Abstract
Over recent years several examples of randomly switching methyltransferases, associated with Type III restriction‐modification (R‐M) systems, have been described in pathogenic bacteria. In every case examined, changes in simple DNA sequence repeats result in variable methyltransferase expression and result in global changes in gene expression, and differentiation of the bacterial cell into distinct phenotypes. These epigenetic regulatory systems are called phasevarions, phase‐variable regulons, and are widespread in bacteria, with 17.4% of Type III R‐M system containing simple DNA sequence repeats. A distinct, recombination‐driven random switching system has also been described in Streptococci in Type I R‐M systems that also regulate gene expression. Here, we interrogate the most extensive and well‐curated database of R‐M systems, REBASE, by searching for all possible simple DNA sequence repeats in the hsdRMS genes that encode Type I R‐M systems. We report that 7.9% of hsdS, 2% of hsdM, and of 4.3% of hsdR genes contain simple sequence repeats that are capable of mediating phase variation. Phase variation of both hsdM and hsdS genes will lead to differential methyltransferase expression or specificity, and thereby the potential to control phasevarions. These data suggest that in addition to well characterized phasevarions controlled by Type III mod genes, and the previously described Streptococcal Type I R‐M systems that switch via recombination, approximately 10% of all Type I R‐M systems surveyed herein have independently evolved the ability to randomly switch expression via simple DNA sequence repeats.
Keywords: bacterial pathogenesis, epigenetics, phase variation, phasevarion, R‐M systems
Abbreviations
- Mod
modification enzyme/methyltransferase
- Res
restriction endonuclease
- R‐M
restriction‐modification system
- SSR
simple sequence repeat
- TRD
target recognition domain
1. INTRODUCTION
Phase variation is the random, high‐frequency reversible switching of gene expression.1 The most common mechanism mediating phase variation of gene expression is slipped‐strand mispairing that occurs in simple sequence repeats (SSRs).1 Rates of phase variation mediated by SSRs are often several orders of magnitude greater than the base mutation rate.1 Many host‐adapted bacterial pathogens contain phase‐variable genes, and these often encode surface associated virulence factors that are subjected to periodic selection, such as iron acquisition systems,2, 3 pili,4 adhesins,5, 6 and lipooligosaccharide biosynthetic genes.7, 8 Several bacterial pathogens also contain mod genes, encoding cytoplasmic Type III DNA methyltransferases, that exhibit phase‐variable expression. We recently characterized the distribution of phase‐variable Type III mod genes in the REBASE database of R‐M systems, and demonstrated that 17.4% of all Type III mod genes contain SSRs.9 ON‐OFF switching of genes encoding Type III methyltransferase expression leads to differential regulation of multiple genes in systems known as phasevarions (phase‐variable regulon; Srikhanta et al 2005). Phasevarions controlled by switching of Type III mod genes have been well‐characterized in a number of host‐adapted bacterial pathogens, such as Haemophilus influenzae,10, 11 Neisseria spp.,12 Helicobacter pylori,13 Moraxella catarrhalis,14, 15 and Kingella kingae 16 (recently reviewed in (17)). Type I R‐M systems comprise three genes, encoding restriction (HsdR), modification (a methyltransferase; HsdM), and target sequence specificity (HsdS) proteins18 (see Figure 1A). The HsdS protein dictates the sequences cleaved and methylated by the HsdR and HsdM subunits, respectively. A functional Type I restriction enzyme consists of the pentamer R2M2S. A trimer made up of M2S is a functional methyltransferase.18 Each HsdS specificity protein is typically made up of two “half” target recognition domains (TRDs; TRD 1 in the 5′ end of the gene, or the 5′ TRD; and TRD 2 in the 3′ end of the gene, or the 3′ TRD); each TRD contributes half to the overall specificity. Therefore, changing a single TRD coding region changes the specificity of the encoded HsdS protein. Variation in hsdS coding sequence has previously been well studied as the basis of differing methyltransferase specificity in Type I R‐M systems,19 and homologous recombination between variable hsdS coding sequences has been shown to generate novel methyltransferase specificities in bacteria.20, 21, 22, 23, 24 A phasevarion controlled by a Type I R‐M system in the major human pathogen Streptococcus pneumoniae has been characterized by a number of groups.25, 26 A phase‐variable Type I R‐M system has also been well characterized in Mycoplasma pulmonis,27, 28 but the effect on gene expression has not been studied. Rather than the ON‐OFF switching seen with Type III R‐M mod genes containing SSRs, the systems described in S. pneumoniae and M. pulmonis shuffle between variable hsdS specificity genes resulting in the expression of multiple different HsdS specificity proteins. This type of phase‐variable Type I R‐M systems have been termed “inverting” Type I systems,29 as the expressed HsdS subunit undergoes phase variation through DNA inversions. Type I R‐M systems that phase vary via changes in the length of SSRs are less well studied. To date, only one hsdM gene and one hsdS gene have been shown to phase vary via SSR tract length changes, both in human adapted pathogens: an hsdM in nontypeable H. influenzae (NTHi), and an hsdS in Neisseria gonorrhoeae. An hsdM gene containing a pentanucleotide SSR tract was identified in H. influenza,30 and we previously noted changes in the length of this tract in multiple nontypeable H. influenzae (NTHi) isolates from paired samples taken from the human nasopharynx and middle ear during cases of otitis media.7 An hsdS gene in N. gonorrhoeae, encoding the NgoAV Type I system,31 contains a poly G tract. Variation in the length of this poly G tract within the hsdS locus changes the reading frame downstream of the poly G tract, and results in either a full‐length or a truncated HsdS protein being produced. The full‐length and truncated HsdS proteins have differing methyltransferase specificities,31 with two of these truncated HsdS subunits combining to participate in sequence recognition32 (see Figure 1A). So rather than an ON‐OFF switch seen in Type III mod genes, variation in SSR length in hsdS genes always produces an active methyltransferase, but one that can switch between two specificities, potentially controlling two unique sets of genes through differential methylation. In order to determine whether Type I systems contain SSRs, and therefore potentially able to control phasevarions, we conducted a survey of all Type I hsdR, hsdM, and hsdS loci for SSRs in the REBASE database of restriction enzymes.33
Figure 1.
