 53. Drew D., Sjöstrand D., Nilsson J., Urbig T., Chin C. N., de Gier J. W., and von Heijne G. (2002) Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. U.S.A. 99, 2690–2695 10.1073/pnas.052018199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bogdanov M., Zhang W., Xie J., and Dowhan W. (2005) Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods 36, 148–171 10.1016/j.ymeth.2004.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Entova S., Billod J. M., Swiecicki J. M., Martin-Santamaría S., and Imperiali B. (2018) Insights into the key determinants of membrane protein topology enable the identification of new monotopic folds. Elife 7, e40889 10.7554/eLife.40889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ray L. C., Das D., Entova S., Lukose V., Lynch A. J., Imperiali B., and Allen K. N. (2018) Membrane association of monotopic phosphoglycosyl transferase underpins function. Nat. Chem. Biol. 14, 538–541 10.1038/s41589-018-0054-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rottig A., and Steinbuchel A. (2013) Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev. 77, 277–321 10.1128/MMBR.00010-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fukuda A., Matsuyama S., Hara T., Nakayama J., Nagasawa H., and Tokuda H. (2002) Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 277, 43512–43518 10.1074/jbc.M206816200 [DOI] [PubMed] [Google Scholar]
- 59. Milo R. (2013) What is the total number of protein molecules per cell volume? a call to rethink some published values. Bioessays 35, 1050–1055 10.1002/bies.201300066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Braun V. (1975) Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415, 335–377 10.1016/0304-4157(75)90013-1 [DOI] [PubMed] [Google Scholar]
- 61. Guo M. S., Updegrove T. B., Gogol E. B., Shabalina S. A., Gross C. A., and Storz G. (2014) MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634 10.1101/gad.243485.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lang M. M., Ingham S. C., and Ingham B. H. (2001) Differentiation of Enterococcus spp. by cell membrane fatty acid methyl ester profiling, biotyping and ribotyping. Lett. Appl. Microbiol. 33, 65–70 10.1046/j.1472-765x.2001.00949.x [DOI] [PubMed] [Google Scholar]
- 63. Rashid R., Cazenave-Gassiot A., Gao I. H., Nair Z. J., Kumar J. K., Gao L., Kline K. A., and Wenk M. R. (2017) Comprehensive analysis of phospholipids and glycolipids in the opportunistic pathogen Enterococcus faecalis. PLoS ONE 12, e0175886 10.1371/journal.pone.0175886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Raines L. J., Moss C. W., Farshtchi D., and Pittman B. (1968) Fatty acids of Listeria monocytogenes. J. Bacteriol. 96, 2175–2177 10.1128/JB.96.6.2175-2177.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ulmschneider M. B., and Sansom M. S. (2001) Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta 1512, 1–14 10.1016/S0005-2736(01)00299-1 [DOI] [PubMed] [Google Scholar]
- 66. Weltzien H. U. (1979) Cytolytic and membrane-perturbing properties of lysophosphatidylcholine. Biochim. Biophys. Acta 559, 259–287 10.1016/0304-4157(79)90004-2 [DOI] [PubMed] [Google Scholar]
- 67. Homma H., Nishijima M., Kobayashi T., Okuyama H., and Nojima S. (1981) Incorporation and metabolism of 2-acyl lysophospholipids by Escherichia coli. Biochim. Biophys. Acta 663, 1–13 10.1016/0005-2760(81)90189-2 [DOI] [PubMed] [Google Scholar]
- 68. Lin Y., Bogdanov M., Lu S., Guan Z., Margolin W., Weiss J., and Zheng L. (2018) The phospholipid-repair system LplT/Aas in Gram-negative bacteria protects the bacterial membrane envelope from host phospholipase A2 attack. J. Biol. Chem. 293, 3386–3398 10.1074/jbc.RA117.001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rock C. O. (1984) Turnover of fatty acids in the 1-position of phosphatidylethanolamine in Escherichia coli. J. Biol. Chem. 259, 6188–6194 [PubMed] [Google Scholar]
- 70. Zheng L., Lin Y., Lu S., Zhang J., and Bogdanov M. (2017) Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Biochim. Biophys. Acta 1862, 1404–1413 10.1016/j.bbalip.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Silva-Rocha R., Martinez-García E., Calles B., Chavarría M., Arce-Rodríguez A., de Las Heras A., Paez-Espino A. D., Durante-Rodriguez G., Kim J., Nikel P. I., Platero R., and de Lorenzo V. (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–675 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Swoboda J. G., Meredith T. C., Campbell J., Brown S., Suzuki T., Bollenbach T., Malhowski A. J., Kishony R., Gilmore M. S., and Walker S. (2009) Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem. Biol. 4, 875–883 10.1021/cb900151k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dobson L., Remenyi I., and Tusnady G. E. (2015) CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 43, W408–412 10.1093/nar/gkv451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cserzö M., Wallin E., Simon I., von Heijne G., and Elofsson A. (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10, 673–676 10.1093/protein/10.6.673 [DOI] [PubMed] [Google Scholar]
- 76. Viklund H., and Elofsson A. (2008) OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24, 1662–1668 10.1093/bioinformatics/btn221 [DOI] [PubMed] [Google Scholar]
- 77. Käll L., Krogh A., and Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 78. Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., Honigschmid P., Schafferhans A., Roos M., Bernhofer M., Richter L., Ashkenazy H., Punta M., Schlessinger A., Bromberg Y., et al. (2014) PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, W337–343 10.1093/nar/gku366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Juretic D., Zoranic L., and Zucic D. (2002) Basic charge clusters and predictions of membrane protein topology. J. Chem. Inf. Comput. Sci. 42, 620–632 [DOI] [PubMed] [Google Scholar]
- 80. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 81. Hofmann K., and Stoffel W. (1993) TMbase: a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374, 166 [Google Scholar]
- 82. Tsirigos K. D., Peters C., Shu N., Kall L., and Elofsson A. (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–407 10.1093/nar/gkv485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described herein are contained within this manuscript.