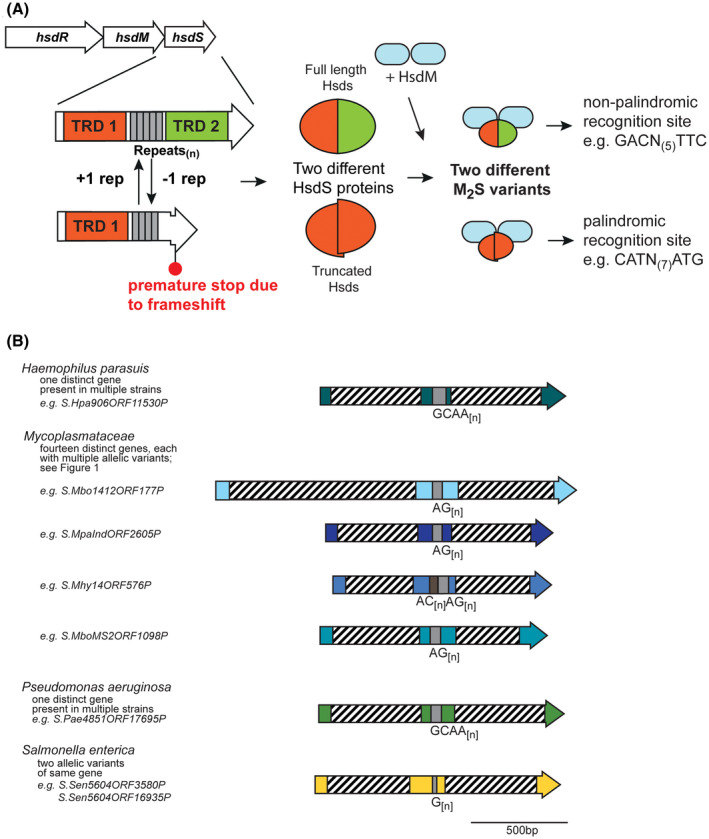
A, Illustration of how phase‐variable switching of hsdS genes occurs. Type I R‐M loci are made up of three genes, encoding a restriction enzyme (hsdR), a methyltransferase (hsdM), and a target sequence specificity protein (hsdS). Each hsdS gene is made up of two target recognition domains (TRDs; TRD 1 in red, and TRD 2 in green), with the SSR tract located between the two TRDs (gray boxes). Loss or gain of repeat units in the SSR tract results in a full‐length hsdS gene being expressed (TRD 1 + 2), which produces a full‐length HsdS protein encoded in a single polypeptide (red/green oval), or a frameshift mutation downstream of the SSR tract, premature transcriptional termination, and results in a truncated HsdS polypeptide (TRD 1 only; red half‐oval). These likely dimerise via the C‐terminal coiled coil region in each truncated HsdS subunit to form a functional HsdS protein. Following oligomerization with an HsdM dimer to form an active methyltransferase, the different HsdS protein subunits result in two different methyltransferase specificities. B, schematic representation of the location of TRD 1 and TRD 2, and the SSR tracts, in a selection of hsdS loci. Colored arrows represent different genes, with color representing homology within each gene if more than one example of this gene is present in REBASE. Hatched boxes represent the locations of the each TRD. The number of different hsdS genes is noted below each species. Unique examples are listed below each individual bacterial species where this hsdS gene is present
2. MATERIALS AND METHODS
2.1. REBASE survey and bioinformatics
We downloaded all genes annotated as Type I loci from REBASE33 (http://rebase.neb.com/rebase/rebase.seqs.html). This consisted of 23,558 hsdS genes, 20,849 hsdM genes, and 18,578 hsdR genes from REBASE on 5 May 2018. We searched for simple sequence repeats computationally with the following restrictions: a minimum of nine repeats for single‐base repeats (eg, AAAAAAAAA), five repeats for two‐base repeats (eg, AGAGAGAGAG), and three repeats for three or more‐base repeats (eg, AGCAGCAGC, AGCTAGCTAGCTAGCT). Cd‐hit,34 with a threshold of 80% nucleotide sequence identity, was used to cluster highly similar sequences together as representative examples. The list of all downloaded genes, all sequences containing one repeat tract, and the list of genes after selection criteria are applied can be found in Supplementary Data 1, 2, and 3 for all hsdS, hsdM, and hsdR, respectively. Phylogenetic analysis was carried out using the multiple sequence alignment program Muscle35 and analyzed by RAxML,36 with a bootstrap value at 1000. The list of unique and representative sequences can be found in Supplementary Data 1‐3 for hsdS, hsdM, and hsdR, respectively. Phylogenetic trees were produced using the data sets in Supplementary Data 1‐3, column E. All individual loci are presented in Supplementary Data 4‐6.
3. RESULTS
In order to identify all phase‐variable Type I hsdS, hsdM, and hsdR genes, we searched the well‐curated restriction enzyme database, REBASE,33 for SSR tracts of DNA. Our search for SSR tracts in the 23,558 hsdS genes, 20,849 hsdM genes, and 18,578 hsdR genes (Supplementary Data 1‐3, column B) showed there were 10,047 hsdS genes, 12,056 hsdM genes, and 13,945 hsdR genes containing at least one SSR tract (Supplementary Data 1‐3, column C). We strictly set our criteria to only select genes for analysis that contained repeat tracts of a length that have previously been shown to lead to high rates of phase variation of the gene containing them.37 For example, mononucleotide SSR tracts of nine bases in length have been shown to phase vary at rates of 1.8‐13.50 × 10−3 38; a tetranucleotide repeat tract consisting of just three repeat units in length phase varied at rates of 0.5‐2.0 × 10−6.39 We also excluded all genes that contained a repeat tract where the repeating unit was divisible by three, that is, those tracts made up of tri‐, hexa‐, and nonanucleotide repeats, as changes in the number of these repeats would not lead to phase‐variable switching of expression. We therefore only defined a Type I hsd gene as “phase‐variable” if the repeat tract in an open reading frame (ORF) was at least nine bases long for mononucleotide repeat tracts (eg, G[9]), five repeats long for dinucleotide repeats (eg, GA[5]), and three repeat units long for tetra‐, penta‐, hexa‐, and octanucleotide repeat tracts (eg, AGCC[3]). After applying our selection criteria for repeat tract length and removing all repeat tracts where the repeat unit is a multiple of 3, we are left with 1838 unique hsdS genes, 416 unique hsdM genes, and 808 unique hsdR genes (Supplementary Data 1‐3, column D). This represents 7.8% of all hsdS genes (1838/23558), 2% of all hsdM genes (416/20849), and 4.3% of all hsdR genes (808/18587).
By clustering genes with >80% identity together, to avoid multiple counting of overrepresented genes in the database, we demonstrate that there are 885 representative phase‐variable hsdS genes, 252 representative phase‐variable hsdM genes, and 314 representative phase‐variable hsdR genes currently annotated in REBASE (Supplementary Data 1‐3, column E). The list of all genes containing phase‐variable SSRs after selection criteria are applied, and all unique phase‐variable representative hsdS, hsdM and hsdR genes are presented in Supplementary Data 1‐3. Phylogenetic trees showing all representative hsdS, hsdM, and hsdR genes are presented in Supplementary Figures 1, 2, 3, respectively (all data from respective Supplementary Data, column E).
Figure 2.
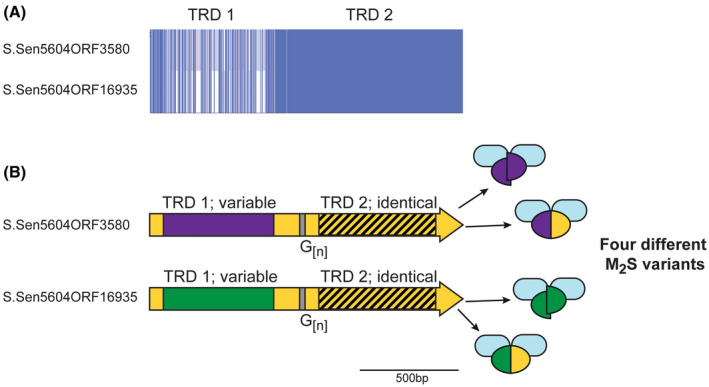
Illustration of the phase‐variable hsdS loci present in Salmonella enterica. Salmonella enterica subsp Enterica, serovar India SA20085604 contains two hsdS loci annotated as containing G[n] tracts—S.Sen5604ORF3580 and S.Sen5604ORF16935. A, Alignment of these two genes was generated using Muscle and viewed using JalView overview feature. B, The region encoding TRD 1 is variable in the two hsdS genes (purple or green boxes) but identical in the region encoding TRD 2 (hatched box, yellow background). The remaining sequence shows high (>95% nucleotide) identity. Variation in the length of the G[n] tracts could potentially result in four different HsdS proteins in a population, represented by different colored ovals (full‐length purple/yellow oval or two half purple ovals from S.Sen5604ORF3580; full‐length green/yellow oval or two half green ovals from S.Sen5604ORF16935), that would combine with an HsdM protein (blue oval) to produce four different M2S methyltransferase variants
Figure 3.
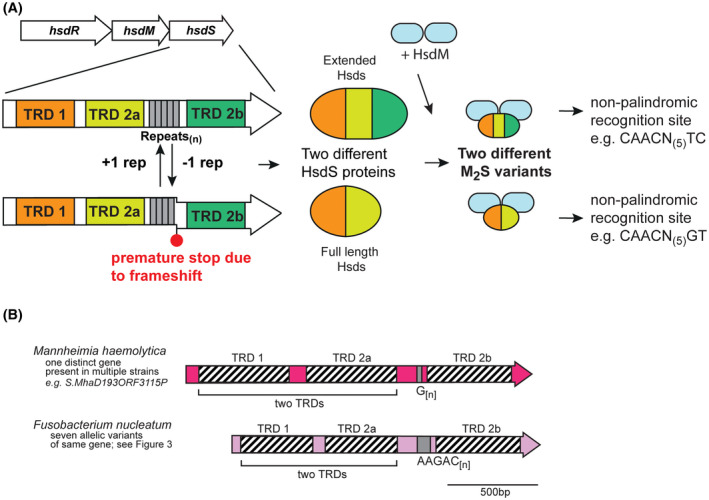
A, Illustration of how phase‐variable switching of extended hsdS genes occurs. Extended hsdS genes contain three separate target recognition domains (TRDs; TRD 1 in orange at the 5′ end, a central TRD 2a in light green, and TRD 2b at the 3′ end in dark green). The SSR tract (gray boxes) is located between TRD 2a and TRD 2b. A frameshift mutation through loss or gain of repeat units in the SSR tract results in TRD 2b being out of frame with the rest of the gene, and expression of a full‐length HsdS protein consisting of TRD 1 + TRD 2a, analogous to that in Figure 1. However, if the SSR tract length results in read‐through to TRD 2b, a protein made up of all three TRDs (TRD 1 + 2a + 2b) is expressed. Following oligomerization with an HsdM dimer to form an active methyltransferase, the different HsdS protein subunits result in two different methyltransferase specificities. B, schematic representation of extended hsdS loci in Mannheimia haemolytica and Fusobacterium nucleatum. Colored arrows represent different genes, with color representing homology within each gene if more than one example of this gene is present in REBASE. Hatched boxes represent the locations of each TRD. The number of different hsdS genes is noted below each species. Unique examples are listed below each individual bacterial species where this hsdS gene is present
3.1. Phase variation of hsdS is more prevalent than hsdM
All previous work with phase‐variable genes in bacteria, including phase‐variable Type III methyltransferases, shows that variation in the length of loci‐encoded SSRs leads to reversible ON‐OFF switching of gene expression.2, 3, 4, 7, 8, 9, 10, 17 However, the sole example of an hsdS gene containing SSRs and showing phase‐variable expression, that of the NgoAV system in N gonorrhoeae strain FA1090,31 demonstrated that rather than ON‐OFF switching, variation in the length of the SSR contained in the encoding hsdS gene lead to production of a full‐length or a truncated HsdS specificity subunit, which resulted in different methyltransferase specificities.31 We observed almost 4 times as many hsdS genes than hsdM containing SSRs (7.8% vs 2%).
3.2. Many bacterial species contain a phase‐variable hsdS gene
Our phylogenetic analysis of the 885 unique representative hsdS genes reveals that a diverse array of species contain an hsdS gene containing a potentially phase‐variable SSR tract (Supplementary Figure 1). As with previous work investigating SSR tracts in Type III mod genes, we observe that phase‐variable hsdS genes are present in both pathogenic and nonpathogenic organisms, including a diverse array in environmental organisms and opportunistic pathogens. For example, we observe a switching between a full‐length HsdS protein and a truncated HsdS protein in an hsdS gene containing a tetranucleotide GCAA[n] repeat in the porcine pathogen Haemophilus parasuis (Figure 1B). The hsdS gene is conserved in all examples containing SSRs, with the SSR tract ranging in length from GCAA[7‐29], which is highly indicative of a phase‐variable mode of expression. No methyltransferase specificity is reported for this HsdS protein in REBASE, but we predict switching between two different specificities dependent on expression of a full‐length HsdS (TRD 1 + TRD 2) or a truncated HsdS (TRD 1 only) due to the number of GCAA[n] repeats present (Figure 1A).
We observe a large group of hsdS genes containing AG[n] SSR tracts in the Mycoplasmataceae (examples in Figure 1B). Our analysis demonstrates that at least 14 different hsdS genes with AG[n] repeat tracts are present in the Mycoplasmatacae (Supplementary Figure [Link], [Link]). Some strains contain R‐M systems with multiple hsdS genes containing AG[n] repeat tracts, for example, Mycoplasma bovirhinis strain HAZ141_2 encodes the adjacent genes S.Mbo1412ORF128 (MBVR141_0128) and S.Mbo1412ORF129 (MBVR141_0129) that both contain AG[n] repeat tracts of eight units in length. Mycoplasma dispar strain GS01 contains a ~9‐kb region (genes CSW10_02740‐CSW10_02780) encoding nine adjacent hsdS genes, each containing a variable number of AG[n] repeats. The resulting diversity of methyltransferase specificities resulting from these multiple, phase‐variable hsdS genes certainly merits further investigation.
We identify two examples of SSR tract containing hsdS genes in a single strain of the human gastric pathogen Salmonella enterica subsp Enterica, serovar India SA20085604 (Figure 1B). These two genes (LFZ16_03585 encoding S.Sen5604ORF3580 and LFZ16_16930 encoding S.Sen5604ORF16935) are not located near each other in the genome. Both these hsdS genes contain mononucleotide G[12] tracts. Our sequence analysis (Figure 2) of these two hsdS genes shows they contain different TRD 1 sequences, but identical TRD 2 sequences, meaning variation in length of the G[12] tract will produce different truncated HsdS proteins (variable TRD 1 only) and different full‐length HsdS proteins (Figure 2), as also illustrated for other examples described above (Figure 1A). Different hsdS genes (by varying TRD 1) means the encoded HsdS proteins will recognize and methylate different sequences, and therefore potentially regulate different phasevarions. Analysis of the hsdM and hsdR genes of these two operons show they are 98% and 100% identical, respectively, and have likely arisen by duplication of the entire Type I system, followed by acquisition of a new TRD 2 in one of the systems.
3.3. The opportunistic human pathogen Pseudomonas aeruginosa contains a phase‐variable hsdS gene that appears to cluster with drug‐resistant strains
Our analysis of hsdS genes containing SSRs revealed a single representative example of a phase‐variable hsdS in the opportunistic human pathogen Pseudomonas aeruginosa. P. aeruginosa is a significant nosocomial pathogen, can cause a range of skin and soft tissue infections, and is a major cause of lung infections in individuals with cystic fibrosis.40 Examination of all sequences in REBASE revealed that a phase‐variable hsdS gene is present in a number of different strains of P. aeruginosa. The individual examples deposited in REBASE contain a GCAA[n] SSR tract of between 3 and 16 repeats. Alignment of these hsdS regions showed they are identical apart from the length of the GCAA[n] repeat tract, indicating the encoded full‐length HsdS proteins will methylate the same sequences, and the resulting truncated HsdS protein will methylate the same sequences, although the full‐length and truncated versions will have different specificities. BLAST analysis for homologues in publically available databases showed that many strains containing this phase‐variable hsdS gene appeared to be either antibiotic resistant,41, 42 or were clinical isolates.43 It is intriguing that a phase‐variable hsdS may be associated with these phenotypes, and further work is required to investigate this hypothesis.
3.4. Mannheimia haemolytica and Fusobacterium nucleatum contain extended phase‐variable hsdS genes
We observed a mononucleotide G[n] tract containing hsdS gene in the bovine pathogen Mannheimia haemolytica. Our analysis of this species shows that this hsdS gene shows high‐sequence conservation in all examples, and therefore encodes a methyltransferase with the same specificity. Our analysis of this system shows that the “S1” TRD annotated in REBASE actually encodes two separate TRDs (Figure 3; TRD 1 and TRD 2a) that would encode a full‐length HsdS protein. Therefore, phase variation of the G[n] tract would actually lead to a full‐length HsdS containing two TRDs (1 + 2a), and a larger “extended” HsdS containing three separate TRDs (1 + 2a + 2b; illustration in Figure 3A). A G[12] tract leads to expression of this extended HsdS protein (TRDs 1 + 2a + 2b; eg, S.MhaD193ORF3115), whereas G[10] and G[11] length tracts leads to an hsdS gene containing just TRD 1 + 2a (eg, TRD 1 + 2a encoding S1.Mha807AORF10585 and TRD 2b encoding S2.Mha807AORF10585, with the S2‐annotated gene containig G[11]). The methyltransferase specificity of the HsdS proteins has been solved; the HsdS protein containing TRDs 1 and 2a methylates the sequence 5′‐CAACN(4)GT, whereas the extended HsdS protein (1 + 2a + 2b) methylates the sequence 5′‐CAACN(5)TC. Therefore, it appears that 5′‐CAAC is the motif recognized by TRD 1, with TRD 2a and TRD 2b likely recognizing 5′‐GT and 5′‐TC, respectively. It is intriguing to speculate that TRD 2b somehow replaces TRD 2a in the final folded protein when the three TRD HsdS protein is expressed, but this would require substantial experimental confirmation. Therefore, production of two distinct methyltransferase activities due to phase variation of a G[n] tract results in expression of a two‐TRD or three‐TRD HsdS protein, each with a different specificity.
A similar system exists in Fusobacterium nucleatum (Figure 3B). F. nucleatum is a Gram‐negative human oral bacterium44 that is involved in the pathogenesis of periodontal disease, and is implicated in preterm births44, 45 and cancer.46, 47 Our examination of hsdS genes containing SSR tracts showed that a diverse group of hsdS genes exists in F. nucleatum, all containing a AAGAC[n] repeat tract (Figure 3B and Figure 4). Individual examples contained between 5 and 12 AAGAC repeats, with 12 repeats giving an extended HsdS protein containing three TRDs (TRDs 1 + 2a + 2b), and 10 or 11 repeats producing a full‐sized HsdS (TRD 1 + TRD 2a) only. The AAGAC[n] repeat tract is always located between TRD 2a and TRD 2b, hence when the number of AAGAC[n] repeats is inframe with TRD 2b, an extended HsdS protein is produced containing three separate TRDs. Sequence analysis of all the systems of this extended HsdS gene found in F. nucleatum (Figure 4) shows there are nine different TRD 1 variants, eight separate TRD 2a variants, and two different TRD 2b variants. The extent of the combinations of these three TRDs in larger sequence databases will reveal how much differential methylation exists in F. nucleatum.
Figure 4.
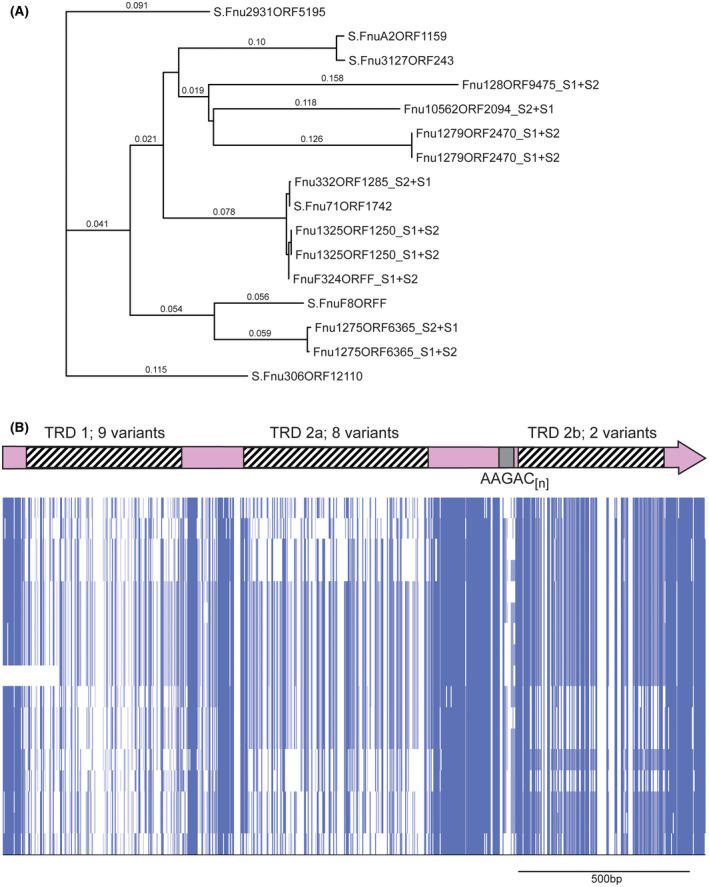
hsdS genes containing AAGAC[n] tracts in Fusobacterium nucleatum. A, a phylogenetic tree was produced by aligning sequences using Muscle, and phylogeny analyzed by RAxML. Where the individual gene is annotated with an “S” prefix, this gene contains an AAGAC[n] repeat tract length where the S1 and S2 regions are in frame, encode the extended HsdS polypeptide, and annotated as a full‐length hsdS gene in REBASE. Where the annotation contains an “S1 + S2” suffix, the AAGAC[n] repeat tract length means that the TRD at the 3′ end of the gene (annotated as TRD 2b here) is out of frame with the 5′ end of the gene encoding TRD 1 and TRD 2a, and annotated as two separate truncated hsdS genes (S1 made up of TRD 1 + TRD 2a, and S2 made up of TRD 2b) in REBASE; B, alignments of the entire hsdS region present in REBASE showing this variation is due to the presence of multiple allelic variants of each of the three TRDs. Sequences were aligned in Muscle, and viewed using JalView overview feature
Both these examples of phase‐variable “extended” hsdS genes shows that an extra method of generating methyltransferase variation appears to have evolved. Phase variation of the described hsdS genes in M. haemolytica and F. nucleatum results in a full‐length HsdS protein, or an extended three TRD containing HsdS protein. Like the situation described in Figure 1, where phase variation of hsdS genes leads to two different methyltransferase specificities, phase variation of these extended hsdS genes also results in two different methyltransferase specificities, but encoded in a single HsdS subunit (expression of extended HsdS protein), rather than dimerization of a two “half” HsdS proteins (truncated HsdS protein).
3.5. Host‐adapted bacterial pathogens contain uncharacterized phase‐variable hsdM genes
Applying the same methodology to search for SSRs in hsdM genes, we observe a number of examples of phase‐variable Type I HsdM methyltransferases in host‐adapted bacterial pathogens. For example, multiple strains of the bovine pathogen Mannheimia haemolytica encode an hsdM gene which contains an ACAGC repeat tract (ACAGC[10‐40]). The human respiratory pathogen H. influenzae encodes an hsdM gene which contains a CGAGA repeat tract (CGAGA[3‐10]). The specificity of this methyltransferase has been solved using PacBio SMRT sequencing: multiple strains of M. haemolytica contain the motif (GACN(5)TTC) (link to entry in REBASE http://rebase.neb.com/cgi-bin/seqget?M.Mha183VI) or (GAAN(5)GTC) (link to entry in REBASE http://rebase.neb.com/cgi-bin/seqget?M.Mha186V) assigned to this HsdM protein, which is the same sequence, just in the opposite orientation; the locus M.Hin375IV in H. influenzae strain 375 has been shown to methylate GGYAN(6)TGA (link to entry in REBASE http://rebase.neb.com/cgi-bin/seqget?M.Hin375IV).
These examples are particularly interesting as both organisms also contain phase‐variable Type III mod genes,9, 10, 48 with M. haemolytica also containing a phase‐variable hsdS gene that contains a mononucleotide G[n] tract (eg, S.MhaD193ORF3115), as described above. The G[n] tract containing hsdS gene in M. haemolytica identified in the hsdS analysis section above is part of a different Type I R‐M system to the system containing a phase‐variable hsdM gene described in this section.
3.6. hsdS and hsdM genes contain SSR tracts in different regions of their coding sequence
Our analysis of hsdM genes that contain SSRs showed that the location of most SSRs is in the 5′ region of the coding sequence, whereas hsdS genes containing SSRs always contain the SSR tract between the two TRD coding sequences, that is, in the middle of the coding sequence. We predict that variation in length of SSRs located in hsdM genes likely leads to ON‐OFF switching, akin to that seen with Type III mod genes9, 17 (Figure 5). Conversely, variation in the length of SSR tracts in hsdS genes leads to full‐length hsdS gene expression, or truncated hsdS gene expression if the SSR tract length leads to a frameshift and premature transcriptional termination. For example, SSR tracts in hsdS genes invariably occur between two TRDs, which leads to expression of multiple HsdS variants, be they full length two TRD proteins, truncated HsdS subunits which then dimerise to form a functional HsdS protein, or an extended three TRD containing HsdS protein (Figure 1 and Figure 3). SSR variation has been demonstrated to result in a switch between specificities due to expression of a full‐length HsdS or truncated HsdS has been demonstrated in N. gonorrhoeae,31 and full and extended versions of HsdS proteins are expressed by M. haemolytica commensurate with SSR tract variation (this work). Thus, phase variation of hsdS genes has evolved to result in expression of multiple variable methyltransferases, whereas hsdM loci switch ON or OFF, akin to that seen with Type III mod genes9 (Figure 5).
Figure 5.
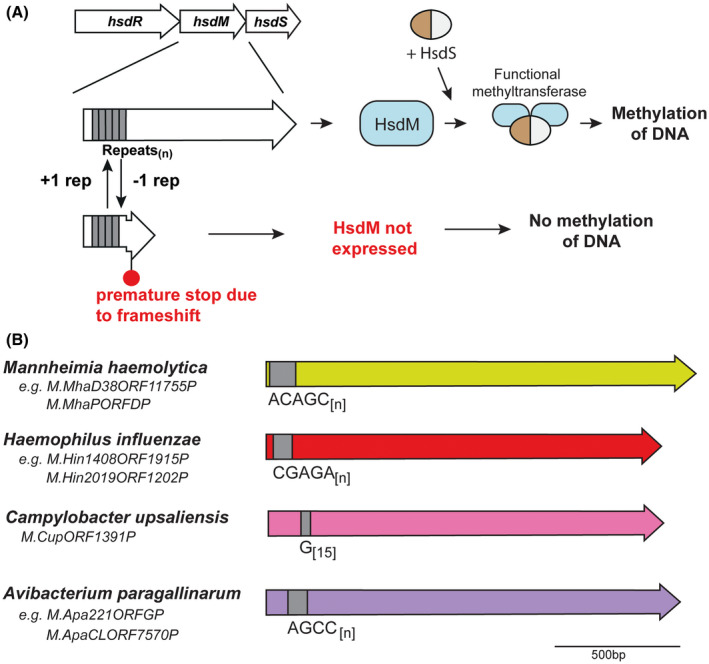
A, Illustration of how phase‐variable switching of hsdM genes occurs. Variation in the length of the SSR tract located in the hsdM ORF results in biphasic ON‐OFF switching of the hsdM gene, which results in expression of a functional HsdM protein and ensuing methyltransferase activity dependent on the HsdS subunit present, or no methyltransferase activity as the hsdM gene is nonfunctional due to a frameshift and premature transcriptional termination, with no resulting methyltransferase activity; B, schematic representation of hsdM loci containing SSR tracts. Colored arrows represent different genes, with color representing homology within each gene if more than one example of this gene is present in REBASE. Unique examples are listed below each individual bacterial species where this hsdM gene is present
3.7. Many bacteria contain an hsdR gene containing short SSR tracts, or SSRs that may not lead to phase‐variable expression
We obtained a total of 808 unique hsdR genes containing a repeat tract that is capable of phase variation, representing 4.3% of all hsdR genes in REBASE. These 808 examples clustered (>80% identity) into 314 representative examples (Supplementary Figure 3; Supplementary Data 3). As with the hsdS and hsdM genes containing phase‐variable repeat tracts, these hsdR genes are widespread in the bacterial domain, comprising environmental and pathogenic organisms. Many Helicobacter spp. (poly C tracts) and Mycoplasmataceae (dinucleotide tracts) are represented, which is not surprising given the prevalence of these types of repeats in these organisms,49 with many R‐M systems in Mycoplasmas previously identified as containing SSRs.50 Many environmental organisms contain hsdR genes with short SSR tracts; for example, our analysis describes hsdR genes containing SSRs in diverse environmental bacterial species such as NinC115ORF23709 from Nitratireductor indicus, a deep sea bacterium isolated from the Indian Ocean51 and MosTT16ORF11850 from Moraxella osloensis, a symbiont of nematode worm Phasmarhabditis hermaphrodita.52 We also identify a number of phase‐variable hsdR genes in unusual or emerging pathogens such as EleORF768 from Eggerthella lenta, an anaerobic Gram‐positive bacilli associated with abdominal sepsis, and Slu143ORF9045 from Staphylococcus lugdunensis, a member of the Staphylococci that has been known to cause septic arthritis.53 However, the majority of the SSR tracts found in hsdR genes were very short (four or fewer repeat units), likely leading to vary low rates of phase variation. In addition, in examples where a single gene is present in multiple different strains, there is no variation in repeat number observed between all the examples present. Taking all hsdR genes as an example, out of 14 702 hsdR genes containing at least one SSR tract, 13 822 of these (94.0%) contain an SSR where the repeat unit is divisible by three (n/3), that is, tri‐, hexa‐, and nonanucleotide repeat tracts. These repeat tracts are potentially variable, but their variation would likely not lead to ON‐OFF switching or variable expression, as no frameshift would occur with variation in SSR tract length. However, the resulting amino acid repeats they encode may have important functional implications, as seen with other amino acid repeat containing proteins such as the RTX family of toxins produced by Gram‐negative bacteria,54 Leucine Rich Repeat (LRP) proteins,55, 56 and HsdS proteins.57, 58 Loss or addition of units in these repeat tracts where the repeating unit is divisible by three would not lead to phase‐variable expression,49 with less selective pressure likely exerted against expansion of tracts of this length as they do not lead to frameshifts and loss of expression.59 However, our findings demonstrate a high abundance of trinucleotide repeat tracts in open reading frames. Similar figures exist for hsdM (12 347 hsdM genes contain at least one repeat tract, 11 929 contain a n/3 repeat tract = 96.6%; 11 383 hsdS genes contain at least one repeat tract, 9517 contain an n/3 repeat tract = 83.6%)
4. DISCUSSION
This is the first time, to our knowledge, that a systematic study has been carried out to identify Type I R‐M systems that contain SSRs capable of mediating phase‐variable expression, and thereby have the potential to control phasevarions (phase‐variable regulons). Every case where a Type III mod gene has been identified containing varying SSR tract lengths has subsequently been shown to control a phasevarion.10, 11, 12, 13, 14, 15, 60 Our analysis demonstrates a number of phase‐variable hsdS genes present in species where phase variation via SSRs has never before been observed. For example, we identify hsdS genes containing SSRs in the human pathogens F. nucleatum and P. aeruginosa. The example in P. aeruginosa is particularly novel, as this species is a major opportunistic pathogen, with particularly significant disease burdens in immunocompromized and cystic fibrosis sufferers.61 The presence of a phasevarion in P. aeruginosa may complicate vaccine development against this organism, via providing a mechanim for altered expression of surface antigens, as is common on other species containing phasevarions.17 Therefore, further investigation of the role of the phase‐variable methyltransferase in P. aeruginosa certainly merits further investigation.
We also identified hsdS and hsdM genes that are apparently phase‐variable in strains with already identified phase‐variable Type III mod genes (eg, M haemolytica, NTHi) in the same genome. Organisms containing multiple phase‐variable Type III mod genes are well characterized, such as Neisseria meningitidis, which can encode modA, modB, and modD,12, 62 and H. pylori, encoding modH,13 and recently identified modJ and modL,9 but this is the first time strains have been identified that encode SSR‐containing Type I and Type III methyltransferases. If the Type I methyltransferases identified in this study are also phase‐variably expressed, in addition to the already characterized phase‐variable Type III methyltransferases in these organisms (H. influenzae and M. haemolytica), this would add a further level of complexity to the study of gene regulation in these species. For example, it is possible that some strains of M. haemolytica could contain up to three independently switching methyltransferases that control phasevarions, as this species contains separate Type I hsdM and hsdS loci that contain SSRs (this study), and a Type III mod gene that contains SSRs.9, 63 The SSR tracts in these two hsdM genes have both been identified previously,7, 30, 63 and their identification in this survey validates our search methodology.
As well as identifying new phase‐variable Type I R‐M systems in a range of bacterial pathogens, our phylogenetic analysis demonstrates that many commensal and environmental bacterial species contain potentially phase‐variable hsd genes. In these cases, examples are often limited to one or two strains of these bacterial species. This could imply that there is less selective pressure to generate phenotypic diversity in these organisms as they exist in a more predicatable environment and use the conventional “sense and respond” gene regulation paradigm of adaptability, that is, these organisms contain many more two‐component sensor‐regulator pairs than small genome pathogens that contain multiphase‐variable methyltransferases.49, 64 It remains to be comprehensively demonstrated that the hsdM and/or hsdS genes that contain SSRs control phasevaions, and how such plasticity of gene expression would be advantageous in a changing environment that cannot be dealt with via coventional “sense and respond” gene regulation strategies. Increased phenotypic diversity is an obvious advantage of phasevarions, particularly in small genome, host‐adapted pathogens; perhaps the increased variablity generated by phasevarions provides a number of additional advantages during adaptation to changing environmental conditions that these organisms may encounter, or that cannot be sensed by conventional means.1
One obvious advantage to methyltransferase variation is resistance to phage. R‐M systems are generally thought of as primitive bacterial immune systems, that protect bacteria from foreign‐incoming DNA, typically from bacteriophage infection.65 Variation in methyltransferase specificity would protect against “escape” phage whose DNA has been methylated at a different sequence to that recognized by the phase‐variable R‐M system expressed by the incoming bacterial cell. The presence of phase‐variable methyltransferases in diverse environmental bacteria, as well as bacterial pathogens, could be explained by increased resistance to phage in the strains containing these systems. For example, in addition to controlling phasevarions, phase variation of the SpnD39III system in S. pneumoniae has been shown to alter resistance to phage.66 Variation in methyltransferases has also be shown to protect against incoming foreign DNA/phage in numerous diverse bacterial species such as Lactococcus lactis 24 Enterococcus faecium 67 Mycoplasma pulmonis 28 and H. influenzae.30 Whether the systems described in this study have evolved to provide resistance to phage, regulate phasevarions, or both, remains to be elucidated.
One limitation of our study, and one that will be particularly interesting to investigate, is that phase variation of an SSR tract between two TRD encoding regions will alter the length of the spacing between the two TRDs. The region between the two TRDs also contains a coiled‐coil domain,68 which in the examples described above occurs immediately before the SSR tract. This coiled‐coil region contains an amino acid TAEL(n) repeat tract.68 It has previously been well described that variation in length of the coiled coil domain itself leads to two different methyltransferase specificities. For example, the Type I R‐M systems EcoR124I and EcoR124II vary in just one TAEL repeat in the coiled‐coil domain separating the two TRDs, which leads to two different methyltransferase specificities—GAAN(6)RTCG (EcoR124I) and GAAN(7)RTCG (EcoR124II).57, 58 We hypothesize that variation in the length of an SSR tract immediately adjacent to the encoded amino acid TAEL repeat would have a similar effect on spacing and therefore specificity, but this would require significant additional analysis and further biochemical analysis to prove this, and is perhaps beyond the scope of this particular study. An analysis of SSR tracts that are divisible by three would add considerable weight to this extra analysis.
Another interesting finding of our study is the discovery of phase‐variable “extended” HsdS proteins that contain three separate TRDs. As noted in our results section, it is tempting to speculate that overall specificity of these three TRD extended HsdS proteins is the result of competition between the centrally encoded TRD (TRD 2a) and the TRD encoded at the 3′ end of the gene (TRD 2b). The sequence and structure of HsdS proteins has been demonstrated to generate HsdS variablility,69, 70 and it would be a worth while follow up study to determine the extent of these “extended” hsdS genes in the bacterial domain, and to perform studies to determine the exact mechanism that results in the multiple different specificities determined for these HsdS proteins.
Our estimates of the prevalence of phase‐variable Type I loci is likely to be an underestimate for a number of reasons. For example, our strict selection criteria likely excludes many examples of genes with short mono‐ and dinucleotide repeat tracts (repeat tracts of less than nine nucleotides long for mononucleotide repeat tracts and less than five repeat units long for dinucleotide repeat tracts); mononucleotide repeat tracts have been shown to phase vary at rates of at least 0.65 × 10−3 in Campylobacter jejuni 38 and a repeat tract of G[7] leads to phase variation of the pptA gene of N meningitidis at a rate of 1 × 10−2.71 Our analysis of only full‐length annotated genes likely leads us to miss many hsdM and hsdR genes that contain SSRs that are phase varied OFF, and which lead to genes being annotated as out‐of‐frame. However, this problem likely does not occur for hsdS genes as they are annotated as functional, albeit truncated, HsdS specificity proteins in REBASE despite containing an SSR where read‐through is prevented to the 3′‐TRD. The use of short read next‐generation sequencing (NGS) technologies that rely on mapping short (<200 bp) reads to reference genomes, and assembling areas where SSRs are present often leads to an underestimation of the repeat tract length due to collapsing the tract down by the assembly software, or alignment to multiple places in the genome72 as assembly software cannot distinguish between sequences.73 This can be particularly problematic in bacterial genomes as the same repeating element may be present in multiple different genes,1 and we have previously discussed these issues at length.9 As such, these sequences need to be confirmed by methods that can accurately discern the sequence of SSR tracts, such as PacBio SMRT long‐read sequencing technology.74, 75
It is interesting to note that nearly all hsdR genes containing a di‐ to nonanucleotide SSR tract that we defined as phase‐variable are very short. For instance, all examples containing tetra‐ and pentanucleotide repeat tracts contain just three repeat units (eg, AATT[3]; CCGGA[3]), and only 6 of 4490 unique examples where the repeat is a dinucleotide (eg, GC[n]) have more than five repeat units. This suggests that while these repeat tracts are theoretically able to expand and contract leading to phase‐variable expression, the lack of examples of variable repeat number in homologues in different strains of the species containing these hsdR genes suggests that these genes do in fact not phase vary, as theoretically, there would be no advantage to gene regulation and adaptability commensurate with switching expression of a restriction enzyme.
It is intriguing that a higher proportion Type III mod genes have evolved to control phasevarions (17.8%;9) than Type I R‐M systems (9.8%; 7.8% of hsdS genes and 2% of hsdM genes; this study). However, when the average length of the DNA sequence recognized by the TRD of these systems is considered, this disparity is perhaps not surprising: Type III Mod methyltransferases recognize and methylate 4/5bp nonpalindromic target sequences (eg, CCGAA; CGAG,76 whereas Type I HsdS proteins need at least a 6‐bp sequence correctly separated for successful recognition (eg, CATN(7/8)ATG68). Therefore, methylation of a 4/5‐bp sequence by a Type III Mod would occur at much higher frequency in the genome than at a >6‐bp sequence by a Type I HsdM2S, which would considerably increase the chances of advantageous epigenetic regulation events occurring with phase‐variable expression of Type III mod genes than Type I hsdMS genes. However, the exact evolutionary selection pressures that have occurred to favor SSR expansion in Type III mod genes over Type I hsdMS genes remain to be explored.
In summary, we identify that almost 7.8% of hsdS genes and almost 2% of hsdM genes (ie, ~10% of all Type I R‐M systems) contain a phase‐variable SSR tract, and consequently are likely to control a phasevarion. A broad array of bacterial species encode SSR‐containing hsd genes, ranging from bacterial pathogens already identified to contain a phase‐variable methyltransferase, to newly identified loci in important opportunistic pathogens and environmental organisms. It appears that phase‐variable methyltransferases have evolved in two separate types of R‐M system on multiple occassions, and that this method of generating phenotypic plasticity is a common contingency strategy that is widely distributed throughout the bacterial domain. Our current study has identified many new bacterial species that contain phasevarions, including many well‐studied bacterial pathogens. The extra level of biological and genetic diversity imparted by phasevarions is clearly a widely utilized mechanism. The identification of the potential for phase‐variable expression of these methyltransferases, and by implication, the ability to control phasevarions, will have a major impact on the study of bacterial virulence, the development of novel therapies and vaccines against the wide variety of important human and animal pathogens that use this epigenetic mechanism of pleiotropic phase variation.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
M. Jennings and J Atack designed research; J. Atack and M. Jennings analyzed data; all authors performed research; J. Atack, M. Jennings, and Y. Zhou wrote the paper; C. Guo performed the search of REBASE.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the Australian National Health and Medical Research Council (NHMRC) Program Grant 1071659 and Principal Research Fellowship 1138466 to MPJ; Project Grants 1099279 to JMA; and 1121629 to YZ; Australian Research Council (ARC) Discovery Projects 170104691 to MPJ, 180100976 to JMA, and 180102060 to YZ. Funding for open access charge: National Health and Medical Research Council, Australia.
Atack JM, Guo C, Yang L, Zhou Y, Jennings MP. DNA sequence repeats identify numerous Type I restriction‐modification systems that are potential epigenetic regulators controlling phase‐variable regulons; phasevarions. The FASEB Journal. 2020;34:1038–1051. 10.1096/fj.201901536RR
John M. Atack and Chengying Guo co‐first author.
REFERENCES
- 1. Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Ann Rev Genet. 2006;40:307‐333. [DOI] [PubMed] [Google Scholar]
- 2. Ren Z, Jin H, Whitby PW, Morton DJ, Stull TL. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin‐haptoglobin binding protein genes of Haemophilus influenzae . J Bacteriol. 1999;181:5865‐5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richardson AR, Stojiljkovic I. HmbR, a hemoglobin‐binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J Bacteriol. 1999;181:2067‐2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blyn LB, Braaten BA, Low DA. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 1990;9:4045‐4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atack JM, Winter LE, Jurcisek JA, Bakaletz LO, Barenkamp SJ, Jennings MP. Selection and counter‐selection of Hia expression reveals a key role for phase‐variable expression of this adhesin in infection caused by non‐typeable Haemophilus influenzae . J Infect Dis. 2015;212:645‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dawid S, Barenkamp SJ, St. Geme, JW . Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc Natl Acad Sci U S A. 1999;96:1077‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fox KL, Atack JM, Srikhanta YN, et al. Selection for phase variation of LOS biosynthetic genes frequently occurs in progression of non‐typeable Haemophilus influenzae infection from the nasopharynx to the middle ear of human patients. PLoS ONE. 2014;9:e90505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poole J, Foster E, Chaloner K, et al. Analysis of nontypeable Haemophilus influenzae phase variable genes during experimental human nasopharyngeal colonization. J Infect Dis. 2013;208:720‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atack JM, Yang Y, Seib KL, Zhou Y, Jennings MP. A survey of Type III restriction‐modification systems reveals numerous, novel epigenetic regulators controlling phase‐variable regulons; phasevarions. Nucleic Acids Res. 2018;46:3532–3542. 10.1093/nar/gky1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Atack JM, Srikhanta YN, Fox KL, et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non‐typeable Haemophilus influenzae . Nat Commun. 2015;6 10.1038/ncomms8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A. 2005;102:5547‐5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srikhanta YN, Dowideit SJ, Edwards JL, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria . PLoS Pathog. 2009;5:e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srikhanta YN, Gorrell RJ, Steen JA, et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori . PLoS ONE. 2011;6:e27569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blakeway LV, Power PM, Jen FE, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014;28:5197‐5207. [DOI] [PubMed] [Google Scholar]
- 15. Seib KL, Peak IR, Jennings MP. Phase variable restriction‐modification systems in Moraxella catarrhalis . FEMS Immunol Med Mic. 2002;32:159‐165. [DOI] [PubMed] [Google Scholar]
- 16. Srikhanta YN, Fung KY, Pollock GL, Bennett‐Wood V, Howden BP, Hartland EL. Phasevarion regulated virulence in the emerging paediatric pathogen Kingella kingae . Infect. Immun. 2017;85:e00319 ‐00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Atack JM, Tan A, Bakaletz LO, Jennings MP, Seib KL. Phasevarions of bacterial pathogens: methylomics sheds new light on old enemies. Trends Microbiol. 2018;26:715‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roberts RJ, Belfort M, Bestor T, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805‐1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gubler M, Braguglia D, Meyer J, Piekarowicz A, Bickle TA. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction‐modification enzymes. EMBO J. 1992;11:233‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gann AA, Campbell AJ, Collins JF, Coulson AF, Murray NE. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987;1:13‐22. [DOI] [PubMed] [Google Scholar]
- 21. Bullas LR, Colson C, Van Pel A. DNA restriction and modification systems in Salmonella. SQ, a new system derived by recombination between the SB system of Salmonella typhimurium and the SP system of Salmonella potsdam. J Gen Microbiol. 1976;95:166‐172. [DOI] [PubMed] [Google Scholar]
- 22. Fuller‐Pace FV, Bullas LR, Delius H, Murray NE. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci U S A. 1984;81:6095‐6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagaraja V, Shepherd JC, Bickle TA. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985;316:371‐372. [DOI] [PubMed] [Google Scholar]
- 24. O'Sullivan D, Twomey DP, Coffey A, Hill C, Fitzgerald GF, Ross RP. Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis . Mol Microbiol. 2000;36:866‐875. [DOI] [PubMed] [Google Scholar]
- 25. Manso AS, Chai MH, Atack JM, et al. A random six‐phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun. 2014;5 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oliver MB, Basu Roy A, Kumar R, Lefkowitz EJ, Swords WE. Streptococcus pneumoniae TIGR4 phase‐locked opacity variants differ in virulence phenotypes. mSphere. 2017;2 10.1128/mSphere.00386-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sitaraman R, Dybvig K. The hsd loci of Mycoplasma pulmonis: organization, rearrangements and expression of genes. Mol Microbiol. 1997;26:109‐120. [DOI] [PubMed] [Google Scholar]
- 28. Dybvig K, Sitaraman R, French CT. A family of phase‐variable restriction enzymes with differing specificities generated by high‐frequency gene rearrangements. Proc Natl Acad Sci. 1998;95:13923‐13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Ste Croix M, Vacca I, Kwun MJ, et al. Phase‐variable methylation and epigenetic regulation by type I restriction–modification systems. FEMS Microbiol Rev. 2017;41:S3‐S15. [DOI] [PubMed] [Google Scholar]
- 30. Zaleski P, Wojciechowski M, Piekarowicz A. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology. 2005;151:3361‐3369. [DOI] [PubMed] [Google Scholar]
- 31. Adamczyk‐Poplawska M, Lower M, Piekarowicz A. Deletion of one nucleotide within the homonucleotide tract present in the hsdS gene alters the DNA sequence specificity of Type I restriction‐modification system NgoAV. J Bacteriol. 2011;193:6750‐6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacWilliams MP, Bickle TA. Generation of new DNA binding specificity by truncation of the type IC EcoDXXI hsdS gene. EMBO J. 1996;15:4775‐4783. [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298‐D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li W, Godzik A. Cd‐hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658‐1659. [DOI] [PubMed] [Google Scholar]
- 35. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792‐1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics. 2014;30:1312‐1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox EC. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135‐156. [DOI] [PubMed] [Google Scholar]
- 38. Bayliss CD, Bidmos FA, Anjum A, et al. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res. 2012;40:5876‐5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farabaugh PJ, Schmeissner U, Hofer M, Miller JH. Genetic studies of the lac repressor. J Mol Biol. 1978;126:847‐863. [DOI] [PubMed] [Google Scholar]
- 40. Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159‐173. [DOI] [PubMed] [Google Scholar]
- 41. Boyle B, Fernandez L, Laroche J, et al. Complete genome sequences of three Pseudomonas aeruginosa isolates with phenotypes of polymyxin B adaptation and inducible resistance. J Bacteriol. 2012;194:529‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Belkum A, Soriaga LB, LaFave MC, et al. Phylogenetic distribution of CRISPR‐cas systems in antibiotic‐resistant Pseudomonas aeruginosa . mBio. 2015;6 10.1128/mBio.01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jeraldo P, Cunningham SA, Quest D, et al. Draft genome sequences of nine Pseudomonas aeruginosa strains, including eight clinical isolates. Genome Announcements. 2015;3:e01154 ‐01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han YW. Fusobacterium nucleatum: a commensal‐turned pathogen. Curr Opin Microbiol. 2015;23:141‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun. 2004;72:2272‐2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shang F‐M, Liu H‐L. Fusobacterium nucleatum and colorectal cancer: a review. World Journal of Gastrointestinal Oncology. 2018;10:71‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kostic A, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe. 2013;14:207‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fox KL, Srikhanta YN, Jennings MP. Phase variable type III restriction‐modification systems of host‐adapted bacterial pathogens. Mol Microbiol. 2007;65:1375‐1379. [DOI] [PubMed] [Google Scholar]
- 49. Mrazek J, Guo X, Shah A. Simple sequence repeats in prokaryotic genomes. Proc Natl Acad Sci U S A. 2007;104:8472‐8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brocchi M, Vasconcelos ATRd, Zaha A. Restriction‐modification systems in Mycoplasma spp. Genetics and Molecular Biology. 2007;30:236‐244. [Google Scholar]
- 51. Lai Q, Yu Z, Yuan J, Sun F, Shao Z. Nitratireductor indicus sp. nov., isolated from deep‐sea water. Int J Syst Evol Microbiol. 2011;61:295‐298. [DOI] [PubMed] [Google Scholar]
- 52. An R, Sreevatsan S, Grewal PS. Moraxella osloensis gene expression in the slug host Deroceras reticulatum . BMC Microbiol. 2008;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mei‐Dan O, Mann G, Steinbacher G, Ballester SJ, Cugat RB, Alvarez PD. Septic arthritis with Staphylococcus lugdunensis following arthroscopic ACL revision with BPTB allograft. Knee Surg Sports Traumatol Arthrosc. 2008;16:15‐18. [DOI] [PubMed] [Google Scholar]
- 54. Frey J. The role of RTX toxins in host specificity of animal pathogenic Pasteurellaceae . Vet Microbiol. 2011;153:51‐58. [DOI] [PubMed] [Google Scholar]
- 55. Hu Y, Huang H, Hui X, et al. Distribution and evolution of yersinia leucine‐rich repeat proteins. Infect Immun. 2016;84:2243‐2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loimaranta V, Hytönen J, Pulliainen AT, et al. Leucine‐rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J Biol Chem. 2009;284:18614‐18623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Price C, Shepherd JC, Bickle TA. DNA recognition by a new family of type I restriction enzymes: a unique relationship between two different DNA specificities. EMBO J. 1987;6:1493‐1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Price C, Lingner J, Bickle TA, Firman K, Glover SW. Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes. J Mol Biol. 1989;205:115‐125. [DOI] [PubMed] [Google Scholar]
- 59. Power PM, Sweetman WA, Gallacher NJ, et al. Simple sequence repeats in Haemophilus influenzae . Infect Genet Evol. 2009;9:216‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seib KL, Jen FE, Tan A, et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6‐adenine DNA methyltransferases of Neisseria meningitidis . Nucleic Acids Res. 2015;43:4150‐4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hoiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663‐1674. [DOI] [PubMed] [Google Scholar]
- 62. Seib KL, Pigozzi E, Muzzi A, et al. A novel epigenetic regulator associated with the hypervirulent Neisseria meningitidis clonal complex 41/44. FASEB J. 2011;25:3622‐3633. [DOI] [PubMed] [Google Scholar]
- 63. Highlander SK, Garza O. The restriction‐modification system of Pasteurella haemolytica is a member of a new family of type I enzymes. Gene. 1996;178:89‐96. [DOI] [PubMed] [Google Scholar]
- 64. Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol Rev. 2014;38:119‐141. [DOI] [PubMed] [Google Scholar]
- 65. Vasu K, Nagaraja V. Diverse functions of restriction‐modification systems in addition to cellular defense. Microbiol Mol Biol Rev. 2013;77:53‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Furi L, Crawford LA, Rangel‐Pineros G, Manso AS, De Ste Croix M, Haigh RD, et al. Methylation warfare: interaction of pneumococcal bacteriophages with their host. J Bacteriol. 2019;201:e00370-19 10.1128/JB.00370-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huo W, Adams HM, Trejo C, Badia R, Palmer KL. A Type I restriction‐modification system associated with Enterococcus faecium subspecies separation. Appl Environ Microbiol. 2019;85:e02174‐02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Loenen WAM, Dryden DTF, Raleigh EA, Wilson GG. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42:20‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cowan GM, Gann AA, Murray NE. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989;56:103‐109. [DOI] [PubMed] [Google Scholar]
- 70. Gough JA, Murray NE, Brenner S. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1‐19. [DOI] [PubMed] [Google Scholar]
- 71. Jen FEC, Warren MJ, Schulz BL, et al. Dual pili post‐translational modifications synergize to mediate meningococcal adherence to platelet activating factor receptor on human airway cells. PLoS Pathog. 2013;9:e1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reinert K, Langmead B, Weese D, Evers DJ. Alignment of next‐generation sequencing reads. Annu Rev Genomics Hum Genet. 2015;16:133‐151. [DOI] [PubMed] [Google Scholar]
- 73. Treangen TJ, Salzberg SL. Repetitive DNA and next‐generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Koren S, Harhay GP, Smith TP, et al. Reducing assembly complexity of microbial genomes with single‐molecule sequencing. Genome Biol. 2013;14:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Eid J, Fehr A, Gray J, et al. Real‐time DNA sequencing from single polymerase molecules. Science. 2009;323:133‐138. [DOI] [PubMed] [Google Scholar]
- 76. Rao DN, Dryden DT, Bheemanaik S. Type III restriction‐modification enzymes: a historical perspective. Nucleic Acids Res. 2014;42:45‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


